Abstract
The accurate identification of antitumor T cell receptors (TCRs) represents a major challenge for the engineering of cell-based cancer immunotherapies. By mapping 55 neoantigen-specific TCR clonotypes (NeoTCRs) from 10 metastatic human tumors to their single-cell transcriptomes, we identified signatures of CD8+ and CD4+ neoantigen-reactive tumor-infiltrating lymphocytes (TILs). Neoantigen-specific TILs exhibited tumor-specific expansion with dysfunctional phenotypes, distinct from blood-emigrant bystanders and regulatory TILs. Prospective prediction and testing of 73 NeoTCR signature–derived clonotypes demonstrated that half of the tested TCRs recognized tumor antigens or autologous tumors. NeoTCR signatures identified TCRs that target driver neoantigens and nonmutated viral or tumor-associated antigens, suggesting a common metastatic TIL exhaustion program. NeoTCR signatures delineate the landscape of TILs across metastatic tumors, enabling successful TCR prediction based purely on TIL transcriptomic states for use in cancer immunotherapy.
Genetic engineering to redirect the antigen specificity of autologous patient immune cells against tumors using T cell receptors (TCRs) and chimeric antigen receptors (CARs) has been effective for the treatment of certain cancer types (1-4). A major challenge in developing engineered cell therapies against the solid epithelial cancers that are responsible for 90% of cancer deaths (5) is the targeting of tumor-specific antigens without destruction of normal cells (6-8). Tumor neoantigens derived from nonsynonymous somatic cancer mutations presented on human leukocyte antigen (HLA) molecules provide tumor specificity for T cell therapies while obviating toxicities associated with targeting normal tissues (9). Tumor neoantigens encoded by somatic mutations present in known cancer driver genes or private mutations specific to individual cancers have emerged as major antigenic targets of CD8+ and CD4+ T cells in immune checkpoint blockade (ICB) and in adoptive cell therapy (ACT) (9-13). Identification of TCRs capable of recognizing tumor neoantigens can lead to the development of cell-based immunotherapies for patients with metastatic solid malignancies. Neoantigen-reactive TCRs may also help to elucidate factors involved with the generation of antitumor immune responses and provide biomarkers that are useful in monitoring antitumor immune responses to ICB and ACT.
Conventional means of identifying tumor-reactive T cells and their cognate TCRs have generally relied on ex vivo T cell functional assays or, when the minimal epitope is known, single-cell sorting of T cells from bulk populations using antigen-specific HLA multimers and then reconstructing the TCRs expressed by the sorted T cells (14-18). Approaches to identify cancer-reactive T cells and their receptors from resected tumors that rely on T cell function can be impaired as a result of tumor-mediated T cell exhaustion and dysfunction (19, 20). Expression of cell surface protein markers of T cell activation and dysfunction can provide a tool for isolating tumor-reactive, neoantigen-specific tumor-infiltrating lymphocytes (TILs) and their receptors from tumors and peripheral blood, although these approaches are often hampered by nonspecific enrichment of irrelevant bystander T cells in tumors (21-26).
Analysis of cellular states can provide insight into the specificity and function of T cells (17, 18). Transcriptomic and epigenetic states of antigen-specific CD8+ T cells from chronic viral infections and tumors have been well established in murine models, but their relevance to the states of human T cells in metastatic tumor samples is unclear (27-29). Recent studies using single-cell transcriptomic profiling of tumors without in vitro culture have demonstrated the heterogeneity of human TIL states (20, 30-32) and explored the phenotypes associated with immunotherapy response in patients (33-36). However, the lack of focus on cancer antigen-specific human CD8+ and CD4+ TILs in single-cell studies has complicated our understanding of the relevant TIL population involved in antitumor immunity within tumor sites. An additional challenge to the identification of antitumor TCRs is characterized by our recent finding that although natural immune reactions do indeed occur in metastatic epithelial cancer, T cell recognition of such cancers is largely skewed toward private neoantigens specific to each cancer (11).
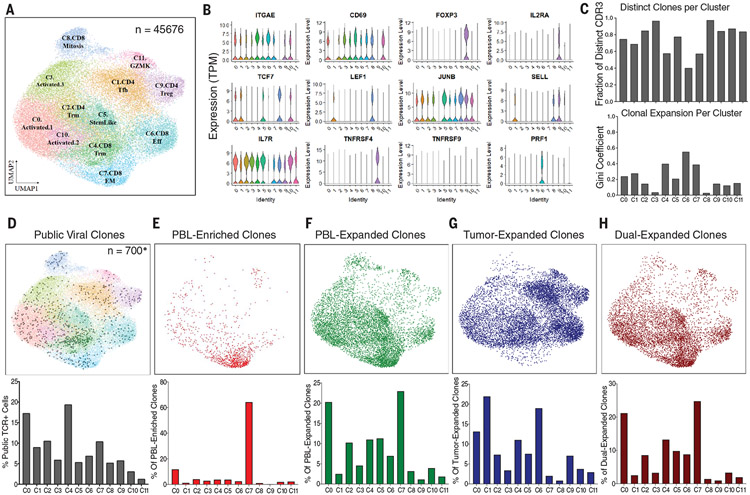
In this study, we evaluated the transcriptomic profiles of neoantigen-reactive TILs within archival metastatic tumor samples from patients to gain insights into intratumoral T cell states. To this end, we performed single-cell RNA sequencing (scRNA-seq) and T cell receptor sequencing (TCR-seq) on CD8+ and CD4+ T cells within metastatic cancers from 10 patients across multiple solid tumor types that included breast, melanoma, colon, and rectal cancers (table S1). We performed unsupervised clustering of 45,676 TILs from tumor digests by using uniform manifold approximation and projection (UMAP) to define 12 distinct transcriptional clusters (Fig. 1A). Patient TILs were distributed across each of the transcriptional phenotypic states regardless of tumor type and whether the patient received prior ICB (table S2 and fig. S1), although some TIL state differences could be attributed to the site of metastasis, as one cluster (C8) was predominantly populated by cells from the sole lymph node metastasis–derived melanoma sample (fig. S1, A to D).
