Abstract
Objectives
To assess the performance of statistical methods used to compare the effectiveness between drugs in an observational setting in the presence of attrition.
Methods
In this simulation study, we compared the estimations of low disease activity (LDA) at 1 year produced by complete case analysis (CC), last observation carried forward (LOCF), LUNDEX, non-responder imputation (NRI), inverse probability weighting (IPW) and multiple imputations of the outcome. All methods were adjusted for confounders. The reasons to stop the treatments were included in the multiple imputation method (confounder-adjusted response rate with attrition correction, CARRAC) and were either included (IPW2) or not (IPW1) in the IPW method. A realistic simulation data set was generated from a real-world data collection. The amount of missing data caused by attrition and its dependence on the ‘true’ value of the data missing were varied to assess the robustness of each method to these changes.
Results
LUNDEX and NRI strongly underestimated the absolute LDA difference between two treatments, and their estimates were highly sensitive to the amount of attrition. IPW1 and CC overestimated the absolute LDA difference between the two treatments and the overestimation increased with increasing attrition or when missingness depended on disease activity at 1 year. IPW2 and CARRAC produced unbiased estimations, but IPW2 had a greater sensitivity to the missing pattern of data and the amount of attrition than CARRAC.
Conclusions
Only multiple imputation and IPW2, which considered both confounding and treatment cessation reasons, produced accurate comparative effectiveness estimates.
Keywords: arthritis, rheumatoid, epidemiology, outcome assessment, health care
Key messages.
What is already known about this subject?
Attrition, defined as an increased selection due to participants leaving the study, is a source of bias for comparative effectiveness research, but it is often not acknowledged.
Several imputation methods exist to account for attrition in comparative effectiveness research with observational data, but comparison of these methods is lacking
What does this study add?
This study provides an extensive comparison of the statistical methods used to compare treatment effectiveness in an observational setting in the presence of attrition.
Omitting to consider confounding, treatment cessation or dropouts leads to biased estimation of effectiveness. The present article provides advices, methods and code to report effectiveness properly.
How might this impact on clinical practice or future developments?
This work suggests that comparative effectiveness results in observational data may have large over or underestimation depending on the method used.
Introduction
In rheumatology, as in other specialties, randomised controlled trials are the gold standard when evaluating treatment efficacy. However, because of the highly selected populations, their conclusions are difficult to generalise to routine clinical practice. For this reason among others, evaluation of the effectiveness of treatments in the real-world patient population is needed.1 Comparative effectiveness between treatments when using observational real-world data requires overcoming several difficulties.
In addition to confounding, a recurring difficulty is missing data.2 The data used to define drug effectiveness can be missing at the follow-up time of interest, while patients are still on treatment, and the missing data may have to be imputed to avoid selection bias,3 using proper imputation methods.4 5 But the data of interest can also be missing because of attrition (an increasing selection due to participants leaving the study).6–8 Considering effectiveness among those remaining on therapy after a certain set of time (complete case analysis; CC9) is known to be a source of bias,3 10 because it excludes from the analysis patients who stopped the drug for an adverse event or lack of effect, thus resulting in a selection bias in favour of responders.11 Although ‘intention to treat’ analysis intends to avoid this bias in controlled trials,12 there is no consensus regarding how this should be handled in an observational study.13 Several statistical methods exist allowing to account for both attrition and confounding, such as inverse probability weighting (IPW)7 14 or multiple imputation (MI) of the outcome.15 16 In rheumatic diseases cohort studies, a popular method to account for potential attrition bias is the LUNDEX index.17 Analogously to non-responder imputation (NRI),18 it corrects for the attrition bias by assuming that all patients stopping their treatment are non-responders. It, therefore, multiplies the estimation of effectiveness by the estimate of each drug survival, which may result in underestimating true drug effectiveness.
Depending on how it is handled, attrition may lead to biased conclusions in comparative effectiveness research. Therefore, the characterisation and comparison of existing statistical methods are needed. The reasons to stop the treatment are often available in registers and, to our knowledge, are generally not employed in standard statistical analyses when accounting for attrition.8
The aim of this research is, thus, to perform an extensive simulation study to compare the ability of different statistical approaches to account for missing data caused by attrition, when studying comparative effectiveness using rheumatology observational data.
Methods
Our study simulates a comparative effectiveness study of two treatments in patients with potentially different baseline characteristics and attrition rates. In this simulation, we compare the effectiveness of tumour necrosis factor inhibitors (TNFi) versus a biologic disease-modifying antirheumatic drugs (bDMARD) with another mode of action (OMA) in patients with rheumatoid arthritis (RA) using the clinical disease activity index (CDAI) definition of low disease activity (LDA) at 1-year follow-up as outcome. We adjust for the following characteristics at treatment start (hereafter referred to as baseline covariates): disease duration, concomitant treatment with conventional synthetic DMARD (csDMARD), the number of previous bDMARDs (prev_bDMARD), and CDAI (CDAI0). A collection of registers is used to generate a single simulation dataset with all CDAI values at 1 year. The simulation study consists in generating, in four iterative steps, missing data of CDAI at 1 year caused by attrition to then compare how different statistical methods estimate effectiveness. The sensitivity of the results is studied by changing the amount and the pattern of the missing data.
