MULTIPLE DIMENSIONS OF THE MUTATION RATE
Antibiotic resistance can be achieved by horizontal acquisition of resistance genes (carried by plasmids or transposons), by recombination of foreign DNA into the chromosome, or by mutations in different chromosomal loci (15). In studies of molecular evolutionary biology, the term mutation rate is applied to estimations of the rate (per generation) of mutation per nucleotide, per locus, or, eventually, for the whole genome, and selectively favorable, unfavorable, or neutral mutations are considered. Differing with this concept, the frequency of mutation measures all the mutants present in a given population, irrespective of whether the mutation events occurred early or late during the growth of the population. In this respect, the frequency of mutants is a cross section of the bacterial population at a given time and reflects not only the mutation rate but also the history of the population before selection is applied.
In the case of antibiotic resistance, the mutation rate is frequently defined as the in vitro frequency at which detectable mutants arise in a bacterial population in the presence of a given antibiotic concentration. Note that we are recording the number of mutant cells and not the number of mutation events. Therefore, we are recording only the selectively favorable mutations for the bacteria that lead to a visible antibiotic resistance phenotype. Such a determination is widely considered an important task for the prognosis of the emergence of antibiotic-resistant bacteria. In the scientific jargon regarding antibiotics, a “mutation rate” is frequently presented in a characteristically naive way that can sometimes be understood as an intrinsic property of a new antimicrobial drug in its interaction with the target bacteria, with a “low mutation rate” being considered an advantage of this drug over its competitors. “This drug induces (?) a low mutation rate” is a familiar but fully mistaken expression. The intention of this review is to reconsider the multiple dimensions of the concept of “mutation rate” from the perspective of reaching a better understanding of its intrinsic complexity and trying to encourage research for more advanced methods of predicting the emergence of mutational resistance to antibiotics.
From the pioneering works of Luria and Delbrück (35), it became clear that evaluation of mutation rates is not easy. In fact, the methods for distinguishing the value of the observed frequency of mutants from the real mutation rate are not easy to apply (33, 35), and fluctuation tests for analysis of the presence of jackpots of preexisting mutants in the tested populations have been developed. In the case of antibiotic resistance, the problem is complicated by the fact that the phenotype does not always reflect the same genotypes in all selected mutants, because mutations in different genes can produce similar antibiotic resistance phenotypes. As an example, when a quinolone resistance mutation rate is determined, this rate is actually the result of the combination of the mutation rates of the genes that encode the synthesis of GyrA, GyrB, ParA, ParC, and several different multidrug resistance (MDR) systems (26, 40). In this respect, the calculated “phenotypic” mutation rate is the result of several different “genotypic” mutation events. In fact, mutations in different loci produce different changes in MICs, and stable maintenance of heterogeneous antibiotic resistance expression classes in bacterial populations is a well-known phenomenon (19).
In recent years, an explosion of published work has demonstrated that the mutation process in bacterial populations is not a static event. On the contrary, a complex network of factors influence the rate and type of mutants that can be selected under antibiotic selective pressure. Mutation rates can largely change for a given antibiotic depending on its concentration during selection (30). Physiological conditions such as the availability of a given carbon source (27) or, in general, bacterial stress (21, 70) may regulate the mutation rate in bacteria. Furthermore, the existence of mutations that produce mutator phenotypes in bacteria (32, 72) and the capability of some antibiotics to increase mutability (37, 61) greatly complicate studies of the effects of population dynamics on the emergence of antibiotic-resistant mutants in bacteria. These elements of variability severely challenge the possibility of predicting the “real” mutation rate just by simple experimental procedures like those frequently used in laboratory experiments.
In the present work we analyze the most relevant factors which influence the emergence and selection of antibiotic resistant mutants in bacterial populations (Table 1). Note that throughout this article we use the term “antibiotic resistance” for any inheritable increase in MICs, irrespective of whether the change is small or large. Such a definition qualifies as “resistant” many organisms that are conventionally considered as having low levels of susceptibility or even being entirely “susceptible” according to National Committee for Clinical Laboratory Standards-type breakpoint-based criteria. Indeed, we (4) and others think that those low-level mutations (see below) have an important role in the final emergence of clinically relevant antibiotic-resistant mutants in bacterial populations.
TABLE 1.
Factors that increase antibiotic resistance mutation rates
| Factors |
|---|
| Unstable sequences surrounding relevant bases for the resistance phenotype |
| Long distance of the R genea to the origin of replication |
| Large number of sites in the R gene that can give rise to a resistance and permissive phenotype |
| Large variety of R genes |
| Low or high copy number of each R gene, if the mutation is recessive or dominant, respectively |
| Few independent antibiotic targets or target access pathways |
| Several independent protective mechanisms |
| Multiple cooperative targets for antibiotic action or cooperative access pathways |
| Few cooperative elements in target protection mechanisms |
| Bacteria under stress (starvation, antibiotic stress, pathogenic stress) |
| Contingency (hypermutable) R genes |
| Transposable elements in the bacterium |
| Bacteria with hypermutable (mutator) phenotype |
| Low level of expression of bacterial programmed cell death |
| Preexisting low-level R gene mutations |
| Low biological cost of R gene mutations |
| Antibiotic concentrations in the selective window |
| Low antibiotic concentrations or short time of exposure |
| Slow killing ability of the antibiotic |
| Small phenotypic lag for the expression of R gene mutations |
| Compartmentalized (structured) physical structure of the selective habitat |
R gene, genes in which mutations produce antibiotic resistance.
MUTABILITY AND MUTATION RATES
The probability that a mutation will give an antibiotic resistance phenotype (mutability) influences the mutation rate. Mutability will depend on the structure and the number of the genes in which mutations can produce a selectable phenotype.
Gene structure.
