Abstract
The pathogenesis‐related 1 (PR1) proteins are members of the cross‐kingdom conserved CAP superfamily (from Cysteine‐rich secretory protein, Antigen 5, and PR1 proteins). PR1 mRNA expression is frequently used for biotic stress monitoring in plants; however, the molecular mechanisms of its cellular processing, localization, and function are still unknown. To analyse the localization and immunity features of Arabidopsis thaliana PR1, we employed transient expression in Nicotiana benthamiana of the tagged full‐length PR1 construct, and also disrupted variants with C‐terminal truncations or mutations. We found that en route from the endoplasmic reticulum, the PR1 protein transits via the multivesicular body and undergoes partial proteolytic processing, dependent on an intact C‐terminal motif. Importantly, only nonmutated or processing‐mimicking variants of PR1 are secreted to the apoplast. The C‐terminal proteolytic cleavage releases a protein fragment that acts as a modulator of plant defence responses, including localized cell death control. However, other parts of PR1 also have immunity potential unrelated to cell death. The described modes of the PR1 contribution to immunity were found to be tissue‐localized and host plant ontogenesis dependent.
Keywords: extracellular proteins, multivesicular bodies, pathogenesis‐related 1, plant immunity, vesicular trafficking
Transiently expressed AtPR1 progresses from the endoplasmic reticulum via ARA6/7‐positive multivesicular bodies toward the cell wall; its effect on immunity is dependent on its C‐terminus integrity and host plant developmental stage.
1. INTRODUCTION
In plants, the pathogenesis‐related 1 (PR1) protein family is a highly conserved member of 17 pathogenesis‐related protein families in total (reviewed in Stintzi et al. [1993] and van Loon and van Strien [1999]). PR1 and PR1‐like proteins play important roles beyond the plant kingdom, as they are present in fungi, insects, and vertebrates, including humans. Together they form a superfamily of secreted proteins with various functions, named CAP (from Cysteine‐rich secretory protein [CRISP], Antigen 5, and PR1 proteins; Gibbs et al., 2008). Even though PR1 is considered to be an antifungal and antioomycete protein, the mechanism for PR1 action is still enigmatic, in contrast to other PR proteins whose function has been elucidated (Agrios, 2005; Joshi et al., 2021).
Plant PR1s are encoded by a family of highly multiplied genes. In the genome of Arabidopsis thaliana, there are 22 PR1‐type genes, primarily arranged in clusters of genes, coding for polar, acidic, or basic type proteins (review in van Loon et al., 2006). Only one of them, A. thaliana PR1 (AtPR1, At2g14610), is activated by pathogens, insects, or chemical treatments, whereas the other PR1‐type genes are constitutively expressed in roots and pollen (van Loon et al., 2006).
The first studies showed PR1 protein level up‐regulation in tobacco and tomato on viral infection, including by tobacco mosaic virus (TMV; Antoniw & Pierpoint, 1978; Camacho Henriquez & Sänger, 1982; van Loon & van Kammen, 1970). However, subsequent PR1‐related experiments concentrated more on its mRNA expression, which became a commonly used reporter of salicylic acid (SA)‐activated defence responses in plants, the hypersensitivity response, and systemic acquired resistance (SAR; Bowling et al., 1994; Jung & Hwang, 2000; Hooft van Huijsduijnen et al., 1987; Somssich et al., 1986). Several studies reported the antifungal and antibacterial acitivity of PR1 (Lincoln et al., 2018; Niderman et al., 1995; Rauscher et al., 1999; Woloshuk et al., 1991). Recently, tobacco and tomato PR1 protein sterol‐binding activity has been found critical for its antimicrobial activity (Gamir et al., 2017). The overexpression of pepper basic PR1 homolog in tobacco enhances plant resistance against pathogenic bacteria and oomycetes (Sarowar et al., 2005). Likewise, the silencing of PR1 genes in tobacco led to increased apoplastic β‐(1,3)‐glucanase activity, decreased callose deposition, and higher susceptibility to Phytophthora (Riviere et al., 2008). As part of the counterattack strategy of pathogens, PR1s are targeted by effectors of pathogens, for example, during host infection by Parastagonospora nodorum (Breen et al., 2016) or by Blumeria graminis f. sp. hordei (Zhang et al., 2012).
The PR1 protein homolog of tomato (P14a), when overexpressed in root tissue, suppresses the plant cell death triggered by the fungal toxin fumonisin B1, as do the human and Ancylostoma PR1 orthologs, indicating a cross‐kingdom cell death‐related function (Lincoln et al., 2018). In tobacco plants expressing a P14a‐GFP (green fluorescent protein) fusion construct, and after inoculation with Pseudomonas syringae pv. tabaci, the up‐regulation of the respective mRNA was general, but the fusion protein expression could be observed only in lesion margins, where cell death and bacterial spread were arrested, indicating posttranscriptional regulation. This is in agreement with the observation of PR1 occurrence in the cell death‐limiting zone in A. thaliana demonstrated by Betsuyaku et al. (2018).
Recently, an 11 amino acid C‐terminal CAP‐derived peptide (CAPE) fragment of tomato PR1b containing the characteristic PxGNxxxxxPY motif has been discovered to function as a defence signalling peptide (Chen et al., 2014). Tomato plants presprayed with CAPE exhibited increased resistance to the bacterial pathogen P. syringae pv. tomato DC3000, and also reduced Spodoptera litura larval growth and weight. AtCAPE1 has been found to function as a negative regulator of salt stress responses in Arabidopsis and as a positive regulator of tomato immunity (Chien et al., 2015). In accordance with this, the interaction of wheat TaPR1 with the effector SnTox3 from P. nodorum hinders the release of the TaCAPE1 peptide from TaPR1, thus preventing the activation of host plant defence responses (Sung et al., 2021).
There is very little data on the cellular trafficking of plant PR1s. Histochemical and proteomic approaches have confirmed a presence of PR1 in the vacuole, cell wall, and exudates (Boudart et al., 2005; Carella et al., 2016; Dixon et al., 1991). In Arabidopsis, PR1 protein tagged with mCherry accumulates in the apoplastic space of the epidermis of seedlings, with only a weak signal detectable in the vacuolar lumen (van Loon et al., 2006). However, in the loss‐of‐function mutant of KEEP ON GOING, a RING E3 ligase involved in abscisic acid signalling during growth and development, the vacuolar PR1‐mCherry signal appeared much stronger (Gu & Innes, 2012). PR1 trafficking and secretion depend on the vesicular pathway regulators SNARE SYP132 or exocyst complex (Du et al., 2015; Kalde et al., 2007). PR1‐GFP expressed transiently in Nicotiana benthamiana leaves colocalizes only partially with Golgi markers and much more prominently with the late endosome/multivesicular body (MVB) FYVE marker, indicative of either unconventional secretion or enhanced degradation‐related pathway. This peculiar localization is dependent on the presence of the PR1 C‐terminus (Pečenková et al., 2017). The Phytophthora brassicae effector RxLR24 affects the intracellular localization of PR1 through interference with secretory pathway‐related RABA GTPases, leading to the accumulation of PR1 in the endoplasmic reticulum (ER) (Tomzcinska et al., 2018).
In this study, we created several tagged A. thaliana PR1 variants and tested them for their localization and immunity‐related functional assays in N. benthamiana. We also assessed the capacity of PR1 to spread to neighbouring cells. We conclude that PR1 undergoes simultaneous partial processing and trafficking dependent on the intact C‐terminal motif. Depending on plant age, the PR1 protein or its fragments can act as both positive or negative, and intracellular or extracellular modulators of local plant defence responses.
2. RESULTS
2.1. PR1‐GFP localizes to multivesicular bodies and extracellular space
We have previously described a highly dynamic dsRed‐FYVE‐ and PR1‐GFP‐positive compartment with architecture similar to MVBs in N. benthamiana (Pečenková et al., 2017). Here, using the MVB markers RFP‐ARA6 and RFP‐ARA7 (Ebine et al., 2011; Jia et al., 2013), we observed their colocalization with PR1‐GFP transiently expressed in N. benthamiana, thus further confirming the identity of the PR1‐GFP compartment as MVBs (Figure 1a). Notably, our data suggest a differential distribution of PR1‐GFP in distinct MVB subpopulations. PR1‐GFP had 82% signal overlap with RFP‐ARA6, and 57% signal overlap with ARA7, which suggests preferred localization in the MVB subpopulation reported to be targeted, besides vacuole, also to the plasma membrane (PM) (Ebine et al., 2011). Plasma membrane‐proximal localization of the PR1‐positive compartment was also observable in A. thaliana seedlings overexpressing PR1 (Figure S1a).
FIGURE 1.
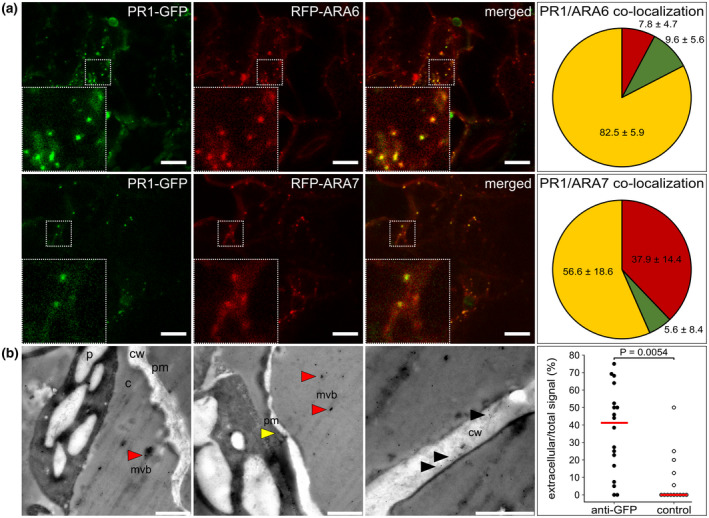
PR1‐GFP (green fluorescent protein) localization in multivesicular bodies (MVBs) and extracellular space. (a) Examples of colocalization of PR1‐GFP with RFP‐ARA6 and RFP‐ARA7 transiently co‐expressed in Nicotiana benthamiana leaves. Insets depict a magnified view of the signal in the dashed squares. Pie charts represent the quantification of colocalization that was performed on at least 10 regions (50 × 50 μm) out of five different scans (n = 329 total analysed objects for ARA6 and n = 141 for ARA7). Red sector, red fluorescent protein (RFP) signal only; green sector, GFP signal only; yellow sector, colocalizing RFP/GFP signals, values indicate mean ± SD. Bars = 10 μm. (b) Immunogold electron microscopy using anti‐GFP and secondary antibodies carrying 10 nm gold particles. Micrographs show different parts of the cells where gold particles are observable: MVB‐like (mvb, red arrowheads), localizations adjacent to the plasma membrane (pm, yellow arrowhead), and cell wall space (cw, black arrowheads; c, cytoplasm; p, plastid). Bars = 1 μm. Panel on the right shows a graph of the portion of PR1‐GFP with extracellular signal (black dots) in comparison to the control experiment (white dots) with an unspecific secondary antibody signal only (18 and 14 images analysed, n = 320 and 76 particles, respectively). Individual data points and median are shown. The p value was calculated with the two‐sided permutation t test
To better resolve this relation of MVB‐ and PM‐ PR1‐GFP localization, we conducted immunogold electron microscopy analysis of N. benthamiana leaves transiently expressing PR1‐GFP employing an anti‐GFP antibody (Figure 1b). In addition to the previously found intracellular localization, we observed PR1‐GFP in MVB‐like compartments adjacent to the PM and outside in the extracellular space, that is, either in proximity to MVB/PM or in more distant cell wall regions indicative of the ability of PR1‐GFP to reach the apoplastic space. However, it was not possible to resolve by this approach whether it occurs via a canonical secretory pathway or via fusion of these MVBs with the PM.
