Abstract
Taxonomy
Bacteria; Phylum Proteobacteria; Class Gammaproteobacteria; Order Lysobacterales (earlier synonym of Xanthomonadales); Family Lysobacteraceae (earlier synonym of Xanthomonadaceae); Genus Xanthomonas; Species X. hortorum; Pathovars: pv. carotae, pv. vitians, pv. hederae, pv. pelargonii, pv. taraxaci, pv. cynarae, and pv. gardneri.
Host range
Xanthomonas hortorum affects agricultural crops, and horticultural and wild plants. Tomato, carrot, artichoke, lettuce, pelargonium, ivy, and dandelion were originally described as the main natural hosts of the seven separate pathovars. Artificial inoculation experiments also revealed other hosts. The natural and experimental host ranges are expected to be broader than initially assumed. Additionally, several strains, yet to be assigned to a pathovar within X. hortorum, cause diseases on several other plant species such as peony, sweet wormwood, lavender, and oak‐leaf hydrangea.
Epidemiology and control
X. hortorum pathovars are mainly disseminated by infected seeds (e.g., X. hortorum pvs carotae and vitians) or cuttings (e.g., X. hortorum pv. pelargonii) and can be further dispersed by wind and rain, or mechanically transferred during planting and cultivation. Global trade of plants, seeds, and other propagating material constitutes a major pathway for their introduction and spread into new geographical areas. The propagules of some pathovars (e.g., X. horturum pv. pelargonii) are spread by insect vectors, while those of others can survive in crop residues and soils, and overwinter until the following growing season (e.g., X. hortorum pvs vitians and carotae). Control measures against X. hortorum pathovars are varied and include exclusion strategies (i.e., by using certification programmes and quarantine regulations) to multiple agricultural practices such as the application of phytosanitary products. Copper‐based compounds against X. hortorum are used, but the emergence of copper‐tolerant strains represents a major threat for their effective management. With the current lack of efficient chemical or biological disease management strategies, host resistance appears promising, but is not without challenges. The intrastrain genetic variability within the same pathovar poses a challenge for breeding cultivars with durable resistance.
Useful websites
https://gd.eppo.int/taxon/XANTGA, https://gd.eppo.int/taxon/XANTCR, https://gd.eppo.int/taxon/XANTPE, https://www.euroxanth.eu, http://www.xanthomonas.org, http://www.xanthomonas.org/dokuwiki
Keywords: bacterial blight, carrot, dandelion, leaf spots, lettuce, pelargonium, tomato, Xanthomonas hortorum
This pathogen profile summarizes the knowledge on the seven pathovars of Xanthomonas hortorum that cause bacterial spot and/or blight on 65 plant species in 15 families, including agricultural, horticultural, and wild plants.

1. INTRODUCTION
The seven pathovars of Xanthomonas hortorum collectively affect 65 plant species in 15 botanical families, including agricultural crops (e.g., tomato, carrot, lettuce), horticultural plants (e.g., pelargonium), and wild plants (e.g., dandelion). This pathogen profile gives the first comprehensive summary of X. hortorum biology, including a history of its taxonomy and an account of its broad host range, and of its distribution and epidemiology, emphasizing intrapathovar differences. The genomics work done on this species is also summarized, with a special focus on pathogen–host interactions. Most previous literature on this pathogen deals with X. hortorum as a homogenous entity. This pathogen profile highlights, for each section, intrapathovar similarities and differences, thus providing a nuanced and detailed look into the complexity of X. hortorum.
2. TAXONOMY UPDATE
The taxonomic history of the different X. hortorum pathovars is long and complex (Figure 1), like that of the genus Xanthomonas. The earliest reports of diseases caused by X. hortorum date back to the 1890s, with the reports describing bacterial leaf spot and blight disease of English ivy in 1894 in Germany (Lindau, 1894), and bacterial blight of geraniums and bacterial leaf spot of lettuce in Massachusetts, USA, in 1898 and 1907, respectively (Stone, 1907; Stone & Smith, 1898). The first proper taxonomic description of the species causing bacterial leaf spot of lettuce, referred to as Bacterium vitians (Brown, 1918), was published in 1918 (Figure 1). In the following years, B. hederae and B. pelargonii were isolated from diseased English ivy (Arnaud, 1920) and diseased geraniums (Brown, 1923), respectively. B. vitians, B. hederae, and B. pelargoni were then reclassified in the genus Phytomonas (Bergey et al., 1923; Burkholder & Guterman, 1932). Phytomonas carotae (originally proposed as “Pseudomonas carotae”) was characterized as the bacterium responsible for bacterial blight of carrot (Kendrick, 1934).
FIGURE 1.
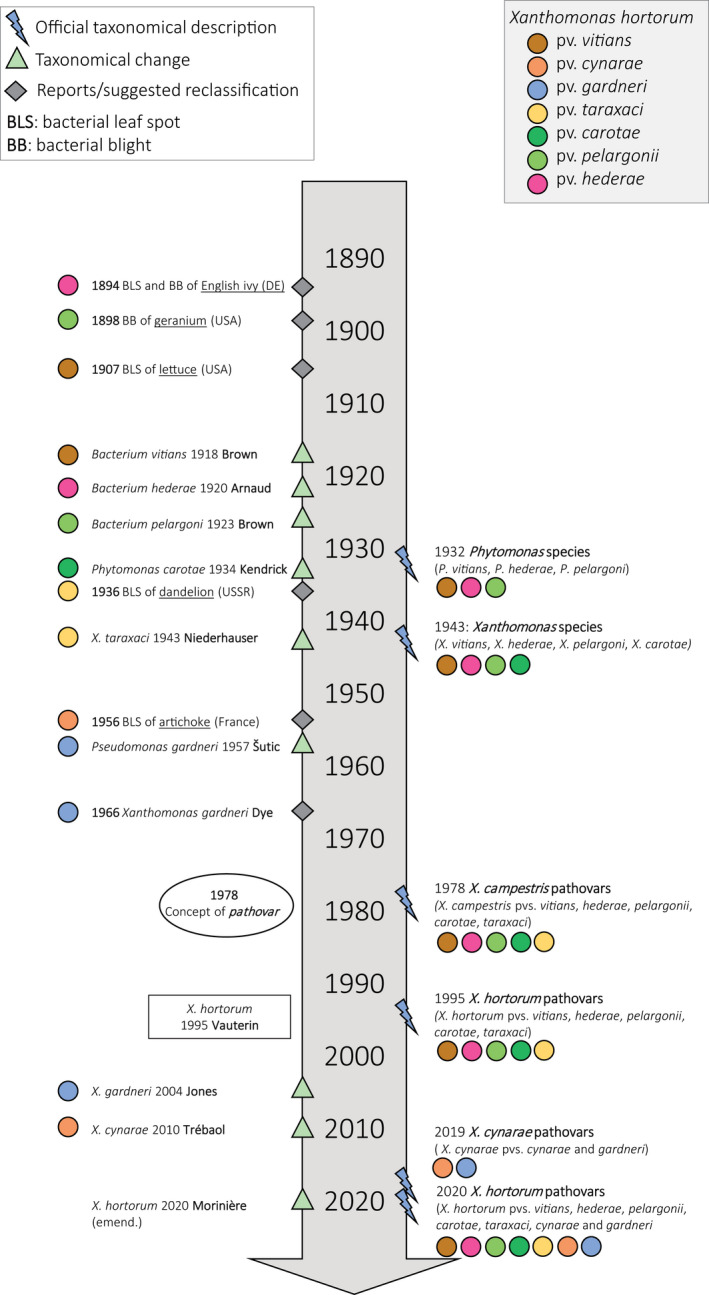
The taxonomical history of Xanthomonas hortorum, outlining official taxonomical descriptions and changes, as well as first reports or suggested reclassifications of the various pathovars
Subsequently, the four species were transferred to the genus Xanthomonas as X. hederae, X. carotae, X. pelargonii, and X. vitians (Dowson, 1943; Starr & Burkholder, 1942). Concurrently, the bacterium responsible for the bacterial blight of Russian dandelion, first reported in the USSR (Sigrianski, 1936), was designated as X. taraxaci (Niederhauser, 1943). Those five pathogens were considered to be individual Xanthomonas species until the introduction of the infrasubspecific epithet “pathovar” (Young et al., 1978), followed by the publication of the first Approved Lists in 1980 (Skerman et al., 1980). Many Xanthomonas species, including X. hederae, X. carotae, X. pelargonii, X. vitians, and X. taraxaci, could only be distinguished by their host range and were thus transferred as pathovars of the polytypic species X. campestris (Young et al., 1978).
Based on DNA–DNA hybridization (DDH) (Palleroni & Bradbury, 1993; Vauterin et al., 1995), these five X. campestris pathovars were classified as pathovars of the new species X. hortorum (Figure 1). The pathotype strain CFBP 5858T (= LMG 733T = NCPPB 939T) of X. hortorum pv. hederae was designated as the species’ type strain. The taxonomical status of “X. hortorum pv. vitians” was unclear, and two variants were distinguished: the former pathotype strain, which is nonpathogenic on lettuce, was labelled “type A”, while “X. hortorum pv. vitians”, pathogenic on lettuce, was designated as “type B”.
A group of strains causing bacterial spot of tomato and pepper (Solanum lycopersicum and Capsicum annuum) was originally named “Pseudomonas gardneri” (Šutic, 1957). Some years later, it was suggested to be part of genus Xanthomonas (Dye, 1966), but it was not formally described as X. gardneri until the beginning of the 21st century (Jones et al., 2004). The taxonomical history of the Xanthomonas strains causing bacterial spot of tomato and pepper has been thoroughly reviewed (Osdaghi et al., 2021; Potnis et al., 2015).
Strains associated with bacterial bract spot of artichoke (Cynara scolymus) were first reported in the 1950s as members of the Xanthomonas genus (Ridé, 1956), yet the official species description as X. cynarae was only provided in 2000 (Trébaol et al., 2000). Although a few phylogenetic studies demonstrated the high genetic relatedness between X. hortorum, X. cynarae, and X. gardneri (Parkinson et al., 2009; Young et al., 2008), they were only recently formally accepted as the same taxonomic entity (Morinière et al., 2020; Timilsina et al., 2019).
Genomic, phenotypic, and pathogenicity analyses were first used to prove the synonymy of X. cynarae and X. gardneri and reclassify them as pathovars of X. cynarae (Timilsina et al., 2019). In that same study, X. hortorum and X. cynarae were acknowledged to be paraphyletic species but were kept separate, based on previous wet‐lab DDH results. However, only the type strain of X. hortorum was included in the 2019 study. A comprehensive analysis revisited the taxonomy of those strains, and included all type, pathotype, or representative strains of X. hortorum and X. cynarae (Morinière et al., 2020). Standard genome‐to‐genome comparison parameters, such as average nucleotide identity (ANI), in silico DDH (isDDH), and tetranucleotide frequencies (Tetra), between X. hortorum and X. cynarae fell into the transition zone of the species boundary (Morinière et al., 2020), a concept described previously (Richter & Rosselló‐Móra, 2009; Rosselló‐Móra & Amann, 2015). Phylogenetic reconstructions suggested a continuous evolution and diversification of pathovars and phenotypic data did not reveal stable diagnostic traits allowing distinction between X. cynarae and X. hortorum strains. X. cynarae was then suggested to be a later heterotypic synonym of X. hortorum and both species were combined into an extended X. hortorum species including seven pathovars (Figure 2): X. hortorum pvs hederae, pelargonii, vitians, carotae, taraxaci, cynarae, and gardneri (Morinière et al., 2020).
FIGURE 2.
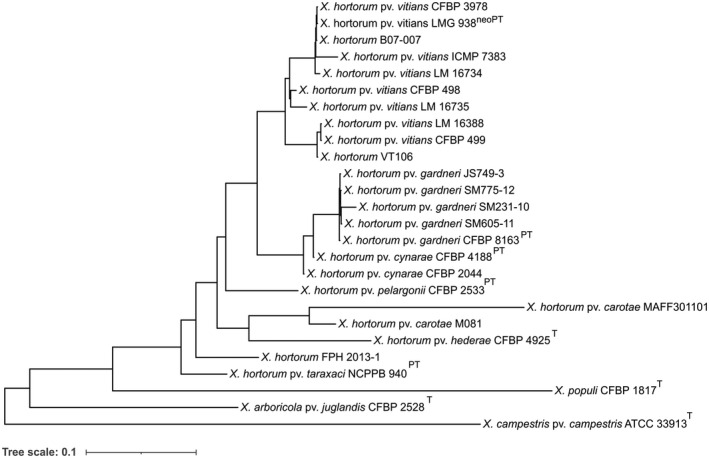
Whole‐genome phylogeny of representative Xanthomonas hortorum strains. The tree was constructed using PhyloPhlAn v. 0.40 (Segata et al., 2013) as previously described in Morinière et al. (2020)
3. HOST RANGE
Making a distinction between natural and experimental hosts of plant‐pathogenic bacteria is important to better understand the extent of their host range (Bull & Koike, 2015). The natural host range of a pathogen consists of naturally infected plants (i.e., in nonexperimental settings), and is the criterion for pathovar identification and classification (Dye et al., 1980). The experimental host range includes plants that show symptoms after artificial inoculation. Its scope depends on the choice of plant species and of inoculation procedures. The experimental host range provides invaluable information on the pathogen's potential to adapt to new host plants (Jacques et al., 2016).
Each X. hortorum pathovar has its own natural host range and the experimental host ranges of multiple pathovars have been studied. Additionally, many unassigned strains within X. hortorum have also been isolated from multiple different plants (e.g., wheat, peony, and hydrangea). Most of the reported natural hosts of X. hortorum belong to the Geraniaceae, Araliaceae, and Asteraceae families, while most of the reported experimental hosts of the pathogen belong to Asteraceae (Table 1). X. hortorum affects more than 65 plant species in 15 botanical families, as summarized in Table 1.
TABLE 1.
The natural and experimental host range of Xanthomonas hortorum pathovars and unassigned strains
| X. hortorum pv. | Isolated from a | Host range type b | Disease c | References | ||
|---|---|---|---|---|---|---|
| Family | Plant genus | Plant species | ||||
| carotae | Apiaceae | Daucus | carota | N | BLB | Kendrick (1934); Myung et al. (2014); du Toit et al. (2014) |
| cynarae | Asteraceae | Cynara | scolymus | N | BBS | Trébaol et al. (2000) |
| Solanaceae | Capsicum | annuum | E | NA | Timilsina et al. (2019) | |
| gardneri | Asteraceae | Cynara | scolymus | E | NA | Timilsina et al. (2019) |
| Euphorbiaceae | Euphorbia | heterophylla | N | BS | Araújo et al. (2015) | |
| Solanaceae | Solanum | lycopersicum | N | BS | Jones et al. (2004); Quezado‐Duval et al. (2004); Timilsina et al. (2019) | |
| Capsicum | annuum | N | BS | Jones et al. (2004); Timilsina et al. (2019) | ||
| Solanum | americanum | E | BS | Araújo et al. (2015) | ||
| Nicandra | physaloides | E | BS | Araújo et al. (2015) | ||
| Brassicaceae | Arabidopsis | thaliana | E | NA | Cândido et al. (2008) | |
| hederae | Araliaceae | Hedera | helix | N | BLS | Arnaud (1920); Trantas et al. (2016) |
| canariensis | N | BLS | Suzuki et al. (2002) | |||
| nepalensis (var. sinensis) | N | BLS | Zhang et al. (2015) | |||
| rhombea | E | NA | Suzuki et al. (2002) | |||
| colchica | E | NA | Leyns et al. (1984) | |||
| Schefflera | atinophylla | N | BLS | Chase (1984); Norman et al. (1999); Tolba (2017) | ||
| arboricola | N | BLS | Chase (1984); Norman et al. (1999) | |||
| Fatsia | japonica | N | BLS | Chase (1984) | ||
| Polyscias | spp. | N | BLS | Norman et al. (1999) | ||
| Plerandra | elegantissima | E | NA | Chase (1984) | ||
| pelargonii | Geraniaceae | Pelargonium | capitatum | N | BB | Knauss and Tammen (1964) |
| peltatum | N | BB | Starr et al. (1955) | |||
| quercifolium | N | BB | Knauss and Tammen (1964) | |||
| radens | N | BB | Knauss and Tammen (1964) | |||
| scandens | N | BB | Knauss and Tammen (1964) | |||
| zonale | N | BB | Leyns et al. (1984) | |||
| × domesticum | N | BB | Stapp (1958) | |||
| × fragrans | N | BB | Knauss and Tammen (1964) | |||
| × hortorum | N | BB | Starr et al. (1955) | |||
| × ignescens | N | BB | Knauss and Tammen (1964) | |||
| Geranium | maculatum | N | BB | Stapp (1958) | ||
| pratense | N | BB | Starr et al. (1955) | |||
| sanguineum | N | BB | Starr et al. (1955) | |||
| sylvaticum | N | BB | Stapp (1958) | |||
| Euphorbiaceae | Euphorbia | pulcherrima | E | NA | Rockey et al. (2015) | |
| taraxaci | Taraxacum | kok‐saghyz | N | BLS | Niederhauser (1943) | |
| vitians | Asteraceae | Lactuca | sativa | N | BLS | Brown (1918); Morinière et al. (2020) |
| serriola | N | BLS | Toussaint et al. (2012); Morinière et al. (2020) | |||
| biennis | E | NA | Toussaint et al. (2012) | |||
| Taraxacum | officinale | E | NA | Toussaint et al. (2012); Morinière et al. (2020) | ||
| Sonchus | oleraceus | E | NA | Toussaint et al. (2012) | ||
| asper | E | NA | Toussaint et al. (2012) | |||
| Artemisia | biennis | E | NA | Toussaint et al. (2012) | ||
| Matricaria | discoidea | E | NA | Toussaint et al. (2012) | ||
| Arctium | minus | E | NA | Toussaint et al. (2012) | ||
| Gnaphalium | uliginosum | E | NA | Toussaint et al. (2012) | ||
| Ambrosia | artemisiifolia | E | NA | Toussaint et al. (2012) | ||
| Galinsoga | quadriradiata | E | NA | Toussaint et al. (2012) | ||
| Senecio | vulgaris | E | NA | Toussaint et al. (2012) | ||
| Solanaceae | Nicotiana | tabacum | E | NA | Toussaint et al. (2012) | |
| Solanum | lycopersicum | E/N? | BLS | Sahin et al. (2003); Al‐Saleh et al. (2011); Morinière et al. (2020) | ||
| Capsicum | annuum | E | NA | Sahin et al. (2003); Al‐Saleh et al. (2011) | ||
| “nigromaculans” | Arctium | lappa | N | BLS | Parkinson et al. (2009); Dehghan‐Niri and Rahimian (2016) | |
| Unassigned | Asteraceae | Artemisia | annua | N | BLS | Ssekiwoko et al. (2009) |
| Cichorium | intybus | N | BLS | Zacaroni et al. (2012) | ||
| Calendula | officinalis | NA | NA | Parkinson et al. (2009) | ||
| Lamiaceae | Lavandula | dentata | N | BLS | Koike et al. (1995) | |
| angustifolia | N | BLS | Koike et al. (1995); Roberts and Parkinson (2014) | |||
| × intermedia | N | BLS | Rotondo et al. (2020) | |||
| × ginginsii | E | NA | Rotondo et al. (2020) | |||
| Oleaceae | Olea | europaea | NA | NA | Young et al. (2010) | |
| Primulaceae | Primula | vulgaris | N | BLS | Nejad et al. (2012) | |
| Hydrangeaceae | Hydrangea | quercifolia | N | BLS | Cottyn et al. (2021); Uddin et al. (1996) | |
| arborescens | N | BLS | Cottyn et al. (2021) | |||
| Paeoniaceae | Paeonia | spp. | N | BB | Oliver et al. (2012); Klass et al. (2019) | |
| Poaceae | Triticum | sp. | E | NA | Egorova et al. (2014) | |
| Poaceae | Hordeum | vulgare | E | NA | Egorova et al. (2014) | |
| Poaceae | Secale | cereale | E | NA | Egorova et al. (2014) | |
| Poaceae | Avena | sativa | E | NA | Egorova et al. (2014) | |
| Lauraceae | Persea | americana | NA | NA | Parkinson et al. (2009) | |
To ensure consistent botanical taxonomy, plant species nomenclature was checked on the World Flora Online database (WFO, 2021).
