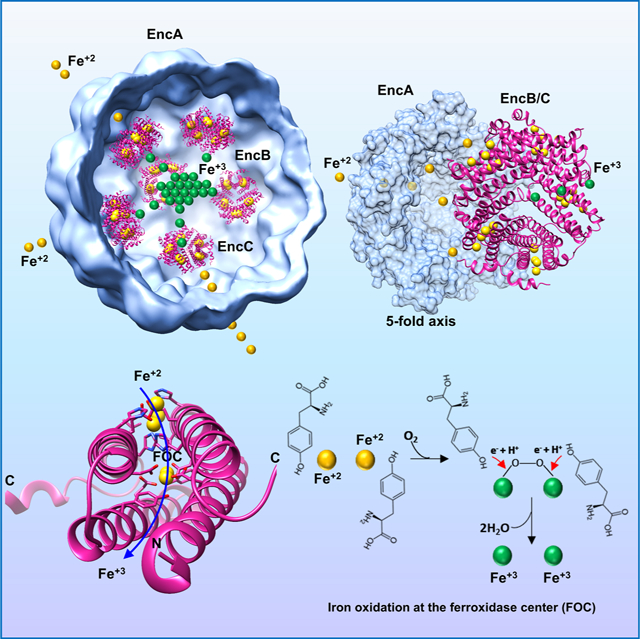
SUMMARY
Encapsulins are bacterial organelle-like cages involved in various aspects of metabolism, especially protection from oxidative stress. They can serve as vehicles for a wide range of medical applications. Encapsulin shell proteins are structurally similar to HK97 bacteriophage capsid protein and their function depends on the encapsulated cargos. The Myxococcus xanthus encapsulin system comprises EncA and three cargos: EncB, EncC and EncD. EncB and EncC are similar to bacterial ferritins which can oxidize Fe+2 to less toxic Fe+3. We analyzed EncA, EncB and EncC by cryo-EM and X-ray crystallography. Cryo-EM shows that EncA cages can have T=3 and T=1 symmetry and that EncA T=1 has a unique protomer arrangement. Also, we define EncB and EncC binding sites on EncA. X-ray crystallography of EncB and EncC reveals conformational changes at the ferroxidase center and additional metal binding sites, suggesting a mechanism for Fe oxidation and storage within the encapsulin shell.
eTOC
Eren et al., characterize M. xanthus ferritin-like proteins EncB and EncC. They show conformational changes at the di-iron binding sites along with additional peripheral iron binding sites in their crystal structures. Based on their findings the authors propose an iron transport and storage mechanism in the M. xanthus encapsulin system.
Graphical Abstract
INTRODUCTION
Encapsulins are protein-based nanocompartments that have been identified in a variety of bacterial and archaeal species (Gabashvili et al., 2020, Giessen, 2016, Nichols et al., 2017). They are involved in a number of metabolic pathways including mitigation of oxidative stress, iron mineralization and storage, anaerobic ammonium oxidation, catabolism and sulfur metabolism (Ahmad et al., 2011, Contreras et al., 2014, Kartal et al., 2013, McHugh et al., 2014, Nichols et al., 2021, Rahmanpour and Bugg, 2013, Sugano et al., 2007). Encapsulins are comprised of a single protomer protein which self-assembles into icosahedral shells that possess the bacteriophage Hong Kong 97 (HK97)-like fold (Sutter et al., 2008, Wikoff et al., 2000). The HK97-like fold is widespread among viruses and has been observed in the capsid proteins of several other tailed bacteriophages, some archaeal phages like HSTV-1 and herpesviruses (Bamford et al., 2005, Fokine et al., 2005, Pietila et al., 2013). Recent bioinformatical analyses propose that encapsulins and HK97-like viruses share a common ancestor (Andreas and Giessen, 2021). As is common with viral capsids, encapsulin protomers can assemble into icosahedrons of different sizes ranging from 20–42 nm. The smallest encapsulins have been identified in Thermotoga marritima, Mycolicibacterium hassiacum, Synechococcus elongatus and Mycobacterium smegmatis (Loncar et al., 2020, Nichols et al., 2021, Sutter et al., 2008, Tang et al., 2021, Wiryaman and Toor, 2021). These encapsulins are formed by 60 protomers with T=1 symmetry. Mid-size encapsulins formed by 180 protomers have been identified in Myxococcus xanthus and Pyrococcus furiosus with T=3 symmetry (Akita et al., 2007, McHugh et al., 2014). The largest encapsulin is found in Quasibacillus thermotolerans with 240 protomers arranged with T=4 symmetry (Giessen et al., 2019).
The function of an encapsulin is determined by the function of its cargo protein. To date, a number of different cargo proteins have been associated with encapsulins including ferritin-like-proteins (FLPs), dye-decolorizing peroxidase (DyP), iron-mineralizing encapsulin-associated firmicute (IMEF), hemerythrin and rubrerythrin (Ahmad et al., 2011, Contreras et al., 2014, Giessen and Silver, 2017, He et al., 2016, McHugh et al., 2014, Rahmanpour and Bugg, 2013). These cargo proteins contain a short (10–40 aa) disordered peptide at their C-termini. The latter serves as a targeting peptide (TP) and delivers the cargo to the interior of the encapsulin before assembly is completed. TPs are generally rich in alanine, proline and glycine residues and usually include a specific anchor sequence like Gly-Ser-Leu, which interacts with a conserved N-terminal helix on the interior surface of the encapsulin. Alternatively, a cargo protein lacking the TP can be fused to the encapsulin shell protein as in the case of P. furiosus (Akita et al., 2007). Also, TPs that are located in the N-terminus of the cargo protein can be found in some encapsulin-cargo systems (Giessen and Silver, 2017).
The M. xanthus encapsulin system comprises the shell subunit EncA and three cargo proteins namely EncB, EncC and EncD (McHugh et al., 2014). Previous studies have shown that EncA and its cargo(s) are involved in mitigation of oxidative stress through ferroxidase activity leading to the oxidation, mineralization, and sequestration of iron inside an EncA shell (McHugh et al., 2014). Deletion of the EncA gene makes M. xanthus highly susceptible to hydrogen peroxide-induced oxidative stress and results in phenotypic growth defects (Kim et al., 2019, McHugh et al., 2014). Previous characterization of native EncA encapsulin by energy dispersive X-ray spectroscopy, inductively coupled mass spectrometry (ICP-MS) and scanning transmission electron microscopy (STEM) has revealed that the electron-dense core of encapsulin particles is highly enriched for iron and phosphorus and can store up to 30,000 iron atoms. (McHugh et al., 2014). Analysis of cargo protein sequences shows that EncB (UniProt ID MXAN_3557) and EncC (UniProt ID MXAN_4464) are homologous to ferritin-like proteins with diiron catalytic sites. On the other hand, EncD (UniProt ID MXAN_2410) is an unclassified protein with no known activity. Here we present a detailed structural study of both EncA and its cargo proteins EncB and EncC.
Our results reveal an another type of M xanthus EncA shell produced by heterologous assembly: it has a T=1 protomer arrangement, distinct from the T=3 architecture of native encapsulins (McHugh et al., 2014). In addition, we show additional Fe binding sites in the high-resolution crystal structure of EncB, one of which corresponds to the “site-C” of previously characterized ferritin nanocages (Ebrahimi et al., 2016, Hempstead et al., 1994, Pfaffen et al., 2013, Stillman et al., 2001, Tatur et al., 2007). We also define structural differences in Fe coordination between EncB and EncC ferroxidase centers (FOCs). Based on these observations, we propose a mechanism for Fe storage mechanism for M. xanthus encapsulin system.
RESULTS
EncA can form nanocompartments with T=1 and T=3 symmetry in the absence of a cargo
The first structure of EncA encapsulin was obtained at 4.6 Å resolution (PDB ID: 4PT2, EMD-5917) using particles isolated from wild-type M. xanthus (McHugh et al., 2014). This structure showed some inner density attributable to cargo proteins at a radius of ~24 Å which did not follow the icosahedral symmetry of the shell, probably due to the partial occupancy of the binding sites. In order to better characterize cargo binding to the shell we used a recombinant E. coli expression system which allowed us to express the shell with a single cargo at a time, as compared to the native system where all the cargo proteins were expressed simultaneously (Figure S1). The majority (~94%) of EncA encapsulins purified from M. xanthus were 32 nm in diameter and arranged in T=3 symmetry (McHugh et al., 2014) whereas with E. coli expression products we observed empty EncA shells of two sizes. About 64% of these particles were similar to the previously described T=3 encapsulin and about 36% were ~18 nm in diameter and arranged in T=1 symmetry (Figure 1A and Figure S2A). In E. coli the T=1 encapsulins only formed in the absence of a co-expressed cargo protein (Figure S2). As T=1 encapsulins from M. xanthus were a minority species (only ~6%) it can be presumed that the smaller encapsulin size cannot accommodate M. xanthus cargo proteins at high occupancy.
Figure 1.
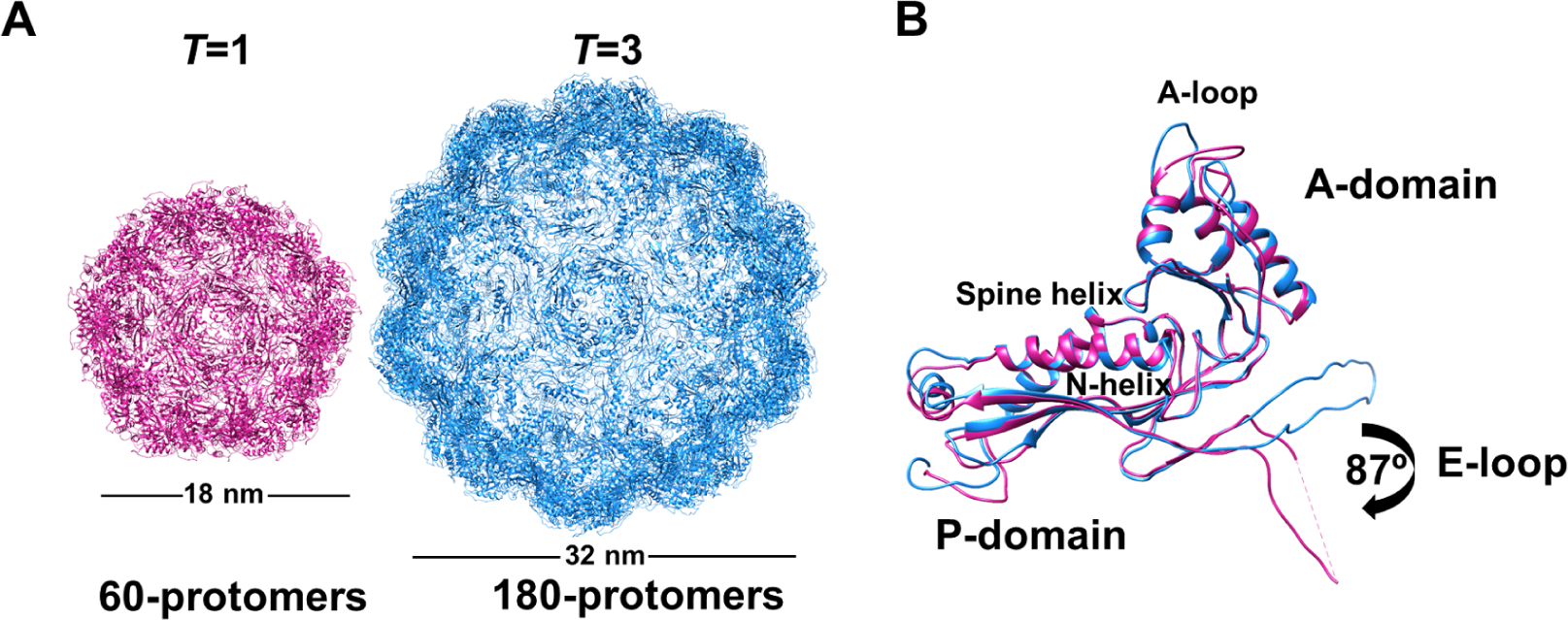
Structural comparison of assembled encapsulin shells from M. xanthus. (A) Icosahedral EncA shell arranged in T=1 formed from 60 protomers (pink) and T=3 from 180 protomers (blue). (B) Superimposed structures of EncA monomers from a T=1 shell (pink) and T=3 shell (blue) showing a conformational rotation in the E-loop. Both structures display a classic HK97 fold with spine α-helix, A- and P-domains with β-hairpin loop (E-loop). See also Figure S2, Figure S3 and Table 1.
