Abstract
Background
A better understanding of the tumor immune microenvironment (TIME) will facilitate the development of prognostic biomarkers and more effective therapeutic strategies in patients with lung cancer. However, little has been reported on the comprehensive evaluation of complex interactions among cancer cells, immune cells, and local immunosuppressive elements in the TIME.
Methods
Whole-exome sequencing and RNA sequencing were carried out on 113 lung cancers. We performed single sample gene set enrichment analysis on TIME-related gene sets to develop a new scoring system (TIME score), consisting of T-score (tumor proliferation), I-score (antitumor immunity) and S-score (immunosuppression). Lung cancers were classified according to a combination of high or low T-score, I-score, and S-scores (eight groups; G1-8). Clinical and genomic features, and immune landscape were investigated among eight groups. The external data sets of 990 lung cancers from The Cancer Genome Atlas and 76 melanomas treated with immune checkpoint inhibitors (ICI) were utilized to evaluate TIME scoring and explore prognostic and predictive accuracy.
Results
The representative histological type including adenocarcinoma and squamous cell carcinoma, and driver mutations such as epidermal growth factor receptor and TP53 mutations were different according to the T-score. The numbers of somatic mutations and predicted neoantigens were higher in Thi (G5-8) than Tlo (G1-4) tumors. Immune selection pressure against neoantigen expression occurred only in Thi and was dampened in Thi/Ilo (G5-6), possibly due to a reduced number of T cells with a high proportion of tumor specific but exhausted cells. Thi/Ilo/Shi (G5) displayed the lowest immune responses by additional immune suppressive mechanisms. The T-score, I-score and S-scores were independent prognostic factors, with survival curves well separated into eight groups with G5 displaying the worst overall survival, while the opposite group Tlo/Ihi/Slo (G4) had the best prognosis. Several oncogenic signaling pathways influenced on T-score and I-scores but not S-score, and PI3K pathway alteration correlated with poor prognosis in accordance with higher T-score and lower I-score. Moreover, the TIME score predicted the efficacy of ICI in patients with melanoma.
Conclusion
The TIME score capturing complex interactions among tumor proliferation, antitumor immunity and immunosuppression could be useful for prognostic predictions or selection of treatment strategies in patients with lung cancer.
Keywords: Tumor Microenvironment; Lung Neoplasms; Immunologic Surveillance; Antigens, Neoplasm
Background
Lung cancer continues to be a leading cause of cancer death worldwide. Despite tremendous advances in surgery, chemotherapy, radiation and targeted therapy, the survival rate is still less than 50% at 5 years. The recent development of immune checkpoint inhibitors (ICI) has revolutionized the treatment of lung cancers,1 but still only a few patients respond to monotherapy or combination therapy.2–4 It is difficult to predict these responders before treatment, a difficulty compounded by the complexity of the immune system precluding the use of a single biomarker. Nonetheless, tumor programmed death-ligand 1 (PD-L1) expression and tumor mutation burden (TMB) have recently been used as biomarkers for ICI.5 A better understanding of the tumor immune microenvironment (TIME) is required to facilitate the identification of prognostic and predictive biomarkers, and for developing more effective therapeutic strategies for patients with lung cancer.
Some time ago, an evaluation of immune system status in the tumor based on the concept of the ‘immune contexture’, was proposed by Galon et al. This is an assessment of immune cell types in the TIME, designated the ‘Immunoscore’6 which quantifies in situ T cell infiltrates and thus incorporates the effects of the host immune response. It has been internationally validated in colorectal cancer and provides better prognostic information than the current tumor-based tumor-node-metastasis (TNM) staging system.7 It may thus be beneficial for cancer classification and improving prognostic accuracy also in other cancer types. Taking this concept further, the ‘Immunogram’ approach proposed by Blank et al8 displays the immune status more comprehensively in each individual patient using radar plots. Immunogram scores based on RNA sequencing (RNA-seq) data and the application of single sample gene set enrichment analysis (ssGSEA) to qualify parameters related to antitumor immunity have been applied to various different cancers.9 10
The TIME commonly contains elements that may severely impair immune responses against the cancer cells. Tumor cells rapidly outgrow their blood supply, leading to hypoxia in the TIME, which in turn promotes immunosuppression.11 Cancer-associated fibroblasts (CAFs) also play a critical role in tumor initiation and progression via their potent immunosuppressive activity.12 The epithelial‐mesenchymal transition (EMT) was also shown to be associated with increased expression of diverse immune inhibitory factors and with an enrichment of immune suppressive cells.13 Overcoming these obstacles to elicit more effective antitumor immune responses is critical for achieving improved clinical outcomes.
In the present study, comprehensive RNA expression profiling of the dynamic interactions in the TIME were evaluated in individual patients with lung cancer. We designed a new scoring system for the TIME and classified patients based on tumor proliferation, antitumor immunity and immunosuppression. We assessed the clinical and genomic characteristics, and immunological features among the patients classified by the TIME score in our Aichi Cancer Center (ACC) cohort and validated its prognostic veracity in the lung cancer cohort in the The Cancer Genome Atlas (TCGA) data set. Using another external melanoma data set, we further investigated whether it could predict therapeutic efficacy of anti-programmed cell death protein-1 (PD-1) therapy.
Methods
Patients and data sets
We accessed a total of 113 tumor samples of patients with lung cancer who underwent surgery at ACC, Ichinomiya Nishi Hospital, Mie Prefectural General Medical Center, Mie-Chuo Medical Center, Anjo Kosei Hospital, Tosei Hospital, Mie University Graduate School of Medicine, and The Jikei University School of Medicine between April 2019 and July 2020. All procedures were in accordance with the ethical standards of the institutions and with the 1964 Helsinki declaration and its later amendments, or comparable ethical standards. Written informed consent was obtained from all individual participants included in the study.
TCGA data set for patients with lung cancer was accessed through the Genomic Data Commons data portal (http://www.cbioportal.org/) as of April 2019. We also utilized the cohorts of melanoma patients treated with ICIs (GSE78220, GSE91061).14 15
DNA and RNA extraction
Tumors and adjacent normal tissues were obtained immediately after surgical resection and stored in RNAlater RNA Stabilization Reagent (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Lymphoprep (Axis-Shield Poc AS, Oslo, Norway) and cryopreserved until use. Genomic DNA was extracted from tumor tissues and matched adjacent normal tissues or PBMCs using either AllPrep DNA/RNA Mini Kits or DNeasy Mini Kits (Qiagen, Hilden, Germany). Total RNA was extracted from tumor tissues using either AllPrep DNA/RNA Mini Kits or RNeasy Mini Kits (Qiagen).
Whole exome sequencing and RNA-seq
For whole exome sequencing (WES) (n=95), sequencing libraries of genomic DNA from tumors and matched normal tissues or PBMCs were prepared using the SureSelect Human All Exon V.6 probe (Agilent Technologies) following the manufacturer’s protocols. The enriched libraries were sequenced as 150 bp paired-end reads using the NovaSeq (Illumina, San Diego, California, USA) at Veritas Genetics (Danvers, Massachusetts, USA). The quality control check including the removal of adapter sequences and low quality reads was conducted using Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The mean coverage of all protein-coding sequences by WES was 116×. Exome reads were independently mapped to the human genome (GRCh38/hg38) using the Burrows-Wheeler Aligner (BWA). Putative somatic mutations were detected using the EBCall (Empirical Bayesian mutation Calling) algorithm.16
For RNA-seq (n=113), an RNA-seq library was prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, Massachusetts, USA) following the manufacturer’s protocols. The enriched libraries were sequenced as 150 bp paired-end reads using NovaSeq (Illumina) at Veritas Genetics. RNA-seq data were analyzed using Salmon17 and RNA reads were mapped to the reference genome (GRCh38/hg38) using BWA. Gene expression values were calculated as transcripts per million using GENCODE V.33.
