Abstract
Big data is defined as being large, varied or frequently updated, and usually generated from real-world interaction. With the unprecedented availability of big data, comes an obligation to maximise its potential for healthcare improvements in treatment effectiveness, disease prevention and healthcare delivery. We review the opportunities and challenges that big data brings to gastroenterology. We review its sources for healthcare improvement in gastroenterology, including electronic medical records, patient registries and patient-generated data. Big data can complement traditional research methods in hypothesis generation, supporting studies and disseminating findings; and in some cases holds distinct advantages where traditional trials are unfeasible. There is great potential power in patient-level linkage of datasets to help quantify inequalities, identify best practice and improve patient outcomes. We exemplify this with the UK colorectal cancer repository and the potential of linkage using the National Endoscopy Database, the inflammatory bowel disease registry and the National Health Service bowel cancer screening programme. Artificial intelligence and machine learning are increasingly being used to improve diagnostics in gastroenterology, with image analysis entering clinical practice, and the potential of machine learning to improve outcome prediction and diagnostics in other clinical areas. Big data brings issues with large sample sizes, real-world biases, data curation, keeping clinical context at analysis and General Data Protection Regulation compliance. There is a tension between our obligation to use data for the common good and protecting individual patient’s data. We emphasise the importance of engaging with our patients to enable them to understand their data usage as fully as they wish.
Keywords: health service research, statistics, clinical trials
Key points.
With unprecedented availability of big data comes an obligation to use it to improve healthcare.
Big data analyses can complement traditional research methodology, and in some cases holds distinct advantages over conventional trials.
Linkage of datasets is challenging but greatly increases their potential power to improve outcomes.
Artificial intelligence and machine-learning algorithms can improve diagnostics and outcomes prediction.
Real-world data need good data curation and an understanding of the clinical context.
We need to engage with our patients, so they understand how we are using their data to improve healthcare.
Definition of big data and sources in healthcare
The term ‘big data’ is generally defined in the information technology industry as data that is high volume (large in size), high velocity (fast moving data updated frequently) or high variety (different sources or data types).1 Big data is often generated from real-world interaction rather than in experiments or trials, and often collected for purposes other than research. Big data already influences many aspects of modern life, from Google search history to mobile phone geolocation. The European Commission further defined big data in healthcare as large, routinely or automatically collected datasets which are electronically captured or stored. These datasets are reusable with multiple purposes, and comprise the fusion or connection of existing databases with the purpose of improving health and health system performance.2 Big data in healthcare and clinical research brings many opportunities and challenges, summarised in table 1.
Table 1.
Opportunities and challenged of big data in clinical research
| Characteristic | Opportunities | Challenges |
| High volume | Large sample sizes and high statistical power | Data may be too large to store and process on a single computer, achievable with cloud computing. |
| High velocity | Can generate timely, relevant research | Risk of getting swamped with new data. |
| High variety | Potential to use novel sources of data, for example, images, smart devices, genomics | May need conversion into a usable format e.g. free text from medical notes to structured data. |
| Real world | Reflects real-world patients and clinical practice | Data often messy with missing data, needs lots of work to make research ready. |
| Not collected for research | Costs less to collect data | May not contain all the information you want, outcome data may be unavailable and not adjudicated. |
With unprecedented volumes of data captured and available to clinicians and researchers, there is a moral obligation to maximise its potential to improve healthcare.3 The applications of big data in healthcare are wide-ranging, and include improvements in the effectiveness and quality of treatments (through earlier disease interventions, reducing errors and understanding causality), disease prevention (through predicting outcomes and understanding global infections), and healthcare delivery (through disseminating evidence and directly involving patients).2 Advances in digitalisation of biomedical data, particularly with genomics and better understanding of cancer pathways, is already developing the field of personalised medicine.4
In this narrative review, we will outline sources of big data in gastroenterology and how these can improve care and research, the power of database linkage, understanding machine learning (ML) and practical issues big data raises which we must overcome to use these essential resources to their fullest potential for our patients.
Sources of data to improve healthcare
The digitisation of healthcare is increasing rapidly and there are currently four main sources of healthcare big data:
Administrative data.
Electronic medical records (EMRs).
Registries.
Patient-generated data.
Administrative data
Administrative data refer to data collected to manage healthcare systems or monitor the health of a population. This focuses on capturing healthcare activity and is similar in format to industrial big data, as used by companies such as Google, which tends to arise from individuals’ incidental digital interaction in exchange for a service. In healthcare this low density, real time information can be used as a surrogate marker for activity and allows predictions of trends and outcomes for these services. These may have very large sample sizes with millions of patients, with examples such as hospital admission rates, general practitioner (GP) attendances and Hospital Episode Statistics (HES) data.
