Abstract
Objectives
To perform an exploratory study to identify a JDM serum metabolic profile that differs from healthy controls (HCs) and responds to immunosuppressive treatment.
Methods
Blood was collected from 9 HCs and 10 patients diagnosed with probable (n = 4) or definite (n = 6) JDM based on the criteria of Bohan and Peter for myositis, with 7 of the 10 providing longitudinal samples following initiation of treatment; these patients comprised the treatment-naïve cohort. Sera underwent mass spectroscopy–based measurements of targeted metabolic intermediates, including 15 amino acids, 45 acylcarnitines (ACs), 15 ceramides and 29 sphingomyelins. Principal components analysis reduced metabolites into smaller sets of factors each comprised of correlated metabolic intermediates. Factor scores and metabolite concentrations were compared with HCs using two-sample t-tests while treatment effects were evaluated using paired t-tests.
Results
Of eight principal components analysis–derived metabolite factors (one AC, two amino acids, three sphingosine and two ceramide), two were significantly associated with JDM: one AC factor containing mostly long-chain ACs (P = 0.049) and one ceramide factor (P < 0.01). For 12 individual ACs, mostly long chain, and three ceramides, concentrations were significantly greater for JDM than HCs. Factors based on these individual metabolites showed decreasing scores with treatment (P = 0.03 and P < 0.01, respectively).
Conclusion
While additional validation is needed, these lipids have potential as JDM serum diagnostic and/or treatment biomarkers. Additionally, the significant association of long-chain ACs and ceramides with JDM offers insights regarding pathogenesis, implicating dysregulation of mitochondrial fatty acid β-oxidation.
Keywords: JDM, metabolomics, biomarkers, principal components analysis, fatty acid oxidation, acylcarnitines, ceramide, insulin resistance
Rheumatology key messages.
JDM is a rare inflammatory disease affecting 2–3/106 children per year.
Metabolic studies in JDM are limited and there are no studies comparing treatment-naïve to control subjects.
A unique serum metabolomic profile consisting of acylcarnitines and ceramides was identified to be responsive to immunosuppression.
Introduction
JDM is a rare, autoimmune, vasculopathic syndrome characterized primarily by inflammation of skeletal muscle, leading to impaired function identified clinically as weakness, decreased endurance and fatigability, most notably of proximal muscle groups. Despite advances in identifying factors involved in establishing the circumstances that result in JDM, many aspects of its pathogenesis remain elusive. Optimal approaches to diagnosing, staging and treating this disease are yet to be determined such that JDM remains a life- and organ-threatening disease with significant morbidities [1]. Further complicating efforts to develop a treatment strategy is the considerable heterogeneity of disease presentation, response to treatment and course, with this variability reflected in the lack of specific and consistent biomarkers [2].
Our current understanding of JDM implicates pathogenic mechanisms involving cellular pathways that elicit both immune and non-immune responses, with skeletal muscle a primary target of these mechanisms resulting in the clinical, histopathologic and biochemical features of JDM [3, 4]. Skeletal muscle, with its insulin-dependent mitochondria-rich myocytes, is a primarily energy-consuming tissue that plays a major role in maintaining metabolic homeostasis through its utilization of glucose, free fatty acids (FAs) and amino acids (AAs) [5–7]. To date, magnetic resonance spectroscopy has been the most extensively used modality to assess altered skeletal muscle mitochondrial bioenergetics and metabolites in the juvenile myopathies, including JDM [8–10] and PM [10]. Application of a newer, more sensitive ‘omics-based’ platform is warranted to gain new insights into the pathogenesis and allow identification of metabolic signatures. As high-throughput technologies have evolved, omics platforms have been employed in the search for biomarkers in multiple classes of disease, including neoplastic, infectious and inflammatory [11–13]. Most omic investigations in JDM have involved genomic and proteomic analyses [14–16].
The amalgamation of systems biology approaches and omics technologies has transformed the ability to quantify low molecular weight molecules using metabolomics; energy-yielding substrates and their metabolic intermediates can be measured using a targeted metabolomics approach. The strength of a metabolomic analysis vs other omics lies in its capacity to acquire a ‘snapshot’ of the physiology of a cell or tissue by evaluating the levels of small molecule intermediaries and end products of metabolic pathways. Altered lipid profiles have been identified by metabolomic analysis in inherited and acquired conditions, as well as chronic inflammatory diseases [13, 17–21]. In a metabolomic analysis of serum from adult DM and PM patients taken at baseline compared with healthy controls (HCs), as well as longitudinally following treatment, altered lipid profiles were identified consistent with dysregulated lipid metabolism, with a response of profiles to immunosuppressive treatment [22]. To the best of our knowledge, no study has been published characterizing the serum metabolic profile of patients with treatment-naïve (TN) JDM compared with HCs or that followed the TN cohort after treatment initiation to measure treatment effect on the metabolic profile. To that end, in this pilot study we compared serum targeted metabolic profiles of probable and definite JDM with those of HCs. In addition, to characterize the impact of therapy we measured metabolic changes after treatment initiation. Together, our results indicate JDM exhibits a treatment-responsive dysregulation of mitochondrial β-oxidation of long-chain FAs, suggesting novel diagnostic and treatment response biomarkers.
Methods
Ethics approval
Research on human subjects was conducted in accordance with the Declaration of Helsinki, using protocols approved by the Institutional Review Boards of Duke University. All patients or their parents gave fully informed written consent to participate and provide biologic samples.