Fig. 1. Transcriptomic landscape of TILs from surgically resected metastatic human cancers.
(A) UMAP of single-cell transcriptome data from 45,676 T cells sorted from 10 archival tumor specimens. Cluster names are based on names of published scRNA signatures with highest correlations (table S5). (B) Violin plots comparing gene expression of common T cell memory and activation markers across UMAP clusters from (A). TPM, transcripts per million. (C) (Top) Bar graph showing the fraction of T cells in each cluster that express distinct CDR3s. (Bottom) Bar graph showing Gini coefficients of CDR3 dispersion throughout clusters. Clusters 4, 6, and 7 have the lowest fraction of distinct CDRs and the highest Gini coefficients, indicating greater average clonal expansion and dispersion. (D to H) (Top) Projection of TCR clonotypes of a given class onto a transcriptomic T cell map. (Bottom) Bar graph showing frequency breakdown by cluster of cells in the corresponding plot. (D) Projection of public TCRs reactive to CMV, EBV, and influenza A (from VDJdb) onto a transcriptomic map from archival patients (n = 700), with the highest frequency in CD8+ TILRM C4. (E) Projection of cells expressing CDR3s that are peripheral blood (PBL)–enriched (i.e., found at a greater frequency in the PBL than in TILs; n = 1115). The majority of PBL-enriched clones are found in the CD8+ TILEM C7 state. (F) Projection of cells expressing CDR3s that are PBL expanded (i.e., found at a frequency greater than twice the limit of detection in the PBL; n = 8566). PBL-expanded clones are found in most phenotypic states but are largely absent in the CD4+ C1 dysfunctional differentiated cluster, CD8+C8.Mitosis, CD4+C9.Treg, and C11.GZMK. (G) Projection of cells expressing CDR3s that are tumor expanded but not PBL expanded (i.e., occurring multiple times in tumors but not found at a frequency greater than twice the limit of detection in the PBL; n = 10,631). Tumor-expanded clones are found in most phenotypic states but are largely absent in C7 and C8. (H) Projection of cells expressing CDR3s that are dual expanded (i.e., occurring multiple times in tumors and found at a frequency greater than twice the limit of detection in the PBL; n = 6436). Dual-expanded clones are found most frequently in C0, C4, C5, and C7.
We performed differential gene expression analysis to compare the TIL phenotypes and identify gene sets that represent each transcriptional cluster (table S3). We scored cells by single-cell gene set enrichment analysis (scGSEA) of our cluster-specific markers and more than 100 gene signatures from other recently published scRNA studies (29-39) and performed cluster correlation analysis to identify TIL cluster phenotypes (Fig. 1A and tables S4 and S5). Phenotypic states ranged from activated TILs (clusters C0, C3, and C10) to resident-memory TILs (TILRM clusters C2 and C4) and CD4+ regulatory TILs (TILTreg cluster C9) (Fig. 1, A and B). We also found that clusters C1 and C6 shared genes with dysfunctional CD4+ and CD8+ TIL populations respectively described by other recent single-cell profiling of melanoma and bladder cancer (table S5) (30, 31). A pseudotime trajectory superimposed on the TIL UMAP indicated that the cells within the C6 CD8+ and C1 CD4+ clusters, along with a subset of activated CD4 and CD8 T cells in C0, represented the most-differentiated TILs within these cell populations (fig. S1E).
By integrating 23,712 TCR clonotypes within the 12 TIL phenotypic clusters using complementarity-determining region 3β (CDR3β) TCR-seq analysis, we found that the majority of TIL clones were unexpanded singletons (83.2% across all clusters), consistent with previous reports (40). Oligoclonal TIL expansion was found mostly within the differentiated dysfunctional CD8+ TIL cluster C6, resident-memory CD8+ TILRM cluster C4, and effector-memory CD8+ TILEM cluster C7, consistent with the idea of clonal expansion after T cell activation and differentiation (Fig. 1C). Previous studies indicated that the tumor microenvironment consists of bystander T cells of viral specificities (23, 34). Analyzing public CDR3β TCR clonotypes reactive to common viral antigens from influenza, cytomegalovirus (CMV), and Epstein-Barr virus (EBV) (41), we identified 700 T cells that contain bystander viral TCRs distributed across multiple cellular states (table S6), with the majority of them in activated C0 TIL (17.3%), CD8+ TILRM C4 (19.4%), and CD8+ TILEM C7 (10.4%) clusters (Fig. 1D).
In parallel, we studied the T cell repertoire in each patient’s peripheral blood from the time of tumor resection and coupled it with the TIL scRNA-seq analysis to better understand T cell expansion within tumors. TIL clonotypes that were enriched in the peripheral blood relative to tumors (PBL-enriched clones) were found preferentially in the CD8+ TILEM C7 state (Fig. 1E), which also contained a large proportion of public viral T cells (Fig. 1D). TIL clonotypes expanded in peripheral blood (PBL-expanded clones) were found within activated C0, TILRM C2 and C4, C5, and CD8+ C7 TILEM states but were largely absent in C1, C6, and C9 clusters containing tumor-expanded clones (Fig. 1F). By contrast, TCR clonotypes enriched in tumors (tumor-expanded clones) were mostly enriched in CD8+ C6 and CD4+ C1 dysfunctional differentiated clusters (Fig. 1G). Furthermore, clones that were expanded in both tumors and peripheral blood (dual-expanded clones) were abundant in TIL states C0 and C7 (Fig. 1H). These results indicate that cellular states of tumor-expanded clones (C1 and C6) are distinct from blood-emigrant TILs within metastatic tumors. Additionally, our data suggest that TIL states containing dual-expanded clones (C0 and C7) are largely populated by irrelevant bystander TILs (including viral-specific TILs) that were expanded in blood before arrival at the tumor site.