N, natural host; E, experimental host; NA, not applicable.
Disease type is only mentioned in the event of a natural host. BLS, bacterial leaf spot; BBS, bacterial bract spot; BLB, bacterial leaf blight; BS, bacterial spot; BB, bacterial blight; NA, not applicable.
X. hortorum pv. hederae is primarily known as a pathogen of English ivy (Hedera helix) (Arnaud, 1920; Trantas et al., 2016), but has also been isolated from diseased plants belonging to other Hedera species (Table 1). Some X. hortorum pv. hederae strains are pathogenic on several other plants of the Araliaceae family (e.g., Schefflera spp.) in natural ecosystems (Table 1). The experimental host range of X. hortorum pv. hederae includes false aralia (Plerandra elegantissima), Japanese ivy (Hedera rhombea) (Suzuki et al., 2002), and Persian ivy (Hedera colchica) (Leyns et al., 1984), but X. hortorum strains have not been reported on those plants in natural conditions. Another X. hortorum pathovar, pv. pelargonii, naturally occurs on a wide range of plant species from the genera Geranium and Pelargonium in the Geraniaceae family (Table 1). Some strains of X. hortorum pv. pelargonii cause mild symptoms on poinsettia (Euphorbia pulcherrima) in experimental conditions (Rockey et al., 2015).
X. hortorum pv. vitians is a pathogen of cultivated lettuce (Lactuca sativa) and probably infects its closest wild relative, the prickly lettuce (Lactuca serriola) (Morinière et al., 2020; Toussaint et al., 2012). This pathovar can be pathogenic on diverse weeds from the Asteraceae family (Table 1) (Toussaint et al., 2012). In greenhouse infection tests, several strains were weakly pathogenic on tomato and two pepper cultivars (C. annuum ‘Marengo’, a sweet pepper, and C. annuum ‘Cayenne Long Slim’, a cayenne pepper) (Al‐Saleh et al., 2011; Morinière et al., 2020; Sahin et al., 2003).
To our knowledge the only known hosts of X. hortorum pvs carotae and taraxaci are their respective initial hosts of isolation. X. hortorum pv. carotae is pathogenic on wild carrot (Daucus carota) and its cultivated subspecies (D. carota subsp. sativus) (Kendrick, 1934; Myung et al., 2014; Temple et al., 2013), while Russian dandelion (Taraxacum kok‐saghyz) is the only reported host of X. hortorum pv. taraxaci (Niederhauser, 1943) (Table 1).
The only recorded natural host of X. hortorum pv. cynarae is artichoke (Cynara scolymus) (Trébaol et al., 2000) and the pathogen also caused leaf spot symptoms in infiltrated C. annuum pepper leaves (Timilsina et al., 2019) (Table 1). X. hortorum pv. gardneri is one of the four xanthomonads responsible for bacterial spot of tomato and pepper, alongside X. euvesicatoria pv. euvesicatoria, X. euvesicatoria pv. perforans, and X. vesicatoria (Jones et al., 2004; Osdaghi et al., 2021; Potnis et al., 2015). X. hortorum pv. gardneri strains were isolated from spot symptoms on tomato and pepper (Jones et al., 2004), as well as the weed plant Euphorbia heterophylla (Araújo et al., 2015). Some X. hortorum pv. gardneri strains are pathogenic on tomato or pepper, while other strains are pathogenic on both (Potnis et al., 2015). Moreover, greenhouse inoculations with X. hortorum pv. gardneri resulted in limited necrosis on artichoke leaves (Timilsina et al., 2019), in chlorotic spots on Arabidopsis thaliana (Cândido et al., 2008), and in leaf lesions on American black nightshade (Solanum americanum) and apple of Peru (Nicandra physaloides) (Araújo et al., 2015).
Several unclassified X. hortorum strains cause disease on other plant species such as peony (Paeonia spp.) (Klass et al., 2019; Oliver et al., 2012) and sweet wormwood (Artemisia annua) (Ssekiwoko et al., 2009) (Table 1). Strains causing an unknown disease of lavender (Lavandula dentata, L. angustifolia, and L. × intermedia) were first identified as X. campestris (Koike et al., 1995), but reclassified as X. hortorum based on sequence data (Roberts & Parkinson, 2014; Rotondo et al., 2020). Strains reported as closely related to X. hortorum are sometimes unavailable in public or private strain collections, as is the case for angular leaf spot disease of oak‐leaf hydrangea (Hydrangea quercifolia) observed in Georgia, USA (Uddin et al., 1996). Recently, similar strains were reported from leaf spot symptoms on hydrangea in Flemish (Belgium) nurseries (Cottyn et al., 2021). Greater burdock (Arctium lappa) is also likely to be a natural host of some unassigned X. hortorum strains (Dehghan‐Niri & Rahimian, 2016).
Other studies suggesting that some strains belong to X. hortorum have not addressed Koch's postulates. As such, it is unclear whether those strains belong to the species. For example, many X. hortorum strains were isolated from seed lots of several Poaceae plants in Russia (e.g., wheat, Triticum sp.; barley, Hordeum vulgare; rye, Secale cereale; and oat, Avena sativa) (Table 1). Infiltration of bacterial suspension through leaves induced vascular or local necrotic lesions in the corresponding Poaceae species (Egorova, 2015; Egorova et al., 2014). Two multilocus sequence analysis (MLSA) studies reported that strains belonging to X. hortorum have been isolated from diseased olive (Olea europaea) (Young et al., 2010), avocado (Persea americana), and pot marigold (Calendula officinalis) (Parkinson et al., 2009) (Table 1).
4. DISEASE SYMPTOMS
The pathovars of X. hortorum can cause bacterial spot and/or bacterial blight on numerous plant species. X. hortorum pvs hederae, taraxaci, and vitians cause bacterial leaf spot on ivy (Figure 3a), dandelion (Figure 3e), and lettuce (Figure 3g), respectively. X. hortorum pvs carotae and pelargonii cause bacterial blight on carrot (Figure 3b) and geranium (Figure 3f), respectively. The symptoms of X. hortorum pv. gardneri can be observed on tomato (Figure 3c) and/or pepper (Figure 3d), depending on the strains, while X. hortorum pv. cynarae causes bacterial spot on artichoke bracts (Figure 3h). The disease symptoms caused by all these pathogens share common characteristics but also have some subtle differences.
FIGURE 3.
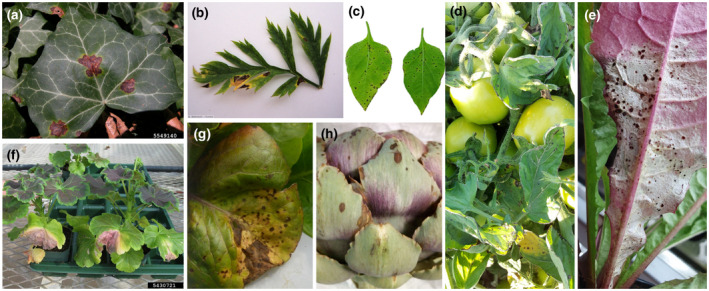
Xanthomonas hortorum pathovars on various hosts. (a) English ivy leaf infected by X. hortorum pv. hederae. Courtesy of Forestry Images and the Penn State Department of Plant Pathology & Environmental Microbiology Archives. (b) X. hortorum pv. carotae symptoms on a carrot leaf. Photograph courtesy of E‐phytia and Benoît Mériaux. (c) X. hortorum pv. gardneri symptoms on pepper (cv. Early Carl Wonder) leaves, 14 days postinoculation (dpi) with X. hortorum pv. gardneri Xg965. Photograph provided by Neha Potnis. (d) Field infection of tomato plant by X. hortorum pv. gardneri. Photograph provided by Eduardo Bernal. (e) Diseased dandelion leaf 12 dpi after inoculation with X. hortorum pv. taraxaci LM 16389 (= CFBP 8644). Photograph provided by Lucas Morinière. (f) X. hortorum pv. pelargonii on geranium (Pelargonium spp.). Photograph courtesy of Forestry Images and Nancy Gregory (University of Delaware). (g) Close‐up of field infection of a lettuce leaf by X. hortorum pv. vitians. Photograph provided by Lucas Morinière. (h) Infection of artichoke head by X. hortorum pv. cynarae. Photograph courtesy of Johan Van Vaerenbergh
Diseases caused by X. hortorum pathovars are characterized by round, water‐soaked lesions on the abaxial surface of leaves (capitulum artichoke bracts, in the case of X. hortorum pv. cynarae) and are usually the first symptoms observed (Norman et al., 1999; Potnis et al., 2015; Pruvost et al., 2010; Ridé, 1956; Rockey et al., 2015; Schornack et al., 2008; Trébaol et al., 2000). These small water‐soaked leaf spots rapidly expand to form angular necrotic lesions.
The presence of a chlorotic halo around spots or lesions is pathovar‐ or plant/cultivar‐dependent. For example, a chlorotic halo is present around lesions caused by X. hortorum pvs pelargonii, hederae, and taraxaci, but its presence varies in angular leaf spot caused by X. hortorum pvs carotae, vitians, and gardneri (Daughtrey & Wick, 1995; Gilbertson, 2002; Myung et al., 2014; Nameth et al., 1999; Pruvost et al., 2010). In advanced infection stages, lesions and spots usually turn dark in colour (brown to black) on plant parts affected by X. hortorum pvs pelargonii, hederae, carotae, gardneri, and vitians. They can also coalesce (e.g., in the presence of X. hortorum pvs hederae, gardneri, and vitians), giving a papery appearance to leaves affected by X. hortorum pv. vitians (Bull & Koike, 2005). In final infection stages, leaves usually harden and dry and, in the case of leaves affected by X. hortorum pv. hederae, a red‐purple margin might appear on their upper surface (Suzuki et al., 2002).
Some very particular leaf symptoms are associated with certain X. hortorum pathovars. For example, X. hortorum pv. pelargonii can cause leaf margin wilting and V‐shaped necrotic areas, depending on the plant species and cultivar (Daughtrey & Wick, 1995). The affected areas eventually drop off, and black stem rot occurs in case of a systemic infection. When the infection expands to the roots, it results in overall wilt and gradual plant death, but no decay or soft rot is observed (Daughtrey & Benson, 2005; Manulis et al., 1994).
Furthermore, leaves are not the only plant parts affected by X. hortorum. X. hortorum pv. gardneri affects tomato fruits, on which it causes characteristic star‐shaped lesions with a raised, scabby appearance (Potnis et al., 2015). On unripe tomato fruits, symptoms look like water‐soaked or slightly raised pale‐green spots, sometimes surrounded by greenish‐white halos. On tomato sepals, symptoms consist of brown lesions, which can turn necrotic; stem lesions are narrow, elongated, and raised (Potnis et al., 2015). X. hortorum pv. hederae occasionally affects stems and petioles (Suzuki et al., 2002), and X. hortorum pv. carotae causes disease on petioles, peduncles, stems, flowers, and leaflets (Gilbertson, 2002). Lesions caused by X. hortorum pvs carotae and vitians can be V‐shaped (Gilbertson, 2002; Sahin, 1997; Scott & Dung, 2020; du Toit et al., 2014).
5. GEOGRAPHIC DISTRIBUTION AND IMPORTANCE
X. hortorum includes a pathovar causing the most devastating bacterial disease of geranium (pv. pelargonii) (Manulis et al., 1994; Munnecke, 1954), an internationally regulated seedborne pathovar affecting carrot (pv. carotae) (Scott & Dung, 2020), and a pathovar reported in most lettuce‐growing areas (pv. vitians) (Sahin, 1997). Furthermore, X. hortorum pv. gardneri, in addition to three other Xanthomonas spp., is a major pathogen on tomato and/or pepper (Jones et al., 2004; Osdaghi et al., 2021; Potnis et al., 2015). Diseases caused by X. hortorum pathovars have been reported in more than 40 countries across all continents except Antarctica (Figure 4), either as one‐time reports or as frequent reoccurrences. One‐time reports do not necessarily mean that the diseases are not recurring or currently present. Examples of frequent reoccurrences include X. hortorum pv. carotae in Canada and the USA (EPPO, 2021), X. hortorum pv. vitians in Canada and France (Morinière et al., 2020; Toussaint, 1999; Toussaint et al., 2012), and X. hortorum pv. gardneri in the USA and Brazil (Araújo et al., 2012, 2015, 2017; Potnis et al., 2015; Quezado‐Duval et al., 2004), the second and sixth largest tomato producers in 2019, respectively, by gross production value (FAOSTAT, 2021).
FIGURE 4.
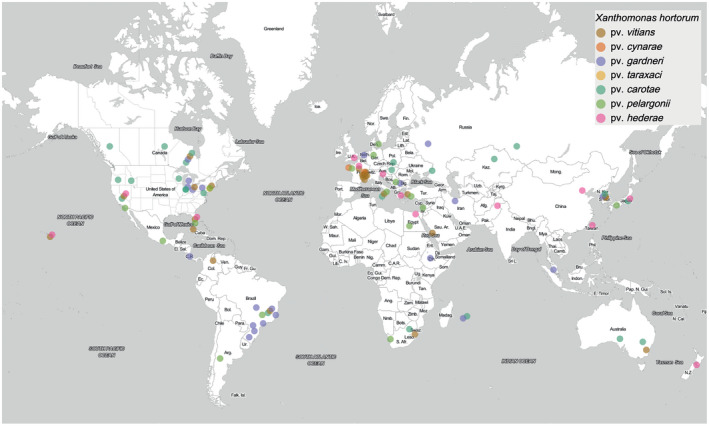
Distribution of the seven Xanthomonas hortorum pathovars. Map from the ggmap R package (Kahle et al., 2019) and data adapted from the European and Mediterranean Plant Protection Organization (EPPO). Location is an approximation based on literature available
Data on the economic impact of X. hortorum is not available for all its pathovars, as there are no reports documenting the cost of the damage caused by X. hortorum pvs cynarae, taraxaci, and hederae. When available, economic impact reports are not recent but are nonetheless informative about the scope of the importance of X. hortorum. For example, bacterial blight of geranium caused by X. hortorum pv. pelargonii is the most devastating bacterial pathogen of geranium and can lead to total geranium loss when environmental conditions are most favourable to this pathogen (Balaž et al., 2016; Manulis et al., 1994; Munnecke, 1954; Nameth et al., 1999). Most carrot seed growers in the Pacific Northwest region of the USA, an important region for US carrot seed production, consider X. hortorum pv. carotae detrimental to seed quality (Dr Jeremiah Dung, The Oregon State University, March 2021, personal communication). In Montreal, Canada, losses due to X. hortorum pv. vitians have led to complete destruction of lettuce fields (Toussaint, 1999). In Florida, USA, the pathovar caused an estimated loss of $4 million from the early to mid‐1990s (Robinson et al., 2006), and also caused substantial economic losses in California and Ohio, USA (Carisse et al., 2000; Sahin, 1997). Losses due to X. hortorum pv. gardneri were estimated to cost the Midwestern US tomato‐processing industry $7–8 million (Ma, 2015; Ma et al., 2011).
X. hortorum pv. gardneri is one of the four xanthomonads causing bacterial spot of tomato and pepper, and multiple reports have studied population structure shifts of those four species, especially in the USA and Brazil (Araújo et al., 2015, 2017; Egel et al., 2018; Pereira et al., 2011). In the USA, most early reported incidences of bacterial spot disease on tomato and pepper were caused by X. euvesicatoria pv. euvesicatoria but a population shift to X. hortorum pv. gardneri has been reported in published work (Egel et al., 2018; Ma, 2015; Ma et al., 2011) and personal communications (Dr Francesca Rotondo and Dr Sally A. Miller, The Ohio State University, March 2021, personal communication). However, recent surveys for tomato and pepper bacterial spot in Brazil have shown a limited presence of X. hortorum pv. gardneri (Araújo et al., 2017).
6. EPIDEMIOLOGY
In general, X. hortorum pathovars thrive in warm, wet, and humid environments in fields and greenhouses (Dye, 1967; Gardner & Kendrick, 1921; Kendrick, 1934; Manulis et al., 1994; Strider, 1985; du Toit et al., 2005; Toussaint, 1999). During inoculation trials, X. hortorum pv. gardneri had a higher virulence at 20°C when compared to other bacterial spot pathogens of tomato and pepper (Araújo et al., 2011), and was more prevalent than other bacterial spot xanthomonads at higher altitudes (Araújo et al., 2017). Furthermore, X. hortorum pv. vitians has an optimal infection temperature of around 23°C (Robinson et al., 2006).
The pathovars colonize the plants through natural openings (e.g., hydathodes, stomata) or wounds (Bernal & Francis, 2021; Dougherty et al., 1974; Ridé, 1956; Schwartz et al., 2017). After gaining entry, they infect the plant vascular system (Barak et al., 2002; Munnecke, 1954). Mesophyll colonization is also possible for X. hortorum pvs pelargonii (Barel et al., 2015) and vitians (authors’ unpublished data). Infections of X. hortorum pvs pelargonii and vitians can sometimes be symptomless (Barak et al., 2002; McPherson & Preece, 1978).
