Conspectus
There is a need for affordable, point-of-care devices to ease the high burden of disease in low-resource settings. The past decade of work on paper-based fluidic devices has resulted the invention of many paper-based biosensors for disease detection. However, a challenge still remains in detecting pathogenic biomarkers from complex human samples without specialized laboratory equipment. Our research has focused on the development of affordable technologies to extract and detect nucleic acids in clinical samples with minimal equipment. Here we describe methods for paper-based extraction, amplification and detection of nucleic acids. This Account provides an overview of our latest technologies developed to detect an array of diseases in low-resource settings.
Graphical Abstract
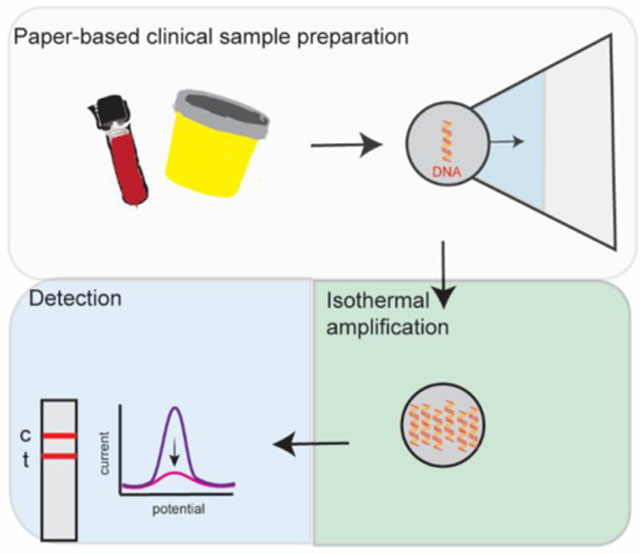
Introduction
Infectious disease prevalence is highest in low-resource settings (LRS) due to a lack of trained personnel, money and infrastructure needed to carry out lab-based molecular diagnostics. The Klapperich Lab focuses on building low-cost point-of-care diagnostics, that don’t require specialized equipment, for use in low-resource settings. These tests are largely comprised of paperfluidic devices that are made to enable extraction, amplification and detection of nucleic acids. While these platforms can be modified to detect any disease, we have focused our efforts on detecting malaria,1,2 influenza3 and various sexually transmitted infections.4–8 We develop technologies that aim to satisfy the World Health Organization’s ASSURED (Affordable, Sensitive, Specific, User friendly, Rapid and Robust, Equipment free and Deliverable to end users) criteria for point-of-care devices.9 Broadly speaking, our technology features paper-based extraction of nucleic acids from clinical samples,1,3,4,6,7 isothermal amplification of target nucleic acids to eliminate thermal cycling requirements,1–8,10 and either lateral flow2–4,6–8 or electrochemical5 detection of nucleic acids. Recently, we have explored CRISPR-based detection to increase assay specificity.5
Paperfluidic sample preparation
In order to first detect a nucleic acid target specific to a pathogen, in most cases that nucleic acid must be extracted from a clinical sample. In high-resource settings, this extraction is perfomed using a multistep protocol requiring centrifugation or a vacuum manifold. To eliminate the need for specialized equipment, we have built paperfluidic systems for the extraction of nucleic acids from various clinical sample types, including nasopharyngeal swabs,11 cervical swabs,12 urethral and vaginal swabs,6 urine4,8 and whole blood.1
Paper-based extraction of H1N1 RNA from nasopharyngeal swabs was demonstrated using a polyethersulfone (PES) membrane attached to a wicking pad (Fig 1).3 The sample was mixed with a lysis buffer that contained Glycoblue coprecipitant (Thermofisher, Lexington, MA) for visualization of the pelleted NA. When the mixture is applied to the PES membrane, the Glycoblue and RNA precipitate is captured by the PES membrane while the liquid phase is wicked away by the absorbent pad. During the initial proof-of-concept demonstration of using PES for RNA extraction, RNA was eluted from the PES membrane via centrifugation. The paper-based extraction yields were comparable to gold-standard benchtop extraction methods.
Figure 1:
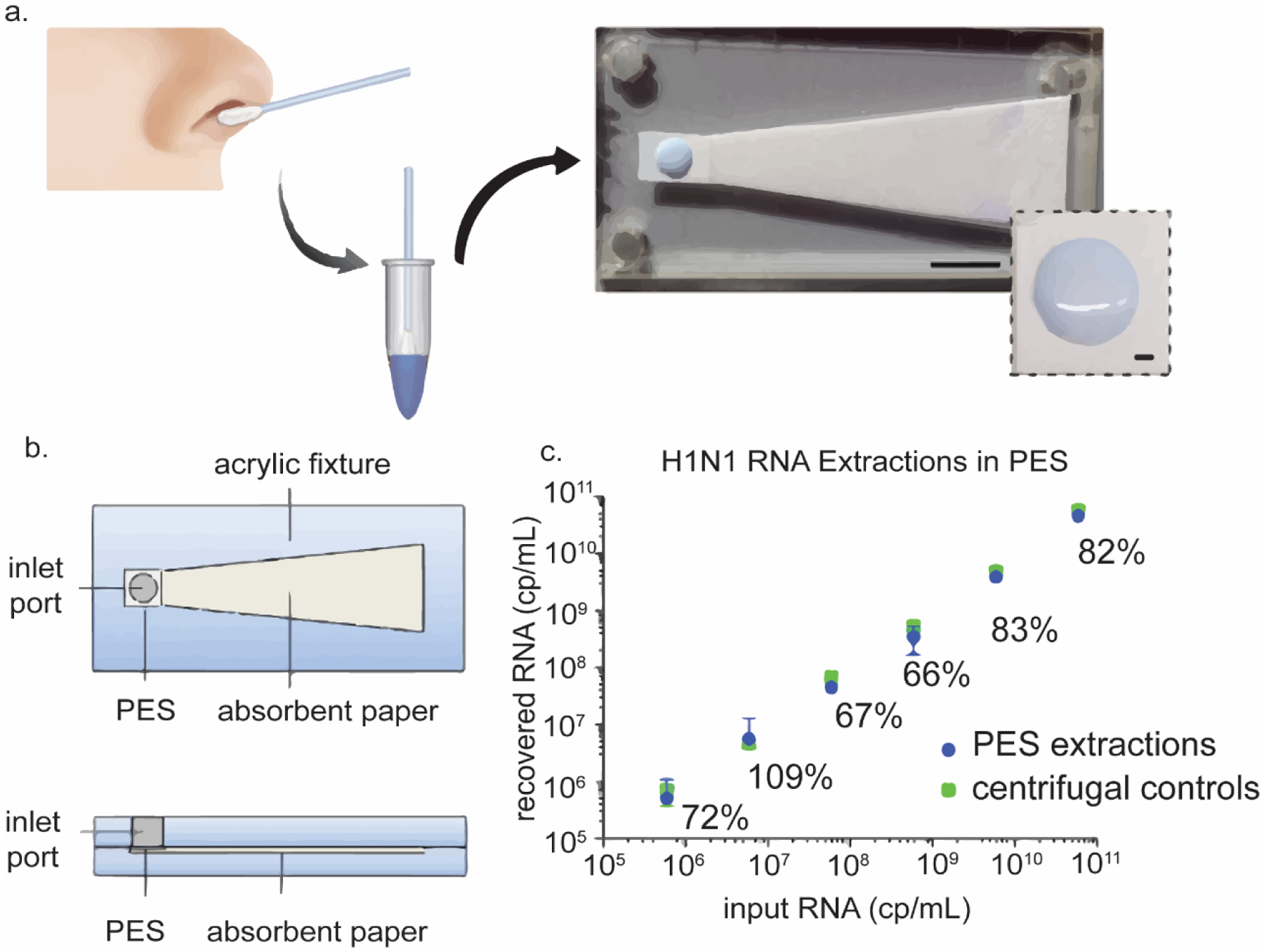
Paperfluidic H1N1 RNA extraction. (a) Nasopharyngeal swabs are lysed and applied to a PES membrane attached to a wicking pad. (b) CAD diagram of paper-based extraction device. (c) RNA recovery extracted with PES is comparable to centrifuge controls. Reproduced from reference (3). Reproduced with permission. Copyright American Chemical Society 2015.
