Abstract
Simple Summary
Ras homologous A (RHOA), a signal mediator and a GTPase, is associated with the progression of gastric cancer (GC). We present novel RHOA inhibitors designed for greater anti-GC potency by means of lead optimization. The RHOA → BIRC5 signaling circuit was found to be a new therapeutic strategy for regulating GC proliferation and migration.
Abstract
Ras homologous A (RHOA), a signal mediator and a GTPase, is known to be associated with the progression of gastric cancer (GC), which is the fourth most common cause of death in the world. Previously, we designed pharmacologically optimized inhibitors against RHOA, including JK-136 and JK-139. Based on this previous work, we performed lead optimization and designed novel RHOA inhibitors for greater anti-GC potency. Two of these compounds, JK-206 and JK-312, could successfully inhibit the viability and migration of GC cell lines. Furthermore, using transcriptomic analysis of GC cells treated with JK-206, we revealed that the inhibition of RHOA might be associated with the inhibition of the mitogenic pathway. Therefore, JK-206 treatment for RHOA inhibition may be a new therapeutic strategy for regulating GC proliferation and migration.
Keywords: gastric cancer, RHOA, JK-206, pharmacogenomics, Rhosin, transcriptomics
1. Introduction
Gastric cancer (GC) is the fifth most prevalent and fourth most lethal cancer worldwide [1]. Although the first-line treatment for GC is endoscopic resection or surgery, the overall 5-year survival rate for GC is only 30% in the United States [2]. Early detection is critical for favorable outcomes and minimizing residual disease in GC; however, an endoscopic screening test for GC is not routinely performed in most Western countries. Consequently, a new and effective therapeutic option (e.g., trastuzumab [3]) for GC is emerging. Personalized medicine may be the best option for improving survival. Nevertheless, in clinical trials, most targeted therapies for GC have been disappointing [3,4,5].
Ras homologous A (RHOA), a small GTPase of the Rho family, plays crucial roles in oncogenic processes, including proliferation, migration, and invasion [6]. In addition, RHOA mediates epithelial–mesenchymal transition and is a destabilizer of microtubules [7]. Previously, we reported that RHOA is the hub mediator of GC progression and a biomarker and therapeutic target for GC [8,9]. Consequently, we designed RHOA inhibitors and evaluated their in vitro efficacy. We previously found that among JK-122–125, JK-122 showed superior suppression of cell viability [8]. In a follow-up study, we identified that JK-136 and JK-139 inhibited cell migration through the lead optimization of JK-122 [10]. Despite the potential of RHOA as a therapeutic target for GC, small-molecule RHOA inhibitors are underexplored [8,10] and the anticancer mechanism of RHOA inhibitors remain to be further elucidated.
In this study, we aimed to design novel RHOA inhibitors by means of the lead optimization of JK-136/JK-139 and elucidate the pathways involved in their anti-cancer mechanisms in GC using transcriptome-based network analysis.
2. Materials and Methods
2.1. Cell Culture
The following human GC cell lines were used within 6 months of resuscitation: AGS (ATCC, Mansfield, VA, USA), SNU216, SNU601, SNU668 (KCLB), and MKN1 (Riken, Tokyo, Japan). The cells were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA) and maintained at 37 °C and 5% CO2. Cell line identities were validated using short tandem repeat profiling (ATCC).
2.2. Synthesis of Hydrazide Derivates
All reactions that were sensitive to air or moisture were conducted under nitrogen. All reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Tokyo Chemical Company (Tokyo, Japan). All anhydrous solvents were distilled over CaH2, P2O5, or Na/benzophenone prior to the reaction, unless otherwise stated. Analytical thin-layer chromatography (TLC) was performed using commercial precoated TLC plates (Silica Gel 60, F254) purchased from Merck KGaA (Darmstadt, Germany). Spots were detected by viewing under UV light (254 nm) or colorizing with charring after dipping in phosphomolybdic acid in ethanol or potassium permanganate aqueous solution. Flash column chromatography was performed on Silica Gel 60 (~0.040–0.063 mm, 230–400 mesh, Merck). Infrared spectra were recorded on an Agilent Cary 670 FTIR instrument (Agilent Technologies, Santa Clara, CA, USA). 1H NMR spectra (DMSO-d6) were recorded on an Agilent 400-MR spectrometer (400 MHz; Agilent Technologies). The chemical shifts are reported in parts per million (δ) units relative to the solvent peak. The 1H NMR data are reported as peak multiplicities: s for singlet; d for doublet; dd for doublet of doublets; ddd for doublet of doublet of doublets; t for triplet; pseudo t for pseudo triplet; brs for broad singlet; and m for multiplet. The coupling constants are reported in Hertz. 13C NMR spectra (DMSO-d6) were recorded on an Agilent 400-MR spectrometer (100 MHz; Agilent Technologies). Mass spectra were recorded using an electrospray ionization source in methylene chloride or methanol.
The general procedure for the synthesis of hydrazides was as follows: a mixture of 4-phenoxybenzaldehyde for JK-201~214 (1 mmol), 6-(benzo[d][1,3]dioxol-5-yl)picolinaldehyde for JK-301–314 (1 mmol), and an appropriate hydrazide compound (1 mmol) in methanol (or ethanol) was stirred at room temperature or heated under reflux for 0.5 h–4 d. The progress of the reaction was monitored using TLC. After completion of the reaction, the contents were cooled to room temperature and poured into ice-cold water (5 mL) while stirring. The solid was filtered, dried, and purified by recrystallization using methanol (or ethanol) to obtain the hydrazide products JK-201–214 and JK-301–314. For detailed analytical data of the compounds, please refer to Method S1.
2.3. Cell Viability Assays
Cells were seeded at a density of 3000/well in 96-well plates and exposed to various concentrations of RHOA inhibitors and DMSO as control for 72 h. The effects of JK-201–214 and JK-301–314 on cell viability were assessed using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS; Promega, Madison, WI, USA) assay. The half-maximal inhibitory concentration (IC50) was defined as the dose required for 50% cell growth inhibition compared to the DMSO control. IC50 values represent the means of triplicate wells from three independent experiments for each drug concentration.
2.4. Migration Assay
AGS (1 × 105), MKN1 (2 × 105), SNU216 (2 × 105), SNU668 (2 × 105), SNU620 (2 × 105), and SNU601 (2 × 105) cells were seeded into Boyden chambers (0.8-μm pore size; Corning Life Sciences, Corning, NY, USA). Medium containing FBS was used as a chemoattractant in the bottom chamber. Migrated cells were stained using a Diff Quick Staining Kit (Thermo Fisher Scientific, Waltham, MA, USA) and photographed under an inverted microscope. The number of migrated cells in three microscopic fields per well was compared to that of control cells, yielding the average %migration.
2.5. Docking Simulations
In silico docking simulations of RHOA with JK inhibitors were performed using AutoDock Vina [11]; the three-dimensional structure of RHOA was obtained from the Protein Data Bank (accession number 1X86). The structure of RHOA consists of two chains: the Rho guanine-nucleotide exchange factor (chain A) and the RHOA structure (chain B). For the docking simulation, the RHOA structure was obtained from the complex. Two JK structures (JK-206 and -312) were drawn and minimized using the Marvin program (ChemAxon; http://www.chemaxon.com, accessed on 30 December 2021; v5.11.4, 2012). For the docking simulation, the pockets in the RHOA complex were searched using the Pck pocket detection program (http://schwarz.benjamin.free.fr/Work/Pck/home.htm, accessed on 30 December 2021). Five pockets greater than 10.0 Å3 in volume were found in the complex. The docking simulations were focused on these pockets, which consisted of 30 pocket residues. Two JK structures were placed in 30 pocket residues and run ten times with different random seeds; subsequently, 300 simulations were carried out. The box size with a length of 15 Å was used to prevent the inhibitors from drifting from the center of the pocket residue. With 300 docking poses, clustering based on the center of mass was performed to categorize these docking structures using Chemistry at HARvard Macromolecular Mechanics [12]. The compounds were ranked according to the following parameters: the lowest energy of the group, the largest number of the group, and the lowest energy conformation in the group.