The two primary sources of inoculum of X. hortorum pathovars are seeds and cuttings, although they can be disseminated through other means as well (e.g., insects, rain, and irrigation water) and can survive on weeds, crop debris, or in soils. Seed is a main source of inoculum for bacterial spot and blight caused by X. hortorum pvs carotae, gardneri, and vitians (Barak et al., 2001, 2002; Kendrick, 1934; Kuan, 1985; Mtui et al., 2010; Sahin & Miller, 1997; du Toit et al., 2005, 2014). Contaminated seed, stecklings, or seedlings may initiate an epidemic in grower fields (McDonald & Linde, 2002; du Toit et al., 2005), which could result in a nonnormal pathogen distribution, as observed for X. hortorum pv. carotae populations (Scott & Dung, 2020). This can pose a challenge to the development of detection methods and durable resistant cultivars.
X. hortorum pvs hederae and pelargonii are mainly transmitted by infected cuttings (Chittaranjan & De Boer, 1997; Norman et al., 1999) because flowers such as geraniums are commonly vegetatively propagated by cuttings. Historically, propagating facilities were inadvertently responsible for distributing infected symptomless plant material (Nameth et al., 1999).
Crop residues can allow X. hortorum pvs carotae and vitians to overwinter for several months or until the following growing season (Christianson et al., 2015; Sahin et al., 2003). X. hortorum pv. carotae can persist in infected carrot foliage on soil for up to a year (Gilbertson, 2002). X. hortorum pv. vitians can survive in crop debris for up to 1 month, in both summer and winter months (Barak et al., 2001; Fayette et al., 2018). X. hortorum pvs vitians and gardneri can survive epiphytically or infect weeds, respectively (Araújo et al., 2015; Barak et al., 2001; Fayette et al., 2018). Soil or crop debris also act as an important inoculum source for X. hortorum pv. carotae, where it can survive for up to 3 months (Kendrick, 1934), and for pv. pelargonii, which can survive in soils for up to a year (Gilbertson, 2002). Survival in weeds, plant residues, and soils can serve as a secondary inoculum source in the presence of favourable hosts and environmental conditions (Gitaitis & Walcott, 2007). If bacterial populations are high, they can re‐emerge from inside the plant tissue and serve as a secondary inoculum on the plant itself or on nearby hosts.
X. hortorum pvs gardneri, carotae, and vitians are also disseminated by wind or rain, or mechanically transferred during planting and cultivation (Potnis et al., 2015; du Toit et al., 2005). X. hortorum pv. carotae has been observed in aerosolized debris generated by carrot seed threshers during field operations (du Toit et al., 2005). X. hortorum pv. pelargonii can be transmitted by greenhouse whiteflies (Trialeurodes vaporariorum) (Bugbee & Anderson, 1963), and insects were noted to be vectors for X. hortorum pv. carotae but no details (insect genus or species) were given (Gilbertson, 2002).
7. IDENTIFICATION AND DETECTION
Visual symptom assessment is the first step to detect a suspected X. hortorum infection and subsequent identification is based on pathogen isolation. X. hortorum strains are readily isolated from infected plant tissue using serial dilution plating. Growth media used can be nonselective (e.g., nutrient agar, sucrose peptone, or yeast‐dextrose‐calcium carbonate [YDC] agar) or semiselective (Saddler & Bradbury, 2015). Irrespective of medium type, X. hortorum colonies are yellow, mucoid, and convex (Saddler & Bradbury, 2015).
Phenotypic profiles of this species, analysed using phenotype microarrays (e.g., Biolog, OMNILOG), remain too variable to provide an accurate identification at the species level (Akhtar & Aslam, 1990; Bouzar et al., 1999; Mirik et al., 2018; Morinière et al., 2020; Myung et al., 2010; Stoyanova et al., 2014; Trébaol et al., 2000; Uddin et al., 1996). Pathovars cannot be distinguished from one another by using such phenotypic profiling as no stable, discriminative traits exist (Morinière et al., 2020; Trébaol et al., 2000). Even though pathovar classification depends on host pathogenicity (see Taxonomy update), the identification of X. hortorum pathovars should not solely rely on the host range. Indeed, some strains of this species can naturally infect hosts other than their original host of isolation (see Host range).
SDS‐PAGE protein profiling and later DDHs (Stefani et al., 1994; Vauterin et al., 1991, 1995) were used to identify X. hortorum pv. vitians “type B”, revealing the existence of aberrant strains (Table 2). Even though fatty acid profiling did not provide identification among pathovars and often remains inaccurate at the species level (Barak & Gilbertson, 2003; Mirik et al., 2018; Sahin et al., 2003; Ssekiwoko et al., 2009; Uddin et al., 1996), it still distinguished between X. hortorum pv. vitians “type B” and the unusual isolates (Sahin et al., 2003). Furthermore, a panel of 16 xanthomonad‐specific monoclonal antibodies (Table 2), used in enzyme‐linked immunosorbent assays (ELISAs), distinguished two serovars of X. hortorum pv. vitians isolates (Sahin et al., 2003).
TABLE 2.
Non‐DNA and DNA‐based identification methods for Xanthomonas hortorum pathovars. The detection targets, taxonomical level of detection, and primer sequence availability, when applicable, are also reported
| Detection | Targeted pathovar | Specific for the targeted pathovar (antibody, primer, or probe name) b | Reference(s) | Comments c | |||
|---|---|---|---|---|---|---|---|
| method | Type | Target(s) | Taxonomical level a | ||||
| ELISA | Non‐DNA | Polyclonal antibodies | Pathovar | pv. pelargonii | Yes | Balaž et al. (2016) | Commercial ELISA kit |
| ELISA | Non‐DNA | Monoclonal antibodies | Xanthomonas species | pv. pelargonii | Yes (MAb Xpel‐1) | Benedict et al. (1990) | NA |
| ELISA | Non‐DNA | Monoclonal antibodies | Pathovar | pv. pelargonii | Yes (McAb 2H5) | Chittaranjan and De Boer (1997) | NA |
| ELISA | Non‐DNA | Monoclonal antibodies | Xanthomonas species/pathovars | pv. vitians | No | Sahin et al. (2003) | Pattern‐based discrimination |
| SDS‐PAGE | Non‐DNA | Various proteins | Xanthomonas species/pathovars | various: pvs vitians (cluster 7d), hederae (cluster 7e), pelargonii (cluster 12) | No | Vauterin et al. (1991) | Pattern‐based discrimination |
| SDS‐PAGE | Non‐DNA | Various proteins | Xanthomonas species/pathovars | pv. vitians | No | Stefani et al. (1994) | Pattern‐based discrimination |
| SDS‐PAGE | Non‐DNA | Various proteins | Pathovar | pv. gardneri | No | Quezado‐Duval et al. (2004) | Pattern‐based discrimination |
| DDH | DNA | DNA homology groups | Xanthomonas species/pathovars | various: pvs pelargonii, hederae, vitians (group 2) | No | Vauterin et al. (1995) | Clustering‐based discrimination |
| MLSA/MLST | DNA | Various housekeeping genes, including gyrB | Xanthomonas species | various pvs | No | Parkinson et al. (2007); Young et al. (2008); Parkinson et al. (2009) | Clustering‐based discrimination |
| MLSA/MLST | DNA | Various housekeeping genes, including gyrB | Pathovar | pv. vitians | No | Fayette et al. (2016) | Clustering‐based discrimination |
| PCR | DNA | RAPD fragments | Pathovar | pv. carotae | Yes (3S/3SR and 9B/9BR) | Meng et al. (2004) | D.L.: 22 fg (3S) and 2 pg (9B) |
| PCR | DNA | CDS or intergenic regions | Pathovar | pv. carotae | Yes (XhcPP02, PP03, PP04, and PP05) | Kimbrel et al. (2011) | NA |
| PCR | DNA | Based on the target of XhcPP02 (Kimbrel et al., 2011) | Pathovar | pv. carotae | Yes (Xhc‐q2) | Temple et al. (2013) | NA |
| PCR | DNA | AFLP fragments | BSX | pv. gardneri | Yes (Bs‐XgF/Bs‐XgR) | Koenraadt et al. (2009); Pereira et al. (2011) | NA |
| PCR | DNA | 1.2 kb DNA‐fragment | Pathovar | pv. pelargonii | Yes | Manulis et al. (1994); Chittaranjan and De Boer (1997) | NA |
| PCR | DNA | ERIC, REP regions | Pathovar | pv. pelargonii | No | Sulzinski et al. (1995) | Pattern‐based discrimination |
| PCR | DNA | ERIC fragment | Pathovar | pv. pelargonii | Yes (XcpMl/XcpM2) | Sulzinski et al. (1996); Sulzinski et al. (1997); Sulzinski et al. (1998); Sulzinski (2001) | NA |
| PCR | DNA | RAPD fragments | Pathovar | pv. vitians | Yes (B162) | Barak et al. (2001) | NA |
| PCR | DNA | BOXA, ERIC, REP, and 16S‐23S rDNA regions | Xanthomonas species/pathovars | pv. vitians | No | Sahin et al. (2003) | Pattern‐based discrimination |
| PCR | DNA | SNP‐based | Pathovar | pv. vitians | No | Hébert et al. (2021) | C t values |
| Multiplex PCR | DNA | ERIC fragment | X. hortorum pv. pelargonii and Ralstonia solanacearum | pv. pelargonii | Yes (DG1/DG2) | Glick et al. (2002) | NA |
| PCR and multiplex PCR | DNA | AFLP fragments | BSX | pv. gardneri | Yes (Bs‐XgF/Bs‐XgR) | Araújo et al. (2012) | D.L.: DNA: 50 pg/µl; bacterial suspension: 5 × 104 cfu/ml (100 bacterial cells per reaction). |
| Real‐time PCR | DNA | ERIC fragment | Pathovar | pv. pelargonii | Yes (XhqF/XhqR) | Farahani and Taghavi (2016) | D.L.: 200 fg |
| Multiplex real‐time PCR | DNA | lepA | BSX +Clavibacter michiganensis subsp. michiganensis, Pseudomonas syringae pv. tomato | pv. gardneri | No | Peňázová et al. (2020) | Cy5 was used for multiplex PCR that targeted BSX together |
| Multiplex real‐time PCR | DNA | hrpB7 primers and probes | BSX | pv. gardneri | Yes, in combination with probe (FP2/RP2) | Strayer, Jeyaprakash, et al. (2016) | D.L.: 5 × 105 cfu/ml |
| LAMP | DNA | Based on PCR product of 9B primer set (Meng et al., 2004) | Pathovar | pv. carotae | Yes (Lace primer set) | Temple and Johnson (2009); Temple et al. (2013) | NA |
| LAMP | DNA | hrpB6 | Pathovar | pv. gardneri | Yes | Stehlíková et al. (2020) | D.L.: 1 pg/µl |
| RPA | DNA | hrpB6 | Pathovar | pv. gardneri | No (XGF/XGR) | Strayer‐Scherer et al. (2019) | D.L.: 5 × 106 cfu/ml; amplified X. hortorum pv. cynarae CFBP 2044 |
BSX, the Xanthomonas species causing bacterial spot of tomato and pepper: X. hortorum pv. gardneri, X. vesicatoria, and X. euvesicatoria pvs euvesicatoria and perforans.
Primer sequences are available in all the DNA‐based detection methods.
D.L., reported detection limits; NA, not available.
Antibodies were used to detect X. hortorum pv. pelargonii. Pathovar‐specific monoclonal antibodies (Benedict et al., 1990; Chittaranjan & De Boer, 1997) and polyclonal antibodies (Balaž et al., 2016; Mirik et al., 2018) were successfully used for serological identification of this pathogen (Table 2), using commercial double‐antibody sandwich ELISA kits (LOEWE Biochemica GmbH and Agdia).
Several DNA‐based molecular assays have been developed over recent decades to identify and detect X. hortorum strains (Table 2). The available methods are limited to four of the seven pathovars, as diagnostics methods are unavailable for X. hortorum pvs hederae, taraxaci, and cynarae at the time of writing. Several PCR detection protocols are available to amplify DNA from many Xanthomonas species, including one or more X. hortorum pathovars, by targeting the 16S rRNA gene (Maes, 1993), the hrp gene cluster (Leite et al., 1994), or gumD, fyuA, and the internal transcribed spacer (ITS) (Adriko et al., 2014). However, these general protocols are usually not implemented for the identification and detection of X. hortorum pathovars. Instead, targeted assays allowing specific detection and identification at the pathovar level are often preferred.
The first targeted detection DNA‐based assays were mostly derived from DNA fingerprint methods, and were used to study the genetic diversity of X. hortorum pathovars (Barak & Gilbertson, 2003; Hamza et al., 2012; Sahin et al., 2003; Sulzinski, 2001). For example, a PCR for X. hortorum pv. carotae was developed from random amplified polymorphic DNA (RAPD) analysis (Meng et al., 2004). Similarly, diagnostic PCR tests for X. hortorum pv. pelargonii were developed from specific DNA fragments identified by RAPD analysis, enterobacterial repetitive intergenic consensus (ERIC) PCR or repetitive extragenic palindromic (REP) PCR (Chittaranjan & De Boer, 1997; Manulis et al., 1994; Sulzinski, 2001; Sulzinski et al., 1995, 1996, 1997, 1998). A multiplex PCR scheme for the simultaneous detection of X. hortorum pv. pelargonii and members of the Ralstonia solanacearum species complex, the second major bacterial pathogen of geranium, was developed from one of the two previously identified ERIC‐PCR fragments (Glick et al., 2002). The same molecular region was used to develop a real‐time quantitative PCR (qPCR) assay, allowing quantification of the pathogen (Farahani & Taghavi, 2016).
For X. hortorum pv. gardneri, a marker identified using amplified fragment polymorphism (AFLP) was initially used to design a diagnostic PCR assay (Koenraadt et al., 2009). The assay was later adapted into a multiplex PCR targeting four Xanthomonas species associated with tomato bacterial spot (Araújo et al., 2012). A multiplex TaqMan qPCR assay differentiating these four species was based on hrpB7, a less conserved gene within the hrpB operon (Strayer, Jeyaprakash, et al., 2016). A multiplex qPCR detecting these four Xanthomonas species, as well as tomato pathogens Clavibacter michiganensis subsp. michiganensis and Pseudomonas syringae pv. tomato, has been recently developed by targeting lepA (Peňázová et al., 2020).
Two X. hortorum pv. vitians‐specific primer pairs, 9308B and B162, were developed from RAPD fragments (Barak et al., 2001), but their specificity to the target varied. Primer pair 9308B failed to amplify isolates recovered from lettuce and weeds around lettuce fields. On the other hand, primer pair B162 successfully detected X. hortorum pv. vitians strains isolated over a 7‐year period (Barak et al., 2001).
Partial gene sequence of gyrB offers a sufficient resolution for the identification of xanthomonad isolates at the species level (Parkinson et al., 2007, 2009). MLSA is preferred to single‐gene (e.g., gyrB) to outline the precise phylogeny of X. hortorum (Morinière et al., 2020). However, MLSA schemes are based on different partial gene sequences, (sub)sets of partial genes, and trimming settings, which complicates the analysis by not allowing proper comparison between studies (Catara et al., 2021). The sequencing of the first draft genome of X. hortorum pv. carotae in 2011 allowed the first use of comparative genomics to develop two new diagnostics assays to detect this pathovar (Kimbrel et al., 2011; Temple et al., 2013). The first assay used a TaqMan qPCR, whereas the second relied on loop‐mediated isothermal amplification (LAMP) (Temple et al., 2013). The latter method showed superior performance compared to qPCR because of its robustness in the presence of inhibitors, and its rapidity, versatility, and usefulness in facilities with limited resources (Kimbrel et al., 2011). Both assays were also the first ones to be used as viability assays (i.e., detection of viable bacterial cells) with a xanthomonad, by including a propidium monoazide treatment prior to DNA extraction.
Two other isothermal amplification methods were recently published for the in‐field detection of X. hortorum pv. gardneri (Table 2), with an emphasis on differentiation from the other xanthomonad species responsible for tomato bacterial spot. The first method is based on recombinase polymerase amplification (RPA) and targets hrcN (hrpB) (Strayer‐Scherer et al., 2019). The second, based on LAMP, targets partial hrpB gene sequence (Stehlíková et al., 2020).
8. GENOMICS
The genome of X. hortorum pv. gardneri ATCC 19865PT (Potnis et al., 2011) was the first X. hortorum genome publicly available in the National Center for Biotechnology Information (NCBI) database (accessed March 2021). At the time of writing, 35 X. hortorum genomes have been deposited in the database, and most (46%) were submitted in 2020 (Table 3). Their completeness varies, and 74% (n = 26) are incomplete (i.e., assembled at the scaffold or contig levels; Table 3). Eight of the nine complete X. hortorum genomes contain at least one plasmid sequence. The average genome size of X. hortorum is 5.26 Mb (4.92–5.68 Mb). The average G + C content is of 63.6% (63.2%–63.9%), and the average predicted coding sequence (CDS) number is 4260 (Table 3). The average size of X. hortorum plasmids is 86.90 kb (29.56–224.70 kb), and three plasmids in X. hortorum pvs vitians and gardneri genomes are larger than 100 kb (Table 3). The average G + C content of plasmids is 60.30% (58.07%–62.18%), with an average of 94 predicted CDS.
TABLE 3.