The DNA structure of a gene is relevant for mutability. If all positions have equivalent possibilities for mutation, long genes might be more prone to mutation than small ones. However, the size of the gene is not the main factor that is relevant for its mutability, because not every mutation in a gene that encodes an antibiotic target results in resistance. Resistance occurs only by mutations which are at the same time permissive (that are nonlethal or, in broader terms, that do not lead to an unacceptable decrease in fitness) and able to produce a resistance phenotype. The probability that an effective resistance mutation will emerge will then be proportional to the number of such positions. For instance, in Escherichia coli changes in at least seven positions in the gyrA gene that result in a quinolone resistance phenotype have been observed, but changes in only three positions in the parC gene that result in a resistance phenotype have been observed (26). It could be expected that the mutation rate for the gyrA gene will be higher than that for the parC gene, and indeed, gyrA mutants are more frequently found than parC mutants. On the contrary, in Streptococcus pneumoniae, quinolone resistance results from changes in two positions of gyrA but results from changes in five positions in parC, and parC mutants are more frequently found. In the same organism, the numbers of positions in the gene that encodes PBP 2×, mutations of which can produce β-lactam resistance, are 13 for cefotaxime resistance but only 1 for penicillin resistance (25). That may contribute, together with the different numbers of targets for both antibiotics (see Cooperative Mutations below), to the higher frequency of mutation toward cefotaxime resistance compared with that toward penicillin resistance. Also, streptomycin resistance results from a change in one or a very few nucleotides of the gene that encodes ribosomal protein S12 (9), and the spontaneous mutation rate is low. The stability of the sequences that surround the nucleotides involved in the mutant phenotype are also highly relevant for mutability. It has been described that the specific mutability for a single base can vary by more than 10,000-fold (17), and long stretches of repeated bases are prone to deletions or insertions by means of slip-strand mispairing (12). Even the position of the gene in the bacterial chromosome affects the mutation rate, since genes farther away from the origin of replication of the chromosome of Enterobacteriaceae can have a mutation rate approximately two times that of genes near this origin (71).
Gene multiplicity.
Since an antibiotic resistance phenotype can be due to mutations in different bacterial loci, the emergence of an antibiotic-resistant mutant will be a function of an overall mutability value that results from the combination of the independent mutability values for these genes. Classical genetic analysis indicates that when mutations in either one or another gene can produce antibiotic resistance, the overall mutability will be the sum of the independent mutability values. On the contrary, when mutations in both genes are required to reach an antibiotic resistance phenotype, the overall mutability will be the product of the independent mutability values for each gene. Let us consider two genes independently involved in an antibiotic resistance phenotype and with independent mutability values of 10−8. If mutations in any of the genes can render an antibiotic resistance phenotype (see Independent Mutations below), the overall mutability will be 2 × 10−8; thus, it is only a little higher than that in the case in which only one gene involved in the resistance phenotype has a mutation. On the contrary, if mutations in both genes are required for resistance (see Cooperative Mutations below), the overall mutability will be 10−16, which is much lower than that in the case in which only one gene is required.
ANTIBIOTIC-CELL INTERACTIONS AND MUTATION RATES
To inhibit their bacterial targets, antibiotics need to cross the cellular envelopes and in some cases (such as 5-nitroimidazoles [28] or pyrazinamide [67]) to be activated by bacterial enzymes before they can gain access to those targets. Bacteria have protection determinants against the antibiotic effect. These include antibiotic-inactivating enzymes (such as β-lactamases [11]) and MDR efflux pumps (56).
Three main types of intrinsic genes (which preexist in the genome of the wild-type susceptible population) are relevant for the emergence of antibiotic-resistant mutants: (i) genes involved in the synthesis and cell positioning of the antibiotic target; mutations in these genes can be denominated target-structural mutations; (ii) genes involved in the access of the antibiotic to the target (including those required for activation of the formerly inactive antibiotic), which are needed for the biochemical access of the antibiotic (see above); mutations in these genes are named target-access mutations; and (iii) genes involved in the protection of the target from the drug, including detoxification by antibiotic-modifying enzymes or efflux of the antibacterial compounds; mutations which activate the expression of those genes (see below) are named target-protection mutations.
A variety of genes can be involved in antibiotic resistance either because there are several different target, access, or protection pathways for the antibiotic in the bacterial cell or because each pathway requires the expression of several genes. Even for a single target, access, protection pathway, several genes can be involved in its synthesis, because it may be constituted by several subunits and/or its expression might be regulated. Regulation is especially relevant in the case of protection determinants, because they are frequently downregulated under standard growing conditions, and antibiotic resistance can be achieved by means of up mutations either in the promoter sequences or the genes that encode the regulatory proteins of these systems. Depending on the specific antibiotic-bacterium interaction at a given antibiotic concentration, antibiotic resistance can result in some cases from single gene mutations (independent mutations), whereas in other cases mutations in several genes (cooperative mutations) are required.
Independent mutations.
If there are a variety of different gene mutations which independently result in antibiotic resistance, the mutation rate will be the sum of the independent mutability values. In the case of target structural mutations, the most salient example is the presence of several subunits (encoded by different genes) in the target. For instance, mutations in the genes that encode either the GyrA or the GyrB subunit of topoisomerase II result in phenotypes of resistance to some quinolones (54). A similar effect is expected to occur in target access mutations. If several genes are required for access of the antibiotic to its target, mutations in each of these genes will produce an antibiotic resistance phenotype. In the case of target protection mutations, the presence of multiple independent antibiotic detoxification mechanisms in the same bacterial cell increases the mutation rate of the bacteria, because mutations that lead to the activation of any of them will increase the MIC of the antibiotic. For instance, low-level quinolone resistance in Pseudomonas aeruginosa can be achieved by means of mutations which activate the expression of different MDR determinants (30).
Cooperative mutations.
