Abstract
Although the carbapenem-hydrolyzing β-lactamase (CHβL) BlaB-1 is known to be in Chryseobacterium meningosepticum NCTC 10585, a second CHβL gene, blaGOB-1, was cloned from another C. meningosepticum clinical isolate (PINT). The G+C content of blaGOB-1 (36%) indicated the likely chromosomal origin of this gene. Its expression in Escherichia coli DH10B yields a mature CHβL with a pI of 8.7 and a relative molecular mass of 28.2 kDa. In E. coli, GOB-1 conferred resistance to narrow-spectrum cephalosporins and reduced susceptibility to ureidopenicillins, broad-spectrum cephalosporins, and carbapenems. GOB-1 had a broad-spectrum hydrolysis profile including penicillins and cephalosporins (but not aztreonam). The catalytic efficiency for meropenem was higher than for imipenem. GOB-1 had low amino acid identity with the class B CHβLs, sharing 18% with the closest, L-1 from Stenotrophomonas maltophilia, and only 11% with BlaB-1. Most of the conserved amino acids that may be involved in the active site of CHβLs (His-101, Asp-103, His-162, and His-225) were identified in GOB-1. Sequence heterogeneity was found for GOB-1-like and BlaB-1-like β-lactamases, having 90 to 100% and 86 to 100% amino acid identity, respectively, among 10 unrelated C. meningosepticum isolates. Each isolate had a GOB-1-like and a BlaB-1-like gene. The same combination of GOB-1-like and BlaB-1-like β-lactamases was not found in two different isolates. C. meningosepticum is a bacterial species with two types of unrelated chromosome-borne class B CHβLs that can be expressed in E. coli and, thus, may represent a clinical threat if spread in gram-negative aerobes.
Chryseobacterium meningosepticum is the most clinically important human pathogen among the Chryseobacterium and Flavobacterium genera. It is responsible for neonatal meningitis, with a mortality of up to 50% (17). C. meningosepticum is also found in pneumonia (J. Fujita, Y. Hata, and S. Irino, Letter, Lancet 335:544, 1990) and endocarditis (7, 52) in immunocompromised patients.
C. meningosepticum (formerly known as Flavobacterium meningosepticum) belonged to the Flavobacterium genus until 1994. Since then, it has been reclassified and belongs now to the Chryseobacterium genus, like Chryseobacterium indologenes and Chryseobacterium gleum (56).
C. meningosepticum is naturally resistant to most β-lactams, including carbapenems (16). A carbapenem-hydrolyzing β-lactamase, (CHβL) BlaB (BlaB-1), from C. meningosepticum NCTC 10585 (CIP 6058) has been described (46). This enzyme belongs to the Ambler class B metallo-β-lactamase group (2), with a broad substrate profile, a relative molecular mass of 26 kDa, and a pI value of 8.5 (46). Recently, in the same species, Ambler class A extended-spectrum β-lactamases have also been characterized (6, 45). These extended-spectrum β-lactamases are inhibited by clavulanic acid, cefoxitin, moxalactam, and imipenem, and their substrate profile does not include carbapenems.
Metalloenzymes usually have a broad spectrum of hydrolysis, except for CphA-1 from Aeromonas hydrophila (31, 49), and are resistant to clinically available β-lactamase inhibitors (8). Within the last few years, metallo-β-lactamases IMP-1, VIM-1, and VIM-2 have been identified as chromosome, plasmid, and/or integron located in several pathogens, such as Acinetobacter baumannii (14), Alcaligenes xylosoxydans, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Serratia marcescens (3, 21, 22, 26, 30, 36, 42). IMP-1 is widespread in Japan (50, 51). The origin of these CHβLs remains, however, unknown.
Our preliminary experiment using isoelectric focusing (IEF) electrophoresis revealed a heterogeneity of pI values in C. meningosepticum isolates. Thus, characterization of the β-lactamase content of C. meningosepticum initiated with the class A ESBLs was continued (6). We report the molecular and biochemical characterization of the CHβL GOB-1 that was weakly related to any class B CHβLs, including BlaB-1. Additionally, sequence analysis of the CHβL genes of 10 C. meningosepticum isolates revealed that each isolate possessed a combination of both types of CHβLs. A combination of two naturally occurring CHβL genes in the same bacterial species had not been reported previously.
MATERIALS AND METHODS
Bacterial strains.
C. meningosepticum PINT was isolated at the Raymond Poincaré hospital (Garches, France). C. meningosepticum AMA and GEO were isolated at the Bicêtre hospital (Le Kremlin-Bicêtre, France), both from tracheoalveolar aspirations. C. meningosepticum AB1572 and H01J100 were from Brita Bruun (11), and reference strains C. meningosepticum CIP 6057 (NCTC 10016), CIP 6058 (NCTC 10585), CIP 6059 (NCTC 10586), CIP 7830 (NCTC 11305), and CIP 79.5 (NCTC 11306) were from the Pasteur Institute (Paris, France). The C. meningosepticum isolates and reference strains were epidemiologically unrelated (data not shown).
Escherichia coli DH10B and rifampin-resistant E. coli JM109 were used for cloning and conjugation assays, respectively, and have been described previously (40, 41). C. meningosepticum isolates were identified as previously described (6, 39, 56). All strains were stored at −70°C in Trypticase soy (TS) broth supplemented with 15% glycerol until testing.