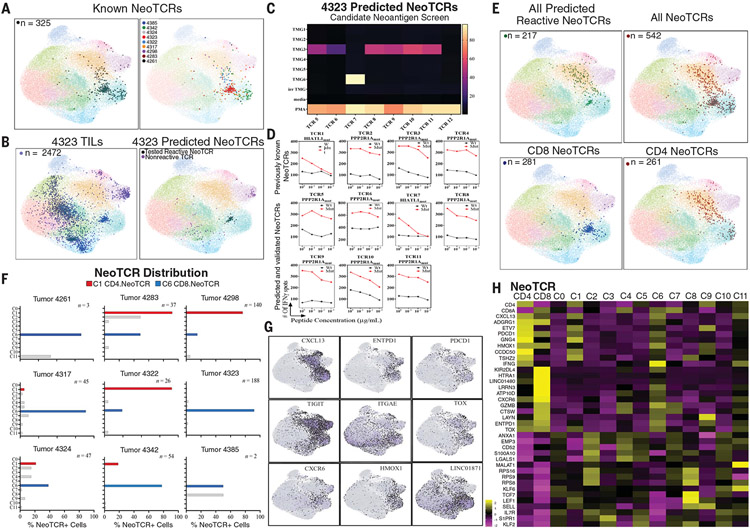
Given that multiple clonally expanded TIL states were attributable to blood-emigrant bystander T cells, we then sought to define cellular states of antitumor neoantigen-specific TILs. To this end, our group had previously used our tandem minigene and peptide screening platform (9, 10) to identify functional CD8+ and CD4+ neoantigen-specific TILs from 9 of 10 archival patients in the scRNA-seq dataset (11, 42, 43). Once these reactive TILs were identified, we performed fluorescence-activated cell sorting of activated 4-1BB+ neoantigen-reactive TILs, followed by TCR reconstruction (14, 26, 44) (fig. S2). We experimentally defined 14 CD8+ and 17 CD4+ neoantigen TCR clonotypes (NeoTCRs) against neoantigens encoded by patient cancer mutations, which also included driver genes such as PIK3CA, KRAS, and TP53 (fig. S2, A and B, and table S7). Within the scRNA-seq data across all samples, we identified 325 single T cells (0.9% of all cells) that expressed these 31 experimentally verified NeoTCRs and backprojected them onto the TIL UMAP (Fig. 2A and fig. S2B). The vast majority of NeoTCR+ TILs (84.3%) were distributed between just two clusters: CD4+ C1 and the CD8+ C6 differentiated dysfunctional states from the nine patients (Fig. 2A). Of the CD4+ clonotypes, 91.3% were found in C1 or C6; 78.5% of CD8+ clonotypes were found in C6 (table S7).
Fig. 2. Neoantigen-reactive CD4 and CD8 T cells exist in common dysfunctional states.
(A) UMAP of all archival T cells overlaid with known neoantigen-reactive T cells from all patients (left; n = 325) and individually by patient (right; see also table S7). (B) (Left) Projection of all T cells from tumor 4323 (n = 2472) onto a transcriptomic map of T cells from all patients. (Right) Projection of newly predicted, experimentally tested neoantigen-reactive TCR-bearing cells from tumor 4323 (tested reactive NeoTCR, black; n = 53) and cells bearing TCRs that were negative in our screening assay (nonreactive TCRs, purple; n = 76). (C) Heatmap of TCR screening of predicted NeoTCRs from tumor 4323, showing reactivity of seven of eight TCRs to tandem minigenes 3 or 6 (TMG3 or TMG6). The right axis, which shows the percentage of activated T cells, indicates those expressing 4-1BB, as determined by flow cytometry. irr TMG, irrelevant TMG (i.e., TMG expressing candidate neoantigens not present in tumor 4323); PMA, phorbol 12-myristate 13-acetate and ionomycin. (D) Peptide titration curves of TMG-reactive TCRs from tumor 4323, as measured by interferon-γ (IFNγ) ELISpot. (Top row) TCRs 1 to 4 were previously known to be reactive to either HIATL1mut (p.G380V) or PPP2R1Amut (p.L432S). (Bottom rows) Newly identified TCRs showing selectivity for the same neoantigens over the corresponding wild-type peptides. Wt, wild type; Mut, mutant. (E) (Top left) Projection of newly identified NeoTCR-expressing cells (n = 217) onto a transcriptomic map from all samples. (Top right) Projection of all known NeoTCRs, including previously known NeoTCRs [from (A)] and newly identified NeoTCR clones (n = 542). (Bottom left) Projection of all known CD8-restricted NeoTCRs, showing high abundance in C6 (n = 281). (Bottom right) Projection of all known CD4-restricted NeoTCRs, showing high abundance in C1 (n = 261). (F) Bar graphs showing distribution of all known NeoTCR-expressing cells across clusters by patient. Gray bars indicate T cell clones seen outside of NeoTCR clusters C1 and C6. (G) Gene expression of representative highly expressed genes from NeoTCR-expressing cells relative to other cells, projected onto a transcriptomic map. (H) Heatmap showing gene expression of the most differentially expressed genes from CD4+ NeoTCR-expressing cells (NeoTCR CD4) and CD8+ NeoTCR-expressing cells (NeoTCR CD8) relative to all clusters. A subset of select stemness and memory genes is shown for comparison.
