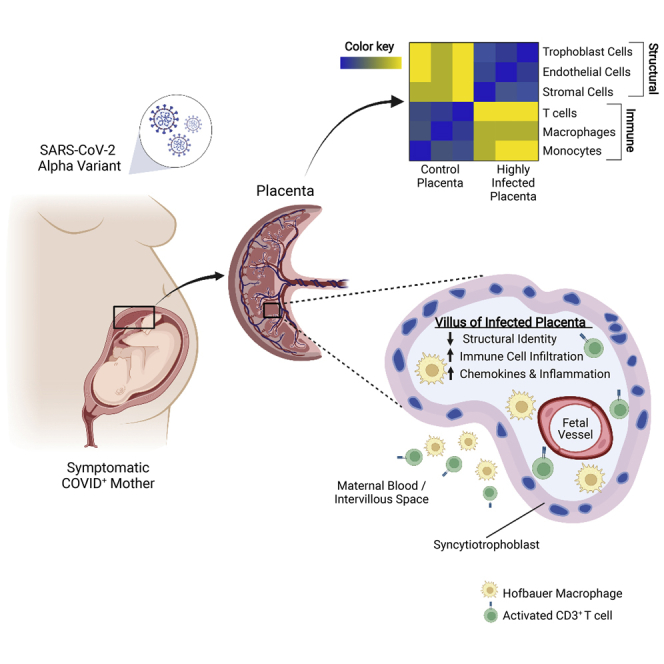
Summary
The effect of SARS-CoV-2 infection on placental function is not well understood. Analysis of placentas from women who tested positive at delivery showed SARS-CoV-2 genomic and subgenomic RNA in 22 out of 52 placentas. Placentas from two mothers with symptomatic COVID-19 whose pregnancies resulted in adverse outcomes for the fetuses contained high levels of viral Alpha variant RNA. The RNA was localized to the trophoblasts that cover the fetal chorionic villi in direct contact with maternal blood. The intervillous spaces and villi were infiltrated with maternal macrophages and T cells. Transcriptome analysis showed an increased expression of chemokines and pathways associated with viral infection and inflammation. Infection of placental cultures with live SARS-CoV-2 and spike protein-pseudotyped lentivirus showed infection of syncytiotrophoblast and, in rare cases, endothelial cells mediated by ACE2 and Neuropilin-1. Viruses with Alpha, Beta, and Delta variant spikes infected the placental cultures at significantly greater levels.
Subject areas: Health sciences, Immunology, Transcriptomics
Graphical abstract
Highlights
-
•
SARS-CoV-2 RNA is detected in 22 out 52 placentas from COVID-positive women
-
•
Infected placentas show extensive infiltration of maternal immune cells
-
•
Infected placentas show increased expression of chemokines and inflammation markers
-
•
Pseudovirus with variant spikes infects placental cultures at higher levels
Health sciences; Immunology; Transcriptomics
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has already taken a devastating toll with the global virus caseload surpassing 490 million infections and more than 6.1 million deaths worldwide. In severe cases, SARS-CoV-2 causes a respiratory illness, whose defining features are an imbalanced inflammatory host response, reduced innate antiviral defenses and an inflammatory “cytokine storm,” endothelial damage, coagulopathies, and thrombosis in several tissues from infected patients (Blanco-Melo et al., 2020).
The effect of SARS-CoV-2 infection during pregnancy on the health of the mother and baby is not fully understood. In the large majority of pregnancies, babies are delivered uninfected when tested by nasopharyngeal swabs and without apparent effect. However, pregnant women with symptomatic SARS-CoV-2 infections are more likely to be admitted to intensive care and the maternal death rates are statistically higher when compared with non-pregnant infected women (Zambrano et al., 2020). Whereas preterm deliveries occur more often in women with suspected or confirmed SARS-CoV-2 infection, no increase in stillbirths or early neonatal deaths was found (Mullins et al., 2021). Prospective and retrospective studies showed that pregnant women infected with SARS-CoV-2 are at an increased risk of adverse events, including higher rates of cesarean section and increased postpartum complications (Marín Gabriel et al., 2020; Prabhu et al., 2020; Woodworth et al., 2020). Vertical transmission from mother to fetus has been reported in a few cases (Alamar et al., 2020; Hecht et al., 2020; Facchetti et al., 2020; Taglauer et al., 2020; Vivanti et al., 2020; Woodworth et al., 2020), although most studies have not detected viral transmission to the fetus (Baergen and Heller, 2020; Cribiù et al., 2021; Della Gatta et al., 2020; Edlow et al., 2020; Kimberlin and Stagno, 2020; Mourad et al., 2021; Penfield et al., 2020; Prabhu et al., 2020; Salvatore et al., 2020; Schwartz, 2020).
The effect of SARS-CoV-2 on placental function is not well understood. Several studies have detected infection of placentas in women who tested positive for the virus at or before delivery. In some cases, the placenta displayed signs of inflammation with increased vascular malperfusion indicative of thrombi within fetal vessels (Baergen and Heller, 2020; Prabhu et al., 2020; Shanes et al., 2020; Vivanti et al., 2020) and infiltration of maternal immune cells (Debelenko et al., 2021; Facchetti et al., 2020; Garrido-Pontnou et al., 2021; Hosier et al., 2020; Lu-Culligan et al., 2021; Morotti et al., 2021). Whether the inflammation resulted from the infection of maternal tissue or the placenta itself is unclear and may depend on the gestational age of the fetus at the time of maternal infection. Several viruses are known to affect placental function and virus-associated inflammation during pregnancy, resulting in chronic cardiovascular disease, diabetes, and obesity later in life (Burton et al., 2016).
SARS-CoV-2 uses ACE2 (Angiotensin-converting enzyme 2) as the primary receptor (Hoffmann et al., 2020) and Neuropilin-1 (NRP1) as a co-receptor (Cantuti-Castelvetri et al., 2020; Daly et al., 2020) in concert with the two proteinases TMPRSS2 (Transmembrane protease serine 2) (Hoffmann et al., 2020) and CTSL (Ou et al., 2020) and the pro-protein convertase FURIN (Shang et al., 2020), amongst others (Daniloski et al., 2021; Schneider et al., 2021; Wang et al., 2021; Wei et al., 2021) for cell entry. All of the entry receptors are expressed at significant levels in the first and second trimester placentas. At term, the entry cofactors are expressed at lower levels in the placenta and the chorioamniotic membranes at the maternal–fetal interface (Baston-Buest et al., 2011; Li et al., 2020; Lu-Culligan et al., 2021; Pique-Regi et al., 2020; Singh et al., 2020; Taglauer et al., 2020). Whether alternative mechanisms are exploited by SARS-CoV-2 to enter the placenta is not known.
SARS-CoV-2 variants that have emerged over the past year are defined by the World Health Organization (WHO) as variants of concern or interest. Four variants of concern have been identified (Alpha (B.1.1.7), Beta (B1.351), Gamma (P.1), and Delta (B.1.617.2)) that contain mutations in the spike protein receptor binding domain and result in increased rates of transmission and lethality (World Health Organization, 2021) (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/). Whether the variants pose an increased risk of the infection of the placenta and adverse effects on placentation and pregnancies is unknown.
In this study, we analyzed placentas from women who tested positive for SARS-CoV-2 at the time of delivery to determine the extent of infection and impact on the inflammatory state of the placental tissues. Of the 22 analyzed placentas, 2 containing high levels of SARS-CoV-2 RNA were both infected with the Alpha (B.1.1.7) variant. These contained the highest levels of viral RNA and both resulted in adverse outcomes for the fetus. The placentas showed infiltration of maternal immune cells and an active immune response at the maternal–fetal interface. Ex vivo infection of placental cultures showed an infection of syncytiotrophoblast. Viruses pseudotyped by the variants of concern spike protein infected placental cells with a greater efficiency than the parental, non-variant spike protein. These results suggest that a direct infection of placental cells can occur and the variants may have increased propensity to infect this critical tissue during pregnancy.
Results
Clinical presentations of SARS-CoV-2 positive mothers, fetal outcomes, and placental pathologies
To understand the effects of SARS-CoV-2 infection during pregnancy, we assembled a panel of placental tissues from 52 women who had been identified as positive for SARS-CoV-2 at the time of admission for delivery at NY Presbyterian Hospital-Weill Cornell (H_1-H_2, P_1-P_20, N1-N30; Table 1). All placental samples were obtained from the Department of Pathology and most of them displayed placental pathologies or were obtained from mothers with clinical presentations. The infection status was determined by qRT-PCR from nasopharyngeal swabs. As controls, placental tissues were obtained from four women who tested negative for SARS-CoV-2 (C_1-C_4), and four SARS-CoV-2 negative women with placental inflammatory pathologies (I_1-I_4) that included acute chorioamnionitis (ACA) and chronic villitis of unknown etiology (CVUE) (Table 1).
Table 1.
Clinical presentations of SARS-CoV-2 positive mothers, fetal outcomes, and placental pathologies
 |
Mat Age: maternal age (years), Gest Age: gestational age (weeks), Mother COVID +/−: mother tested RT-PCR positive (+) or negative (−) at birth, COVID-19 Symptoms +/−: patient noted to have mild symptoms unless otherwise noted as “severe” (+) or was noted to be asymptomatic (−); NA if notes had no documentation of COVID-19 symptoms, DFM: decreased fetal movement, NICU: neonate intensive care unit, MCI: massive chronic intervillositis, IUFD: intra-uterine fetal demise, MFI: maternal floor infarction, PPH: postpartum hemorrhage, CHI: chronic histiocytic intervillositis, ACA: acute chorioamnionitis, T2D: type 2 diabetes, Mec: meconium, IVT: intervillous thrombi, VUE: villitis of unknown etiology, BMI: body mass index, TAB: therapeutic abortion, D&C: dilation and curettage, IDA: iron deficiency anemia, ITP: immune thrombocytic purpura, HTN: hypertension, CIN1: Cervical Intra-epithelial Neoplasia Grade 1, PCS: Pelvic Congestion Syndrome, PEC: preeclampsia SF: severe features, GBS: Group B Streptococcus+, Di/Di: dichorionic/diamniotic, GDM: Gestational Diabetes Mellitus, PTL: pre-term labor, PAPP-A: pregnancy-associated Plasma Protein A, UCTD: undifferentiated connective tissue disorder, PROM: premature rupture of membranes, OUD: opoid use disorder, HCV: Hepatitis C Virus+, PIH: pregnancy-induced/gestational hypertension. Gray shaded rows show the fetal demise/NICU admission.
The pregnant women ranged in age from 16 to 51 years, with the majority in their 20s and 30s. Two pregnancies resulted in intra-uterine fetal demise (IUFD) (H_2, P_10). One fetus that was delivered preterm at 25 weeks of gestation was admitted to the neonatal intensive care unit (NICU) where the infant remained for several months (H_1). All neonates tested negative for SARS-CoV-2 by nasopharyngeal swabs at 24-h post-delivery. Among the placentas delivered from mothers who tested positive for SARS-CoV-2, 33% (17 cases) presented with fetal vascular malperfusion (FVM), 23% (12 cases) displayed maternal vascular malperfusion (MVM), and 8% (4 cases) overlapped for both pathologies. None of the healthy control placentas from SARS-CoV-2 negative mothers displayed FVM or MVM (Table 1).
SARS-CoV-2 viral RNA presence in the placenta
To determine the extent of SARS-CoV-2 infection in the placenta of infected mothers, we isolated RNA from formalin-fixed paraffin embedded (FFPE) sections from all placental samples and measured genomic and replicating viral RNA by qRT-PCR using primers against SARS-CoV-2-N. The presence of a PCR amplicon of the correct size on melting curves and agarose gels confirmed the positive and negative results (Figure 1A and data not shown). Genomic and subgenomic viral RNA was detected in 22 of the 52 placentas from SARS-CoV-2 positive mothers (42%) (Table 1). The presence of SARS-CoV-2 RNA in the placenta was not correlated with the observed FVM (10 displayed FVM out of 22 positive placentas; Table 1).
Figure 1.
SARS-CoV-2 virus is present in placentas from infected mothers and results in inflammatory responses
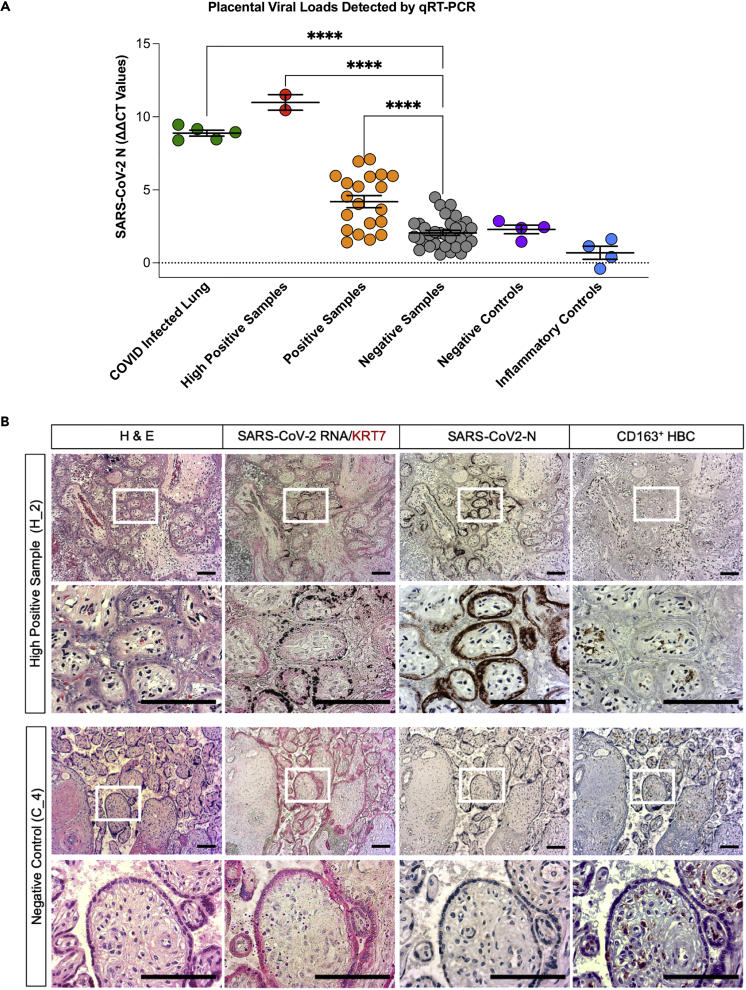
(A) Graph showing ΔΔCT values of RNA samples isolated from FFPE patient placenta slides from the different patient cohorts included in this study. Positive control lung samples (n = 5), high positive samples (n = 2), and positive samples (n = 20) are significantly higher than negative samples (n = 30). Negative (n = 4) and inflammatory controls (n = 4). Statistical analysis was performed using a one-way ANOVA and adjusted for by multiple comparisons test using the Benjamini–Hochberg FDR method ∗∗∗∗p value < 0.0001. Data are presented as mean ± SEM.