Creation of the simulation dataset
An original data set composed of a collaboration of RA registers including TNFi and OMA, including >45 000 treatment courses19 (see table 1) was used to construct a realistic open cohort simulation data set composed of 10 000 treatment courses. The variables of this simulation data set were treatment (TNFi or OMA), CDAI value at treatment start and at 12 months (CDAI0, CDAI12, respectively), all confounders cited above and the treatment status, which indicates the last available status about the patient’ treatment. It is equal to ‘ongoing’ if the patient is still on treatment at the time of data extraction and to the reason for treatment cessation otherwise (see online supplemental material for more details).
Table 1.
Characteristics of the patients under treatment in the initial real-world register collection
|
|
Other mode of actions | TNF inhibitor |
| N | 6067 | 40 767 |
| Disease duration (median (IQR) in year) | 9.8 (4.6, 17.9) | 7.4 (2.9, 14.6) |
| Treatment duration (median (IQR) in year) | 1.2 (0.5, 2.7) | 1.7 (0.6, 4.3) |
| Number of previous bDMARD (number (%)) | ||
| 0 | 1451 (23.9) | 23 016 (56.5) |
| 1 | 2019 (33.3) | 12 269 (30.1) |
| 2 | 1383 (22.8) | 3896 (9.6) |
| 3+ | 1214 (20.0) | 1586 (3.9) |
| Concomitant csDMARD (number (%)) | ||
| MTX | 2982 (49.2) | 20 062 (49.2) |
| MTX +other | 68 (1.1) | 1080 (2.6) |
| None | 1907 (31.4) | 15 047 (36.9) |
| Other | 1110 (18.3) | 4578 (11.2) |
| CDAI0 (median (IQR)) | 23.4 (16.5, 32.0) | 23.0 (13.9, 33.5) |
| CDAI12 (median (IQR)) | 10.0 (5.0, 17.3) | 7.0 (2.9, 14.0) |
| Treatment status (number (%)) | ||
| Ongoing | 3336 (55.0) | 24 968 (61.2) |
| Stopped for adverse event | 459 (7.6) | 1876 (4.6) |
| Stopped for ineffectiveness | 748 (12.3) | 3369 (8.3) |
| Stopped for pregnancy | 5 (0.1) | 53 (0.1) |
| Stopped for remission | 62 (1.0) | 189 (0.5) |
| Stopped for other | 313 (5.2) | 1089 (2.7) |
| Stopped for unspecified reason | 1144 (18.9) | 9223 (22.6) |
| Missing CDAI12 (number (%)) | 1680 (27.7) | 9415 (23.1) |
| Number (%) of patients stopping after 1 year (number (%)) | 1017 (16.8) | 6081 (14.9) |
| LDA at 12 month (number (%)) | 1811 (51.4) | 15 873 (63.7) |
Number of observations (N), number and proportion of patients having 0, 1, 2, 3 and more (3+) previous biological DMARD, of patient having methotrexate alone (MTX), methotrexate with other csDMARD (MTX +other), at least an csDMARD other than MTX (other) or no concomitant synthetic DMARD treatment (none), median (IQR) value of baseline CDAI (CDAI0) and CDAI at 12 months (CDAI12).
CDAI, Clinical Disease Activity Index; csDMARD, conventional synthetic disease-modifying antirheumatic drugs; TNF, tumour necrosis factor.
annrheumdis-2021-221477supp001.pdf (964KB, pdf)
The simulation data set was constructed to have the same proportion of treatments (OMA and TNFi) as the whole data collection. For each treatment group, CDAI0, disease duration, prev_bDMARD, csDMARD and the treatment status were independently randomly drawn from the entire collection. The treatment duration values in the simulation data set were then randomly sampled while matching exactly the treatment, the treatment status, and prev_bDMARD. CDAI12 was then randomly sampled from the entire data set while matching exactly the treatment, the treatment status, prev_bDMARD, and the categories of CDAI0 (see online supplemental figure 1 for a graphical representation of the data generation). By doing so, we recreated the association between CDAI12 and the reason to discontinue using the reasons to discontinue that happened between 12 and 36 months in the original register collection. Our simulation, thus, imposed this same association between CDAI12 and discontinuations before 12 months. In the resulting data set for the 12-month follow-up, more than 55% of the treatment status are ‘ongoing’ (table 2).
Table 2.