Antimicrobial agents and MIC determinations.
The antimicrobial agents used in this study have been described (41). MICs were determined by an agar dilution technique on Mueller-Hinton agar (Sanofi-Diagnostics Pasteur) with an inoculum of 104 CFU per spot (34). The plates were incubated at 35°C for 18 h before MIC determinations were performed as previously described (34).
Cloning experiments, PCR amplifications, and recombinant plasmids.
Genomic DNAs were extracted as described previously (35). Fragments from Sau3AI partially digested genomic DNA from C. meningosepticum PINT were cloned in pBK-CMV phagemid (Stratagene, Ozyme, Amsterdam, The Netherlands) (Table 1) and expressed in E. coli DH10B as previously described (35). Antibiotic-resistant colonies were selected onto amoxicillin (30 μg/ml) and kanamycin (30 μg/ml) containing TS agar plates.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| pBK-CMV phagemid | Neomycin and kanamycin resistant | Stratagene |
| pPCRScript Cam SK | Chloramphenicol resistant | Stratagene |
| pBS2 | A 2.4-kb DNA fragment from C. meningosepticum PINT that contained blaGOB-1 in the BamHI site of pBK-CMV | This study |
| pBS3 | Entire blaGOB-1 gene in the SrfI site of pPCRScript Cam SK | This study |
| pBS4 | Entire blaBlaB-1 in the SrfI site of pPCRScript Cam SK | This study |
Recombinant plasmid DNA was obtained from 100-ml TS broth cultures grown overnight in the presence of amoxicillin (30 μg/ml) at 37°C. Plasmid DNAs were recovered by using Qiagen columns (Qiagen, Courtaboeuf, France) before restriction digest analyses.
16S rDNA fragments were amplified by PCR using the universal 16S RNA primers 5′-AGAGTTTGATCHTGGYTYAGA-3′ and 5′-ACGGYTACCTTGTTACGACTTC-3′, where Y is C or T and H is A, C, or T (4), and genomic DNAs of C. meningosepticum isolates as the template. Primers used to amplify blaGOB-1-like genes were primer 1 (5′-GCTATGAGAAATTTTGCTACACTG-3′) or primer 3 (5′-GGAGTGGTAAAAGATGAAATGTGC-3′) and primer 2 (5′-TCATACTTATTTATCTTGGG-3′) (Fig. 1).
FIG. 1.
Nucleotide sequence of a 2,384-bp DNA fragment of recombinant plasmid pBS2 carrying blaGOB-1 and the 117 bp of the 3′ end of the endo-beta-N-acetylglucosaminidase gene of C. meningosepticum PINT. The deduced amino acid sequences are given in a single-letter code. The start and stop codons of the blaGOB-1 gene and the stop codon of the endo-beta-N-acetylglucosaminidase gene are in bold. The vertical arrow indicates the peptide leader cleavage site in E. coli as determined by N-terminal sequencing. The putative −35 and −10 sequences of the putative promoter and ribosome binding site (RBS) for blaGOB-1 are underlined. Primers 1, 2, and 3 used to PCR amplify blaGOB-1-like genes from other C. meningosepticum isolates are indicated by an arrow.
In order to establish a comparison of MICs of β-lactams for E. coli DH10B harboring either blaGOB-1 or blaBlaB-1, PCR products of blaGOB-1 from C. meningosepticum PINT were obtained using primers 2 and 3, and those for blaBlaB-1 from C. meningosepticum CIP 6058 were obtained using primers 4 (5′-GTGAATGTAGCAGAGTGTTAATG-3′) and primer 5 (5′-GTTGTCTGGTTAAGCGTTCG-3′) located at the 5′ and the 3′ end of blaBlaB-1 (Table 1) (46). Each PCR fragment was cloned into the same pPCR-Script CamSK vector (Stratagene) and electrotransformed into E. coli DH10B.
Conjugation assays, plasmid content, and Southern hybridization.
Plasmid DNA extractions of C. meningosepticum isolates were attempted according to two different methods (18, 24). Direct transfer of resistance genes into in vitro-obtained rifampin-resistant E. coli JM109 was attempted by liquid and solid conjugation assays and by electroporation of the putative plasmid DNA suspension into E. coli DH10B (41). Transconjugants and electroporants were selected on TS agar plates containing either rifampin (200 μg/ml) and amoxicillin (30 μg/ml) or amoxicillin, respectively. Southern hybridizations were performed using a 0.8% electrophoresis gel containing unrestricted genomic DNAs of C. meningosepticum isolates and a PCR-prepared internal probe for blaGOB-1 (see below). Visualization was made using the ECL nonradioactive hybridization kit as described by the manufacturer (Amersham Pharmacia Biotech, Orsay, France).
DNA sequencing and protein analysis.