A switch between T=3 and T=1 has only been observed for M. xanthus EncA so far. In the previous M. xanthus encapsulin studies the T=1 EncA has not been structurally characterized (McHugh et al., 2014). We solved the cryo-EM structures of EncA T=3 and T=1 encapsulins each at 3.4 Å resolution (Figure 1A and Table 1). EncA monomers showed the typical HK97 fold and consisted of a P-domain (peripheral domain), A-domain (axial domain) and E-loop (extension loop) (Figure 1B). Structural alignment of EncA from T=3 and T=1 encapsulins showed a conformational rotation of the E-loop by ~90° (Figure 1B and Figure S3A). A similar E-loop rotation of ~60° was observed when the structure of T. maritima T=1 encapsulin was compared to the structure of P. furiosus T=3 encapsulin. (Gabashvili et al., 2020, Sutter et al., 2008). In T=1 encapsulins E-loops are responsible for tight two-fold interactions that are not observed in larger particles such as T=3 encapsulins and bacteriophages. The E-loop rotation in T=1 compared to T=3 protomers results in the formation of additional β-sheet interactions with the neighboring monomer, creating a tighter packing (Nichols et al., 2017). In M. xanthus the tighter packing between the EncA monomers resulted in a substantial increase in dimer interface area in T=1 in comparison to T=3 (1058 Å2 vs. 308 Å2, respectively; Figure S3A). The E-loop formed additional hydrophobic interactions and hydrogen bonds with the neighboring P-domain resulting in a −14.6 kcal/mol increase in the calculated binding energy (PISA (Krissinel and Henrick, 2007)) between EncA monomers (Figure S3B and C).
Table 1.
Cryo-EM data collection, processing, and refinement statistics
| Deposition ID | EMD-24815 | EMD-24814 | EMD-24816 | EMD-24832 |
|---|---|---|---|---|
| PDB: 7S21 | PDB: 7S20 | PDB: 7S2T | PDB: 7S4Q | |
| Data collection | ||||
| Magnification | 130,000 | 130,000 | 130,000 | 130,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Total dose (e−/Å) | 66 | 66 | 46 | 46 |
| Frame rate | 4 frames/sec | 4 frames/sec | 5 frames/sec | 5 frames/sec |
| Defocus range (μm) | −0.6 to −2.6 | −0.6 to −2.6 | −0.6 to −2.6 | −0.6 to −2.6 |
| Pixel size | 1.06 | 1.06 | 0.537 | 0.537 |
| Data processing | ||||
| Symmetry | I1 | I1 | I1 | I1 |
| Initial particle number | 20706 | 20706 | 8139 | 10357 |
| Final particle number | 5183 | 9309 | 7961 | 8512 |
| Resolution (Å) | 3.4 | 3.4 | 3.5 | 3.1 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Refinement | ||||
| Map sharpening B factor (Å2) | −90 | −90 | −90 | −90 |
| Model composition | ||||
| Non-hydrogen atoms | 2041 | 6364 | 6677 | 6659 |
| Protein residues | 265 | 825 | 862 | 862 |
| B factors (Å 2 ) | ||||
| Protein | 86.41 | 79.31 | 97.95 | 102.02 |
| R.m.s. deviations | ||||
| Bond length (Å) | 0.006 | 0.005 | 0.007 | 0.008 |
| Bond angles (°) | 0.765 | 0.729 | 0.903 | 0.928 |
| Validation | ||||
| Model-to-map fit CC protein | 0.84 | 0.80 | 0.80 | 0.81 |
| MolProbity score | 2.17 | 2.30 | 2.36 | 2.35 |
| Clashscore | 9.87 | 12.95 | 14.00 | 14.00 |
| Sidechain outliers (%) | 0.00 | 0.3 | 0.9 | 0.3 |
| Ramachandran | ||||
| Favored (%) | 85.44 | 83.88 | 81.88 | 82.59 |
| Allowed (%) | 14.56 | 16.12 | 18.00 | 17.41 |
| Disallowed (%) | 0.00 | 0.00 | 0.12 | 0.00 |
Encapsulins have been classified into four distinct families based on the cargo proteins they contain (Andreas and Giessen, 2021). Among these, only Family 1 and Family 2 encapsulins have been characterized. Family 1 is comprised of the classical encapsulins of varying sizes which are involved in the protection of the organism against oxidative stress (Andreas and Giessen, 2021). M. xanthus EncA is included in this family. Family 2 encapsulins are mostly involved in sulfur metabolism (Andreas and Giessen, 2021). Structures of encapsulins from Family 1 with T=1, T=3 and T=4 symmetry have been solved (Almeida et al., 2021). In contrast, only one structure has been determined for the Family 2 encapsulins, while no structure is available for Family 3 and Family 4 encapsulins (Almeida et al., 2021). Structural comparisons show that the N-terminus of Family 2 shell proteins including the N-terminal helix (N-helix) extends towards the outside of the capsid, in contrast to Family 1 shell proteins where the N-terminus is sequestered inside the encapsulin shell (Andreas and Giessen, 2021). EncA T=1 encapsulin is slightly smaller in size (18 nm) in comparison to other naturally occurring T=1 encapsulins from Family 1 (which range from 20–24 nm), and Family 2 (26.5 nm) (Figure S4A) (Nichols et al., 2021). Structural alignment of EncA protomers with the Family 1 protomers at the 2-fold-axis showed that one of the subunits mostly overlapped (root mean square deviation [RMSD] of 1.16 Å), whereas the other subunit was rotated by about 45° resulting in an overall RMSD of 6.0 Å between the dimers (Figure S4B). A similar alignment with the Family 2 S. elongatus encapsulin PCC 7942 protomers showed an even more pronounced rotation of one of the subunits by about 60° resulting in an overall RMSD of 15 Å between the dimers (Figure S4C). The differences in the subunit arrangements between EncA and the other T=1 encapsulins resulted from the different structural alignment of the E-loop between the protomers (Figure S4D). As such, EncA showed a unique T=1 assembly in comparison to the previously characterized Family 1 and Family 2 T=1 encapsulins. The EncA T=1 shell has an inner volume of 8.71 × 105 Å3. In contrast the EncA T=3 shell has an inner volume of 3.09 × 106 A3 which corresponds to ~3.5 times more storage space. The volume difference can mean that T=3 encapsulins might be able to store more Fe than T=1 encapsulins.
Binding site of TP on EncA
To date only a few encapsulins with cargo proteins have been characterized structurally. These structures show that in most cases the cargo is not symmetrically arranged within the icosahedral shell likely due to low occupancy or conformational flexibility created by the N/C-terminus linker, which makes it hard to resolve the cargo density (Giessen et al., 2019, LaFrance et al., 2021, McHugh et al., 2014, Ross et al., 2021, Tang et al., 2021, Xiong et al., 2020). In the cases where the cargo density could be resolved, the encapsulin shells had considerably higher resolution in comparison to the cargo (Giessen et al., 2019, LaFrance et al., 2021, Ross et al., 2021, Tang et al., 2021). Among all encapsulin structures deposited in the Protein Data Bank (PDB) only two show well defined TP binding sites on the shell protein namely Q. thermotolerans T=4 encapsulin (PDB ID: 6NJ8) and T. maritima T=1 encapsulin (PDB ID: 3DKT) (Giessen et al., 2019, Sutter et al., 2008). M. xanthus cargo proteins EncB, EncC and EncD vary in size and share only marginal sequence similarity (23.4%−30.1%). However, their C-terminal TP is highly conserved with the consensus sequence LTVGSLRR (in EncD the last residue is G, Figure S5) (McHugh et al., 2014). Although, EncB and EncC share only 30% identity both are homologous to bacterial ferritin-like proteins with diiron binding sites.
To obtain the cryo-EM structure of EncA with its cargo proteins we co-expressed EncA with either EncB or EncC using a dual-expression plasmid in E. coli. In both cases, the cargo protein expression was much higher in comparison to EncA (Figure S1B and C). On the other hand, the cargo incorporated into the shell was only about 30–40% of EncA as judged from the SDS-PAGE gels, suggesting that the binding sites were not fully occupied (Figure S1B and C). This is similar to the native M. xanthus encapsulin system where the total cargo incorporation was calculated as ~30% (EncA-72%, EncB-7%, EncC-15% and EncD-6%) (McHugh et al., 2014). We solved EncA-EncB and EncA-EncC structures at 3.5 Å and 3.2 Å resolution, respectively, after applying icosahedral (I) symmetry (Figure 2A and Table 1). Both structures had T=3 symmetry and showed clear densities for the TPs and some additional residues (Figure 2). The TP densities were similar at the 5-, 3- and 2-fold axes in the icosahedral reconstructions and we did not observe any substantial differences between the TPs located at the pentameric and hexameric facets (Figure 2B and Figure S6A and B). The binding sites were located at the P-domain in a hydrophobic cleft formed between the N-terminal helix and the 3-stranded-β-sheet (Figure 3A and B). Analysis of the binding sites revealed that the TP-shell interactions were a combination of electrostatic and hydrophobic interactions (Figure 3C). The arginine residues at the tip were more flexible in comparison to the rest of the conserved residues as determined by the B-factors (Figure S6C) and pointed towards a highly negatively charged patch where the side chains could form hydrogen bonds and electrostatic interactions with multiple residues in any orientation (Figure 3B and C). The residues LTVG were more rigid forming a curved region in the loop which extended towards the hydrophobic cleft on the shell protein (Figure 3B and Figure S6C). The non-conserved residues that were visible in the structure showed more flexibility, as determined by the B-factors, which is not surprising as that region has minimal interaction with the shell and extends towards the interior of the encapsulin (Figure 3A and Figure S6C).
Figure 2.
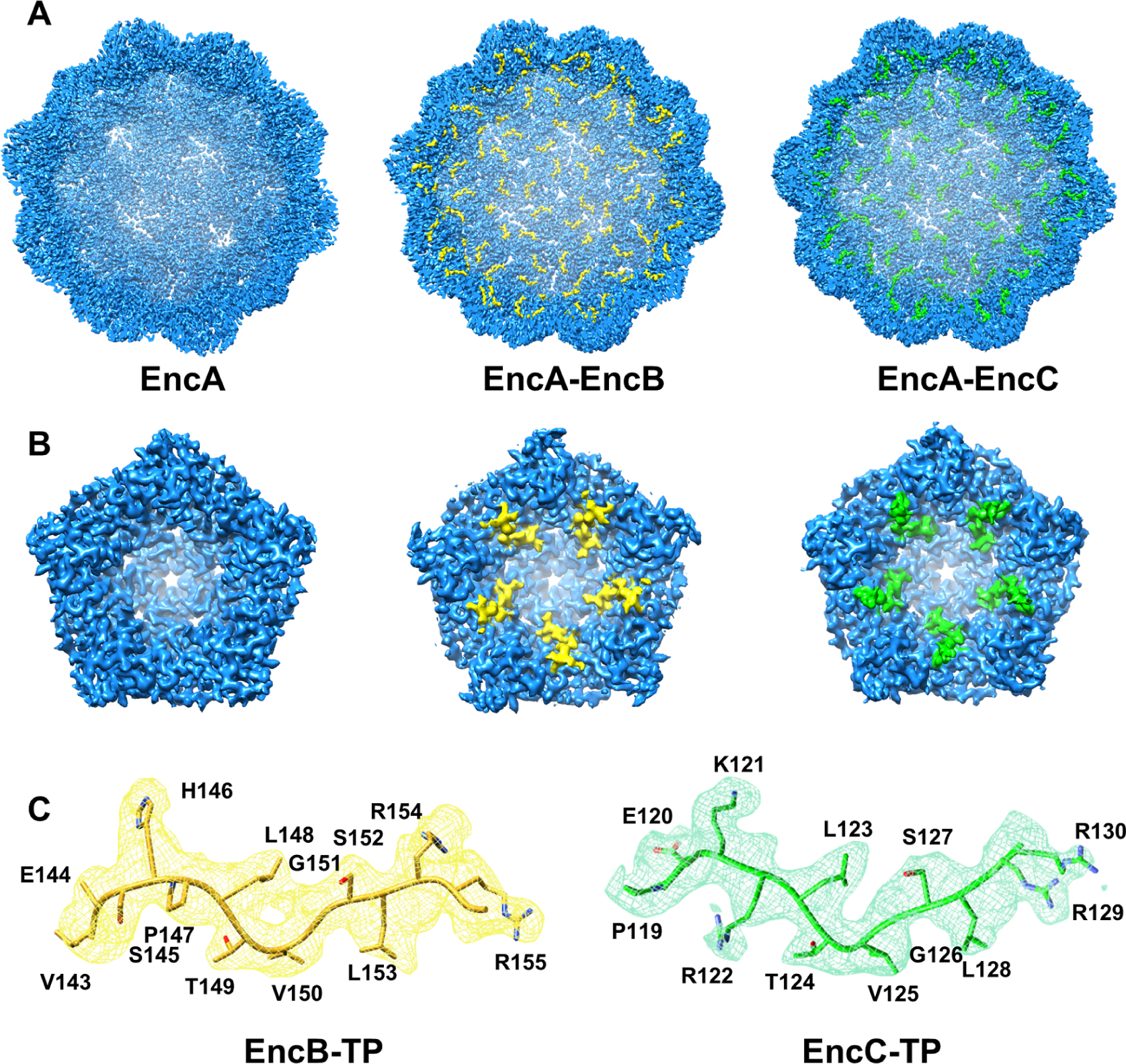
Locations of TP binding sites on the EncA shell. (A) Cryo-EM maps of empty EncA (left), EncA-EncB (middle) and EncA-EncC (right) viewed from inside the shell. (B) Close-up views of TP densities as viewed from inside the shell along a 5-fold axis: EncA (left), EncA-EncB (middle) and EncA-EncC (right). EncA shell density is shown in blue. EncB TP density is shown in yellow and EncC TP density is shown in green. (C) Atomic models of EncB TP (yellow cartoon) and EncC TP (green cartoon) built inside the cryo-EM densities (displayed as mesh) in matching colors. See also Table 1, Figure S5 and Figure S6.