Administrative big data has a role in service quality evaluation and can inform commissioning and patient choice. A recent paper reviewed hospital admission data in inflammatory bowel disease (IBD) to assess trends over a 10-year period, demonstrating a reduction in admission length and the commissioning impact of admissions for elective medical therapies.5 Administrative clinical coding data has the risk of misclassification, for example, cholangiocarcinoma coding error contributed to a reported rise in intrahepatic tumours.6
EMRs and the problems with traditional data
EMRs and Healthcare Information Systems refers to data collected to manage patients’ healthcare. These are generated at the point of patient care and can include clinical episodes, diagnostic tests including clinicopathological results and therapeutic interventions. Electronic healthcare data is largely underutilised with a paucity of evidence of real-world application.7
Assimilating medical data from medical notes, letters and reports requires collection and analysis of high-density data that is often retrospective.8 Natural language processing (NLP), a development of ML, can be used to analyse and extract information from unstructured text. In the USA, NLP was recently demonstrated to be effective at extracting quality indicators from unstructured colonoscopy procedure reports.9 However, clinical documents are naturally fraught with human errors and omissions, and analysing and interpreting such data without its original clinical context poses challenges.3 Unstructured clinical data may have questionable data validity, and non-standardised heterogeneous data sources, requiring high curation and management resources. To improve health quality, we need to be able to structure and analyse this complex medical data—this process often involves narrowing broad clinical meanings (referred to as Ballung concepts) into pinpoint diagnoses: this may aid data analysis, but removes nuanced description of clinical phenomena which do not always reach set criteria.
An example of a traditional big clinical dataset which improves the effectiveness and quality of treatment is the National Cancer Registration and Analysis Service (NCRAS) housed in Public Health England.10 This uses semiautomatic data collection from multidisciplinary team meetings and extraction from text-based pathology and radiology reports. This is in part automated to reduce duplications, however mostly extracted by trained Cancer Registration Officers. This allows for a rich data set to allow supporting of service provision, clinical audit, commissioning planning of services, public health and epidemiological research.
Registries
Registries are records about a health condition within a specific population. These may be set up specifically for research, such as biobanks, but might be set up for non-research activity, such as quality registries. Healthcare registry data may be more detailed, structured and research focused, therefore, generally better suited to automatically generated data points and may be well structured to fit into relational databases. With rapid digitisation across healthcare, registry data can be prospectively collected and analysed automatically in real time.7 Such registries can be used for assessment of quality in specific disease management, looking for significant variation in disease management and outcomes. Analysis of big data permits quantification of these disparities and facilitates the elimination of unwarranted variation in quality.
The UK’s National Endoscopy Database (NED) demonstrates real-time automated capture of clinical data directly from endoscopy reporting systems (ERS), avoiding double entry of data.11 The standardised datapoints used by all ERSs create a rich centralised database with granular details. This allows quality assurance with the automatic calculation of key performance indicators (KPIs) for both individual endoscopists and their organisations, to monitor and improve endoscopy quality. This reduces the burden of paper audit and feedback on endoscopists and endoscopy unit leads. Assessing the impact of automated feedback on colonoscopy KPIs is the topic of the NED Automated Performance Reports to Improve Quality Outcomes Trial (APRIQOT), a national clinical trial currently underway.12 NED currently does not analyse free-text comments losing some clinical descriptive details and nuanced Ballung concepts; while this may enrich analyses in the future, current systems based on NLP of free-text data are not yet effective, with a study of free-text reports capturing measures of performance, finding that 13 of the 20 measures were inaccurate.13
At a national level, NED provides an accurate overview of endoscopy workload and workforce activity, to facilitate planning. For example, in 2019, we were able to assess that 50% of the endoscopy workforce was not performing the minimum number of annual procedures.14 The value of this real-time data was demonstrated recently during the coronavirus pandemic: NED quickly allowed national organisations to see the effect of the pandemic on weekly national endoscopy activity, monitoring its recovery and the potential burden of reduced cancer diagnostics to help plan resuming services.15
Patient-generated data
We have discussed the challenge of capturing clinical context; similar nuanced variables which are difficult to capture are patient-related experiences, values and preferences. Without these we lose the patient perspective of disease. These are challenging to code as quantitative variables for analysis.16 Technologies associated with big data offer solutions of directly involving patients with data entry and using big data platforms to directly provide information to patients. Patient-related outcomes measures (PROMs) and measuring what matters to patients is central in driving improvement in patient care, and are increasingly being used in healthcare research and in service development.17
Patient generated data typically uses apps or devices and is a growing area of data, although not established so far. Electronic surveys, social media and smartphone apps and devices are all potential sources of data. An example of PROMs being used in big data is the UK IBD registry.18 This registry combines hospital record data from physicians, who are describe the phenotype of IBD accurately, however, they use PROMs and patient experience to provide data on how active their disease is and its impact. As a growing network, the registry has faced difficulties with missing data, however they have started to use this patient and clinical data to create KPIs based on patient experience for units in prescribing of biological medications.19
Big data and clinical research
Although randomised controlled trials (RCTs) are considered the gold standard method to determine the value of treatments, they are expensive and take a long time from inception to publication. Further, there is growing concern that trials may not be entirely representative of the patient population at large, which can have implications for how results should be translated into practice.
