Abstract
Background
Renal cell carcinomas (RCC) are a heterogeneous group of neoplasms. Recent genome and transcriptome studies revealed characteristic genomic aberrations and molecular features of different histologic RCC subtypes, including chromophobe renal cell carcinoma (ChRCC).
Objective
To characterize the gene expression and biomarker signatures associated with for ChRCC.
Design, setting, and participants
We performed integrative RNA sequencing analysis from 1,049 RCC specimens from The Cancer Genome Atlas and in-house studies. Our workflow identified genes relatively enriched in ChRCC, including FOXI1, RHCG, and a novel long non-coding RNA (lncRNA), LINC01187. We assessed the expression pattern of FOXI1 and RHCG protein by immunohistochemistry (IHC) and LINC01187 mRNA by RNA in situ hybridization (RNA-ISH) in whole tissue sections representing a cohort of 197 RCC cases, including both primary and metastatic tumors.
Outcome Measurements and Statistical Analysis
The FOXI1 and RHCG IHC staining, as well as the LINC01187 RNA-ISH staining were evaluated in each case for intensity, pattern and localization of expression.
Results and Limitations
All primary and metastatic classic ChRCCs demonstrated homogeneous positive labeling for FOXI1, RHCG proteins and LINC01187 transcript. Unclassified RCC with oncocytic features, oncocytoma, and hybrid oncocytic tumor (HOT), as well as all but two cases of eosinophilic ChRCC also stained positive. Importantly, metastatic and primary RCC of all other subtypes showed no staining for FOXI1 and RHCG proteins and LINC01187 transcript. In normal kidney, FOXI1, RHCG, and LINC01187 were detected in the distal nephron segment, specifically in the intercalated cells. Two cases of eosinophilic ChRCC with focal expression of FOXI1 and LINC01187, and Golgi-like RHCG staining were found to contain MTOR gene mutations upon DNA sequencing.
Conclusions
We demonstrate a pipeline for identification and validation of RCC subtype specific biomarkers that can aid in the confirmation of cell of origin and may further aid accurate classification and diagnosis of renal tumors.
Patient summary
FOXI1, RHCG, and LINC01187 are lineage-specific signature genes for ChRCC.
Keywords: renal cell carcinoma, chromophobe, classification, hybrid oncocytic tumor, next generation sequencing, immunohistochemistry, RNA in situ hybridization
1. Introduction
Renal cell carcinoma (RCC) is the most common malignancy of the adult kidney. [1] With the advance of clinical, histological and molecular characterization, RCC classification has expanded from 4 subtypes in the 1997 Heidelberg classification [2] to 12 distinct subtypes in the 2016 World Health Organization (WHO) classification of renal tumors. [3] Recent comprehensive genome and transcriptome studies revealed characteristic genomic aberrations of different histologic RCC subtypes and have elucidated the molecular subtypes therein. [4–7]
Chromophobe renal cell carcinoma (ChRCC) accounts for approximately 5% of renal malignant neoplasms. [8] They are subdivided into two histologic categories: the classic ChRCC with large cells with prominent cell borders, pale to eosinophilic cytoplasm, and raisinoid nuclei with perinuclear halos; and the eosinophilic variant of ChRCC with acinar architecture, abundant eosinophilic cytoplasm and focal nuclear wrinkling with subtle perinuclear clearing/halos. [9] In the vast majority of cases, these tumors can be classified based on morphology alone. However, some cases show morphologic overlap with other entities including clear cell RCC, MiT family translocation RCC, and renal oncocytoma. [10, 11] The pan-RCC study from The Cancer Genome Atlas (TCGA) demonstrated that approximately 6–10% of the tumors included in the clear cell RCC, papillary RCC, and chromophobe RCC cohorts were initially misclassified. [5]
In order to gain a better understanding of the molecular features, gene expression signatures, and cell-of-origin of RCC subtypes, especially the rare RCC subtypes, we performed integrative analysis of RNAseq data from 1,049 RCC specimens assembled by combining TCGA, in-house rare RCC subtype RNAseq and normal human kidney single cell RNAseq (scRNA-seq) studies. In this report, we focused on characterization of chromophobe renal cell carcinoma and related oncocytic neoplasms. Our workflow identified Forkhead box I1 protein (FOXI1), Rh family C glycoprotein (RHCG) and a novel long noncoding RNA (lncRNA) LINC01187 as lineage-specific genes in ChRCC. The normal human kidney scRNA-seq data showed that FOXI1, RHCG, and LINC01187 genes are expressed in the intercalated cells of distal nephron, indicating the potential cell-of-origin of ChRCC. Next, we performed in-depth characterization of the expression pattern for FOXI1, RHCG, and LINC01187 using either immunohistochemistry or RNA in situ hybridization. Our findings showed relatively high specificity and sensitivity of expression of these genes in oncocytic renal tumors including ChRCC, especially in the metastatic setting, thus indicating their potential as biomarkers to aid the diagnostic categorization of RCC. We also confirmed their expression in the distal tubule of normal human kidney.
2. Patients and methods
2.1. Patients
This study was performed under Institutional Review Board-approved protocols (with waiver of informed consent). A search of the Michigan Medicine surgical pathology database identified consecutive cases of classic ChRCC (n= 33), eosinophilic ChRCC (n=10), unclassified RCC with oncocytic features (n= 6), hybrid oncocytic tumor (n=7), and oncocytoma (n=18). For comparison, we included cases of clear cell RCC (6 World Health Organization/International Society of Urological Pathology [WHO/ISUP] grade 2, 10 WHO/ISUP grade 3, 14 WHO/ISUP grade 4), clear cell papillary RCC (n=5), mucinous tubular and spindle cell carcinoma (MTSCC, n=1), papillary RCC (6 type 1, 4 type 2). While all the previous cases reflect resection specimens, biopsy material from 4 cases of ChRCC were also included. All available ChRCC metastases were retrieved (18 sites from 5 patients), as well as clear cell RCC (15 sites from 15 patients) and papillary RCC metastases (5 sites from 3 patients) from various sites. All cases were clinically diagnosed in Michigan Medicine surgical pathology through morphologic assessment and immunohistochemical workup as deemed necessary; all cases were re-reviewed by two study pathologists with expertise in genitourinary pathology (SLS and RM) for histopathologic assessment and diagnostic confirmation.
2.2. Biomarker Nomination
RNAseq data was integrated from a combined cohort of major RCC subtypes (clear cell, papillary, and ChRCC) from TCGA and rare RCC specimens from the Michigan Center for Translational Pathology (n=1049 in total). The updated classification of TCGA RCC cases was used [5], and samples annotated as “mixed” were excluded. Raw sequencing reads were aligned to the GRCh38 reference genome using STAR [12]; aligned reads that overlap with annotated genes according to Gencode v23 [13] were then counted using featureCounts [14]. A scaling normalization scheme (TMM) was applied to all samples to adjust sequencing depth [15]. Genes with no or low expression, defined as having an median of <1.5 reads per kilobase of transcript per million mapped reads (RPKM), were removed prior to differential expression (DE) analysis. DE analyses were performed with limma [16] on voom-transformed count data [17]. Systematic differences between two data sources (in-house and TCGA) were adjusted by including data source as a covariate in the linear model. [18, 19] To identify ChRCC-specific genes, we made pairwise comparison between ChRCC and all other subtypes using fitting contrast models. Differentially expressed genes shared in all pairs of comparison (Benjamini-Hochberg adjusted p<0.05 and median fold-change>=2) were selected as candidates of ChRCC biomarkers. The candidate gene expression in the intercalated cells (presumed cell-of-origin) was examined using an in-house single cell sequencing data set (unpublished). A cancer-specific biomarker was defined as a gene expressed in a given cancer subtype with very low or no expression in any nephron segment. A lineage-specific biomarker was defined as a gene expressed in both a given cancer subtype and certain nephron segments. As a part of the enterprise, we built a webportal (“Renaissance”) with easy data visualization to facilitate nomination of lineage-specific or cancer-specific biomarkers for specific subtypes of RCC.
2.3. Immunohistochemistry
Immunohistochemistry (IHC) was performed on representative whole formalin fixed, paraffin embedded (FFPE) 5 micron-thick tissue sections. Heat induced epitope retrieval was performed with FLEX TRS Low pH Retrieval buffer (6.10; Dako, Carpinteria, CA) for 20 minutes. After peroxidase and protein blocking, the rabbit polyclonal FOXI1 antibody (1:250; Atlas Antibodies, Stockholm, Sweden) or rabbit polyclonal RHCG antibody (1:4000; LSBio, Seattle, WA) was applied to the sections and incubated at room temperature for 60 minutes. The FLEX + Rabbit EnVision System (Dako, Carpinteria, CA) and DAB chromogen were used for detection. Immunopositivity was defined as brown pigmentation in the membrane/cytoplasm (for RHCG) or nucleus (for FOXI1) of tumor cells. The staining was independently assessed by two pathologists (SLS and RM) in each case for presence and pattern of expression.
2.4. RNA in situ Hybridization
RNA in situ hybridization (RNA-ISH) was performed on 4 micron-thick FFPE tissue sections using the RNAscope 2.5 HD Brown kit (Advanced Cell Diagnostics, Newark, CA) and a target probe against human LINC01187 (532311). RNA quality was evaluated in all cases using a positive control probe against human peptidylprolyl isomerase B (PPIB). Assay background was monitored using a negative control probe (DapB). After deparaffinization, hydrogen peroxide pretreatment and target retrieval, tissue sections were permeabilized using protease and hybridized with target probe in the HybEZ oven for 2 hours at 40°C. After two washes, the samples were processed for a series of signal amplification steps. Chromogenic detection was performed using DAB, counterstained with 50% Gill’s Hematoxylin I (Fisher Scientific, Rochester, NY).
