Abstract
The MYC oncogene encodes for the MYC protein and is frequently dysregulated across multiple cancer cell types, making it an attractive target for cancer therapy. MYC overexpression leads to MYC binding at active enhancers, resulting in a global transcriptional amplification of active genes. Because super-enhancers are frequently dysregulated in cancer, we hypothesized that MYC preferentially invades into super-enhancers and alters the cancer genome organization. To that end, we performed ChIP-seq, RNA-seq, circular chromosome conformation capture (4C-seq), and Spike-in Quantitative Hi-C (SIQHiC) on the U2OS osteosarcoma cell line with tetracycline-inducible MYC. MYC overexpression in U2OS cells modulated histone acetylation and increased MYC binding at super-enhancers. SIQHiC analysis revealed increased global chromatin contact frequency, particularly at chromatin interactions connecting MYC binding sites at promoters and enhancers. Immunofluorescence staining showed that MYC molecules formed punctate foci at these transcriptionally active domains after MYC overexpression. These results demonstrate the accumulation of overexpressed MYC at promoter–enhancer hubs and suggest that MYC invades into enhancers through spatial proximity. At the same time, the increased protein–protein interactions may strengthen these chromatin interactions to increase chromatin contact frequency. CTCF siRNA knockdown in MYC-overexpressed U2OS cells demonstrated that removal of architectural proteins can disperse MYC and abrogate the increase in chromatin contacts. By elucidating the chromatin landscape of MYC-driven cancers, we can potentially target MYC-associated chromatin interactions for cancer therapy.
Dysregulation of the MYC oncogene, which encodes for the MYC transcription factor, is common in cancer. Elevated mRNA and protein expression of MYC is seen across most cancer types in The Cancer Genome Atlas (TCGA) data set (Schaub et al. 2018), making it an attractive target for cancer therapy. However, there are a multitude of causes for MYC dysregulation, including focal amplification of the MYC gene (Beroukhim et al. 2010), aberrations in the web of oncogenic signaling pathways regulating MYC (Kress et al. 2015), and the acquisition of new enhancer regulatory elements in spatial proximity to the MYC gene locus through deletions and translocations (Affer et al. 2014). The MYC protein itself is notoriously termed as “undruggable,” because of the unstructured nature of the protein and the lack of enzymatic binding sites or prominent pockets for small molecule inhibitor binding (McKeown and Bradner 2014).
MYC usually binds to the promoters of actively transcribing genes at physiological levels. Upon overexpression, MYC not only increases binding at active promoters but “invades” into active enhancers as well, resulting in a global transcriptional amplification of active genes (Lin et al. 2012). Canonical MYC binding sites become saturated and MYC occupies noncanonical binding sites with lower affinity, resulting in an up-regulation of genes associated with malignant transformation (Walz et al. 2014). Enhancer invasion has also been observed in the MYC family member MYCN (Zeid et al. 2018).
At present, the mechanism underlying MYC enhancer invasion has not been well studied. Enhancers recruit transcription factors to activate linearly distant gene promoters via long-range chromatin interactions (Carter et al. 2002; Plank and Dean 2014). Recent studies have defined a category of enhancers known as super-enhancers: clusters of enhancers with exceptionally high enrichment of transcriptional activators (Hnisz et al. 2013; Lovén et al. 2013; Whyte et al. 2013). Super-enhancers are associated with a more extensive network of chromatin interactions to multiple distant target genes (Cao et al. 2017), to serve as regulatory hubs to govern processes important to cell identity (Whyte et al. 2013). This led us to question whether MYC enhancer invasion might preferentially occur at super-enhancers and if this might alter the cancer genome architecture.
In this study, we aimed to investigate the immediate changes to the enhancer and chromatin interaction landscapes after MYC overexpression, particularly at super-enhancers, using Spike-In Quantitative Hi-C (SIQHiC), a modified Hi-C approach that normalizes chromatin interactions by cell count.
Results
MYC overexpression leads to increased MYC binding at super-enhancers
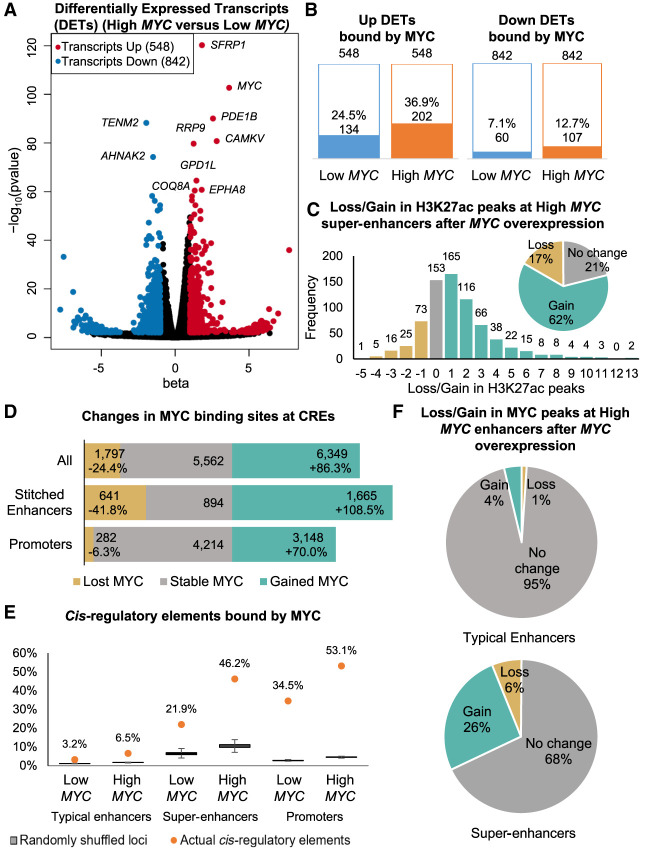
We profiled the MYC enhancer binding landscape changes associated with MYC overexpression using the U2OS osteosarcoma cell line inserted with a tetracycline-inducible MYC cassette, because these cells are not addicted to MYC expression and have been used in previous studies to investigate the effects of MYC overexpression (Walz et al. 2014; Lorenzin et al. 2016). Doxycycline treatment for 30 h increased MYC gene expression by about 35× compared to vehicle treatment, together with an increase in MYC protein expression (Supplemental Fig. S1A–C). Hereafter, we refer to vehicle-treated and doxycycline-treated U2OS cells as Low MYC cells and High MYC cells, respectively. RNA-seq identified 548 significantly up-regulated and 842 significantly down-regulated transcripts after MYC overexpression (Fig. 1A), in line with previous observations by Walz et al. (2014). MYC is known to repress a subset of genes, for example, through its association with ZBTB17 (also known as MIZ1) (Walz et al. 2014). Although down-regulated transcripts outnumbered up-regulated transcripts (Fig. 1B), only 12.7% of the down-regulated transcripts were direct targets of MYC, indicating that direct repression was not the main mechanism for down-regulation. Most of these down-regulated transcripts were lowly expressed, with reads per kilobase per million (RPKM) values <1. Because MYC overexpression had been shown to increase total RNA content per cell (Lovén et al. 2012), transcripts that were up-regulated to a smaller extent than the increase in total RNA content would appear to be down-regulated (Kress et al. 2015). In contrast to the down-regulated transcripts, 24.5% of the up-regulated transcript promoters were bound by MYC at endogenous MYC expression levels and increased to 36.9% after MYC overexpression (Fig. 1B).
Figure 1.
MYC overexpression leads to differential transcript expression, differential H3K27ac signal, and increased MYC binding at super-enhancers. (A) Significantly up-regulated (red) and down-regulated (blue) transcripts after MYC overexpression (|beta| > 1, FDR < 0.05, Wald test). Top 10 significantly regulated transcripts are labeled. (B) Number of differentially expressed transcripts (DETs) bound by MYC in Low MYC and High MYC cells. (C) Bar graph and pie chart showing gain and loss of H3K27ac peaks at super-enhancers after MYC overexpression. (D) Bar graph showing gain and loss of MYC binding sites at stitched enhancers and promoters after MYC overexpression. (E) Proportion of typical enhancers, super-enhancers, and promoters being bound by MYC compared to randomly shuffled coordinates. The proportion of MYC-bound CREs are shown as orange dots. Box plots show 1000 iterations of MYC occupancy at random genomic loci of the same size and on the same chromosome as the actual CREs. (F) Pie charts showing gain and loss of MYC ChIP-seq peaks at typical enhancers (top) and super-enhancers (bottom) after MYC overexpression.
We performed a gene set enrichment analysis (GSEA) to determine the pathways dysregulated by MYC overexpression. Previously published gene sets of MYC-regulated genes were significantly enriched in our data set (Supplemental Fig. S1D). Consistent with previous literature (Eilers and Eisenman 2008), MYC overexpression activated pathways involved in cell proliferation, including ribosome biogenesis, translation, mitochondrial biogenesis, and splicing (Supplemental Fig. S1E). MYC overexpression activated the MAPK14 pathway, which is implicated in osteoblast motility (Supplemental Fig. S1E; Rodríguez-Carballo et al. 2016), and down-regulated cell adhesion genes (Supplemental Fig. S1F), suggesting a shift toward a cell migration phenotype.
We performed H3K27ac ChIP-seq to identify active cis-regulatory elements (CREs) in Low MYC and High MYC cells. H3K27ac peaks within 2.5 kb of transcription start sites were labeled as active promoter peaks, whereas the remaining peaks were labeled as enhancer peaks. We identified super-enhancers using a similar approach to ROSE (rank ordering of super-enhancers) (Lovén et al. 2013; Whyte et al. 2013). Briefly, enhancer peaks within 12.5 kb of each other were stitched together, and super-enhancers were separated from typical enhancers based on their H3K27ac ChIP-seq signal (Methods; Supplemental Fig. S2A). In total, we identified ∼28,500 stitched enhancers in both Low MYC and High MYC cells, of which ∼7300 stitched enhancers were lost and gained after MYC overexpression (Supplemental Fig. S2C). We identified 890 super-enhancers in the Low MYC cells and 725 super-enhancers in the High MYC cells (Supplemental Fig. S2E). Although many stitched enhancers were gained or lost, almost all High MYC super-enhancers overlapped with Low MYC stitched enhancers (Fig. 1C), indicating that super-enhancers remain highly acetylated and novel super-enhancers were not activated after MYC overexpression. Sixty-two percent of the High MYC super-enhancers gained between 1 and 13 H3K27ac peaks after MYC overexpression (Fig. 1C), with 15% merging multiple Low MYC stitched enhancers (Supplemental Fig. 2F), resulting in super-enhancers with significantly more constituent H3K27ac peaks (Supplemental Fig. 2G).