2.6. Gene Expression and Differentially Expressed Gene (DEG) Analyses
Following JK-206, -312, and DMSO treatments on AGS, MKN1, and SNU601 cells, total cellular mRNA was isolated using RNeasy kits (Qiagen, Valencia, CA, USA), reverse transcribed, and hybridized to the Affymetrix GeneChip Human 2.0 ST Array (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
We performed DEG analysis using a microarray of GC cells treated with JK-206/-312 versus DMSO. For GC cell samples treated with DMSO, we used three samples (GEO accession numbers: GSM3984792, GSM3984796, and GSM3984800) from our previous dataset (GEO accession number: GSE135068) [10].
To identify the functional contexts of DEGs, we investigated their biological pathways using Molecular Signatures Database v7.4 [13] and Gene Set Enrichment Analysis v4.1.0 [14] based on the hallmark gene set [13] for group-wise pathway analyses, and Gene Set Variation Analysis (GSVA) [15] for a sample-wise pathway analysis. In the context analysis, an adjusted p value (false discovery rate (FDR)) was used for statistical significance.
2.7. Validation of mRNA Expression Patterns of DEGs in Independent Transcriptome Datasets
To validate the mRNA expression patterns of DEGs in publicly available independent datasets, we inspected the expression levels of RHOA and DEGs using publicly available transcriptome datasets, GSE135068 [10] and the expression level dataset of the Cancer Cell Line Encyclopedia (CCLE) [16], GSE110237 [17], and GSE83913 [18].
2.8. Protein–Protein Interaction (PPI) Network Construction
To explore the interactions between our RHOA inhibitors and perturbed gene sets in GC cells treated with the JK series of compounds versus DMSO, we constructed a PPI network with the resultant DEGs identified from the gene set analysis and participant genes of the RHOA signaling pathway. The RHOA signaling pathway genes were manually selected from our previous review [19]. The PPI network was constructed using the STRING [20] and Biological General Repository for Interaction Datasets (BioGRID) databases [21]. Gene ontology analysis was performed using the STRING database to investigate the functional role of the PPI network. An adjusted p value (FDR) < 0.05 was considered statistically significant for this investigation. An independent GC dataset, GSE36968 [22], was used to inspect gene expression patterns among DEGs in the PPI network between GC tumor tissues (tumor group) and normal tissues adjacent to the tumor (control group) using DEG and correlation analyses. Statistical tests between the two groups were performed using the two-sided paired t-test. Statistical significance was set at p < 0.05. Correlation analysis was performed using the Pearson correlation coefficient (PCC). PCC > |0.5| and p < 0.05 were considered statistically significant.
2.9. Statistics
The DEGs satisfied the following conditions: (i) the cut-off value of fold-change (FC) was greater than 1.3 or less than 1/1.3; and (ii) statistical tests among GC cell groups were performed using the two-sided paired t-test, with statistical significance at p value < 0.05.
An adjusted p value (FDR) < 0.05 was considered statistically significant.
3. Results
3.1. Design and Synthesis of RHOA Inhibitors
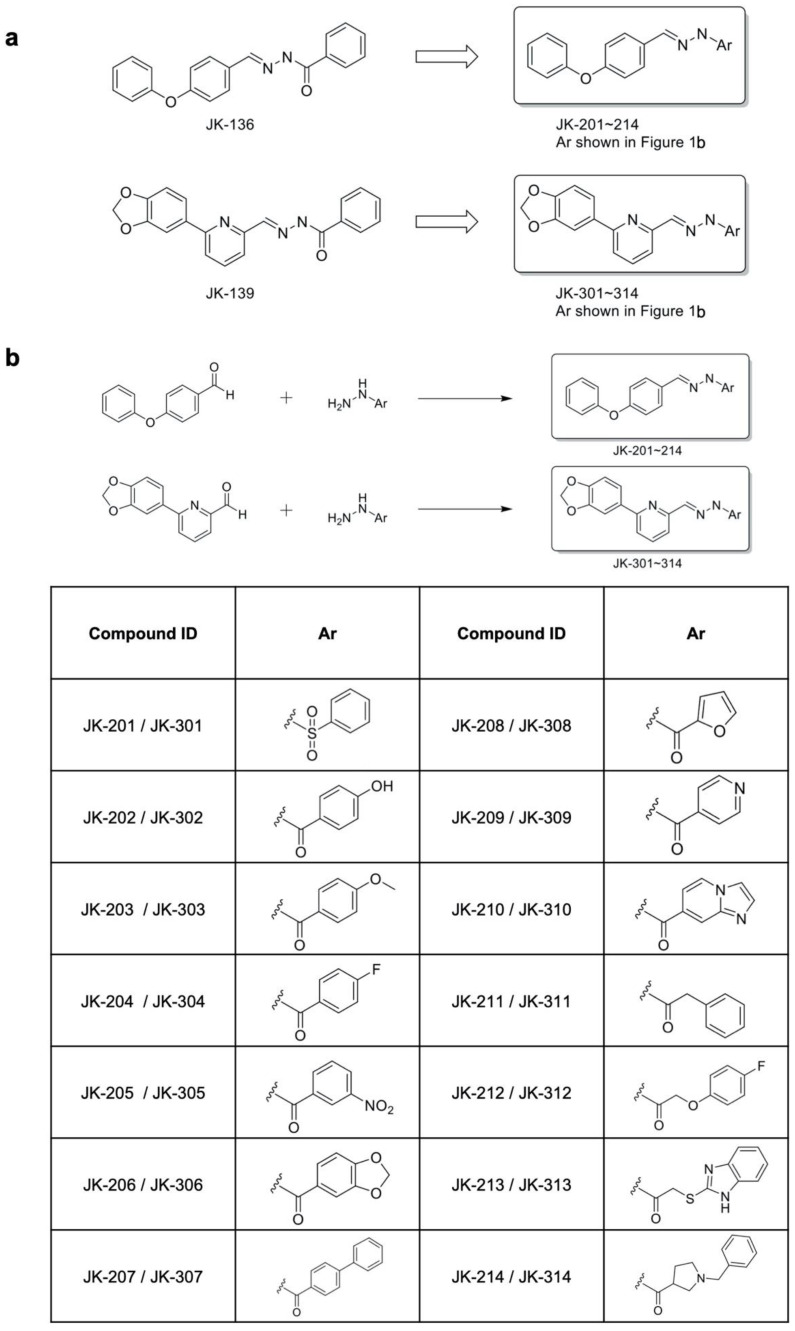
We designed and synthesized a series of new anti-GC compounds of varying sizes, with various substituents, and with different electronic effects on phenyl-ring and heterocyclic systems for the structural modification of the benzoyl group in JK-136 and JK-139. In our previous work [8], we performed the biological evaluation of the five different compounds of an RHOA inhibitor of our own design (JK-121, -122, -123, -124, and -125) to show different chemical properties. Out of the five compounds, the compound with the benzoyl group (JK-122) exhibited the best growth inhibition in in vitro assays of GC cells and the best binding affinity to RHOA. Based on these previous results, we assumed the benzoyl group to be the necessary skeletal group of the compound and decided to introduce other functional groups on it for further optimization of our novel RHOA inhibitors. The synthetic strategy for the hydrazides JK-201–214 and JK-301–314 is displayed in Figure 1.
Figure 1.
Design and synthesis of Ras homologous A (RHOA) inhibitors. (a) Rational design of novel RHOA inhibitors. (b) Synthesis of hydrazide derivatives. Reagents and conditions: methanol or ethanol, room temperature or reflux, 0.5 h–4 d.
