Abstract
Human immunodeficiency virus type 1 (HIV-1) isolates resistant to (−)-β-d-dioxolane-guanosine (DXG), a potent and selective nucleoside analog HIV-1 reverse transcriptase (RT) inhibitor, were selected by serial passage of HIV-1LAI in increasing drug concentrations (maximum concentration, 30 μM). Two independent selection experiments were performed. Viral isolates for which the DXG median effective concentrations (EC50s) increased 7.3- and 12.2-fold were isolated after 13 and 14 passages, respectively. Cloning and DNA sequencing of the RT region from the first resistant isolate identified a K65R mutation (AAA to AGA) in 10 of 10 clones. The role of this mutation in DXG resistance was confirmed by site-specific mutagenesis of HIV-1LAI. The K65R mutation also conferred greater than threefold cross-resistance to 2′,3′-dideoxycytidine, 2′,3′-dideoxyinosine, 2′,3′-dideoxy-3′-thiacytidine, 9-(2-phosphonylmethoxyethyl)adenine, 2-amino-6-chloropurine dioxolane, dioxolanyl-5-fluorocytosine, and diaminopurine dioxolane but had only marginal effects on 3′-azido-3′-deoxthymidine (AZT) susceptibility. However, when introduced into a genetic background for AZT resistance (D67N, K70R, T215Y, T219Q), the K65R mutation reversed the AZT resistance. DNA sequencing of RT clones derived from the second resistant isolate identified the L74V mutation, previously reported to cause ddI resistance. The L74V mutation also decreased the AZT resistance when the mutation was introduced into a genetic background for AZT resistance (D67N, K70R, T215Y, T219Q) but to a lesser degree than the K65R mutation did. These findings indicate that DXG and certain 2′,3′-dideoxy compounds (e.g., ddI) can select for the same resistance mutations and thus may not be optimal for use in combination. However, the combination of AZT with DXG or its orally bioavailable prodrug (−)-β-d-2,6-diaminopurine-dioxolane should be explored because of the suppressive effects of the K65R and L74V mutations on AZT resistance.
Breakthrough replication of drug-resistant viral variants is a major obstacle in the search for effective treatments for human immunodeficiency virus type 1 (HIV-1) infection. Resistant variants have emerged with each Food and Drug Administration-approved class of antiretroviral agents, including nucleoside analogue reverse transcriptase (RT) inhibitors, nonnucleoside RT inhibitors, and protease inhibitors. Additionally, variants resistant to combinations of antiretroviral agents have been reported (2, 5). Problems with drug toxicities and suboptimal pharmacokinetics have also hindered the development of effective treatments for HIV-1 infection.
In response to these shortcomings, a variety of new antiretroviral compounds have been sought and discovered. Testing of each of these in clinical trials is neither economically feasible nor practical, making in vitro analysis a vital aspect of preclinical evaluation of drug candidates. Many previous studies have demonstrated that in vitro selection and characterization of drug-resistant viral variants is a useful means of identifying the genetic and biochemical mechanisms of resistance to specific compounds (24), as well as determining the time course for the development of resistance (14). This knowledge can help identify the most promising antiretroviral agents for clinical development. Additionally, in vitro studies can identify drug candidates for combination therapy by characterizing cross-resistance mutations as well as suppressor mutations that reverse phenotypic resistance (6, 14, 15).
(−)-β-d-Dioxolane-guanosine (DXG) was recently synthesized and licensed for clinical development (10). DXG is a purine nucleoside analog with the natural d configuration and is both a potent and selective inhibitor of HIV-1 and hepatitis B virus in vitro (4, 8, 10, 22, 26). An unique feature of DXG is the substitution of a dioxolane ring for the sugar moiety found in natural nucleosides (Fig. 1). The triphosphorylated form of DXG is recognized as a substrate by HIV-1 RT and is subsequently incorporated into DNA, resulting in chain termination because DXG lacks a 3′-OH group necessary for chain extension (Fig. 1) (8). The median effective concentration (EC50) of DXG is ∼27 nM in activated peripheral blood mononuclear cells (PBMCs), making it the most potent purine nucleoside analog against HIV-1 in these cells. In addition, DXG has no apparent cytotoxicity up to 100 μM. One potential limitation of DXG is its limited aqueous solubility (5); however, promising orally bioavailable prodrugs of DXG are available. Specifically, (−)-β-d-2,6-diaminopurine-dioxolane (DAPD) and (−)-β-d-2-amino-6-chloropurine dioxolane (ACPD) have favorable pharmacokinetic properties and undergo rapid biotransformation to DXG in vivo (4, 22). DAPD is in clinical development, and thus, this study was undertaken to determine the potential for HIV-1 to develop resistance to DXG.