Sequencing of the 2.4-kb cloned DNA fragment of recombinant plasmid pBS2, of 16S rDNA fragments, and of PCR products that contained blaBlaB-1-like and blaGOB-1-like genes was performed using an ABI 373 sequencer (Applied Biosystems, Foster City, Calif.). The nucleotide and deduced protein sequences were analyzed with software available over the Internet at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov.) and at Pedro's BioMolecular Research Tools website (http://www.fmi.ch/biology/research_tools.html.), and hydrophobicity analysis of the N-terminal region of the open reading frame (ORF) was performed as described (http://genome.cbs.dtu.dk./services/SignalP/ [25]). Multiple nucleotide or protein sequence alignments were carried out using the program ClustalW (http://www2.ebi.ac.uk/clustalw). A dendrogram of GOB-1 β-lactamase was derived from the multiple sequence alignment by a parsimony method using the phylogeny package PAUP (Phylogenetic Analysis Using Parsimony) version 3.0 (53).
β-Lactamase extraction.
A culture of E. coli DH10B (pBS2) was grown overnight at 37°C in 4 liters of TS broth containing kanamycin (30 μg/ml) and amoxicillin (30 μg/ml). Bacterial suspensions were pelleted, resuspended in 40 ml of 20 mM Tris-HCl buffer (pH 8), disrupted by sonification (three times at 50 W for 30 s using a Vibra Cell 75022 Phospholyser [Bioblock, Illkirch, France]), and centrifuged for 1 h at 48,000 × g at 4°C. Nucleic acids were precipitated by addition of 0.2 M spermine (7% [vol/vol]) (Sigma, Saint-Quentin Fallavier, France) overnight at 4°C. This suspension was ultracentrifuged at 100,000 × g for 1 h at 4°C, and the supernatant contained the β-lactamase extract.
β-Lactamase purification.
The β-lactamase extract from E. coli DH10B (pBS2) was filtered through a 0.45-μm-pore-size filter (Millipore, Saint-Quentin-en-Yvelines, France) prior its loading onto a preequilibrated Q-Sepharose column (Amersham Pharmacia Biotech). The enzyme which was recovered in the flowthrough was then dialyzed overnight at 4°C against 50 mM phosphate buffer, pH 7. The enzymatic fraction was then loaded onto a preequilibrated S-Sepharose column (Amersham Pharmacia Biotech). The enzyme was eluted by a linear NaCl gradient (0 to 1 M) in phosphate buffer (pH 7). The β-lactamase was eluted at a concentration of 170 mM NaCl. The fraction containing the β-lactamase activity was dialyzed overnight against 30 mM cacodylate buffer, pH 6.5, containing 50 μM ZnCl2. The specific activities of the β-lactamase extract and of the purified β-lactamase from E. coli DH10B (pBS2) were compared using 100 μM of imipenem as substrate as previously described (40).
N-terminal sequencing and isoelectric focusing.
In order to determine the site for cleavage of the mature protein of GOB-1 β-lactamase, the purified enzyme was submitted to an Edman analysis (19) at the laboratory for protein microsequencing at the Pasteur Institute, Paris, France. Purified enzyme and marker proteins were subjected to sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis (20 mA, 5 h, room temperature). Proteins were then electrotransferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore) by using the Mini Protean II transfer cell (8 by 7.3 cm) (Bio-Rad) in 50 mM Tris–50 mM borate buffer (pH 8.7) at room temperature (3.5 V/cm, overnight). The membrane was then rinsed in distilled water and stained with a solution made of 0.05% Coomassie brilliant blue R-250 in methanol and water (50:50 [vol/vol]) for 5 min. The membrane was then destained in methanol and water (50:40 [vol/vol]) and acetate and water (10:40 [vol/vol]). The protein band was then excised with a razor blade and allowed to air dry. The amino-terminal sequence of the β-lactamase was determined with an automated Edman sequencer on a model 473A gas phase sequencer (Applied Biosystems).
The purified enzyme from a culture of E. coli DH10B (pBS2) and β-lactamase extracts from cultures of 10 C. meningosepticum isolates were subjected to analytical IEF on an ampholine polyacrylamide gel with a pH of 3.5 to 9.5 (Ampholine PAG plate; Amersham Pharmacia Biotech) for 90 min at 1,500 V, 50 mA, and 30 W. The focused β-lactamases were detected by overlaying the gel with 1 mM nitrocefin (Oxoid, Paris, France) or with an iodine-starch agar gel containing 0.5% (wt/vol) of imipenem (31) in 100 mM phosphate buffer (pH 7.0). These gels were preincubated with or without 100 mM EDTA (ethylenediaminetetraacetic acid) and with or without 5 mM clavulanic acid (38). The pI values were determined and compared to those of known β-lactamases.
Kinetic measurements and relative molecular mass determination.
Purified β-lactamase was used for kinetic measurements performed at 30°C in 30 mM cacodylate (pH 6.5) supplemented with 50 μM ZnCl2 as described previously (46). The rates of hydrolysis were determined with a Pharmacia ULTROSPEC 2000 spectrophotometer and were computer analyzed using the SWIFT II software (Amersham Pharmacia Biotech).
Km and kcat values were determined by analyzing the β-lactam hydrolysis under initial rate conditions by using the Eadie-Hoffstee linearization of the Michaelis-Menten equation as previously described (13, 41).
Various concentrations of EDTA or clavulanic acid were preincubated with the enzyme for 10 min at 30°C before testing the rate of imipenem hydrolysis. The 50% inhibitory concentration (IC50) of these inhibitors was then determined.