JK-201 and JK-301 were designed to introduce a sulfonyl group instead of a carbonyl group in the hydrazide spacers of JK-136 and JK-139. JK-202–207 and JK-302–307 possess structural diversity with different substituents in the phenyl ring of these compounds. In particular, JK-206 and JK-306 were designed to contain oxygen, forming a 1,3-dioxole ring connected to their phenyl rings. JK-208–210 and JK-308–310 were designed to form aryl heterocyclic systems. We further extended the spacer length with methylene or pyrrolidine moieties with more flexible hydrazide functional group systems, leading to the design of JK-211–214 and JK-311–314.
For the synthesis of the target hydrazides JK-201–214, commercially available 4-phenoxybenzaldehyde was treated with appropriate hydrazide reagents in methanol or ethanol at room temperature or under reflux conditions. The reaction of 6-(1,3-benzodioxol-5-yl)-2-pyridinecarbaldehyde with appropriate hydrazide reagents in methanol or ethanol at room temperature or heating conditions resulted in the creation of the hydrazide products JK-301–314.
3.2. Selection of RHOA Inhibitors
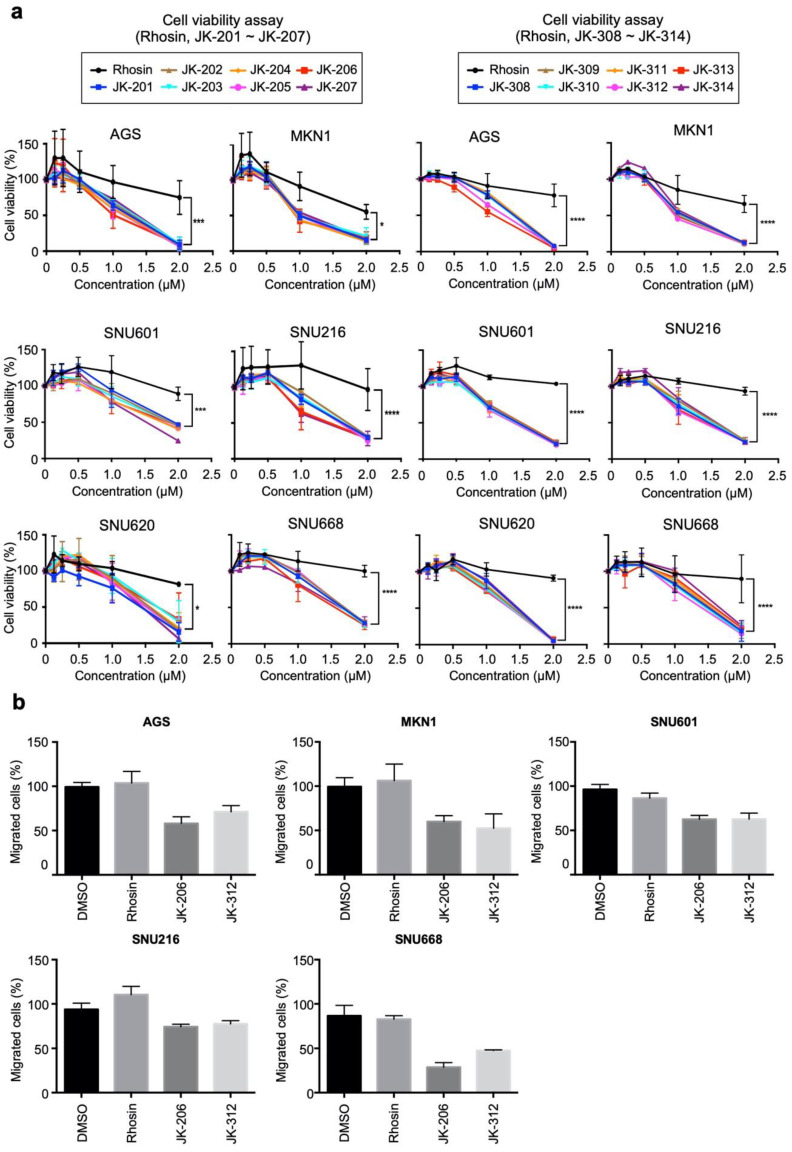
We evaluated the IC50 values of the final RHOA inhibitors (Rhosin (Millipore, Burlington, MA, USA), a nonclinical RHOA inhibitor [23], JK201–214, and JK301–314) against GC cell lines (Figure 2a and Figure S1). The designed compounds, including JK-206 and JK-312, exhibited IC50 values of less than 2.5 µM (Figure 2a and Figure S1). Overall, all compounds showed substantially lower cell viability compared with Rhosin, and JK-206 and JK-312 showed the best cell viability inhibition potency (Figure 2a and Figure S1). In our previous study [8], we assessed the dependency of RHOA signaling in cell lines. RHOA expression levels varied in most GC cells as AGS, MKN1, and SNU601 cells had low, moderate, and high RHOA expression, respectively. Performing a RHOA gene mutation through cancer.sanger.ac.uk (accessed on 30 December 2021), it was found that AGS has a p.E64del (amino acid) mutation on RHOA, but the MKN1 and SNU601 cell lines have no mutation on RHOA. Based on these results, we strategically selected candidate compounds based on the drug treatment results in AGS, MKN1, and SNU601 cells. We determined the efficacy of the compounds using cell density assays after treatment with 2 µM of each compound. In AGS and SNU601 cells, JK-206 exhibited the best inhibitory performance compared to other JK-200 series compounds and JK-312 showed the best inhibitory performance compared to other JK-300 series compounds (Figure S2). In migration assays, JK-206 and -312 remarkably inhibited wound healing in AGS, MKN1, SNU601, SNU216, and SNU668 cells (Figure 2b). Based on these results, we selected JK-206 and JK-312 for further experimentation.
Figure 2.
RHOA inhibitors suppressed cell growth and migration in gastric cancer (GC) cells. (a) The GC cell lines AGS, MKN1, SNU601, SNU216, SNU620, and SNU668 were treated with 15 small-molecule candidates including Rhosin (see also Figure S1). (b) The migration assay showed significant inhibition of wound healing with treatment with both JK-206 and JK-312 (* p value < 0.05, *** p value < 0.001, **** p value < 0.0001).
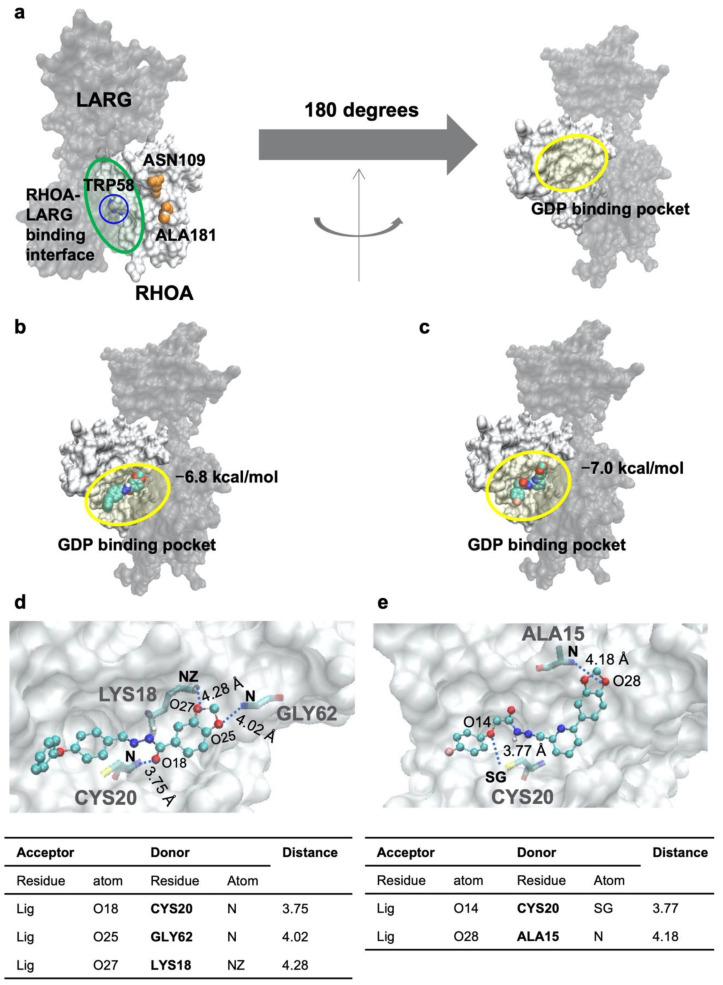
3.3. Docking Simulations Indicated That JK-206 and -312 Bind with the Key Residue of RHOA
Docking simulations of RHOA with JK-206 and -312 were performed. The RHOA protein’s molecular representation is described on Figure 3a. The lowest energy conformers were found, as shown in Figure 3b,c. The docking energies of JK-206 and -312 are −6.8 and −7.0 kcal/mol, respectively, indicating that the inhibitors can tightly bind to the RHOA structure. The binding pocket in RHOA that interacts with JK-206 and -312 is the GDP-binding site on which the original ligand, GDP, of RHOA interacts. The docking of the JK compounds on GDP-binding pocket can compete against the original ligand, GDP. The competition inhibition may prevent the activation of the interaction between RHOA and LARG. The hydrogen bond patterns between the inhibitor and RHOA are shown in Figure 3d,e. A detailed analysis of the hydrogen bond is shown below the hydrogen bond patterns.