FIG. 1.
Chemical structure of DXG.
MATERIALS AND METHODS
Chemicals.
DXG, 2′,3′-dideoxycytidine (ddC), 3′-azido-3′-deoxythymidine (AZT), 3′-deoxy-2′,3′-didehydrothymidine (D4T), 2′,3′-dideoxy-3′-thiacytidine (3TC), ACPD, DAPD (10), 9-(2-phosphonylmethoxyethyl)adenine (PMEA), and β-d-dioxolanyl-5-fluorocytosine (d-FDOC) were synthesized by our group and were provided by R. F. Schinazi. 2′,3′-Dideoxyinosine (ddI) and phosphonoformate (PFA) were purchased from Sigma Chemical Company, St. Louis, Mo. The compounds were prepared as 10 or 30 mM stock solutions in sterile water or dimethyl sulfoxide as appropriate and were stored at −20°C. Stock solutions were diluted to the appropriate concentrations with RPMI 1640 medium (Whittaker MA Bioproducts, Walkersville, Md.) immediately before use.
Cells.
MT-2 cells (AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health; contributed by D. Richman) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (JRH Biosciences, Lenexa, Kans.), 2 μM l-glutamine, 10 mM HEPES buffer, and antibiotics (50 IU of penicillin per ml, 50 μg of streptomycin per ml). PBMCs were isolated from HIV-1-seronegative donors and were activated with phytohemagglutinin (10 μg/ml; Difco Labs, Detroit, Mich.) for 3 days prior to infection with HIV-1. At the time of infection, PBMCs were transferred to and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 μM l-glutamine, 10% interleukin-2 (Cellular Products, Buffalo, N.Y.), and antibiotics.
Viruses.
Stock preparations of HIV-1LAI were prepared as described previously (21). Briefly, MT-2 cells were electroporated with 10 μg of plasmid DNA that encodes the HIV-1LAI proviral clone. Culture supernatants were harvested 5 to 7 days after infection at the peak viral cytopathic effect (CPE). Prior to selection for DXG-resistant virus, the plasmid-derived virus was passed 10 times as cell-free virus in MT-2 cells. The infectivity of the virus preparation was determined by threefold endpoint dilution in MT-2 cells (six cultures per dilution), and the 50% tissue culture infective dose was calculated by use of the equation of Reed and Muench (23).
Selection of resistant viruses.
Resistant virus was selected in two independent experiments by serial passage of HIV-1LAI in MT-2 cells in increasing concentrations of DXG. MT-2 cells (1.0 × 106) were pretreated with DXG for 2 h prior to inoculation with HIV-1LAI. Virus replication was measured by daily monitoring of viral cytopathic effects (CPEs). At a +2 CPE (more than two syncytia per field at a ×100 magnification), the culture fluids were clarified by centrifugation (200 × g for 10 min) and the supernatant was harvested. A 0.1-ml aliquot of the harvested supernatant was subsequently used to initiate a new passage in fresh MT-2 cells. Virus was passaged two to three times for each concentration, depending on how rapidly the virus grew in the presence of the drug at a particular concentration. The selective pressure was increased from an initial DXG concentration of 2.5 μM to a final concentration of 30 μM after 13 (selection 1) or 14 (selection 2) passages. After every two passages, the log10 reduction in viral infectivity in 30 μM DXG was determined to monitor the passaged virus for a reduction in susceptibility to DXG (19).
Antiviral susceptibility determinations.
Virus susceptibility to DXG and other compounds was determined in MT-2 cells as described previously (19). Briefly, MT-2 cells were infected at a multiplicity of 0.01 50% tissue culture infective dose per cell in the presence of serial drug dilutions. Each dilution was tested in triplicate. Culture supernatants were harvested 4 or 7 days after infection and were assayed for p24 antigen production by a commercial assay (DuPont, NEN Products, Wilmington, Del.). Virus susceptibility to DXG was also determined in PBMCs as described previously (25).
Cloning and DNA sequencing of HIV-1 RT.
