Abstract
This study presents the results of SARS-CoV-2 surveillance in sewage water of 11 municipalities and marine bioindicators in Galicia (NW of Spain) from May 2020 to May 2021. An integrated pipeline was developed including sampling, pre-treatment and biomarker quantification, RNA detection, SARS-CoV-2 sequencing, mechanistic mathematical modeling and forecasting. The viral load in the inlet stream to the wastewater treatment plants (WWTP) was used to detect new outbreaks of COVID-19, and the data of viral load in the wastewater in combination with data provided by the health system was used to predict the evolution of the pandemic in the municipalities under study within a time horizon of 7 days. Moreover, the study shows that the viral load was eliminated from the treated sewage water in the WWTP, mainly in the biological reactors and the disinfection system. As a result, we detected a minor impact of the virus in the marine environment through the analysis of seawater, marine sediments and, wild and aquacultured mussels in the final discharge point of the WWTP.
Keywords: SARS-CoV-2, COVID-19, Wastewater-based epidemiology, Virus variants, Viral load, Predictive model, Stochastic SIR model
Graphical abstract

1. Introduction
The world is experiencing a pandemic caused by the virus SARS-CoV-2, firstly detected in December 2019 in China (WHO, 2020). The rapid spread of the virus has evidenced the need for the development of tools to massively detect the presence of the virus in local communities that, combined with individual screening methods, would contribute to SARS-CoV-2 surveillance. Diagnostic methods based on qRT-PCR assay to amplify the genetic material of the virus were set up to confirm the disease in patients and asymptomatic population (Corman et al., 2020). Various studies confirmed the presence of SARS-CoV-2 in feces of patients with COVID-19 (Chen et al., 2020; Wu et al., 2020; Xiao et al., 2020). As a result, the genetic material of the virus can be detected in the wastewater in varying concentrations, from 20 to 3 × 106 copies/L (Foladori et al., 2020; Lastra et al., 2022) depending on number of infected individuals, viral load in feces and dilution in the wastewater. Recent studies detected the presence of different variants of the virus, including Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron, in wastewater (Pérez-Cataluña et al., 2021; Wolfe et al., 2022).
The possibility of using the wastewater epidemiology approach for COVID-19 was raised soon after the onset of the pandemics (Ahmed et al., 2020; Bivins et al., 2020; Medema et al., 2020; Wurtzer et al., 2020). Wastewater monitoring has been proven already as a viable method to track markers of human activity including drugs, pharmaceuticals, pathogen microorganisms and virus (LaTurner et al., 2021). The first report on detection of SARS-CoV-2 RNA in wastewater was released in March 2020 by Medema and coauthors, in which fragments of the genetic material of SARS-CoV-2 were identified in several sewage samples from The Netherlands in the beginning of March 2020, right after the first case of COVID-19 human infection was detected in the country. The complete version of this study was published by Medema et al. (2020) and includes a quantitative analysis by qRT-PCR. This study points to sewage surveillance as a potential tool for early warning of (re-) emergence or increase of infections by SARS-CoV-2.
During 2020 and the beginning of 2021, efforts were made worldwide to develop methods and procedures to analyze sewage water as a simple and fast tool to know the prevalence of the virus in the community. The viral load in sewage water was used to detect COVID-19 outbreaks and track the evolution of the infected population (Larsen and Wigginton, 2020), with the possibility to detect hot spots sampling the sewage network or monitor specific areas, facilities, or schools (Fielding-Miller et al., 2021; Gibas et al., 2021; Haak et al., 2022) and different studies confirmed wastewater monitoring as a convenient complementary approach to COVID-19 surveillance and testing strategies (Peccia et al., 2020; Foladori et al., 2020; Kitajima et al., 2020; Randazzo et al., 2020).
After increasing evidences of the capacities of sewage surveillance, the European Commission has highlighted the importance of the surveillance of SARS-CoV-2 and its variants in wastewaters as a rapid and cost-effective analysis, as well as a reliable source of information about the transmission of SARS-CoV-2 in the population (European Commission, 2021). Wastewater monitoring should be considered as a complementary tool for surveillance of SARS-CoV-2 and other pathogens in the population.
The protocols for detection of the genetic material in wastewater and its quantification have been increasingly optimized (Ahmed et al., 2022) since the beginning of the pandemic. The screening of SARS-CoV-2 variants in complex matrices, such as urban wastewaters, can be very challenging (Martin et al., 2020; Crits-Christoph et al., 2021; Graber et al., 2021; La Rosa et al., 2021). However, multiple efforts were made during 2021 to consolidate the sequencing of SARS-CoV-2 ARN in wastewater samples, and its usefulness for detecting variants in circulation has been seen (Islam et al., 2021; Izquierdo-Lara et al., 2021; Hillary et al., 2021; Fontenele et al., 2021). Different studies have applied next-generation sequencing approaches to the viral RNA obtained from the sewage samples during the wastewater surveillance protocols. Many of these sequencing studies have been benefited from the ARTIC Network (https://artic.network/ncov-2019) amplicon library, which amplifies the whole viral genome with 400 bps amplicons as a previous step before sequencing. This approach works correctly with sewage samples if the genetic material is not degraded below the size threshold. Sewage sequencing protocols have allowed the detection of SARS-CoV-2 variants of concern in a certain area before these results were obtained in clinical samples (Jahn et al., 2021). Sequencing of sewage samples can be used as well for detecting novel SNVs (single nucleotide variants) or recent mutations that are not yet widespread and take longer to be detected in clinical samples (Fontenele et al., 2021). The mutations that arise from the analysis of sewage samples can be compared with clinical samples from the same regions in order to reveal if there are regionally prevalent variants or if some variants are being imported to a concrete region (Martin et al., 2020; Crits-Christoph et al., 2021).
In this paper we present a methodology developed for sewage water surveillance of SARS-CoV-2 in Galicia (NW of Spain) through 11 wastewater treatment plants (WWTPs) located in medium-sized municipalities with no discharge from hospitals. The pipeline of the method, illustrated in the graphical abstract, integrates: wastewater sampling in the WWTPs of study (at the inlet, outlet effluent and final discharge points), sampling from marine sediment and bioindicators, pre-treatment and biomarker quantification, RNA detection by RT-qPCR, sequencing of SARS-CoV-2 in wastewater samples, data management through a digital platform and epidemics forecasting by a predictive mechanistic model. This work provides new evidence of wastewater RT-qPCR analysis being a reliable method for early detection of SARS-CoV-2 outbreaks in the community as it has been validated in previous studies (Randazzo et al., 2020) and, in combination with the data reported by the health system, for monitoring and prediction of the evolution of the pandemics at the municipal-level. This work contributes a number of relevant distinguishing features with respect previously published studies, from which we highlight the following aspects: the evaluation of the fate of the virus in wastewater and marine environments, the assessment of the efficiency of the WWTPs in the removal of the genetic material of the virus, and the development of a mechanistic model that, combining data from the health system, shows predictive capacity in forecasting the evolution of the pandemics at the municipality level. Besides, the integral approach including the detection of variants from wastewater samples is a significant source of information for the monitoring of the impact of the pandemic.
2. Materials and methods
2.1. WWTP sampling
The detection of the genetic material of the SARS-CoV-2 virus was carried out in wastewater treatment plants (WWTP) in Galicia (NW Spain) during one year, from May 2020 to May 2021 (Fig. 1 ). Eleven WWTPs with different treatment technologies were selected from medium-sized municipalities with a connected population between 2000 and 23,000 inhabitants (Table 1 ). The selected plants had no wastewater discharge from hospitals that could increase the viral load in the wastewater, falsifying the measured viral load in the sewage water generated by the infected population. Some samples from Nigrán and Baiona WWTPs (Fig. 1), collected in March and April 2020, were also analyzed for the presence of SARS-CoV-2 genetic material. Four additional WWTPs were included in the study in June–August 2020 upon request of regional health authorities to help in the control of COVID-19 outbreaks (these WWTPs were identified as A, B, C and D in Fig. 1).