We conclude that the PR1‐positive compartments are MVBs, and that the PR1‐GFP can be secreted into the cell wall space.
2.2. PR1‐GFP trafficking to MVBs does not involve endocytosis
Furthermore, we wanted to assess if the observed PR1 MVB‐related signal is cell‐autonomous, or whether it at least partially originates from an uptake from the extracellular space and/or neighbouring cells. Because there was no effect of the inhibition of endocytosis in the creation of PR1‐positive MVB (Figure 2a), nor colocalization with endocytic dye FM4‐64 (Figure S1b), we sought other evidence that the PR1‐compartment is, or is not, cell autonomous, employing the co‐expression of PR1‐GFP and cytoplasmic SEC10‐mCherry exocyst core subunit as a reference from the same expression vector in N. benthamiana leaves (Figure 2b). We looked for directly transformed cells containing both GFP and mCherry signals, with neighbouring cells with penetrated GFP signal only (Figure 2b). Such cases were indeed found, and the PR1signal imported into the neighbouring cells had vesicle‐like characteristics (Figure 2b). This phenomenon indicates the intercellular trafficking of PR1‐GFP protein and possible paracrine mode of action (e.g., Ge et al., 2019). However, it should be stressed that cells with supposedly imported PR1‐GFP signals represented a minor fraction of fluorescent cells, and the GFP signal intensity was relatively low. To check if the PR1‐GFP protein might spread to more distant cells from the infiltration site, we performed western blot analyses with leaf tissue distant from the infiltrated parts. However, we never detected any PR1 protein spread using either anti‐GFP for PR1‐GFP or anti‐hemagglutinin (anti‐HA) for HA‐tagged constructs created for this purpose (data not shown).
FIGURE 2.
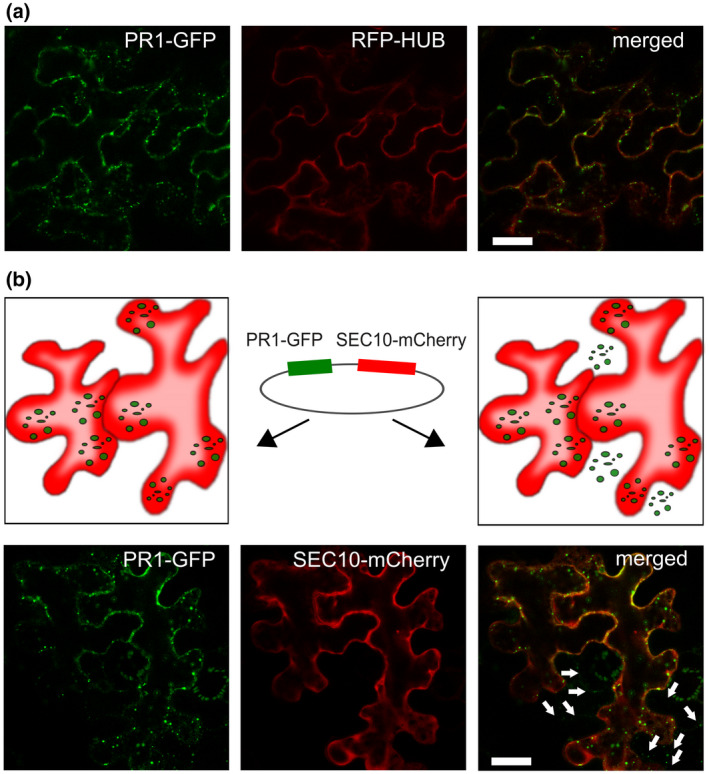
The PR1‐GFP (green fluorescent protein) trafficking is not dependent on endocytosis. (a) Insensitivity of the PR1‐GFP multivesicular body (MVB) compartment formation to co‐expressed endocytic inhibitor RFP‐HUB construct in Nicotiana benthamiana leaves. Bar = 10 μm. (b) Paracrine cell‐to‐cell spread of PR1‐GFP. Upper row: schematic of a situation in leaf epidermal cells indicative of the two‐construct expression in one cell (on the left) and cell‐to‐cell trafficking of one protein (PR1‐GFP) to the neighbouring cell (on the right). Middle: a schematic presentation of a vector encoding both PR1‐GFP and cytoplasmic construct SEC10‐mCherry as a reference. Lower row: cells that have only GFP signal (white arrow) and are neighbours to cells with both GFP and RFP signals. The penetrated signal was found to be of vesicular type (white arrows). Bar = 10 μm
Our findings suggest that the PR1 in N. benthamiana epidermis probably executes its function in the extracellular space, because no prominent spread from one cell interior to another was observed. A minor portion of the secreted PR1 can, by an undescribed mechanism, spread from cells where it is synthesized directly to the neighbouring cells; both modes are indicative of its short‐distance tissue‐localized activity.
2.3. C‐terminus‐dependent localization and partial proteolysis of PR1‐GFP
Next, we wanted to assess what are the sequence requirements for the observed PR1 trafficking. Based on the suggested processing of the C‐terminal CAPE peptides of PR1‐like family proteins, and on previous findings on C‐terminus‐dependent PR1 localization (Chen et al., 2014; Chien et al., 2015; Pečenková et al., 2017), we set about checking in more detail the localization of constructs PR1‐GFP (full‐length PR1, further referred to as FL) and a 50 amino acid‐truncated version PR1ΔC (D50 variant; Pečenková et al., 2017), and also the newly created GFP‐tagged PR1 variant with the presumed cleavage site YDPR mutated into AAAA (4A variant), and another lacking the last 11 amino acids, thus mimicking suggested PR1 C‐terminal processing (PR1Δ11; D11; see also Figure 3 for the schematic representation of constructs). We tested whether these truncations and mutations affect the protein structure. Our comparison of ab initio modelled 3D protein structures (without GFP) of all variants suggests that the overall conformation would not be altered dramatically. The conformation of D50 is the most distinct/deviated from FL (Figure 3a), while the conformation for version 4A and D11 is affected only by very minor changes.
FIGURE 3.
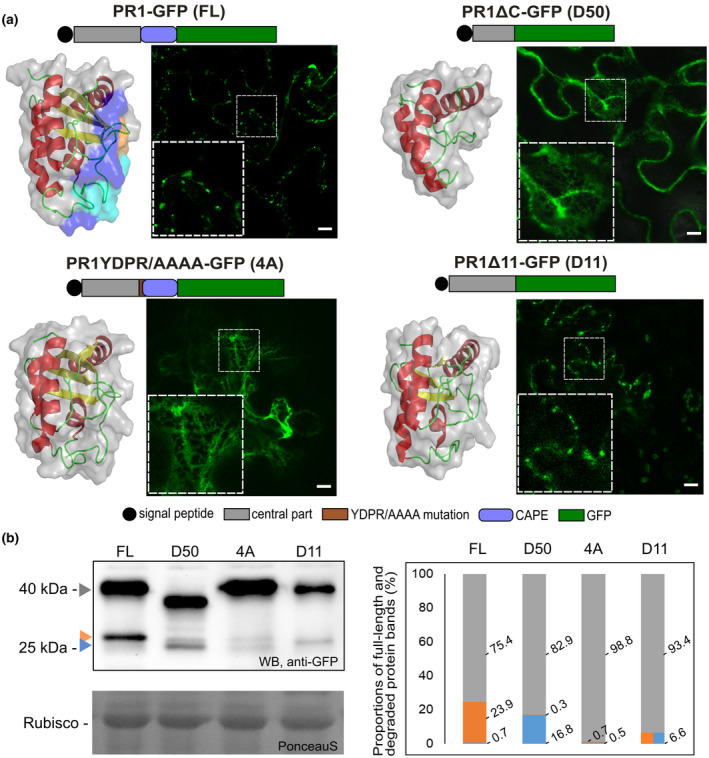
Localization and expression patterns of PR1 and its truncated and mutated variants tagged with green fluorescent protein (GFP). (a) Schematic representation and intracellular localizations in transiently transformed Nicotiana benthamiana leaves and three‐dimensional models are presented for each of the construct variants. In full‐length wild‐type (FL) models, orange shows a region differing from mutated 4A, cyan a missing part in D11, and light blue a missing part in D50. Confocal images show either vesicular type of localization (FL and D11) or endoplasmic reticulum (ER)‐retained (D50 and 4A) localization of PR1 variants. FL, full‐length PR1‐GFP; D50, PR1ΔC‐GFP; 4A, PR1YDPR/AAAA‐GFP; D11, PR1Δ11‐GFP. Insets depict a magnified view of the signal in the dashed squares. Bars = 10 μm. (b) Western blot analysis of PR1 variant expression using anti‐GFP antibody. The whole fusion protein bands (c.40 kDa) are indicated with a grey arrowhead. Lower bands are indicative of proteolysis, the blue arrowhead points to the smaller band corresponding to c.27 kDa free GFP, and the orange arrowhead points to the slightly bigger band with additional amino acids from PR1 C‐terminus, c.29 kDa. On the right: graph of the intensity distributions for two whole fusion constructs (grey bars) and cleaved bands (orange bars and blue bars for 29 and 27 kDa bands, respectively) for each of the variants. In the case of D11, the proteolysis gave only one GFP band of weight between 29 and 27 kDa, indicative of alternative proteolysis (orange–blue bar). The quantification shows enhanced proteolysis resulting in the strongest upper 29 kDa band in FL, especially in comparison to the D50 variant with the most prominent unspecific 27 kDa GFP band. The 4A construct was the least proteolysed
We analysed the localization of variants in N. benthamiana leaf epidermis, and we observed that the YDPR motif is crucial for the protein exit from ER (Figure 3a). In variant 4A, the protein was arrested within the ER, similar to the truncated variant D50. The pronounced difference in localization of FL and 4A could be explained by the effect of mutation on either the protein structure or on the supposed cleavage motif. The cleavage‐mimicking variant D11 exited the ER similarly to the FL, suggesting that the cleavage of CAPE peptide and progression in trafficking might be coupled. To further relate PR1 processing with observed localization, western blot analysis was performed. Using GFP‐specific antibody, we could detect the full‐length fusion proteins, and also the smaller GFP‐positive bands indicative of nonspecific GFP cleavage (c.27 kDa); in addition, we could detect a slightly larger one that corroborates the specific processing occurring near the C‐terminal end of PR1 (c.29 kDa; Figure 3b). For each of the constructs, the proteolysis was quantified as a distribution of the three bands in percentages and compared among variants (Figures 3b and S2). The proteolysis was partial for all constructs, but there was a prominent proteolysis of the FL (the least portion of the whole length band, 75.4%) as a consequence of the most prominent specific (23.9%) and minimal nonspecific cleavage (0.7%). The 4A construct was the least processed (98.8% for the whole fusion protein band) while the D50 construct, in accordance with the 3D structure prediction, was the most prominently nonspecifically proteolysed construct (16.6% of the smaller GFP band). These observations are consistent with the comparative analysis of conserved motifs in CAP proteins and the predicted importance of the YPDR motif for proteolysis (Chen et al., 2014; Chien et al., 2015).
Collectively, our data show the crucial role of the C‐terminal motif YDPR for the progression of PR1 trafficking from ER to MVBs, as well as the coincidental partial protein processing.