HIV-1 RT was cloned from infected cell lysates as described previously (17, 18). The full-length RT-coding region was amplified by PCR and was ligated into a PCRII TA cloning vector (InVitrogen, San Diego, Calif.). Escherichia coli INVαF′ was subsequently transfected with the vector, and transformants were identified via EcoRI digestion. Plasmid DNA from transformants was purified (Qiagen Inc., Chatsworth, Calif.) and was sequenced as described previously (19, 21).
Production of mutant recombinant HIV-1.
Generation of mutant recombinant HIV-1 was performed as described previously (18). Mutant RT was generated via oligonucleotide-directed mutagenesis and was ligated into the pXXHIV-1LAI cloning vector. Cloning was facilitated by the presence of two silent restriction sites at the 5′ and 3′ ends (XmaI and XbaI, respectively) of the pXXHIV-1LAI RT (17, 18). Infectious mutant recombinant HIV-1 was generated by electroporating MT-2 cells with mutated pXXHIV-1LAI clones. The presence of the desired mutations was verified by DNA sequencing.
Statistical analysis.
The Wilcoxon rank sum test for independent sample means was used to test the significance of the observed differences in mean EC50s. One-tailed tests were used in comparing the EC50s for wild-type and recombinant viruses as the hypothesis tested was that the EC50s for recombinant viruses were greater than the EC50s for wild-type viruses.
Molecular modeling of DXG.
Calculations were performed on a Silicon Graphics Indy workstation with SYBYL, version 6.1a, software (Tripos Associates, St. Louis, Mo.). ddI, DAPD, and DXG were constructed by using the routines available in SYBYL. The geometries of the molecules were optimized, and charges were generated by using PM3 hamiltonian in a MOPAC interface. A grid search was performed on each molecule with 300 increments about the rotatable bonds C-2→N-1→C-1′→O-4′ and C-3′→C-4′→C-5′→O-5′. The global minimum conformation was further relaxed. Energy minimizations were performed by using the Tripos force field with MOPAC charges by the conjugate gradient method. These molecules were superimposed for comparisons. The molecules were fit by use of the “fit atoms” option in SYBYL by using N-1, C-2, N-3, C-4, N-7, C-8, N-9, and C-1′ atoms in such a way that the root mean square deviation is minimized.
RESULTS
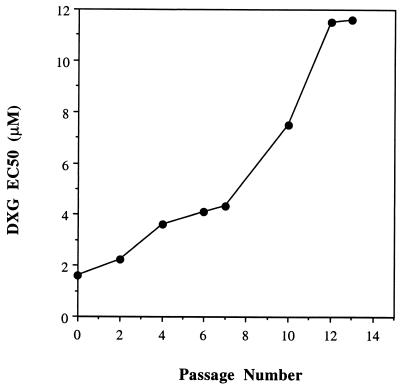
DXG-resistant HIV-1LAI isolates were selected in vitro in two separate experiments by serial passage of virus in MT-2 cells in escalating drug concentrations. In the first experiment, the EC50 of DXG increased only about twofold following the first seven passages (Fig. 2). By passage 10, however, a fivefold increase in the EC50 was apparent compared to the EC50 for control HIV-1LAI passaged in the absence of drug. The level of DXG resistance reached a maximum of 7.3-fold by passage 13 and did not increase further with subsequent passages. In the second selection experiment, DXG-resistant virus emerged at a rate similar to that observed in the first selection, reaching its maximal resistance level of 12.2-fold by passage 14 (data not shown).
FIG. 2.
In vitro resistance to DXG.
Genotypic analysis.
To determine the molecular basis for DXG resistance, the RT region (amino acids 1 to 560) of proviral DNA was amplified by PCR and was cloned from the infected cells of passage 13 (selection 1) and passage 14 (selection 2). As a control, proviral DNA was amplified and cloned from cells infected with virus passaged in the absence of drug. DNA sequencing identified the presence of a lysine (K)-to-arginine (R) mutation at position 65 (AAA to AGA) in 10 of 10 RT clones derived from passage 13 infected cells (selection 1). The K65R mutation was not detected in any of the control clones (0 of 10). Sequence analysis of RT clones derived from the second selection (passage 14 cells) revealed a leucine (L)-to-valine (V) mutation at position 74 (ATT to AGT) in 8 of 10 RT clones but in none of the control clones (0 of 10). No other mutations in the RT region were found in more than one clone from either selection experiment.
Susceptibilities of recombinant viruses.
