Abstract
Simple Summary
RAS mutation is the most frequent oncogenic alteration in human cancers and KRAS is the most frequently mutated, notably in non-small cell lung carcinomas (NSCLC). Various attempts to inhibit KRAS in the past were unsuccessful in these latter tumors. However, recently, several small molecules (AMG510, MRTX849, JNJ-74699157, and LY3499446) have been developed to specifically target KRAS G12C-mutated tumors, which seems promising for patient treatment and should soon be administered in daily practice for non-squamous (NS)-NSCLC. In this context, it will be mandatory to systematically assess the KRAS status in routine clinical practice, at least in advanced NS-NSCLC, leading to new challenges for thoracic oncologists.
Abstract
KRAS mutations are among the most frequent genomic alterations identified in non-squamous non-small cell lung carcinomas (NS-NSCLC), notably in lung adenocarcinomas. In most cases, these mutations are mutually exclusive, with different genomic alterations currently known to be sensitive to therapies targeting EGFR, ALK, BRAF, ROS1, and NTRK. Recently, several promising clinical trials targeting KRAS mutations, particularly for KRAS G12C-mutated NSCLC, have established new hope for better treatment of patients. In parallel, other studies have shown that NSCLC harboring co-mutations in KRAS and STK11 or KEAP1 have demonstrated primary resistance to immune checkpoint inhibitors. Thus, the assessment of the KRAS status in advanced-stage NS-NSCLC has become essential to setting up an optimal therapeutic strategy in these patients. This stimulated the development of new algorithms for the management of NSCLC samples in pathology laboratories and conditioned reorganization of optimal health care of lung cancer patients by the thoracic pathologists. This review addresses the recent data concerning the detection of KRAS mutations in NSCLC and focuses on the new challenges facing pathologists in daily practice for KRAS status assessment.
Keywords: lung cancer, KRAS, molecular biology, algorithms, samples
1. Introduction
The most recent global report on the epidemiology of cancer diseases states that lung cancer has the highest mortality among 36 cancer types considered, and it is the second most frequently diagnosed cancer type around the world [1]. Therefore, in 2020, based on data collected from 185 countries, the number of diagnosed cases was estimated at more than 2.2 million cases (11.4% of all cancers), while mortality was around 1.8 million cases (18.0%) [1,2]. It is well known that the mortality of NSCLC is strongly associated with late diagnosis [1,3,4]. Among the lung cancer histological subtypes, non-small cell lung carcinoma (NSCLC) is the most frequent histological subtype, representing at least 85% of all cases [5]. The therapeutic strategy of advanced-stage or metastatic NSCLC, notably of non-squamous (NS)-NSCLC, has evolved dramatically in the last few years, thanks to the development of new targeted therapies and immune checkpoint inhibitors (ICIs) administered alone or in association with chemotherapy and targeted therapies [6,7,8]. These different therapies have significantly increased the overall survival of NS-NSCLC patients [9,10]. Therefore, several therapeutic molecules for routine clinical practice were rapidly granted marketing authorization by the Food and Drug Administration (FDA) in the US and by the European Medicines Administration (EMA) in Europe [11,12,13]. Different drugs target genomic alterations in EGFR, ALK, ROS1, BRAF, and NTRK, or more recently in RET and MET [11,12,13]. In the case of absence of oncogenetic addiction, ICIs are now widely used as significant alternative therapeutic options [7,8]. Moreover, besides the different promising ICIs, new molecules are currently being evaluated in many clinical trials and recent results suggest the coming soon administration of additional targeted therapies for NS-NSCLC in daily practice [10,12,14,15,16,17]. Among these new drugs, some target KRAS mutations, particularly the KRAS G12C mutation [18,19,20,21]. The development of these therapies raises a lot of hope and expectations in thoracic oncology since the KRAS mutations, which have been considered for a long time as non-targetable, are identified in more than 30% of NSCLC patients, at least in Europe and in the US, depending on the different populations [15,22,23,24,25,26,27]. However, among the KRAS-mutated NSCLC, a KRAS G12C mutation has been identified in 30% to 50% of cases and currently represents the only targetable mutation associated with different clinical trials or allowing the treatment of patients with one of the KRAS G12C inhibitors (sotorasib), which has been recently approved by the FDA/EMA for second line treatment based on phase II results (CodeBreak 100 clinical trial) [23,25,26,28,29,30]. It is noteworthy that other different solid tumors can have a KRAS mutation, notably a KRAS G12C mutation, such as some colon and pancreatic carcinomas [31,32]. In this regard, a recent clinical trial [CodeBreaK100 (NCT03600883)] demonstrated the efficacy of the sotorasib in pancreatic carcinoma mutated for KRAS G12C [33].
KRAS has been added to the list of genes that certainly must be characterized for advanced-stage NS-NSCLC at diagnosis, progression, and recurrence [8]. One of the consequences is a modification of the diagnostic algorithms used by most of the clinical and molecular pathology laboratories. So, the challenge will now be to set up and use specific and sensitive molecular testing for KRAS status assessment, considering notably both the quality and quantity of the DNA that is extracted from tissue, cytological, and blood samples, but also the turnaround time (TAT) to send the report to the physician.
After a brief reminder of the recent data concerning the biology and epidemiology of KRAS mutations in NSCLC, notably of the KRAS G12C mutation, as well as their impact on patient prognosis, this review reports the strategic issues and new challenges faced in the assessment of the KRAS status in daily practice by the thoracic pathologists.
2. KRAS Biology and the Signaling Pathways Induced by KRAS Mutations
A substantial number of studies related to the biology of KRAS have been performed and are being published almost every day, notably into lung cancer, highlighting the fact that this topic represents a huge stake in thoracic oncology. We report here some of the major and specific knowledge in the field, mainly considering the signaling pathways induced in lung cancer cells by the KRAS mutation. KRAS mutations affect cell biology since, in a non-mutated KRAS cell, the KRAS protein, which is a small guanosine triphosphate (GTPase), connects cell membrane growth factor receptors to intracellular signaling pathways and many transcription factors, thus playing a pivotal role in a variety of cellular processes [34,35,36]. The KRAS gene produces two isoforms (KRAS-4A and KRAS-4B), each of which has a distinct C-terminus and hence might cause various membrane interactions within different lipid raft interactions. The two isoforms coexist in cells, and although KRAS-4B is the most common, both can cause lung cancer in mice. However, even in the absence of the KRAS-4B isoform, KRAS-4A has been shown to cause metastatic lung adenocarcinomas in vivo [37]. The binding of ligands to their transmembrane receptors, usually receptor tyrosine kinases, activates the upstream signaling pathways of KRAS, as does recruitment of docking proteins, such as GRB2, in association with RAS-specific GEFs, which enhance KRAS activation. Of note, only certain KRAS mutant lung cancers (those expressing both galectin-3 and integrin αvβ3) rely on a process of anchorage-independent growth, showing the close relationship between membrane signaling and KRAS dependence in lung cancer [38,39]. Just upstream of KRAS, SOS1 is a guanine exchange factor (GEF) that binds and activates the GDP-bound RAS family of proteins at its catalytic binding site, promoting GDP to GTP exchange. In addition to its catalytic site, SOS1 can engage GTP-bound KRAS at an allosteric site that enhances its GEF function. SOS1 depletion or specific genetic inactivation of its GEF function has been shown to reduce the survival of KRAS-mutated tumor cells [40,41].
KRAS downstream signaling has been extensively investigated over recent decades and depends on the tissue of origin [42,43]. Four major routes downstream of KRAS have been characterized: the RAF/MEK/ERK MAPK, PI3K/AKT, RALGEF/TBK1 and RAF1-MAPK independent pathways. The RAF/MEK MAPK pathway activates transcription factors via ERK nuclear translocation to stimulate the expression of diverse genes involved in cell proliferation, survival, differentiation, and cell-cycle regulation. While this pathway is normally activated in response to growth factors, mutations in EGFR, KRAS, and BRAF maintain a residual activity and drive oncogenic addiction. The PI3K/AKT pathway also plays a key role in RAS-mediated tumorigenesis, in which AKT phosphorylation leads to the activation of major downstream targets such as the mammalian target of rapamycin (mTOR), forkhead box O (FOXO), or nuclear factor (NF)-κB, which stimulate survival, cell-cycle progression, metabolism, and invasion. While studied mainly in other KRAS dependent cancers, the importance of the RAL/TBK1 pathway in RAS-induced lung cancer was confirmed in an RNAi screen to identify lethal partners of oncogenic KRAS [44]. Given the importance of tumor immunogenicity in the response to ICIs, TBK1 seems to play a central role since it contributes to innate immunity by activating interferon regulatory factor 3/7 (IRF3/7), thereby inducing type 1 interferon gene expression and supporting cell growth and self-renewal [45,46,47]. The unexpected requirement for RAF1 in KRAS-driven NSCLC has been identified in mouse models as a new and important independent pathway. RAF1 has been shown to be important for the initiation and progression of NSCLC [48]. Interestingly, elimination of RAF1 expression led to a drastic reduction in lung tumors without affecting the activity of the MAPK pathways, illustrating a new pivotal downstream route of regulation. c-RAF ablation induced regression of advanced Kras/Trp53 mutant lung adenocarcinoma by a mechanism independent of MAPK signaling [48]. Of note, the four non-exhaustive pathways cited above can be activated differently depending on the mutation, either heterozygous or homozygous, or on the expressed KRAS isoform, in synergy with the KRAS wild-type allele and other RAS family members, i.e., NRAS and HRAS [49,50,51]. Therefore, even if a lot of knowledge has been obtained over the last few years, further research is still needed to define a comprehensive scenario of the context of activation of downstream pathways of KRAS in lung tumors.
The depletion or accumulation of different metabolites, which results from aberrant cancer metabolism, has different ramifications for the function of surrounding immune cells, and thus has an impact on the tissue microenvironment. To manage the metabolic challenges imposed by this microenvironment, cancer cells can participate in many cooperative metabolic interactions that support tumor growth and invasion, as well as competitive metabolic interactions that induce tumor survival by limiting nutrient availability to the antitumor immune system [34,52]. Taken together, increasing our knowledge of the different effector pathways downstream of KRAS, such as dysregulated metabolism, might lead in the future to the enhancement of many therapeutic interventions both alone and in combination with direct inhibition of KRAS. Additionally, a recent study from Kobayashi and colleagues nicely provides new insights into the biological role of silent mutations in KRAS and their potential to be translated into novel therapies [53]. Briefly, synonymous single-nucleotide polymorphisms (sSNPs) that alter programmed translational speed affect expression and function of the encoded protein [54,55]. The sSNPs do not result in the change of the amino acid sequence because these nucleotide changes usually occur in the third base of a codon. This often leads to the conclusion that a lack of protein sequence alteration will not have any functional consequences [54,56,57]. However, sSNPs can affect the expression of neighboring genes, but also the mRNA splicing, stability, andstructure, as well as protein function and folding. Synergistic advances in next-generation sequencing have led to the identification of sSNPs associated with disease penetrance [55]. Therefore, over 50 human non-tumoral and tumoral diseases have been associated with these synonymous mutations [54,56,57]. So, it is noteworthy that these types of mutations in KRAS were proven to be crucial for protein and mRNA expression and thus can lead to amplification and overexpression of this gene. As a result of this sSNP, it was demonstrated that the cell proliferation and metastasis can be enhanced, and the resistance to targeted therapeutics can be increased [53]. This discovery presents an opportunity for new treatments targeting KRAS in NSCLC.
3. Epidemiology of KRAS Mutations in Non-Small Cell Lung Carcinoma
Depending on the population and the series, the frequency of KRAS mutations in lung cancer patients is variable, identified in around 20% to 40% of lung adenocarcinomas and in around 5% to 7% of lung squamous cell lung carcinomas [58,59,60,61,62,63,64,65]. These mutations are much more frequent in Caucasian and Hispanic populations (from 25% to 35%) than in Asian populations (including Chinese and Indian patients) and in Iranians (globally from 9% to 13%) [59,61,62,63,65,66,67,68]. Of interest, a recent study identified a KRAS mutation in 24% of NSCLC Chinese patients [69]. KRAS mutations are more frequent in smokers (30%) than in non-smoker (10%) populations [61,62,63,65,66,67,68]. Other epidemiological factors have been associated with these mutations (gender, age, histological subtypes of lung adenocarcinoma, histological grading, tumor stage), leading to some discrepant results [60,62,63,70,71,72,73].
Different KRAS mutation subtypes have been defined in NSCLC [60]. KRAS mutations occur in the GTP-binding domain of the protein, mainly affecting codons 12 and 13 in exon 2 and codon 61 in exon 3 [64]. However, the majority of these mutations are substitutions of glycine in codon 12: G12C (40−50% of all KRAS mutations), G12V (11−26%), G12D (6–17%), G12A (6–12%), and G12S (2–5%) [60,62,74,75,76]. Other KRAS mutations include different rare mutations in codon 12 [G12R (1.7%)], codon 13 [G13C (6–12%); G13D (3–9%)], and codon 61 [Q61L (1–2%), Q61H (4%)]. Exceptional mutations in exon 4 affecting codon 146 have also been reported in less than 1% of cases, notably in metastatic NS-NSCLC. KRAS G12C and KRAS G12V are more frequent in smokers while KRAS G12D and KRAS G12S are more frequent in non-smoker patients [77]. The proportion of KRAS G12C-mutated subtypes is slightly lower across a series of Asian patients in comparison with Caucasian patients [61].
KRAS mutations can occur as a result of NSCLC progression treated with different tyrosine kinase inhibitors (TKIs) [78,79]. Therefore, a KRAS G12C mutation is noted in around 1% of patients showing tumor progression after first-line treatment with TKIs [79].
4. Impact of the KRAS Status on the Prognosis of Non-Small Cell Lung Carcinoma Patients
It is noteworthy that many contradictory studies into the prognostic value of KRAS mutations in NSCLC patients have been published [80,81,82]. Therefore, when examining these studies and drawing conclusions, the prognostic role of a KRAS mutation in lung cancer is still debatable. This is probably firstly due to the different methodologies used for molecular testing (notably the sensitivity and specificity of the different approaches), but also to variable patient inclusion criteria (including age, gender, and ethnicity) to assess the prognostic impact of these KRAS mutations. Moreover, the prognosis of KRAS-mutated NS-NSCLC can vary depending on the disease stage [83,84,85]. Recent studies have shown a higher risk of tumor recurrence after surgical resection in early-stage cancers with KRAS G12C mutations in comparison to other KRAS-mutated tumors and non-KRAS-mutated tumors [83,84,85]. Thus, it is certainly of strong interest to look systematically for the KRAS mutational status in resected lung specimens.
Some studies integrated the relationship between the KRAS mutation and other oncogenic mutations, the PD-L1 expression, and/or the tumor mutational burden (TMB) [74,86,87]. These different biological criteria can certainly have a strong impact on the behavior of KRAS-mutated tumors. Thus, according to their evaluation, this may explain some discrepancies among studies related to the impact of KRAS mutations on disease prognosis. Therefore, some initial series only evaluated the KRAS status without looking for different associated genomic alterations, notably in STK11, KEAP1, and/or TP53 [88]. However, more than 50% of the KRAS mutated-NSCLC show a concurrent genomic alteration on one or more other genes (mainly, TP53, STK11, KEAP1, CDKN2A, AKT1, PI3KA, BRAF) [89,90,91]. The most common co-mutations are observed in TP53 (35–40%) and STK11 (12–20%) [89,90,91,92]. The prognosis of NS-NSCLC patients may also vary according to the subtype of the KRAS mutation. Therefore, the prognostic impact of KRAS G12C is debatable in certain series [83,84,85,93]. A recent study showed that KRAS G12C is mutually exclusive compared to known actionable driver mutations, but non-driver co-mutations are common (STK11, 21.5%; KEAP1, 7.0%; TP53, 48.0%) [94]. KRAS G12C mutations can also be associated with ERBB2 amplifications and ERBB4 mutations [73]. KRAS G12V mutations are frequently associated with PTEN mutations, while KRAS G12D mutations are associated with PDGFRA mutations [90]. TP53 mutations on certain exons, such as exon 8, have been associated with worse prognosis in certain series of NSCLC patients [95]. Moreover, prognosis may even be worse in the case of TP53, STK11, and KEAP1 co-mutations [59,63,76,82,88,96,97,98,99,100,101,102,103,104,105]. An experimental study demonstrated that NRF2 activation, in association with a loss in STK11 and KRAS activation, can be associated with a negative prognosis and tumor progression, and can lead to resistance to immunotherapy [106]. Globally, the presence of several co-mutations in association with the KRAS mutation leads to more aggressive tumors and chemo-resistance. Moreover, the association between KRAS mutations and a high level of gene copy number variation of KRAS can be an indicator of worse prognosis [107].
