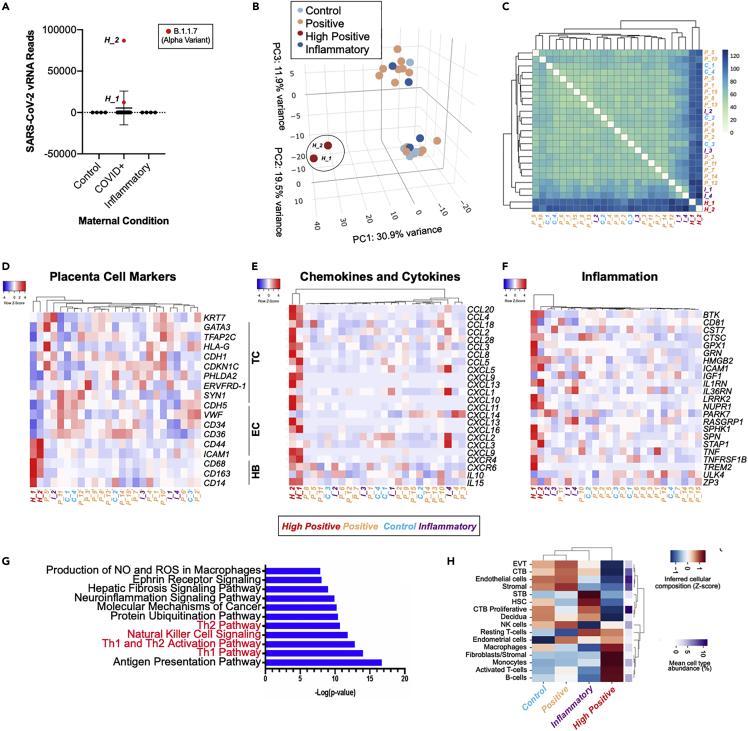
Figure 6.
Transcriptional analysis of term placentas of COVID-19 patients
(A) SARS-CoV-2 viral RNA FPKM levels in placentas from healthy control pregnancies (n = 4), COVID+ mothers (n = 17), and placentas with non-COVID related inflammatory pathologies (n = 4). B.1.1.07 Alpha variant (red) was detected in the two high positive placentas (H_1, H_2).
(B) PCA analysis of gene expression profiles in control samples (C, n = 4), high positive samples (H, n = 2), positive samples (P, n = 15), and inflammatory samples (I, n = 4).
(C) Expression heatmap of sample-to-sample distances for the overall gene expression.
(D–F) Heatmap from RNA-seq data of placentas from control samples (C, n = 4), high positive samples (H, n = 2), positive samples (P, n = 15), and inflammatory samples (I, n = 4) showing placental cell marker gene expression (D), chemokine and cytokine gene expression (E), and inflammatory associated gene expression (F).
(G) Ingenuity pathway analysis (IPA) depicting the top canonical biological pathways affected when comparing high positive SARS-CoV-2 placentas with control placentas. TC, trophoblast cells; EC, endothelial cells; HB, Hofbauer cells.
(H) Heatmap of cellular deconvolution estimating the relative abundance of cell types at the maternal–fetal interface. Mean values per group were the Z-score transformed per row. The original mean relative proportion of each cell type is displayed on the right.
See also Table S2; Figures S4 and S5.