To confirm the suspected roles of the K65R and L74V mutations in DXG resistance, each mutation was introduced individually by site-specific mutagenesis into a wild-type proviral clone (HIV-1LAI). Additionally, each mutation was introduced into a genetic background for AZT resistance (67N, 70R, 215Y, 219Q) to assess the effect of each mutation on AZT susceptibility. Table 1 summarizes the susceptibilities of the K65R mutant recombinant viruses to DXG and AZT assayed in MT-2 cells (inhibition of p24 antigen production). The K65R mutation conferred 8.7-fold DXG resistance, which was similar to the level of DXG resistance observed for virus isolated in the first selection experiment (7.3-fold). A similar level of DXG resistance (8.0-fold) was observed when the K65R mutant was assayed in PBMCs (data not shown). The K65R mutation had a minimal effect on AZT susceptibility in a wild-type background, increasing the AZT EC50 by only 3.3-fold (Table 1). By contrast, the K65R mutation markedly increased susceptibility to AZT when the mutation was introduced into an AZT resistance background, reducing the level of AZT resistance from 47.7- to 1.3-fold (P < 0.05). The AZT resistance mutations had little effect on the level of DXG resistance conferred by the K65R mutation, reducing this from 8.7- to 5.0-fold (P < 0.05).
TABLE 1.
DXG-AZT mutational interactions
| Virus | DXG-susceptible viruses
|
AZT-susceptible viruses
|
||
|---|---|---|---|---|
| EC50 (μM)a | Fold resistanceb | EC50 (μM) | Fold resistance | |
| HIV-1LAI | 0.28 ± 0.04 | 0.003 ± 0.000 | ||
| HIV-1LAI K65R | 2.41 ± 0.35 | 8.7 | 0.010 ± 0.003 | 3.3 |
| HIV-1LAI L74V | 1.17 ± 0.26 | 4.2 | 0.006 ± 0.002 | 1.9 |
| HIV-1LAI 4×AZTc | 0.58 ± 0.17 | 2.1 | 0.14 ± 0.04 | 47.7 |
| HIV-1LAI 4×AZT + K65R | 1.40 ± 0.16 | 5.0 | 0.004 ± 0.0004 | 1.3 |
| HIV-1LAI 4×AZT + L74V | 1.04 ± 0.14 | 3.7 | 0.019 ± 0.006 | 6.3 |
EC50s were determined via the p24 antigen assay in MT-2 cells (see Materials and Methods). Values are means ± standard errors (n = 3 or 4 experiments).
Fold resistance relative to the EC50 for the wild-type virus.
AZT-resistant strain with mutations 67N, 70R, 215Y, and 219Q.
The L74V mutation conferred ∼4.2-fold DXG resistance when introduced into the HIV-1LAI clone. This was ∼3-fold lower DXG resistance than that observed for virus isolated in the second selection experiment (12.2-fold increase in the EC50). Tenfold resistance was observed when the L74V mutant virus was assayed in PBMCs (data not shown). Similar to K65R, the L74V mutation did not reduce susceptibility to AZT in a wild-type background (P = 0.19) (Table 1). The L74V mutation did, however, reduce the level of AZT resistance from 47.7- to 6.3-fold when the mutation was introduced into an AZT resistance background (P < 0.05), although this reduction was less than that observed with the K65R mutation (Table 1). The AZT resistance mutations had essentially no effect on the level of DXG resistance conferred by the L74V mutation (4.2- to 3.7-fold).
Cross-resistance of the K65R mutant.
The K65R recombinant virus was tested for susceptibility to different antiretroviral agents in MT-2 cells to assess the level of cross-resistance conferred by this mutation. Table 2 shows that the K65R mutation significantly reduced the susceptibility to the other dioxolane derivatives tested (d-ACPD and d-DAPD), as well as to ddI, ddC, PMEA, d-FDOC, PFA, and 3TC. The K65R mutation had minimal effects on susceptibility to AZT (3.3-fold increase) (Table 1).
TABLE 2.
Cross-resistance of HIV-1LAI encoding K65R
| Antiviral agent | Mean EC50 (μM)a | Fold resistanceb |
|---|---|---|
| ddC | 6.6 ± 1.9 | 3.1 |
| ddI | 14.2 ± 3.6 | 3.1 |
| D4T | 18.4 ± 6.3 | 3.0 |
| 3TC | >60 | >14.2 |
| PFA | 66.3 ± 3.2 | 3.0 |
| d-ACPD | 12.9 ± 3.9 | 3.7 |
| d-DAPD | 48.1 ± 8.9 | 4.5 |
| d-FDOC | 2.6 ± 0.6 | 7.7 |
| PMEA | >60 | >4.3 |
EC50s were determined by measurement of inhibition of p24 antigen production in MT-2 cells (see Materials and Methods). Values are means ± standard errors (n = 3 or 4 experiments).