Figure 3.
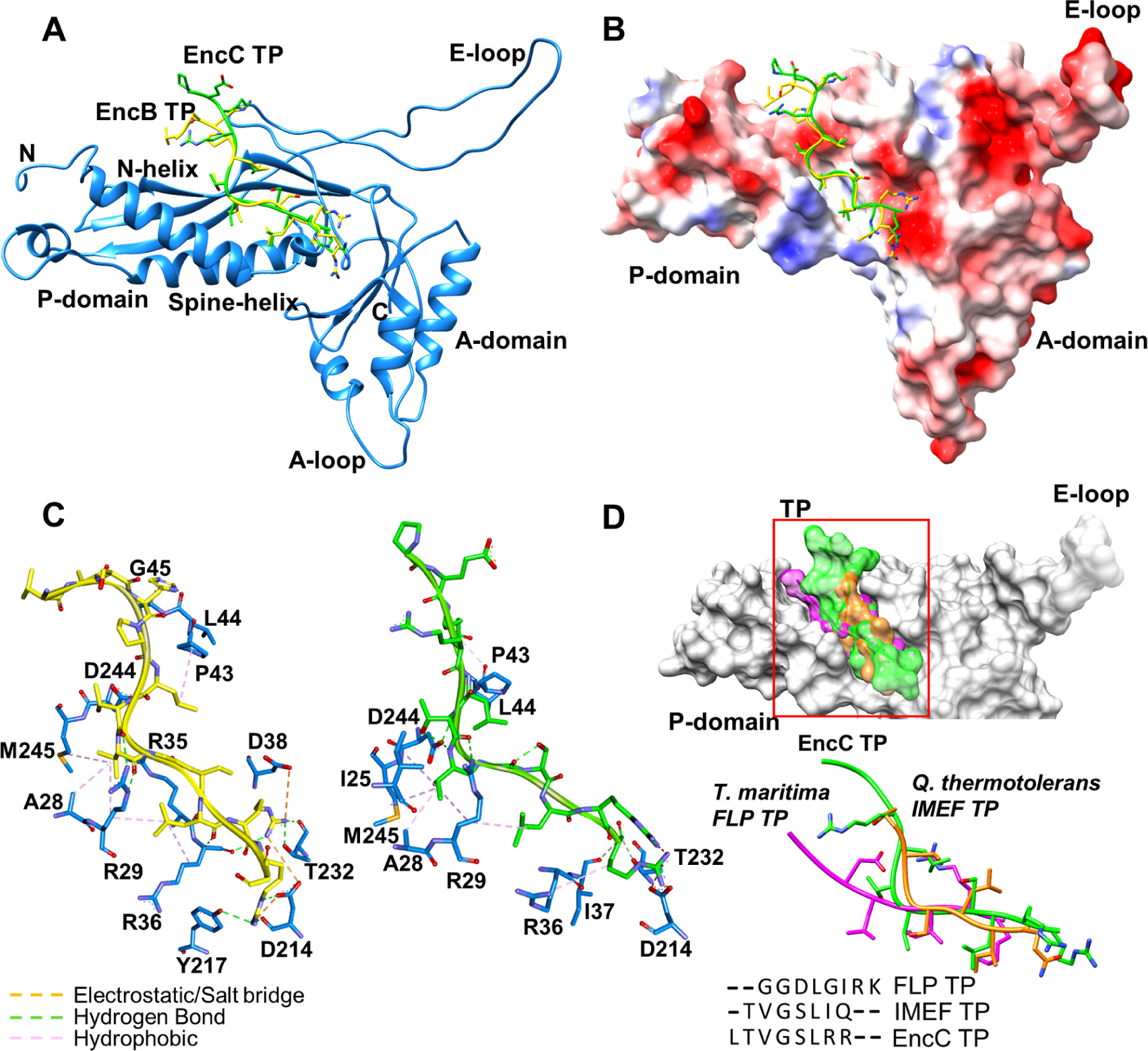
Locations of TP binding sites on an EncA monomer. (A) EncB TP (yellow) and EncC TP (green) located on EncA (blue). (B) EncB TP and EncC TP shown on EncA surface colored according to the electrostatic surface potential (blue: electropositive, red: electronegative and white: neutral). (C) Close-up views of EncB TP (left) and EncC TP (right) binding pockets. Electrostatic interactions and salt bridges are represented as orange dashes, hydrogen bonds are shown as green dashes and hydrophobic interactions are shown as pink dashes. (D) Structural comparison of TPs from different organisms. Surface representations of TPs from M. xanthus EncC (green), T. maritima FLP (magenta) and Q. thermotolerans IMEF (orange) located on shell protein (gray) are shown on top. Superimposed structures and sequence alignment of TPs are shown at the bottom. See also Figure S5 and Figure S6.
Encapsulin shell protein folding is highly conserved among all the structurally characterized encapsulins despite sequence diversity (Nichols et al., 2017). We compared the conserved TP of M. xanthus cargo proteins to the TPs of T. maritima FLP and Q. thermotolerans IMEF (Giessen et al., 2019, Sutter et al., 2008). All the TPs targeted a similar area on the shell (Figure 3D). There was very little sequence conservation among the M. xanthus EncB/C and T. maritima FLP TPs (33% sequence identity) (Figure 3D). By contrast, Q. thermotolerans TP was highly homologous to EncB/C TPs (71% sequence identity). EncC TP showed a conformation similar to the IMEF TP even though the conserved residues did not overlap in the superimposed structures, likely due to the different binding residues of the shell proteins. In comparison, FLP TP adopted a different conformation which extended towards the N-helix (Figure 3D). Analysis of TP interactions showed greater interface areas (630/640, 570 and 470 Å2 for EncB/C TP, FLP TP and IMEF TP, respectively) and higher binding energies (−9/−10, −5 and −6 kcal/mol for EncB/C TP, FLP TP and IMEF TP, respectively) for EncB and EncC in comparison to FLP and IMEF.
Cargo binding to EncA
Different binding modes of cargo to T=1 and T=4 encapsulins have been observed (Giessen et al., 2019, LaFrance et al., 2021, Nichols et al., 2021, Ross et al., 2021, Tang et al., 2021). In M. smegmatis two dodecameric hexamers of DyP with a 2-fold axis of symmetry stretch across the interior of the encapsulin (Tang et al., 2021). In Q. thermotolerans the IMEF cargo binds as dimers most likely at the 2-fold hexameric capsomers (Giessen et al., 2019). In T. maritima the decameric cargo protein with 5-fold symmetry preferentially binds to the pentameric axis of the icosahedral shell (LaFrance et al., 2021). In H. ochraceum four decameric EncFtn are arranged with tetrahedral symmetry within the icosahedral shell where two of them are located at the 5-fold axes and the other two are found between the 5-fold and 3-fold axes (Ross et al., 2021). During reconstructions we observed strong inner densities within the EncA-EncB and EncA-EncC shells which disappeared during high resolution icosahedral reconstructions indicating that the cargo within the encapsulin did not follow icosahedral symmetry and was rotationally averaged during symmetry-imposed processing. This result was in agreement with the previous reconstruction of the EncA-cargo from M. xanthus (McHugh et al., 2014). In order to resolve the cargo density within the encapsulin shells we subtracted the icosahedral shell from the EncA-EncC particles and employed a focused classification. The final reconstruction showed an ~8.0 Å resolution cargo density matching the size of the decameric EncC at the 5-fold-axis (Figure 4). The cargo was located ~3.4 nm away from the shell and ~2.4–2.7 nm away from the tip of the resolved TP. We could not obtain a density for the rest of the C-terminus in any of our reconstructions. Here it should be noted that only a small fraction of encapsulin particles (24%) showed cargo densities around the 5-fold axes indicating low occupancy in these areas either due to the shell containing a low number of cargos and/or some cargo being asymmetrically localized inside the shell. In the natural M. xanthus encapsulin there are 3 cargo proteins with different sizes where the length of their C-terminal disordered region differs significantly suggesting a true symmetry of the cargo within the shell is improbable (Figure S5). We also attempted to localize the cargo inside the EncA-EncB shells. However, the number of particles that showed resolvable cargo densities were much lower in comparison to EncA-EncC therefore we decided not to characterize these particles any further.
Figure 4.
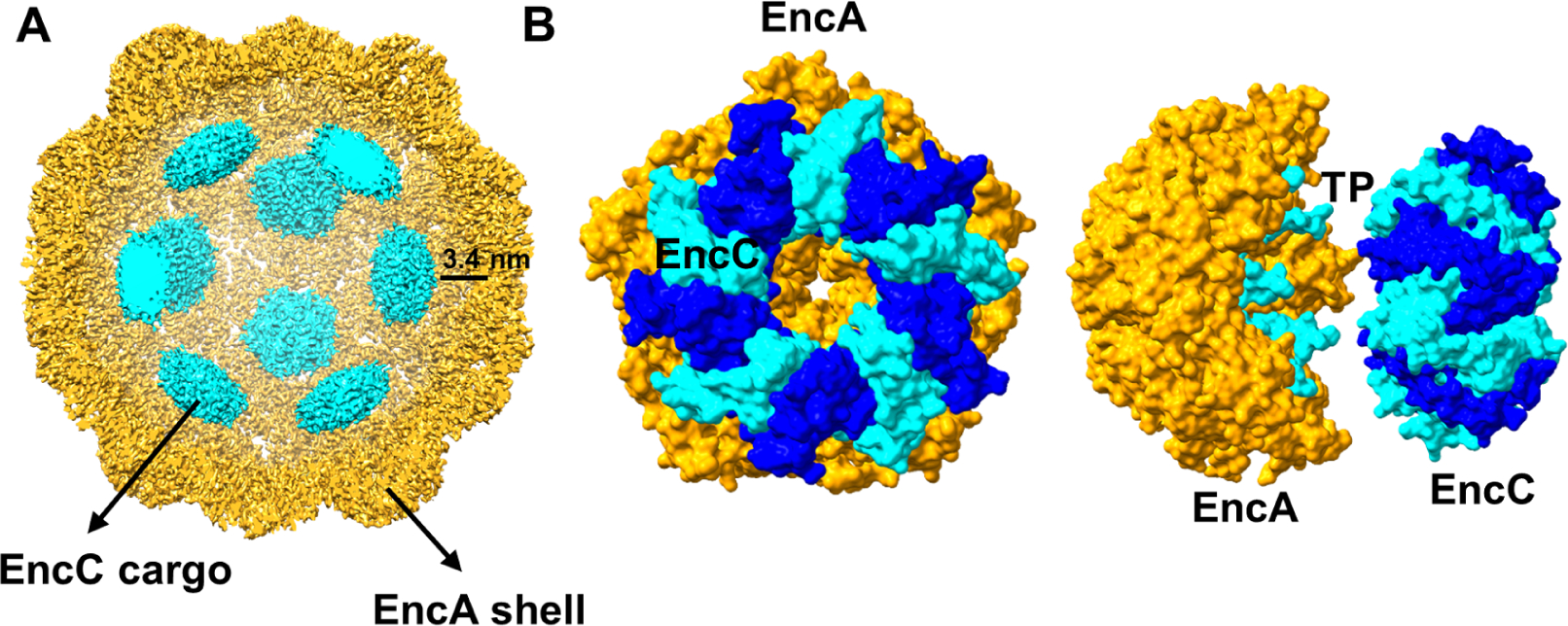
Cargo location inside the EncA shell. (A) Inside view of EncC-loaded EncA. An 8 Å map of cargo (cyan) was combined with the 3.2 Å map of the shell (yellow). (B) Surface representations of the pentameric EncA and decameric EncC structures docked into the cryo-EM density as viewed from front (left) and side (right). The individual monomers of EncC are colored in blue and cyan while EncA is colored in orange. The TP binding to the EncA pentamer is also shown in cyan and visible in the side-view.
High resolution structures of EncB and EncC
Previous M. xanthus encapsulin studies have shown that the system acts as an “iron-megastore” that can store up to 10 times more Fe atoms in comparison to the classical bacterial ferritins and protects the bacteria from oxidative stress (McHugh et al., 2014). This storage capacity is due to the ability of the ferritin-like cargo proteins, EncB and EncC, to oxidize Fe+2 to Fe+3 using a conserved catalytically active FOC. In order to obtain high-resolution structures of the cargo proteins we crystallized both EncB and EncC as decamers with 1.86 Å and 2.49 Å resolutions, respectively (Figure 5A, B and Table 2). Both EncB and EncC decamers were formed by 5 repeating units of 2 antiparallel α-helices which formed a dimerization interface required for ferroxidase activity (Figure 5A, B and C). The observed EncB decamers were 7.0 nm wide and 5.0 nm thick while the EncC decamers were 8.5 nm wide and 5.0 nm thick with more visible C-termini (Figure 5A and B). However, these are not the final particle sizes since both structures lacked densities for some residues located at the N- and C-termini, either due to proteolytic cleavage during crystallization or disorder. The visible N-termini of alternating subunits extended towards the center of the decameric ring, while the C-termini were arranged around the circumference. In that position, the C-termini can freely interact with the outer encapsulin shell. Both proteins had folds similar to the previously defined encapsulated ferritins Rhodospirillum rubrum Rru_A0973, Haliangium ochraceum EncFtn and Pyrococcus furiosus PFC_05175 where the decamers resemble toroids with D5 symmetry (Figure 5D) (He et al., 2019, Piergentili et al., 2020). This architecture is very different from classical bacterial ferritins which form 24-meric nanocages and do not require an outer shell to store iron (Tosha et al., 2012). Electrostatic potential map analysis of EncB showed an overall even distribution of the charges with localized negative areas concentrated around the middle of the dimerization interface as viewed from outside (Figure 5E). On the other hand, EncC was mostly negatively charged on the outside and inside of the α-helical core regions forming the toroidal particles while the C-terminus regions remained mostly neutral with positively charged patches (Figure 5F).