To eliminate all centrifugation steps in a sample-extraction process, a PES membrane was next incorporated into a fully integrated device for the extraction, amplification and detection of human papillomavirus (HPV) DNA from cervical swabs (Fig 2).12 Again, the sample was first mixed with a lysis buffer that contained Glycoblue coprecipitant. Upon application to the PES membrane, the DNA-Glycoblue precipitate was captured by the PES membrane while the liquid phase was wicked away by the absorbent pad. After a series of ethanol washes to remove impurities, the DNA of interest was amplified directly on the PES membrane by the addition of an aqueous solution containing the amplification reagents. After amplification, the DNA was eluted onto a lateral flow strip for visual detection. This technology was also used to detect Neisseria gonorrhoeae (NG) DNA from vaginal and urethral swabs to demonstrate the versatility of this platform.6
Figure 2:
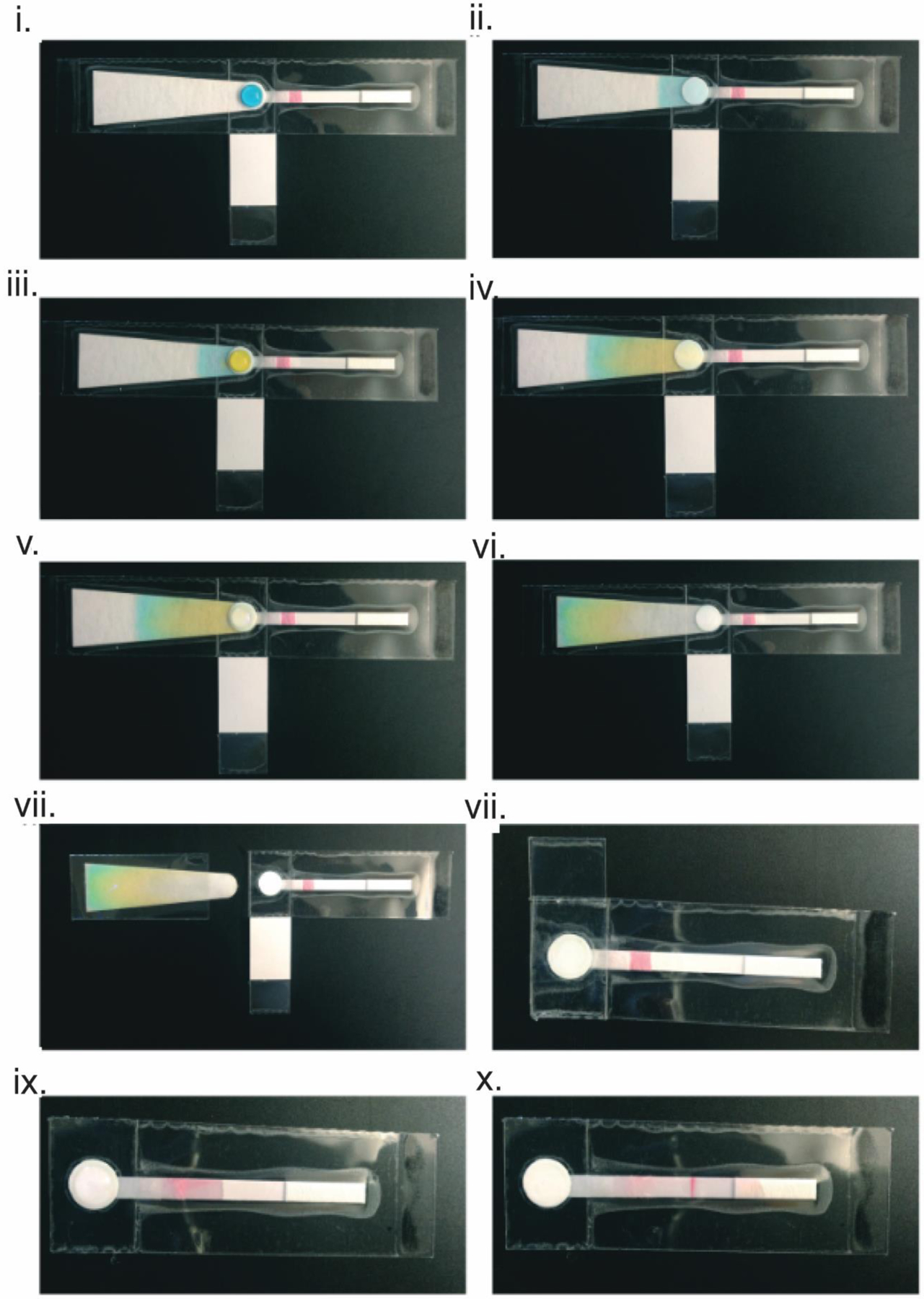
Fully integrated paperfluidic device. (i) A lysed sample, represented with blue dye, is applied to the PES membrane. (ii) The liquid phase of the sample is wicked away by the absorbent pad via capillary action while the solid phase remains on the PES membrane. (iii) The PES membrane is washed with 70% ethanol, represented here with yellow dye. (iv) The ethanol is wicked away by the wicking pad, removing impurities on the PES membrane. (v) A final wash with 100% ethanol, represented using water, is applied to the PES membrane. (vi) The final wash solution wicks through the absorbent pad, removing all impurities to leave behind purified DNA on the PES membrane. (vii) The absorbent pad is removed. (viii) The LAMP reagents are applied to the PES membrane and the bottom end of the chip is folder over to cover the membrane containing the purified DNA and LAMP reagents. (ix) After amplification, the covering on the membrane is removed as is the hydrophobic barrier between the PES membrane and the lateral flow strip. Water is applied to the PES membrane to elute the purified DNA. (x) The eluted DNA wicks to the right through the lateral flow strip. Reproduced from reference (7) with permission. Copyright Royal Society of Chemistry 2016.