Fig. 1.
Wastewater treatment plants (WWTP) sampled for monitoring the presence of SARS-CoV-2 in wastewater and marine environment.
Table 1.
Characteristics of the selected WWTP for the SARS-CoV-2 monitoring.
| WWTP | Served population | Population equivalent (p.e.) | design flow (m3/day) |
peak flow (m3/h) |
secondary treatment | tertiary treatment | Discharge point | |
|---|---|---|---|---|---|---|---|---|
| 1 | Baiona | 12,090 | 36,000 | 7314 | 690 | Biologic -Act. Sludge | N and P removal + chlorination | In the sea |
| 2 | Nigrán | 17,038 | 70,000 | 19,600 | 1469 | Biologic -Act. Sludge | N removal + UV | In the sea |
| 3 | Gondomar | 11,067 | 24,000 | 6720 | 562 | Biologic -Act. Sludge | N removal + UV | River |
| 4 | Cambados | 20,841 | 48,000 | 12,000 | 750 | Biologic -Act. Sludge | N removal + chlorination | In the sea |
| 5 | Moraña | 1672 | 6225 | 1680 | 168 | Biologic -Act. Sludge | N removal + chlorination | River |
| 6 | Porto do Son | 4054 | 12,764 | 3192 | 319 | Biologic -Act. Sludge | UV | In the sea |
| 7 | Muros | 4697 | 9000 | 1500 | 150 | Biologic -Act. Sludge | N removal + microfiltration + UV | In the sea |
| 8 | Melide | 5725 | 15,000 | 4310 | 431 | Biologic -Act. Sludge | N removal | River |
| 9 | Ares | 22,850 | 52,000 | 13,278 | 1500 | Biologic -Act. Sludge | N and P removal + chlorination | In the sea |
| 10 | Cedeira | 5146 | 10,395 | 3119 | 312 | Biologic -Act. Sludge | microfiltration + UV | River |
| 11 | Noia | 14,000 | 20,000 | 5000 | 500 | Biologic -Fixed bed | N removal + UV | River mouth |
| C | Burela | 9566 | 14,700 | 2320 | 214 | None (Physicochemical treatment) | None | In the sea |
Sampling was carried out in the WWTPs as well as in the discharge point in the marine environment. In the WWTPs, 1 or 2 samples per week were taken in the raw water in the inlet stream to the plant (identified as M1), the effluent of the secondary settling tank (M2) and the final discharge effluent after disinfection (M3). Samples of thickened sludge were also collected (M5) to determine if the increasing solid concentration in the sludge would allow for more sensitive detection of the genetic material of the virus. In the marine environment, samples were taken every two weeks in the final discharge point, and identified as follows: samples of seawater (M4), marine sediment (BIOIND-S) and bioindicators: wild mussels (BIOIND) and aquaculture mussels (BIOIND-A). The authors paid special attention to the detection of SARS-CoV-2 in mussels due to their extraordinary capacity for filtering water and the possibility of concentrating viral RNA, increasing the probability of the detection of the viral genetic material.
Samples M1-M3 were collected in the WWTPs as 24 h-composite samples using an automatic sampling system (Teledyne ISCO, model 3700 full size, USA). M4 samples were collected in the discharge point of the WWTP, 1 m below the sea surface. M5 samples were collected as grab sample directly from the sludge settling tank. 1 L of all the samples (M1-M5) were collected and stored in amber glass bottles and kept at 4 °C during storage and transportation to the laboratory (maximum 24 h). In the laboratory, the samples (M1-M5) were filtered through a 20–25 μm cellulose filter (Whatman®- Grade 4) to remove coarse suspended particles. The filtered sample was divided into two aliquots: one for wastewater concentration, RNA isolation, and detection by qPCR; and the other aliquot for biomarkers determination by liquid chromatography.
2.2. Biomarkers quantification
Caffeine (1,3,7-trimethylxanthine) was selected as a biomarker in the sewage water for each WWTP included in this study. The biomarker was used to normalize the viral load variability due to dilution and floating population effects. Caffeine was determined by HPLC using an Agilent 1260 Infinity II (Agilent Technologies, CA, USA) chromatograph equipped with a gradient quaternary pump, oven, autosampler and a UV–vis diode array detector. 10 mL of the filtered wastewater samples were spiked with 4 μg of caffeine and then determined by HPLC as described by Moret et al. (2012). The spiked caffeine samples were eluted at 1.0 mL·min−1 and separated on a 4.6 × 100 mm (4 μm) Poroshell 120EC-C18 column at 25 °C. The mobile phase consisted of 0.1% phosphoric acid aqueous solution (eluent A) and acetonitrile (eluent B). Elution was held for 6 min in isocratic mode, using 80% of eluent A and 20% of eluent B. 50 μL injection volume was used and caffeine was detected in the UV range at 274 nm. All the samples were analyzed in triplicate to assure the precision and reproducibility of the results with a coefficient of variation below 10%.
2.3. Wastewater concentration
Although several methods of concentration of samples were initially evaluated, the adsorption-precipitation protocol with aluminum chloride (AlCl3), previously described in Randazzo et al. (2019), was selected because of better quality and quantity of RNA obtained. Briefly, 150 mL of water was transferred into a beaker of 200 mL and 75 μL of the enveloped rhabdovirus SVCV (105 TCID50/mL) (an enveloped RNA virus) was inoculated to each water sample as a concentration control. The SVCV virus was selected because it is an enveloped virus like the SARS-CoV-2, and it is frequently used in our research group. The pH of each sample was adjusted to 6.0 and 0.9 N AlCl3 solution was added to the sample at 1:100 ratio. The pH was again readjusted to 6.0 and samples were mixed at room temperature in an orbital shaker at 150 rpm during 15 min. Then, samples were centrifuged at 1700g for 20 min. in a Sorval ST Plus Series centrifuge (Thermo Scientific, USA) and the pellet was resuspended in 10 mL of 3% beef extract at pH 7.5 and transferred to 15 mL centrifuge tubes (Falcon). Samples were mixed again at room temperature in an orbital shaker at 200 rpm for 10 min. and centrifuged at 1900g for 30 min. in the Allegra™ X-22R Centrifuge (Beckman Coulter). Finally, the pellet was resuspended in 1 mL 1× PBS and samples were stored at −20 °C until RNA isolation.
2.4. RNA isolation
Three procedures were used to extract RNA from the concentrated sewage water, seawater, marine sediment and bioindicator (mussels) samples. First 884 samples of concentrated sewage water, seawater and sediment were processed using the QIAmp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. 140 μL of concentrated samples were used for this purpose. Rest of the concentrated sewage, seawater and sediment samples were processed using Maxwell RSC Pure Food GMO and Authentication kit (Promega, Madison, USA) with a slight modification. Briefly, 300 μL of concentrated sewage water, seawater and sediment samples were mixed vigorously with 400 μL of CTAB (Cetyl Trimethyl Ammonium Bromide) and 40 μL of proteinase K and incubated 10 min. at 60 °C. After centrifugation at 16000g for 10 min., the supernatant was mixed with 300 μL of lysis buffer and transferred to the Maxwell® Instrument Cartridge where RNA was automatically isolated in 80 μL of elution buffer. The purpose of using the Maxwell RSC Pure food GMO and Authentication kit was the reduction of time effort for RNA extractions, since this procedure was automatized.
