Abstract
Recently, reverse genetics systems of plant negative‐stranded RNA (NSR) viruses have been developed to study virus–host interactions. Nonetheless, genetic rescue of plant NSR viruses in both insect vectors and monocot plants is very limited. Northern cereal mosaic virus (NCMV), a plant cytorhabdovirus, causes severe diseases in cereal plants through transmission by the small brown planthopper (SBPH, Laodelphax striatellus) in a propagative manner. In this study, we first developed a minireplicon system of NCMV in Nicotiana benthamiana plants, and then recovered a recombinant NCMV virus (rNCMV‐RFP), with a red fluorescent protein (RFP) insertion, in SBPHs and barley plants. We further used rNCMV‐RFP and green fluorescent protein (GFP)‐tagged barley yellow striate mosaic virus (rBYSMV‐GFP), a closely related cytorhabdovirus, to study superinfection exclusion, a widely observed phenomenon in dicot plants rarely studied in monocot plants. Interestingly, cellular superinfection exclusion of rBYSMV‐GFP and rNCMV‐RFP was observed in barley leaves. Our results demonstrate that two insect‐transmitted cytorhabdoviruses are enemies rather than friends at the cellular level during coinfections in plants.
Keywords: cytorhabdovirus, monocot plants, NCMV, plant negative‐stranded RNA viruses, reverse genetics systems, superinfection exclusion (SIE)
The recombinant NCMV‐RFP virus was rescued in barley plants and insect vectors to reveal cellular superinfection exclusion of two cytorhabdoviruses, NCMV‐RFP and BYSMV‐GFP, in barley.
1.
Plant negative‐stranded RNA (NSR) viruses cause severe diseases in dicot and monocot plants worldwide. Most plant NSR viruses are heavily reliant on specific insect vectors for long‐distance transmission in fields. Plant NSR viruses have caused significant yield losses and reduced quality of economically important crops. The reverse genetics system of plant viruses is a powerful technique for investigating virus biology, virus–host interactions, and biotechnological control of plant disease (Zang et al., 2020). Although reverse genetics systems of plant positive‐strand RNA viruses have been established for more than 30 years, recovery of plant NSR viruses from cDNA clones has only been achieved in recent years (Zang et al., 2020). The first reverse genetic systems of plant NSR viruses, Sonchus yellow net nucleorhabdovirus (SYNV), including the minireplicon (MR) system and agroinfiltration of full‐length infectious cDNA clones, was established in dicot plants (Ganesan et al., 2013; Wang et al., 2015). We used insect injection and transmission to recover a cytorhabdovirus, barley yellow striate mosaic virus (BYSMV), from full‐length cDNA clones in insect vectors (small brown planthopper [SBPH], Laodelphax striatellus) and monocot plants (barley, Hordeum vulgare) (Fang et al., 2019; Gao et al., 2019). Subsequently, tomato spotted wilt virus, an economically important, segmented NSR virus, was rescued in Nicotiana benthamiana plants through codon optimization and agroinfiltration (Feng et al., 2020). More recently, rose rosette virus, with a seven‐segmented NSR genome, was recovered in N. benthamiana plants and roses by airbrush delivery (Verchot et al., 2020). Although reverse genetic systems for these plant NSR viruses have been successfully established, recovery of insect‐transmitted NSR viruses in their insect vectors and host plants is still limited and remains to be developed extensively for examining the basis of the complicated virus–host plant–insect vector interactions.
Northern cereal mosaic virus (NCMV), a cytorhabdovirus, causes severe disease in barley and wheat plants, and occurs mainly in Japan, Korea, and China (Lee & Shikata, 1977; Murayama & Lu, 1967; Yang et al., 2018). NCMV contains a negative‐stranded RNA genome consisting of 13,222 nucleotides (Tanno et al., 2000). The NCMV genome encodes five structural proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and polymerase (L). In addition, the NCMV genome encodes five accessory proteins: P3, P4, P5, P6, and P9 in the order 3′‐N‐P‐P3‐P4‐P5‐P6‐M‐G‐P9‐L‐5′ (Tanno et al., 2000). NCMV and BYSMV are phylogenetically closely related cytorhabdoviruses with similar genome organization and sequence identity (Yan et al., 2015). In addition, the two cytorhabdoviruses share similar host ranges in the family Poaceae, and both are transmitted by SBPHs (Cao et al., 2018). Recently, we developed an antigenomic minireplicon system of BYSMV (BYSMV‐agMR) and studied the intracellular movement of the BYSMV P protein (Fang et al., 2019). We rescued the full‐length BYSMV from cDNA clones and developed versatile delivery platforms based on BYSMV vectors in cereal plants (H. vulgare) and SBPHs (Gao et al., 2019). We used the reverse genetics system of BYSMV to uncover the functions of host factors such as carbon catabolite repression 4 and casein kinase 1 in cytorhabdovirus infection (Gao et al., 2020; Zhang et al., 2020). Despite increasing progress in plant NSR virus studies recently, a reverse genetics system for NCMV remains to be developed for highlighting the biological features of NCMV, as well as its interaction with other NSR viruses.