Figure 3.
Molecular docking simulations and hydrogen bond patterns. (a) Molecular representation of the RHOA–LARG complex. The RHOA structure is illustrated using a white surface model. The interacting partner protein, LARG (Rho guanine nucleotide exchange factor 12), is shaded based on a black transparency surface model. The two structures are interacting on the RHOA–LARG-binding interface circled by transparent green in the left figure. The TRP58 is located on the RHOA–LARG-binding surface. The channel is formed between ASN109 and ALA181, which are illustrated by orange spheres. The GDP-binding pocket (transparent yellow circle in the right figure) exists on the opposite side (180 degrees’ rotation of the principal axis of the proteins). The docking poses (b) JK-206 and (c) JK-312 are drawn on the RHOA. The inhibitors are also drawn using space-filling models colored by atom types (oxygen: red; nitrogen: blue; carbon: cyan). The hydrogen-bond-interacting patterns of (d) JK-206 and (e) JK-312 are drawn. The inhibitors are drawn using ball-and-stick models colored by atom types. The interacting residues are drawn using a stick model colored by atom types. The interacting hydrogen bonds are drawn using dot lines with the bonding distances, with the Angstrom as the unit. Detailed information on the hydrogen bonds is tabulated below the hydrogen-bond-interacting pattern.
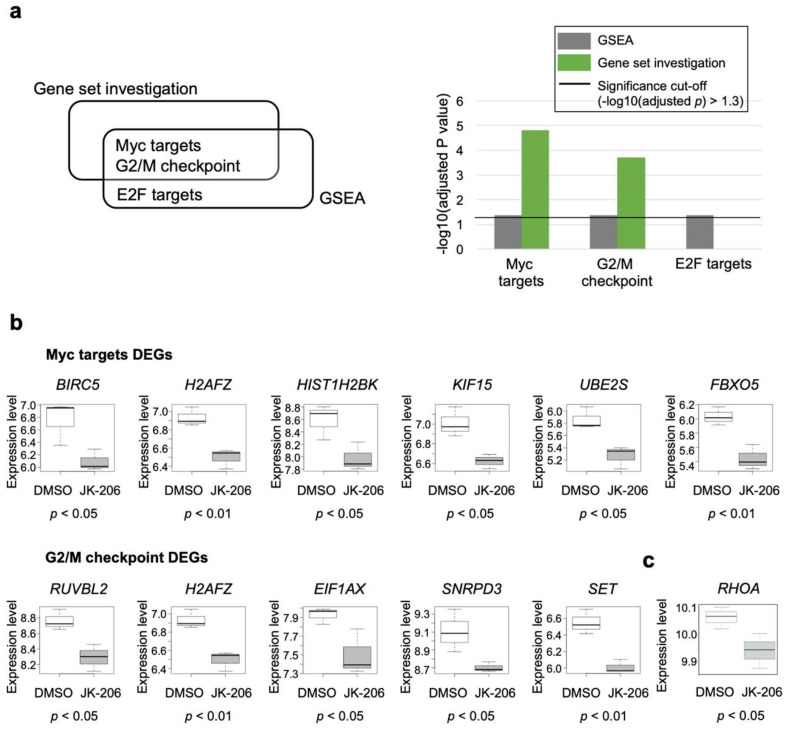
3.4. Gene Set Analysis Revealed That JK-206 Treatment Perturbed Myc Targets and G2/M Checkpoint
We performed gene set analyses using gene expression datasets from GC cells (AGS, MKN1, and SNU601). GC cells were treated with the selected RHOA inhibitors JK-206 and -312 (experimental groups) and DMSO (control group). Table S1 includes a list of DEGs that are common and uncommon to JK-206- and JK-312-treated (versus DMSO) GC cells. JK-206 treatment of GC cells showed that Myc targets (p value < 0.0001 from the gene set investigation) and G2/M checkpoint (p value < 0.0001 from the gene set investigation) gene sets were significantly enriched compared to the control group (Figure 4a). Of the common genes between the Myc targets and the G2/M checkpoint gene sets, 10 DEGs were identified in JK-206-treated GC cells compared to the control group, and all of them were downregulated (Figure 4b). The expression levels of RHOA were also decreased significantly (p < 0.05) after the inhibition of RHOA by JK-206 treatment compared to DMSO treatment in GC cells (Figure 4c). The genes relating to Myc targets (i.e., BIRC5, H2AFZ, HIST1H2BK, KIF15, UBE2S, and FBXO5; Figure S3a) and G2/M checkpoint (i.e., RUVBL2, H2AFZ, EIF1AX, SNRPD3, and SET; Figure S3b) were down-regulated in JK-312-treated GC cells compared to DMSO-treated GC cells. However, the gene set analysis failed in JK-312-treated GC cells compared to the control group and we could not obtain statistically significant gene sets. We additionally performed a sample-specific pathway-activity analysis for the expression data of JK-312-treated cells, instead of using the GSEA (the group-wise pathway enrichment analysis). In the results of the sample-wise pathway analysis conducted using the gene set variation analysis (GSVA) [15], it was found that JK-312-treated GC cell lines were also suppressed in hallmark gene sets of Myc targets, G2/M checkpoint, and E2F targets such as JK-206-treated GC cell lines compared to DMSO-treated GC cell lines (Figure S4).
Figure 4.
Gene set analysis of GC cells treated with JK-206 and -312 compared to those treated with dimethyl sulfoxide (DMSO). (a) Significantly detected gene sets of gene set analyses. (b) Expression profiles of the differentially expressed genes (DEGs) in the two gene sets (Myc targets and G2/M checkpoint). (c) Expression pattern of RHOA in JK-206- vs. DMSO-treated GC cells.
3.5. Validation of mRNA Expression Patterns of DEGs Related to Myc Targets and G2/M Checkpoint in Independent Datasets
To determine whether Myc-target genes and G2/M-checkpoint related genes were suppressed due to the RHOA inhibition, we inspected publicly available independent datasets. For validating the expression levels of RHOA, Myc-target genes, and G2/M-checkpoint-related genes, we re-visited and inspected the transcriptome data (GEO accession: GSE135068) [10] from our previous study of other RHOA inhibitors (JK-136 and -139). We found that, upon RHOA inhibition by JK-136 and -139, most of the expression levels of RHOA and DEGs related to Myc targets and G2/M checkpoint (depicted in Figure 4b) were consistently decreased (Figure S5).
