Abstract
Genome-wide association studies (GWAS) have been highly informative in discovering disease-associated loci but are not designed to capture all structural variations in the human genome. Using long-read sequencing data, we discovered widespread structural variation within SINE-VNTR-Alu (SVA) elements, a class of great ape-specific transposable elements with gene-regulatory roles, which represents a major source of structural variability in the human population. We highlight the presence of structurally variable SVAs (SV-SVAs) in neurological disease–associated loci, and we further associate SV-SVAs to disease-associated SNPs and differential gene expression using luciferase assays and expression quantitative trait loci data. Finally, we genetically deleted SV-SVAs in the BIN1 and CD2AP Alzheimer's disease–associated risk loci and in the BCKDK Parkinson's disease–associated risk locus and assessed multiple aspects of their gene-regulatory influence in a human neuronal context. Together, this study reveals a novel layer of genetic variation in transposable elements that may contribute to identification of the structural variants that are the actual drivers of disease associations of GWAS loci.
Discovering the genetic variation underlying human diseases is a common goal in human genetics, and the rapid increase of genome-wide association studies (GWAS) has generated a vast catalog of single-nucleotide polymorphisms (SNPs) associated with specific traits and diseases (MacArthur et al. 2017). In most cases, GWAS do not identify the genetic variation that drives the trait but use SNPs as markers to highlight trait-associated loci through linkage disequilibrium (LD) (Edwards et al. 2013). This calls for elaborate post-GWAS analysis to shed light on the genes and mechanisms involved in specific traits (Backman et al. 2021; Mortezaei and Tavallaei 2021). A comprehensive view of the genetic structural variants that exist within loci containing trait-associated SNPs is an essential first step to assessing how these variants may lead to disease susceptibility on both genetic and functional levels (Eichler 2019).
One source of structural variation that has not been sufficiently considered comes from transposable elements (TEs), which together constitute >42% of the human genome (Smit 1999; International Human Genome Sequencing Consortium 2001; Audano et al. 2019; Linthorst et al. 2020). Although the vast majority of TEs do not alter coding regions of our genome, some TE classes harbor strong gene regulatory potential that can directly affect gene expression levels (Jacobs et al. 2014; Wang et al. 2014; Chuong et al. 2016; Fuentes et al. 2018; Pontis et al. 2019). The TE-mediated regulatory effect on genes is highly tissue-specific and has been shown to be particularly prominent in a neuronal environment (Jacob-Hirsch et al. 2018; Trizzino et al. 2018; Pontis et al. 2019; Miao et al. 2020; Sundaram and Wysocka 2020). TEs are activated during aging, neurodegeneration, and neurological diseases, but whether this is a cause or a consequence of the disease pathology remains unknown in many cases (Frank et al. 2005; Li et al. 2013; Van Meter et al. 2014; Guo et al. 2018; Shpyleva et al. 2018).
Only TEs of the Alu, L1, and SVA (SINE-VNTR-Alu) families can still actively spread through the genome, and new insertions cause variation between individuals in the form of presence/absence TE-insertional polymorphisms (Kazazian et al. 1988; Batzer et al. 1991; Brouha et al. 2003; Ostertag et al. 2003). TEs can alter gene regulation in the locus in which they insert, such that the presence or absence of a TE can lead to inter-individual differences in gene expression. There are approximately 60,475 Alu, 10,018 L1, and 6417 SVA TE-insertional polymorphisms known, with new insertions occurring every 40, 63, and 63 births, respectively (Feusier et al. 2019; Collins et al. 2020). Some of these new insertions have been linked to diseases (Makino et al. 2007; Hancks and Kazazian 2016; Sekar et al. 2016; Payer et al. 2017; Payer and Burns 2019; Pfaff et al. 2021). Next to presence/absence TE polymorphisms, structural variation within fixed TEs (TE insertions observed in all individuals in the human population) has also been reported (Savage et al. 2013, 2014), although the prevalence of this type of structural variation has remained elusive. The repetitive nature of TEs increases the propensity for unequal crossover events or DNA polymerase slippage during meiosis, for which variable number of tandem repeats (VNTRs) are especially susceptible (Brookes 2013). SVA elements harbor unusually large VNTRs as their internal segment and have a unique sequence composition compared to other VNTRs in our genome. The structural variation in VNTRs is particularly interesting because they are often associated with gene-regulatory functions, and many genes have accrued VNTRs as essential regulatory elements for their expression (International Human Genome Sequencing Consortium 2001; Fondon et al. 2008).
It is becoming increasingly clear that gene-regulatory properties of TEs were co-opted during evolution, leading to the integration of TEs as novel gene-regulatory elements in preexisting gene expression networks (Cordaux and Batzer 2009; Chuong et al. 2016). As such, TEs have become an integral part of normal human gene regulation. Because our genome has become dependent on TEs for specific aspects of gene regulation, structural variation within fixed TEs could account for inter-individual differences in temporal or spatial aspects of gene expression. Despite the possible roles structurally variable TEs may play in human health or disease, this level of structural variation has remained largely undocumented. This is mainly a result of technical limitations associated with the highly repetitive DNA sequences within TEs, which makes identifying structural variations in TEs using short-read sequencing strategies extremely challenging. The development of long-read sequencing techniques provides, for the first time, the opportunity to accurately assess the level of structural variation (Eichler 2019). This allows for the evaluation of possible associations between disease susceptibility and specific structural variations found in fixed TEs in our genome (Audano et al. 2019; Chaisson et al. 2019; Sulovari et al. 2019; Ewing et al. 2020; Ebert et al. 2021; Porubsky et al. 2021).
In this study we discovered that SVA retrotransposons, a great ape-specific class of TEs, constitutes a major source of hidden genetic variation that is not taken into account by conventional genetic case-control studies. We set out to investigate the biological consequences of structural variability in SVAs, focusing on SV-SVAs in Alzheimer's disease (AD)– and Parkinson's disease (PD)–associated GWAS loci. We assessed the gene-regulatory influence of SV-SVAs in a human neuronal context by genetic deletion of SVAs in three disease-associated loci. Our findings highlight the importance of careful mapping of structural variations within fixed TEs in the human population and argue for their inclusion in complex trait genetics as a layer of genetic variation that may, in some cases, confer the actual disease susceptibility to a GWAS-identified locus.
Results
SVAs are a major contributor to inter-individual structural variation
In our search for structural variations in TEs that may be associated with differential susceptibility to neurological disorders, we first determined which TEs are likely to play a regulatory role in human neuronal gene expression. We specifically focused on TEs active in neuronal cells, because previous research has associated aberrant TE activation with several neurological disorders (Frank et al. 2005; Li et al. 2013; Van Meter et al. 2014; Bragg et al. 2017; Guo et al. 2018; Shpyleva et al. 2018). To detect TEs that may have a regulatory influence on nearby genes in neuronal cells, we performed chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) for the enhancer-associated marker EP300 in human embryonic stem cell (hESC)-derived cortical organoids (Eiraku et al. 2008; Visel et al. 2009). Highest neuronal EP300 enrichment in TEs was observed for the active classes of TEs in our genome, including LINEs and all members of the SVA family (Fig. 1A,B). These data support previously published findings in which histone ChIP-seq showed SVAs become active in a neuronal environment (Pontis et al. 2019).
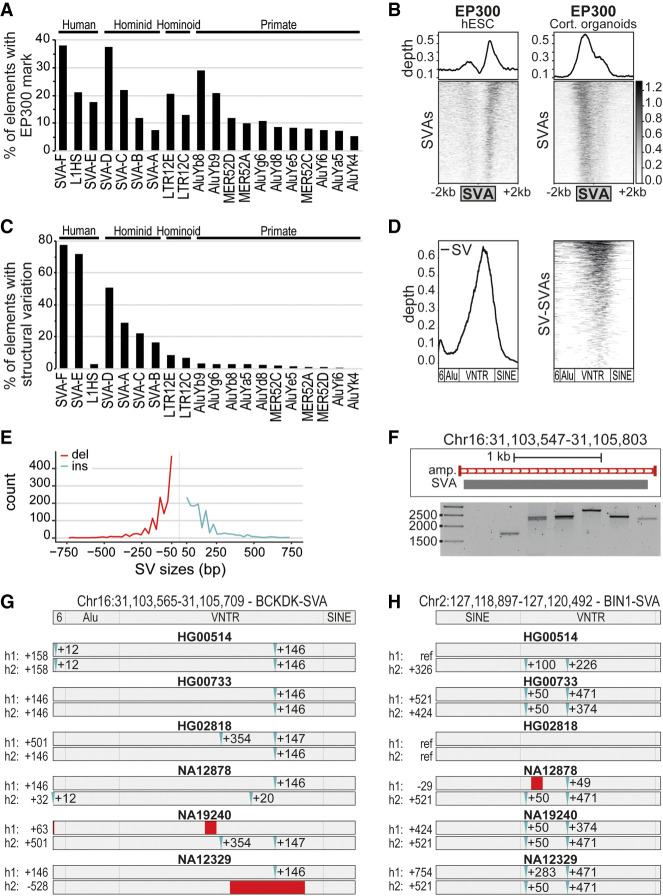
Figure 1.
SVAs are a major contributor to inter-individual structural variation. (A) Percentage of transposable elements with EP300 enhancer mark in cortical organoids; the top 20 enriched elements are shown. (B) Coverage heatmaps at full-length SVAs (GRCh37) in hESCs and cortical organoids for EP300 (hESCs: average of two replicates; cortical organoids: average of two biological and two technical replicates). Bottom gray box: average size SVAs. (C) Percentage of “full-length” TEs per class with structural variation based on Audano et al. (2019), grouped by the species they originated in. (D) Relative abundance of structural variation (left) and corresponding coverage heatmap (right) showing that most structural variation resides in the VNTR region of SVAs. Approximate SVA structure is shown below. (E) Distribution of structural variation (SV) sizes for insertions (ins) and deletions (del) in SVAs. (F) Example of structural variants for SVA in PCR-amplified region Chr 16: 31,103,547–31,105,803 (GRCh38 assembly). PCR-amplified region shown in red. (G,H) Schematic overview of SV-SVAs in phased assemblies of Ebert et al. (2021) of listed genomes for specified regions with approximate size shown. Estimated location of insertions (blue) and deletions (red) compared to reference genome.
For the top 20 neuronally active TE classes, we analyzed inter-individual structural variations. We used data from the recently published “patched human reference genome assembly” based on 15 human genomes sequenced by Pacific Biosciences (PacBio) long-read sequencing (Audano et al. 2019). In this assembly, structural variants of 50 bp or greater identified in any of the 15 individuals were included as alternate loci to improve the representation of allelic diversity in the reference assembly. Whereas a low level of structural variation was identified for almost all classes, we found an extraordinarily high level of structural variation in SVA elements (Fig. 1C). Almost half of full-length SVAs were structurally variable in the human population. The biggest contribution came from SVA-D, SVA-E, and SVA-F elements, representing the evolutionarily youngest classes of SVAs, which contain the largest VNTR region. This region was highly enriched for structural variations, together with the 5′ region, which contains a hexamer repeat (Fig. 1D; Supplemental Fig. S1A). Most variations observed in SVAs were between 50 and 200 bp (Fig. 1E), but ∼20% of the variations were >200 bp in size.