If mutations in several different genes are required for antibiotic resistance, the overall mutability will be the product of the independent mutability values. For instance, the presence in the bacterial cell of several effective (lethal or inhibitory) targets, each one of which is capable of an independent interaction with the antibiotic, ensures that single mutations (target structural mutations) will infrequently arise, as the bacteria should be killed because of the availability of other points of antibiotic action. The spontaneous rate of mutation to cefotaxime resistance in S. pneumoniae is higher than that for penicillin resistance because cefotaxime has one less penicillin-binding protein target (24). Another example of this situation is the requirement in both gyrA and grlA for the acquisition of a phenotype of resistance to some fluoroquinolones in Staphylococcus aureus (16). Also, if the antibiotic can be independently and effectively transported through multiple independent access routes, mutation in all of the access pathways (target access mutations) will be required to acquire an antibiotic resistance phenotype. If multiple cooperative protection elements are required to effectively protect the target, mutations that result in the expression of all of them (target protection mutations) will be required for an antibiotic resistance phenotype and the overall mutability will decrease. In this respect, it has been shown that both the chromosomal β-lactamase and the MDR system MexAB-OprM are required for the full protection of P. aeruginosa from β-lactam antibiotics (45).
For any specific antibiotic, a combination of target, access, and protection pathway genes is involved in its interaction with the bacterial cell. Mutations in one or another of those genes may produce low-level resistance, whereas high-level resistance frequently requires mutations in more than one gene. In this respect, low antibiotic concentrations will select independent mutations, whereas high antibiotic concentrations should preferentially select cooperative mutations (see The Antibiotic Selective Process and Mutation Rates). As an example, low-level quinolone resistance can be acquired by independent mutations in different genes (see above), whereas high-level fluoroquinolone resistance frequently requires successive mutations in gyrA and grlA (53) and in regulatory sequences of efflux pumps as well.
Gene copy number.
In the case of target-structural mutations in high-copy-number genes, mutations in one of the copies of the gene will produce a mixed population of target molecules. Some of them will correspond to the mutated allele of the target, and the majority will correspond to the wild-type allele. If the mutant target is not functionally dominant, many wild-type targets will still remain in the bacteria, thus masking the effect of the mutation. A high copy number of the gene will reduce the observed emergence of mutants. For instance, mutations in rrn genes, which encode 23S rRNA, probably occur at a similar rate in E. coli, S. aureus, Mycobacterium, or Helicobacter pylori. In practice, such mutations, which lead to a macrolide resistance phenotype (55, 73), are detectable only in the last two organisms, because they have only one or two available copies of the rrn genes (65, 73). The converse (increased mutation rate) should happen if the mutant allele for antibiotic resistance is dominant over the wild-type one, because mutations in one or another of the copies that encode the target gene will produce an antibiotic resistance phenotype. In the case of dominance, a large number of gene copies will increase mutability. This point is particularly relevant in the case of target protective mutations. The presence of multiple copies of a detoxification gene frequently produces an antibiotic resistance phenotype due to a gene-dosing effect (43). An example of this is plasmid-encoded β-lactamases (41). However, if the amount of the product of the gene is not enough to fully protect the target or mutations are required to change the substrate specificity, the presence of multiple copies of the protection gene might increase the overall mutability, as mutations in one or another copy will render an antibiotic resistance phenotype. That may have occurred during the evolution of extended-spectrum β-lactamases (7, 51).
BACTERIAL PHYSIOLOGICAL BACKGROUND AND MUTATION RATES
Adaptive mutation.
The mutation process has classically been studied in actively dividing bacteria (33, 35), as it was assumed that mutations occur as the consequence of errors during the DNA replication process. However, more recent work has demonstrated that mutation also occurs in nondividing cells (22, 36, 64, 69). These mutation events are the basis of the so-called adaptive mutation (or stress-induced mutagenesis) (63). A special feature of stress-induced mutagenesis is the fact that the mutation rate can increase over time by several orders of magnitude for cells under starvation conditions (50, 69).
In all of the models of adaptive mutation analyzed so far, studies have been performed by using nonlethal selection, because it was assumed that a lethal selector will kill bacteria long before they enter in starvation. Perhaps for this reason regulation of the emergence of antibiotic-resistant mutants under bacterial stress has been poorly studied. Nonetheless, some recent work has begun to fill this gap. It has been demonstrated that quinolones, which are able to induce the SOS mutagenic response (57), increase the rate of emergence of resistance to these drugs in E. coli (62). The emergence of MDR mutants increases in P. aeruginosa under antibiotic challenge (1). E. coli exposed to streptomycin displays a hypermutable phenotype (61). Finally, the mutation rate of Salmonella enterica serotype Typhimurium for rifampin resistance increases under starvation conditions (27). These observations indicate that bacterial growth conditions have a dramatic effect on the mutation rate. Analysis of several model systems have demonstrated that stress-enhanced bacterial mutation is a regulated phenomenon. The main factors in this process are stress-responsive error-prone DNA polymerases V (umuCD) and IV (dinB), which transiently increase the rate of mutation (58). Bacteria growing in vivo are frequently under stress (20) because they are starved, under antibiotic treatment, or challenged by the need to colonize novel environments under the inhibitory effects of host defense mechanisms. We then suggest that the frequencies of mutation are probably much higher in the course of an infective process than those that have been determined by in vitro analysis.
Gene-specific regulation of antibiotic resistance mutation rate.