Patients and samples
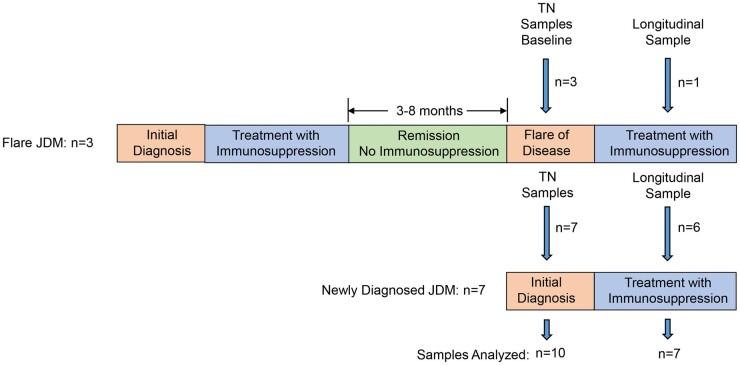
Participants included 10 patients diagnosed with probable (n = 4) or definite (n = 6) JDM using the criteria of Bohan and Peter for myositis [23]. The diagnosis of probable JDM was based on meeting clinical and laboratory criteria alone while those fulfilling criteria for definite disease had EMG and/or skeletal muscle biopsy consistent with JDM. These 10 patients constituted the TN group (Fig. 1). Seven of the ten were newly diagnosed and had not received prior immunosuppression. The remaining three were previously diagnosed with, and treated for, JDM and had experienced a disease flare after a clinical remission off medications for a period ranging from 8 to 53 months. Baseline samples were obtained from all 10 patients within the TN group. Longitudinal samples were available for six newly diagnosed and one flare patient; the time from baseline to longitudinal sample attainment ranged from 3 to 8 months.
Fig. 1.
Schematic representation of disease course and sample collection in 10 JDM patients comprising the TN cohort
Ten gender- and age-matched (±12 months) controls without inflammatory musculoskeletal disease or a history of chronic disease were recruited from Duke paediatric clinics presenting for well-child visits (n = 5) and from the Duke paediatric rheumatology clinic, where they were diagnosed with non-inflammatory arthralgias secondary to mechanical factors, i.e. joint hypermobility (n = 5). All were assessed for acute illness and were found to be healthy at the time of sample acquisition.
Venous blood was collected in serum separator tubes from patients and HCs. After standing for 30 min, samples were centrifuged at 1200 rpm for 15 min at room temperature for serum separation. Samples were stored at −80°C until they were analysed; no samples had undergone a freeze–thaw cycle until use for this study. One control sample was lost in processing.
Serum metabolomic analysis
The first analysis compared metabolic profiles from TN sera vs HC sera. The second, a longitudinal analysis, included seven patients from the initial analysis; one of the seven (patient 7 in Tables 1 and 3) was the only flare patient with a longitudinal sample available at the time of analysis. Stable isotope dilution and mass spectrometry (MS) techniques were used to measure 104 small molecule metabolites using a targeted MS-based metabolic platform including AAs (n = 15), acylcarnitines (ACs; n = 45), ceramides (n = 15) and sphingomyelins (n = 29), metabolites related to skeletal muscle metabolism. A targeted approach was applied, as it allows for absolute quantification of metabolite concentrations. Analyses were performed in a single batch in the Duke Molecular Physiology Core. AA and AC measures were made by flow injection tandem MS as previously described [24, 25]. Data were acquired using a Micromass Quattro Micro liquid chromatography–MS system running MassLynx 4.0 software (Waters, Milford, MA, USA). Ceramides and sphingomyelins were extracted as described previously [26, 27] and analysed by flow injection tandem MS for precursors of m/z 264 and m/z 184, respectively, using a Xevo TQS mass spectrometer (Waters).
Table 1.
Demographic, clinical and pathologic characteristics of studied JDM patients
| Patient number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Sex | F | F | F | F | F | F | F | F | F | M |
| Age, years | 4 | 7 | 16 | 16 | 3 | 4 | 14 | 15 | 14 | 14 |
| TN or flare | TN | TN | TN | TN | TN | TN | Flare | TN | Flare | Flare |
| Disease duration, months | 5 | 4 | 6 | 4 | 3 | 4 | 0.5 a | 3 | 0.75 a | 0.75 a |
| Definite or probable JDM | Probable | Definite | Definite | Definite | Probable | Definite | Definite | Definite | Probable | Probable (patient amyopathic) |
| Medications during disease courseb | S/I/P/M/H | S/I/P/M/H | S/I/P/M/R | S/I/P/M/R | S/I/P/M | S/I/P/M | S/I/P/M | P/M/H | P/M | P/M/H |
| Medications initiated with flare | — | — | — | — | — | — | S/I/P/M | — | P/M | P/M/H |
| BMI/weight (kg) prior to treatment | 14.8/16.1 | 14.8/23 | 29/75.1 | 18.5/45.3 | 15.8/15.8 | 21/26.7 | 21.4/50.5 | 18.6/49 | 15/36.3 | 16.9/46.2 |
| BMI/weight (kg) at longitudinal sample | 15.3/19.2 | 15.8/25.4 | 39.5/104.1 | 17.4/43.1 | 18.8/20.1 | 28.8/30.7 | 20.1/52.8 | NA | NA | NA |
| Muscle biopsy | ND | D | D | D | ND | D | D | D | ND | ND |
| Biopsy findingsc | ND | M, P, T | M, P | M, P | ND | M | M, P, T | P, T, MI | ND | ND |
| MRI | D, ab | ND | D, ab | D, ab | D, ab | D, ab | D, ab | D, ab | D, ab | D, nl |
| EMG | ND | ND | D, ab | ND | ND | D, ab | D, ab | ND | ND | ND |
| Rash present | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CK, U/L | 80 | 132 | 49 | 5759 | 191 | 6668 | 25 | 1277 | 96 | 182 |
| Myositis-specific antibody | UD | UD | UD | MI-2 | PM/SCL | HMG-CoA | UD | NXP2 | TIF-1ɣ | UD |
D: done; ND: not done; nl: normal; ab: abnormal; CK: creatinine kinase; UD: undetected.