TIME score
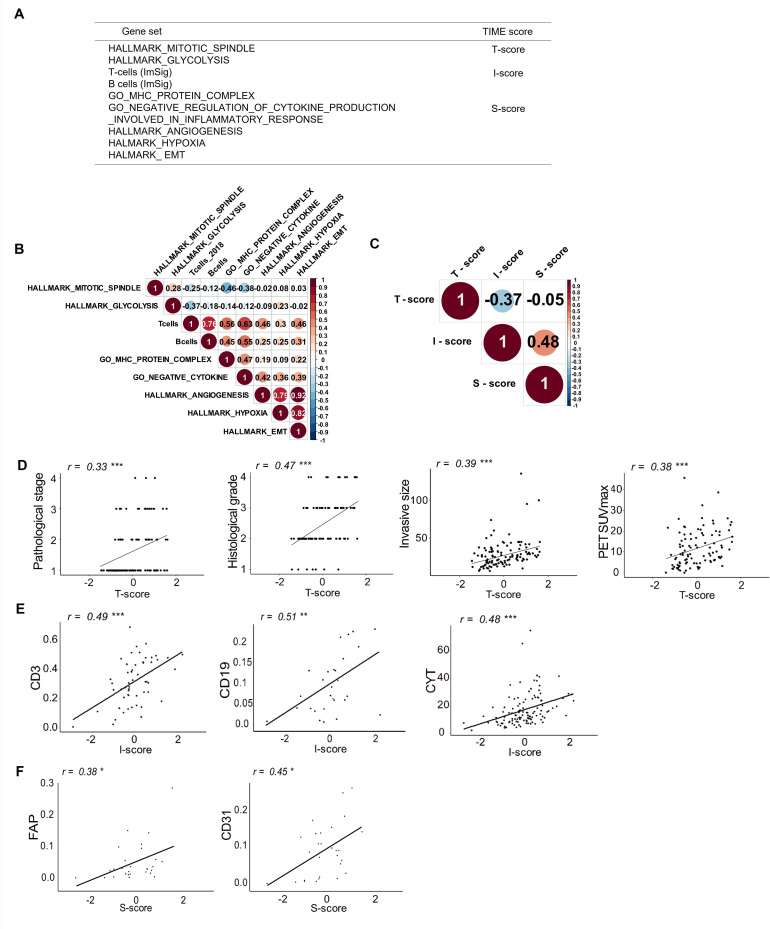
Based on the idea that the status of the TIME can be described by factors relevant to the tumor proliferation (T-factor), antitumor immunity (I-factor) and immunosuppression (S-factor),18–20 we selected nine representative gene sets related to those factors in the molecular signature database (MsigDB V.7.1) or gene sets previously verified and reported21 (figure 1A). In clinical practice, tumor proliferative activity is often evaluated by Fluorine-18-fluoro-2-deoxy-D-glucose (FDG)-positron emission tomography (PET) to visualize glucose metabolism, including aerobic glycolysis (also known as the Warburg effect) in proliferating tumor cells. FDG-PET is a reliable tool for the accurate diagnosis and the prediction of overall survival (OS) in patients with lung cancer. Cytotoxic chemotherapeutics targeting the apparatus of mitosis (eg, Taxanes or Vinca alkaloids) have been commonly used for patients with lung cancer. Therefore, we selected gene sets related to glycolysis/glucogenesis and cell division (mitosis) for T-score, rather than each specific pathway involved in cell proliferation such as p53, WNT or MAPK. For I-factor, we selected the gene sets related to the underpinnings of adaptive immunity including lymphocytes (CD8+ and CD4+ T cells and B cells) and MHC I and II peptide complex, rather than focusing on specific immune effector or suppressive cells. For S-factor, we selected several representative suppressive factors that have been previously reported in lung cancer.11–13 The individual RNA expression score for ssGSEA was calculated using the gene set variation analysis program22 on R package V.4.0.1. After z-score normalization of the ssGSEA score on each gene set, we calculated the mean values of z-scores of gene sets allocated to T-factors, I-factors or S-factors and scored as T-scores, I-scores or S-scores (figure 1A and online supplemental figure S1A). The median values of each score were used as cut-off points to categorize high or low T-scores, I-scores, and S-scores in each cohort.
Figure 1.
TIME scoring using RNA sequencing data and associations with clinical features and experimental data in patients with lung cancer. (A) Gene sets and TIME scores used in this study. (B) Correlogram showing correlations between each gene signature of the TIME factors in the Aichi Cancer Center (ACC) cohort. The size of the circles indicates the degree of correlation. (C) Correlogram showing correlations between each TIME score (T-score, I-score and S-score) in the ACC cohort. Association of clinicopathological findings or experimental data with T-score (D), I-score (E) and S-score (F) are shown. CYT, cytolytic activity; epithelial‐mesenchymal transition; FAP, fibroblast activation protein; PET, positron emission tomography; SUVmax, maximum standardized uptake value; TCGA, The Cancer Genome Atlas; TIME, tumor immune microenvironment. *p<0.05; **p<0.01; ***p<0.001.
jitc-2021-003765supp002.pdf (5.2MB, pdf)
Cell-type quantification and deconvolution
Quantification and deconvolution of different cell types of the TIME from RNA-seq data were done by xCell (http://xcell.ucsf.edu),23 quanTIseq (https//icbi-med.ac.at/quantiseq)24 and CIBERSORT (https//cibersort.standord.edu).25
HLA typing and HLA class I epitope binding prediction
HLA types were determined from the WES data of normal genomic DNA using the HLA typing software Omixon target HLA (Omixon). Predicted candidate neoantigens derived from missense mutations on individual HLA alleles are estimated using Neoantimon (https://github.com/hase62/Neoantimon).26 We counted the number of neoantigen peptides with IC50 value <500 nmol/L and corresponding wild-type peptide with >500 nmol/L as the predicted neoepitope. We defined a missense mutation capable of generating one or more HLA-A, HLA-B, or HLA-C–restricted neoepitopes as a predicted neoantigen (p-neoAg). In addition, we evaluated the RNA expression data using bam-readcount (https://github.com/genome/bam-readcount) and calculated the number of expressed neoantigen epitopes (e-neoAgs) in p-neoAgs as previously reported.27
Flow cytometry
Fresh tumor digests (FTD) from tumor tissues were prepared using the gentleMACS tumor dissociator (Miltenyi Biotec, Auburn, California, USA), according to the manufacturer’s instructions, and were cryopreserved until use. Cryopreserved FTD were thawed in Roswell Park Memorial Institute (RPMI) 1640 mediumand then stained after blocking Fc receptors using Human BD Fc Block (BD BioSciences, San Jose, California, USA). The following monoclonal antibodies were used: Brilliant Violet (BV)421-labeled CD3, Allophycocyanin (APC)-labeled CD4, Fluorescein isothiocyanate (FITC)-labeled CD8, BV711-labeled CD103, Phycoerythrin(PE)-labeled CD39, BV786-labeled PD-1, BV650-labeled Tim-3 and PE-labeled CD31 (all from BioLegend, San Diego, California, USA), and APC-labeled fibroblast activation protein (FAP) (R&D Systems, Minneapolis, Minnesota, USA). SYTOX AADvanced Dead Cell Stain Kit (Thermo Fisher, Waltham, Massachusetts, USA) or Zombie NIR Fixable Viability Kit (BioLegend) was used to exclude dead cells. Stained cells were analyzed using an LSRFortessa flow cytometer (BD BioSciences) and the data processed using FlowJo V.10.0.7 (FlowJo, LLC, Ashland, Oregon, USA). Gating strategies in this study are shown in online supplemental figure S2.