Characteristics of the patients in the simulated dataset
|
|
Other mode of action | TNF inhibitor |
| N | 1295 | 8705 |
| Disease duration (median (IQR) in year) | 9.7 (4.5, 17.8) | 7.3 (2.9, 14.3) |
| Treatment duration (median (IQR) in year) | 1.2 (0.5, 2.7) | 1.6 (0.5, 4.2) |
| Number of previous bDMARD (number (%)) | ||
| 0 | 326 (25.2) | 4973 (57.1) |
| 1 | 409 (31.6) | 2618 (30.1) |
| 2 | 294 (22.7) | 775 (8.9) |
| 3+ | 266 (20.5) | 339 (3.9) |
| Concomitant csDMARD (number (%)) | ||
| MTX | 611 (47.2) | 4330 (49.7) |
| MTX +other | 18 (1.4) | 248 (2.8) |
| None | 427 (33.0) | 3143 (36.1) |
| Other | 239 (18.5) | 984 (11.3) |
| CDAI0 (median (IQR)) | 23.2 (16.4, 32.4) | 23.0 (14.0, 33.3) |
| CDAI12 (median (IQR)) | 10.2 (5.3, 17.0) | 8.0 (3.4, 15.1) |
| treatment status (number (%)) | ||
| Ongoing | 740 (57.1) | 5355 (61.5) |
| Stopped for adverse event | 95 (7.3) | 407 (4.7) |
| Stopped for ineffectiveness | 159 (12.3) | 755 (8.7) |
| Stopped for pregnancy | 1 (0.1) | 11 (0.1) |
| Stopped for remission | 17 (1.3) | 41 (0.5) |
| Stopped for other | 57 (4.4) | 215 (2.5) |
| Stopped for unspecified reason | 226 (17.5) | 1921 (22.1) |
| LDA | 639 (49.3) | 5210 (59.9) |
Number of observations (N), number and proportion of patients having 0, 1, 2, 3 and more (3+) previous biological DMARD, of patient having methotrexate alone (MTX), methotrexate with other csDMARD (MTX +other), at least an csDMARD other than MTX (other) or no concomitant synthetic DMARD treatment (none), median (IQR) value of age, body mass index (BMI), baseline CDAI (CDAI0) and CDAI at 12 months (CDAI12).
CDAI, Clinical Disease Activity Index; csDMARD, conventional synthetic disease-modifying antirheumatic drugs; LDA, low disease activity; TNF, tumour necrosis factor.
Generation of missing data
Data can be missing completely at random when the probability of missing is independent of observed and unobserved data, missing at random (MAR) when the probability of missing depends on observed data, and missing not at random (MNAR) when the probability of missing depends on unobserved data. The patients whose CDAI values at 12 months were set to missing in the simulated data set were chosen by sampling from the patients having a treatment status different from ‘ongoing’ with the conditional probability of having a missing CDAI12 value in the original real data. These probabilities were extracted from a generalised linear model applied on the initial ‘real-world’ register collection estimating the probability of having a missing outcome as a function of the treatment, prev_bDMARD, csDMARD, CDAI0, CDAI12 and the treatment status. Before applying this generalised linear model, all predictors were imputed using MI by chained equation (mice) with 40 iteration, 40 samples and predictive mean matching.
Different ways of deleting data were applied, leading to different types of missing patterns for the different estimation methods considered (see table 3):
Table 3.
Missingness conditions for the simulation, 1 being the reference condition
| Condition | Attrition for OMA (%) | Attrition for TNFi (%) | CDAI12 OR for OMA | CDAI12 OR for TNFi | Missingness for CC, LOCF, LUNDEX, NRI, IPW1 | Missingness for IPW2 and CARRAC |
| 1 | 20 | 20 | 1 | 1 | MNAR | MAR |
| 2 | 10 | 20 | 1 | 1 | MNAR | MAR |
| 3 | 15 | 20 | 1 | 1 | MNAR | MAR |
| 4 | 25 | 20 | 1 | 1 | MNAR | MAR |
| 5 | 30 | 20 | 1 | 1 | MNAR | MAR |
| 6 | 10 | 10 | 1 | 1 | MNAR | MAR |
| 7 | 15 | 15 | 1 | 1 | MNAR | MAR |
| 8 | 25 | 25 | 1 | 1 | MNAR | MAR |
| 9 | 30 | 25 | 1 | 1 | MNAR | MAR |
| 10 | 20 | 20 | 1.07 | 1 | MNAR | MNAR for OMA, MAR for TNFi |
| 11 | 20 | 20 | 1.14 | 1 | MNAR | MNAR for OMA, MAR for TNFi |
| 12 | 20 | 20 | 1.07 | 1.07 | MNAR | MNAR |
| 13 | 20 | 20 | 1.14 | 1.14 | MNAR | MNAR |
Attrition for OMA of TNFi indicates the percentage of missing CDAI at 12 months for the other mode of action treatment (OMA) of the reference treatment TNFi, CDAI12 OR are the OR of the CDAI value at 12 months (CDAI12) in the generalised linear model predicting missingness of CDAI12 used to create missing values in the simulation. A CDAI12 OR of 1.07 or 1.14 implies that the odds of having a CDAI12 missing is multiplied by 2 or by 4 for an increase of 10 points of CDAI12. The column ‘Missingness for CC, LOCF, LUNDEX, NRI, IPW1’ indicates if the missing data are MNAR or MAR for the methods CC, LOCF, LUNDEX, NRI, IPW not accounting for the reasons for treatment cessation in the attrition weights (IPW1). The column ‘Missingness for IPW2 and CARRAC’ does the same for IPW accounting for the reasons for treatment cessation in the attrition weights (IPW2) and CARRAC.
CARRAC, confounder-adjusted response rate with attrition correction; CC, complete case analysis; CDAI, Clinical Disease Activity Index; IPW, inverse probability weighting; LDA, low disease activity; LOCF, last observation carried forward; MAR, missing at random; MNAR, missing not at random; NRI, non-responder imputation; OMA, another mode of action; TNF, tumor necrosis factor.
A reference scenario (missingness condition 1) with 30% treatment cessation in both treatments and no association between effectiveness at 1 year and treatment cessation.