Figure 5.
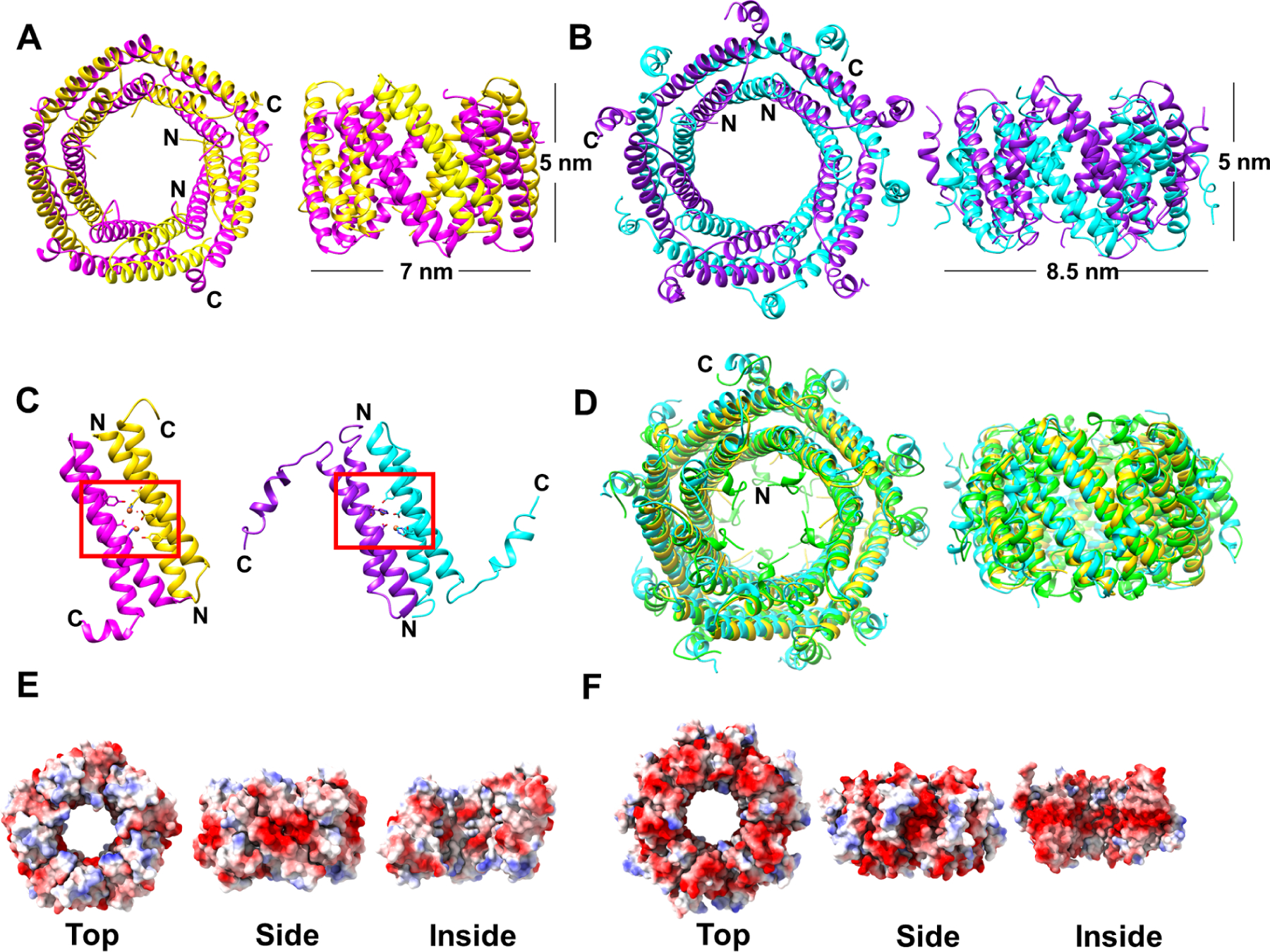
Crystal structures of EncB and EncC decamers. (A) EncB crystal structure in P21 space group is shown from top (left) and side view (right). The α-helical pairs forming the catalytic dimer interface are shown in magenta and yellow. (B) EncC crystal structure in C2 space group is shown from top (left) and side view (right). The α-helical pairs forming the catalytic dimer interface are shown in purple and cyan. The observed dimensions of the decamers are indicated in the side views. (C) The location of FOCs formed by the dimer pairs of EncB (left) and EncC (right) are indicated by red boxes. (D) Structural alignment of EncB (yellow), EncC (cyan) and R. rubrum encapsulated ferritin Rru_A0973 (PDB ID: 5DA5, green) is shown from top (left) and side view (right). (E) Electrostatic surface representation of EncB decamers as viewed from top, side and inside. (F) Electrostatic surface representation of EncC decamers as viewed from top, side and inside. Electrostatic surface potential is colored in blue for electropositive, red for electronegative and white for neutral areas. See also Table2.
Table 2.
X-Ray Diffraction Data Collection and Refinement Statistics
| Deposition ID | EncB | EncB | EncC |
|---|---|---|---|
| PDB-7S5C | PDB-7S5K | PDB-7S8T | |
| Data collection | |||
| Beamline | APS-22-ID-D | APS-22-ID-D | APS-22-ID-D |
| Wavelength | 1.0 | 1.0 | 1.0 |
| Space group | P21 | C2 | C2 |
| Cell dimensions | |||
| α,ß,γ (Å) | 63.8 68.4 83.4 | 127.5 44.4 128 | 148.7 47.8 135.3 |
| a,b,c (°) | 90.0 103.6 90.0 | 90.0 114.1 90.0 | 90 90.1 90 |
| Resolution range (Å) | 43.34–1.86 (1.93–1.86) | 63.44–1.95 (2.02–1.95) | 37.77–2.49 (2.58–2.49) |
| Redundancy | 6.2 (3.2) | 6.6 (5.5) | 6.6 (6.3) |
| Completeness (%) | 95.4 (71.5) | 98.1 (90.9) | 96.3 (92.9) |
| I/σ | 12.8 (1.4) | 7.6 (1.5) | 12.5 (6.7) |
| Rmeas | 0.091 (0.87) | 0.20 (0.95) | 0.16 (0.54) |
| CC1/2 | 1.00 (0.59) | 1.00 (0.35) | 1.00 (0.95) |
| No. of unique reflections | 56065 (4286) | 47414 (4395) | 32528 (3099) |
| Refinement | |||
| Resolution (Å) | 1.86 | 1.95 | 2.49 |
| No. of reflections used | 54973 (3934) | 47324 (4349) | 32498 (3097) |
| Reflections used for R-free | 1970 (145) | 1995 (181) | 1994 (192) |
| Rwork/Rfree | 0.18/0.22 | 0.21/0.24 | 0.32/0.34 |
| No. of atoms | |||
| Protein/water/metal | 5117/354/30 | 5230/146/26 | 6341/86/7 |
| RMSDs | |||
| Bond lengths (Å) and angles (°) | 0.01/1.41 | 0.01/1.19 | 0.006/1.08 |
| Ramachandran (%) | |||
| Favored/allowed/outliers | 99.4/0.6/0.0 | 98.7/1.3/0.0 | 93.4/5.5/1.1 |
| Rotamer outliers (%) | 0.0 | 0.7 | 1.0 |
| Average B-factor | 38.4 | 39.1 | 57.4 |
| Protein/water/metal | 37.8/44.5/55.4 | 38.9/41.6/70.3 | 57.5/48.6/58.7 |
Statistics for the highest-resolution shell are shown in parenthesis
EncB and EncC iron binding sites
Similar to classical ferritins and diiron-carboxylate proteins, both cargos contained a duplicated motif consisting of two consecutive helices and a conserved EXXH motif (Figure 5C) (Nordlund and Eklund, 1995). The diiron binding site of EncC was very similar to the previously defined binding sites of Rru_A0973 (PDB ID: 5DA5) and PFC_05175 (PDB ID: 5N5E) (Figure S7A) (He et al., 2016, He et al., 2019). In the EncC structure one state of Fe coordination (state 1) could be observed where Fe atoms were tightly coordinated by 3 glutamic acid residues and a histidine residue forming a 5-coordinate Fe+2 complex (Figure 6A and Figure S7A). In this state, nearby conserved tyrosine residues were located in close proximity (3.3– 4.0 Å) to each Fe+2 (Figure 6A). A conserved tyrosine near the iron binding site has been commonly observed in bacterial ferritins (Hall et al., 2002, Honarmand Ebrahimi et al., 2015, Nordlund and Eklund, 1995). This tyrosine residue has been suggested to form an intermediate tyrosyl radical that is involved in direct conversion of O2 to H2O whereas the coordinating histidine residues are likely involved in electron transfer paths (Nordlund and Eklund, 1995). Interestingly, the crystal structure of EncB showed a second state (state 2) of the FOC which has not been previously observed in the encapsulated ferritin structures that show Fe atoms at their FOC (Figure 6B and Figure S7B). In this state, the E28 (E17 in EncC) residues on both sides of the FOC shifted away from the Fe atom with the rotation of the side chains by 180° in comparison to the EncC E17 residues (Figure S7B and C). This rotation increased the distance between the E28 carboxylate groups and the Fe from 2.7A to 4.1 and 4.4 Å (Figure S7C). Also, in EncB, the distances between both Y35 (Y24 in EncC) residues and the Fe atoms decreased to only 2.6 Å due to conformational shifts of the tyrosine residues and/or the Fe atoms (Figure 6B and S7C). These changes brought the Fe atoms from a high-affinity 5-coordinated state to a lower affinity 4-coordinated state.
Figure 6.
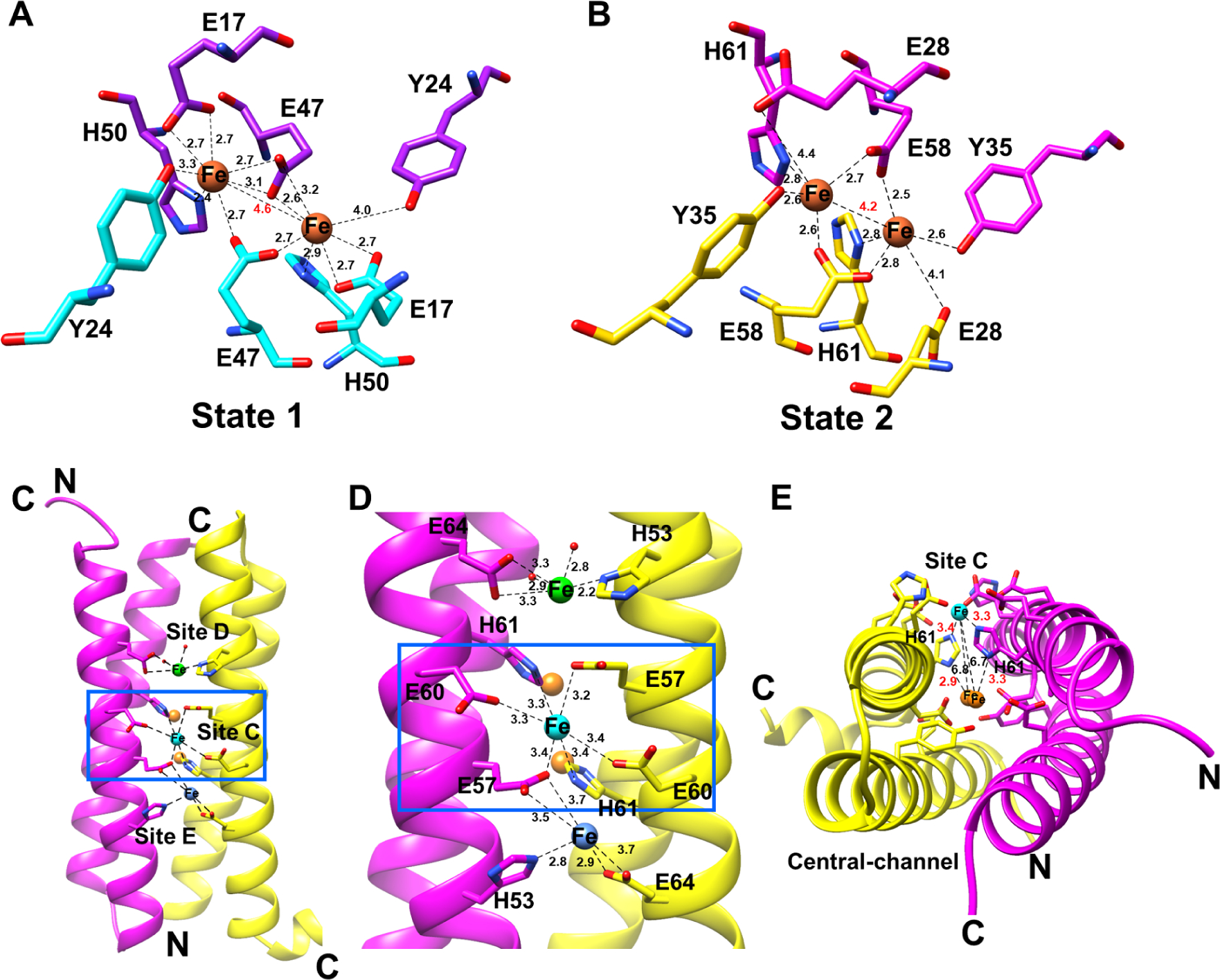
EncB and EncC ferroxidase centers (FOCs). (A) Diiron binding site of EncC showing Fe+2 coordination in “state 1” which is the equivalent of iron coordination shown in other encapsulated ferritin structures. (B) Diiron binding site of EncB showing Fe+2 coordination in “state 2”. Coordinating residues in the FOC from each α-helix are shown in purple and cyan for EncC, and magenta and yellow in EncB. Fe atoms are shown as orange spheres. Distances between Fe atoms and the coordinating residues are shown as black dashed lines. A red label shows the distance between two Fe atoms. (C) Additional metal binding sites formed by the glutamic acid-histidine ladder in EncB as viewed from outside. EncB monomers are colored in magenta and yellow. The “site C” Fe atom is shown as a cyan sphere, while the loosely coordinated Fe atoms in “site D” and “site E” are shown as green and blue spheres, respectively. (D) Close-up view of the EncB additional metal binding sites. Distances between Fe atoms and coordinating residues are shown as black dashes. (E) EncB site C and FOC as observed along the axis of the α-helices. Site C Fe is colored in cyan while the FOC Fe atoms are colored in orange. Distances are shown as black dashes. Distances between the bridging histidine residues and the Fe atoms are labeled in red. Distances between the site C Fe and FOC Fe atoms are labeled in black. See also Figure S7 and Figure S8.