Tumor-infiltrating lymphocyte expansion and ELISA
Surgically resected tumors were cut into pieces and cultured in RPMI 1640 medium containing 100 U/mL penicillin, 100 µg/mL streptomycin, 2 mmol/l-glutamine and 10% fetal calf serum along with recombinant human interleukin (IL)-2 (6000 IU/mL) (PeproTech, Cranbury, New Jersey, USA) in 6–12 wells of 24-well plates to expand tumor-infiltrating lymphocytes (TILs). Half of the medium with IL-2 was replaced every 3–4 days for 2 weeks. Expanded TILs (1×105) from each well were co-cultured with FTD (1×105) in 96-well plates overnight, after which interferon (IFN)-γ production was measured using human IFN-γ ELISA kits (Thermo Fisher) according to the manufacturer’s instructions.
Statistical analysis
All clinical data were analyzed using R V.4.0.1 (R Development Core Team, Vienna, Austria). Clinical data were analyzed using Fisher’s exact test or χ2 test for categorical variables and Mann-Whitney U test, Kruskal-Wallis test or Welch t-test for continuous variables. Correlations were calculated using Spearman rank correlation testing. OS was evaluated using the Kaplan-Meier method by the log-rank test. The Cox proportional hazards model was used for multivariate analysis. The area under curve for predicting ICI responses in melanoma cohort was calculated using nine gene sets as mentioned above. We regarded a probability level of 0.05 as statistically significant.
Results
Patients’ characteristics
In total, 113 cases of lung cancer were analyzed in this study. The clinicopathological characteristics of these patients are summarized in table 1.
Table 1.
Clinicopathological features of 113 patients with lung cancer
| Characteristic | N=113 |
| Age | 71 (64, 76) |
| Sex | |
| Female | 41 (36%) |
| Male | 72 (64%) |
| Smoking index (pack-year) | 30 (3, 48) |
| Histology | |
| Adenocarcinoma | 76 (67%) |
| Squamous cell carcinoma | 23 (20%) |
| Others | 14 (13%) |
| Histological grade | |
| 1 | 7 (6.8%) |
| 2 | 55 (53%) |
| 3 | 29 (28%) |
| 4 | 12 (12%) |
| Unknown | 10 |
| Invasive tumor size (mm) | 26 (18, 34) |
| T factor | |
| 1 | 52 (46%) |
| 2 | 51 (45%) |
| 3 | 6 (5.3%) |
| 4 | 4 (3.5%) |
| N factor | |
| 0 | 81 (72%) |
| 1 | 14 (12%) |
| 2 | 14 (12%) |
| X | 3 (2.7%) |
| M factor | |
| 0 | 110 (97.3%) |
| 1 | 3 (2.7%) |
| Pathological stage | |
| I | 71 (63%) |
| II | 23 (20%) |
| III | 16 (14%) |
| IV | 3 (2.7%) |
| Median (IQR); n (%) |
TIME scoring and association with clinical features and flow cytometry and gene expression data
To evaluate complex interactions among cancer cells, immune cells, and local immunosuppressive elements in the TIME, we tried to develop a new scoring system based on the three major factors defined here as tumor proliferation, antitumor immunity and immunosuppression. We focused on nine gene sets related to those TIME factors as shown in figure 1A and online supplemental table S1). We performed ssGSEA using bulk RNA-seq data from the 113 lung tumor samples and enrichment scores were z-score normalized. We allocated z-scores of gene sets related to mitotic spindle and glycolysis to tumor factors (T-score). T cells and B cells,21 as well as MHC protein complexes were allocated to antitumor immune factors (I-score). Finally, inhibition of inflammatory cytokine production, angiogenesis, hypoxia, and EMT were allocated to immunosuppressive factors (S-score) (figure 1A). All data for the process of TIME scoring are shown in online supplemental table S2). The correlation coefficient of each ssGSEA score for the gene sets and T-scores, I-scores and S-scores were calculated as shown in figure 1B and C. The score for T-cells was found to be correlated with the B-cell score and the GO_MHC_PROTEIN_COMPLEX in the I-score (Spearman’s rank correlation coefficient, r=0.76 and 0.56, respectively, both p<0.001, figure 1B). The scores for HALLMARK_ANGIOGENESIS, HALLMARK_HYPOXIA and HALLMARK_EMT were strongly correlated with each other in the S-score (r=0.79, 0.92, and 0.82; p<0.001; figure 1B). The score for GO_NEGATIVE_REGULATION_OF_CYTOKINE_PRODUCTION_INVOLVED_IN_INFLAMMATORY_RESPONSE in the S-score was moderately correlated with the scores for T cells, B cells and GO_MHC_PROTEIN_COMPLEX in the I-score (r=0.63, 0.55 and 0.47, respectively; p<0.001, figure 1B). T-scores and I-scores were negatively correlated (Spearman’s rank correlation coefficient, r=−0.37, p<0.001), whereas I-scores and S-scores were positively correlated (r=0.48, p<0.001, figure 1C).
jitc-2021-003765supp001.xlsx (54.5KB, xlsx)
We next investigated associations between each TIME score and clinicopathological features or flow cytometry and gene-expression data. The T-score was positively correlated with pathological stage, histological grade, invasive lesion size, and the maximum standardized uptake value on FDG-PET (Spearman’s rank correlation coefficient, r=0.33, 0.47, 0.39 and 0.38, respectively; all p<0.001; figure 1D). The I-score was correlated with the proportions of CD3+ cells and CD19+ cells in FTD from tumor tissues detected by flow cytometry (r=0.49 and 0.51; p<0.001, respectively; figure 1E and online supplemental figure S2). The I-score was also correlated with cytolytic activity (CYT) as estimated from the levels of expression of messenger RNA for GZMA and PRF1, which are associated with cytotoxic T cell infiltration and activity28 (r=0.48; p<0.001; figure 1E). The correlations between I-score and different immune cell scores calculated by cell-type deconvolution and quantification pipelines29 were shown in online supplemental figure S3. Finally, the S-score was correlated with the frequency of cells positive for FAP, which is associated with CAFs, and for CD31+ endothelial cells by flow cytometry (r=0.38 and 0.45; p=0.024 and 0.012, respectively; figure 1F). The S-score was also correlated with the abundance of M2 macrophages, stromal and endothelial cells inferred from xCell,23 and the expression of anti-inflammatory cytokine genes (TGFB1 and IL-10) and immune checkpoint molecule genes (CTLA4, PDCD1 (PD-1), HAVCR2 (Tim-3) and LAG3) (online supplemental figure S4). These data suggest that T-scores, I-scores, and S-scores based on TIME are consistent with experimental data and are likely to reflect clinical features in patients with lung cancer.
We also tested another combination of similar gene sets from gene ontology instead of hallmark regarding T-scores or S-scores. However, those scores were equal to or lower in terms of correlation coefficients with clinical features or flow cytometry data (online supplemental figure S1B, C), thus we continued to use gene sets described in figure 1A hereafter.