To test the dependence of the observed comparative effectiveness on the amount of attrition, the proportion of missing CDAI12 was set to 10%, 15%, 20%, 25% and 30% in OMA only, and then in both treatments.
To assess the sensitivity of the estimators to MNAR data, we changed the association between CDAI12 values and missingness of CDAI12 caused by attrition by modifying the OR for CDAI12 yielded by the generalised linear model predicting the missing values of CDAI12 (ie, the effect of CDAI12 values on the odds of having a missing CDAI12). It was set to 1 for the reference treatment, and to 1.07 or 1.14 (probability of having a CDAI12 missing multiplied by 2 or 4 for an increase in 10 points of CDAI12) for OMA only, and then in both treatments.
For treatment courses with CDAI12 set to missing, the treatment durations were imputed with plausible values (treatment duration between 0 and 12 months) using MI with predictive mean matching, including CDAI0, disease duration and treatment as covariates.
Simulation
For each condition, the simulation consisted in generating 1000 samples with missing data caused by treatment cessation and estimating the difference in LDA proportion between the two treatments with different statistical methods.
We report bias as the difference between the value estimated in the simulation sample and the true value. The true value is defined here as the LDA rate given by the treatment effect of a linear model predicting LDA on the complete simulation data set (ie, before inducing missing values) adjusting for baseline covariates. In addition, we estimate coverage as the percentage of CIs in the simulation samples which included the true value.
All code to generate simulation data, estimate measures of LDA, and generate the manuscript figures and tables is available in a repository.20 All the simulation, statistical analysis and figures were made in R V.4.1.0,21 using the library ipw 22 for IPW, mice 23 for MI with chained equation, geepack 24 for the generalised estimating equations.
Methods to estimate the LDA
The dataset analysed contained one line per patient’s treatment course. All methods used a generalised linear model with Huber-White robust standard errors predicting a binary outcome as a function of the baseline covariates (referred hereafter as the adjusted model).25–27 In line with Cheung and co-authors26 27, we use a Gaussian identity link, and the coefficient for treatment provides the increase of LDA rate compared with the reference treatment. The following estimation methods were considered.
Complete case analysis
CC consists of restricting the analysis only to available data. We considered here the adjusted estimation of the LDA difference between treatments.
Last observation carried forward
All the missing CDAI12 values were set to the last available value of CDAI, which could be the baseline value. The LDA rate is then estimated with the adjusted model.
LUNDEX
LDA rate (PLDA-LUNDEX) is estimated by the proportion of patients reaching LDA ( ) obtained by the adjusted model multiplied by the Kaplan Meyer estimates of the drug survival at the time of outcome evaluation17 :
| (equation 1) |
CIs were calculated by bootstrap using the quantiles of 1000 samples.
Non-responder imputation
All missing values caused by attrition were set to a non-responder value. The LDA proportion was then estimated using the adjusted model.
Inverse probability weighting 1 and 2
The inverse probability weights for treatment ( ) and for attrition ( ) were computed using a generalised linear model. Weights for treatment ( ) included the baseline covariates. Weights for censoring ( ) included the same baseline covariates in the first version of this method (IPW1), or additionally included the treatment status (IPW2). The LDA proportion was then computed using the adjusted model with weights equal to .
Multiple imputation
Missing disease activity values at 12 months were imputed using MI using chained equations (mice) with the predictive mean matching algorithm, with baseline covariates and the treatment status.
For each of the imputed samples, the LDA difference between treatments was calculated with the adjusted model. The overall estimate and its SE are then pooled from the 10 mice samples using Rubin’s rule.28 The overall method is hereafter named confounder-adjusted response rate with attrition correction (CARRAC).
Results
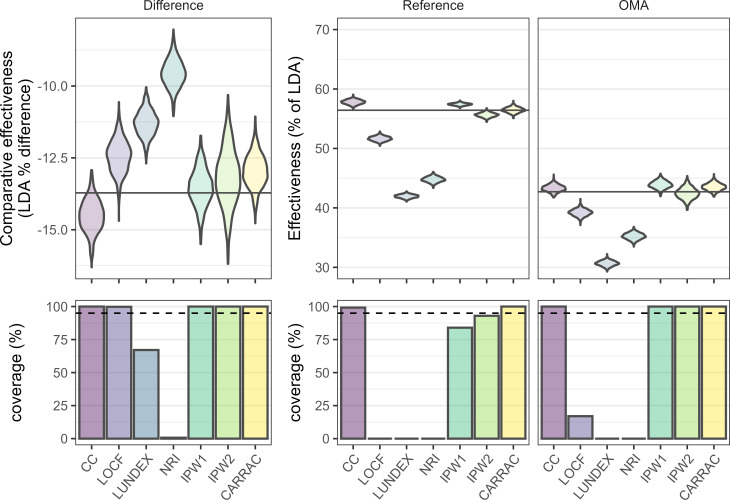
In the reference scenario, CARRAC and IPW2 provided almost unbiased LDA for each treatment (figure 1), thereby estimating almost unbiased LDA difference for these two methods. CC and IPW1 overestimated LDA in both treatments. As their overestimations were similar in OMA and in the reference treatment, the absolute LDA difference was almost unbiased. Last observation carried forward (LOCF), LUNDEX and NRI strongly underestimated LDA in both treatments. Because the underestimation was much lower in OMA, the absolute LDA difference was underestimated by these methods. The coverage was 95% or above in individual treatments only with CARRAC, but due to their smaller bias when considering the LDA difference, the two IPW methods and CC retained good coverage for the comparative effectiveness.