Next, we looked into MYC occupancy at CREs. MYC overexpression led to a loss of 1797 MYC binding sites and a gain of 6349 MYC binding sites, of which 3148 binding sites were gained at promoters and 1665 at stitched enhancers (Fig. 1D). The proportion of MYC binding sites at stitched enhancers increased from 17.6% to 21.4%, whereas the proportion of MYC binding sites at promoters remained similar (61.5%–61.8%) (Supplemental Fig. 3A), recapitulating previous observations of enhancer invasion. The 21.9% of super-enhancers overlapping with MYC binding sites increased to 46.2% after MYC overexpression (Fig. 1E). and 26% of super-enhancers gained MYC binding sites after MYC overexpression, compared to only 4% of typical enhancers (Fig. 1F; Supplemental Fig. S3B,C). H3K27ac ChIP-seq profiles of MYC binding sites show that MYC binds directly adjacent to constituent H3K27ac peaks and not randomly between the constituent peaks within the super-enhancers (Supplemental Fig. S3D). To test whether the preferential binding of MYC at super-enhancers was due to size, we randomly shuffled the genomic intervals of CREs within the same chromosome and overlapped them with MYC binding sites. Significantly more actual super-enhancers overlapped with MYC binding sites than 1000 iterations of randomly shuffled genomic loci (Fig. 1E), indicating that MYC preferentially binds at super-enhancers regardless of their size. Together, these results indicate that overexpressed MYC preferentially accumulates at H3K27ac peaks in super-enhancers.
We further compared the MYC binding sites that were pre-existing, lost, or gained after MYC overexpression. Pre-existing MYC binding sites at promoters and enhancers consistently had the highest H3K27ac and MYC ChIP-seq signals, whereas gained MYC binding sites had lower signals (Supplemental Fig. S3E–G), indicating that open chromatin demarcated by H3K27ac is required for MYC binding. Lost promoter MYC binding sites had high H3K27ac signal comparable to pre-existing and gained MYC binding sites (Supplemental Fig. S3G). This suggests that H3K27ac is required but insufficient for MYC binding at promoters, and other factors are involved in the recruitment of MYC, such as binding site affinity and recruitment by other transcription factors (Lorenzin et al. 2016).
We performed motif analysis at MYC ChIP-seq peaks to ascertain whether there may be different chromatin factors recruiting MYC to promoters and enhancers. Promoter MYC binding sites were more enriched for other transcription factor binding motifs compared to typical enhancer and super-enhancer MYC binding sites (Supplemental Fig. S4A,B). Thirty-five motifs occurred at >50% of promoter MYC binding sites, whereas no single motif occurred at >50% of enhancer MYC binding sites (Supplemental Fig. S4A,B). Top enriched motifs at promoters included known promoter binding factors such as the KLF/SP family of transcription factors, which contain guanine/cytosine (G/C) rich sequences. Similarly, top enriched motifs at enhancers contained G/C rich sequences, such as MAZ, ZBTB17, and EGR2. Chromatin interactions frequently connect CpG rich promoters and enhancers together (Pachano et al. 2021), and these interactions may be mediating the recruitment of MYC from promoter binding sites to enhancers. Gained MYC binding sites were slightly less enriched for transcription factor motifs than pre-existing MYC binding sites (Supplemental Fig. S4F), whereas lost MYC binding sites were poorly enriched for transcription factor motifs, suggesting that MYC is likely recruited to these binding sites by other transcription factors (Supplemental Fig. S4G). However, the same transcription factor motifs were enriched for in lost, pre-existing, and gained MYC binding sites, indicating that recruitment of supraphysiological MYC to gained binding sites by new interactors is unlikely (Supplemental Fig. S4E).
Spike-in Quantitative Hi-C reveals increased global chromatin contact frequency per cell after MYC overexpression
In traditional Hi-C techniques, cross-sample normalization is based on the assumption that chromatin interactions are largely stable across biological conditions (Lun and Smyth 2015; Stansfield et al. 2018). However, perturbation of factors involved in chromatin organization such as CTCF and cohesin can result in global changes in chromatin contact frequency. We hypothesized that increased global MYC binding at CREs not only increases transcription of associated genes but also stabilizes chromatin interactions at these genes, resulting in similar global changes in chromatin contact frequency.
Cell-count normalization techniques have been developed for RNA-seq (Lovén et al. 2012) and ChIP-seq (Orlando et al. 2014) to account for global changes in transcription and occupancy, respectively. Cell-count normalized transcription analyses had previously revealed a global increase in transcription after MYC overexpression (Lovén et al. 2012; Nie et al. 2012). These methods involve mixing a control sample from an orthogonal species into the experimental human samples at a fixed cell number ratio. The recently published Absolute Quantification of Architecture Hi-ChIP protocol (AQuA-HiChIP) (Gryder et al. 2019) utilizes the same strategy of spiking in mouse cells into human samples, running on the assumption that contact frequency for untreated mouse cells should be the same across samples.
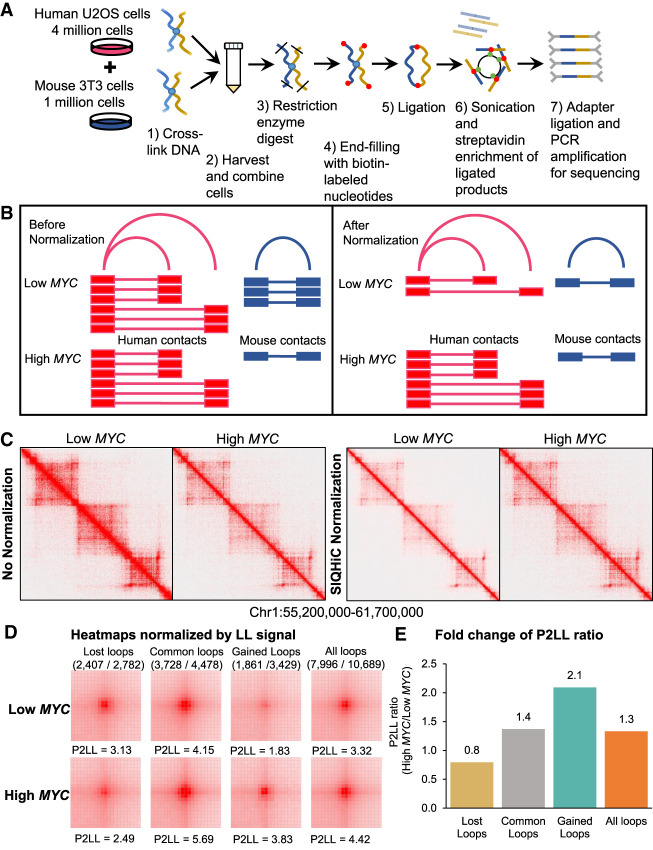
In our study, we adapted the AQuA-HiChIP protocol (Gryder et al. 2019) to perform Spike-In Quantitative Hi-C on Low MYC and High MYC cells in duplicate, to elucidate the global changes in chromatin contact frequency. In brief, mouse 3T3 cells were cross-linked and mixed into each sample of human cells at a ratio of 1:4, before continuing with the Arima Hi-C kit protocol (Fig. 2A; Methods), preserving this cell number ratio throughout all subsequent steps of library preparation. If global chromatin contact frequency is unchanged, we expect to obtain a similar human to mouse chromatin contact ratio (HMR) across all samples. Hence, the relative difference between the HMR of different samples reflects the differences in global chromatin contact frequency and can be used to normalize the samples (Fig. 2B).
Figure 2.
SIQHiC normalization reveals increased chromatin contact frequency per cell after MYC overexpression. (A) Brief overview of the SIQHiC workflow. (B) Cartoon illustrating Hi-C contacts before (left) and after (right) SIQHiC normalization. SIQHiC normalization scaled down the Low MYC contacts such that the number of mouse contacts in both conditions was the same, thereby revealing an increase in human chromatin contacts. (C) Hi-C matrix heat maps of a region on Chromosome 1. Left panel: No normalization. Right panel: SIQHiC normalization. (D) Aggregate Peak Analysis (APA) plots at 5-kb resolution showing the aggregate signal of “Lost,” “Common,” and “Gained” chromatin loop sets in Low MYC and High MYC cells identified using the nonnormalized Hi-C contact matrices. (P) Peak signal at the center pixel, (LL) average signal of the 3 × 3 square at the lower left corner of the APA plot, representing local background, (P2LL) ratio of P to LL. APA color scales were normalized by the LL signal. Loop sets were filtered to remove short loops near the diagonal (shown above each APA plot; numerator: number of filtered loops; denominator: total number of loops). (E) Fold change of P2LL ratio between High MYC and Low MYC cells.
Ambiguous read pairs mapping to both species consistently made up a small percentage of the total reads per sample (∼0.3%) and were discarded (Supplemental Table S1). Using SIQHiC, we observe a 2.1- to 4.4-fold increase in HMR after MYC overexpression, suggesting an equivalent increase in global chromatin contacts (Supplemental Table S2). Hi-C chromatin interaction heat maps of Low MYC and High MYC cells appear to show a decrease in chromatin interactions after MYC overexpression (Fig. 2C, left). After SIQHiC normalization, we observed an increase in chromatin interactions instead (Fig. 2D, right). As expected, Low MYC duplicates had similar HMRs (33.4 and 36.4), reflecting consistent chromatin contact frequencies at physiological MYC levels (Supplemental Table S2). Conversely, High MYC duplicates had higher and more variable HMRs (146.9 and 78.1) after doxycycline induction of MYC expression. (Supplemental Table S2).