2.4. C‐terminus‐dependent extracellular and vacuolar trafficking destinations of PR1‐RFP variants
Because the anticipated trafficking destination for PR1 is the extracellular space, we also employed an alternative fluorescent tag, red fluorescent protein (RFP), which is not sensitive to the low pH of the cell wall (see Figure 4 for a schematic representation of constructs). Besides FL PR1‐RFP, we again prepared truncated versions of PR1 protein tagged with RFP, and we observed their localization. The extracellular signal was robust for RFP‐tagged FL and D11 variants in infiltrated N. benthamiana leaves. For D50 and 4A variants, no extracellular fluorescent signal was detected; instead vacuolar and ER localizations were found, similar to corresponding GFP‐tagged constructs (a comparison of the GFP‐ and RFP‐tagged constructs for each of the variants is shown in Figure 4b). In the case of RFP‐tagged variants, observation of intracellular structures was prevented by the overintensive extracellular signal. Confirmation of the extracellular/apoplastic signal for FL and D11 by western blotting is shown in Figure S3.
FIGURE 4.
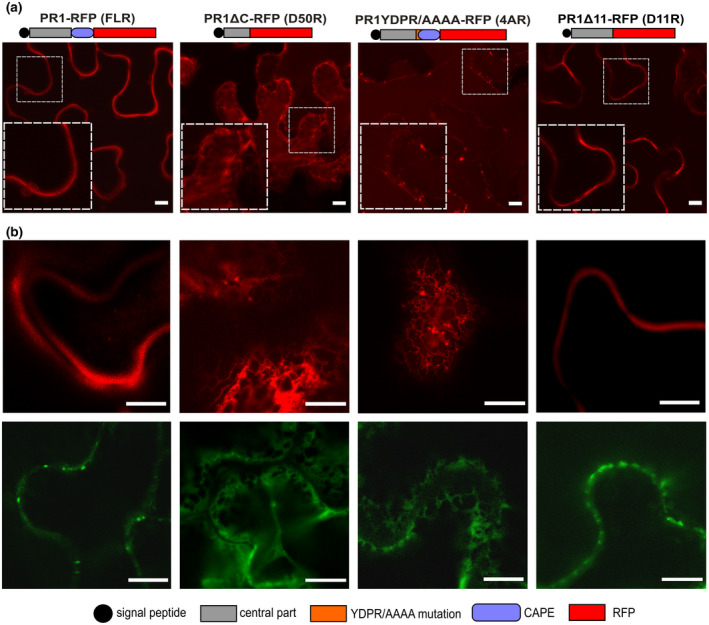
Subcellular localizations of red fluorescent protein (RFP)‐tagged PR1 variants. (a) Schematic representation of RFP‐tagged PR1 variants and their localization in infiltrated Nicotiana benthamiana leaves. Insets depict a magnified view of the signal in the dashed squares. A strong extracellular signal for FL and D11 variants, and endoplasmic reticulum and vacuolar localization of D50 and 4A was observed. In all cases a minor portion of the vesicular signal was detected as well. Bars = 10 μm. (b) The representative images showing corresponding localizations of FL, D50, 4A, and D11 variants tagged with RFP (upper row) and GFP (lower row). Bars = 100 μm
In addition to the confirmation of GFP‐ and RFP‐tagged PR1 variants localization congruency (overlapping, and due to the GFP pH sensitivity also complementary), these results confirm that in N. benthamiana epidermis only the wild‐type construct or the one that lacks the C‐terminus downstream from the supposed cleavage site reaches the extracellular space.
2.5. Trafficking of the GFP and RFP double‐tagged FL PR1 to MVB and extracellular space
To follow the intact nonmutated FL PR1 trafficking route, we had to take into consideration its partial C‐terminal proteolysis; therefore, the reported localization could also be assigned to either specifically or nonspecifically released C‐terminal tags. To simultaneously observe the cellular trafficking of putative proteolytically processed parts of PR1, we prepared a construct carrying GFP placed between the signal sequence and N‐terminus of the secreted PR1 part and RFP on its C‐terminus (for schematic see Figure 5a). We then followed the localization in a transient expression assay in N. benthamiana leaves and also in Nicotiana tabacum pollen tubes. These experiments confirmed and extended observations acquired using single‐tagged constructs. PR1 protein with both tags was present in both ER and MVB. Because the construct transit from ER to MVB was found to be coupled with C‐terminus processing, the presence of both tags in MVB is indicative of a common trafficking route of either the whole protein or both processed protein parts. Nevertheless, some accumulation of GFP‐labelled N‐terminus on the ER, and even in the cytoplasm and nucleus, is indicative of a partial separate N‐terminus trafficking pathway, which is also supported by the absolute absence of RFP signal from these compartments (Figure 5b). Unlike the PR1 with RFP tag only, the RFP‐part of the double‐tagged construct was also present in the vacuole (Figure 5b). Interestingly, in the pollen, both PR1 N‐ and C‐termini reached the vacuole and extracellular space; there was also a small portion of exclusively GFP‐positive cytoplasmic signal, supporting previous observations of GFP‐containing N‐terminal part leakage from the ER to the cytoplasm (Figure 5b). The accumulation of vacuolar and cytoplasmic signals for this construct may also be the consequence of slower and partially compromised protein processing due to the prominent fusion protein enlargement by the two fluorescent tags. The schematic conclusion for observed localizations, compiled with those for single GFP and RFP tags, is summarized in Figure 5c.
FIGURE 5.
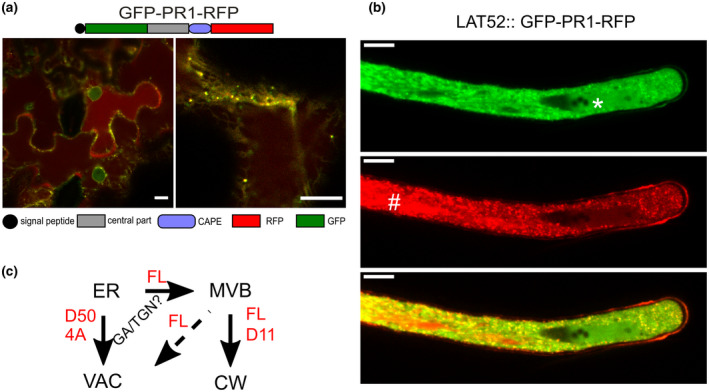
Double‐tagged full‐length PR1 construct carrying green fluorescent protein (GFP) placed between the signal sequence and N‐terminus of the secreted part and red fluorescent protein (RFP) on the C‐terminus transiently expressed in Nicotiana benthamiana leaves (a) or in germinating pollen of Nicotiana tabacum (b). The protein is present in both endoplasmic reticulum (ER) and multivesicular bodies (MVBs) (some vesicles are exclusively marked with a single tag), as well as in the extracellular space where the RFP tag dominates due to its pH insensitivity. Separate signals are observed for GFP in the nucleus and RFP in the vacuole, also in the case of pollen tube (nucleus marked by an asterisk and vacuole by a hash). Bars = 10 μm. (c) Scheme of PR1 trafficking. The full‐length PR1 progresses from ER through the secretory pathway via MVBs and subsequently to the extracellular space or vacuole. An involvement of the transit via the Golgi apparatus is not well supported by these observations. The constructs with prominent deletion (D50) or with severely mutated YDPR site (4A) are retained on the ER and/or reach the vacuole. The progress of PR1 through the trafficking pathways is coupled to the recognition and partial proteolysis of the YDPR motif, based on the fact that the D11 construct can reach the extracellular space as well
To conclude, these experiments with differentially labelled N‐ and C‐ termini tagged FL PR1 further prove its trafficking path from the ER via MVB and mostly to the extracellular space; in addition, compartments marked by only one fluorophore support the existence of the partial proteolytic processing.
2.6. PR1 variants differently modulate host plant defence in an age‐dependent manner
Next, we tested whether the full‐length PR1 and truncated and mutated variants differ in the immunity function in N. benthamiana leaves, in addition to their different subcellular localization and final destinations. To this end, we employed two well‐established pathogen model systems, tobacco mosaic virus (TMV) and nonvirulent bacterium P. syringae pv. tomato DC3000 hrcC (further referred to as hrcC; this strain is differentially selectable from Agrobacterium because it carries resistance to chloramphenicol). The variant localizations and the comparability of protein levels were verified by confocal microscopy and western blotting, respectively, and were found to be always consistent with the representative image in Figure 3.
We co‐expressed PR1 variants with pathogenic TMV carrying free RFP as a reporter of infection and quantified the virus spread (Figure 6a,b). In agreement with previous data on PR1 role in immunity, the expression of the full‐length PR1 construct significantly improved plant resistance towards TMV while truncated/mutated variants compromised plant immunity in adult N. benthamiana plants (Figure 6b). Interestingly, in young plants, the spread of TMV was impaired in the presence of D50 or D11 variants and enhanced with FL and especially 4A variants.
FIGURE 6.
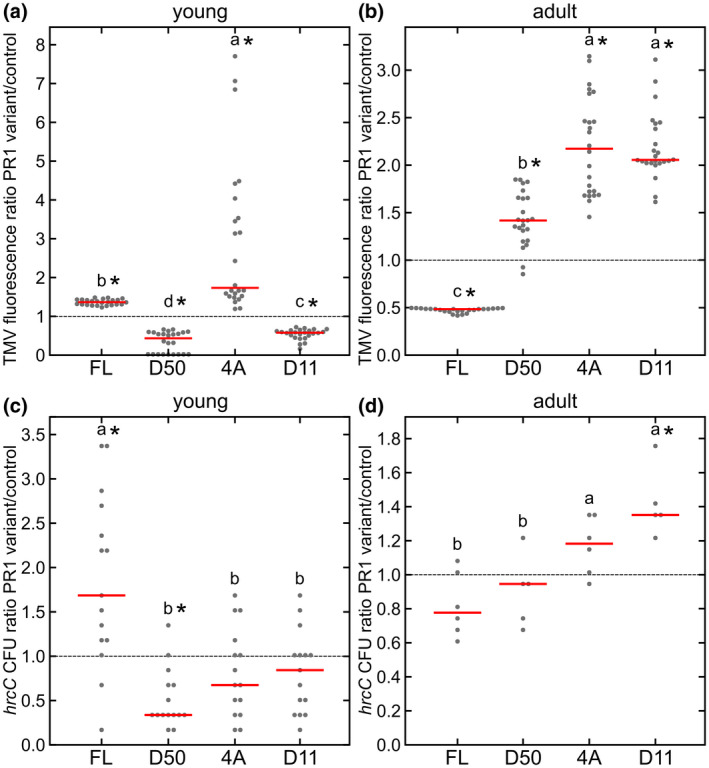
The analysis of green fluorescent protein (GFP)‐tagged PR1 variant influence on Nicotiana benthamiana immunity. Tobacco mosaic virus (TMV) spread quantified based on red fluorescent protein (RFP) in young (a) and adult (b) plants. Results for each variant are presented as TMV RFP fluorescence normalized to intensities found for control construct (value 1, dashed line); n = 24 for each of the constructs. Susceptibility of infiltrated leaves to Pseudomonas syringae pv. tomato DC3000 hrcC for young (c) and adult (d) plants for each of the variants, normalized by the control experiment with Agrobactrium tumefaciens with hrcC (value 1); n = 14 for young plants (left) and n = 6 for adult plants (right). Individual data points and median are shown. Values significantly different from control experiments are marked with asterisks, as determined by analysis of variance with post hoc Tukey HSD calculator. Different lower case letters indicate significant differences between variants
The same general trends were found for co‐inoculation of PR1 variants with P. syringae hrcC. The defence of younger plants was unexpectedly worse for the FL variant, while the D50 variant significantly improved the plant´s resistance (Figure 6c,d). In the case of older N. benthamiana plants transiently expressing variants, FL PR1 seemed to slightly improve resistance compared to control combination of Agrobacterium and hrcC (set to value 1), but were not significantly different from the D50 truncated variant (Figure 6d). Consistently, in both TMV and hrcC experiments, the FL and processing‐mimicking variant D11 effects on immunity were in opposition to each other.