Figure 1.
Distribution of the effectiveness measured by the low disease activity (LDA) proportion (upper panels) and the associated coverage (percentage of CIs in the simulation samples which include the true value, lower panels) for each treatment (reference and another mode of action (OMA)—other modes of action, middle and right panels) and for the difference between the treatments (difference, left panels). The methods analysed are complete case analysis (CC), last observation carried forward (LOCF), LUNDEX, non-responder imputation (NRI), inverse probability weighting accounting for the reasons for treatment cessation in the attrition weights (IPW2) or not (IPW1), and confounder-adjusted response rate with attrition correction by reason for drug cessation (CARRAC). The widths of the violins are fixed, so the area of the violin does not represent the number of counts. The true value is represented as a black horizontal line.
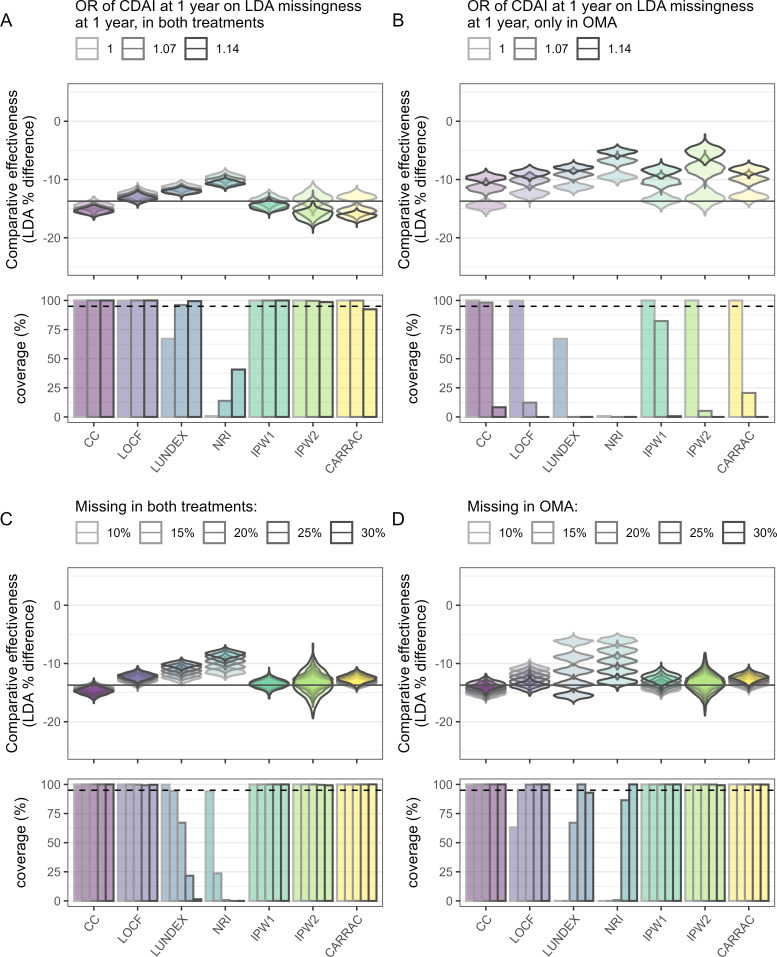
When increasing the association between missingness and the true value of effectiveness at 1 year for both treatments (figure 2A and online supplemental figure 2), LUNDEX, NRI, IPW1, LOCF and CC estimations were almost unaffected, while IPW2 and CARRAC started to overestimate the absolute LDA difference. When the association between missingness of CDAI12 and CDAI12 values existed only in one treatment (figure 2B and online supplemental figure 3), all methods overestimated the difference in LDA, leading to inadequate coverage.
Figure 2.
Low disease activity (LDA) difference between treatments and the associated coverage (percentage of CIs in the simulation samples that include the true value) of the reference situation (condition 1) and when having missing not at random in both treatments (condition 12, A), when having missing not at random in one treatment (conditions 10 and 11, B), when having a changing proportion of missing due to attrition in both treatments conditions (conditions 6–9, C) and when having a changing proportion of missing due to attrition in only one treatment (conditions 2–5, D). The methods analysed are complete case analysis (CC), last observation carried forward (LOCF), LUNDEX, non-responder imputation (NRI), inverse probability weighting accounting for the reasons for treatment cessation in the attrition weights (IPW2) or not (IPW1) and confounder-adjusted response rate with attrition correction by reason for drug cessation (CARRAC). The widths of the violins are fixed, so the area of the violin do not represent the number of counts. The true value is represented as a black horizontal line.
When changing the amount of missingness in both treatments, CARRAC, CC and IPW1 provided unchanged LDA difference estimation when compared with the reference situation (figure 2C and online supplemental figure 4). IPW2 estimations remained approximately unbiased, but their dispersion increased significantly. The bias yielded by NRI and LUNDEX, and to a lesser extent by LOCF, was increased when the amount of missing data increased equally in both treatments.