In addition to two different iron coordination states, we detected a glutamic acid-histidine ladder along the periphery of the two α-helices forming the FOC (Figure 6C). The ladder was formed by 3 glutamic acid residues and 2 histidine residues from each dimer. The EncB structure showed that this ladder acted as a secondary Fe binding center (Figure 6C and D). Superimposed structures of all the EncB dimers showed that one Fe atom consistently was found at the center of the ladder and was coordinated by four glutamic acid residues and two histidine residues (Figure 6D). We determined that this site corresponds to the “site C” of the bacterial ferritins (Honarmand Ebrahimi et al., 2015, Ebrahimi et al., 2016, Hempstead et al., 1994, Pfaffen et al., 2013, Stillman et al., 2001, Tatur et al., 2007). Similar to bacterial ferritins, EncB site C was located ~7.0 Å from the FOC (Figure 6E) (Pfaffen et al., 2013). One histidine residue (H61) was bridging between the site C and the FOC, coordinating Fe atoms in both sites (Figure 6E). We observed two more potential Fe binding sites at the top and bottom of the ladder (which we named as site D and site E, respectively) where we could locate Fe atoms in some of the dimers (Figure 6C and 6D). Four out of five of the ladder residues and all of the site C residues were also conserved in EncC, Rru_A0973 and Hoch_3836 (Figure S7D and S8A). On the other hand, three out of five ladder residues were conserved in PFC_05175 (Figure S8A). The ladder has an electronegative nature which can potentially attract the positively charged Fe+2 atoms (Figure S8B). Therefore, we propose that this region may form the “gateway” for Fe+2 atoms for some encapsulated ferritins. The site C Fe+2 entry has also been proposed for some bacterial ferritins (Ebrahimi, 2015). Interestingly, Piergentili et al. identified another Fe binding site on the opposite side of FOC located at the central-channel facing side for Rru_A0973 (Piergentili et al., 2020). The authors hypothesized that this site would be the Fe “entry” site as opposed to that proposed in this study. However, this central channel entry theory could not be readily applied to some of the other encapsulated ferritins including EncB and EncC in which one of the Fe coordinating residues is changed to an arginine (R27) and an alanine (A16), respectively (Figure S8A and C). In the study carried out by Piergentili et al., glutamic to alanine mutations resulted in abrogation of metal binding to the site, providing evidence that this site cannot be an Fe binding site for EncC (Piergentili et al., 2020). In the EncB structure the orientation of glutamic acid and arginine side chains do not support Fe coordination where the glutamic acid side chains point away from the binding site (Figure S8C). The presence of the arginine residues at that site creates a neutral to electropositive surface area at the binding site as opposed to the highly electronegative glutamic acid-histidine ladder (Figure S8C). Therefore, we conclude that there are likely different Fe “entry” sites for different encapsulated ferritins and in some cases Fe+2 movement to the FOC is from outside-in and in others it is from inside-out of the central channel.
Both EncB and EncC were crystallized in the presence of Ca ions. We observed Ca binding sites in close proximity to the FOC in the high-resolution structures of EncB (Figure S8D). The Ca ions were separated by 3.2 Å and they were located at 3.3–3.8 Å from the nearby Fe atoms (Figure S8D). They were coordinated by T31 and E58 from each of the α-helices and a H2O molecule (Figure S8D). Interestingly, a Ca binding site formed by threonine, glutamic acid and H2O and located between the iron entry channel and the FOC has been described for the plant ferritin SFER4 (Masuda et al., 2010). This site was described as a metal transit site as the mutation of the glutamic acid resulted in a delay in the oxidation of the iron. It is possible that this site may act in a similar manner in EncB, i.e., as a Fe+3 transit site for the exit of the molecule from the FOC. The Ca binding site is located between the FOC and the central channel of the decamer which supports the leaving of Fe+3 through the central channel.
Analysis of EncA shell pores
It has been proposed that the negatively charged pores located at the 5-fold axes of the shell allow the entry of positively charged Fe+2 to the lumen of the encapsulin (He et al., 2016). In that communication, Fe+2 was suggested to gain access to the metal binding sites within the central channel of the cargo protein prior to oxidation within the FOC and then released as Fe+3 (He et al., 2016). We analyzed the pores of EncA at the 2-fold, 3-fold and 5-fold axes (Figure S9). Our results showed that the pores at the 5-fold-axis and 3-fold axis were open while the pores at the 2-fold axis remained closed. The surrounding environment of the 5-fold-axis pore had electronegative patches on the outside and more neutral charges on the inside, while the smaller 3-fold-axis pore area was highly electropositive on the outside and highly electronegative on the inside (Figure S9). The electropositive environment is created by arginine residues (R261) located at the tip of the E-loops surrounding the periphery of the pore and other arginine residues located in close proximity to the pore. It is unlikely that the positively charged Fe+2 atoms will diffuse towards the arginine residues. Therefore, we conclude that the 5-fold-axis pore is the most likely pore for Fe+2 entry. Our comparisons of EncA, EncA-EncB and EnA-EncC 5-fold-axis pores reveals a dynamic pore where the pore size varies between 6 Å for EncA to ~9 Å for EncA-EncB and ~10 Å EncA-EncC (Figure 7). The pore periphery contains a threonine (T200) and a glycine (G201) residue while the pore is surrounded by a tyrosine (Y197), glutamic (E198) and a lysine (K199). The electronegative charge is controlled by the positioning of the glutamic and tyrosine residues while the lysine side chain remains mostly buried. Our structures show that the pore size can vary by at least 3 Å between a “closed state” and “semi-open” state. The pore size is likely controlled by the influx of the cations and/or some other small molecules that might require a larger opening for the entry into the encapsulin. In the case of Fe+2 side entry to the FOC, and Fe+3 exit via the central-channel of the decamer, the latter need not be aligned with a symmetry axis, showing how the asymmetrically arranged cargo can access Fe+2 diffusing from the 5-fold-axis pores.
Figure 7.

Analysis of the EncA pores at the 5-fold-axis. Close-up views of the (A) EncA (blue), (B) EncA-EncB (pink) and (C) EncA-EncC (orange) shell at the 5-fold-axis showing the pores. Top panels show the cartoon representations where the pore-surrounding residues are shown as stick models. Bottom panels show the surface representations of the shell proteins. The sizes of pores, in Å, are shown in the centers of the upper panels. See also Figure S9.
DISCUSSION
Encapsulins constitute a growing family of nanocompartments that have been identified in aerobic and anaerobic microbes found in terrestrial and aquatic environments as well as bacterial and archaeal extremophiles. Although encapsulin research has gained momentum only recently, encapsulins have already been applied in various scientific fields (Efremova et al., 2021, Gabashvili et al., 2020, Giessen, 2016, Jones and Giessen, 2021, Sigmund et al., 2018). Their small size, self-assembly properties, stability across a wide range of temperature and pH, and ease of genetic modification facilitates their use for targeted drug/label delivery. It has been shown that encapsulins can be heterologously expressed in mammalian cells without causing cell toxicity thereby expanding their potential uses in the medical field (Sigmund et al., 2018). Encapsulins can be engineered to encapsulate both endogenous and exogenous cargos. The interior space of these nanocages can be used to house nanoreactors for the synthesis of biomimetic nanomaterials (Diaz et al., 2021), and cargo containers that would enable them to be used as cancer cell trackers and in MRI imaging (Efremova et al., 2021, Sigmund et al., 2018). The exterior surfaces of encapsulins can be used to display targeting ligands and antigenic molecules for the construction of targeted theragnostic nanoparticles and nano-vaccines (Choi et al., 2016, Choi et al., 2021). Cargo loading can be achieved by appending N/C-terminal TPs to the cargo(s). On the other hand, surface modification can be done through the engineering of the shell protein loops. Despite the current and potential of future applications of encapsulins in various fields, the structural characterization of known or newly discovered encapsulin systems has been very limited. Although, computational techniques and modeling studies can aid in the design of genetically modified encapsulin systems for biotechnological applications, the lack of structural information still acts as a barrier to such applications. For this reason, we carried out a comprehensive structural study of the M. xanthus EncA encapsulin-cargo system. Our structures show well defined EncB and EncC TP binding sites on EncA. Structural comparisons with the other TP-shell pairs show that even widely differing TPs can target the similar sites on the shell proteins which makes it easier to choose the right encapsulin cargo-TP pair for engineering. Also, we show that the same encapsulin protein can assemble with either T=1 or T=3 symmetry when expressed recombinantly. This in turn might aid in loading the cargo into a nanocage of defined size in a controlled manner.
The metal binding capabilities of naturally existing encapsulin proteins are also of particular importance as these cargoes can be altered to bind different metals including heavy metals. Encapsulins have very high metal storage capabilities. For instance, M. xanthus can store up to ~30,000 Fe atoms within its shell as opposed to the ferritins which can store up to 4500 Fe atoms (McHugh et al., 2014). As such, encapsulins can be engineered to bind to toxic heavy metals for environmental cleanup. In fact, it has been shown that the active site Fe+2 of ferritins can readily be replaced by proxy metals including Zn+2, Ca+2, Mg+2, Co+2 and Cu+2 (Ebrahimi, 2015, Theil, 2013). Alternatively, the metal binding abilities of encapsulins can be altered to develop better medical imaging techniques. Although, metal binding and storage properties of bacterial ferritins have been extensively characterized less information is available for the encapsulated ferritins. To date, all the encapsulated ferritin structures have shown a unique iron coordination environment at the FOC. In comparison, our high-resolution EncB structures show conformational changes at the FOC emphasizing the importance of the conserved tyrosine residues. Also, we observe alternative Fe binding sites located at the sides of the decameric cargo proteins one of which corresponds to the site C of bacterial ferritins.
Different mechanisms have been proposed for the reduction of the Fe+2 at the FOC of bacterial ferritins (Honarmand Ebrahimi et al., 2017, Honarmand Ebrahimi et al., 2015). In the first model, the site C is not occupied by an extra Fe+2 and the two Fe+2 ions in the ferroxidase center are oxidized via a peroxodiferric intermediate which eventually forms two Fe+3 and H2O2. In the second model, site C is occupied by Fe+2 and a conserved tyrosine residue acts as an e− + H+ donor to the peroxidiferric intermediates forming Fe+3, Fe+4 (the presence of Fe+4 species at the FOC still remains hypothetical) and H2O molecules in addition to a tyrosyl radical. The high-valency Fe+4 oxidizes nearby Fe+2 releasing another H2O molecule and Fe+3. The tyrosyl radical is then reduced by the addition of another e− + H+ donor (Honarmand Ebrahimi et al., 2017). Although, this model falls in line with the extra metal sites we observe and the movement of the conserved tyrosine residues towards the Fe atoms at the FOC, it is not fully applicable to the encapsulated ferritins. First, the model is based on a single conserved tyrosine residue at the FOC. Second the model does not explain how the site C Fe+2, which is located 6.8 Å away from the FOC Fe atoms, can act as an e− donor.