We further studied TIL states from tumor 4323, a rectal cancer liver metastasis, which had four previously defined NeoTCRs (tumor 4323 NeoTCRs 1 to 4) recognizing two somatic neoantigens HIATL1mut (p.G380V) and PPP2R1Amut (p.L432S) (table S7). Tumor 4323 CD8+ TILs were broadly distributed across TILEM C7, TILRM C4, and C6, whereas CD4+ cells were in TILRM C2 and TILTreg C9 (Fig. 2B). The four dominant NeoTCRs that represent 140 single TILs were found almost exclusively in the differentiated CD8+ C6 cluster (91.4%; Fig. 2A). Because dysfunctional and differentiated TIL states could be driven by chronic T cell–tumor neoantigen interaction, we hypothesized that other subdominant clones within the differentiated CD8+ C6 cluster state might also be neoantigen reactive. We therefore reconstructed and tested eight additional TCR clonotypes from tumor 4323 TILs within cluster C6 (table S7, fig. S2B, and materials and methods). Seven of the eight newly predicted TCRs were also found to be reactive against either HIATL1mut or PPP2R1Amut neoantigens (Fig. 2, C and D), and 45 of 48 single cells expressing the newly identified NeoTCRs were found in the CD8+ C6 dysfunctional differentiated cluster. Beyond the ability to capture previously unknown NeoTCRs, CD8+ C6 also demonstrated selectivity for neoantigen reactivity, because other dominant TCR clonotypes from tumor 4323 TILs that had previously screened negative for reactivity against candidate neoantigens (“dominant nonreactive TCRs”) were largely found in the CD8+ C4 TILRM and CD4+ C9 TILTreg states (Fig. 2B and table S7).
We extended this analysis by reconstructing and experimentally screening candidate TCR clonotypes from other tumor samples that populated the CD4+ C1 and CD8+ C6 dysfunctional differentiated clusters against their autologous tumor mutation–encoded candidate neoantigens. Including those predicted from tumor 4323, we successfully defined 23 NeoTCR clonotypes (expressed by 217 TILs, median: 9 T cells per clonotype, range: 1 to 34) targeting 10 neoantigens (Fig. 2E and table S7). Although many of the newly identified NeoTCRs targeted the same tumor neoantigens defined by in vitro expanded TIL screening, we also identified NeoTCRs targeting previously unknown neoantigens that were not defined by prior screening of cultured TILs (e.g., 10 novel NeoTCRs against four newly identified neoantigens from tumor 4298; Fig. 2E and table S7). Combined backprojection of all 54 (31 previously known plus 23 newly identified) NeoTCR clonotypes representing 542 individual TILs showed that, irrespective of patient tumor histology, clusters C1 and C6 contained the majority (86.5%) of all neoantigen-reactive TILs (Fig. 2F and fig. S3A), suggesting a shared neoantigen-specific TIL program within metastatic cancers. Among 21 CD8+ NeoTCRs expressed by 281 cells, 81.1% of cells were present in C6. Of the 31 CD4+ NeoTCR clones expressed by 261 cells, 60.5% of cells were located within C1, whereas 89.3% were within either C1 or C6. Notably, five CD4+ NeoTCR clones found in C6 were not observed in C1 (table S7), potentially representing CD4 cytotoxic T lymphocytes, as previously observed (31). Overall, the frequency of CD8+ NeoTCRs increased 22-fold in C6 compared with the overall frequency, and the frequency of CD4+ NeoTCRs increased fivefold in C1 compared with the overall frequency (fig. S3B). Consistent with our prior study showing that less differentiated T cell states lack enrichment of neoantigen-reactive TILs (45), only 7.4% of neoantigen-reactive TILs were found in tumor-resident memory states CD4+TILRM (C2) and CD8+TILRM (C4) or the stemlike C5 (Fig. 2F and fig. S3A). Additionally, fewer than 1.5% of neoantigen-reactive TILs were found among dual-expanded clonotypes previously reported to be associated with immunotherapy response (34), suggesting that only a minority of dual-expanded TILs consist of tumor-relevant clonotypes (Fig. 1H).
Restricting the gene expression analysis to only verified NeoTCR+ TILs within the archival samples enabled us to develop molecular profiles of CD4+ and CD8+ neoantigen-reactive TILs (table S8). CD4+ and CD8+ NeoTCR-expressing cells shared expression of multiple genes—including those encoding the cytokine CXCL13; tissue-homing protein CXCR6; inhibitory markers TIGIT, PD1 (PDCD1), CD39 (ENTPD1), and LAG3; and TOX, a regulator of T cell exhaustion (Fig. 2, G and H, and table S8)—which have all been previously reported to be expressed by dysfunctional T cells within the tumor microenvironment (21-23, 29-31). CD4+ and CD8+ NeoTCR-expressing cells also shared expression of several genes that were not previously reported to be associated with intratumoral dysfunctional T cells, including ADGRG1, HMOX1, LINC01871, DUSP4, and ACP5 (Fig. 2H and table S8). CD4+ and CD8+ NeoTCR cells also shared down-regulation of stemness and memory genes IL7R, CD44, and KLF2; lactate dehydrogenase LDH1; annexin ANXA1; the calcium channel S100A10; and cell cycle regulator RGCC (table S8). Additionally, CD8+ NeoTCR TILs expressed effector T cell genes GZMA, GZMB, GZMK, IFNG, and PRF1; exhaustion markers LAG3, HAVCR2 (TIM3), and ENTPD1 (CD39); and tissue-residency marker ITGAE (CD103), which have been previously reported to enrich for neoantigen-specific TILs (Fig. 2H and table S8) (21-23, 46).
Pathway analysis of gene networks highlighted several upstream regulators of genes expressed by NeoTCR4+ and NeoTCR8+ cells (table S9). These gene modules implicated a TCR- dependent up-regulation of genes within both CD4+ and CD8+ NeoTCR cells (CD28, TCR, and IL2), in concert with cytokine-encoding genes IFNA1/2, IFNB1, IL27, IL4, and IL6 that are involved in follicular T helper (TFH) cell maintenance and activation (47, 48). The high expression of CXCL13 by both CD4+ and CD8+ NeoTCRs, in addition to TFH cell activation through type 1 interferons (47, 48) and the implication of B cell receptors in the CD8+ NeoTCR signature (table S9), suggests that antitumor neoantigen-specific TIL activation might involve tertiary lymphoid structures within metastatic deposits of human tumors (49, 50).