Fold resistance relative to the EC50 for the parental virus.
Computer modeling.
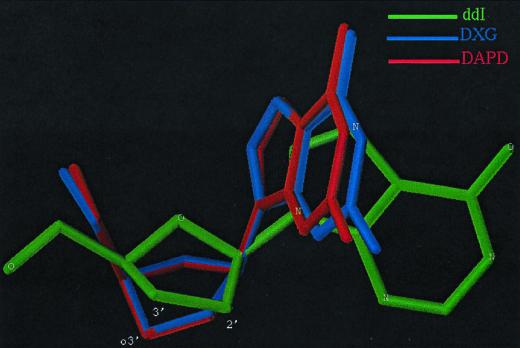
Figure 3 shows the results of structural modeling of DXG, DAPD, and ddI. The pictures of the fit of the atoms show conformational differences among ddI, DAPD, and DXG. DAPD and DXG are closely related when the pseudorotation angle and other torsional angles are compared. However, ddI has a sugar conformation different from those of DAPD and DXG. The longer bond length of C-2′→O-3′ in DAPD and DXG than that of C-2′→C-3′ in ddI and the electronic effects of O-3′ play a key role in providing the unique conformations of DAPD and DXG.
FIG. 3.
Computer modeling of DXG, DAPD, and ddI.
DISCUSSION
HIV-1 variants resistant to DXG, a nucleoside analog RT inhibitor, were selected twice by serial passage of HIV-1LAI in increasing concentrations of compound. After 13 and 14 passages, variants with 7.3- and 12.2-fold increases in EC50s, respectively, were isolated. DNA sequencing and site-specific mutagenesis demonstrated that the K65R mutation was responsible for the DXG resistance observed in the first selection experiment. The K65R mutation also conferred cross-resistance to ddI, ddC, 3TC, PMEA, PFA, d-ACPD, d-FDOC, and d-DAPD but produced little change in AZT susceptibility. The K65R mutation was found to reverse the AZT resistance when the mutation was introduced into an AZT resistance background by ∼37-fold. These results are similar to those reported by Zhang et al. (30), who found that the K65R mutation resulted in resistance to ddC and cross-resistance to ddI but no change in susceptibility to AZT.
DNA sequence analysis of resistant virus from the second selection identified a leucine-to-valine substitution at codon 74. An L74V recombinant virus exhibited 4.2-fold resistance, which was ∼3-fold less than the level of resistance observed in the selected virus (12.2-fold). This difference was reproducible in three separate experiments and thus was not likely due to assay variation. Because no other mutations were consistently found in RT clones derived from this selection, mutations outside of the gene for RT, such as in the envelope gene or another HIV gene, could have contributed to the observed DXG resistance in the selected virus. The L74V mutation was also found to reduce the level of AZT resistance when the mutation was introduced into an AZT resistance background by ∼8-fold. A study by St. Clair et al. (28) identified the L74V mutation in ddI-resistant virus isolated from patients who had changed from AZT to ddI therapy and determined that L74V conferred cross-resistance to ddC and increased susceptibility to AZT. Our finding that DXG selects for the K65R and L74V mutations in vitro has been confirmed in recent work by Borroto-Esoda et al. [K. Borroto-Esoda, J. Mewshaw, D. Wakefield, B. Hooper, J. Jeffrey, P. Furman, and B. McCreedy, Antivir. Ther. 4(Suppl. 1), abstr. 3, 1999] and Gu et al. (8). Borroto-Esoda et al. [Antivir. Ther. 4(Suppl. 1), abstr. 3, 1999] performed similar selection experiments with DAPD, a prodrug of DXG. Their initial selection with DAPD isolated the K65R mutant, whereas a second selection isolated the L74V mutant. Gu et al. (8) have shown that virus with the K65R, L74V, or M184V mutation exhibits two- to fivefold resistance to DXG or DAPD. The M184V mutation was not selected in our study, but low-level DXG cross-resistance from this mutation is not surprising because it is well known that it confers low-level cross-resistance to ddI and ddC. According to the recent “closed” RT structure published by Huang et al. (9), K65R, L74V, and M184I/V are each located in the region of RT that interacts with the incoming nucleotide. Therefore, mutations at each of these positions could potentially change the interaction of RT with DXG triphosphate.