(B) Brightfield microscopy images of a representative COVID high positive placental sample H_2 and a representative negative control placental sample C_4. Slides were stained by H&E, in situ hybridization for SARS-CoV-2-RNA and counterstained for syncytial trophoblast marker cytokeratin (KRT7, red), and by immunohistochemistry for SARS-CoV-2-N protein (brown) as well as for CD163-positive Hofbauer cells (HBC). Scale bars = 100 μm.
See also Figure S1.
Viral RNA was not detected in the placenta samples obtained from SARS-CoV-2 negative healthy mothers and uninfected mothers with unrelated inflammatory pathologies (Table 1). This provided five distinct cohorts for this study: high positive samples (H_1, H_2; ddCT value >9), positive samples (P_1, P_3-P_4, P_9-P_15; ddCT >4.5, and P_2, P_5–P_8, P_16–P_20, borderline positive; presence of amplicon and ddCT <4.5), negative samples (N1–N30; ddCT <4.5 and no amplicon), and negative controls (C_1-C_4) and inflammatory controls (I_1-I_4) (Figure 1A and Table 1).
Strikingly, the three pregnancies from SARS-CoV-2 positive mothers that resulted in IUFD or admission of the neonate to the NICU-delivered placentas that were highly positive (H_1 and H_2) or positive (P_10) for SARS-CoV-2 (Table 1, grey shaded rows).
Placental syncytiotrophoblast are the primary target for SARS-CoV-2 infection of pregnant females at term
We next investigated placental pathologies and identified the cells in the placental chorionic villi that were infected by SARS-CoV-2. Adjacent placental sample sections (10 microns apart) were stained with hematoxylin and eosin (H&E) and examined for the presence of replicating viral RNA by in situ hybridization and for the presence of SARS-CoV-2 nucleocapsid protein (SARS-CoV-2 N) by immunohistochemistry. Hofbauer cells (HBC) of the placenta and maternal monocytes/macrophages were visualized by immunohistochemistry with an anti-CD163 antibody. Subgenomic viral RNA was readily detected by in situ hybridization in the high positive samples H_1 and H_2 and sporadically in positive samples but not in negative controls (Figures 1B and S1, and not shown). The presence of viral RNA was restricted to the Keratin-7 (KRT7)-positive trophoblast layers that anatomically cover the chorionic villi structures. These consist of an outer layer of syncytiotrophoblast that faces the maternal blood (MB) space and a cytotrophoblast layer underneath (Figures 1B and S1). Similarly, the expression of the SARS-CoV-2 N protein was detected in adjacent sections within the same villi. The N protein was localized to the syncytiotrophoblast layers in the high positive placental samples (Figures 1B and S1).
Histological examination of the H&E-stained sections showed massive chronic intervillositis (MCI) in placenta H_1, and chronic histiocytic intervillositis (CHI) in placenta H_2, consistent with the pathology reported in Table 1. The intervillous spaces showed extensive infiltration of maternal immune cells, perivillous fibrin deposition, and clots with erythrocytes, mononuclear cells, and fibrin deposition (Figures 1B and S1); chorionic villi were collapsed along with evidence of ischemia. The syncytial trophoblast layer of the high positive samples had significantly fewer nuclei and displayed evidence of trophoblast necrosis suggesting a loss of structural integrity within the fetal compartment of the infected placenta. In contrast, none of the negative samples or negative controls displayed massive chronic intervillositis (MCI), chronic histiocytic intervillositis (CHI), or infiltration of maternal immune cells.
Immune cell infiltration into intervillous spaces and invasion of chorionic villi
To determine the localization of maternal immune cell populations in SARS-CoV-2-infected placentas, monocytes/macrophages, T cells, and natural killer cells (NK) in the FFPE tissue slides from high positive and negative control samples were stained for CD163, CD3, CD4, and CD56, respectively. Intravilllous HBC and intervillous maternal macrophages were not infected with SARS-CoV-2 as evidenced by the absence of SARS-CoV-2 RNA and N protein in adjacent placental tissue sections (Figure 1B). In the high positive placental samples H_1 and H_2, immunostaining for CD163 showed massive infiltration of CD163-positive monocytes/macrophages into the intervillous space as well as increased numbers of HBC within the chorionic fetal villi compared with the uninfected negative controls (Figures 2D, 2H, and 2I). In addition, a significantly higher number of CD163-positive HBC was detected in the chorionic plate (CP) and a slight increase in the number of macrophages in the maternal decidua (Figure 2I).
Figure 2.
Immune cell infiltration of CD3+ and CD4+ T cells, CD56+ NK, and CD163+ Hofbauer cells in a high positive placenta from a COVID-infected mother versus a normal healthy control placenta
(A–D) Representative brightfield images of IHC stained FFPE placental tissue sections showing infiltration within maternal blood space (MB), fetal intravillous space (FV), chorionic plate (CP), and maternal decidua (MD) from high positive sample H_2.
(E–H) Representative images of the same regions within the placenta from a non-infected negative control, C_3. Sections were stained for CD3 (A and E), CD4 (B and F), CD56 (C and G), CD163 (D and F). Scale bars = 100 μm.
(I) Quantification of CD3+ T cells, CD4+ helper T cells, CD56+ NK cells, and CD163 + macrophages/HBC detectable by DAB staining across the indicated regions of placental tissue listed along the top X axis. The number of images analyzed for each antibody are listed in Table S1. Values represent the percentage of DAB positive nuclei out of total number of cells based on nuclear detection by a hematoxylin signal. The grey bar in the boxplots represents the median, and the inner, colored boxes represent the interquartile range (25th and 75th percentiles). Statistical analysis was performed using a two-tailed Mann–Whitney U-test and adjusted for multiple testing with the Benjamini–Hochberg FDR method. ∗p < 0.05, ∗∗p < 0.01.
Significantly increased levels of CD3-positive T cells were detected within the MB space, within the fetal chorionic villi (FV), and in the CP of the high positive samples. In contrast, most of the CD3-positive T cells in the negative control sample were restricted to the maternal decidua (MD) (Figures 2A, 2E, 2I, S2A, and S2B). The localization of infiltrating CD4-positive T helper cells followed the same trend (Figures 2B, 2F, and 2I). CD56-positive NK cells were mainly localized to the maternal decidua in both of the high positive samples and negative control samples (Figures 2C, 2G, and 2I). Whereas few NK cells were detected in regions other than the maternal decidua by CD56 staining, there was a reduction in the presence of NK cells in both the MB space and the fetal villi of the infected samples relative to the uninfected controls. In contrast, increased numbers of CD56-positive NK cells were observed in the maternal decidua in the high positive placental samples compared with the negative control (Figures 2C, 2G, and 2I). The increase in infiltrating macrophages and T cells into the fetal regions of the placenta may be an indicator for the observed adverse outcome of pregnancy (Table 1).
Taken together, our results show that the syncytiotrophoblast is the primary target for SARS-CoV-2 infection of the placenta. In response to infection in the high positive placentas, there was a massive migration of maternal monocyte/macrophages and T cells and a retention of CD56-positive NK cells within the maternal decidua, caused either by SARS-CoV-2 infection of the mother, the placenta, or both.
Placental explants and cell clusters are permissive for pseudotype virus
The susceptibility of placental cells to support SARS-CoV-2 replication is not clear. Virus entry requires cell surface ACE2 on the target cell and TMPRSS2 and cathepsins that serve to process the spike protein (Hoffmann et al., 2020). In addition, NRP1 is thought to play a role as a co-receptor for virus entry in some cell-types (Cantuti-Castelvetri et al., 2020; Daly et al., 2020). Spike protein pseudotyped lentiviruses provide an accurate and sensitive means to determine the susceptibility of cells to SARS-CoV-2 entry (Tada et al., 2020). We used this approach to determine the susceptibility of the placental cells to support SARS-CoV-2 entry. Fresh placental isolates from SARS-CoV-2 negative mothers were obtained shortly following delivery. The fetal CP and maternal decidua were then removed and samples containing terminal, intermediate and stem chorionic villi were used to establish placental villi explant cultures (Figure 3C). To allow the passage of small cell clusters, additional placental cell clusters were prepared by enzymatic digestion of the chorionic villi followed by filtration (Figure 3D). The cultures were then infected with a dual green fluorescent protein/nanoluciferase (GFP.Nluc) reporter lentiviral vector pseudotyped by the D614G SARS-CoV-2 spike (S) (Tada et al., 2020). Vesicular stomatitis virus G protein (VSV-G) pseudotyped lentivirus, which infects cells with high infectivity using a ubiquitous cell surface receptor, was used as a control. The results showed that the placental cultures were susceptible to infection by the D614G pseudotype. The infectivity was 2.7-fold lower on average than of the VSV-G pseudotype, indicative of a relatively efficient entry mediated by the spike protein (Figures 3A, 3B, and 3E). The explant cultures were approximately five-fold less infectable as compared with the clusters and single cells, likely owing to the reduced surface accessibility in the explants. The addition of the HIV reverse transcriptase inhibitor nevirapine (NVP) confirmed that the luciferase activity was primarily owing to virus entry and not carry-over from residual virions (Figure 3A).
Figure 3.
Placental explants and cell clusters infected by SARS-CoV-2 S protein pseudotyped lentiviruses
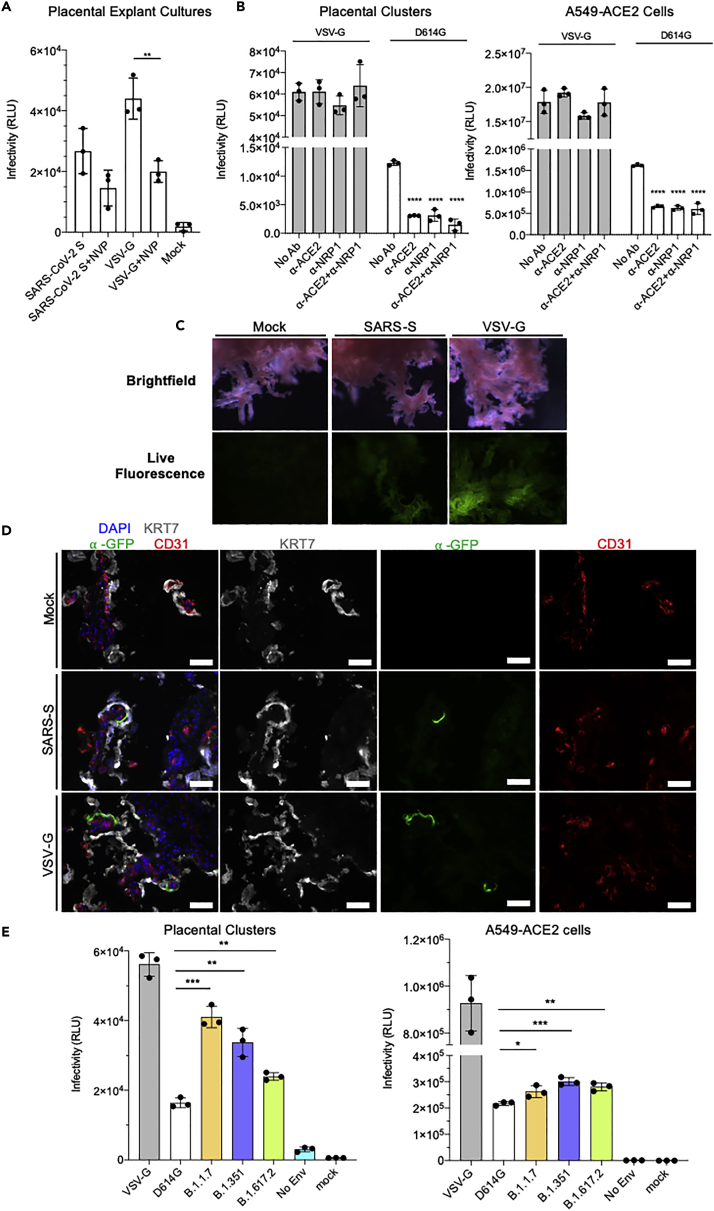
(A) Graph showing relative luminescence units (RLU) from placental explant cultures 72 hpi after infection with lentivirus pseudotyped with SARS-CoV-2 spike or VSV-G protein with or without the addition of reverse transcriptase inhibitor Nevirapine (NVP). Statistical significance was determined by the two-tailed unpaired t-test (∗p ≤ 0.05, ∗∗p ≤ 0.005). Data are presented as mean ± SEM.
(B) Graphs showing RLU from infected isolated primary placental cell clusters (left) and infected A549-ACE2 cells (right) 72 hpi with the addition of blocking antibodies against ACE2 and NRP1. Statistical significance was determined by the two-tailed unpaired t-test (∗∗p ≤ 0.005, ∗∗∗p ≤ 0.001). Data are presented as mean ± SEM.