We reasoned that the shared neoantigen-specific TIL gene signatures might enable us to prospectively predict antitumor, neoantigen-specific TCRs purely on the basis of TIL transcriptomic states. We first tested the sensitivity and specificity of versions of potential NeoTCR4 and NeoTCR8 signatures containing various lengths of gene lists to correctly call known NeoTCRs from the archival specimens (table S10). From these analyses, we found that a 40-gene version of the putative NeoTCR4 signatures and a 243-gene version of the putative NeoTCR8 signatures performed with the highest sensitivity and specificity, and we established these as NeoTCR4 and NeoTCR8 signatures, respectively [area under the curve (AUC) of receiver operator characteristic (ROC) > 0.9; table S10, tab 2]. Additionally, we compared effector and dysfunctional gene sets consisting of different minimal combinations of classical T cell activation or dysfunction markers (ENTPD1, CXCL13, PDCD1, ITGAE, TIGIT, TOX, LAG3, HAVCR2, and GZMK; table S10, tab 3) for their ability to capture CD4+ and CD8+ NeoTCRs. Gene sets that contain CXCL13 performed well in capturing CD4+ NeoTCR cells (AUC ≤ 0.8) but were inferior (AUC ≤ 0.8) to the NeoTCR8 signature in predicting CD8+ NeoTCRs. Notably, versions of the NeoTCR4 and NeoTCR8 signatures that intentionally excluded these nine genes demonstrated high sensitivity and specificity (NeoTCR4 AUCs > 0.8 and NeoTCR8 AUCs > 0.9; table S10, tab 4). These data together suggest that the TIL dysfunctional program identified here is not limited to just a few known exhaustion or activation genes but also includes several genes with yet-unidentified functions that together establish a recurrent dysfunctional transcriptional module within neoantigen-specific TILs from human cancers.
Correlation analyses of the NeoTCR4 and NeoTCR8 gene signatures demonstrated that the NeoTCR4 signature was most similar to those of the lung cancer TIL signatures Caushi. CD4.Tfh.2 [Pearson’s correlation coefficient (r) = 0.725] and Wu.CD4.IL6ST (Pearson’s r = 0.612), whereas the NeoTCR8 signature was most similar to those of exhausted CD8 cells observed in basal and squamous cell carcinoma (Yost.CD8.Exhausted, Pearson’s r = 0.766) and melanoma (Oliveira.TTE, Pearson’s r = 0.747) (table S5) (34, 36, 38, 39). These data suggest that the neoantigen-reactive CD4+ and CD8+ dysfunctional TIL states that we identified within colorectal cancer, breast cancer, and melanoma also exist in other tumor histologies. To investigate whether NeoTCR signatures can distinguish bystander viral-reactive TILs from tumor viral antigen–reactive TILs, we performed scRNA and TCR-seq on TILs from a lymph node metastasis from human papilloma virus 16 (HPV16)–positive anal squamous cell cancer (tumor 4397). From the tumor, there was one previously known CD8+ TCR reactive against HPV16 antigen E4 (the only HPV antigen expressed by the tumor; fig. S4, A to C). Despite being observed only once among the 941 sequenced T cells, when scored by scGSEA, the E4-reactive TCR (TCR1) was the fourth-highest NeoTCR8 signature–expressing cell (fig. S4D), which suggests that antitumor viral antigen–specific TILs are similar to neoantigen-reactive dysfunctional TIL states, and that antitumor T cells in metastatic tumors may exist in shared dysfunctional states regardless of antigen class.
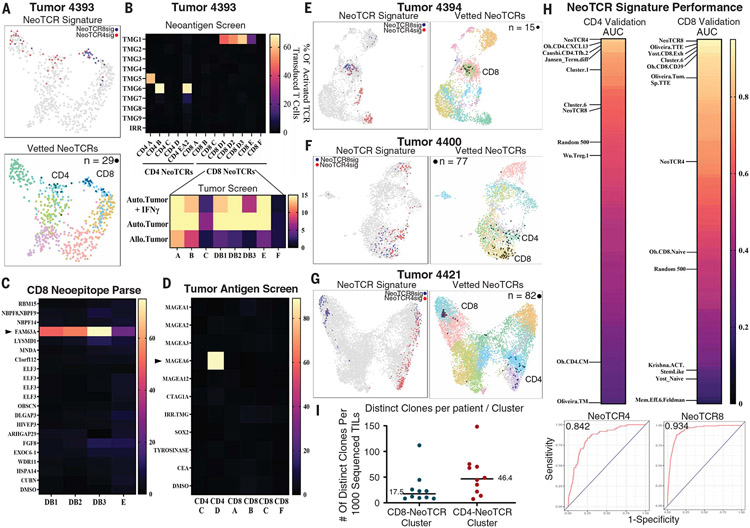
We then tested the utility of NeoTCR signatures to prospectively predict antitumor neoantigen-reactive TILs from four independent tumor samples from patients with metastatic colon adenocarcinoma. We performed scRNA-seq and scTCR-seq on patient TILs from single-cell suspensions of surgically resected metastatic tumors and scored each individual T cell using NeoTCR8 and NeoTCR4 signatures to identify putative antitumor NeoTCR TIL states (fig. S5 and table S4). From 630 TILs sequenced from tumor 4393, we reconstructed eight candidate CD8+ TCRs and six candidate CD4+ TCRs using NeoTCR signatures and subsequently screened TCR-transduced T cells for their response to 156 autologous candidate neoantigens, nine candidate tumor-associated antigens (table S11 and materials and methods), and a patient-derived xenograft (PDX) tumor line (Fig. 3, A to D, and fig. S6). The results of the screening assays indicated that four of the candidate CD8+ TCRs recognized a FAM63Amut (p.D460N) neoepitope and the autologous PDX, three CD8+ TCRs recognized the autologous PDX tumor but not any screened candidate neoantigens, one CD4+ TCR recognized a FUT1mut (p.343_344del) neoepitope, two CD4+ TCRs recognized a PCNTmut (p.P2122L) neoepitope, and one CD4+ TCR recognized the highly expressed nonmutated tumor-associated antigen (TAA) MAGEA6 (Fig. 3, B to D, and table S7). Backprojection of these 11 NeoTCRs indicated that they represented 29 single cells (or ~4.6%) of the single-cell map from tumor 4393 TILs (Fig. 3A).