Genome metrics of representative Xanthomonas hortorum strains
| Organism | Submission year | Strain | GenBank assembly accession | Assembly level | Contigs/scaffolds | Size (Mb) | GC (%) | N50 (bp) | CDS a | Plasmids (bp) |
|---|---|---|---|---|---|---|---|---|---|---|
| X. hortorum | 2017 | B07‐007 | GCA_002285515.1 | Complete | 2 | 5.25 | 63.6 | 5,175,249 | 4241 | pB07007 (75,655 bp) |
| X. hortorum | 2019 | VT 106 | GCA_008728175.1 | Complete | 2 | 5.15 | 63.7 | 5,101,806 | 4135 | pVT106 (44,015 bp) |
| X. hortorum | 2020 | FPH2013‐1 | GCA_011305375.1 | Scaffold | 70 | 5.22 | 63.7 | 190,176 | 4187 | – |
| X. hortorum pv. carotae | 2013 | M081 | GCA_000505565.1 | Chromosome | 1 | 5.05 | 63.7 | 5,052,399 | 8446 | – |
| X. hortorum pv. carotae | 2020 | MAFF 301101 | GCA_015726835.1 | Contig | 423 | 5.10 | 63.7 | 20,618 | 3972 | – |
| X. hortorum pv. cynarae | 2018 | CFBP 4188PT | GCA_002939985.1 | Scaffold | 102 | 5.06 | 63.7 | 145,505 | 4060 | – |
| X. hortorum pv. cynarae | 2020 | CFBP 2044 | GCA_903978235.1 | Complete | 2 | 5.12 | 63.7 | 5,079,002 | 8640 | CFBP2044_p40 (40,232 bp) |
| X. hortorum pv. gardneri | 2015 | SM234‐10 | GCA_001009295.1 | Scaffold | 179 | 5.33 | 63.5 | 49,361 | 4337 | – |
| X. hortorum pv. gardneri | 2015 | SM605‐11 | GCA_001009325.1 | Scaffold | 158 | 5.34 | 63.5 | 61,942 | 4327 | – |
| X. hortorum pv. gardneri | 2015 | SM775‐12 | GCA_001009625.1 | Scaffold | 176 | 5.25 | 63.6 | 59,240 | 4244 | – |
| X. hortorum pv. gardneri | 2016 | JS749‐3 | GCA_001908755.1 | Complete | 3 | 5.42 | 63.5 | 5,158,913 | 4373 | pJS749‐3.1 (211,336 bp), pJS749‐3.2 (45,952 bp) |
| X. hortorum pv. gardneri | 2020 | CFBP 8163PT | GCA_012922265.1 | Contig | 121 | 5.15 | 63.7 | 125,969 | 4151 | – |
| X. hortorum pv. hederae | 2018 | CFBP 4925T | GCA_002940005.1 | Scaffold | 313 | 5.32 | 63.8 | 42,684 | 4299 | – |
| X. hortorum pv. pelargonii | 2020 | CFBP 2533PT | GCA_012922215.1 | Contig | 94 | 5.21 | 63.8 | 134,256 | 4176 | – |
| X. hortorum pv. taraxaci | 2020 | NCPPB 940PT | GCA_903978185.1 | Complete | 2 | 5.03 | 63.8 | 4,999,567 | 8628 | NCPPB940_p30 (29,567 bp) |
| X. hortorum pv. vitians | 2020 | LM 16735 | GCA_012922125.1 | Contig | 138 | 5.19 | 63.7 | 118,265 | 4192 | – |
| X. hortorum pv. vitians | 2020 | LMG 938neoPT | GCA_012922135.1 | Contig | 119 | 5.03 | 63.8 | 141,718 | 4036 | – |
| X. hortorum pv. vitians | 2020 | LM 16388 | GCA_012922175.1 | Contig | 121 | 5.07 | 63.7 | 115,523 | 4063 | – |
| X. hortorum pv. vitians | 2020 | CFBP 3978 | GCA_012922195.1 | Contig | 131 | 5.13 | 63.7 | 141,718 | 4137 | – |
| X. hortorum pv. vitians | 2020 | CFBP 499 | GCA_012922335.1 | Contig | 132 | 5.17 | 63.7 | 118,955 | 4161 | – |
| X. hortorum pv. vitians | 2020 | LM 16734 | GCA_014338485.1 | Complete | 2 | 5.27 | 63.7 | 5,213,310 | 4223 | pLM16734 (57,250 bp) |
| X. hortorum pv. vitians | 2020 | CFBP 498 | GCA_903978195.1 | Complete | 4 | 5.68 | 63.2 | 5,365,193 | 4654 | CFBP498_p224 (224,704 bp), CFBP498_p47 (47,063 bp), CFBP498_p41 (41,583 bp) |
| X. hortorum pv. vitians | 2016 | ICMP 7383 | GCA_001908775.1 | Complete | 4 | 5.63 | 63.3 | 5,313,102 | 4511 | pICMP7383.1 (203,385 bp), pICMP7383.2 (61,840 bp), pICMP7383.3 (47,122 bp) |
The number of CDS is a direct output from NCBI and three numbers appear to overestimate the actual number.
The essential genome (genes required for growth and survival, irrespective of environmental conditions; Koonin, 2000) of X. hortorum pv. vitians LM 16734 was recently characterized through saturated transposon insertion sequencing (Morinière et al., 2021) and included 370 protein‐coding genes. These genes were mostly associated with critical cellular processes (e.g., translation, energy production, lipid transport), with 355 and 334 of them conserved within X. hortorum and the Xanthomonadaceae family, respectively.
8.1. Lipo‐ and exo‐polysaccharides
Lipopolysaccharides (LPSs) are involved in biofilm formation and protecting pathogens from their environment (Corsaro et al., 2001; Newman et al., 2002, 2007). X. hortorum pv. gardneri ATCC 19865PT has a 17.7 kb ancestral‐type LPS gene cluster, like that of X. campestris pv. campestris ATCC 33913T (Potnis et al., 2011). The LPS gene clusters of X. hortorum pvs cynarae CFBP 4188PT and gardneri ATCC 19865PT were highly syntenic, but different to that of X. hortorum pv. hederae CFBP 4925T (Timilsina et al., 2019). X. hortorum pvs carotae M081, cynarae CFBP 4188PT, gardneri ATCC 19865PT, and hederae CFBP 4925T possess wzm and wzt homologs (Kimbrel et al., 2011; Potnis et al., 2011; Timilsina et al., 2019), involved in the transport of LPS band A in Pseudomonas aeruginosa (Rocchetta & Lam, 1997).
Exopolysaccharides (EPSs) are involved in xanthan biogenesis, and they protect xanthomonad pathogens from environmental stress (Kakkar et al., 2015; Kamoun & Kado, 1990; Sutherland, 1993). The EPS gene cluster of X. hortorum pvs carotae and vitians is arranged similarly to that of X. campestris pv. campestris and contains all 12 genes from the gumB‐gumM cluster (Kimbrel et al., 2011; Morinière et al., 2021). Unlike in other Xanthomonas species (Katzen et al., 1998; Kim et al., 2008), only the mutations of gumE, gumI, and gumJ were lethal in X. hortorum pv. vitians LM 16734 (Morinière et al., 2021). The presence of a tRNA gene flanking the cluster in some Xanthomonas genomes suggests a horizontal transfer acquisition (Lu et al., 2008). However, no evidence of insertion elements was found in the EPS gene cluster of X. hortorum pvs carotae and vitians (Kimbrel et al., 2011; Morinière et al., 2021).
8.2. Secretion systems
Secretion systems and their effector proteins are crucial determinants of virulence in the Xanthomonas genus (Büttner & Bonas, 2010). There are two types of type II secretion system (T2SS) clusters within Xanthomonas: the T2SS‐xps, directly involved in virulence, and the T2SS‐xcs, which has seemingly no direct virulence function (Szczesny et al., 2010). The pathotype strains of X. hortorum pvs hederae, gardneri, and cynarae, in addition to strain B07‐007, have complete T2SS‐xps (xpsD‐xpsN) and T2SS‐xcs (xcsC‐xcsN) clusters (Alvarez‐Martinez et al., 2021; Timilsina et al., 2020). Unlike T2SS‐xps, the T2SS‐xcs cluster is not conserved within Xanthomonas spp. (Timilsina et al., 2020) but is conserved between the four X. hortorum strains.
The type III secretion system (T3SS) delivers effector proteins that, in turn, can suppress or trigger plant defence mechanisms (Büttner, 2016; White et al., 2009). The T3SS is found in most Xanthomonas strains, including X. hortorum (Timilsina et al., 2020). The T3SS of X. hortorum pv. gardneri ATCC 19865PT is a mosaic hrp cluster, with elements like that of X. campestris pv. campestris ATCC 33913T, but also including novel effectors (see Molecular host–pathogen interactions) (Potnis et al., 2011). X. hortorum pv. carotae M081 has a complete hrp cluster and is predicted to be functional (Kimbrel et al., 2011). Furthermore, a recent study reported that the T3SS hrp cluster in X. hortorum pv. gardneri ATCC 19865PT, cynarae CFBP 4188PT, hederae CFBP 4925T, and carotae M081 are similar, with some differences in the two 20 kb regions flanking the cluster (Merda et al., 2017).
The type IV secretion system (T4SS) is involved in protein transfer as well as bacterial conjugation (Guglielmini et al., 2014; Lawley et al., 2003; Llosa et al., 2002). X. hortorum pv. gardneri ATCC 19865PT has two plasmidborne and one chromosomal T4SS clusters (Potnis et al., 2011). The chromosomal cluster of X. hortorum pv. gardneri is complete and similar to that of X. campestris pv. campestris ATCC 33913T. One of the two X. hortorum pv. gardneri plasmidborne clusters is 98% and 89% identical to the T4SS clusters of Burkholderia multivorans ATCC 17616 and Acidovorax avenae subsp. citrulli AAC001, respectively. The other plasmidborne cluster is similar to the one found in X. vesicatoria ATCC 35937T and X. euvesicatoria pv. perforans 91‐118 (Potnis et al., 2011). The presence of a T4SS cluster in X. hortorum pv. carotae M081 was suggested by the detection of virB genes scattered over three different contigs but its functionality was inconclusive (Kimbrel et al., 2011).
The type V secretion system (T5SS) is responsible for the secretion of various proteins, including adhesins, which are important for host colonization as they are among the first contact points between pathogen and host (Meuskens et al., 2019). The members of T5SS are autotransporters, with the exception of type 5b, which is formed of two proteins (Guérin et al., 2017). In Xanthomonas spp., T5SS clusters belong to categories 5a, 5b, and 5c (Alvarez‐Martinez et al., 2021). X. hortorum pv. gardneri ATCC 19865PT and X. hortorum B07‐007 have three types of T5SS (types 5a, 5b, and 5c) (Alvarez‐Martinez et al., 2021).
The type VI secretion system (T6SS) is mostly responsible for bacterial antagonism, thus playing an important role in competition (Bayer‐Santos et al., 2019; Russell et al., 2014). In Xanthomonas spp., three subclasses of T6SSs have been reported (T6SS‐I, T6SS‐II, and T6SS‐III) (Alvarez‐Martinez et al., 2021; Bayer‐Santos et al., 2019; Timilsina et al., 2020). X. hortorum pvs hederae WHRI 7744, gardneri ATCC 19865PT, and cynarae CFBP 4188PT do not possess T6SS‐I and T6SS‐III clusters. A complete T6SS‐II cluster was detected in X. hortorum pv. hederae WHRI 7744, but not in strains CFBP 4188PT and ATCC 19865PT (Timilsina et al., 2020). However, two different studies reported that no T6SS was found in X. hortorum pv. gardneri and X. hortorum (unspecified pathovar). In one study, strain numbers were not specified (Bayer‐Santos et al., 2019), while in the other, the two strains were X. hortorum pv. gardneri ATCC 19865PT and X. hortorum B07‐007 (Alvarez‐Martinez et al., 2021).
8.3. Copper resistance and homeostasis
Copper resistance is attributed to the acquisition of a copper resistance gene cluster through horizontal gene transfer (Behlau et al., 2008; Bender et al., 1990; Cooksey, 1994). Copper resistance is usually plasmid encoded (Stall et al., 1986) and can thus be acquired via conjugation by other bacteria (Basim et al., 1999). Because copper‐based solutions have been extensively used for controlling bacterial spot diseases, with recommendations going back to the 1920s (Abrahamian et al., 2020; Higgins, 1922; Obradovic et al., 2008), copper‐resistant strains pose a challenge for disease management (see Disease control and management).
X. hortorum pv. gardneri strains differed in their response to copper. For example, strain ATCC 19865PT has copLAB homologs on the chromosome (cohLAB) and is homeostatic to copper, growing in copper concentrations up to 75 mg/L (Potnis et al., 2011). In contrast, strains JS749‐3 and ICMP 7383 have plasmidborne copLAB and copMGCDF genes (Richard, Boyer, et al., 2017; Richard, Ravigné, et al., 2017), as well as cusAB/smmD systems, involved in heavy metal efflux resistance and originally described in Stenotrophomonas maltophilia (Crossman et al., 2008). Strains JS749‐3 and ICMP 7383 are copper‐resistant and can grow in copper concentrations up to 470 mg/L (Richard, Ravigné, et al., 2017).
9. MOLECULAR HOST–PATHOGEN INTERACTIONS
The interactions of Xanthomonas species with their plant hosts involve the coordinated expression of various virulence factors (e.g., quorum sensing, effectors, avirulence genes) (Alvarez‐Martinez et al., 2021; Ryan et al., 2011; Timilsina et al., 2020). Quorum sensing, a chemical communication mechanism allowing bacteria to regulate group behaviours in response to stimulus, involves the production, release, and detection of auto‐inducers (Bassler, 1999; von Bodman et al., 2003; Miller & Bassler, 2001; Ng & Bassler, 2009; Whitehead et al., 2001). In Xanthomonas, the production and sensing of diffusible signal factors (DSF, e.g., α,β‐unsaturated fatty acids) (Wang et al., 2004) are regulated by genes within the regulation of the pathogenicity factors (rpf) cluster.
Knocking out rpfF and rpfC in X. hortorum pv. pelargonii Xhp305 altered in planta motility, decreased disease severity on pelargonium plants, and disrupted the plant colonization pattern (Barel et al., 2015). The resulting inability of X. hortorum pv. pelargonii to switch back and forth between biofilm and planktonic lifestyles is thus DSF‐dependent (Barel et al., 2015), and this shift is essential for pathogenicity (He & Zhang, 2008). Furthermore, in the rpfF and rpfC mutants, genes gumM, pilC, and pilT were down‐regulated compared to the wild type, suggesting that gumM expression and biofilm production, and the type 4 pilus apparatus are DSF‐dependent in X. hortorum pv. pelargonii (Barel et al., 2015).
Effectors are used by Xanthomonas species to trigger or suppress host defence mechanisms. Repertoires of effectors (effectomes) have been suggested to play a role determining host specificity (Hajri et al., 2009). Within X. hortorum, effector‐related work is mainly focused on pv. gardneri, but there are also reports on pv. carotae strains (more information below). The T3SS of X. hortorum pv. gardneri ATCC 19865PT was associated with hrpW (Potnis et al., 2011), a gene predicted to encode a pectate lyase (White et al., 2009), involved in plant tissue maceration and rotting (Collmer & Keen, 1986). The function of effector gene xopZ2, located downstream of hrpW, was suggested by avrBs2 reporter gene fusion (Potnis et al., 2011). Other T3SS effectors (T3Es) in X. hortorum pv. gardneri strains were also reported: XopAM, XopAO (homolog of AvrRpm1 from P. syringae), XopAQ (homolog of Rip6/Rip11 from R. solanacearum), and XopAS (homolog of HopAS1 from P. syringae). Effectors XopAM and XopAO were demonstrated to be dependent on the T3SS using the AvrBs2 reporter system (Potnis et al., 2011). In addition, four novel T3Es were reported in multiple field strains of X. hortorum pv. gardneri: a second XopE2 paralog, in addition to XopJ and two predicted effectors, named T3EP and PTP, with homologs in R. solanacearum and X. campestris pv. campestris, respectively (Schwartz et al., 2015).
Effector AvrHah1, a transcription activator‐like (TAL) effector of the AvrBs3/PthA family (Schornack et al., 2008), was the first characterized effector of X. hortorum pv. gardneri. AvrHah1 was able to trigger a Bs3‐dependent hypersensitive response (HR) on pepper plants (Schornack et al., 2008). Gain‐of‐function experiments with a X. euvesicatoria pv. euvesicatoria strain revealed that avrHah1 is responsible for enhanced water‐soaking in pepper leaves, a phenotype typical for the compatible interaction of X. hortorum pv. gardneri, the donor pathogen (Schornack et al., 2008). The virulence function of AvrHah1, triggering enhanced water‐soaking in its known hosts tomato, pepper, and Nicotiana benthamiana, was attributed to the movement of water into the infected apoplast (Schwartz et al., 2017). Gene avrBs7 was also identified in X. hortorum pv. gardneri as another avirulence gene as its product triggered an HR in Capsicum baccatum var. pendulum (Potnis et al., 2012). When the corresponding single dominant resistance gene Bs7 was introgressed into C. annuum ‘Early Calwonder’ (ECW), the resulting near‐isogenic line ECW‐70R was resistant to strains harbouring avrBs7.
Twenty‐one candidate T3E genes were identified in X. hortorum pv. carotae M081, and the products of two of them, AvrBs2 and XopQ, were found to elicit effector‐triggered immunity (Kimbrel et al., 2011). Using Agrobacterium‐mediated transient expression of avrBs2 from X. hortorum pv. carotae in transgenic N. benthamiana triggered an HR in a Bs2‐dependent manner. In contrast, no phenotypes were visible in wild‐type N. benthamiana lacking Bs2 on delivery of the same DNA construct. Transient expression of xopQ also resulted in strong and rapid HRs in most of the infiltrated leaves of wild‐type Nicotiana tabacum, perhaps mediated by another resistance gene. These observations indicated a possibility for resistance gene‐mediated control of X. hortorum pv. carotae (Kimbrel et al., 2011). A core Xanthomonas effectome of nine effectors, including AvrBs2, XopQ, and XopZ previously described, was reported in the tested strains of a study including X. hortorum B07‐007 and X. hortorum pv. gardneri ATCC 19865PT.
10. DISEASE CONTROL AND MANAGEMENT
An integrated control programme that focuses on excluding, reducing, or eradicating the pathogen, in combination with various methods like biological control and host resistance breeding, is the most suitable to manage bacterial spot pathogens like X. hortorum (Agrios, 2005; Marin et al., 2019). Preventing X. hortorum infections by excluding the pathogen from its hosts is crucial, especially because global trade of plants, seeds, and other propagating material plays an important role in the dissemination of this species (see Epidemiology).
Because X. hortorum pv. gardneri is locally present in the territory of the European and Mediterranean Plant Protection Organization (EPPO), the pathogen is on the EPPO A2 list and is recommended for regulation as a quarantine pest (EPPO, 2021). Since 2020, the EU (European Union) Plant Health Law regulates this pathovar as a nonquarantine pest (RNQP; Picard et al., 2018) on seeds, propagating, and planting material of tomato and peppers as well as propagating material of ornamental peppers (EU Commission, 2019). Other Regional Plant Protection Organizations (RPPOs) can implement regional phytosanitary regulations. For example, X. hortorum pv. carotae is considered an A1 plant pest by the Caribbean Plant Protection Commission (CPPC) and is under strict quarantine control there. Certification programmes propose requirements for production of disease‐free plants. For example, certification scheme EPPO PM4/3 outlines various testing methods for X. hortorum pv. pelargonii on different propagation materials (nuclear, basic stock, and certified cuttings). Because cool temperatures during propagation suppress symptom expression, methods to detect low pathogen numbers in asymptomatic tissue are thus crucial (see Identification and detection).
Physical or chemical treatment of the planting material can decrease pathogen inoculum (Janse & Wenneker, 2002). Hot‐water seed treatment reduced X. hortorum pvs carotae and gardneri infections. However, hot‐water seed treatment can sometime be unsuitable. For example, a treatment at 50°C for 2 h of lettuce seeds against X. hortorum pv. vitians significantly reduced seed germination (Carisse et al., 2000). Some chemical seed treatments against this pathovar, such as soaking in 1% sodium hypochlorite for 5 min (Carisse et al., 2000), in 3% hydrogen peroxide for 5 min, or in suspensions of copper hydroxide plus mancozeb (Pernezny et al., 2002), were more effective in reducing seed contamination than others (copper hydroxide alone, benzoyl peroxide, or calcium peroxide). The seed treatments described above are limited to university or extension research and, to the best of our knowledge, are not found in official documents by the National Seed Health System (NSHS) or the International Seed Federation (ISF‐ISHI). The use of seed treatments remains at the discretion of seed production companies, which must indicate what treatment was used on each seed lot.