Baseline samples were obtained from all patients; longitudinal samples were available from those patients indicated by bold number.
Onset of flare to treatment in months.
Medications during disease course: S: solumedrol; I: IVIG; P: prednisone; M: methotrexate; R: rituximab; H: hydroxychloroquine.
Skeletal muscle biopsy findings: M: major histocompatibility complex I upregulation; P: perifascicular atrophy; T: tubuloreticular inclusions; MI: microinfarction.
Table 3.
Longitudinal decline in AC and ceramide component scores following treatment
| Baseline AC analysis | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7a |
|---|---|---|---|---|---|---|---|
| Baseline ceramide analysis | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7a |
| Months to longitudinal sampling | 8.0 | 5.5 | 3.0 | 4.5 | 4.0 | 4.0 | 8.0 |
| Baseline sample score | 0.117 | 0.410 | 1.743 | 2.236 | 0.894 | −−0.322 | −0.370 |
| Follow-up sample score | −0.937 | −1.264 | −1.315 | −0.890 | −0.500 | 0.058 | −0.457 |
| Months to longitudinal sampling | 8.0 | 5.5 | 2.0 | 4.5 | 4.0 | 4.0 | 8.0 |
| Baseline | −0.652 | −0.505 | 1.542 | 1.140 | −0.592 | −0.152 | 1.721 |
| Follow-up | −0.706 | −1.746 | 0.266 | −0.487 | −1.256 | −0.332 | −0.298 |
In baseline analysis of the AC data (Table 2), 12 AC concentrations were significantly greater in the TN group. In longitudinal analyses, a single PCA-derived factor explaining 51% of the variation in these ACs decreased by 1.43 .s.d. (P = 0.03) during treatment. Mean decline of −1.43 s.d. (P = 0.03).
Baseline ceramide analysis revealed three of four concentrations higher in the TN group at baseline (C24:1, d18-1-C18 and d18-1-C24-1). These loaded on a single PCA-derived factor. In longitudinal analyses, this factor, which explained 78% of the variance, decreased by 0.98 s.d. (P < 0.01) during treatment. Mean decline of −0.97 s.d. (P < 0.01).
Flare patient.
Statistical analysis
To identify metabolite associations and reduce multiple comparisons we employed principal components analysis (PCA) on log-transformed, standardized metabolite measurements separately to reduce the dimensionality of data to a limited number of principal component (PC) factors. This technique has been used previously, including a study by Kraus et al. [19] based on 739 observations considering the same metabolites presented here [20]. Individual metabolites with a factor load of ≥0.6 are reported as composing a given factor. Interpretable factors (those accounting for a large amount of the variance) as well as individual metabolites were assessed for their association with JDM using t-tests. In additional analyses we used regression controls for baseline BMI and change in BMI to assess whether any associations between JDM and our outcome variables were due to BMI.
Results
Patient characteristics
Table 1 shows the clinical and pathologic features of TN patients. Patients’ ages ranged from 4 to 16 years at diagnosis (mean 7.8). All patients had rash at diagnosis; one patient’s disease course (patient 10) has been amyopathic. Myositis-specific antibodies were identified in five patients (MI-2, PM/SCL, HMG-CoA, NXP2 and TIF-1ɣ); this 50% myositis-specific antibody positivity is consistent with published values [28, 29]. Muscle biopsy was performed in six patients and EMG in three who had also been biopsied.
Immunomodulatory medications received during the JDM disease course up to procurement of longitudinal serum samples are listed in Table 1. Prior to experiencing a flare of their disease, patients 7, 9 and 10 had been weaned from either methotrexate (patients 7 and 9) or methotrexate/hydroxychloroquine (patient 10) following at least 24 months of clinical remission on these medications alone. Two of the flare patients (patients 7 and 9) also received methylprednisolone and prednisone; additionally, patient 9 received IVIG. Flare patients had been off all immunomodulatory medications for at least 8 months prior to developing flare symptoms.
At longitudinal serum sampling, there were seven patient samples available from the TN cohort: six newly diagnosed patients (patients 1–6) and one flare patient (patient 7). All longitudinal samples were obtained at a time when patient disease activity was improving on medication as assessed clinically and by laboratory measures.
No patients or HC subjects were on glucocorticoid treatment for another disease (i.e. asthma) or other immunosuppression at the time of serum sampling. Due to the presence of one obese subject in the control group, mean BMI was higher among HCs compared with TN patients (23.7 vs 18.5; P = 0.11). Mean BMI increased from 19.7 to 22.5 (P = 0.15) at the time of the longitudinal sample during treatment.
Baseline metabolomics analysis, TN vs HC serum
Eight PCA-derived metabolite factors were identified from serum and included one AC factor, two AA factors, three sphingosine factors and two ceramide factors. Of these eight factors, two—one AC and one ceramide factor—were significantly associated with JDM.
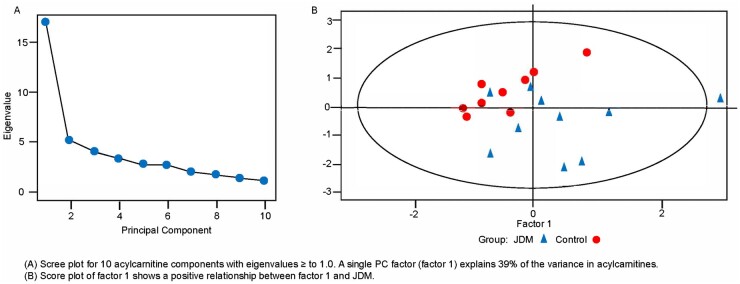
A single PCA factor (PC factor 1) explained 39% of the variance in ACs and was composed of 26 ACs that loaded ≥0.6 on PC factor 1 (Table 2, in bold text). These included 14 long-chain ACs (LCACs; 14–20 carbon atoms), 8 medium-chain ACs (8–12 carbon atoms) and 4 short-chain ACs (2–6 carbon atoms). PC factor 1 scores were significantly greater in the TN group compared with the HC group by 0.89 s.d. (P = 0.049). Fig. 2A shows the eigenvalues for the 10 AC components with eigenvalues ≥1. The score plot (Fig. 2B) shows a positive relationship between PC factor 1 and JDM. Statistically controlling for BMI did not change this positive relationship between JDM and PC factor 1. As BMI was unrelated to factor 1, it was dropped from our final models.