In cases where the detection of a pathogen from whole blood is needed, the process requires more washing to remove inhibitors of downstream amplification techniques. To deal with whole blood samples, we built a flexible, paper-based device for nucleic acid sample preparation from blood (SNAPflex) (Fig 3). The device resembles the integrated paperfluidic device made by Rodriguez et al., but instead uses a glass fiber membrane to capture the nucleic acids. Similar to the previously published paperfluidic device,13 the sample is first mixed with lysis buffer that contains Glycoblue precipitant. The nucleic acid-glycoblue precipitate is again left behind on the glass fiber membrane while the solution phase is wicked away with the absorbent pad. This device has been used to extract HIV RNA and malaria DNA from whole blood for either immediate use or long-term storage of the extracted nucleic acid.
Figure 3:
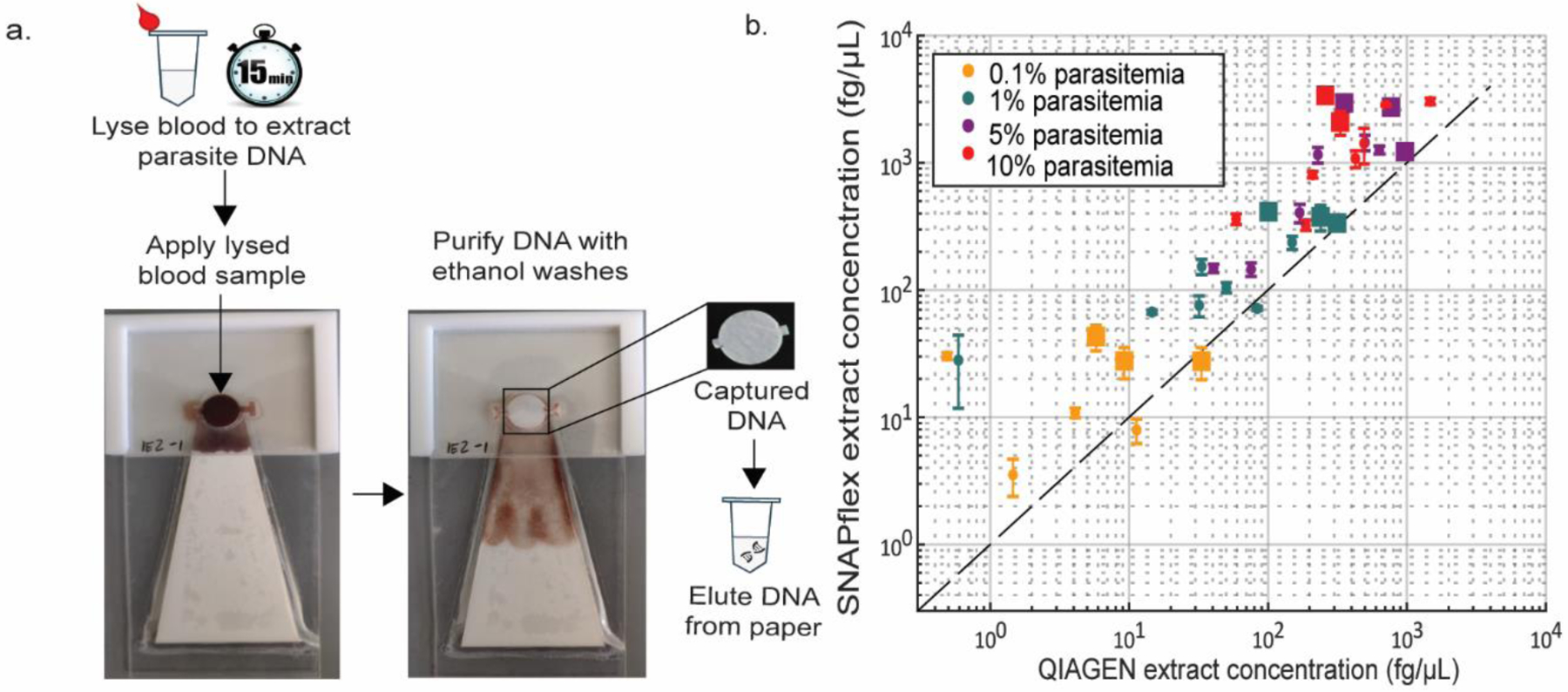
SNAPflex extraction of P. falciparum DNA using a glass fiber membrane. (a) lysed blood is applied to a glass fiber membrane. The liquid phase is wicked away by the absorbent pad. A series of ethanol washes purfies the DNA captured on the membrane. The membrane containing captured DNA is removed and used to elute the DNA. (b) SNAPflex extraction performs better than QIAGEN extraction. Adapted from refs (1) and (2). Reproduced with permission. Copyright Royal Society of Chemistry 2020 (1) and American Chemical Society 2021 (2).
When detection of low concentrations of pathogenic DNA in large volumes, such as urine, DNA must first be concentrated from large volumes of the sample. This can be done using solid-phase extraction techniques, such as using a porous polymer monolith to extract Chlamydia trachomatis (CT) DNA from clinical samples.8 However, the solid phase extraction of DNA requires chaotropic agents and nonpolar solvents that inhibit downstream nucleic acid amplification techniques (NAATs). An alternative to DNA extraction via solid phase extraction is a DNA enrichment strategy where the negatively charged DNA is captured by a positively charged substrate.4 We have demonstrated Trichomonas vaginalis (TV) DNA capture using a chitosan-modified paper membrane, which acquires a positive charge at a slightly acidic pH, from large urine samples without using inhibitors of downstream NAAT.4 As shown in Figure 4, the chitosan-functionalized paper is placed in a syringe that processes large volumes of urine. As the urine passes through the membrane, DNA is captured by the positively charged chitosan-functionalized membrane.