Based on our EncB and EncC crystal structures, we propose a modified version of the second model (Figure 8). In this model, two conserved tyrosine residues act as e− + H+ donors to the peroxidiferric intermediates simultaneously forming two Fe+4 intermediates instead of a Fe+3 and Fe+4. As the Fe+2 at the site C is located ~7.0 Å away from the nearest Fe atom at the FOC in EncB it is highly unlikely that it can directly act as an e− donor to any of the Fe+4 molecules. Therefore, we propose that site C Fe+2 transfers the e− through an electron transfer chain formed by the two histidine residues (H61) bridging the site C to the FOC without the need for any significant conformational changes. According to our proposed model, first one Fe+2 located at the site C acts as the e− donor through one of the histidine residues and upon its oxidation to Fe+3 it is replaced by a second Fe+2 that acts as the second e− donor through the other histidine (Figure 8). This model, similar to that proposed previously, requires the reduction of the tyrosyl radical by a redox partner for the completion of the cycle (Honarmand Ebrahimi et al., 2017). According to our model the Fe+3 at site C should leave from the peripheral region. On the other hand, it is not clear whether Fe+3 molecules at the FOC move through alternative D and E sites or move into the central channel of the decamer. A surface representation of the EncB dimer shows that the central channel is partially blocked from the inside by two arginine residues (R27, Figure S8B). However, there are two narrow openings above and below the arginines that can give passage to the Fe molecules. Also, it is possible that the arginine side chains can move to open and close the passage to the channel.
Figure 8.
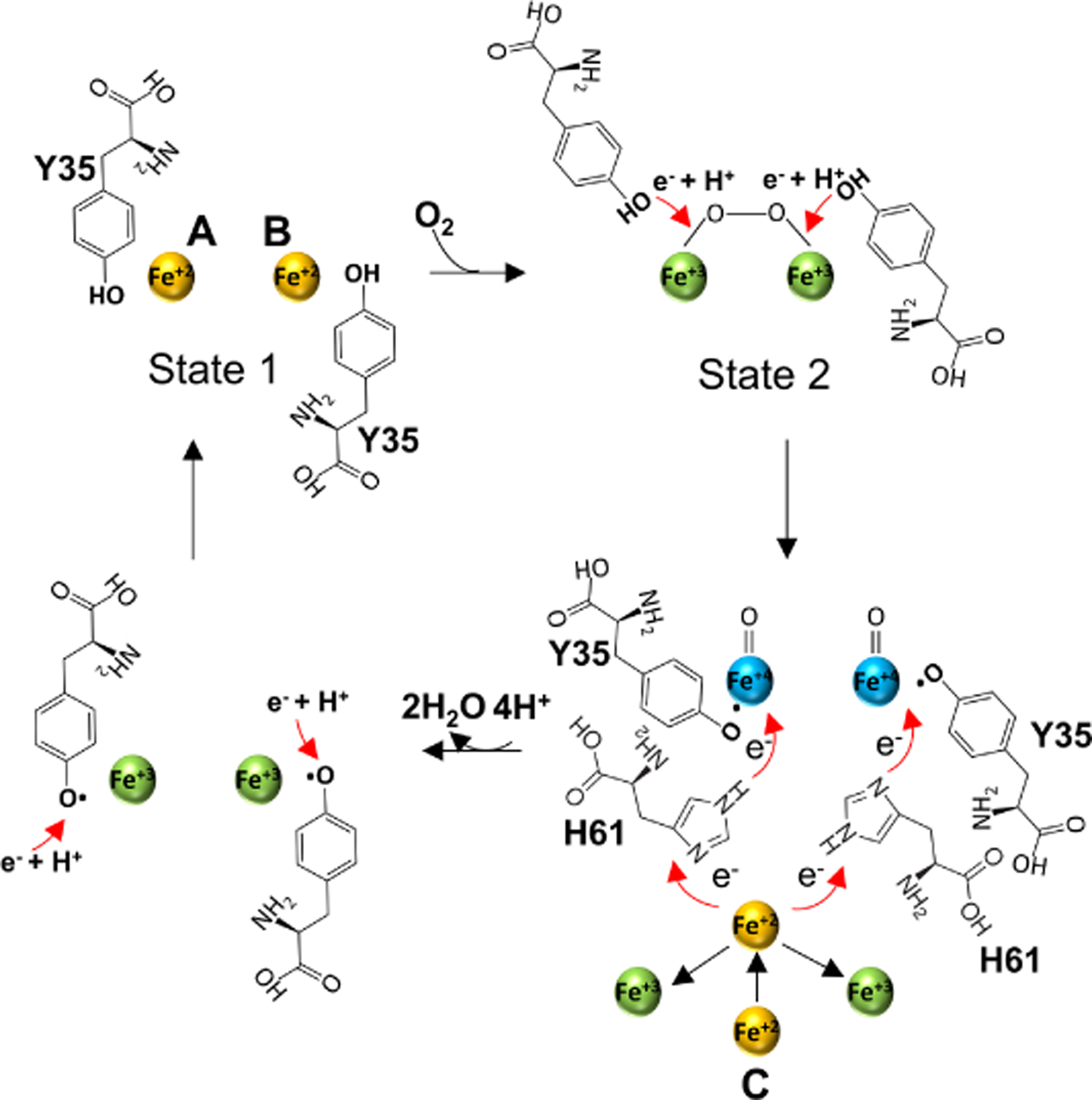
Proposed mechanism of Fe+2 oxidation at the FOC of EncB. In step 1, the 2 Fe+2 atoms in the “high-affinity” binding state (state 1) interact with O2 to form a peroxodiferric intermediate. In step 2, the two conserved Y35 move towards the peroxodiferric intermediate to donate e− + H+ to the Fe+2 atoms forming two tyrosyl radicals and two hypothetical Fe+4 intermediates with an unknown oxidation state. In step 3, one of the two Fe+4 is reduced to Fe+3 through an electro-transfer chain formed by the site C Fe+2 and H61 releasing a H2O molecule. The oxidized site C Fe+3 leaves the site and is replaced by a second Fe+2 which in turn reduces the second Fe+4 releasing a second H2O. In step 4 the tyrosyl radical is reduced by an unidentified redox partner followed by the addition of a proton. Finally, Fe+3 atoms at the FOC are displaced by the incoming Fe+2. See also Figure S10.
Our observation of dynamic pores in the encapsulin shells suggests that the pores can open and close enabling the “selective” entry of the metals and/or small molecules. The dynamic nature of the pores shows that the encapsulins can be engineered to serve as selective channels through which substrates of interest such as drugs could enter the shell or the mass transport could be enhanced by designing enlarged pores (Williams et al., 2018b).
Finally, we propose a simple model for Fe storage in the lumen of the encapsulin (Figure S10). In this model Fe+2 enters the encapsulin through the “open” pores located at the 5-fold-axis. Upon entry Fe+2 diffuses to the negatively charged iron binding sites formed by the glutamic acid-histidine ladder. From these sites Fe+2 is transported to the FOC where it is oxidized to Fe+3. Fe+3, which has a lower affinity for the FOC Fe binding sites, moves towards the central-channel and is displaced by another Fe+2. Fe+3 then exits the central-channel to enter the lumen of the encapsulin where it is stored in a crystalline mineral form, likely in the form of mineral ferrihydrite, [FeO(OH)]8[FeO(H2PO4)] (Hiemstra and Zhao, 2016).
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Paul T. Wingfield (wingfiep@mail.nih.gov).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Maps and models have been deposited in the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (accession numbers: EncA (T=1) EMD-24815, EncA (T=3) EMD-24814, EncA-EncB EMD-24816 and EncA-EncC EMD-24832). Models and crystal structures have been deposited in the Protein Data Bank, https://www.ebi.ac.uk/pdbe/ (PDB ID codes 7S21 EncA (T=1), 7S20 EncA (T=3), 7S2T EncA-EncB, 7S4Q EncA-EncC, 7S5C EncB-P21, 7S5K EncB-C2, 7S8T EncC).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL and SUBJECT DETAILS
For protein expression T7 Express Crystal Competent E. coli derived from BL21 strain was used. The bacteria were transformed with pETDuet-1 plasmids containing M. xanthus EncA, EncA and EncB or EncA and EncC genes. Cells were initially grown in Luria Broth (LB) medium containing 0.1 mg/ml ampicillin at 37°C and induced at 20°C with 1 mM IPTG. Expression and purification were done as described in Method Details.
METHOD DETAILS
Expression and purification of proteins
EncA (MXAN_3556) was cloned into the first cloning site of pETDuet-1 (GenScript) between the BamH1-Not1 restriction sites. The first cloning site of pETDuet-1 contains a N-terminal His-tag and adds a short linker sequence (SQDP) between the His-tag and the first residue of the EncA protein which likely makes the N-terminus His-tag more accessible. For co-expression, EncA was cloned into pETDuet-1 BamH1-Not1 sites and EncB (MXAN_3557) was cloned into the second cloning site between the Nde1-Xho1 restriction sites of the same expression plasmid, with the addition of a 6-His-tag at the N-terminus and a stop codon at the C-terminus to generate the EncA-EncB-pETDuet-1 plasmid. Similarly, EncC (MXAN_4464) with an N-terminal His-tag was cloned into the second cloning site between the Nde1-Xho1 restriction sites of EncA-pETDuet-1 plasmid to generate the EncA-EncC-pETDuet-1 plasmid. In this design the N-terminus His-tags of the cargo proteins are not accessible when they are encapsulated by EncA. However, the excess cargo that is not encapsulated can be captured by a nickel column. T7 Express Crystal Competent E. coli (New England Biolabs) transformed with the expression plasmids were grown overnight to a cell density of OD600 1.5 in 1 L flasks at 37 °C in the presence of 0.1 mg/ml ampicillin. The cells were induced with 1 mM IPTG and grown at 20 °C for 24h. The cells were collected by centrifugation at 5000 rpm for 20 min at 4 °C using a Beckman Coulter Avanti J30I refrigerated centrifuge (Beckman Coulter). Cells were then resuspended in 40 ml ice cold 50 mM Tris pH 8.0, 200 mM NaCl buffer supplemented with cOmplete EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN), 1% Triton-X100, 1% Tween-20 and 0.5 mg/ml lysozyme. The cells were sonicated with 20 bursts of 10 sec followed by intervals of 30 sec cooling at 60 kHz. Cell debris was removed by spinning the lysate at 12,500 rpm for 30 min at 4 °C (Beckman Coulter). The cleared supernatant for each preparation was incubated with 2 ml of HisPur Ni-NTA resin (ThermoFisher) for 20 min at room temperature. The cleared lysate-Ni-NTA resin was then loaded onto an empty gravity flow column. After the lysate flowed through the column, the column was washed with 20 ml of 50 mM Tris pH 8.5, 200 mM NaCl, 25 mM imidazole. The proteins were eluted with 10 ml of 50 mM Tris pH 8.5, 200 mM NaCl, 300 mM imidazole. The eluted proteins were concentrated to 5 ml using Amicon Ultra-15 centrifugal filter concentrator with 30 kDa cutoff (MilliporeSigma). The proteins were applied to a HiLoad Superdex 200 16/600 column (Cytiva) for size exclusion chromatography equilibrated with 20 mM HEPES pH 7.3, 150 mM NaCl. Eluates corresponding to EncA, EncA-EncB, EncA-EncC, EncB and EncC peaks were collected and further analyzed by SDS-PAGE using 4–20% Tris-Glycine gels (Invitrogen).
Sample preparation and data collection
Three microliters of the samples at 0.5 mg/ml were applied to Quantifoil R1.2/1.3 300 mesh Cu grids coated with 2 nm ultrathin carbon (Electron Microscopy Sciences, Protochips, Inc.) which were previously plasma-cleaned for 12 s with an argon-oxygen mixture (25% oxygen) in a model 1020 plasma cleaner (Fischione). Using a Leica EM GP plunger (Leica Microsystems), excess liquid was blotted 5–6 s after a 60 s wait time (95% humidity, 4°C) and grids were flash frozen in liquid ethane.
Data collection was performed with a Titan Krios G3 cryo-electron microscope (ThermoFisher) operated at 300 kV at the MICEF (NIH). For EncA without cargo micrographs were recorded as dose-fractionated movies with a Gatan K2 Summit direct electron detector operated in counting mode at a nominal magnification of x130,000 (calibrated pixel size of 1.06 Å). The total dose for each exposure was 73 e−/Å2 where the total exposure time was 10 s fractionated into 40 frames with 0.25 s exposure time for each frame. The nominal defocus range used was −0.6 to −2.6 μm. For EncA-EncB and EncA-EncC micrographs were recorded as dose-fractionated movies with a Gatan K2 Summit direct electron detector operated in super-resolution mode at a nominal magnification of x130,000 (calibrated pixel size of 0.537 Å). The total dose for each exposure was 46 e−/Å2 where the total exposure was 6 s fractionated into 30 frames with 0.2 s exposure time for each frame. For EncA, EncA-EncB and EncA-EncC samples 2403, 7843 and 7715 movies were acquired, respectively, using SerialEM (Mastronarde, 2005).