The prognosis of patients with KRAS-mutated tumors is associated with the level of PD-L1 expression in tumor cells [63,108,109,110,111,112]. Thus, KRAS-mutated tumors with a high level of PD-L1 expression demonstrated worse prognosis [63,108,109,110,111,112]. Moreover, some studies showed that PD-L1 expression is also linked to TP53 mutations and tobacco exposure [113,114]. PD-L1 expression seems to be variable according to the subtype of the KRAS mutation [108,115]. Independently of the tobacco status, the TMB is higher in KRAS mutations than in wild-type KRAS NS-NSCLC [116]. However, the TMB level is lower in KRAS G12D mutated tumors than in other KRAS-mutated tumors [86]. This could be explained by the fact that KRAS G12D is most often identified in non-smoker patients [86].
5. The KRAS G12C Mutation Is a New Target for Promising Therapy in Advanced-Stage or Metastatic Non-Small Cell Lung Carcinoma
It was demonstrated several years ago that KRAS-mutated NSCLCs are more chemo-resistant than KRAS wild-type NSCLCs [117]. Moreover, KRAS-mutated NSCLC showed no response to TKIs targeting EGFR mutations [118]. So, KRAS mutations have been considered until recently to not be “druggable” with some targeted therapies [26,119]. Initially, different strategies focused on direct targeting of KRAS proteins and downstream inhibition of KRAS effector pathways [120,121]. However, most of these strategies were associated with a lack of tumor responsiveness [120,121]. Therefore, the identification of a hidden pocket adjacent to switch II in GDP-bound KRAS suddenly renewed great interest in the direct targeting of the mutant KRAS proteins [122]. So, different molecules were rapidly developed, in particular those targeting KRAS G12C mutated NSCLC (Figure 1) [105,106]. However, the initial KRAS G12C inhibitors were not sufficiently potent for further development for humans [105]. More recently, novel covalent small-molecule inhibitors of KRAS G12C mutant proteins, notably AMG 510 (sotorasib) and MRTX849 (adagrasib), provided initially promising results [123,124,125,126,127,128]. Therefore, after the positive results obtained in vitro and in vivo on inhibition of growth of different KRAS G12C mutated cells, new drugs are currently under evaluation on patients in different clinical trials and should soon be administered in daily practice to advanced or metastatic NS-NSCLC patients showing a KRAS G12C mutation [30,129,130]. Thus, promising results have been obtained in phase I and phase II clinical trials with sotorasib and adagrasib targeting KRAS G12C mutated NSCLC showing 37% and 45% overall response rate (ORR), respectively. In these studies, KRAS and STK11 co-mutations confer a better response to targeted therapies [23,25,30,123,131,132,133,134,135]. As mentioned previously, only one drug (sotorasib) has been FDA and EMA approved as second line therapy in KRAS G12C mutated tumors [23,28,29]. Caution in interpretation is necessary until the advantages of overall survival of some other direct inhibitors of mutant proteins are confirmed in large-scale phase III randomized trials. In addition, many clinical trials are currently being developed to evaluate additional KRAS G12C inhibitors, such as LY3499446 (clinicaltrials.gov identifier: NTC04165031) and JNJ-746999157 (clinicaltrials.gov identifier: NTC0400630) and other KRAS mutation-specific inhibitors in NSCLC patients (Table 1) [136,137,138]. In fact, the different KRAS inhibitors can be classified in different therapeutic families, including KRAS mutation-specific inhibitors, pan-KRAS inhibitors, downstream KRAS inhibitors, upstream KRAS inhibitors, and finally, some inhibitors targeting cellular metabolism and autophagy [139].
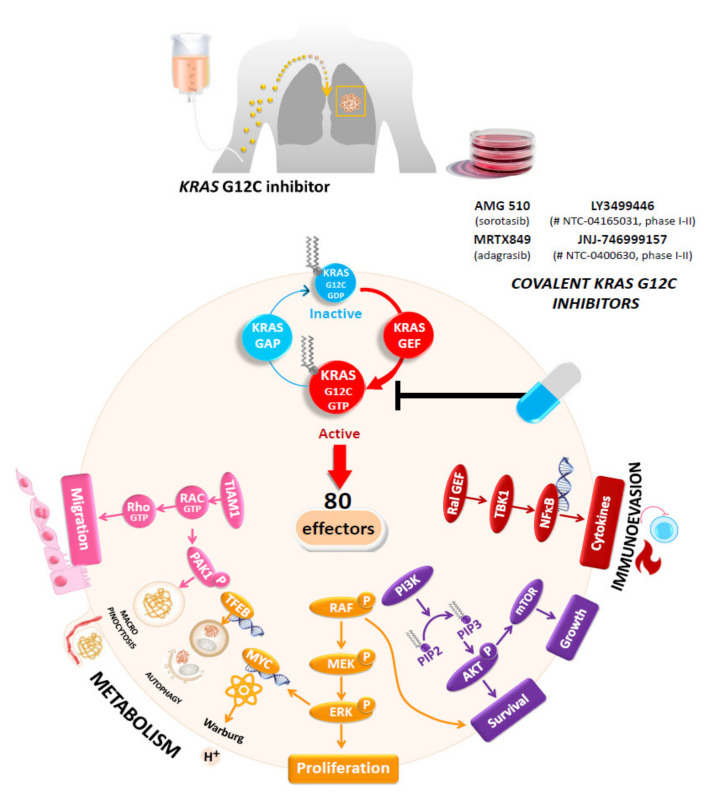
Figure 1.
The promise of KRAS G12C inhibitors. Compared with wild-type KRAS, cysteine 12 (C12) mutations destroy the GTPase activity of KRAS and lock it in the GTP-bound state. Once activated, Ras signals through 80 effectors, thus activating many different signaling pathways involved in cell proliferation, survival, metabolism, and migration. The best-characterized pathways are the MAPK (Raf-MEK-ERK), PI3K/AKT, and Ral pathways. In contrast, the small molecule drugs sotorasib and adagrasib can form a covalent bond with C12 in the KRAS-G12C protein, causing KRAS to take on an inactive state.
Table 1.
A few examples of ongoing clinical trials based on inhibitors targeting KRAS in non-small cell lung carcinoma.
| Therapeutic Family | ||
|---|---|---|
| Clinical trial | NTC04165031 | LY3499446 |
| NTC0400630 | JNJ-746999157 | |
| Pan-KRAS inhibitors | NCT04000529 | TNO155 + ribocicid or + spartalizumab |
| NCT04916236 | RMC-4630 + LY3214996 | |
| NCT04111458 | BI 1701963 + trametinib | |
| NCT03114319 | TNO155 alone | |
| NCT04045496 | JAB-3312 | |
| Dowstream KRAS inhibitors | NCT03681483 | RO516766 |
| NCT03284502 | HM95573 + cobimetinib | |
| NCT02974725 | LXH254 + LTT462 | |
| NCT04620330 | VS-6766 + defactinib | |
| Upstream + downstream KRAS inhibitors | NCT02230553 | Trametinib + lapatinib |
| NCT03704688 | Trametinib + poniotinib | |
| NCT04967079 | Trametinib + aniotinib | |
| Futibatinib + binimetinib |
6. Resistance Mechanisms Induced in Non-Small Cell Lung Carcinomas by Specific Inhibitors Targeting the KRAS G12C Mutation
More than 50% of the patients treated with sotorasib and adagrasib do not show significant tumor reduction when treated with these specific KRAS G12C inhibitors [140,141,142,143]. Moreover, resistance to these molecules occurred relatively early, most of the time a few months following the treatment initiation [139]. Therefore, recent clinical data showed that the duration of response for the great majority after sotorasib treatment is quite short, since the median progression-free survival is around 6 months [23]. These different results stress the urgent need to better characterize the resistance pathways that can be induced by the KRAS G12C inhibitors [139,142]. In fact, different mechanisms of primary and secondary resistances occurring in KRAS-mutated NSCLC patients treated with these inhibitors can explain these clinical results (Figure 2) [141,143,144,145,146,147,148,149,150,151,152,153,154,155]. Most of these resistance mechanisms are driven by genomic alterations (KRAS mutation or amplification or mutations on other genes) or by different bypass mechanisms [143,145,147]. So, many mechanisms of resistances are due to genetic alterations in the nucleotide exchange function or adaptive mechanisms in downstream pathways or in expressed KRAS G12C [143,148,156,157]. Thus, KRAS G12C can stay in an active state as a result of activation of the EGFR or AURKA signaling pathways [157]. This can be mediated by PTPN11/SHP2 recruiting SOS1 to promote conversion to an active state [158].
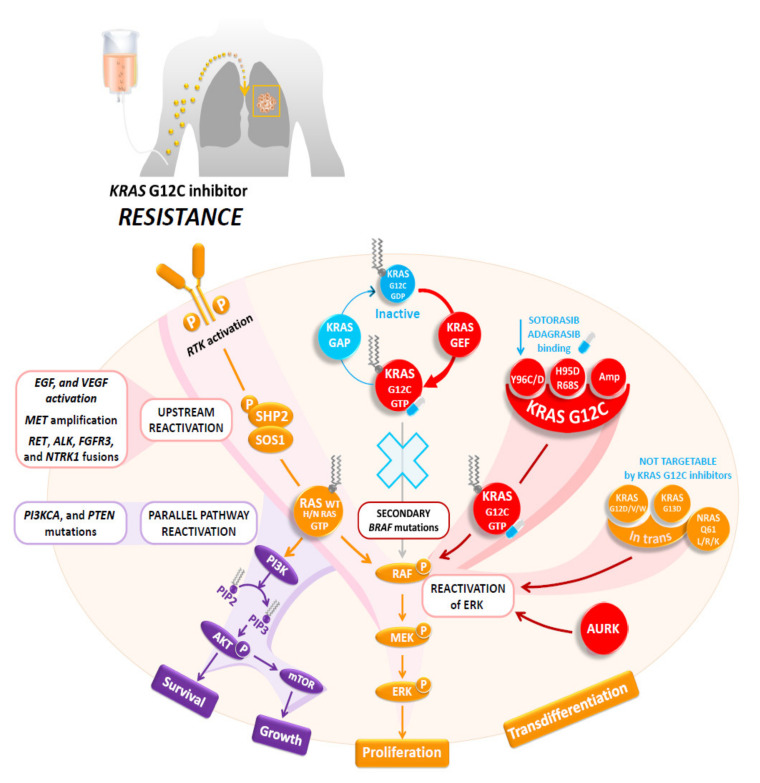
Figure 2.
Multifaceted resistance mechanisms to KRAS G12C inhibitors include secondary mutations of KRAS G12C that potentiate its nucleotide exchange (Y40A, N116H, or A146V), impair the inherent GTPase activity (A59G, Q61L, or Y64A), or impact other MAPK effectors (such as BRAF mutations). Innate or acquired resistance can also occur through a high RTK activity (EGFR or MET) on the cell surface, which bypasses the KRAS G12C inhibition and activates wild-type N/H-RAS via upstream SHP2 and AURKA activation. Ultimately, this restores the MAPK pathway independently of KRAS G12C and leads to activation of parallel oncogenic pathways such as the PI3K/AKT pathway. Alternatively, KRAS amplification or diminished degradation up-regulates KRAS G12C to an excessive level that does not bind the inhibitors.
Primary or intrinsic resistance mechanisms are often associated with the fact that not all KRAS mutant cells depend on KRAS activation to maintain their viability, and this can be maintained despite ablation of the KRAS mutant protein, highlighting the heterogeneity of the different KRAS-mutated tumor cell subclones [159]. In this regard, it was shown that activation of the downstream effectors (i.e., ERK and AKT) was not suppressed after KRAS knockdown. The RAS–RAF–ERK pathway has multiple independent mechanisms that maintain signaling in its active state while being selectively targeted. Therefore, intrinsic resistance relies on this as a fundamental pathway for cell survival [143].
The predominant secondary resistance mechanisms are linked to the onset of new KRAS mutations [145,147]. One major new mutation, the Y96D mutation, occurs at a relevant position for sotorasib and adagrasib binding, leading to resistance to both inhibitors [141,149]. Other KRAS mutations, such as R86S and H95D, have been identified in association with secondary resistance to KRAS G12C inhibitors [141]. In addition, acquired mutations can be identified alone or in association with acquired mutations in the switch II pocket [141]. So, G12D, G12V, and G12W in trans can also be detected at tumor progression [141,143]. Consequently, different clones can present resistance by impeding the binding of drugs to KRAS G12C mutated cells and other tumor cells acquire resistance through mutations that are not targetable by KRAS G12C inhibitors. As already described for tumors rearranged for ALK and treated with crizotinib, amplification in the KRAS G12C allele can also be an independently acquired resistance mechanism to KRAS G12C inhibitors [143,145,147]. Off-target mechanisms of secondary resistance, occurring on different genes, are also possible [126,128,130]. So, in a similar manner to that described for many TKIs (notably those targeting EGFR mutations or ALK rearrangements), secondary resistance associated to KRAS G12C inhibitors can be linked to MET amplification [143,147,152,160,161]. A RET fusion can be a mechanism of secondary resistance since RET kinase domain activation leads to oncogenic signaling of the PI3K-AKT, JNKs, and BRAF–MEK–ERK pathways, thus promoting cell survival and tumor promotion [162]. Other fusions have been reported to be associated with resistance to adagrasib such as ALK and FGFR3 fusions, or more rarely, BRAF and NTRK1 fusions [140,143,147]. Polyclonal activation of different RAS isoforms, associated with different NRAS mutations (such as Q61L, Q61R, and Q61K) can play an important role in secondary mechanisms of resistance to KRAS G12C inhibitors [143,145]. Other mutations of BRAF, PI3KCA, and PTEN can be associated with this resistance [145,147]. In addition, an important off-target mechanism of resistance to adagrasib is histological transformation, such as squamous cell carcinoma transformation [141]. From a practical point of view, early identification of some of these different mechanisms of resistance can allow alternative treatments to NS-NSCLC harboring a KRAS G12C mutation to be proposed. Finally, among one of the recently identified mechanisms, BCL6 expression seems to be an important cause of resistance [163]. As consequences of these different mechanisms of resistance, many clinical trials are ongoing or are going to soon start combining a KRAS G12C inhibitor and different other therapeutic agents, some of which will later be detailed in the perspectives section [164]. Briefly, as an example, the combination therapies currently programed in the CodeBreak 101 clinical trials include the association of soratosib with different molecules such as AMG 404, trametinib, RMC-4630, afatinib, pembrolizumab, panitumumab, carboplatin, pemetrexed, and docetaxel, atezolizumab, everolimus, palbociclib, MVASI® (bevacizumab-awwb), TNO155, FOLFIRI, and loperamide [164].
Abbreviations: RTK: receptor tyrosine-kinase, KRAS: Kirsten rat sarcoma, GAP: GTPase activating proteins, GEF: guanine nucleotide exchange factors, SOS: son of sevenless, AURKA: Aurora kinase B, SOS: son of sevenless, SHP2: Src homology region 2 domain-containing phosphatase-2, Ral: Ras-like, NF-kB: nuclear factor-kB, RAF: RAF proto-oncogene serine/threonine-protein kinase, MEK: Mitogen-activated protein kinase kinase, ERK: extracellular signal-regulated kinase.