To validate these findings and to assess if any additional structural variation in SVA elements was not correctly captured, we performed an additional BLAST-alignment-based analysis on five of the PacBio genome assemblies (Supplemental Table S1). We focused on SVA sequences present on Chromosome 1 in the GRCh38 assembly as a proof of principle and included small structural variations (20–50 bp difference in SVA size) that were not included in previous analyses (Audano et al. 2019). We observed structural variation within 49.1% of full-length SVAs present on Chr 1, a slight increase over what was discovered in our first analysis. Structural variation was primarily observed in the center (VNTR) and 5′ (hexamer) region of the SVA. The distribution of structural variation between SVA classes was comparable to previous analysis, with most SV-SVA elements belonging to the youngest types.
To rule out sequencing or assembly errors leading to the observed variation, we selected five human-specific SVAs in which to validate the structural variations by PCR analysis. We confirmed structural variations within all five studied SVAs in a platform of 236 human genomic DNA samples (Fig. 1F; Supplemental Fig. S1B–G) and observed differences in size up to 1000 bp. For the SV-SVAs most extensively analyzed (PCR on 81 and 236 genomes), we found additional variations not observed in the initially sequenced human genomes. We therefore used recently published phased genome assemblies (Ebert et al. 2021) and were able to trace all variants observed by PCR analysis in the haplotype-specific sequencing data (Fig. 1F–H; Supplemental Fig. S1B). This suggests the actual variability in SVAs may be even higher in the population than we found based on the 15 unphased PacBio genomes (Audano et al. 2019). Together these data reveal that SVA elements are a major source of structural variation between individuals.
SV-SVAs reside in gene-regulatory regions
We next determined whether SV-SVAs have the potential to influence gene expression. Previous studies have shown that SVAs have strong gene regulatory potential and are identified as cis-regulatory elements by epigenetic marks (Savage et al. 2013, 2014; Jacobs et al. 2014; Bragg et al. 2017; Trizzino et al. 2017; Pontis et al. 2019). In addition, SVAs are enriched in open and active chromatin regions in numerous tissues and senescent cells, and show active epigenetic marks in the brain (De Cecco et al. 2013; Trizzino et al. 2018; Pontis et al. 2019). In further support of the regulatory potential of SVA elements, we showed that an SVA-F element upstream of a minimal promoter induced an 11.5× increase in luciferase activity relative to the empty vector control in mouse ESCs (mESCs; P = 0.00227, two-sided t-test with Bonferroni correction) (Fig. 2A). mESCs were used to fully assess the SVA's regulatory potential using an approach described in Jacobs et al. (2014), in which SVA activity is measured in a cellular model system that lacks KRAB zinc finger proteins, a family of endogenous SVA repressors found in primates. A much lower but still significant enhancement of luciferase activity by the SVA-F element was also observed under conditions of KRAB zinc finger-mediated repression by the recently identified SVA repressor ZNF91 (P = 0.00441, two-sided t-test with Bonferroni correction) (Fig. 2A; Jacobs et al. 2014; Haring et al. 2021).
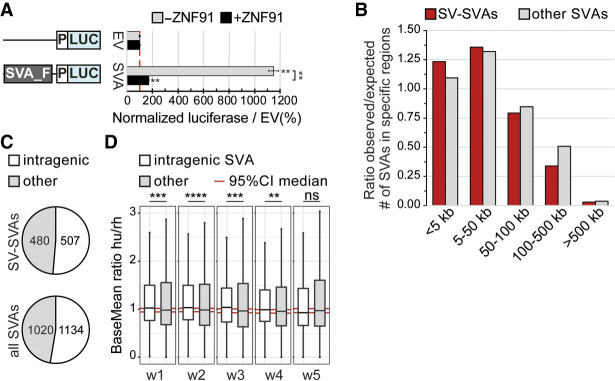
Figure 2.
SV-SVAs reside in gene-regulatory regions. (A) Luciferase activity of construct without (EV) and with an SVA element (SVA_F) upstream of a minimal promoter (P) in mESCs. N3n9, two-sided t-test with Bonferroni correction, (**) P < 0.01. Error bars: SEM. (B) Distribution of SV-SVAs (red) and non-SV-SVAs (gray) per distance to TSS. Only SVAs > 1 kb are shown. (C) Number of SV-SVAs and non-SV-SVAs that are intragenic is comparable (χ2(1, N = 2154) = 1.10, P = 0.29). Only SVAs > 1000 bp are shown. (D) Box plots showing base mean expression ratio (human/rhesus) for transcripts with an intragenic SVA in humans (white; 1151) and without (gray; 23,296) in ESC-derived cortical organoids of 1- to 5-wk old. Red line shows 95% CI of 10,000× bootstrapped median of transcripts without an SVA with sample size of 1151. Wilcoxon rank-sum test: (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01, ns = not significant.
SVAs have also been shown to influence gene expression through insertion near or within genes (Jacobs et al. 2014; Bragg et al. 2017; Trizzino et al. 2018; Haring et al. 2021). We found that 82% of SV-SVAs are located <50 kb from a transcription start site (TSS) (Fig. 2B), 1.33× more than expected by random distribution throughout the genome (P < 2.2 × 10−16, χ2 test). This is in line with previous research showing a higher than expected number of SVAs in genic regions (Savage et al. 2013). SVAs in active chromatin regions are known to preferentially reside in gene bodies, suggesting they may function as intronic regulatory elements (Trizzino et al. 2018). We found that a high percentage (51.4%) of SV-SVAs were intragenic, which was comparable to non-SV-SVAs (P = 0.29, χ2 test) (Fig. 2C). This supports findings from previous work, in which additionally a positive correlation between SVA number and gene transcription was found (Gianfrancesco et al. 2019). To further analyze the intronic regulatory effect of SVAs on a genome-wide level, we compared the expression levels of genes with and without an intragenic SVA in cortical organoids derived from human and rhesus macaque embryonic stem cells (Field et al. 2019). Notably, rhesus macaques, like all non-great-ape species, do not contain any SVAs in their genome. Human genes with an intragenic SVA displayed a modest but significant increase in expression levels compared to the non-SVA ortholog in rhesus. This difference was not observed for genes that lack intragenic SVAs in both human and rhesus (Fig. 2D; Supplemental Fig. S2). These data provide support for an intronic regulatory role of SVAs. The location of SV-SVAs in close proximity to or within genes is consistent with the hypothesis that these SVAs may have gene regulatory potential, and that structural variability within these elements could differentially regulate nearby genes.
Disorder-associated loci identified by GWAS are rich in SV-SVAs
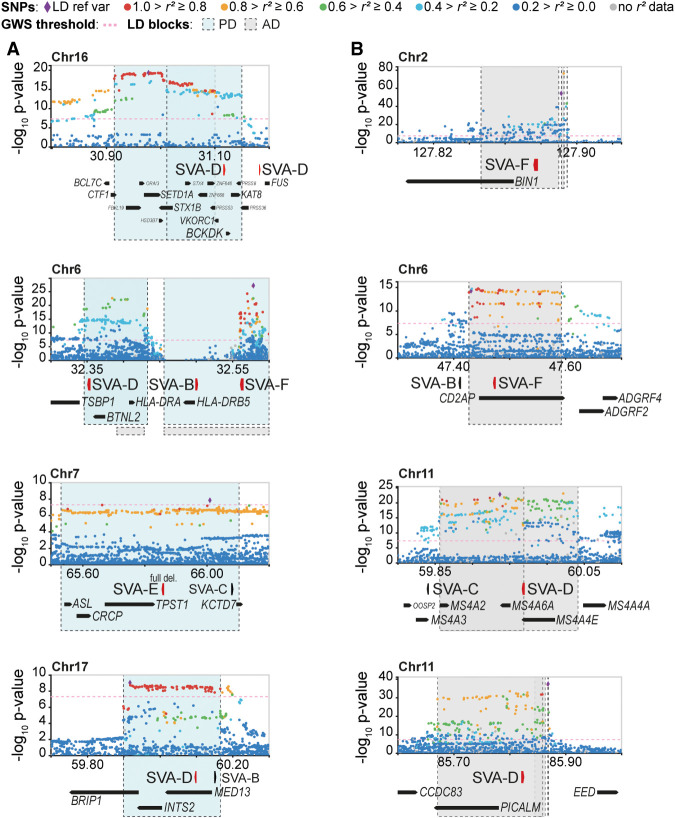
Previous research showed that insertional polymorphisms of TEs are in LD with trait-associated SNPs, constituting a potential causative genetic variant for numerous human phenotypes (Payer et al. 2017). We extended this approach beyond TE-insertional polymorphisms, asking whether we could detect SV-SVAs in LD with established neurological disorder-associated SNPs, which could thus have possible involvement in the development of these conditions. Using reported SNPs from the NHGRI-EBI catalog as source, we considered a variety of complex diseases with a presumed major genetic component and searched for SVAs within disorder-associated LD blocks. We determined the presence of SVAs in disorder-associated LD blocks in AD, PD, bipolar disorder, amyotrophic lateral sclerosis (ALS), autism spectrum disorder (ASD), multiple sclerosis (MS), and schizophrenia. AD-associated LD blocks contained 13 SVAs (in 12 out of 94 LD blocks) and PD-associated LD blocks contained 23 SVAs (in 19 out of 114 LD blocks). A substantial number of SVAs were also found in LD blocks associated with schizophrenia (69 SVAs in 59 out of 470 blocks), ASD (38 SVAs in 28 out of 178 blocks), bipolar disorder (36 SVAs in 21 out of 151 blocks), and MS (19 SVAs in 13 out of 198 blocks). Only one SVA was found in ALS-associated LD blocks, but the number of identified ALS-associated loci was also low (15 loci). The number of SVAs located within LD blocks was higher than the number expected by random distribution of the elements for PD (2.6×, P < .00001, χ2 test), ASD (2.1×, P < .00001), bipolar disorder (2.3×, P < .00001), schizophrenia (1.6×, P = .000106), and MS (1.9×, P = .00555), but not for AD (1.7×, P = .0545), neuroblastoma (1.7×, P = .586), or ALS (1.3×, P = .868).
We further focused our investigation on PD- and AD-associated loci, because aging is the greatest risk factor for those diseases, and the epigenetic changes associated with aging can uncover the hidden regulatory potential of TEs (Li et al. 2013; Guo et al. 2018). For PD, 23 SVAs were located within 19 LD blocks, of which nine SVAs were structurally variable and one showed insertional polymorphisms in the population (Fig. 3A; Supplemental Fig. S3A). SV-SVAs were also found intronic or close to the TSS of three other well-studied PD-associated genes: NURR1, SYT11, and PARK7 (Supplemental Fig. S4A; Jankovic et al. 2005; Simón-Sánchez et al. 2009; Nalls et al. 2014, 2019). For AD, the 13 SVAs found within those LD blocks were located near highly validated AD risk genes including BIN1, PICALM, and CD2AP (Fig. 3B; Hamza et al. 2010; Jansen et al. 2019; Schwartzentruber et al. 2021). Eight of these SVAs were found to be structurally variable (Fig. 3B; Supplemental Fig. S3B). SV-SVAs were also found intronic or close to the TSS of five other well-studied AD-associated genes: MS4A1, BACE1, PSEN1, DMXL1, and SPRED2 (Supplemental Fig. S4B; Sherva et al. 2014; Ma et al. 2015; Yu et al. 2016; Kelleher and Shen 2017; Schwartzentruber et al. 2021). The GWAS upon which the LD blocks were based were performed without accurate knowledge of the level of structural variation in TEs. Our analysis suggests that like any other structural variation near the tag SNPs, structurally variable SVAs need to be considered as candidate causal factors for the disease risk contained within these loci. This emphasizes the need to assess the gene-regulatory roles of these specific SV-SVAs in more detail.