RNA from bivalve samples (bioindicators) was isolated using the Maxwell RSC simply RNA Tissue Kit (Promega, Madison, USA) attending the manufacturer's instructions. Briefly, bivalves were carefully extracted from the shell. A small fragment from gill, mantle, gonad and digestive gland of 3 bivalve samples were pooled in a single sample and homogenized in 200 μL of chilled 1-Thioglycerol/Homogenization solution. After homogenization, 200 μL of lysis buffer were added to the samples and mixed vigorously for 15 s. Then, 400 μL of the samples were transferred to the Maxwell® Instrument Cartridge and RNA was automatically isolated in 50 μL of nuclease free water.
The concentration and purity of the isolated RNA were determined by a NanoDrop™ 1000 spectrophotometer (NanoDrop Technologies, Inc., DE, USA). RNA was kept at −80 °C until further use.
2.5. SARS-CoV-2 detection by qPCR assay
Before the SARS-CoV-2 detection, the viral concentration methodology was validated by quantitative PCR using the SVCV primers N-SVCV-For and N-SVCV-rev of SVCV rhabdovirus (Table 2 ). The results of this validation have been detailed in the supplementary material S1.
Table 2.
Primers and probes used for the SARsCoV-2 detection by RT-qPCR.
| Primer name | Target gene | Sequence (5′-3′) | Reference |
|---|---|---|---|
| nCOV_N1 F | Nucleocapsin (N) | GACCCCAAAATCAGCGAAAT | CDC assay |
| nCOV_N1 R | TCTGGTTACTGCCAGTTGAATCTG | ||
| nCOV_N1 P | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | ||
| nCOV_N2 F | TTACAAACATTGGCCGCAAA | ||
| nCOV_N2 R | GCGCGACATTCCGAAGAA | ||
| nCOV_N2 P | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | ||
| ESarbeco_F | Envelope (E) | ACAGGTACGTTAATAGTTAATAGCGT | Corman et al., 2020 |
| ESarbeco_R | ATATTGCAGCAGTACGCACACA | ||
| Esarbeco_P1 | FAM-ACACTAGCCATCCTTACTGCGCTTCGQSY7 | ||
| N-SVCV-For | Nucleoprotein (N) | TGAGGTGAGTGCTGAGGATG | López-Muñoz et al., 2010 |
| N-SVCV-Rev | CCATCAGCAAAGTCCGGTAT |
SARS-CoV-2 RNA was detected by TaqMan real-time RT-PCR using the kit GoTaq® Probe 1-Step RT-qPCR System, 12.5 mL (Promega, Winsconsin, US). Two oligonucleotide primers and probes designed by the US Centers for Disease Control and Prevention (CDC) were used to target two regions of the nucleocapsid gene (N), as well as the primers and probe that target the gene E (Table 2). For each target gene, each sample was analyzed by qPCR in two technical replicates (6 qPCRs per sample) using a StepOnePlus Real-time PCR System (Applied Biosystems, USA). Each reaction mix (20 μL) contained 10 μL GoTaq® Probe qPCR Master Mix with dUTP (2×), supplemented with CXR Reference Dye, 30 μM according to manufacturing protocols, 0.4 μL GoScript™ RT Mix for 1-Step RT-qPCR, 1 μL of each primer pair (10 μM), 0.5 μL probe (10 μM), 2.1 μL of Ultrapure™ distilled water (Invitrogen) and 5 μL of each RNA sample. The thermal cycling conditions were 45 °C for 15 min. Followed by a preheating at 95 °C for 2 min. and 45 cycles amplification at 95 °C for 3 s, and 55 °C for 30 s. Standard curves were added on each qPCR plate. They were composed by seven serial dilutions (1:10) of synthetic RNA transcripts of the genetic targets with a known concentration (200,000 copies/μL):2019-nCoV_N_Positive Control and 2019-nCoV_E_Positive Control (IDT-Technologies, USA). Results were obtained with the StepOne Software v2.3. A sample was considered positive if Ct values with a correct amplification plot were obtained in at least two target genes. From each sample, Ct values of each technical replicate were transformed to the number of genomic copies/reaction PCR from the equation of each standard curve obtained for each target gene. The minimum coefficient of determination allowed on each standard curve was R2 > 0.8. Then, the average of copies/reaction obtained for each sample were extrapolated to copies/L. The efficiency and sensitivity of each pair of primers (N1, N2 and E) were estimated and detailed in Supplementary material S2 and the effect of possible PCR inhibition was estimated and detailed in Supplementary material S3.
2.6. Sequencing of SARS-CoV-2 in sewage samples
Five samples of interest were selected for ARTIC amplicon-based sequencing of SARS-CoV-2 in order to identify SARS-CoV-2 variants. Sewage samples were selected based on qPCR Cts for genes N and E below the threshold of 30. The only exception was a sample selected based on its nature (sampled from a Mink farm in which the propagation of the virus to the animals was reported). This sample was characterized by lower Cts but it was also included. The second criterion was the sampling time. Samples were selected from the months of December 2020 and January 2021, which were characterized by the maximum levels of viral spread in Galicia (NW Spain). Selected samples belong to the WWTP in the towns of Baiona, Melide and Noia (Fig. 1).
Starting from the concentrated samples and the RNA extraction explained in sections “2.4. RNA isolation” and “2.5. SARS-CoV-2 detection by qPCR assay”, selected samples were submitted to reverse transcription to cDNA using the High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems) following the manufacturer instructions. The obtained cDNA was amplified using the ARTIC primers set V.3. (Integrated DNA Technologies, USA) developed by the ARTIC Network and which cover the entire genome of SARS-CoV-2 with amplicons of 400 bps (https://github.com/artic-network/artic-ncov2019). Two PCR reactions were performed for every sample, each one with one Pool of ARTIC primers. PCR reactions consisted in 12 μL of DreamTaq Master Mix, 4 μL of the respective ARTIC Primer Pool (10 μM), 2.5 μL cDNA and 6.5 μL of Nuclease free water for a total reaction volume of 25 μL. Cycling conditions were 1× (98 °C, 30s), 30× (98 °C, 15 s; 65 °C, 5 min). After this, PCR products were merged for each sample and submitted to purification. Samples quality and concentration were measured and sequencing libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina). Finally, amplicon sequencing libraries were sequenced in an Illumina NovaSeq6000 equipment (Macrogen, Korea).
2.7. Bioinformatic analysis
Reads were quality-checked using the FastQC tool on Omics Box (Biobam; https://www.biobam.com/omicsbox). Paired reads were trimmed in the QIAGEN CLC Genomics Workbench 20.0 (https://digitalinsights.qiagen.com/). Quality trimming was performed removing the first 20 nts of each read and applying a quality threshold of 0.01 (Phred = 20). Nextera adapter sequences were trimmed as well. Trimmed reads were mapped against a SARS-CoV-2 reference genome (ID: NC_045512). Mapping parameters were set to Match score = 1, Mismatch cost = 2, Insertion cost = 3, Deletion cost = 3, Length Fraction = 0.5, Similarity Fraction = 0.8. Average mapping coverage was calculated for each sample based on the mapped reads sequencing depth (considering the bases of the mapped reads and the bases that conform the reference genome). Mapped reads tracks were generated and analyzed for variant detection using the basic variant detection tool of the CLC Genomics Workbench 20.0.1. The variant analysis parameters were set to Ploidy = 1, Nonspecific read matches were ignored, Minimum coverage = 10, Minimum variant reads count = 2, Minimum frequency = 5%. Frequency was calculated considering the number of reads carrying the mutation in comparison with every read covering the position. Only mutations that caused changes in the protein sequence were considered for further analyses. Known mutations that define variants of current concern were retrieved from the Covariants database (CoVariants.org). Moreover, novel mutations non-defined in previously described SARS-CoV-2 variants of concern were retrieved from the sequencing data with stricter filters (Minimum coverage = 100, Quality >30, Variant reads >10 and Frequency > 5%).