7. Impact of KRAS Mutations on the Efficiency of Immunotherapy
Previous studies demonstrated that the outcome of advanced or metastatic NS-NSCLC patients treated with ICIs was better for tumors with KRAS mutations compared to the outcome with wild-type KRAS tumors [114]. This can probably be explained by a higher overall level of PD-L1 expression in KRAS-mutated tumors [87]. Therefore, PD-L1 expression is relevant for prediction of the efficacy of ICIs for patients with KRAS-mutated tumors more than in patients with other types of NSCLC [87,115,165,166,167]. In the study by Jeanson et al., the sensitivity of ICIs appears to be associated more with the expression of PD-L1 by the tumor cells than with the different types of KRAS mutations [115]. In the study by Lauko et al., the response to ICIs of NSCLC patients with a brain metastasis was higher in KRAS-mutated tumors [168]. In a recent study that enrolled patients with PD-L1 positive tumors, pembrolizumab monotherapy did not significantly improve the outcome of KRAS-mutated tumors compared with chemotherapy for patients with wild-type disease [169]. So, in addition to the KRAS mutational status, some authors propose to take into consideration not only the expression of PD-L1 but also other biological parameters such as the density of CD8 positive lymphocytes infiltrating the tumor cells when assessing the response to ICIs [165]. Moreover, the ICI response with or without platinum-based chemotherapy or with or without anti-CTL4 treatment is also dependent on the presence of TP53, STK11, and/or KEAP1 co-mutations [170,171,172,173]. Thus, the association of these different genomic alterations leads to variable immune signatures [170,171,172,173,174]. Most of the studies showed that the association of STK11 and KEAP1 mutations with a KRAS mutation leads to ICIs resistance [104,174]. KRAS and TP53 mutations are frequently associated with NSCLC [62,88,172]. These mutated NSCLC seem to present a long-term response to ICIs [175]. Finally, different subgroups of tumors can be distinguished, certain tumors showing a KEAP1 mutation in the absence of a TP53 mutation lead to an immune desert, while both KEAP1 and TP53 associated mutations show a rich immune infiltrate [172,176]. Other genes of interest such as SMARCA4 should also be studied, since SMARCA4 and KRAS co-mutations lead to a weak response to ICIs [177].
Currently, the first-line treatment in daily practice of patients with KRAS-mutated tumors associates ICIs (pembrolizumab) and chemotherapy or immunotherapy (pembrolizumab) alone for tumors expressing PD-L1 in more than 50% of tumor cells [7,178]. A therapeutic strategy using specific KRAS inhibitors alone might be administered in the near future as first-line treatment for tumors expressing PD-L1 in less than 50% of tumor cells [23,30,129,130]. In this regard, a clinical trial (CodeBreak 201) has recently started in KRAS G12C mutated aNS-NSCLC for sotorasib administration in tumors with less than 1% of PD-L1 expression and/or STK11 mutation [179]. Association of this targeted therapy with immunotherapy could also be proposed [22]. For this treatment it is mandatory that no genomic alteration in EGFR, ALK, ROS1, BRAF, and NTRK is detected.
Some studies showed that STK11 mutations are less frequent in all KRAS-mutated tumors than in non KRAS-mutated tumors [180]. However, the KRAS G12C mutation can be associated with STK11 and/or KEAP1 mutations and show a low response to immunotherapy [89,91,92,104,181,182]. Therefore, in the case of these co-mutations, specific KRAS G12C inhibitors may be a favorable approach as first-line therapy. Another approach focused on combined treatment that can associate KRAS G12C inhibitor molecules and immunotherapy [22]. This combined therapy may shift the balance away from an immuno-suppressive tumor microenvironment to allow effective anti-tumor immunity [22,183]. In this regard, it seems highly appropriate to rapidly set up for late-stage NS-NSCLC patients reflex testing looking for the status of KRAS, STK11, and KEAP1 before administrating any first-line therapy. However, we have to keep in mind that some NS-NSCLC showing KRAS and STK11 mutations also respond well to immunotherapy, highlighting that other currently uncertain biological mechanisms should also be identified and evaluated in the near future [184].
8. Biological Samples and Molecular Testing
KRAS mutations, notably the KRAS G12C mutation, can be identified from tissue samples (bronchial biopsies, transthoracic biopsies, and more rarely biopsies from a metastatic site or a surgically resected sample), from different cytological and fluid samples (bronchial aspirates, fine needle aspiration, endobronchial ultrasound and transbronchial needle aspiration, bronchoalveolar lavage, pleural and cerebrospinal fluid) and from blood [15,105,185,186,187,188,189]. These mutations are currently being characterized using targeted sequencing (RT-PCR and droplet digital PCR) or next-generation sequencing (NGS) approaches [189,190]. Moreover, liquid biopsy is certainly a very promising tool to use at baseline but also at progression in patients treated with specific inhibitors of KRAS G12C mutations to track new KRAS mutations, notably the Y96D mutation, as well as other mutations such as the R86S and H95D mutations [141]. When considering the increasing number of genes to be evaluated at diagnosis or at tumor progression, NGS must be used as a priority. However, setting up and using NGS systematically can still be associated with a number of constraints, which will be detailed below [191].
9. Opportunities and Challenges for the Thoracic Pathologists
Different consensus expert opinions on the use of testing to select treatments targeting KRAS inhibitors did not recommend KRAS-mutation testing as a daily practice stand-alone assay [13,188]. In recent years, some laboratories have developed KRAS testing if EGFR, ALK, and ROS1 genomic alterations are not detected in late-stage NSCLC. However, considering the increasing number of genes of interest for targeted therapy for these tumors, it seems today that this strategy should be abandoned. In this regard, KRAS mutations must be considered with a larger multigene panel, including many other genes of interest, notably EGFR, ALK, ROS1, BRAF, NTRK, RET, MET, and HER2 [13]. In parallel, copy number variation, especially affecting KRAS, also needs to be assessed. As discussed above, several genes of interest (such TP53, STK11, and KEAP1) should ideally be concomitantly evaluated in addition to the KRAS status [91,104]. At the same time, it is mandatory to assess PD-L1 expression in tumor cells, and if possible, also in immune cells. In this regard, different constraints can arise and present many challenges for a pathology and molecular laboratory (Table 2). Thus, according to the sample size and type, and/or the percentage of tumor cells, the different technologies must be well controlled to make a pathological and a molecular testing report in a short TAT. The accessibility to NGS in daily routine practice and thus assessment of the KRAS status is variable from one laboratory to another and from one country to another [191].
Table 2.
Challenges associated with routine clinical testing of KRAS mutations in NSCLC for a pathology laboratory. NGS, next generation sequencing; IHC, immunohistochemistry; IVDR, in vitro drug regulation.
| Challenges |
|---|
|
9.1. Tracking the False-Negative and False-Positive Results
The characterization of the different genomic alterations such as mutations, rearrangements, or amplifications, requires extracted nucleic acid (DNA/RNA) that is not degraded and is obtained in a sufficient quantity for the sequencing method of choice [192]. In this respect, the NGS approaches are becoming more and more sensitive and are now in competition with targeted sequencing methods for use in daily practice. The molecular pathologist needs to select the method of molecular testing depending on the budget and on the hospital organization. Some technologies, such as those based on amplicon-based library preparation, can be more effective than hybrid capture-based NGS library preparation in the case of small tissue lung biopsies and/or a low percentage of tumor cells [174]. The identification of KRAS mutations using NGS requires 10–15 ng or 40–50 ng of DNA, respectively, for amplicon-based or hybrid capture technologies [193]. Large panels (more than 300 genes) need a higher amount of DNA than medium-sized panels (from 20 to 50 genes). For multigene panels, KRAS is sequenced with deep coverage (>500 x), where the specific depth of coverage depends on whether the assay is a hybrid capture or an amplicon-based approach, as well as whether the values are de-duplicated or not. Globally, the hot spots at codons 12, 13, and 61 can be routinely detected down to at least a 5% variant-allele frequency calling [194].
DNA degradation can provide false-negative or false-positive results, notably in the context of assessment of KRAS mutations [167,176]. These false results can be due to poor management of one or several steps of the pre-analytical phase [176,195]. For example, a long period of cold ischemia, a long or short fixative time, can have consequences on false-negative molecular results due to DNA degradation or false-positive molecular results due to DNA deamination [176,195].
KRAS mutations present on tumor cells must be distinguished from KRAS mutations associated with clonal hematopoiesis on indeterminate potential (CHIP) [196,197,198]. CHIP is defined as a “hematologic malignancy-associated somatic mutation” in blood or bone marrow that is not associated with other diagnostic criteria for a hematological malignancy [197,199]. CHIP occurs frequently in the elderly and presents pitfalls for KRAS mutation assessment performed with circulating free DNA (cf-DNA) [196,197,198]. In this respect, NGS approaches that employ the simultaneous analysis of cf-DNA and matched DNA blood cells can confirm the presence of somatic mutations and eliminate false identification of the somatic mutation as tumor-specific [197,199]. Moreover, since pancreatic and colon cancers and other solid tumors such as low-grade serous ovarian and endometrial carcinomas show KRAS mutations, the presence of this mutation detected with cf-DNA may indicate the co-existence of a carcinoma other than the diagnosed NS-NSCLC [200]. It is noteworthy that a KRAS mutation can also be identified in certain non-malignant diseases and theoretically, can be found with cf-DNA, mostly when using very sensitive approaches [201,202,203,204]. For advanced NSCLC, some studies have documented the presence of ct-DNA in about 85% of cases, highlighting that a negative result with a blood sample does not eliminate the presence of the KRAS mutation in tissue samples [205]. Thus, naive patients with slow-growing lung cancers may be at risk of having a negative result. In fact, negative results with plasma must also be considered as inconclusive, leading to a tissue biopsy [206].
9.2. Algorithm Testing: How to Be Optimal?
Molecular testing for KRAS status assessment currently varies according to the country and to the institution and different organizations [207,208,209,210]. If testing follows the international guidelines, NGS needs to be done at diagnosis to test at least EGFR, ALK, ROS1, BRAF, NTRK, RET, and MET [13,211]. Therefore, in this context, it seems much easier, more cost-effective, and less tissue-consuming to use an NGS approach than to do multiple RT-PCR analyses with or without immunohistochemistry and fluorescent in situ studies (i.e., for ALK and ROS1). Considering that other genes of interest (notably KRAS and HER2) are present in the majority of NGS panels, it is obvious that NGS is the better approach for systematically gaining access to the KRAS status for NS-NSCLC [209]. However, NGS at baseline or even at progression and at recurrence is not currently possible in all countries [191,212]. Moreover, one drawback of doing some molecular tests, notably when evaluating the KRAS status, is that the therapeutic molecule targeting KRAS G12C is not yet available in routine clinical practice and is currently mandatory only for inclusion into clinical trials using KRAS inhibitors. Thus, most of the physicians do not ask for KRAS mutation assessments in daily practice.
We also need to distinguish between bespoke molecular testing, when the pathologist is simply waiting for the physician to request evaluation of the KRAS status, and reflex molecular testing, performed by the pathologist for all diagnoses of NS-NSCLC, even without knowing the disease stage at the time of diagnosis and whatever the sample type (tissue biopsies, cytological sample, surgically resected specimen, etc.). It is noteworthy that even if KRAS inhibitors are most likely first administered for advanced-stage NSCLC showing a KRAS G12C mutation, these drugs will probably be indicated in the future for early-stage NSCLC [213]. The latter strategy will be associated with new challenges for the pathologists, also including the use of NGS in this indication.
9.3. Should a Liquid Biopsy Be Integrated into KRAS Status Assessment for NS-NSCLC?
In the absence of tissue or cytological material at diagnosis, the KRAS status can be evaluated using a liquid biopsy (LB) [186]. During tumor progression or recurrence, notably in patients treated with different anti-EGFR or anti-ALK TKIs, it is now well-admitted that a LB can be performed systematically to look for different druggable genomic alterations and this can include the detection of a KRAS G12C mutation [206]. In case of an inconclusive result, and if possible, depending on the patient status and tumor site, a tissue re-biopsy should be done to track resistance mechanisms [206]. NGS approaches can routinely use a LB and thus provide access to the KRAS status and associated genomic alterations [15,190,214,215,216,217]. However, NGS with a LB is currently not often available in most laboratories, in Europe at least, and is set up for routine testing in only a few comprehensive cancer centers [191,215,218]. Therefore, many laboratories use targeted sequencing methods, such as RT-PCR or ddPCR [219,220,221]. Interestingly, a recent study demonstrated that the amount of cf-DNA was higher when some genomic alterations such as KRAS were identified in tumors, in contrast to a lower amount of cf-DNA associated with some other mutations detected on EGFR or TP53 [222]. However, the interest in a tissue re-biopsy is currently progressing since a number of mechanisms of resistance (such as gene amplification and rearrangement) are found much more easily or only found (such as histological transformation and epithelial-mesenchymal transformation) in tissue samples and not in blood samples [206]. However, LBs will hopefully be more developed and requested by physicians in the future too, since this non-invasive approach for KRAS status evaluation and of other genomic alteration is repeatable and does not need hospitalization for invasive procedures. However, there are still some gaps to fill in this setting in order to better use LBs in daily practice, but this method is certainly also pivotal to look for resistance mechanisms occurring under treatment, in particular with KRAS G12C inhibitors. In this setting, LBs definitively allows an easy monitoring of the disease [218].
9.4. To Be Able to Deliver the Results in a Turnaround Time That Follows International Guidelines
As for the other genomic alterations of interest, the TAT to report the KRAS status to the physician needs to be mastered in routine clinical practice. According to the international guidelines, this TAT must be no more than 10 working days [13,211]. New considerations have now been set up for reflex NGS panel testing for any newly diagnosed NS-NSCLC [223]. Moreover, this can be performed despite the initial level of information concerning the disease staging status. This option opens great opportunities to considerably reduce the TAT and should enable genotype-directed targeted therapies, including KRAS inhibitors, in an efficient and certainly cost-effective manner, not only for advanced stages, but also in near-treatment strategies for early stages of NSCLC.
9.5. Sampling and Timing to Detect the Mechanisms of Resistance Associated with Targeted Therapies of KRAS-Mutated Lung Cancers
As mentioned previously, the biological mechanisms leading to therapeutic resistance to different drugs targeting KRAS mutations, notably KRAS G12C, can be intrinsic and present at baseline or be acquired at progression under therapy. The issue for the pathologist concerns rapid identification of this mechanism in order to switch the therapy to another molecule [146]. Therefore, the questions that can arise are: (i) what are the best samples for characterization of these mechanisms? and (ii) when should characterizations be done? Some of the resistant mechanisms are certainly easier to assess with a tissue biopsy (such as a MET amplification) while other mechanisms may be identified using a liquid biopsy and/or a tissue biopsy (such as new KRAS mutations or BRAF mutations) [144,145,147]. Moreover, as different mechanisms of resistance occur at progression in patients treated with different TKIs, early detection of this mechanism can be done with blood samples by establishing monitoring, even before the radiological onset of tumor progression [160,161]. However, other mechanisms of resistance, notably histological transformation, can be assessed only with a tissue biopsy.
9.6. Other Challenges
KRAS mutations are rarely detected in lung squamous cell carcinomas (LSCC) (between 4% to 8%), however, evaluation of the status in LSCC may be required in the near future, resulting in an increase in the workload and an increase in the budget dedicated to molecular biology testing, which should be taken into consideration [74,224].
As previously mentioned, it is very important to combine the status of KRAS mutations and other possible biomarkers to better predict the response to immunotherapy. Among the latter, one challenge was implementing the TMB results for patient care, knowing the multiple issues surrounding the use of NGS for TMB evaluation [225,226,227,228]. As an example, the US FDA approved pembrolizumab for the treatment of cancer patients with TMB >10 mutations/megabase (mut/Mb) [229]. Although the approval provides a novel therapeutic option for cancer patients, it raises major concerns [230]. Therefore, a TMB of 10 mut/Mb is certainly an arbitrary cut-off for immunotherapy treatment selection. Additionally, TMB-High, defined by >10 mut/Mb, fails to predict improved ICIs response across different cancer types [230]. Therefore, the reproducibility of TMB cut-off became the focal point for ICI treatment [230]. Thus, despite initially some promising results, the “TMB story” was quite disappointing for many reasons, leading to the fact that this biomarker is not use in routine clinical practice, at least currently in Europe [228]. Conversely, as already mentioned above, to combine the status of KRAS and additional biomarkers such as STK11, KEAP1, and TP53 is certainly of stronger importance to immunotherapy response prediction [102,103,104].