Increasing the amount of missingness in OMA but not in TNFi decreased the absolute LDA difference between the treatments provided by CC and IPW1, while the LDA proportion difference yielded by LOCF, LUNDEX and NRI increased strongly (figure 2D and online supplemental figure 5). IPW2 stayed unbiased but started to produce more dispersed estimations when the percentage of missingness was 30%. CARRAC estimations of LDA difference slightly underestimated the absolute true difference when increasing the amount of attrition but retained coverage for all missingness conditions.
Discussion
Using a simulation data set generated from a real-world data collection, the present simulation study addresses the issue that some patients may stop the treatment not because it does not work but for some complex reasons including adverse events, remission, etc, which could be taken into account to accurately assess effectiveness. We, therefore, investigated several methods used to compare the response rates of two treatments in presence of confounding and attrition. We focused on attrition, by manipulating the missingness pattern to create greater treatment discontinuation in one treatment versus the other or by increasing the risk of treatment discontinuation. Because observational studies usually use adjusted models to estimate the causal effect of treatments, we used as the true estimate the difference in LDA proportion based on an adjusted model applied on the simulation data set without missing data.
We first observed that the bias in the difference of effectiveness was always lower than the strongest bias observed for the effectiveness estimated in individual treatments. This will always be the case if the estimation of effectiveness is biased in the same direction in both treatments. As expected, methods including a model for attrition and based on MIs and IPW performed well29 30 as long as missingness was dependent only on known covariates (MAR data), but not on unmeasured information (MNAR data). Although an important part of the treatment cessation reasons was unknown, this information was still valuable when estimating effectiveness in the presence of attrition. Including the treatment status in the calculation of the censoring weights permitted IPW to estimate more precisely LDA for each treatment. On the other hand, it led to a higher sensibility to the amount of attrition and to the association between missingness and effectiveness value. This result highlights the importance of model specification for the missingness pattern and the difference between IPW and MI. For instance, the model used for missingness in IPW did not include an interaction term between treatment and variables predicting effectiveness at 1 year (such as baseline disease activity for instance), thus misspecifying the differential effect introduced in the data. MI using predictive mean matching, in the CARRAC method, is less sensitive to misspecification because the model defines the distribution of missing data, which has less variation between the treatments.
CC and IPW1 were biased in each individual treatment, indicating a persistent association between CDAI12 values and their missingness. This association, reflecting the fact that patients remaining on treatment tend to have a better response than those stopping,31 caused the LDA estimated by CC in each treatment to increase with the amount of missing data at 1 year or with the increase in association between CDA12 values and their missingness. IPW1 did so to a smaller extent, as its model partially accounts for the attrition. When the amount of attrition was similar in the two treatments, the biases were similar, and these two methods correctly estimated the difference in LDA. But when the missingness differed between the two treatments, these methods yielded strongly biased estimations of the differential effectiveness.
LOCF, LUNDEX, and NRI underestimated LDA in each treatment. LOCF assumes that the missing values of CDAI are identical to the last available values, although disease activity is known to globally decrease with time.32 33 LUNDEX and NRI considered all patients without information at 1 year as non-responders, although studies show that treatment cessation is not only due to ineffectiveness34 but also due to various other reasons, such as adverse events, pregnancy, or even remission. Therefore, when increasing the proportion of missing values, the LDA proportion yielded by NRI and LUNDEX converge to 0%, which is the LDA when all data at 1 year are missing, and the one provided by LOCF converge to an intermediate value comprised between the baseline proportion of LDA and the one at 1 year.
Three main groups of methods with different ways of handling missing data caused by attrition emerged from our discussion: those including a model for attrition (IPW2 and CARRAC), those considering patients who stopped treatment as non-responders (LUNDEX and NRI) or as keeping the same disease activity in time (LOCF) and those not adjusting for attrition (IPW1 and CC). Within each group, these methods did not handle confounding the same way either, thus causing a residual difference between them.
Strength and limitations
The use of data stemming from a real register to generate a simulation data set is a strength of this study, as it allowed to test the statistical methods on close to real-world data. The large variety of methods studied and the release of the code on an open-access repository are also assets to the present work, which guarantee the accessibility of the methods. These methods are also generalisable for any disease or treatments, where treatments may be stopped for different reasons. However, as in every clinical study, the quality of the imputation will depend on model specification, so careful thought should be given to the covariates included in the model for each particular case. Nevertheless, a limitation of any simulation study is that results depend on the model used to generate the data. In the case of this study, our simulation design favours both CARRAC and IPW2, as they are the only ones making use of the treatment status used to generate the data. Thus, we may underestimate the impact of model misspecification. Another limitation is the use of only baseline disease activity, baseline confounders, and reason for stopping to estimate response rate. Though this choice corresponds to the reality of registry data, which usually have few intermediate visits, further models including repeated measurements could be informative. Finally, we used the association between the disease activity at 1 year and the reasons to stop in the future to recreate in the simulation data set the link between the reasons to stop before 1 year and the value of CDAI a patient would have had at 1 year. This procedure may underestimate the real association between reasons for cessation and disease activity. Therefore, the difference between the statistical methods presented here may be more pronounced in real applied analysis.