Management of epiphytic X. hortorum (see Epidemiology) is challenging. Suppression methods of epiphytic X. hortorum pv. carotae on carrot foliage include sanitation and the use of drip irrigation to avoid wetting the phyllosphere during seed maturation (du Toit et al., 2005). Crop rotations or fallow periods could be used to eliminate contamination in plant debris by overwintering pathovars (Barak et al., 2001). In addition, good weed control and removing diseased plants can reduce inoculum amount (Barak et al., 2001; Toussaint et al., 2012), keeping in mind that some X. hortorum pathovars can survive epiphytically on weeds, and even infect them (see Epidemiology). Another good practice for decreasing the risk of disseminating X. hortorum pv. pelargonii involves not growing perennial Geranium spp. near greenhouse facilities producing Pelargonium spp. (Nameth et al., 1999).
Foliar applications of copper‐based bactericides have been used for X. hortorum pvs carotae and vitians with variable efficacy depending on various factors (e.g., application time, disease development stage, and climate) (Bull & Koike, 2005; du Toit et al., 2005). Copper‐based applications are unsustainable as they have adverse environmental effects, and because copper‐induced resistant strains are problematic for sustainable, long‐term control (Fishel, 2005; Husak, 2015; Willis & Bishop, 2016). For example, copper‐induced resistance was reported in strains of X. hortorum pv. gardneri (Abbasi et al., 2015; Khanal et al., 2020). The use of nanoparticles was studied to manage copper‐resistant and/or copper‐tolerant strains. For example, silver (Ag) nanoparticles merged in a double‐stranded DNA–graphene oxide matrix (Ag‐dsDNA‐GO) exhibited antibacterial activity against copper‐tolerant X. hortorum pv. gardneri strains (Strayer, Oscoy, et al., 2016).
Biological control solutions against some X. hortorum pathovars have been proposed. For example, alternative nontoxic compounds that induce a systemic acquired resistance (SAR) in the host plant can provide a more environmentally sustainable approach to disease management than pesticides. The compound acibenzolar‐S‐methyl (ASM), a benzothiadiazole, released in Europe as Bion (Syngenta Ltd) and in the United States as Actigard 50WG (Syngenta Crop Protection Inc.), has shown positive results for controlling bacterial spot caused by X. hortorum pvs gardneri, pelargonii, and carotae (regarding the latter, the application was only successful on seeds) (Blainski et al., 2018; Pontes et al., 2016; Simmons et al., 2010; Yellareddygari et al., 2013). However, a limitation of ASM is its adverse effect on tomato growth and yield, which may be attributed to the energy cost associated with resistance induction (Romero et al., 2001).
Treating geranium leaves with methyl jasmonate inhibited multiplication of X. hortorum pv. pelargonii (Zhang, Grefer, et al., 2009) and spraying leaves with EPSs from Lactiplantibacillus plantarum triggered a local induced resistance in tomato leaves against X. hortorum pv. gardneri (Blainski et al., 2018). When tested on agar plates, various essential oils inhibited X. hortorum pathovars. Origanum compactum (oregano) and Thymus vulgaris (thyme) inhibited X. hortorum pv. pelargonii (Kokoskova & Pavela, 2006), and three oregano species (O. acutidens, O. rotundifolium, and O. vulgare) seemed to inhibit the growth of X. hortorum pvs pelargonii and vitians (Dadasoglu et al., 2011). Geraniin, a tannin extracted from sugar maple, resulted in high mortality of X. hortorum pv. vitians bacterial cells when tested by plate counting (Delisle‐Houde et al., 2021).
Two P. syringae pv. syringae isolates, 422 and 17‐049, decreased the colonization of X. hortorum pv. carotae on carrot leaves (Belvoir et al., 2019). Less virulent quorum‐sensing mutants that elicit plant SAR might have a potential in management of X. hortorum pv. pelargonii (Barel et al., 2015). Bacteriophages applied as foliar sprays provided significant control of the disease caused by X. hortorum pv. gardneri in field trials (Balogh et al., 2003; Flaherty et al., 2000). However, bacteriophage effectiveness strongly depended on UV radiation and other environmental factors that affect their persistence in the phyllosphere (Iriarte et al., 2007). In greenhouse trials on potted geraniums, daily foliar sprays of bacteriophages significantly reduced disease caused by X. hortorum pv. pelargonii (Flaherty et al., 2001).
11. HOST RESISTANCE
Resistance breeding research against X. hortorum has been focused on tomato, pepper, lettuce, carrot, and pelargonium, and multiple plant cultivars showed moderate to high resistance against the various pathovars. Wild tomatoes have a broad‐spectrum resistance against multiple X. hortorum pv. gardneri strains (Liabeuf et al., 2015). Screening of S. lycopersicum and Solanum pimpinellifolium germplasm using HR identified partially resistant S. lycopersicum lines, as well as three S. pimpinellifolium accessions (LA2533, LA1936, and PI 128,216) resistant against the pathovar. This resistance may be controlled by one to four loci with moderate heritability. The S. pimpinellifolium lines were also resistant under field conditions (Liabeuf et al., 2015). Another wild tomato genotype, S. lycopersicum var. cerasiforme line PI 114490, possessed broad‐spectrum resistance to multiple xanthomonad pathogens, including X. hortorum pv. gardneri. Resistance in PI 114490 is quantitatively inherited, and QTL‐3 locus and allele at QTL‐11 are major contributors of resistance against X. hortorum pv. gardneri (Bernal et al., 2020).
A mutation in DMR6 (downy mildew resistance 6) in Arabidopsis, conferring broad‐spectrum resistance to various Xanthomonas and Pseudomonas phythopathogens, was tested in tomato (Thomazella et al., 2016). The stable transgenic tomato plants were resistant against X. hortorum pv. gardneri and were not compromised in their growth and development.
In pepper, two dominant resistance genes, Bs3 and Bs7, are known to confer resistance against X. hortorum pv. gardneri strains carrying avirulence genes avrHah1 and avrBs7, respectively (Potnis et al., 2012; Schornack et al., 2008). However, the plasmidborne nature of both avirulence genes suggests vulnerability to resistance breakdown, so they have not been further considered in breeding programmes. Screening of core pepper germplasm collection against X. hortorum pv. gardneri revealed that more than 40 PI lines of C. baccatum in greenhouse conditions and multiple PI lines of C. annuum showed promising resistance levels (Potnis et al., 2012). A total of 20 significant single nucleotide polymorphisms (SNPs), co‐located within 150 kb of 92 unique genes, were recently identified against the pathovar (Potnis et al., 2019).
Regarding X. hortorum pv. vitians, different lettuce genotypes (L. sativa) show differential responses to the pathogen. For example, romaine and butterhead lettuce cultivars are among the highly susceptible ones (Pernezny et al., 1995). Moderately resistant cultivars include both green‐leaf (e.g., Waldmann's Green and Grand Rapids) and red‐leaf (e.g., Red Line) cultivars (Carisse et al., 2000), although other studies have noted their susceptibility (Bull et al., 2007). Such discrepancy could be due to differences in the experimental setups of the studies, such as using different strains for the pathogenicity tests. Other moderately resistant cultivars include Little Gem and Reine des Glaces (Batavia crisphead) (Bull et al., 2007). These two latter cultivars were deemed to be promising in breeding resistant cultivars against X. hortorum pv. vitians (Hayes, Trent, Mou, et al., 2014; Hayes, Trent, Truco, et al., 2014). However, undesirable traits (e.g., small size and low yield) associated with cv. Little Gem are of concern. Furthermore, this cultivar has also shown variable resistance in separate studies, making it an unattractive candidate (Bull et al., 2007; Lu & Raid, 2013). This difference could be due either to virulence dissimilarities at the strain level or to host susceptibility variation as a result of different environmental conditions at the cultivar evaluation locations (Lu & Raid, 2013). In addition, resistance of cv. Reine des Glaces was also highly dependent on environmental conditions (Bull et al., 2007).
Genetic maps of various wild lettuce species like L. serriola, L. saligna, and L. virosa, have revealed multiple genes conferring broad resistance (McHale et al., 2009; Truco et al., 2013). However, L. saligna and L. virosa have compatibility issues, making hybridization difficult. The broad resistance in wild lettuce species have yet to be tested against X. hortorum pv. vitians.
The high genetic variability of the pathogen population is a challenge for breeding cultivars with durable resistance. Resistance against MLSA‐based groups B, D, and E of X. hortorum pv. vitians was identified to be controlled by a single dominant locus, Xanthomonas resistance 1 (Xar1), in the Batavia heirloom cv. La Brillante (Bull et al., 2016; Hayes, Trent, Mou, et al., 2014; Hayes, Trent, Truco, et al., 2014). Two other cultivars, Little Gem and Pavane, carry Xar1 alleles and are resistant to Californian isolates of X. hortorum pv. vitians. Another locus identified as X. campestris vitians resistance (Xcvr) was found in the same linkage group (LG2) during the mapping of a PI 358001‐1 × Tall Guzmaine population. The durability of Xar1 and Xcvr resistances in cv. La Brillante and PI 358001‐1 raised concerns because of the high variability in the pathogen population (Hayes, Trent, Mou, et al., 2014; Hayes, Trent, Truco, et al., 2014). Major and minor quantitative trait loci (QTLs) controlling this resistance were identified and co‐located in the same region of LG2 as previously identified with Xar1 and Xcvr (Sandoya et al., 2019).
A germplasm screening of carrot species (e.g., PI lines, public inbred lines, commercial cultivars, and wild varieties) indicated four PI lines (PI 263601, PI 418967, PI 432905, and PI 432906) and two of the wild relatives, Ames 7674 and SS10 OR, were the most resistant against X. hortorum pv. carotae (Christianson et al., 2015). The resistant PI lines are promising for use in commercial breeding programmes (Christianson et al., 2015).
In the genus Pelargonium, only a small number of pelargonium and geranium species have been screened for resistance against X. hortorum pv. pelargonii based on symptom expression alone after pathogen inoculation (Griesbach & Olbricht, 2002; Zhang, Sairam, et al., 2009). Five resistant pelargonium species were identified (Griesbach & Olbricht, 2002; Zhang, Sairam, et al., 2009), but most commercially important cultivars of Pelargonium zonale hybrids were highly susceptible (Griesbach & Olbricht, 2002; Zhang, Sairam, et al., 2009).
12. RESEARCH PERSPECTIVES
Several advances improving our understanding of the X. hortorum species have recently been published. However, some knowledge gaps remain, mainly related to extent of host range, detection, and control methods, including host resistance. Given the broad and diverse host range described for this species, it is likely that unreported hosts remain to be identified in various ecosystems. Further investigation of the natural and experimental host ranges of X. hortorum could provide insight into its evolutionary history and determine if plant domestication influenced host specialization of the pathovars of X. hortorum.
The recent increased availability of genomic data for X. hortorum will help in the identification of novel isolates from new natural hosts through establishing quick, field‐deployable detection methods. Such tools will also be very beneficial for phytosanitary control, especially as prevention strategies are preferred to formulation applications and are less costly than containment and eradication measures.
There is a significant need to conduct a comprehensive comparative genomics analysis of this species, especially in view of the recent taxonomical changes. Because plasmids offer a potentially large source of variation in the species, determining the plasmid content of strains and their contribution to pathogenicity is highly relevant. The recent application of a TnSeq analysis in X. hortorum pv. vitians paves the way to functional genomics analysis of other X. hortorum members. Aside from providing insights into essential bacterial genes in different in vitro and in planta conditions, TnSeq would also considerably improve our understanding of X. hortorum biology.
Most important commercial varieties are still highly susceptible to diseases caused by X. hortorum. The inefficiency of application‐based control strategies further consolidates host resistance as a promising area for devising practical and durable disease control solutions against X. hortorum. Because nonhost resistance is more durable than host resistance, screening more nonhost species for their disease response to X. hortorum could uncover broad, nonhost resistance genes against the pathovars. Furthermore, exploring recent advancements in the field of host resistance against bacteria, such as CRISPR/Cas9‐mediated gene mutations, also sound promising for breeding X. hortorum‐resistant plant cultivars. However, as highlighted throughout this pathogen profile, the high genetic variability of these phytopathogens affecting several plant families represents a real challenge for long‐term resistance.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
This article is based on work from COST Action CA16107 EuroXanth, supported by COST (European Cooperation in Science and Technology). N.C.D. was supported by grant no. IZCOZ0_177064 from the Swiss National Science Foundation (SNSF). L.M. was funded by a grant from the French Ministry of Higher Education, Research and Innovation. J.M.J. acknowledges the support by SCRI (grant no. 2020‐51181‐32154) from the USDA National Institute of Food and Agriculture and from Ohio Department of Agriculture Specialty Crops Block Grant (AGR‐SCG‐19‐03). J.M.J. and E.B. are also grateful to support from USDA NIFA award no. 2018‐67013‐28490 through the NSF/NIFA Plant Biotic Interactions Program. N.P. acknowledges support by the SCRI (grant no. 2019‐51181‐30010) from the USDA National Institute of Food and Agriculture. J.F.P. acknowledges support from the Department of Life Sciences and Facility Management of the Zurich University of Applied Sciences (ZHAW) in Wädenswil. The authors thank John Bennett (ZHAW) for critically proofreading the manuscript.
Dia, N.C. , Morinière, L. , Cottyn, B. , Bernal, E. , Jacobs, J.M. , Koebnik, R. , et al (2022) Xanthomonas hortorum – beyond gardens: Current taxonomy, genomics, and virulence repertoires. Molecular Plant Pathology, 23, 597–621. 10.1111/mpp.13185
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- Abbasi, P.A. , Khabbaz, S.E. , Weselowski, B. & Zhang, L. (2015) Occurrence of copper‐resistant strains and a shift in Xanthomonas spp. causing tomato bacterial spot in Ontario. Canadian Journal of Microbiology, 61, 753–761. [DOI] [PubMed] [Google Scholar]
- Abrahamian, P. , Sharma, A. , Jones, J.B. & Vallad, G.E. (2020) Dynamics and spread of bacterial spot epidemics in tomato transplants grown for field production. Plant Disease, 105, 566–575. [DOI] [PubMed] [Google Scholar]
- Adriko, J. , Mbega, E.R. , Mortensen, C.N. , Wulff, E.G. , Tushemereirwe, W.K. , Kubiriba, J. et al. (2014) Improved PCR for identification of members of the genus Xanthomonas . European Journal of Plant Pathology, 138, 293–306. [Google Scholar]
- Agrios, G.N. (2005) Plant pathology. Burlington, MA: Elsevier Academic Press. [Google Scholar]
- Akhtar, M.A. & Aslam, M. (1990) Xanthomonas campestris pv. hederae causing bacterial leaf spot of Schefflera at Islamabad. Pakistan Journal of Agricultural Research, 11, 290–292. [Google Scholar]
- Al‐Saleh, M.A. , Ibrahim, Y.E. , Abo‐Elyousr, K.A.M. & Alibrahim, J.S. (2011) Population dynamics of Xanthomonas campestris pv. vitians on different plant species and management of bacterial leaf spot of lettuce under greenhouse conditions. Crop Protection, 30, 883–887. [Google Scholar]
- Alvarez‐Martinez, C.E. , Sgro, G.G. , Araujo, G.G. , Paiva, M.R.N. , Matsuyama, B.Y. , Guzzo, C.R. et al. (2021) Secrete or perish: The role of secretion systems in Xanthomonas biology. Computational and Structural Biotechnology Journal, 19, 279–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo, E.R. , Costa, J.R. , Ferreira, M.A.S.V. & Quezado‐Duval, A.M. (2012) Simultaneous detection and identification of the Xanthomonas species complex associated with tomato bacterial spot using species‐specific primers and multiplex PCR. Journal of Applied Microbiology, 113, 1479–1490. [DOI] [PubMed] [Google Scholar]
- Araújo, E.R. , Costa, J.R. , Ferreira, M.A.S.V. & Quezado‐Duval, A.M. (2017) Widespread distribution of Xanthomonas perforans and limited presence of X. gardneri in Brazil. Plant Pathology, 66, 159–168. [Google Scholar]
- Araújo, E.R. , Costa, J.R. , Pontes, N.C. & Quezado‐Duval, A.M. (2015) Xanthomonas perforans and X. gardneri associated with bacterial leaf spot on weeds in Brazilian tomato fields. European Journal of Plant Pathology, 143, 543–548. [Google Scholar]
- Araújo, E.R. , Pereira, R.C. , Ferreira, M.A.S.V. , Café‐Filho, A.C. , Moita, A.W. & Quezado‐Duval, A.M. (2011) Effect of temperature on pathogenicity components of tomato bacterial spot and competition between Xanthomonas perforans and X. gardneri . Acta Horticulturae, 914, 39–42. [Google Scholar]
- Arnaud, G. (1920) Une maladie bactérienne du lierre (Hedera helix L.). Comptes Rendus Hebdomadaires des Séances de l’Académie des Sciences, 171, 121–122. [Google Scholar]
- Balaž, J. , Ivanović, Ž. , Davidović, A. , Iličić, R. , Janse, J. & Popović, T. (2016) Characterization of Xanthomonas hortorum pv. pelargonii isolated from geranium in Serbia. Plant Disease, 100, 164–170. [DOI] [PubMed] [Google Scholar]
- Balogh, B. , Jones, J.B. , Momol, M.T. , Olson, S.M. , Obradovic, A. , King, P. et al. (2003) Improved efficacy of newly formulated bacteriophages for management of bacterial spot on tomato. Plant Disease, 87, 949–954. [DOI] [PubMed] [Google Scholar]
- Barak, J.D. & Gilbertson, R.L. (2003) Genetic diversity of Xanthomonas campestris pv. vitians, the causal agent of bacterial leafspot of lettuce. Phytopathology, 93, 596–603. [DOI] [PubMed] [Google Scholar]
- Barak, J.D. , Koike, S.T. & Gilbertson, R.L. (2001) Role of crop debris and weeds in the epidemiology of bacterial leaf spot of lettuce in California. Plant Disease, 85, 169–178. [DOI] [PubMed] [Google Scholar]
- Barak, J.D. , Koike, S.T. & Gilbertson, R.L. (2002) Movement of Xanthomonas campestris pv. vitians in the stems of lettuce and seed contamination. Plant Pathology, 51, 506–512. [Google Scholar]
- Barel, V. , Chalupowicz, L. , Barash, I. , Sharabani, G. , Reuven, M. , Dror, O. et al. (2015) Virulence and in planta movement of Xanthomonas hortorum pv. pelargonii are affected by the diffusible signal factor (DSF)‐dependent quorum sensing system. Molecular Plant Pathology, 16, 710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basim, H. , Stall, R.E. , Minsavage, G.V. & Jones, J.B. (1999) Chromosomal gene transfer by conjugation in the plant pathogen Xanthomonas axonopodis pv. vesicatoria . Phytopathology, 89, 1044–1049. [DOI] [PubMed] [Google Scholar]
- Bassler, B.L. (1999) How bacteria talk to each other: regulation of gene expression by quorum sensing. Current Opinion in Microbiology, 2, 582–587. [DOI] [PubMed] [Google Scholar]
- Bayer‐Santos, E. , de Ceseti, L.M. , Farah, C.S. & Alvarez‐Martinez, C.E. (2019) Distribution, function and regulation of type 6 secretion systems of Xanthomonadales. Frontiers in Microbiology, 10, 1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlau, F. , Belasque, J. , Bergamin Filho, A. , Graham, J.H. , Leite, R.P. & Gottwald, T.R. (2008) Copper sprays and windbreaks for control of citrus canker on young orange trees in southern Brazil. Crop Protection, 27, 807–813. [Google Scholar]
- Belvoir, T. , Scott, J.C. & Dung, J.K. (2019) Identification and screening of bacteria for the biocontrol of Xanthomonas hortorum pv. carotae in carrot seed crops. APS Annual Meeting. St Paul, MN: Plant Health 2019.