The assessment and identification of some somatic variants of KRAS mutations that are pathogenic (oncogenic) and function could be of strong interest to including patients with these variants into clinical trials [231,232,233]. Along with the development of high-covering NGS panels, the underlying point is whether a NSCLC with an unknown variant of KRAS should or should not be included in a precision oncology trial. Well-established pathogenic and benign variants are quite easily recognized, but there is still an urgent need to broaden the classification of a variant of interest from the unknown significance category, from either the potentially benign to the potentially pathogenic (oncogenic) categories, to support the inclusion of a patient in a clinical trial [231,232,233].
The choice of algorithm has a strong impact on the workload of the technicians, the engineers, and the pathologists, including for the post-analytical phase. It is also important to say that most of the molecular testing is usually done on demand by physicians. In this regard, the establishment of reflex testing by the pathologist should be done under a signed agreement with both partners.
As for other genomic alterations associated with a possible targeted therapy, the different molecular biology tests used in a hospital laboratory for KRAS status assessment need to be included in different programs of external quality assurance and obtain accreditation and validation [234,235].
Finally, the pathologists should also set up, in collaboration with the physician, a standby state of the scientific literature concerning different novel technical developments, molecule targets, drug developments, mechanisms of resistance etc., in relation to lung cancer, and more specifically to the different topics concerning KRAS in NSCLC. One major challenge will certainly be the ability to rapidly transfer different new biomarkers of responsiveness, resistance and/or toxicity related to the different molecules targeting the KRAS mutations to daily practice. This could also be done through different advanced training programs.
10. Perspectives
The current efficacy of the new KRAS G12C inhibitors highlights the urgent need to systematically identify NSCLC patients who can benefit from these therapies alone or in combination with other drugs. In this new situation, pathologists have the responsibility to test for KRAS mutations, notably reporting KRAS G12C pathogenic variants. This should be done as a routine test using a comprehensive up-front molecular gene panel and before first-line therapy. Accumulated data from preclinical and clinical studies showed that KRAS G12C-targeted therapeutics as single agents are inevitably thwarted by drug resistance [143,145,147]. Thus, despite initial promising results, current KRAS specific inhibitors all induce many mechanisms of resistance. Therefore, new developments are ongoing, notably those based on biochemical analyses and some in vitro screening [236,237,238]. A promising strategy to optimize KRAS G12C inhibitor therapy concerns combination treatments with other therapeutic agents, the identification of which is empowered by the insightful appreciation of compensatory signaling pathways or evasive mechanisms, such as those that attenuate immune responses [137,239]. In this regard, many clinical trials are currently open in order to assess efficacy of combination treatments [164]. These new therapeutic strategies associate or will associate different combinations of molecules, including some SHP2 inhibitors or MEK inhibitors together with KRAS G12C specific inhibitors [130,240,241,242,243,244,245]. Other molecules targeting KRAS mutations have been tested or are ongoing, such as those targeting mTOR, IGF1R, FASN, or EGLN1 in association with inhibitors targeting KRAS G12C mutations, or those that modify glutamine metabolism [246,247]. Moreover, new drugs targeting other KRAS subtype mutations, such as KRAS G12D, are under evaluation in vitro or in clinical trials [246,248,249]. Pan-KRAS drugs have also the potential to address a broad range of patient populations, including KRAS G12D-, KRAS G12V-, KRAS G13D-, KRAS G12R-, and KRAS G12A-mutant or KRAS wild-type-amplified cancers, as well as cancers with acquired resistance to KRAS G12C inhibitors [246].
11. Conclusions
There is a strong necessity to look exhaustively for the different mutations in KRAS in NSCLC patients. In this regard, many studies and initiatives are currently ongoing to improve the different aspects related to KRAS-mutated lung cancers [250]. However, the histopathologist will continue to play a major role in molecular testing, notably in thoracic oncology [251]. Therefore, the pathologist is more than ever a pivotal actor in this setting and will very soon be at the front line of identification of KRAS mutations in NS-NSCLC in routine clinical practice.
Acknowledgments
The author thanks Christiane Brahimi-Horn for editing this manuscript, the Conseil Départemental des Alpes Maritimes, the “Ligue Départementale de Lutte contre le Cancer des Alpes Maritimes”, the “Canceropôle PACA”, the French Government (Agence Nationale de Recherche, ANR) through the ‘Investments for the Future’ LABEX SIGNALIFE (ANR-11-LABX-0028-01 and IDEX UCAJedi ANR-15-IDEX-01) and [AD-ME project R19162DD]; CANC’AIR Genexposomic project; DREAL PACA, ARS PACA, Région Sud, INSERM cancer; the “Institut National du Cancer” (INCa) Plan Cancer, ITMO-Cancer; Children Medical Safety Research Institute (CMSRI, Vaccinophagy project R17033DJA).
Author Contributions
Conceptualization, P.H.; review of the literature, C.B., V.H., P.B., M.I., B.M. and P.H. writing—original draft preparation, C.B., P.B., M.I. and P.H.; writing—review and editing, C.B., V.H., P.B., M.I., B.M. and P.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The author declares no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Duma N., Santana-Davila R., Molina J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019;94:1623–1640. doi: 10.1016/j.mayocp.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson A.G., Tsao M.S., Beasley M.B., Borczuk A.C., Brambilla E., Cooper W.A., Dacic S., Jain D., Kerr K.M., Lantuejoul S., et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022;17:362–387. doi: 10.1016/j.jtho.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Peng L., Wang Z., Stebbing J., Yu Z. Novel immunotherapeutic drugs for the treatment of lung cancer. Curr. Opin. Oncol. 2022;34:89–94. doi: 10.1097/CCO.0000000000000800. [DOI] [PubMed] [Google Scholar]
- 7.Reck M., Remon J., Hellmann M.D. First-Line Immunotherapy for Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2022;40:586–597. doi: 10.1200/JCO.21.01497. [DOI] [PubMed] [Google Scholar]
- 8.Tan A.C., Tan D.S.W. Targeted Therapies for Lung Cancer Patients with Oncogenic Driver Molecular Alterations. J. Clin. Oncol. 2022;40:611–625. doi: 10.1200/JCO.21.01626. [DOI] [PubMed] [Google Scholar]
- 9.Howlader N., Forjaz G., Mooradian M.J., Meza R., Kong C.Y., Cronin K.A., Mariotto A.B., Lowy D.R., Feuer E.J. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N. Engl. J. Med. 2020;383:640–649. doi: 10.1056/NEJMoa1916623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melosky B., Wheatley-Price P., Juergens R.A., Sacher A., Leighl N.B., Tsao M.S., Cheema P., Snow S., Liu G., Card P.B., et al. The rapidly evolving landscape of novel targeted therapies in advanced non-small cell lung cancer. Lung Cancer. 2021;160:136–151. doi: 10.1016/j.lungcan.2021.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Hanna N.H., Robinson A.G., Temin S., Baker S., Jr., Brahmer J.R., Ellis P.M., Gaspar L.E., Haddad R.Y., Hesketh P.J., Jain D., et al. Therapy for Stage IV Non-Small-Cell Lung Cancer with Driver Alterations: ASCO and OH (CCO) Joint Guideline Update. J. Clin. Oncol. 2021;39:1040–1091. doi: 10.1200/JCO.20.03570. [DOI] [PubMed] [Google Scholar]
- 12.Kazdal D., Hofman V., Christopoulos P., Ilié M., Stenzinger A., Hofman P. Fusion-positive non-small cell lung carcinoma: Biological principles, clinical practice, and diagnostic implications. Genes Chromosomes Cancer. 2022;61:244–260. doi: 10.1002/gcc.23022. [DOI] [PubMed] [Google Scholar]
- 13.Mosele F., Remon J., Mateo J., Westphalen C.B., Barlesi F., Lolkema M.P., Normanno N., Scarpa A., Robson M., Meric-Bernstam F., et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020;31:1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 14.Hofman P., Barlesi F. Companion diagnostic tests for treatment of lung cancer patients: What are the current and future challenges? Expert Rev. Mol. Diagn. 2019;19:429–438. doi: 10.1080/14737159.2019.1611426. [DOI] [PubMed] [Google Scholar]
- 15.Hofman P. What Is New in Biomarker Testing at Diagnosis of Advanced Non-Squamous Non-Small Cell Lung Carcinoma? Implications for Cytology and Liquid Biopsy. J. Mol. Pathol. 2021;2:147–172. doi: 10.3390/jmp2020015. [DOI] [Google Scholar]
- 16.Imyanitov E.N., Iyevleva A.G., Levchenko E.V. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit. Rev. Oncol. Hematol. 2021;157:103194. doi: 10.1016/j.critrevonc.2020.103194. [DOI] [PubMed] [Google Scholar]
- 17.Rebuzzi S., Zullo L., Rossi G., Grassi M., Murianni V., Tagliamento M., Prelaj A., Coco S., Longo L., Bello M.D., et al. Novel Emerging Molecular Targets in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2021;22:2625. doi: 10.3390/ijms22052625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corral de la Fuente E., Olmedo Garcia M.E., Gomez Rueda A., Lage Y., Garrido P. Targeting KRAS in Non-Small Cell Lung Cancer. Front. Oncol. 2022;11:792635. doi: 10.3389/fonc.2021.792635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Hu L., Xu C. Kirsten rat sarcoma inhibitors in clinical development against nonsmall cell lung cancer. Curr. Opin. Oncol. 2022;34:66–76. doi: 10.1097/CCO.0000000000000808. [DOI] [PubMed] [Google Scholar]
- 20.Thein K.Z., Biter A.B., Hong D.S. Therapeutics Targeting Mutant KRAS. Annu. Rev. Med. 2021;72:349–364. doi: 10.1146/annurev-med-080819-033145. [DOI] [PubMed] [Google Scholar]
- 21.Xu Q., Zhang G., Liu Q., Li S., Zhang Y. Inhibitors of the GTPase KRAS(G12C) in cancer: A patent review (2019–2021) Expert Opin. Ther. Pat. 2022;5:1–31. doi: 10.1080/13543776.2022.2032648. [DOI] [PubMed] [Google Scholar]
- 22.Adderley H., Blackhall F.H., Lindsay C.R. KRAS-mutant non-small cell lung cancer: Converging small molecules and immune checkpoint inhibition. eBioMedicine. 2019;41:711–716. doi: 10.1016/j.ebiom.2019.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong D.S., Fakih M.G., Strickler J.H., Desai J., Durm G.A., Shapiro G.I., Falchook G.S., Price T.J., Sacher A., Denlinger C.S., et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullard A. KRAS’s undruggability cracks? Nat. Rev. Drug. Discov. 2019;18:488. doi: 10.1038/d41573-019-00102-y. [DOI] [PubMed] [Google Scholar]
- 25.Reck M., Carbone D.P., Garassino M., Barlesi F. Targeting KRAS in non-small-cell lung cancer: Recent progress and new approaches. Ann. Oncol. 2021;32:1101–1110. doi: 10.1016/j.annonc.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Salgia R., Pharaon R., Mambetsariev I., Nam A., Sattler M. The improbable targeted therapy: KRAS as an emerging target in non-small cell lung cancer (NSCLC) Cell Rep. Med. 2021;2:100186. doi: 10.1016/j.xcrm.2020.100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie M., Xu X., Fan Y. KRAS-Mutant Non-Small Cell Lung Cancer: An Emerging Promisingly Treatable Subgroup. Front. Oncol. 2021;11:672612. doi: 10.3389/fonc.2021.672612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. [(accessed on 5 March 2022)]. Available online: https://www.ema.europa.eu/en/medicines/human/summaries-opinion/lumykras.
- 29. [(accessed on 6 March 2022)]; Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-sotorasib-kras-g12c-mutated-nsclc.
- 30.Skoulidis F., Li B.T., Dy G.K., Price T.J., Falchook G.S., Wolf J., Italiano A., Schuler M., Borghaei H., Barlesi F., et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N. Engl. J. Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prior I.A., Hood F.E., Hartley J.L. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan M.B., Corcoran R.B. Therapeutic strategies to target RAS-mutant cancers. Nat. Rev. Clin. Oncol. 2018;15:709–720. doi: 10.1038/s41571-018-0105-0. [DOI] [PubMed] [Google Scholar]
- 33. [(accessed on 12 March 2022)]. Available online: https://ascopubs.org/doi/abs/10.1200/JCO.2022.40.36_suppl.360490.