In-host increased mutability is a general situation that may affect several different bacterial genes. However, regulation of mutability also has a gene-specific component. It has been pointed out that bacteria have two different sets of genes: housekeeping genes, which are relevant for basic bacterial metabolism and structure and that mutate at an expected low frequency, and contingency genes (52), which are important for bacterial adaptability to changing environments and that are highly mutable. Indeed, very high rates of mutation may indicate not a real mutation but, rather, some programmed recombination event. Examples of such contingency genes are those involved in phase changes. Phase variation is a long-studied phenomenon by which bacteria can rapidly change their immunogenic characteristics and hence evade host defenses. This process is characterized by switching between high and low levels of expression of a particular group of genes. Switching is the consequence of specific DNA rearrangements in the genes for regulatory proteins or the promoters of the genes involved in the phase change. DNA regions involved in switching present, then, a high mutation rate, which is specific for these regions and which is tightly regulated (68, 76). Few of these systems of specific regulation of mutagenesis have been analyzed so far in relation to antibiotic resistance. For instance, switching between phenotypes of chloramphenicol resistance and susceptibility with frequencies in the range 10−4 to 10−5 have been described for two isolates of Proteus mirabilis (13) and Agrobacterium radiobacter (42). In both cases a switch in the expression of a chloramphenicol acetyltransferase (which is silent in the susceptible populations) was responsible for the emergence of a resistant population.
Hypermutable (mutator) strains.
Recent work has shown that many bacterial populations harbor a proportion of cells with a mutator phenotype. These cells have a mutation rate that is increased from 10 to 50 up to 10,000 times (48), generally as a consequence of a defective methyl-directed mismatch repair system. Because such highly mutable bacteria can rapidly emerge in a previously homogeneous population, the overall mutation rate will increase. The mutator phenotype allows bacteria to develop a large variability of alleles that can evade (by mutation) stressful environments during the infective process or antibiotic treatment (32, 46). Selection of mutator alleles by antibiotic therapy with different drugs can be the basis for the emergence of some phenotypes of multiple antibiotic resistance among bacteria with mutations in more than one target (32). Mutator bacteria are not expected to be fixed in the majority of bacterial populations as they tend to be less fit than organisms with stable genomes in nonstressful environments. In fact, the emergence of hypermutable cells may be modulated by systems of programmed cell death in bacteria. The same stress conditions that trigger the appearance of a mutator phenotype may also regulate the expression of antitoxin-toxin systems in bacteria which play a role in programmed bacterial cell death (49). The expected consequence is suicide and a consequent decrease in the bacterial population, reducing the possibility that a mutant will emerge or that the mutation will be fixed (see below). Nevertheless, a subpopulation of mutators may be maintained in bacterial populations challenged by fluctuating (stressful and nonstressful) environments. A succession of environmental bottlenecks is not an unexpected situation in the natural history of many bacterial organisms and may be particularly frequent among pathogenic or epidemigenic bacterial clones.
In addition to mutator genes, other elements may contribute to the general intrinsic mutability of bacterial genomes. For instance, transposable elements also play a role by increasing the rate of mutation (29), and the transposition event can be regulated by the environment, including the presence of antibiotics (75). In such a way, the bacterial genome structure, together with the presence of mutations in genes involved in DNA repair, and the growing conditions are major determinants for the general mutability of bacterial populations.
Preexisting low-level antibiotic resistance mutations.
As stated before, the presence of low-level antibiotic resistance determinants is frequently required, in combination with other mechanisms, to produce a high-level antibiotic resistance phenotype. A direct effect of preexisting low-level antibiotic resistance mutations will then be a net increase in the mutation rate when the selection is performed under high antibiotic concentrations. However, it seems that the presence of low-level antibiotic resistance mechanisms might favor the emergence of clinically relevant antibiotic-resistant bacteria even when the antibiotic selective concentration allows the growth of mutants with mutations in a single gene. This hypothesis is backed by works from Markham (38) and Markham and Neyfakh (39), in which the investigators demonstrate that reserpine, an inhibitor of the MDR efflux pumps from S. aureus and S. pneumoniae, drastically reduces the mutation rate, leading to quinolone-resistant mutants in these bacterial species (38, 39). Similarly, tetracycline-selected marO or marR E. coli mutants have a 1,000-fold higher rate of mutation to fluoroquinolone resistance (14). This situation may also apply for antibiotics with an inoculum effect, since the presence of bacteria growing at a high density can be considered a transient phenotypic mechanism of low-level antibiotic resistance (60). The implication of this fact for the development of antibiotic resistance in the course of an infection is clear: the presence of low-level antibiotic resistance determinants in bacteria will increase the possibility of the emergence of a clinically relevant antibiotic resistance phenotype. In the same respect, the presence of local accumulations of bacteria (or, eventually, biofilm formation) will favor the emergence of antibiotic-resistant mutants as the consequence of the inoculum effect.
THE ANTIBIOTIC SELECTIVE PROCESS AND MUTATION RATES
Once a mutation that potentially might render an antibiotic resistance phenotype has occurred, the bacterium carrying the mutated allele must compete with the wild-type ancestor bacterial population. The outcome of the competition process depends on its relative fitness, defined as the efficiency of multiplication of the mutant cell compared with that of the wild-type ancestor population (18). Thus, not only the mutability but the selective process as well will have a relevant effect on the final mutation rate values.
Bacterial fitness.
Antibiotic resistance changes bacterial fitness (2) usually by decreasing its value (6, 34), which is generally known as the biological cost of resistance, but in some cases, fitness is increased (8). In time, this reduction can be compensated by mutations in other loci of the bacterial chromosome (5, 10, 66), so that the antibiotic-resistant bacteria can present a level of fitness equal to or even higher than those of the original wild-type strains. If the mutant bacterium has a lower level of fitness than the wild type, it will be cleared from the bacterial population during growth prior to selection and the mutation rate will be underestimated. On the contrary, a higher level of fitness of the mutant bacteria will produce an overestimation of the mutation rate. We should remind readers that in vitro-determined bacterial fitness does not necessary equal in vivo fitness. In this respect, it has recently been described that highly fluoroquinolone-resistant Salmonella strains might be counterselected in the field (23). Also, different fitness-compensating mutations are selected when antibiotic-resistant bacterial populations are growing in vivo compared with those selected in in vitro experiments (5). These differences highlight the need for very careful interpretation of the results obtained with current in vitro models for the emergence of antibiotic-resistant mutants in bacterial populations.