SARS-CoV-2 genomes sequenced from Galician patients were downloaded from the GISAID database (Elbe and Buckland-Merrett, 2017), from the beginning of the pandemic (December 2020) until the end of this study (27/04/2021). A total of 500 genomes were downloaded and used for further analyses. Mutations were analyzed in comparison to the Wuhan reference using the Nextclade tool (https://clades.nextstrain.org). The frequency of each mutation was calculated considering the number of genomes in which each mutation appeared compared to the total number of genomes. A comparative analysis was performed to study which mutations where shared between Galician clinical samples and the studied sewage samples.
Genomes were also downloaded from GISAID for clinical samples collected in Portugal and the Spanish Autonomous Regions during the months of December 2020 and January 2021 (a total of 865 and 1862 genomes respectively were considered). The same comparative analysis between clinical and sewage samples in the same winter period was performed.
The online tool covidcg (covidcg.org) was used in order to determine the worldwide distribution of several mutations found in the sewage samples. Specifically, mutations that were not shared with clinical samples, i.e. mutations exclusively identified in sewage and not related to specific variants of interest so far, were analyzed using this tool.
2.8. Predictive mathematical model
2.8.1. Model identification
A mathematical model of the SIR type is developed (Model-1 in the supplementary information S4) with states: S, I, R, Iu, Io including as observables the number of infected persons reported by the health system (Io) and the total number of infected persons (I) estimated from the viral copies detected in wastewater samples (see supplementary information for details). The data of the number of new infected persons per day in each municipality were obtained by SERGAS (Galician Health Service).
The municipalities included in this study are small-medium size, between 2000 and 23,000 inhabitants (Table 1). For this population range, the effects of stochasticity in the dynamics are significant, and therefore they need to be taken into account. In a stochastic regime, the dynamics corresponding to Model-1 are described by a Chemical Master Equation (CME) (Pájaro et al., 2019), simulated here by means of the Gillespie's Stochastic Simulation Algorithm (SSA), which provides exact realizations of the CME (Gillespie, 2007).
2.8.2. Model calibration
The model calibration was performed using two different time course data sets (viral number of copies in wastewater and number of infected persons reported by the health system). Data were partitioned in periods of maximum 7 days in which the restriction levels can be considered to be constant. The model has only two parameters for calibration. We obtained 4 parameter vectors, corresponding to 4 different levels of restrictions. Interestingly, there were no significant variations among the parameters among the different WWTPs.
2.8.3. Process variability monitoring
In order to assess the variability of the process due to the effects of dilution and floating population, we monitor two auxiliary variables: the concentration of caffeine in the sample (previously reported as a reliable biomarker), and the daily cell-phone mobility data for the municipalities in the study (provided by Nommon Solutions and Technologies, Spain).
2.8.4. Model predictions
Predictions are made by solving the Model-1 equations by stochastic simulation (105 realizations of the SSA algorithm) with initial conditions being the number per copies in wastewater at the starting date of interest (t0) and the number of accumulated infected persons calculated from the daily new infected persons in the municipality (14 days), provided by the health system. The parameter vector is fixed according to the level of population restrictive policy in the period of interest (values obtained at the calibration stage). From the 105 realizations of the SSA algorithm, the mean and standard deviation of the mean for predicted states were computed, in order to provide confidence intervals for the forecast values.
3. Results
3.1. Presence of SARS-CoV-2 in WWTPs and marine bioindicators
Sampling and analysis of SARS-CoV-2 in 11 WWTP and their marine environment were carried out from May 2020 to May 2021. Besides, 3 additional samples from Nigrán (M1 in March 12, 2020) and Baiona WWTP (M1 and M2 samples in April 28, 2020) were also included in this study. Overall, 1342 samples (sewage water, seawater, marine sediment and mussels) were taken and analyzed. It represents 24.5 samples per week on average during 13 months.
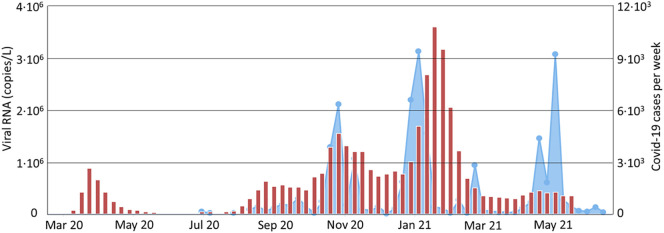
The presence of the genetic material of the virus SARS-CoV-2 in the inlet stream to the WWTP (M1 samples) is shown in Fig. 2 with the active COVID-19 cases reported by the regional health authorities from Galicia region. The vertical bars show the percentage of positive samples versus the total samples analyzed per month in the 11 WWTP in this study. The presence of viral RNA in the raw wastewater (M1 samples) followed a similar trend to the profile active cases of COVID-19 in the region, confirming that the analysis of viral RNA in the WWTP raw water can be used to monitor the evolution of the pandemic in each of the 11 municipalities and the region as a whole. It is important to note the peaks in positive M1 samples in Nov-2020 and Jan-2021 corresponding with the 3rd and 4th COVID-19 waves registered in Galicia and Spain.
Fig. 2.
Presence of viral RNA of SARS-CoV-2 in the WWTP inlet stream in the 11 municipal WWTP expressed as percentage of positive analysis (bars) and the total active cases in the Galicia region (line) reported by the public health system.
Fig. 3 shows the average viral load in M1 samples in the 11 WWTP and the weekly new cases for the total period of the study in the 11 municipalities. This figure shows that the viral load signal in M1 samples peaks 7–10 days in advance. The detection in advance is evident in the two more intense waves of COVID-19 in Galicia in November 2020 and January 2021. The evolution of the viral load in the raw wastewater (M1) can be observed in Baiona WWTP (Table 3 ). The viral load was decreasing during Nov-2020 as the 3rd wave active cases decreased in the area. During Dec-2020, the active cases remained constant in the region, but the viral load in M1 (raw water in the WWTP inlet) was steadily increasing until the end of the month (29 Dec 2020) when the viral load was about 20 times higher than that in the beginning of the month. These results anticipated the explosion of COVID-19 cases in the early January 2021 and the 4th wave.
Fig. 3.
Weekly average viral load in the inlet stream to the WWTP of the 11 municipalities (line plot with shaded area) and the new weekly infections reported from the public health system in Galicia region.
Table 3.