Conclusion
Correct estimation of effectiveness requires considering confounding, treatment cessation and dropouts. While CARRAC and IPW can produce proper estimates, methods omitting one of those, such as LUNDEX, NRI or CC, yield biased estimation, depending on the amount of attrition in the treatments. While the choice of methods is important, and some methods make stronger assumptions than others, model specification remains crucial. Careful justification of the model used for both missingness and confounding is necessary to obtain trustworthy and accurate results.
Acknowledgments
The authors thank all the rheumatologists and patients who participated in the registries used to initially create the simulation dataset.
Footnotes
Handling editor: Josef S Smolen
Twitter: @denis_mongin, @delcourvoisier
Contributors: DM performed the simulation study, the data analysis, participated in the interpretation of the results, and designed and wrote the manuscript; KL participated in the study design and contributed to the interpretation of the results; AF contributed to the interpretation of the results; TF contributed to the study design and to the interpretation of the results; DSC designed the study, contributed to the implementation of the simulation study and the data analysis, participated in the interpretation of the results and is the garantor of this work. All authors revised critically the manuscript and approved its current version.
Funding: This project was supported by an unrestricted grant from AbbVie.
Competing interests: None declared.
Patient and public involvement statement: Because the present work is a methodological simulation study, it was not appropriate to involve patients or the public in its design, conduct or reporting.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. The simulated dataset and all the code used to generate the data, perform the simulation study and create the figures and tables have been made openly available in the following repository: https://gitlab.com/dmongin/comparative-effectiveness. Concerning the original register collection used to create the simulation dataset, all the data belong to the registries. Anonymised data can be shared as long as agreements are made with all participating registries.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Because the study is a retrospective study using completely deidentified data, it was exempted from ethical approval. Approval of each local ethical committee for the collection of clinical data in each register was obtained for the register collection used to create the simulation dataset.
References
- 1. Guo Q, Wang Y, Xu D, et al. Rheumatoid arthritis: pathological mechanisms and modern pharmacologic therapies. Bone Res 2018;6:1–14. 10.1038/s41413-018-0016-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Courvoisier D, Lauper K, Bergstra SA, et al. Op0199 points to consider when analysing and reporting comparative effectiveness research with observational data in rheumatology. Ann Rheum Dis 2020;79:124–5. 10.1136/annrheumdis-2020-eular.1162 [DOI] [PubMed] [Google Scholar]
- 3. van der Heijden GJMG, Donders ART, Stijnen T, et al. Imputation of missing values is superior to complete case analysis and the missing-indicator method in multivariable diagnostic research: a clinical example. J Clin Epidemiol 2006;59:1102–9. 10.1016/j.jclinepi.2006.01.015 [DOI] [PubMed] [Google Scholar]
- 4. Schafer JL. Analysis of incomplete multivariate data. CRC Press, 1997. [Google Scholar]
- 5. Mongin D, Lauper K, Turesson C, et al. Imputing missing data of function and disease activity in rheumatoid arthritis registers: what is the best technique? RMD Open 2019;5:e000994. 10.1136/rmdopen-2019-000994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nunan D, Aronson J, Bankhead C. Catalogue of bias: attrition bias. BMJ Evid Based Med 2018;23:21–2. 10.1136/ebmed-2017-110883 [DOI] [PubMed] [Google Scholar]
- 7. Howe CJ, Cole SR, Lau B, et al. Selection bias due to loss to follow up in cohort studies. Epidemiology 2016;27:91–7. 10.1097/EDE.0000000000000409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lauper K, Kedra J, De Wit M, et al. Op0198 a Systematic Review to Inform the Eular Points to Consider When Analysing and Reporting Comparative Effectiveness Research with Observational Data in Rheumatology. Ann Rheum Dis 2020;79:123.2–4. 10.1136/annrheumdis-2020-eular.882 [DOI] [PubMed] [Google Scholar]
- 9. Eekhout I, de Boer RM, Twisk JWR, et al. Missing data: a systematic review of how they are reported and handled. Epidemiology 2012;23:729–32. 10.1097/EDE.0b013e3182576cdb [DOI] [PubMed] [Google Scholar]
- 10. Markusse IM, Landewé R, Wolterbeek R, et al. Linear extrapolation of missing radiographic change scores in clinical trials does not spuriously overestimate group radiographic changes in rheumatoid arthritis. Rheumatology 2016;55:1295–300. 10.1093/rheumatology/kew030 [DOI] [PubMed] [Google Scholar]
- 11. Horwitz RI, Viscoli CM, Berkman L, et al. Treatment adherence and risk of death after a myocardial infarction. Lancet 1990;336:542–5. 10.1016/0140-6736(90)92095-Y [DOI] [PubMed] [Google Scholar]
- 12. Newell DJ. Intention-to-treat analysis: implications for quantitative and qualitative research. Int J Epidemiol 1992;21:837–41. 10.1093/ije/21.5.837 [DOI] [PubMed] [Google Scholar]