MYC overexpression weakens TAD insulation and strengthens a set of chromatin loops
Given that MYC overexpression increased chromatin contact frequency, we wanted to know whether MYC overexpression reshapes the global chromatin landscape. A/B compartment analysis showed few instances of compartment switching between Low MYC and High MYC cells, with Chromosome 8 shown as an example in Supplemental Figure S5A. However, MYC overexpression weakened insulation at topologically associating domain (TAD) boundaries (Supplemental Fig. S5B), resulting in a loss of 2912 TAD boundaries (Supplemental Fig. S5C). Lost and common TAD boundaries showed similar loss of insulation, as seen by the reduced amplitude of TAD separation score, whereas insulation at gained boundaries remained relatively unchanged (Supplemental Fig. S5B). This indicates that inactive compartments were not activated by MYC overexpression, but chromatin contact frequency increased interactions across TAD boundaries.
Next, we identified 7266 and 7910 significant chromatin loops in Low MYC and High MYC cells, respectively, with approximately 60% of these loops common between both conditions (Supplemental Fig. S5D). We performed an aggregate plot analysis (APA) to show the aggregate signal of the chromatin loops that were “gained” and “lost” after MYC overexpression, or “common” between both conditions. In this analysis, the 105-kb × 105-kb contact matrices surrounding the midpoints of all loops in each loop set are overlapped such that the center pixel (P) of the APA plot shows the aggregate signal of the midpoints of these loops. The lower left corner of the plot (LL) represents the contact frequency for shorter interactions in the vicinity of the loop set and gives an indication of the local random interaction frequencies. Without SIQHiC normalization, APA plots show decreased peak signal enrichment (P) at chromatin loops after MYC overexpression (Supplemental Fig. S6A,B). Using SIQHiC to normalize for cell count revealed a 2.2- and 3.3-fold increase in peak signal enrichment (P) at common and gained loop sets instead (Supplemental Fig. S6C,D). Using the ratio of signal at P to the average signal at LL (P2LL) to normalize peak signals against local background interactions, we observed a smaller increase in chromatin contact frequency at common and gained loop sets compared to SIQHiC normalization (1.4-fold and 2.1-fold, respectively) (Fig. 2D,E), indicating that MYC overexpression increased local random interactions but to a lesser extent than loop interactions. Taken together, APA analysis showed that MYC overexpression increased global chromatin contact frequency as a whole but strengthened these chromatin loops in particular.
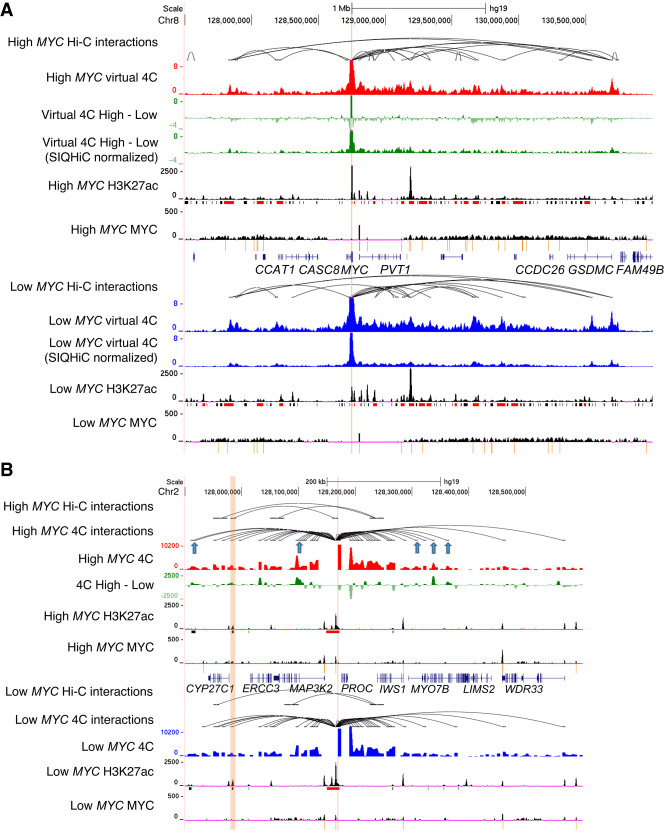
We zoomed in on the MYC gene locus because MYC is known to be regulated by super-enhancers through chromatin interactions (Shi et al. 2013; Herranz et al. 2014; Zhang et al. 2016; Bahr et al. 2018). We generated virtual 4C plots from our Hi-C data, using the MYC gene promoter as the viewpoint, to visualize the chromatin interactions at this locus. Without normalization, virtual 4C showed a general decrease in chromatin interaction frequencies (Fig. 3A). However, SIQHiC normalization of the Hi-C reads revealed a general increase in virtual 4C signal instead (Fig. 3A).
Figure 3.
MYC overexpression increases chromatin contact frequency and chromatin loops at super-enhancers. (A) Genome browser view of High MYC and Low MYC cells at the MYC locus. Tracks show Hi-C chromatin loops, virtual 4C signal of interactions at the MYC promoter (red and blue tracks, respectively), H3K27ac and MYC ChIP-seq signal. The difference between the virtual 4C signals of High MYC and Low MYC cells are shown with and without SIQHiC normalization (green tracks). Super-enhancers (red) and typical enhancers (black) are shown as bars below the H3K27ac ChIP-seq tracks. MYC binding peaks are shown as yellow bars below the MYC ChIP-seq tracks. (B) 4C-seq of chromatin interactions at a randomly selected MYC-bound super-enhancer near the PROC gene on Chromosome 2. Tracks show Hi-C chromatin loops, 4C-seq signal (red and blue tracks), difference between 4C-seq signal of High MYC and Low MYC cells (green), H3K27ac and MYC ChIP-seq signal. Gained Hi-C chromatin loop is highlighted in orange. Blue arrows show additional gained chromatin interactions identified using 4C-seq.
Because Hi-C assays the entire genome, it requires very high sequencing depth for sufficient resolution and limits chromatin loop detection within a specific region of interest (Babu and Fullwood 2015; Sati and Cavalli 2017). Hence, we performed circular chromosome conformation capture (4C-seq) at a randomly selected MYC bound super-enhancer near the PROC gene which gained a chromatin loop to the CYP27C1 gene after MYC overexpression. 4C-seq showed that the gained Hi-C chromatin loop was already present in Low MYC cells, but MYC overexpression increased the interaction frequency of this loop (Fig. 3B). 4C-seq also identified additional chromatin loops at the MYC-bound super-enhancer locus that were not significant in the Hi-C data (Fig. 3B, blue arrows). Taken together, MYC overexpression strengthens a set of gained chromatin loops.
SIQHiC normalization shows reduced chromatin contact frequency after CTCF siRNA knockdown
To validate our SIQHiC normalization approach, we transfected High MYC cells with either small interfering RNA (siRNA) targeting CTCF (siCTCF) or nontargeting siRNA (siControl) and performed SIQHiC in duplicate. siCTCF knockdown reduced CTCF expression by 80% compared to siControl (Supplemental Fig. S7A). SIQHiC normalization showed that global chromatin contact frequency in siCTCF cells was reduced by 55% compared to siControl cells (Supplemental Tables S3, S4). After CTCF siRNA knockdown, 74.3% (4633/6236) of TAD boundaries from siControl cells remained (Supplemental Fig. S7B), but 47.1% (5624/11,929) of chromatin loops were lost (Supplemental Fig. S7C). TAD boundaries lost after CTCF siRNA knockdown showed a decrease in insulation, but insulation at the common and gained TAD boundaries remained mostly unchanged (Supplemental Fig. S7D). These results are similar to previous reports of insulation preservation at selected TAD boundaries when CTCF is incompletely depleted (Zuin et al. 2014; Khoury et al. 2020). Without normalization, siCTCF appeared to selectively decrease chromatin contact frequency at siControl-unique chromatin loops and increase chromatin contact frequency at a small set of siCTCF-unique loops, whereas contact frequency at common loops remained unchanged (Supplemental Fig. S7C,E,F), similar to results from Zuin et al. (2014) and Kubo et al. (2021). After applying SIQHiC normalization, we observed a global decrease in chromatin contact frequency at all loop sets instead (Supplemental Fig. S7G,H), indicating that the siCTCF unique loops were not being induced after CTCF depletion but rather reduced to a lesser extent compared to siControl unique and common loops. This demonstrates the necessity for cell-count normalization when perturbing chromatin architectural factors.
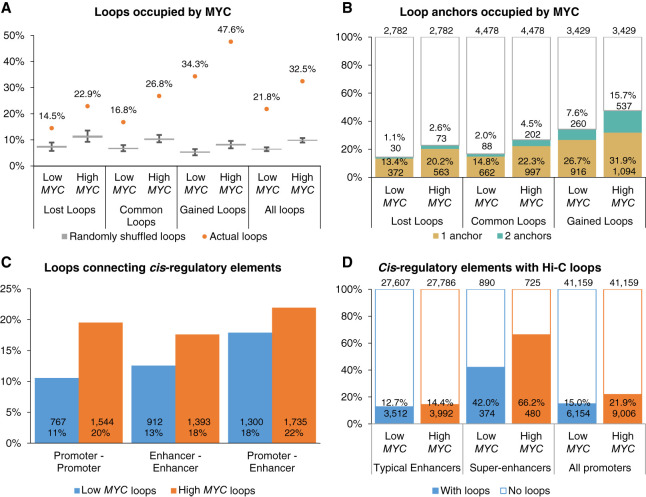
Chromatin loops tend to connect MYC binding sites at super-enhancers
We overlapped all Hi-C loop anchors with MYC binding sites and found that the proportion of all chromatin loops with MYC binding sites increased from 21.8% to 32.5% after MYC overexpression (Fig. 4A). Gained loops were particularly enriched in MYC binding, with 47.6% of these loops occupied by MYC in High MYC cells, which was considerably higher than the proportion of randomly shuffled chromatin loop coordinates overlapping with MYC binding sites (Fig. 4A). Notably, more gained loops were occupied by MYC at physiological MYC levels (34.3%) compared to common (16.8%) and lost loops (14.3%) (Fig. 4A). Additionally, 7.6% of the gained loops had MYC binding at both anchors at physiological MYC levels and increased to 15.7% after MYC overexpression (Fig. 4B). APA analysis revealed a 1.54-fold increase in chromatin contact frequency at loops connecting MYC binding sites, compared to a 1.32-fold increase at loops with no MYC binding (Supplemental Fig. S8A). Gene Ontology analysis showed that MYC-bound genes at lost chromatin loops were associated with negative regulation of transcription (Supplemental Fig. S8B). MYC-bound genes at gained chromatin loops were associated with previously shown MYC dysregulated pathways such as transcription, translation, and cell–cell adhesion (Supplemental Figs. S1E,F, S8C). These results suggest that chromatin interactions connecting MYC-bound genomic loci are preferentially strengthened after MYC overexpression.