Distribution of representative histology and driver mutations according to TIME scores
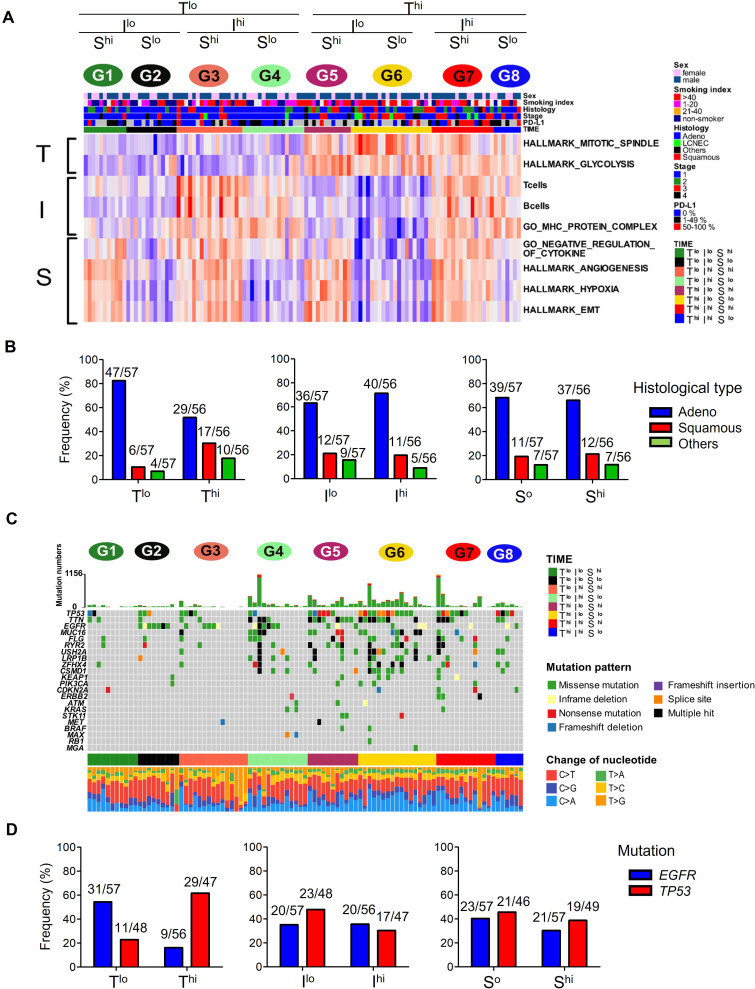
To investigate dynamic interactions among T-factors, I-factors, and S-factors, the 113 patients with lung cancer were stratified according to a combination of high or low of T-score (T), I-score (I), and S-score (S). For this, patients were divided into eight groups as follows: Group 1 (G1). Tlo/Ilo/Shi, Group 2 (G2). Tlo/Ilo/Slo, Group 3 (G3). Tlo/Ihi/Shi, and Group 4 (G4). Tlo/Ihi/Slo, Group 5 (G5). Thi/Ilo/Shi, Group 6 (G6). Thi/Ilo/Slo, Group 7 (G7). Thi/Ihi/Shi, Group 8 (G8). Thi/Ihi/Slo (online supplemental tables S2 and S3). The heatmaps of each gene set normalized by z-score and clinical data in all eight groups are shown in figure 2A. Immune cell composition and immune-related molecule gene expression in eight groups were also shown in online supplemental figure S5.
Figure 2.
Histological and mutational analyses and the TIME score. (A) Heatmap of each gene set normalized by z-score of 113 lung cancers in eight groups (G1-8) stratified according to high or low of T-scores, I-scores and S-scores. Top bars show sex, smoking status, histology, stage, and PD-L1 TPS. (B) The prevalence of lung cancer histological type according to T-score, I-score and S-score. (C) Representative driver gene mutations and types of mutations present in the eight groups. (D) The frequencies of representative driver gene mutations (EGFR and TP53 mutations) according to T-scores, I-scores and S-scores. EGFR mutations were evaluated by both WES and target sequencing data (113 cases), and TP53 mutations were by WES only (95 cases). EMT, epithelial‐mesenchymal transition; EGFR, epidermal growth factor receptor; PD-L1, programmed death-ligand 1; TIME, tumor immune microenvironment; TPS, Tumor Proportion Score; WES, whole exome sequencing.
We first examined associations between each TIME score and the typical histological type of the cancer. Adenocarcinomas were present at a higher rate in G1-4 (Tlo; 47/57, 82.5%) than in G5-8 (Thi; 29/56, 51.8%) (χ2 test, p<0.01; figure 2B left). In contrast, there were more squamous cell carcinomas in the Thi groups (17/56, 30.4%) than in the Tlo groups (6/57, 10.5%) (p<0.05). However, there were no significant differences in the frequencies of these histotypes according to their Ilo and Ihi or Slo and Shi status (figure 2B middle and right). Next, recurrent driver mutations were investigated, based on the WES data (figure 2C) and target sequencing data. Epidermal growth factor receptor (EGFR) mutations were present in the Tlo groups more frequently than in the Thi groups (31/57, 54.4%-versus-9/56, 16.1%; p<0.01; figure 2D left). On the other hand, TP53 mutations were more frequent in the Thi than Tlo groups (29/47, 61.7%-versus-11/48, 22.9%, p<0.01). However, again, there were no differences in the frequencies of mutations in either EGFR or TP53 between Ilo and Ihi or between Slo and Shi groups (figure 2D middle and right). Accordingly, the T-score was significantly lower in EGFR mutant lung tumors compared with those with wild-type EGFR (online supplemental figure S6A), but was significantly higher in tumors with TP53 mutations, and the I-score was significantly lower in tumors with TP53 mutations (online supplemental figure S6B).
Single nucleotide variants, neoantigens and immune selection pressure
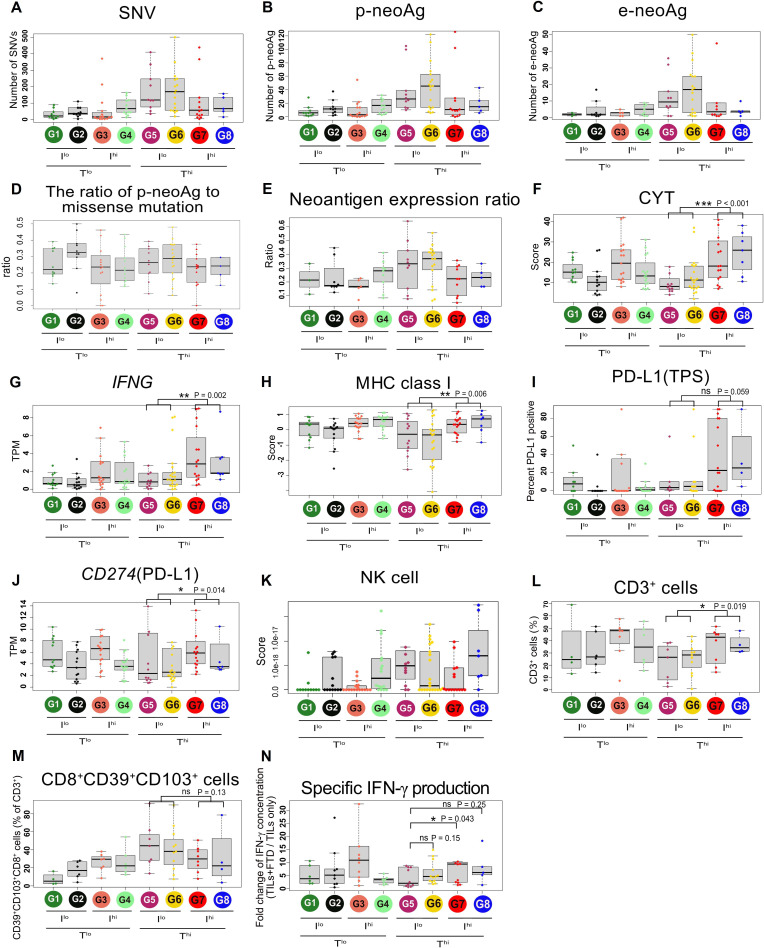
Mutation-derived neoantigens are thought to be key targets for antitumor immune responses and immunoediting.30 31 Therefore, we next investigated the numbers of single nucleotide variants (SNVs), p-neoAgs and e-neoAgs in the eight groups (figure 3A–C). The numbers of SNVs and p-neoAgs were higher in the Thi than Tlo groups (Welch’s t-test, p=0.010 and 0.036, respectively; online supplemental figure S7A, B). However, there were no significant differences in the numbers of e-neoAgs between the two groups (p=0.23; Online supplemental figure S7C). Immune selection pressure may eliminate tumor subclones that e-neoAgs or affect the expression of neoantigens thorough the suppression of transcripts that contain neoantigens.32 It has been shown that certain immunogenic cancers exhibit preferential depletion of neoantigenic mutations within the totality of mutations in the tumor.28 33 34 To test whether the same may apply in the subgroup of lung cancer, we compared the ratio of observed to expected neoantigen28 and the ratio of neoantigen per somatic mutation as previously reported.34 However, there were no significant differences among the groups (online supplemental figure S8A-D and figure 3D). Next, we compared the ratio of e-neoAgs to p-neoAgs which we designated the ‘neoantigen expression ratio’ as previously reported27 (figure 3E). Although this ratio was modestly higher in the Thi than Tlo group (p=0.036; online supplemental figure S8E), additional incorporation of the I-score status in this analysis revealed a highly significantly lower ratio in the Thi/Ihi group (G7,8) than in the Thi/Ilo group (G5,6) (p=0.007; online supplemental figure S8F). In contrast, this ratio was not lower in the Tlo/Ihi (G3,4) than in the Tlo/Ilo (G1,2) group (p=0.89). These results suggest that immune selection pressure against neoantigen messenger RNA (mRNA) expression is likely to occur only in the Thi group (G5-8) with high neoantigen loads, and that it might be dampened in the Thi/Ilo group (G5,6).