Image processing
Super-resolution movies were binned over 1.074 × 1.074 pixels. Movie alignment and CTF estimations were carried out using cisTEM (Grant et al., 2018). A total of 20706 particles were picked from EncA micrographs using automatic picking feature. After 2D classification 9309 particles were selected for T=3 EncA and 5183 particles were selected for T=1 EncA to generate ab-initio 3D models and for auto refinement using cisTEM. A final round of manual refinement was carried out for each structure with a soft mask. The resolution calculation was based on the gold standard Fourier Shell Correlation (FSC) at the 0.143 criterion. From EncA-EncB micrographs initially 8139 particles were picked manually for EncA-EncB and 7961 particles were selected after 2D classification for ab-initio model building and subsequent auto and manual refinement steps. From EncA-EncC micrographs 10357 particles were picked manually for EncA-EnC and 8512 particles were selected after 2D classification for ab-initio model building and subsequent auto and manual refinement steps. Cryo-EM data collection, refinement, and validation statistics are given in Table 1. MolProbity was used for structure validation (Williams et al., 2018a).
Determination of cargo location
EncA-EncC micrographs were re-processed using RELION-3.1 (Scheres, 2012). Movie alignment was carried out by MotionCor2 (Zheng et al., 2017). A total of 20313 particles were picked using the auto-pick function. After 2D classification 13454 particles were picked for further processing. Initially a 3D classification and an initial model building followed by 3D auto-refinement was performed. Then the shell density was subtracted from the original particle images and a focused 3D classification without image alignment was done. Only one class with 3165 particles showed clear densities that had similar size and shape with the cargo. Particles from this class were selected and refined to obtain the final structure. In order to perform the particle subtraction and focused classification, a 60 Å low pass filtered full mask of the encapsulin shell and cargo was created with soft edges using the RELION mask creation tool. To generate the empty shell mask the volume eraser tool of UCSF Chimera was used to remove the inner density belonging to the cargo proteins (Pettersen et al., 2004). Then the empty shell mask was subtracted from the full mask to obtain a mask of only the cargo proteins without the shell.
Cryo-EM model building and refinement
Model building into the cryo-EM maps was initiated by fitting the previously solved cryo-EM structure of EncA (PDB ID: 4PT2) into the EncA (T=3 and T=1), EncA-EncB and EncA-EncC maps using UCSF Chimera and real-space refinement in Phenix (Adams et al., 2010). The resulting structures was rebuilt in Coot (Emsley et al., 2010) followed by multiple cycles of real-space refinement in Phenix. The TP models were built manually into the extra densities observed in EncA-EncB and EncA-EncC maps that were not observed in EncA map and subjected to multiple rounds of refinement in Phenix.
Crystallization of cargo proteins
EncB and EncC proteins were concentrated to 15 mg/ml Amicon Ultra-15 centrifugal filter concentrator with 10 kDa cutoff. Crystals were obtained using the sitting drop vapor diffusion technique by mixing 1 μl of protein with 1 μl of mother liquor. Both EncB and EncC crystals appeared in 20% PEG 550 MME, 10% PEG 20000, 100 mM Tris-Bicine pH 8.5, 30 mM CaCl2 and 5 mM FeCl2 in 1 week. Crystals were protected with 30% MPD before flash-freezing in liquid nitrogen. All data collections were carried out at the Advanced Photon Source (APS, Lemont, IL) beamline 22-ID-D using the high-speed Eiger X 16M detector. Data were collected from multiple crystals in each case and the final structures were obtained using data from best diffracting crystals.
Model building and refinement for crystals
Data processing and integration was done with XDS and scaling was done with XSCALE (Kabsch, 2010). Molecular replacement was carried out with Phaser (McCoy et al., 2007) using PDB 5N5E as a search model for both structures. Final structures were obtained after multiple rounds of model building with Coot and refinement with Phenix Refine. For refinement, XYZ coordinates, real-space, individual B-factors, occupancies and NCS strategies were applied. We solved the crystal structures of EncB decamers in space groups P21 and C2 at 1.86 Å and 1.95 Å resolution, respectively. Both structures showed similar features, so we refer to the P21 structure due to its higher resolution for the remainder of the text. In these crystal structures EncB amino acids 14–80 were visible All the screened EncC crystals showed pseudo merohedral twinning. The EncC crystals were arranged in C2 space group mimicking C222 symmetry and the resolution was 2.5 Å. The crystal data was refined with the twin law -h, -k, l for the detwinning of the data. The crystal structure showed densities for amino acids 1–79. The crystallographic data and refinement statistics are listed in Table 2.
Model visualization and analysis
The structures were visualized using either UCSF Chimera or UCSF ChimeraX (Goddard et al., 2018). Protein interfaces were analyzed using PDBePISA (Krissinel and Henrick, 2007). The theoretical binding energies were calculated using PDBePISA.
QUANTIFICATION and STATISTICAL ANALYSIS
Statistical data in Table 1 for the cryo-EM structures and in Table 2 for the X-ray crystal structures calculated were obtained from the outputs of Phenix, MolProbity and the wwwPDB oneDep validation server (https://validate-rcsb-2.wwpdb.org).
Supplementary Material
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| T7 express crystal competent E. coli | New England Biolabs | Cat# C3022 |
| Chemicals, peptides, and recombinant proteins | ||
| LB broth base | Invitrogen | Cat# 12780-052 |
| Ampicillin | Fisher Scientific | Cat# BP1760-25 |
| Isopropyl-beta-D-thiogalactopyranoside | MP Biomedicals | Cat# 102101 |
| Tris base | Fisher Scientific | Cat# BP152-1 |
| Sodium Chloride | KD Medical | Cat# RGF-3270 |
| cOmplete EDTA-free protease inhibitor cocktail | Roche | Cat# 11873580001 |
| Triton-X100 | MP Biomedicals | Cat# 194854 |
| Tween-20 | Sigma-Aldrich | Cat# P-1379 |
| Lysozyme | Chemical Dynamics Corporation | Cat# 56-3660-00 |
| HisPur Ni-NTA resin | ThermoFisher | Cat# 88221 |
| Imidazole | Sigma-Aldrich | Cat# I3386-500G |
| HEPES | Acros Organics | Cat# 215001000 |
| 4–20% SDS-PAGE gels | Invitrogen | Cat# XV04200PK20 |
| O2-Ar gas (25–75%) | Roberts Oxygen Company | R0120508012 |
| Ethane gas | Matheson Tri Gas | 5003838 |
| PEG 550 MME | Hampton Research | HR2-611 |
| PEG 20000 | Hampton Research | HR2–609 |
| Bicine | Fluka | Cat# 14871 |
| MPD | Hampton Research | HR2–627 |
| Calcium chloride | Fisher Scientific | Cat# C-79 |
| Iron chloride | Sigma-Aldrich | Cat# 372870 |
| Deposited data | ||
| EncA (T=1) cryo-EM map | This paper | EMD-24815 |
| EncA (T=3) cryo-EM map | This paper | EMD-24814 |
| EncA-EncB cryo-EM map | This paper | EMD-24816 |
| EncA-EncC cryo-EM map | This paper | EMD-24832 |
| EncA (T=1) structure | This paper | PDB: 7S21 |
| EncA (T=3) structure | This paper | PDB: 7S20 |
| EncA-EncB structure | This paper | PDB: 7S2T |
| EncA-EncC structure | This paper | PDB: 7S4Q |
| EncB crystal structure (space group P21) | This paper | PDB: 7S5C |
| EncB crystal structure (space group C2) | This paper | PDB: 7S5K |
| EncC crystal structure (space group C2) | This paper | PDB: 7S8T |
| M. xanthus encapsulin protein EncA | McHugh et al., 2014 | PDB: 4PT2, EMD-5917 |
| T. maritima FLP | Sutter et al., 2008 | PDB: 3DKT |
| Q. thermotolerans IMEF | Giessen et al., 2019 | PDB: 6NJ8, EMD-9383 |
| M. smegmatis encapsulin | Tang et al., 2021 | PDB: 7BOJ, EMD-30130 |
| T. maritima encapsulin | Xiong et al., 2020 | PDB: 7K5W, EMD-22617 |
| M. hassiacum encapsulin | Loncar et al., 2020 | PDB: 6I9G |
| S. elongatus encapsulin | Nichols et al., 2021 | PDB: 6X8M, EMD-22094 |
| Rru_A0973 | He et al., 2016 | PDB: 5DA5 |
| PFC_05175 | He et al., 2019 | PDB: 5N5E |
| Hoch_3836 | He et al., 2019 | PDB: 5N5F |
| Recombinant DNA | ||
| EncA-pETDuet-1 | This paper | N/A |
| EncA-EncB-pETDuet-1 | This paper | N/A |
| EncA-EncC-pETDuet-1 | This paper | N/A |
| Software and algorithms | ||
| SerialEM | Mastronarde, 2005 | https://bio3d.colorado.edu/SerialEM/ |
| cisTEM | Grant et al., 2018 | https://cistem.org |
| MolProbity | Williams et al., 2018a | http://molprobity.biochem.duke.edu |
| RELION-3.1 | Scheres, 2012 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/ucsf-software |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| Phenix | Adams et al., 2010 | https://phenix-online.org |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| XDS | Kabsch, 2010 | https://xds.mr.mpg.de |
| XSCALE | Kabsch, 2010 | https://xds.mr.mpg.de/html_doc/xscale_program.html |
| Phaser | McCoy et al., 2007 | https://www-structmed.cimr.cam.ac.uk/phaser_obsolete/ |
| UCSF ChimeraX | Goddard et al., 2018 | https://www.cgl.ucsf.edu/chimerax/ |
| PDBePISA | Krissinel and Henrick, 2007 | https://www.ebi.ac.uk/pdbe/pisa/ |
| Other | ||
| Beckman Coulter Avanti J30I refrigerated centrifuge | Beckman Coulter | https://www.beckman.com/centrifuges |
| Leica EM GP Plunger | Leica Microsystems | https://www.leica-microsystems.com |
| Plasma Cleaner Model 1020 | Fischione | https://www.fischione.com |
| Quantifoil R1.2/1.3 300 mesh Cu grids coated with 2 nm ultrathin carbon | Electron Microscopy Sciences, Protochips, Inc. | Cat# Q310CR1.3–2nm |
HIGHLIGHTS.