Figure 4.
Chromatin interactions are enriched at super-enhancers and gained chromatin loops tend to connect MYC binding sites. (A) Percentage of Hi-C chromatin loops occupied by MYC compared to randomly shuffled loop coordinates. Percentage of MYC-bound chromatin loops are shown as orange dots. Box plots show 1000 iterations of MYC occupancy at random genomic loci of the same size and on the same chromosome as the actual chromatin loops. (B) Percentage of chromatin loop anchors occupied by MYC. (C) Percentage of Hi-C chromatin loops connecting cis-regulatory elements. (D) Percentage of cis-regulatory elements with Hi-C chromatin loops.
Next, we wanted to know whether chromatin interactions are preferentially altered at the enhancers invaded by MYC. We annotated the Hi-C loops according to their overlap with promoters and stitched enhancers. MYC overexpression increased chromatin looping between promoters (11%–20%), between stitched enhancers (13%–18%), and between promoters and stitched enhancers (18%–22%) (Fig. 4C). In particular, the proportion of CRE loops within the lost loop set was similar to the common loop set, whereas the proportion of CRE loops within the gained loop set was markedly higher (Supplemental Fig. S9A), showing that chromatin interactions are preferentially strengthened between CREs. Consistent with previous research (Cao et al. 2017), a markedly higher proportion of super-enhancers (42.0%) was associated with Hi-C loops compared to typical enhancers (12.7%) and promoters (15.0%), and MYC overexpression further increased the proportion of super-enhancers with loops to 66.2% (Fig. 4D). MYC-bound CREs were more associated with chromatin loops compared to non-MYC-bound CREs, particularly after MYC overexpression (Supplemental Fig. S9B). Because super-enhancers are larger than typical enhancers, we compared the constituent H3K27ac peaks within typical and super-enhancers. Similarly, more MYC-bound super-enhancer constituent H3K27ac peaks overlapped with Hi-C loops (27.3%–39.9%) compared to MYC-bound typical enhancer constituents (13.2%–21.2%) and promoter loci (15.4%–28.0%) (Supplemental Fig. S9B,C).
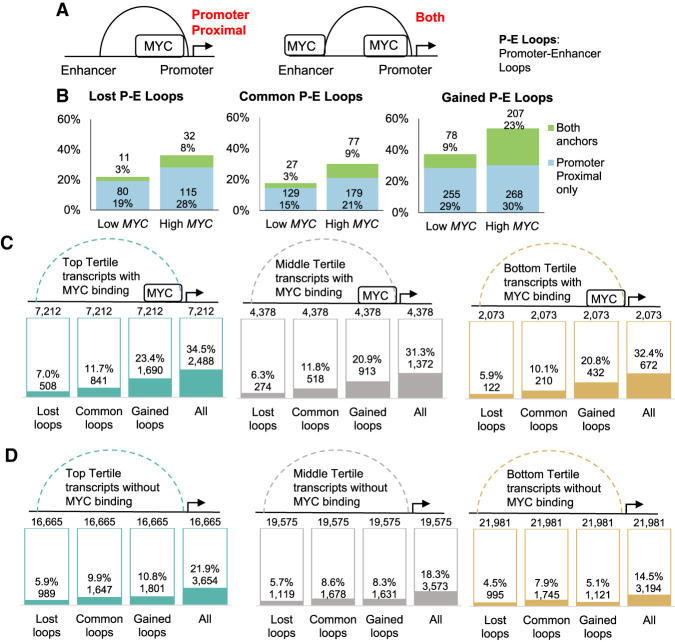
Because MYC binds at both promoters and enhancers, we looked at promoter–enhancer chromatin loops (P-E loops) in detail. We overlapped P-E loops with MYC binding sites and categorized the loops as being bound proximal to the promoter loop anchor or bound at both loop anchors (Fig. 5A). MYC overexpression increased the proportion of P-E loops with only proximal MYC binding sites from 16% to 26%, whereas P-E loops bound at both anchors increased from 3% to 16% (Supplemental Fig. S9D). Although lost P-E loops gained MYC binding sites at the promoter proximal loop anchor after MYC overexpression (22%–36%), few of these loci gained MYC at the distal anchor as well (3%–8%) (Fig. 5B). In contrast, gained P-E loops acquired more distal MYC binding sites (9%–23%) (Fig. 5B). Taken together, our results suggest that MYC overexpression preferentially strengthens chromatin interactions between MYC binding sites at promoters and enhancers.
Figure 5.
MYC is enriched at both anchors of gained promoter–enhancer chromatin loops regardless of transcriptional activity. (A) Promoter–enhancer chromatin loops are categorized as having promoter-proximal MYC binding or MYC binding at both loop anchors. (B) Percentage of lost (left), common (middle), and gained (right) promoter–enhancer chromatin loops occupied by MYC at the promoter proximal anchor or at both anchors. (C,D) Percentage of (C) MYC-bound and (D) non-MYC-bound transcripts with Hi-C chromatin loops. Transcripts were stratified into top (green), middle (gray), and bottom (yellow) tertiles based on transcript expression after MYC overexpression.
MYC overexpression increases chromatin contact frequency regardless of transcription activity
Previous studies have linked transcription activity with the formation of chromatin interactions, possibly through cohesin binding and positioning (Busslinger et al. 2017; Isoda et al. 2017; Heinz et al. 2018) and cis-regulatory element mobility (Gu et al. 2018; Nagashima et al. 2019). Because MYC overexpression leads to a global increase in transcription, we wanted to know whether the observed increase in chromatin interactions is a result of this increase in transcriptional activity. First, we compared significantly up- and down-regulated transcripts (P < 0.05, abs(beta) > 1) with nonregulated transcripts (P > 0.95, transcripts per million > 1). Hi-C loops are not preferentially gained or lost at MYC regulated transcripts compared to MYC nonregulated transcripts (Supplemental Fig. S10A). However, more MYC-bound transcripts gained Hi-C loops (Supplemental Fig. S10B) compared to transcripts without MYC binding (Supplemental Fig. S10C), indicating that MYC binding contributed to chromatin looping regardless of differential transcript expression.
When transcripts were stratified into tertiles based on transcript expression, top tertile transcript promoters overlapped better with Hi-C loops compared to the bottom tertile (25.7% and 16.1%, respectively), demonstrating that transcriptional activity positively correlates with chromatin looping (Supplemental Fig. S11A). However, we found that MYC-bound transcripts were more associated with gained Hi-C loops compared to transcripts without MYC, regardless of expression levels (Fig. 5C,D). Transcripts with pre-existing or gained MYC binding after MYC overexpression were more associated with gained Hi-C loops regardless of expression levels (Supplemental Fig. S11B,C), indicating that the direct binding of MYC increases chromatin contact frequency regardless of the level of transcriptional activity.
In contrast, among transcripts that lost MYC binding after MYC overexpression, bottom and middle tertile transcripts more associated with lost Hi-C loops and less associated with gained Hi-C loops, whereas top tertile transcripts were associated with gained Hi-C loops (Supplemental Fig. S11D), demonstrating the contribution of transcriptional activity to chromatin contact frequency when MYC is not present. Together, these results show that transcriptional activity correlates with chromatin interactions but the direct binding of MYC also increases chromatin contact frequency through other mechanisms.
Chromatin contacts can be disrupted by removing CTCF
Because chromatin interactions were connecting MYC binding sites together at these regions of high transcriptional activity, we wanted to know whether the overexpressed MYC was being spatially constrained at these regions or merely diffused throughout the nucleus. Immunofluorescence staining showed that MYC formed discrete punctate spots within the nucleus after MYC overexpression, overlapping regions with high RNA polymerase II signal (Supplemental Fig. S12). These punctate spots were lost after CTCF siRNA knockdown, indicating that these foci can be disrupted by removing chromatin architectural factors (Supplemental Fig. S12).