Table 2.
HC and TN means on acylcarnitine factor 1 and concentrations (N = 45)
| Variable name | Concentration, μM, meana |
P-value | Variable name | Concentration, μM, meana |
P-value | ||
|---|---|---|---|---|---|---|---|
| HCs | JDM (TN) | HCs | JDM (TN) | ||||
| Factor 1 | −0.470 | 0.423 | 0.049* | C12:1 | −0.312 | 0.281 | 0.206 |
| C2b | −0.361 | 0.325 | 0.140 | C12-OH/C10-DC | −0.319 | 0.287 | 0.196 |
| C3 | −0.340 | 0.306 | 0.166 | C14 | −0.406 | 0.365 | 0.094 |
| C4/Ci4 | −0.170 | 0.153 | 0.480 | C14:1 | −0.390 | 0.351 | 0.108 |
| C4-OH | −0.500 | 0.450 | 0.040* | C14:2 | −0.212 | 0.191 | 0.395 |
| C4-DC/Ci4-DC | −0.372 | 0.335 | 0.120 | C14:1-OH | −0.146 | 0.131 | 0.562 |
| C5 | −0.073 | 0.066 | 0.760 | C14-OH/C12-DC | −0.155 | 0.140 | 0.536 |
| C5:1 | 0.187 | −0.168 | 0.456 | C16 | −0.440 | 0.396 | 0.060 |
| C5-DC | 0.078 | −0.070 | 0.758 | C16-OH/C14-DC | −0.513 | 0.462 | 0.030* |
| C5-OH/C3-DC | −0.514 | 0.463 | 0.029* | C16:1 | −0.484 | 0.436 | 0.041* |
| C6 | −0.571 | 0.514 | 0.013* | C16:1-OH/C14:1-DC | −0.430 | 0.387 | 0.074 |
| C7-DC | −0.086 | 0.077 | 0.734 | C16:2 | −0.255 | 0.229 | 0.306 |
| C8 | −0.210 | 0.189 | 0.400 | C18 | −0.672 | 0.605 | 0.002** |
| C8:1 | 0.332 | −0.299 | 0.177 | C18-OH/C16-DC | −0.524 | 0.472 | 0.030* |
| C8:1-DC | 0.053 | −0.048 | 0.833 | C18:1 | −0.640 | 0.576 | 0.004** |
| C8-OH/C6-DC | −0.038 | 0.034 | 0.880 | C18:1-DC | −0.546 | 0.491 | 0.020* |
| C8:1-OH/C6:1-DC | 0.547 | −0.492 | 0.019* | C18:1-OH/C16:1-DC | −0.533 | 0.480 | 0.020* |
| C10 | −0.154 | 0.139 | 0.540 | C18:2 | −0.283 | 0.255 | 0.240 |
| C10:1 | −0.019 | 0.017 | 0.942 | C18:2-OH | −0.196 | 0.177 | 0.433 |
| C10:2 | 0.226 | −0.204 | 0.364 | C20 | −0.363 | 0.326 | 0.130 |
| C10:3 | 0.377 | −0.339 | 0.122 | C20-OH/C18-DC | −0.513 | 0.461 | 0.029* |
| C10-OH/C8-DC | −0.096 | 0.086 | 0.703 | C20:4 | 0.077 | −0.069 | 0.760 |
| C12 | −0.250 | 0.225 | 0.315 | C22 | −0.181 | 0.163 | 0.690 |
Twenty-six of 45 acylcarnitine concentrations (in bold text) loaded >0.6 on a PCA-derived factor (PC factor 1) composed primarily of LCACs. PC factor 1 scores were significantly greater in the TN patients compared with the HCs (P = 0.049). Twelve individual mostly LCAC concentrations were significantly higher in the TN group.
Mean concentrations are based on data logged to the base 10 then standardized.
Acylcarnitines loading ≥0.60 are in bold.
P < 0.05,
P < 0.01.
Fig. 2.
Scree and score plots of serum acylcarnitines in subjects, JDM vs HCs
(A) Scree plot for 10 acylcarnitine components with eigenvalues ≥1.0. A single PC factor (factor 1) explains 39% of the variance in acylcarnitines. (B) Score plot of PC factor 1 shows a positive relationship between PC factor 1 and JDM.
Individual assessments indicated that 12 ACs had significantly greater concentrations in the TN group, with standardized mean differences >0.8. These included two short-chain ACs (C4-OH and C5-OH/C3-DC), two medium-chain ACs (C6 and C8:1-OH/C6:1-DC) and eight LCACs (C16-OH/C14-DC, C16:1, C18, C18-OH/C16-DC, C18:1, C18:1-DC, C18:1-OH/C16:1-DC and C20-OH/C18-DC). Eight of these also loaded ≥0.6 on PC factor 1, with six of the eight being LCACs.