Stained slides were examined under 100x and 200x magnification for RNA-ISH signals in tumor cells and adjacent benign kidney tissues by four study participants including three pathologists (SLS, XW, RM, and RM). The RNA-ISH assay stains each RNA molecule/transcript as an individual brown, punctate dot. The number of dots per cell was counted and expression level was evaluated according to the RNAscope scoring criteria as follow: score 0 = no staining or <1 dot per 10 cells, score 1 = 1–3 dots per cell, score 2 = 4–9 dots per cell and non or very few dot clusters, score 3 = 10–15 dots per cell and <10% dots in clusters, score 4 = >15 dots per cell and > 10% dots are in clusters. The H-score was calculated for each examined tissue section as the sum of the percentage of cells with score 0–4 [(A%x0)+(B%x1)+(C%x2)+(D%x4)+(E%x4), A+B+C+D+E=100], using previously published scoring criteria. [3, 20]
3. Results
3.1. Nomination of Cancer-specific and Lineage-specific Biomarkers in ChRCC
Our group constructed a workflow which includes bioinformatic analysis of RNAseq data from clinical RCC samples to identify ChRCC-specific biomarkers (Figure 1A). Comparison of RNAseq data from ChRCC with the other RCC subtypes and normal kidney tissues identified 2,751 up- and 2,107 down-regulated genes (>2 fold-change and adjusted p <0.05, Figure 1B). To further identify ChRCC-specific biomarkers, we performed pairwise comparisons with clear cell RCC, papillary RCC and several rare subtypes. A total of 1,268 genes were nominated as potential ChRCC biomarkers by filtering for genes with significant up-regulation (> 2 fold-change and adjusted p <0.05). Next, these genes were further ranked by a scoring scheme considering both expression level and significance. Using this strategy, Forkheadbox protein I1 (FOXI1), the lncRNA (LINC01187), Rh C glycoprotein (RHCG), and HEPACAM family member 2 (HEPACAM2) genes were ranked as the top candidates in our list (Figure 1C). These biomarkers were annotated as lineage-specific genes, as they demonstrated restricted expression mainly within the intercalated cells in normal kidney tissue as observed in our single cell sequencing data (Figure 1D). In addition, we also identified cancer specific genes such as KLK15, LRRTM1 and lncRNA LINC00588 that were expressed only in ChRCC but absent in other RCC subtypes and normal renal cell types (Figure 1E). These lineage- and cancer-specific genes were considered to have potential for visual labeling of ChRCC as they were consistently elevated in TCGA ChRCC cases which were initially diagnosed as other subtypes based on histologic evaluation (Figure 1F). We also performed geneset enrichment analysis (GSEA) of differentially expressed genes between ChRCC vs other tumors and ChRCC versus benign tissues (Supplementary Figure 1). We observed negative enrichment of epithelial-mesenchymal transition as the most significant event.
Figure 1.
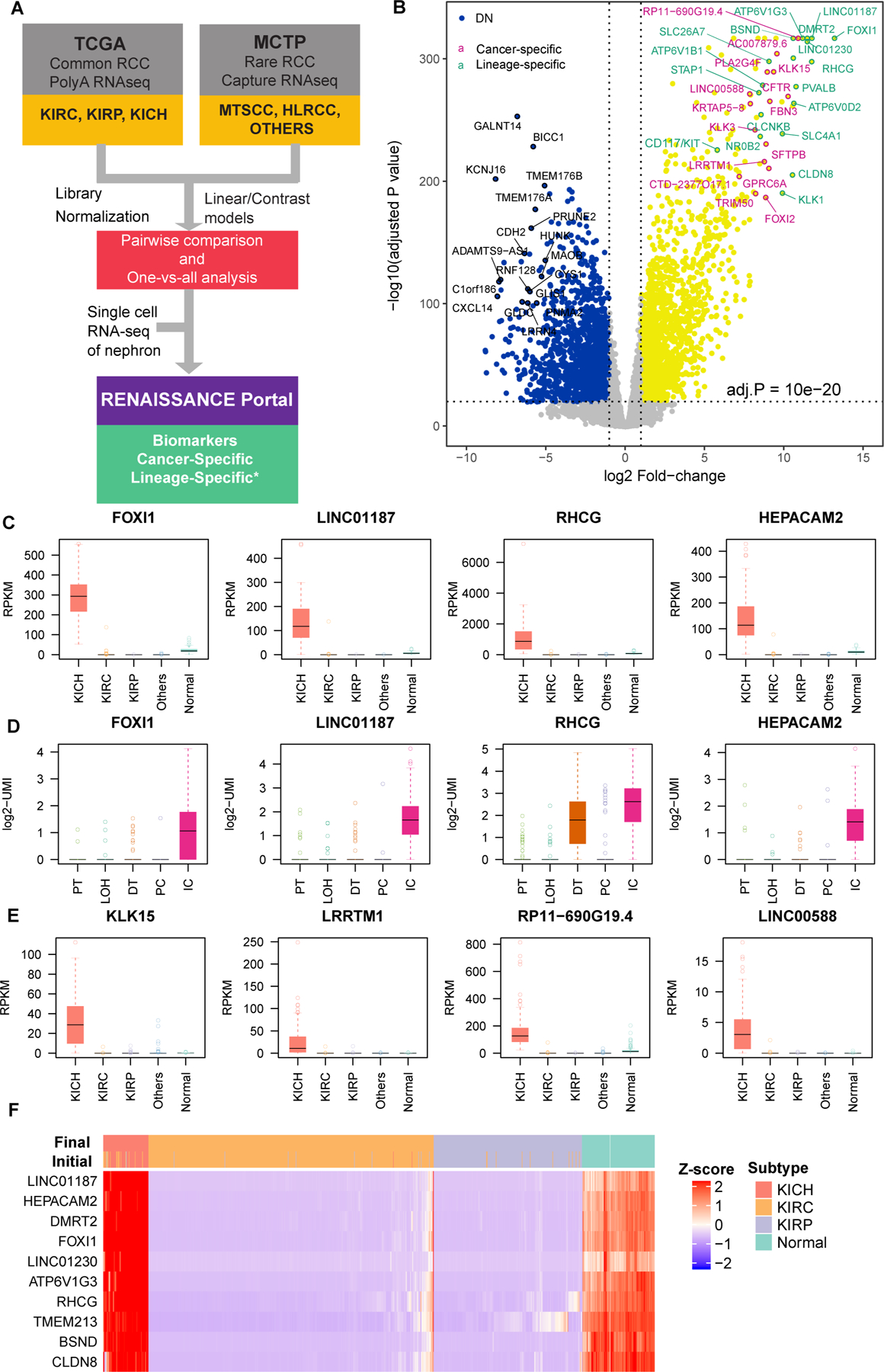
Nomination of cancer and lineage biomarkers of ChRCC. A) Schematic of the gene analysis pipeline to determine cancer specific and lineage specific expression in ChRCC. B) Volcano plot represents significantly differentially expressed genes in ChRCC compared to normal and other RCC subtypes. Selected top up- and down-regulated genes are circled and labeled. C) Expression of top ChRCC lineage specific genes across major RCC subtypes. KICH = ChRCC, KIRC = CCRCC, KIRP = PRCC. D) Expression of top ChRCC lineage specific genes across major epithelial cell types in human kidney (from single cell sequencing). PT = proximal tubule, LOH = loop of Henle, DT = distal tubule, PC = principle cell, IC = intercalated cell. E) Expression of top ChRCC cancer specific genes across major RCC subtypes. F) Nominated biomarkers are highly specific to ChRCC. Initial and final diagnoses (after re-evaluation) of TCGA RCC cases are annotated on the top of the heatmap. Cancer samples with high expression of nominated biomarkers were almost exclusively ChRCC, including cases with confusing histology (i.e., cases were re-assigned to different subtypes).
Based on single cell RNA sequencing data from normal human kidney, FOXI1 and LINC01187 expression were found to be highly restricted to intercalated cells among the renal tubular cell types (Figure 1D); this phenomenon is also faithfully reflected in the staining pattern we observed in the benign kidney regions adjacent to these tumors (Figure 2), thereby validating our nomination as lineage-specific biomarkers. Next, to investigate the cancer specificity, we systematically evaluated the expression of FOXI1 and RHCG at the protein level by IHC in a renal neoplasm cohort comprised of 197 samples and LINC01187 by RNA-ISH in a similar cohort containing 162 samples representing various histologic subtypes (Figures 3–6, Supplementary Figures 2–8).
Figure 2.