Fig. 3. NeoTCR signatures enable prospective identification of neoantigen-, tumor-associated antigen-, and tumor-reactive CD4 and CD8 TCRs from newly resected tumors.
(A) (Top) Transcriptomic map of 630 TILs sequenced from tumor 4393 (colon). Cells that score in the 95th percentile of NeoTCR4 and NeoTCR8 signatures are highlighted. (Bottom) Experimentally verified CD4+ and CD8+ NeoTCR-expressing cells (n = 29) backprojected onto a transcriptomic map (black). (B) (Top) Screening of eight predicted candidate CD8+ TCRs and six candidate CD4+ TCRs against peptide pools and TMGs that represent 156 tumor mutations identified four TMG- and peptide pool–reactive CD8+ TCRs and three TMG- and peptide pool–reactive CD4+ TCRs. TMGs are shown for clarity. (Bottom) Screening of CD8+ TCRs against corresponding autologous tumors demonstrated selective reactivity toward autologous (Auto.) tumors relative to allogeneic (Allo.) tumors by 7 of 8 TCRs. The right axis, which shows the percentage of activated TCR transduced cells, refers to cells expressing 4–1BB as determined by flow cytometry. (C) Reactivity deconvolution data of the TMG-reactive CD8+ TCRs against constituent peptides of their reactive TMGs for newly identified TCRs from tumor 4393. CD8+ TCRs reactive to TMG1 (TCRs DB1, DB2, DB3, and E) demonstrated reactivity to FAM63Amut (p.D460N). (D) Screening of neoantigen–nonreactive TCRs from tumor 4393 against autologous dendritic cells expressing TAA RNA or pulsed with TAA peptide pools identified tumor 4393 CD4 TCR D as reactive toward MAGEA6. (E) (Left) Transcriptomic map of 2972 TILs sequenced from tumor 4394 (colon). Cells that score in the 95th percentile of NeoTCR4 and NeoTCR8 signatures are highlighted. (Right) Experimentally verified CD8+ NeoTCR-expressing cells (n = 15) backprojected onto a transcriptomic map. (F) (Left) Transcriptomic map of 2559 TILs sequenced from tumor 4400 (colon). Cells that score in the 95th percentile of NeoTCR4 and NeoTCR8 signatures are highlighted. (Right) Experimentally verified CD4+ and CD8+ NeoTCR-expressing cells (n = 77) backprojected onto a transcriptomic map. (G) (Left) Transcriptomic map of 10,049 TILs sequenced from tumor 4421 (colon). Cells that score in the 98th percentile of NeoTCR4 and NeoTCR8 signatures are highlighted. (Right) Experimentally verified CD4+ and CD8+ NeoTCR-expressing cells (n = 82) backprojected onto a transcriptomic map. (H) AUC scores showing the sensitivity and specificity of NeoTCR4, NeoTCR8, and published signatures in calling verified tumor- and neoantigen-reactive TCRs from prospective tumor samples. scGSEA was performed on T cells from samples 4393, 4394, 4400, and 4421, and ROC curves were generated to compare signature sensitivity and specificity. AUC values of all signatures were ranked by their ability to call CD4+ NeoTCRs (top left) and CD8+ NeoTCRs (top right). Selected signatures of interest are highlighted. (Bottom) ROC curves of highest-scoring signatures for CD4 (NeoTCR4; left) and CD8 (NeoTCR8; right) NeoTCRs. (I) Dot plots showing numbers of clones present in the C1 NeoTCR4 and C6 NeoTCR8 states per tumor from Fig. 1A, normalized to 1000 sequenced T cells for each tumor. Bars denote median values.
From tumor 4394, we synthesized and tested 12 CD8+ TCRs from the candidate CD8+ NeoTCR cluster against 185 autologous candidate neoantigens (Fig. 3E; no CD4+ clonotypes were tested because the CD4+ NeoTCR cluster comprised highly polyclonal singletons). We identified two CD8+ TCRs against the driver neoantigen KRASmut (p.G12V) and one CD8+ TCR that was nonreactive against screened mutations but did recognize an autologous PDX line (Fig. 3E and table S7). These three NeoTCRs represented 15 cells (0.5% of 2972 TILs) (Fig. 3E). From tumor 4400, we prospectively predicted 15 CD8+ TCRs and 14 CD4+ TCRs on the basis of NeoTCR4 and NeoTCR8 signatures (Fig. 3F). Neoantigen screening against 485 autologous candidate neoantigens and three highly expressed TAAs (table S11) led to the identification of five CD4+ TCRs that mediate recognition of KRASmut (p.G13D driver neoepitope) and nine CD8+ TCRs that were reactive with the tumor PDX line but did not recognize the screened neoantigen candidates (Fig. 3F and table S7). These 14 antitumor TCRs were expressed in 77 cells, representing 3.0% of the total cells captured (Fig. 3F). From tumor 4421, we prospectively predicted 7 CD8+ TCRs and 11 CD4+ TCRs on the basis of candidate NeoTCR clusters and tested them against 239 autologous candidate neoantigens and an autologous tumor organoid (Fig. 3G and fig. S6). Two CD8+ TCRs recognized the ANO9mut p.151_154del neoepitope, one recognized the FLNBmut p.H2539Q neoepitope and organoid, and two recognized the UHRF2mut (p.R212P) neoepitope and organoid, whereas two CD8+ TCRs were exclusively organoid reactive (table S7). Two of the 11 CD4+ TCRs were also neoantigen reactive to ESRP1mut (p.E180D) and THOC6mut (p.R116H). These nine reactive TCRs represented 82 cells (0.8% of 10,049 TILs; Fig. 3G).