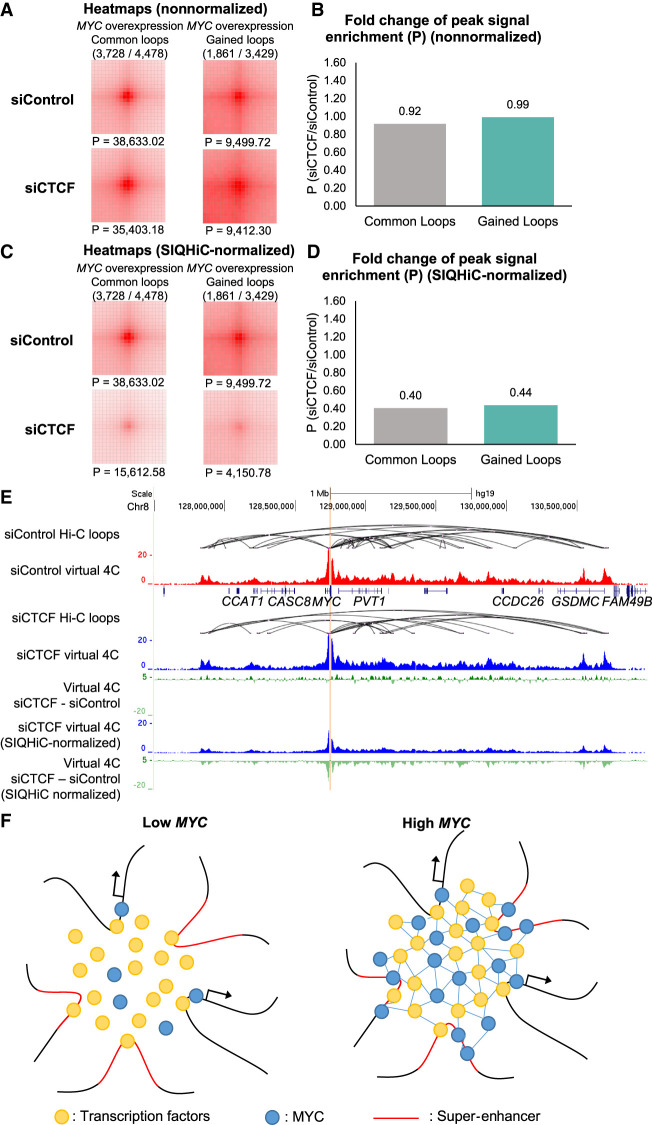
To confirm this, we looked at the siCTCF-mediated changes in chromatin interaction frequencies at the common and gained loops after MYC overexpression. SIQHiC-normalized APA analysis showed similar reductions in chromatin contact frequency at common and gained loops, which was not evident in the nonnormalized APA plots (Fig. 6A–D). SIQHiC-normalized virtual 4C plots of interactions at the MYC promoter illustrated the decrease in chromatin contact frequency (Fig. 6E), recapitulating previous observations in CTCF-depleted SEM cells (Hyle et al. 2019). Although CTCF depletion reduced MYC expression in SEM cells (Hyle et al. 2019), MYC was not down-regulated in siCTCF cells because of exogenous MYC expression from the tetracycline-inducible system (Supplemental Fig. S7A). While maintaining high MYC expression, we show that reduction of chromatin architectural proteins such as CTCF can reduce chromatin interactions at MYC foci and disperse MYC. Taken together, we propose a model where MYC accumulates at canonical promoter binding sites, binds to noncanonical binding sites at spatially adjacent super-enhancers, and forms protein–protein interactions with other transcription factors to stabilize chromatin interactions at these domains (Fig. 6F).
Figure 6.
Chromatin contacts can be disrupted through CTCF siRNA knockdown. (A) siControl and siCTCF Aggregate Peak Analysis plots at 5-kb resolution showing the nonnormalized aggregate signal at “Common” and “Gained” chromatin loops previously identified in High MYC cells. (P) Peak signal at the center pixel. (B) Fold change of nonnormalized peak signal enrichment (P) at common and gained loops between siControl and siCTCF cells. (C) Same APA analysis as in A but with SIQHiC normalization applied. (D) Fold change of SIQHiC-normalized peak signal enrichment (P) at common and gained loops between siControl and siCTCF cells. (E) Genome browser view showing siControl and siCTCF virtual 4C signal of interactions at the MYC promoter (red and blue tracks, respectively). The difference between the siControl and siCTCF virtual 4C signals are shown with and without SIQHiC normalization (green tracks). (F) Proposed model of overexpressed MYC accumulating at canonical promoter binding sites, binding to spatially adjacent super-enhancers, and forming protein–protein interactions with other transcription factors to stabilize chromatin interactions within the domain.
Discussion
MYC overexpression has long been described as a key driving force for oncogenesis in multiple cell types, resulting in tumors that become highly dependent on elevated MYC expression (Gabay et al. 2014; Bradner et al. 2017), but it is still unclear how overexpressed MYC shifts from regulating its canonical target genes to activate oncogenes. Recent studies have shown that overexpressed MYC invades into distal enhancer regulatory elements to differentially regulate gene expression (Lin et al. 2012; Nie et al. 2012; Sabò et al. 2014). Because cancer cells frequently acquire novel, cancer-specific super-enhancer signatures that are associated with cancer initiation and maintenance (Ooi et al. 2016; Tsang et al. 2019; Raisner et al. 2020), we wondered whether MYC binds preferentially to super-enhancers to perform its oncogenic functions. In this paper, we use the U2OS cell line with a tetracycline-inducible MYC system, which has been widely used for the study of transcriptional regulation by MYC (Walz et al. 2014; Lorenzin et al. 2016; de Pretis et al. 2017; Nie et al. 2020) and is a reasonable model for establishing a parsimonious understanding of MYC enhancer invasion. We show that overexpressed MYC indeed invades into super-enhancers in particular and leads to changes in the enhancer landscape and chromatin interactome. In particular, super-enhancers gained more constituent H3K27ac peaks and increased enrichment of MYC binding after MYC overexpression.
Although there was a loss and gain of more than 7000 stitched enhancers after MYC overexpression, we observed no de novo super-enhancer formation at H3K27ac unmarked loci. This is consistent with results from Poli et al. (2018) using TERT-immortalized human mammary epithelial (IMEC) cells, where cancer-associated de novo enhancers were not activated in MYC-overexpressed IMEC cells but marked by H3K27ac only after deriving secondary mammospheres. Earlier studies have shown that MYC does not bind to closed chromatin (Guccione et al. 2006; Lin et al. 2012; Soufi et al. 2012), suggesting that MYC invades into active super-enhancers but is unable to activate silent oncogenic super-enhancers by itself.
Because super-enhancers are associated with abundant long-range chromatin interactions and high interaction frequencies (Schmitt et al. 2016; Beagrie et al. 2017; Cao et al. 2017), we wanted to know whether MYC super-enhancer invasion alters the three-dimensional chromatin interactome. Using SIQHiC, a modified Hi-C protocol normalizing for cell count between samples, we observed increased global chromatin contact frequency after MYC overexpression. Chromatin interactions at MYC binding sites were preferentially strengthened and connected MYC binding sites together, notably between promoters and enhancers. Kieffer-Kwon et al. (2017) previously showed that MYC up-regulation during B cell activation is accompanied by a twofold increase in chromatin loops, particularly the formation of B cell-associated chromatin loops. In this work, MYC overexpression alone strengthened pre-existing loops but did not increase the total number of chromatin loops, suggesting that other chromatin factors are required to reconfigure the chromatin interaction landscape in cancer as well as in B cell activation.
Previous studies have shown that CTCF siRNA knockdown does not deplete CTCF protein completely and leads to limited disruption of TAD boundaries (Zuin et al. 2014; Khoury et al. 2020), and this was recapitulated in our study. In contrast, degron-mediated CTCF protein degradation resulted in widescale weakening of TAD insulation (Nora et al. 2017; Kubo et al. 2021). Although CTCF siRNA knockdown had limited effects on TAD structure, SIQHiC-normalization revealed a global decrease in chromatin contact frequency at all chromatin loop sets, demonstrating the importance of cell count normalization in analyzing the perturbation of chromatin architectural factors.
Previous research has shown that active transcription may play a role in maintaining the three-dimensional genome organization (Busslinger et al. 2017; Isoda et al. 2017; Gu et al. 2018; Heinz et al. 2018; Nagashima et al. 2019). Here, we show that transcriptional activity does correlate with chromatin interactions. Although MYC overexpression increases global transcription of active genes, we observed increased chromatin contact frequency regardless of transcription activity, indicating that direct binding of MYC contributes to chromatin interactions via other mechanisms.
MYC overexpression is a common event in oncogenesis across multiple cancers but cannot alone induce malignant transformation. Our results suggest that other cancer-initiating mutations are required to reconfigure the epigenetic and chromatin interaction landscape, with MYC playing a role in maintaining these epigenetic alterations by stabilizing these transient changes in chromatin interactions. These cancers would in turn become reliant on MYC and other participants of these chromatin interactions. If MYC expression is inhibited, we suspect that the reduction in protein–protein interactions will reduce these aberrant chromatin interactions.
Alternatively, these chromatin interactions can be perturbed by inhibiting other architectural factors involved. In this work, CTCF knockdown reduced chromatin contact frequency and dispersed MYC, supporting this line of investigation. A recent study has identified curaxins as a class of anticancer drugs that can disrupt chromatin looping by intercalating with DNA and interfering with CTCF binding (Kantidze et al. 2019), without causing DNA damage (Gasparian et al. 2011; Kantidze et al. 2019; Lu et al. 2021). Curaxins also exert anticancer activity through trapping of the histone chaperone FACT complex on chromatin (Gasparian et al. 2011). Crucially, both chromatin looping disruption and FACT complex trapping led to reduced expression of MYC family genes (Carter et al. 2015; Kantidze et al. 2019; Wang et al. 2020). Although complete degradation of CTCF is lethal in normal cells (Moore et al. 2012; Kemp et al. 2014), curaxins were well tolerated in xenograft models across multiple cancer cell types (Dermawan et al. 2016; Kim et al. 2016; Barone et al. 2017) and recently completed phase I clinical trials (Sarantopoulos et al. 2020), making it a promising drug against MYC-addicted cancers.
Taken together, our manuscript has demonstrated that MYC overexpression leads to MYC invasion into super-enhancers, and SIQHiC uncovered an increase in chromatin interactions at these regions. We have also shown the utility of SIQHiC in investigating chromatin architectural proteins such as CTCF. Our results lay the groundwork for further research to elucidate the role of MYC in maintaining cancer-specific chromatin interactions in established cancers and ways to therapeutically target these chromatin interactions. We anticipate that SIQHiC will continue to be useful for uncovering changes in chromatin contact frequency when perturbing chromatin architectural factors and using drugs such as curaxins.
Methods
Cell lines
U2OS osteosarcoma cells (ATCC HTB-96) with a doxycycline-inducible MYC system were kindly provided by Elmar Wolf's lab from Universität Würzburg, Germany. Cell lines were authenticated by STR profiling, and mycoplasma testing was performed using the MycoAlert PLUS Mycoplasma Detection kit (Lonza). U2OS cells were grown in DMEM supplemented with 10% tetracycline-free fetal bovine serum (Clontech), 100 U/mL penicillin, and 100 µg/mL streptomycin. MYC expression was induced in U2OS cells by the addition of 1 µg/mL doxycycline (Clontech) for 30 h. CTCF siRNA knockdown was performed using ON-TARGETplus SMARTpool Human CTCF (10664) siRNA (Dharmacon L-020165-00-0005). Cells were transfected with 5 nM CTCF siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer's instructions, with medium replacement after 24 h, and cells were harvested after 48 h.