Figure 4.
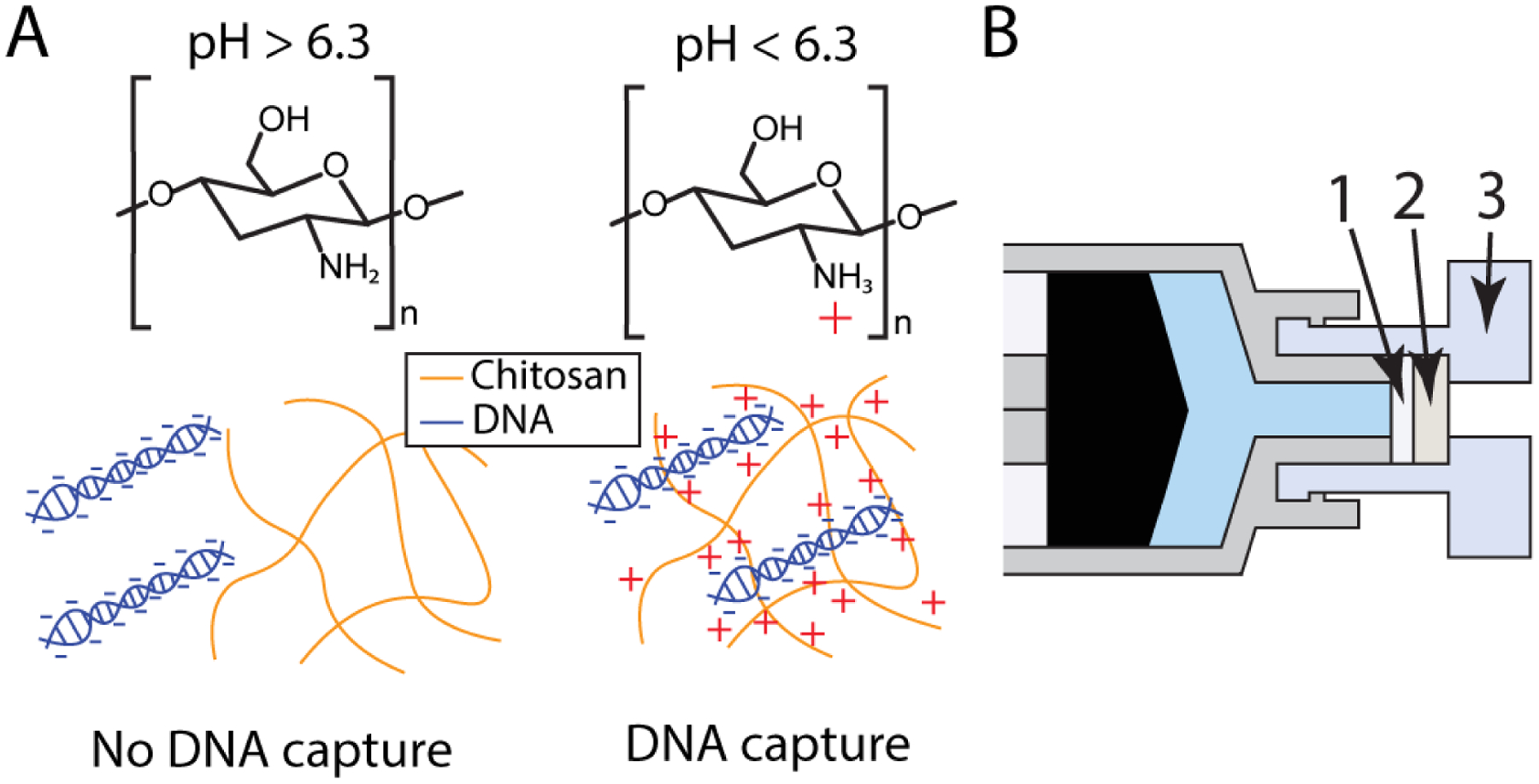
(a) Chitosan aquires a positive charge at slightly acidic pHs, allowing it to capture negatively charged DNA. (b) A chitosan-functionalized membrane (1) is placed in front of a cellulose backing membrane (2) in a female Luer cap (3). The sample is pushed through the chitosan-functionalized membrane with the syringe. The filter is then removed and used as a template for a TV tHDA assay. Reproduced from ref (4) with permission. Copyright Royal Society of Chemistry 2020.
Isothermal amplification
Once pathogenic DNA or RNA of interest has been isolated, for most detection methods, it needs to be amplified. In high-resource settings, this is done with the polymerase chain reaction (PCR), which requires expensive equipment for thermocycling. In order to obviate the need for a thermal cycler, a number of isothermal amplification techniques have been developed. These techniques can amplify DNA at a constant temperature, necessitating only a water bath or an inexpensive resistive heating element. These techniques have been extensively reviewed.14 We have predominately used thermophilic helicase-dependent amplification (tHDA)4,6,8,10 and loop-mediated isothermal amplification (LAMP).3,5,7,10 Our lab also engineered a novel isothermal amplification technique called Iso-IMRS, which achieves a sensitivity similar to that of PCR.2
HDA assays employ helicase enzymes to unwind DNA in preparation for replication via DNA polymerase.15,16 Unlike PCR, they do not require successive heating steps to de-hybridize the DNA and can be performed at a single temperature. After DNA unwinding, single stranded binding protein (SSBP) in the reaction mixture stabilizes the DNA and allows the primers to bind. Once both of the primers have bound to the template, DNA polymerase will begin the replication process. Early versions of the protocol used a helicase and polymerase able to perform amplification at 37°C. Later, a thermostable helicase was used, in conjunction with Bst DNA polymerase, to enable amplification at a higher temperature (65°C), thereby improving the reaction efficiency and reducing the formation of non-specific products. HDA assays that employ the thermostable helicase are called tHDA assays. We have used tHDA assays to amplify Chlamydia trachomatis DNA,8 Neisseria gonorrhoeae DNA,6 and Trichomonas vaginalis DNA4.