Viral load in the raw wastewater to the Baiona WWTP anticipating the 3rd COVID-19 wave in Jan-2021.
| Date | Viral load (copies/L) |
||
|---|---|---|---|
| N1 Gene | N2 Gene | E Gene | |
| 14-Nov-20 | 7.30 × 104 | ||
| 17-Nov-20 | 2.13 × 104 | ||
| 25-Nov-20 | 8.93 × 103 | ||
| 26-Nov-20 | 1.06 × 104 | ||
| 2-Dec-20 | 6.86 × 103 | ||
| 14-Dec-20 | 3.64 × 104 | ||
| 29-Dec-20 | 1.30 × 105 | 2.59 × 105 | 3.90 × 104 |
| 4-Jan-21 | 2.87 × 105 | 5.16 × 105 | 1.89 × 105 |
| 9-Jan-21 | 1.98 × 105 | 2.22 × 105 | 4.37 × 105 |
| 16-Jan-21 | 3.60 × 105 | 2.90 × 105 | 4.91 × 105 |
| 20-Jan-21 | 1.06 × 105 | 3.58 × 104 | 7.32 × 104 |
| 25-Jan-21 | 5.59 × 104 | 3.68 × 104 | 2.96 × 104 |
| 30-Jan-21 | 3.77 × 104 | 2.12 × 104 | 1.86 × 103 |
| 31-Jan-21 | 4.52 × 104 | 2.73 × 104 | 6.26 × 103 |
It is important to note from Fig. 2, Fig. 3 that there was a decreasing of the COVID-19 impact in March–May 2021 (with ~5% to ~30% of fully vaccinated people, respectively) but the decreasing of new and active cases did not correspond with the reduction of the viral load in the WWTP raw water.
The analysis of the genetic material of SARS-CoV-2 in the discharge effluent (M3), the effluent from the secondary settling tank (M2) and the concentrated sludge (M5) in the WWTP showed very low impact. Only 1% of M3, 3% of M2 and 4% of M5 samples were positive. Moreover, there is not a clear relation of high viral load in the raw wastewater and the detection of the viral RNA in the discharge effluent of other intermediate points in the plant. These results suggest that the biological treatment and the disinfection of wastewater (with chlorine or UV light, Table 1) in the sewage plant were able to degrade and/or remove the genetic material of the virus from the wastewater. Conversely, in the physicochemical plants (e.g., Burela WWTP, Fig. 1) the viral RNA was detected in the raw water and in intermediate sampling points in the plant, suggesting that chemical flocculation and physical separations were not completely effective in removing the viral RNA from the wastewater (with the exception of Melide WWTP in which there is no specific disinfection treatment, only a tertiary treatment consisting on N removal).
The presence of the viral RNA in the marine environment was mainly limited to the discharge points of the Cambados and Muros WWTP (Table 4 ). The viral RNA was not detected on marine sediment; it was only detected in mussels (bioindicator) and seawater. It is believed that the presence of the viral RNA in the marine environment was associated with uncontrolled discharge points and wastewater bypass in the plants. Moreover, the viral RNA was detected mainly in the marine environment in July–November in 2020 but not in January 2021 when the highest viral loads (over 106 copies/mL) were measured in the raw water to the WWTP during the 4th COVID-19 wave. As we will discuss later, here it is important to remark that RT-qPCR detection cannot be used to assess infectivity.
Table 4.
Impact of SARS-CoV-2 in the sea (Positive samples / total number of samples).
| WWTP | Baiona | Nigrán | Gondomar | Cambados | Muros | Ares |
|---|---|---|---|---|---|---|
| M4 | 0/2 | 1/11 | 0/10 | 4/52 | 2/13 | 0/2 |
| BioInd | 0/9 | 0/19 | 1/19 | 1/10 | −/− | 0/1 |
| BioInd-A | −/− | 0/1 | −/− | 4/42 | −/− | −/− |
| BioInd-S | −/− | 0/11 | 0/10 | 0/11 | 0/13 | 0/2 |
| Total | 0/11 | 1/42 | 1/39 | 9/115 | 2/26 | 0/5 |
3.2. Forecast of the evolution of the pandemics by municipalities using a predictive mathematical model with wastewater and clinical data
The mathematical model developed in this study (Model-1) shows a good predictive capacity allowing us to forecast the evolution of the pandemics (number of infected persons, observed and unobserved by the health system) in all the municipalities within a horizon of 7 days. In Fig. 4 we include, as representative illustration of the model outcome, the predictions for three different municipalities (Ares, Melide, Baiona) at different time periods. The figures, data and software codes for the whole study are available online at https://github.com/DIMCoVAR/Model-1. The model predictions (mean and standard deviations) for total number of infected (blue) and observed number of infected (black) are depicted in Fig. 4 together with the real data obtained from wastewater samples (blue circles) and health system (black squares). From the 105 realizations of the SSA algorithm, the mean (solid blue line) and standard deviation (dotted blue line) are computed. Note that, as expected from a stochastic process, the confidence interval increases with time, making longer term predictions impractical, as it has been reported in previous studies (Castro et al., 2020).
Fig. 4.
Predictions of the evolution of the epidemics obtained by the SIR-modified stochastic model (Model-1) in the municipalities of Ares, Melide and Baiona at different starting dates. Blue lines are the mean (solid) and standard deviation (dashed) of the total infected persons as predicted by the model. Black lines are the mean (solid) and standard deviation (dashed) of the number of infected observed by the health system as predicted by the model. Real data from water samples and health system are represented by blue circles and black squares, respectively.
3.3. SARS-CoV-2 sequencing in sewage samples
Sequencing process allowed us to obtain about 18 million reads per sample. After the trimming, more than 17 million reads were retained for further analyses in all the samples. Trimmed reads were mapped to the SARS-CoV-2 reference genome (ID: NC_045512), being possible to obtain a successful number of reads of SARS-CoV-2 in 4 out of 5 sequenced sewage samples. It is noteworthy that the sample in which an insufficient coverage of reads was obtained was the worst sample in terms of CT values, the one obtained from a Mink farm. With the other samples from 3 Galician towns (Baiona, Melide and Noia) it was possible to obtain successful sequencing results, obtaining an average base coverage of 4098× (all the details about the sequencing, trimming and mapping processes are shown in Table 5 ).
Table 5.
Information covering the sequencing process and the variant analyses.
