To the Editor —
Biologists are important stakeholders in genomic data, both as data generators and as users of genomic data resources. Tools to efficiently visualize and analyze multi-omic data are increasingly important to ensure accessibility and usability of genomic data for all biologists1. We developed the gEAR portal (http://umgear.org) to enable these functionalities (Fig. 1). The development of visualization tools for multi-omic biological data has not kept pace with the generation of transcriptomic and epigenetic data. Repositories allow users to deposit large datasets, but do little in the way of analysis. Therefore, the user must download the raw data and perform analyses locally, which is both time-consuming and out of the reach of many biologists2. While some web-based resources are now available to facilitate multi-omic data analysis3, these resources are typically designed for specific domains, do not enable viewing data spatially overlaid onto a broad set of anatomical structures, and do not allow interrogation of multiple datasets at one time.
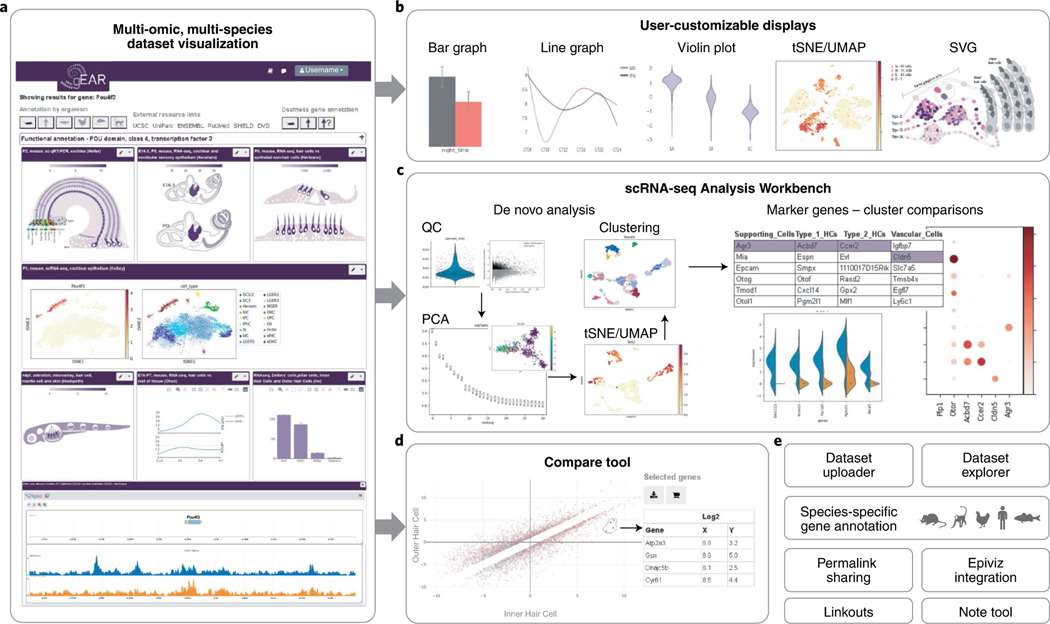
Fig. 1 |. gEAR overview.
a, gEAR result page example. b, Users can select from a variety of custom displays. c, The scRNA-seq Analysis Workbench allows de novo analysis of a dataset; or, for datasets uploaded with expression metadata, analysis can start from marker gene identification and cluster comparison. d, The Compare tool identifies differentially expressed genes to generate gene carts. e, Additional features.
The gEAR portal provides a single platform for data deposition, display, analysis and interrogation, with a focus on allowing users to customize the site and their own data displays. The gEAR portal supports uploaded datasets for a range of organisms, including human, mouse, rat, zebrafish, chicken and marmoset. Multiple datasets can be displayed side-by-side within a display profile, showing not only a user’s own data in a visual form of choice, but also allowing the user to select from hundreds of previously uploaded public datasets to display alongside their own for cross-experimental validation and discovery. Integration of the Epiviz4 tool allows epigenetic modifications to be viewed in the context of gene expression. The portal is also designed to display a gene’s species-specific annotation and links out to information on other systems such as the UCSC Genome Browser5, CCDS6, Ensembl7, OMIM8, and a variety of domain-specific resources. Further features available via drop-down menus include links to the raw data in the Gene Expression Omnibus (GEO)2, the associated publication, a tool to compare gene expression across conditions, a tool to store and export gene lists for downstream analysis of differentially expressed genes, and a single-cell RNA-seq Analysis Workbench.