- 13. Bell ML, Fiero M, Horton NJ, et al. Handling missing data in RCTs; a review of the top medical journals. BMC Med Res Methodol 2014;14:118. 10.1186/1471-2288-14-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cole SR, Hernán MA. Constructing inverse probability weights for marginal structural models. Am J Epidemiol 2008;168:656–64. 10.1093/aje/kwn164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Floden L, Bell ML. Imputation strategies when a continuous outcome is to be dichotomized for responder analysis: a simulation study. BMC Med Res Methodol 2019;19:161. 10.1186/s12874-019-0793-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Demirtas H. Practical advice on how to impute continuous data when the ultimate interest centers on Dichotomized outcomes through Pre-Specified thresholds. Commun Stat Simul Comput 2007;36:871–89. 10.1080/03610910701418424 [DOI] [Google Scholar]
- 17. Kristensen LE, Saxne T, Geborek P. The LUNDEX, a new index of drug efficacy in clinical practice: results of a five-year observational study of treatment with infliximab and etanercept among rheumatoid arthritis patients in southern Sweden. Arthritis Rheum 2006;54:600–6. 10.1002/art.21570 [DOI] [PubMed] [Google Scholar]
- 18. Moore AR, Straube S, Eccleston C, et al. Estimate at your peril: imputation methods for patient withdrawal can bias efficacy outcomes in chronic pain trials using responder analyses. Pain 2012;153:265–8. 10.1016/j.pain.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 19. Courvoisier DS, Chatzidionysiou K, Mongin D, et al. The impact of seropositivity on the effectiveness of biologic anti-rheumatic agents: results from a collaboration of 16 registries. Rheumatology 2021;60:820–8. 10.1093/rheumatology/keaa393 [DOI] [PubMed] [Google Scholar]
- 20. Mongin D, Lauper K, Finckh A. Data from: accounting for missing data caused by drug cessation in observational comparative effectiveness research: a simulation study. Novembre 23, 2021. Available: https://gitlab.com/dmongin/comparative-effectiveness [DOI] [PMC free article] [PubMed]
- 21. R Core Team . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2019. https://www.R-project.org [Google Scholar]
- 22. van der der WM, Geskus RB. ipw: an R package for inverse probability weighting. Journal of Statistical Software 2011;43:1–23. 10.18637/jss.v043.i13 [DOI] [Google Scholar]
- 23. van BS, Groothuis-Oudshoorn K. Mice: multivariate imputation by Chained equations in R. Journal of Statistical Software 2011;45:1–67. 10.18637/jss.v045.i03 [DOI] [Google Scholar]
- 24. Højsgaard S, Halekoh U, Yan J. The R package geepack for generalized estimating equations. Journal of Statistical Software 2005;15:1–11. 10.18637/jss.v015.i02 [DOI] [Google Scholar]
- 25. Frisell T, Dehlin M, Di Giuseppe D, et al. Comparative effectiveness of abatacept, rituximab, tocilizumab and TNFi biologics in RA: results from the nationwide Swedish register. Rheumatology 2019. 10.1093/rheumatology/key433. [Epub ahead of print: 21 Jan 2019]. [DOI] [PubMed] [Google Scholar]
- 26. Cheung YB. A modified least-squares regression approach to the estimation of risk difference. Am J Epidemiol 2007;166:1337–44. 10.1093/aje/kwm223 [DOI] [PubMed] [Google Scholar]
- 27. Zou G. A modified poisson regression approach to prospective studies with binary data. Am J Epidemiol 2004;159:702–6. 10.1093/aje/kwh090 [DOI] [PubMed] [Google Scholar]
- 28. Rubin DB. Multiple imputation for nonresponse in surveys. John Wiley & Sons, 2004. [Google Scholar]
- 29. Jones M, Mishra GD, Dobson A. Analytical results in longitudinal studies depended on target of inference and assumed mechanism of attrition. J Clin Epidemiol 2015;68:1165–75. 10.1016/j.jclinepi.2015.03.011 [DOI] [PubMed] [Google Scholar]
- 30. Seaman SR, White IR. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res 2013;22:278–95. 10.1177/0962280210395740 [DOI] [PubMed] [Google Scholar]
- 31. Iannaccone CK, Fossel A, Tsao H, et al. Factors associated with attrition in a longitudinal rheumatoid arthritis registry. Arthritis Care Res 2013;65:1183–9. 10.1002/acr.21940 [DOI] [PubMed] [Google Scholar]
- 32. Courvoisier DS, Alpizar-Rodriguez D, Gottenberg JE, et al. Rheumatoid arthritis patients after initiation of a new biologic agent: trajectories of disease activity in a large multinational cohort study. EBioMedicine 2016;11:302–6. 10.1016/j.ebiom.2016.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aletaha D, Funovits J, Keystone EC, et al. Disease activity early in the course of treatment predicts response to therapy after one year in rheumatoid arthritis patients. Arthritis Rheum 2007;56:3226–35. 10.1002/art.22943 [DOI] [PubMed] [Google Scholar]
- 34. Verstappen M, van Mulligen E, de Jong PHP, et al. DMARD-free remission as novel treatment target in rheumatoid arthritis: a systematic literature review of achievability and sustainability. RMD Open 2020;6:e001220. 10.1136/rmdopen-2020-001220 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2021-221477supp001.pdf (964KB, pdf)
Data Availability Statement
Data are available upon reasonable request. The simulated dataset and all the code used to generate the data, perform the simulation study and create the figures and tables have been made openly available in the following repository: https://gitlab.com/dmongin/comparative-effectiveness. Concerning the original register collection used to create the simulation dataset, all the data belong to the registries. Anonymised data can be shared as long as agreements are made with all participating registries.