- 34.Kerk S.A., Papagiannakopoulos T., Shah Y.M., Lyssiotis C.A. Metabolic networks in mutant KRAS-driven tumours: Tissue specificities and the microenvironment. Nat. Rev. Cancer. 2021;21:510–525. doi: 10.1038/s41568-021-00375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H.J., Lee H.N., Jeong M.S., Jang S.B. Oncogenic KRAS: Signaling and Drug Resistance. Cancers. 2021;13:5599. doi: 10.3390/cancers13225599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukhopadhyay S., Vander Heiden M.G., McCormick F. The Metabolic Landscape of RAS-Driven Cancers from biology to therapy. Nat. Cancer. 2021;2:271–283. doi: 10.1038/s43018-021-00184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmón M., Paniagua G., Lechuga C.G., Fernández-García F., Zarzuela E., Álvarez-Díaz R., Musteanu M., Guerra C., Caleiras E., Muñoz J., et al. KRAS4A induces metastatic lung adenocarcinomas in vivo in the absence of the KRAS4B isoform. Proc. Natl. Acad. Sci. USA. 2021;118:e2023112118. doi: 10.1073/pnas.2023112118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seguin L., Kato S., Franovic A., Camargo M.F., Lesperance J., Elliott K.C., Yebra M., Mielgo A., Lowy A.M., Husain H., et al. An integrin β₃-KRAS-RalB complex drives tumour stemness and resistance to EGFR inhibition. Nat. Cell Biol. 2014;16:457–468. doi: 10.1038/ncb2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seguin L., Camargo M.F., Wettersten H.I., Kato S., Desgrosellier J.S., von Schalscha T., Elliott K.C., Cosset E., Lesperance J., Weis S.M., et al. Galectin-3, a Druggable Vulnerability for KRAS-Addicted Cancers. Cancer Discov. 2017;7:1464–1479. doi: 10.1158/2159-8290.CD-17-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freedman T.S., Sondermann H., Friedland G.D., Kortemme T., Bar-Sagi D., Marqusee S., Kuriyan J. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc. Natl. Acad. Sci. USA. 2006;103:16692–16697. doi: 10.1073/pnas.0608127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeng H.H., Taylor L.J., Bar-Sagi D. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat. Commun. 2012;3:1168. doi: 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayers J.R., Torrence M.E., Danai L.V., Papagiannakopoulos T., Davidson S.M., Bauer M.R., Lau A.N., Ji B.W., Dixit P.D., Hosios A.M., et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart A., Coker E.A., Pölsterl S., Georgiou A., Minchom A.R., Carreira S., Cunningham D., O’Brien M.E., Raynaud F.I., de Bono J.S., et al. Differences in Signaling Patterns on PI3K Inhibition Reveal Context Specificity in KRAS-Mutant Cancers. Mol. Cancer Ther. 2019;18:1396–1404. doi: 10.1158/1535-7163.MCT-18-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C., et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chien Y., Kim S., Bumeister R., Loo Y.M., Kwon S.W., Johnson C.L., Balakireva M.G., Romeo Y., Kopelovich L., Gale M., Jr., et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 46.Ou Y.H., Torres M., Ram R., Formstecher E., Roland C., Cheng T., Brekken R., Wurz R., Tasker A., Polverino T., et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol. Cell. 2011;41:458–470. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry A.K., Chow E.K., Goodnough J.B., Yeh W.C., Cheng G. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J. Exp. Med. 2004;199:1651–1658. doi: 10.1084/jem.20040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanclemente M., Francoz S., Esteban-Burgos L., Bousquet-Mur E., Djurec M., Lopez-Casas P.P., Hidalgo M., Guerra C., Drosten M., Musteanu M., et al. c-RAF Ablation Induces Regression of Advanced Kras/Trp53 Mutant Lung Adenocarcinomas by a Mechanism Independent of MAPK Signaling. Cancer Cell. 2018;33:217–228.e4. doi: 10.1016/j.ccell.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 49.Hammond D.E., Mageean C.J., Rusilowicz E.V., Wickenden J.A., Clague M.J., Prior I.A. Differential reprogramming of isogenic colorectal cancer cells by distinct activating KRAS mutations. J. Proteome Res. 2015;14:1535–1546. doi: 10.1021/pr501191a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tahir R., Renuse S., Udainiya S., Madugundu A.K., Cutler J.A., Nirujogi R.S., Na C.H., Xu Y., Wu X., Pandey A. Mutation-Specific and Common Phosphotyrosine Signatures of KRAS G12D and G13D Alleles. J. Proteome Res. 2021;20:670–683. doi: 10.1021/acs.jproteome.0c00587. [DOI] [PubMed] [Google Scholar]
- 51.Varshavi D., Varshavi D., McCarthy N., Veselkov K., Keun H.C., Everett J.R. Metabolic characterization of colorectal cancer cells harbouring different KRAS mutations in codon 12, 13, 61 and 146 using human SW48 isogenic cell lines. Metabolomics. 2020;16:51. doi: 10.1007/s11306-020-01674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyssiotis C.A., Kimmelman A.C. Metabolic interactions in the tumor microenvironment. Trends Cell Biol. 2017;27:863–875. doi: 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi Y., Chhoeu C., Li J., Price K.S., Kiedrowski L.A., Hutchins J.L., Hardin A.I., Wei Z., Hong F., Bahcall M., et al. Silent mutations reveal therapeutic vulnerability in RAS Q61 cancers. Nature. 2022;603:335–342. doi: 10.1038/s41586-022-04451-4. [DOI] [PubMed] [Google Scholar]
- 54.Hunt R.C., Simhadri V.L., Iandoli M., Sauna Z.E., Kimchi-Sarfaty C. Exposing synonymous mutations. Trends Genet. 2014;30:308–321. doi: 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Sharma Y., Miladi M., Dukare S., Boulay K., Caudron-Herger M., Groß M., Backofen R., Diederichs S. A pan-cancer analysis of synonymous mutations. Nat. Commun. 2019;10:1–14. doi: 10.1038/s41467-019-10489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brest P., Lapaquette P., Souidi M., Lebrigand K., Cesaro A., Vouret-Craviari V., Mari B., Barbry P., Mosnier J.F., Hébuterne X., et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat. Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 57.Sauna Z.E., Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 58.Acker F., Stratmann J., Aspacher L., Nguyen N.T.T., Wagner S., Serve H., Wild P.J., Sebastian M. KRAS Mutations in Squamous Cell Carcinomas of the Lung. Front. Oncol. 2021;11:788084. doi: 10.3389/fonc.2021.788084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.El Osta B., Behera M., Kim S., Berry L.D., Sica G., Pillai R.N., Owonikoko T.K., Kris M.G., Johnson B.E., Kwiatkowski D.J., et al. Characteristics and Outcomes of Patients with Metastatic KRAS-Mutant Lung Adenocarcinomas: The Lung Cancer Mutation Consortium Experience. J. Thorac. Oncol. 2019;14:876–889. doi: 10.1016/j.jtho.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guibert N., Ilie M., Long E., Hofman V., Bouhlel L., Brest P., Mograbi B., Marquette C.H., Didier A., Mazieres J., et al. KRAS Mutations in Lung Adenocarcinoma: Molecular and Epidemiological Characteristics, Methods for Detection, and Therapeutic Strategy Perspectives. Curr. Mol. Med. 2015;15:418–432. doi: 10.2174/1566524015666150505161412. [DOI] [PubMed] [Google Scholar]
- 61.Jia Y., Jiang T., Li X., Zhao C., Zhang L., Zhao S., Liu X., Qiao M., Luo J., Shi J., et al. Characterization of distinct types of KRAS mutation and its impact on first-line platinum-based chemotherapy in Chinese patients with advanced non-small cell lung cancer. Oncol. Lett. 2017;14:6525–6532. doi: 10.3892/ol.2017.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y., Li H., Zhu J., Zhang Y., Liu X., Li R., Zhang Q., Cheng Y. The Prevalence and Concurrent Pathogenic Mutations of KRAS (G12C) in Northeast Chinese Non-small-cell Lung Cancer Patients. Cancer Manag. Res. 2021;13:2447–2454. doi: 10.2147/CMAR.S282617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma Z., Zhang Y., Deng C., Fu F., Deng L., Li Y., Chen H. The prognostic value of Kirsten rat sarcoma viral oncogene homolog mutations in resected lung adenocarcinoma differs according to clinical features. J. Thorac. Cardiovasc. Surg. 2022;163:e73–e85. doi: 10.1016/j.jtcvs.2020.05.097. [DOI] [PubMed] [Google Scholar]
- 64.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Si X., Pan R., Ma S., Li L., Liang L., Zhang P., Chu Y., Wang H., Wang M., Zhang X., et al. Genomic characteristics of driver genes in Chinese patients with non-small cell lung cancer. Thorac. Cancer. 2021;12:357–363. doi: 10.1111/1759-7714.13757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cavagna R., Escremim de Paula F., Sant’Anna D., Santana I., da Silva V.D., da Silva E.C.A., Bacchi C.E., Miziara J.E., Dias J.M., De Marchi P., et al. Frequency of KRAS p.Gly12Cys Mutation in Brazilian Patients with Lung Cancer. JCO Glob. Oncol. 2021;7:639–645. doi: 10.1200/GO.20.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das B.R., Bhaumik S., Ahmad F., Mandsaurwala A., Satam H. Molecular spectrum of somatic EGFR and KRAS gene mutations in non small cell lung carcinoma: Determination of frequency, distribution pattern and identification of novel variations in Indian patients. Pathol. Oncol. Res. 2015;21:675–687. doi: 10.1007/s12253-014-9874-7. [DOI] [PubMed] [Google Scholar]
- 68.Fathi Z., Mousavi S.A.J., Roudi R., Ghazi F. Distribution of KRAS, DDR2, and TP53 gene mutations in lung cancer: An analysis of Iranian patients. PLoS ONE. 2018;13:e0200633. doi: 10.1371/journal.pone.0200633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou F., Huang Y., Cai W., Li J., Su C., Ren S., Wu C., Zhou C. The genomic and immunologic profiles of pure pulmonary sarcomatoid carcinoma in Chinese patients. Lung Cancer. 2021;153:66–72. doi: 10.1016/j.lungcan.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 70.Cai L., Chen Y., Tong X., Wu X., Bao H., Shao Y., Luo Z., Wang X., Cao Y. The genomic landscape of young and old lung cancer patients highlights age-dependent mutation frequencies and clinical actionability in young patients. Int. J. Cancer. 2021;149:883–892. doi: 10.1002/ijc.33583. [DOI] [PubMed] [Google Scholar]
- 71.Deng C., Zheng Q., Zhang Y., Jin Y., Shen X., Nie X., Fu F., Ma X., Ma Z., Wen Z., et al. Validation of the Novel International Association for the Study of Lung Cancer Grading System for Invasive Pulmonary Adenocarcinoma and Association with Common Driver Mutations. J. Thorac. Oncol. 2021;16:1684–1693. doi: 10.1016/j.jtho.2021.07.006. [DOI] [PubMed] [Google Scholar]
- 72.Nassar A.H., Adib E., Kwiatkowski D.J. Distribution of KRAS (G12C) Somatic Mutations across Race, Sex, and Cancer Type. N. Engl. J. Med. 2021;384:185–187. doi: 10.1056/NEJMc2030638. [DOI] [PubMed] [Google Scholar]
- 73.Saleh M.M., Scheffler M., Merkelbach-Bruse S., Scheel A.H., Ulmer B., Wolf J., Buettner R. Comprehensive Analysis of TP53 and KEAP1 Mutations and Their Impact on Survival in Localized- and Advanced-Stage NSCLC. J. Thorac. Oncol. 2021;17:76–88. doi: 10.1016/j.jtho.2021.08.764. [DOI] [PubMed] [Google Scholar]
- 74.Judd J., Abdel Karim N., Khan H., Naqash A.R., Baca Y., Xiu J., VanderWalde A.M., Mamdani H., Raez L.E., Nagasaka M., et al. Characterization of KRAS Mutation Subtypes in Non-small Cell Lung Cancer. Mol. Cancer Ther. 2021;20:2577–2584. doi: 10.1158/1535-7163.MCT-21-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malapelle U., Passiglia F., Cremolini C., Reale M.L., Pepe F., Pisapia P., Avallone A., Cortinovis D., De Stefano A., Fassan M., et al. RAS as a positive predictive biomarker: Focus on lung and colorectal cancer patients. Eur. J. Cancer. 2021;146:74–83. doi: 10.1016/j.ejca.2021.01.015. [DOI] [PubMed] [Google Scholar]
- 76.Sebastian M., Eberhardt W.E.E., Hoffknecht P., Metzenmacher M., Wehler T., Kokowski K., Alt J., Schütte W., Büttner R., Heukamp L.C., et al. KRAS G12C-mutated advanced non-small cell lung cancer: A real-world cohort from the German prospective, observational, nation-wide CRISP Registry (AIO-TRK-0315) Lung Cancer. 2021;154:51–61. doi: 10.1016/j.lungcan.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 77.Riely G.J., Marks J., Pao W. KRAS mutations in non-small cell lung cancer. Proc. Am. Thorac. Soc. 2009;6:201–205. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 78.Du X., Yang B., An Q., Assaraf Y.G., Cao X., Xia J. Acquired resistance to third-generation EGFR-TKIs and emerging next-generation EGFR inhibitors. Innovation. 2021;2:100103. doi: 10.1016/j.xinn.2021.100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Serna-Blasco R., Sánchez-Herrero E., Sanz-Moreno S., Rodriguez-Festa A., García-Veros E., Casarrubios M., Sierra-Rodero B., Laza-Briviesca R., Cruz-Bermúdez A., Mielgo-Rubio X., et al. KRAS p.G12C mutation occurs in 1% of EGFR-mutated advanced non-small-cell lung cancer patients progressing on a first-line treatment with a tyrosine kinase inhibitor. ESMO Open. 2021;6:100279. doi: 10.1016/j.esmoop.2021.100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aviel-Ronen S., Blackhall F.H., Shepherd F.A., Tsao M.S. K-ras mutations in non-small-cell lung carcinoma: A review. Clin. Lung Cancer. 2006;8:30–38. doi: 10.3816/CLC.2006.n.030. [DOI] [PubMed] [Google Scholar]
- 81.Karachaliou N., Mayo C., Costa C., Magrí I., Gimenez-Capitan A., Molina-Vila M.A., Rosell R. KRAS mutations in lung cancer. Clin. Lung Cancer. 2013;14:205–214. doi: 10.1016/j.cllc.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 82.Mascaux C., Iannino N., Martin B., Paesmans M., Berghmans T., Dusart M., Haller A., Lothaire P., Meert A.P., Noel S., et al. The role of RAS oncogene in survival of patients with lung cancer: A systematic review of the literature with meta-analysis. Br. J. Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finn S.P., Addeo A., Dafni U., Thunnissen E., Bubendorf L., Madsen L.B., Biernat W., Verbeken E., Hernandez-Losa J., Marchetti A., et al. Prognostic Impact of KRAS G12C Mutation in Patients with NSCLC: Results from the European Thoracic Oncology Platform Lungscape Project. J. Thorac. Oncol. 2021;16:990–1002. doi: 10.1016/j.jtho.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 84.Jones G.D., Caso R., Tan K.S., Mastrogiacomo B., Sanchez-Vega F., Liu Y., Connolly J.G., Murciano-Goroff Y.R., Bott M.J., Adusumilli P.S., et al. KRAS G12C Mutation Is Associated with Increased Risk of Recurrence in Surgically Resected Lung Adenocarcinoma. Clin. Cancer Res. 2021;27:2604–2612. doi: 10.1158/1078-0432.CCR-20-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith S.A., van Berkel V.H. Commentary: Every detail matters-Understanding the impact of KRAS mutations. J. Thorac. Cardiovasc. Surg. 2022;163:e88–e89. doi: 10.1016/j.jtcvs.2020.06.024. [DOI] [PubMed] [Google Scholar]
- 86.Gao G., Liao W., Ma Q., Zhang B., Chen Y., Wang Y. KRAS G12D mutation predicts lower TMB and drives immune suppression in lung adenocarcinoma. Lung Cancer. 2020;149:41–45. doi: 10.1016/j.lungcan.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 87.Schoenfeld A.J., Rizvi H., Bandlamudi C., Sauter J.L., Travis W.D., Rekhtman N., Plodkowski A.J., Perez-Johnston R., Sawan P., Beras A., et al. Clinical and molecular correlates of PD-L1 expression in patients with lung adenocarcinomas. Ann. Oncol. 2020;31:599–608. doi: 10.1016/j.annonc.2020.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goulding R.E., Chenoweth M., Carter G.C., Boye M.E., Sheffield K.M., John W.J., Leusch M.S., Muehlenbein C.E., Li L., Jen M.H., et al. KRAS mutation as a prognostic factor and predictive factor in advanced/metastatic non-small cell lung cancer: A systematic literature review and meta-analysis. Cancer Treat. Res. Commun. 2020;24:100200. doi: 10.1016/j.ctarc.2020.100200. [DOI] [PubMed] [Google Scholar]
- 89.Aredo J.V., Padda S.K., Kunder C.A., Han S.S., Neal J.W., Shrager J.B., Wakelee H.A. Impact of KRAS mutation subtype and concurrent pathogenic mutations on non-small cell lung cancer outcomes. Lung Cancer. 2019;133:144–150. doi: 10.1016/j.lungcan.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scheffler M., Ihle M.A., Hein R., Merkelbach-Bruse S., Scheel A.H., Siemanowski J., Brägelmann J., Kron A., Abedpour N., Ueckeroth F., et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J. Thorac. Oncol. 2019;14:606–616. doi: 10.1016/j.jtho.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 91.Skoulidis F., Heymach J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arbour K.C., Jordan E., Kim H.R., Dienstag J., Yu H.A., Sanchez-Vega F., Lito P., Berger M., Solit D.B., Hellmann M., et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018;24:334–340. doi: 10.1158/1078-0432.CCR-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lei L., Wang W.X., Yu Z.Y., Liang X.B., Pan W.W., Chen H.F., Wang L.P., Fang Y., Wang M., Xu C.W., et al. Real-World Study in Advanced Non-Small Cell Lung Cancer with KRAS Mutations. Transl. Oncol. 2020;13:329–335. doi: 10.1016/j.tranon.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spira A.I., Tu H., Aggarwal S., Hsu H., Carrigan G., Wang X., Ngarmchamnanrith G., Chia V., Gray J.E. A retrospective observational study of the natural history of advanced non-small-cell lung cancer in patients with KRAS p.G12C mutated or wild-type disease. Lung Cancer. 2021;159:1–9. doi: 10.1016/j.lungcan.2021.05.026. [DOI] [PubMed] [Google Scholar]
- 95.Liu Y., Xu F., Wang Y., Wu Q., Wang B., Yao Y., Zhang Y., Han-Zhang H., Ye J., Zhang L., et al. Mutations in exon 8 of TP53 are associated with shorter survival in patients with advanced lung cancer. Oncol. Lett. 2019;18:3159–3169. doi: 10.3892/ol.2019.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Corsini E.M., Ripley R.T. Commentary: KRAS-mutant lung adenocarcinomas—A work in progress. J. Thorac. Cardiovasc. Surg. 2022;163:e87–e88. doi: 10.1016/j.jtcvs.2020.06.037. [DOI] [PubMed] [Google Scholar]