For another validation, since the RHOA inhibitors (JK-206, -312, -136, and -139) demonstrated down-regulation of RHOA and the DEGs (depicted in Figure 4b), we assumed that there were positive correlations between RHOA and the DEGs. In 13 GC cell lines from CCLE [16], we calculated Pearson’s correlation, and found that most of the correlations were positive (Figure S6a). We also inspected other publicly available transcriptomic datasets for RHOA inhibition by siRNA and knockout in gastric and prostate cancer cell lines (GEO accessions GSE110237 and GSE83913, respectively) [17,18]. Dataset GSE110237 was derived from experiments of RHOA knockdown in GC cells [17] and dataset GSE83913 was derived from experiments of RHOA knockout in prostate cancer cells [18]. We confirmed that most of the DEGs were also down-regulated after knockdown of RHOA in GC cell lines and after knockout of RHOA in prostate cancer cell lines (Figure S6b,c).
3.6. Network Construction Revealed That JK-206 Perturbed Microtubule Formation and the Cell Cycle
In this study, since the group-wise and sample-specific pathway analyzes showed consistencies in GC cell lines treated with JK-206, we performed further investigation only on JK-206-treated GC cells compared to the control group. JK-206 was designed and optimized to inhibit RHOA expression. To explore the biological mechanism perturbed by JK-206 treatment in GC cells, we constructed PPI networks using the 10 genes (Figure 4b) and RHOA signaling pathway genes as inputs in the STRING [20] and BioGRID [21] databases. In the PPI network (Figure 5a), the RHOA signaling pathway and the 10 DEGs (Figure 4b) out of the common genes between the two gene sets (Myc targets and G2/M checkpoint) were found to interact with each other through RAC1 and CDK4. Most of the gene expression levels in the RHOA signaling pathway were also downregulated (Figure 5a).
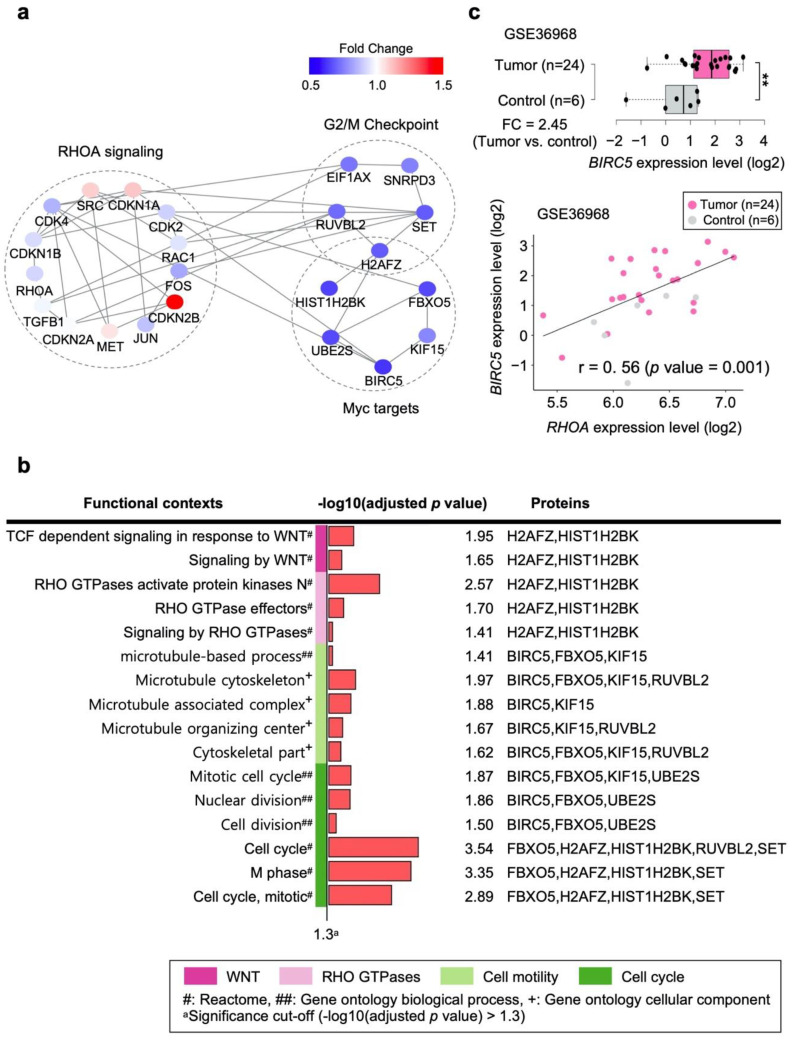
Figure 5.
Protein-protein interaction (PPI) network construction and functional context analysis of the network of GC cells treated with JK-206 compared to those treated with DMSO. (a) PPI networks constructed from RHOA signaling and DEGs of GC cells treated with JK-206 compared to those treated with DMSO. (b) Functional contexts of GC cells treated with JK-206 compared to those treated with DMSO in the PPI network. (c) Differentially expressed BIRC5 between tumor and control groups and correlation between BIRC5 and RHOA in the independent GC patient dataset (GEO accession number: GSE36968). r: Pearson’s correlation coefficient. ** p value < 0.01.
The functional contexts of JK-206 treatment in GC cells were found to involve WNT signaling, Rho GTPases, cell motility, and the cell cycle (Figure 5b). The gene expression levels of microtubules (i.e., baculoviral IAP repeat-containing 5 (BIRC5), F-Box protein 5 (FBXO5), RuvB-like AAA+ ATPase 2 (RUVBL2), and kinesin family member 15 (KIF15)) were downregulated in JK-206-treated GC cells compared to the control group (Figure 4b and Figure 5b). Therefore, JK-206 treatment had an inhibitory effect on microtubule activity in GC cells, which is in agreement with our migration assay results (Figure 2b). Similarly, our previously reported RHOA inhibitors (i.e., JK-136 and -139) regulate actin polymerization and cell migration [10]. Similar to the functional contexts identified in this study, cell-cycle-related gene sets (i.e., M phase and nuclear division) were identified in the functional context analysis in our previous study [10]. These gene sets comprised the protein products H2A.Z variant histone 1 (H2AFZ), HIST1H2BK, SET nuclear proto-oncogene (SET), UBE2S, FBXO5, BIRC5, and RUVBL2 of downregulated DEGs (Figure 4b and Figure 5b). In line with these results, JK-206 treatment exerted an inhibitory effect on cell-cycle-related activity against GC cells.
To determine whether the expression levels of the genes identified in the PPI network were replicated in a GC patient dataset, we performed DEG and correlation analyses with the 23 genes in Figure 5a using an independent GC patient dataset (GEO accession number: GSE36968) [22]. We assumed that the downregulated genes in the PPI networks (Figure 5a) were upregulated in the tumor versus control groups in the dataset GSE36968. In fact, BIRC5 was significantly upregulated in the tumor group compared with the control group (FC = 2.45, p value < 0.01; Figure 5c) and its expression level showed a significantly positive correlation with that of RHOA (r = 0.56, p value = 0.001; Figure 5c). Furthermore, the expression levels of RAC1, CDK2, RUVBL2, and MET (shown in Figure 5a) were upregulated in the tumor group compared to the control group and showed a noticeably positive correlation with the expression level of RHOA (Figure S7). Considering the positive expression correlation of BIRC5, RAC1, CDK2, RUVBL2, and MET with RHOA in the GC patient dataset, the downregulation of the genes with JK-206 treatment in the GC cell models indicates that RHOA signaling may be involved in upstream regulation of genes in GC.
4. Discussion
In this study, we designed and synthesized a series of new RHOA inhibitors to evaluate and improve their biological activity [10]. Although the RHOA signaling pathway is considered a potential therapeutic target for GC, the effects of RHOA inhibitors on cancer cells are yet to be elucidated. Among the new compounds we studied, JK-206 could effectively inhibit cancer cell survival and migration. To determine the mechanism of action of JK-206, we analyzed transcriptomic data of GC cell lines treated with JK-206 compared to those treated with DMSO. We found that JK-206 suppressed biological contexts associated with WNT, Rho GTPases, Myc targets, G2/M checkpoint, and microtubule-related gene sets. In our previous study, we confirmed that the RHOA signaling pathway plays a crucial role in GC progression [9]. RHOA interacts with c-Myc, resulting in synergistic reinforcement of the expression of cancer phenotypes and behaviors favoring cancer development, including cell migration, invasion, and metastasis [24,25].