LAMP assays employ 4–6 primers that target between 6–8 regions of the target DNA and a strand-displacing DNA polymerase to amplify a target gene.17,18 Typically these reactions run at ~65°C. Initially, the assay was designed to incorporate four primers that targeted six regions of the gene; the addition of two more primers was added later to accelerate the reaction.19 The primers are designed such that the amplicons self-hybridize into a dumbbell structure; these dumbbell structures serve as additional points of initiation for the primers, leading to exponential amplification. LAMP is known to be resistant to common PCR inhibitors that are found in complex sample matrices.14 We have used LAMP reactions to amplify HPV DNA.5,7,10 We have also developed a method to characterize LAMP amplicons during assay development using fluorescent primers.20 We used reverse-transcriptase LAMP (RT-LAMP) to reverse-transcribe H1N1 RNA to DNA prior to amplifying the cDNA of interest.3
A novel isothermal amplification technique to detect P. falciparum genomic DNA was engineered in our laboratory.2 A collaborator devised a computational method to design PCR primers by finding identical multirepeat sequences (IMRS) in the genome. Primers designed to bind to these regions result in more amplification than those that only bind to one location on the genome. Using these primers, they made an assay requiring thermocycling with improved sensitivity compared to PCR.21 We developed an isothermal version of this assay, called iso-IMRS to detect P. falciparum DNA. This assay utilizes one forward primer which binds to 52 sites in the genome and one reverse primer which binds to 55 sites in the genome. Due to the fact that the primers target repeat sequences, iso-IMRS product results in amplicons of differing sizes. Iso-IMRS exhibits similar sensitivity to qPCR and successfully amplified P. falciparum DNA from human saliva and blood samples.
CRISPR Cas12a-based detection
One drawback of HDA and LAMP assays is that they often suffer from false-positive results. In PCR, the denaturation step reduces the impact of off target primer binding events. Isothermal techniques do not have this natural “reset” and accumulate non-specific binding events throughout the reaction time. The relatively long primers used in HDA make them prone to primer dimer formation15 while the large number of primers used in LAMP can lead to self-annealing of the primers, leading to nonspecific amplification.12 We coupled a LAMP reaction with a CRISPR-Cas12a assay to improve assay specificity.5 CRISPR-Cas12a is a RNA-guided enzyme that, upon binding to a specific sequence of double-stranded DNA, exhibits random endonuclease behavior (Fig 5).22 This target-activated endonuclease activity can be monitored fluorescently,22–25 visually via lateral flow strips24,26,27 or electrochemically.5,28,29 The CRISPR-Cas12a enzyme does not exhibit this endonuclease activity in the absence of its target sequence. Therefore, the Cas12a does not detect off target amplification products that have been generated in a LAMP reaction.5 Cas12a is one of many CRISPR enzymes that have been used for pathogen diagnostics; CRISPR-based detection of diseases has been extensively reviewed.30 We chose to use Cas12a to detect HPV DNA because HPV is a DNA virus and the substrate for Cas12a is double-stranded DNA. Other Cas enzymes act on different substrates, such as RNA or ssDNA; these enzymes and their roles in diagnostics have been extensively reviewed.30
Figure 5.
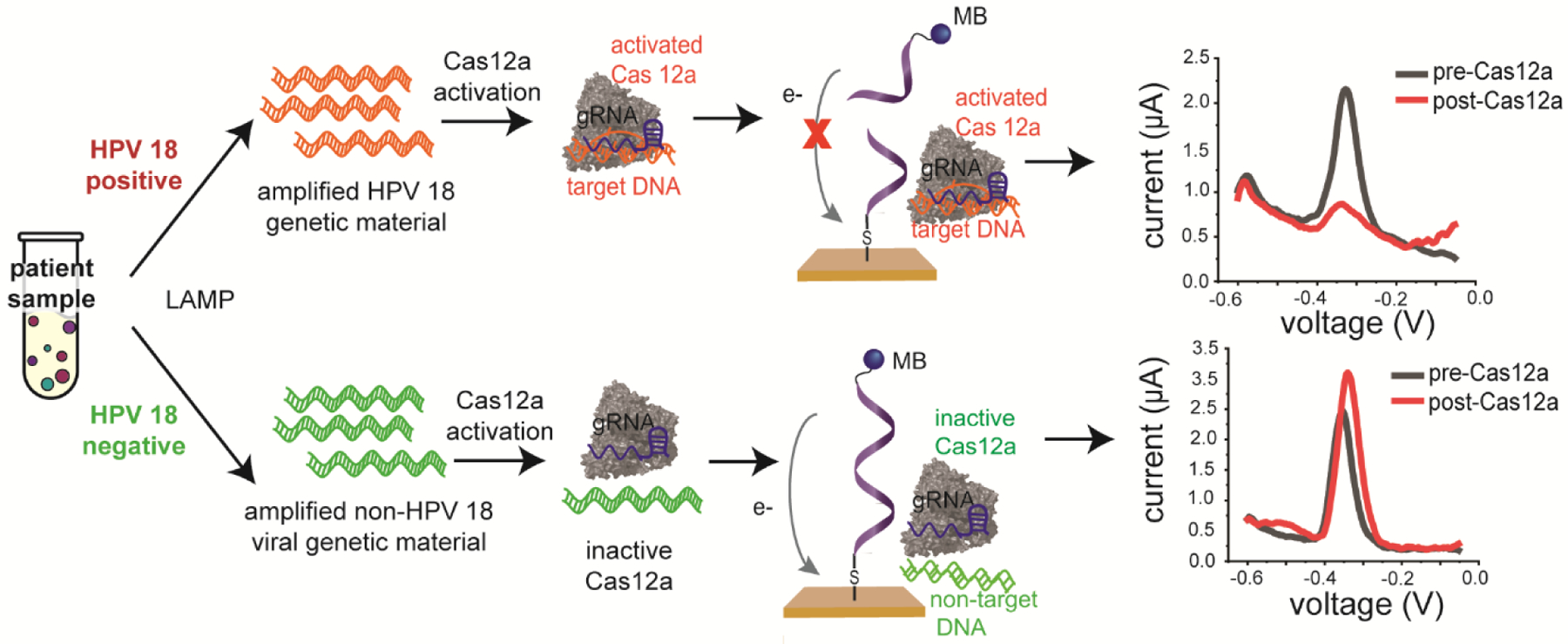
Cas12a is engineered to target HPV 18 DNA. In the presence of target DNA, Cas12a exhibits random endonuclease activity that degrades the single-stranded reporter DNA on the electrodes, resulting in a decrease in signal from the methylene blue (MB). In the presence of non-target DNA, Cas12a does not become activated and does not degrade the reporter DNA on the electrode. Reproduced from reference (5) with permission. Copyright American Chemical Society 2021.
Readout mechanisms
Once the gene of interest has been amplified, it must be detected. There are numerous readout mechanisms to detect amplified DNA, including colorimetric, fluorescent and electrochemical.31 These transduction modalities in POC biosensors has been extensively reviewed.32 Colorimetric readouts, including lateral flow readouts, are commonly used in POC diagnostics because they are easy to read without instrumentation. Fluorescent readout mechanisms offer increased sensitivity compared to colorimetric readouts, but results can be impaired by the auto fluorescence of other molecules or substrates in the reaction. Electrochemical readouts are favorable for POC diagnostics due to their portability, affordability and quantitative nature. Our technological development utilizes lateral-flow and electrochemical readouts.