Figure 3.
SV-SVAs reside in Parkinson's and Alzheimer's disease–associated LD blocks. (A,B) Regional SNP association plots with SV-SVAs (red) shown in LD blocks of PD (blue) (A) and AD (gray) (B). The associated SNPs (AD; de Rojas et al. 2021, PD; Nalls et al. 2019) are plotted with their respective meta-analysis genome-wide significant P-values (GWS [Genome-wide significance], P < 5 × 10−8; as −log10 values) and are distinguished by linkage disequilibrium (r2) of nearby SNPs on a blue to red scale, from r2 = 0 to 1, based on pairwise r2 values from the 1000 Genomes Phase3 (ALL) reference panel. Gene annotations: NCBI RefSeq Select database. Assembly GRCh37, scale in Mb.
SVA variants are associated with disease-risk SNPs and have differential gene regulatory potential
Based on the regulatory potential of SVAs and their presence in neurological disease-associated loci, we hypothesized that structural variation within SVAs could be a causal factor in disease risk for a number of GWAS-identified trait loci. To test this, we first focused on two human-specific SV-SVAs in loci associated with AD and PD. One of these SVAs was chosen as an example of an SVA nearby a gene body and the other of an SVA in a gene-poor region. Structural variants of these SVAs were identified by PCR, and the presence of the disease-associated tag SNP was determined by Sanger sequencing. We focused on individuals homozygous for specific SVA structural variants to ensure a correct association between the risk SNP and the presence of a certain SVA variant. The first SV-SVA we selected is located 2.7 kb from the TSS of the gene encoding for branched chain ketoacid dehydrogenase kinase (BCKDK) (Figs. 3A, 4A,B). This gene has an exonic SNP, rs14235, for which the minor (risk) allele is associated with a 1.17× to 1.36× increase in mean Lewy body count (P < 0.041–0.0026) (Nalls et al. 2014; Heckman et al. 2017). The putative mechanism of involvement of BCKDK in PD development is not entirely clear, but the nearby gene KAT8, which resides in the same LD block, may influence PD by modulating autophagic flux (Chang et al. 2017). We identified multiple structural variants for the nearby SVA by PCR analysis (Fig. 1F; Supplemental Fig. S1F) and analyzed rs14235 in 76 individuals homozygous for each structural variant of the nearby SVA. There was a significant relationship between the SNP alleles and the SVA structural variants (P < 2.2 × 10−16, Fisher's exact test). The minor (risk) allele was exclusively observed in individuals containing the SVA variant with either −528 bp or +146 bp structural variations, whereas all individuals with the reference variant and the +500 bp SVA variant were homozygous for the ancestral (major) allele (Fig. 4B). This indicates a strong link between specific structural variants of the SVA in this locus and the disease-associated haplotype identified by GWAS. This makes the SVA near BCKDK a potential candidate for carrying the actual mechanistic cause for the association of the minor (risk) allele of rs14235 with increased Lewy body count and increased risk of PD.
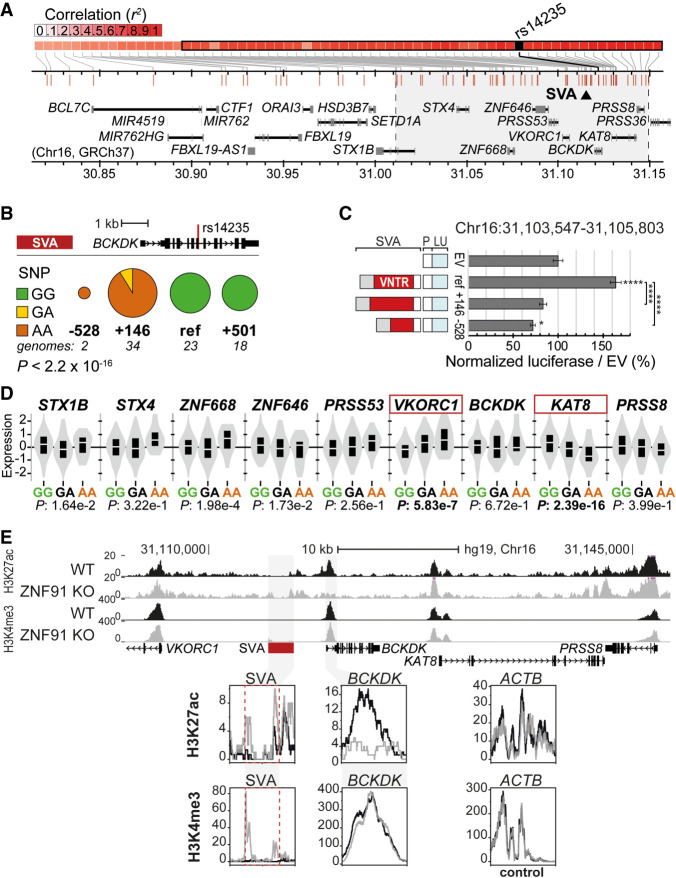
Figure 4.
Structurally variable SVA near BCKDK links to a disease-associated SNP and has the potential to differentially regulate nearby genes. (A) Overview of LD block for rs14235, with area r2> 0.8 highlighted in gray. Approximate location of SVA marked with black triangle. (B) rs14235 genotyping analysis for individuals homozygotic for BCKDK-SVA variants −600 (n = 2), ref (n = 23), +150 (n = 34), and +500 (n = 18). (Ancestral allele) G; (risk allele) A. Fisher's exact test: P < 2.2 × 10−16. (C) Schematic overview of luciferase constructs (P = minimal promoter, LU = luciferase gene) with BCKDK-SVA variants (Chr 16: 31,103,547–31,105,803 GRCh38), with corresponding luciferase activity in transfected mESCs. N3n9, except BCKDK-SVA ref (n = 8). One-way ANOVA with Tukey's multiple comparison, (****) P < 0.0001, (*) P < 0.05. Error bars: SEM. (D) Analysis of eQTL data in cortex for rs14235 for genes within the LD block with r2 > 0.8. Normalized expression is shown. Genes considered significant are shown in a red box. (E) KO of the SVA repressor ZNF91 lowers H3K27ac at the promoter of BCKDK and increases H3K4me3 methylation at the SVA near BCKDK in hESCs. ACTB shown as control enhancer region. (Top) Overview of locus, (bottom) magnification of regions of interest.
We next examined whether each of the SVA structural variants associated with the minor (risk) allele showed different gene regulatory potential. Both SVA variants (+146 bp and −528 bp) that consistently occur in conjunction with the minor allele showed significantly reduced gene regulatory potential compared to the reference variant/ancestral allele (P < 0.0001, two-way ANOVA, Tukey's multiple comparison) (Fig. 4C). The difference in reporter gene expression between the +146 bp variant and the reference SVA was still observed following the overexpression of ZNF91, a factor mediating strong repression of SVAs in a human-cellular context (P = 0.0274, two-way ANOVA, Tukey's multiple comparison) (Supplemental Fig. S5A). This shows that structural variations within the SVA near BCKDK harbor highly differential gene regulatory potential, and each of these SVA variants may therefore have different effects on nearby gene expression.
We next determined whether the presence of different SVA structural variants could also influence gene expression in vivo. Based on the knowledge that the rs14235 SNP allele was significantly associated with the structural variant of the nearby SVA, we compared eQTL data for rs14235 in brain tissue. We examined expression data from cortex and substantia nigra, two well-studied brain areas involved in PD, and found rs14235 was significantly associated with VKORC1 and KAT8 expression in cortex (Fig. 4D). KAT8 expression in the cortex was decreased in the presence of the minor (risk) allele for SNP rs14235. KAT8 expression in substantia nigra displayed a trend in the same direction (Supplemental Fig. S5C). These data are consistent with the observed repressive regulatory effect of the two SVA variants that coincide with the minor (risk) allele (Fig. 4B–D). Individuals with the minor (risk) allele also showed decreased expression of KAT8, ZNF646, PRSS36, and RP11-196G in other brain areas. No association of rs14235 with BCKDK expression levels was found. To further assess the regulatory effect of the SVA in this locus, we asked whether ectopic activation of the SVA would lead to changes in epigenetic marks in the BCKDK locus. We previously showed that in hESCs carrying a ZNF91 genetic deletion, SVAs become epigenetically activated (Haring et al. 2021). This is also the case for the SVA near BCKDK, which showed an increase in H3K4me3 in ZNF91 knockout (KO) hESCs (Fig. 4E). We also found epigenetic alterations at the BCKDK promoter, indicating that in human ESCs, the ectopic activation of the upstream SVA affected the epigenetic state of the BCKDK promoter.