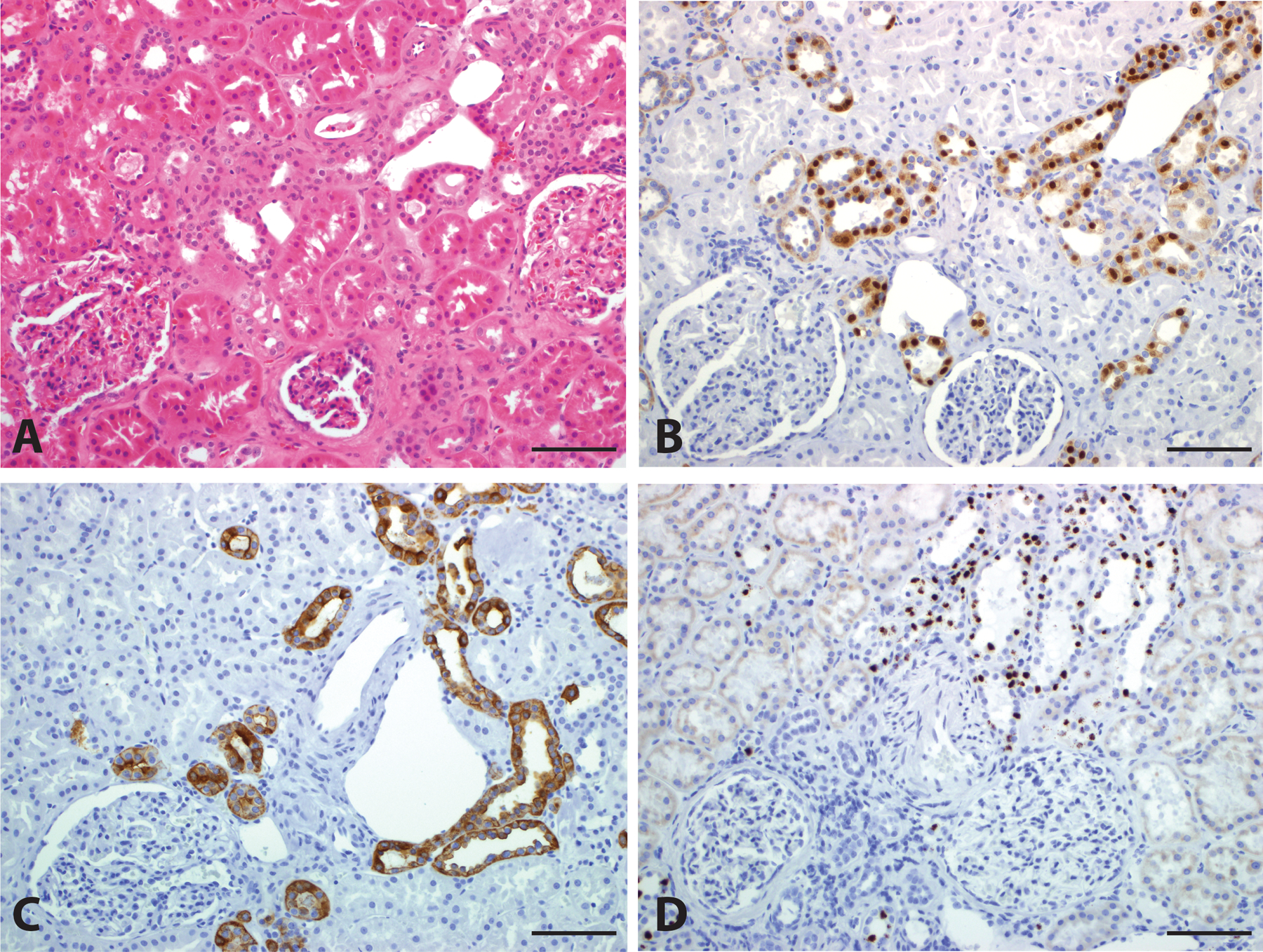
Biomarker Expression in Normal Kidney. Normal kidney tissue (A, H&E, 200x) demonstrates nuclear staining for FOXI1 (B, 200x), circumferential membranous staining for RHCG (C, 200x), and high level nuclear expression of LINC01187 (D, 200x) in distal tubules. Scale bar = 100 microns.
Figure 3.
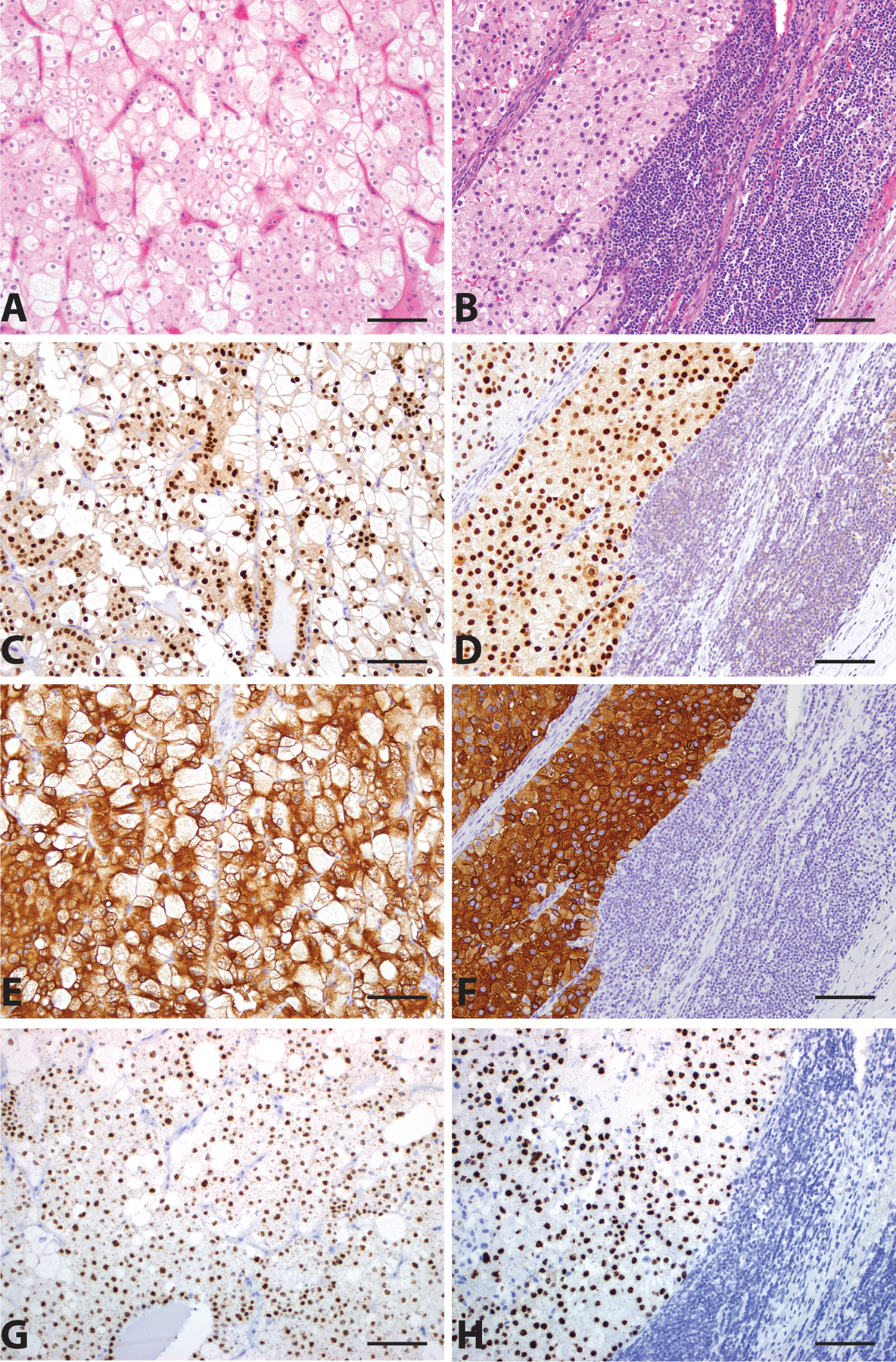
Classic Chromophobe Renal Cell Carcinoma (ChRCC). Primary (A, H&E, 200x) and metastatic (B, H&E, 200x) ChRCC demonstrate strong nuclear staining for FOXI1 (C and D, 200x), circumferential membranous staining for RHCG (E and F, 200x), and high level nuclear LINC01187 expression (G and H, 200x). Scale bar = 100 microns.
Figure 6.