- 97.Di Federico A., De Giglio A., Parisi C., Gelsomino F. STK11/LKB1 and KEAP1 mutations in non-small cell lung cancer: Prognostic rather than predictive? Eur. J. Cancer. 2021;157:108–113. doi: 10.1016/j.ejca.2021.08.011. [DOI] [PubMed] [Google Scholar]
- 98.Krishnamurthy N., Goodman A.M., Barkauskas D.A., Kurzrock R. STK11 alterations in the pan-cancer setting: Prognostic and therapeutic implications. Eur. J. Cancer. 2021;148:215–229. doi: 10.1016/j.ejca.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Izar B., Zhou H., Heist R.S., Azzoli C.G., Muzikansky A., Scribner E.E., Bernardo L.A., Dias-Santagata D., Iafrate A.J., Lanuti M. The prognostic impact of KRAS, its codon and amino acid specific mutations, on survival in resected stage I lung adenocarcinoma. J. Thorac. Oncol. 2014;9:1363–1369. doi: 10.1097/JTO.0000000000000266. [DOI] [PubMed] [Google Scholar]
- 100.Kadota K., Sima C.S., Arcila M.E., Hedvat C., Kris M.G., Jones D.R., Adusumilli P.S., Travis W.D. KRAS Mutation Is a Significant Prognostic Factor in Early-stage Lung Adenocarcinoma. Am. J. Surg. Pathol. 2016;40:1579–1590. doi: 10.1097/PAS.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim Y.T., Kim T.Y., Lee D.S., Park S.J., Park J.Y., Seo S.J., Choi H.S., Kang H.J., Hahn S., Kang C.H., et al. Molecular changes of epidermal growth factor receptor (EGFR) and KRAS and their impact on the clinical outcomes in surgically resected adenocarcinoma of the lung. Lung Cancer. 2008;59:111–118. doi: 10.1016/j.lungcan.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 102.Mograbi B., Heeke S., Hofman P. The Importance of STK11/LKB1 Assessment in Non-Small Cell Lung Carcinomas. Diagnostics. 2021;11:196. doi: 10.3390/diagnostics11020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ricciuti B., Arbour K.C., Lin J.J., Vajdi A., Vokes N., Hong L., Zhang J., Tolstorukov M.Y., Li Y.Y., Spurr L.F., et al. Diminished efficacy of PD-(L)1 inhibition in STK11- and KEAP1-mutant lung adenocarcinoma is impacted by KRAS mutation status. J. Thorac. Oncol. 2022;17:399–410. doi: 10.1016/j.jtho.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Skoulidis F., Goldberg M.E., Greenawalt D.M., Hellmann M.D., Awad M.M., Gainor J.F., Schrock A.B., Hartmaier R.J., Trabucco S.E., Gay L., et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wahl S.G.F., Dai H.Y., Emdal E.F., Berg T., Halvorsen T.O., Ottestad A.L., Lund-Iversen M., Brustugun O.T., Førde D., Paulsen E.E., et al. The Prognostic Effect of KRAS Mutations in Non-Small Cell Lung Carcinoma Revisited: A Norwegian Multicentre Study. Cancers. 2021;13:4294. doi: 10.3390/cancers13174294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singh A., Daemen A., Nickles D., Jeon S.M., Foreman O., Sudini K., Gnad F., Lajoie S., Gour N., Mitzner W., et al. NRF2 Activation Promotes Aggressive Lung Cancer and Associates with Poor Clinical Outcomes. Clin. Cancer Res. 2021;27:877–888. doi: 10.1158/1078-0432.CCR-20-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fung A.S., Karimi M., Michiels S., Seymour L., Brambilla E., Le-Chevalier T., Soria J.C., Kratzke R., Graziano S.L., Devarakonda S., et al. Prognostic and predictive effect of KRAS gene copy number and mutation status in early stage non-small cell lung cancer patients. Transl. Lung Cancer Res. 2021;10:826–838. doi: 10.21037/tlcr-20-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Falk A.T., Yazbeck N., Guibert N., Chamorey E., Paquet A., Ribeyre L., Bence C., Zahaf K., Leroy S., Marquette C.H., et al. Effect of mutant variants of the KRAS gene on PD-L1 expression and on the immune microenvironment and association with clinical outcome in lung adenocarcinoma patients. Lung Cancer. 2018;121:70–75. doi: 10.1016/j.lungcan.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 109.Nadal E., Chen G., Prensner J.R., Shiratsuchi H., Sam C., Zhao L., Kalemkerian G.P., Brenner D., Lin J., Reddy R.M., et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J. Thorac. Oncol. 2014;9:1513–1522. doi: 10.1097/JTO.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 110.Renaud S., Falcoz P.E., Schaëffer M., Guenot D., Romain B., Olland A., Reeb J., Santelmo N., Chenard M.P., Legrain M., et al. Prognostic value of the KRAS G12V mutation in 841 surgically resected Caucasian lung adenocarcinoma cases. Br. J. Cancer. 2015;113:1206–1215. doi: 10.1038/bjc.2015.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shepherd F.A., Domerg C., Hainaut P., Jänne P.A., Pignon J.P., Graziano S., Douillard J.Y., Brambilla E., Le Chevalier T., Seymour L., et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J. Clin. Oncol. 2013;31:2173–2181. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tao L., Miao R., Mekhail T., Sun J., Meng L., Fang C., Guan J., Jain A., Du Y., Allen A., et al. Prognostic Value of KRAS Mutation Subtypes and PD-L1 Expression in Patients with Lung Adenocarcinoma. Clin. Lung Cancer. 2021;22:e506–e511. doi: 10.1016/j.cllc.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 113.Calles A., Liao X., Sholl L.M., Rodig S.J., Freeman G.J., Butaney M., Lydon C., Dahlberg S.E., Hodi F.S., Oxnard G.R., et al. Expression of PD-1 and Its Ligands, PD-L1 and PD-L2, in Smokers and Never Smokers with KRAS-Mutant Lung Cancer. J. Thorac. Oncol. 2015;10:1726–1735. doi: 10.1097/JTO.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 114.Dong Z.Y., Zhong W.Z., Zhang X.C., Su J., Xie Z., Liu S.Y., Tu H.Y., Chen H.J., Sun Y.L., Zhou Q., et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 115.Jeanson A., Tomasini P., Souquet-Bressand M., Brandone N., Boucekine M., Grangeon M., Chaleat S., Khobta N., Milia J., Mhanna L., et al. Efficacy of Immune Checkpoint Inhibitors in KRAS-Mutant Non-Small Cell Lung Cancer (NSCLC) J. Thorac. Oncol. 2019;14:1095–1101. doi: 10.1016/j.jtho.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 116.Liu C., Zheng S., Jin R., Wang X., Wang F., Zang R., Xu H., Lu Z., Huang J., Lei Y., et al. The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett. 2020;470:95–105. doi: 10.1016/j.canlet.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 117.Garassino M.C., Marabese M., Rusconi P., Rulli E., Martelli O., Farina G., Scanni A., Broggini M. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann. Oncol. 2011;22:235–237. doi: 10.1093/annonc/mdq680. [DOI] [PubMed] [Google Scholar]
- 118.Zer A., Ding K., Lee S.M., Goss G.D., Seymour L., Ellis P.M., Hackshaw A., Bradbury P.A., Han L., O’Callaghan C.J., et al. Pooled Analysis of the Prognostic and Predictive Value of KRAS Mutation Status and Mutation Subtype in Patients with Non-Small Cell Lung Cancer Treated with Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors. J. Thorac. Oncol. 2016;11:312–323. doi: 10.1016/j.jtho.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 119.Moore A.R., Rosenberg S.C., McCormick F., Malek S. RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 2020;19:533–552. doi: 10.1038/s41573-020-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lindsay C.R., Jamal-Hanjani M., Forster M., Blackhall F. KRAS: Reasons for optimism in lung cancer. Eur. J. Cancer. 2018;99:20–27. doi: 10.1016/j.ejca.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 122.Janes M.R., Zhang J., Li L.S., Hansen R., Peters U., Guo X., Chen Y., Babbar A., Firdaus S.J., Darjania L., et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell. 2018;172:578–589.e17. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 123.Lindsay C.R., Garassino M.C., Nadal E., Öhrling K., Scheffler M., Mazières J. On target: Rational approaches to KRAS inhibition for treatment of non-small cell lung carcinoma. Lung Cancer. 2021;160:152–165. doi: 10.1016/j.lungcan.2021.07.005. [DOI] [PubMed] [Google Scholar]
- 124.Briere D.M., Li S., Calinisan A., Sudhakar N., Aranda R., Hargis L., Peng D.H., Deng J., Engstrom L.D., Hallin J., et al. The KRAS(G12C) Inhibitor MRTX849 Reconditions the Tumor Immune Microenvironment and Sensitizes Tumors to Checkpoint Inhibitor Therapy. Mol. Cancer Ther. 2021;20:975–985. doi: 10.1158/1535-7163.MCT-20-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Canon J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D., Gaida K., Holt T., Knutson C.G., Koppada N., et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 126.Fell J.B., Fischer J.P., Baer B.R., Blake J.F., Bouhana K., Briere D.M., Brown K.D., Burgess L.E., Burns A.C., Burkard M.R., et al. Identification of the Clinical Development Candidate MRTX849, a Covalent KRAS(G12C) Inhibitor for the Treatment of Cancer. J. Med. Chem. 2020;63:6679–6693. doi: 10.1021/acs.jmedchem.9b02052. [DOI] [PubMed] [Google Scholar]
- 127.Lanman B.A., Allen J.R., Allen J.G., Amegadzie A.K., Ashton K.S., Booker S.K., Chen J.J., Chen N., Frohn M.J., Goodman G., et al. Discovery of a Covalent Inhibitor of KRAS(G12C) (AMG 510) for the Treatment of Solid Tumors. J. Med. Chem. 2020;63:52–65. doi: 10.1021/acs.jmedchem.9b01180. [DOI] [PubMed] [Google Scholar]
- 128.Hallin J., Engstrom L.D., Hargis L., Calinisan A., Aranda R., Briere D.M., Sudhakar N., Bowcut V., Baer B.R., Ballard J.A., et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Naim N., Moukheiber S., Daou S., Kourie H.R. KRAS-G12C covalent inhibitors: A game changer in the scene of cancer therapies. Crit. Rev. Oncol. Hematol. 2021;168:103524. doi: 10.1016/j.critrevonc.2021.103524. [DOI] [PubMed] [Google Scholar]
- 130.Palma G., Khurshid F., Lu K., Woodward B., Husain H. Selective KRAS G12C inhibitors in non-small cell lung cancer: Chemistry, concurrent pathway alterations, and clinical outcomes. NPJ Precis. Oncol. 2021;5:98. doi: 10.1038/s41698-021-00237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Arbour K.C., Rizvi H., Plodkowski A.J., Hellmann M.D., Knezevic A., Heller G., Yu H.A., Ladanyi M., Kris M.G., Arcila M.E., et al. Treatment Outcomes and Clinical Characteristics of Patients with KRAS-G12C-Mutant Non-Small Cell Lung Cancer. Clin. Cancer Res. 2021;27:2209–2215. doi: 10.1158/1078-0432.CCR-20-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Blair H.A. Sotorasib: First Approval. Drugs. 2021;81:1573–1579. doi: 10.1007/s40265-021-01574-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Köhler J., Jänne P.A. If Virchow and Ehrlich Had Dreamt Together: What the Future Holds for KRAS-Mutant Lung Cancer. Int. J. Mol. Sci. 2021;22:3025. doi: 10.3390/ijms22063025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sunaga N., Miura Y., Kasahara N., Sakurai R. Targeting Oncogenic KRAS in Non-Small-Cell Lung Cancer. Cancers. 2021;13:5956. doi: 10.3390/cancers13235956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang S.S., Nagasaka M. Spotlight on Sotorasib (AMG 510) for KRAS (G12C) Positive Non-Small Cell Lung Cancer. Lung Cancer. 2021;12:115–122. doi: 10.2147/LCTT.S334623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nagasaka M., Li Y., Sukari A., Ou S.I., Al-Hallak M.N., Azmi A.S. KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne? Cancer Treat. Rev. 2020;84:101974. doi: 10.1016/j.ctrv.2020.101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nagasaka M., Potugari B., Nguyen A., Sukari A., Azmi A.S., Ou S.I. KRAS Inhibitors- yes but what next? Direct targeting of KRAS- vaccines, adoptive T cell therapy and beyond. Cancer Treat. Rev. 2021;101:102309. doi: 10.1016/j.ctrv.2021.102309. [DOI] [PubMed] [Google Scholar]
- 138.Uprety D., Adjei A.A. KRAS: From undruggable to a druggable Cancer Target. Cancer Treat. Rev. 2020;89:102070. doi: 10.1016/j.ctrv.2020.102070. [DOI] [PubMed] [Google Scholar]
- 139.Zhang J., Zhang J., Liu Q., Fan X.X., Leung E.L., Yao X.J., Liu L. Resistance looms for KRAS G12C inhibitors and rational tackling strategies. Pharmacol. Ther. 2022;229:108050. doi: 10.1016/j.pharmthera.2021.108050. [DOI] [PubMed] [Google Scholar]
- 140.Akhave N.S., Biter A.B., Hong D.S. Mechanisms of Resistance to KRAS(G12C)-Targeted Therapy. Cancer Discov. 2021;11:1345–1352. doi: 10.1158/2159-8290.CD-20-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Awad M.M., Liu S., Rybkin I.I., Arbour K.C., Dilly J., Zhu V.W., Johnson M.L., Heist R.S., Patil T., Riely G.J., et al. Acquired Resistance to KRASG12C Inhibition in Cancer. N. Engl. J. Med. 2021;384:2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reita D., Pabst L., Pencreach E., Guérin E., Dano L., Rimelen V., Voegeli A.C., Vallat L., Mascaux C., Beau-Faller M. Direct Targeting KRAS Mutation in Non-Small Cell Lung Cancer: Focus on Resistance. Cancers. 2022;14:1321. doi: 10.3390/cancers14051321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao Y., Murciano-Goroff Y.R., Xue J.Y., Ang A., Lucas J., Mai T.T., Da Cruz Paula A.F., Saiki A.Y., Mohn D., Achanta P., et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature. 2021;599:679–683. doi: 10.1038/s41586-021-04065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Addeo A., Banna G.L., Friedlaender A. KRAS G12C Mutations in NSCLC: From Target to Resistance. Cancers. 2021;13:2541. doi: 10.3390/cancers13112541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Blaquier J.B., Cardona A.F., Recondo G. Resistance to KRASG12C Inhibitors in Non-Small Cell Lung Cancer. Front. Oncol. 2021;11:787585. doi: 10.3389/fonc.2021.787585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Colombo M., Ndembe G., Broggini M. KRAS Targeting and Resistance: Anticipating the Expectable. J. Thorac. Oncol. 2021;16:1239–1241. doi: 10.1016/j.jtho.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 147.Dunnett-Kane V., Nicola P., Blackhall F., Lindsay C. Mechanisms of Resistance to KRASG12C Inhibitors. Cancers. 2021;13:151. doi: 10.3390/cancers13010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hata A.N., Shaw A.T. Resistance looms for KRASG12C inhibitors. Nat. Med. 2020;26:169–170. doi: 10.1038/s41591-020-0765-z. [DOI] [PubMed] [Google Scholar]
- 149.Koga T., Suda K., Fujino T., Ohara S., Hamada A., Nishino M., Chiba M., Shimoji M., Takemoto T., Arita T., et al. KRAS Secondary Mutations That Confer Acquired Resistance to KRAS G12C Inhibitors, Sotorasib and Adagrasib, and Overcoming Strategies: Insights from In Vitro Experiments. J. Thorac. Oncol. 2021;16:1321–1332. doi: 10.1016/j.jtho.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 150.Liu J., Kang R., Tang D. The KRAS-G12C inhibitor: Activity and resistance. Cancer Gene Ther. 2021 doi: 10.1038/s41417-021-00383-9. [DOI] [PubMed] [Google Scholar]
- 151.Moore A.R., Malek S. The promise and peril of KRAS G12C inhibitors. Cancer Cell. 2021;39:1059–1061. doi: 10.1016/j.ccell.2021.07.011. [DOI] [PubMed] [Google Scholar]
- 152.Suzuki S., Yonesaka K., Teramura T., Takehara T., Kato R., Sakai H., Haratani K., Tanizaki J., Kawakami H., Hayashi H., et al. KRAS Inhibitor Resistance in MET-Amplified KRAS (G12C) Non-Small Cell Lung Cancer Induced By RAS- and Non-RAS-Mediated Cell Signaling Mechanisms. Clin. Cancer Res. 2021;27:5697–5707. doi: 10.1158/1078-0432.CCR-21-0856. [DOI] [PubMed] [Google Scholar]
- 153.Tanaka N., Lin J.J., Li C., Ryan M.B., Zhang J., Kiedrowski L.A., Michel A.G., Syed M.U., Fella K.A., Sakhi M., et al. Clinical Acquired Resistance to KRAS(G12C) Inhibition through a Novel KRAS Switch-II Pocket Mutation and Polyclonal Alterations Converging on RAS-MAPK Reactivation. Cancer Discov. 2021;11:1913–1922. doi: 10.1158/2159-8290.CD-21-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tang D., Kroemer G., Kang R. Oncogenic KRAS blockade therapy: Renewed enthusiasm and persistent challenges. Mol. Cancer. 2021;20:128. doi: 10.1186/s12943-021-01422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tsai Y.S., Woodcock M.G., Azam S.H., Thorne L.B., Kanchi K.L., Parker J.S., Vincent B.G., Pecot C.V. Rapid idiosyncratic mechanisms of clinical resistance to KRAS G12C inhibition. J. Clin. Investig. 2022;132:e155523. doi: 10.1172/JCI155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ryan M.B., Fece de la Cruz F., Phat S., Myers D.T., Wong E., Shahzade H.A., Hong C.B., Corcoran R.B. Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS(G12C) Inhibition. Clin. Cancer Res. 2020;26:1633–1643. doi: 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xue J.Y., Zhao Y., Aronowitz J., Mai T.T., Vides A., Qeriqi B., Kim D., Li C., de Stanchina E., Mazutis L., et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature. 2020;577:421–425. doi: 10.1038/s41586-019-1884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ruess D.A., Heynen G.J., Ciecielski K.J., Ai J., Berninger A., Kabacaoglu D., Görgülü K., Dantes Z., Wörmann S.M., Diakopoulos K.N., et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat. Med. 2018;24:954–960. doi: 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 159.Singh A., Greninger P., Rhodes D., Koopman L., Violette S., Bardeesy N., Settleman J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hofman P. Detecting Resistance to Therapeutic ALK Inhibitors in Tumor Tissue and Liquid Biopsy Markers: An Update to a Clinical Routine Practice. Cells. 2021;10:168. doi: 10.3390/cells10010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Reita D., Pabst L., Pencreach E., Guérin E., Dano L., Rimelen V., Voegeli A.C., Vallat L., Mascaux C., Beau-Faller M. Molecular Mechanism of EGFR-TKI Resistance in EGFR-Mutated Non-Small Cell Lung Cancer: Application to Biological Diagnostic and Monitoring. Cancers. 2021;13:4926. doi: 10.3390/cancers13194926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guo J., Liu Y., Lv J., Zou B., Chen Z., Li K., Feng J., Cai Z., Wei L., Liu M., et al. BCL6 confers KRAS-mutant non-small-cell lung cancer resistance to BET inhibitors. J. Clin. Investig. 2021;131:e133090. doi: 10.1172/JCI133090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. [(accessed on 6 March 2022)]; Available online: https://www.clinicaltrials.gov/ct2/show/NCT04185883.