Antibiotic concentrations.
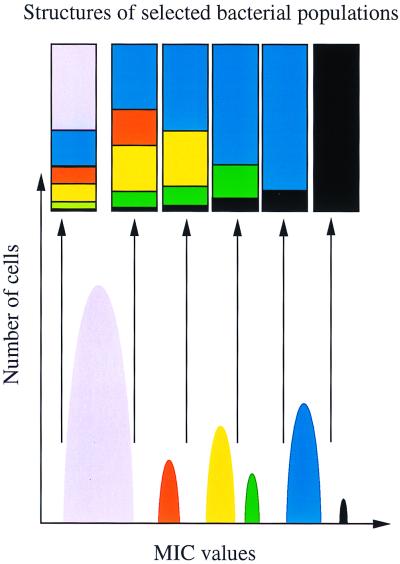
The concentration of the selector has an important role in the rate of mutation to antibiotic resistance. For instance, the rate of selection of low-level quinolone-resistant mutants in P. aeruginosa can range from 1.2 × 10−6 to 4 × 10−10, depending on the type and concentration of the quinolone used for selection (30). As stated earlier, a phenotype of increased antibiotic resistance may arise from mutations in different genes. These mutations, however, may provide quite different levels of antibiotic resistance. In fact, heterogeneous expression of antibiotic resistance (even by changes in the same target gene) is a well-known phenomenon (19). At low selector concentrations, mutations in any of those genes can effectively protect the bacteria from the action of the antibiotic and thus be selectable. However, once the antibiotic concentration rises, the number of selectable mutants decreases (Fig. 1). At certain antibiotic concentrations, combinations of mutations in more than one gene might be required to provide the resistance phenotype (see above), so that at high selector concentrations, a sharp decrease in the mutation rate will occur.
FIG. 1.
Patchwork structure of bacterial populations selected at different antibiotic concentrations. The different bacterial populations that can be obtained upon selection with different concentrations of the same antibiotic are shown. Six different phenotypes, each one corresponding to mutations either in different loci or in different positions in the same locus, that render different MICs are indicated. The rectangles show the compositions of the bacterial populations selected with the antibiotic concentration fixed by the arrow at the bottom. The black box at the bottom of all rectangles contains the same amount of bacterial cells in all populations. Note that for any population maximum selection is obtained at selective antibiotic concentrations close to the MIC for the population.
Another important point (3) is that the probability that a specific type of mutant will emerge is expected to have a maximum at one particular antibiotic concentration close to the MIC for the organism (Fig. 1). For instance, a specific antibiotic concentration may be sufficient to decrease the growth rate or to suppress the original ancestor population but may not be sufficient to affect the resistant variant population. Beyond this concentration, antibiotic concentrations may be able to reduce or suppress in an equivalent way the growth of both susceptible and variant populations, and therefore, no selection for the variant is expected to occur. The same applies when the antibiotic concentration is below the level to which both populations are susceptible. Therefore, the selection of a particular antibiotic-resistant variant may happen only in a narrow range of drug concentrations that define a selective window. The conclusion is that the observed mutation rate is very sensitive to changes in drug concentration, and different rates and types of mutants may be obtained in a discontinuous way along the range of concentrations. On the other hand, as the selective effect of the drug may depend (as for β-lactam antibiotics) on the time of exposure, this period of time may be critical to yield one or another mutation rate.
Finally, the dynamics of the antibiotic action on the bacterial cell may modify the mutation rate. If the bacterial population is not killed effectively by a given antibiotic, the cells are maintained under stress, which may increase the mutation rate (see above). On the contrary, some potentially effective resistance mutations may have a phenotypic lag; that is, the resistance phenotype is not immediately evident after mutation but appears some time later. If at this time the bacterium is rapidly killed by the drug, the resistance phenotype will never arise.
Physical structure of selective habitat.
We have seen that both mutability and fitness are relevant for the emergence of antibiotic-resistant bacteria. However, the capability of a bacterium to compete with other partners depends on the environment in which bacteria grow. Structured and compartmentalized environments (such as surfaces) allow bacteria to occupy different niches and thus do not compete with each other. Under these circumstances, all possible alleles in the population capable of surviving the selective pressure will grow. The effect of the physical structure of the culture medium on the variability of bacterial populations has been studied for bacterial phenotypes other than antibiotic resistance phenotypes (31, 59). The results obtained indicate that growth on structured habitats increases the variability of bacterial populations and accelerates their evolution in response to environmental inputs. The habitat colonized by bacteria during an infection is frequently heterogeneous, and bacteria are frequently attached to surfaces or are inside host cells. Since these types of environments (structured habitats) allow the emergence of higher degrees of different phenotypes, more types of antibiotic-resistant mutants are probably selected during an infection than during in vitro tests with liquid cultures. Actually, it has been shown that P. aeruginosa strains isolated from the sputum of each cystic fibrosis patient present a high degree of variability both in morphotypes and in antibiotic susceptibility profiles (47, 77). Finally, the size of the selective habitat and the total bacterial population size may also influence the mutation rate. The larger the population, the more likely the fixation of hypermutable alleles (74) which highly affect the mutation rate (see before). The increased in-host evolution of bacterial populations as a consequence of both a higher mutation rate and selection in large, densely populated structured habitats might then have a major role in the emergence of antibiotic-resistant mutants during the infective process.