(C) Brightfield and live fluorescence microscopy images of cultured placental explants, mock-infected (Mock, left column), 72 hpi with either SARS-CoV-2-S pseudotyped lentivirus (SARS-CoV-2-S, center column) or VSV-G pseudotyped lentivirus (VSV-G, right column).
(D) Fluorescence microscopy images on sections from mock-infected (Mock, top row), SARS-CoV-2-S pseudotyped lentivirus-infected (SARS-CoV-2-S, center row), or VSV-G pseudotyped lentivirus-infected (VSV-G, bottom row) explants, stained for the GFP reporter (green) syncytial trophoblast marker, cytokeratin (KRT7, grey), endothelial marker CD31 (red), and DAPI nuclear stain (blue). Scale bars = 500 μm.
(E) Graphs showing RLU from placental cell clusters (left) and A549-ACE2 cells (right) infected with lentivirus pseudotyped by VSV-G (control), wild-type SARS-CoV-2 spike (D614G), Alpha variant (B.1.1.7), Beta variant (B.1.351), and Delta variant (B.1.617.2) spike. Lentivirus without a pseudotyped enveloped protein was included as a control (No Env). Statistical analysis was performed using one-way ANOVA (∗p value < 0.05, ∗∗p value < 0.005, ∗∗∗p value < 0.001). Statistical significance was determined by the two-tailed unpaired t-test (∗∗p ≤ 0.005, ∗∗∗p ≤ 0.001). Data are presented as mean ± SEM.
See also Figure S3.
ACE2 and TMPRSS2 are expressed in the placenta at low levels in the third trimester (Ouyang et al., 2021; Pique-Regi et al., 2020; Singh et al., 2020); however, syncytiotrophoblast expresses the alternative receptor, NRP1 (Arad et al., 2017; Baston-Buest et al., 2011; Cantuti-Castelvetri et al., 2020; Daly et al., 2020). To determine the relative roles of ACE2 and NRP1 in SARS-CoV-2 entry in the placenta, placental cell clusters were treated with anti-ACE2 or anti-NRP1 blocking antibodies before infection with the pseudotyped virus. Both antibodies decreased the infectivity of the D614G spike protein pseudotyped lentivirus (anti-ACE2, 3.5-fold; anti-NRP1, 3.1-fold). The combination of the two antibodies further decreased infectivity by two-fold, resulting in a 7.3-fold reduction compared with untreated controls (Figure 3B, left panel; Figure S3A). For comparison, we tested the lung cell-line A549-ACE2. Whereas the overall infectability of the cells was much higher than the primary placental cells, the antibodies decreased the infection to a similar extent (anti-ACE2, 2.6-fold; anti-NRP1: 2.9-fold) consistent with an earlier report (Tada et al., 2020), and the combination resulted in no further decrease (Figure 3B, right panel). The antibodies did not block the infection by the VSV-G pseudotype demonstrating specificity of the antibodies (Figure 3B, left and right panels; Figure S3A). The results suggest that SARS-CoV-2 viral entry of the placental tissue is dependent on both ACE2 and NRP1, and that infectivity is increased in the presence of both receptors simultaneously.
To determine the cell types targeted for infection, placental explant cultures were infected with the spike protein pseudotypes and then visualized by fluorescence microscopy for the GFP reporter. Live GFP fluorescence could be visualized in the infected explant cultures and was more robust for the VSV-G pseudotype-infected cultures (Figure 3C). To determine the identity of the infected cells, the explant cultures were processed for immunofluorescence by staining with an antibody against the trophoblast marker KRT7/Cytokeratin and the endothelial marker CD31. The results showed GFP-positive cells in small patches of the syncytiotrophoblast on the outer perimeter of the chorionic villi but not in endothelial cells (Figure 3D). The VSV-G pseudotype showed a similar pattern, typically with larger clusters of GFP-positive cells by live cell fluorescence microscopy of the infected explant cultures. No GFP-positive cells were found in mock-infected explant cultures (Figure 3D).
Increased infection of placental cell clusters by viruses with variant spike proteins
SARS-CoV-2 variants containing mutated spike proteins have been shown to spread with increased transmissibility and replicate at higher virus loads in vivo. This raises the possibility that the variants might pose a greater risk to the pregnant mother and the developing fetus. To test the ability of the variants to infect placental cells, we infected placental cultures with lentiviruses pseudotyped by Alpha (B1.1.7), Beta (B1.351), and Delta (B.1.617.2) variants of concern spike proteins (Tada et al., 2020, 2021a, 2021b, 2022). The results showed that the variant pseudotypes infected the placental clusters with increased infectivity relative to the earlier D614G pseudotype. Infection by the Alpha pseudotype was increased 2.7-fold, on average, whereas Beta and Delta were increased 2.2-fold and 2.1-fold, respectively (Figure 3E, left panel; Figure S3B). The variants did not show the same increase upon infection of A549-ACE2 cells demonstrating that the increases were specific to placental cells and not caused by a general increased infectivity of the variants (Figure 3E, right panel).
Primary placental cell clusters are permissive to live SARS-CoV-2
To further determine the susceptibility of placental cells to SARS-CoV-2, we infected primary human placental cell clusters ex vivo. Placentas isolated from healthy term deliveries were digested into cell clusters of approximately 50–100 cells, plated on Matrigel-coated plates, and infected with live SARS-CoV-2 (MOI = 1). The cells were collected 24-h post-infection (hpi) and analyzed by qRT-PCR and immunofluorescence staining.
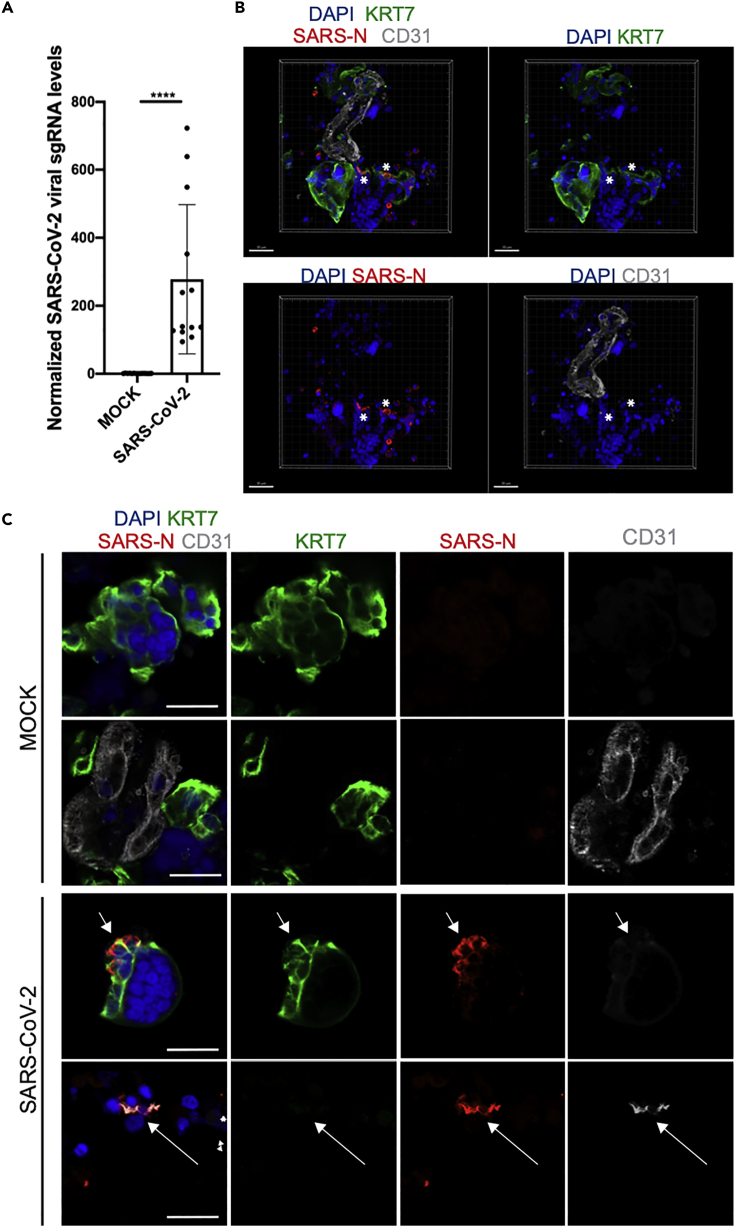
qRT-PCR quantification using primers targeting subgenomic N transcripts showed the presence of SARS-CoV-2 viral RNA in the placental clusters, demonstrating the presence of active virus infection (Figure 4A). To determine the infected cell types in the placental cell clusters, we performed immunostaining for the SARS-CoV-2 N protein and confocal imaging. This analysis showed infected KRT7-positive trophoblast cells (Figures 4B and 4C; Video S1) and, in addition, the presence of a small number of infected CD31+ endothelial cells (Figure 4C).
Figure 4.
Primary human placenta cells can be infected with SARS-CoV-2 ex vivo
(A) qRT-PCR analysis of relative viral N subgenomic RNA expression in primary placental cell clusters infected with SARS-CoV-2 ex vivo (MOI = 1) at 24 hpi and normalized to ACTB levels (mean ± SD; n = 12 from four independent experiments; Student’s t test, ∗∗∗∗p < 0.0001).
(B) Three-dimensional reconstruction of confocal imaging of primary placental cell clusters infected with SARS-CoV-2 ex vivo (MOI = 1) at 24 hpi, stained for trophoblast marker KRT7 (green), SARS-N (red), endothelial marker CD31 (grey), and DAPI (blue). Scale bar = 30 μm.
(C) Confocal imaging of primary placental cell clusters infected with MOCK (top rows) or SARS-CoV-2 (MOI = 1, bottom rows) ex vivo at 24 hpi, stained for trophoblast marker KRT7 (green), SARS-N (red), endothelial marker CD31 (grey), and DAPI (blue). Arrows indicate the presence of SARS-N nucleocapsid protein in trophoblast and endothelial cells. Scale bar = 20 μm.
See also Video S1.
Video shows isolated primary placental clusters infected with live SARSCoV-2 virus (MOI = 1, bottom rows) ex vivo at 24 hpi, that were then stained for trophoblast marker KRT7 (green), SARS-N (red), endothelial marker CD31 (grey), and DAPI (blue). Scale bar = 50 μm.
To determine the transcriptional changes induced by SARS-CoV-2 infection, we analyzed the transcriptome of ex vivo infected placental cell clusters at 24 hpi by RNA-seq. Alignment to the viral genome detected SARS-CoV-2 viral RNA levels in SARS-CoV-2-infected samples (Figure 5A). Principal component analysis (PCA) demonstrated that the infected samples occupy a distinct transcriptional space compared with mock-infected control samples (Figure 5B). Strong similarity among SARS-CoV-2-infected samples with distinct separation from mock-infected samples was observed in the heatmap of sample-to-sample distances (Figure 5C).
Figure 5.
SARS-CoV-2 infection of ex vivo placental explants demonstrates a robust inflammatory response
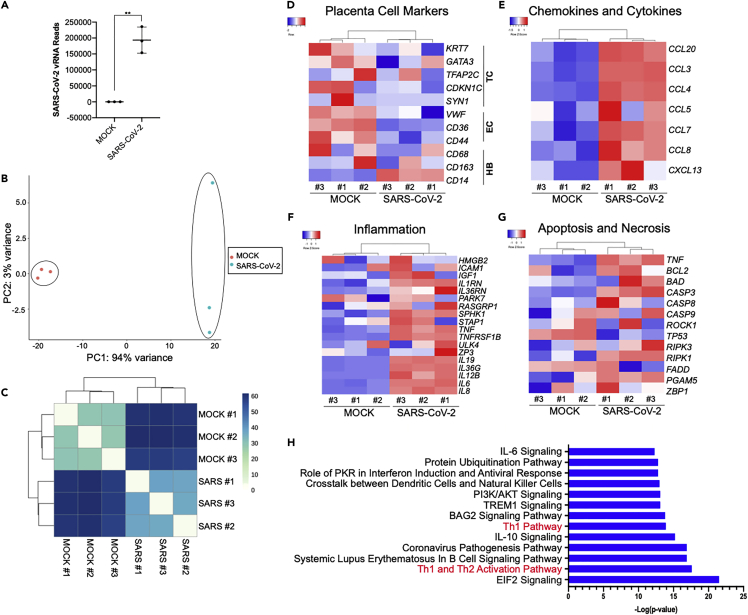
(A) SARS-CoV-2 viral RNA FPKM levels in mock and SARS-CoV-2-infected placental cell clusters. Data are presented as mean ± SD. n = 3 biological replicates, Student’s t test (∗∗p = 0.0012).
(B) PCA analysis of gene expression profiles in mock infected samples (n = 3) and SARS-CoV-2-infected samples (n = 3 biological replicates).
(C) Expression heatmap of sample-to-sample distances for the overall gene expression.
(D–G) Heatmap from RNA-seq data of showing placental cell marker gene expression (D), chemokine and cytokine gene expression (E), inflammatory associated gene expression (F), and cell death marker gene expression (G).
(H) IPA depicting the top canonical biological pathways affected when comparing SARS-CoV-2-infected placenta cell clusters with mock infected controls.
TC, trophoblast cells; EC, endothelial cells; HB, Hofbauer cells.
To determine the effects of SARS-CoV-2 infection on the expression of placental cell-type specific markers genes, differential expression analysis was performed on the RNA-seq data. The analysis showed decreased expression of trophoblast cell markers KRT7, GATA3, CDKN1C and endothelial cell markers VWF, CD36, CD44, and increased the expression of the Hofbauer cell marker CD14 (Figure 5D). In addition, a strong inflammatory response was seen in infected samples with dramatic increases in the expression of chemokine, proinflammatory cytokines (Figure 5E) and other inflammation-related genes (Figure 5F), a finding consistent with inflammatory responses to SARS-CoV-2 infection in other tissues (Han et al., 2021; Tang et al., 2021; Yang et al., 2020, 2021). In addition, a trend for increased expression of cell death-related genes is observed in SARS-CoV-2-infected placental cell clusters, suggesting an acute apoptotic response to infection (Figure 5G).