- Bender, C.L. , Malvick, D.K. , Conway, K.E. , George, S. & Pratt, P. (1990) Characterization of pXV1OA, a copper resistance plasmid in Xanthomonas campestris pv. vesicatoria . Applied and Environmental Microbiology, 56, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict, A.A. , Alvarez, A.M. & Pollard, L.W. (1990) Pathovar‐specific antigens of Xanthomonas campestris pv. begoniae and X. campestris pv. pelargonii detected with monoclonal antibodies. Applied and Environmental Microbiology, 56, 572–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergey, D.H. , Harrison, F.C. , Breed, R.S. , Hammer, B.W. & Huntoon, F.M. (1923) Bergey’s manual of determinative bacteriology. Baltimore, MD: Williams and Wilkins. [Google Scholar]
- Bernal, E. & Francis, D.M. (2021) Processing tomato germplasm with improved resistance to bacterial spot. HortScience, 56, 519–520. [Google Scholar]
- Bernal, E. , Liabeuf, D. & Francis, D.M. (2020) Evaluating quantitative trait locus resistance in tomato to multiple Xanthomonas spp. Plant Disease, 104, 423–429. [DOI] [PubMed] [Google Scholar]
- Blainski, J.M.L. , da Rocha Neto, A.C. , Schimidt, E.C. , Voltolini, J.A. , Rossi, M.J. & Di Piero, R.M. (2018) Exopolysaccharides from Lactobacillus plantarum induce biochemical and physiological alterations in tomato plant against bacterial spot. Applied Microbiology and Biotechnology, 102, 4741–4753. [DOI] [PubMed] [Google Scholar]
- von Bodman, S.B. , Bauer, W.D. & Coplin, D.L. (2003) Quorum sensing in plant‐pathogenic bacteria. Annual Review of Phytopathology, 41, 455–482. [DOI] [PubMed] [Google Scholar]
- Bouzar, H. , Jones, J.B. , Stall, R.E. , Louws, F.J. , Schneider, M. , Rademaker, J.L.W. et al. (1999) Multiphasic analysis of xanthomonads causing bacterial spot disease on tomato and pepper in the Caribbean and Central America: evidence for common lineages within and between countries. Phytopathology, 89, 328–335. [DOI] [PubMed] [Google Scholar]
- Brown, N.A. (1918) Some bacterial diseases of lettuce. Journal of Agricultural Research, 13, 367–388. [Google Scholar]
- Brown, N.A. (1923) Bacterial leaf spot of geranium in the eastern United States. Journal of Agricultural Research, 23, 361–372. [Google Scholar]
- Bugbee, W.M. & Anderson, N.A. (1963) Whitefly transmission of Xanthomonas pelargonii and histological examination of leafspots of Pelargonium hortorum . Phytopathology, 53, 177–178. [Google Scholar]
- Bull, C.T. , Goldman, P. , Hayes, R. , Madden, L. & Koike, S. (2007) Genetic diversity of lettuce for resistance to bacterial leaf spot caused by Xanthomonas campestris pv. vitians . Plant Health Progress, 8, 11. [Google Scholar]
- Bull, C.T. & Koike, S.T. (2005) Evaluating the efficacy of commercial products for management of bacterial leaf spot on lettuce. Plant Health Progress, 6, 3. Available at: https://apsjournals.apsnet.org/doi/10.1094/PHP‐2005‐1121‐01‐RS [Accessed 24th September 2021]. [Google Scholar]
- Bull, C.T. & Koike, S.T. (2015) Practical benefits of knowing the enemy: modern molecular tools for diagnosing the etiology of bacterial diseases and understanding the taxonomy and diversity of plant‐pathogenic bacteria. Annual Review of Phytopathology, 53, 157–180. [DOI] [PubMed] [Google Scholar]
- Bull, C. , Trent, M. & Hayes, R. (2016) Three races of Xanthomonas campestris pv. vitians causing bacterial leaf spot on lettuce identified. Science to Practice, Vol. 106. APS Annual Meeting. St Paul, MN: American Phytopathological Society, pp. 437‐P. https://www.apsnet.org/meetings/Documents/2016_meeting_abstracts/aps2016_244.htm [Accessed 18th January 2022]. [Google Scholar]
- Burkholder, W.H. & Guterman, C.E.F. (1932) Synergism in a bacterial disease of Hedera helix . Phytopathology, 22, 781–784. [Google Scholar]
- Büttner, D. (2016) Behind the lines–actions of bacterial type III effector proteins in plant cells. FEMS Microbiology Reviews, 40, 894–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. & Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiology Reviews, 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Cândido, E.S. , Pereira, J.L. , Quezado‐Duval, A.M. , Noronha, E.F. , Krüger, R.H. & Quirino, B.F. (2008) Xanthomonas gardneri exoenzymatic activity towards plant tissue. World Journal of Microbiology and Biotechnology, 24, 163–170. [Google Scholar]
- Carisse, O. , Ouimet, A. , Toussaint, V. & Philion, V. (2000) Evaluation of the effect of seed treatments, bactericides, and cultivars on bacterial leaf spot of lettuce caused by Xanthomonas campestris pv. vitians . Plant Disease, 84, 295–299. [DOI] [PubMed] [Google Scholar]
- Catara, V. , Cubero, J. , Pothier, J.F. , Bosis, E. , Bragard, C. , Đermić, E. et al. (2021) Trends in molecular diagnosis and diversity studies for phytosanitary regulated Xanthomonas . Microorganisms, 9, 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, A.R. (1984) Xanthomonas campestris pv. hederae causes a leaf spot of five species of Araliaceae . Plant Pathology, 33, 439–440. [Google Scholar]
- Chittaranjan, S. & De Boer, S.H. (1997) Detection of Xanthomonas campestris pv. pelargonii in geranium and greenhouse nutrient solution by serological and PCR techniques. European Journal of Plant Pathology, 103, 555–563. [Google Scholar]
- Christianson, C.E. , Jones, S.S. & du Toit, L.J. (2015) Screening carrot germplasm for resistance to Xanthomonas hortorum pv. carotae . HortScience, 50, 341–350. [Google Scholar]
- Collmer, A. & Keen, N.T. (1986) The role of pectic enzymes in plant pathogenesis. Annual Review of Phytopathology, 24, 383–409. [Google Scholar]
- Cooksey, D.A. (1994) Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiology Reviews, 14, 381–386. [DOI] [PubMed] [Google Scholar]
- Corsaro, M.M. , De Castro, C. , Molinaro, A. & Parrilli, M. (2001) Structure of lipopolysaccharides from phytopathogenic Gram‐negative bacteria. Recent Research Developments in Phytochemistry, 119–138. [Google Scholar]
- Cottyn, B. , De Paepe, B. , Dia, N.C. , Van Malderghem, C. , Pothier, J.F. , & Van Vaerenbergh, J. (2021) First report of bacterial leaf spot of Hydrangea in retail nurseries in Belgium caused by strains assigned to a new Xanthomonas hortorum clade. New Disease Reports, 43, e12008. [Google Scholar]
- Crossman, L.C. , Gould, V.C. , Dow, J.M. , Vernikos, G.S. , Okazaki, A. , Sebaihia, M. et al. (2008) The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biology, 9, R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadasoglu, F. , Aydin, T. , Kotan, R. , Cakir, A. , Ozer, H. , Kordali, S. et al. (2011) Antibacterial activities of extracts and essential oils of three Origanum species against plant pathogenic bacteria and their potential use as seed disinfectants. Journal of Plant Pathology, 271–282. [Google Scholar]
- Daughtrey, M.L. & Benson, D.M. (2005) Principles of plant health management for ornamental plants. Annual Review of Phytopathology, 43, 141–169. [DOI] [PubMed] [Google Scholar]
- Daughtrey, M.L. & Wick, R.L. (1995) Geraniums IV. The grower’s manual. In: White, J.W. (Ed.) Vascular wilt diseases. Chicago, IL: Ball Publishing, pp. 237–242. [Google Scholar]
- Dehghan‐Niri, M. & Rahimian, H. (2016) First report of Xanthomonas gardneri causing bacterial leaf spot on burdock in Iran. Journal of Plant Pathology, 98, 677. [Google Scholar]
- Delisle‐Houde, M. , Blais, M. , Tweddell, R.J. & Rioux, D. (2021) Antibacterial activity of geraniin from sugar maple leaves: an ultrastructural study with the phytopathogen Xanthomonas campestris pv. vitians . Journal of Plant Pathology, 103, 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty, D.E. , Powell, C.C. & Larsen, P.O. (1974) Epidemiology and control of bacterial leaf spot and stem rot of Pelargonium hortorum . Phytopathology, 64, 1081–1083. [Google Scholar]
- Dowson, W.J. (1943) On the generic names Pseudomonas, Xanthomonas and Bacterium for certain bacterial plant pathogens. Transactions of the British Mycological Society, 26, 4–14. [Google Scholar]
- Dye, D.W. (1966) Cultural and biochemical reactions of additional Xanthomonas species. New Zealand Journal of Science, 9, 913–919. [Google Scholar]
- Dye, D.W. (1967) Bacterial spot of ivy caused by Xanthomonas hederae (Arnaud, 1920) Dowson 1939, in New Zealand. New Zealand Journal of Science, 10, 481–485. [Google Scholar]
- Dye, D.W. , Bradbury, J.F. , Goto, M. , Hayward, A.C. , Lelliott, R.A. & Schroth, M.N. (1980) International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Review of Plant Pathology, 59, 153–168. [Google Scholar]
- Egel, D.S. , Jones, J.B. , Minsavage, G.V. , Creswell, T. , Ruhl, G. , Maynard, E. et al. (2018) Distribution and characterization of Xanthomonas strains causing bacterial spot of tomato in Indiana. Plant Health Progress, 319–321. [Google Scholar]
- Egorova, M.S. (2015) Species diversity and diagnostic methods for phytopathogenic bacteria of the genus Xanthomonas affecting plants of the Poaceae family . PhD thesis, Moscow, University of Moscow.
- Egorova, M.S. , Mazurin, E.S. , Polityko, V.A. , & Ignatov, A.N. (2014) Diversity of phytopathogenic bacteria of genus Xanthomonas isolated from Poaceae plants in Russia. RUDN Journal of Agronomy and Animal Industries, 4, 47–53. [Google Scholar]
- EPPO (2021) EPPO global database. Available at: https://gd.eppo.int/ [Accessed 25th April 2021]. [Google Scholar]
- EU Commission . (2019) Commission Implementing Regulation (EU) 2019/2072 of 28 November 2019 establishing uniform conditions for the implementation of Regulation (EU) 2016/2031 of the European Parliament and the Council, as regards protective measures against pests of plants, and repealing Commission Regulation (EC) No 690/2008 and amending Commission Implementing Regulation (EU) 2018/2019. Official Journal, L319, 1–278. [Google Scholar]
- FAOSTAT . (2021) FAOSTAT. Available at: http://www.fao.org/faostat/en/#home [Accessed 30th September, 2021]. [Google Scholar]
- Farahani, A.S. & Taghavi, S.M. (2016) Sensitive and specific detection of Xanthomonas hortorum pv. pelargonii in geranium by real‐time PCR. Journal of Plant Protection Research, 56, 265–269. [Google Scholar]
- Fayette, J. , Jones, J.B. , Pernezny, K. , Roberts, P.D. & Raid, R. (2018) Survival of Xanthomonas campestris pv. vitians on lettuce in crop debris, irrigation water, and weeds in south Florida. European Journal of Plant Pathology, 151, 341–353. [Google Scholar]
- Fayette, J. , Raid, R. , Roberts, P.D. , Jones, J.B. , Pernezny, K. , Bull, C.T. et al. (2016) Multilocus sequence typing of strains of bacterial spot of lettuce collected in the United States. Phytopathology, 106, 1262–1269. [DOI] [PubMed] [Google Scholar]
- Fishel, F. (2005) Pesticide toxicity profile: copper‐based pesticides. EDIS, 11, 1–4. [Google Scholar]
- Flaherty, J.E. , Harbaugh, B.K. , Jones, J.B. , Somodi, G.C. & Jackson, L.E. (2001) H‐mutant bacteriophages as a potential biocontrol of bacterial blight of geranium. HortScience, 36, 98–100. [Google Scholar]
- Flaherty, J.E. , Somodi, G.C. , Jones, J.B. , Harbaugh, B.K. & Jackson, L.E. (2000) Control of bacterial spot on tomato in the greenhouse and field with H‐mutant bacteriophages. HortScience, 35, 882–884. [Google Scholar]
- Gardner, M.W. & Kendrick, J.B. (1921) Bacterial spot of tomato. Journal of Agricultural Research, 21, 123–156. [Google Scholar]
- Gilbertson, R.L. (2002) Bacterial leaf blight of carrot. In: Davis, R.M. & Raid, R.N. (Eds.) Compendium of umbelliferous crop diseases. St Paul, MN: American Phytopathological Society, pp. 11–12. [Google Scholar]
- Gitaitis, R. & Walcott, R. (2007) The epidemiology and management of seedborne bacterial diseases. Annual Review of Phytopathology, 45, 371–397. [DOI] [PubMed] [Google Scholar]
- Glick, D.L. , Coffey, C.M. & Sulzinski, M.A. (2002) Simultaneous PCR detection of the two major bacterial pathogens of geranium. Journal of Phytopathology, 150, 54–59. [Google Scholar]
- Griesbach, E. & Olbricht, K. (2002) Resistance to Xanthomonas hortorum pv. pelargonii in the genus Pelargonium . Journal of Plant Diseases and Protection, 109, 553–568. [Google Scholar]
- Guérin, J. , Bigot, S. , Schneider, R. , Buchanan, S.K. & Jacob‐Dubuisson, F. (2017) Two‐partner secretion: combining efficiency and simplicity in the secretion of large proteins for bacteria–host and bacteria–bacteria interactions. Frontiers in Cellular and Infection Microbiology, 7, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini, J. , Néron, B. , Abby, S.S. , Garcillán‐Barcia, M.P. , de la Cruz, F. & Rocha, E.P.C. (2014) Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Research, 42, 5715–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajri, A. , Brin, C. , Hunault, G. , Lardeux, F. , Lemaire, C. , Manceau, C. et al. (2009) A «Repertoire for Repertoire» hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas . PLoS One, 4, e6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza, A.A. , Robene‐Soustrade, I. , Jouen, E. , Lefeuvre, P. , Chiroleu, F. , Fisher‐Le Saux, M. et al. (2012) Multilocus sequence analysis‐ and amplified fragment length polymorphism‐based characterization of xanthomonads associated with bacterial spot of tomato and pepper and their relatedness to Xanthomonas species. Systematic and Applied Microbiology, 35, 183–190. [DOI] [PubMed] [Google Scholar]
- Hayes, R.J. , Trent, M.A. , Mou, B. , Simko, I. , Gebben, S.J. & Bull, C.T. (2014) Baby leaf lettuce germplasm enhancement: developing diverse populations with resistance to bacterial leaf spot caused by Xanthomonas campestris pv. vitians . HortScience, 49, 18–24. [Google Scholar]
- Hayes, R.J. , Trent, M.A. , Truco, M.J. , Antonise, R. , Michelmore, R.W. & Bull, C.T. (2014) The inheritance of resistance to bacterial leaf spot of lettuce caused by Xanthomonas campestris pv. vitians in three lettuce cultivars. Horticulture Research, 1, 14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y.‐W. & Zhang, L.‐H. (2008) Quorum sensing and virulence regulation in Xanthomonas campestris . FEMS Microbiology Reviews, 32, 842–857. [DOI] [PubMed] [Google Scholar]
- Hébert, P.‐O. , Laforest, M. , Xu, D. , Ciotola, M. , Cadieux, M. , Beaulieu, C. et al. (2021) Genotypic and phenotypic characterization of lettuce bacterial pathogen Xanthomonas hortorum pv. vitians populations collected in Quebec, Canada. Agronomy, 11, 2386. [Google Scholar]
- Higgins, B.B. (1922) The bacterial spot of pepper. Pythopathology, 12, 501–516. [Google Scholar]
- Husak, V.V. (2015) Copper and copper‐containing pesticides: metabolism, toxicity and oxidative stress. Journal of Vasyl Stefanyk Precarpathian National University, 2, 39–51. [Google Scholar]
- Iriarte, F.B. , Balogh, B. , Momol, M.T. , Smith, L.M. , Wilson, M. & Jones, J.B. (2007) Factors affecting survival of bacteriophage on tomato leaf surfaces. Applied and Environmental Microbiology, 73, 1704–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques, M.‐A. , Arlat, M. , Boulanger, A. , Boureau, T. , Carrère, S. , Cesbron, S. et al. (2016) Using ecology, physiology, and genomics to understand host specificity in Xanthomonas . Annual Review of Phytopathology, 54, 163–187. [DOI] [PubMed] [Google Scholar]
- Janse, J.D. & Wenneker, M. (2002) Possibilities of avoidance and control of bacterial plant diseases when using pathogen‐tested (certified) or ‐treated planting material. Plant Pathology, 51, 523–536. [Google Scholar]
- Jones, J.B. , Lacy, G.H. , Bouzar, H. , Stall, R.E. & Schaad, N.W. (2004) Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Systematic and Applied Microbiology, 27, 755–762. [DOI] [PubMed] [Google Scholar]
- Kahle, D. , Wickham, H. , Jackson, S. & Korpela, M. (2019) Package ‘ggmap’. Available at: https://github.com/dkahle/ggmap [Accessed 13th January 2022].