| Sample ID | 929 | 963 | 971 | 989 | 996 |
|---|---|---|---|---|---|
| Location | Noia | Melide | A Baña | Baiona | Melide |
| Sampling date | 28/12/2020 | 10/01/2021 | 15/01/2021 | 16/01/2021 | 17/01/2021 |
| Sample type | M1 (sewage inlet) |
M1 (sewage inlet) |
Mink farm |
M1 (sewage inlet) |
M1 (sewage inlet) |
| Ct gene N | 26.78 | 26.18 | 34.18 | 29.21 | 27.64 |
| Ct gene E | 27.62 | 27.39 | 35.69 | 29.50 | 28.34 |
| ARTIC amplification, Illumina Nextera sequencing and SARS-CoV-2 mapping | |||||
| Total reads | 18,523,470 | 18,580,114 | 18,249,222 | 19,551,208 | 18,051,722 |
| Trimmed reads | 17,639,592 | 18,068,128 | 17,473,150 | 19,073,506 | 17,352,478 |
| Mapped reads | 1,378,004 | 1,400,298 | 3,500 | 83,004 | 2,165,954 |
| Average coverage/depth | 5,646.63 X | 5,731.26 X | 3.87 X | 339.34 X | 8,771.89 X |
| Aminoacidic mutations | 73 | 70 | 28 | 68 | 100 |
| Mutations - unique mutations for a certain variant are highlighted in bold letters- |
S:A222V N:A220V ORF1b:V767L |
S:A222V N:A220V |
S:S477N S:N501Y S:A570D S:D614G |
S:A222V N:A220V ORF8:Q27* |
|
| Variants likely present -variants detected with unique mutations non shared by other strains are highlighted in bold letters- |
20E (EU1) - Spain 20A.EU2 - Europe |
20E (EU1) - Spain |
20A.EU2 - Europe 20I/501Y.V1 - B.1.1.7 – UK 20H/501Y.V2 - B.1.351 – South Africa 20J/501Y.V3 – P.1 – Brazil 20B/S.484K - P.2 – Brazil 20C/S.452R - B.1.427/9 – California 20C/S.484K - B.1.526 – New York 20A/S.484K - B.1.525 |
20E (EU1) - Spain 20I/501Y.V1 - B.1.1.7 – UK |
|
| Novel mutations - coverage>100, freq>5%, variant reads>10, quality>30 | 24 | 27 | 9 | 52 | |
| Novel mutations found in Galician clinical samples | 3 | 5 | 7 | ||
3.4. Basic variants analysis
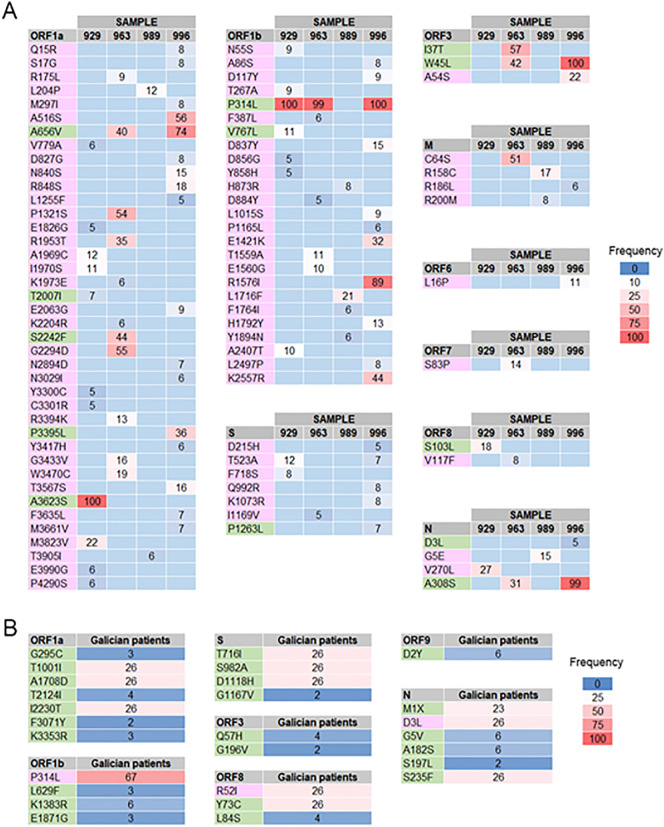
The analysis of variants displayed an important number of mutations contained in our samples, ranging from 68 to 100 single amino acidic changes (Table 5). Among all these mutations, it is worth mentioning the presence of non-synonymous nucleotide changes that have defined new virus lineages of concern. These mutations, as well as the lineages that they define are displayed in Table 5. Specifically, mutations of Spike (A222V) and N gene (A220v) that are defining mutations of 20E (EU1) clade, initially expanded in Spain and spread widely across Europe, were detected in a very high frequency (88%–100%) in Noia (929) and Melide (963 and 996) samples (Fig. 5A). Moreover, mutations defining another clade spread across Europe and named as 20A.EU2 have been also found in two samples, Noia (929) and Baiona (989). These defining mutations are located in the ORF1b (V767L) and Spike (S477N) and showed a frequency of 10%–20% (Fig. 5A). The analysis allowed to find as well other important mutations of Spike that determine relevant lineages in terms of virus spread: N501Y (mutation shared by B.1.1.7, B.1.351 and P1 lineages, commonly known as UK, South African and Brazilian variants respectively) and D614G (shared by all the new variants of concern). In fact, this last mutation is one of the most frequently represented as well as showing the highest read coverage. These mutations were detected in the 989 sample from Baiona. In addition to the aforementioned mutations shared by several lineages of clinical concern that have been appeared throughout the pandemic, two additional mutations characteristic of the UK variant (B.1.1.7) were found in two different samples, 989-Baiona and 996-Melide. More specifically, S:A570D and ORF8:Q27* that are represented in the set of reads with a frequency of 17% and 40% respectively (Fig. 5A).
Fig. 5.
A. Identification of known mutations from different SARS-Cov-2 variants of current concern in sewage samples. Mutations are represented in the genomic position in which they occur and named after the amino acidic change they cause in the protein sequence. Samples, read coverage or depth and the frequency of occurrence are represented for each mutation. The variants of concern that are characterized by the found mutations are also indicated. B. The most common mutations associated with currently described variants are represented for 500 SARS-Cov-2 genomes from Galician clinical samples. Mutations are divided by gene and the frequency is represented using a color scale. Frequency here indicates the percentage of genomes in which a certain mutation appears (only mutations present in 10 or more genomes were considered). Mutations highlighted in rose were also detected in the studied sewage samples.
After identifying the known mutations defining linages of concern that were found in the sewage samples, a comparative analysis was performed with viral genomes from patients. A set of SARS-CoV-2 genomes from 500 Galician patients collected since the beginning of the pandemic until the end of April 2021 were analyzed in order to pool the set of mutations present in the population. Fig. 6B shows the most common mutations (those that appear at least in 10 out of 500 genomes) present in the population that were associated with the aforementioned lineages of concern. Note that some of the most common mutations from sewage, are also found in patients (S:A222V, S:N501Y, S:A570D, S:D614G, ORF8:Q27* and N:A220V) (Fig. 5A).
Fig. 6.
A. Novel mutations identified in the sewage samples (coverage>100, freq>5%, variant reads>10, quality>30). Mutations are divided by gene and the frequency is represented using a color scale. The frequency of each mutation represents the number of variant reads related to the total number of reads which cover the position. B. The most common mutations not associated with currently described variants are represented for 500 SARS-Cov-2 genomes from Galician clinical samples. Mutations are divided by gene and the frequency is represented using the same color scale. Frequency here indicates the percentage of genomes in which a certain mutation appears (only mutations present in 10 or more genomes were considered). A correspondence between novel mutations shared by clinical and sewage samples is indicated: rose (sewage samples) and green (clinical samples).
The information associated with viral variants in the sewage samples uncovers the mutation pool of the overall population, as seen with the matching results. We have been able to find relatively high number of amino acidic variations that until now have not been associated with relevant variants as those presented above. A total number of 87 of these novel mutations were identified in sewage samples, with the employed filtering criteria (Fig. 6A). Some of these mutations showed remarkably high frequencies and were found in several samples, as is the case of ORF1a:A656V, ORF1b:P314L, ORF3:W45L or N:A308S (Fig. 6A). The most relevant case, ORF1b:P314L was also found in the Galician patients in a very high frequency (Fig. 6A), which, together with the previous results, shows a remarkable correlation between sewage samples and patients from the same geographic area.
Certainly, many of the mutations appear at low frequencies. In order to determine if the sample size of 500 Galician genomes was not large enough, the same analysis was performed with genomes from the rest of Spain and Portugal. After the comparison, it became clear that the Galician genome database represents quite accurately the mutation pool of the Iberian Peninsula, since the results are practically identical to those already described.
Focusing on the less frequent mutations, Spike mutations not associated with variants of concern and which appeared in sewage samples and not in patients, were analyzed in detail. The places and dates in which they had been detected were analyzed (Supplementary material S5, Fig. S4). Despite that these mutation had not been detected in Galicia until the present sewage samples, it was seen that S:D215H was a relatively old mutation with a great presence especially in the UK. The other Spike mutations previously undetected in Galicia are much more recent and their number of worldwide sequences is still starting to rise since the beginning of 2021. Sewage samples can be useful to detect new mutations that in the future may be of great importance in clinical samples as well.