From the four prospective patients, 37 of 73 (50.7%) predicted TCRs were reactive to tumors, neoantigens, or TAAs (Table 1). Among predicted CD8 TCRs, this frequency was 60.9%, and predicted CD4 TCRs were positive in our screen 35.4% of the time (Table 1). Notably, though our coculture assays of predicted CD4 TCRs did not show any direct antitumor reactivity, TCR recognition of tumor material through cross-presentation via antigen-presenting cells has not been evaluated. We did not identify any significant gene expression differences between TIL-expressing TCRs that were neoantigen and/or tumor reactive compared with TIL-containing TCRs that were nonreactive in our screens, nor did we find significant differences between gene expression profiles of neoantigen-specific TCR-expressing cells relative to exclusively PDX tumor–reactive TCR-containing cells that did not recognize screened candidate neoantigens (fig. S6).
Table 1. Summary of prospectively predicted TCRs from four patient tumor TILs to identify antitumor, neoantigen-reactive TCRs using the NeoTCR gene signature.
“Tumor” refers to direct organoid or PDX reactivity. NeoAg, neoantigen; TAA, tumor-associated antigen.
| Prospective prediction of TCRs reactive with tumor | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD8 | CD4 | All | |||||||||||
| Tumor | Tested | Tumor and NeoAg-reactive |
NeoAg only | Tumor only | Total | Tested | NeoAg-reactive | TAA-reactive | Tumor only | Total | Tested | Total | % |
| 4393 | 8 | 4 | 0 | 3 | 7 | 6 | 3 | 1 | 0 | 4 | 14 | 11 | 79 |
| 4394 | 12 | 2 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 12 | 3 | 25 |
| 4400 | 15 | 0 | 0 | 9 | 9 | 14 | 5 | 0 | 0 | 5 | 29 | 14 | 48 |
| 4421 | 7 | 3 | 2 | 2 | 7 | 11 | 2 | 0 | 0 | 2 | 18 | 9 | 50 |
| All | 42 | 9 | 2 | 15 | 26 61.9% |
31 | 10 | 1 | 0 | 11 35.4% |
73 | 37 | 50.7 |
To assess the sensitivity and specificity of the NeoTCR signatures and compare them to other published T cell signatures in terms of their ability to successfully identify neoantigen-reactive TCRs, we performed scGSEA on individual cells and performed ROC analysis of the ability of given signatures to call CD4+ NeoTCRs, CD8+ NeoTCRs, and public viral TCRs from the archival specimens (training sets; table S12 and fig. S7) and CD4+ NeoTCRs and CD8+ NeoTCRs from the prospective specimens (validation sets; Fig. 3H and table S12). In both the training and validation sets, the NeoTCR4 and NeoTCR8 signatures demonstrated the highest combination of sensitivity and specificity for CD4+ and CD8+ NeoTCRs, respectively (Fig. 3H, fig. S7, and table S12). We also identified TIL phenotypic states from other studies that performed well in our validation sets; namely, the bladder cancer–derived CD4.CXCL13 (31) and non–small cell lung cancer–derived CD4.Tfh.2 (39) signatures for CD4+ NeoTCRs, and terminally exhausted signatures from melanoma (38) and skin carcinoma (36) for CD8+ NeoTCRs. Although the studies did not examine the neoantigen reactivity of CD4+ T cells, the conserved phenotypic states shared with the validated CD4+ NeoTCRs identified here likely indicate that those samples also contain CD4+ neoantigen-reactive T cells. Notably, signatures from stemlike T cells associated with ACT response (Krishna.ACT.StemLike) (45) and ICB response (CD8_G, Mem.Eff) (33) were especially poor at highlighting NeoTCRs, consistent with the majority of antitumor T cells being in a dysfunctional state in progressing metastatic cancer (Fig. 3H and table S12) (45). Finally, the relative median AUC values for the NeoTCR4 and NeoTCR8 signatures were low for public virus-reactive T cells, suggesting high specificity of this dysfunctional program for tumor antigen-specific TILs (table S12 and fig. S7). Combining scRNA data from all 15 sequenced tumors, we estimated that a median of 17.5 clonotypes will be present within a given CD8+ NeoTCR cluster, and 46.4 clonotypes will be present within a given CD4+ NeoTCR cluster for every 1000 TILs sequenced per patient (Fig. 3I), which markedly enhances the landscape of possible antitumor T cell clonotypes within solid tumors.
In this Report, we have identified shared gene expression profiles of neoantigen-specific CD4+ and CD8+ T cells within metastatic solid human cancers. Neoantigen-specific TILs largely exhibited tumor-specific clonal expansion, with only limited overlap with dually expanded TILs found in the peripheral blood at the level of our sequencing depth (33, 34). Our results support prior high-dimensional phenotyping studies showing that tumor-reactive T cells are enriched within differentiated dysfunctional cellular states (23, 30, 31), with very few stemlike antitumor T cells (38, 45). We leveraged the NeoTCR dysfunctional signatures to identify antitumor TCRs with limited TIL material, in some cases identifying even an apparently unexpanded clonotype within the NeoTCR clusters as neoantigen reactive (e.g., tumor 4323 TCR10). Using signatures derived from relatively few neoantigen-reactive clones (54 clonotypes expressed by 542 cells), more than half of all prospectively tested TCRs expressing NeoTCR4 or NeoTCR8 signatures in this analysis were neoantigen and/or tumor reactive, which suggests a high degree of tumor specificity in T cells that exhibit NeoTCR states. These signatures offer the potential to identify antitumor TCRs without the need for functional screening of candidate neoantigens. Further, as the roles of neoantigen-specific CD4+ T cells in establishing an antitumor response by supporting the activation of cytolytic CD8+ T cells have come into view (51, 52), the NeoTCR4 signature identified here will allow for expeditious identification of such CD4+ antitumor TCRs.