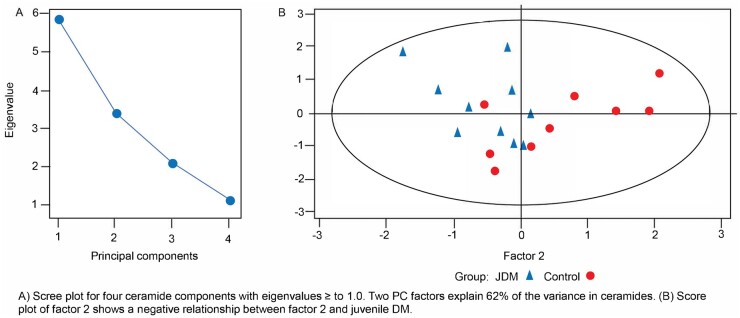
In a PCA of serum ceramides, two PC factors accounted for 62% of the variance (see Supplementary Table S1, available at Rheumatology online). The PC factor 1 score, with high loadings of nine ceramides, was not significantly associated with TN patients vs HCs. PC factor 2, with high loadings of four C26-related ceramides, was negatively associated with TN group membership (P < 0.01). Fig. 3A shows eigenvalues for the four ceramide components with eigenvalues ≥1, while the score plot in Fig. 3B shows a negative relationship between PC factor 2 and JDM. In individual assessments, the concentrations of ceramides C23, C24:1 and C24 (d18:1/24:1) were higher in the TN group (P < 0.05) and C18 (d18:1/C18:0) trended in the same direction (P < 0.059, in bold text in Supplementary Table S1, available at Rheumatology online). Controlling for BMI did not change the negative relationship between TN and PC factor 2 and BMI was again dropped from our final models.
Fig. 3.
Scree and score plots of serum ceramides in subjects, JDM vs HCs
(A) Scree plot for four ceramide components with eigenvalues ≥1.0. Two PC factors explain 62% of the variance in ceramides. (B) Score plot of PC factor 2 shows a negative relationship between PC factor 2 and JDM.
Three PC factors explained 77% of the variance noted in sphingosine items (see Supplementary Table S2, available at Rheumatology online). PC factor 1 contained 22 of 32 sphingosines loading ≥0.6. Sphingosines C35:3, C43:3, C45 and C45:1 loaded ≥0.6 on PC factor 2, while sphingosines C44:1 and C44:2 loaded ≥0.6 on PC factor 3. No significant associations were present between sphingosine factors and JDM. Five individual TN sphingosine concentrations—C35:3, C38:1, C42:1, C43:1 and C43:3—were significantly less than in HCs, with mean differences >0.80. Sphingosine concentrations of C37:2, C40:1, C45 and C45:1 were also lower and trending towards significance.
Two AA PC factors explained 54% of the AA variance; however, neither of these factor scores differed for JDM and HCs. Similarly, none of the individual AAs were related to JDM in individual t-tests. As neither the sphingosine nor the AA factors were associated with JDM, we do not present scree or score plots for these analyses.
Longitudinal analysis of serum acylcarnitines and ceramides
Longitudinal data were available for 7 of 10 members of the TN group (6 new diagnosis and 1 flare); longitudinal samples were not collected for HCs. In baseline analysis of the AC data, 12 AC concentrations (C4-OH, C5-OH/C3-DC, C6, C8:1-OH/C6:1-DC, C16-OH/C14-DC, C16-1, C18, C18-0H/C16-DC, C18-1, C18-1-DC, C18-1-OH/C16-1-DC and C20-OH/C18-DC) were significantly greater in the TN group. In longitudinal analyses, a single PCA-derived factor explaining 51% of the variation in these ACs decreased by 1.43 s.d. (P = 0.03) during treatment (Table 3). Factor scores declined for six of seven samples, including the only flare sample (patient 7). For the ceramides, three of four concentrations noted to be higher in the TN group at baseline [C24:1, C18 (d18:1/18:0) and C24:1 (d18:1/24:1)] loaded on a single PCA-derived factor. In longitudinal analyses, this factor, which explained 78% of the variance, decreased by 0.98 s.d. (P < 0.01) during treatment. As with the ACs, factor scores declined for six of seven samples.
Discussion
In this cross-sectional and longitudinal pilot analysis we provide preliminary evidence that serum from patients with TN JDM differs from HCs on two PCA-based metabolic profiles at diagnosis. Further, these metabolic differences decline with treatment. At diagnosis, TN patients exhibited greater scores for an AC factor (AC factor 1; Table 2) composed mainly of LCAC items and greater concentrations of 12 individual AC species, again, mainly LCACs, as compared with HCs. TN patients had lower scores for a ceramide factor (factor 2; see Supplementary Table S1, available at Rheumatology online) and greater concentrations of four individual ceramide species that did not load on factor 2. Longitudinal analyses indicated that TN as compared with HC differences in ACs and ceramides declined with immunosuppressive treatment. As such, this profile has promise as a minimally invasive diagnostic and treatment-response biomarker. To our knowledge this is the first report of targeted serum metabolic profiles in JDM prior to and during treatment. In addition to their potential as biomarkers, these AC and ceramide metabolites may inform the pathogenesis of JDM, as they represent the physiologic state of a patient with new-onset disease prior to treatment.
In this study we found greater concentrations of ACs in patients compared with HCs, principally even chain number LCACs, indicating a disruption of early stages of mitochondrial FA β-oxidation. ACs are fatty acyl lipids and perform a principal role regulating the balance of skeletal muscle carbohydrate and lipid metabolism. As such, ACs are considered indicators of the state of mitochondrial FA β-oxidation, as they reflect cellular fluctuations in substrate supply and flux limitation of specific enzymes [30–33]. FAs are thioesterified to acetyl coenzyme A to form their cognate fatty acyl CoA esters; this step is required for FAs to enter a metabolic pathway. Mitochondrial oxidation of long-chain fatty acyl CoA (≥14 carbon atoms) requires the long-chain acyl CoA L-carnitine be transferred to L-carnitine. The resulting long-chain AC is transported into the mitochondrial matrix by the carnitine shuttle (see Supplementary Fig. S1, available at Rheumatology online). Within the mitochondria, long-chain FAs are converted to non-oxidized acyl-CoAs and these acyl-CoAs undergo chain shortening during successive rounds of β-oxidation. The acyl-CoA ester is converted to acetyl CoA molecules by shortening of the acyl-CoA by two carbons per cycle [32, 34]. When mitochondrial FA β-oxidation is disrupted, non-oxidized acyl-CoA esters of various chain lengths accumulate. In this situation, acyl-CoAs and their chain-shortened derivatives can be transferred back to L-carnitine, forming the corresponding ACs [32, 34]. This allows for transport of these ACs out of the mitochondria and eventually from the cell where they may undergo alternative pathways of oxidation; various ACs and their metabolites may be detected in the blood and/or urine [34, 35].