CONCLUSIONS AND PERSPECTIVES
In this review, we have stated that the “mutation rate” is not a simple characteristic of a specific bacterial species-antibiotic association. On the contrary, the probability of the emergence of antibiotic-resistant mutants is a complex phenomenon, as previously recognized by others (44), in which the physiology, the genetics, the antibiotic-bacterium dynamics, and the historical behavior of bacterial populations, together with the physical structure of the selective medium, play major roles. We must assume that the mutation rate determined under conventional laboratory conditions probably differs greatly from that in vivo at the site of infection. In such a way, more than a single mutation rate, bacterial populations may have multiple different mutation rates. The time has arrived to face this complexity. We hope that the analysis presented in this review may contribute to an understanding of the factors influencing these mutation rates (Table 1) with the aim of predicting a theoretical rate of emergence of resistant mutants for a given antibiotic challenge by the use of appropriate mathematical models. To implement the models, more precise experimental data are urgently required. Before the introduction of a new antimicrobial agent in clinical practice, it is advisable to know in advance (phase I trials) the possibilities of development of mutational resistance in different bacterial pathogens. Analysis of both natural and hypermutable populations under challenge with a variety of close antibiotic concentrations (starting very near the MIC), and preferably with compartmentalized (solid) media, will provide important data that can be used to predict the frequency and type of resistant variants that may arise during therapy. Current advances in genomics and bacterial physiology should provide the basis for the precise determination of some of the quantitative data required for the proposed analysis. We encourage the development of such important pieces of research.
ACKNOWLEDGMENTS
We thank Marc Lipsitch, from the Harvard Medical School, Bruce Levin, from Emory University, José Pérez-Martín, from the Centro Nacional de Biotecnología, and Jesús Blázquez, from the Ramón y Cajal Hospital, for fruitful criticisms, discussions, and comments on draft versions of the manuscript.
REFERENCES
- 1.Alonso A, Campanario E, Martinez J L. Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology. 1999;145:2857–2862. doi: 10.1099/00221287-145-10-2857. [DOI] [PubMed] [Google Scholar]
- 2.Andersson D I, Levin B R. The biological cost of antibiotic resistance. Curr Opin Microbiol. 1999;2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 3.Baquero F, Negri M C. Selective compartments for resistant microorganisms in antibiotic gradients. Bioessays. 1997;19:731–736. doi: 10.1002/bies.950190814. [DOI] [PubMed] [Google Scholar]
- 4.Baquero F, Negri M C, Morosini M I, Blazquez J. The antibiotic selective process: concentration-specific amplification of low-level resistant populations. Ciba Found Symp. 1997;207:93–105. doi: 10.1002/9780470515358.ch7. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkman J, Nagaev I, Berg O G, Hughes D, Andersson D I. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287:1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 6.Bjorkman J, Hughes D, Andersson D I. Virulence of antibiotic-resistant Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:3949–3953. doi: 10.1073/pnas.95.7.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blazquez J, Morosini M I, Negri M C, Gonzalez-Leiza M, Baquero F. Single amino acid replacements at positions altered in naturally occurring extended-spectrum TEM beta-lactamases. Antimicrob Agents Chemother. 1995;39:145–149. doi: 10.1128/aac.39.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blot M, Hauer B, Monnet G. The Tn5 bleomycin resistance gene confers improved survival and growth advantage on Escherichia coli. Mol Gen Genet. 1994;242:595–601. doi: 10.1007/BF00285283. [DOI] [PubMed] [Google Scholar]
- 9.Bonny C, Montandon P E, Martin S M, Stutz E. Analysis of streptomycin-resistance of Escherichia coli mutants. Biochim Biophys Acta. 1991;1089:213–219. doi: 10.1016/0167-4781(91)90010-j. [DOI] [PubMed] [Google Scholar]
- 10.Bottger E C, Springer B, Pletschette M, Sander P. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat Med. 1998;4:1343–1344. doi: 10.1038/3906. [DOI] [PubMed] [Google Scholar]
- 11.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bzymek M, Saveson C J, Feschenko V V, Lovett S T. Slipped misalignment mechanisms of deletion formation: in vivo susceptibility to nucleases. J Bacteriol. 1999;181:477–482. doi: 10.1128/jb.181.2.477-482.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charles I G, Harford S, Brookfield J F, Shaw W V. Resistance to chloramphenicol in Proteus mirabilis by expression of a chromosomal gene for chloramphenicol acetyltransferase. J Bacteriol. 1985;164:114–122. doi: 10.1128/jb.164.1.114-122.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen S P, McMurry L M, Hooper D C, Wolfson J S, Levy S B. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies J E. Origins, acquisition and dissemination of antibiotic resistance determinants. Ciba Found Symp. 1997;207:15–27. [PubMed] [Google Scholar]
- 16.Deplano A, Zekhnini A, Allali N, Couturier M, Struelens M J. Association of mutations in grlA and gyrA topoisomerase genes with resistance to ciprofloxacin in epidemic and sporadic isolates of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:2023–2025. doi: 10.1128/aac.41.9.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drake J W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elena S F, Lenski R E. Test of synergistic interactions among deleterious mutations in bacteria. Nature. 1997;390:395–398. doi: 10.1038/37108. [DOI] [PubMed] [Google Scholar]