Differences in in vitro selection methods have been reported to yield different resistance mutations (12). In this study, however, the same selection methods yielded two different resistance mutations (K65R and L74V). As mentioned above, HIV-1LAI was passaged 10 times in the absence of DXG prior to the start of the selection procedures, which likely increased the genetic heterogeneity of the starting virus population. Kinjerski and Buckheit (11) have reported that the genetic composition of the starting virus population may result in the selection of different genetic variants in vitro. In our study, the isolation of two distinct resistant mutants may have been a chance event as a result of the presence of different resistant quasispecies in the starting virus population or as a result of evolution under drug selective pressure.
That ddI, ddC, and DXG select similar resistance mutations suggests that the triphosphate forms of these analogs interact similarly with the active site of RT. Although the three compounds differ from each other in their nucleoside base compositions, they are all β-d enantiomers and contain similar although not identical sugar components. ddI and ddC are both 2′,3′-dideoxy nucleosides, whereas DXG is a 1,3-dioxolane nucleoside. Computer modeling suggests that the triphosphate forms of DXG and ddI (ddATP) have distinct structural configurations (Fig. 3), although these differences are apparently not sufficient to alter the resistance mutations that are selected. By contrast, AZT selects different resistance mutations than DXG or ddI, probably as a result of the large 3′ azido group of AZT. These observations indicate that the base component of a nucleoside has little influence on the resistance mutations that are selected, whereas the sugar component can be an important determinant (e.g., ddI and DXG versus AZT). These findings are supported by the “closed” RT structure published by Huang et al. (9), which showed that the large 3′ azido group of AZT is likely responsible for the selection of mutations on the “rear face” of RT. Conversely, the lack of 3′ substituents on compounds such as DXG, ddI, and ddC is likely responsible for the selection of mutations on the “front face” of RT.
While the suppressive effects of the L74V and M184V mutations have been documented, this property of the K65R mutation has not been previously recognized. Mutations selected by nonnucleoside RT inhibitors (L100I [3], Y181C [13, 14]) and the foscarnet resistance mutations (W88G, E89K, L92I, Q161L [29]), have also been shown to suppress AZT resistance when introduced into an AZT resistance background. The K65R and L74V mutations confer resistance to 2′,3′-dideoxy (7, 16, 27, 28, 30) and dioxolane nucleoside RT inhibitors, while they suppress the phenotypic effects of AZT resistance mutations. This is a significant finding given that DXG and AZT target the same site of HIV-1 RT but appear to have antagonistic resistance mutations. As mentioned above, the lack of AZT cross-resistance conferred by K65R and L74V is likely the result of the different interactions of the two compounds with RT. The suppression of AZT resistance by these mutations, however, is more difficult to explain. Gu et al. (7, 9) reported that the K65R mutation confers resistance to ddC and noted that positions 65 to 71 of RT play important roles in RT binding of deoxynucleoside triphosphate substrates. Additionally, Sluis-Cremer et al. [N. Sluis-Cremer, D. Arion, N. Kaushik, H. Lim, and M. A. Parniak, Antivir. Ther. 4(Suppl. 1), abstr. 29, 1999] recently reported that the K65R mutation results in a 10-fold decrease in the binding of ddCTP to HIV-1 RT. Given that K65R is an important site for deoxynucleoside triphosphate binding [Sluis-Cremer et al., Antivir. Ther. 4(Suppl. 1), abstr. 29, 1999], it is possible that mutations at this position in RT interfere with the process of pyrophosphorolysis, a proposed mechanism of AZT resistance (1, 20). There is in fact some evidence to suggest that a K65R mutant is less able than wild-type virus to undergo pyrophosphorolysis [Sluis-Cremer et al., Antivir. Ther. 4(Suppl. 1), abstr. 29, 1999], thus potentially explaining how this mutation reverses AZT resistance. Additional studies on the interactions between the AZT resistance mutations and K65R and L74V would provide more insight into this observation. Of note is that the AZT resistance background used in this study did not include M41L. St Clair et al. (28) showed that while L74V could reverse the AZT resistance caused by T215Y, this effect was lost in the presence of the M41L mutation. Because we did not include the M41L mutation in our studies, it is unknown whether M41L would have a similar effect on the reversal of the AZT resistance caused by K65R.