The circulating metabolic profile we identified for TN patients supports a model whereby disruptions in skeletal muscle mitochondrial β-oxidation lead to an increase in acyl CoAs that, impeded from further β-oxidation, are converted to other metabolites including ceramides and diacylglycerols or transferred back to L-carnitine to form ACs, some of which are oxidized in the liver by ω-oxidation, a ‘rescue’ pathway [34–36]. In addition to increased circulating ACs, we report an increase in specific ceramides [C23, C24:1, C24:1 (d18:1/24:1) and C18:0 (d18:1/18:0)] and dicarboxyacylcarnitines (C8:1-OH/C6:1-DC, C16-OH/C14-DC and C18:1-OH/C16:1-DC), all of which decrease following initiation of immunosuppression, suggesting a return of homeostatic metabolism. Our results are consistent with findings in an adult DM and PM study evaluating the lipid profiles from newly diagnosed patients as compared with HCs in which the reported profiles are altered compared with HCs and respond to immunosuppressive treatment [19]. They are also consistent with earlier metabolic studies in JDM using P31 magnetic resonance spectroscopy in which subjects with JDM were found to have decreased levels of adenosine triphosphate, suggesting that the acetyl-CoA produced by mitochondrial FA β-oxidation is not available for oxidative phosphorylation [15].
The conditions that lead to dysregulation and disruption of mitochondrial FA β-oxidation in skeletal muscle have been most widely investigated in obesity, type 2 diabetes mellitus and cardiovascular disease [11, 13, 17, 30]. Common factors cited as instigating and perpetuating derangement are lipotoxicity and insulin resistance [37]. Lipotoxicity is the storage of excess lipid in non-adipose tissue, including skeletal muscle, and can lead to changes in metabolic flux that have deleterious cellular effects, including activation of endoplasmic reticulum (ER) stress responses, inflammation and insulin resistance; these lipids include fatty acyl-CoAs and ceramide [37–39]. Conversely, ER stress is a critical regulator of lipid biosynthesis [38, 40]. In JDM, activation of ER stress responses is considered a non-immune mechanism of skeletal muscle injury [12]. Pro-inflammatory cytokines, including IL-6 and TNF-α, play a role in metabolic disease through their effects on signalling pathways leading to insulin resistance [13, 37]. These cytokines play a significant pro-inflammatory role in JDM pathogenesis [7, 41] such that it could be hypothesized that inflammation affects mitochondrial function in JDM.
The value of this pilot study is 2-fold. First, we identify a metabolic profile comprised of lipids that discriminate individuals with TN JDM from HCs and, as indicated in the longitudinal analyses, demonstrate a treatment response in patients with reductions in ACs and ceramides. Second, our results inform disease pathogenesis, opening new avenues for investigation. Nevertheless, several weaknesses should be mentioned. This analysis is exploratory and based on a small number of patients with a rare, clinically and pathogenically heterogeneous disease. We did not include other inflammatory disease conditions as controls; our results may not be specific to JDM and, in additional studies, other inflammatory disease controls are warranted. However, in a metabolomic analysis of juvenile-onset SLE, no specific AC or ceramide profile was identified [42]. Additionally, we did not employ specific disease measures to determine disease status at the time of the longitudinal samples.
There are variables that affect metabolism, including diet and fed/fasting status, pubertal status and medications, for which we did not specifically control. In this study, as all samples were randomly collected with respect to fed/fasting state and regular diet, we expect these variables to influence metabolites similarly for both groups (JDM and HCs). In the absence of a controlled fed/fasting state or diet, overall metabolite variation will be much greater, likely increasing the chance of a type 2 error (failing to uncover significant associations) while reducing the chance of a type 1 error (false positive). Furthermore, using regression controls for baseline BMI and change in BMI, it was determined that associations between JDM and our outcome variables were not due to BMI and BMI did not influence our longitudinal results. Pubertal status was known for all study individuals except for controls seen in the clinic (n = 5); therefore, 5 of 10 were matched for pubertal status and all were age- and gender-matched. Prior to diagnosis, study patients were not on immunosuppressive medications or medications known to alter FA metabolism, including those indicated for treatment of diabetes (type 1 or 2), depression, seizure disorders or inflammatory disease. During treatment they received a variety of immunosuppressive medications, including glucocorticoids and subcutaneous methotrexate. It is well known that cellular responses to inflammation include shifts in metabolic pathways in immune effector cells as well as tissue cells such as hepatocytes and myocytes [43]. The effects of immunosuppression on metabolic changes are likely dose and duration dependent and controlling for these variables will be important when investigating the value of metabolic biomarkers in assessing disease activity, medication response and medication toxicity [44, 45]. Although we did not control for these variables, our objective in this study was to see if there was a change in baseline signatures following treatment and to use this data to inform future metabolic studies in JDM where these factors will be studied.