Big data can complement clinical trials by increasing their efficiency and cost-effectiveness through informing power calculations, site selection and point-of-care randomisation, and improving efficiency of data capture.8 Big data is well suited to hybrid RCTs, which includes both traditional and pragmatic clinical trial elements. It begins with randomisation to different intervention groups, with some data collected through standardised RCT procedures, but the remaining data collected through routine healthcare visits via EMRs and administrative data.20
Big data is also well suited to phase IV trials assessing safety and adverse events of medicines and interventions, particularly rate events. An example this is the National Health Service (NHS) Bowel Cancer Screening Programme, which has a large and rich database of bowel screening colonoscopy procedures and histological data. This is manually entered by an expert group of trained bowel screening practitioners. Analysing 163 000 procedures from this large database found significant correlations between the rare event of postpolypectomy bleeding and polyps being located in the caecum (but not elsewhere in the proximal colon), which was later confirmed in a large systematic review and meta-analysis.21 22
For rare and chronic diseases prospective cohorts utilising standardised big data collection can assess treatments and determine predictive factors related to outcomes unfeasible in traditional trials. This was recently demonstrated in the PROTECT trial assessing paediatric IBD.23
The effectiveness for some clinical interventions is difficult to test in RCTs, such as the organisation of clinical services. Big data analysis can provide a solution to these problems by providing cost-effective, timely, population-wide analyses. However, big data’s large nature has the risk of creating statistical noise, with data showing significant correlation between variables, without hypothesis-driven causal reasoning or clinical significance. Although hypothesis-generation can be synergistic with traditional research methods to test the generated hypothesis,8 without an RCT a lack of a causal structure can limit interpretation of data, and a theoretical framework linking cause to effect should underwrite any trial design or analysis methodology.16 When an RCT is not suited the ‘target trial’ methodology is recommended to assess for causal inference from large observational databases, and outlines a framework for comparative effectiveness.24 We also recommend some basic do’s and don’ts of big data causal inference research (table 2).
Table 2.
Do’s and Don’ts of big data causal inference research
| Do | Don’t |
| Prespecify study design and analysis in the study protocol and statistical analysis plan. | Mine data for interesting results |
| Register the study for example, on ClinicalTrials.gov | |
| Use tools like GitLAb to manage and share programming codes.45 | Use hypothesis tests without a good reason |
| Find a good team, data science is a team sport needing clinical, epidemiological, statistical, and programming skills. | |
| Follow the EQUATOR network reporting guidelines.46 | Assume that patient and public involvement is too difficult or less important. |
| Publish disappointing/negative findings |
EQUATOR, enhancing the quality and transparency of heath research.
Big data also has a role in disseminating research findings and developing translational medicine in larger populations. An example of this is the Liver Investigation Testing Marker Utility in Steatohepatitis project.25 This brings together clinicians and scientists from academic centres across Europe with the common goal to develop, validate and qualify biomarkers in non-alcoholic fatty liver disease. This international group has identified 55 biomarkers with requirements for technical and clinical validation with the aim of translating this research to monitor response to treatment and progression in non-alcoholic fatty liver disease.