We then tested how much of the described immunity effects of FL PR1 is executed by the C‐terminal CAPE peptide alone. For this purpose, we tested a construct carrying the PR1 signal peptide (amino acids 1–26) fused to the N‐terminus of GFP, and the PR1 C‐terminal 15 amino acids (i.e., 11 amino acids of CAPE preceded by the cleavage motif; variant SGC) fused to the C‐terminus. The construct carrying the signal peptide of PR1 fused to the N‐terminus of GFP was used as an additional control (variant SG; see also Figure S4a for schematic representation). SGC, the CAPE‐containing variant, significantly compromised the defence of young plants and significantly improved adult plant defence (Figure S4b), unlike the SG variant (presented in comparison to control plants treated with Agrobacterium/hrcC), which is also consistent with immunity features of construct D11.
We also tested whether the effect of PR1 variants containing cleavable CAPE (FL, SGC) on the defence in young plants may be related to the changes in cell death. Indeed, the co‐expression of CAPE‐containing PR1 variants with hrcC led to a significant decrease of trypan blue‐positive cells, indicating CAPE‐dependent suppression of cell death (Figure S4c; no differences were observed for adult plants expressing construct variants, data not shown). Because the presence of the synthetic CAPE peptide had no inhibitory effect on hrcC growth in vitro (Figure S4d), we could assign the effect of the CAPE on plant cell death to the modification of the plant defence responses.
To summarize, our assays of TMV spread and P. syringae hrcC propagation in infiltrated N. benthamiana leaves confirm the immunity‐modulating function for all of the GFP‐tagged PR1 variants. The positive or negative effect of PR1 depends on the plant developmental stage, that is, in younger plants PR1 may contribute to bacterial progress, probably due to cell death suppression. Cell death suppression is not an exclusive PR1 function, because C‐terminus truncated and mutated variants were also capable of contributing differentially to plant immunity.
3. DISCUSSION
In this work, we performed a comparative analysis of localization, trafficking, proteolysis, and immunity effects of A. thaliana PR1 and its truncated/mutated variants transiently expressed in N. benthamiana. We employed a transient overexpression approach to mimic the highly enhanced PR1 expression and overload of the secretory pathway that occurs during biotic stresses (Jelitto‐Van Doren et al., 1999). Our data document the complex trafficking of PR1 in N. benthamiana epidermis mainly to the two cellular compartments: cell wall/apoplast and, in case of functional and structural disturbances (by truncation, mutation or N‐terminally fused tag), also to the vacuole (schematically summarized in Figure 5c). From the behaviour of all tested truncated and mutated variants, we conclude that the progress from ER to MVB (GFP) and extracellular space (RFP) is tightly related to the presence of the YD motif in the supposed YD‐PR cleavage site, with its upstream PR1 amino acid context. The motif intactness may be required for proper protein processing, as concluded from the localization of D11 comparable to FL; on the other hand, the quantity of proteins trafficked from the ER to MVBs does not reflect the quantity of the processed portion. Variants with differently deleted N‐termini should be further employed to verify the importance of the YDPR motif and upstream context, and possibly the importance of disulphide bonds for the structure and trafficking. Because all these observations are valid for N. benthamiana epidermal cells, it should be examined if they are also applicable to A. thaliana.
The preference of PR1 for co‐localization with plant‐specific RabF GTPase ARA6 subpopulation of MVBs is in agreement with the suggested defence‐related function that might also involve a preferential ARA6 activity targeting to the apoplast over ARA7‐regulated vacuolar transport/import (Bourdais et al., 2019; Ebine et al., 2011; Inada et al., 2016; Ito et al., 2016). The absence of PR1 co‐localization with FM4‐64 and insensitivity of MVB‐localized PR1 to endocytosis inhibition (Figure S1b) demonstrate that PR1 localization to MVBs is not a result of retrograde transport of the secreted PR1. However, we cannot rule out that a proportion of the PR1 follows the canonical secretory route (Pečenková et al., 2017), which is supported by the partial colocalization of PR1 with Golgi apparatus markers (Pečenková et al., 2017) and by the dependence of PR1 trafficking on Golgi SNAREs BET12 and MEMB12 in specific plant cell types (Chung et al., 2018).
Our results obtained from PR1 co‐immunoprecipitation using overexpressing A. thaliana seedlings confirm the strong association of the overproduced protein with ER‐related chaperone CDC48 that, besides its up‐regulation by treatment with pathogens, is also related to ER stress, as well as to MVB loading in yeast (Kama et al., 2018; Figure S5). It remains to be clarified whether the PR1‐positive MVB compartment is related to ER bodies reported as a resident PR1 intracellular compartment, and a source for apoplastic PR1 in response of Arabidopsis to the nonadapted pathogenic fungus Colletotrichum gloeosporioides (Watanabe et al., 2013). The observed protein accumulation in either ER bodies or MVBs could be also interpreted as an artefact produced by protein overexpression (Saberianfar & Menassa, 2017). Nevertheless, our results reproducibly pinpoint the strict rules of this protein trafficking that are affected by the disruption of protein motifs, which might also reflect the changes of localization patterns occurring on pathogen attack (Nakano et al., 2014; Tomczynska et al., 2018).
We could not confirm the actual physical contact of PR1‐positive MVBs with the PM and extracellular space using electron microscopy, despite repeatedly observing their tight appression in vivo. Although contact sites between MVBs and the PM have been recently shown in an artificial bimolecular fluorescence complementation assay, so far there is a lack of evidence for direct crosstalk of endogenous MVBs and the PM (Hansen & Nielsen, 2017; Tao et al., 2019).
We found evidence of partial proteolytic processing, which is consistent with findings by Chen et al. (2014), Breen et al. (2017) and Sung et al. (2021). It remains to be further elucidated if this is a consequence of excessive protein expression and exhaustion of processing machinery, or whether it really reflects a more limited need for CAPE peptide than for the full‐length protein. In addition, in all of our western blot analyses, we often noticed bands of proteins with increased size, indicative of multimerization, which has been confirmed in the case of homologs of PR1 in other species (Lu et al., 2013; Sheng, Olrichs, Geerts, Kaloyanova, et al., 2019; Shen, Olrichs, Geerts, Li, et al., 2019). The phenomenon of dimerization and formation of sodium dodecyl sulphate‐resistant complexes remains to be further elaborated.
The bacterial immunity assays showed a clear trend for FL and D11 constructs to have opposite effects on plant immunity, indicating the importance of CAPE presence for immunity function. We speculate that, in accordance with previous findings (Betsuyaku et al., 2018; Lincoln et al., 2018), the CAPE contributes to young plant susceptibility by cell death suppression, thus providing a more favourable environment for bacterial amplification. In older plants, the effect of cell death suppression may not be strong enough to suppress the endogenous age‐related activation of senescence. However, PR1 may also have a cell‐death‐independent immunity function. For instance, both FL and 4A variants suppressed the cell death spread in young plants, but unlike FL the 4A suppressed bacterial accumulation; these discrepancies remain to be further analysed. Our immunity assays also revealed that the PR1 C‐terminus (CAPE) is not the only PR1 fragment responsible for its activities. N‐terminal and central parts of PR1 are also important and capable of affecting plant defence, which is best demonstrated by the immunostimulatory activity of the D50 variant in younger plants. Interestingly, regardless of plant age and TMV or P. syringae hrcC infection, the intracellular localization of PR1 remained unchanged (data not shown). It should be noted that the results of these experiments must be interpreted with caution due to the fact that the combinations of applied pathogens may also have unexpected effects on endogenous immunity. Our reverse transcription PCR analysis showed that the levels of endogenous N. benthamiana PR1a were up‐regulated at 48 hours postinfection (hpi) even by the immunologically inert control construct SG; similarly, 48 hpi for hrcC were required for NbPR1a up‐regulation in adult plants, confirming the importance of PR1‐defence in anti‐Pseudomonas salicylic acid and/or age‐related immunity (Figure S6; Kus et al., 2002). In addition, there were no differences in range of NbPR1a activation among constructs. Our observations are in agreement with Pruss et al. (2008) who have shown that Agrobacterium tumefaciens activates defence in N. benthamiana, which should be further addressed by separate studies.
There are no reliable Arabidopsis T‐DNA insertional knockout mutants available and, if so, the study of a single mutant would be inconclusive due to the existence of an additional 21 PR1‐like proteins and their putative redundancy. Our extensive evolutionary tree reconstruction revealed that among them, a jasmonate‐responsive AtPRB1 (Santamaria et al., 2001) and several other pollen‐ and abiotic stress‐related proteins (Figure S7, in blue letters), are the most closely related to AtPR1, which may be relevant for future studies.
To conclude, PR1 trafficking from the ER to the cell wall or vacuole is coupled to the YDPR motif intactness, which is also possibly important for the C‐terminal‐dependent proteolysis. The full‐length protein, as well as its processed peptides, executes local defence‐related apoplastic signalling, the outcome of which depends on the plant age and may include cell death containment.
4. EXPERIMENTAL PROCEDURES
4.1. Plant material
N. benthamiana plants were propagated in a greenhouse in soil, under 16/8 h light/dark conditions and ambient temperature. The seeds of A. thaliana Col‐0 were surface‐sterilized and plated onto half‐strength Murashige and Skoog medium (1/2×MS). The plants were propagated in vitro for 7 days (23°C, 16/8 h light/dark) and used for experiments or transferred to Jiffy tablets and cultivated in a growth chamber (23°C, 14/10 h light/dark) for 5–6 weeks.
4.2. Cloning procedure
For preparation and cloning of new PR1 (At2g14610) construct variants, a previously described clone of full‐length PR1 (Pečenková et al., 2017) was used as a template. A list of primers enabling cloning into pENTR3C, pTNT (for in vitro translation system), and pHD223 (carrying pollen‐specific Lat52 promoter, Klahre et al., 2006) is presented in Table S1. Standard restriction/ligation cloning procedures were used. Cloned constructs were transferred from an Entry vector by a recombinant LR reaction of Gateway cloning (Invitrogen) into pGWB2 (for CaMV 35S promoter‐driven expression) and pGWB5 (for GFP‐fusion and 35S‐driven expression) vectors (Nakagawa et al., 2007), and pK7RWG2 for RFP‐fusion variants (Karimi et al., 2002). To prepare the construct for the endocytic inhibitor (RFP‐HUB), first AtHUB was amplified and cloned together with the ubiquitin promoter and RFP into the binary vector pHD71 (kindly provided by Benedikt Kost, Erlangen). Construct CDC48b‐RFP was obtained from Manfred Heinlein (Strasbourg) (Niehl et al., 2012), constructs RFP‐ARA6 and RFP‐ARA7 from Falco Kruger (Heidelberg), and SEC10‐mCherry construct for recloning into the pFRETgc‐2in1‐NN multicistronic vector (Hecker et al., 2015) was obtained from Jitka Ortmannová (Prague).
4.3. Transient expression in N. benthamiana
Prepared constructs were used for the transformation of Agrobacterium tumefaciens GV3101. Overnight cultures were diluted in the infiltration buffer (for microscopic analysis and bacterial assays in 150 mM MgSO4, 100 μM acetosyringone) and adjusted to OD600 0.1. After incubation on the bench at room temperature for 2 h, these cultures were used for infiltration by syringe on the abaxial N. benthamiana leaf surface. For the immunity assays, young plants at the stage of two to four true leaves (c.2 weeks old) and adult plants in the stage eight to 10 true leaves (c.4 weeks old) were used.