Lateral-flow readout
A common readout for POC devices for use in LRS is the lateral-flow readout. One of the first examples of a lateral flow readout is a pregnancy test. As shown in Fig 6, a lateral flow strip consists of a membrane that contains antibodies against the antigen of interest. In order to detect amplified nucleic acids using a lateral flow strips, the primers used to amplify the DNA are tagged with either a small molecular that acts as an antigen, such as FAM or DIG, or biotin. FAM and DIG are often used because antibodies to these two molecules are commercially available and used routinely in molecular biology. The assay is designed to generate amplicons that are dual labeled with the antigen and the biotin.33 When the sample is applied to the lateral flow strip, the antigen binds to the antibodies on the strip while the biotinylated side of the amplicon binds to streptavidin-coated gold nanoparticles present at the end of the test strip that is dipped in the sample of interest. The aggregation of the gold nanoparticles on the antibody line results in a red line that is visible to the naked eye. We have used lateral flow readouts to detect amplicons generated by HDA,4,6 LAMP3,7 and iso-IMRS.2
Figure 6:
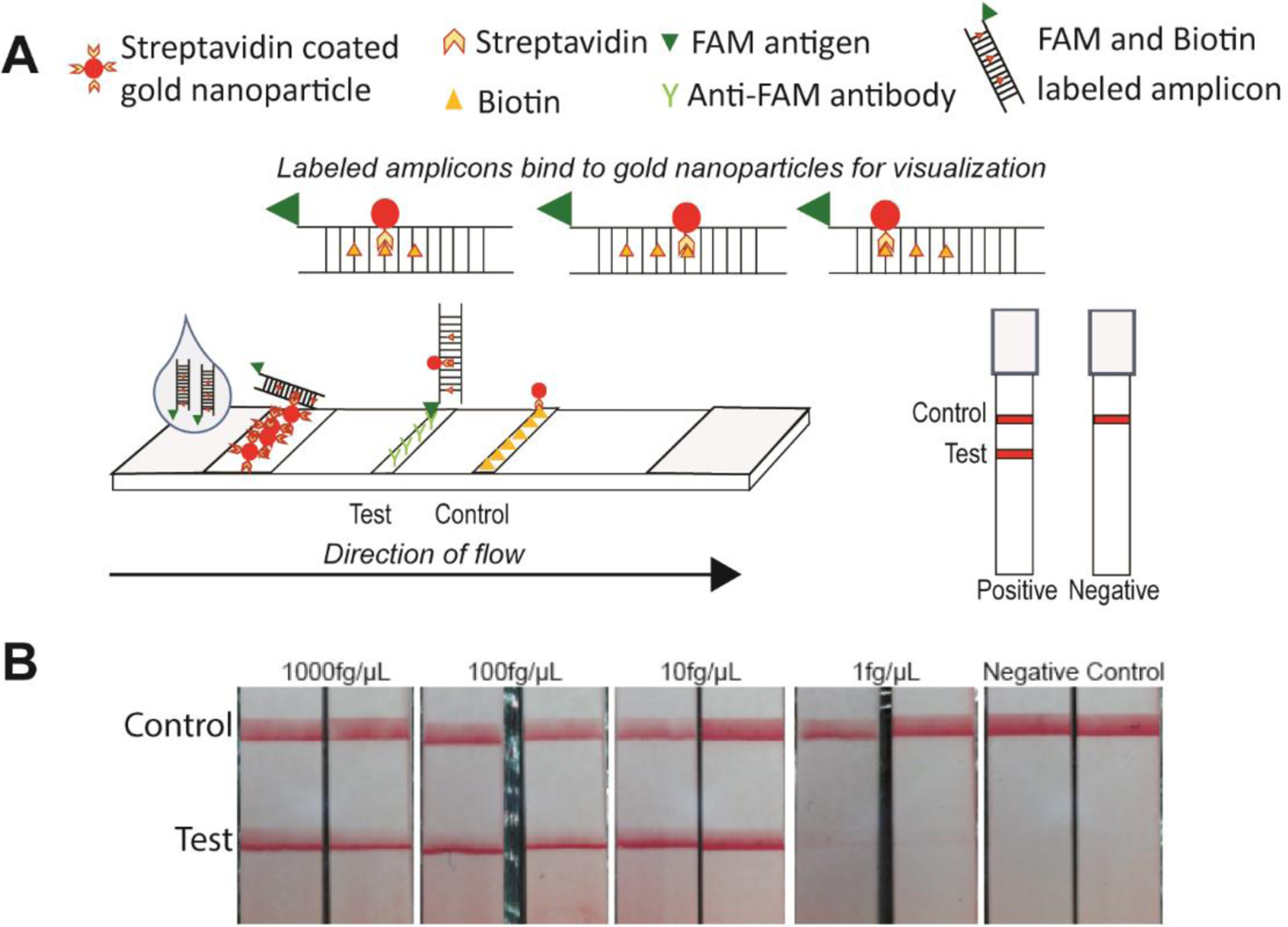
Lateral flow detection of amplicons. (a) Lateral flow strips designed to capture labeled amplicons. (b) Visual readout of iso-IMRS amplicons. Reproduced from reference (2) with permission. Copyright American Chemical Society 2021.