- Kakkar, A. , Nizampatnam, N.R. , Kondreddy, A. , Pradhan, B.B. & Chatterjee, S. (2015) Xanthomonas campestris cell–cell signalling molecule DSF (diffusible signal factor) elicits innate immunity in plants and is suppressed by the exopolysaccharide xanthan. Journal of Experimental Botany, 66, 6697–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. & Kado, C.I. (1990) Phenotypic switching affecting chemotaxis, xanthan production, and virulence in Xanthomonas campestris . Applied and Environmental Microbiology, 56, 3855–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen, F. , Ferreiro, D.U. , Oddo, C.G. , Ielmini, M.V. , Becker, A. , Pühler, A. et al. (1998) Xanthomonas campestris pv. campestris gum mutants: effects on xanthan biosynthesis and plant virulence. Journal of Bacteriology, 180, 1607–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick, J.B. (1934) Bacterial blight of carrot. Journal of Agricultural Research, 49, 493–510. [Google Scholar]
- Khanal, S. , Hind, S.R. & Babadoost, M. (2020) Occurrence of copper‐resistant Xanthomonas perforans and X. gardneri in Illinois tomato fields. Plant Health Progress, 21, 338–344. [Google Scholar]
- Kim, J.G. , Taylor, K.W. , Hotson, A. , Keegan, M. , Schmelz, E.A. & Mudgett, M.B. (2008) XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas‐infected tomato leaves. The Plant Cell, 20, 1915–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrel, J.A. , Givan, S.A. , Temple, T.N. , Johnson, K.B. & Chang, J.H. (2011) Genome sequencing and comparative analysis of the carrot bacterial blight pathogen, Xanthomonas hortorum pv. carotae M081, for insights into pathogenicity and applications in molecular diagnostics. Molecular Plant Pathology, 12, 580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass, T.L. , Long, J.J. , Summers, J.A. , Roman‐Reyna, V. , Koebnik, R. , Jacobs, J.M. et al. (2019) First report of bacterial blight of peony caused by Xanthomonas hortorum in Ohio. Plant Disease, 103, 2940. [Google Scholar]
- Knauss, J.F. & Tammen, J. (1964) The resistance of Pelargonium to Xanthomonas pelargonii (Brown) Starr and Burkholder. Phytopathology, 54, 128. [Google Scholar]
- Koenraadt, H. , van Betteray, B. , Germain, R. , Hiddink, G. , Jones, J.B. & Oosterhof, J. (2009) Development of specific primers for the molecular detection of bacterial spot of pepper and tomato. Acta Horticulturae, 808, 99–102. [Google Scholar]
- Koike, S.T. , Tjosvold, S.A. , Cooksey, D.A. & Azad, H.R. (1995) A bacterial leaf disease of lavender caused by Xanthomonas campestris . Plant Disease, 79, 859. [Google Scholar]
- Kokoskova, B. & Pavela, R. (2006) Effectivity of essential oils against Xanthomonas hortorum pv. pelargonii, the causal agent of bacterial blight on geranium. Mitteilungen‐Biologischen Bundesanstalt für Land und Forstwirtschaft, 408, 77. [Google Scholar]
- Koonin, E.V. (2000) How many genes can make a cell: the minimal‐gene‐set concept. Annual Review of Genomics and Human Genetics, 1, 99–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan, T.‐L. (1985) Detection of Xanthomonas campestris pv. carotae in carrot seed. Plant Disease, 69, 758–760. [Google Scholar]
- Lawley, T.D. , Klimke, W.A. , Gubbins, M.J. & Frost, L.S. (2003) F factor conjugation is a true type IV secretion system. FEMS Microbiology Letters, 224, 1–15. [DOI] [PubMed] [Google Scholar]
- Leite, R.P. , Minsavage, G.V. , Bonas, U. & Stall, R.E. (1994) Detection and identification of phytopathogenic Xanthomonas strains by amplification of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria . Applied and Environmental Microbiology, 60, 1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyns, F. , De Cleene, M. , Swings, J.‐G. & De Ley, J. (1984) The host range of the genus Xanthomonas . The Botanical Review, 50, 308–356. [Google Scholar]
- Liabeuf, D. , Francis, D.M. & Sim, S.C. (2015) Screening cultivated and wild tomato germplasm for resistance to Xanthomonas gardneri . Acta Horticulturae, 1069, 65–70. [Google Scholar]
- Lindau, G. (1894) Der Epheukrebs. Zeitschrift für Pflanzenkrankheiten, 4, 1–3. [Google Scholar]
- Llosa, M. , Gomis‐Rüth, F.X. , Coll, M. & de la Cruz, F. (2002) Bacterial conjugation: a two‐step mechanism for DNA transport. Molecular Microbiology, 45, 1–8. [DOI] [PubMed] [Google Scholar]
- Lu, H. , Patil, P. , Van Sluys, M.‐A. , White, F.F. , Ryan, R.P. , Dow, J.M. et al. (2008) Acquisition and evolution of plant pathogenesis‐associated gene clusters and candidate determinants of tissue‐specificity in Xanthomonas . PLoS One, 3, e3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. & Raid, R. (2013) A novel screening method for evaluation of lettuce germplasm for bacterial leaf spot resistance. HortScience, 48, 171–174. [Google Scholar]
- Ma, X. (2015) Characterization and management of bacterial leaf spot of processing tomato in Ohio. PhD thesis, Ohio, Ohio State University. [Google Scholar]
- Ma, X. , Lewis Ivey, M.L. & Miller, S.A. (2011) First report of Xanthomonas gardneri causing bacterial spot of tomato in Ohio and Michigan. Plant Disease, 95, 1584. [DOI] [PubMed] [Google Scholar]
- Maes, M. (1993) Fast classification of plant‐associated bacteria in the Xanthomonas genus. FEMS Microbiology Letters, 113, 161–165. [Google Scholar]
- Manulis, S. , Valinsky, L. , Lichter, A. & Gabriel, D.W. (1994) Sensitive and specific detection of Xanthomonas campestris pv. pelargonii with DNA primers and probes identified by random amplified polymorphic DNA analysis. Applied and Environmental Microbiology, 60, 4094–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, V.R. , Ferrarezi, J.H. , Vieira, G. & Sass, D.C. (2019) Recent advances in the biocontrol of Xanthomonas spp. World Journal of Microbiology and Biotechnology, 35, 72. [DOI] [PubMed] [Google Scholar]
- McDonald, B.A. & Linde, C. (2002) The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica, 124, 163–180. [Google Scholar]
- McHale, L.K. , Truco, M.J. , Kozik, A. , Wroblewski, T. , Ochoa, O.E. , Lahre, K.A. et al. (2009) The genomic architecture of disease resistance in lettuce. Theoretical and Applied Genetics, 118, 565–580. [DOI] [PubMed] [Google Scholar]
- McPherson, G.M. & Preece, T.F. (1978) Bacterial blight of Pelargonium: movement, symptom production and distribution of Xanthomonas pelargonii (Brown) Starr & Burkholder in Pelargonium × hortorum Bailae, following artificial inoculation. In: Proceedings of the 4th International Conference on Plant Pathogenic Bacteria, Angers.
- Meng, X.Q. , Umesh, K.C. , Davis, R.M. & Gilbertson, R.L. (2004) Development of PCR‐based assays for detecting Xanthomonas campestris pv. carotae, the carrot bacterial leaf blight pathogen, from different substrates. Plant Disease, 88, 1226–1234. [DOI] [PubMed] [Google Scholar]
- Merda, D. , Briand, M. , Bosis, E. , Rousseau, C. , Portier, P. , Barret, M. et al. (2017) Ancestral acquisitions, gene flow and multiple evolutionary trajectories of the type three secretion system and effectors in Xanthomonas plant pathogens. Molecular Ecology, 26, 5939–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuskens, I. , Saragliadis, A. , Leo, J.C. & Linke, D. (2019) Type V secretion systems: an overview of passenger domain functions. Frontiers in Microbiology, 10, 1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.B. & Bassler, B.L. (2001) Quorum sensing in bacteria. Annual Review of Microbiology, 55, 165–199. [DOI] [PubMed] [Google Scholar]
- Mirik, M. , Aysan, Y. & Baysal‐Gurel, F. (2018) Bacterial spot and blight diseases of ornamental plants caused by different Xanthomonas species in Turkey. Plant Protection Science, 54, 240–247. [Google Scholar]
- Morinière, L. , Burlet, A. , Rosenthal, E.R. , Nesme, X. , Portier, P. , Bull, C.T. , et al. (2020) Clarifying the taxonomy of the causal agent of bacterial leaf spot of lettuce through a polyphasic approach reveals that Xanthomonas cynarae Trébaol et al. 2000 emend. Timilsina et al. 2019 is a later heterotypic synonym of Xanthomonas hortorum Vauterin et al. 1995. Systematic and Applied Microbiology, 43, 126087. [DOI] [PubMed] [Google Scholar]
- Morinière, L. , Lecomte, S. , Gueguen, E. & Bertolla, F. (2021) In vitro exploration of the Xanthomonas hortorum pv. vitians genome using transposon insertion sequencing and comparative genomics to discriminate between core and contextual essential genes. Microbial Genomics, 7, 000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mtui, H.D. , Bennett, M.A. , Maerere, A.P. , Miller, S.A. , Kleinhenz, M.D. & Sibuga, K.P. (2010) Effect of seed treatments and mulch on seedborne bacterial pathogens and yield of tomato (Solanum lycopersicum Mill.) in Tanzania. Journal of Animal and Plant Sciences, 8, 1006–1015. [Google Scholar]
- Munnecke, D.E. (1954) Bacterial stem rot and leaf spot of Pelargonium . Phytopathology, 44, 627–632. [Google Scholar]
- Myung, I.S. , Moon, S.Y. , Jeong, I.H. , Lee, S.W. , Lee, Y.H. & Shim, H.S. (2010) Bacterial leaf spot of iceberg lettuce caused by Xanthomonas campestris pv. vitians type B, a new disease in South Korea. Plant Disease, 94, 790. [DOI] [PubMed] [Google Scholar]
- Myung, I.S. , Yoon, M.J. , Lee, J.Y. , Kim, G.D. , Lee, M.H. , Hwang, E.Y. et al. (2014) First report of bacterial leaf blight of carrot caused by Xanthomonas hortorum pv. carotae in Korea. Plant Disease, 98, 275. [DOI] [PubMed] [Google Scholar]
- Nameth, S.T. , Daughtrey, M.L. , Moorman, G.W. & Sulzinski, M.A. (1999) Bacterial blight of geranium: a history of diagnostic challenges. Plant Disease, 83, 204–212. [DOI] [PubMed] [Google Scholar]
- Nejad, M.A. , Rahimian, H. & Tohidfar, M. (2012) Identification of the causal agent of a leaf spot of primula by dnaK and rpoD gene sequences in Mazandaran. Iranian Journal of Plant Pathology, 48, e287. [Google Scholar]
- Newman, M.‐A. , Dow, J.M. , Molinaro, A. & Parrilli, M. (2007) Invited review: priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. Journal of Endotoxin Research, 13, 69–84. [DOI] [PubMed] [Google Scholar]
- Newman, M.‐A. , von Roepenack‐Lahaye, E. , Parr, A. , Daniels, M.J. & Dow, J.M. (2002) Prior exposure to lipopolysaccharide potentiates expression of plant defenses in response to bacteria. The Plant Journal, 29, 487–495. [DOI] [PubMed] [Google Scholar]
- Ng, W.‐L. & Bassler, B.L. (2009) Bacterial quorum‐sensing network architectures. Annual Review of Genetics, 43, 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhauser, J.S. (1943) A bacterial leaf spot and blight of the Russian dandelion. Phytopathology, 33, 959–961. [Google Scholar]
- Norman, D.J. , Chase, A.R. , Stall, R.E. & Jones, J.B. (1999) Heterogeneity of Xanthomonas campestris pv. hederae strains from araliaceous hosts. Phytopathology, 89, 646–652. [DOI] [PubMed] [Google Scholar]
- Obradovic, A. , Jones, J.B. , Balogh, B. & Momol, M.T. (2008) Integrated management of tomato bacterial spot. In: Ciancio, A. & Mukerji, K.G. (Eds.) Integrated management of diseases caused by fungi, phytoplasma and bacteria. Dordrecht, Netherlands: Springer, pp. 211–223. [Google Scholar]
- Oliver, C.L. , Cai, R. , Vinatzer, B.A. , Bush, E.A. & Hansen, M.A. (2012) First report of bacterial spot of peony caused by a Xanthomonas sp. in the United States. Plant Disease, 96, 581. [DOI] [PubMed] [Google Scholar]
- Osdaghi, E. , Sharma, A. , Goss, E. , Abrahamian, P. , Newberry, E. , Potnis, N. et al. (2021) A centenary for bacterial spot of tomato and pepper. Molecular Plant Pathology, 22, 1500–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleroni, N.J. & Bradbury, J.F. (1993) Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. International Journal of Systematic and Evolutionary Microbiology, 43, 606–609. [DOI] [PubMed] [Google Scholar]
- Parkinson, N. , Aritua, V. , Heeney, J. , Cowie, C. , Bew, J. & Stead, D. (2007) Phylogenetic analysis of Xanthomonas species by comparison of partial gyrase B gene sequences. International Journal of Systematic and Evolutionary Microbiology, 57, 2881–2887. [DOI] [PubMed] [Google Scholar]
- Parkinson, N. , Cowie, C. , Heeney, J. & Stead, D. (2009) Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. International Journal of Systematic and Evolutionary Microbiology, 59, 264–274. [DOI] [PubMed] [Google Scholar]
- Peňázová, E. , Dvořák, M. , Ragasová, L. , Kiss, T. , Pečenka, J. , Čechová, J. et al. (2020) Multiplex real‐time PCR for the detection of Clavibacter michiganensis subsp. michiganensis, Pseudomonas syringae pv. tomato and pathogenic Xanthomonas species on tomato plants. PLoS One, 15, e0227559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, R.C. , Araújo, E.R. , Ferreira, M.A.S.V. & Quezado‐Duval, A.M. (2011) Occurrence of Xanthomonas species causing bacterial spot in fresh market tomato fields in Brazil. Acta Horticulturae, 914, 61–64. [Google Scholar]
- Pernezny, K. , Nagata, R. , Raid, R.N. , Collins, J. & Carroll, A. (2002) Investigation of seed treatments for management of bacterial leaf spot of lettuce. Plant Disease, 86, 151–155. [DOI] [PubMed] [Google Scholar]
- Pernezny, K. , Raid, R.N. , Stall, R.E. , Hodge, N. & Collins, J. (1995) An outbreak of bacterial spot of lettuce in Florida caused by Xanthomonas campestris pv. vitians . Plant Disease, 79, 359–360. [Google Scholar]
- Picard, C. , Afonso, T. , Benko‐Beloglavec, A. , Karadjova, O. , Matthews‐Berry, S. , Paunovic, S.A. et al. (2018) Recommended regulated non‐quarantine pests (RNQPs), associated thresholds and risk management measures in the European and Mediterranean region. EPPO Bulletin, 48, 552–568. [Google Scholar]
- Pontes, N.D.C. , Nascimento, A.D.R. , Golynski, A. , Maffia, L.A. , Rogério de Oliveira, J. & Quezado‐Duval, A.M. (2016) Intervals and number of applications of Acibenzolar‐S‐Methyl for the control of bacterial spot on processing tomato. Plant Disease, 100, 2126–2133. [DOI] [PubMed] [Google Scholar]
- Potnis, N. , Branham, S.E. , Jones, J.B. & Wechter, W.P. (2019) Genome‐wide association study of resistance to Xanthomonas gardneri in the USDA pepper (Capsicum) collection. Phytopathology, 109, 1217–1225. [DOI] [PubMed] [Google Scholar]
- Potnis, N. , Krasileva, K. , Chow, V. , Almeida, N.F. , Patil, P.B. , Ryan, R.P. et al. (2011) Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics, 12, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potnis, N. , Minsavage, G. , Smith, J.K. , Hurlbert, J.C. , Norman, D. , Rodrigues, R. et al. (2012) Avirulence proteins AvrBs7 from Xanthomonas gardneri and AvrBs1.1 from Xanthomonas euvesicatoria contribute to a novel gene‐for‐gene interaction in pepper. Molecular Plant‐Microbe Interactions, 25, 307–320. [DOI] [PubMed] [Google Scholar]
- Potnis, N. , Timilsina, S. , Strayer, A. , Shantharaj, D. , Barak, J.D. , Paret, M.L. et al. (2015) Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Molecular Plant Pathology, 16, 907–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruvost, O. , Boyer, C. , Robène‐Soustrade, I. , Jouen, E. , Saison, A. , Hostachy, B. et al. (2010) First report of Xanthomonas hortorum pv. carotae causing bacterial leaf blight of carrot in Mauritius. Plant Disease, 94, 1069. [DOI] [PubMed] [Google Scholar]
- Quezado‐Duval, A.M. , Leite Júnior, R.P. , Truffi, D. & Camargo, L.E.A. (2004) Outbreaks of bacterial spot caused by Xanthomonas gardneri on processing tomato in Central‐West Brazil. Plant Disease, 88, 157–161. [DOI] [PubMed] [Google Scholar]
- Richard, D. , Boyer, C. , Lefeuvre, P. , Canteros, B.I. , Beni‐Madhu, S. , Portier, P. et al. (2017) Complete genome sequences of six copper‐resistant Xanthomonas strains causing bacterial spot of solaneous plants, belonging to X. gardneri, X. euvesicatoria, and X. vesicatoria, using long‐read technology. Genome Announcements, 5, e01693‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, D. , Ravigné, V. , Rieux, A. , Facon, B. , Boyer, C. , Boyer, K. et al. (2017) Adaptation of genetically monomorphic bacteria: evolution of copper resistance through multiple horizontal gene transfers of complex and versatile mobile genetic elements. Molecular Ecology, 26, 2131–2149. [DOI] [PubMed] [Google Scholar]
- Richter, M. & Rosselló‐Móra, R. (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences of the United States of America, 106, 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridé, M. (1956) Sur une maladie nouvelle de l’artichaut (Cynara scolymus L.). Comptes Rendus Hebdomadaires des Séances de l’Académie des Sciences, 243, 174–177. [Google Scholar]
- Roberts, S.J. & Parkinson, N.M. (2014) A bacterial leaf spot and shoot blight of lavender caused by Xanthomonas hortorum in the UK. New Disease Reports, 30, 1. [Google Scholar]
- Robinson, P.E. , Jones, J.B. & Pernezny, K. (2006) Leaf spot of lettuce: relationship of temperature to infection and potential host range of Xanthomonas campestris pv. vitians . Plant Disease, 90, 465–470. [DOI] [PubMed] [Google Scholar]
- Rocchetta, H.L. & Lam, J.S. (1997) Identification and functional characterization of an ABC transport system involved in polysaccharide export of A‐band lipopolysaccharide in Pseudomonas aeruginosa . Journal of Bacteriology, 179, 4713–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey, W. , Potnis, N. , Timilsina, S. , Hong, J.C. , Vallad, G.E. , Jones, J.B. et al. (2015) Multilocus sequence analysis reveals genetic diversity in xanthomonads associated with Poinsettia production. Plant Disease, 99, 874–882. [DOI] [PubMed] [Google Scholar]
- Romero, A.M. , Kousik, C.S. & Ritchie, D.F. (2001) Resistance to bacterial spot in bell pepper induced by acibenzolar‐S‐methyl. Plant Disease, 85, 189–194. [DOI] [PubMed] [Google Scholar]
- Rosselló‐Móra, R. & Amann, R. (2015) Past and future species definitions for Bacteria and Archaea. Systematic and Applied Microbiology, 38, 209–216. [DOI] [PubMed] [Google Scholar]
- Rotondo, F. , Testen, A.L. , Horvat, M.M. , Roman‐Reyna, V. , Klass, T.L. , Jacobs, J.M. et al. (2020) First report of Xanthomonas hortorum causing bacterial leaf spot of lavender (Lavandula × intermedia) in Ohio. Plant Disease, 105, 484. [Google Scholar]
- Russell, A.B. , Peterson, S.B. & Mougous, J.D. (2014) Type VI secretion system effectors: poisons with a purpose. Nature Reviews Microbiology, 12, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R.P. , Vorhölter, F.‐J. , Potnis, N. , Jones, J.B. , Van Sluys, M.‐A. , Bogdanove, A.J. et al. (2011) Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nature Reviews Microbiology, 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Saddler, G.S. & Bradbury, J.F. (2015) Xanthomonas. In: Rainey, F. , Whitman, W.B. , Kämpfer, P. , Trujillo, M. , Chun, J. , DeVos, P. , Hedlund, B. & Dedysh, S. (Eds.) Bergey’s manual of systematics of Archaea and Bacteria. Chichester, UK: John Wiley & Sons, pp. 1–53. [Google Scholar]
- Sahin, F. (1997) Detection, identification and characterization of strains of Xanthomonas campestris pv. vesicatoria by traditional and molecular methods, and resistance in Capsicum species to Xanthomonas campestris pv. vesicatoria pepper race 6. PhD thesis, Ohio, The Ohio State University. [Google Scholar]
- Sahin, F. , Abbasi, P.A. , Ivey, M.L. , Zhang, J. & Miller, S.A. (2003) Diversity among strains of Xanthomonas campestris pv. vitians from lettuce. Phytopathology, 93, 64–70. [DOI] [PubMed] [Google Scholar]
- Sahin, F. & Miller, S.A. (1997) Identification of the bacterial leaf spot pathogen of lettuce, Xanthomonas campestris pv. vitians, in Ohio, and assessment of cultivar resistance and seed treatment. Plant Disease, 81, 1443–1446. [DOI] [PubMed] [Google Scholar]
- Sandoya, G.V. , Maisonneuve, B. , Truco, M.J. , Bull, C.T. , Simko, I. , Trent, M. et al. (2019) Genetic analysis of resistance to bacterial leaf spot in the heirloom lettuce cultivar Reine des Glaces. Molecular Breeding, 39, 1–11. [Google Scholar]
- Schornack, S. , Minsavage, G.V. , Stall, R.E. , Jones, J.B. & Lahaye, T. (2008) Characterization of AvrHah1, a novel AvrBs3‐like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytologist, 179, 546–556. [DOI] [PubMed] [Google Scholar]
- Schwartz, A.R. , Morbitzer, R. , Lahaye, T. & Staskawicz, B.J. (2017) TALE‐induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proceedings of the National Academy of Sciences of the United States of America, 114, e897–e903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, A.R. , Potnis, N. , Timilsina, S. , Wilson, M. , Patané, J. , Martins, J. et al. (2015) Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Frontiers in Microbiology, 6, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, J.C. & Dung, J.K.S. (2020) Distribution of Xanthomonas hortorum pv. carotae populations in naturally infested carrot seed lots. Plant Disease, 104, 2144–2148. [DOI] [PubMed] [Google Scholar]
- Segata, N. , Börnigen, D. , Morgan, X.C. & Huttenhower, C. (2013) PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nature Communications, 4, 2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrianski, A.M. (1936) Rubber bearers. In Spravochnik agronoma po borʹbe s bolezniami selʹskokhoziaistvennykh rastenii [Handbook for the agronomist on the control of diseases of agricultural plants]. Moscow: Selʹkhozgiz, pp. 270–275. [Google Scholar]
- Simmons, R. , Wu, B.‐M. & Johnson, K. (2010) Evaluating Actigard® for controlling Xanthomonas blight in carrot seed crops.