The power of linkage
Currently, disparate datasets about all aspects of patient care are available across the UK but access for researchers to link and exploit them is limited, and repetitive applications to link datasets for individual projects results in significant duplication of effort. Achieving reliable, patient-level linkage of all routine datasets specific diseases would help to quantify inequalities and identify processes and procedures associated with best practice and improved patient outcomes. Linkage of datasets has its own challenges, including consent, privacy and linkage error particularly if patient identifiers are pseudonymised at source.26
An example is the UK COloRECTal cancer Repository, which aims to quantify the characteristics of, and any variation in, colorectal cancer and its management across the UK, by working in partnership with all relevant UK data providers to create a single virtual research repository of all the UK colorectal cancer data, ensuring robust quality assurance and standardised processing to make the data readily available to the research community and other relevant users. The resulting cancer intelligence has huge potential and will help promote early diagnosis, optimise treatments, improve the efficiency and cost-effectiveness of NHS services, ensure patient-centred care and, ultimately, improve outcomes. This model allows strict information governance while also allowing ready access to those with the necessary approvals, thereby increasing efficiency and reducing duplication.
The potential for new informative outputs from linkage of individual data-rich datasets is also substantial, for example:
Linkage of endoscopy datasets to cancer outcomes datasets to permit more detailed analyses of factors influencing poor clinical outcomes.
Linkage of endoscopy datasets, like NED, to other datasets, such as NCRAS, the UK IBD registry and English HES data would permit automated capture and analysis of endoscopic complications and post-colonoscopy colorectal cancers.
Precision medicine-linking genotype and clinical data will permit the investigation of colorectal cancer treatment response and outcome in relation to genetics and tumour biology.
Data linkage can also allow clinical process and outcome information to be linked to social care, consumer, social media, housing, pollution, energy, environment, transport and many other datasets. The analytical possibilities are huge. For example, linking NHS bowel cancer screening programme data with a socioeconomic market segmentation tool allowed exploration and hypothesis generation of the impact of socioeconomic profiles in variations in bowel cancer screening uptake, and the development of a prospective trial assessing the impact of public health engagement strategies on uptake of bowel screening in different socioeconomic groups.27 28
Machine learning and artificial intelligence
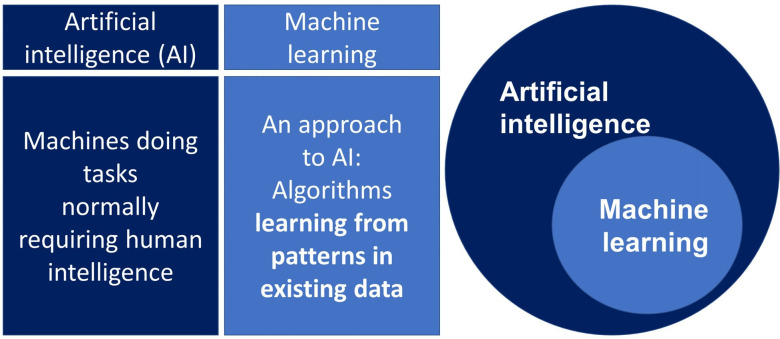
ML describes a set of methods that use algorithms to learn from observations in data or from interactions in the real world. ML methods are well suited to identifying patterns in complex data and are now the most commonly used approach to developing artificial intelligence systems, which aims to use computers to automate human tasks (figure 1). ML may be supervised ML, in which a model is trained using curated clinical data with specific known outcomes, or unsupervised ML which does not have predefined outcomes, and is used as an exploratory tool to find patterns or clusters within datasets.29
Figure 1.
Machine learning and AI.
ML algorithms are closely related to statistical methods used widely in medical research and epidemiology but are well suited to data with very large numbers of variables, or where the interaction or combination of variables is informative. Examples of this type of data includes unstructured data like images, text or speech, and ML algorithms are now the dominant method for automating the analysis of these types of data, such as interpreting and classifying medical images.