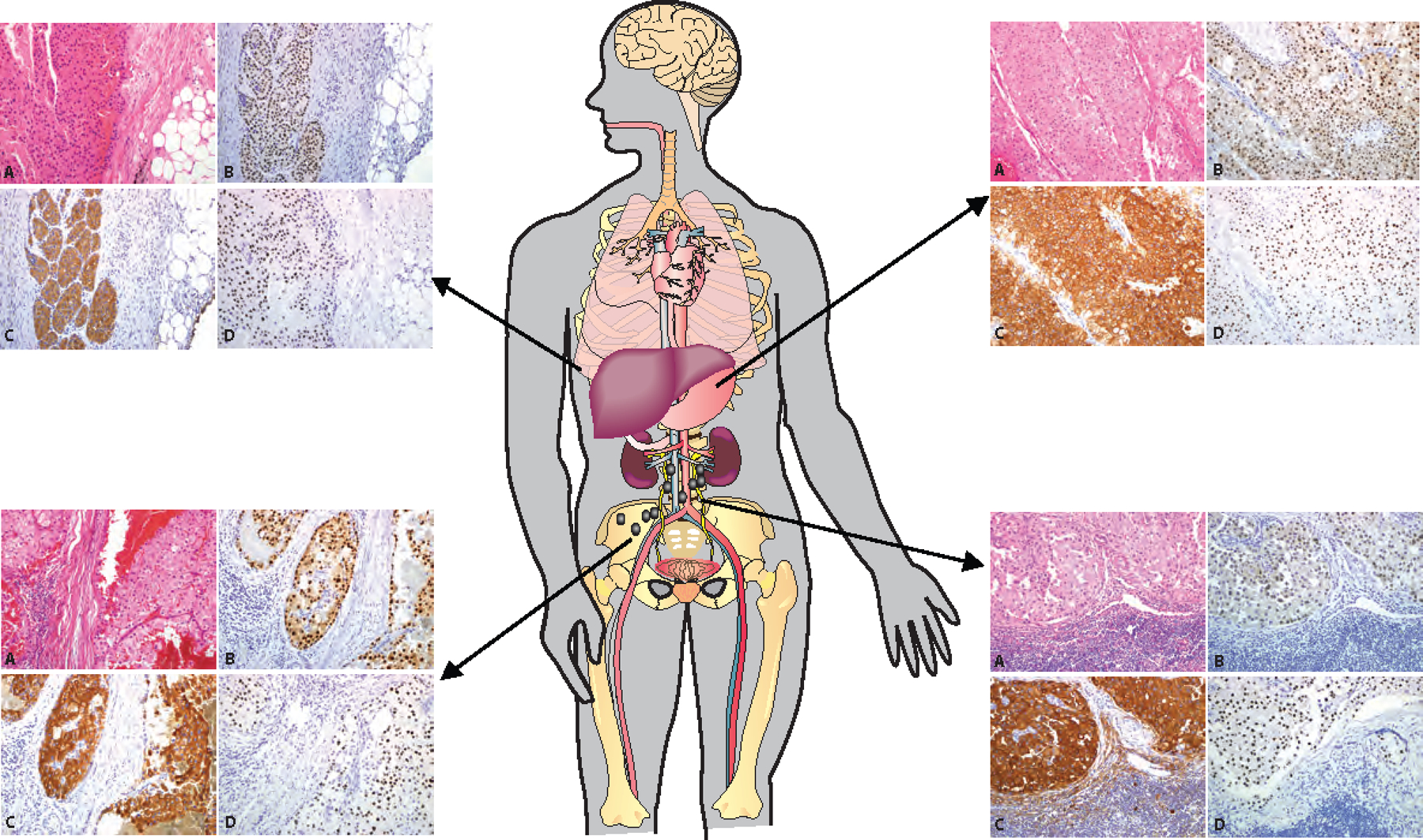
Biomarker Expression in Metastatic ChRCC. Positive expression of FOXI1, RHCG, and LINC01187 by metastatic ChRCC at four different anatomic sites, including omentum, right psoas muscle, stomach, and retroperitoneal lymph nodes. (A, H&E, 100x; B, FOXI1 IHC 100x; C, RHCG IHC 100x; D, LINC01187 RNAISH 100x)
3.2. Nuclear expression of the transcription factor FOXI1 is enriched in ChRCC
Both primary and metastatic ChRCC (Figure 3A–3D) demonstrated diffuse and strong positive nuclear staining for FOXI1 protein by IHC on whole tissue sections. In contrast, other RCC subtypes such as clear cell RCC (Figure 4A–4D), show no visible staining, in either primary (Figures 4A and 4C) or metastatic sites (Figures 4B and 4D).
Figure 4.
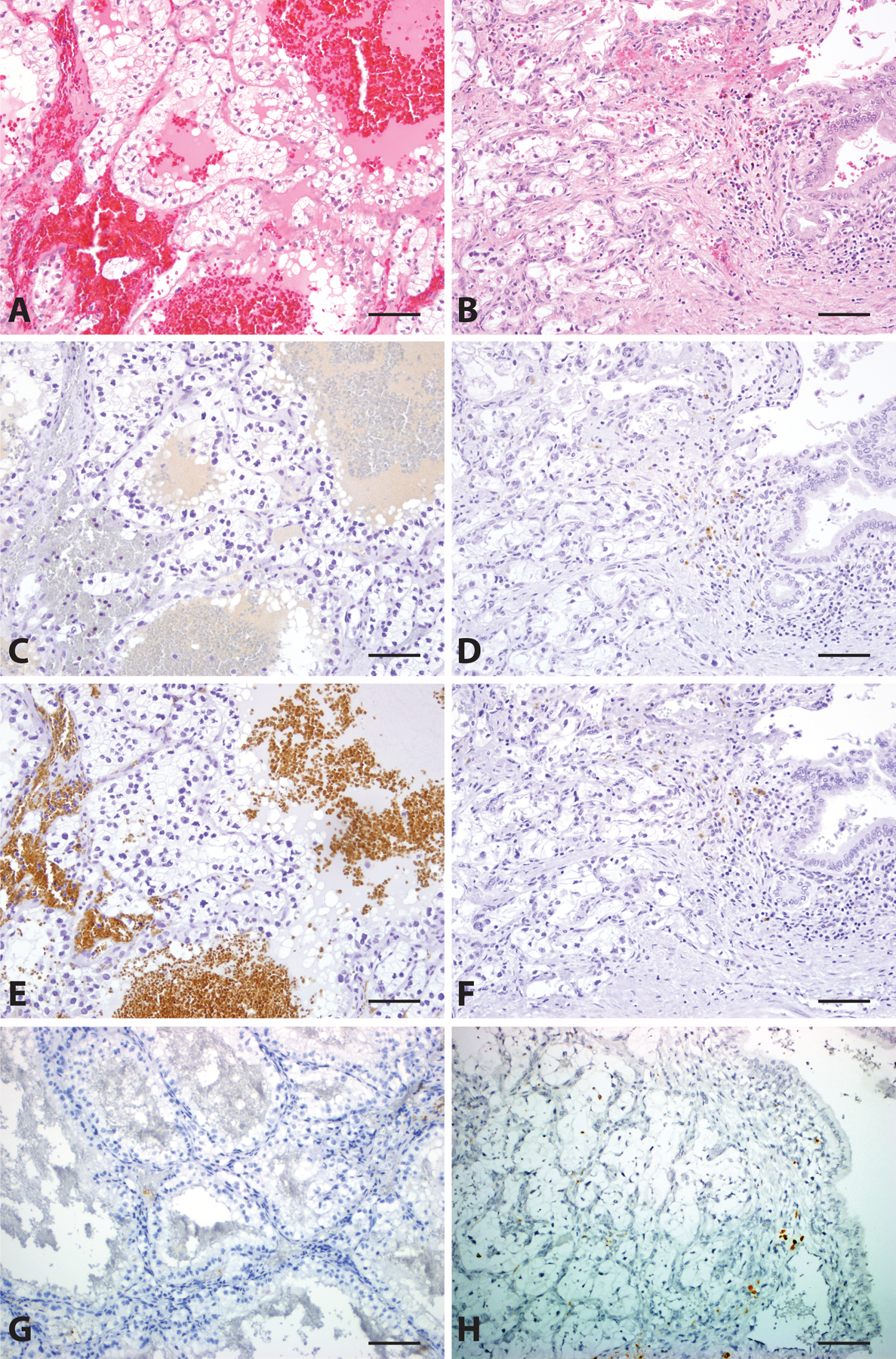
Clear Cell Renal Cell Carcinoma (CCRCC). Primary (A, H&E, 200x) and metastatic (B, H&E, 200x) CCRCC are negative for FOXI1 (C and D, 200x), RHCG (E and F, 200x), and LINC01187 (G and H, 200x) expression. Scale bar = 100 microns.
It was intriguing to note that FOXI1 protein staining in sarcomatoid ChRCC was present only in the epithelioid tumor cell component (Supplementary Figures 2A and 2C), including those epithelioid cells entrapped by spindled cells within the sarcomatoid component (Supplementary Figures 2B and 2D), while the high-grade spindle cell component was predominantly negative for FOXI1 staining. All cases of eosinophilic ChRCC (Supplementary Figures 3A and 3C) also demonstrated positive FOXI1 staining. To evaluate some of the main oncocytic entities which fall within the clinical spectrum and differential diagnosis of ChRCC, we performed further interrogation which demonstrated that renal oncocytoma (Supplementary Figures 3B and 3D), and unclassified RCC with oncocytic features also showed positive FOXI1 staining. Interestingly, all evaluated hybrid oncocytic tumors (HOT), a renal tumor most frequently encountered in patients with the Birt-Hogg-Dubé syndrome, demonstrated a dual phenotype in which only one tumor epithelial cell population was positive for FOXI1 staining (Supplementary Figures 4A–4D), thus simulating a checkered staining pattern on low-power evaluation.
In this cohort, no primary (Figures 4A and 4C) or metastatic (Figures 4B and 4D) clear cell RCC, clear cell papillary RCC, primary (Supplementary Figures 5A and 5C) or metastatic (Supplementary Figures 5B and 5D) papillary RCC, FH-deficient RCC, collecting duct carcinoma, or MTSCC showed FOXI1 staining. 3/8 (37.5%) primary MiT family translocation RCCs showed very weak and focal nuclear staining for FOXI1 in scattered cells (Supplementary Figures 6A– 6F); the only case of metastatic translocation-associated RCC stained in this cohort was completely negative for FOXI1 staining. (Table 1)
Table 1.