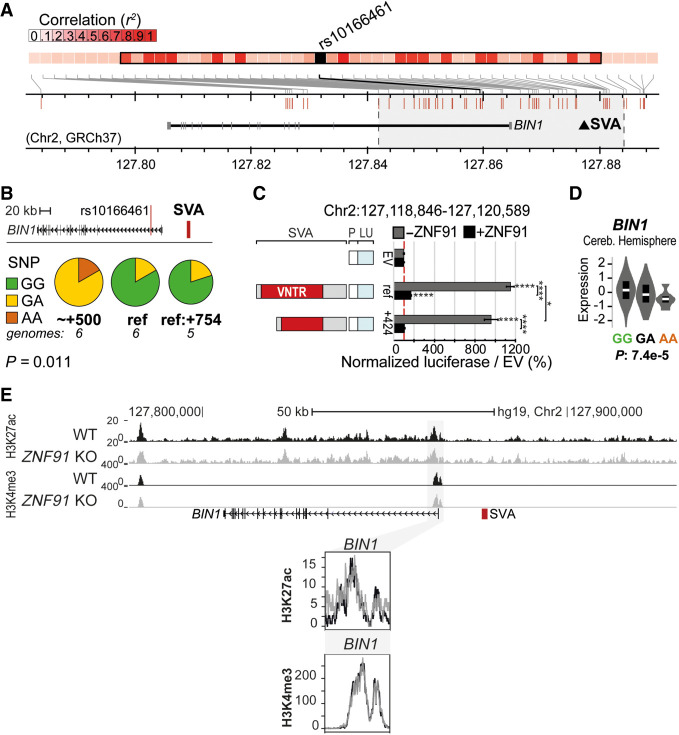
The second SV-SVA we analyzed is located 12 kb upstream of the TSS of the AD-associated gene BIN1. The upstream region of this gene has been named the second-most important susceptibility locus in late-onset AD (LOAD) (http://www.alzgene.org). BIN1 colocalizes and interacts with tau protein, and SNPs upstream of BIN1 have been linked to increased expression of the gene and risk for LOAD (Seshadri et al. 2010; Carrasquillo et al. 2011; Hu et al. 2011; Lambert et al. 2011, 2013; Lee et al. 2011; Wijsman et al. 2011; Chapuis et al. 2013; Masoodi et al. 2013). The upstream region of BIN1 contains a structurally variable SVA (Figs. 3B, 5A,B) (Chr 2: 127,118,897–127,120,492, GRCh38), located just 17 kb from the AD-associated risk SNP rs10166461 (allele effect −0.2636, P = 3.82 × 10−6) (Beecham et al. 2014). In a similar PCR validation to determine the association of SVA variants with the AD-associated SNP rs10166461, we found that the risk allele was more often observed in individuals carrying the +424 bp or +521 bp (∼+500) SVA variant, and although the number of sequenced individuals was limited, we found a significant association between SNP allele and SVA variant (P = 0.011, Fisher's exact test) (Fig. 5B). Structural variants of this SVA associated with the minor (risk) allele showed significant differential regulatory potential in the luciferase reporter assay (P < 0.05, two-way ANOVA, Tukey's multiple comparison) (Fig. 5C). In the presence of the SVA repressor ZNF91 this difference was not maintained. Furthermore, eQTL analysis revealed that individuals carrying the risk allele of rs10166461 displayed significantly lower expression of BIN1, but this effect was only observed in the cerebellum (Fig. 5D). Despite the apparent tissue-specific association between rs10166461 and changes in BIN1 expression, the direction of the gene expression effect corresponded with the results of our luciferase assays, in which the +424 bp variant (most often present in individuals with the risk allele of rs10166461) had a significantly lower regulatory potential than the reference SVA variant (Fig. 5C). Consistent with the cerebellum-specific association of rs10166461 with differences in BIN1 expression, in cortical organoids we found no significant alterations for H3K4me3 and H3K27ac histone marks in the BIN1 locus under ZNF91 KO conditions (Fig. 5E). We also examined eQTL data for other SV-SVAs in AD and PD loci. Significant differential expression associations for SNPs within the LD blocks overlapping these SVAs were observed for numerous genes, including CD2AP, HLA-DRB1/5/6, PICALM, MS4A6A, MS4A4E, and APOE (see Supplemental Table S2). Together, these data show the relationship between SV-SVAs and GWAS-identified risk SNPs and suggest that these SVA variants could potentially be involved in disease susceptibility through gene regulation in these loci. Further functional research on specific loci remains necessary to fully understand which and how SV-SVAs may contribute to disease development.
Figure 5.
Structurally variable SVA near BIN1 links to a disease-associated SNP and has the potential to differentially regulate nearby genes. (A) Overview of LD block for rs10166461, with area r2 > 0.8 highlighted in gray. Approximate location of SVA marked with black triangle. (B) rs10166461 genotyping analysis for individuals homozygotic for BIN1-SVA variants ref (n = 6) and +424 or +521 (∼+500, n = 6) and heterozygotic for ref and +754 (n = 5). Ancestral allele = G, risk allele = A. Fisher's exact test: P = 0.0108. (C) Schematic overview of luciferase constructs (P = minimal promoter, LU = luciferase gene) with BIN1-SVA variants, with corresponding luciferase activity in transfected mESCs with and without ZNF91. Two-way ANOVA with Tukey's multiple comparison. (****) P < 0.0001, (*) P < 0.05. Error bars: SEM. (D) Analysis of eQTL data in cerebellar hemisphere for rs10166461 and BIN1. (E) KO of the SVA repressor ZNF91 does not influence H3K27ac and H3K4me3 at the promoter of BIN1 in hESCs. (Top) Overview of locus, (bottom) magnification of regions of interest.
Genetic deletion of SVA elements in AD/PD loci alters the epigenome and nearby gene expression
To further investigate the gene-regulatory role of the SVAs in the BCKDK and BIN1 loci, we engineered genetic deletions of the SVAs in hESCs using CRISPR-Cas9 technology. BIN1-SVA KO and BCKDK-SVA KO hESCs were subsequently directed into a neuronal fate by generating cortical organoids to analyze the regulatory effect of the SVAs in a neuronal context (Fig. 6; Supplemental Figs. S6, S7). Five weeks after the onset of neuronal differentiation, organoids were harvested for RNA-seq and ChIP-seq. We supplemented a transcriptomic profile of the loci in the respective SVA KO cells with ChIP-seq profiles of H3K4me3 and H3K27ac epigenetic marks to identify genes that may be under regulatory influence of the SVA.
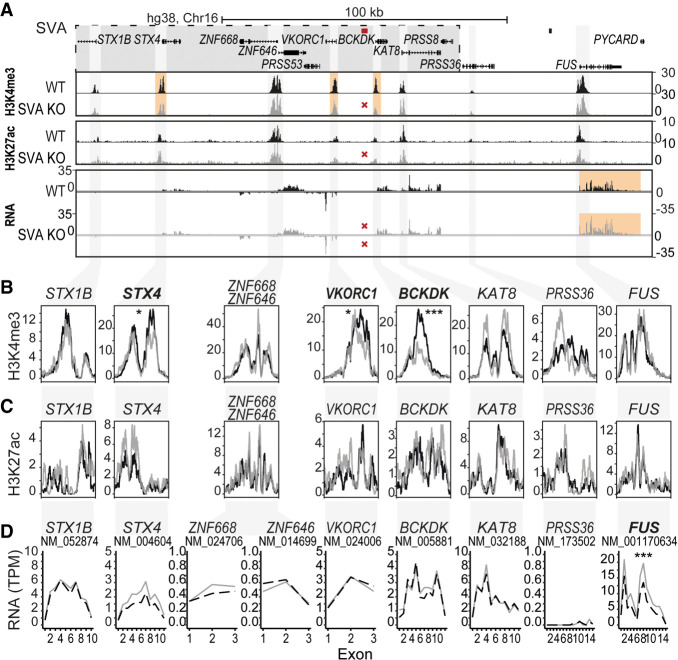
Figure 6.
SVA deletion alters the epigenome and nearby gene expression. (A) Overview of locus. LD block shown in gray box, location of SVA removed by CRISPR-Cas9 KO shown in red. (B–D) Magnification of genes within a 200 kb region of deleted SVA. (B) H3K4me3 ChIP-seq, the mean of three replicates is shown. (C) H3K27ac ChIP-seq, the mean of three replicates is shown. (D) Mean expression of three replicates shown per exon for transcripts. Adjusted P-value of DESeq2 is shown for the whole transcript and ChIP peaks. (***) P < 0.001, (*) P < 0.05. Genes reaching statistical significance are indicated in bold.
Genetic deletion of the SVA located in the BCKDK locus resulted in a statistically significant reduction in H3K4me3 at the promoters of BCKDK, VKORC1, and STX4 (Fig. 6A,B; Supplemental Table S3). Although we observed an increase of H3K4me3 signal in part of the promoter of KAT8 and PRSS36, this result did not reach the threshold of significance. No significant changes were observed for H3K27ac, which was detected at a much lower level overall (Fig. 6C; Supplemental Table S3). Despite the clear changes in histone marks, the RNA expression levels of BCKDK, VKORC1, STX4, or KAT8 were not significantly altered in BCKDK-SVA KO neurons. Instead, the deletion of the SVA in the BCKDK locus resulted in a strong increase in expression of the gene encoding RNA-binding protein FUS (FUS; adjusted P = 1.53 × 10−11, DESeq2), located ∼75 kb from the deletion site (Fig. 6D). Although no direct link has been reported for FUS with PD, this gene is linked to other neurodegenerative diseases such as ALS, frontotemporal dementia (FTD), and essential tremor (ET) (Kwiatkowski et al. 2009; Vance et al. 2009; Mackenzie et al. 2010; Wu et al. 2013; Deng et al. 2014). The expression of FUS was previously shown to be controlled by an SVA directly upstream of the FUS gene, which further supports our current observations (Savage et al. 2014). None of the other 61 genes in a window of 1 Mb upstream of and downstream from the SVA showed differential expression upon SVA deletion, with the exception of SRCAP, located 404 kb from the SVA (Supplemental Fig. S8). Removal of the SVA 12 kb from the TSS of BIN1 did not result in detectable epigenetic and transcriptional changes in the locus in the context of the cortex model system we used (Supplemental Fig. S6), but this may reflect the selective cerebellum-specific association of BIN1 expression with the risk allele and its associated SVA variant (Fig. 5D). Therefore, use of a model system for cerebellum rather than cortex may be important for investigating the effect of SVA KO in the BIN1 locus.
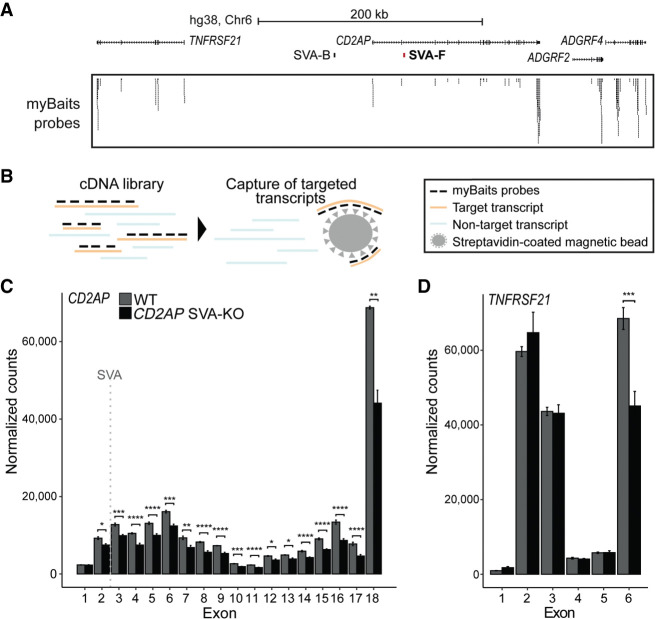
In a final approach, we aimed to investigate the gene-regulatory role of an intronic SVA in CD2AP, another AD-associated gene (Fig. 3B). We engineered a genetic deletion of the SVA in hESCs using CRISPR-Cas9 (Fig. 7A) and generated cortical organoids from the CD2AP-SVA KO hESCs. We performed targeted mRNA enrichment before RNA-seq (RNA CaptureSeq) to specifically enrich for transcripts derived from the CD2AP locus and 38 other unrelated control genes, allowing us to track the expression levels of each of the CD2AP exons separately. This was important because intronic SVAs may differentially influence the expression level of exons upstream of and downstream from the SVA, as is observed for a pathogenic intronic SVA that leads to X-linked dystonia-parkinsonism (Bragg et al. 2017). Furthermore, this RNA CaptureSeq approach increased the overall resolution of the selected gene transcripts, allowing for a more accurate assessment of the consequences of removal of the intronic SVA for CD2AP gene expression (Fig. 7B). DESeq2 analysis comparing gene expression in CD2AP-SVA KO cortical organoids to unedited control organoids revealed significantly lower expression levels for all CD2AP exons downstream from the SVA insertion site (Fig. 7C). Of the two exons upstream of the SVA, exon 1 was not significantly affected by SVA removal, and only a modest reduction was observed for exon 2. Although the final exon of the nearby gene TNRFSF21 also showed a difference in expression between KO and unedited controls, a consistent change in expression levels for TNRFSF21 was not observed (Fig. 7D). No significant differences in expression levels between CD2AP-SVA KO and unedited control organoids were observed for the vast majority of control genes residing in other genetic loci included in the RNA CaptureSeq analysis (Supplemental Table S4).