The use of artificial intelligence algorithm techniques to identify colorectal polyps from high definition while light colonoscopy images is currently being trialled in the UK with the GI-genius system. The system was trained on 1.5 million images of polyps manually annotated by expert endoscopists and has a near 100% real time sensitivity for detection of clinically relevant polyps, however, only one in every two lesions detected was rated as adenomatous by centralised pathology. This adjunct may help reduce the limitation of human visual perception, fatigue, distraction and variable alertness during examination, but risk prompting removal of benign distal hyperplastic polyps and increasing the false-positive rate at colonoscopy.30
Other potential applications for ML in gastroenterology include developing predictive algorithms that can be used to predict outcomes or support in diagnosis. These algorithms typically use the large volumes of data avilable in electronic healthcare records, detecting patterns in the underlying data that can be used to make predictions about patients’ futures. In hepataitis B, ML algorithms using genomic and clinical data have developed a model to determine viral variants which predicted HBV e antigen status to facilitate clinical decision making.31
Gastroenterology as a specialty is potentially well placed to making use of ML in this way, since the management of many gastro-intestinal conditions (eg, IBD) requires multiple labotory tests and imaging over time that can provide the type of deep, variable-rich datasets well suited to ML methods. An early example of this is an Israeli and UK study looking at the risk of colorectal cancer from blood work, age and sex.32 Using decision trees and cross-validation techniques, the authors generated a prediction model for colorectal cancer in primary care who’s area under the curve outperformed both a standard linear statistical model and iron deficiency anaemia management guideline criteria. ML is, however, limited by the requirement for data of sufficient quality and volume to be useful. For example, in developing an algorithm to automatically detect tumours on CT scan images, a typical ML algorithm would need to be trained using imaging data that has been read by a radiologist and labelled with the correct diagnosis. Labelled data are often time consuming or expensive to collect, and any errors or biases in the training data are carried through into the predictions made by the algorithm. ML is also less useful for analyses where the challenge is to answer a causal questions, such as is X drug more effective or associated with fewer adverse events than Y drug. ML algorithms should generally be restricted to making predictions rather than explaining why things happen.
Issues with big data
Traditional empirical science using hypothesis-testing is designed to filter out noise, demonstrate correlation and hypothesise causations which are believed through their replication and corroboration. Big data embraces this noise with real-world unfiltered data, with the potential to see underlying patterns at a large scale. However, this can bring problems of scale:
High volume brings a low threshold for significance.
Due to the size of data, many analyses may reach statistical significance without substantial clinical relevance (eg, demonstration of a statistically significant relative risk of 1.03). While such results might warrant further research to understand potential causation, it should not necessarily be interpreted as sufficient to warrant any change in clinical guidance.
High variety and volume can lead to measuring too much.
In measuring and testing increasingly large numbers of variable and comparisons, the problem of multiplicity arises, with an increased risk of finding statistical significance by happenstance and erroneously rejecting a null hypothesis (type 1 error).
Specific to KPIs, there is also a temptation to ‘measure everything’ simply because data are available. While this presents an excellent opportunity to refine KPIs and strengthen their evidence base, it should not be used as a thoughtless and counterproductive opportunity to create ever-increasing numbers of KPIs, increasing complexity rather than reducing it.
High variety has variable quality and needs context, curation and General Data Protection Regulation (GDPR) compliance.
While one of the advantages of big data is its potential to cut through the noise to see underlying patterns, large-scale data also increase the problems of methodological issues with data quality, data inconsistency and instability: the ‘rubbish in, rubbish out’ phenomenon. The annotation, curation and linkage of datasets is complex work, and fraught with difficulty and missed opportunity if data miners are unaware of the clinical contexts and wider goals of the project. Unsupervised ML models have been used to discover latent infectious disease using social media and demonstrates the risk of misinterpretation from curation without theoretical understanding.33 Google Influenza Trends accurately predicted outbreaks of influenza in the USA 7 days before the Center for Disease Control, using Google search terms for influenza symptoms. However, a few winters later the previously swift and accurate theory-free and data-rich model overstated the outbreak by a factor of two. Unsupervised ML was able to find statistical patterns of correlation in the data, but without understanding causation; theory-free analysis of correlations is fragile.34
With the ethical obligation to engage in the curation of a big dataset, there are challenges of data ownership, security and privacy.35 The European Union and UK GDPR set out principles of data lawfulness, purpose limitation, data minimisation, accuracy, storage limitation, confidentiality and accountability.36 37 This has sparked international debate on the use of big data in research and healthcare. The scale of the dynamic flow of data between organisations and across international borders has led to improvements in health, but has increased privacy-related harms, as relying on individual specific consent to minimise harm is increasingly unfeasible.38 As GDPR excludes anonymised data, increasingly big data is stored and shared as aggregated or pseudonymised data which theoretically cannot be traced back to individual subjects, which poses challenges for data linkage.8
Real-world data have real-world errors.