Superinfection exclusion (SIE) is a well‐known phenomenon in which the first established infections of many viruses prevent the subsequent reinfection by the same or closely related viruses (Laureti et al., 2020). For human and animal viruses, including hepatitis C virus, West Nile virus, vesicular stomatitis virus, human immunodeficiency virus, herpesviruses, and poxviruses, SIE has been shown to prevent secondary infections in single cells (Kobiler et al., 2010; Liao et al., 2017; Nethe et al., 2005; Ramirez et al., 2010; Schaller et al., 2007; Simon et al., 1990; Tscherne et al., 2007; Zou et al., 2009). SIE has also been shown to occur at a cellular level for many plant positive‐stranded viruses, like citrus tristeza virus, tobacco mosaic virus, turnip crinkle virus, plum pox virus, soilborne wheat mosaic virus, and apple latent spherical virus (Bergua et al., 2014; Dietrich & Maiss, 2003; Folimonova, 2012; Julve et al., 2013; Miyashita & Kishino, 2010; Takahashi et al., 2007; Zhang et al., 2017, 2019). More recently, it was reported that the matrix protein of SYNV, a plant NSR virus, mediated SIE by regulating virion assembly (Zhou et al., 2019). However, due to the lack of reverse genetic systems, reported research on the phenomenon of SIE between two different cytorhabdoviruses in monocot plants has lagged behind those reported in dicot plants. In this study, we first developed a minireplicon of NCMV and then rescued the full‐length NCMV from full‐length cDNA in SBPHs and barley plants. Based on the NCMV and BYSMV infectious cDNA clones, recombinant viruses NCMV‐RFP and BYSMV‐GFP expressing red and green fluorescent proteins, respectively, were used to study SIE of these closely related cytorhabdoviruses in monocot plants.
To develop an antigenomic minireplicon (agMR) of NCMV, we generated a reporter cassette based on the NCMV antigenome through substituting the open reading frames (ORFs) of NCMV N and P with the green fluorescent protein (GFP) and red fluorescent protein (RFP) ORFs, respectively. In addition, the intergenic sequences (ISs) between the NCMV P and P3 genes were inserted between the GFP and RFP ORFs, resulting in the pNCMV‐agMR plasmid (Figure 1a). The ORFs of the NCMV N, P, and L genes were individually introduced into the pGD vector for ectopic protein expression, which is essential for initiation of plant rhabdovirus infections (Figure 1b). To suppress host RNA silencing responses and improve accumulation of viral core proteins, three viral RNA silencing suppressor genes (VSRs; tomato bushy stunt virus p19, barley stripe mosaic virus γb, and tobacco etch virus HC‐Pro) were inserted into the pGD vector (Figure 1c) described previously (Fang et al., 2019; Gao et al., 2019).
FIGURE 1.
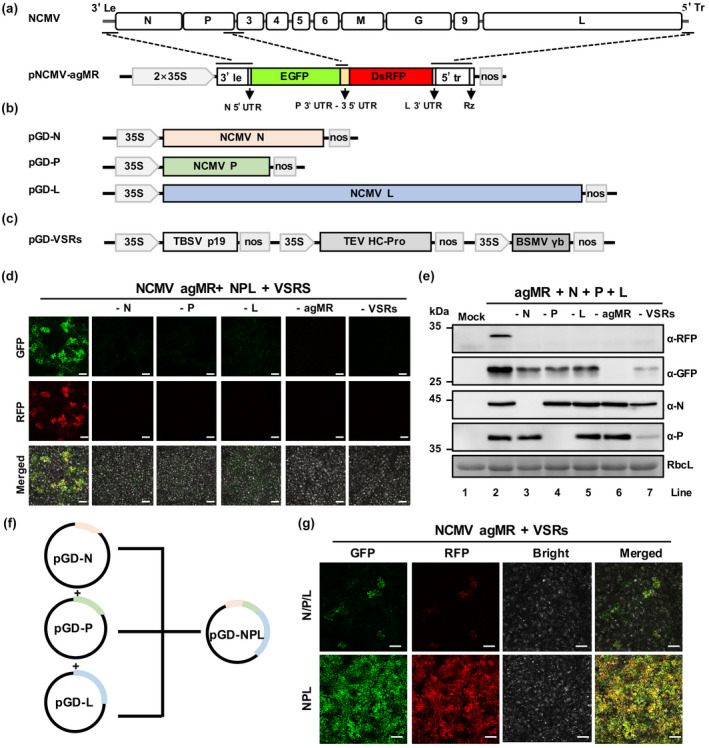
Construction of a northern cereal mosaic virus (NCMV) antigenomic minireplicon system in Nicotiana benthamiana leaves. (a) Schematic diagram showing a binary construct to express NCMV antigenomic minireplicon. Le, leader. Tr, trailer; UTR, untranslated region; P 3′ UTR‐3 5′ UTR, intergenic sequences including P 3′ UTR, intergenic sequence, and 3 5′ UTR; Rz, ribozyme sequence; nos, nopaline synthase terminator. (b) Schematic diagrams showing binary constructs to express the NCMV N, P, and L proteins. (c) Schematic diagram showing a binary construct to express the viral RNA silencing suppressor genes (VSRs) tomato bushy stunt virus p19, barley stripe mosaic virus γb, and tobacco etch virus HC‐Pro. (d) green fluorescent protein (GFP) and red fluorescent protein (RFP) foci in N. benthamiana infiltrated with different combinations of NCMV agMR system at 6 days postinoculation (dpi) seen with a fluorescence microscope. Scale bars, 200 μm. (e) Western blot analyses of expression of RFP, GFP, NCMV N and P proteins in leaves shown in (d) with specific antibodies against RFP, GFP, NCMV N and P proteins, respectively. (f) Schematic diagram of representative binary constructs to express NCMV N, P, and L, singly or together. (g) GFP and RFP foci in N. benthamiana infiltrated with different combinations of NCMV agMR system at 6 dpi. Scale bars, 200 μm