Figure 3.
Neoantigens, immune responses and immunoediting. The number of SNVs (A), predicted neoantigens (p-neoAgs) (B), and expressed neoantigens (e-neoAgs) (C), the ratio of p-neoAg to missense mutation (D), and the neoantigen expression ratio (e-neoAg/p-neoAg) (E) in the eight groups. (F) and G) Cytolytic activity score (CYT) and IFN-γ TPM. (H) MHC class I score calculated based on HLA-A, HLA-B, and HLA-C TPM. PD-L1 TPS (I) and CD274 (PD-L1) TPM (J) in the eight groups. NK cell score inferred from xCell (K). Percentages of CD3+ (L) and CD39+CD103+CD8+ cells (M) in FTD detected by flow cytometry. (N) Fold-change of IFN-γ concentration (TILs plus FTD/TILs only). IFN, interferon; FTD, fresh tumor digests; NK, natural killer; ns, not significant; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; SNVs, single nucleotide variants; TIL, tumor-infiltrating lymphocytes; TPM, transcripts per million; TPS, Tumor Proportion Score. *p<0.05; **p<0.01; ***p<0.001.
Influence of TILs and immune response on the TIME score
We particularly focused on the Thi (G5-8) groups where immune selection pressure against neoantigens was likely to have been strongest. The CYT score and IFN-γ mRNA were lower in the Thi/Ilo groups (G5,6) compared with Thi/Ihi (G7,8) (Mann-Whitney U test, p<0.001 and 0.002, respectively; figure 3F and G and online supplemental figure S8G, H). The patterns of expression of MHC class I and PD-L1, which are induced by IFN-γ, were similar to IFN-γ expression (figure 3H–J). The inverse correlation of MHC class I expression and natural killer cell score as seen in ‘missing self’ recognition was not observed (figure 3K). Furthermore, tumor specific, but exhausted CD8+ T cells expressing CD39 and CD103 cell surface markers35 tended to be higher in Thi/Ilo (G5,6), although the frequency of CD3+ cells is rather low in these groups (figure 3L and M and online supplemental figure S8I, J). We next investigated IFN-γ production from cultured TILs challenged with autologous tumor cells (online supplemental figure S9). The concentration of IFN-γ in the Ilo groups (G5,6) tended to be lower than in the Ihi groups (G7,8), but this difference was not statistically significant (p=0.15; online supplemental figure S8K. Thi/Ilo/Shi (G5) exhibited the lowest IFN-γ production among the Thi groups (Mann-Whitney U test, vs G6, G7, G8, p=0.15, 0.043 and 0.25, respectively; figure 3N). Taken together, limited numbers and possible exhaustion of TILs as well as decreased tumor MHC class I expression might limit immune selection against neoantigens in the Thi/Ilo G5 and G6 groups. The lowest immune response in the Thi/Ilo/Shi G5 group might be attributable to additional immunosuppressive mechanisms.
TIME scores of lung cancers in the TCGA cohort and their association with survival
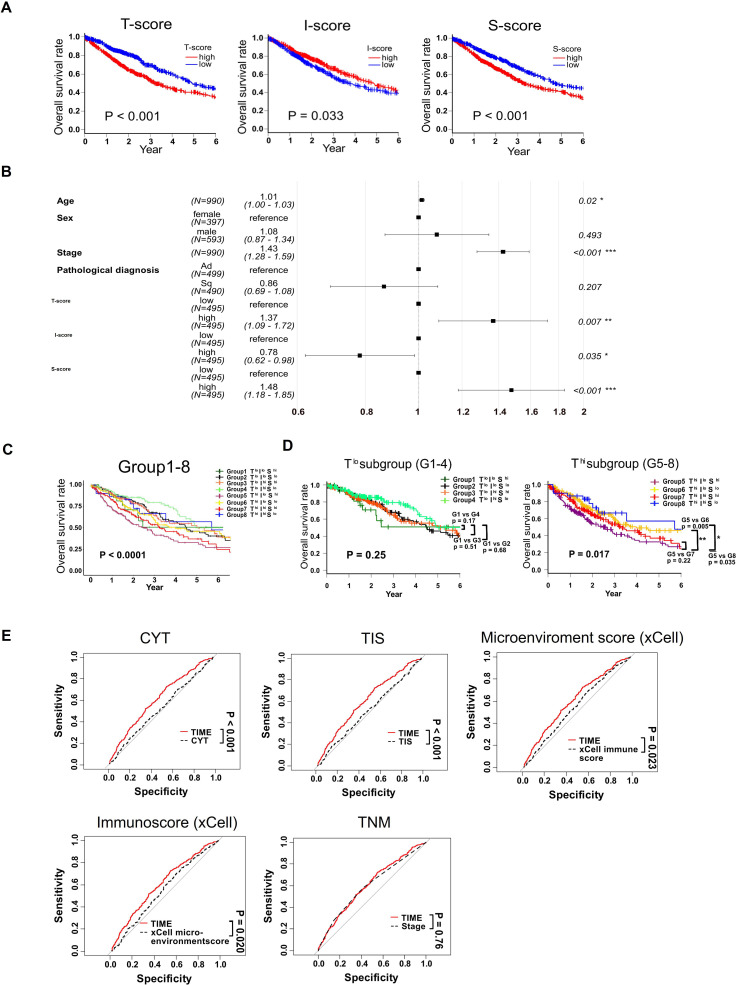
We next examined the prognostic impact of the TIME score. Because our survival follow-up (median 1.5 years) was too short to evaluate prognosis for patients predominantly with stage I and II disease in our own cohort (83%, table 1), we utilized the TCGA cohort with long-term follow-up data from 990 patients with lung cancer regardless of differences in some clinical features such as histology (online supplemental table S4). We performed the same analyses as for the ACC cohort (online supplemental figure S10). As shown in figure 4A, OS indicated that the T-score, I-score, and S-score were significant prognostic factors (Thi: HR 1.62 (1.33–1.97); p<0.001, Ihi: HR 0.81 (0.66–0.98); p=0.033, Shi: HR 1.45 (1.19–1.77); p<0.001). In addition, the Cox proportional hazards model adjusted by age, sex, pathological stage, and histology indicated that Thi and Shi were independent unfavorable prognostic factors and that Ihi was an independent favorable prognostic factor (Cox proportional hazards model, T-score: HR, 1.37 (1.09–1.72); p=0.007, I-score: HR, 0.78 (0.62–0.98); p=0.035, S-score: HR, 1.48 (1.18–1.85); p<0.001; figure 4B). OS was clearly different in the eight groups (log-rank test, p<0.0001; figure 4C) whereby the Thi/Ilo/Shi G5 group had the worst prognosis, while the inverse group Tlo/Ihi/Slo (G4) had the best prognosis.
Figure 4.