Electrochemical readout
Electrochemical readouts are favorable for point-of-care diagnostics due to their sensitivity, portability and affordability. A common electrochemical biosensor is the glucose meter. We used an electrochemical platform to monitor HPV-activated Cas12a endonuclease activity.5 Importantly, we used novel gold leaf electrodes that cost nearly one order of magnitude less ($0.50/electrode) than the most inexpensive version of their commercially available counterparts (~$4.00/electrode). While gold is a common substrate for electrochemical biosensors due its ease of modification with thiolated molecules,34 it is often expensive and requires specialized equipment to fabricate. We made gold electrodes using an equipment-free fabrication method that can be done in low-resource settings at an affordable cost (Fig 7). The gold leaf is inexpensive because it is very thin. Our work demonstrates an important step forward in being able to bring gold-based electrochemical biosensors to low-resource settings.35
Figure 7:
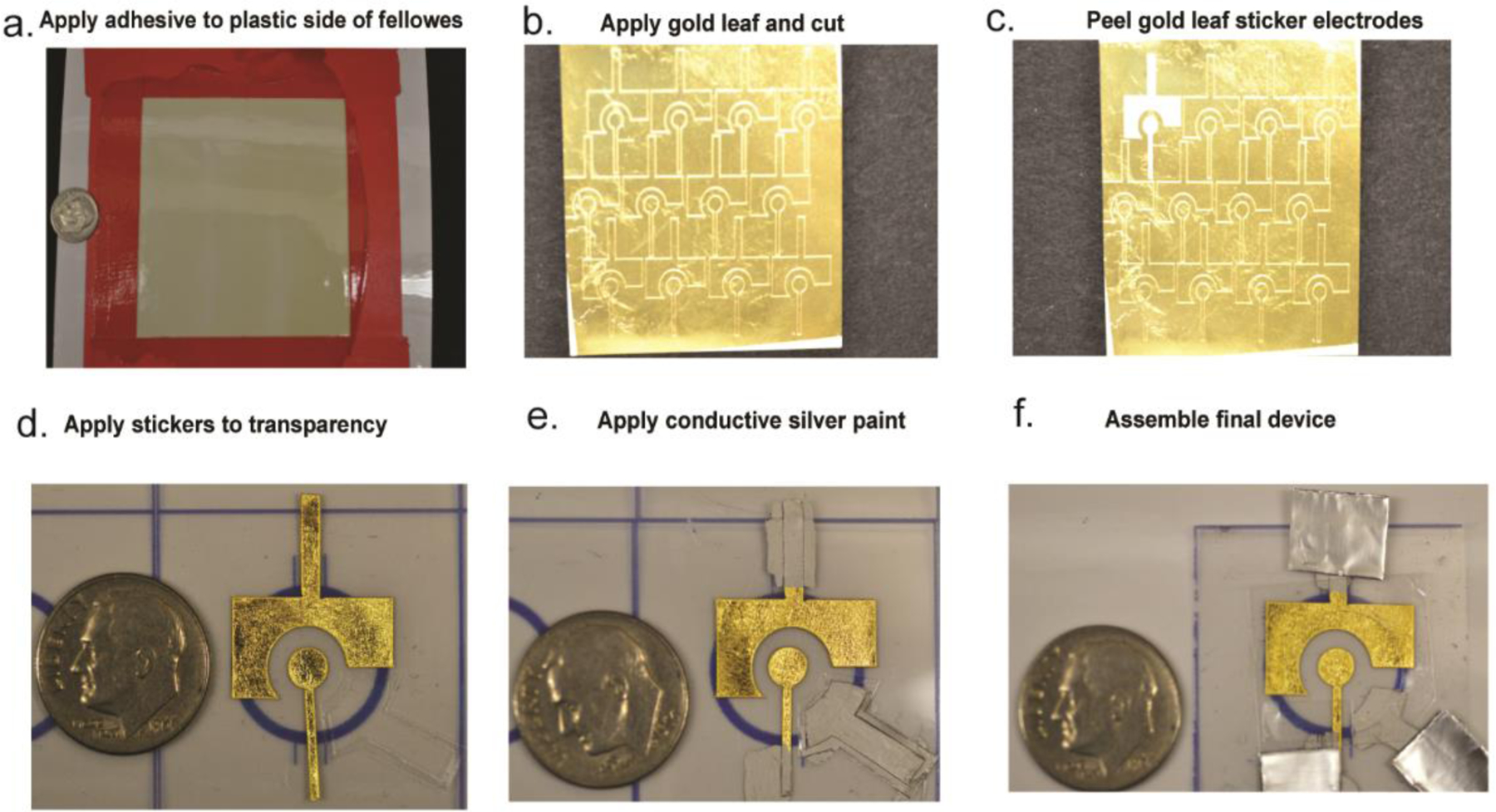
Equipment-free fabrication of gold leaf electrodes. Reproduced from reference (5) with permission. Copyright American Chemical Society 2021.
Biography
Marjon Zamani, Ph.D. is Postdoctoral Associate in Chemical Engineering at the Massachusetts Institute of Technology. In May 2021, she received her Ph.D. in Biomedical Engineering from Boston University in Dr. Klapperich’s lab. She builds electrochemical biosensors for point-of-care diagnostics.
Ariel L. Furst, Ph.D. is a Raymond (1921) & Helen St. Laurent Career Development Professor of Chemical Engineering at the Massachusetts Institute of Technology. She did her postdoctoral fellowship at UC Berkeley and obtained her Ph.D. in Chemistry from the California Institute of Technology. She develops electrochemical biosensors for clinical diagnostics.
Catherine M. Klapperich, Ph.D. is the Vice Chair of Biomedical Engineering at Boston University, Professor of Biomedical Engineering, Materials Science Engineering and Mechanical Engineering at Boston University and the Director of the Precision Diagnostics Center at Boston University. Her work focuses on building paperfluidic devices for the extraction, amplification and detection of nucleic acids in low-resource settings.