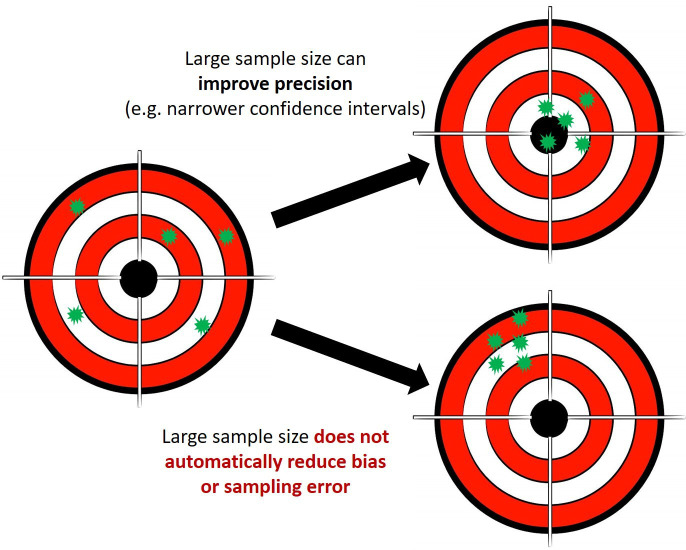
Bias and sampling errors are not necessarily reduced by increasing the sample size, (figure 2) and there is a risk that biases in datasets are reproduced by algorithms in their prospective application. For example, social injustices leading to potential disparities or under-representation of ethnic and population groups in healthcare datasets can be reinforced during analyses and lead to potential harm: during risk stratification for hypertrophic cardiomyopathy in the USA using targeted genetic testing, the genetic data used had over-representation of white Americans. This led to multiple patients from an African or unspecified ancestry, having genetic results misclassified as pathogenic. After wider genomic analysis of a more diverse population, these were all reclassified as benign.39
Figure 2.
Big data and the risk of bias.
Patient and clinician generated datasets are at risk of reporting bias through distortion of presented data due to selective disclosure. The phenomenon of gaming statistics at an organisation and individual level has been described when clinicians face targets.40 Automation of data collection through EMRs may reduce this.
Real-world data have real-world patients who should be engaged with.
There is an ethical obligation to use big data for the common good. Just as physicians have a moral obligation to learn from their own experiences, we have an obligation to use EMRs with easily accessible healthcare data to conduct analyses to improve care.35 This can create tension with protecting an individual patient’s data if not managed properly.
The failed care data project demonstrated that problems with data management and communication can lead to conflicting legal duties in protecting patients’ data. This centralisation effort of health and social care data in 2013 was judged as being flawed by the National Data Guardian in its inadequacy of explaining the benefits of data-sharing and allowing the possibility that personal data might be accessed by commercial companies. Problems with systems required for patients to opt-out and unclear criteria for accessing the collected health data posed risks to the trust between patients and GPs. This created a power struggle between patients, GPs, Health and Social Care Information Centre, the government and data purchasers.41 This highlights the need for clear data access, management and sharing protocols and the importance of easy to use and dynamic consent processes.
Polls show there is public support for sharing patient data for medical research (77%), however, there are very low levels of awareness of how the NHS uses healthcare data.42 43 Data breaches by companies and social media are well reported,44 and similar incidents in healthcare could lead to mistrust of healthcare data collection. The key message from the Caldicott report was the importance of dialogue with the public, we owe it to citizens to enable them to understand data usage as fully as they wish, and ensure information about how data is accessed, by whom, and for what purposes is available.41
Conclusion
We have described how big data can complement and offer distinct advantages to traditional research methods. Through the linkage of datasets and increasing sophistication of data analysis using ML the prevalence of big data in research and clinical practice will continue to increase. Statistical pragmatism, investment in data curation and engaging patients in understanding the use of their data are highlighted as important factors in the development of big data in gastroenterology.
Footnotes
Twitter: @drjamiec, @Rutter_Matt
Contributors: JC wrote the definitions, Sources, research and issues sections of the manuscript, compiled others’ section and submitted the paper. JC addressed reviewers comments. BB wrote the machine learning and artificial intelligence section, contributed to the definitions and issues sections, and reviewed drafts. EM contributed to the power of linkage section and reviewed drafts. MR wrote the power of linkage section, significantly edited the entire manuscript and reviewed drafts.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Provenance and peer review: Commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not required.
References
- 1. Beyer MA, Laney D. The importance of ‘big data’: a definition. Stamford, CT Gart 2012.
- 2. Habl C, Renner A-T, Bobek J. Study on big data in public health, telemedicine and healthcare 2016.