The results of this study yield several important conclusions. First, there are common patterns of resistance to DXG and 2′,3′-dideoxynucleoside analogs, which suggest that some of these compounds should not be used in combination with each other. Second, that DXG, ddI, and ddC select for similar resistance mutations indicates that the nucleoside base component has little effect on the resistance mutations selected for by a given nucleoside analogue RT inhibitor. Third, the lack of cross-resistance to AZT and the reversal of AZT resistance by DXG resistance mutations provide strong rationales for the use of these compounds in combination with one another.
ACKNOWLEDGMENTS
This work was supported by grants from the Medical Research Service of the U.S. Department of Veterans Affairs (to R.F.S. and J.W.M.), the U.S. Department of Defense (to R.F.S. and J.W.M.), and the National Institutes of Health (grant RO1 AI-25899 [to C.K.C. and R.F.S.] and grant AI-41980 [to R.F.S.]).
REFERENCES
- 1.Arion D, Kaushik N, McCormick S, Borkow G, Parniak M A. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 2.Balzarini J, Karlsson A, Perez-Perez M J, Camarasa M J, Tarpley W G, De Clercq E. Treatment of human immunodeficiency virus type 1 (HIV-1)-infected cells with combinations of HIV-1-specific inhibitors results in a different resistance pattern than does treatment with single-drug therapy. J Virol. 1993;67:5353–5359. doi: 10.1128/jvi.67.9.5353-5359.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckheit R W, Kinjerski T L, Fliakas-Boltz V, Russell J D, Stup T L, Pallansch L A, Brouwer W G, Dao D C, Harrison W A, Schultz R J, Bader J P, Yang S S. Structure-activity and cross-resistance evaluations of a series of human immunodeficiency virus type 1-specific compounds related to oxathiin carboxanilide. Antimicrob Agents Chemother. 1995;39:2718–2727. doi: 10.1128/aac.39.12.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Boudinot F D, Chu C K, McClure H M, Schinazi R F. Pharmacokinetics of (−)-β-d-2-aminopurine dioxolane and (−)-β-d-2-amino-6-chloropurine dioxolane, and their antiviral metabolite (−)-β-d-dioxolane guanine in rhesus monkeys. Antimicrob Agents Chemother. 1996;40:2332–2336. doi: 10.1128/aac.40.10.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condra J H, Schleif W A, Blahy O M, Gabryelski L, Graham D, Quintero J, Rhodes A, Robbins H, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K, Deutsch P, Emini E. In vivo emergence of HIV-1 to variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 6.De Clercq E. HIV resistance to reverse transcriptase inhibitors. Biochem Pharmacol. 1994;47:155–169. doi: 10.1016/0006-2952(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 7.Gu Z, Gao Q, Fang H, Salomon H, Parniak M A, Goldberg E, Cameron J, Wainberg M A. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistance to 2′3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1994;38:275–281. doi: 10.1128/aac.38.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu Z, Wainberg M A, Nguyen-BA N, L'Heureux L, de Muys J-M, Bowlin T L, Rando R F. Mechanism of action and in vitro activity of 1′,3′-dioxolanylpurine nucleoside analogues against sensitive and drug-resistant human immunodeficiency virus type 1 variants. Antimicrob Agents Chemother. 1999;43:2376–2382. doi: 10.1128/aac.43.10.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 10.Kim H O, Schinazi R F, Shanmuganathan K, Cannon D L, Alves A J, Jeong L S, Beach J W, Chu C K. 1,3-Dioxolanylpurine nucleosides (2R, 4R) and (2R, 4S) with selective anti-HIV-1 activity in human lymphocytes. J Med Chem. 1993;36:30–37. doi: 10.1021/jm00053a004. [DOI] [PubMed] [Google Scholar]
- 11.Kinjerski T L, Buckheit R W., Jr The role of genotypic heterogeneity in wild type virus populations on the selection of non-nucleoside reverse transcriptase inhibitor resistant viruses. Antivir Res. 1997;33:109–115. doi: 10.1016/s0166-3542(96)01008-x. [DOI] [PubMed] [Google Scholar]
- 12.Kleim J P, Winkler I, Rosner M, Kirsch R, Rubsamen-Waigmann H, Paessens A, Riess G. In vitro selection for different mutational patterns in the HIV-1 reverse transcriptase using high and low selective pressure of the nonnucleoside reverse transcriptase inhibitor HBY 097. Virology. 1997;231:112–118. doi: 10.1006/viro.1997.8513. [DOI] [PubMed] [Google Scholar]