Protein extraction and Western blot
Cells were lysed in RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific) for 30 min at 4°C, with vortexing every 10 min. Cell lysate was centrifuged to remove cell debris. Protein concentration was measured using the Pierce BCA Protein Assay kit (Thermo Fisher Scientific). Fifty micrograms of protein was loaded into a 12% SDS-PAGE gel for electrophoresis and transferred onto a PVDF membrane. The membrane was blocked with 5% nonfat milk in Tris- buffered saline with 0.1% Tween 20 (TBST) for 1 h at room temperature, followed by incubation with primary antibodies overnight at 4°C. The following primary antibodies were used: anti-MYC (Abcam ab32072; diluted 1:1000) and anti-vinculin (Sigma-Aldrich V9131; diluted 1:200). The membrane was washed three times with TBST, followed by incubation with either mouse (Cell Signalling Technologies 7076; diluted 1:5000) or rabbit (Cell Signalling Technologies 7074; diluted 1:5000) HRP-conjugated secondary antibodies for 1 h at room temperature. After washing three times with TBST, bands were imaged using Clarity Western ECL Substrate (Bio-Rad) on the ImageQuant LAS 500 (GE Healthcare).
Immunofluorescence staining
Cells were grown on a coverslip, fixed in 4% formaldehyde for 15 min, and permeabilized with 0.2% Triton X-100 in Tris-buffered saline (TBS-TX100). Cells were then blocked with 1% bovine serum albumin (BSA) in 0.2% TBS-TX100 for 3 h at room temperature and incubated with primary antibodies against MYC (Abcam ab32072) or RNA polymerase II (BioLegend 664906) for 2 h at room temperature. Coverslips were washed three times with 0.2% TBS-TX100, incubated with antimouse secondary antibodies conjugated with Alexa Fluor 555 (Life Technologies A21422) or antirabbit secondary antibodies conjugated with Alexa Fluor 488 (Life Technologies A11034) for 1 h at room temperature, followed by incubation with DAPI (Sigma-Aldrich) for 10 min. Coverslips were mounted onto slides and imaged on the Nikon A1R confocal microscope at 100× magnification.
Reverse transcription and quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from U2OS cells with or without MYC induction in triplicate, using the RNeasy Mini kit (QIAGEN). RNA was reverse-transcribed into complementary DNA using the qScript cDNA Synthesis kit (Quantabio). Quantitative PCR was performed on the QuantStudio 5 (Thermo Fisher Scientific) using the GoTaq qPCR Mastermix (Promega), with TBP as the housekeeping control. Primer sequences are listed in Supplemental Table S5.
Circular chromosome conformation capture experimental procedure
4C-seq was performed in duplicate on U2OS cells with or without MYC induction as previously described (Cao et al. 2017). Forty million cells were crosslinked in PBS supplemented with 1% formaldehyde for 10 min at room temperature, followed by quenching with 0.25 M glycine for 5 min. Cells were washed three times with PBS supplemented with 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich). Nuclei were isolated by lysing cells in 4C lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 5 mM EDTA, 0.5% Igepal CA-630) for 10 min at 4°C, followed by homogenization using a dounce homogenizer (Wheaton) (50 strokes using Pestle B).
Nuclei were permeabilized at 37°C with the addition of 0.3% SDS for 1 h, followed by 2% Triton-X 100 for 1 h. Primary enzyme digestion with HindIII-HF (NEB) was done for 18 h at 37°C, before proximity ligation in dilute conditions with T4 DNA Ligase (Thermo Fisher Scientific) overnight at 16°C. Crosslinking was reversed with the addition of 55 µg/mL Proteinase K (Ambion) for 4 h at 65°C and overnight at 37°C, and treated with RNase A for 1 h at 37°C. DNA was purified using phenol/chloroform extraction followed by ethanol precipitation to yield the 3C library. Secondary enzyme digestion with DpnII (NEB) was done on the 3C library overnight at 37°C, followed by proximity ligation and de-crosslinking as described above. 4C libraries were generated for each viewpoint through nested inverse-PCR using Phusion DNA Polymerase (Thermo Fisher Scientific), with primers listed in Supplemental Table S5. DNA fragments between 200 and 1000 bp were isolated from the 4C libraries by gel excision after running the 4C libraries on a 4%–20% gradient TBE gel (Thermo Fisher Scientific). The gel slices were shredded and macerated in Tris EDTA buffer overnight at 37°C, and DNA was precipitated using ethanol. The 4C libraries were multiplexed and single-end 1 × 150-bp sequencing was performed on the Illumina MiSeq.
RNA-seq experimental procedure
Total RNA was extracted from U2OS cells with or without MYC induction in duplicate, using the RNeasy Mini kit (QIAGEN). Ribosomal RNA depletion and library preparation were performed using the TruSeq Stranded Total RNA LT kit (Illumina) according to the manufacturer's protocol. Paired-end 2 × 100-bp sequencing was performed on the Illumina HiSeq 2500.
ChIP-seq experimental procedure
H3K27ac and MYC ChIP-seq were performed in duplicate on U2OS cells with or without MYC induction. Cells were fixed in 1% formaldehyde and sonicated using the truChIP Chromatin Shearing kit (Covaris) on the ME220 Focused-Ultrasonicator (Covaris) according to the manufacturer's protocol. H3K27ac and MYC-bound DNA were immunoprecipitated using anti-H3K27ac (Abcam ab4729, Lot: GR150367-1) and anti-MYC (Santa Cruz Biotechnology sc-764, Lot: H1712), with anti-IgG (Santa Cruz Biotechnology sc-2027, Lot: H2615) as the negative control. Three and one-half micrograms of each antibody was rotated with 15 µL of Dynabeads Protein G magnetic beads (Invitrogen) overnight at 4°C and washed three times with beads wash buffer (0.1% Triton X-100 in PBS) to remove unbound antibodies. Antibody-bound beads were combined with sonicated chromatin from five million cells and rotated overnight at 4°C. Beads were then washed three times with Shearing Buffer D3 (Covaris), once with high salt washing buffer (50 mM HEPES, pH 7.5, 350 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS), once with lithium chloride wash buffer (10 mM Tris, pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate), and once with Tris-EDTA buffer. Chromatin was eluted from the magnetic beads in 100 µL elution buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 1% SDS) supplemented with 2 µL of 0.5 mg/mL RNase A (QIAGEN) for 2 h at 55°C. Chromatin was then de-crosslinked with the addition of 2 µL of 20 mg/mL Proteinase K (Ambion) for 4 h at 55°C and overnight at 37°C. For total input, chromatin from 500,000 cells was treated with RNase A and de-crosslinked similarly. DNA was purified using the MinElute PCR Purification kit (QIAGEN). ChIP-seq libraries were prepared using the ThruPLEX DNA-seq kit (Rubicon) and sequenced paired-end 2 × 100 bp on the HiSeq 2500 (Illumina).
SIQHiC experimental procedure
SIQHiC was performed in duplicate on U2OS cells with or without MYC induction, with the addition of untreated mouse 3T3 cells. Human U2OS cells and mouse 3T3 cells were counted using the Countess II automated cell counter (Thermo Fisher Scientific) and fixed with 2% formaldehyde using the Arima Hi-C kit (Arima Genomics). One million fixed mouse 3T3 cells were added to each sample of four million fixed human U2OS cells before subsequent steps of restriction enzyme digest, biotin end filling, and ligation using the Arima-HiC kit (Arima Genomics) according to the manufacturer's protocol. Libraries were prepared using the KAPA Hyper-Prep kit (KAPA), according to the Arima-HiC kit protocol. SIQHiC libraries were sequenced paired-end 2 × 150-bp on the HiSeq 4000 (Illumina).
Genome assembly
4C-seq, ChIP-seq, RNA-seq, and SIQHiC libraries were all mapped to the hg19 human genome assembly, in line with previous published data sets on MYC enhancer invasion that were analyzed using this assembly. We do not expect significant differences when the libraries are mapped to the newer GRCh38 genome assembly because a previous analysis of TCGA data had shown significant concordance between hg19 and GRCh38 versions (Gao et al. 2019).
4C-seq data processing
4C-seq libraries were mapped to the hg19 genome using BWA-MEM (Li 2013) version 0.7.5a-r405 using default settings, and significant 4C interactions were identified using the r3Cseq pipeline (q < 0.05) (Thongjuea et al. 2013) on the CSI NGS portal (An et al. 2020) (https://csibioinfo.nus.edu.sg/).
ChIP-seq data processing
ChIP-seq libraries were mapped to the hg19 genome using BWA-MEM (Li 2013) version 0.7.5a-r405 using default settings. PCR duplicates were removed using SAMtools (Li et al. 2009) version 1.7, and reads falling within the ENCODE consensus blacklisted regions (The ENCODE Project Consortium 2012) were removed using BEDTools (Quinlan and Hall 2010) version 2.26.0. ChIP-seq signals were visualized using deepTools (Ramírez et al. 2014) version 3.2.1 (bamCompare ‐‐normalizeUsing RPKM ‐‐operation subtract -bs 1). BAM files of biological replicates were merged before calling ChIP-seq peaks using MACS2 (Zhang et al. 2008) version 2.1.2. H3K27ac ChIP-seq peaks within 2 kb of transcription start sites were labeled as active promoter peaks whereas the rest were labeled as enhancer peaks.
Super-enhancers were called as described previously (Cao et al. 2017). Briefly, H3K27ac enhancer peaks within 12.5 kb of each other were stitched together and ranked based on H3K27ac enrichment. The point where a line with slope 1 is tangential to the ranked H3K27ac enrichment curve was chosen as a cutoff to separate super-enhancers from typical enhancers.
To identify differential H3K27ac peaks, BAM files from all replicates and conditions were merged and a common list of H3K27ac peaks was called using MACS2. Read counts for the common list of H3K27ac peaks were obtained using Rsubread (Liao et al. 2019). Differential H3K27ac peaks were identified using DESeq2 (Love et al. 2014) version 1.24.0 (padj < 0.01 and padj < 0.01, respectively).