Prognostic impact of the TIME score in TCGA cohort. (A) Kaplan-Meier curves for OS according to high or low T-scores, I-scores and S-scores. (B) Forrest plot to evaluate OS in TCGA lung cancer using multivariate Cox hazard proportional model analysis. (C) Kaplan-Meier curves of OS. (D) Kaplan-Meier curves of OS in Tlo (G1-4) and Thi (G5-8) groups separately. (E) The ROC of prediction for OS of TIME score is shown comparing to CYT, cytolytic activity; TIS, Tumor Inflammatory Score, microenvironment score (xCell), immunoscore (xCell) and TNM classification. *p<0.05; **p<0.01; ***p<0.001. IFN, interferon; ns, not significant; OS, overall survival; ROC, receiver operator characteristic; TCGA, The Cancer Genome Atlas; TIME, tumor immune microenvironment; TNM, tumor, node, metastasis.
Because the level of immune pressure was different between Tlo (G1-4) and Thi (G5-8), we next performed subgroup analyses and found that OS was similar among all Tlo groups (log-rank test, p=0.25). However, there was a statistically significant difference in the Thi groups (p=0.017, figure 4D), largely due to the S-score rather than the I-score (online supplemental figure S11). Thus, G5 (Thi/Ilo/Shi) patients had a poor prognosis compared with G6 (Thi/Ilo/Slo) or G8 (Thi/Ihi/Slo) (p=0.005 and p=0.035, respectively), but not to G7 (Thi/Ihi/Shi) (p=0.22). These results suggest that immunosuppression is more likely to affect the prognosis of patients in all Thi groups (G5-8).
Finally, we tested the performance of TIME score comparing with other previously reported prognostic scores or factors. As shown in figure 4E, the receiver operator characteristic (ROC) of prediction for OS of TIME score was better than those of CYT, TIS (Tumor Inflammatory Score), microenvironment score (xCell) or immunoscore (xCell), but comparable and not superior to that of traditional TNM classification.
Oncogenic signaling pathway, TIME score, and prognosis in lung cancers
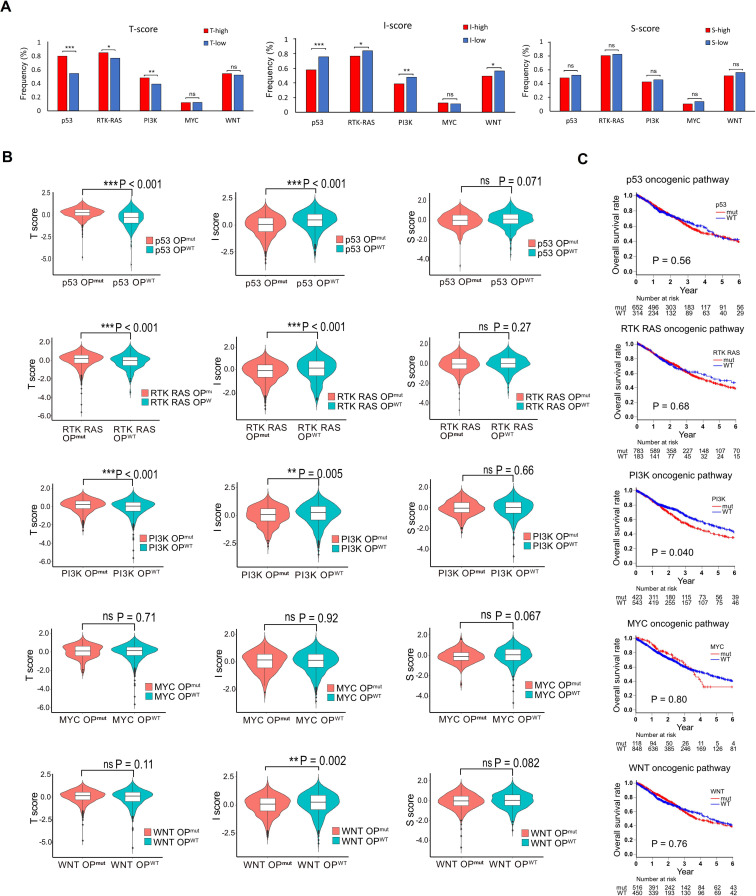
It has been known that common drivers of tumorigenesis modulate the tumor immune milieu.36 37 Therefore, we evaluated the alterations in oncogenic signaling pathway according to the Sanchez-Vega’s methods38 and investigated the correlation with T-scores, I-scores and S-scores in lung cancers. As shown in figure 5A, higher mutational rates in genes involved in oncogenic signaling pathways were observed in Thi group (p53, RTK-RAS, and PI3K) compared with Tlo group. In contrast, the higher mutational rate was observed in Ilo group (p53, RTK-RAS, PI3K and WNT). There were no differences between high and low of S-score. Accordingly, T-score was significantly higher, and I-score was significantly lower in the groups with those alterations (figure 5B and online supplemental figure S12). We then investigated the impact of pathway alterations on prognosis in lung cancer patients. Patients with PIK3 pathway altered tumors alone displayed poor prognosis (figure 5C). These results reveal that PI3K pathway alteration correlates with poor prognosis of patients with lung cancer in accordance with higher T-score and lower I-score.
Figure 5.
Oncogenic pathway, tumor immune microenvironment score and prognosis in lung cancers. (A) The impact of oncogenic signaling pathways on T-scores, I-scores and S-scores in lung cancers are examined. The fraction of samples with at least one alteration detected in five signaling pathways are shown in low or high T-scores, I-scores and S-scores. (B) and C), T-scores, I-scores and S-scores (B) and overall survival (C) are compared between the cases with and without signaling pathway alterations. *p<0.05; **p<0.01; ***p<0.001; ns, not significant.
The possibility of TIME score to predict the response to ICI
We attempted to test whether the TIME score predicts responses to ICI, but an appropriate cohort of patients with lung cancer treated with ICI was not available. Because TIME scoring in various different cancers revealed that lung cancer and melanoma had similar patterns of T-scores, I-scores and S-scores (online supplemental figure S13) and analogous prognoses based thereon (online supplemental figure S14). Thus, despite of totally different entities from lung cancer, we utilized the data set of 76 patients with melanoma treated with ICI (GSE78220, GSE91061). A heatmap of 76 cases is shown in online supplemental figure S15A. There was no significant difference in OS (log-rank test, p=0.12; online supplemental figure S15B), although the disease control rate tended to be lower in the Thi/Ilo/Shi group (G5) than in G4, G6 or G7 (3/10 vs 8/13, 8/12, or 7/12; Fisher’s exact test, p=0.21, 0.19 and 0.23, respectively; online supplemental figure S15C). Finally, we investigated whether the TIME score predicts objective response rates (ORRs). We generated ROC curves for TIME scores, (TIS; reported to be a predictive biomarker for response to ICI treatment39 40), or for CD274 (PD-L1) expression. The resulting area under the curve (AUC) of TIME score tended to be higher than that of TIS (0.71 vs 0.58, p=0.14) or CD274 expression (0.71 vs 0.52, p=0.073) (online supplemental figure S15D). Kaplan-Meier survival curves for the two groups (high vs low score using optimal cut-off level for the predictivity of the ORR) revealed that survival was superior for patients in the TIME high score group (log-rank test, p=0.015), but that the TIS or PD-L1 expression were not informative (log-rank test, p=0.29 and p=0.94, respectively; online supplemental figure S15E).
Discussion
To achieve a better understanding of the TIME in lung cancer, here we establish a new model to evaluate the TIME and its association with prognosis. Components of the TIME were assessed in three categories, namely tumor proliferation, antitumor immunity and immunosuppression, dichotomized into high and low as T-scores, I-scores and S-scores, respectively. After verifying these scores against the clinicopathological findings and experimental data, we showed that the histological type and driver mutations in lung cancers were different in the different categories of T-score, but were not influenced by I-scores or S-scores. Thus, adenocarcinomas and tumors with EGFR mutations were largely clustered in Tlo groups (G1-4), while squamous cell carcinoma and cancers with TP53 mutations were predominantly found in the Thi groups (G5-8). Given that the T-score representing gene activity relevant to cell division and metabolism, it might be that adenocarcinoma and EGFR-mutant tumors tend to be more indolent, while squamous cell carcinoma and TP53-mutated tumors possess more aggressive properties.