Summary of FOXI1 expression in the validation cohort.
| Tumor Type | Positive Cases in Primary Tumors | Positive cases in Metastatic Tumors |
|---|---|---|
| Classic ChRCC | 32/32 (100%) | 18/18 (100%) |
| Sarcomatoid ChRCC (spindled cells) | 1/3 (33%) *focal in 1 |
0/1 (0%) |
| ChRCC with marked cellular atypia | 4/4 (100%) *patchy in 1, weak in another |
N/A |
| EosinophilicChRCC | 10/10 (100%) *patchy/focal in 3 |
N/A |
| Hybrid oncocytic tumor (HOT) | 7/7 (100%) *one of the two cell populations only |
N/A |
| RCC, unclassified (oncocytic) | 6/6 (100%) | N/A |
| Oncocytoma | 18/18 (100%) | N/A |
| Clear Cell RCC | 0/30 (0%) | 0/17 (0%) |
| Papillary RCC | 0/11 (0%) | 0/8 (0%) |
| Clear Cell Papillary RCC | 0/5 (0%) | N/A |
| MTSCC | 0/1 (0%) | N/A |
| FH-deficient RCC | 0/1 (0%) | N/A |
| Collecting Duct Carcinoma | 0/3 (0%) | 0/1 (0%) |
| Translocation RCC | 3/8 (37.5%) *weak in 3 |
0/1 (0%) |
| RCC, unclassified | 1/6 (0%) *focal blush in 1 |
0/6 (0%) |
3.3. Membranous expression pattern of RHCG in ChRCC
The staining patterns we observed with RHCG IHC performed on whole tissue sections can be grouped into three categories: circumferential membranous staining (pattern 1; Supplementary Figure 7A), apical cup-like staining (pattern 2; Supplementary Figure 7B), and Golgi-like/secretory staining (pattern 3; Supplementary Figure 7C).
Diffuse and strong circumferential membranous RHCG staining was identified in primary and metastatic ChRCC (Figure 3E and 3F), while primary and metastatic clear cell RCC were negative for RHCG staining (Figure 4E and 4F). Sarcomatoid ChRCC showed circumferential membranous RHCG staining in the epithelioid tumor cell component (Supplementary Figure 2E), including those epithelioid cells entrapped by spindled cells within the sarcomatoid component (Supplementary Figure 2F). Eosinophilic ChRCC cases showed either Golgi-like/secretory (6/10, 60%; Supplementary Figure 3E) or apical cup-like (4/10, 40%) RHCG staining. All cases of oncocytoma (Supplementary Figure 3F) and low-grade oncocytic unclassified RCC demonstrated apical cup-like RHCG staining; the same was true of one tumor epithelial cell population in HOTs, again leading to a low-power checkered appearance to this tumor similar to the FOXI1 staining described above for this tumor type (Supplementary Figures 4E–4F).
In this cohort, no primary (Figure 4E) or metastatic (Figure 4F) clear cell RCC, clear cell papillary RCC, primary (Supplementary Figure 5E) or metastatic (Supplementary Figure 5F) papillary RCC, FH-deficient RCC, collecting duct carcinoma, or MTSCC showed RHCG staining. Half (4/8) of the MiT family translocation RCC cases showed patchy membranous RHCG staining (Supplementary Figures 6G–6I); this finding only showed partial concordance with focal weak nuclear FOXI1 staining. The case of metastatic MiT family translocation RCC was negative for RHCG. A detailed summary is displayed in Table 2.
Table 2.
Summary of RHCG expression in the validation cohort.
| Tumor Type | Positive Cases in Primary Tumors | Positive cases in Metastatic Tumors |
|---|---|---|
| Classic ChRCC | 32/32 (100%) Circumferential membranous |
18/18 (100%) Circumferential membranous |
| Sarcomatoid ChRCC (spindled cells) | 0/3 (0%) | 0/1 (0%) |
| ChRCC with marked cellular atypia | 4/4 (100%) Circumferential membranous, patchy – 2 Apical cup-like – 1 Circumferential membranous and Golgi-like/secretory – 1 |
N/A |
| EosinophilicChRCC | 10/10 (100%) Apical cup-like – 4 Golgi-like/secretory – 6 |
N/A |
| Hybrid oncocytic tumor (HOT) | 7/7 (100%) Apical cup-like in one cell population (checkered) |
N/A |
| RCC, unclassified (oncocytic) | 6/6 (100%) Apical cup-like |
N/A |
| Oncocytoma | 18/18 (100%) Apical cup-like |
N/A |
| Clear Cell RCC | 0/30 (0%) | 0/17 (0%) |
| Papillary RCC | 0/11 (0%) | 0/8 (0%) |
| Clear Cell Papillary RCC | 0/5 (0%) | N/A |
| MTSCC | 0/1 (0%) | N/A |
| FH-deficient RCC | 0/1 (0%) | N/A |
| Collecting Duct Carcinoma | 0/3 (0%) | 0/1 (0%) |
| Translocation RCC | 4/8 (50%) Circumferential membranous, all focal/patchy |
0/1 (0%) |
| RCC, unclassified | 0/6 (0%) | 0/6 (0%) |
Nuclear expression of the lncRNA biomarker LINC01187 is highly enriched in ChRCC
To validate our in-silico nomination, we performed LINC01187 RNA-ISH on whole tissue sections across 167 RCC tumor samples (including 34 classic ChRCC, 8 eosinophilic ChRCC, and 5 metastatic ChRCC cases from 18 metastatic sites). In our final analysis, we considered a total of 162/167 (97%) cases. Five cases were excluded due to insufficient staining of the positive control gene.
High level LINC01187 nuclear expression was seen in both primary (mean H-score = 371; range = 247 to 398) and metastatic ChRCC (mean H-score = 368; range = 308 to 400) (Figure 3G and 3H), while primary and metastatic clear cell RCC (Figure 4G and 4H) showed no expression (H-score = 0 in all 10 primary and 15 metastatic cases studied).
Sarcomatoid ChRCC showed high level LINC01187 expression in the classic ChRCC epithelioid tumor cell component (Supplementary Figure 2G), but not in the spindle cell component of the sarcomatoid regions (Supplementary Figure 2H). Almost all eosinophilic ChRCC (6/8) had high LINC01187 expression (mean H-score = 254; range = 250 to 386 in positive cases, H-score = 4 in two negative cases) (Supplementary Figure 3G). Other oncocytic tumors including unclassified RCC with oncocytic features and oncocytomas (Supplementary Figure 3H) also showed high LINC01187 expression (mean H-score = 377; range = 189 to 400), suggesting that these tumors originate from a common nephron segment. All HOTs showed a dual phenotype, with just one population of cells demonstrating high level LINC01187 expression, rendering a checkered pattern as mentioned above similar to our observations with FOXI1 and RHCG staining (Supplementary Figures 4E–4F). All 4 ChRCC biopsy samples demonstrated high LINC01187 expression within the tumor (Supplementary Figure 3I–3L).
Importantly, in this cohort, no LINC01187 RNA expression was detected in primary clear cell RCC (Figure 4G), clear cell papillary RCC, papillary RCC (Supplementary Figure 5G), MiT family translocation RCC (Supplementary Figure 6J–6L), collecting duct carcinoma, or MTSCC samples (H-score = 0 in all 32 primary case). All the evaluated metastatic non-ChRCC cases, including clear cell RCC (Figure 4H) and papillary RCC (Supplementary Figure 5H), as well as one metastatic sarcomatoid ChRCC (H-score =0 in all 19 metastatic samples), showed complete absence of LINC01187 expression. (Table 3)
Table 3.
Summary of LINC01187 expression in the validation cohort.
| Primary Tumors | Metastatic Tumors | |||
|---|---|---|---|---|
| Positive Cases | Average H-Score | Positive Cases | Average H-Score | |
| Classic ChRCC | 34/34 (100%) | 371.2 | 18/18 (100%) | 367.8 |
| Sarcomatoid ChRCC (spindled cells) | 0/3 (0%) | 0 | 0/1 (0%) | 0 |
| Eosinophilic ChRCC | 6/8 (75%) | 254.0 | 0 | N/A |
| Hybrid oncocytic tumor (HOT) | 7/7 (100%) | 307.3 | 0 | N/A |
| RCC, unclassified (oncocytic) | 6/6 (100%) | 346.0 | 0 | N/A |
| Oncocytoma | 18/18 (100%) | 386.7 | 0 | N/A |
| Clear Cell RCC | 0/10 (0%) | 0 | 0/15 (0%) | 0 |
| Papillary RCC | 0/11 (0%) | 0 | 0/5 (0%) | 0 |
| Clear Cell Papillary RCC | 0/5 (0%) | 0 | 0 | N/A |
| MTSCC | 0/1 (0%) | 0 | 0 | N/A |
| Collecting Duct Carcinoma | 0/3 (0%) | 0 | 0 | N/A |
| Translocation RCC | 0/4 (0%) | 0 | 0 | N/A |
| RCC, unclassified | 0/6 (0%) | 1.3 | 0/7 (0%) | 0.3 |
Finally, to further characterize the expression pattern of ChRCC biomarkers in tumor and normal kidney tissues, we performed FOXI1 and LINC01187 dual RNA-ISH staining in classic ChRCC. These results confirm the co-expression of FOXI1 and LINC01187 in the majority of classic ChRCC tumor epithelial cells and in the intercalated cells in adjacent normal kidney tissues. This finding further supports the notion that ChRCC originates from the distal nephron segment (Figure 5).
Figure 5.