Ingenuity pathway analysis (IPA) of the genes enriched in SARS-CoV-2-infected samples compared with mock-infected controls highlighted canonical biological pathways associated with viral infection and immune response, including the Th1 and Th2 activation pathways, Coronavirus pathogenesis pathway, and IL-10 and IL-6 signaling (Figure 5H). The pathway with the most prominent alteration in the infected placental cell clusters was EIF2 signaling, a pathway previously identified as the top pathway changed in SARS-CoV-2-infected human pancreatic islets (Tang et al., 2021).
Placentas infected by SARS-CoV-2 at term show induction of inflammation genes
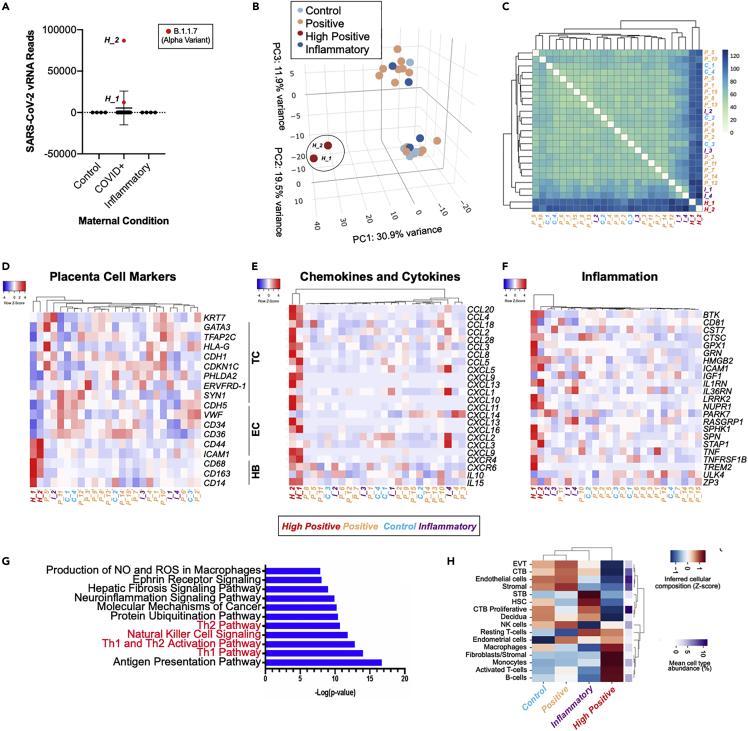
SARS-CoV-2 infection has been shown to induce major transcriptional alterations in infected cells. To determine the host response of the human placenta to SARS-CoV-2 infection, we analyzed the transcriptome of placentas from COVID-19 positive and COVID-19 negative mothers shown in Table 1 and Figure 1A by RNA-seq. Of the COVID-19 positive mothers, two placentas were high positive (high positive samples, H_1, H_2) and 15 were positive (positive samples, P_1-P_15) for SARS-CoV-2 RNA by qRT-PCR (Figure 1A, Table S2). Of the COVID-19 negative mothers, four were healthy controls (control samples, C_1-C_4) and four were placentas with non-COVID related inflammatory pathologies (inflammatory samples, I_1-I_4) (Figure 1A, Table S2). Alignment to the viral genome detected SARS-CoV-2 viral RNA levels in the two high positive samples analyzed (11.1%; H_1 and H_2), and both samples were identified as the Alpha (B.1.1.7) variant (Figure 6A). Furthermore, in both high positive samples, SARS-CoV-2 viral RNA reads can be detected across the whole genome (Figure S4A). PCA demonstrated that the high positive samples H_1 and H_2 occupy a distinct transcriptional space compared with the negative controls (C) and the inflammatory samples (I), as well from the positive samples (P) (Figure 6B). Clustering of an expression heatmap of sample-to-sample distances demonstrated a high degree of similarity between the two high positive samples (H_1 and H_2) and a broad separation from all other samples, whereas two of the inflammatory samples (I_1 and I-4) and two positive samples (P_5 and P_10) displayed a moderate degree of similarity (Figure 6C).
Figure 6.
Transcriptional analysis of term placentas of COVID-19 patients
(A) SARS-CoV-2 viral RNA FPKM levels in placentas from healthy control pregnancies (n = 4), COVID+ mothers (n = 17), and placentas with non-COVID related inflammatory pathologies (n = 4). B.1.1.07 Alpha variant (red) was detected in the two high positive placentas (H_1, H_2).
(B) PCA analysis of gene expression profiles in control samples (C, n = 4), high positive samples (H, n = 2), positive samples (P, n = 15), and inflammatory samples (I, n = 4).
(C) Expression heatmap of sample-to-sample distances for the overall gene expression.
(D–F) Heatmap from RNA-seq data of placentas from control samples (C, n = 4), high positive samples (H, n = 2), positive samples (P, n = 15), and inflammatory samples (I, n = 4) showing placental cell marker gene expression (D), chemokine and cytokine gene expression (E), and inflammatory associated gene expression (F).
(G) Ingenuity pathway analysis (IPA) depicting the top canonical biological pathways affected when comparing high positive SARS-CoV-2 placentas with control placentas. TC, trophoblast cells; EC, endothelial cells; HB, Hofbauer cells.
(H) Heatmap of cellular deconvolution estimating the relative abundance of cell types at the maternal–fetal interface. Mean values per group were the Z-score transformed per row. The original mean relative proportion of each cell type is displayed on the right.
See also Table S2; Figures S4 and S5.
To determine the transcriptional changes in the placenta in response to virus infection, differential expression analysis was performed on the four groups based on their category and number of viral reads detected by RNA-seq (Table S2), using three comparisons: (1) high positive versus control samples, to determine the effects of maternal COVID infection coupled with high viral reads in the placenta; (2) positive versus control samples to analyze the effects of maternal COVID infection without viral reads detected by RNA-seq in the placenta; and (3) inflammatory versus control samples, to determine the effects of other non-COVID inflammatory pathologies on the placenta.
Expression and cluster analysis of placental cell type specific markers showed an increase in the expression of HBC macrophage marker genes CD68, CD163, CD14 and a decrease in the expression of trophoblast markers KRT7, TFAP2C, HLA-G, PHLDA2 and endothelial specific markers CDH5, VWF, CD36, CD44 in the high positive samples compared with all other samples (Figure 6D), similar to what was found in ex vivo infected human placenta cell clusters (Figure 5D). No major differences were observed in expression levels of apoptosis- or necrosis-related genes (Figure S4B), suggesting that the downregulation of placenta cell specific markers is owing to the loss of cell identity rather than cell death. The loss of cellular identity has also been observed in several SARS-CoV-2-infected tissues (Omer et al., 2021; Tang et al., 2021). High positive placental samples show a strong upregulation of chemokines and cytokines that is more robust than that found in inflammatory control samples (Figure 6E) and is consistent with lung autopsy samples from COVID-19 patients (Han et al., 2021). Similar to ex vivo infected placental cell clusters, the high positive placental samples also show an increased expression of inflammatory-related genes (Figures 5F and 6F). Significantly decreased expression of several placental trophoblast and endothelial cell markers (TFAP2C, VWF), increased the expression of Hofbauer cell markers (CD68, CD163), and of chemokine and cytokine genes (CXCL10, CCL20, IL15) and inflammation markers (NURP1, TNFRSF1B) was validated by qRT-PCR of the high positive samples (Figure S5).
IPA of the genes enriched in the high positive placental samples compared with controls from uninfected mothers highlighted canonical biological pathways associated with viral infection and immune response, including antigen presentation, Th1 and Th2 immune cell activation, and natural killer cell signaling (Figure 6G).
The top canonical pathways altered in the inflammatory samples as compared with healthy controls included EIF2 signaling and mitochondrial dysfunction (Figure S4C). Interferon signaling and hypercytokinemia/hyperchemokinemia that are known to play a critical role in the pathogenesis of influenza were the major pathways enriched in the positive samples compared with controls (Figure S4D). Importantly, the IPA analysis of ex vivo infected placental cell clusters (Figure 5H) and of the high positive placental samples (Figure 6G) showed significant changes in Th1 and Th2 signaling pathways as well as pathways involving natural killer cells, consistent with the quantitative analysis of T cells and NK cells by immunohistochemistry (Figure 2I). These changes were specific to the infected placentas as they were not detected in placentas with other inflammatory pathologies (Figures 6G and S4C).
To estimate the abundances of inflammatory cell types within the placental samples, we performed cellular deconvolution by CIBERSORTx (Newman et al., 2019) using a cell-type reference specific to term placentas (Pique-Regi et al., 2020). The analysis showed that control and positive samples displayed a similar pattern of inferred relative cell proportions, whereas the inflammatory and high positive samples were more dissimilar (Figure 6H). Specifically, the high positive samples showed an increase in the relative abundance of T cells, monocytes and macrophages, B cells, and fibroblasts in comparison with control samples. Interestingly, there was also a decrease in the pattern of signatures related to specialized placental cell types such as cytotrophoblasts (CTB), extravillous trophoblast (EVT), endothelial cells, and decidual cells. These data are consistent with the immune cell infiltration we found in the placenta (Figure 2). Furthermore, the data suggest that SARS-CoV-2 infection of placental tissue may have a profound effect on the cellular identity and tissue integrity of the placental tissue. Taken together, our findings identify an inflammatory phenotype in SARS-CoV-2 positive placentas, most pronounced in the high positive samples, that is complemented by a loss of chorionic villi cell identity.
Discussion
The placenta is a vital organ that provides the gestational interface between mother and fetus. Viral infections can result in placental dysfunction, leading to pregnancy complications with increased morbidity and mortality for the mother and fetus (John and Hemberger, 2012; Maltepe et al., 2010; Rossant and Cross, 2001) and can developmentally program the fetus for a chronic disease later in life (Burton et al., 2016). Such effects are notable for Zika virus infection where trophoblasts serve as a route of mother-to-fetus infection (Tan et al., 2019).
Although the large majority of infants born to SARS-CoV-2-infected mothers do not become infected, our results show that SARS-CoV-2 RNA can frequently be detected in the placenta. In placental tissues that were highly infected, the virus was primarily localized to the syncytiotrophoblast and the placenta was infiltrated with maternal immune cells with the transcriptional hallmarks of inflammation. Our findings do not directly show the presence of live virus in the placenta although virus is detected at least in some pregnancies. In support of this, the two highly infected placentas showed high levels of viral RNA; placental cells were infected by the pseudotyped and live SARS-CoV-2; placental samples stained for nucleocapsid protein; and RNA-seq showed high copy numbers of viral RNA across the entire virus genome.
Within a cohort of placental samples from mothers who tested positive for SARS-CoV-2 RNA at delivery, 22 of 52 (42%) had detectable genomic and replicating viral RNA, a frequency higher than that found in previous studies (Debelenko et al., 2021; Hecht et al., 2020; Facchetti et al., 2020; Lu-Culligan et al., 2021). The increased frequency may be because most of the placental samples we studied were from deliveries in which the mother was infected late in pregnancy and the mothers or newborns presented with clinical pathologies.
The two placental samples that we identified with high levels of viral RNA were both infected with the SARS-CoV-2 Alpha variant, one of which resulted in fetal demise and the other was a preterm delivery that required extended care in the NICU. Both mothers presented with symptomatic COVID-19, and tested positive for SARS-CoV-2 RNA and antiviral IgM and IgG antibody, indicating a recent infection (Kubiak et al., 2021; Prabhu et al., 2021). In contrast, only one out of 20 pregnancies of SARS-CoV-2 positive mothers with medium virus content in the placenta (P_10) and one out of 30 pregnancies of SARS-CoV-2 positive mothers with no virus detected in the placenta (N15) resulted in IUFD. This may have been triggered by poorly controlled maternal type 2 diabetes (P_10) or by anencephaly of the fetus (N15). Infants born from COVID-19 positive mothers with placentas high positive or positive for the virus tested negative for SARS-CoV-2 by PCR on nasal swabs, demonstrating the protective role of the placenta in preventing vertical transmission (Cribiù et al., 2021). Despite the protection of the newborn from infection, it remains possible that infection of the placenta could have long-term effects on the infant, suggesting the importance of continued monitoring of the child for developmental defects.
In placentas with high virus content, viral RNA and antigen were detected in a large fraction of the syncytiotrophoblast, the outer cell layer covering the fetal chorionic villi situated at the interphase to MB. In those placentas, virus was not detected in the fetal HBC or stromal and endothelial cells within the chorionic villi or outside the villi. Other studies have also provided evidence for SARS-CoV-2 infection in the placental syncytiotrophoblast (Alamar et al., 2020; Facchetti et al., 2020; Hecht et al., 2020; Hosier et al., 2020; Mulvey et al., 2020; Penfield et al., 2020; Taglauer et al., 2020; Vivanti et al., 2020). However, two reports on a preterm placenta and a placenta from a newborn with vertically transmitted SARS-CoV-2 detected SARS-CoV-2 RNA in HBC and stromal cells inside the villi and in maternal macrophages and epithelial cells at the maternal–fetal interface (Facchetti et al., 2020; Verma et al., 2021). In those cases, it is possible that infection occurred at an earlier gestational stage that allowed for viral spread beyond the syncytiotrophoblast layer.