The TMB is generally high in lung cancer and melanoma, dues to exposure to carcinogens or ultraviolet radiation.41 These cancers are also known to be more sensitive to ICI than many other solid tumors.42 Neoantigens derived from tumor-specific mutations are now acknowledged as major targets of antitumor immune responses.43–45 Thus, neoantigen immune selection occurs in the process of tumor escape27 31 32 or in developing ICI resistance.46 Decreased neoantigen expression by promotor methylation under immune selection pressure has been reported as a mechanism of escape in lung cancer evolution.32 In our study, the numbers of SNVs and predicted neoantigens were higher in the Thi groups and neoantigen immune selection appeared to occur in Thi/Ihi groups (G7,8), but not Tlo/Ihi groups (G3,4). These findings suggest that tumor-specific T cell responses against abundant neoantigens may be induced in patients in the Thi groups, thus resulting in neoantigen immunoselection. On the other hand, immune responses may not be sufficiently powerful to elicit immunoselection in the Tlo groups with a low neoantigen load.
We investigated the power of our method of assessing the TIME on prognosis using the TCGA cohort over a follow-up period of up to 7 years and found that T-scorehi, I-scorelo and S-scorehi were all independent unfavorable prognostic factors. T-scorehi might contribute to poor prognosis not only because of accelerated cell division and glycolysis in tumor cells but also increased immunosuppression due to the hyperactivation of the cell cycle program.47 Furthermore, specific driver mutations not only exert on intrinsic influence on the fate of tumor cells but can have profound effects on host immune system.36 37 These notions are supported by our data that the I-score was negatively correlated with the T-score. Immune cells contributing to the I-score represent adaptive immune cells, both T cells and B cells. A strong T cell infiltration has been reported to be associated with good clinical outcome in many different tumor types including lung cancer.48 In addition, it has been reported that the presence of B cells in the tumor is also associated with better immune responses and clinical outcome in lung cancer.49 Expression of HLA and β2-microglobulin as well as tumor antigens are critical components to induce adaptive immune responses; thus, their loss leads to tumor escape from immunity.27 50 51 The S-score is constituted by factors that include inflammatory cytokine inhibition, angiogenesis, hypoxia, and EMT, which are known to suppress antitumor immunity and are related to poor prognosis.11 39 52 Thus, we analyzed OS by combining high or low of T-score, I-scores and S-scores, resulting in stratification of the patients into eight groups. Of these, the Thi/Ilo/Shi group (G5) showed the worst prognosis.
There are many therapies currently approved for patients with lung cancer, including agents that target oncogenic driver mutations, as well as ICI that enhance antitumor responses. Briefly, molecular targeting therapies such as tyrosine kinase inhibitors will usually be selected as first-line therapy in patients with druggable mutations as seen in the Tlo (G1-4) groups. On the other hand, patients without such druggable mutations but harboring abundant mutations in the Thi (G5-8) groups may be more likely to respond to immune-modulatory therapies. Recently, ICI therapy has resulted in survival benefits especially for patients whose tumors have a high TMB, or PD-L1 expression.5 However, the predictive capacity of either of these markers is low under most circumstances. As was the case with patients in the G5 group (Thi/Ilo/Shi) in this study, immune responses were clearly hampered by multiple mechanisms, despite sufficient numbers of neoantigens available to be targeted by the immune system. In fact, we showed that patients with melanomas in group G5 were resistant to ICI. Therefore, to amplify the I-factor, strategies such as adoptive T cell transfer therapy or cancer vaccination to change ‘cold’ into ‘hot’ tumors by replenishing or enhancing T cells in the tumor53 54 might be needed before subsequent ICI could effectively enhance T cell function.55 To regulate high T-factors in this group, a combination of direct cytotoxic chemotherapy or treatment with inhibitors of cyclin-dependent kinases56 may be required to control aggressive tumor cells. We revealed that tumors with PI3K pathway alteration in accordance with higher T-score and lower I-score were associated with poor prognosis. Therefore, PI3K inhibitor might improve clinical outcome of patients with lung cancer by enhancing antitumor immune responses as well as regulating tumor cell proliferation.57 Additionally, anti-angiogenic agents58 or agents targeting tumor-associated fibroblasts59 might also be required to reduce S-factor parameters to enhance the induction of effective immune responses. The same is true for the other seven groups; optimized drug combinations and personalized therapies based on each individual TIME will be needed to offer the best probability of prolonged survival.
There are several limitations to this study. First, this scoring model does not cover all the hallmarks of cancer,60 and may thus miss some aspects of the TIME in lung cancer. Second, because our survival data were too immature to assess early-stage lung cancers, we used the TCGA cohort to evaluate OS despite the presence of some features different from our cohort. Third, we utilized the melanoma cohort for prediction of ICI response because the data sets of both clinical information and RNA-seq from patients with ICI-treated lung cancer were not available. Fourth, the TIME score was evaluated using a single tumor region and thus tumor heterogeneity within the same patients32 was not considered in this study.
Despite these limitations, we developed a new evaluation model based on TIME factors associated with prognosis in lung cancer. Because bulk WES and RNA-seq data, as used in this study, can be generated from small biopsy samples, this scoring system may be useful for evaluating longitudinal changes in the TIME of individual patients and tumors for further prognostic predictions or selection of treatment strategies in patients with lung cancer.
Acknowledgments
We are grateful to all the patients who participated in this study. We acknowledge the excellent technical assistance of Seiko Shibata and Masami Iwano for the preparation of tumor tissue specimens and Yuki Abe for assisting with the analysis of patient information. The super-computing resource was provided by Human Genome Center, the Univ. of Tokyo (http://sc.hgc.jp/shirokane.html).
Footnotes
Contributors: HM conducted the study, accepted full responsibility for the study, had access to all the data and the final decision to submit for publication. SShi, YT, HKu, and HM designed the project. SShi, YT, HKo, TM, YS, ADO, MM and RN performed the experiments. SShi and RY performed bioinformatics analyses. HT, TN, NS, KM, SSho, YS, SH, HS, FW, KA, KF, SK, MT, and TO provided human clinical samples. SShi, DM and HM analyzed the results and wrote the manuscript. All authors reviewed the manuscript. The final version was approved to be submitted by all authors.
Funding: This work was supported by the Aichi Cancer Center Joint Research Project on Priority Areas (HM) and was also supported in part by the Japan Society for the Promotion of Science KAKENHI grant numbers 20K09187 (YT) and 19H03528 (HM), and the Japanese Respiratory Foundation (SShi).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data are available upon reasonable request. The data set generated and analyzed during the current study are available in the Japanese Genotype-Phenotype Archive under accession number DRA012560.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
The study protocol was approved by the institutional ethical review board of Aichi Cancer Center (2018-2-20). Participants gave informed consent to participate in the study before taking part.