To rescue the NCMV agMR system, Agrobacterium harbouring the plasmids pNCMV agMR, pGD‐N, ‐P, ‐L, and ‐VSRs were coinfiltrated into N. benthamiana leaves. Six days postinfiltration (dpi), GFP and RFP fluorescence was observed in the infiltrated regions of N. benthamiana leaves (Figure 1d). In contrast, GFP and RFP fluorescence was not observed in N. benthamiana leaves without expression of any one of N, P, L, agMR, or VSRs (Figure 1d). Consistently, western blotting analyses showed that accumulation of GFP and RFP was only detected in infiltrated leaves expressing all components (Figure 1e). In the agMR system, efficient expression of the reporter genes requires delivery of all the DNA structures into single plant cells. To improve the recovery efficiency of the NCMV agMR, the N, P, and L expression cassettes were inserted into a single plasmid designated pGD‐NPL (Figure 1f). As expected, pGD‐NPL dramatically increased foci numbers compared with the separate pGD‐N, pGD‐P, and pGD‐L plasmids (Figure 1g). These results demonstrate that we have successfully established an efficient NCMV minireplicon system in N. benthamiana leaves.
The successful establishment of the NCMV‐agMR system indicates that all the core proteins and VSRs function to assemble infectious virus replication complexes (Figure 1). Accordingly, we next generated a full‐length construct of the NCMV antigenome with an RFP reporter gene insertion to monitor virus infection. To this end, the RFP gene flanked by the P/P3 gene junction sequences was amplified and inserted between the NCMV N and P genes (Figure 2a). To rescue rNCMV‐RFP in vivo, Agrobacterium harbouring plasmids pNCMV‐RFP, pGD‐NPL, and pGD‐VSRs were coinfiltrated into N. benthamiana leaves. At 12 dpi, RFP foci were observed in the cytoplasm of infiltrated leaves (Figure 2b), indicating that rNCMV‐RFP was recovered in single cells of N. benthamiana leaves.
FIGURE 2.
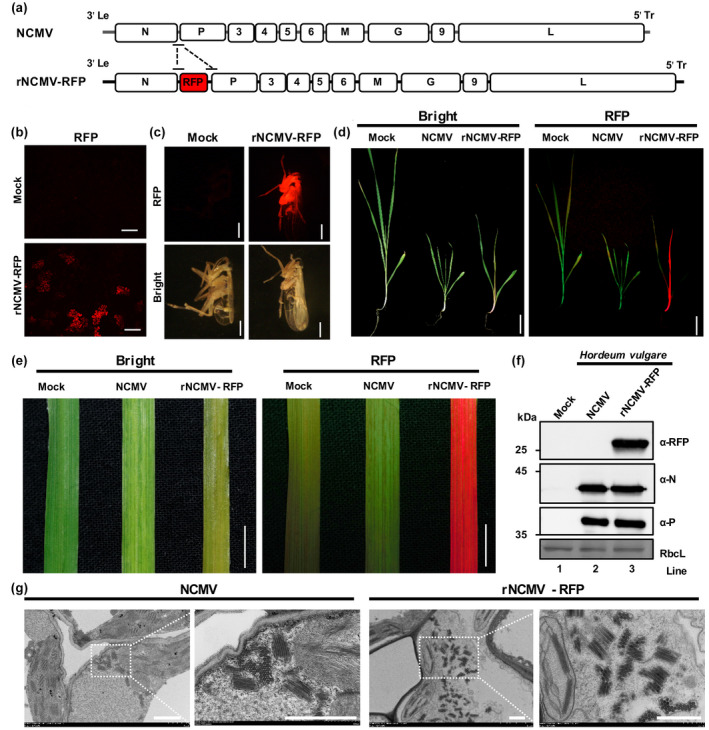
Recovery of infectious northern cereal mosaic virus (NCMV) in Nicotiana benthamiana leaves, small brown planthoppers (SBPHs, Laodelphax striatellus), and barley plants (Hordeum vulgare). (a) Schematic diagram of the full‐length northern cereal mosaic virus‐RFP (NCMV‐RFP) complementary DNA (cDNA) containing duplicate P/3 gene junctions flanking the red fluorescent protein (RFP) gene between the N and P of NCMV antigenome cDNA. (b) RFP foci in N. benthamiana infiltrated with NCMV‐RFP, NCMV nucleoprotein (N),phosphoprotein(P),and the large RNA polymerase(L), and viral RNA silencing suppressors (VSRs) at 12 days postinoculation (dpi) seen with a fluorescence microscope. Scale bars, 200 μm. (c) Images of SBPHs infected by NCMV‐RFP. At 10 dpi, the SBPHs were observed with a fluorescence stereomicroscope. Scale bars, 1 mm. (d) Symptoms on NCMV‐ and recombinant NCMV virus (rNCMV‐RFP)‐infected barley plants. Scale bars, 1 cm. (e) RFP fluorescence in barley leaves resulting from systemic infection with NCMV and rNCMV‐RFP. Scale bars, 1 cm. (f) Western blot analysis of NCMV proteins and RFP expression in plants shown in (d) with specific antibodies against NCMV N, P, and RFP proteins. (g) Electron micrographs of thin sections of barley leaves infected with NCMV and rNCMV‐RFP