Infection of the placenta was followed by extensive infiltration of maternal immune cells. In placentas with a high virus load, the maternal space and fetal chorionic villi contained large numbers of macrophages, T lymphocytes, increased numbers of CD3-positive and CD4-positive T cells in the fetal CP, as well as higher numbers of CD56-positive NK cells restricted to the maternal decidua. The source of the NK cells was not clear. They could have resulted from the proliferation of uterine NK cells residing at the maternal–fetal interface or infiltration of NK cells from the maternal circulation. No viral RNA or protein was detected in immune cells after careful and rigorous analysis, suggesting immune cells are not a viral source in the placenta. Other studies have reported on infiltration of maternal immune cells into the placenta in the case of live-borne and stillborn neonates that tested positive for SARS-CoV-2 (Facchetti et al., 2020; Garrido-Pontnou et al., 2021; Kirtsman et al., 2020; Vivanti et al., 2020). Our study, for the first time, reports on extensive immune cell infiltration in placentas with high levels of the Alpha variant virus without vertical transmission of the virus. Consistent with other reports, the two high positive samples showed intervillous infiltration by inflammatory immune cells, chronic histiocytic intervillositis with trophoblast necrosis, intervillous space collapse, and increased perivillous fibrin deposition (Debelenko et al., 2021; Facchetti et al., 2020; Garrido-Pontnou et al., 2021; Linehan et al., 2021; Lu-Culligan et al., 2021; Marton et al., 2021; Verma et al., 2021). The two heavily infected placentas H_1 and H_2 in our study were both infected with the Alpha variant. These placentas were delivered in the early Spring 2021, whereas the other infected placentas were delivered before December 2020, before the widespread presence of the Alpha variant in the US (Alpert et al., 2021).
Transcriptome analysis of placental tissue and of acutely infected placental cell clusters revealed the increased expression chemokines, cytokines, and inflammatory-related genes. Overall loss of placental cell lineages identities was detected in the high positive samples, similar to what has been reported for several SARS-CoV-2-infected tissues (Omer et al., 2021; Tang et al., 2021). Moreover, a previous study reported an increase in immunopathology gene expression in one severe SARS-CoV-2 positive placenta but not in placental tissue with a low virus load (Cribiù et al., 2021). In our study, placental samples with low virus content displayed altered interferon signaling pathways as detected by IPA; however, they showed only minor differences in their gene expression patterns compared with negative control samples. A recent single-cell RNA-seq study by Lu-Culligan et al. on two virus-negative placentas from women with severe COVID-19 revealed robust immune activation in the placenta, including the upregulation of interferon-stimulated genes and of genes indicative of mediating innate-to-adaptive immune cell communication in response to maternal COVID-19 (Lu-Culligan et al., 2021). In contrast, the high virus levels detected in our study of two placentas infected with the Alpha variant, in combination with the observed up-regulation of immune response genes and infiltration of immune cells into maternal and fetal placental compartments support the hypothesis that these are in response to the infection of the mother and the presence of SARS-COV-2 in the placenta.
To determine the ability of placental cells to support SARS-CoV-2 infection, we tested the infectability of ex vivo placental explant and cell cluster culture models both with live SARS-CoV-2 virus and with spike protein pseudotyped lentiviruses. Term placentas express low levels of ACE2 and TMPRSS2 (Ouyang et al., 2021; Pique-Regi et al., 2020; Singh et al., 2020;), as well as the entry cofactor NRP1 (Cantuti-Castelvetri et al., 2020; Daly et al., 2020). NRP1 is expressed at the maternal–fetal interface in decidual cells and syncytiotrophoblast where it acts as a co-receptor for vascular endothelial growth factor (VEGF) on endothelial cells and plays an important role both in pregnancy and the immune system (Arad et al., 2017; Baston-Buest et al., 2011). In addition, SARS-CoV-2 infection has been reported to increase (Lu-Culligan et al., 2021) or decrease placental ACE2 expression, causing dysregulation of the renin-angiotensin system (Verma et al., 2021). We found that live and pseudotyped virus targeted the syncytiotrophoblast and, in rare cases, endothelial cells. The infection was partially blocked by anti-ACE2 and anti-NRP1 antibodies, suggesting the role of both proteins in virus entry into placental cells. The antibody treatment did not completely prevent infection of the placental cells suggesting the possibility of a yet unidentified entry cofactor, although we could not rule-out the possibility that the antibodies were not completely efficient at blocking virus entry. Alpha, Beta, and Delta variant pseudotypes infected the placental cell clusters with increased efficiency, raising concern that the variants may present an increased risk to the developing fetus. Such a concern was further supported by our finding that the two placentas that contained high levels of virus were both infected by the Alpha variant and were associated with a strong inflammatory response and adverse outcome of pregnancy.
This study focused on the effect of maternal infection late in pregnancy on the placenta. It will be important, in addition, to study the effect of infections during the earlier stages of pregnancy where the impact on placental health could be greater. Such a possibility is of particular importance with the high prevalence of the Delta variant that replicates with increased virus loads in vivo (Lopez Bernal et al., 2021; Luo et al., 2021) and has an increased ability to infect placental cells. The increased frequency with which variants of concern appear to infect the placenta should be considered by clinicians advising their patients on vaccination during pregnancy. In addition, the increased expression of SARS-CoV-2 entry cofactors in the first and second trimester may increase the susceptibility of the placenta to infection. An infection at these earlier stages could affect placental development and morphogenesis resulting in an increased risk of placental pathologies and clinical outcomes for mother and fetus. Our study highlights the importance of monitoring the health of COVID-19 mothers and their infants in order to identify possible long-term clinical impacts of SARS-CoV-2 infection during pregnancy.
Limitations of the study
The present study used a limited cohort of women who tested positive for SARS-CoV-2 at delivery, and all placental samples were obtained from Pathology. It will be important to investigate the impact of SARS-CoV-2 maternal infection during the first and second trimester on placental development and on the health of the newborns. With the exception of two placentas from mothers infected with the Alpha variant, whose pregnancies resulted in adverse outcomes for the fetuses and contained high virus levels, all placentas in this study were from mothers infected with the original SARS-CoV-2. Given that viruses with variant spike proteins infected placental cultures at significantly greater levels, it will be important to study the effect of the recent SARS-CoV-2 variants Delta and Omicron on the placenta and its long-term effects on the health of mothers and their newborns.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Cytokeratin | Agilent Dako | Agilent Cat# Z0622; RRID:AB_2650434 |
| anti-rabbit alkaline phosphatase | Jackson ImmunoResearch | Jackson ImmunoResearch Labs Cat# 111-055-003; RRID:AB_2337947 |
| Sheep anti-human Neuropilin-1 | R&D Systems | R and D Systems Cat# AF3870; RRID:AB_884367 |
| anti-ACE2 | Agilent | AdipoGen Cat# AG-20A-0032; RRID:AB_2490114 |
| Rabbit anti-SARS-CoV-2 nucleocapsid (N) antibody | GeneTex | GeneTex Cat# GTX635679; RRID:AB_2888553 |
| Rabbit anti-CD163 | Novus Biologicals | Novus Cat# NBP1-49776-1ml; RRID:AB_10011892 |
| Anti-CD3 | Leica Biosystems | Leica Biosystems Cat# PA0554; RRID:AB_10554454 |
| Anti-CD4 | Leica Biosystems | Leica Biosystems Cat# PA0371; RRID:AB_10554438 |
| Anti-CD56 | Leica Biosystems | Leica Biosystems Cat# PA0192; RRID:AB_10554753 |
| Sheep anti-human CD31/PECAM1 | R&D Systems | R and D Systems Cat# AF806; RRID:AB_355617 |
| Chicken anti-GFP | Abcam | Abcam Cat# ab13970; RRID:AB_300798 |
| Anti-KRT7 | Agilent Dako | Agilent Cat# M701829-2; RRID:AB 2134589 |
| AlexaFluor488-donkey anti-chicken, AlexaFluor594-donkey anti-sheep, AlexaFluor647-donkey anti-rabbit |
Jackson ImmunoResearch | Jackson ImmunoResearch Labs Cat# 703-545-155; RRID:AB_2340375, Jackson ImmunoResearch Labs Cat# 713-585-003; RRID:AB_2340747, Jackson ImmunoResearch Labs Cat# 711-585-152; RRID:AB_2340621 |
| AlexaFluor488-donkey-anti-mouse, AlexaFluor568-donkey-anti-rabbit, AlexaFluor647-donkey-anti-sheep |
ThermoFisher | Thermo Fisher Scientific Cat# A32766; RRID:AB_2762823, Thermo Fisher Scientific Cat# A10042; RRID:AB_2534017, Thermo Fisher Scientific Cat# A21448; RRID:AB_10374882 |
| Bacterial and virus strains | ||
| SARS-CoV-2 isolate USA-WA1/2020 | Center for Disease Control and Prevention, obtained through BEI Resources NIAID, NIH | NR-52281 |
| SARS-CoV-2 S D614G pseudotyped reporter virus SARS-CoV-2 S B1.1.7 pseudotyped reporter virus SARS-CoV-2 S B1.351 pseudotyped reporter virus SARS-CoV-2 S B.1.617.2 pseudotyped reporter virus VSV-G pseudotyped reporter virus |
(Tada et al., 2020, 2021b, 2022) | N/A |
| Biological samples | ||
| FFPE blocks of placental samples | Department of Pathology and Laboratory Medicine, WCM | N/A |
| Fresh placental samples | WCM under IRB exempt approval IRB #20-07022453 |
N/A |
| Critical commercial assays | ||
| RNeasy FFPE kit | Qiagen | Cat# 73604 |
| LunaScript® RT SuperMix Kit | New England Biolabs | Cat# E3010L |
| Luna® Universal qPCR Master Mix | New England Biolabs | Cat# #M3003 |
| EnzMet kit | Nanoprobes, Yaphank NY | Cat# 111-055-003 |
| RBC Lysis Buffer | Biolegend | Cat# 420301 |
| Human umbilical cord dissociation kit | Millitenyi Biotec | Cat# 130-105-737 |
| Nano-Glo® Luciferase Assay System | Promega | Cat# N1120 |
| SuperScript III Platinum SYBR Green One-Step qRT–PCR Kit | Invitrogen | Cat# 11736059 |
| ImmPRESS Reagent kit | Vector Laboratories | Cat# MPX-2402 |
| TruSeq Stranded Total RNA Library Prep Kit | Illumina | Cat# RS-122-2103 |
| Quick-RNA FFPE Miniprep | Zymo Research | Cat# R1008 |
| Experimental models: Cell lines | ||
| Vero E6 | ATCC | #CRL-1586 |
| A549-ACE2 | (Tada et al., 2021b) | N/A |
| Software and algorithms | ||
| GraphPad Prism software | Graphpad.com | N/A |
| IDT OligoAnalyzer | Integrated DNA Technologies | N/A |
| Stardist version 0.7.1 | (Schmidt et al., 2018) | N/A |
| Labelme version 4.5.9 | https://github.com/wkentaro/labelme | N/A |
| Imaris software | Bitplane | N/A |
| cutadapt v1.18 | (Kechin et al., 2017) | N/A |
| STAR v2.5.2b | (Dobin et al., 2013) | N/A |
| HTSeq-count v 0.11.2 | (Anders et al., 2015) | N/A |
| DESeq2 v1.26.0 | (Love et al., 2014) | N/A |
| Heatmapper | (Babicki et al., 2016) | N/A |
| Source code for analysis of IHC data | This paper | N/A |
| Deposited data | ||
| RNA-seq data of patient placentas and ex vivo placental clusters have been deposited in the GEO repository database. | This paper | GEO: GSE181238 |
| IHC data have been deposited in the zenodo repository database. | This paper | https://doi.org/10.5281/zenodo.5182825 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Heidi Stuhlmann (hes2011@med.cornell.edu)
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human placental samples
Placental samples from SARS-CoV-2 positive women and controls were obtained at delivery by the Department of Pathology and Laboratory Medicine at Weill Cornell Medicine (WCM)/NY Presbyterian Hospital. Maternal and gestational age of the patients are provided in Table 1.
Placental explant and cell cluster cultures
Fresh, de-identified placentas from SARS-CoV-2-negative mothers were collected within 0.5–2 h post-delivery from Labor & Delivery at WCM/NYP under an approved IRB exempt protocol (#20-07022453, WCM). Tissue samples were dissected by removing the fetal chorionic plate and any remaining maternal decidual tissue. Primary explant cultures and cell clusters were cultured in DMEM/F12 medium supplemented with 10% FBS and 100 U/mL penicillin, 100 μg/mL streptomycin and 0.25 μg/mL amphotericin B (Massimiani et al., 2019).
Cell lines
Vero E6 (African green monkey [Chlorocebus aethiops] kidney) and A549 (adenocarcinomic human alveolar basal epithelial cell line)-ACE2 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin and 100 μg/mL streptomycin, and maintained at 37°C with 5% CO2.
Methods details
Placental samples
All placental samples from SARS-CoV-2 positive women and controls were obtained at delivery by the Department of Pathology and Laboratory Medicine at Weill Cornell Medicine. All women admitted for delivery were tested by nasal swabs for acute SARS-CoV-2 infection by qRT-PCR, and serologically for previous infection at Weill Cornell Medicine Department of Pathology and Laboratory Medicine. Infants were tested for SARS-CoV-2 at birth and 1 week of age by nasal swabs and RT-PCR. Placental samples were fixed for 48 h in formalin and then processed and embedded into formalin fixed paraffin embedded (FFPE) blocks by the pathology department. FFPE placental samples from 4 healthy women who tested negative for SARS-CoV-2 as controls and an additional 4 samples with inflammation pathologies obtained from SARS-CoV-2 negative patients were also included in the study. Unstained sections and H&E sections of the FFPE blocks were performed at the Weill Cornell Clinical & Translational Science Center (CTSC) core facility. Additional H&E staining was performed by the Weill Cornell Histology core facility.