Through DEG and gene set analyses, 10 DEGs were found to be associated with two gene sets related to cell motility and cycle (i.e., Myc targets and G2/M checkpoint) in GC cells treated with JK-206 compared to those treated with DMSO (Figure 4b). These genes are involved in apoptosis inhibition (BIRC5) [26], oncogene activation (H2AFZ and SET) [27,28], protein degradation regulation (FBXO5 and UBE2S) [29,30], and carcinogenesis (RUVBL2 and eukaryotic translation initiation factor 1A X-linked (EIF1AX)) [31,32]. In our networks, the inhibition of RHOA signaling by JK-206 treatment may induce the suppression of Myc targets through downregulation of BIRC5, H2AFZ, HIST1H2BK, KIF15, UBE2S, and FBXO5 in GC cells. KIF15 promotes cell mitosis and structural assembly [33,34] and its high gene expression is associated with poor prognosis in GC patients; on the contrary, the suppression of its expression inhibits GC cell progression, promotion of apoptosis, and cell cycle arrest [35]. In multiple GC dataset analyses, H2AFZ was identified as a crucial hub gene in the GC co-expression network, which indicated that H2AFZ has high degrees of interaction with multiple participant genes and may function as a control node in the GC co-expression network [36]. BIRC5, also known as survivin, is an apoptosis inhibitor [37] and acts as a resistance factor to anticancer therapies [38,39]. BIRC5 is a molecular marker of the poor prognosis in lung, pancreatic, and breast cancers [40]. It is known to increase lymph node metastasis [41] and is associated with poor clinical outcomes in GC [42]. Increased expression of BIRC5 is a risk factor for cancer progression and poor outcomes in breast cancer [43]. Though BIRC5 has been validated as a target of cancer drugs [44], the number of BIRC5 inhibitors available for clinical testing is limited [45]. As BIRC5 participates in large cellular networks, pathway inhibitors [46] have been proposed as promising alternatives to a single protein inhibitor [45]. In this study, BIRC5 was remarkably suppressed in GC cells when RHOA was inhibited by the RHOA inhibitor JK-206 (Figure 4b). BIRC5 was found to interact with the RHOA signaling pathway (Figure 5a) and was associated with mitotic activity (Figure 5b). In the independent GC dataset, BIRC5 was highly expressed in the tumor group compared to the control group, and its expression level was substantially correlated with that of RHOA (Figure 5c). Therefore, JK-206 could inhibit RHOA and, thus, suppress BIRC5.
Further investigation of BIRC5, H2AFZ, HIST1H2BK, KIF15, UBE2S, and FBXO5 is crucial to broaden the understanding of cancer invasion and migration, and to promote the discovery and evaluation of new therapeutic targets.
RHOA activity increases in pre-anaphase mitotic cells and participates in G2/M transition through mitotic cell rounding and de-adhesion [47]. The inhibition of RHOA leads to the inhibition of G2/M transition via the interference of mitotic cell rounding and de-adhesion [47,48]. In this study, JK-206 treatment dysregulated the G2/M checkpoint in GC cells. Disturbance of the G2/M checkpoint induces G2/M arrest, genomic instability, and subsequently, cell apoptosis [49]. JK-206 treatment may drive G2/M checkpoint arrest or catastrophic mitosis, resulting in subsequent apoptosis, which was confirmed by a notable decrease in cell viability in the cell viability assay (Figure 2a and Figure S1). Modifications of apoptosis- and cell-cycle-related pathways were also derived using our previously designed RHOA inhibitors (JK-136 and -139) [10]. Cross-talk between the RHOA signaling pathway and G2/M checkpoint is yet to be elucidated. Among the participant genes of the G2/M checkpoint gene set, RUVBL2, EIF1AX, and SET are known to play important roles in the cross-talk between RHOA signaling and cell cycle activity [50,51,52]. RUVBL2 is associated with poor prognosis in multiple types of cancer [52,53,54]. RUVBL2 is involved in DNA replication and its inhibition results in cancer cell death via cell cycle (i.e., S-phase) arrest and subsequent replication catastrophe in non-small cell lung cancer [52]. EIF1AX stimulates cell proliferation by inducing cell cycle transition (i.e., G1/S) via the inhibition of p21 expression, resulting in poor prognosis in breast cancer patients [51]. SET regulates the G2/M transition by modulating CDK1 [50], and it results in tumor progression and poor clinical outcomes in GC [55]. Therefore, SET is considered a potential therapeutic target [50,56,57]. In future studies, the specific roles of identified DEGs in the G2/M checkpoint gene set (i.e., RUVBL2, H2AFZ, EIF1AX, SNRPD3, and SET) need to be investigated to understand apoptosis or mitotic catastrophe induced by G2/M checkpoint alterations and discover new therapeutic targets.
The migration of cancer cells requires the reformation of microtubules, and RHOA is essential for cell motility, adhesion, and the regulation of actin and microtubules [58]. In our PPI network construction, JK-206 was found to be associated with the microtubule formation and function in GC cells, which was also confirmed by the remarkable reduction in cell migration in the cell migration assay (Figure 2b). We hypothesize that the downregulation of BIRC5, FBXO5, KIF15, and RUVBL2 led to the perturbation of microtubule dynamics with JK-206 treatment in GC cells. Considering the biological roles of microtubules in the cell cycle, JK-206-induced perturbation of microtubule dynamics may have affected cell cycle transition, especially at the G2/M checkpoint. Inhibitors also serve as chemical probes to reveal biological mechanisms [59]. JK-206 showed good efficacy in the in vitro viability assay (Figure 2a,b), and through network analysis, we showed an association between RHOA and BIRC5 in the transcriptome dataset of JK-206-treated GC cell lines. We only performed in vitro and in silico assays to measure the efficacy of JK-206; however, the discrepancy between in vitro and in vivo experiments might be observed due to GC heterogeneity, as we previously reported [10]. Further studies based on diverse GC cell lines and xenograft models are needed.
There are limitations. In this study, the mRNA and protein expressions of RHOA and the DEGs by JK-206 were not investigated through RT-PCR and western blotting. Thus, our conclusions should be carefully interpreted.
In this study, a new RHOA inhibitor, JK-206, substantially decreased both cell viability and migration in GC cells (Figure 2a,b). Through systematic analysis of the gene expression dataset, we identified that JK-206 treatment led to suppression of the expression levels of oncogenic genes, which play key roles in apoptosis, carcinogenesis, and protein degradation. JK-206 treatment affected pathways related to the G2/M checkpoint, Myc target, and microtubule dynamics. Thus, the impact of JK-206 on multiple oncogenic pathways represents a promising therapeutic potential for GC as a pathway inhibitor.
5. Conclusions
In conclusion, we identified the optimized compound JK-206 as an RHOA inhibitor in GC. This study not only supports our previous extensive studies regarding RHOA as a therapeutic target in GC [8,10,60], but also suggests RHOA-mediated mitogenic pathway regulation as a new therapeutic strategy for GC.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14071604/s1, Figure S1: RHOA inhibitors suppress cell growth and migration in GC.; Figure S2: Cell viability assay at 2 μM treatment; Figure S3: Expression patterns of the DEGs (depicted in Figure 4b) in JK-206, JK-312, and DMSO treatments in GC cells; Figure S4: JK-206 and JK-312 treatments in GC cells down-regulated hallmark gene sets of Myc targets, G2/M checkpoint, and E2F targets compared to the DMSO treatment in GC cells; Figure S5: Expression patterns of the DEGs (depicted in Figure 4b) by another RHOA inhibitors in GC cells; Figure S6: Correlations with RHOA and DEGs (depicted in Figure 4b) and expression patterns of the DEGs by knockdown and knockout of RHOA in GC and prostate cancer cells; Figure S7: Differentially expressed genes in association with RHOA expression level in an independent GC dataset (GSE36968); Table S1: A list of DEGs common and uncommon to JK-206 treated (versus DMSO treated) and JK-312 treated (versus DMSO treated) GC cells; Method S1: Synthesis of hydrazide derivatives.