4.4. Confocal microscopy
For live‐cell imaging, either infiltrated parts of N. benthamiana leaves (48 hpi) or 5–7‐day‐old seedlings were used for observation under a Zeiss LSM880 confocal microscope using 63× oil immersion objective. For cell wall visualization, propidium iodide (Sigma‐Aldrich) 50 μg/ml, dissolved in water, was used. Plasma membrane and endosomes were visualized using 5 μM FM4‐64 (Invitrogen). For Brefeldin A (BFA) treatment, seedlings were transferred from agar plates into liquid 1/2×MS medium supplemented with 50 μm BFA (Sigma‐Aldrich) and incubated for the indicated time at room temperature. Excitation wavelengths used were 488 nm for GFP and 561 nm for RFP, mCherry, FM4‐64 and propidium iodide. The images were analyzed using Zen 2.1 Software (Carl Zeiss GmbH) and Fiji/ImageJ (Schindelin et al., 2012). Pollen transformation and the microscopic analysis of pollen tube growth was performed according to Pejchar et al. (2020).
4.5. Immunogold labelling and transmission electron microscopy
The immunogold labelling for transmission electron microscopy was performed according to Vancová et al. (2019), with modifications. Briefly, freshly harvested material was fixed in a 4% paraformaldehyde + 0.1% glutaraldehyde (c.2 h at 4°C), dehydrated through an ascending ethanol series (30%–100% ethanol). Samples were embedded in LR White resin. Thin sections (70 nm) were cut on a Reichert–Jung Ultracut E Ultramicrotome and used on Formvar/carbon‐coated Ni grids. The sections were treated with blocking buffer composed of 1% fish skin gelatin and 0.05% Tween 20 (Sigma‐Aldrich) dissolved in phosphate‐buffered saline, and then incubated for 1–2 h with anti‐GFP antibodies (AS153001; Agrisera) diluted 1:50. Secondary antibody conjugates (anti‐rabbit 10 nm gold; Aurion) were diluted 1:50 in blocking buffer and incubated with samples for 1 h at room temperature. Controls for nonspecific binding of the secondary antibody were performed by omitting the primary antibody. The sections were analysed and photographed using a JEM‐1011 electron microscope with a Megaview III camera and analysed in ImageJ (Schindelin et al., 2012).
4.6. SDS‐PAGE and western‐blot
Total protein extracts were performed from leaf discs (6 mm diameter, four discs per leaf) of infiltrated N. benthamiana, or from 4–6‐week‐old A. thaliana rosette leaves, using either phospate‐buffered saline or extraction buffer as described in Hála et al. (2008). Usually, 12% polyacrylamide gels were used. Proteins were blotted onto a nitrocellulose membrane or polyvinylidene difluoride membrane (quality of transfer and load control was checked by Ponceau S staining) and blocked overnight at 4°C with 5% nonfat dry milk in phosphate‐buffered saline. Primary antibodies anti‐PR1 (AS10687; dilution 1:1000; Agrisera), anti‐GFP (AS152987; dilution 1:1000; Agrisera), anti‐HA (26183; dilution 1:1000; Thermofisher Scientific), and anti‐RFP (W4021; 1:5000; MBL) were incubated with the membranes for 3 h at room temperature in the blocking solution. Horseradish peroxidase‐conjugated antibodies (anti‐rabbit and anti‐mouse; Promega) were applied followed by chemiluminescent ECL detection (Amersham) by the Bio‐Rad documentation system. Using the Gel Analysis function of ImageJ, signal intensities for protein bands were determined for each construct from five or six different samples (each sample was prepared from two to four leaf discs from one or two infiltrated leaves).
4.7. Virus inoculation
Fully expanded leaves of N. benthamiana were agroinfiltrated as described in Cerovska et al. (2012), using TMV expressing GFP (GenBank accession number KF981446). On each plant, three fully developed leaves were infiltrated with 200 µl of Agrobacterium suspension (OD600 = 0.2 in infiltration solution 10 mM 2‐(N‐morpholino)ethanesulfonic acid, 100 μM acetosyringone, 10 mM MgCl2). Duplicate samples (1 cm) were collected from each plant under UV illumination to ensure processing of only virus‐infected tissue. Leaf tissue was homogenized in 400 μl of phosphate‐buffered saline using ceramic beads and a FastPrep 24 instrument, and total protein content was measured using total protein assay (BioRad). Samples were then equilibrated to 1 mg/ml total protein concentration and GFP fluorescence measured using an Infinite F200 instrument (Tecan).
4.8. Bacterial sensitivity assays
For assessment of a construct impact on plant immunity, 24 hpi with Agrobacterium carrying PR1 variants, P. syringae pv. tomato DC3000 mutant strain hrcC (donated by Chris Staiger, West Lafayette, USA; Yuan & He, 1996) in OD600 = 0.05 (in distilled water), was infiltrated, into two or three leaves from two or three plants for each construct. After 24 h, an equal number of leaf discs was excised from infiltrated leaves for each variant and the surface was sterilized with 70% ethanol and homogenized using Bead Ruptor Elite (steel beads 5 mm, two cycles of 10 s, 4.5 m/s; Omni International). A series of dilutions was plated onto Luria–Bertani (LB) medium (Duchefa) with rifampicin (25 µg/ml) and chloramphenicol (34 µg/ml) and left for approximately 30 h until the colonies became visible and countable. The number of colonies (colony‐forming units, cfu) was normalized to the corresponding control (Agrobacterium/hrcC, further on abbreviated as “A”, approximately 9.2 × 105 cfu/cm2 for young plants [two‐ to four‐leaf stage] and 1.5 × 105 cfu/cm2 for adult plants in stages prior to flowering). The expression levels of construct variants from leaf discs were verified as being comparable by western blot (as in section 2.6).
4.9. Trypan blue staining
To visualize cell death spread, N. benthamiana leaves were boiled for 3 min in a 1:1 (vol/vol) mixture of ethanol:trypan blue staining solution (10 ml lactic acid, 10 ml glycerol, 10 g phenol, and 10 mg trypan blue, dissolved in 10 ml distilled water). The leaves were then destained in 2.5 g/ml chloral hydrate in distilled water.
4.10. The construction of 3D models of PR1 variants
The structural models for the wild‐type and all studied PR1 deletion/mutation variants were built independently using the Robetta ab initio algorithm (Webb & Sali, 2016). We chose this approach to avoid the template‐based topological constraints that typically occur in homology modelling, which would mask the potential structural defects in the mutated variants. To validate the ability of the ab initio approach to predict the correct structure, the structural model for the PR1 FL variant was also independently constructed using homology modelling with tomato and yeast PR1 orthologs as templates (pdb ids 1CFE and 5ETE, respectively). The best models from both approaches were very similar, with backbone root‐mean‐square‐deviation (RMSD) values <3 Å.
4.11. RNA isolation and reverse transcription PCR
RNA was isolated from young and adult N. benthamiana leaves (always pooled infiltrated parts from two leaves) 48 hpi with Agrobacterium/constructs and 24 hpi with hrcC, additionally from leaves at 48 hpi with hrcC. The RNeasy Plant kit (Qiagen) was used according to the manufacturer's instructions. The obtained RNA (500 ng to 1 μg) was subjected to reverse transcription using an oligo(dT) primer (Transcriptor High Fidelity cDNA Synthesis Kit; Roche) according to the manufacturer's instructions. PCR was performed using OneTaq DNA polymerase (NEB) in 25–28 cycles. The primers used for reverse transcription PCR analysis were designed according to Wu et al. (2017) (EF1α‐F: 5′‐ATGATTACTGGTACCTCCCG‐3′, EF1α‐R: 5′‐ACCTAGCCTTGGAATACTG‐3′, PR1a‐F: 5′‐CGTTGAGATGTGGGTCAATG‐′ and PR1a‐R: 5′‐CCTAGCACATCCAACACGAA‐3′). Similar results were obtained for each of the three repetitions of the RNA isolation experiment.
4.12. Apoplastic fluid isolation
The method of apoplastic fluid isolation from infiltrated N. benthamiana was performed according to Joosten (2012) with modifications. Briefly, two full‐size infiltrated leaves were cut off and immersed in 50 ml of cold distilled water with plant protease inhibitors (Sigma). Vacuum was applied for 2–5 min, then, after vacuum release, the leaves were surface‐dried with paper, wrapped in Parafilm sheets, and placed petal side up in a centrifuge tube. After centrifugation (1000 × g, 10 min, 4°C), around 300 μl of apoplastic fluid was gained and proteins precipitated overnight with acetone on −20°C, centrifuged and diluted in 50 µl of water. Typically, 0.1 mg/ml of protein was recovered per leaf.
4.13. Assessment of antibacterial CAPE peptide activity
P. syringae hrcC was grown overnight on plates containing LB medium (with antibiotics rifampicin and chloramphenicol as described above) at 28°C. Bacteria were resuspended in liquid LB and diluted to final OD600 0.001. Synthetic C9 peptide (Vidia; Vestec) was added at final concentrations of 100, 10, 1, and 0 µM and incubated at 28°C for 2 h with slight agitation (80 rpm). The reaction was carried in three parallels in multiwell plates. Serial dilutions were spread onto LB plates with rifampicin (25 µg/ml) and chloramphenicol (34 µg/ml), and left for approximately 30 h until the colonies became visible and countable.
[Correction added on 15 February 2022, after first online publication: the section 4.13 heading has been updated in this version.]
4.14. Co‐immunoprecipitation
Arabidopsis seedlings (1 g of 5–7‐day‐old) were used for the co‐immunoprecipitation of proteins. Extracts were prepared in Sec6/8 buffer (Hála et al., 2008; 20 mM HEPES, pH 6.8, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 0.5% Tween 20; supplemented with 1× protease inhibitor cocktail; Sigma‐Aldrich) giving a concentration of protein of 5–15 mg/ml, and used for incubation with an in vitro synthesized bait. Protein complexes with GFP‐tagged baits were isolated using a µMACS GFP‐tagged protein isolation kit (Miltenyi Biotec), while those with HA‐tagged baits were isolated with a Pierce HA‐Tag Magnetic IP/Co‐IP Kit (Thermo Scientific), with co‐incubation with total protein extract prepared from A. thaliana adult rosette leaves, in both cases according to the manufacturer's instructions. Eluates were resolved on a 12% polyacrylamide gel and further processed by mass spectrometry.
4.15. Mass spectrometry
Each of the lanes with resolved eluates was cut into four bands. After in‐gel digestion with trypsin, eluted peptides were identified using UHPLC Dionex Ultimate3000 RSLC nano (Dionex) connected to an ESI‐Q‐TOF Maxis Impact mass spectrometer (Bruker). Measurements were carried out in positive ion mode with precursor ion selection in the range 400–1400 mass‐to‐charge ratio; up to 10 precursor ions were selected for fragmentation from each mass spectrum. Peak lists were extracted from raw data by Data Analysis v. 4.1 (Bruker Daltonics) and uploaded to the data management system Proteinscape (Bruker Daltonics). For protein identification, the Mascot server (v. 2.4.1; Matrix Science) was used with a custom‐made database containing A. thaliana sequences (downloaded from Uniprot website on 11 October 2016; 33,705 sequences) complemented with common laboratory contaminants. The following parameters were set during searches: enzyme trypsin, one allowed missed cleavage, tolerance 10 ppm in MS mode and 0.05 Da in MS/MS mode, carbamidomethylation of cysteines was set as a fixed modification and oxidation of methionines as a variable modification, Mascot decoy search was used to calculate false discovery rate (FDR). Identified proteins were filtered so that final FDR was 1%. The obtained Mascot results were also devoid of statistically supported, false‐positive interactors obtained with GFP tag alone (Ortmannová et al., 2021).