The relative molecular mass of the purified β-lactamase was determined by gel filtration using a 1.6- by 47-cm column packed with Superdex 75 (Amersham Pharmacia Biotech) equilibrated and eluted with phosphate buffer (pH 7) containing 150 mM NaCl. Each elution peak was tested for β-lactamase activity by using nitrocefin as substrate. The peak that showed the highest β-lactamase activity was linearly plotted against the logarithm of the molecular masses of the standard proteins (Amersham Pharmacia Biotech) to determine the relative molecular mass of the purified β-lactamase.
Nucleotide sequence accession numbers.
The nucleotide and deduced β-lactamase amino acid sequences reported in this work have been assigned to the GenBank and EMBL databases under the accession no. AF189290 to AF189305 and AF090141. The nucleotide sequences of the 16S rDNAs have been assigned to the accession no. AF207070 to AF207079.
RESULTS
Cloning and sequence analysis of blaGOB-1.
Partially Sau3AI-digested genomic DNA from C. meningosepticum PINT was cloned into the BamHI site of pBK-CMV. Three recombinant E. coli DH10B clones were obtained. One of them, harboring pBS2 (the smallest insert [2.4 kb]), was selected for further studies.
DNA sequence analysis of the 2,384-bp insert of pBS2 revealed an ORF of 873 bp, encoding a 290-amino-acid preprotein (Fig. 1). Putative −35 (TTGAAA) and −10 (TTTATT) promoter regions and a ribosome binding site (AAAACA) were found along with a putative ATG initiation codon at position 243 (Fig. 1).
The G+C content of this ORF was 36%, which lies close to the G+C ratio found for other C. meningosepticum genes recorded in the EMBL and GenBank sequence database (36.1 to 41.6%). The codon usage of this ORF was also similar to those calculated for the set of these C. meningosepticum genes (data not shown). From the sequencing data, one would expect the first 18 amino acids of this ORF, which contains numerous hydrophobic residues found by hydrophobicity analysis, to be the leader peptide (Fig. 1). This was indeed the case, since Edman analysis (nine cycles) determined the N-terminal sequence of the purified protein from a culture of E. coli DH10B (pBS2) cells as being QVVK. The cleavage site of the leader peptide was therefore deduced to be just after the alanine residue at position 18 (Fig. 1).
Further DNA sequence analysis of the downstream region of this ORF identified the 3′ end terminal sequence of an endo-beta-N-acetylglucosaminidase F1 gene (Fig. 1) (54).
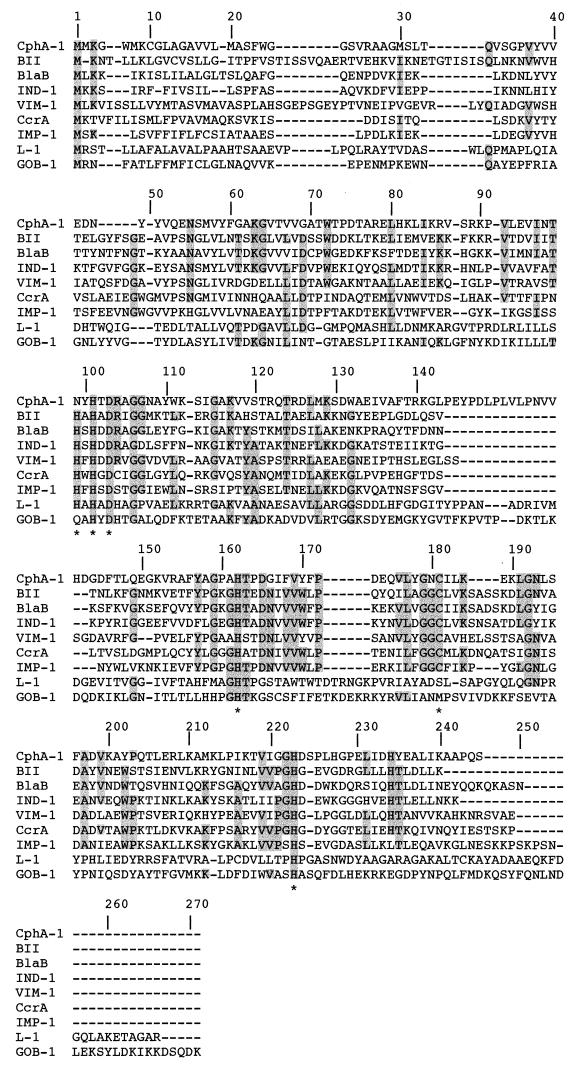
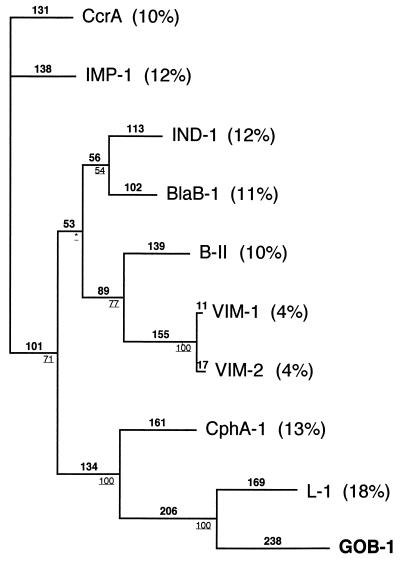
The mature protein (named GOB-1 for class B β-lactamase of C. meningosepticum) expressed in E. coli DH10B cells had a relative molecular mass determined by gel filtration to be 28.2 kDa. His-101, Asp-103, His-162, and His-225 identified by biochemical analysis or by crystal structure analysis as interacting with a Zn2+ cofactor in Bacillus cereus 569H/9 enzyme or in CcrA were found in GOB-1 (Fig. 2) (10, 12). However, the histidine residue at position 99 found in most class B CHβLs was changed for a glutamine residue in GOB-1 (Fig. 2). The comparison of GOB-1 with other class B β-lactamases revealed only weak identity (Fig. 3). The highest percentage of identity was with L-1 from Stenotrophomonas maltophilia (18%) and only 11% with BlaB-1 from C. meningosepticum.