NCMV is transmitted by SBPHs and our previous studies have demonstrated BYSMV can infect SBPHs by injection of insects with an extract of virus‐infected plant leaves (Gao et al., 2019; Qiao et al., 2022; Tanno et al., 2000). Accordingly, crude extract of rNCMV‐RFP‐infected N. benthamiana leaves was injected into second‐instar nymphs and the injected SBPHs were reared on fresh rice seedlings. At 10 dpi, RFP fluorescence was observed in five out of 106 surviving SPBHs (Figure 2c). Immunoblotting analyses showed accumulation of RFP, NCMV N, and P proteins in the injected SBPHs (Figure S1). To confirm that the RFP observed at 10 dpi was due to multiplication of rNCMV‐RFP, and not due to the injected extract per se, we examined the SBPHs immediately after injection with crude extract from rNCMV‐RFP‐infected barley plants (Figure S2). As shown in Figure S2b, the RFP fluorescence intensity of the SPBHs observed immediately after injection was much lower than the SPBHs infected with rNCMV‐RFP at 10 dpi, indicating that rNCMV‐RFP was successfully rescued in its insect vector.
To determine whether rNCMV‐RFP could be transmitted from infected SBPHs to barley plants, we transferred rNCMV‐RFP‐ or wild‐type NCMV‐infected SBPHs to healthy barley plants for a 3‐day infection access period. At 10 dpi, barley plants infected by either rNCMV‐RFP or wild‐type NCMV exhibited dwarf symptoms compared with mock‐treated plants (Figure 2d, left panel), while RFP fluorescence was only observed in leaves systemically infected by rNCMV‐RFP, rather than those infected by NCMV (Figure 2d,e). Western blotting analysis showed that abundant RFP accumulated in leaves infected systemically by rNCMV‐RFP, but not in those of wild‐type NCMV‐infected plants (Figure 2f) whereas both N and P proteins were detected in all the infected plants (Figure 2f). In addition, a large number of bacilliform particles were observed in the cytoplasm of barley stem and leaf cells infected by NCMV or rNCMV‐RFP (Figure 2g; Figure S3). In conclusion, these results demonstrate that rNCMV‐RFP was successfully rescued in barley plants.
The SIE phenomenon, in which a primary viral infection prevents subsequent viral infection by closely related viruses, has been observed in a broad range of bacterial, plant, and animal viruses (Bergua et al., 2014; Tatineni & French, 2016). However, most of the studies have mainly focused on the SIE by the same virus (Bergua et al., 2014; Whitaker‐Dowling et al., 1983; Zhang et al., 2017). Studies of SIE between two different viruses are limited. Tatineni and French (2016) showed that two wheat viruses, wheat streak mosaic virus (WSMV) and Triticum mosaic virus (TriMV), could efficiently coinfect the same wheat cell, suggesting that there was no SIE phenomenon between these two divergent viruses. NCMV and BYSMV are two closely related members of the genus Cytorhabdovirus with 62% amino acid identity of their polymerase (L) proteins (Yan et al., 2015). The GFP‐tagged BYSMV (rBYSMV‐GFP) has been rescued in our previous studies (Gao et al., 2019). To test whether these two viruses could coinfect barley plants, rNCMV‐RFP and rBYSMV‐GFP were used to examine the nature of coinfection by NCMV and BYSMV in a susceptible barley cultivar, Golden Promise (Figure 3a). Five rNCMV‐RFP‐infected SBPHs and five rBYSMV‐GFP‐infected SBPHs were transferred to a healthy barley seedling to coinoculate rNCMV‐RFP and rBYSMV‐GFP. The upper noninoculated leaves were examined for systemic infection by rNCMV‐RFP and rBYSMV‐GFP at 10 dpi (Figure 3b,c). Barley leaves observed at the whole‐organism level indicated the presence of both GFP and RFP in the same barley leaves (Figure 3b). However, cellular‐level observation showed that rNCMV‐RFP and rBYSMV‐GFP could hardly be detected within the same cell (Figure 3c). Western blot analyses revealed that the proteins NCMV N, BYSMV N, GFP, and RFP were expressed in the expected combinations (Figure 3d). We also observed that two BYSMV variants tagged with the two different fluorescent proteins exhibited SIE in the barley leaves (Figure S4). Taken together, these results reveal that NCMV and BYSMV exhibit SIE at the cellular level, although they coinfected one whole barley plant.