4.16. Fluorescence resonance energy transfer analysis
Analysis was performed on leaves of N. benthamiana expressing PR1‐GFP and CDC48b‐RFP either individually or in combination. The quantification of fluorescence resonance energy transfer was performed according to Feige et al. (2005) using five to 10 images for each of the two controls and the PR1‐GFP and CDC48b‐RFP combination. The statistical analysis was performed in Excel using the t test as verification of statistical significance.
4.17. Phylogenetic analysis
The tree was constructed based on the alignment of all proteins containing CAP domains of selected species. Sequences were aligned using MUSCLE and manually trimmed. Phylogenetic analysis was performed using the neighbour‐joining method with 500 bootstrap replicates.
Supporting information
FIGURE S1 PR1‐GFP localization in seedlings of Arabidopsis thaliana. (a) Localization of PR1‐GFP in root cells in a typical MVB‐like compartment (left panel). Bar = 10 μm. Root cells of A. thaliana expressing PR1‐GFP (green) and propidium iodide (PI; 10 μg/ml) stained cell walls (red; middle panel). Bar = 5 μm. PR1‐GFP and PI signals zoomed and visualized using processed three‐dimensional reconstruction of Z stacks acquired with airy scan mode of confocal microscopy (right panel). Bar = 1 μm. (b) Minor co‐localization of the PR1‐positive compartment with endocytotic dye FM4‐64 (1 μg/ml) stained compartment (left panel, pointed by arrowheads). PR1‐GFP compartment is not affected by BFA (50 μM) and only partially co‐localizes with FM4‐64 stained BFA bodies (middle panel, arrowheads); PR1‐GFP and FM4‐64 stained BFA bodies depicted in more detail using higher magnification (right panel). Bars = 10 μm
FIGURE S2 Examples of the western blot analysis performed with leaf extracts from infiltrated Nicotiana benthamiana, using GFP‐specific antibody. (a) Detection of the full‐length fusion proteins and also the smaller GFP‐positive band. FL, full‐length PR1‐GFP; D50, PR1ΔC‐GFP; 4A, PR1YDPR/AAAA‐GFP; D11, PR1Δ11‐GFP on the left. Western blot of the GFP control constructs with signal peptide (SP)‐GFP only (variant SG) and SP‐GFP‐CAPE (variant SGC) with CNYDPRGNYVNEKPY peptide on C‐end (SGC) on the right. (b) Two different exposition times of western blot for SG, FL, D50, 4A, D11, and SGC constructs, showing the difference in expression levels of SG and SGC constructs versus PR1 variants; the red arrowhead points to the probable bands of GFP with fused PR1 C‐terminus
FIGURE S3 Confirmation by western blot for extracellular/apoplastic signal for FL and D11. The upper row shows total cell extracts from Nicotiana benthamiana expressing RFP‐tagged PR1 variants: full‐length, FLR (PR1‐RFP), D50R (PR1ΔC‐RFP), 4AR (PR1YDPR/AAAA‐RFP), and D11R (PR1Δ11‐RFP). The lower rows show the apoplastic fluid from leaves infiltrated with the same constructs. The antibody recognizes cleaved RFP tag only for FLR and D11R, in agreement with confocal observations. However, the presence of their uncleaved variants in apoplastic fluid is detectable using anti‐PR1 (Agrisera; unspecific recognition of bands of c.50 and 30 kDa is observable for all variants). Ponceau S staining shows minor contamination with cytoplasmic RuBisCO, equal for all analysed samples. The FLR variant is prone to the formation of SDS‐resistant higher molecular weight multimers. The prominent band seen between 25 and 35 kDa with anti‐PR1 antibody (Agrisera) is a product of unspecific interaction observable with the extracts enriched for proteins from the cell wall and plasma membrane fractions from both Arabidopsis and Nicotiana
FIGURE S4 PR1C‐terminal CAPE peptide effect on localization and immunity. (a) Schematic representation of signal peptide (SP)‐GFP (variant SG) and SP‐GFP‐CAPE (variant SGC) fusion proteins, their intracellular localizations, and effects on antibacterial plant immunity in transiently transformed Nicotiana benthamiana leaves. Both constructs demonstrate endoplasmic reticulum (ER)‐retained localization (with supposed cleavage and further trafficking of CAPE in case of SGC). Bars = 10 μm. (b) Graph shows susceptibility toward hrcC, expressed as ratios of cfu quantified for SG and SGC versus control A (Agrobacterium tumefaciens with P. syringae hrcC; value 1), counted for young and adult plants. Peptide presence significantly enhances susceptibility in younger plants and resistance in adult plants (n = 6). Significantly different values determined by analysis of variance (ANOVA) with post hoc Tukey HSD are marked with asterisks. Bars = 10 μm. (c) Trypan blue staining intensities of each of employed PR1 variants, expressed as a ratio of stained area toward the total analysed infiltrated area. Representative images of trypan blue stained leaves of N. benthamiana expressing PR1 variants are shown above the graph for each of the variants, as well as the control Agrobacterium/hrcC. Different small letters indicate significant differences between variants determined by ANOVA with post hoc Tukey HSD calculator (n = 6). Infiltrated variants: A, Agrobacterium/hrcC infiltrated control; FL, full‐length PR1‐GFP; D50, PR1ΔC‐GFP; 4A, PR1YDPR/AAAA‐GFP; D11, PR1Δ11‐GFP; SG, SP‐GFP; SGC, SP‐GFP‐CAPE. Bar = 100 μm. (d) cfu of hrcC cultivated in liquid medium in the presence of different concentrations of synthetic peptide CAPE. The peptide has no significant influence on in vitro growth of hrcC bacteria
FIGURE S5 Detection of proteins possibly interacting with PR1 using the co‐immunoprecipitation method. (a) Table 1. Several putative protein interactors found in co‐immunoprecipitation experiments with PR1‐GFP overexpressed in Arabidopsis thaliana. (b) Verification of PR1‐CDC48 interaction using fluorescence resonance energy transfer (FRET) analysis. Analysis was performed on leaves of Nicotiana benthamiana co‐expressing PR1‐GFP with CDC48b‐RFP. Bars = 20 μm. The graph shows the quantification of FRET analysis (according to Feige et al., 2005) performed using five to 10 images for each of the two controls and the PR1–GFP and CDC48b–RFP combination. The significant positive FRET was detected in case of PR1‐GFP and CDC48b‐RFP
FIGURE S6 Reverse transcription (RT)‐PCR analysis of NbPR1a up‐regulation. (a) The RT‐PCR was performed for mock, hrcC‐infiltrated, and leaves infiltrated with hrcC together with control construct SG, construct SGC, as well as FL and D50 constructs which have opposite effects on plant immunity. EF1‐a gene was used as a housekeeping control. (b) Appearance of plants 24 hr postinoculation (hpi): different constructs at this stage do not cause differences in symptoms development on sites of infiltration. (c) RT‐PCR 48 hpi with hrcC in comparison to mock infiltration; NbPR1a is up‐regulated in adult leaves. (d) The appearance of lesions in 5 days postinoculation with hrcC
FIGURE S7 Phylogenetic tree of PR1‐like proteins. The phylogenetic tree was constructed based on the alignment using all proteins containing CAP domains of selected species. Sequences were aligned using MUSCLE and manually trimmed. Phylogenetic analysis has been performed using the neighbour‐joining method with 500 bootstrap replicates. CAP domain‐containing proteins are present throughout whole land plants evolution with subsequent repeated gene amplification. Based on our evolutionary tree, PR1‐like proteins might have originally pollen‐ and abiotic‐stress‐related functions which later on evolved also to a defence function (deduced based on RNA expression pattern [TAIR, Genevestigator], no function was assigned to other closely related paralogs). The most closely related to AtPR1 is AtPRB1, which participates in jasmonate‐related defence (Santamaria et al., 2001). Ambtr, Amborella trichopoda; Arath, Arabidopsis thaliana; Brara, Brassica rapa; Capgr, Capsella grandiflora; Poptr, Populus trichocarpa; Theca, Theobroma cacao; Vitvi, Vitis vinifera
TABLE S1 List of primers used for cloning
ACKNOWLEDGEMENTS
The authors would like to thank to Falco Kruger (Heidelberg) for providing RFP‐ARA6 and RFP‐ARA7 constructs, Manfred Heinlein (Strasbourg) for the CDC48b‐RFP construct, Kateřina Malinská for confocal microscopy support, Jana Šťovíčková for technical assistance, Jitka Ortmannová for providing the SEC10‐mCherry construct in 2in1 vector, Ludmila Businská for N. benthamiana plant cultivation and propagation, Miroslav Hyliš for sample preparation guidance and transmission electron microscopy service, Roman Pleskot for help with figure design, and Vladimír Pečenka for critical reading of the manuscript.
Pečenková, T. , Pejchar, P. , Moravec, T. , Drs, M. , Haluška, S. , Šantrůček, J. , et al (2022) Immunity functions of Arabidopsis pathogenesis‐related 1 are coupled but not confined to its C‐terminus processing and trafficking. Molecular Plant Pathology, 23, 664–678. 10.1111/mpp.13187
Funding information
This research was funded by the Ministry of Education, Youth and Sports of the Czech Republic (MŠMT) project and European Regional Development Fund project CZ.02.1.01/0.0/0.0/16_019/0000738 “Centre for Experimental Plant Biology”. Light microscopy was done at the Imaging Facility of the Institute of Experimental Botany AS CR supported by the MEYS CR (LM2018129 Czech‐Bioimaging) and IEB AS CR. The authors have no conflicts of interest to declare
Contributor Information
Tamara Pečenková, Email: pecenkova@ueb.cas.cz.