4. Discussion
The pandemic of the SARS-CoV-2 spread rapidly in Europe since February 2020 with a significant impact in Spain. The restrictive policies upon the population issued by the government resulted in a minimum impact of COVID-19 during May–June 2020. Therefore, at that moment the detection and control of the new outbreaks was of utmost importance for the health authorities. The analysis of the sewage water for the virus and its genetic material achieved widespread importance (Ahmed et al., 2020; Randazzo et al., 2020; Cervantes-Avilés et al., 2021). In that context, our results confirmed the capacity of the sewage water surveillance to follow the evolution of the pandemic in the studied community, through SARS-CoV-2 sewage monitoring of a number of representative municipalities (medium sized and without discharges from hospitals). In addition, and in contrast to other research conducted on the presence of SARS-CoV-2 in sewage, our study aimed also at exploring the detection of the virus in the marine environment and the capacity of the WWTP to remove the virus (Galicia is well known for its fish, shellfish and aquaculture activities). Our results confirmed the capacity of the biological reactors and the disinfection system in WWTP to eliminate the virus (Balboa et al., 2021; Serra-Compte et al., 2021). The impact in the marine environment was of minor importance and the detection of the virus in seawater and wild and aquacultured mussels can be associated to uncontrolled discharge of wastewater and alleviation from the sewage network. In terms of bioindicators, recently Le Guernic et al. (2022) also detected the presence of SARS-CoV-2 genetic material in zebra mussels exposed to raw and treated wastewater confirming these mollusks as good environmental indicators that can be used for active surveillance of pathogenic microorganisms in environmental waters. It is important to remark here that, the RT-qPCR analysis in this work targets specific RNA gene sequences and it cannot be used to assess infectivity. However, several studies exploring SARS-CoV-2 stability and replication in the environment supported that, after wastewater treatment, the release of infective viral particles is unlikely (Rimoldi et al., 2020; Westhaus et al., 2021).
We observed that the average viral load signal over all the WWPTs peaked before the health system reported cases in the community in the most important waves in the course of the study. This observation is coherent with an anticipative capacity of the sewage signal. The effect of the vaccination program may explain the relatively high concentrations of viral RNA in the wastewater despite the decreasing amount of cases detected by the national health system (as a percentage of infected individuals who are vaccinated and with less symptoms might go unnoticed) in March–May 2021 (with ~5% and ~ 30% of the population fully vaccinated, respectively). Bivins and Bibby (2021) measured lower concentrations of viral genetic material in the wastewater after the massive vaccination in a college campus (closed community).
To the best of our knowledge, this is the first study in which wastewater samples and data from the health system are combined to successfully predict the number of active cases of COVID-19 using a mechanistic mathematical model. The inclusion of wastewater samples endows the model with higher anticipative capacity. Moreover, the model is demonstrated to be robust, and therefore easily extrapolated to other WWTP of municipalities in the same population range (the low number of parameters of the model makes very easy to re-calibrate the model for its use in very different locations and or scenarios). As key factors for the success (predictive capacity) and robustness of the model, we highlight the following: i) the evolution of the pandemics is evaluated locally at the level of municipalities, ii) we take into account the effects of stochasticity, such that the confidence intervals for the predictions are automatically generated, iii) the predictions are made for time horizons of 7 days.
As the pandemic progressed, the occurrence of mutations and new virus lineages caused much concern due to the increased infectivity of the virus (Korber et al., 2020; Plante et al., 2021; Volz et al., 2021a). As this fact conditions the control of the pandemic, being able to find mutations of interest as early as possible using sewage seems very interesting. The most advanced mass sequencing methods have allowed to obtain the sequence of the virus in several countries in the world (Crits-Christoph et al., 2021; Fontenele et al., 2021; Jahn et al., 2021). Despite the difficulty of sequencing given the high degradation of the sample and the fragmentation of the viral genome, it is possible to detect specific mutations that provide guidance on the variants that may be present in the population. In the present work, linage-defining mutations as well as shared mutations by the most infective lineages acknowledged at the time of study, were identified. The constant emergence of new viral variants casts doubts on whether a mutation will invariably define a single lineage. However, sequencing results allow the identification of specific mutations compatible with arising lineages of concern at the time the samples were taken. Specifically, some mutations defining the B.1.1.7 lineage at the time of sequencing were identified. This variant, also known as Alpha variant, spread worldwide by the end of 2020 (Volz et al., 2021b), a time compatible with the detection of some of these mutations in our samples dated December 2020/January 2021. Moreover, it is important to highlight that two of the most frequent mutations with high coverage in our analysis are ORF1b:P314L and S:D614G, that are commonly found to be co-occurring and that can greatly increase the infectivity of the virus (Ogawa et al., 2020). Remarkably, methods based on RT qPCR for mutation monitoring have been developed in order to detect and monitor specific SARS-CoV-2 variants (see for example Lee et al., 2021; Wurtzer et al., 2022).
Furthermore, it could be evidenced in this work a marked agreement between the most frequent mutations detected in sewage and patients of the same geographical area, which constitutes another evidence of the potential of sewage analysis to represent the evolution of the pandemic. Novel mutations are appearing continually. In sewage samples we have detected mutations which appeared recently and had not been detected in the geographical area of study. Novel mutations that the virus is adopting may end up having great relevance, so early detection in sewage could be very useful.
5. Conclusions
This study confirmed that the analysis of the genetic material of SARS-CoV-2 in the sewage water in the WWTP inlet stream is a sensitive and practical method to detect new outbreaks of COVID-19 and to evaluate the evolution of the pandemic in the community.
As part of the integral approach for SARS-CoV-2 surveillance, this study developed a mechanistic model to predict the number of infected people in municipalities of small-medium size based on the viral load in the sewage water and the data of infections from the public health system. Our results confirm that the model is robust and has predictive capacity, being capable of forecasting the evolution of the pandemics in the municipalities time horizon of seven days. We highlight as key aspects for the predictive capability and robustness of the model that i) is local (municipality-based), ii) it takes into account the stochastic nature of the process.
Moreover, the analysis of variants showed a marked agreement between the most frequent mutations detected in sewage and patients of the same geographical area.
The treatment of wastewater in biological reactors and the subsequent disinfection favors the elimination of the virus from treated sewage water. The study of the presence of virus in the marine environment in the discharge point of the WWTP revealed a minor impact of the SARS-CoV-2 in seawater, marine sediment and, wild and aquacultured mussels.
Code availability
Code and figures available at https://github.com/DIMCoVAR/Model-1
Data availability
ARCTIC amplicon library sequenced reads are deposited in the NCBI SRA repository, accessible through the Bioproject PRJNA783202.
CRediT authorship contribution statement
Beatriz Novoa: Conceptualization, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. Raquel Ríos-Castro: Methodology, Investigation. Irene Otero-Muras: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. Susana Gouveia: Investigation, Resources. Adrián Cabo: Investigation, Resources. Amaro Saco: Formal analysis, Investigation. Magalí Rey-Campos: Formal analysis, Investigation. Manuel Pájaro: Methodology, Formal analysis, Software, Validation, Writing – review & editing. Noelia Fajar: Data curation, Formal analysis, Investigation, Validation. Raquel Aranguren: Investigation. Alejandro Romero: Investigation. Antonella Panebianco: Investigation. Lorena Valdés: Investigation. Pedro Payo: Conceptualization, Resources, Funding acquisition. Antonio A. Alonso: Conceptualization, Methodology, Funding acquisition. Antonio Figueras: Conceptualization, Formal analysis, Investigation, Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing. Claudio Cameselle: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors gratefully acknowledge the support of this work through the project “DIMCoVAR” funded in the program “Fondo Supera COVID” of the CRUE (Spanish Universities)-Santander Foundation. The authors also thank Aguas de Galicia and Consellería de Sanidade - Xunta de Galicia for their support and funding of this study. The IIM-CSIC group is also funded by projects 0474_BLUEBIOLAB from EU FEDER Program Interreg Spain-Portugal and IN607B 2019/01 from Consellería de Economía, Emprego e Industria (GAIN), Xunta de Galicia. MP acknowledges support from grant FJC2019-041397-I funded by MCIN/AEI/10.13039/501100011033.