- 3. Goodman KW. Ethics, medicine, and information technology. Cambridge University Press, 2015. [Google Scholar]
- 4. GS O, Kuznetsov VA. Big genomics and clinical data analytics strategies for precision cancer prognosis. Sci Rep 2016;6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahmad A, Laverty AA, Alexakis C, et al. Changing nationwide trends in endoscopic, medical and surgical admissions for inflammatory bowel disease: 2003-2013. BMJ Open Gastroenterol 2018;5:e000191. 10.1136/bmjgast-2017-000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khan SA, Emadossadaty S, Ladep NG, et al. Rising trends in cholangiocarcinoma: is the ICD classification system misleading us? J Hepatol 2012;56:848–54. 10.1016/j.jhep.2011.11.015 [DOI] [PubMed] [Google Scholar]
- 7. Mehta N, Pandit A. Concurrence of big data analytics and healthcare: a systematic review. Int J Med Inform 2018;114:57–65. 10.1016/j.ijmedinf.2018.03.013 [DOI] [PubMed] [Google Scholar]
- 8. Hulsen T, Jamuar SS, Moody AR, et al. From big data to precision medicine. Front Med 2019;6:34. 10.3389/fmed.2019.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laique SN, Hayat U, Sarvepalli S, et al. Application of optical character recognition with natural language processing for large-scale quality metric data extraction in colonoscopy reports. Gastrointest Endosc 2021;93:750–7. 10.1016/j.gie.2020.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Henson KE, Elliss-Brookes L, Coupland VH, et al. Data resource profile: National cancer registration dataset in England. Int J Epidemiol 2020;49:16–16h. 10.1093/ije/dyz076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. TJW L, Siau K, Esmaily S. Development of a national automated endoscopy database: the United Kingdom national endoscopy database (NED). United Eur Gastroenterol J 2019;7:798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Catlow J, Sharp L, Kasim A, et al. The National endoscopy database (NED) automated performance reports to improve quality outcomes trial (APRIQOT) randomized controlled trial design. Endosc Int Open 2020;8:E1545–52. 10.1055/a-1261-3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mehrotra A, Dellon ES, Schoen RE, et al. Applying a natural language processing tool to electronic health records to assess performance on colonoscopy quality measures. Gastrointest Endosc 2012;75:1233–9. e14. 10.1016/j.gie.2012.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. NED - National Endoscopy Database. Jt. Advis. Gr. GI Endosc. Available: https://ned.jets.nhs.uk/kpi/ [Accessed 31 May 2020].
- 15. Rutter MD, Brookes M, Lee TJ, et al. Impact of the COVID-19 pandemic on UK endoscopic activity and cancer detection: a national endoscopy database analysis. Gut 2021;70:1–7. 10.1136/gutjnl-2020-322179 [DOI] [PubMed] [Google Scholar]
- 16. Chin-Yee B, Upshur R. Three problems with big data and artificial intelligence in medicine. Perspect Biol Med 2019;62:237–56. 10.1353/pbm.2019.0012 [DOI] [PubMed] [Google Scholar]
- 17. Calvert M, Kyte D, Price G, et al. Maximising the impact of patient reported outcome assessment for patients and society. BMJ 2019;364:k5267. 10.1136/bmj.k5267 [DOI] [PubMed] [Google Scholar]
- 18. UK IBD Registry - IBD Registry. Available: https://ibdregistry.org.uk/ [Accessed 29 Jun 2020].
- 19. Shawihdi M, Cummings F, Bloom S. PTH-126 audit of biological therapy for inflammatory bowel disease: results from the UK IBD registry. In: Gut. BMJ 2019:A97.2–8. [Google Scholar]
- 20. Zhu M, Sridhar S, Hollingsworth R, et al. Hybrid clinical trials to generate real-world evidence: design considerations from a sponsor's perspective. Contemp Clin Trials 2020;94:105856. 10.1016/j.cct.2019.105856 [DOI] [PubMed] [Google Scholar]
- 21. Rutter MD, Nickerson C, Rees CJ, et al. Risk factors for adverse events related to polypectomy in the English bowel cancer screening programme. Endoscopy 2014;46:90–7. 10.1055/s-0033-1344987 [DOI] [PubMed] [Google Scholar]
- 22. Jaruvongvanich V, Prasitlumkum N, Assavapongpaiboon B, et al. Risk factors for delayed colonic post-polypectomy bleeding: a systematic review and meta-analysis. Int J Colorectal Dis 2017;32:1399–406. 10.1007/s00384-017-2870-0 [DOI] [PubMed] [Google Scholar]
- 23. Hyams JS, Davis Thomas S, Gotman N, et al. Clinical and biological predictors of response to standardised paediatric colitis therapy (protect): a multicentre inception cohort study. Lancet 2019;393:1708–20. 10.1016/S0140-6736(18)32592-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hernán MA, Robins JM. Using big data to emulate a target trial when a randomized trial is not available. Am J Epidemiol 2016;183:758–64. 10.1093/aje/kwv254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LITMUS project.. Available: https://litmus-project.eu/ [Accessed 29 Jun 2020].