Martin Potocký, Email: potocky@ueb.cas.cz.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Agrios, G.N. (2005) How plants defend themselves against pathogens. In: Agrios, G.N. (Ed.), Plant Pathology. Academic Press, 207–248. [Google Scholar]
- Antoniw, J.F. & Pierpoint, W.S. (1978) Purification of a tobacco leaf protein associated with resistance to virus infection. Biochemical Society Transactions, 6, 248–250. [DOI] [PubMed] [Google Scholar]
- Betsuyaku, S. , Katou, S. , Takebayashi, Y. , Sakakibara, H. , Nomura, N. & Fukuda, H. (2018) Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector‐triggered immunity in Arabidopsis thaliana . Plant and Cell Physiology, 59, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudart, G. , Jamet, E. , Rossignol, M. , Lafitte, C. , Borderies, G. , Jauneau, A. et al. (2005) Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: identification by mass spectrometry and bioinformatics. Proteomics, 5, 212–221. [DOI] [PubMed] [Google Scholar]
- Bourdais, G. , McLachlan, D.H. , Rickett, L.M. , Zhou, J. , Siwoszek, A. , Häweker, H. et al. (2019) The use of quantitative imaging to investigate regulators of membrane trafficking in Arabidopsis stomatal closure. Traffic, 20, 168–180. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A. , Guo, A. , Cao, H. , Gordon, A.S. , Klessig, D.F. & Dong, X. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. The Plant Cell, 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen, S. , Williams, S.J. , Outram, M. , Kobe, B. & Solomon, P.S. (2017) Emerging insights into the functions of pathogenesis‐related protein 1. Trends in Plant Science, 22, 871–879. [DOI] [PubMed] [Google Scholar]
- Breen, S. , Williams, S.J. , Winterberg, B. , Kobe, B. & Solomon, P.S. (2016) Wheat PR‐1 proteins are targeted by necrotrophic pathogen effector proteins. The Plant Journal, 88, 13–25. [DOI] [PubMed] [Google Scholar]
- Camacho Henriquez, A. & Sänger, H.L. (1982) Analysis of acid‐extractable tomato leaf proteins after infection with a viroid, two viruses and a fungus and partial purification of the “pathogenesis‐related” protein p14. Archives of Virology, 74, 181–196. [DOI] [PubMed] [Google Scholar]
- Carella, P. , Merl‐Pham, J. , Wilson, D.C. , Dey, S. , Hauck, S.M. , Vlot, A.C. et al. (2016) Comparative proteomics analysis of phloem exudates collected during the induction of systemic acquired resistance. Plant Physiology, 171, 1495–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerovska, N. , Hoffmeisterova, H. , Moravec, T. , Plchova, H. , Folwarczna, J. , Synkova, H. et al. (2012) Transient expression of Human papillomavirus type 16 L2 epitope fused to N‐ and C‐terminus of coat protein of Potato virus X in plants. Journal of Biosciences, 37, 125–133. [DOI] [PubMed] [Google Scholar]
- Chen, Y.L. , Lee, C.Y. , Cheng, K.T. , Chang, W.H. , Huang, R.N. , Nam, H.G. et al. (2014) Quantitative peptidomics study reveals that a wound‐induced peptide from PR‐1 regulates immune signaling in tomato. The Plant Cell, 26, 4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, P.S. , Nam, H.G. & Chen, Y.R. (2015) A salt‐regulated peptide derived from the CAP superfamily protein negatively regulates salt‐stress tolerance in Arabidopsis . Journal of Experimental Botany, 66, 5301–5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, K.P. , Zeng, Y. , Li, Y. , Ji, C. , Xia, Y. & Jiang, L. (2018) Signal motif‐dependent ER export of the Qc‐SNARE BET12 interacts with MEMB12 and affects PR1 trafficking in Arabidopsis . Journal of Cell Science, 131, jcs202838. [DOI] [PubMed] [Google Scholar]
- Dixon, D.C. , Cutt, J.R. & Klessig, D.F. (1991) Differential targeting of the tobacco PR‐1 pathogenesis‐related proteins to the extracellular space and vacuoles of crystal idioblasts. The EMBO Journal, 10, 1317–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y. , Mpina, M.H. , Birch, P.R.J. , Bouwmeester, K. & Govers, F. (2015) Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiology, 169, 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine, K. , Fujimoto, M. , Okatani, Y. , Nishiyama, T. , Goh, T. , Ito, E. et al. (2011) A membrane trafficking pathway regulated by the plant‐specific RAB GTPase ARA6. Nature Cell Biology, 13, 853–859. [DOI] [PubMed] [Google Scholar]
- Feige, J.N. , Sage, D. , Wahli, W. , Desvergne, B. & Gelman, L. (2005) PixFRET, an ImageJ plug‐in for FRET calculation that can accommodate variations in spectral bleed‐throughs. Microscopy Research and Technique, 68, 51–58. [DOI] [PubMed] [Google Scholar]
- Gamir, J. , Darwiche, R. , van't Hof, P. , Choudhary, V. , Stumpe, M. , Schneiter, R. et al. (2017) The sterol‐binding activity of PATHOGENESIS‐RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. The Plant Journal, 89, 502–509. [DOI] [PubMed] [Google Scholar]
- Ge, Z. , Zhao, Y. , Liu, M.‐C. , Zhou, L.‐Z. , Wang, L. , Zhong, S. et al. (2019) LLG2/3 are co‐receptors in BUPS/ANX‐RALF signaling to regulate Arabidopsis pollen tube integrity. Current Biology, 29, 3256.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, G.M. , Roelants, K. & O’Bryan, M.K. (2008) The CAP superfamily: cysteine‐rich secretory proteins, antigen 5, and pathogenesis‐related 1 proteins – roles in reproduction, cancer, and immune defense. Endocrine Reviews, 29, 865–897. [DOI] [PubMed] [Google Scholar]
- Gu, Y. & Innes, R.W. (2012) The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. The Plant Cell, 24, 4717–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hála, M. , Cole, R. , Synek, L. , Drdová, E. , Pečenková, T. , Nordheim, A. et al. (2008) An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. The Plant Cell, 20, 1330–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, L.L. & Nielsen, M.E. (2017) Plant exosomes: using an unconventional exit to prevent pathogen entry?. Journal of Experimental Botany, 69, 59–68. [DOI] [PubMed] [Google Scholar]
- Hecker, A. , Wallmeroth, N. , Peter, S. , Blatt, M.R. , Harter, K. & Grefen, C. (2015) Binary 2in1 vectors improve in planta (co)localization and dynamic protein interaction studies. Plant Physiology, 168, 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft van Huijsduijnen, R.A.M. , Kauffmann, S. , Brederode, F.T. , Cornelissen, B.J.C. , Legrand, M. , Fritig, B. et al. (1987) Homology between chitinases that are induced by TMV infection of tobacco. Plant Molecular Biology, 9, 411–420. [DOI] [PubMed] [Google Scholar]
- Inada, N. , Betsuyaku, S. , Shimada, T.L. , Ebine, K. , Ito, E. , Kutsuna, N. et al. (2016) Modulation of plant RAB GTPase‐mediated membrane trafficking pathway at the interface between plants and obligate biotrophic pathogens. Plant and Cell Physiology, 57, 1854–1864. [DOI] [PubMed] [Google Scholar]
- Ito, E. , Uemura, T. , Ueda, T. & Nakano, A. (2016) Distribution of RAB5‐positive multivesicular endosomes and the trans‐Golgi network in root meristematic cells of Arabidopsis thaliana . Plant Biotechnol, 33, 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C. & van Kammen, A. (1970) Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacum var. “Samsun” and “Samsun NN”. II. Changes in protein constitution after infection with tobacco mosaic virus. Virology, 40, 199–211. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. & Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annual Review of Phytopathology, 44(1), 135–162. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. & van Strien, E.A. (1999) The families of pathogenesis‐related proteins, their activities, and comparative analysis of PR‐1 type proteins. Physiological and Molecular Plant Pathology, 55, 85–97. [Google Scholar]
- Jelitto‐Van Dooren, E.P.W.M. , Vidal, S. & Denecke, J. (1999) Anticipating endoplasmic reticulum stress: a novel early response before pathogenesis‐related gene induction. The Plant Cell, 11, 1935–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, T. , Gao, C. , Cui, Y. , Wang, J. , Ding, Y. , Cai, Y. et al. (2013) ARA7(Q69L) expression in transgenic Arabidopsis cells induces the formation of enlarged multivesicular bodies. Journal of Experimental Botany, 64, 2817–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, M.H.A.J. (2012) Isolation of apoplastic fluid from leaf tissue by the vacuum infiltration‐centrifugation technique. Methods in Molecular Biology, 835, 603–610. [DOI] [PubMed] [Google Scholar]
- Joshi, V. , Joshi, N. , Vyas, A. & Jadhav, S.K. (2021) Pathogenesis‐related proteins: role in plant defense. In: Jogaiah, S. (Ed.), Biocontrol agents and secondary metabolites. Woodhead Publishing, pp. 573–590. [Google Scholar]
- Jung, H.W. & Hwang, B.K. (2000) Isolation, partial sequencing, and expression of pathogenesis‐related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria . Molecular Plant‐Microbe Interactions, 13, 136–142. [DOI] [PubMed] [Google Scholar]
- Kalde, M. , Nühse, T.S. , Findlay, K. & Peck, S.C. (2007) The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis‐related protein 1. Proceedings of the National Academy of Sciences of the United States of America, 104, 11850–11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kama, R. , Gabriely, G. , Kanneganti, V. , Gerst, J.E. & Gilmore, R. (2018) Cdc48 and ubiquilins confer selective anterograde protein sorting and entry into the multivesicular body in yeast. Molecular Biology of the Cell, 29, 948–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M. , Inzé, D. & Depicker, A. (2002) GATEWAYTM vectors for Agrobacterium‐mediated plant transformation. Trends in Plant Science, 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Klahre, U. , Becker, C. , Schmitt, A.C. & Kost, B. (2006) Nt‐RhoGDI2 regulates Rac/Rop signaling and polar cell growth in tobacco pollen tubes. The Plant Journal, 46, 1018–1031. [DOI] [PubMed] [Google Scholar]
- Kus, J.V. , Zaton, K. , Sarkar, R. & Cameron, R.K. (2002) Age‐related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae . The Plant Cell, 14, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, J.E. , Sanchez, J.P. , Zumstein, K. & Gilchrist, D.G. (2018) Plant and animal PR1 family members inhibit programmed cell death and suppress bacterial pathogens in plant tissues. Molecular Plant Pathology, 19, 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. , Faris, J.D. , Sherwood, R. & Edwards, M.C. (2013) Dimerization and protease resistance: new insight into the function of PR‐1. Journal of Plant Physiology, 170, 105–110. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Kurose, T. , Hino, T. , Tanaka, K. , Kawamukai, M. , Niwa, Y. et al. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering, 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nakano, R.T. , Yamada, K. , Bednarek, P. , Nishimura, M. & Hara‐Nishimura, I. (2014) ER bodies in plants of the Brassicales order: biogenesis and association with innate immunity. Frontiers in Plant Science, 5, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niderman, T. , Genetet, I. , Bruyère, T. , Gees, R. , Stintzi, A. , Legrand, M. et al. (1995) Pathogenesis‐related PR‐1 proteins are antifungal: isolation and characterization of three 14‐kilodalton proteins of tomato and of a basic PR‐1 of tobacco with inhibitory activity against Phytophthora infestans . Plant Physiology, 108, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehl, A. , Amari, K. , Gereige, D. , Brandner, K. , Mély, Y. & Heinlein, M. (2012) Control of Tobacco mosaic virus movement protein fate by CELL‐DIVISION‐CYCLE protein48. Plant Physiology, 160, 2093–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmannová, J. , Sekereš, J. , Kulich, I. , Šantrůček, J. , Dobrev, P. , Žárský, V. et al. (2021) The EXO70B2 exocyst subunit contributes to papillae and encasement formation in anti‐fungal defence in Arabidopsis . Journal of Experimental Botany, 73, 742–755. [DOI] [PubMed] [Google Scholar]
- Pečenková, T. , Pleskot, R. & Žárský, V. (2017) Subcellular localization of Arabidopsis pathogenesis‐related 1 (PR1) protein. International Journal of Molecular Sciences, 18(4), 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchar, P. , Sekereš, J. , Novotný, O. , Žárský, V. & Potocký, M. (2020) Functional analysis of phospholipase Dδ family in tobacco pollen tubes. The Plant Journal, 103, 212–226. [DOI] [PubMed] [Google Scholar]