SARS-CoV-2 detection in RNA from FFPE placental sections by qRT-PCR
Total RNA samples were prepared from FFPE placental tissue sections, followed by DNaseI treatment using a RNeasy FFPE kit (Qiagen). To quantify for replicating virus, subgenomic viral nucleocapsid RNA copy numbers were determined by qRT-PCR. cDNA was generated by using a RT SuperMix Kit (LunaScript®) and SARS-CoV-2 N and cellular glyceraldehyde phosphate dehydrogenase (GAPDH) transcripts were quantified by qRT-PCR with a Luna® Universal qPCR Master Mix (NEB) on a CFX384 Touch Real-Time PCR Detection System (BioRad).
qRT-PCR graphs were generated using GraphPad Prism software.
RNA in situ hybridization to detect SARS-CoV-2 RNA on FFPE placental sections
Probe design
Oligonucleotide probes were synthesized with a 20–25 nucleotides complementarity to SARS-CoV-2 genomic RNA. Sequences were chosen to minimize off-target hybridization to cellular transcripts using NCBI BLAST. IDT OligoAnalyzer (Integrated DNA Technologies) was used to identify probe pairs with similar thermodynamic properties, melting temperature of 45–60°C, GC content of 40%–55%, and low self-complementary. The 3′ end of probes used for proximity ligation signal amplification was designed with a sequence partially complementary to the 61 bp long backbone and partially complementary to the 21 bp insert as described previously (Yang et al., 2020).
Tissue viral RNA staining pretreatment
Sections of FFPE placental samples were deparaffinized by 2 × 5 min treatments with 100% xylene at room temperature. The slides were rinsed twice for 1 min each in 100% ethanol at room temperature and then air dried. Endogenous peroxidase activity was quenched by treating for 10 min with 0.3% hydrogen peroxide at room temperature followed by washing with DEPC treated water. The samples were incubated 15 min at 95–100 °C in antigen retrieval solution (ACDBio), rinsed in DEPC-treated water, dehydrated for 3 min in 100% ethanol at room temperature and air dried. Tissue sections were permeabilized for 30 min at 40°C using RNAscope protease plus solution (ACDBio) and rinsed in DEPC-treated water.
SARS-CoV-2 RNA detection by probes using proximity ligation
Hybridization was performed overnight at 40°C in hybridization buffer [2× SSC, 20% formamide (Thermo Fischer Scientific, Waltham, MA, USA), 2.5% (vol/vol) polyvinylsulfonic acid, 20 mM ribonucleoside vanadyl complex (New England Biolabs, Ipswich, MA, USA), 40 U/mL RNasin (Promega, Madison, WI, USA), 0.1% (vol/vol) Tween 20 (Sigma Aldrich), 100 μg/mL salmon sperm DNA (Thermo Fisher Scientific), 100 μg/mL yeast RNA (Thermo Fisher Scientific)]. DNA probes were dissolved in DEPC-treated water and added at a final concentration of 100 nM (Integrated DNA Technologies, Coralville, IA, ISA). Samples were washed briefly and incubated in 2× SSC, 20% formamide, 40 U/mL RNasin at 40 °C and then washed four times (5 min each) in PBS, 0.1% (vol/vol) Tween 20, and 4 U/mL RNasin (Promega, Madison, WI, USA). The slides were then incubated at 37°C with 100 nM insert/backbone oligonucleotides in PBS containing 1× SSC, 0.1% (vol/vol) Tween 20, 100 μg/mL salmon sperm DNA (Thermo Fisher Scientific), 100 μg/mL yeast RNA (Thermo Fisher Scientific), 40 U/mL RNasin. After four washes, tissues were incubated at 37 °C with 0.1 U/μL T4 DNA ligase (New England Biolabs, Ipswich, MA, USA) in 50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT, 250 μg/mL BSA, 0.05% Tween 20, 40 U/mL RNasin, followed by incubation with 0.1 U/μL phi29 DNA polymerase in 50 mM Tris-HCl, 10 mM MgCl2, 10 mM (NH4)2SO4, 250uM dNTPs, 1 mM DTT, 0.05% Tween 20, 40 U/mL RNasin pH 7.5 at 30 °C. The slides were washed and endogenous biotin was blocked for 1 h using Avidin/Biotin blocking kit (Vector laboratories, Burlingame, CA, USA). Rolling cycle amplicons were identified using a biotin labeled DNA probe at a concentration of 5 nM at 37 °C in PBS containing 1× SSC, 0.1% Tween 20, 100 μg/mL salmon sperm DNA, 100 μg/mL yeast RNA. After washing, samples were incubated for 60 min with 1:100 diluted streptavidin-HRP (Thermo Fisher Scientific) in PBS at room temperature followed by washing. Labeling was accomplished using EnzMet kit (Nanoprobes). The slides were further labeled overnight with rabbit anti-cytokeratin 1:250 (Dako Z0622) at 4°C. After washing, the samples were incubated with anti-rabbit alkaline phosphatase antibody (1:1000, Jackson Immunoresearch, Baltimore, PA, USA) and stained using Fast Red substrate kit (Abcam, Cambridge, MA, USA). The samples were counterstained with Hematoxylin (Vector laboratories, Burlingame, CA, USA) and mounted in Permount (Fischer Scientific).
Placental explant and cell cluster cultures
Fresh, de-identified placentas from SARS-CoV-2-negative mothers were collected within 0.5–2 h post-delivery from Labor & Delivery at WCM/NYP under an approved IRB exempt protocol (#20-07022453, Weill Cornell Medicine.) Tissue samples were dissected by removing the fetal chorionic plate and any remaining maternal decidual tissue. Primary explant cultures (1 cm × 1 cm × 2 cm) containing terminal, intermediate and stem chorionic villi were further dissected, washed in ice-cold 1× PBS to remove maternal blood, and plated into 48-well plastic dishes in DMEM/F12 medium supplemented with 10% FBS and 100 U/mL penicillin, 100 μg/mL streptomycin and 0.25 μg/mL amphotericin B, as previously described (Massimiani et al., 2019).
Placental cell clusters were prepared from fresh chorionic villi tissue samples by mincing with scissors and 10 blade scalpels and then digested with 0.2 mg/mL collagenase/0.8U/mL dispase (Roche) and recombinant DNAse I (Sigma) in MACS buffer (PBS/2mM EDTA, 0.5% bovine serum albumin (BSA)) at 42°C with agitation by pipetting with a 5 mL stripette. The digested tissue was filtered through 100 μm filters (Corning), and red blood cells (RBC) were removed using RBC Lysis Buffer (Biolegend). The clusters were washed in MACS buffer and viability was determined by Trypan Blue (GIBCO) exclusion. The cultures were plated onto Matrigel-coated 96-well dishes and μ-slide 8-well chamber slides (ibidi GmbH, Germany) at confluent density in DMEM/F12 supplemented with 10% FBS and penicillin/streptomycin/fungizone and then incubated for 24 h at 37 °C in 5% CO2 prior to infection with pseudotyped virus.
For infection with live SARS-CoV-2, sections of fresh chorionic villi were minced with sterile scalpels, digested for 7 min in Accutase (Innovative Cell Technologies) or isolated using a human umbilical cord dissociation kit (Millitenyi Biotec) and then filtrated through a 100 μm cell strainer (Falcon) to obtain cell clusters of ∼50–100 cells. Red blood cells were lysed using RBC Lysis Buffer (Biolegend), washed with PBS-0.5% BSA, and resuspended in culture medium (DMEM, 10%FBS, 1% Pen-Strep-Glutamax). Cell viability was determined with Trypan blue exclusion (Gibco). The cell clusters were plated on Matrigel-coated plates (Corning, hESC-qualified) at 4 × 105 per well in 24-well plates or 3 × 104 per well in glass-like polymer bottom 96-well plates (CellVis).
SARS-CoV-2 propagation and titration
SARS-CoV-2 isolate USA-WA1/2020 (NR-52281) was provided by the Center for Disease Control and Prevention (CDC) and obtained through BEI Resources, NIAID, NIH and propagated on Vero E6 cells in DMEM supplemented with 2% FBS, 4.5 g/L D-glucose, 4 mM L-glutamine, 10 mM non-essential amino acids, 1 mM sodium pyruvate and 10 mM HEPES using a passage-2 stock of virus. Three days post-infection, the culture supernatant was harvested and the virus was concentrated by filtering for 20 min through a 100 k-Da Amicon Ultra 15 centrifugal filter (Millipore Sigma) at ∼4000 rpm. The virus was resuspended and infectious titers were determined by plaque assay in Vero E6 cells in Minimum Essential Media supplemented with 2% FBS, 4 mM L-glutamine, 0.2% BSA, 10 mM HEPES and 0.12% NaHCO3 and 0.7% agar. All MOI values were based on titer determined from plaque assays on Vero E6 cells. Work with live SARS-CoV-2 was performed in the CDC/USDA-approved biosafety level-3 (BSL-3) facility of the Icahn School of Medicine at Mount Sinai in accordance with institutional biosafety requirements.
Infection of ex vivo placental cultures
Infection of explants and placental clusters with pseudotyped lentivirus
Lentiviruses encoding dual GFP/nanoluciferase reporter genes and pseudotyped by SARS-CoV-2 spike (S) protein D614G, B1.1.7 (Alpha variant), B1.351 (Beta variant), B.1.617.2 (Delta variant), or by VSV-G were prepared as previously described (Tada et al., 2020, 2021b, 2022). The viruses were concentrated 10-fold by ultracentrifugation and titers were quantified by reverse transcriptase assay. Placental explant cultures and cell clusters were infected with 10 μL SARS-CoV-2 S or VSV-G pseudotyped lentivirus (Tada et al., 2020) and lysed at 72 h post infection (hpi). Luciferase activity was measured using a Nano-Glo Assay Kit (Promega) and read on an Envision microplate luminometer (Perkin Elmer). For antibody blocking experiments, placental cell clusters were pretreated for 30 min with anti-NRP1 mAb (R&D Systems) or anti-ACE2 mAb (Agilent) prior to infection.
Infection of placental clusters with live SARS-CoV-2
Placental cell clusters were infected with SARS-CoV-2 at day-1 at a MOI = 1, or mock-infected as described (Yang et al., 2020). At 24 hpi, cells were washed with PBS and lysed in in TRIzol (Invitrogen) for RNA analysis or fixed for 60 min at room temperature in 4% formaldehyde for immunofluorescence staining. All work involving live SARS-CoV-2 was performed in the CDC/USDA-approved BSL-3 facility of the Icahn School of Medicine at Mount Sinai in accordance with institutional biosafety requirements.
qRT-PCR for viral load of SARS-CoV-2 infected placental clusters
Total RNA was extracted using Trizol followed by ezDNAse treatment (Thermo Fisher Scientific) per manufacturer’s instructions. To quantify viral replication, measured by the accumulation of subgenomic N transcripts, one-step quantitative real-time PCR was performed using a Super-Script III Platinum SYBR Green One-Step qRT–PCR Kit (Invitrogen) with primers specific for TRS and beta-actin (ACTB) as an internal reference, as previously described (Yang et al., 2020). Reactions were analyzed on a QuantStudio 6 Flex Real Time PCR Instrument (Applied Biosystems). The delta-delta-cycle threshold (ΔΔCT) was determined relative to ACTB and mock-infected samples. Graphs were generated using GraphPad Prism software.
Immunohistochemistry of FFPE placental sections
Immunohistochemistry (IHC) of FFPE slides was done using an ImmPRESS Reagent kit (Vector Laboratories). The slides were dewaxed for 45 min at 55°C and then rehydrated using xylenes followed by a standard ethanol gradient. For antigen retrieval, the slides were treated in a steamer for 35 min at pH 6.1 in sodium citrate buffer, blocked for 1 h at room temperature in 2.5% horse serum (Vector laboratories) and then incubated overnight at 4°C in a humid chamber with primary antibodies (SARS-CoV-2-N, GeneTex, 1:100; CD163, Novus Biologicals, 1:250) diluted in 1% BSA/0.1% Triton X- PBS (PBST). The slides were treated with 3% hydrogen peroxide (Sigma H1009) at room temperature, washed 3 times with 0.1% PBST and then incubated for 1 h at room temperature with ImmPRESS anti-rabbit peroxidase conjugated antibody (Vector Laboratories). Unbound antibody was removed by rinsing with 0.1% PBST with final wash in PBS. The signals were developed using freshly prepared DAB substrate (Vector Labs) and counterstained with Hematoxylin (RICCA Chemical Company). The stained slides were dehydrated using an increasing ethanol gradient, treated with xylenes, and mounted with Permount solution (Thermo Fisher Scientific). For cell quantification analysis, additional IHC staining was performed by the Weill Cornell Medicine Center for Translational Pathology using primary antibodies against CD3 (Leica), CD4 (Leica), CD56 (Leica), and CD163 (Leica). Brightfield images were acquired using a Zeiss microscope (Carl Zeiss, Germany).
Analysis of IHC data
IHC images were decomposed into a hematoxylin and a diaminobenzidine intensity channel using a preset color space for the stains from the scikit-image (Van Der Walt et al., 2014) (version 0.18.2). The hematoxylin channel was normalized to the unit space after capping the intensity to the 3rd and 98th percentiles and used for segmentation with Stardist version 0.7.1 (Schmidt et al., 2018) with the 2D_versatile_fluo pretrained model. The intensity of each nucleus in both hematoxylin and diaminobenzidine channels was calculated by a mean reduction for each nucleus. As the IHC signal does not linearly reflect the stoichiometry of the chemical reaction, the signal was discretized into positive and negative fractions per image. Specifically, intensity values across images were normalized by subtracting the location and dividing by the standard deviation of each whole image (Z-score), and nuclei were declared positive for diaminobenzidine signal if values for the markers CD3, CD4, CD56 and CD163 were above zero (Figures S2C–S2F). For CD163 staining in maternal blood space, analysis of the blood space around fetal villi was restricted by creating polygonal masks using the labelme program (https://github.com/wkentaro/labelme, version 4.5.9). Statistical testing was performed between control and COVID-19 placenta donors with a two-sided Mann-Whitney U test, and adjusted for multiple comparisons with the Benjamini–Hochberg FDR method using pingouin, version 0.3.12 (Vallat, 2018).