- 26. Hagger-Johnson G, Harron K, Fleming T, et al. Data linkage errors in hospital administrative data when applying a pseudonymisation algorithm to paediatric intensive care records. BMJ Open 2015;5:8118. 10.1136/bmjopen-2015-008118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gavens L, Whiteley L, Belencsak A, et al. Market segmentation tools provide insights into demographic variations in bowel cancer screening uptake. J Epidemiol Community Health 2019;73:778–85. 10.1136/jech-2018-211085 [DOI] [PubMed] [Google Scholar]
- 28. Smith SG, Wardle J, Atkin W, et al. Reducing the socioeconomic gradient in uptake of the NHS bowel cancer screening programme using a simplified supplementary information leaflet: a cluster-randomised trial. BMC Cancer 2017;17:1–9. 10.1186/s12885-017-3512-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med Res Methodol 2019;19:64. 10.1186/s12874-019-0681-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hassan C, Wallace MB, Sharma P, et al. New artificial intelligence system: first validation study versus experienced endoscopists for colorectal polyp detection. Gut 2020;69:799–800. 10.1136/gutjnl-2019-319914 [DOI] [PubMed] [Google Scholar]
- 31. Mueller-Breckenridge AJ, Garcia-Alcalde F, Wildum S, et al. Machine-learning based patient classification using hepatitis B virus full-length genome quasispecies from Asian and European cohorts. Sci Rep 2019;9:1–12. 10.1038/s41598-019-55445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kinar Y, Kalkstein N, Akiva P, et al. Development and validation of a predictive model for detection of colorectal cancer in primary care by analysis of complete blood counts: a binational retrospective study. J Am Med Inform Assoc 2016;23:879–90. 10.1093/jamia/ocv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim S, Tucker CS, Kumara S. An unsupervised machine learning model for discovering latent infectious diseases using social media data. J Biomed Inform 2017;66:82–94. 10.1016/j.jbi.2016.12.007 [DOI] [PubMed] [Google Scholar]
- 34. Harford T. Big data: a big mistake? Signif 2014;11:14–19. 10.1111/j.1740-9713.2014.00778.x [DOI] [Google Scholar]
- 35. Goodman KW, Goodman KW. Biomedical research, from genomes to populations: big data and the growth of knowledge. In: Ethics, medicine, and information technology. Cambridge University Press, 2015: 121–46. [Google Scholar]
- 36. General Data Protection Regulation (GDPR) compliance guidelines. Available: https://gdpr.eu/ [Accessed 25 Apr 2021].
- 37. Guide to the UK general data protection regulation (UK GDPR) | ICO. Available: https://ico.org.uk/for-organisations/guide-to-data-protection/guide-to-the-general-data-protection-regulation-gdpr/ [Accessed 25 Apr 2021].
- 38. Knoppers BM, Thorogood AM. Ethics and big data in health. Curr Opin Syst Biol 2017;4:53–7. 10.1016/j.coisb.2017.07.001 [DOI] [Google Scholar]
- 39. Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016;375:655–65. 10.1056/NEJMsa1507092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bevan G, Hood C. What’s measured is what matters: Targets and gaming in the English public health care system. Public Adm 2006;84:517–38. 10.1111/j.1467-9299.2006.00600.x [DOI] [Google Scholar]
- 41. National Data Guardian for Health and Care . Review of data security, consent and Opt-Outs national data guardian, 2016. [DOI] [PubMed] [Google Scholar]
- 42. Patient data in research | Wellcome. Available: https://wellcome.org/what-we-do/our-work/our-policy-work-using-patient-data-research [Accessed 11 Jan 2021].
- 43. Clemence M, Gilby N, Shah J. Wellcome trust monitor wave 2 tracking public views on sicence, biomedical research and science education 2013.
- 44. Data breaches - BBC News. Available: https://www.bbc.co.uk/news/topics/c0ele42740rt/data-breaches [Accessed 23 Apr 2021].
- 45. GitLab . The DevOps lifecycle with GitLab. GitLab, 2019. Available: https://about.gitlab.com/stages-devops-lifecycle/ [Accessed 10 Jan 2021].
- 46. Equator . The EQUATOR network | Enhancing the QUAlity and transparency of health research. Equator Resour. Cent., 2020. Available: https://www.equator-network.org/ [Accessed 10 Jan 2021].