- 165.Noordhof A.L., Damhuis R.A.M., Hendriks L.E.L., de Langen A.J., Timens W., Venmans B.J.W., van Geffen W.H. Prognostic impact of KRAS mutation status for patients with stage IV adenocarcinoma of the lung treated with first-line pembrolizumab monotherapy. Lung Cancer. 2021;155:163–169. doi: 10.1016/j.lungcan.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 166.Shirasawa M., Yoshida T., Shimoda Y., Takayanagi D., Shiraishi K., Kubo T., Mitani S., Matsumoto Y., Masuda K., Shinno Y., et al. Differential Immune-Related Microenvironment Determines Programmed Cell Death Protein-1/Programmed Death-Ligand 1 Blockade Efficacy in Patients with Advanced NSCLC. J. Thorac. Oncol. 2021;16:2078–2090. doi: 10.1016/j.jtho.2021.07.027. [DOI] [PubMed] [Google Scholar]
- 167.Sun L., Hsu M., Cohen R.B., Langer C.J., Mamtani R., Aggarwal C. Association Between KRAS Variant Status and Outcomes with First-line Immune Checkpoint Inhibitor-Based Therapy in Patients with Advanced Non-Small-Cell Lung Cancer. JAMA Oncol. 2021;7:937–939. doi: 10.1001/jamaoncol.2021.0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lauko A., Kotecha R., Barnett A., Li H., Tatineni V., Ali A., Patil P., Mohammadi A.M., Chao S.T., Murphy E.S., et al. Impact of KRAS mutation status on the efficacy of immunotherapy in lung cancer brain metastases. Sci. Rep. 2021;11:18174. doi: 10.1038/s41598-021-97566-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Herbst R.S., Lopes G., Kowalski D.M. Associated of KRAS mutation status with response to prembrolizumab monotherapy given as fist-line therapy for PD-L1 positive advanced nonsquamous NSCLC in keynote; Proceedings of the ESMO Immuno-Oncology Congress; Geneva, Switzerland. 11–14 December 2019. [Google Scholar]
- 170.Gu M., Xu T., Chang P. KRAS/LKB1 and KRAS/TP53 co-mutations create divergent immune signatures in lung adenocarcinomas. Ther. Adv. Med. Oncol. 2021;13:17588359211006950. doi: 10.1177/17588359211006950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pathak R., Salgia R. Near-Complete Response to Combined Pembrolizumab and Platinum-Doublet in a Patient with STK11/KRAS Mutated Advanced Lung Adenocarcinoma. Clin. Lung Cancer. 2021;23:E137–E139. doi: 10.1016/j.cllc.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 172.Pelosi G. KEAP1 and TP53 (Co)mutation in Lung Adenocarcinoma: Another Bullet for Immunotherapy? J. Thorac. Oncol. 2021;16:1979–1983. doi: 10.1016/j.jtho.2021.10.004. [DOI] [PubMed] [Google Scholar]
- 173.Pore N., Wu S., Standifer N., Jure-Kunkel M., de Los Reyes M., Shrestha Y., Halpin R., Rothstein R., Mulgrew K., Blackmore S., et al. Resistance to Durvalumab and Durvalumab plus Tremelimumab Is Associated with Functional STK11 Mutations in Patients with Non-Small Cell Lung Cancer and Is Reversed by STAT3 Knockdown. Cancer Discov. 2021;11:2828–2845. doi: 10.1158/2159-8290.CD-20-1543. [DOI] [PubMed] [Google Scholar]
- 174.Pavan A., Bragadin A.B., Calvetti L., Ferro A., Zulato E., Attili I., Nardo G., Dal Maso A., Frega S., Menin A.G., et al. Role of next generation sequencing-based liquid biopsy in advanced non-small cell lung cancer patients treated with immune checkpoint inhibitors: Impact of STK11, KRAS and TP53 mutations and co-mutations on outcome. Transl. Lung Cancer Res. 2021;10:202–220. doi: 10.21037/tlcr-20-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Frost N., Kollmeier J., Vollbrecht C., Grah C., Matthes B., Pultermann D., von Laffert M., Lüders H., Olive E., Raspe M., et al. KRAS(G12C)/TP53 co-mutations identify long-term responders to first line palliative treatment with pembrolizumab monotherapy in PD-L1 high (50%) lung adenocarcinoma. Transl. Lung Cancer Res. 2021;10:737–752. doi: 10.21037/tlcr-20-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Scalera S., Mazzotta M., Corleone G., Sperati F., Terrenato I., Krasniqi E., Pizzuti L., Barba M., Vici P., Gallo E., et al. KEAP1 and TP53 Frame Genomic, Evolutionary, and Immunologic Subtypes of Lung Adenocarcinoma with Different Sensitivity to Immunotherapy. J. Thorac. Oncol. 2021;16:2065–2077. doi: 10.1016/j.jtho.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 177.Liu L., Ahmed T., Petty W.J., Grant S., Ruiz J., Lycan T.W., Topaloglu U., Chou P.C., Miller L.D., Hawkins G.A., et al. SMARCA4 mutations in KRAS-mutant lung adenocarcinoma: A multi-cohort analysis. Mol. Oncol. 2021;15:462–472. doi: 10.1002/1878-0261.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Planchard D., Popat S., Kerr K., Novello S., Smit E.F., Faivre-Finn C., Mok T.S., Reck M., Van Schil P.E., Hellmann M.D., et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018;4:iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 179. [(accessed on 8 March 2022)]; Available online: https://www.clinicaltrials.gov/ct2/show/NCT04933695.
- 180.Gibert J., Clavé S., Hardy-Werbin M., Taus Á., Rocha P., Longarón R., Piquer G., Chaib I., Carcereny E., Morán T., et al. Concomitant genomic alterations in KRAS mutant advanced lung adenocarcinoma. Lung Cancer. 2020;140:42–45. doi: 10.1016/j.lungcan.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 181.Amanam I., Mambetsariev I., Gupta R., Achuthan S., Wang Y., Pharaon R., Massarelli E., Koczywas M., Reckamp K., Salgia R. Role of immunotherapy and co-mutations on KRAS-mutant non-small cell lung cancer survival. J. Thorac. Dis. 2020;12:5086–5095. doi: 10.21037/jtd.2020.04.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Davis A.P., Cooper W.A., Boyer M., Lee J.H., Pavlakis N., Kao S.C. Efficacy of immunotherapy in KRAS-mutant non-small-cell lung cancer with comutations. Immunotherapy. 2021;13:941–952. doi: 10.2217/imt-2021-0090. [DOI] [PubMed] [Google Scholar]
- 183.Van Maldegem F., Downward J. Mutant KRAS at the Heart of Tumor Immune Evasion. Immunity. 2020;52:14–16. doi: 10.1016/j.immuni.2019.12.013. [DOI] [PubMed] [Google Scholar]
- 184.Nadal E., Heeke S., Benzaquen J., Vilariño N., Navarro A., Azuara D., Hofman P. Two Patients with Advanced-Stage Lung Adenocarcinoma with Radiologic Complete Response to Nivolumab Treatment Harboring an STK11/LKB1 Mutation. JCO Precis. Oncol. 2020;11:1238–1245. doi: 10.1200/PO.20.00174. [DOI] [PubMed] [Google Scholar]
- 185.De Kock R., Knoops C., Baselmans M., Borne B.V.D., Brunsveld L., Scharnhorst V., Deiman B. Sensitive cell-free tumor DNA analysis in supernatant pleural effusions supports therapy selection and disease monitoring of lung cancer patients. Cancer Treat. Res. Commun. 2021;29:100449. doi: 10.1016/j.ctarc.2021.100449. [DOI] [PubMed] [Google Scholar]
- 186.Merker J.D., Oxnard G.R., Compton C., Diehn M., Hurley P., Lazar A.J., Lindeman N., Lockwood C.M., Rai A.J., Schilsky R.L., et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 187.Nacchio M., Sgariglia R., Gristina V., Pisapia P., Pepe F., De Luca C., Migliatico I., Clery E., Greco L., Vigliar E., et al. KRAS mutations testing in non-small cell lung cancer: The role of Liquid biopsy in the basal setting. J. Thorac. Dis. 2020;12:3836–3843. doi: 10.21037/jtd.2020.01.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Veluswamy R., Mack P.C., Houldsworth J., Elkhouly E., Hirsch F.R. KRAS G12C-Mutant Non-Small Cell Lung Cancer: Biology, Developmental Therapeutics, and Molecular Testing. J. Mol. Diagn. 2021;23:507–520. doi: 10.1016/j.jmoldx.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 189.Vessies D.C.L., Greuter M.J.E., van Rooijen K.L., Linders T.C., Lanfermeijer M., Ramkisoensing K.L., Meijer G.A., Koopman M., Coupé V.M.H., Vink G.R., et al. Performance of four platforms for KRAS mutation detection in plasma cell-free DNA: ddPCR, Idylla, COBAS z480 and BEAMing. Sci. Rep. 2020;10:8122. doi: 10.1038/s41598-020-64822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Roosan M.R., Mambetsariev I., Pharaon R., Fricke J., Husain H., Reckamp K.L., Koczywas M., Massarelli E., Bild A.H., Salgia R. Usefulness of Circulating Tumor DNA in Identifying Somatic Mutations and Tracking Tumor Evolution in Patients with Non-small Cell Lung Cancer. Chest. 2021;160:1095–1107. doi: 10.1016/j.chest.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Horgan D., Curigliano G., Rieß O., Hofman P., Büttner R., Conte P., Čufer T., Gallagher W.M., Georges N., Kerr K., et al. Identifying the Steps Required to Effectively Implement Next-Generation Sequencing in Oncology at a National Level in Europe. J. Pers. Med. 2022;12:72. doi: 10.3390/jpm12010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hofman P. The challenges of evaluating predictive biomarkers using small biopsy tissue samples and liquid biopsies from non-small cell lung cancer patients. J. Thorac. Dis. 2019;11((Suppl. 1)):S57–S64. doi: 10.21037/jtd.2018.11.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bruno R., Fontanini G. Next Generation Sequencing for Gene Fusion Analysis in Lung Cancer: A Literature Review. Diagnostics. 2020;10:521. doi: 10.3390/diagnostics10080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Legras A., Barritault M., Tallet A., Fabre E., Guyard A., Rance B., Digan W., Pecuchet N., Giroux-Leprieur E., Julie C., et al. Validity of Targeted Next-Generation Sequencing in Routine Care for Identifying Clinically Relevant Molecular Profiles in Non-Small-Cell Lung Cancer: Results of a 2-Year Experience on 1343 Samples. J. Mol. Diagn. 2018;20:550–564. doi: 10.1016/j.jmoldx.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 195.Ilie M., Hofman P. Pitfalls in lung cancer molecular pathology: How to limit them in routine practice? Curr. Med. Chem. 2012;19:2638–2651. doi: 10.2174/092986712800493002. [DOI] [PubMed] [Google Scholar]
- 196.Bauml J., Levy B. Clonal Hematopoiesis: A New Layer in the Liquid Biopsy Story in Lung Cancer. Clin. Cancer Res. 2018;24:4352–4354. doi: 10.1158/1078-0432.CCR-18-0969. [DOI] [PubMed] [Google Scholar]
- 197.Hu Y., Ulrich B.C., Supplee J., Kuang Y., Lizotte P.H., Feeney N.B., Guibert N.M., Awad M.M., Wong K.K., Jänne P.A., et al. False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin. Cancer Res. 2018;24:4437–4443. doi: 10.1158/1078-0432.CCR-18-0143. [DOI] [PubMed] [Google Scholar]
- 198.Shlush L.I. Age-related clonal hematopoiesis. Blood. 2018;131:496–504. doi: 10.1182/blood-2017-07-746453. [DOI] [PubMed] [Google Scholar]
- 199.Razavi P., Li B.T., Brown D.N., Jung B., Hubbell E., Shen R., Abida W., Juluru K., De Bruijn I., Hou C., et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019;25:1928–1937. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Indini A., Rijavec E., Ghidini M., Cortellini A., Grossi F. Targeting KRAS in Solid Tumors: Current Challenges and Future Opportunities of Novel KRAS Inhibitors. Pharmaceutics. 2021;13:653. doi: 10.3390/pharmaceutics13050653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Anglesio M.S., Papadopoulos N., Ayhan A., Nazeran T.M., Noë M., Horlings H.M., Lum A., Jones S., Senz J., Seckin T., et al. Cancer-Associated Mutations in Endometriosis without Cancer. N. Engl. J. Med. 2017;376:1835–1848. doi: 10.1056/NEJMoa1614814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chang J.C., Montecalvo J., Borsu L., Lu S., Larsen B.T., Wallace W.D., Sae-Ow W., Mackinnon A.C., Kim H.R., Bowman A., et al. Bronchiolar Adenoma: Expansion of the Concept of Ciliated Muconodular Papillary Tumors with Proposal for Revised Terminology Based on Morphologic, Immunophenotypic, and Genomic Analysis of 25 Cases. Am. J. Surg. Pathol. 2018;42:1010–1026. doi: 10.1097/PAS.0000000000001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sasaki E., Masago K., Fujita S., Hanai N., Yatabe Y. Frequent KRAS and HRAS mutations in squamous cell papillomas of the head and neck. J. Pathol. Clin. Res. 2020;6:154–159. doi: 10.1002/cjp2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wittersheim M., Heydt C., Hoffmann F., Büttner R. KRAS mutation in papillary fibroelastoma: A true cardiac neoplasm? J. Pathol. Clin. Res. 2017;3:100–104. doi: 10.1002/cjp2.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zill O.A., Banks K.C., Fairclough S.R., Mortimer S.A., Vowles J.V., Mokhtari R., Gandara D.R., Mack P.C., Odegaard J.I., Nagy R.J., et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin. Cancer Res. 2018;24:3528–3538. doi: 10.1158/1078-0432.CCR-17-3837. [DOI] [PubMed] [Google Scholar]
- 206.Rolfo C., Mack P.C., Scagliotti G.V., Baas P., Barlesi F., Bivona T.G., Herbst R.S., Mok T.S., Peled N., Pirker R., et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018;13:1248–1268. doi: 10.1016/j.jtho.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 207.Hofman P., Rouleau E., Sabourin J.C., Denis M., Deleuze J.F., Barlesi F., Laurent-Puig P. Predictive molecular pathology in non-small cell lung cancer in France: The past, the present and the perspectives. Cancer Cytopathol. 2020;128:601–610. doi: 10.1002/cncy.22318. [DOI] [PubMed] [Google Scholar]
- 208.Vigliar E., Iaccarino A., Sciortino M., De Luca C., Malapelle U., Bellevicine C., Troncone G. Molecular predictive testing in precision oncology: The Italian experience. Cancer Cytopathol. 2020;128:622–628. doi: 10.1002/cncy.22290. [DOI] [PubMed] [Google Scholar]