Funding for open access charge: Universidade de Vigo/CISUG.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.155140.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Mueller J.F.… First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Shanks O.C.… Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Bibby K. Wastewater surveillance during mass COVID-19 vaccination on a college campus. Environ. Sci. Technol. Lett. 2021;8:792–798. doi: 10.1021/acs.estlett.1c00519. [DOI] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Bibby K.… Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Castro M., Ares S., Cuesta J.A., Manrubia S. The turning point and end of an expanding epidemic cannot be precisely forecast. PNAS. 2020;117:26190–26196. doi: 10.1073/pnas.2007868117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Avilés P., Moreno-Andrade I., Carrillo-Reyes J. Approaches applied to detect SARS-CoV-2 in wastewater and perspectives post-COVID-19. J. Water Process Eng. 2021;40 doi: 10.1016/j.jwpe.2021.101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen L., Deng Q., Cheng Z.… The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Drosten C.… Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph A., Kantor R.S., Olm M.R., Nelson K.L.… Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio. 2021;12 doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob.Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Commission HERA Incubator: anticipating together the threat of COVID-19 variants. 2021. https://ec.europa.eu/info/sites/default/files/communication-hera-incubator-anticipating-threat-covid-19-variants_en.pdf
- Fielding-Miller R., Karthikeyan S., Gaines T., Knight R.… . 2021. Wastewater and surface monitoring to detect COVID-19 in elementary school settings: the Safer at School Early Alert project. medRxiv 2021.10.19.21265226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know?A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenele R.S., Kraberger S., Hadfield J., Varsani A.… High-throughput sequencing of SARS-CoV-2 in wastewater provides insights into circulating variants. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Munir M. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782 doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D.T. Stochastic simulation of chemical kinetics. Annu. Rev. Phys. Chem. 2007;58:35–55. doi: 10.1146/annurev.physchem.58.032806.104637. [DOI] [PubMed] [Google Scholar]
- Graber T.E., Mercier É., Bhatnagar K., Delatolla R.… Near real-time determination of B.1.1.7 in proportion to total SARS-CoV-2 viral load in wastewater using an allele-specific primer extension PCR strategy. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak L., Delic B., Li L., Guarin T., Mazurowski L., Dastjerdi N.G., Dewan A., Pagilla K. Spatial and temporal variability and data bias in wastewater surveillance of SARS-CoV-2 in a sewer system. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary L.S., Farkas K., Maher K.H., Jones D.L.… Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam A., Sayeed M.A., Kalam M.A., Hassan M.M.… Molecular epidemiology of SARS-CoV-2 in diverse environmental samples globally. Microorganisms. 2021;9:1696. doi: 10.3390/microorganisms9081696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Lara R., Elsinga G., Heijnen L., de Graaf M.… Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerg. Infect. Dis. 2021;27:1405–1415. doi: 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K., Dreifuss D., Topolsky I., Beerenwinkel N. 2021. Detection of SARS-CoV-2 variants in Switzerland by genomic analysis of wastewater samples. medRxiv 2021.01.08.21249379. [DOI] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Rose J.B.… SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Montefiori D.C.… Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Suffredini E.… Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastra A., Botello J., Pinilla A., Flores J.… SARS-CoV-2 detection in wastewater as an early warning indicator for COVID-19 pandemic.Madrid region case study. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTurner Z.W., Zong D.M., Kalvapalle P., Stadler L.B.… Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guernic A., Palos Ladeiro M., Boudaud N., Geffard A.… First evidence of SARS-CoV-2 genome detection in zebra mussel (Dreissena polymorpha) J. Environ. Manag. 2022;301 doi: 10.1016/j.jenvman.2021.113866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Imakaev M., Armas F., McElroy K.A., Gu X., Duvallet C., Chandra F., Chen H., Leifels M., Mendola S., Floyd-O’Sullivan R. Quantitative SARS-CoV-2 alpha variant B. 1.1. 7 tracking in wastewater by allele-specific RT-qPCR. Environ.Sci.Technol.Lett. 2021;8(8):675–682. [Google Scholar]
- López-Muñoz A., Roca F.J., Sepulcre M.P., Meseguer J., Mulero V. Zebrafish larvae are unable to mount a protective antiviral response against waterborne infection by spring viremia of carp virus. Dev. Comp. Immunol. 2010;34:546–552. doi: 10.1016/j.dci.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Martin J., Klapsa D., Wilton, Majumdar M. Tracking SARS-CoV-2 in sewage: evidence of changes in virus variant predominance during COVID-19 pandemic. Viruses. 2020;12 doi: 10.3390/v12101144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Moret S., Hidalgo M., Sanchez J.M. Simple and fast methods based on solid-phase extraction coupled to liquid chromatography with UV detection for the monitoring of caffeine in natural, and wastewater as marker of anthropogenic impact. ISRN Chromatogr. 2012;2012 doi: 10.5402/2012/487138. [DOI] [Google Scholar]
- Ogawa J., Zhu W., Tonnu N., Singer O., Hunter T., Ryan (Firth) A.L., Pao G.M. 2020. The D614G mutation in the SARS-CoV2 Spike protein increases infectivity in an ACE2 receptor dependent manner. bioRxiv 2020.07.21.214932. [DOI] [Google Scholar]
- Pájaro M., Otero-Muras I., Vázquez C., Alonso A.A. Transient hysteresis and inherent stochasticity in gene regulatory networks. Nat. Commun. 2019;10:4581. doi: 10.1038/s41467-019-12344-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Omer S.B.… Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Chiner-Oms Á., Cuevas-Ferrando E., Sánchez G. 2021. Detection of genomic variants of SARS-CoV-2 circulating in wastewater by high-throughput sequencing. medRxiv. [DOI] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Shi P.-Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Piqueras J., Evtoski Z., Sastre G., Sancho R., Gonzalez C., Sánchez G. Interlaboratory comparative study to detect potentially infectious human enteric viruses in influent and effluent waters. Food Environ. Virol. 2019;11:350–363. doi: 10.1007/s12560-019-09392-2. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Compte A., González S., Arnaldos M., Litrico X.… Elimination of SARS-CoV-2 along wastewater and sludge treatment processes. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Connor T.R. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Ferguson N.M.… Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany - suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Naming the coronavirus disease (COVID-19) and the virus that causes it [WWW Document] 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it URL. (accessed 10.28.21)
- Wolfe M., Hughes B., Duong D., Chan-Herur V., Wigginton K.R., White B.J., Boehm A.B. 2022. Detection of SARS-CoV-2 variant Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron in wastewater settled solids using mutation-specific assays is associated with regional detection of variants in clinical samples. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Huang X.… Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill. 2020;25(50) doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Levert M., Cluzel N., Almayrac J.L., Charpentier C., Masnada S., Gillon-Ritz M., Mouchel J.M., Maday Y., Boni M. SARS-CoV-2 genome quantification in wastewaters at regional and city scale allows precise monitoring of the whole outbreaks dynamics and variants spreading in the population. Sci. Total Environ. 2022;810 doi: 10.1016/j.scitotenv.2021.152213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Zhao J.… Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
ARCTIC amplicon library sequenced reads are deposited in the NCBI SRA repository, accessible through the Bioproject PRJNA783202.