References
- 1.Kolluri N, Albarran N, Fan A, et al. SNAPflex: A paper-and-plastic device for instrument-free RNA and DNA extraction from whole blood. Lab Chip. 2020;20(18):3386–3398. doi: 10.1039/d0lc00277a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolluri N, Kamath S, Lally P, et al. Development and Clinical Validation of Iso-IMRS: A Novel Diagnostic Assay for P. falciparum Malaria. Anal Chem. 2021;93(4):2097–2105. doi: 10.1021/acs.analchem.0c03847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez NM, Linnes JC, Fan A, Ellenson CK, Pollock NR, Klapperich CM. Paper-Based RNA Extraction, in Situ Isothermal Amplification, and Lateral Flow Detection for Low-Cost, Rapid Diagnosis of Influenza A (H1N1) from Clinical Specimens. Anal Chem. 2015;87(15):7872–7879. doi: 10.1021/acs.analchem.5b01594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbohm JM, Robson JM, Singh R, et al. Rapid electrostatic DNA enrichment for sensitive detection of: Trichomonas vaginalis in clinical urinary samples. Anal Methods. 2020;12(8):1085–1093. doi: 10.1039/c9ay02478f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zamani M, Robson JM, Fan A, Bono MS, Furst AL, Klapperich CM. Electrochemical Strategy for Low-Cost Viral Detection. ACS Cent Sci. Published online 2021. doi: 10.1021/acscentsci.1c00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horst AL, Rosenbohm JM, Kolluri N, et al. A paperfluidic platform to detect Neisseria gonorrhoeae in clinical samples. Biomed Microdevices. 2018;20(2):35. doi: 10.1007/s10544-018-0280-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez NM, Wong WS, Liu L, Dewar R, Klapperich CM. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip. 2016;16(4):753–763. doi: 10.1039/c5lc01392e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linnes JC, Fan A, Rodriguez NM, Lemieux B, Kong H, Klapperich CM. Paper-based molecular diagnostic for Chlamydia trachomatis. doi: 10.1039/c4ra07911f [DOI] [PMC free article] [PubMed]
- 9.Smith S, Korvink JG, Mager D, Land K. The potential of paper-based diagnostics to meet the ASSURED criteria. RSC Adv. 2018;8(59):34012–34034. doi: 10.1039/C8RA06132G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linnes JC, Rodriguez NM, Liu L, Klapperich CM. Polyethersulfone improves isothermal nucleic acid amplification compared to current paper-based diagnostics. Biomed Microdevices. 2016;18(2):30. doi: 10.1007/s10544-016-0057-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez NM, Linnes JC, Fan A, Ellenson CK, Pollock NR, Klapperich CM. Paper-Based RNA Extraction, in Situ Isothermal Amplification, and Lateral Flow Detection for Low-Cost, Rapid Diagnosis of Influenza A (H1N1) from Clinical Specimens. Anal Chem. 2015;87(15):7872–7879. doi: 10.1021/acs.analchem.5b01594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez NM, Wong WS, Liu L, Dewar R, Klapperich CM. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip. 2016;16(4):753–763. doi: 10.1039/c5lc01392e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez NM, Wong WS, Liu L, Dewar R, Klapperich CM. A Fully Integrated Paperfluidic Molecular Diagnostic Chip for the Extraction, Amplification, and Detection of Nucleic Acids from Clinical Samples. Lab Chip. 2015;16(4):753–763. doi: 10.1039/c5lc01392e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asadi R, Mollasalehi H. The mechanism and improvements to the isothermal amplification of nucleic acids, at a glance. Anal Biochem. Published online May 21, 2021:114260. doi: 10.1016/j.ab.2021.114260 [DOI] [PubMed] [Google Scholar]
- 15.Barreda-García S, Miranda-Castro R, de-los-Santos-Álvarez N, Miranda-Ordieres AJ, Lobo-Castañón MJ. Helicase-dependent isothermal amplification: a novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal Bioanal Chem. 2018;410(3):679–693. doi: 10.1007/s00216-017-0620-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5(8):795–800. doi: 10.1038/sj.embor.7400200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becherer L, Borst N, Bakheit M, Frischmann S, Zengerle R, Von Stetten F. Loop-mediated isothermal amplification (LAMP)-review and classification of methods for sequence-specific detection. Anal Methods. 2020;12(6):717–746. doi: 10.1039/c9ay02246e [DOI] [Google Scholar]
- 18.Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):e63. doi: 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16(3):223–229. doi: 10.1006/mcpr.2002.0415 [DOI] [PubMed] [Google Scholar]
- 20.Landaverde L, Wong W, Hernandez G, Fan A, Klapperich C. Method for the elucidation of LAMP products captured on lateral flow strips in a point of care test for HPV 16. Anal Bioanal Chem. 2020;412(24):6199–6209. doi: 10.1007/s00216-020-02702-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raju LS, Kamath S, Shetty MC, et al. Genome Mining–Based Identification of Identical Multirepeat Sequences in Plasmodium falciparum Genome for Highly Sensitive Real-Time Quantitative PCR Assay and Its Application in Malaria Diagnosis. J Mol Diagnostics. 2019;21(5):824–838. doi: 10.1016/j.jmoldx.2019.04.004 [DOI] [PubMed] [Google Scholar]
- 22.Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science (80-). 2018;360(6387):436–439. doi: 10.1126/science.aar6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding X, Yin K, Li Z, et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-18575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsou JH, Leng Q, Jiang F. A CRISPR Test for Detection of Circulating Nuclei Acids. Transl Oncol. 2019;12(12):1566–1573. doi: 10.1016/j.tranon.2019.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganguli A, Mostafa A, Berger J, et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(37):22727–22735. doi: 10.1073/pnas.2014739117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Ji P, Fan H, et al. CRISPR/Cas12a technology combined with immunochromatographic strips for portable detection of African swine fever virus. Commun Biol. 2020;3(1):62. doi: 10.1038/s42003-020-0796-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolbasov DV, Zou W, Bai J, et al. Cas12a-Based On-Site and Rapid Nucleic Acid Detection of African Swine Fever. Published online 2019. doi: 10.3389/fmicb.2019.02830 [DOI] [PMC free article] [PubMed]
- 28.Dai Y, Somoza RA, Wang L, et al. Exploring the Trans-Cleavage Activity of CRISPR Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew Chemie Int Ed. Published online 2019. doi: 10.1002/anie.201910772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D, Yan Y, Que H, et al. CRISPR/Cas12a-Mediated Interfacial Cleaving of Hairpin DNA Reporter for Electrochemical Nucleic Acid Sensing. ACS Sensors. 2020;5(2):557–562. doi: 10.1021/acssensors.9b02461 [DOI] [PubMed] [Google Scholar]
- 30.Jolany Vangah S, Katalani C, Booneh HA, Hajizade A, Sijercic A, Ahmadian G. CRISPR-Based Diagnosis of Infectious and Noninfectious Diseases. Biol Proced Online. 2020;22(1):1–14. doi: 10.1186/s12575-020-00135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi JR. Development of Point-of-Care Biosensors for COVID-19. Front Chem. 2020;8(May). doi: 10.3389/fchem.2020.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pashchenko O, Shelby T, Banerjee T, Santra S. A Comparison of Optical, Electrochemical, Magnetic, and Colorimetric Point-of-Care Biosensors for Infectious Disease Diagnosis. ACS Infect Dis. 2018;4(8):1162–1178. doi: 10.1021/acsinfecdis.8b00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamphee H, Chaiprasert A, Prammananan T, Wiriyachaiporn N, Kanchanatavee A, Dharakul T. Rapid molecular detection of multidrug- resistant tuberculosis by PCR-nucleic acid lateral flow immunoassay. PLoS One. 2015;10(9):1–17. doi: 10.1371/journal.pone.0137791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wink T, Van Zuilen SJ, Bult A, Van Bennekom WP. Tutorial Review Self-Assembled Monolayers for Biosensors. [DOI] [PubMed]
- 35.Newsham E, Richards-Kortum R. CRISPR-Based Electrochemical Sensor Permits Sensitive and Specific Viral Detection in Low-Resource Settings. ACS Cent Sci. Published online 2021:926–928. doi: 10.1021/acscentsci.1c00555 [DOI] [PMC free article] [PubMed] [Google Scholar]