Author Contributions
Data curation, M.B. and H.J.E.; Formal analysis, M.B., S.P. and J.-H.K.; Funding acquisition, S.P. and S.N.; Methodology, Y.H.K., J.L. and S.N.; Project administration, S.N.; Resources, H.J.E.; Software, M.B., S.P. and H.-Y.L.; Supervision, J.L. and S.N.; Validation, H.J.E. and Y.H.K.; Visualization, S.P.; Writing—original draft, M.B., S.P. and J.-H.K.; Writing—review and editing, J.L. and S.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Gachon University Gil Medical Center (grant number FRD2019-11-02 (2) to S.N.), the National Research Foundation of Korea (NRF-2020R1F1A1069206 to S.N.; NRF-2020R1I1A1A0106589212 to S.P.), and the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program (KGM5362111 to J.L.). The funders and sponsors had no role in study design; collection, management, analysis, and interpretation of data; writing of the report; or the decision to submit the report for publication. Neither did they have ultimate authority over any of these activities.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are publicly available. The microarray dataset in this study was deposited in the NCBI GEO repository (GEO accession GSE188908) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188908, accessed on 30 December 2021). For GC-cell-treated DMSO, three samples (GSM3984792, GSM3984796, and GSM3984800) were downloaded from our previously deposited dataset, GSE135068 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135068, accessed on 30 December 2021).
Conflicts of Interest
H.J.E. is an employee from Corestem Inc. The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Karimi P., Islami F., Anandasabapathy S., Freedman N.D., Kamangar F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol. Biomark. Prev. 2014;23:700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bang Y.J., Van Cutsem E., Feyereislova A., Chung H.C., Shen L., Sawaki A., Lordick F., Ohtsu A., Omuro Y., Satoh T., et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 4.Iqbal S., Goldman B., Fenoglio-Preiser C.M., Lenz H.J., Zhang W., Danenberg K.D., Shibata S.I., Blanke C.D. Southwest Oncology Group study S0413: A phase II trial of lapatinib (GW572016) as first-line therapy in patients with advanced or metastatic gastric cancer. Ann. Oncol. 2011;22:2610–2615. doi: 10.1093/annonc/mdr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lordick F., Kang Y.K., Chung H.C., Salman P., Oh S.C., Bodoky G., Kurteva G., Volovat C., Moiseyenko V.M., Gorbunova V., et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): A randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:490–499. doi: 10.1016/S1470-2045(13)70102-5. [DOI] [PubMed] [Google Scholar]
- 6.Sahai E., Marshall C.J. RHO-GTPases and cancer. Nat. Rev. Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 7.Nakaya Y., Sukowati E.W., Wu Y., Sheng G. RhoA and microtubule dynamics control cell-basement membrane interaction in EMT during gastrulation. Nat. Cell Biol. 2008;10:765–775. doi: 10.1038/ncb1739. [DOI] [PubMed] [Google Scholar]
- 8.Chang H.R., Nam S., Lee J., Kim J.H., Jung H.R., Park H.S., Park S., Ahn Y.Z., Huh I., Balch C., et al. Systematic approach identifies RHOA as a potential biomarker therapeutic target for Asian gastric cancer. Oncotarget. 2016;7:81435–81451. doi: 10.18632/oncotarget.12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nam S., Chang H.R., Kim K.T., Kook M.C., Hong D., Kwon C.H., Jung H.R., Park H.S., Powis G., Liang H., et al. PATHOME: An algorithm for accurately detecting differentially expressed subpathways. Oncogene. 2014;33:4941–4951. doi: 10.1038/onc.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J.H., Park S., Lim S.M., Eom H.J., Balch C., Lee J., Kim G.J., Jeong J.H., Nam S., Kim Y.H. Rational design of small molecule RHOA inhibitors for gastric cancer. Pharm. J. 2020;20:601–612. doi: 10.1038/s41397-020-0153-6. [DOI] [PubMed] [Google Scholar]
- 11.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks B.R., Brooks C.L., 3rd, Mackerell A.D., Jr., Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanzelmann S., Castelo R., Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikari N., Serizawa A., Tanji E., Yamamoto M., Furukawa T. Analysis of RHOA mutations and their significance in the proliferation and transcriptome of digestive tract cancer cells. Oncol. Lett. 2021;22:735. doi: 10.3892/ol.2021.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alkasalias T., Alexeyenko A., Hennig K., Danielsson F., Lebbink R.J., Fielden M., Turunen S.P., Lehti K., Kashuba V., Madapura H.S., et al. RhoA knockout fibroblasts lose tumor-inhibitory capacity in vitro and promote tumor growth in vivo. Proc. Natl. Acad. Sci. USA. 2017;114:E1413–E1421. doi: 10.1073/pnas.1621161114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nam S., Kim J.H., Lee D.H. RHOA in Gastric Cancer: Functional Roles and Therapeutic Potential. Front. Genet. 2019;10:438. doi: 10.3389/fgene.2019.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K., O’Donnell L., Oster S., Theesfeld C., Sellam A., et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y.H., Liang H., Liu X., Lee J.S., Cho J.Y., Cheong J.H., Kim H., Li M., Downey T.J., Dyer M.D., et al. AMPKalpha modulation in cancer progression: Multilayer integrative analysis of the whole transcriptome in Asian gastric cancer. Cancer Res. 2012;72:2512–2521. doi: 10.1158/0008-5472.CAN-11-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang X., Marchioni F., Sipes N., Evelyn C.R., Jerabek-Willemsen M., Duhr S., Seibel W., Wortman M., Zheng Y. Rational design of small molecule inhibitors targeting RhoA subfamily Rho GTPases. Chem. Biol. 2012;19:699–710. doi: 10.1016/j.chembiol.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan C.H., Lee S.W., Li C.F., Wang J., Yang W.L., Wu C.Y., Wu J., Nakayama K.I., Kang H.Y., Huang H.Y., et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat. Cell Biol. 2010;12:457–467. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauzeau V., Berenjeno I.M., Citterio C., Bustelo X.R. A transcriptional cross-talk between RhoA and c-Myc inhibits the RhoA/Rock-dependent cytoskeleton. Oncogene. 2010;29:3781–3792. doi: 10.1038/onc.2010.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altieri D.C. Survivin and apoptosis control. Adv. Cancer Res. 2003;88:31–52. doi: 10.1016/s0065-230x(03)88303-3. [DOI] [PubMed] [Google Scholar]
- 27.Yang H.D., Kim P.J., Eun J.W., Shen Q., Kim H.S., Shin W.C., Ahn Y.M., Park W.S., Lee J.Y., Nam S.W. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget. 2016;7:11412–11423. doi: 10.18632/oncotarget.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Switzer C.H., Cheng R.Y., Vitek T.M., Christensen D.J., Wink D.A., Vitek M.P. Targeting SET/I(2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene. 2011;30:2504–2513. doi: 10.1038/onc.2010.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tekcham D.S., Chen D., Liu Y., Ling T., Zhang Y., Chen H., Wang W., Otkur W., Qi H., Xia T., et al. F-box proteins and cancer: An update from functional and regulatory mechanism to therapeutic clinical prospects. Theranostics. 2020;10:4150–4167. doi: 10.7150/thno.42735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L., Liang Y., Li P., Liang Q., Sun H., Xu D., Hu W. Oncogenic Activities Of UBE2S Mediated By VHL/HIF-1alpha/STAT3 Signal Via The Ubiquitin-Proteasome System In PDAC. Onco Targets Ther. 2019;12:9767–9781. doi: 10.2147/OTT.S228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnamoorthy G.P., Davidson N.R., Leach S.D., Zhao Z., Lowe S.W., Lee G., Landa I., Nagarajah J., Saqcena M., Singh K., et al. EIF1AX and RAS Mutations Cooperate to Drive Thyroid Tumorigenesis through ATF4 and c-MYC. Cancer Discov. 2019;9:264–281. doi: 10.1158/2159-8290.CD-18-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rousseau B., Menard L., Haurie V., Taras D., Blanc J.F., Moreau-Gaudry F., Metzler P., Hugues M., Boyault S., Lemiere S., et al. Overexpression and role of the ATPase and putative DNA helicase RuvB-like 2 in human hepatocellular carcinoma. Hepatology. 2007;46:1108–1118. doi: 10.1002/hep.21770. [DOI] [PubMed] [Google Scholar]