- 13.Larder B A. 3′-Azido-3′-deoxythymidine resistance suppressed by a mutation conferring human immunodeficiency virus type 1 resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1992;36:2664–2669. doi: 10.1128/aac.36.12.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larder B A. Viral resistance and the selection of antiretroviral combinations. J Acquired Immune Defic Syndr. 1995;10:S28–S33. [PubMed] [Google Scholar]
- 15.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 16.Martin J L, Wilson J E, Haynes R L, Furman P A. Mechanism of resistance of human immunodeficiency virus type 1 to 2′,3′-dideoxyinosine. Proc Natl Acad Sci USA. 1993;90:6135–6139. doi: 10.1073/pnas.90.13.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellors J W, Im G J, Tramontano E, Winkler S R, Medina D J, Dutschman G E, Bazmi H Z, Piras G, Gonzalez C J, Cheng Y C. A single conservative amino acid substitution in the reverse transcriptase of human immunodeficiency virus-1 confers resistance to (+)-(5S)-4,5,6,7- tetrahydro-5-methyl-6-(3-methyl-2-butenyl)imidazo[4,5,1-jk][1,4]benzodiazepin-2(1H)-thione (TIBO R82150) Mol Pharmacol. 1992;43:11–16. [PubMed] [Google Scholar]
- 18.Mellors J W, Dutschman G E, Im B J, Tramontano E, Winkler S R, Cheng Y-C. In vitro selection and molecular characterization of human immunodeficiency virus type 1 resistant to non-nucleoside inhibitors of reverse transcriptase. Mol Pharmacol. 1992;41:446–451. [PubMed] [Google Scholar]
- 19.Mellors J W, Bazmi H Z, Schinazi R F, Roy B M, Hsiou Y, Arnold E, Wir J, Mayers D L. Novel mutations in reverse transcriptase of human immunodeficiency virus type 1 reduce susceptibility to foscarnet in laboratory and clinical isolates. Antimicrob Agents Chemother. 1995;39:1087–1092. doi: 10.1128/aac.39.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer P R, Matsuura S E, So A G, Scott W A. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc Natl Acad Sci USA. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen M H, Schinazi R S, Shi C, Goudgaon N M, NcKenna P M, Mellors J W. Resistance of human immunodeficiency virus type 1 to acyclic 6-phenylselenenyl and 6-phenylthiopyrimidines. Antimicrob Agents Chemother. 1994;38:2409–2414. doi: 10.1128/aac.38.10.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajagopalan P, Boudinot F D, Chu C K, Tennant B C, Baldwin B H, Schinazi R F. Pharmacokinetics of (−)-β-d-2,6-diaminopurine dioxolane and its metabolite, dioxolane guanosine, in woodchucks (Marmota monax) Antivir Chem Chemother. 1996;7:65–70. [Google Scholar]
- 23.Reed L J, Muench H. A simple method for estimating fifty percent end points. Am J Hyg. 1938;27:493–496. [Google Scholar]
- 24.Richman D D. Resistance of clinical isolates of human immunodeficiency virus to antiretroviral agents. Antimicrob Agents Chemother. 1993;37:1207–1213. doi: 10.1128/aac.37.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schinazi R F, Sommadossi J-P, Saalmann V, Cannon D L, Xie M-Y, Hart G, Smith G C, Hahn E F. Activity of 3′-azido-3′-deoxythymidine nucleotide dimers in primary lymphocytes infected with human immunodeficiency virus type 1. Antimicrob Agents Chemother. 1990;34:1061–1067. doi: 10.1128/aac.34.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schinazi R F, McClure H M, Boudinot F D, Xiang Y, Chu C K. Development of (−)-β-d-2,6-diaminopurine dioxolane as a potential antiviral agent. Antivir Res. 1994;23:81. [Google Scholar]
- 27.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1997;5(8):129–142. [Google Scholar]
- 28.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Resistance to DDI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 29.Tachedjian G, Mellors J, Bazmi H, Birch C, Mills J. Zidovudine resistance is suppressed by mutations conferring resistance of human immunodeficiency virus type 1 to foscarnet. J Virol. 1996;70:7171–7181. doi: 10.1128/jvi.70.10.7171-7181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang D, Caliendo A M, Eron J J, DeVore E M, Kaplan J C, Hirsch M S, D'Aquila R T. Resistance to 2′,3′-dideoxycytidine conferred by a mutation in codon 65 of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1994;38:282–287. doi: 10.1128/aac.38.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]