FIGURE 3.
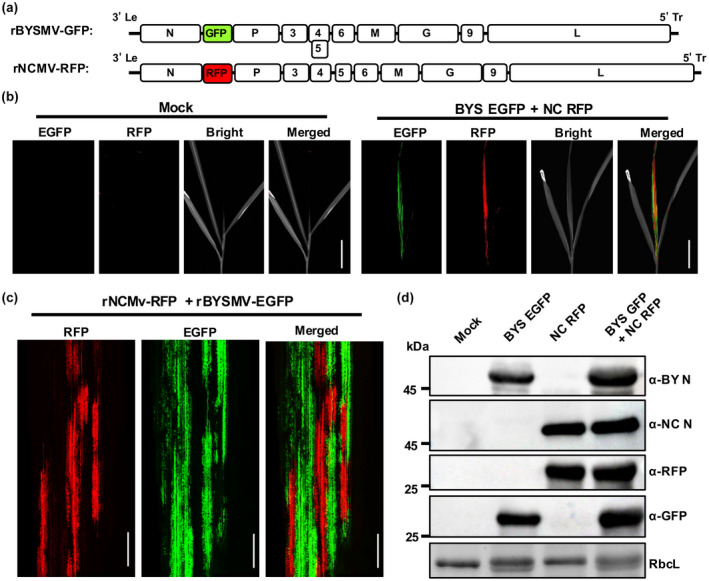
Superinfection exclusion (SIE) of recombinant northern cereal mosaic virus (rNCMV‐RFP) and GFP‐tagged barley yellow striate mosaic virus (rBYSMV‐GFP) in barley plants. (a) Schematic diagram of the genome of rNCMV‐RFP and rBYSMV‐GFP. (b) Coinfection of barley plants by rNCMV‐RFP and rBYSMV‐GFP at 10 days postinoculation. Scale bars, 3 cm. (c) SIE between rNCMV‐RFP and rBYSMV‐GFP in barley at the cellular level. Scale bars, 1 μm. (d) Western blot analysis of red fluorescent protein (RFP), green fluorescent protein (GFP), NCMV N, and BYSMV N expression in plants shown in (b) with specific antibodies against RFP, GFP, NCMV N, and BYSMV N proteins, respectively
Although NCMV has been reported for more than 70 years, studies about NCMV are limited in morphological and molecular functions due to the lack of a reverse genetic system. Here, we first developed an MR system of NCMV in N. benthamiana plants (Figure 1), and then recovered a recombinant NCMV virus (rNCMV‐RFP) in SBPHs and barley plants (Figure 2). The successful development of an MR and a full‐length reverse genetic system has provided a powerful tool to study the replication and pathogenesis of NCMV. Due to the classical discontinuous transcription and well‐defined transcription units of rhabdoviruses, rhabdovirus genomes are able to maintain multiple insertions of transcription units (Gao et al., 2019; Jackson et al., 2005; Xu et al., 2022). The full‐length reverse genetic system of NCMV permits stable foreign protein expression in barley plants and SBPHs, and rapid screening of gene functions in host plants and insect vectors.
The data above suggest that NCMV and BYSMV, two closely related NSR plant viruses, exhibit SIE at the cellular level (Figure 3). Therefore, cross‐protection could be used to control virus transmission in the field, which means insulating plants from a disease‐causing virus by simply preinoculating them with a mild variant of the same virus (Guo et al., 2020). A large number of plant‐infecting viruses are transmitted by insect vectors to their host plants and many of the insect vectors transmit several different virus species (Moreno & Lopez‐Moya, 2019). For instance, SBPHs are vectors of both NCMV and BYSMV. It would be interesting to determine whether NCMV and BYSMV can coinfect and be transmitted by one single SBPH. It remains to be determined whether the SIE between NCMV and BYSMV in plants shares similar features in insect vectors. Vector transmission is an essential step in the infection cycle of most plant viruses (Whitfield et al., 2015). Thus, clarifying the phenomenon and mechanism of SIE in insect vectors among various plant viruses could be useful to control the spread and epidemiology of the viruses.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
X.F. performed the majority of the experiments, assisted by J.Q., Y.Z., Q.G., D. G. Y.Y., X.L, and W.X. X.F. and X.W. analysed the data. X.W and X.F. wrote the manuscript, assisted by Y.W. All authors discussed the results and commented on the manuscript.