FIG. 2.
Multiple-sequence alignment of amino acid sequence of GOB-1 from C. meningosepticum PINT isolate with those of eight class B CHβLs. Sequence comparison was performed first by aligning the proteins by using the ClustalW program. Then, adjustments were made to reduce the number of gaps and to maintain alignment of the putative active residues of the active sites. The origins of metallo-β-lactamases are as follows: CphA-1 from A. hydrophila AE036 (31), BII from B. cereus 5/B/6 (29), BlaB (BlaB-1) from C. meningosepticum CIP 6058 (NCTC 10585) (46), IND-1 from C. indologenes (5), VIM-1 from P. aeruginosa VR-143/97 (28), CcrA from B. fragilis TAL 3636 (44), IMP-1 from S. marcescens TN9106 (36), and L-1 from S. maltophilia IID1275 (57). Amino acids that were identical for at least five out of nine aligned amino acid sequences are shaded in grey. Stars refer to conserved amino acids identified by crystal structure determination as interacting in the binding to the Zn2+ cofactor or to the water molecule in the B. cereus 569H/9 enzyme or in CcrA (12, 20). The numbering scheme refers to the CcrA enzyme (44). Dashes indicate gaps introduced to optimize the alignment.
FIG. 3.
Dendrogram obtained for nine representative CHβLs calculated with ClustalW followed by adjustments to reduce the number of gaps and to maintain alignment of the residues identified in the active sites of some CHβLs. Branch lengths are to scale and proportional to the number of amino acid changes. The percentages at the branching point (bold and underlined) refer to the number of times a particular node was found in 100 bootstrap replications (the stars indicate uncertainty of nodes with bootstrap values of less than 50%). The distance along the vertical axis has no significance. BlaB-1 (BlaB) and GOB-1 were from C. meningosepticum, IND-1 was from C. indologenes, CphA-1 was from A. hydrophila, L-1 was from S. maltophilia, BII was from B. cereus, VIM-1 and VIM-2 were from P. aeruginosa, CcrA was from B. fragilis, and IMP-1 was from S. marcescens. Percent amino acid identities to GOB-1 are indicated in parentheses.
β-Lactam resistance phenotype and plasmid analysis.
The MICs of β-lactams for C. meningosepticum PINT showed that it was resistant to all tested β-lactams except piperacillin, as previously reported (Table 2) (6, 16). Similar MICs (within a two-dilution range) were obtained for the C. meningosepticum isolates except for C. meningosepticum H01J100, for which MICs of all β-lactams were lower (data not shown).
TABLE 2.
MICs of β-lactams for C. meningosepticum PINT, E. coli DH10B (pBS3), E. coli DH10B (pBS4), and the E. coli DH10B reference strain
| β-Lactam | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| C. meningosepticum PINT | E. coli DH10B (pBS3)a | E. coli DH10B (pBS4)b | E. coli DH10B | |
| Amoxicillin | 256 | 64 | 128 | 4 |
| Ticarcillin | 256 | 64 | 256 | 4 |
| Piperacillin | 32 | 2 | 4 | 1 |
| Cephalothin | 512 | 32 | 16 | 2 |
| Cefepime | 32 | 0.06 | 0.03 | 0.03 |
| Cefoxitin | 32 | 16 | 2 | 1 |
| Cefpirome | 32 | 0.5 | 0.06 | 0.06 |
| Ceftazidime | 256 | 16 | 0.5 | 0.5 |
| Cefotaxime | 64 | 0.25 | 0.12 | 0.12 |
| Aztreonam | >512 | 0.25 | 0.25 | 0.25 |
| Moxalactam | 64 | 1 | 0.12 | 0.12 |
| Imipenem | 32 | 0.5 | 0.5 | 0.12 |
| Meropenem | 16 | 0.12 | 0.12 | 0.06 |
E. coli DH10B (pBS3) expressed GOB-1.
E. coli DH10B (pBS4) expressed BlaB-1.
Recombinant plasmids pBS3 and pBS4 were constructed by cloning the ORF of blaGOB-1 and blaBlaB-1 in plasmid pPCR-Script Cam SK, respectively, without the putative promoter regions of these β-lactamase genes (Table 1). E. coli DH10B (pBS3) showed a decreased susceptibility to all β-lactams except to aztreonam (Table 2), thus indicating that blaGOB-1 was involved at least partially in the resistance to carbapenems of C. meningosepticum PINT. MICs of penicillins were higher against E. coli DH10B (pBS4) than those against E. coli DH10B (pBS3), while the opposite was found for cephalosporins (Table 2). Both recombinant E. coli strains remained fully susceptible to aztreonam. MICs of carbapenems were similar against E. coli DH10B (pBS3) and E. coli DH10B (pBS4), although the amino acid identity of GOB-1 and BlaB-1 was low.