M. xanthus encapsulin EncA T=1 and T=3 structures are determined at 3.4 Å resolution EncB and EncC cargo protein binding sites on EncA are identified
EncB and EncC structures are determined at 1.86 and 2.49 Å resolution, respectively Multiple iron binding sites on cargo proteins are characterized
Acknowledgments
This work utilized the NIH Multi-Institute Cryo-EM Facility (MICEF) and the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). The authors thank Huaibin Wang for technical support on the NIH MICEF Titan Krios Electron Microscope. This research was supported by the Intramural Research Program of the NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Footnotes
Declaration of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- ADAMS PD, AFONINE PV, BUNKOCZI G, CHEN VB, DAVIS IW, ECHOLS N, HEADD JJ, HUNG LW, KAPRAL GJ, GROSSE-KUNSTLEVE RW, MCCOY AJ, MORIARTY NW, OEFFNER R, READ RJ, RICHARDSON DC, RICHARDSON JS, TERWILLIGER TC & ZWART PH 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr, 66, 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AHMAD M, ROBERTS JN, HARDIMAN EM, SINGH R, ELTIS LD & BUGG TD 2011. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry, 50, 5096–107. [DOI] [PubMed] [Google Scholar]
- AKITA F, CHONG KT, TANAKA H, YAMASHITA E, MIYAZAKI N, NAKAISHI Y, SUZUKI M, NAMBA K, ONO Y, TSUKIHARA T & NAKAGAWA A 2007. The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J Mol Biol, 368, 1469–83. [DOI] [PubMed] [Google Scholar]
- ALMEIDA AV, CARVALHO AJ & PEREIRA AS 2021. Encapsulin nanocages: Protein encapsulation and iron sequestration. Coordination Chemistry Reviews, 448, 214188. [Google Scholar]
- ANDREAS MP & GIESSEN TW 2021. Large-scale computational discovery and analysis of virus-derived microbial nanocompartments. Nat Commun, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAMFORD DH, GRIMES JM & STUART DI 2005. What does structure tell us about virus evolution? Curr Opin Struct Biol, 15, 655–63. [DOI] [PubMed] [Google Scholar]
- CHOI B, MOON H, HONG SJ, SHIN C, DO Y, RYU S & KANG S 2016. Effective Delivery of Antigen-Encapsulin Nanoparticle Fusions to Dendritic Cells Leads to Antigen-Specific Cytotoxic T Cell Activation and Tumor Rejection. ACS Nano, 10, 7339–50. [DOI] [PubMed] [Google Scholar]
- CHOI H, EOM S, KIM HU, BAE Y, JUNG HS & KANG S 2021. Load and Display: Engineering Encapsulin as a Modular Nanoplatform for Protein-Cargo Encapsulation and Protein-Ligand Decoration Using Split Intein and SpyTag/SpyCatcher. Biomacromolecules, 22, 3028–3039. [DOI] [PubMed] [Google Scholar]
- CONTRERAS H, JOENS MS, MCMATH LM, LE VP, TULLIUS MV, KIMMEY JM, BIONGHI N, HORWITZ MA, FITZPATRICK JA & GOULDING CW 2014. Characterization of a Mycobacterium tuberculosis nanocompartment and its potential cargo proteins. J Biol Chem, 289, 18279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIAZ D, VIDAL X, SUNNA A & CARE A 2021. Bioengineering a Light-Responsive Encapsulin Nanoreactor: A Potential Tool for In Vitro Photodynamic Therapy. ACS Appl Mater Interfaces, 13, 7977–7986. [DOI] [PubMed] [Google Scholar]
- EBRAHIMI KH, BILL E, HAGEDOORN PL & HAGEN WR 2016. Spectroscopic evidence for the role of a site of the di-iron catalytic center of ferritins in tuning the kinetics of Fe(ii) oxidation. Mol Biosyst, 12, 3576–3588. [DOI] [PubMed] [Google Scholar]
- EBRAHIMI KH, HAGEDOORN PL, HAGEN RW 2015. Unity in the Biochemistry of the Iron-Storage Proteins Ferritin and Bacterioferritin. Chem Rev, 115, 295–326. [DOI] [PubMed] [Google Scholar]
- EFREMOVA MV, BODEA SV, SIGMUND F, SEMKINA A, WESTMEYER GG & ABAKUMOV MA 2021. Genetically Encoded Self-Assembling Iron Oxide Nanoparticles as a Possible Platform for Cancer-Cell Tracking. Pharmaceutics, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMSLEY P, LOHKAMP B, SCOTT WG & COWTAN K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOKINE A, LEIMAN PG, SHNEIDER MM, AHVAZI B, BOESHANS KM, STEVEN AC, BLACK LW, MESYANZHINOV VV & ROSSMANN MG 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A, 102, 7163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GABASHVILI AN, CHMELYUK NS, EFREMOVA MV, MALINOVSKAYA JA, SEMKINA AS & ABAKUMOV MA 2020. Encapsulins-Bacterial Protein Nanocompartments: Structure, Properties, and Application. Biomolecules, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIESSEN TW 2016. Encapsulins: microbial nanocompartments with applications in biomedicine, nanobiotechnology and materials science. Curr Opin Chem Biol, 34, 1–10. [DOI] [PubMed] [Google Scholar]
- GIESSEN TW, ORLANDO BJ, VERDEGAAL AA, CHAMBERS MG, GARDENER J, BELL DC, BIRRANE G, LIAO M & SILVER PA 2019. Large protein organelles form a new iron sequestration system with high storage capacity. Elife, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIESSEN TW & SILVER PA 2017. Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat Microbiol, 2, 17029. [DOI] [PubMed] [Google Scholar]
- GODDARD TD, HUANG CC, MENG EC, PETTERSEN EF, COUCH GS, MORRIS JH & FERRIN TE 2018. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci, 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRANT T, ROHOU A & GRIGORIEFF N 2018. cisTEM, user-friendly software for single-particle image processing. Elife, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALL DR, HADDEN JM, LEONARD GA, BAILEY S, NEU M, WINN M & LINDLEY PF 2002. The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 A, respectively. Acta Crystallogr D Biol Crystallogr, 58, 70–80. [DOI] [PubMed] [Google Scholar]
- HE D, HUGHES S, VANDEN-HEHIR S, GEORGIEV A, ALTENBACH K, TARRANT E, MACKAY CL, WALDRON KJ, CLARKE DJ & MARLES-WRIGHT J 2016. Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE D, PIERGENTILI C, ROSS J, TARRANT E, TUCK LR, MACKAY CL, MCIVER Z, WALDRON KJ, CLARKE DJ & MARLES-WRIGHT J 2019. Conservation of the structural and functional architecture of encapsulated ferritins in bacteria and archaea. Biochem J, 476, 975–989. [DOI] [PubMed] [Google Scholar]
- HEMPSTEAD PD, HUDSON AJ, ARTYMIUK PJ, ANDREWS SC, BANFIELD MJ, GUEST JR & HARRISON PM 1994. Direct observation of the iron binding sites in a ferritin. FEBS Lett, 350, 258–62. [DOI] [PubMed] [Google Scholar]
- HIEMSTRA T & ZHAO W 2016. Reactivity of ferrihydrite and ferritin in relation to surface structure, size, and nanoparticle formation studied for phosphate and arsenate. Environ. Sci.: Nano, 3, 1265–1279. [Google Scholar]
- HONARMAND EBRAHIMI K, BILL E, HAGEDOORN PL & HAGEN WR 2017. Spectroscopic evidence for the presence of a high-valent Fe(IV) species in the ferroxidase reaction of an archaeal ferritin. FEBS Lett, 591, 1712–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONARMAND EBRAHIMI K, HAGEDOORN PL & HAGEN WR 2015. Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem Rev, 115, 295–326. [DOI] [PubMed] [Google Scholar]
- JONES JA & GIESSEN TW 2021. Advances in encapsulin nanocompartment biology and engineering. Biotechnol Bioeng, 118, 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KABSCH W 2010. Xds. Acta Crystallogr D Biol Crystallogr, 66, 125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARTAL B, DE ALMEIDA NM, MAALCKE WJ, OP DEN CAMP HJ, JETTEN MS & KELTJENS JT 2013. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev, 37, 428–61. [DOI] [PubMed] [Google Scholar]
- KIM D, CHOI J, LEE S, HYUN H, LEE K & CHO K 2019. Mutants defective in the production of encapsulin show a tan-phase-locked phenotype in Myxococcus xanthus. J Microbiol, 57, 795–802. [DOI] [PubMed] [Google Scholar]
- KRISSINEL E & HENRICK K 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol, 372, 774–97. [DOI] [PubMed] [Google Scholar]
- LAFRANCE B, CASSIDY-AMSTUTZ C, NICHOLS RJ, OLTROGGE LM, NOGALES E & SAVAGE DF 2021. The encapsulin from Thermatoga maritima is a flavoprotein with a symmetry matched ferritin-like cargo protein. bioRxiv, 441214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LONCAR N, ROZEBOOM HJ, FRANKEN LE, STUART MCA & FRAAIJE MW 2020. Structure of a robust bacterial protein cage and its application as a versatile biocatalytic platform through enzyme encapsulation. Biochem Biophys Res Commun, 529, 548–553. [DOI] [PubMed] [Google Scholar]
- MASTRONARDE DN 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol, 152, 36–51. [DOI] [PubMed] [Google Scholar]
- MASUDA T, GOTO F, YOSHIHARA T & MIKAMI B 2010. Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin. J Biol Chem, 285, 4049–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCOY AJ, GROSSE-KUNSTLEVE RW, ADAMS PD, WINN MD, STORONI LC & READ RJ 2007. Phaser crystallographic software. J Appl Crystallogr, 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCHUGH CA, FONTANA J, NEMECEK D, CHENG N, AKSYUK AA, HEYMANN JB, WINKLER DC, LAM AS, WALL JS, STEVEN AC & HOICZYK E 2014. A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J, 33, 1896–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NICHOLS RJ, CASSIDY-AMSTUTZ C, CHAIJARASPHONG T & SAVAGE DF 2017. Encapsulins: molecular biology of the shell. Crit Rev Biochem Mol Biol, 52, 583–594. [DOI] [PubMed] [Google Scholar]
- NICHOLS RJ, LAFRANCE B, PHILLIPS NR, RADFORD DR, OLTROGGE LM, VALENTIN-ALVARADO LE, BISCHOFF AJ, NOGALES E & SAVAGE DF 2021. Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism. Elife, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORDLUND P & EKLUND H 1995. Di-iron-carboxylate proteins. Curr Opin Struct Biol, 5, 758–66. [DOI] [PubMed] [Google Scholar]
- PETTERSEN EF, GODDARD TD, HUANG CC, COUCH GS, GREENBLATT DM, MENG EC & FERRIN TE 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem, 25, 1605–12. [DOI] [PubMed] [Google Scholar]
- PFAFFEN S, ABDULQADIR R, LE BRUN NE & MURPHY ME 2013. Mechanism of ferrous iron binding and oxidation by ferritin from a pennate diatom. J Biol Chem, 288, 14917–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PIERGENTILI C, ROSS J, HE D, GALLAGHER KJ, STANLEY WA, ADAM L, MACKAY CL, BASLÉ A, WALDRON KJ, CLARKE DJ & MARLES-WRIGHT J 2020. Dissecting the structural and functional roles of a putative metal entry site in encapsulated ferritins. J Biol Chem, 295, 15511–15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PIETILA MK, LAURINMAKI P, RUSSELL DA, KO CC, JACOBS-SERA D, HENDRIX RW, BAMFORD DH & BUTCHER SJ 2013. Structure of the archaeal head-tailed virus HSTV-1 completes the HK97 fold story. Proc Natl Acad Sci U S A, 110, 10604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAHMANPOUR R & BUGG TD 2013. Assembly in vitro of Rhodococcus jostii RHA1 encapsulin and peroxidase DypB to form a nanocompartment. FEBS J, 280, 2097–104. [DOI] [PubMed] [Google Scholar]
- ROSS J, MCIVER Z, LAMBERT T, PIERGENTILI C, GALLAGHER KJ, BIRD JE, CRUICKSHANK FL, ZARAZÚA-ARVIZU E, HORSFALL LE, WALDRON KJ, WILSON MD, MACKAY CL, BASLÉ A, CLARKE DJ & MARLES-WRIGHT J 2021. Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment. bioRxiv, 439977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHERES SH 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol, 180, 519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIGMUND F, MASSNER C, ERDMANN P, STELZL A, ROLBIESKI H, DESAI M, BRICAULT S, WORNER TP, SNIJDER J, GEERLOF A, FUCHS H, HRABE DE ANGELIS M, HECK AJR, JASANOFF A, NTZIACHRISTOS V, PLITZKO J & WESTMEYER GG 2018. Bacterial encapsulins as orthogonal compartments for mammalian cell engineering. Nat Commun, 9, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STILLMAN TJ, HEMPSTEAD PD, ARTYMIUK PJ, ANDREWS SC, HUDSON AJ, TREFFRY A, GUEST JR & HARRISON PM 2001. The high-resolution X-ray crystallographic structure of the ferritin (EcFtnA) of Escherichia coli; comparison with human H ferritin (HuHF) and the structures of the Fe(3+) and Zn(2+) derivatives. J Mol Biol, 307, 587–603. [DOI] [PubMed] [Google Scholar]
- SUGANO Y, MURAMATSU R, ICHIYANAGI A, SATO T & SHODA M 2007. DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family: ASP171 replaces the distal histidine of classical peroxidases. J Biol Chem, 282, 36652–8. [DOI] [PubMed] [Google Scholar]
- SUTTER M, BOEHRINGER D, GUTMANN S, GUNTHER S, PRANGISHVILI D, LOESSNER MJ, STETTER KO, WEBER-BAN E & BAN N 2008. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol, 15, 939–47. [DOI] [PubMed] [Google Scholar]
- TANG Y, MU A, ZHANG Y, ZHOU S, WANG W, LAI Y, ZHOU X, LIU F, YANG X, GONG H, WANG Q & RAO Z 2021. Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin. Proc Natl Acad Sci U S A, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TATUR J, HAGEN WR & MATIAS PM 2007. Crystal structure of the ferritin from the hyperthermophilic archaeal anaerobe Pyrococcus furiosus. J Biol Inorg Chem, 12, 615–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THEIL EC 2013. Ferritin: the protein nanocage and iron biomineral in health and in disease. Inorg Chem, 52, 12223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOSHA T, BEHERA RK, NG HL, BHATTASALI O, ALBER T & THEIL EC 2012. Ferritin protein nanocage ion channels: gating by N-terminal extensions. J Biol Chem, 287, 13016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIKOFF WR, LILJAS L, DUDA RL, TSURUTA H, HENDRIX RW & JOHNSON JE 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science, 289, 2129–33. [DOI] [PubMed] [Google Scholar]
- WILLIAMS CJ, HEADD JJ, MORIARTY NW, PRISANT MG, VIDEAU LL, DEIS LN, VERMA V, KEEDY DA, HINTZE BJ, CHEN VB, JAIN S, LEWIS SM, ARENDALL WB 3RD, SNOEYINK J, ADAMS PD, LOVELL SC, RICHARDSON JS & RICHARDSON DC 2018a. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci, 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS EM, JUNG SM, COFFMAN JL & LUTZ S 2018b. Pore Engineering for Enhanced Mass Transport in Encapsulin Nanocompartments. ACS Synth Biol, 7, 2514–2517. [DOI] [PubMed] [Google Scholar]
- WIRYAMAN T & TOOR N 2021. Cryo-EM structure of a thermostable bacterial nanocompartment. IUCrJ, 8, 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIONG X, SUN C, VAGO FS, KLOSE T, ZHU J & JIANG W 2020. Cryo-EM Structure of Heterologous Protein Complex Loaded Thermotoga Maritima Encapsulin Capsid. Biomolecules, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHENG SQ, PALOVCAK E, ARMACHE JP, VERBA KA, CHENG Y & AGARD DA 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods, 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Maps and models have been deposited in the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (accession numbers: EncA (T=1) EMD-24815, EncA (T=3) EMD-24814, EncA-EncB EMD-24816 and EncA-EncC EMD-24832). Models and crystal structures have been deposited in the Protein Data Bank, https://www.ebi.ac.uk/pdbe/ (PDB ID codes 7S21 EncA (T=1), 7S20 EncA (T=3), 7S2T EncA-EncB, 7S4Q EncA-EncC, 7S5C EncB-P21, 7S5K EncB-C2, 7S8T EncC).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