- 209.Kerr K.M., Bibeau F., Thunnissen E., Botling J., Ryška A., Wolf J., Öhrling K., Burdon P., Malapelle U., Büttner R. The evolving landscape of biomarker testing for non-small cell lung cancer in Europe. Lung Cancer. 2021;154:161–175. doi: 10.1016/j.lungcan.2021.02.026. [DOI] [PubMed] [Google Scholar]
- 210.Thunnissen E., Weynand B., Udovicic-Gagula D., Brcic L., Szolkowska M., Hofman P., Smojver-Ježek S., Anttila S., Calabrese F., Kern I., et al. Lung cancer biomarker testing: Perspective from Europe. Transl. Lung Cancer Res. 2020;9:887–897. doi: 10.21037/tlcr.2020.04.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lindeman N.I., Cagle P.T., Aisner D.L., Arcila M.E., Beasley M.B., Bernicker E.H., Colasacco C., Dacic S., Hirsch F.R., Kerr K., et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol. 2018;13:323–358. doi: 10.1016/j.jtho.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 212.Vavala T., Malapelle U., Veggiani C., Ludovini V., Papotti M., Leone A., Graziano P., Minari R., Bono F., Sapino A., et al. Molecular profiling of advanced non-small cell lung cancer in the era of immunotherapy approach: A multicenter Italian observational prospective study of biomarker screening in daily clinical practice. J. Clin. Pathol. 2021;75:207339. doi: 10.1136/jclinpath-2020-207339. [DOI] [PubMed] [Google Scholar]
- 213.Chaft J.E., Rimner A., Weder W., Azzoli C.G., Kris M.G., Cascone T. Evolution of systemic therapy for stages I-III non-metastatic non-small-cell lung cancer. Nat. Rev. Clin. Oncol. 2021;18:547–557. doi: 10.1038/s41571-021-00501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gragnano G., Nacchio M., Sgariglia R., Conticelli F., Iaccarino A., De Luca C., Troncone G., Malapelle U. Performance evaluation of a fully closed real-time PCR platform for the detection of KRAS p.G12C mutations in liquid biopsy of patients with non-small cell lung cancer. J. Clin. Pathol. 2021:207416. doi: 10.1136/jclinpath-2021-207416. [DOI] [PubMed] [Google Scholar]
- 215.Heitzer E., van den Broek D., Denis M.G., Hofman P., Hubank M., Mouliere F., Paz-Aeres L., Schuuring E., Sültmann H., Vainer G., et al. Recommendations for a practical implementation of circulating tumor DNA mutation testing in metastatic non-small cell lung cancer. ESMO Open. 2022;7:e100399. doi: 10.1016/j.esmoop.2022.100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hofman P. Next-Generation Sequencing with Liquid Biopsies from Treatment-Naïve Non-Small Cell Lung Carcinoma Patients. Cancers. 2021;13:2049. doi: 10.3390/cancers13092049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Holm M., Andersson E., Osterlund E., Ovissi A., Soveri L.M., Anttonen A.K., Kytölä S., Aittomäki K., Osterlund P., Ristimäki A. Detection of KRAS mutations in liquid biopsies from metastatic colorectal cancer patients using droplet digital PCR, Idylla, and next generation sequencing. PLoS ONE. 2020;15:e0239819. doi: 10.1371/journal.pone.0239819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Malapelle U., Tiseo M., Vivancos A., Kapp J., Serrano M.J., Tiemann M. Liquid Biopsy for Biomarker Testing in Non-Small Cell Lung Cancer: A European Perspective. J. Mol. Pathol. 2021;2:255–273. doi: 10.3390/jmp2030022. [DOI] [Google Scholar]
- 219.Heeke S., Benzaquen J., Hofman V., Ilié M., Allegra M., Long-Mira E., Lassalle S., Tanga V., Salacroup C., Bonnetaud C., et al. Critical Assessment in Routine Clinical Practice of Liquid Biopsy for EGFR Status Testing in Non-Small-Cell Lung Cancer: A Single-Laboratory Experience (LPCE, Nice, France) Clin. Lung Cancer. 2020;21:56.e8–65.e8. doi: 10.1016/j.cllc.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 220.Heeke S., Hofman V., Benzaquen J., Otto J., Tanga V., Zahaf K., Allegra M., Long-Mira E., Lassalle S., Marquette C.H., et al. Detection of EGFR Mutations from Plasma of NSCLC Patients Using an Automatic Cartridge-Based PCR System. Front. Pharmacol. 2021;12:657743. doi: 10.3389/fphar.2021.657743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lin C.C., Huang W.L., Wei F., Su W.C., Wong D.T. Emerging platforms using liquid biopsy to detect EGFR mutations in lung cancer. Expert Rev. Mol. Diagn. 2015;15:1427–1440. doi: 10.1586/14737159.2015.1094379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lam V.K., Zhang J., Wu C.C., Tran H.T., Li L., Diao L., Wang J., Rinsurongkawong W., Raymond V.M., Lanman R.B., et al. Genotype-Specific Differences in Circulating Tumor DNA Levels in Advanced NSCLC. J. Thorac. Oncol. 2021;16:601–609. doi: 10.1016/j.jtho.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Miller T.E., Yang M., Bajor D., Friedman J.D., Chang R.Y.C., Dowlati A., Willis J.E., Sadri N. Clinical utility of reflex testing using focused next-generation sequencing for management of patients with advanced lung adenocarcinoma. J. Clin. Pathol. 2018;71:1108–1115. doi: 10.1136/jclinpath-2018-205396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Joshi A., Mishra R., Desai S., Chandrani P., Kore H., Sunder R., Hait S., Iyer P., Trivedi V., Choughule A., et al. Molecular characterization of lung squamous cell carcinoma tumors reveals therapeutically relevant alterations. Oncotarget. 2021;12:578–588. doi: 10.18632/oncotarget.27905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Fenizia F., Wolstenholme N., Fairley J.A., Rouleau E., Cheetham M.H., Horan M.P., Torlakovic E., Besse B., Al Dieri R., Tiniakos D.G., et al. Tumor mutation burden testing: A survey of the International Quality Network for Pathology (IQN Path) Virchows Arch. 2021;479:1067–1072. doi: 10.1007/s00428-021-03093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Heeke S., Hofman P. Tumor mutational burden assessment as a predictive biomarker for immunotherapy in lung cancer patients: Getting ready for prime-time or not? Transl. Lung Cancer Res. 2018;7:631–638. doi: 10.21037/tlcr.2018.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Heeke S., Benzaquen J., Hofman V., Long-Mira E., Lespinet V., Bordone O., Marquette C.H., Delingette H., Ilié M., Hofman P. Comparison of Three Sequencing Panels Used for the Assessment of Tumor Mutational Burden in NSCLC Reveals Low Comparability. J. Thorac. Oncol. 2020;15:1535–1540. doi: 10.1016/j.jtho.2020.05.013. [DOI] [PubMed] [Google Scholar]
- 228.Sholl L.M., Hirsch F.R., Hwang D., Botling J., Lopez-Rios F., Bubendorf L., Mino-Kenudson M., Roden A.C., Beasley M.B., Borczuk A., et al. The Promises and Challenges of Tumor Mutation Burden as an Immunotherapy Biomarker: A Perspective from the International Association for the Study of Lung Cancer Pathology Committee. J. Thorac. Oncol. 2020;15:1409–1424. doi: 10.1016/j.jtho.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.FDA FDA Approves Pembrolizumab for Adults and Children with TMB- H Solid Tumors. [(accessed on 3 March 2022)]; Available online: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors.
- 230.Prasad V., Addeo A. The FDA approval of pembrolizumab for patients with TMB >10 mut/Mb: Was it a wise decision? No. Ann. Oncol. 2020;31:1112–1114. doi: 10.1016/j.annonc.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 231.Horak P., Griffith M., Danos A.M., Pitel B.A., Madhavan S., Liu X., Chow C., Williams H., Carmody L., Barrow-Laing L., et al. Standards for the classification of pathogenicity of somatic variants in cancer (oncogenicity): Joint recommendations of Clinical Genome Resource (ClinGen), Cancer Genomics Consortium (CGC), and Variant Interpretation for Cancer Consortium (VICC) Genet. Med. 2022 doi: 10.1016/j.gim.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A., et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Suybeng V., Koeppel F., Harlé A., Rouleau E. Comparison of Pathogenicity Prediction Tools on Somatic Variants. J. Mol. Diagn. 2020;22:1383–1392. doi: 10.1016/j.jmoldx.2020.08.007. [DOI] [PubMed] [Google Scholar]
- 234.Long E., Hofman V., Ilie M., Washetine K., Lespinet V., Bonnetaud C., Bordone O., Gavric-Tanga V., Gaziello M.C., Lassalle S., et al. Accreditation of the activity of molecular pathology according to ISO 15189: Key steps to follow and the main potential pitfalls. Ann. Pathol. 2013;33:12–23. doi: 10.1016/j.annpat.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 235.Long-Mira E., Washetine K., Hofman P. Sense and nonsense in the process of accreditation of a pathology laboratory. Virchows Arch. 2016;468:43–49. doi: 10.1007/s00428-015-1837-1. [DOI] [PubMed] [Google Scholar]
- 236.Kim J., Lee H.M., Cai F., Ko B., Yang C., Lieu E.L., Muhammad N., Rhyne S., Li K., Haloul M., et al. The hexosamine biosynthesis pathway is a targetable liability in KRAS/LKB1 mutant lung cancer. Nat. Metab. 2020;2:1401–1412. doi: 10.1038/s42255-020-00316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lee H., Cai F., Kelekar N., Velupally N.K., Kim J. Targeting PGM3 as a Novel Therapeutic Strategy in KRAS/LKB1 Co-Mutant Lung Cancer. Cells. 2022;11:176. doi: 10.3390/cells11010176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mathieu M., Steier V., Fassy F., Delorme C., Papin D., Genet B., Duffieux F., Bertrand T., Delarbre L., Le-Borgne H., et al. KRAS G12C fragment screening renders new binding pockets. Small GTPases. 2022;13:225–238. doi: 10.1080/21541248.2021.1979360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Asimgil H., Ertetik U., Çevik N.C., Ekizce M., Doğruöz A., Gökalp M., Arık-Sever E., Istvanffy R., Friess H., Ceyhan G.O., et al. Targeting the undruggable oncogenic KRAS: The dawn of hope. JCI Insight. 2022;7:e153688. doi: 10.1172/jci.insight.153688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lu H., Liu C., Velazquez R., Wang H., Dunkl L.M., Kazic-Legueux M., Haberkorn A., Billy E., Manchado E., Brachmann S.M., et al. SHP2 Inhibition Overcomes RTK-Mediated Pathway Reactivation in KRAS-Mutant Tumors Treated with MEK Inhibitors. Mol. Cancer Ther. 2019;18:1323–1334. doi: 10.1158/1535-7163.MCT-18-0852. [DOI] [PubMed] [Google Scholar]
- 241.Fedele C., Ran H., Diskin B., Wei W., Jen J., Geer M.J., Araki K., Ozerdem U., Simeone D.M., Miller G., et al. SHP2 Inhibition Prevents Adaptive Resistance to MEK Inhibitors in Multiple Cancer Models. Cancer Discov. 2018;8:1237–1249. doi: 10.1158/2159-8290.CD-18-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Fedele C., Li S., Teng K.W., Foster C.J.R., Peng D., Ran H., Mita P., Geer M.J., Hattori T., Koide A., et al. SHP2 inhibition diminishes KRASG12C cycling and promotes tumor microenvironment remodeling. J. Exp. Med. 2021;218:e20201414. doi: 10.1084/jem.20201414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hofmann M.H., Gmachl M., Ramharter J., Savarese F., Gerlach D., Marszalek J.R., Sanderson M.P., Kessler D., Trapani F., Arnhof H., et al. BI-3406, a Potent and Selective SOS1-KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition. Cancer Discov. 2021;11:142–157. doi: 10.1158/2159-8290.CD-20-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Molina-Arcas M., Moore C., Rana S., van Maldegem F., Mugarza E., Romero-Clavijo P., Herbert E., Horswell S., Li L.S., Janes M.R., et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci. Transl. Med. 2019;11:eaaw7999. doi: 10.1126/scitranslmed.aaw7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ning W., Yang Z., Kocher G.J., Dorn P., Peng R.W. A Breakthrough Brought about by Targeting KRAS(G12C): Nonconformity Is Punished. Cancers. 2022;14:390. doi: 10.3390/cancers14020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hofmann M.H., Gerlach D., Misale S., Petronczki M., Kraut N. Expanding the Reach of Precision Oncology by Drugging All KRAS Mutants. Cancer Discov. 2022 doi: 10.1158/2159-8290.CD-21-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Reggiani F., Sauta E., Torricelli F., Zanetti E., Tagliavini E., Santandrea G., Gobbi G., Damia G., Bellazzi R., Ambrosetti D., et al. An integrative functional genomics approach reveals EGLN1 as a novel therapeutic target in KRAS mutated lung adenocarcinoma. Mol. Cancer. 2021;20:63. doi: 10.1186/s12943-021-01357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sakamoto K., Kamada Y., Sameshima T., Yaguchi M., Niida A., Sasaki S., Miwa M., Ohkubo S., Sakamoto J.I., Kamaura M., et al. K-Ras(G12D)-selective inhibitory peptides generated by random peptide T7 phage display technology. Biochem. Biophys. Res. Commun. 2017;484:605–611. doi: 10.1016/j.bbrc.2017.01.147. [DOI] [PubMed] [Google Scholar]
- 249.Wang X., Allen S., Blake J.F., Bowcut V., Briere D.M., Calinisan A., Dahlke J.R., Fell J.B., Fischer J.P., Gunn R.J., et al. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS(G12D) Inhibitor. J. Med. Chem. 2021;65:3123–3133. doi: 10.1021/acs.jmedchem.1c01688. [DOI] [PubMed] [Google Scholar]
- 250.Nissley D.V., McCormick F. RAS at 40: Update from the RAS Initiative. Cancer Discov. 2022 doi: 10.1158/2159-8290.CD-21-1554. [DOI] [PubMed] [Google Scholar]
- 251.Ooft M.L., Boissiere J., Sioletic S. The histopathologist is essential in molecular pathology quality assurance for solid tumours. Virchows Arch. 2021;479:1263–1265. doi: 10.1007/s00428-021-03226-y. [DOI] [PubMed] [Google Scholar]