- Pruss, G.J. , Nester, E.W. & Vance, V. (2008) Infiltration with Agrobacterium tumefaciens induces host defense and development‐dependent responses in the infiltrated zone. Molecular Plant‐Microbe Interactions, 21, 1528–1538. [DOI] [PubMed] [Google Scholar]
- Rauscher, M. , Ádám, A.L. , Wirtz, S. , Guggenheim, R. , Mendgen, K. & Deising, H.B. (1999) PR‐1 protein inhibits the differentiation of rust infection hyphae in leaves of acquired resistant broad bean. The Plant Journal, 19, 625–633. [DOI] [PubMed] [Google Scholar]
- Rivière, M.P. , Marais, A. , Ponchet, M. , Willats, W. & Galiana, E. (2008) Silencing of acidic pathogenesis‐related PR‐1 genes increases extracellular β‐(1→3)‐glucanase activity at the onset of tobacco defence reactions. Journal of Experimental Botany, 59, 1225–1239. [DOI] [PubMed] [Google Scholar]
- Saberianfar, R. & Menassa, R. (2017) Protein bodies: how the ER deals with high accumulation of recombinant proteins. Plant Biotechnology Journal, 15, 671–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria, M. , Thomson, C.J. , Read, N.D. & Loake, G.J. (2001) The promoter of a basic PR1‐like gene, AtPRB1, from Arabidopsis establishes an organ‐specific expression pattern and responsiveness to ethylene and methyl jasmonate. Plant Molecular Biology, 47, 641–652. [DOI] [PubMed] [Google Scholar]
- Sarowar, S. , Young, J.K. , Eui, N.K. , Ki, D.K. , Byung, K.H. , Islam, R. et al. (2005) Overexpression of a pepper basic pathogenesis‐related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Reports, 24, 216–224. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. et al. (2012) Fiji: an open‐source platform for biological‐image analysis. Nature Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, J. , Olrichs, N.K. , Geerts, W.J. , Kaloyanova, D.V. & Helms, J.B. (2019) Metal ions and redox balance regulate distinct amyloid‐like aggregation pathways of GAPR‐1. Scientific Reports, 9, 15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, J. , Olrichs, N.K. , Geerts, W.J. , Li, X. , Rehman, A.U. , Gadella, B.M. et al. (2019) Zinc binding regulates amyloid‐like aggregation of GAPR‐1. Bioscience Reports, 39, BSR20182345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich, I.E. , Schmelzer, E. , Bollmann, J. & Hahlbrock, K. (1986) Rapid activation by fungal elicitor of genes encoding “pathogenesis‐related” proteins in cultured parsley cells. Proceedings of the National Academy of Sciences of the United States of America, 83, 2427–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A. , Heitz, T. , Prasad, V. , Wiedemann‐Merdinoglu, S. , Kauffmann, S. , Geoffroy, P. et al. (1993) Plant “pathogenesis‐related” proteins and their role in defense against pathogens. Biochimie, 75, 687–706. [DOI] [PubMed] [Google Scholar]
- Sung, Y.C. , Outram, M.A. , Breen, S. , Wang, C. , Dagvadorj, B. , Winterberg, B. et al. (2021) PR1‐mediated defence via C‐terminal peptide release is targeted by a fungal pathogen effector. New Phytologist, 229, 3467–3480. [DOI] [PubMed] [Google Scholar]
- Tao, K. , Waletich, J.R. , Wise, H. , Arredondo, F. & Tyler, B.M. (2019) Tethering of multi‐vesicular bodies and the tonoplast to the plasma membrane in plants. Frontiers in Plant Science, 10, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczynska, I. , Stumpe, M. & Mauch, F. (2018) A conserved RxLR effector interacts with host RABA‐type GTPases to inhibit vesicle‐mediated secretion of antimicrobial proteins. The Plant Journal, 95, 187–203. [DOI] [PubMed] [Google Scholar]
- Vancová, M. , Bílý, T. , Nebesářová, J. , Grubhoffer, L. , Bonnet, S. , Park, Y. et al. (2019) Ultrastructural mapping of salivary gland innervation in the tick Ixodes ricinus . Scientific Reports, 9, 6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S. , Shimada, T.L. , Hiruma, K. & Takano, Y. (2013) Pathogen infection trial increases the secretion of proteins localized in the endoplasmic reticulum body of Arabidopsis . Plant Physiology, 163(2), 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, B. & Sali, A. (2016). Comparative protein structure modeling using MODELLER. Current Protocols in Protein Science, 2016, 5.6.1–5.6.37. [DOI] [PubMed] [Google Scholar]
- Woloshuk, C.P. , Meulenhoff, J.S. , Sela‐Buurlage, M. , van den Elzen, P.J.M. & Cornelissen, B.J.C. (1991) Pathogen‐induced proteins with inhibitory activity toward Phytophthora infestans . The Plant Cell, 3, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L. , Wu, H. , Chen, L. , Zhang, H. & Gao, X. (2017) Induction of systemic disease resistance in Nicotiana benthamiana by the cyclodipeptides cyclo (l‐Pro‐l‐Pro) and cyclo (d‐Pro‐d‐Pro). Molecular Plant Pathology, 18, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J. & He, S.Y. (1996) The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. Journal of Bacteriology, 178, 6399–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W.J. , Pedersen, C. , Kwaaitaal, M. , Gregersen, P.L. , Mørch, S.M. , Hanisch, S. et al. (2012) Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Molecular Plant Pathology, 13, 1110–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 PR1‐GFP localization in seedlings of Arabidopsis thaliana. (a) Localization of PR1‐GFP in root cells in a typical MVB‐like compartment (left panel). Bar = 10 μm. Root cells of A. thaliana expressing PR1‐GFP (green) and propidium iodide (PI; 10 μg/ml) stained cell walls (red; middle panel). Bar = 5 μm. PR1‐GFP and PI signals zoomed and visualized using processed three‐dimensional reconstruction of Z stacks acquired with airy scan mode of confocal microscopy (right panel). Bar = 1 μm. (b) Minor co‐localization of the PR1‐positive compartment with endocytotic dye FM4‐64 (1 μg/ml) stained compartment (left panel, pointed by arrowheads). PR1‐GFP compartment is not affected by BFA (50 μM) and only partially co‐localizes with FM4‐64 stained BFA bodies (middle panel, arrowheads); PR1‐GFP and FM4‐64 stained BFA bodies depicted in more detail using higher magnification (right panel). Bars = 10 μm
FIGURE S2 Examples of the western blot analysis performed with leaf extracts from infiltrated Nicotiana benthamiana, using GFP‐specific antibody. (a) Detection of the full‐length fusion proteins and also the smaller GFP‐positive band. FL, full‐length PR1‐GFP; D50, PR1ΔC‐GFP; 4A, PR1YDPR/AAAA‐GFP; D11, PR1Δ11‐GFP on the left. Western blot of the GFP control constructs with signal peptide (SP)‐GFP only (variant SG) and SP‐GFP‐CAPE (variant SGC) with CNYDPRGNYVNEKPY peptide on C‐end (SGC) on the right. (b) Two different exposition times of western blot for SG, FL, D50, 4A, D11, and SGC constructs, showing the difference in expression levels of SG and SGC constructs versus PR1 variants; the red arrowhead points to the probable bands of GFP with fused PR1 C‐terminus
FIGURE S3 Confirmation by western blot for extracellular/apoplastic signal for FL and D11. The upper row shows total cell extracts from Nicotiana benthamiana expressing RFP‐tagged PR1 variants: full‐length, FLR (PR1‐RFP), D50R (PR1ΔC‐RFP), 4AR (PR1YDPR/AAAA‐RFP), and D11R (PR1Δ11‐RFP). The lower rows show the apoplastic fluid from leaves infiltrated with the same constructs. The antibody recognizes cleaved RFP tag only for FLR and D11R, in agreement with confocal observations. However, the presence of their uncleaved variants in apoplastic fluid is detectable using anti‐PR1 (Agrisera; unspecific recognition of bands of c.50 and 30 kDa is observable for all variants). Ponceau S staining shows minor contamination with cytoplasmic RuBisCO, equal for all analysed samples. The FLR variant is prone to the formation of SDS‐resistant higher molecular weight multimers. The prominent band seen between 25 and 35 kDa with anti‐PR1 antibody (Agrisera) is a product of unspecific interaction observable with the extracts enriched for proteins from the cell wall and plasma membrane fractions from both Arabidopsis and Nicotiana
FIGURE S4 PR1C‐terminal CAPE peptide effect on localization and immunity. (a) Schematic representation of signal peptide (SP)‐GFP (variant SG) and SP‐GFP‐CAPE (variant SGC) fusion proteins, their intracellular localizations, and effects on antibacterial plant immunity in transiently transformed Nicotiana benthamiana leaves. Both constructs demonstrate endoplasmic reticulum (ER)‐retained localization (with supposed cleavage and further trafficking of CAPE in case of SGC). Bars = 10 μm. (b) Graph shows susceptibility toward hrcC, expressed as ratios of cfu quantified for SG and SGC versus control A (Agrobacterium tumefaciens with P. syringae hrcC; value 1), counted for young and adult plants. Peptide presence significantly enhances susceptibility in younger plants and resistance in adult plants (n = 6). Significantly different values determined by analysis of variance (ANOVA) with post hoc Tukey HSD are marked with asterisks. Bars = 10 μm. (c) Trypan blue staining intensities of each of employed PR1 variants, expressed as a ratio of stained area toward the total analysed infiltrated area. Representative images of trypan blue stained leaves of N. benthamiana expressing PR1 variants are shown above the graph for each of the variants, as well as the control Agrobacterium/hrcC. Different small letters indicate significant differences between variants determined by ANOVA with post hoc Tukey HSD calculator (n = 6). Infiltrated variants: A, Agrobacterium/hrcC infiltrated control; FL, full‐length PR1‐GFP; D50, PR1ΔC‐GFP; 4A, PR1YDPR/AAAA‐GFP; D11, PR1Δ11‐GFP; SG, SP‐GFP; SGC, SP‐GFP‐CAPE. Bar = 100 μm. (d) cfu of hrcC cultivated in liquid medium in the presence of different concentrations of synthetic peptide CAPE. The peptide has no significant influence on in vitro growth of hrcC bacteria
FIGURE S5 Detection of proteins possibly interacting with PR1 using the co‐immunoprecipitation method. (a) Table 1. Several putative protein interactors found in co‐immunoprecipitation experiments with PR1‐GFP overexpressed in Arabidopsis thaliana. (b) Verification of PR1‐CDC48 interaction using fluorescence resonance energy transfer (FRET) analysis. Analysis was performed on leaves of Nicotiana benthamiana co‐expressing PR1‐GFP with CDC48b‐RFP. Bars = 20 μm. The graph shows the quantification of FRET analysis (according to Feige et al., 2005) performed using five to 10 images for each of the two controls and the PR1–GFP and CDC48b–RFP combination. The significant positive FRET was detected in case of PR1‐GFP and CDC48b‐RFP
FIGURE S6 Reverse transcription (RT)‐PCR analysis of NbPR1a up‐regulation. (a) The RT‐PCR was performed for mock, hrcC‐infiltrated, and leaves infiltrated with hrcC together with control construct SG, construct SGC, as well as FL and D50 constructs which have opposite effects on plant immunity. EF1‐a gene was used as a housekeeping control. (b) Appearance of plants 24 hr postinoculation (hpi): different constructs at this stage do not cause differences in symptoms development on sites of infiltration. (c) RT‐PCR 48 hpi with hrcC in comparison to mock infiltration; NbPR1a is up‐regulated in adult leaves. (d) The appearance of lesions in 5 days postinoculation with hrcC
FIGURE S7 Phylogenetic tree of PR1‐like proteins. The phylogenetic tree was constructed based on the alignment using all proteins containing CAP domains of selected species. Sequences were aligned using MUSCLE and manually trimmed. Phylogenetic analysis has been performed using the neighbour‐joining method with 500 bootstrap replicates. CAP domain‐containing proteins are present throughout whole land plants evolution with subsequent repeated gene amplification. Based on our evolutionary tree, PR1‐like proteins might have originally pollen‐ and abiotic‐stress‐related functions which later on evolved also to a defence function (deduced based on RNA expression pattern [TAIR, Genevestigator], no function was assigned to other closely related paralogs). The most closely related to AtPR1 is AtPRB1, which participates in jasmonate‐related defence (Santamaria et al., 2001). Ambtr, Amborella trichopoda; Arath, Arabidopsis thaliana; Brara, Brassica rapa; Capgr, Capsella grandiflora; Poptr, Populus trichocarpa; Theca, Theobroma cacao; Vitvi, Vitis vinifera
TABLE S1 List of primers used for cloning
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