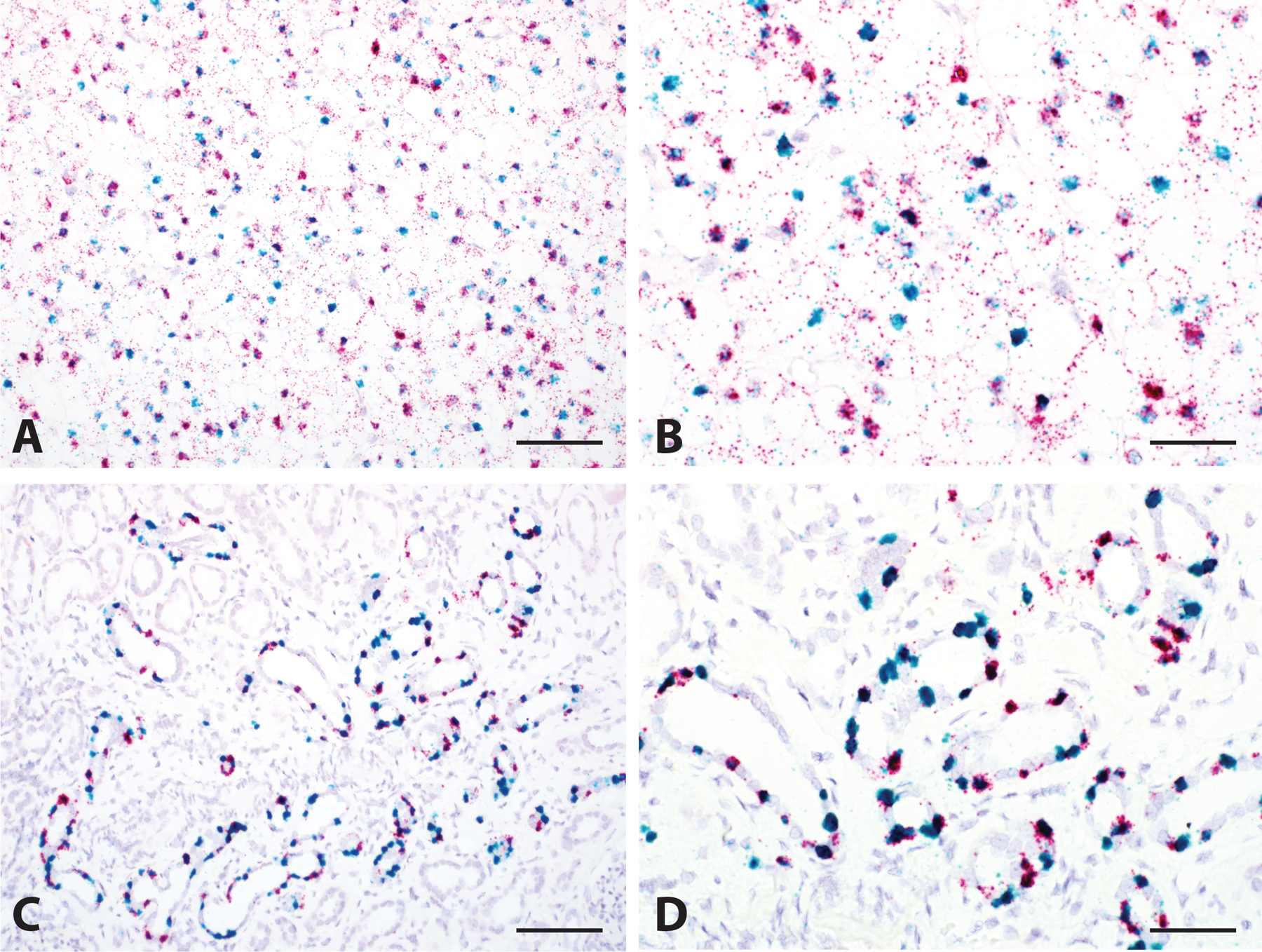
Dual RNA in situ Hybridization for FOXI1 and LINC01187. Co-expression of FOXI1 (Red) and LINC01187 (Green) was seen in the majority of classic ChRCC tumor cells (A, 200x and B, 400x), as well as in the intercalated cells in normal kidney tissue (C, 200x and D, 400x). Scale bar = 100 microns for A and C, 50 microns for B and D.
3.4. Uniform expression of biomarkers across multiple ChRCC metastatic sites
To evaluate the uniformity and intra-patient heterogeneity (if any) of biomarker expression across multiple metastatic sites, we assessed FOXI1, RHCG, and LINC01187 staining in metastatic ChRCC tumors from 18 different metastatic sites collected from 5 patients. All tumor samples demonstrated diffuse and strong FOXI1 staining, circumferential membranous RHCG staining, and high LINC01187 expression (H-score > 300). Figure 6 shows the staining observed in metastatic ChRCC samples from omentum, right psoas muscle, stomach, and retroperitoneal lymph nodes. This representative data illustrates the uniform spatial and temporal expression of FOXI1, RHCG proteins and LINC01187 transcripts.
3.5. Biomarker evaluation identifies morphologic mimics of eosinophilic ChRCC containing distinct molecular aberrations
During the evaluation of FOXI1, RHCG and LINC01187 expression, we made an interesting observation that 2 of 10 eosinophilic ChRCC cases in our RCC cohort showed very low level nuclear FOXI1 staining and were largely negative for LINC01187 expression (Supplementary Figure 8A–8D and 8G–8H). These tumors demonstrated diffuse Golgi-like/secretory RHCG staining (Supplementary Figure 8E–8F). The above findings suggested that these two cases may likely be a distinct disease subtype where a distinct molecular process is associated with eosinophilic ChRCC or a renal tumor that morphologically closely mimics eosinophilic ChRCC. To investigate this further, we subjected matched tumor and adjacent benign kidney samples from these two cases to whole exome sequencing to identify mutations and copy number pattern, as well as transcriptome sequencing to study the global gene expression pattern. It has been well-documented that ChRCCs contain the characteristic recurrent one copy loss of several chromosomes including 1, 2, 6, 10, 13, 17. Based on our copy number variation (CNV) analysis, these two index cases were diploid for these chromosomes and lacked the typical ChRCC copy loss pattern (Supplementary Figure 9). On the other hand, intriguingly, both cases contained hotspot missense activating mutations (p.S2215Y, p.L2427Q) in the MTOR gene within the kinase domain. We revisited the TCGA database and identified 3 additional cases that were initially diagnosed as ChRCC or PRCC and reclassified after sequencing analysis as mixed RCC with MTOR mutations in the kinase domain. RNAseq data from these two cases also confirmed lack of expression of FOXI1 and LINC01187 genes.
4. Discussion
In this study, we described a pipeline for nominating RCC subtype specific biomarkers using RNAseq data, followed by experimental validation in clinical specimens. Our results demonstrated high enrichment of FOXI1, RHCG, and LINC01187 in classic and eosinophilic variants of ChRCC, as well as metastatic ChRCC, indicating the preserved spatial and temporal expression in metastatic ChRCC. These biomarkers are also expressed in oncocytic renal neoplasms including unclassified RCC with oncocytic features, HOTs, and oncocytomas, but not in clear cell RCC, papillary RCC, clear cell papillary RCC, collecting duct carcinoma or MTSCC tumor in either primary or the metastatic setting. Some MiT family translocation RCC cases show very weak/focal FOXI1 staining, and patchy RHCG staining, but no LINC01187 expression.
FOXI1 is a transcription factor required for the differentiation of intercalated cells in the distal renal tubules. Mouse studies have shown that FOXI1 is a master regulator of vacuolar H+-ATPase proton pump subunits in the kidney, inner ear, and epididymis. [21] Lindgren et al. reported the enriched gene expression of putative FOXI1 transcriptional targets in the intercalated cells of the distal nephron, and in ChRCC. [22]
RHCG transports ammonia and is expressed in sites that are important for ammonia secretion. [23] Liu et al. report that based on Northern blot studies, the major organs with RHCG expression in humans include fetal kidney and adult kidney, testis, brain, placenta, pancreas, and prostate. [24] Multiple groups have reported that in the normal human kidney, there is apical and basolateral RHCG expression in the distal convoluted tubule, connecting segment, initial collecting tubule, and throughout the collecting duct. [23] Han et al. and Brown et al. report that RHCG is localized to alpha-intercalated cells in the human kidney. RHCG also stains red blood cells, likely due to the similar structure to erythroid Rh proteins, so care should be taken to verify which cells are staining, particularly in cases of highly vascular renal neoplasms.
The function of the lncRNA LINC01187 has not been characterized yet. Expression of both FOXI1 and LINC01187 were very specific to ChRCC in our pan-RCC RNAseq data analysis. In addition, the expression of these two genes shows a strong correlation across TCGA tumors (n=10,000 tumors representing more than 30 different tumor types) RNAseq data. The nuclear localization and selective co-expression pattern of these genes among human normal and cancer cell types suggest either a transcriptional co-regulation or functional relationship between these two genes. This is further supported by the fact that the lncRNA LINC01187 is located ~80 kilobases 3’ of FOXI1 on human chromosome 5q region. A recent study on FOXA1 activation in advanced prostate cancer reported co-expression of FOXA1 and lncRNA FOXMIND that are adjacent to each other on chromosome 14q. The duplications and translocations within the FOXA1-FOXMIND locus resulted in overexpression of FOXA1 driven by the regulatory element FOXMIND. [25]
In adjacent benign kidney, FOXI1 and LINC01187 are co-expressed in a restricted cell population (the intercalated cells of distal tubules), indicating the potential cell-of-origin of ChRCC and the lineage-specificity of these biomarkers. Previous studies have suggested that ChRCC arises from the intercalated cell of the collecting duct system. [26, 27] Shinmura et al. selected 3 genes (including BSND and ATP6V1G3) with specific transcriptional expression in ChRCC using data from the TCGA. [28] Strong diffuse expression of both proteins was seen in 23/23 ChRCC, 13/14 renal oncocytoma, 0/153 clear cell RCC, and 0/10 papillary RCC. Han et al. described apical and basolateral RHCG expression in the distal convoluted tubule, connecting segment, initial collecting tubule, and throughout the collecting duct. RHCG was expressed by 2/2 ChRCC and 2/2 renal oncocytoma but not clear cell RCC (0/5) or papillary RCC (0/4). [27] To our knowledge, the current study describes the most comprehensive analysis of potential lineage-specific biomarkers for ChRCC evaluated in multiple RCC subtypes and renal oncocytoma. Our work helps characterize a common cell-of-origin for diverse oncocytic renal tumors including classic and eosinophilic ChRCC and renal oncocytoma. Further studies are needed to better characterize how a single cell type transforms into neoplastic entities that are diverse at the cellular, physiologic, and genomic levels. Robinson et al. have reported that STAT3 regulated gene PIM1 is only upregulated in ChRCC but not in CCRCC or PRCC, while MYC, VIM, ICAM1 and ITGB4 genes were downregulated only in ChRCC. [29] Our analysis recaptures the differential expression of these genes. In addition, our pan-RCC analysis also indicates PIM1 is upregulated in a small subset of CCRCC and PRCC samples.