Plasmid analysis and attempts to transfer the β-lactam resistance markers from C. meningosepticum to E. coli failed, thus suggesting the likely chromosomal origin of blaGOB-1.
Biochemical properties of GOB-1.
IEF analysis revealed that E. coli DH10B (pBS2) produced only one β-lactamase activity with a pI value of 8.7. This pI value did not correspond to the pI value of 8.3 found for the carbapenem-hydrolyzing activity identified in C. meningosepticum PINT.
Specific activity prior to and after purification enabled us to determine the 400-fold purification factor for GOB-1 from E. coli DH10B (pBS2). The specific activity of the purified enzyme was 73.2 μmol · min−1 · mg of protein−1.
Kinetic parameters of GOB-1 revealed a broad spectrum of hydrolysis with a strong activity against meropenem, compared to that against imipenem (Table 3). GOB-1 β-lactamase has a strong activity against amoxicillin, benzylpenicillin, piperacillin, and extended-spectrum cephalosporins. Hydrolysis of aztreonam was not detectable. The hydrolytic activity of GOB-1 β-lactamase was inhibited by EDTA (IC50, 25 μM) but not by class A β-lactamase inhibitors, such as clavulanic acid (IC50, >10 mM). GOB-1 was therefore classified the functional CHβL group 3a according to the Bush classification (9, 43).
TABLE 3.
Kinetic parameters of β-lactam antibiotics for the purified carbapenem-hydrolyzing β-lactamase GOB-1
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (μM−1·s−1)a |
|---|---|---|---|
| Benzylpenicillin | 109 | 204 | 1.87 |
| Amoxicillin | 1,024 | 357 | 0.35 |
| Ticarcillin | 147 | 76.3 | 0.52 |
| Piperacillin | 169 | 279 | 1.66 |
| Cephalothin | 24 | 16.1 | 0.67 |
| Cefepime | 52.6 | 10.7 | 0.20 |
| Cefoxitin | 46.8 | 11.9 | 0.25 |
| Cefuroxime | 26.9 | 26.3 | 0.98 |
| Ceftazidime | 71.4 | 54.5 | 0.76 |
| Cefotaxime | 51.1 | 43.7 | 0.85 |
| Aztreonam | NDb | <0.5 | —c |
| Moxalactam | 78.8 | 10.4 | 0.13 |
| Imipenem | 60 | 39.4 | 0.66 |
| Meropenem | 5.4 | 29.2 | 5.34 |
Standard deviations were within 10%.
ND, not determinable.
—, the hydrolysis parameter could not be calculated.
Distribution of GOB-1-like and BlaB-like β-lactamases and 16S rDNA sequencing.
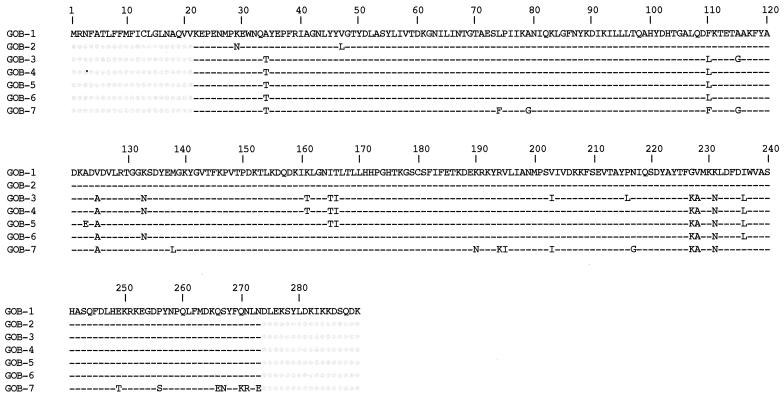
EDTA-inhibited activities obtained by comparison of pI values with or without EDTA were heterogeneous for the 10 C. meningosepticum isolates (Table 4). Only one EDTA-inhibited hydrolysis activity was detected for C. meningosepticum PINT, AMA, 7830, 79.5, CIP 6057, AB1572, and H01J100 isolates. Three isolates produced two EDTA-inhibited activities (Table 4). Additionally, clavulanic acid-inhibited β-lactamase activities varied from one isolate to the other (Table 4). Southern hybridization experiments using nonrestricted genomic DNA of C. meningosepticum isolates and a PCR-amplified 731-bp fragment internal to blaGOB-1 as a probe yielded a hybridization signal that corresponded to the chromosomal band (data not shown), showing that each C. meningosepticum isolate possessed a chromosomally located blaGOB-1-like gene. PCR fragments of GOB-1- and BlaB-1-like genes of 10 C. meningosepticum isolates (except for a blaGOB-1-like gene from C. meningosepticum CIP 7830 that failed to yield a PCR-positive result) were sequenced on both strands. Sequences for entire blaBlaB-1-like genes and a partial portion of the blaGOB-1-like genes yielding 252 out of 290 amino acids were obtained using the designed PCR primers. The deduced amino acid sequences revealing heterogeneity among GOB-1-like β-lactamases having 90 to 100% amino acid identity (Fig. 4).