Our results appear to implicate multiple pathways involved in maintaining metabolic and biosynthetic homeostasis in the face of JDM. Further delineation of systems involved in initiating dysregulated lipid metabolism as well as those leading to the deleterious consequences of JDM will benefit from a combined lipidomic/transcriptomic analysis. Interrogation of genes involved in lipid metabolism, oxidative phosphorylation, ER stress and stress responses, apoptosis and mitophagy will add to our knowledge of pathogenesis. To better understand the value of our metabolic profile, it is important to perform a lipidomic study in a larger cohort of juvenile myositis patients controlling for covariates considered to affect metabolism and disease activity. A second study would ideally include patients in remission off medication and those with active disease on treatment, with analyses using disease activity measures to further correlate treatment effect to disease stage. Other inflammatory diseases (i.e. systemic lupus) should be included as disease controls. This study will require a large number of individuals; it will, by necessity, be a collaborative study.
In summary, this pilot study has provided evidence of an altered lipid profile in TN patients with JDM compared with HCs that shows a significant response to immunosuppressive therapy in longitudinal analyses. As this is an exploratory study and we have not adjusted for multiple comparisons, our findings are provisional and will require corroboration. Our data show an increase in ACs and ceramides that we propose are derived from excess lipid present in skeletal muscle due to dysfunction and dysregulation of lipid metabolism, a potential mechanism of disease pathogenesis in JDM.
Supplementary Material
Acknowledgements
The authors thank Ca’ Lecia Fleming for her excellent handling of patient samples and work obtaining patient consent and clinical data collection. We also would like to thank Drs Michael Muehlbauer and Tim Koves for their expertise in preparing samples for analysis.
J.A.D., K.M.H., A.M.R. and D.S.P. were involved in the conception and design of the study. L.L. analysed and interpreted the data. J.A.D. and K.M.H. contributed to the interpretation of the results. J.A.D. wrote the first draft and K.M.H., L.L., D.S.P. and A.M.R. made critical revisions to the manuscript. O.I. participated in the acquisition of data and laboratory studies and reviewed the manuscript. All authors read and approved the final manuscript.
Funding: This work was supported by the Cure Juvenile Myositis Foundation.
Disclosure statement: The authors declare no conflicts of interest.
Data availability statement
The data that support the findings of this study are available from the corresponding author on request.
Supplementary data
Supplementary data are available at Rheumatology online.
Contributor Information
Jeffrey A Dvergsten, Department of Pediatrics, Duke Children’s Hospital, Duke University Medical Center.
Ann M Reed, Department of Pediatrics, Duke Children’s Hospital, Duke University Medical Center.
Lawrence Landerman, Department of Pediatrics, Duke Children’s Hospital, Duke University Medical Center.
David S Pisetsky, Department of Medicine and Immunology, Duke University Medical Center and Research Service, Durham VA Medical Center.
Olga Ilkayeva, Department of Medicine, Duke Molecular Physiology Institute, Duke School of Medicine, Durham, NC, USA.
Kim M Huffman, Department of Medicine, Duke Molecular Physiology Institute, Duke School of Medicine, Durham, NC, USA.
References
- 1. Feldman BM, Rider LG, Reed AM, Pachman LM. Juvenile dermatomyositis and other idiopathic inflammatory myopathies of childhood. Lancet 2008;371:2201–12. [DOI] [PubMed] [Google Scholar]
- 2. Wu Q, Wedderburn LR, McCann LJ. Juvenile dermatomyositis: latest advances. Best Pract Res Clin Rheumatol 2017;31:535–57. [DOI] [PubMed] [Google Scholar]
- 3. Nagaraju K, Lundberg IE. Polymyositis and dermatomyositis: pathophysiology. Rheum Dis Clin N Am 2011;37:159–71. [DOI] [PubMed] [Google Scholar]
- 4. Wienke J, Deakin CT, Wedderburn LR, van Wijk F, van Royen-Kerkhof A. Systemic and tissue inflammation in juvenile dermatomyositis: from pathogenesis to the quest for monitoring tools. Front Immunol 2018;9:2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marette A, Liu Y, Sweeney G. Skeletal muscle glucose metabolism and inflammation in the development of the metabolic syndrome. Rev Endocr Metab Disord 2014;15:299–305. [DOI] [PubMed] [Google Scholar]
- 6. Kuzmiak-Glancy A, Willis WT. Skeletal muscle fuel selection occurs at the mitochondrial level. J Exp Biol 2014;217:1993–2003. [DOI] [PubMed] [Google Scholar]
- 7. Yaribeygi H, Farrokhi FR, Butler AE, Sahebkar A. Insulin resistance: review of the underlying molecular mechanisms. J Cell Physiol 2019;234:8152–61. [DOI] [PubMed] [Google Scholar]
- 8. Park JH, Kari S, King LE, Olsen NJ. Analysis of 31P MR spectroscopy data using artificial neural networks for longitudinal evaluation of muscle disease: dermatomyositis. NMR Biomed 1998;11:245–56. [DOI] [PubMed] [Google Scholar]
- 9. Park JH, Niermann KJ, Ryder NM et al. Muscle abnormalities in juvenile myositis patients. Arthritis Rheum 2000;43:2359–67. [DOI] [PubMed] [Google Scholar]
- 10. Chung Y-L, Rider LG, Bell JD et al. Muscle metabolites, detected in urine by proton spectroscopy, correlate with disease damage in juvenile idiopathic inflammatory myopathies. Arthritis Rheum 2005;53:565–70. [DOI] [PubMed] [Google Scholar]
- 11. Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci 2019;20:4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ludwig KR, Hummon AB. Mass spectrometry for the discovery of biomarkers of sepsis. Mol Biosyst 2017;13:648–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rinaldo P, Cowan TM, Matern D. Acylcarnitine profile analysis. Genet Med 2008;10:151–6. [DOI] [PubMed] [Google Scholar]
- 14. Miller FW, Chen W, O’Hanlon TP et al. Genome-wide association study identifies HLA 8.1 ancestral haplotype alleles as major genetic risk factors for myositis phenotypes. Genes Immun 2015;16:470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bilgic H, Ytterberg SR, Amin S et al. Interleukin-6 and type I interferon-regulated genes and chemokines mark disease activity in dermatomyositis. Arthritis Rheum 2009;60:3436–46. [DOI] [PubMed] [Google Scholar]