Supporting information
FILE S1. Experimental procedures
FIGURE S1. Western blot analyses of expression of the NCMV N, P and red fluorescent protein (RFP) proteins in small brown planthoppers shown in Figure 1c with specific antibodies against NCMV N, P, and RFP proteins
FIGURE S2. Red fluorescent protein (RFP) fluorescence of small brown planthoppers (SBPHs, Laodelphax striatellus) after microinjection with crude extracts of mock‐ or rNCMV‐RFP‐infected barley leaves. (a) Crude extracts from barley leaves infected by rNCMV‐RFP. (b) Images of SBPHs injected with crude extracts shown in (a). The SBPHs were observed immediately after injection seen with a fluorescence stereomicroscope. Scale bars, 1 mm
FIGURE S3. Electron micrographs of thin sections stems of barley plants infected with NCMV and rNCMV‐RFP at 12 days postinoculation
FIGURE S4. Coinfection and superinfection exclusion (SIE) of rBYSMV‐RFP and rBYSMV‐GFP in barley plants. (a) Schematic diagram of the genome of rBYSMV‐RFP and rBYSMV‐GFP. (b) Coinfection of barley by rBYSMV‐RFP and rBYSMV‐GFP at 10 days postinoculation. Scale bars, 3 cm. (c) SIE between rBYSMV‐RFP and rBYSMV‐GFP in barley at the cellular level. Scale bars, 1 μm. (d) Western blot analysis of BYSMV N, red fluorescent protein (RFP), and green fluorescent protein (GFP) expression in plants shown in (b) with specific antibodies against BYSMV N, RFP, and GFP proteins, respectively
TABLE S1. Primers used in this study
ACKNOWLEDGEMENTS
We thank Professor Jialin Yu, Dawei Li, Chenggui Han, and Yongliang Zhang for their helpful discussion. We thank Professor Hongqin Miao and Dianping Di for providing the NCMV‐infected wheat plants. This work was supported by grants from the Natural Science Foundation of China 31872920 (X.B.W.) and 32102150 (Q.G.), as well as the China Postdoctoral Science Foundation 2021T140713 (Q.G.).
Fang, X.‐D. , Qiao, J.‐H. , Zang, Y. , Gao, Q. , Xu, W.‐Y. , Gao, D.‐M. et al. (2022) Developing reverse genetics systems of northern cereal mosaic virus to reveal superinfection exclusion of two cytorhabdoviruses in barley plants. Molecular Plant Pathology, 23, 749–756. 10.1111/mpp.13188
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Bergua, M. , Zwart, M.P. , El‐Mohtar, C. , Shilts, T. , Elena, S.F. & Folimonova, S.Y. (2014) A viral protein mediates superinfection exclusion at the whole‐organism level but is not required for exclusion at the cellular level. Journal of Virology, 88, 11327–11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Q. , Xu, W.‐Y. , Gao, Q. , Jiang, Z.‐H. , Liu, S.‐Y. , Fang, X.‐D. et al. (2018) Transmission characteristics of barley yellow striate mosaic virus in its planthopper vector Laodelphax striatellus . Frontiers in Microbiology, 9, 1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, C. & Maiss, E. (2003) Fluorescent labelling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. Journal of General Virology, 84, 2871–2876. [DOI] [PubMed] [Google Scholar]
- Fang, X.‐D. , Yan, T. , Gao, Q. , Cao, Q. , Gao, D.‐M. , Xu, W.‐Y. et al. (2019) A cytorhabdovirus phosphoprotein forms mobile inclusions trafficked on the actin/ER network for viral RNA synthesis. Journal of Experimental Botany, 70, 4049–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, M. , Cheng, R. , Chen, M. , Guo, R. , Li, L. , Feng, Z. et al. (2020) Rescue of tomato spotted wilt virus entirely from complementary DNA clones. Proceedings of the National Academy of Sciences of the United States of America, 117, 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folimonova, S.Y. (2012) Superinfection exclusion is an active virus‐controlled function that requires a specific viral protein. Journal of Virology, 86, 5554–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan, U. , Bragg, J.N. , Deng, M. , Marr, S. , Lee, M.Y. , Qian, S. et al. (2013) Construction of a Sonchus yellow net virus minireplicon: a step toward reverse genetic analysis of plant negative‐strand RNA viruses. Journal of Virology, 87, 10598–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q. , Xu, W.‐Y. , Yan, T. , Fang, X.‐D. , Cao, Q. , Zhang, Z.‐J. et al. (2019) Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytologist, 223, 2120–2133. [DOI] [PubMed] [Google Scholar]
- Gao, Q. , Yan, T. , Zhang, Z.‐J. , Liu, S.‐Y. , Fang, X.‐D. , Gao, D.‐M. et al. (2020) Casein kinase 1 regulates cytorhabdovirus replication and transcription by phosphorylating a phosphoprotein serine‐rich motif. The Plant Cell, 32, 2878–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Q. , Zhang, S. , Sun, R. , Yao, X. , Zhang, X.‐F. , Tatineni, S. et al. (2020) Superinfection exclusion by p28 of Turnip crinkle virus is separable from its replication function. Molecular Plant‐Microbe Interactions, 33, 364–375. [DOI] [PubMed] [Google Scholar]