References
- 1.Doroshow DB, Sanmamed MF, Hastings K, et al. Immunotherapy in non-small cell lung cancer: facts and hopes. Clin Cancer Res 2019;25:4592–602. 10.1158/1078-0432.CCR-18-1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced Nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627–39. 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015;373:123–35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst RS, Baas P, Kim D-W, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540–50. 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 5.Alborelli I, Leonards K, Rothschild SI, et al. Tumor mutational burden assessed by targeted NGS predicts clinical benefit from immune checkpoint inhibitors in non-small cell lung cancer. J Pathol 2020;250:19–29. 10.1002/path.5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960–4. 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 7.Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018;391:2128–39. 10.1016/S0140-6736(18)30789-X [DOI] [PubMed] [Google Scholar]
- 8.Blank CU, Haanen JB, Ribas A. The "cancer immunogram". Science 2016;352:658–60. [DOI] [PubMed] [Google Scholar]
- 9.Karasaki T, Nagayama K, Kuwano H, et al. An Immunogram for the Cancer-Immunity cycle: towards personalized immunotherapy of lung cancer. J Thorac Oncol 2017;12:791–803. 10.1016/j.jtho.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi Y, Kushihara Y, Saito N, et al. A novel scoring method based on RNA-seq immunograms describing individual cancer-immunity interactions. Cancer Sci 2020;111:4031–40. 10.1111/cas.14621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salem A, Asselin M-C, Reymen B. Targeting hypoxia to improve non-small cell lung cancer outcome. J Natl Cancer Inst 2018;110. 10.1093/jnci/djx160. [Epub ahead of print: 01 01 2018]. [DOI] [PubMed] [Google Scholar]
- 12.Xiang H, Ramil CP, Hai J, et al. Cancer-Associated fibroblasts promote immunosuppression by inducing ROS-generating monocytic MDSCs in lung squamous cell carcinoma. Cancer Immunol Res 2020;8:436–50. 10.1158/2326-6066.CIR-19-0507 [DOI] [PubMed] [Google Scholar]
- 13.Lou Y, Diao L, Cuentas ERP, et al. Epithelial-Mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res 2016;22:3630–42. 10.1158/1078-0432.CCR-15-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 2017;171:934–49. 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiraishi Y, Sato Y, Chiba K, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res 2013;41:e89. 10.1093/nar/gkt126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans E. "Salmon". Midwives Chron 1972;86:118–9. [PubMed] [Google Scholar]
- 18.Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (time) for effective therapy. Nat Med 2018;24:541–50. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa A, Kieffer Y, Scholer-Dahirel A, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 2018;33:463–79. 10.1016/j.ccell.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 20.Liberzon A, Birger C, Thorvaldsdóttir H, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nirmal AJ, Regan T, Shih BB, et al. Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol Res 2018;6:1388–400. 10.1158/2326-6066.CIR-18-0342 [DOI] [PubMed] [Google Scholar]
- 22.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 2013;14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. 10.1186/s13059-017-1349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finotello F, Mayer C, Plattner C, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-Seq data. Genome Med 2019;11:34. 10.1186/s13073-019-0638-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasegawa T, Hayashi S, Shimizu E, et al. Neoantimon: a multifunctional R package for identification of tumor-specific neoantigens. Bioinformatics 2020;36:4813–6. 10.1093/bioinformatics/btaa616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nejo T, Matsushita H, Karasaki T, et al. Reduced neoantigen expression revealed by longitudinal Multiomics as a possible immune evasion mechanism in glioma. Cancer Immunol Res 2019;7:1148–61. 10.1158/2326-6066.CIR-18-0599 [DOI] [PubMed] [Google Scholar]
- 28.Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finotello F, Rieder D, Hackl H, et al. Next-Generation computational tools for interrogating cancer immunity. Nat Rev Genet 2019;20:724–46. 10.1038/s41576-019-0166-7 [DOI] [PubMed] [Google Scholar]
- 30.Yarchoan M, Johnson BA, Lutz ER, et al. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer 2017;17:209–22. 10.1038/nrc.2016.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012;482:400–4. 10.1038/nature10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenthal R, Cadieux EL, Salgado R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature 2019;567:479–85. 10.1038/s41586-019-1032-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mlecnik B, Bindea G, Angell HK, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016;44:698–711. 10.1016/j.immuni.2016.02.025 [DOI] [PubMed] [Google Scholar]
- 34.Matsushita H, Hasegawa K, Oda K, et al. The frequency of neoantigens per somatic mutation rather than overall mutational load or number of predicted neoantigens per se is a prognostic factor in ovarian clear cell carcinoma. Oncoimmunology 2017;6:e1338996. 10.1080/2162402X.2017.1338996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duhen T, Duhen R, Montler R, et al. Co-Expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat Commun 2018;9:2724. 10.1038/s41467-018-05072-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wellenstein MD, de Visser KE. Cancer-Cell-Intrinsic mechanisms shaping the tumor immune landscape. Immunity 2018;48:399–416. 10.1016/j.immuni.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 37.Kalbasi A, Ribas A. Tumour-Intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol 2020;20:25–39. 10.1038/s41577-019-0218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 2018;173:321–37. 10.1016/j.cell.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damotte D, Warren S, Arrondeau J, et al. The tumor inflammation signature (TIS) is associated with anti-PD-1 treatment benefit in the CERTIM pan-cancer cohort. J Transl Med 2019;17:357. 10.1186/s12967-019-2100-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500–1. 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srivastava PK. Neoepitopes of cancers: looking back, looking ahead. Cancer Immunol Res 2015;3:969–77. 10.1158/2326-6066.CIR-15-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schumacher TN, Hacohen N. Neoantigens encoded in the cancer genome. Curr Opin Immunol 2016;41:98–103. 10.1016/j.coi.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 45.Ward JP, Gubin MM, Schreiber RD. The role of neoantigens in naturally occurring and therapeutically induced immune responses to cancer. Adv Immunol 2016;130:25–74. 10.1016/bs.ai.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anagnostou V, Smith KN, Forde PM, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov 2017;7:264–76. 10.1158/2159-8290.CD-16-0828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Stanger BZ. Cell cycle regulation meets tumor immunosuppression. Trends Immunol 2020;41:859–63. 10.1016/j.it.2020.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298–306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- 49.Germain C, Gnjatic S, Tamzalit F, et al. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am J Respir Crit Care Med 2014;189:832–44. 10.1164/rccm.201309-1611OC [DOI] [PubMed] [Google Scholar]
- 50.Gettinger S, Choi J, Hastings K, et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov 2017;7:1420–35. 10.1158/2159-8290.CD-17-0593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsushita H, Hasegawa K, Oda K, et al. Neoantigen load and HLA-class I expression identify a subgroup of tumors with a T-cell-inflamed phenotype and favorable prognosis in homologous recombination-proficient high-grade serous ovarian carcinoma. J Immunother Cancer 2020;8:e000375. 10.1136/jitc-2019-000375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soltermann A, Tischler V, Arbogast S, et al. Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin Cancer Res 2008;14:7430–7. 10.1158/1078-0432.CCR-08-0935 [DOI] [PubMed] [Google Scholar]
- 53.Keskin DB, Anandappa AJ, Sun J, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 2019;565:234–9. 10.1038/s41586-018-0792-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anandappa AJ, Wu CJ, Ott PA. Directing traffic: how to effectively drive T cells into tumors. Cancer Discov 2020;10:185–97. 10.1158/2159-8290.CD-19-0790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ott PA, Hu-Lieskovan S, Chmielowski B, et al. A phase Ib trial of personalized neoantigen therapy plus anti-PD-1 in patients with advanced melanoma, non-small cell lung cancer, or bladder cancer. Cell 2020;183:347–62. 10.1016/j.cell.2020.08.053 [DOI] [PubMed] [Google Scholar]
- 56.O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol 2016;13:417–30. 10.1038/nrclinonc.2016.26 [DOI] [PubMed] [Google Scholar]
- 57.Sun P, Meng L-H. Emerging roles of class I PI3K inhibitors in modulating tumor microenvironment and immunity. Acta Pharmacol Sin 2020;41:1395–402. 10.1038/s41401-020-00500-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vokes EE, Salgia R, Karrison TG. Evidence-Based role of bevacizumab in non-small cell lung cancer. Ann Oncol 2013;24:6–9. 10.1093/annonc/mds608 [DOI] [PubMed] [Google Scholar]
- 59.Sahai E, Astsaturov I, Cukierman E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174–86. 10.1038/s41568-019-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003765supp002.pdf (5.2MB, pdf)
jitc-2021-003765supp001.xlsx (54.5KB, xlsx)
Data Availability Statement
Data are available in a public, open access repository. Data are available upon reasonable request. The data set generated and analyzed during the current study are available in the Japanese Genotype-Phenotype Archive under accession number DRA012560.