In our cohort, two of the cases originally classified as eosinophilic ChRCC showed FOXI1 protein staining in rare cells and very low focal expression of LINC01187. Next generation sequencing of these two cases revealed hotspot missense mutations in the MTOR gene, which encodes a conserved serine/threonine kinase and a key component of both mTORC1 and mTORC2 signaling complexes. This observation demonstrated that there could be different molecular aberrations associated with the same histologic subtype. The mTORC complexes are critical to metabolism and normal/tumor cell growth. [30] Multiple missense mutations at FAT and kinase domains are activating mutations that contribute to the pathogenesis of cancer, including the p.S2215Y mutation. [31] Chen et al. have reported somatic MTOR p.L2427R mutations in two cases of sporadic, previously unclassified, RCC with predominantly nested architecture and eosinophilic and vacuolated cytoplasm. [32] In contrast to our cases, both of these cases from Chen et al.’s cohort demonstrated loss of chromosome 1. It is currently unclear whether these tumors are related to each other. Cancer cases with mTOR activating mutations may respond well to mTOR inhibitor therapies, if/when needed in such cases. [33]
Finally, the loss of expression of both FOXI1 protein and LINC01187 transcript in sarcomatoid ChRCC regions indicates a likely de-differentiated state resulting from the loss of the lineage specific FOXI1 transcription factor. While the mechanism behind this loss remains to be delineated, the current study provides the first glimpse of a putative causal event underlying the clinically aggressive sarcomatoid transformation of ChRCC. A series of 14 sarcomatoid ChRCCs demonstrated that most were identified at an advanced stage, and metastasis and death due to disease were not uncommon. [34]
Currently, the IHC markers most commonly used to support a diagnosis of ChRCC are cytokeratin 7 and CD117 (KIT) [10, 35]. These markers are positive in a variety of tumors from multiple organ systems (Supplementary Figure 10), and therefore may be of limited use for a pathologist attempting to classify a metastatic lesion, especially on core needle biopsy. Based on pan-cancer RNAseq data, CD117 expression can be found in a broader variety of neoplasms than FOXI1. In addition, the cytoplasmic/membranous CD117 staining is at times more difficult to interpret than the nuclear FOXI1 labeling. Based on published data, CD117 staining in ChRCC shows limited sensitivity with a 70–80% detection rate. Interestingly, it has been reported that CD117-negative ChRCC tends to be of advanced stage. [35] Although it was previously not well-understood why ChRCC and other oncocytic renal neoplasms demonstrate immunohistochemical staining for CD117, our scRNA-seq data provides a likely explanation for this phenomenon, as our analysis annotated CD117 as another lineage-specific marker which is expressed by intercalated cells in the distal nephron (Figure 1). Our current study now further expands the list of lineage-specific markers to FOXI1 and LINC01187, along with several other candidates.
Strengths of this study include the use of RNAseq data from both common and rare renal tumors, as well as the use of a large tissue validation cohort including a broad variety of common and rare tumor types. Employing whole tissue sections of tumor for staining allowed a more comprehensive evaluation of each of these markers, including assessment of the extent of intratumoral heterogeneity. Despite the comprehensive analysis of various types of renal tumors, oncocytoma RNAseq data was not available in the TCGA pan-renal cancer dataset. Hence, identification of markers that distinguish the benign oncocytoma from ChRCC are considered to be goals for future investigation.
5. Conclusion
In summary, we demonstrated a pipeline for identification and validation of candidate RCC subtype biomarkers. Our findings demonstrated FOXI1, RHCG, and LINC01187 overexpression is enriched in oncocytic renal neoplasms including primary and metastatic ChRCC. They are also lineage-specific genes labeling intercalated cells of distal tubules in normal nephron. The FOXI1, RHCG, and LINC01187 overexpression detected by IHC and RNA-ISH may serve as a panel of potential diagnostic markers to assist diagnosis of ChRCC, especially in metastatic tumors, or primary renal tumors with unusual morphology or poor differentiation.
Supplementary Material
Supplementary Figure 1. Geneset enrichment analysis of Hallmark pathways with differentially expressed genes from (A) ChRCC versus Others and (B) ChRCC versus Benign tissues. Pathways that showed significant adjusted p-values (<0.05) are shown as bar plots. (NES: normalized enrichment score)
Supplementary Figure 2. Sarcomatoid Chromophobe Renal Cell Carcinoma (ChRCC). Sarcomatoid ChRCC (A, H&E, 100x, and B, H&E, 400x) demonstrates strong staining for FOXI (C, 100x and D, 400x), RHCG (E, 100x and F, 400x), and LINC01187 (G, 100x and H, 400x) only in cells with classic epithelioid morphology. Scale bar = 200 microns for A, C, E, and G; 50 microns for B, D, F, and H.
Supplementary Figure 4. Hybrid Oncocytic Tumor (HOT). HOT (A, H&E, 100x; B, H&E, 400x) demonstrates strong staining for FOXI1 (C, 100x; D, 400x), RHCG (E, 100x; F, 400x), and LINC01187 (G, 100x; H, 400x) limited to one cell population, leading to a checkered appearance. Scale bar = 200 microns for A, C, E, and G; 50 microns for B, D, F, and H.
Supplementary Figure 3. Eosinophilic Chromophobe Renal Cell Carcinoma (Eosinophilic ChRCC) and Oncocytoma. Eosinophilic ChRCC (A, H&E, 200x) and oncocytoma (B, H&E, 200x) both demonstrate strong nuclear expression of FOXI1 (C and D, 200x), RHCG (E and F, 200x), and LINC01187 (G and H, 200x). All oncocytomas showed apical cup-like RHCG staining; some eosinophilic ChRCC demonstrate Golgi-like RHCG staining as seen here. ChRCC biopsy samples show strong nuclear expression of LINC01187 (I-L, 100x). Scale bar = 100 microns for A-H; 200 microns for I-L.
Supplementary Figure 5. Papillary Renal Cell Carcinoma (PRCC). Primary (A, H&E, 200x) and metastatic (B, H&E, 200x) PRCC are negative for FOXI1 (C and D, 200x), RHCG (E and F, 200x), and LINC01187 (G and H, 200x) expression. Scale bar = 100 microns.
Supplementary Figure 7. Patterns of RHCG Expression. Three patterns of RHCG staining were seen, including circumferential membranous staining (A, 200x), apical cup-like staining (B, 200x), and Golgi-like staining (C, 200x). Scale bar = 100 microns.
Supplementary Figure 6. Translocation Renal Cell Carcinoma (t-RCC). Cases of t-RCC (A-C, H&E, 400x) can show weak and patchy nuclear FOXI1 expression (D-F, 400x) and patchy RHCG expression (G-H, 400x), although other cases lack expression of either marker. The case illustrated in the right-hand column is negative for RHCG staining (I, 400x). All cases of t-RCC were negative for LINC01187 (J-L, 400x). Scale bar = 50 microns.
Supplementary Figure 9. Copy number variation analysis in Cases with MTOR Mutation. Both cases have diploid chromosomes and lacked the typical ChRCC copy loss pattern.
Supplementary Figure 8. Biomarker Expression in Cases with MTOR Mutations, Mimicking Eosinophilic Chromophobe Renal Cell Carcinoma. These two cases (A and B, H&E, 200x) demonstrated very focal and weak nuclear staining for FOXI1 (C and D, 200x), despite diffuse Golgi-like RHCG staining (E and F, 200x). Both cases were negative for LINC01187 expression (G and H, 200x). Scale bar = 100 microns.
Supplementary Figure 10. Box plots showing expression of KIT (CD117), FOXI1, LINC01187, and RHCG across TCGA cancer types from multiple organ systems. ACC, Adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, prostate adenocarcinoma; READ, Rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, Testicular Germ Cell Tumors; THCA, thyroid carcinoma; THYM, Thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, Uterine Carcinosarcoma; UVW, Uveal Melanoma.
Acknowledgements
Research reported in this publication was supported by the University of Michigan Anatomic Pathology Funding Committee and the National Cancer Institutes of Health under Award Number P30CA046592 by the use of the following Cancer Center Shared Resource(s): Tissue and Molecular Pathology. We are thankful to Michelle Vinco and Deborah Postiff for tissue procurement and sample processing.