- Jackson, A.O. , Dietzgen, R.G. , Goodin, M.M. , Bragg, J.N. & Deng, M. (2005) Biology of plant rhabdoviruses. Annual Review of Phytopathology, 43, 623–660. [DOI] [PubMed] [Google Scholar]
- Julve, J.M. , Gandía, A. , Fernández‐del‐Carmen, A. , Sarrion‐Perdigones, A. , Castelijns, B. , Granell, A. et al. (2013) A coat‐independent superinfection exclusion rapidly imposed in Nicotiana benthamiana cells by tobacco mosaic virus is not prevented by depletion of the movement protein. Plant Molecular Biology, 81, 553–564. [DOI] [PubMed] [Google Scholar]
- Kobiler, O. , Lipman, Y. , Therkelsen, K. , Daubechies, I. , & Enquist, L.W. (2010) Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nature Communications, 1, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laureti, M. , Paradkar, P.N. , Fazakerley, J.K. & Rodriguez‐Andres, J. (2020) Superinfection exclusion in mosquitoes and its potential as an arbovirus control strategy. Viruses, 12, 1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.H. & Shikata, E. (1977) Occurrence of northern cereal mosaic virus in Korea. Korean Journal of Applied Entomology, 16, 87–92. [Google Scholar]
- Liao, J. , Wei, Q. , Fan, J. , Zou, Y. , Song, D. , Liu, J. et al. (2017) Characterization of retroviral infectivity and superinfection resistance during retrovirus‐mediated transduction of mammalian cells. Gene Therapy, 24, 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita, S. & Kishino, H. (2010) Estimation of the size of genetic bottlenecks in cell‐to‐cell movement of soil‐borne wheat mosaic virus and the possible role of the bottlenecks in speeding up selection of variations in trans‐acting genes or elements. Journal of Virology, 84, 1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, A.B. & Lopez‐Moya, J.J. (2019) When viruses play team sports: mixed infections in plants. Phytopathology, 110, 29–48. [DOI] [PubMed] [Google Scholar]
- Murayama, D. & Lu, Y.‐T. (1967) Some physical properties of northern cereal mosaic virus. Journal of the Faculty of Agriculture, Hokkaido University, 55, 182–190. [Google Scholar]
- Nethe, M. , Berkhout, B. , & van der Kuyl, A.C. (2005) Retroviral superinfection resistance. Retrovirology, 2, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, J.H. , Gao, Q. , Zang, Y. , Fang, X.D. & Wang, X.B. (2022) A versatile expression platform in insects and cereals based on a cytorhabdovirus. Methods in Molecular Biology, 2400, 163–170. [DOI] [PubMed] [Google Scholar]
- Ramirez, S. , Perez‐del‐Pulgar, S. , Carrion, J.A. , Coto‐Llerena, M. , Mensa, L. , Dragun, J. et al. (2010) Hepatitis C virus superinfection of liver grafts: a detailed analysis of early exclusion of non‐dominant virus strains. Journal of General Virology, 91, 1183–1188. [DOI] [PubMed] [Google Scholar]
- Schaller, T. , Appel, N. , Koutsoudakis, G. , Kallis, S. , Lohmann, V. , Pietschmann, T. et al. (2007) Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene‐tagged viral genomes. Journal of Virology, 81, 4591–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, K.O. , Cardamone, J.J. , Whitaker‐Dowling, P.A. , Youngner, J.S. , & Widnell, C.C. (1990) Cellular mechanisms in the superinfection exclusion of vesicular stomatitis virus. Virology, 177, 375–379. [DOI] [PubMed] [Google Scholar]
- Takahashi, T. , Sugawara, T. , Yamatsuta, T. , Isogai, M. , Natsuaki, T. & Yoshikawa, N. (2007) Analysis of the spatial distribution of identical and two distinct virus populations differently labeled with cyan and yellow fluorescent proteins in coinfected plants. Phytopathology, 97, 1200–1206. [DOI] [PubMed] [Google Scholar]
- Tanno, F. , Nakatsu, A. , Toriyama, S. & Kojima, M. (2000) Complete nucleotide sequence of Northern cereal mosaic virus and its genome organization. Archives of Virology, 145, 1373–1384. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. & French, R. (2016) The coat protein and NIa protease of two Potyviridae family members independently confer superinfection exclusion. Journal of Virology, 90, 10886–10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne, D.M. , Evans, M.J. , von Hahn, T. , Jones, C.T. , Stamataki, Z. , McKeating, J.A. et al. (2007) Superinfection exclusion in cells infected with hepatitis C virus. Journal of Virology, 81, 3693–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. , Herath, V. , Urrutia, C.D. , Gayral, M. , Lyle, K. , Shires, M.K. et al. (2020) Development of a reverse genetic system for studying rose rosette virus in whole plants. Molecular Plant‐Microbe Interactions, 33, 1209–1221. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Ma, X. , Qian, ShaSha , Zhou, X. , Sun, K. , Chen, X. et al. (2015) Rescue of a plant negative‐strand RNA virus from cloned cDNA: insights into enveloped plant virus movement and morphogenesis. PLoS Pathogens, 11, e1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker‐Dowling, P. , Youngner, J.S. , Widnell, C.C. & Wilcox, D.K. (1983) Superinfection exclusion by vesicular stomatitis virus. Virology, 131, 137–143. [DOI] [PubMed] [Google Scholar]
- Whitfield, A.E. , Falk, B.W. & Rotenberg, D. (2015) Insect vector‐mediated transmission of plant viruses. Virology, 479–480, 278–289. [DOI] [PubMed] [Google Scholar]
- Xu, W.‐Y. , Fang, X.‐D. , Cao, Q. , Gao, Q. , Gao, D.‐M. , Qiao, J.‐H. et al. (2022) A cytorhabdovirus‐based expression vector in Nilaparvata lugens, Laodelphax striatellus, and Sogatella furcifera . Insect Biochemistry and Molecular Biology, 140, 103703. [DOI] [PubMed] [Google Scholar]
- Yan, T. , Zhu, J.‐R. , Di, D. , Gao, Q. , Zhang, Y. , Zhang, A. et al. (2015) Characterization of the complete genome of Barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology, 478, 112–122. [DOI] [PubMed] [Google Scholar]
- Yang, F. , Zhang, A. , Li, X. , Huo, L. , Di, D. & Miao, H. (2018) Occurrence and alternation of cytorhabdoviruses on wheat in northern China. Bioscience Journal, 34, 1472–1476. [Google Scholar]
- Zang, Y. , Fang, X.‐D. , Qiao, J.‐H. , Gao, Q. & Wang, X.‐B. (2020) Reverse genetics systems of plant negative‐strand RNA viruses are difficult to be developed but powerful for virus–host interaction studies and virus‐based vector applications. Phytopathology Research, 2, 29. [Google Scholar]
- Zhang, S. , Sun, R. , Guo, Q. , Zhang, X.F. & Qu, F. (2019) Repression of turnip crinkle virus replication by its replication protein p88. Virology, 526, 165–172. [DOI] [PubMed] [Google Scholar]
- Zhang, X.‐F. , Sun, R. , Guo, Q. , Zhang, S. , Meulia, T. , Halfmann, R. et al. (2017) A self‐perpetuating repressive state of a viral replication protein blocks superinfection by the same virus. PLoS Pathogens, 13, e1006253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z.‐J. , Gao, Q. , Fang, X.‐D. , Ding, Z.‐H. , Gao, D.‐M. , Xu, W.‐Y. et al. (2020) CCR4, a RNA decay factor, is hijacked by a plant cytorhabdovirus phosphoprotein to facilitate virus replication. eLife, 9, e53753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Sun, K. , Zhou, X. , Jackson, A.O. & Li, Z. (2019) The matrix protein of a plant rhabdovirus mediates superinfection exclusion by inhibiting viral transcription. Journal of Virology, 93, e00680‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, G. , Zhang, B. , Lim, P.‐Y. , Yuan, Z. , Bernard, K.A. , & Shi, P.‐Y. (2009) Exclusion of West Nile virus superinfection through RNA replication. Journal of Virology, 83, 11765–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FILE S1. Experimental procedures
FIGURE S1. Western blot analyses of expression of the NCMV N, P and red fluorescent protein (RFP) proteins in small brown planthoppers shown in Figure 1c with specific antibodies against NCMV N, P, and RFP proteins
FIGURE S2. Red fluorescent protein (RFP) fluorescence of small brown planthoppers (SBPHs, Laodelphax striatellus) after microinjection with crude extracts of mock‐ or rNCMV‐RFP‐infected barley leaves. (a) Crude extracts from barley leaves infected by rNCMV‐RFP. (b) Images of SBPHs injected with crude extracts shown in (a). The SBPHs were observed immediately after injection seen with a fluorescence stereomicroscope. Scale bars, 1 mm
FIGURE S3. Electron micrographs of thin sections stems of barley plants infected with NCMV and rNCMV‐RFP at 12 days postinoculation
FIGURE S4. Coinfection and superinfection exclusion (SIE) of rBYSMV‐RFP and rBYSMV‐GFP in barley plants. (a) Schematic diagram of the genome of rBYSMV‐RFP and rBYSMV‐GFP. (b) Coinfection of barley by rBYSMV‐RFP and rBYSMV‐GFP at 10 days postinoculation. Scale bars, 3 cm. (c) SIE between rBYSMV‐RFP and rBYSMV‐GFP in barley at the cellular level. Scale bars, 1 μm. (d) Western blot analysis of BYSMV N, red fluorescent protein (RFP), and green fluorescent protein (GFP) expression in plants shown in (b) with specific antibodies against BYSMV N, RFP, and GFP proteins, respectively
TABLE S1. Primers used in this study
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