Figure 7.
Intronic SVA deletion alters exon expression of CD2AP gene. (A) Overview of CD2AP locus. Location of SVA removed by CRISPR-Cas9 KO shown in red. Probes from myBaits targeting CD2AP exons. (B) Schematic of capture of targeted transcripts with myBaits probes. (C,D) Normalized mean expression of three replicates shown per exon of CD2AP and the nearby highly expressed gene TNFRSF21. Location of SVA indicated with dashed gray line. Adjusted P-value from DESeq2 shown for each exon. (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01, (*) P < 0.05.
Taken together, these results show that structurally variable SVAs in important neurological disease-associated loci have the potential to affect the surrounding epigenome and/or transcriptome. Although the genetic variants that cause gene dysregulation remain undiscovered for the majority of the GWAS-identified disease-risk loci, our data present structural variation in SVAs and other TEs as novel potential genetic modifiers of gene regulation in these loci.
Discussion
Our study reveals that structural variations in SVAs are previously unknown candidates for causal genetic variants not captured by conventional GWAS. Multiple lines of research indicate that SVAs can influence the expression of nearby genes (De Cecco et al. 2013; Savage et al. 2013, 2014; Jacobs et al. 2014; Bragg et al. 2017; Trizzino et al. 2017, 2018; Pontis et al. 2019; Haring et al. 2021), and the extensive structural variability that we observed in SVAs genome-wide suggests that this influence could be highly variable within the human population. For two SVAs analyzed in detail in this study, we find that specific SVA variants are significantly associated with the presence of nearby risk SNPs identified in GWAS, whereas other variants are not. Although all individuals will carry a certain SVA in a disease-associated locus, the particular structural variation present within this SVA in each individual may have important consequences. In fact, we showed that different SVA structural variants show differential gene regulatory potential in vitro. We propose that inter-individual structural variation in SVA elements may be an important genetic variable that may directly and functionally link to disease risk. Furthermore, in those cases where an SVA influences gene expression in the locus, the structural variation in the SVA may be an even more accurate indicator for disease risk than the risk SNP originally identified by GWAS.
Previous research into structural variation within specific SVAs supports our hypothesis that these variations can be involved in disease. In X-linked dystonia-parkinsonism, the length of a disease-causing SVA insertion in an intron of the TAF1 gene displayed differential gene regulatory potential and correlated with disease onset (Bragg et al. 2017). Two other SV-SVAs near ALS- and PD-associated genes were also shown to have differential regulatory potential, although in these cases the causal relationship to the disease remains unproven (Savage et al. 2013, 2014). Our study reveals multiple SV-SVAs in AD- and PD-associated loci that are potential contributors to disease susceptibility. eQTL data of SNPs in the BCKDK and BIN1 loci showed an association of the disease-risk allele to changes in gene expression. Importantly, for both loci the direction of gene expression change associated with the risk alleles was consistent with our luciferase assay data in which different SVA variants were explored functionally. The two SVA variants associated with the risk allele rs14235 in the BCKDK locus displayed an expansion (+146 bp variant) and deletion (−528 bp variant) relative to the reference; however, both variants showed a significant reduction in gene regulatory potential relative to the reference in the luciferase reporter assay. Potential mechanisms underlying differences in regulation of gene expression between SVA variants remain to be explored in future work.
Removal of BCKDK and CD2AP SV-SVAs produced subtle but significant changes on gene expression and epigenetic landscape, whereas no change was detected upon deletion of the SV-SVA in the BIN1 locus. This is not completely unexpected, because SV-SVAs are only one potential layer of gene regulation among many others. If SV-SVAs produced large changes in gene expression, we might expect the phenotypic consequences of these variations to be evident in early life. On the contrary, it could be the case that the influence of SV-SVAs is highly tissue- or condition-specific and is not easily captured in our model systems. Structural variations in SVAs may be particularly important in age-related neurological diseases, because the epigenetic changes associated with aging could uncover the latent regulatory potential of these TEs (Li et al. 2013; Guo et al. 2018). In that sense, SV-SVAs may contribute to the progressive character of these diseases, because with increased age the differences in regulatory potential of SVA variants could become increasingly more prominent. For the SVA in the PD-associated BCKDK locus, along with an influence on the epigenetic state of nearby gene promoters, we found a possible gene-regulatory effect on FUS, a gene located just outside of the PD-associated LD block. These findings are compatible with the concept that the causal variant within a disease-associated locus could be a gene-regulatory element that influences the expression of genes inside and/or outside of the LD block. Although there is no strong support for a role of FUS dysregulation in PD, FUS has been associated with other neurodegenerative diseases such as ALS, FTD, and ET (Kwiatkowski et al. 2009; Vance et al. 2009; Mackenzie et al. 2010; Wu et al. 2013; Deng et al. 2014). In addition, PINK1 and PARKIN, two well-known PD-associated genes, were found to be genetic modifiers of FUS-induced neurodegeneration (Chen et al. 2016). Given the role of FUS in other neurodegenerative diseases, it is not unlikely that FUS dysregulation, caused by specific SVA variants in the BCKDK locus, is in some way a contributor to the neuropathology in PD, but whether FUS is indeed regulated by the SVA in the BCKDK locus remains to be investigated in detail.
Collectively, this study reveals an extensive level of structural variation in TEs that has escaped detection by SNP arrays and short-read sequencing techniques. Our analysis stresses the importance of accurate mapping and characterization of structural variations in TEs in disease-associated loci. The methods used by GWAS do not identify structural variations owing to their repetitive nature and size, so there may be numerous additional risk loci where the disease association is caused by structural variation in TEs. Whole-genome sequencing (WGS) is perfectly suitable for genotyping insertional polymorphisms, however variations in repetitive sequences are still overlooked. Ebert et al. (2021) describe a complementary method to identify structural variations with haplotype-resolved assemblies from long-read sequencing technology and genotype the identified structural variations on short-read sequencing data for further population analysis. Although this method allows for the discovery of structural variations on short reads, the limiting factor is still highly repetitive DNA regions such as SVAs, thus giving an underrepresentation of the true level of structural variability. Indeed, we found that the poor genotype quality of the specific SVAs of interest in the PanGenie callset of 1000 Genomes samples (Ebert et al. 2021) made for a noninformative SVA-eQTL analysis. In the future, a targeted genotyping of SVAs using an alternative genotyper may be able to improve the SVA genotyping quality using short reads. Alternatively, long-read sequencing may be a better technology to fully characterize structural variations and their association with disease, because it is estimated that ∼83% of insertions are missed with short-read-calling algorithms (Chaisson et al. 2019). Recently haplotype-resolved assemblies have been reported, which allow for more sensitive structural variation discovery by taking into account both alleles for each locus. This strategy outputs a much more accurate representation of the total structural variability in the human genome, indicating that the structural variation data set used in our study is an underrepresentation of the total structural variation in the human population. Although long-read sequencing is expensive and not yet widely used, it is a matter of time before it becomes the standard way to perform genome sequencing. The reliability of genetic disease susceptibility markers and predictions will be significantly enhanced once GWAS include SNPs alongside all other structural variations, regardless of the size of these variants or whether they are located within TEs. Finally, increased awareness of the hidden gene regulatory potential of noncoding DNA elements and the availability of human model systems and genetic engineering to study functional noncoding elements will allow us to functionally interrogate these variants to gain a better understanding of the mechanism of disease. Ultimately, this may contribute to the discovery of novel therapeutic targets and strategies for disease treatment.
Methods
Cell culture and SVA KO
hESCs were cultured and treated as described previously (Haring et al. 2021) to generate KOs of the SVAs located at Chr 2: 127,118,846–127,120,589, Chr 16: 31,103,565–31,105,709, and Chr 6: 47,505,039–47,506,780 (GRCh38) using the CRISPR-Cas9 system (Supplemental Fig. S7B–D). Cortical organoids were grown based on the methods described by Eiraku et al. (2008). See Supplemental Material for details and full methods.
ChIP-seq
ChIP
ChIP on EP300 was performed as described previously (Jacobs et al. 2014), with an excess of EP300 antibody (C-20; sc-585 X; Lot B0211 and Lot E2610). For H3K27ac and H3K4me3 data, ChIP was based on Vermunt et al. (2014), with 5 µg H3K27ac (Abcam ab4729, Lot GR3303561-2) and 6 µg H3K4me3 (Millipore 07-473, Lot 3394198) per sample. ChIP-seq data analysis on EP300 was performed as described previously (Jacobs et al. 2014). For the full procedure, see Supplemental Material.
Library preparation and sequencing
Paired-end indexed ChIP DNA libraries were prepared using Illumina TruSeq ChIP Sample Preparation Kit according to the guidelines, with minor exceptions (see Supplemental Material). For H3K27ac and H3K4me3, 2 × 75 bp paired-end sequencing was performed by MAD: Dutch Genomics Service & Support Provider of the University of Amsterdam using a NextSeq 550 Illumina sequencer. EP300 data were sequenced on an Illumina HiSeq 2000 sequencing device.
EP300 enrichment in TE classes
EP300 summit and peak files of replicates were merged and only peaks of which the summit overlapped with a repeat were kept to increase specificity. These 19,030 peaks were used as input for the TE-analysis pipeline (https://github.com/4ureliek/TEanalysis) (Kapusta et al. 2013) to calculate repeat classes significantly enriched for EP300. For the top 20 enriched elements [log2(observed number of elements overlapping with a EP300 peak/expected)], the percentage of elements overlapping with a peak was reported (obs_hits/nb_of_TE_in_genome). See Supplemental Table S1 for output.
RNA-seq
RNA isolation, library preparation, and sequencing
Between six and nine organoids were collected on day 35 after start of differentiation, rinsed in medium, homogenized in 400 µL TRIzol (Invitrogen) by pipetting, and stored at −80°C for later use. RNA was isolated according to the manufacturer's protocol with DNase I treatment (Sigma-Aldrich) and cleaned-up using the RNA Clean & Concentrator-5 kit (Zymo Research). Libraries were generated with the TruSeq Stranded Total RNA Library Prep (Illumina) kit, and 2 × 75 bp paired-end reads were sequenced by MAD: Dutch Genomics Service & Support Provider of the University of Amsterdam using a NextSeq 550 Illumina sequencer.
RNA CaptureSeq
Samples of CD2AP-SVA KO and unedited control were used for RNA CaptureSeq. RNA isolation of cortical organoids and library preparation were performed as described previously. Enrichment of targeted transcripts was performed with biotinylated probes from myBaits Custom RNA-seq following the manufacturer's standard protocol v.5.00 (Arbor Biosciences, Ref #200320-91) based on Mercer et al. (2012). The cDNA libraries were pooled, and 100 ng was used for capture. After capture, samples were amplified with 14 PCR cycles and purified with 1× AMPure XP beads. The sequencing was performed as described previously.