TABLE 4.
pI values of β-lactamase activity detected in C. meningosepticum isolates and in E. coli DH10B harboring pBS2 (GOB-1) or pBS4 (BlaB-1) and the corresponding GOB-1-like and BlaB-1-like sequences
| Strains | pI values
|
Shared amino acid sequence
|
||
|---|---|---|---|---|
| Clavulanic acid-inhibited β-lactamase activity | EDTA-inhibited β-lactamase activity | GOB-1 like | BlaB-1 like | |
| C. meningosepticum PINT | 7.6 | 8.3 | GOB-1 | BlaB-1 |
| C. meningosepticum CIP 6058 | 8.5 | 7.8, 8.7 | GOB-5 | BlaB-1 |
| C. meningosepticum AMA | 8.5 | 8.3 | GOB-6 | BlaB-2 |
| C. meningosepticum GEO | 7.0 | 7.5, 8.5 | GOB-4 | BlaB-3 |
| C. meningosepticum CIP 7830 | 7.5 | 8 | +a | BlaB-6 |
| C. meningosepticum CIP 6059 | 8.4 | 7.5, 8.5 | GOB-3 | BlaB-3 |
| C. meningosepticum CIP 79.5 | 7.6 | 8.4 | GOB-7 | BlaB-5 |
| C. meningosepticum AB1572 | 7.6 | 8.3 | GOB-1 | BlaB-7 |
| C. meningosepticum H01J100 | 8.1 | 8.3 | GOB-2 | BlaB-8 |
| C. meningosepticum CIP 6057 | 7.6 | 8.6 | GOB-1 | BlaB-4 |
| E. coli DH10B (pBS2) | —b | 8.7 | GOB-1 | — |
| E. coli DH10B (pBS4) | — | 8.5 | — | BlaB-1 |
+, GOB-1-like positive results by hybridization but negative by PCR amplification.
—, not detectable.
FIG. 4.
Amino acid comparison of the GOB-1-like β-lactamases from nine C. meningosepticum isolates. Dashes indicate identical amino acids, and dots indicate undetermined sequences. GOB-1 was from C. meningosepticum PINT, CIP 6057, and AB1572, GOB-2 was from C. meningosepticum HO1J100, GOB-3 was from C. meningosepticum CIP 6059, GOB-4 was from C. meningosepticum GEO, GOB-5 was from C. meningosepticum CIP 6058, GOB-6 was from C. meningosepticum AMA, and GOB-7 was from C. meningosepticum CIP 79.5. Numbering is according to the GOB-1 sequence.
Alignment of the BlaB-1-like sequences of 10 C. meningosepticum isolates also revealed heterogeneity, with 86 to 100% amino acid identity (Fig. 5). The same GOB-1-like or BlaB-1-like sequences were found in several isolates, for example, GOB-1 in C. meningosepticum PINT, AB1572, and CIP 6057 and BlaB-1 in C. meningosepticum PINT and CIP 6058 (Table 4). However, within two given C. meningosepticum isolates, the same combination of GOB-1-like and BlaB-1-like β-lactamases was not found (Table 4). 16S rDNA sequencing identified homogeneous sequences (from 96 to 99% identity) among the studied C. meningosepticum isolates (data not shown, accession numbers available).
FIG. 5.
Amino acid comparison of the BlaB-1-like β-lactamases from 10 C. meningosepticum isolates. Dashes indicate identical amino acids. BlaB-1 was from C. meningosepticum CIP 6058 and PINT, BlaB-2 was from C. meningosepticum AMA, BlaB-3 was from C. meningosepticum GEO and CIP 6059, BlaB-4 was from C. meningosepticum CIP 6057, BlaB-5 was from C. meningosepticum CIP 79.5, BlaB-6 was from C. meningosepticum CIP 7830, BlaB-7 was from C. meningosepticum AB1572, and BlaB-8 was from C. meningosepticum H01J100. Numbering is according to the BlaB-1 sequence.