Motif analysis was performed to find the frequency of HOCOMOCO Human v11 Core motifs (Kulakovskiy et al. 2018) occurring at MYC binding sites using FIMO (Grant et al. 2011) with default parameters.
RNA-seq data processing
Total RNA-seq libraries were mapped to the hg19 genome and read counts for UCSC RefSeq transcripts were obtained using kallisto v0.44.0 (Bray et al. 2016). Differentially expressed transcripts were identified using sleuth v0.30.0 (Pimentel et al. 2017) (fdr < 0.05, |beta|>1) in R (R Core Team 2021). Gene set enrichment analysis was performed using GSEA v4.1.0 (Subramanian et al. 2005). Gene Ontology (GO) functional annotation was performed using DAVID (Huang da et al. 2009).
SIQHiC data processing
SIQHiC libraries were analyzed using the Juicer (v1.5) pipeline (Durand et al. 2016) with some modifications to obtain HIC matrix files. Because SIQHiC libraries include human and mouse DNA, paired reads were separated and mapped as single reads using BWA-MEM (Li 2013) to an artificial reference genome combining hg19 human and mm10 mouse genome sequences. Human and mouse chromosomes were appended with “H” and “M”, respectively, to avoid chromosome name duplication. After mapping, reads were paired together again and split into two files. Paired reads both mapping to human chromosomes were placed together as Human-Human paired reads (H-H), and paired reads both mapping to mouse chromosomes were placed together as Mouse-Mouse paired reads (M-M). Ambiguous read pairs that separately mapped to different species were discarded. H-H and M-M reads were processed using Juicer separately, filtering out duplicates, intrafragment reads, and reads with MAPQ < 30 to generate HIC matrix files for each species. H-H reads of biological replicates were also merged and processed in the same way to generate HIC matrix files for downstream analyses.
The ratio between human and mouse contacts for each sample was calculated. Because the mouse 3T3 cells in each sample come from the same population and were spiked into the human cells at the same ratio of 1:4, we expect to obtain a similar HMR ratio across all samples. Hence, the relative difference between the HMR of different samples (SIQHiC ratio) reflects the changes in global chromatin contact frequency. The SIQHiC ratio was calculated such that samples with lower HMR are normalized against samples with the highest HMR.
Hi-C contact matrices from merged biological replicates were normalized using the SIQHiC ratio by adding a custom normalization vector to the original contact matrix HIC file using the Juicer “addNorm” subroutine, where the magnitude of each bin was the SIQHiC ratio raised to the power of (−0.5). In this way, the Hi-C matrices are cell count-normalized to the sample with the highest HMR to prevent scale-up extrapolation errors.
The SIQHiC normalized matrix for each chromosome was extracted from the HIC file using the Juicer “dump” subroutine, combined together, and finally reassembled into a SIQHiC normalized HIC matrix file using the Juicer “pre” subroutine. Because the SIQHiC normalization vector for the High MYC Hi-C matrix is 1, the SIQHiC normalized matrix is equivalent to the nonnormalized matrix. Hence, only the Low MYC Hi-C matrix was SIQHiC-normalized. Original and SIQHiC-normalized HIC matrices were balanced using the Knight-Ruiz algorithm for subsequent analyses.
Aggregate peak analyses was performed using the Juicer “apa” subroutine using both nonnormalized and SIQHiC-normalized contact matrices using the parameters “-k KR –u –n 30”. APA heat maps were generated using pheatmap v1.0.12 (https://rdrr.io/cran/pheatmap/) in R (R Core Team 2021).
Compartments were identified using the Juicer “eigenvector” and “pearsons” subroutines at 100-kb resolution. Because eigenvalues signs are arbitrary, we correlated eigenvectors with the number of gene promoters within each 100-kb bin, such that gene rich regions were assigned positive eigenvalues.
Topologically associating domain boundaries were identified using the HiCExplorer (Ramírez et al. 2018) “hicFindTADs” subroutine using the parameter “‐‐minDepth 50000 ‐‐maxDepth 500000 ‐‐thresholdComparisons 0.001 ‐‐delta 0.01 ‐‐correctForMultipleTesting fdr”. Lost, common, and gained TAD boundaries were identified by intersecting Low MYC and High MYC TAD boundaries. TAD separation scores of lost, common, and gained TAD boundaries were visualized using deepTools (Ramírez et al. 2014) version 3.2.1.
Loops were called using the Juicer “hiccups” subroutine with parameter “-k KR -m 1024 ‐‐ignore_sparsity”. Lost, common, and gained loops were identified using pgltools (Greenwald et al. 2017) by merging and intersecting Low MYC loops with High MYC loops using a 10-kb intersecting window at each loop anchor. Loop subsets are listed in Supplemental Table S5. Hi-C loops were annotated according to their overlap with MYC binding sites, promoters, typical enhancers, and super-enhancers using pgltools.
Virtual 4C tracks were extracted from the Hi-C matrices by using the Juicer “dump” subroutine, dumping a 5-kb window against its entire chromosome using a 5-kb bin size.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE164777.
Supplementary Material
Acknowledgments
This research is supported by the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education Academic Research Fund Tier 3 grant awarded to Daniel Tenen (MOE2014-T3-1-006). This research is supported by the NRF Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative and a Singapore Ministry of Education Academic Research Fund Tier 2 grant awarded to M.J.F. (MOET2EP30120-0009).
Author contributions: Y.X.S. and M.J.F. contributed to the conception and design of the study. Y.X.S. conducted the experiments and developed the SIQHiC protocol. Y.X.S. and K.C. performed the bioinformatics analyses. Y.X.S. and M.J.F. reviewed the data and wrote the manuscript. All authors reviewed and approved the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276313.121.
Freely available online through the Genome Research Open Access option.
Competing interest statement
M.J.F. declares two patents on methodologies related to ChIA-PET. No other competing interests are declared.
References
- Affer M, Chesi M, Chen WD, Keats JJ, Demchenko YN, Tamizhmani K, Garbitt VM, Riggs DL, Brents LA, Roschke AV, et al. 2014. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 28: 1725–1735. 10.1038/leu.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An O, Tan KT, Li Y, Li J, Wu CS, Zhang B, Chen L, Yang H. 2020. CSI NGS Portal: an online platform for automated NGS data analysis and sharing. Int J Mol Sci 21: 3828. 10.3390/ijms21113828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu D, Fullwood MJ. 2015. 3D genome organization in health and disease: emerging opportunities in cancer translational medicine. Nucleus 6: 382–393. 10.1080/19491034.2015.1106676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, Murison A, Langenfeld K, Petretich M, Scognamiglio R, et al. 2018. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553: 515–520. 10.1038/nature25193 [DOI] [PubMed] [Google Scholar]
- Barone TA, Burkhart CA, Safina A, Haderski G, Gurova KV, Purmal AA, Gudkov AV, Plunkett RJ. 2017. Anticancer drug candidate CBL0137, which inhibits histone chaperone FACT, is efficacious in preclinical orthotopic models of temozolomide-responsive and -resistant glioblastoma. Neuro Oncol 19: 186–196. 10.1093/neuonc/now141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. 2017. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543: 519–524. 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905. 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, Hnisz D, Young RA. 2017. Transcriptional addiction in cancer. Cell 168: 629–643. 10.1016/j.cell.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM. 2017. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544: 503–507. 10.1038/nature22063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Fang Y, Tan HK, Goh Y, Choy JYH, Koh BTH, Tan JH, Bertin N, Ramadass A, Hunter E, et al. 2017. Super-enhancers and broad H3K4me3 domains form complex gene regulatory circuits involving chromatin interactions. Sci Rep 7: 2186. 10.1038/s41598-017-02257-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. 2002. Long-range chromatin regulatory interactions in vivo. Nat Genet 32: 623–626. 10.1038/ng1051 [DOI] [PubMed] [Google Scholar]
- Carter DR, Murray J, Cheung BB, Gamble L, Koach J, Tsang J, Sutton S, Kalla H, Syed S, Gifford AJ, et al. 2015. Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci Transl Med 7: 312ra176. 10.1126/scitranslmed.aab1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pretis S, Kress TR, Morelli MJ, Sabò A, Locarno C, Verrecchia A, Doni M, Campaner S, Amati B, Pelizzola M. 2017. Integrative analysis of RNA polymerase II and transcriptional dynamics upon MYC activation. Genome Res 27: 1658–1664. 10.1101/gr.226035.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermawan JKT, Hitomi M, Silver DJ, Wu Q, Sandlesh P, Sloan AE, Purmal AA, Gurova KV, Rich JN, Lathia JD, et al. 2016. Pharmacological targeting of the histone chaperone complex FACT preferentially eliminates glioblastoma stem cells and prolongs survival in preclinical models. Cancer Res 76: 2432–2442. 10.1158/0008-5472.CAN-15-2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3: 95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. 2008. Myc's broad reach. Genes Dev 22: 2755–2766. 10.1101/gad.1712408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay M, Li Y, Felsher DW. 2014. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 4: a014241. 10.1101/cshperspect.a014241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao GF, Parker JS, Reynolds SM, Silva TC, Wang L-B, Zhou W, Akbani R, Bailey M, Balu S, Berman BP, et al. 2019. Before and after: comparison of legacy and harmonized TCGA Genomic Data Commons’ data. Cell Syst 9: 24–34.e10. 10.1016/j.cels.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparian AV, Burkhart CA, Purmal AA, Brodsky L, Pal M, Saranadasa M, Bosykh DA, Commane M, Guryanova OA, Pal S, et al. 2011. Curaxins: anticancer compounds that simultaneously suppress NF-κB and activate p53 by targeting FACT. Sci Transl Med 3: 95ra74. 10.1126/scitranslmed.3002530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald WW, Li H, Smith EN, Benaglio P, Nariai N, Frazer KA. 2017. Pgltools: a genomic arithmetic tool suite for manipulation of Hi-C peak and other chromatin interaction data. BMC Bioinformatics 18: 207. 10.1186/s12859-017-1621-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Pomella S, Sayers C, Wu XS, Song Y, Chiarella AM, Bagchi S, Chou H-C, Sinniah RS, Walton A, et al. 2019. Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat Genet 51: 1714–1722. 10.1038/s41588-019-0534-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Swigut T, Spencley A, Bauer MR, Chung M, Meyer T, Wysocka J. 2018. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359: 1050–1055. 10.1126/science.aao3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B. 2006. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol 8: 764–770. 10.1038/ncb1434 [DOI] [PubMed] [Google Scholar]
- Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L, et al. 2018. Transcription elongation can affect genome 3D structure. Cell 174: 1522–1536.e22. 10.1016/j.cell.2018.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, Xu L, Castillo-Martin M, Llobet-Navás D, Cordon-Cardo C, et al. 2014. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 20: 1130–1137. 10.1038/nm.3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hyle J, Zhang Y, Wright S, Xu B, Shao Y, Easton J, Tian L, Feng R, Xu P, Li C. 2019. Acute depletion of CTCF directly affects MYC regulation through loss of enhancer–promoter looping. Nucleic Acids Res 47: 6699–6713. 10.1093/nar/gkz462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda T, Moore AJ, He Z, Chandra V, Aida M, Denholtz M, Piet van Hamburg J, Fisch KM, Chang AN, Fahl SP, et al. 2017. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell 171: 103–119.e18. 10.1016/j.cell.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantidze OL, Luzhin AV, Nizovtseva EV, Safina A, Valieva ME, Golov AK, Velichko AK, Lyubitelev AV, Feofanov AV, Gurova KV, et al. 2019. The anti-cancer drugs curaxins target spatial genome organization. Nat Commun 10: 1441. 10.1038/s41467-019-09500-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CJ, Moore JM, Moser R, Bernard B, Teater M, Smith LE, Rabaia NA, Gurley KE, Guinney J, Busch SE, et al. 2014. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep 7: 1020–1029. 10.1016/j.celrep.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury A, Achinger-Kawecka J, Bert SA, Smith GC, French HJ, Luu P-L, Peters TJ, Du Q, Parry AJ, Valdes-Mora F, et al. 2020. Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains. Nat Commun 11: 54. 10.1038/s41467-019-13753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon K-R, Nimura K, Rao SSP, Xu J, Jung S, Pekowska A, Dose M, Stevens E, Mathe E, Dong P, et al. 2017. Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol Cell 67: 566–578.e10. 10.1016/j.molcel.2017.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Neznanov N, Wilfong CD, Fleyshman DI, Purmal AA, Haderski G, Stanhope-Baker P, Burkhart CA, Gurova KV, Gudkov AV, et al. 2016. Preclinical validation of a single-treatment infusion modality that can eradicate extremity melanomas. Cancer Res 76: 6620–6630. 10.1158/0008-5472.CAN-15-2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TR, Sabò A, Amati B. 2015. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer 15: 593–607. 10.1038/nrc3984 [DOI] [PubMed] [Google Scholar]
- Kubo N, Ishii H, Xiong X, Bianco S, Meitinger F, Hu R, Hocker JD, Conte M, Gorkin D, Yu M, et al. 2021. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat Struct Mol Biol 28: 152–161. 10.1038/s41594-020-00539-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Sharipov RN, Fedorova AD, Rumynskiy EI, Medvedeva YA, Magana-Mora A, Bajic VB, Papatsenko DA, et al. 2018. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res 46: D252–D259. 10.1093/nar/gkx1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN].
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Consortium. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2019. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47: e47. 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67. 10.1016/j.cell.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzin F, Benary U, Baluapuri A, Walz S, Jung LA, von Eyss B, Kisker C, Wolf J, Eilers M, Wolf E. 2016. Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife 5: e15161. 10.7554/eLife.15161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. 2012. Revisiting global gene expression analysis. Cell 151: 476–482. 10.1016/j.cell.2012.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Liu C, Liu Y, Luo A, Chen J, Lei Z, Kong J, Xiao X, Zhang S, Wang Y-Z, et al. 2021. Curaxin-induced DNA topology alterations trigger the distinct binding response of CTCF and FACT at the single-molecule level. Biochemistry 60: 494–499. 10.1021/acs.biochem.1c00014 [DOI] [PubMed] [Google Scholar]
- Lun ATL, Smyth GK. 2015. diffHic: a Bioconductor package to detect differential genomic interactions in Hi-C data. BMC Bioinformatics 16: 258. 10.1186/s12859-015-0683-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MR, Bradner JE. 2014. Therapeutic strategies to inhibit MYC. Cold Spring Harb Perspect Med 4: a014266. 10.1101/cshperspect.a014266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JM, Rabaia NA, Smith LE, Fagerlie S, Gurley K, Loukinov D, Disteche CM, Collins SJ, Kemp CJ, Lobanenkov VV, et al. 2012. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS One 7: e34915. 10.1371/journal.pone.0034915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima R, Hibino K, Ashwin SS, Babokhov M, Fujishiro S, Imai R, Nozaki T, Tamura S, Tani T, Kimura H, et al. 2019. Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II. J Cell Biol 218: 1511–1530. 10.1083/jcb.201811090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79. 10.1016/j.cell.2012.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Guo C, Das SK, Chow CC, Batchelor E, Simons SSJ, Levens D. 2020. Dissecting transcriptional amplification by MYC. eLife 9: e52483. 10.7554/eLife.52483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi WF, Xing M, Xu C, Yao X, Ramlee MK, Lim MC, Cao F, Lim K, Babu D, Poon L-F, et al. 2016. Epigenomic profiling of primary gastric adenocarcinoma reveals super-enhancer heterogeneity. Nat Commun 7: 12983. 10.1038/ncomms12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG. 2014. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9: 1163–1170. 10.1016/j.celrep.2014.10.018 [DOI] [PubMed] [Google Scholar]
- Pachano T, Sánchez-Gaya V, Ealo T, Mariner-Faulí M, Bleckwehl T, Asenjo HG, Respuela P, Cruz-Molina S, Muñoz-San Martín M, Haro E, et al. 2021. Orphan CpG islands amplify poised enhancer regulatory activity and determine target gene responsiveness. Nat Genet 53: 1036–1049. 10.1038/s41588-021-00888-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Plank JL, Dean A. 2014. Enhancer function: mechanistic and genome-wide insights come together. Mol Cell 55: 5–14. 10.1016/j.molcel.2014.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli V, Fagnocchi L, Fasciani A, Cherubini A, Mazzoleni S, Ferrillo S, Miluzio A, Gaudioso G, Vaira V, Turdo A, et al. 2018. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat Commun 9: 1024. 10.1038/s41467-018-03264-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R, Bainer R, Haverty PM, Benedetti KL, Gascoigne KE. 2020. Super-enhancer acquisition drives oncogene expression in triple negative breast cancer. PLoS One 15: e0235343. 10.1371/journal.pone.0235343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9: 189. 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rodríguez-Carballo E, Gámez B, Ventura F. 2016. P38 MAPK signaling in osteoblast differentiation. Front Cell Dev Biol 4: 40. 10.3389/fcell.2016.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabò A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. 2014. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511: 488–492. 10.1038/nature13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarantopoulos J, Mahalingam D, Sharma N, Iyer RV, Ma WW, Ahluwalia MS, Johnson S, Purmal A, Shpigotskaya P, Hards A, et al. 2020. Results of a completed phase I trial of CBL0137 administered intravenously (IV) to patients (Pts) with advanced solid tumors. J Clin Oncol 38: 3583. 10.1200/JCO.2020.38.15_suppl.3583 [DOI] [Google Scholar]
- Sati S, Cavalli G. 2017. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma 126: 33–44. 10.1007/s00412-016-0593-6 [DOI] [PubMed] [Google Scholar]
- Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, Zhao W, Zhang X, Ventura A, Liu Y, et al. 2018. Pan-cancer alterations of the MYC oncogene and its proximal network across the Cancer Genome Atlas. Cell Syst 6: 282–300.e2. 10.1016/j.cels.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, et al. 2016. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep 17: 2042–2059. 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe J-S, Minder JL, Mercan F, Wang E, Eckersley-Maslin MA, et al. 2013. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev 27: 2648–2662. 10.1101/gad.232710.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield JC, Cresswell KG, Vladimirov VI, Dozmorov MG. 2018. HiCcompare: an R-package for joint normalization and comparison of HI-C datasets. BMC Bioinformatics 19: 279. 10.1186/s12859-018-2288-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongjuea S, Stadhouders R, Grosveld FG, Soler E, Lenhard B. 2013. r3Cseq: an R/Bioconductor package for the discovery of long-range genomic interactions from chromosome conformation capture and next-generation sequencing data. Nucleic Acids Res 41: e132. 10.1093/nar/gkt373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang FH, Law CT, Tang TC, Cheng CL, Chin DW, Tam WV, Wei L, Wong CC, Ng IO, Wong CM. 2019. Aberrant super-enhancer landscape in human hepatocellular carcinoma. Hepatology (Baltimore, Md) 69: 2502–2517. 10.1002/hep.30544 [DOI] [PubMed] [Google Scholar]
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. 2014. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511: 483–487. 10.1038/nature13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sui Y, Li Q, Zhao Y, Dong X, Yang J, Liang Z, Han Y, Tang Y, Ma J. 2020. Effective inhibition of MYC-amplified group 3 medulloblastoma by FACT-targeted curaxin drug CBL0137. Cell Death Dis 11: 1029. 10.1038/s41419-020-03201-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeid R, Lawlor MA, Poon E, Reyes JM, Fulciniti M, Lopez MA, Scott TG, Nabet B, Erb MA, Winter GE, et al. 2018. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat Genet 50: 515–523. 10.1038/s41588-018-0044-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, Meyerson M. 2016. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet 48: 176–182. 10.1038/ng.3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, Brouwer RWW, van de Corput MPC, van de Werken HJG, Knoch TA, van Ijcken WFJ, et al. 2014. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci 111: 996–1001. 10.1073/pnas.1317788111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.