Data analysis
Data were analyzed on the public Freiburg Galaxy server (Goecks et al. 2010; Afgan et al. 2018) (https://usegalaxy.eu). Reads were trimmed using Trimmomatic (Bolger et al. 2014) version 0.36.5 for paired-end reads, removing adapters (ILLUMINACLIP TruSeq3 paired-end), cutting when average quality per base in a 4-base sliding window was below 20, and dropping reads below a length of 30. Reads were mapped using HISAT2 (Galaxy Version 2.1.0 + galaxy5) (Kim et al. 2019) against the built-in hg38. featureCounts (Galaxy Version 1.6.4 + galaxy2) (Liao et al. 2014) was used to assign reads to NCBI RefSeq hg38 features with -p, -d 75 -D 900 -B -C. Output was used for DESeq2 (Galaxy Version 2.11.40.6 + galaxy1) (Love et al. 2014) using default settings. bamCoverage (Galaxy Version 3.3.2.0.0) from the deepTools2 package (Ramírez et al. 2016) was used to generate coverage tracks (bin size 1) and scale these with a scaling factor based on the number of uniquely assigned reads from featureCounts. Scaled coverage tracks were merged using wiggletools (Zerbino et al. 2014) mean, and wig files transformed to bigWig files using the wigToBigWig script (http://hgdownload.soe.ucsc.edu/admin/exe/). See Supplemental Table S3 for DESeq2 output.
Differential exon usage analysis
To visualize differential exon usage (Fig. 6D; Supplemental Fig. S6B,C), raw RNA-seq data were preprocessed and analyzed with the RNA-seq analysis pipeline (https://github.com/KoesGroup/Snakemake_hisat-DESeq), with the following modification of the featureCounts rule using “exon” as feature type: Snakefile, line 194 featureCounts -p -t exon -g exon_id -T 8 -F GTF -O -M -a {input.gff} -o {output} {input.bams}. Preprocessed reads were mapped to hg38 and reads were assigned to NCBI RefSeq hg38 features. The read count files were used as input for differential exon usage analysis. From here on, analysis was performed in R (version 3.4.1) using the packages dplyr (version 1.0.2; https://cran.r-project.org/package=dplyr), reshape2 (version 1.4.4) (Wickham 2007), and tidyr (version 1.1.2; https://cran.r-project.org/package=tidyr). The NCBI RefSeq hg38 annotation file was modified to contain only exon information and filtered based on canonical transcript IDs and coding region. Read counts file was merged with prefiltered annotation file through a translation table and exon counts normalized to transcript per million. Data were displayed using ggplot2 (version 3.3.3; https://ggplot2.tidyverse.org) (Wickham 2016).
Capture data analysis
RNA CaptureSeq data were preprocessed and analyzed with the RNA-seq analysis pipeline (https://github.com/BleekerLab/snakemake_rnaseq). Preprocessed reads were mapped to hg38, allowing up to 20 multimappers during the alignment. Reads were assigned to Ensembl V104, counting features for both transcripts and exons. From here on, analysis was performed in R (version 4.1.0) using the packages tidyverse (version 1.3.1) (Wickham et al. 2019), reshape2 (version 1.4.4) (Wickham 2007), and plyr (version 1.8.6) (Wickham 2011). The read counts file was used as input for differential exon usage analysis with DESeq2 (version 1.32.0) (Love et al. 2014). To correct for variability in control loci presumably unaffected by SVA KO, size factor was estimated based on all captured genes (Supplemental Table S1) excluding genes located in the CD2AP locus (Fig. 7A). See Supplemental Table S4 for DESeq2 output.
Structural variation analysis
Structural variant data were retrieved from Audano et al. (2019) and alternate loci were removed. For each repeat class of interest, regions with one or both ends outside the repeat that overlapped > 95% with the total repeat length were filtered out to exclude full insertions from the analysis. Transposable elements that showed EP300 (enhancer) marks in cortical organoids were selected, and the coordinates were extracted from RepeatMasker (UCSC Genome Browser hg38). The alternate haplotypes were removed from each TE subclass, and distribution plots were used to select a minimum length per TE subclass (Supplemental Table S1), which was used as criteria to filter the elements. All analyses were performed using GenomicRanges package version 1.36.1 (https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html) in R version 3.6.0 (2019-04-26) (Lawrence et al. 2013; R Core Team 2019). For script, see Rcode_SVsinTEs in Supplemental Code. Because the annotation of SVAs in RepeatMasker for hg38 includes many SVA fragments near one another, we merged these using the join_sva.py code (see Supplemental Code). For Figure 1D, structural variation locations were plotted at SVAs using data of Audano et al. (2019) and the deepTools (Ramírez et al. 2016) package at the Galaxy platform (Goecks et al. 2010; Afgan et al. 2018). To include hexamer repeats not annotated in the RepeatMasker (hg38 assembly) as SVA, simple repeats at the 5′ side of SVAs with a size >1000 bp were merged with the SVA if they were within 20 bp of each other using BEDTools (Quinlan and Hall 2010) merge. The structural variation data were filtered as described previously. Insertions were transformed into deletions for clarity of overlap (insertions were represented as only 1 bp overlap, which were not clearly visible in the heatmap). computeMatrix (Galaxy Version 3.3.2.0.0) was used to prepare the data for profile plotting with the following settings: regions were scaled to the mean size of SVAs containing structural variation (1908 bp), ‐‐binSize 1, ‐‐sortRegions descend, ‐‐sortUsing mean, ‐‐missingDataAsZero True. Heatmap and coverage plots were generated using plotHeatmap (Galaxy Version 3.3.2.0.1) for only SV-SVAs (987). For Figure 1E, sizes of structural variations overlapping with SVAs were plotted in R (R Core Team 2019) using ggplot2 (Wickham 2016) geom_freqpoly() with 60 bins. The lines from 0 to 50 and −50 to 0 bp were manually removed, because no structural variations with these sizes were present in the data. For Figure 1, G and H, BLAST (Altschul et al. 1990) was used to search for the SVAs of interest plus 500 bp flanks in the depicted genomes (Ebert et al. 2021), and MUSCLE (Unipro UGENE) (Okonechnikov et al. 2012) was used for alignment to interpret the results. SVA regions were determined based on the repeat browser (Fernandes et al. 2020). Manually curated results were visualized using Adobe Illustrator CC (Adobe Inc.). See Supplemental Table S1 for sequences and information about data. Analysis of intragenic SVAs was performed on the public European Galaxy server (https://usegalaxy.eu). The hg38 NCBI RefSeq GTF file was filtered for transcripts, converted to BED12 using Convert GTF to BED12 (Galaxy Version 357), sorted using BEDTools (Quinlan and Hall 2010) SortBED (Galaxy Version 2.29.2) and overlapped with SV- and non-SV-SVAs using BEDTools (Quinlan and Hall 2010) ClosestBed (Galaxy Version 2.29.2). TSSs were generated from the hg38 NCBI RefSeq transcript GTF file and the GenomicRanges package (version 1.44.0; https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html) was used to determine locations relative to TSS in R (version 4.1.0) (R Core Team 2019). See Supplemental Table S1 for information. Differences in distribution of SV- and non-SV-SVAs were tested using a χ2 test of independence. For distance to TSS: χ2(4, N = 2154) = 7.4468, P = 0.1141. For intragenic versus non-intragenic: χ2(1, N = 2154) = 1.10, P = 0.29. For number of SV-SVAs within 50 kb of TSS: χ2(1, N = 987) = 172.3, P < 2.2 × 10−16. For number of SVAs in AD/PD LD blocks: PD (23/8.8, χ2 (df: 1) = 23.195, P < .00001), ASD (38/17.8, χ2 (df: 1) = 23.057, P < .00001), bipolar disorder (36/15.6, χ2 (df: 1) = 26.939, P < .00001), schizophrenia (69/43.6, χ2 (df: 1) = 15.018, P = .000106), MS (19/10.2, χ2 (df: 1) = 7.691, P = .00555), AD (13/7.7, χ2 (df: 1) = 3.696, P = .0545), neuroblastoma (1/0.6, χ2 (df: 1) = 0.296, P = .586), and ALS (1/0.8, χ2 (df: 1) = 0.0276, P = .868).
Genome-wide analysis of structural variation within TEs
Structural variations in SVAs were additionally analyzed by aligning reference SVA sequences with SVAs identified in WGS files (GCA_002180035.3_HG00514_prelim_3.0_genomic.fna, HG00733_prelim_1.0, HG01352_prelim_2.1, HG02059_prelim_1.0, NA19240_prelim_3.0), using the te-polymorphisms-analysis.py script (see Supplemental Code). Because this analysis included extensive manual interpretation of output, only SVAs on Chromosome 1 were analyzed in five individuals to confirm high structural variability in SVAs. In short, reference sequence and 500 bp flanking sequences were retrieved from hg38 using the Bio.SeqIO package from Biopython (Cock et al. 2009). lastdb (Kiełbasa et al. 2011) was used to build an index of the WGS files. SVA-flanking sequences with ≥90% similarity to reference were identified in the sequencing contigs using lastal (Kiełbasa et al. 2011), and the sequence in between the flanks with the highest similarity was stored as SVA sequence. Pairwise sequence alignment of reference SVA sequence and SVA sequence identified in contigs was performed using EMBOSS Needle or Stretcher (Myers and Miller 1988) to determine structural variation relative to the reference genome.
LD block analysis
Linkage disequilibrium (LD) blocks (threshold r2 ≥ 0.8) were determined with the SNiPA Block Annotation tool (https://snipa.helmholtz-muenchen.de/) (Arnold et al. 2015) using the 1000 Genomes Project (Phase 3/Version 5, European population, GRCh37 assembly, Ensembl 87) data for SNPs from the GWAS Catalog (P ≤ 10−8) (MacArthur et al. 2017). SNPs for which no LD blocks could be calculated or that were not in LD were filtered out, and duplicate blocks were removed from the list. To minimize bias in calculating the percentage of LD blocks overlapping with an SVA, blocks completely within other blocks were removed. LD blocks were annotated for hg19 and overlapped with the hg19 SVA annotation (see Supplemental Table S1) using the UCSC Genome Browser Table Browser tool. See Supplemental Table S5 for downloaded files and information about LD blocks. Locus overview tracks were plotted using pyGenomeTracks (https://pypi.org/project/pyGenomeTracks/) (Ramírez et al. 2018; Lopez-Delisle et al. 2021) and regional plots were made using LocusZoom (AD) (https://my.locuszoom.org/) (Pruim et al. 2010) or the PD GWAS Locus Browser (https://pdgenetics.shinyapps.io/GWASBrowser/) (Grenn et al. 2020). Data were used from de Rojas et al. (2021) and Nalls et al. (2019). Because of plotting restrictions of LocusZoom, infinite P-values (P = 0) present in the de Rojas et al. meta-analysis for the APOE locus were transformed to 1 × 10−320. Gene annotations were derived from NCBI RefSeq Select database, assembly GRCh37.
eQTL plots
Violin plots were generated for described regions and SNPs using the GTEx eQTL Dashboard tool (https://gtexportal.org/home/eqtlDashboardPage) on May 20, 2020, for the cortex and substantia nigra.
Pairwise linkage disequilibrium
Heatmap matrices were generated using the interactive LDmatrix tool of the LDlink suite (https://ldlink.nci.nih.gov/) (Machiela and Chanock 2015). Data from the European population of the 1000 Genomes Project (Phase 3/Version 5) were used to match Caucasian gDNA samples. See Supplemental Table S1 for SNPs included in image.