The gEAR portal’s specialized workflow for single-cell RNA-seq analysis enables a user to upload their dataset and use the Analysis Workbench to perform a stepwise analysis, including quality control, principal component analysis, clustering, dimensionality reduction and marker gene identification. All analyses within the gEAR portal require no coding knowledge and are available through user-friendly interface controls. Users can also upload precomputed analyses, a feature that is particularly useful when hosting annotated and published datasets. Biologists may upload expression or epigenetic data to gEAR (publicly or privately) and perform a rapid initial analysis. Users then curate their datasets to choose how they should be displayed, selecting from options such as bar or line plots, colorized anatomical scalable vector graphics (SVGs), violin plots, or t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) displays. Data displays can be customized to match the displays in publications. Individual datasets or profiles can be shared privately with collaborators initially, then publicly via a short permalink URL as a primary method of data availability within a publication.
A unique feature of the gEAR portal is the ability to display gene expression data using SVGs of anatomical sites rendered as cartoons, where each cell or structure is colored on the basis of a user-customizable color gradient derived from the dataset expression matrix. This feature allows cell-type-specific data to be rendered in a spatial-transcriptomic-like presentation. Furthermore, in situ spatial transcriptomic data from techniques such as MERFISH9 or Visium10 are naturally supported by the portal. By tracking dataset information and using standardized metadata, the gEAR portal actively supports FAIR (Findability, Accessibility, Interoperability and Reusability) data use principles1 and assists users in tracking source data. We have also added a plug-in framework so that users running their own portals can add additional, domain-specific functionality.
While initially generated to support the hearing research community, the gEAR portal has already been adopted as a key data-sharing and visualization resource for other communities, including atlases of brain cell types as a component of the BRAIN Initiative Cell Census Network (BICCN; http://nemoanalytics.org). Both portals use the abilities of the gEAR portal to compare multi-omic data modalities alongside spatial and physiological data, which gives the user comprehensive and molecular insight into the inner ear and brain in biological context. Users are welcome to create accounts and add their data to either of these or even create a new portal for their own domain of interest using the gEAR framework.
Acknowledgements
We are grateful to the members of the Hearing Restoration Project for serving as the focus group in developing this tool and their continuous feedback. We thank R. Marini for technical assistance. This work was funded by the Hearing Restoration Project (R.H.); R01-DC013817 and R01-DC019370 (R.H.); R24- 96MH114815 (R.H. and O.W.); and R01-GM114267 (H.C.B).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Methods thanks Eran Mukamel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Code availability
The gEAR code is available on GitHub (https://github.com/IGS/gEAR) or Zenodo (https://doi.org/10.5281/zenodo.4818596) and released under the GNU Affero General Public License v3.0.
References
- 1.Wilkinson MD et al. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T. et al. Nucleic Acids Res. 41, D991–D995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cakir B. et al. NAR Genom. Bioinform 2, lqaa052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chelaru F, Smith L, Goldstein N. & Bravo HC Nat. Methods 11, 938–940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haeussler M. et al. Nucleic Acids Res. 47, D853–D858 (2019). (D1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pruitt KD et al. Genome Res. 19, 1316–1323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham F. et al. Nucleic Acids Res. 47, D745–D751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKusick VA Am. J. Hum. Genet 80, 588–604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffitt JR et al. Proc. Natl Acad. Sci. USA 113, 11046–11051 (2016). [Google Scholar]
- 10.Marx V. Nat. Methods 18, 9–14 (2021). [DOI] [PubMed] [Google Scholar]