Immunofluorescence staining for infected placental explants and cell clusters
Pseudovirus infected explant cultures at 72 hpi were drop-fixed overnight at 4°C in 4% paraformaldehyde in PBS containing Ca2+Mg2+. The fixed explants were dehydrated in 30% sucrose in PBS overnight at 4°C and then embedded in optimal cutting temperature compound (OCT) on dry ice and the frozen blocks were sectioned on a cryomicrotome at 10-μm thickness. The sections were blocked for 1 h in 10% donkey serum (Jackson ImmunoResearch labs) in 0.1% PBST. Primary antibodies ((rabbit anti-Cytokeratin, Dako, 1:1000; sheep anti-human CD31, R&D Systems, 1:500); chicken anti-GFP, Abcam, 1:1000) were diluted in 10% donkey serum-0.1% PBST and incubated overnight at 4°C followed by incubation with secondary antibodies (AlexaFlour647-donkey anti-rabbit, AlexaFlour594-donkey anti-sheep, and AlexaFlour488-donkey anti-chicken, Jackson ImmunoResearch labs, 1:500). The clusters were then stained using 4′,6-diamidino-2-phenylindole (DAPI). Slides were mounted with coverslips using ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific). Slides were imaged on a Zeiss fluorescence microscope and analyzed using ImageJ software.
SARS-CoV-2 infected cells were fixed in paraformaldehyde and blocked in 5% normal donkey serum in PBS-0.05% Triton X-0.01% Saponin (PBS-TSP). Primary antibodies (SARS-CoV-2-N, GeneTex, 1:200; KRT7, Agilent Dako, 1:400; CD31/PECAM1, R&D Systems, 1:1000) were incubated overnight at 4°C in blocking buffer, followed by incubation with secondary antibodies (AlexaFluor488-donkey-anti-mouse, AlexaFluor594-donkey-anti-rabbit, AlexaFluor647-donkey-anti-sheep, ThermoFisher, 1:500) in PBS-TSP, and counterstaining with DAPI (Thermo Fisher Scientific). Images were acquired using a Zeiss LSM 800 Confocal microscope and processed using Imaris software (Bitplane).
RNA-sequencing and RNA-seq data analysis
Total RNA extracted from placental cell clusters and FFPE sections was used for RNA-seq analysis. RNA from ex vivo infected placental cell clusters was further purified using RNAClean XP beads (Beckman Coutler). RNA was used to prepare libraries for RNA-seq with the TruSeq Stranded Total RNA Library Prep Kit (Illumina). The libraries were sequenced with pair-end 51 bps on an Illumina NovaSeq6000 sequencer. Sequencing reads were cleaned by trimming adapter sequences and low-quality bases using cutadapt v1.18 (Kechin et al., 2017). Sequences were aligned to the human reference genome (GRCh37) combined with the SARS-CoV-2 genome (NC_045512.2) using STAR v2.5.2b (Dobin et al., 2013). Raw read counts per gene were extracted using HTSeq-count v 0.11.2 (Anders et al., 2015).
A large fraction of viral reads in the ex vivo placental cell clusters infected with SARS-CoV-2 resulted in human gene counts that were much lower than those of the mock samples. For comparison of human genes expression in the ex vivo samples, the raw gene counts in the mock and SARS-CoV-2 infected ex vivo samples were down-sampled such that the human gene counts are comparable amongst these samples.
Differential expression analysis was performed using DESeq2 v1.26.0 (Love et al., 2014). Regularized logarithm transformation to the counts data was applied using the rlog function. Principle component analysis (PCA) was performed with the transformed human gene counts using the plotPCA function. Sample-to-sample distances were determined by applying the R dist function to the transpose of the transformed human gene counts, and hierarchical clustering was done based on the distance using the R hc function. Heatmap plots were performed based on the transformed counts data, and average linkage clustering analysis with Euclidean distance measurement method were generated using Heatmapper (Babicki et al., 2016).
10× Single-cell RNA sequencing fastq files were downloaded from DBGAP (accession PRJNA560990) and aligned using CellRanger software. The aligned counts-data were then processed using Seurat (Butler et al., 2018) and cells were annotated by cell type specified by the markers in the published data source (Pique-Regi et al., 2020). Deconvolution software Bisque (Jew et al., 2020) was used with the reference single cell data sampled to 100 single cell transcriptomes per cell type and the bulk RNAseq of the different cohorts was then deconvoluted using the single cell data as reference. Cell type proportions were inferred and these proportions clustered and plotted as heatmaps.
qRT-PCR analysis was performed on total RNA samples using Quick-RNA FFPE Miniprep (ZYMO Research) according to the manufacturer instructions. cDNA was prepared using LunaScript RT SuperMix Kit (NEB) and quantitative real-time PCR was performed using Luna Universal qPCR Master Mix (NEB) on a cfx384 qPCR instrument (BioRad) with gene specific primers. Delta-delta-cycle threshold (ΔΔCT) was determined relative to the GAPDH RNA internal reference and non-infected placenta samples. Every sample was analyzed in n = 3 technical replicates.
Quantification and statistical analysis
All experiments were in technical triplicates unless otherwise indicated. Statistical analysis was performed using a one-way ANOVA and adjusted for by multiple comparisons test using the Benjamini-Hochberg FDR method (Figure 1), a two-tailed Mann-Whitney U-test and adjusted for multiple testing with the Benjamini-Hochberg FDR method (Figure 2), and a one-way ANOVA and Student’s t test (Figures 3, 4, and 5). ∗ = p value < 0.05, ∗∗ = p value < 0.005, ∗∗∗ = p value < 0.001, ∗∗∗∗ = p value < 0.0001. Data are represented as mean ± SEM.
Acknowledgments
We thank the patients, their families, and healthcare workers fighting the COVID-19 pandemic. We would also like to acknowledge Michael D. Glendenning for his technical assistance with the IHC, the Weill Cornell Medicine Genomics Core, the Weill Cornell Medicine Histology Core, and the Weill Cornell Medicine Center for Translational Pathology. This work was supported by a Weill Cornell Medicine COVID-19 Research Grant (H.S., R.E.S., R.N.B.), the NCI (R01CA234614) and NIAID (2R01AI107301) and NIDDK (R01DK121072) to Department of Medicine, Weill Cornell Medicine (R.E.S.), NIDDK (R01DK119667, R01DK119667-02S1) to S.C. R.E.S. and S.C. are supported as an Irma Hirschl Trust Research Award Scholar. L.B.A. was supported in part by NYSTEM Training grant. L.A.L. is supported by an F32 post-doctoral fellowship from the National Institutes of Health (1F32HD096810-01A1) and a Weill Cornell Medicine Research Assistance for Primary Parents Award. A.F.R. is supported by an NCI T32CA203702 grant. N.R.L was supported by grants from the NIH (DA046100, AI122390, and AI120898). T.T. was supported by the Vilcek/Goldfarb Fellowship Endowment Fund.
Author contributions
Conceptualization and methodology: L.B.A., H.S., L.A.L., R.E.S., Y.B., S.C., R.N.B., Y.J.Y., N.R.L., and B.R.t.; investigation: L.B.A., L.A.L., Y.B., T.T., L.C., A.F.R., S.U., B.C.L., V.C., C.G.J., and H.S.; writing – original draft: L.B.A., H.S., L.A.L., S.C., R.E.S., N.R.L., R.N.B., M.P., and Y.J.Y.; writing – review and editing: L.B.A., H.S., L.A.L., S.C., R.E.S., T.T., N.R.L., A.F.R., Y.J.Y., R.N.B., and B.R.t.O.; funding acquisition: H.S., R.E.S., and S.C.
Declaration of interests
O.E. is a scientific advisor and equity holder in Freenome, Owkin, Volastra Therapeutics, and One Three Biotech. R.E.S. is on the scientific advisory board of Miromatrix Inc and is a consultant and speaker for Alnylam Inc.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104223.
Contributor Information
Shuibing Chen, Email: shc2034@med.cornell.edu.
Robert E. Schwartz, Email: res2025@med.cornell.edu.
Heidi Stuhlmann, Email: hes2011@med.cornell.edu.
Supplemental information
Data and code availability
-
•
RNA-seq data of patient placentas and ex vivo placental clusters were deposited in the GEO repository database and will be available as of the date of publication. The accession number is listed in the key resources table.
-
•
Immunohistochemistry (IHC) analysis data were deposited in the zenodo repository database and will be available as of the date of publication. The accession number is listed in the key resources table.
-
•
The source code for the analysis of IHC data will be available as of the date of publication. The link is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alamar I., Abu-Arja M.H., Heyman T., Roberts D.J., Desai N., Narula P., Dygulska B. A possible case of vertical transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a newborn with positive placental in situ hybridization of SARS-CoV-2 RNA. J. Pediatr. Infect. Dis. Soc. 2020;9:636–639. doi: 10.1093/jpids/piaa109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert T., Brito A.F., Lasek-Nesselquist E., Rothman J., Valesano A.L., Mackay M.J., Petrone M.E., Breban M.I., Watkins A.E., Vogels C.B.F., et al. Early introductions and transmission of SARS-CoV-2 variant B.1.1.7 in the United States. Cell. 2021;184:2595–2604.e13. doi: 10.1016/j.cell.2021.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arad A., Nammouz S., Nov Y., Ohel G., Bejar J., Vadasz Z. The expression of neuropilin-1 in human placentas from normal and preeclamptic pregnancies. Int. J. Gynecol. Pathol. 2017;36:42–49. doi: 10.1097/PGP.0000000000000283. [DOI] [PubMed] [Google Scholar]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baergen R.N., Heller D.S. Placental pathology in covid-19 positive mothers: preliminary findings. Pediatr. Dev. Pathol. 2020;23:177–180. doi: 10.1177/1093526620925569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baston-Buest D.M., Porn A.C., Schanz A., Kruessel J.S., Janni W., Hess A.P. Expression of the vascular endothelial growth factor receptor neuropilin-1 at the human embryo-maternal interface. Eur. J. Obstet. Gynecol. Reprod. Biol. 2011;154:151–156. doi: 10.1016/j.ejogrb.2010.10.018. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton G.J., Fowden A.L., Thornburg K.L. Placental origins of chronic disease. Physiol. Rev. 2016;96:1509–1565. doi: 10.1152/physrev.00029.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., Van Der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribiù F.M., Erra R., Pugni L., Rubio-Perez C., Alonso L., Simonetti S., Croci G.A., Serna G., Ronchi A., Pietrasanta C., et al. Severe SARS-CoV-2 placenta infection can impact neonatal outcome in the absence of vertical transmission. J. Clin. Invest. 2021;131:e145427. doi: 10.1172/JCI145427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debelenko L., Katsyv I., Chong A.M., Peruyero L., Szabolcs M., Uhlemann A.C. Trophoblast damage with acute and chronic intervillositis: disruption of the placental barrier by severe acute respiratory syndrome coronavirus 2. Hum. Pathol. 2021;109:69–79. doi: 10.1016/j.humpath.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Gatta A.N., Rizzo R., Pilu G., Simonazzi G. Coronavirus disease 2019 during pregnancy: a systematic review of reported cases. Am. J. Obstet. Gynecol. 2020;223:36–41. doi: 10.1016/j.ajog.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlow A.G., Li J.Z., Collier A.Y., Atyeo C., James K.E., Boatin A.A., Gray K.J., Bordt E.A., Shook L.L., Yonker L.M., et al. Assessment of maternal and neonatal SARS-CoV-2 viral load, transplacental antibody transfer, and placental pathology in pregnancies during the COVID-19 pandemic. JAMA Netw. Open. 2020;3:e2030455. doi: 10.1001/jamanetworkopen.2020.30455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchetti F., Bugatti M., Drera E., Tripodo C., Sartori E., Cancila V., Papaccio M., Castellani R., Casola S., Boniotti M.B., et al. SARS-CoV2 vertical transmission with adverse effects on the newborn revealed through integrated immunohistochemical, electron microscopy and molecular analyses of Placenta. EBioMedicine. 2020;59:102951. doi: 10.1016/j.ebiom.2020.102951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Pontnou M., Navarro A., Camacho J., Crispi F., Alguacil-Guillén M., Moreno-Baró A., Hernandez-Losa J., Sesé M., Ramón Y.C.S., Garcia Ruíz I., et al. Diffuse trophoblast damage is the hallmark of SARS-CoV-2-associated fetal demise. Mod. Pathol. 2021;34:1704–1709. doi: 10.1038/s41379-021-00827-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J.L., Quade B., Deshpande V., Mino-Kenudson M., Ting D.T., Desai N., Dygulska B., Heyman T., Salafia C., Shen D., et al. SARS-CoV-2 can infect the placenta and is not associated with specific placental histopathology: a series of 19 placentas from COVID-19-positive mothers. Mod. Pathol. 2020;33:2092–2103. doi: 10.1038/s41379-020-0639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosier H., Farhadian S.F., Morotti R.A., Deshmukh U., Lu-Culligan A., Campbell K.H., Yasumoto Y., Vogels C.B., Casanovas-Massana A., Vijayakumar P., et al. SARS-CoV-2 infection of the placenta. J. Clin. Invest. 2020;130:4947–4953. doi: 10.1172/JCI139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jew B., Alvarez M., Rahmani E., Miao Z., Ko A., Garske K.M., Sul J.H., Pietiläinen K.H., Pajukanta P., Halperin E. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 2020;11:1971. doi: 10.1038/s41467-020-15816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John R., Hemberger M. A placenta for life. Reprod. Biomed. Online. 2012;25:5–11. doi: 10.1016/j.rbmo.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Kechin A., Boyarskikh U., Kel A., Filipenko M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 2017;24:1138–1143. doi: 10.1089/cmb.2017.0096. [DOI] [PubMed] [Google Scholar]