Although the identified NeoTCR cells ranged from 0.1 to 9.1% of all TILs within the 14 tumors in which neoantigen reactivity was known, this likely represents an underestimate because we did not synthesize and experimentally determine tumor or mutation specificity for every TCR clonotype within the NeoTCR clusters, nor did we assess tumor or nonmutated TAA reactivity in the archival specimens (Fig. 3I). T cells expressing TCRs that target random mutations, mutations in tumor driver genes (e.g., KRAS, TP53, and PIK3CA), a nonmutated tumor-associated antigen (MAGEA6), an oncogenic viral antigen (HPV16-E4), and autologous tumor-reactive orphan receptors all converged on the same dysfunctional phenotype. Our results are further evidenced by a recent meta-analysis indicating that expression of CXCL13, one of the most differentially expressed genes within the NeoTCR4 and NeoTCR8 signatures, represents an independent variable for predicting responses to ICB (53). The existence of several orphan tumor-reactive receptors that are reactive to autologous tumor material but tested negative against candidate neoantigens within the NeoTCR clusters also implies that the landscape of tumor antigens remains broader than that of the somatic mutations we have tested, as has been recently suggested (54-56). Further evidence for this point is the lack of gene expression differences between NeoTCR signature–expressing cells that tested negative in our screens and those that tested positive. We find it likely that intratumoral T cells that acquire a dysfunctional, clonally expanded phenotype do so because they are reacting to tumor-relevant antigens, although this study did not investigate whether differences in target antigen expression or TCR functional avidity can differentially contribute to T cell dysfunction. We propose that TCRs from cells in the NeoTCR transcriptomic state can be rapidly identified without additional TIL growth, activation, or testing, thus providing opportunities to develop patient-specific neoantigen-targeting TCR immunotherapies against metastatic solid tumors, even when endogenous NeoTCR-expressing cells from those tumors are dysfunctional and/or exhausted. The potential roles of the genes expressed in the NeoTCR signature (including those that have unknown T cell function) in mediating tumor-specific TIL dysfunction in metastatic human cancers also remain to be explored in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Surgery Branch TIL Laboratory and clinical team for generating TIL; we also thank patients enrolled in our clinical protocols. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). We also thank NIDAP for providing additional computational support and the CCR Genomics Core for next-generation sequencing support.
Funding:
This research was supported by the Center for Cancer Research intramural research program of the National Cancer Institute. Support from the CCR Single Cell Analysis Facility was funded by FNLCR contract no. HHSN261200800001E. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Footnotes
Competing interests: F.J.L., S.Kr., R.Y., K.H., J.C.Y., P.F.R., and S.A.R. are listed as inventors on a patent application (US provisional patent application no. 62/992,701, PCT patent application no. PCT/US2021/023240) submitted by NCI that covers the use of NeoTCR signatures to identify antitumor TCRs. The other authors declare no competing interests.
Data and materials availability: All data are available in the main text or the supplementary materials. Sequencing data generated as part of this study are available on dbGaP (the Database of Genotypes and Phenotypes) under accession no. phs002748.v1.p1. Previously published tumor exome and RNA-seq data can be found on dbGaP under accession nos. phs002735.v1.p1 and phs001003.v2.p1, respectively.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Robbins PF et al. , J. Clin. Oncol 29, 917–924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan RA et al. , Science 314, 126–129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.June CH, Sadelain M, N. Engl. J. Med 379, 64–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochenderfer JN, Yu Z, Frasheri D, Restifo NP, Rosenberg SA, Blood 116, 3875–3886 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A, CA Cancer J. Clin 70, 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Morgan RA et al. , Mol. Ther 18, 843–851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkhurst MR et al. , Mol. Ther 19, 620–626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linette GP et al. , Blood 122, 863–871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran E, Robbins PF, Rosenberg SA, Nat. Immunol 18, 255–262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robbins PF et al. , Nat. Med 19, 747–752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkhurst MR et al. , Cancer Discov. 9, 1022–1035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gubin MM et al. , Nature 515, 577–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Rooij N et al. , J. Clin. Oncol 31, e439–e442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkhurst M et al. , Clin. Cancer Res 23, 2491–2505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keskin DB et al. , Nature 565, 234–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altman JD et al. , Science 274, 94–96 (1996).8810254 [Google Scholar]
- 17.Han A, Glanville J, Hansmann L, Davis MM, Nat. Biotechnol 32, 684–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y-C et al. , Mol. Ther 26, 379–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blank CU et al. , Nat. Rev. Immunol 19, 665–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Leun AM, Thommen DS, Schumacher TN, Nat. Rev. Cancer 20, 218–232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duhen T et al. , Nat. Commun 9, 2724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gros A et al. , J. Clin. Invest 124, 2246–2259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simoni Y et al. , Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Yossef R et al. , JCI Insight 3, e122467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheper W et al. , Nat. Med 25, 89–94 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Pasetto A et al. , Cancer Immunol. Res 4, 734–743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wherry EJ et al. , Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Sen DR et al. , Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott AC et al. , Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H et al. , Cell 181, 747 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Oh DY et al. , Cell 181, 1612–1625.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen CS et al. , Nature 576, 465–470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sade-Feldman M et al. , Cell 176, 404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu TD et al. , Nature 579, 274–278 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Miller BC et al. , Nat. Immunol 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yost KE et al. , Nat. Med 25, 1251–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tirosh I et al. , Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira G et al. , Nature 596, 119–125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caushi JX et al. , Nature 596, 126–132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valpione S et al. , Nat. Commun 12, 4098 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagaev DV et al. , Nucleic Acids Res. 48, D1057–D1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin N et al. , Clin. Cancer Res 27, 5084–5095 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y-C et al. , J. Immunother. Cancer 9, e002595 (2021).34321276 [Google Scholar]
- 44.Paria BC et al. , J. Immunother 44, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishna S et al. , Science 370, 1328–1334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghorani E et al. , Nat. Cancer 1, 546–561 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu-Trantien C et al. , JCI Insight 2, 91487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denton AE et al. , J. Exp. Med 216, 621–637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabrita R et al. , Nature 577, 561–565 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Joshi NS et al. , Immunity 43, 579–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alspach E et al. , Nature 574, 696–701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui C et al. , Cell 184, 6101–6118.e13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Litchfield K et al. , Cell 184, 596–614.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang W et al. , Nat. Med 25, 767–775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roudko V et al. , Cell 183, 1634–1649.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gee MH et al. , Cell 172, 549–563.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.