- Skerman, V.B.D. , McGowan, V. & Sneath, P.H.A. (1980) Approved lists of bacterial names. International Journal of Systematic and Evolutionary Microbiology, 30, 225–420. [DOI] [PubMed] [Google Scholar]
- Ssekiwoko, F. , Mulumba, J.W. , Carter, B.A. , Stanford, H. , Parkinson, N. , Kelly, P. et al. (2009) Xanthomonas hortorum pathogenic on Artemisia annua newly reported in Uganda. Plant Pathology, 58, 795. [Google Scholar]
- Stall, R.E. , Loschke, D.C. & Jones, J.B. (1986) Linkage of copper resistance and avirulence loci on a self‐transmissible plasmid in Xanthomonas campestris pv. vesicatoria . Phytopathology, 76, 240–243. [Google Scholar]
- Stapp, C. (1958) Pflanzenpathogene Bakterien. Berlin.
- Starr, M.P. & Burkholder, W.H. (1942) Lipolytic activity of phytopathogenic bacteria determined by means of spirit blue agar and its taxonomic significance. Phytopathology, 32, 598–604. [Google Scholar]
- Starr, M.P. , Volcani, Z. & Munnecke, D.E. (1955) Relationship of Xanthomonas pelargonii and Xanthomonas geranii . Phytopathology, 45, 613–617. [Google Scholar]
- Stefani, E. , Raio, A. , Bazzi, C. & Zoina, A. (1994) Identification and grouping of Xanthomonas campestris pv. vitians using SDS‐PAGE. Phytopathologia Mediterranea, 33, 99–104. [Google Scholar]
- Stehlíková, D. , Beran, P. , Cohen, S.P. & Čurn, V. (2020) Development of real‐time and colorimetric loop mediated isothermal amplification assay for detection of Xanthomonas gardneri . Microorganisms, 8, 1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, G.E. (1907) Bacterial disease of lettuce. Massachusetts Agricultural Experiment Station Annual Report Botany, 19, 163–164. [Google Scholar]
- Stone, G.E. & Smith, R.E. (1898) A disease of the cultivated Geranium . Massachusetts Agricultural Experiment Station Annual Report Botany, 10, 164. [Google Scholar]
- Stoyanova, M. , Vancheva, T. , Moncheva, P. & Bogatzevska, N. (2014) Differentiation of Xanthomonas spp. causing bacterial spot in Bulgaria based on BIOLOG system. International Journal of Microbiology, 2014, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer, A. , Jeyaprakash, A. , Minsavage, G.V. , Timilsina, S. , Vallad, G.E. , Jones, J.B. et al. (2016) A multiplex real‐time PCR assay differentiates four Xanthomonas species associated with bacterial spot of tomato. Plant Disease, 100, 1660–1668. [DOI] [PubMed] [Google Scholar]
- Strayer, A. , Ocsoy, I. , Tan, W. , Jones, J.B. & Paret, M.L. (2016) Low concentrations of a silver‐based nanocomposite to manage bacterial spot of tomato in the greenhouse. Plant Disease, 100, 1460–1465. [DOI] [PubMed] [Google Scholar]
- Strayer‐Scherer, A. , Jones, J.B. & Paret, M.L. (2019) Recombinase polymerase amplification assay for field detection of tomato bacterial spot pathogens. Phytopathology, 109, 690–700. [DOI] [PubMed] [Google Scholar]
- Strider, D.L. (1985) Diseases of floral crops. New York, NY: Praeger. [Google Scholar]
- Sulzinski, M.A. (2001) Differentiation of Xanthomonas campestris pvs pelargonii and hederae by polymerase chain reaction. Journal of Phytopathology, 149, 45–49. [Google Scholar]
- Sulzinski, M.A. , Moorman, G.W. , Schlagnhaufer, B. & Romaine, C.P. (1995) Fingerprinting of Xanthomonas campestris pv. pelargonii and related pathovars using random‐primed PCR. Journal of Phytopathology, 143, 429–433. [Google Scholar]
- Sulzinski, M.A. , Moorman, G.W. , Schlagnhaufer, B. & Romaine, C.P. (1996) Characteristics of a PCR‐based assay for in planta detection of Xanthomonas campestris pv. pelargonii . Journal of Phytopathology, 144, 393–398. [Google Scholar]
- Sulzinski, M.A. , Moorman, G.W. , Schlagnhaufer, B. & Romaine, C.P. (1997) A simple DNA extraction method for PCR‐based detection of Xanthomonas campestris pv. pelargonii in geraniums. Journal of Phytopathology, 145, 213–215. [Google Scholar]
- Sulzinski, M.A. , Schlagnhaufer, B. , Moorman, G.W. & Romaine, C.P. (1998) PCR‐based detection of artificial latent infections of geranium by Xanthomonas campestris pv. pelargonii . Journal of Phytopathology, 146, 111–114. [Google Scholar]
- Sutherland, I.W. (1993) Xanthan. In: Swings, J.G. & Civerolo, E.L. (Eds.) Xanthomonas. Dordrecht, Netherlands: Springer, pp. 363–388. [Google Scholar]
- Šutic, D. (1957) Bakterioze crvenog patlidzana [tomato bacteriosis]. Posebna Izdanja Institut za Zastitu Bilja Beograd [Special Edition Institute for Plant Protection Belgrade], 6, 1–65. [Google Scholar]
- Suzuki, A. , Kusumoto, S. , Horie, H. & Takikawa, Y. (2002) Bacterial leaf spot of ivy caused by Xanthomonas campestris pv. hederae . Journal of General Plant Pathology, 68, 398–400. [Google Scholar]
- Szczesny, R. , Jordan, M. , Schramm, C. , Schulz, S. , Cogez, V. , Bonas, U. et al. (2010) Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria . New Phytologist, 187, 983–1002. [DOI] [PubMed] [Google Scholar]
- Temple, T.N. , du Toit, L.J. , Derie, M.L. & Johnson, K.B. (2013) Quantitative molecular detection of Xanthomonas hortorum pv. carotae in carrot seed before and after hot‐water treatment. Plant Disease, 97, 1585–1592. [DOI] [PubMed] [Google Scholar]
- Temple, T.N. & Johnson, K.B. (2009) Detection of Xanthomonas hortorum pv. carotae on and in carrot with loop‐mediated isothermal amplification (LAMP). Phytopathology, 99, S186. [Google Scholar]
- Thomazella, D.P. de T. , Brail, Q. , Dahlbeck, D. & Staskawicz, B. (2016) CRISPR‐Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad‐spectrum disease resistance. bioRxiv, 064824. [DOI] [PMC free article] [PubMed]
- Timilsina, S. , Kara, S. , Jacques, M.A. , Potnis, N. , Minsavage, G.V. , Vallad, G.E. , et al. (2019) Reclassification of Xanthomonas gardneri (ex Šutič 1957) Jones et al. 2006 as a later heterotypic synonym of Xanthomonas cynarae Trébaol et al. 2000 and description of X. cynarae pv. cynarae and X. cynarae pv. gardneri based on whole genome analyses. International Journal of Systematic and Evolutionary Microbiology, 69, 343–349. [DOI] [PubMed] [Google Scholar]
- Timilsina, S. , Potnis, N. , Newberry, E.A. , Liyanapathiranage, P. , Iruegas‐Bocardo, F. , White, F.F. et al. (2020) Xanthomonas diversity, virulence and plant–pathogen interactions. Nature Reviews Microbiology, 18, 415–427. [DOI] [PubMed] [Google Scholar]
- du Toit, L.J. , Crowe, F. , Derie, M. , Simmons, R. & Pelter, G. (2005) Bacterial blight in carrot seed crops in the Pacific Northwest. Plant Disease, 89, 896–907. [DOI] [PubMed] [Google Scholar]
- du Toit, L.J. , Derie, M.L. , Christianson, C.E. , Hoagland, L. & Simon, P. (2014) First report of bacterial blight of carrot in Indiana caused by Xanthomonas hortorum pv. carotae . Plant Disease, 98, 685. [DOI] [PubMed] [Google Scholar]
- Tolba, I.H. (2017) Bacterial leaf spot of araliaceous plants caused by Xanthomonas campestris pv. hederae in Egypt. Journal of Plant Protection and Pathology, 8, 287–295. [Google Scholar]
- Toussaint, V. (1999) Bacterial leaf spot, a new disease of lettuce in Quebec caused by Xanthomonas campestris pv. vitians . Phytoprotection, 80, 121–125. [Google Scholar]
- Toussaint, V. , Benoit, D.L. & Carisse, O. (2012) Potential of weed species to serve as a reservoir for Xanthomonas campestris pv. vitians, the causal agent of bacterial leaf spot of lettuce. Crop Protection, 41, 64–70. [Google Scholar]
- Trantas, E.A. , Sarris, P.F. , Mpalantinaki, E. , Papadimitriou, M. , Ververidis, F. & Goumas, D.E. (2016) First report of Xanthomonas hortorum pv. hederae causing bacterial leaf spot on ivy in Greece. Plant Disease, 100, 2158. [Google Scholar]
- Trébaol, G. , Gardan, L. , Manceau, C. , Tanguy, J.L. , Tirilly, Y. & Boury, S. (2000) Genomic and phenotypic characterization of Xanthomonas cynarae sp. nov., a new species that causes bacterial bract spot of artichoke (Cynara scolymus L.). International Journal of Systematic and Evolutionary Microbiology, 50, 1471–1478. [DOI] [PubMed] [Google Scholar]
- Truco, M.J. , Ashrafi, H. , Kozik, A. , van Leeuwen, H. , Bowers, J. , Wo, S.R.C. et al. (2013) An ultra‐high‐density, transcript‐based, genetic map of lettuce. G3: Genes, Genomes, Genetics, 3, 617–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin, W. , McCarter, S. & Gitaitis, R. (1996) First report of bacterial leaf spot of oakleaf hydrangea caused by a pathovar of Xanthomonas campestris . Plant Disease, 80, 599. [Google Scholar]
- Vauterin, L. , Hoste, B. , Kersters, K. & Swings, J. (1995) Reclassification of Xanthomonas . International Journal of Systematic and Evolutionary Microbiology, 45, 472–489. [DOI] [PubMed] [Google Scholar]
- Vauterin, L. , Swings, J. & Kersters, K. (1991) Grouping of Xanthomonas campestris pathovars by SDS‐PAGE of proteins. Journal of General Microbiology, 137, 1677–1687. [Google Scholar]
- Wang, L.‐H. , He, Y. , Gao, Y. , Wu, J.E. , Dong, Y.‐H. , He, C. et al. (2004) A bacterial cell–cell communication signal with cross‐kingdom structural analogues. Molecular Microbiology, 51, 903–912. [DOI] [PubMed] [Google Scholar]
- WFO (2021) World flora online. Available at: http://www.worldfloraonline.org [Accessed 27th April 2021]. [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. & Koebnik, R. (2009) The type III effectors of Xanthomonas . Molecular Plant Pathology, 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead, N.A. , Barnard, A.M.L. , Slater, H. , Simpson, N.J.L. & Salmond, G.P.C. (2001) Quorum‐sensing in Gram‐negative bacteria. FEMS Microbiology Reviews, 25, 365–404. [DOI] [PubMed] [Google Scholar]
- Willis, B.E. & Bishop, W.M. (2016) Understanding fate and effects of copper pesticides in aquatic systems. Journal of Geoscience and Environment Protection, 4, 37–42. [Google Scholar]
- Yellareddygari, S.K. , Kumar, K.V.K. , Sudini, H. , Reddy, E.C.S. , Sayyed, R.Z. , Madhuri, K.V.N. et al. (2013) Evaluation of biopesticides for control of Xanthomonas campestris pv. pelargonni in geranium seedlings under greenhouse conditions. In: Reddy, M.S. , Ilao, R.I. , Faylon, P.S. , Dar, W.D. , Sayyed, R. , Sudini, H. , Kumar, K.V.K. & Armanda, A. (Eds.) Recent advances in biofertilizers and biofungicides (PGPR) for sustainable agriculture. Proceedings of 3rd Asian Conference on Plant Growth‐Promoting Rhizobacteria (PGPR) and other Microbials, Manila, Philippines, 21‐24 April, 2013, Asian PGPR Society for Sustainable Agriculture, pp. 491–495. [Google Scholar]
- Young, J.M. , Dye, D.W. , Bradbury, J.F. , Panagopoulos, C.G. & Robbs, C.F. (1978) A proposed nomenclature and classification for plant pathogenic bacteria. New Zealand Journal of Agricultural Research, 21, 153–177. [Google Scholar]
- Young, J.M. , Park, D.C. , Shearman, H.M. & Fargier, E. (2008) A multilocus sequence analysis of the genus Xanthomonas . Systematic and Applied Microbiology, 31, 366–377. [DOI] [PubMed] [Google Scholar]
- Young, J.M. , Wilkie, J.P. , Park, D.‐C. & Watson, D.R.W. (2010) New Zealand strains of plant pathogenic bacteria classified by multi‐locus sequence analysis; proposal of Xanthomonas dyei sp. nov. Plant Pathology, 59, 270–281. [Google Scholar]
- Zacaroni, A.B. , Koike, S.T. , de Souza, R.M. & Bull, C.T. (2012) Bacterial leaf spot of radicchio (Cichorium intybus) is caused by Xanthomonas hortorum . Plant Disease, 96, 1820. [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Grefer, D. , Feasel, J. , Ferencak, M. , Sairam, R.V. & Goldman, S.L. (2009) Exogenous methyl jasmonate inhibits the spread/multiplication of Xanthomonas campestris pv. pelargonii in the leaves of Pelargonium × hortorum . Archives of Phytopathology and Plant Protection, 42, 930–939. [Google Scholar]
- Zhang, S. , Sairam, R.V. , Grefer, D. , Feasel, J. , Ferencak, M. & Goldman, S.L. (2009) Resistance to Xanthomonas campestris pv. pelargonii in geranium and diagnosis of the bacterial blight using polymerase chain reaction. Archives of Phytopathology and Plant Protection, 42, 1109–1117. [Google Scholar]
- Zhang, X.F. , Fu, L.N. , Yang, J. , Li, X. & Ji, G.H. (2015) First report of bacterial leaf spot of Hedera nepalensis var. sinensis caused by Xanthomonas campestris pv. hederae in China. Plant Disease, 100, 1007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