- 19.Figueiredo A M, Ha E, Kreiswirth B N, de Lencastre H, Noel G J, Senterfit L, Tomasz A. In vivo stability of heterogeneous expression classes in clinical isolates of methicillin-resistant staphylococci. J Infect Dis. 1991;164:883–887. doi: 10.1093/infdis/164.5.883. [DOI] [PubMed] [Google Scholar]
- 20.Foley I, Marsh P, Wellington E M H, Smith A W, Brown M R W. General stress response master regulator rpoS is expressed in human infection: a possible role in chronicity. J Antimicrob Chemother. 1999;43:164–165. doi: 10.1093/jac/43.1.164. [DOI] [PubMed] [Google Scholar]
- 21.Foster P L. Adaptive mutation: the uses of adversity. Annu Rev Microbiol. 1993;47:467–504. doi: 10.1146/annurev.mi.47.100193.002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster P L, Trimarchi J M. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giraud E, Brisabois A, Martel J L, Chaslus-Dancla E. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob Agents Chemother. 1999;43:2131–2137. doi: 10.1128/aac.43.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakenbeck R. Beta-lactam-resistant Streptococcus pneumoniae: epidemiology and evolutionary mechanism. Chemotherapy (Basel) 1999;45:83–94. doi: 10.1159/000007170. [DOI] [PubMed] [Google Scholar]
- 25.Hakenbeck R, Kaminski K, van der Linden M, Paik J, Reichmann F, Zähner D. Penicillin-binding proteins in β-lactam-resistant Streptococcus pneumoniae. Microb Drug Resist. 1999;5:91–99. doi: 10.1089/mdr.1999.5.91. [DOI] [PubMed] [Google Scholar]
- 26.Hooper D C. Mechanisms of fluoroquinolone resistance. Drug Resist Updates. 1999;2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 27.Hughes D, Andersson D I. Carbon starvation of Salmonella typhimurium does not cause a general increase of mutation rates. J Bacteriol. 1997;179:6688–6691. doi: 10.1128/jb.179.21.6688-6691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kedderis G L, Argenbright L S, Miwa G T. Mechanism of reductive activation of a 5-nitroimidazole by flavoproteins: model studies with dithionite. Arch Biochem Biophys. 1988;262:40–48. doi: 10.1016/0003-9861(88)90166-x. [DOI] [PubMed] [Google Scholar]
- 29.Kidwell M G, Lisch D R. Transposable elements and host genome evolution. Trends Ecol Evol. 2000;15:95–99. doi: 10.1016/s0169-5347(99)01817-0. [DOI] [PubMed] [Google Scholar]
- 30.Kohler T, Michea-Hamzehpour M, Plesiat P, Kahr A L, Pechere J C. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korona R, Nakatsu C H, Forney L J, Lenski R E. Evidence for multiple adaptive peaks from populations of bacteria evolving in a structured habitat. Proc Natl Acad Sci USA. 1994;91:9037–9041. doi: 10.1073/pnas.91.19.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeClerc J E, Li B, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 33.Lederberg J, Lederberg E M. Replica plating and indirect selection of bacterial mutants. J Bacteriol. 1952;63:399–406. doi: 10.1128/jb.63.3.399-406.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenski R E. The cost of antibiotic resistance from the perspective of a bacterium. Ciba Found Symp. 1997;207:131–140. doi: 10.1002/9780470515358.ch9. [DOI] [PubMed] [Google Scholar]
- 35.Luria S E, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maenhaut Michel G, Shapiro J A. The roles of starvation and selective substrates in the emergence of araB-lacZ fusion clones. EMBO J. 1994;13:5229–5239. doi: 10.1002/j.1460-2075.1994.tb06854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mamber S W, Kolek B, Brookshire K W, Bonner D P, Fung-Tomc J. Activity of quinolones in the Ames Salmonella TA102 mutagenicity test and other bacterial genotoxicity assays. Antimicrob Agents Chemother. 1993;37:213–217. doi: 10.1128/aac.37.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markham P N. Inhibition of the emergence of ciprofloxacin resistance in Streptococcus pneumoniae by the multidrug efflux inhibitor reserpine. Antimicrob Agents Chemother. 1999;43:988–989. doi: 10.1128/aac.43.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markham P N, Neyfakh A A. Inhibition of the multidrug transporter NorA prevents emergence of norfloxacin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:2673–2674. doi: 10.1128/aac.40.11.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez J L, Alonso A, Gomez-Gomez J M, Baquero F. Quinolone resistance by mutations in chromosomal gyrase genes. Just the tip of the iceberg? J Antimicrob Chemother. 1998;42:683–688. doi: 10.1093/jac/42.6.683. [DOI] [PubMed] [Google Scholar]
- 41.Martinez J L, Cercenado E, Rodriguez-Creixems M, Vicente-Perez M F, Delgado-Iribarren A, Baquero F. Resistance to beta-lactam/clavulanate. Lancet. 1987;ii:1473. doi: 10.1016/s0140-6736(87)91180-9. [DOI] [PubMed] [Google Scholar]
- 42.Martinez J L, Martinez-Suarez J, Culebras E, Perez-Diaz J C, Baquero F. Antibiotic inactivating enzymes from a clinical isolate of Agrobacterium radiobacter. J Antimicrob Chemother. 1989;23:283–284. doi: 10.1093/jac/23.2.283. [DOI] [PubMed] [Google Scholar]
- 43.Martinez J L, Baquero F. Epidemiology of antibiotic-inactivating enzymes and DNA probes: the problem of quantity. J Antimicrob Chemother. 1990;26:301–303. doi: 10.1093/jac/26.3.301-a. [DOI] [PubMed] [Google Scholar]
- 44.Mass W K. Mutations to antibiotic resistance. In: Lorian V, editor. Antibiotics in laboratory and medicine. 2nd ed. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 669–682. [Google Scholar]