- Kimberlin D.W., Stagno S. Can SARS-CoV-2 infection Be acquired in utero?: more definitive evidence is needed. JAMA. 2020;23:1788–1789. doi: 10.1001/jama.2020.4868. [DOI] [PubMed] [Google Scholar]
- Kirtsman M., Diambomba Y., Poutanen S.M., Malinowski A.K., Vlachodimitropoulou E., Parks W.T., Erdman L., Morris S.K., Shah P.S. Probable congenital SARS-CoV-2 infection in a neonate born to a woman with active SARS-CoV-2 infection. Cmaj. 2020;192 doi: 10.1503/cmaj.200821. E647–e650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiak J.M., Murphy E.A., Yee J., Cagino K.A., Friedlander R.L., Glynn S.M., Matthews K.C., Jurkiewicz M., Sukhu A.C., Zhao Z., et al. Severe acute respiratory syndrome coronavirus 2 serology levels in pregnant women and their neonates. Am. J. Obstet. Gynecol. 2021;225:73.e1–73.e7. doi: 10.1016/j.ajog.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Chen L., Zhang J., Xiong C., Li X. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PLoS One. 2020;15:e0230295. doi: 10.1371/journal.pone.0230295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan L., O'donoghue K., Dineen S., White J., Higgins J.R., Fitzgerald B. SARS-CoV-2 placentitis: an uncommon complication of maternal COVID-19. Placenta. 2021;104:261–266. doi: 10.1016/j.placenta.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Bernal J., Gower C., Andrews N. Effectiveness of covid-19 vaccines against the B.1.617.2 (delta) variant. Reply. N. Engl. J. Med. 2021;385:e92. doi: 10.1056/NEJMc2113090. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu-Culligan A., Chavan A.R., Vijayakumar P., Irshaid L., Courchaine E.M., Milano K.M., Tang Z., Pope S.D., Song E., Vogels C.B.F., et al. Maternal respiratory SARS-CoV-2 infection in pregnancy is associated with a robust inflammatory response at the maternal-fetal interface. Med (N Y) 2021;2:591–610.e10. doi: 10.1016/j.medj.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C.H., Morris C.P., Sachithanandham J., Amadi A., Gaston D.C., Li M., Swanson N.J., Schwartz M., Klein E.Y., Pekosz A., Mostafa H.H. Infection with the SARS-CoV-2 delta variant is associated with higher recovery of infectious virus compared to the Alpha variant in both unvaccinated and vaccinated individuals. Clin. Infect. Dis. 2021;18:ciab986. doi: 10.1093/cid/ciab986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltepe E., Bakardjiev A.I., Fisher S.J. The placenta: transcriptional, epigenetic, and physiological integration during development. J. Clin. Invest. 2010;120:1016–1025. doi: 10.1172/JCI41211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín Gabriel M.A., Reyne Vergeli M., Caserío Carbonero S., Sole L., Carrizosa Molina T., Rivero Calle I., Cuadrado Pérez I., Álvarez Fernández B., Forti Buratti A., Fernández-Cañadas Morillo A. Maternal, perinatal and neonatal outcomes with COVID-19: a multicenter study of 242 pregnancies and their 248 infant newborns during their first month of life. Pediatr. Infect. Dis. J. 2020;39:e393–e397. doi: 10.1097/INF.0000000000002902. [DOI] [PubMed] [Google Scholar]
- Marton T., Hargitai B., Hunter K., Pugh M., Murray P. Massive perivillous fibrin deposition and chronic histiocytic intervillositis a complication of SARS-CoV-2 infection. Pediatr. Dev. Pathol. 2021;24:450–454. doi: 10.1177/10935266211020723. [DOI] [PubMed] [Google Scholar]
- Massimiani M., Lacko L.A., Burke Swanson C.S., Salvi S., Argueta L.B., Moresi S., Ferrazzani S., Gelber S.E., Baergen R.N., Toschi N., et al. Increased circulating levels of Epidermal Growth Factor-like Domain 7 in pregnant women affected by preeclampsia. Transl. Res. 2019;207:19–29. doi: 10.1016/j.trsl.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morotti D., Cadamuro M., Rigoli E., Sonzogni A., Gianatti A., Parolin C., Patanè L., Schwartz D.A. Molecular pathology analysis of SARS-CoV-2 in syncytiotrophoblast and hofbauer cells in placenta from a pregnant woman and fetus with COVID-19. Pathogens. 2021;10:479. doi: 10.3390/pathogens10040479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourad M., Jacob T., Sadovsky E., Bejerano S., Simone G.S., Bagalkot T.R., Zucker J., Yin M.T., Chang J.Y., Liu L., et al. Placental response to maternal SARS-CoV-2 infection. Sci. Rep. 2021;11:14390. doi: 10.1038/s41598-021-93931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins E., Hudak M.L., Banerjee J., Getzlaff T., Townson J., Barnette K., Playle R., Perry A., Bourne T., Lees C.C. Pregnancy and neonatal outcomes of COVID-19: coreporting of common outcomes from PAN-COVID and AAP-SONPM registries. Ultrasound Obstet. Gynecol. 2021;57:573–581. doi: 10.1002/uog.23619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey J.J., Magro C.M., Ma L.X., Nuovo G.J., Baergen R.N. Analysis of complement deposition and viral RNA in placentas of COVID-19 patients. Ann. Diagn. Pathol. 2020;46:151530. doi: 10.1016/j.anndiagpath.2020.151530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer D., Pleniceanu O., Gnatek Y., Namestnikov M., Cohen-Zontag O., Goldberg S., Friedman Y.E., Friedman N., Mandelboim M., Vitner E., et al. Human kidney spheroids and monolayers provide insights into SARS-CoV-2 renal interactions. J. Am. Soc. Nephrol. 2021;32:2242–2254. doi: 10.1681/ASN.2020111546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Tracking SARS-CoV-2 variants. 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y., Bagalkot T., Fitzgerald W., Sadovsky E., Chu T., Martínez-Marchal A., Brieño-Enríquez M., Su E.J., Margolis L., Sorkin A., Sadovsky Y. Term human placental trophoblasts express SARS-CoV-2 entry factors ACE2, TMPRSS2, and furin. mSphere. 2021;6 doi: 10.1128/mSphere.00250-21. e00250–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield C.A., Brubaker S.G., Limaye M.A., Lighter J., Ratner A.J., Thomas K.M., Meyer J.A., Roman A.S. Detection of severe acute respiratory syndrome coronavirus 2 in placental and fetal membrane samples. Am. J. Obstet. Gynecol. MFM. 2020;2:100133. doi: 10.1016/j.ajogmf.2020.100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R., Romero R., Tarca A.L., Luca F., Xu Y., Alazizi A., Leng Y., Hsu C.D., Gomez-Lopez N. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? Elife. 2020;9:e58716. doi: 10.7554/eLife.58716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu M., Cagino K., Matthews K.C., Friedlander R.L., Glynn S.M., Kubiak J.M., Yang Y.J., Zhao Z., Baergen R.N., Dipace J.I., et al. Pregnancy and postpartum outcomes in a universally tested population for SARS-CoV-2 in New York City: a prospective cohort study. BJOG. 2020;127:1548–1556. doi: 10.1111/1471-0528.16403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu M., Murphy E.A., Sukhu A.C., Yee J., Singh S., Eng D., Zhao Z., Riley L.E., Yang Y.J. Antibody response to coronavirus disease 2019 (COVID-19) messenger RNA vaccination in pregnant women and transplacental passage into cord blood. Obstet. Gynecol. 2021;138:278–280. doi: 10.1097/AOG.0000000000004438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J., Cross J.C. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Salvatore C.M., Han J.Y., Acker K.P., Tiwari P., Jin J., Brandler M., Cangemi C., Gordon L., Parow A., Dipace J., Delamora P. Neonatal management and outcomes during the COVID-19 pandemic: an observation cohort study. Lancet Child. Adolesc. Health. 2020;4:721–727. doi: 10.1016/S2352-4642(20)30235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U., Weigert M., Broaddus C., Myers G. Springer International Publishing; 2018. Cell Detection with Star-Convex Polygons; pp. 265–273. [Google Scholar]
- Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D.A. An analysis of 38 pregnant women with COVID-19, their newborn infants, and maternal-fetal transmission of SARS-CoV-2: maternal coronavirus infections and pregnancy outcomes. Arch. Pathol. Lab. Med. 2020;144:799–805. doi: 10.5858/arpa.2020-0901-SA. [DOI] [PubMed] [Google Scholar]
- Shanes E.D., Mithal L.B., Otero S., Azad H.A., Miller E.S., Goldstein J.A. Placental pathology in COVID-19. Am. J. Clin. Pathol. 2020;154:23–32. doi: 10.1093/ajcp/aqaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Bansal V., Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 2020;32:108175. doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Dcosta B.M., Samanovic M.I., Herati R.S., Cornelius A., Zhou H., Vaill A., Kazmierski W., Mulligan M.J., Landau N.R. Convalescent-phase sera and vaccine-elicited antibodies largely maintain neutralizing titer against global SARS-CoV-2 variant spikes. mBio. 2021;12:e0069621. doi: 10.1128/mBio.00696-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Fan C., Chen J.S., Kaur R., Stapleford K.A., Gristick H., Dcosta B.M., Wilen C.B., Nimigean C.M., Landau N.R. An ACE2 microbody containing a single immunoglobulin fc domain is a potent inhibitor of SARS-CoV-2. Cell Rep. 2020;33:108528. doi: 10.1016/j.celrep.2020.108528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Mulligan M.J., Landau N.R. Partial resistance of SARS-CoV-2 Delta variants to vaccine-elicited antibodies and convalescent sera. iScience. 2021;24:103341. doi: 10.1016/j.isci.2021.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Zhou H., Samanovic M.I., Dcosta B.M., Cornelius A., Herati R.S., Mulligan M.J., Landau N.R. Neutralization of SARS-CoV-2 variants by mRNA and adenoviral vector vaccine-elicited antibodies. Front. Immunol. 2022;13:797589. doi: 10.3389/fimmu.2022.797589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglauer E., Benarroch Y., Rop K., Barnett E., Sabharwal V., Yarrington C., Wachman E.M. Consistent localization of SARS-CoV-2 spike glycoprotein and ACE2 over TMPRSS2 predominance in placental villi of 15 COVID-19 positive maternal-fetal dyads. Placenta. 2020;100:69–74. doi: 10.1016/j.placenta.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Lacko L.A., Zhou T., Tomoiaga D., Hurtado R., Zhang T., Sevilla A., Zhong A., Mason C.E., Noggle S., et al. Pre- and peri-implantation Zika virus infection impairs fetal development by targeting trophectoderm cells. Nat. Commun. 2019;10:4155. doi: 10.1038/s41467-019-12063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Uhl S., Zhang T., Xue D., Li B., Vandana J.J., Acklin J.A., Bonnycastle L.L., Narisu N., Erdos M.R., et al. SARS-CoV-2 infection induces beta cell transdifferentiation. Cell Metab. 2021;33:1577–1591.e7. doi: 10.1016/j.cmet.2021.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallat R. Pingouin: statistics in Python. J. Open Source Softw. 2018;3:1026. [Google Scholar]
- Van Der Walt S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T. scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Joshi C.S., Silverstein R.B., He M., Carter E.B., Mysorekar I.U. SARS-CoV-2 colonization of maternal and fetal cells of the human placenta promotes alteration of local renin-angiotensin system. Med. (N Y) 2021;2:575–590.e5. doi: 10.1016/j.medj.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanti A.J., Vauloup-Fellous C., Prevot S., Zupan V., Suffee C., Do Cao J., Benachi A., De Luca D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020;11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Alfajaro M.M., Deweirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2021;184:76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth K.R., Olsen E.O., Neelam V., Lewis E.L., Galang R.R., Oduyebo T., Aveni K., Yazdy M.M., Harvey E., Longcore N.D., et al. Birth and infant outcomes following laboratory-confirmed SARS-CoV-2 infection in pregnancy - SET-NET, 16 jurisdictions, March 29-October 14, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1635–1640. doi: 10.15585/mmwr.mm6944e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Jaffré F., Nilsson-Payant B.E., Bram Y., Wang P., Zhu J., Zhang T., Redmond D., Houghton S., et al. An immuno-cardiac model for macrophage-mediated inflammation in COVID-19 hearts. Circ. Res. 2021;129:33–46. doi: 10.1161/CIRCRESAHA.121.319060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffré F., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano L.D., Ellington S., Strid P., Galang R.R., Oduyebo T., Tong V.T., Woodworth K.R., Nahabedian J.F., 3rd, Azziz-Baumgartner E., Gilboa S.M., Meaney-Delman D. Update: characteristics of symptomatic women of reproductive age with laboratory-confirmed SARS-CoV-2 infection by pregnancy status - United States, January 22-October 3, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1641–1647. doi: 10.15585/mmwr.mm6944e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video shows isolated primary placental clusters infected with live SARSCoV-2 virus (MOI = 1, bottom rows) ex vivo at 24 hpi, that were then stained for trophoblast marker KRT7 (green), SARS-N (red), endothelial marker CD31 (grey), and DAPI (blue). Scale bar = 50 μm.
Data Availability Statement
-
•
RNA-seq data of patient placentas and ex vivo placental clusters were deposited in the GEO repository database and will be available as of the date of publication. The accession number is listed in the key resources table.
-
•
Immunohistochemistry (IHC) analysis data were deposited in the zenodo repository database and will be available as of the date of publication. The accession number is listed in the key resources table.
-
•
The source code for the analysis of IHC data will be available as of the date of publication. The link is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.