PCR
SVA and SNP amplification
SVAs were amplified from human gDNA samples of patients diagnosed with neurodegenerative disease, which were a kind gift from Coriell Institute Biorepository (DNA panels NDPT088, NDPT087, NDPT083 from the NINDS Repository), and gDNA from a cohort of cognitively healthy centenarians. See Supplemental Material for details and full methods. For primer details, see Supplemental Table S6; for details on identified variants for BCKDK- and BIN1-SVA, see Supplemental Table S1 and Supplemental File S1.
Luciferase assay
Luciferase assays using SVAs cloned upstream of a luciferase reporter were performed in mouse ESCs as described previously (Jacobs et al. 2014). Details of primers used for cloning are provided in Supplemental Table S6. A detailed description and full methods are provided in Supplemental Material.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE167409. For scripts, see Supplemental Codes. Plasmids and cell lines are available on request to the corresponding author.
Supplementary Material
Acknowledgments
This work was supported by a Human Frontier Science Program (HFSP) Career Development Award (CDA00030/2016C) to F.M.J.J. and a European Research Council (ERC) starting grant (ERC-2016-stG-716035) to F.M.J.J. and a grant from Alzheimer Nederland (WE.03-2018-07) to F.M.J.J., H.H., and M.J.T.R. We thank Lindsay Payer and Kathleen Burns for helpful discussions and advice about the project; Evan Eichler, Tobias Marschall, Peter Audano, and Marc Bonder for helpful discussion about SV-eQTL analysis; the National Institute of Neurological Disorders and Stroke (NINDS) Human Genetics Resource Center for gDNA; Gonzalo Congrains Sotomayor for technical assistance with FACS; Cindy Wagemans for technical support during gDNA isolation of SVA KO lines; Sophie Imhof and Elias Brandorff for providing the BIN1-SVA reference plasmid; Sol Katzman for EP300 ChIP-data processing; Wim de Leeuw for technical support with bioinformatics; MAD: Dutch Genomics Service & Support Provider of the University of Amsterdam for sequencing; and the Evolutionary Neurogenomics Group and others at the Swammerdam Institute for Life Sciences (SILS) for helpful discussions.
Author contributions: F.M.J.J. and E.J.v.B. conceptualized the project. F.M.J.J., E.J.v.B., R.L.F.P.G., M.L., and A.D.E. conceived or designed elements of the study. E.J.v.B. and R.L.F.P.G. performed SV-TE/SVA analysis. F.M.J.J. and D.H. generated EP300 data. E.J.v.B., M.L., F.M.J.J., and A.D.E. were involved in the BLAST SV-SVA analysis. E.J.v.B., J.P., P.F.R., A.-F.E.S., and F.M.J.J. performed PCR and SNP genotyping and generated SVA-plasmids. E.J.v.B., R.L.F.P.G., I.C., and E.R.B. were involved in the LD block analysis and visualization. E.J.v.B., R.L.F.P.G., and I.C. were involved in SVA KO experiments. R.L.F.P.G. performed RNA CaptureSeq experiment with the support of J.L.R. and F.T.G.W. for the analysis. E.J.v.B., R.L.F.P.G., and C.M. performed the statistical analyses. F.M.J.J., M.J.T.R., and H.H. acquired funding for the study. E.J.v.B., R.L.F.P.G., F.M.J.J., and C.M. wrote the manuscript with contributions of M.L., A.D.E., M.J.T.R., and H.H. F.M.J.J. supervised the study.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275515.121.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, et al. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46: W537–W544. 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmüller G. 2015. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 31: 1334–1336. 10.1093/bioinformatics/btu779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, et al. 2019. Characterizing the major structural variant alleles of the human genome. Cell 176: 663–675.e19. 10.1016/j.cell.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman JD, Li AH, Marcketta A, Sun D, Mbatchou J, Kessler MD, Benner C, Liu D, Locke AE, Balasubramanian S, et al. 2021. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature 599: 628–634. 10.1038/s41586-021-04103-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Gudi VA, Mena JC, Foltz DW, Herrera RJ, Deininger PL. 1991. Amplification dynamics of human-specific (HS) Alu family members. Nucleic Acids Res 19: 3619–3623. 10.1093/nar/19.13.3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, Corneveaux JJ, Hardy J, Vonsattel JP, Younkin SG, et al. 2014. Genome-wide association meta-analysis of neuropathologic features of Alzheimer's disease and related dementias. PLoS Genet 10: e1004606. 10.1371/journal.pgen.1004606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg DC, Mangkalaphiban K, Vaine CA, Kulkarni NJ, Shin D, Yadav R, Dhakal J, Ton ML, Cheng A, Russo CT, et al. 2017. Disease onset in X-linked dystonia-parkinsonism correlates with expansion of a hexameric repeat within an SVA retrotransposon in TAF1. Proc Natl Acad Sci 114: E11020–E11028. 10.1073/pnas.1712526114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes KJ. 2013. The VNTR in complex disorders: the forgotten polymorphisms? A functional way forward? Genomics 101: 273–281. 10.1016/j.ygeno.2013.03.003 [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HHJ. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci 100: 5280–5285. 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Sando SB, Aasly JO, et al. 2011. Replication of BIN1 association with Alzheimer's disease and evaluation of genetic interactions. J Alzheimers Dis 24: 751–758. 10.3233/JAD-2011-101932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, Gardner EJ, Rodriguez OL, Guo L, Collins RL, et al. 2019. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun 10: 1784. 10.1038/s41467-018-08148-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F, Kerchner GA, Ayalon G, Bingol B, Sheng M, et al. 2017. A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat Genet 49: 1511–1516. 10.1038/ng.3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV, Geller F, Sottejeau Y, Harold D, Dourlen P, et al. 2013. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry 18: 1225–1234. 10.1038/mp.2013.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Deng J, Wang P, Yang M, Chen X, Zhu L, Liu J, Lu B, Shen Y, Fushimi K, et al. 2016. PINK1 and Parkin are genetic modifiers for FUS-induced neurodegeneration. Hum Mol Genet 25: 5059–5068. 10.1093/hmg/ddw310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351: 1083–1087. 10.1126/science.aad5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25: 1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, et al. 2020. A structural variation reference for medical and population genetics. Nature 581: 444–451. 10.1038/s41586-020-2287-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703. 10.1038/nrg2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM. 2013. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12: 247–256. 10.1111/acel.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Gao K, Jankovic J. 2014. The role of FUS gene variants in neurodegenerative diseases. Nat Rev Neurol 10: 337–348. 10.1038/nrneurol.2014.78 [DOI] [PubMed] [Google Scholar]
- de Rojas I, Moreno-Grau S, Tesi N, Grenier-Boley B, Andrade V, Jansen IE, Pedersen NL, Stringa N, Zettergren A, Hernández I, et al. 2021. Common variants in Alzheimer's disease and risk stratification by polygenic risk scores. Nat Commun 12: 3417. 10.1038/s41467-021-22491-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert P, Audano PA, Zhu Q, Rodriguez-Martin B, Porubsky D, Bonder MJ, Sulovari A, Ebler J, Zhou W, Serra Mari R, et al. 2021. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 372: eabf7117. 10.1126/science.abf7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Beesley J, French JD, Dunning AM. 2013. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 93: 779–797. 10.1016/j.ajhg.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE. 2019. Genetic variation, comparative genomics, and the diagnosis of disease. N Engl J Med 381: 64–74. 10.1056/NEJMra1809315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. 2008. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3: 519–532. 10.1016/j.stem.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Ewing AD, Smits N, Sanchez-Luque FJ, Faivre J, Brennan PM, Richardson SR, Cheetham SW, Faulkner GJ. 2020. Nanopore sequencing enables comprehensive transposable element epigenomic profiling. Mol Cell 80: 915–928.e5. 10.1016/j.molcel.2020.10.024 [DOI] [PubMed] [Google Scholar]
- Fernandes JD, Zamudio-Hurtado A, Clawson H, Kent WJ, Haussler D, Salama SR, Haeussler M. 2020. The UCSC repeat browser allows discovery and visualization of evolutionary conflict across repeat families. Mob DNA 11: 13. 10.1186/s13100-020-00208-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feusier J, Watkins WS, Thomas J, Farrell A, Witherspoon DJ, Baird L, Ha H, Xing J, Jorde LB. 2019. Pedigree-based estimation of human mobile element retrotransposition rates. Genome Res 29: 1567–1577. 10.1101/gr.247965.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field AR, Jacobs FMJ, Fiddes IT, Phillips APR, Reyes-Ortiz AM, LaMontagne E, Whitehead L, Meng V, Rosenkrantz JL, Olsen M, et al. 2019. Structurally conserved primate lncRNAs are transiently expressed during human cortical differentiation and influence cell-type-specific genes. Stem Cell Reports 12: 245–257. 10.1016/j.stemcr.2018.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondon JW III, Hammock EAD, Hannan AJ, King DG. 2008. Simple sequence repeats: genetic modulators of brain function and behavior. Trends Neurosci 31: 328–334. 10.1016/j.tins.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Frank O, Giehl M, Zheng C, Hehlmann R, Leib-Mösch C, Seifarth W. 2005. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J Virol 79: 10890–10901. 10.1128/JVI.79.17.10890-10901.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes DR, Swigut T, Wysocka J. 2018. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. eLife 7: e35989. 10.7554/eLife.35989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianfrancesco O, Geary B, Savage AL, Billingsley KJ, Bubb VJ, Quinn JP. 2019. The role of SINE-VNTR-Alu (SVA) retrotransposons in shaping the human genome. Int J Mol Sci 20: 5977. 10.3390/ijms20235977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. 10.1186/gb-2010-11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenn FP, Kim JJ, Makarious MB, Iwaki H, Illarionova A, Brolin K, Kluss JH, Schumacher-Schuh AF, Leonard H, Faghri F, et al. 2020. The Parkinson's Disease Genome-Wide Association Study Locus Browser. Mov Disord 35: 2056–2067. 10.1002/mds.28197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Jeong HH, Hsieh YC, Klein HU, Bennett DA, De Jager PL, Liu Z, Shulman JM. 2018. Tau activates transposable elements in Alzheimer's disease. Cell Rep 23: 2874–2880. 10.1016/j.celrep.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, Yearout D, Kay DM, Doheny KF, Paschall J, Pugh E, et al. 2010. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet 42: 781–785. 10.1038/ng.642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HHJ. 2016. Roles for retrotransposon insertions in human disease. Mob DNA 7: 9. 10.1186/s13100-016-0065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring NL, van Bree EJ, Jordaan WS, Roels JRE, Congrains Sotomayor G, Hey TM, White FTG, Galland MD, Smidt MP, Jacobs FMJ. 2021. ZNF91 deletion in human embryonic stem cells leads to ectopic activation of SVA retrotransposons and up-regulation of KRAB zinc finger gene clusters. Genome Res 31: 551–563. 10.1101/gr.265348.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman MG, Kasanuki K, Diehl NN, Koga S, Soto A, Murray ME, Dickson DW, Ross OA. 2017. Parkinson's disease susceptibility variants and severity of Lewy body pathology. Parkinsonism Relat Disord 44: 79–84. 10.1016/j.parkreldis.2017.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Pickering E, Liu YC, Hall S, Fournier H, Katz E, Dechairo B, John S, Van Eerdewegh P, Soares H, et al. 2011. Meta-analysis for genome-wide association study identifies multiple variants at the BIN1 locus associated with late-onset Alzheimer's disease. PLoS One 6: e16616. 10.1371/journal.pone.0016616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Jacob-Hirsch J, Eyal E, Knisbacher BA, Roth J, Cesarkas K, Dor C, Farage-Barhom S, Kunik V, Simon AJ, Gal M, et al. 2018. Whole-genome sequencing reveals principles of brain retrotransposition in neurodevelopmental disorders. Cell Res 28: 187–203. 10.1038/cr.2018.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245. 10.1038/nature13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic J, Chen S, Le WD. 2005. The role of Nurr1 in the development of dopaminergic neurons and Parkinson's disease. Prog Neurobiol 77: 128–138. 10.1016/j.pneurobio.2005.09.001 [DOI] [PubMed] [Google Scholar]
- Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, Sealock J, Karlsson IK, Hägg S, Athanasiu L, et al. 2019. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat Genet 51: 404–413. 10.1038/s41588-018-0311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470. 10.1371/journal.pgen.1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr., Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. 1988. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332: 164–166. 10.1038/332164a0 [DOI] [PubMed] [Google Scholar]
- Kelleher RJ III, Shen J. 2017. Presenilin-1 mutations and Alzheimer's disease. Proc Natl Acad Sci 114: 629–631. 10.1073/pnas.1619574114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res 21: 487–493. 10.1101/gr.113985.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJJ, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208. 10.1126/science.1166066 [DOI] [PubMed] [Google Scholar]
- Lambert JC, Zelenika D, Hiltunen M, Chouraki V, Combarros O, Bullido MJ, Tognoni G, Fievet N, Boland A, Arosio B, et al. 2011. Evidence of the association of BIN1 and PICALM with the AD risk in contrasting European populations. Neurobiol Aging 32: 756.e11–756.e15. 10.1016/j.neurobiolaging.2010.11.022 [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, et al. 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45: 1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan M, Carey V. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, Jimenez-Velazquez IZ, Rogaeva E, St George-Hyslop PH, Mayeux R. 2011. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol 68: 320–328. 10.1001/archneurol.2010.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Prazak L, Chatterjee N, Grüninger S, Krug L, Theodorou D, Dubnau J. 2013. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci 16: 529–531. 10.1038/nn.3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Linthorst J, Meert W, Hestand MS, Korlach J, Vermeesch JR, Reinders MJT, Holstege H. 2020. Extreme enrichment of VNTR-associated polymorphicity in human subtelomeres: genes with most VNTRs are predominantly expressed in the brain. Transl Psychiatry 10: 369. 10.1038/s41398-020-01060-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Delisle L, Rabbani L, Wolff J, Bhardwaj V, Backofen R, Grüning B, Ramírez F, Manke T. 2021. pyGenomeTracks: reproducible plots for multivariate genomic data sets. Bioinformatics 37: 422–423. 10.1093/bioinformatics/btaa692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Yu JT, Tan L. 2015. MS4A cluster in Alzheimer's disease. Mol Neurobiol 51: 1240–1248. 10.1007/s12035-014-8800-z [DOI] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, et al. 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45: D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ. 2015. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31: 3555–3557. 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Rademakers R, Neumann M. 2010. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol 9: 995–1007. 10.1016/S1474-4422(10)70195-2 [DOI] [PubMed] [Google Scholar]
- Makino S, Kaji R, Ando S, Tomizawa M, Yasuno K, Goto S, Matsumoto S, Tabuena MD, Maranon E, Dantes M, et al. 2007. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am J Hum Genet 80: 393–406. 10.1086/512129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoodi TA, Al Shammari SA, Al-Muammar MN, Alhamdan AA, Talluri VR. 2013. Exploration of deleterious single nucleotide polymorphisms in late-onset Alzheimer disease susceptibility genes. Gene 512: 429–437. 10.1016/j.gene.2012.08.026 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. 2012. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol 30: 99–104. 10.1038/nbt.2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Fu S, Lyu C, Gontarz P, Wang T, Zhang B. 2020. Tissue-specific usage of transposable element-derived promoters in mouse development. Genome Biol 21: 255. 10.1186/s13059-020-02164-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortezaei Z, Tavallaei M. 2021. Recent innovations and in-depth aspects of post-genome wide association study (post-GWAS) to understand the genetic basis of complex phenotypes. Heredity (Edinb) 127: 485–497. 10.1038/s41437-021-00479-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, Miller W. 1988. Optimal alignments in linear space. Comput Appl Biosci 4: 11–17. 10.1093/bioinformatics/4.1.11 [DOI] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, et al. 2014. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet 46: 989–993. 10.1038/ng.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, et al. 2019. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol 18: 1091–1102. 10.1016/S1474-4422(19)30320-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28: 1166–1167. 10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Goodier JL, Zhang Y, Kazazian HHJ. 2003. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet 73: 1444–1451. 10.1086/380207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer LM, Burns KH. 2019. Transposable elements in human genetic disease. Nat Rev Genet 20: 760–772. 10.1038/s41576-019-0165-8 [DOI] [PubMed] [Google Scholar]
- Payer LM, Steranka JP, Yang WR, Kryatova M, Medabalimi S, Ardeljan D, Liu C, Boeke JD, Avramopoulos D, Burns KH. 2017. Structural variants caused by Alu insertions are associated with risks for many human diseases. Proc Natl Acad Sci 114: E3984–E3992. 10.1073/pnas.1704117114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff AL, Bubb VJ, Quinn JP, Koks S. 2021. Reference SVA insertion polymorphisms are associated with Parkinson's disease progression and differential gene expression. NPJ Park Dis 7: 44. 10.1038/s41531-021-00189-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontis J, Planet E, Offner S, Turelli P, Duc J, Coudray A, Theunissen TW, Jaenisch R, Trono D. 2019. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell 24: 724–735.e5. 10.1016/j.stem.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porubsky D, Ebert P, Audano PA, Vollger MR, Harvey WT, Marijon P, Ebler J, Munson KM, Sorensen M, Sulovari A, et al. 2021. Fully phased human genome assembly without parental data using single-cell strand sequencing and long reads. Nat Biotechnol 39: 302–308. 10.1038/s41587-020-0719-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. 2010. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9: 189. 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Savage AL, Bubb VJ, Breen G, Quinn JP. 2013. Characterisation of the potential function of SVA retrotransposons to modulate gene expression patterns. BMC Evol Biol 13: 101. 10.1186/1471-2148-13-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage AL, Wilm TP, Khursheed K, Shatunov A, Morrison KE, Shaw PJ, Shaw CE, Smith B, Breen G, Al-Chalabi A, et al. 2014. An evaluation of a SVA retrotransposon in the FUS promoter as a transcriptional regulator and its association to ALS. PLoS One 9: e90833. 10.1371/journal.pone.0090833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Cooper S, Liu JZ, Barrio-Hernandez I, Bello E, Kumasaka N, Young AMH, Franklin RJM, Johnson T, Estrada K, et al. 2021. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer's disease risk genes. Nat Genet 53: 392–402. 10.1038/s41588-020-00776-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, et al. 2016. Schizophrenia risk from complex variation of complement component 4. Nature 530: 177–183. 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, et al. 2010. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303: 1832–1840. 10.1001/jama.2010.574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Tripodis Y, Bennett DA, Chibnik LB, Crane PK, de Jager PL, Farrer LA, Saykin AJ, Shulman JM, Naj A, et al. 2014. Genome-wide association study of the rate of cognitive decline in Alzheimer's disease. Alzheimers Dement 10: 45–52. 10.1016/j.jalz.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpyleva S, Melnyk S, Pavliv O, Pogribny I, Jill James S. 2018. Overexpression of LINE-1 retrotransposons in autism brain. Mol Neurobiol 55: 1740–1749. 10.1007/s12035-017-0421-x [DOI] [PubMed] [Google Scholar]
- Simón-Sánchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, et al. 2009. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet 41: 1308–1312. 10.1038/ng.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev 9: 657–663. 10.1016/S0959-437X(99)00031-3 [DOI] [PubMed] [Google Scholar]
- Sulovari A, Li R, Audano PA, Porubsky D, Vollger MR, Logsdon GA, Warren WC, Pollen AA, Chaisson MJP, Eichler EE. 2019. Human-specific tandem repeat expansion and differential gene expression during primate evolution. Proc Natl Acad Sci 116: 23243–23253. 10.1073/pnas.1912175116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Wysocka J. 2020. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos Trans R Soc Lond B Biol Sci 375: 20190347. 10.1098/rstb.2019.0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res 27: 1623–1633. 10.1101/gr.218149.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Kapusta A, Brown CD. 2018. Transposable elements generate regulatory novelty in a tissue-specific fashion. BMC Genomics 19: 468. 10.1186/s12864-018-4850-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211. 10.1126/science.1165942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. 2014. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun 5: 5011. 10.1038/ncomms6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermunt MW, Reinink P, Korving J, de Bruijn E, Creyghton PM, Basak O, Geeven G, Toonen PW, Lansu N, Meunier C, et al. 2014. Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep 9: 767–779. 10.1016/j.celrep.2014.09.023 [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858. 10.1038/nature07730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xie G, Singh M, Ghanbarian AT, Raskó T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. 2014. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 516: 405–409. 10.1038/nature13804 [DOI] [PubMed] [Google Scholar]
- Wickham H. 2007. Reshaping data with the reshape package. J Stat Softw 21: 1–20. 10.18637/jss.v021.i12 [DOI] [Google Scholar]
- Wickham H. 2011. The split-apply-combine strategy for data analysis. J Stat Softw 40: 1–29. 10.18637/jss.v040.i01 [DOI] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. https://ggplot2.tidyverse.org. [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. 2019. Welcome to the tidyverse. J Open Source Softw 4: 1686. 10.21105/joss.01686 [DOI] [Google Scholar]
- Wijsman EM, Pankratz ND, Choi Y, Rothstein JH, Faber KM, Cheng R, Lee JH, Bird TD, Bennett DA, Diaz-Arrastia R, et al. 2011. Genome-wide association of familial late-onset Alzheimer's disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS Genet 7: e1001308. 10.1371/journal.pgen.1001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YR, Foo JN, Tan LCS, Chen CM, Prakash KM, Chen YC, Bei JX, Au WL, Chang CW, Wong TY, et al. 2013. Identification of a novel risk variant in the FUS gene in essential tremor. Neurology 81: 541–544. 10.1212/WNL.0b013e31829e700c [DOI] [PubMed] [Google Scholar]
- Yu M, Liu Y, Shen J, Lv D, Zhang J. 2016. Meta-analysis of BACE1 gene rs638405 polymorphism and the risk of Alzheimer's disease in Caucasion and Asian population. Neurosci Lett 616: 189–196. 10.1016/j.neulet.2016.01.059 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Johnson N, Juettemann T, Wilder SP, Flicek P. 2014. WiggleTools: parallel processing of large collections of genome-wide datasets for visualization and statistical analysis. Bioinformatics 30: 1008–1009. 10.1093/bioinformatics/btt737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.