References
- 1.Umer M, Mohib Y, Atif M, Nazim M. Skeletal metastasis in renal cell carcinoma: A review. Ann Med Surg (Lond) 2018;27:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovacs G, Akhtar M, Beckwith BJ, et al. The Heidelberg classification of renal cell tumours. J Pathol 1997;183:131–3. [DOI] [PubMed] [Google Scholar]
- 3.Udager AM, Mehra R. Morphologic, Molecular, and Taxonomic Evolution of Renal Cell Carcinoma: A Conceptual Perspective With Emphasis on Updates to the 2016 World Health Organization Classification. Arch Pathol Lab Med 2016;140:1026–37. [DOI] [PubMed] [Google Scholar]
- 4.Davis CF, Ricketts CJ, Wang M, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014;26:319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen F, Zhang Y, Senbabaoglu Y, et al. Multilevel Genomics-Based Taxonomy of Renal Cell Carcinoma. Cell Rep 2016;14:2476–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ricketts CJ, De Cubas AA, Fan H, et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep 2018;23:313–26 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor AS, Spratt DE, Dhanasekaran SM, Mehra R. Contemporary Renal Tumor Categorization With Biomarker and Translational Updates: A Practical Review. Arch Pathol Lab Med 2019;143:1477–91. [DOI] [PubMed] [Google Scholar]
- 8.Reuter VETS. Adult renal tumors. . Lippincott Williams & Wilkins: Philadelphia, PA, 2010. [Google Scholar]
- 9.Brunelli M, Eble JN, Zhang S, et al. Eosinophilic and classic chromophobe renal cell carcinomas have similar frequent losses of multiple chromosomes from among chromosomes 1, 2, 6, 10, and 17, and this pattern of genetic abnormality is not present in renal oncocytoma. Mod Pathol 2005;18:161–9. [DOI] [PubMed] [Google Scholar]
- 10.Zhao W, Tian B, Wu C, et al. DOG1, cyclin D1, CK7, CD117 and vimentin are useful immunohistochemical markers in distinguishing chromophobe renal cell carcinoma from clear cell renal cell carcinoma and renal oncocytoma. Pathol Res Pract 2015;211:303–7. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy JM, Wang X, Plouffe KR, et al. Clinical and morphologic review of 60 hereditary renal tumors from 30 hereditary renal cell carcinoma syndrome patients: lessons from a contemporary single institution series. Med Oncol 2019;36:74. [DOI] [PubMed] [Google Scholar]
- 12.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Searle S, Frankish A, Bignell A, et al. The GENCODE human gene set. Genome Biology 2010;11. [Google Scholar]
- 14.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 15.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cieslik M, Chugh R, Wu YM, et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res 2015;25:1372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson DR, Wu YM, Lonigro RJ, et al. Integrative clinical genomics of metastatic cancer. Nature 2017;548:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Zhang Y, Chen YB, et al. VSTM2A Overexpression Is a Sensitive and Specific Biomarker for Mucinous Tubular and Spindle Cell Carcinoma (MTSCC) of the Kidney. Am J Surg Pathol 2018;42:1571–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidarsson H, Westergren R, Heglind M, et al. The forkhead transcription factor Foxi1 is a master regulator of vacuolar H-ATPase proton pump subunits in the inner ear, kidney and epididymis. PLoS One 2009;4:e4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindgren D, Eriksson P, Krawczyk K, et al. Cell-Type-Specific Gene Programs of the Normal Human Nephron Define Kidney Cancer Subtypes. Cell Rep 2017;20:1476–89. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong AJ, Halabi S, Eisen T, et al. Everolimus versus sunitinib for patients with metastatic non-clear cell renal cell carcinoma (ASPEN): a multicentre, open-label, randomised phase 2 trial. Lancet Oncol 2016;17:378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Chen Y, Mo R, et al. Characterization of human RhCG and mouse Rhcg as novel nonerythroid Rh glycoprotein homologues predominantly expressed in kidney and testis. J Biol Chem 2000;275:25641–51. [DOI] [PubMed] [Google Scholar]
- 25.Parolia A, Cieslik M, Chu SC, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 2019;571:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda N, Toi M, Hiroi M, Shuin T, Enzan H. Review of renal oncocytoma with focus on clinical and pathobiological aspects. Histol Histopathol 2003;18:935–42. [DOI] [PubMed] [Google Scholar]
- 27.Han KH, Croker BP, Clapp WL, et al. Expression of the ammonia transporter, rh C glycoprotein, in normal and neoplastic human kidney. J Am Soc Nephrol 2006;17:2670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinmura K, Igarashi H, Kato H, et al. BSND and ATP6V1G3: Novel Immunohistochemical Markers for Chromophobe Renal Cell Carcinoma. Medicine (Baltimore) 2015;94:e989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson RL, Sharma A, Bai S, et al. Comparative STAT3-Regulated Gene Expression Profile in Renal Cell Carcinoma Subtypes. Front Oncol 2019;9:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbons JJ, Abraham RT, Yu K. Mammalian target of rapamycin: discovery of rapamycin reveals a signaling pathway important for normal and cancer cell growth. Semin Oncol 2009;36 Suppl 3:S3–S17. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Pham CG, Albanese SK, et al. Mechanistically distinct cancer-associated mTOR activation clusters predict sensitivity to rapamycin. J Clin Invest 2016;126:3526–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YB, Mirsadraei L, Jayakumaran G, et al. Somatic Mutations of TSC2 or MTOR Characterize a Morphologically Distinct Subset of Sporadic Renal Cell Carcinoma With Eosinophilic and Vacuolated Cytoplasm. Am J Surg Pathol 2019;43:121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Battelli C, Cho DC. mTOR inhibitors in renal cell carcinoma. Therapy 2011;8:359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauer SR, Zhou M, Master VA, Osunkoya AO. Chromophobe renal cell carcinoma with sarcomatoid differentiation: a clinicopathologic study of 14 cases. Anal Quant Cytopathol Histpathol 2013;35:77–84. [PubMed] [Google Scholar]
- 35.Kruger S, Sotlar K, Kausch I, Horny HP. Expression of KIT (CD117) in renal cell carcinoma and renal oncocytoma. Oncology 2005;68:269–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Geneset enrichment analysis of Hallmark pathways with differentially expressed genes from (A) ChRCC versus Others and (B) ChRCC versus Benign tissues. Pathways that showed significant adjusted p-values (<0.05) are shown as bar plots. (NES: normalized enrichment score)
Supplementary Figure 2. Sarcomatoid Chromophobe Renal Cell Carcinoma (ChRCC). Sarcomatoid ChRCC (A, H&E, 100x, and B, H&E, 400x) demonstrates strong staining for FOXI (C, 100x and D, 400x), RHCG (E, 100x and F, 400x), and LINC01187 (G, 100x and H, 400x) only in cells with classic epithelioid morphology. Scale bar = 200 microns for A, C, E, and G; 50 microns for B, D, F, and H.
Supplementary Figure 4. Hybrid Oncocytic Tumor (HOT). HOT (A, H&E, 100x; B, H&E, 400x) demonstrates strong staining for FOXI1 (C, 100x; D, 400x), RHCG (E, 100x; F, 400x), and LINC01187 (G, 100x; H, 400x) limited to one cell population, leading to a checkered appearance. Scale bar = 200 microns for A, C, E, and G; 50 microns for B, D, F, and H.
Supplementary Figure 3. Eosinophilic Chromophobe Renal Cell Carcinoma (Eosinophilic ChRCC) and Oncocytoma. Eosinophilic ChRCC (A, H&E, 200x) and oncocytoma (B, H&E, 200x) both demonstrate strong nuclear expression of FOXI1 (C and D, 200x), RHCG (E and F, 200x), and LINC01187 (G and H, 200x). All oncocytomas showed apical cup-like RHCG staining; some eosinophilic ChRCC demonstrate Golgi-like RHCG staining as seen here. ChRCC biopsy samples show strong nuclear expression of LINC01187 (I-L, 100x). Scale bar = 100 microns for A-H; 200 microns for I-L.
Supplementary Figure 5. Papillary Renal Cell Carcinoma (PRCC). Primary (A, H&E, 200x) and metastatic (B, H&E, 200x) PRCC are negative for FOXI1 (C and D, 200x), RHCG (E and F, 200x), and LINC01187 (G and H, 200x) expression. Scale bar = 100 microns.
Supplementary Figure 7. Patterns of RHCG Expression. Three patterns of RHCG staining were seen, including circumferential membranous staining (A, 200x), apical cup-like staining (B, 200x), and Golgi-like staining (C, 200x). Scale bar = 100 microns.
Supplementary Figure 6. Translocation Renal Cell Carcinoma (t-RCC). Cases of t-RCC (A-C, H&E, 400x) can show weak and patchy nuclear FOXI1 expression (D-F, 400x) and patchy RHCG expression (G-H, 400x), although other cases lack expression of either marker. The case illustrated in the right-hand column is negative for RHCG staining (I, 400x). All cases of t-RCC were negative for LINC01187 (J-L, 400x). Scale bar = 50 microns.
Supplementary Figure 9. Copy number variation analysis in Cases with MTOR Mutation. Both cases have diploid chromosomes and lacked the typical ChRCC copy loss pattern.
Supplementary Figure 8. Biomarker Expression in Cases with MTOR Mutations, Mimicking Eosinophilic Chromophobe Renal Cell Carcinoma. These two cases (A and B, H&E, 200x) demonstrated very focal and weak nuclear staining for FOXI1 (C and D, 200x), despite diffuse Golgi-like RHCG staining (E and F, 200x). Both cases were negative for LINC01187 expression (G and H, 200x). Scale bar = 100 microns.
Supplementary Figure 10. Box plots showing expression of KIT (CD117), FOXI1, LINC01187, and RHCG across TCGA cancer types from multiple organ systems. ACC, Adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, prostate adenocarcinoma; READ, Rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, Testicular Germ Cell Tumors; THCA, thyroid carcinoma; THYM, Thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, Uterine Carcinosarcoma; UVW, Uveal Melanoma.