- 33.Vale R.D., Milligan R.A. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 34.Zhong A., Tan F.Q., Yang W.X. Chromokinesin: Kinesin superfamily regulating cell division through chromosome and spindle. Gene. 2016;589:43–48. doi: 10.1016/j.gene.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 35.Tao J., Sun G., Li Q., Zhi X., Li Z., He Z., Chen H., Zhou A., Ye J., Xu G., et al. KIF15 promotes the evolution of gastric cancer cells through inhibition of reactive oxygen species-mediated apoptosis. J. Cell Physiol. 2020;235:9388–9398. doi: 10.1002/jcp.29743. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y., Grabsch H., Ivanova T., Tan I.B., Murray J., Ooi C.H., Wright A.I., West N.P., Hutchins G.G., Wu J., et al. Comprehensive genomic meta-analysis identifies intra-tumoural stroma as a predictor of survival in patients with gastric cancer. Gut. 2013;62:1100–1111. doi: 10.1136/gutjnl-2011-301373. [DOI] [PubMed] [Google Scholar]
- 37.Altieri D.C. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr. Opin. Cell Biol. 2006;18:609–615. doi: 10.1016/j.ceb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Morgillo F., Woo J.K., Kim E.S., Hong W.K., Lee H.Y. Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer Res. 2006;66:10100–10111. doi: 10.1158/0008-5472.CAN-06-1684. [DOI] [PubMed] [Google Scholar]
- 39.Tran J., Master Z., Yu J.L., Rak J., Dumont D.J., Kerbel R.S. A role for survivin in chemoresistance of endothelial cells mediated by VEGF. Proc. Natl. Acad. Sci. USA. 2002;99:4349–4354. doi: 10.1073/pnas.072586399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Duan N., Zhang C., Zhang W. Survivin and Tumorigenesis: Molecular Mechanisms and Therapeutic Strategies. J. Cancer. 2016;7:314–323. doi: 10.7150/jca.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J., Zhu Z., Sun Z., Sun X., Wang Z., Xu H. Survivin gene expression increases gastric cancer cell lymphatic metastasis by upregulating vascular endothelial growth factor-C expression levels. Mol. Med. Rep. 2014;9:600–606. doi: 10.3892/mmr.2013.1858. [DOI] [PubMed] [Google Scholar]
- 42.Liu J.L., Gao W., Kang Q.M., Zhang X.J., Yang S.G. Prognostic value of survivin in patients with gastric cancer: A systematic review with meta-analysis. PLoS ONE. 2013;8:e71930. doi: 10.1371/journal.pone.0083350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van ‘t Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 44.Fesik S.W. Promoting apoptosis as a strategy for cancer drug discovery. Nat. Rev. Cancer. 2005;5:876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 45.Altieri D.C. Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 46.Van der Greef J., McBurney R.N. Innovation: Rescuing drug discovery: In vivo systems pathology and systems pharmacology. Nat. Rev. Drug Discov. 2005;4:961–967. doi: 10.1038/nrd1904. [DOI] [PubMed] [Google Scholar]
- 47.Marchesi S., Montani F., Deflorian G., D’Antuono R., Cuomo A., Bologna S., Mazzoccoli C., Bonaldi T., Di Fiore P.P., Nicassio F. DEPDC1B coordinates de-adhesion events and cell-cycle progression at mitosis. Dev. Cell. 2014;31:420–433. doi: 10.1016/j.devcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maddox A.S., Burridge K. RhoA is required for cortical retraction and rigidity during mitotic cell rounding. J. Cell Biol. 2003;160:255–265. doi: 10.1083/jcb.200207130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bucher N., Britten C.D. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br. J. Cancer. 2008;98:523–528. doi: 10.1038/sj.bjc.6604208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canela N., Rodriguez-Vilarrupla A., Estanyol J.M., Diaz C., Pujol M.J., Agell N., Bachs O. The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J. Biol. Chem. 2003;278:1158–1164. doi: 10.1074/jbc.M207497200. [DOI] [PubMed] [Google Scholar]
- 51.Li Y., Guo L., Ying S., Feng G.H., Zhang Y. Transcriptional repression of p21 by EIF1AX promotes the proliferation of breast cancer cells. Cell Prolif. 2020;53:e12903. doi: 10.1111/cpr.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yenerall P., Das A.K., Wang S., Kollipara R.K., Li L.S., Villalobos P., Flaming J., Lin Y.F., Huffman K., Timmons B.C., et al. RUVBL1/RUVBL2 ATPase Activity Drives PAQosome Maturation, DNA Replication and Radioresistance in Lung Cancer. Cell Chem. Biol. 2020;27:105–121.e14. doi: 10.1016/j.chembiol.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lauscher J.C., Loddenkemper C., Kosel L., Grone J., Buhr H.J., Huber O. Increased pontin expression in human colorectal cancer tissue. Hum. Pathol. 2007;38:978–985. doi: 10.1016/j.humpath.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Breig O., Bras S., Martinez Soria N., Osman D., Heidenreich O., Haenlin M., Waltzer L. Pontin is a critical regulator for AML1-ETO-induced leukemia. Leukemia. 2014;28:1271–1279. doi: 10.1038/leu.2013.376. [DOI] [PubMed] [Google Scholar]
- 55.Yuan X., Zhang T., Zheng X., Zhang Y., Feng T., Liu P., Sun Z., Qin S., Liu X., Zhang L., et al. Overexpression of SET oncoprotein is associated with tumor progression and poor prognosis in human gastric cancer. Oncol. Rep. 2017;38:1733–1741. doi: 10.3892/or.2017.5788. [DOI] [PubMed] [Google Scholar]
- 56.Cristobal I., Garcia-Orti L., Cirauqui C., Cortes-Lavaud X., Garcia-Sanchez M.A., Calasanz M.J., Odero M.D. Overexpression of SET is a recurrent event associated with poor outcome and contributes to protein phosphatase 2A inhibition in acute myeloid leukemia. Haematologica. 2012;97:543–550. doi: 10.3324/haematol.2011.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu H., Gu Y., Wang H., Yin J., Zheng G., Zhang Z., Lu M., Wang C., He Z. Overexpression of PP2A inhibitor SET oncoprotein is associated with tumor progression and poor prognosis in human non-small cell lung cancer. Oncotarget. 2015;6:14913–14925. doi: 10.18632/oncotarget.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ridley A.J. Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castoreno A.B., Eggert U.S. Small molecule probes of cellular pathways and networks. ACS Chem. Biol. 2011;6:86–94. doi: 10.1021/cb1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J.H., Eom H.J., Lim G., Park S., Lee J., Nam S., Kim Y.H., Jeong J.H. Differential effects, on oncogenic pathway signalling, by derivatives of the HNF4 alpha inhibitor BI6015. Br. J. Cancer. 2019;120:488–498. doi: 10.1038/s41416-018-0374-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are publicly available. The microarray dataset in this study was deposited in the NCBI GEO repository (GEO accession GSE188908) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188908, accessed on 30 December 2021). For GC-cell-treated DMSO, three samples (GSM3984792, GSM3984796, and GSM3984800) were downloaded from our previously deposited dataset, GSE135068 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135068, accessed on 30 December 2021).