- 16. Wienke J, Bellutti Enders F, Lim J et al. Galectin-9 and CXCL-10 as biomarkers for disease activity in juvenile dermatomyositis: a longitudinal cohort study and multicohort validation. Arthritis Rheum 2019;71:1377–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aguer C, McCoin CS, Knotts TA et al. Acylcarnitines: potential implications for skeletal muscle insulin resistance. FASEB J 2015;29:336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruiz M, Labarthe F, Fortier A et al. Circulating acylcarnitine profile in human heart failure: a surrogate of fatty acid metabolic dysregulation in mitochondria and beyond. Am J Physiol Heart Circ Physiol 2017;313:H768–81. [DOI] [PubMed] [Google Scholar]
- 19. Kraus WE, Pieper CF, Huffman KM et al. Association of plasma small-molecule intermediate metabolites with age and body mass index across six diverse study populations. J Gerontol A Biol Sci Med Sci 2016;71:1507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunter WG, Kelly JP, McGarrah RW et al. Metabolomic profiling identifies novel circulating biomarkers of mitochondrial dysfunction differentially elevated in heart failure with preserved versus reduced ejection fraction: evidence for shared metabolic impairments in clinical heart failure. J Am Heart Assoc 2016;5:e003190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guma M, Tiziani S, Firestein G. Metabolomics in rheumatic disease: desperately seeking biomarkers. Nat Rev Rheumatol 2016;12:269–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raouf J, Idborg H, Englund P et al. Targeted lipidomics analysis identified altered serum lipid profiles in patients with polymyositis and dermatomyositis. Arthritis Res Ther 2018;20:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bohan A, Peter JB. Polymyositis and dermatomyositis. N Engl J Med 1975;292:344–7. [DOI] [PubMed] [Google Scholar]
- 24. An J, Muoio DM, Shiota M et al. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med 2004;10:268–74. [DOI] [PubMed] [Google Scholar]
- 25. Ferrara CT, Wang P, Neto EC et al. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet 2008;4:e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Merrill AH Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipodomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 2005;36:207–24. [DOI] [PubMed] [Google Scholar]
- 27. Huffman KM, Shah SH, Stevens RD et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care 2009;32:1678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rider LG, Nistala K. The juvenile idiopathic inflammatory myopathies: pathogenesis, clinical and autoantibody phenotypes, and outcomes. J Intern Med 2016;280:24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tansley SL, Simou S, Shaddick G et al. Autoantibodies in juvenile-onset myositis: their diagnostic value and associated clinical phenotype in a large UK cohort. J Autoimmun 2017;84:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab 2012;15:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fahy E, Cotter D, Sud M, Subramaniam S. Lipid classification, structures and tools. Biochim Biophys Acta 2011;1811:637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCoin CS, Knotts TA, Adams SH. Acylcarnitines—old actors auditioning for new roles in metabolic physiology. Nat Rev Endocrinol 2015;11:617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ribel-Madsen A, Ribel-Madsen R, Brons R et al. Plasma acylcarnitine profiling indicates increased fatty acid oxidation relative to tricarboxylic acid cycle capacity in young, healthy low birth weight men. Physiol Rep 2016;4:e12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adeva-Andany MM, Carneiro-Freire N, Seco-Filgueira M, Fernández-Fernández C, Mouriño-Bayolo D. Mitochondrial β-oxidation of saturated fatty acids in humans. Mitochondrion 2019;46:73–90. [DOI] [PubMed] [Google Scholar]
- 35. Wanders RJ, Komen J, Kemp S. Fatty acid omega-oxidation as a rescue pathway for fatty acid oxidation disorders in humans. FEBS J 2011;278:182–94. [DOI] [PubMed] [Google Scholar]
- 36. Ferreira NS, Engelsby H, Neess D et al. Regulation of very-long chain acyl chain ceramide synthesis by acyl-CoA-binding protein. J Biol Chem 2017;292:7588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Luca C, Olefsky JM. Inflammation and insulin resistance. FEBS Lett 2008;582:97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Herpen NA, Schrauwen-Hinderling VB. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol Behav 2008;94:231–41. [DOI] [PubMed] [Google Scholar]
- 39. Han J, Kaufman RJ. The role of ER stress in lipid metabolism and lipotoxicity. J Lipid Res 2016;57:1329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cao X, Lu X-M, Tuo X et al. Angiotensin-converting enzyme 2 regulates endoplasmic reticulum stress and mitochondrial function to preserve skeletal muscle lipid metabolism. Lipids Health Dis 2019;18:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reed AM, Peterson E, Bilgic H et al. Changes in novel biomarkers of disease activity in juvenile and adult dermatomyositis are sensitive biomarkers of disease course. Arthritis Rheum 2012;64:4078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson GA, Waddington KE, Coelewij L et al. Increased apolipoprotein-B:A1 ratio predicts cardiometabolic risk in patients with juvenile onset SLE. EBiomedicine 2021;65:103243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weyand CM, Zeisbrich M, Goronzy JJ. Metabolic signatures of T-cells and macrophages in rheumatoid arthritis. Curr Opin Immunol 2017;46:112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ellero-Simatos S, Szymanska E, Rullman T et al. Assessing the metabolic effects of prednisolone in healthy volunteers using urine metabolic profiling. Genome Med 2012;4:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker B, Bruce I. SLE and metabolic syndrome. Lupus 2013;22:1259–66. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on request.