- 45.Masuda N, Gotoh N, Ishii C, Sakagawa E, Ohya S, Nishino T. Interplay between chromosomal β-lactamase and the MexAB-OprM efflux system in intrinsic resistance to β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:400–402. doi: 10.1128/aac.43.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- 47.Mereghetti L, Marquet-van der Mee N, Loulergue J, Rolland J C, Audurier A. Pseudomonas aeruginosa from cystic fibrosis patients: study using whole cell RAPD and antibiotic susceptibility. Pathol Biol. 1998;46:319–324. [PubMed] [Google Scholar]
- 48.Miller J H. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- 49.Mittenhuber G. Ocurrence of MazEF-like antitoxin/toxin systems in bacteria. J Mol Biotechnol. 1999;1:295–302. [PubMed] [Google Scholar]
- 50.Mittler J E, Lenski R E. New data on excisions of Mu from E. coli MCS2 cast doubt on directed mutation hypothesis. Nature. 1990;344:173–175. doi: 10.1038/344173a0. [DOI] [PubMed] [Google Scholar]
- 51.Morosini M I, Negri M C, Shoichet B, Baquero M R, Baquero F, Blazquez J. An extended-spectrum AmpC-type beta-lactamase obtained by in vitro antibiotic selection. FEMS Microbiol Lett. 1998;165:85–90. doi: 10.1111/j.1574-6968.1998.tb13131.x. [DOI] [PubMed] [Google Scholar]
- 52.Moxon E R, Rainey P B, Nowak M A, Lenski R E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 53.Munoz Bellido J L, Alonso Manzanares M A, Yague Guirao G, Gutierrez Zufiaurre M N, Toldos M C, Segovia Hernandez M, Garcia Rodriguez J A. In vitro activities of 13 fluoroquinolones against Staphylococcus aureus isolates with characterized mutations in gyrA, gyrB, grlA, and norA and against wild-type isolates. Antimicrob Agents Chemother. 1999;43:966–968. doi: 10.1128/aac.43.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura S, Nakamura M, Kojima T, Yoshida H. gyrA and gyrB mutations in quinolone-resistant strains of Escherichia coli. Antimicrob Agents Chemother. 1989;33:254–255. doi: 10.1128/aac.33.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nash K A, Inderlied C B. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob Agents Chemother. 1995;39:2625–2630. doi: 10.1128/aac.39.12.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 57.Phillips I, Culebras E, Moreno F, Baquero F. Induction of the SOS response by new 4-quinolones. J Antimicrob Chemother. 1987;20:631–638. doi: 10.1093/jac/20.5.631. [DOI] [PubMed] [Google Scholar]
- 58.Radman M. Enzymes of evolutionary change. Nature. 1999;401:866–887. doi: 10.1038/44738. [DOI] [PubMed] [Google Scholar]
- 59.Rainey P B, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 60.Reguera J A, Baquero F, Perez-Diaz J C, Martinez J L. Synergistic effect of dosage and bacterial inoculum in TEM-1 mediated antibiotic resistance. Eur J Clin Microbiol Infect Dis. 1988;7:778–779. doi: 10.1007/BF01975047. [DOI] [PubMed] [Google Scholar]
- 61.Ren L, Rahman M S, Humayun M Z. Escherichia coli cells exposed to streptomycin display a mutator phenotype. J Bacteriol. 1999;181:1043–1044. doi: 10.1128/jb.181.3.1043-1044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riesenfeld C, Everett M, Piddock L J, Hall B G. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob Agents Chemother. 1997;41:2059–2060. doi: 10.1128/aac.41.9.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenberg S M, Harris R S, Torkelson J. Molecular handles on adaptive mutation. Mol Microbiol. 1995;18:185–189. doi: 10.1111/j.1365-2958.1995.mmi_18020185.x. [DOI] [PubMed] [Google Scholar]
- 64.Rosenberg S M, Harris R S, Longerich S, Galloway A M. Recombination-dependent mutation in non-dividing cells. Mutat Res. 1996;350:69–76. doi: 10.1016/0027-5107(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 65.Sander P, Prammananan T, Meier A, Frischkorn K, Bottger E C. The role of ribosomal RNAs in macrolide resistance. Mol Microbiol. 1997;26:469–480. doi: 10.1046/j.1365-2958.1997.5811946.x. [DOI] [PubMed] [Google Scholar]
- 66.Schrag S J, Perrot V, Levin B R. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc R Soc London Ser B. 1997;264:1287–1291. doi: 10.1098/rspb.1997.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 68.Seifert H S. Questions about gonococcal pilus phase and antigenic variation. Mol Microbiol. 1996;21:433–440. doi: 10.1111/j.1365-2958.1996.tb02552.x. [DOI] [PubMed] [Google Scholar]
- 69.Shapiro J A. Observations on the formation of clones containing araB-lacZ cistron fusions. Mol Gen Genet. 1984;194:79–90. doi: 10.1007/BF00383501. [DOI] [PubMed] [Google Scholar]
- 70.Shapiro J A. Genome organization, natural genetic engineering and adaptive mutation. Trends Genet. 1997;13:98–104. doi: 10.1016/s0168-9525(97)01058-5. [DOI] [PubMed] [Google Scholar]
- 71.Sharp P M, Shields D C, Wolfe K H, Li W H. Chromosomal location and evolutionary rate variation in enterobacterial genes. Science. 1989;246:808–810. doi: 10.1126/science.2683084. [DOI] [PubMed] [Google Scholar]
- 72.Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon P H, Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 73.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics. 1999;152:485–493. doi: 10.1093/genetics/152.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Torres O R, Korman R Z, Zahler S A, Dunny G M. The conjugative transposon Tn925: enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in Bacillus subtilis and E. faecalis. Mol Gen Genet. 1991;225:395–400. doi: 10.1007/BF00261679. [DOI] [PubMed] [Google Scholar]
- 76.Willems R, Paul A, van der Heide H G, ter Avest A R, Mooi F R. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 1990;9:2803–2809. doi: 10.1002/j.1460-2075.1990.tb07468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolter J M, Kotsiou G, McCormack J G. Mixed morphotype testing of Pseudomonas aeruginosa cultures from cystic fibrosis patients. J Med Microbiol. 1995;42:220–224. doi: 10.1099/00222615-42-3-220. [DOI] [PubMed] [Google Scholar]