DISCUSSION
GOB-1 is a broad-spectrum class B β-lactamase like the previously identified BlaB-1 (BlaB) in C. meningosepticum NCTC 10585 (CIP 6058). Comparison of their kinetic constants revealed that BlaB-1 hydrolyzed benzylpenicillin better than GOB-1. Additionally, a comparison of MICs of β-lactams for E. coli expressing either GOB-1 or BlaB-1 revealed that GOB-1 hydrolyzed ceftazidime and cefoxitin more significantly than BlaB-1 does (hydrolysis constants of BlaB-1 for ceftazidime are not available [46]). GOB-1 β-lactamase hydrolysis of meropenem was greater than that of imipenem. Imipenem is usually hydrolyzed better than meropenem by class B CHβLs with two exceptions, the group 3a B. cereus II enzyme and the group 3b AsbM1 enzyme from A. hydrophila (43). GOB-1, like BlaB-1, conferred only a slight increase in the MICs of carbapenems once its gene was cloned on a multicopy plasmid and expressed in E. coli. Similar results have been found for the CHβLs IMP-1, VIM-1, and VIM-2 (28, 36, 42). These results, together with data for kinetic constants of carbapenems, may indicate that an additional decrease of outer membrane permeability for carbapenems may explain the resistance to carbapenems observed for C. meningosepticum (32). In this regard, the pI value of 8.3 for the EDTA-inhibited β-lactamase identified in C. meningosepticum PINT did not correspond to the pI value of 8.7 for GOB-1 expressed in E. coli DH10B. This result may be explained either by a pI value of 8.3 corresponding to BlaB-1 also found in C. meningosepticum PINT, by a weak or lack of expression of GOB-1, or to differences in leader peptide cleavage in E. coli and in C. meningosepticum. Such a difference in the N-terminal end of the mature protein of GOB-1 may lead to its low concentration in the periplasmic space in E. coli. Whatever the reason is, the low level of resistance to carbapenems conferred by GOB-1 in E. coli may explain its difficulty in being detected once expressed in enterobacterial clinical isolates. Studies of the pI values of the C. meningosepticum isolates revealed that GOB-1-like and BlaB-1-like β-lactamases may not always be expressed since for some C. meningosepticum isolates, only one EDTA-inhibited β-lactamase was evidenced by IEF gel electrophoresis although two CHβL genes had been identified (Table 4). However, since the pI values of GOB-1 and BlaB-1 were very close, one cannot exclude that they cannot be distinguished on the IEF gel.
Several variants of CHβLs have been found in S. maltophilia, A. hydrophila, and Bacteroides fragilis. However, in these species, it was determined that variants from reference CHβLs had 88 to 95% identity (31, 38, 43, 47, 48). To the best of our knowledge, it is the first time that two CHβLs with only 11% amino acid identity were identified in the same bacterial species. The significance of this result remains to be determined. The regulation of these CHβLs, if any, would be of interest as described for the A. hydrophila CHβL (1). It may be hypothesized that CHβLs in C. meningosepticum may counteract the effect of antibiotics produced by this Chryseobacterium species (33, 37). Additionally, the presence of two CHβL genes in C. meningosepticum may be used as a tool for a PCR-based identification of this species.
The amino acid sequence of GOB-1 allowed its classification in the sequence-based subclass B3 of metallo-CHβLs (43) along with L-1, the only other member of this subgroup, whereas BlaB-1 is a member of the subclass B1.
The primary structure of GOB-1 β-lactamase keeps most of the conserved amino acid residues of class B β-lactamases that act in the interaction with the Zn2+ cofactor or with the water molecule located in the active site, as shown for CcrA (58) or for L-1 (55): His-101, Asp-103, His-162, and His-225 (43). However, like CphA-1, GOB-1 lacks the His-99 residue, which is also involved in Zn2+ binding, but possesses instead a glutamine residue (asparagine in CphA) (31). Therefore, the absence of His-99 does not seem to be involved in narrowing the spectrum of GOB-1 (Fig. 2).
GOB-1 β-lactamase, like L-1, lacks a cysteine residue at position 181 that is involved in the interaction with a Zn2+ ion. In L-1, Cys-181 function is replaced by a histidine residue located at position 104 (55). It could be the same for GOB-1, which possesses also a histidine residue at this same position.
While this work was in progress, two BlaB variants were reported in GenBank, BlaC from C. meningosepticum NCTC 10016 and BlaB-2 from C. meningosepticum 97/P/5443. We have also identified BlaC (BlaB-4) from the same C. meningosepticum NCTC 10016 isolate and BlaB-2 from another C. meningosepticum isolate (C. meningosepticum AMA).
Although some genetic variation was identified among BlaB-1-like and GOB-1-like sequences, none of the studied C. meningosepticum isolates could be assigned to a special C. meningosepticum subgroup. Indeed, the C. meningosepticum isolates had 96 to 99% identity, according to the results of 16S rDNA sequencing.
Time will tell if gram-negative aerobes, such as C. meningosepticum, may be a reservoir for diffusion of CHβL genes to opportunistic pathogens. P. aeruginosa and Acinetobacter spp. that share low natural permeability towards most β-lactams are good candidates for expressing these carbapenem resistance genes. Finally, since C. meningosepticum CHβLs provide only a low level of resistance to carbapenems once they are expressed in E. coli, their routine detection in gram-negative clinical pathogens shall be performed at best with PCR-based methods previously described for blaIMP-1 detection among American and Japanese isolates (23, 52).
ACKNOWLEDGMENTS
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche, Université Paris XI, Faculté de Médecine Paris Sud (grant UPRES, JE-2227), and the French network “Les β-lactamases: de l'observation clinique à la structure,” France.
We thank E. Ronco and B. Bruun for the gift of some C. meningosepticum isolates and L. Poirel for precious advice.
REFERENCES
- 1.Alksne L E, Rasmussen B A. Expression of the AsbA1, OXA-12, and AsbM1 beta-lactamases in Aeromonas jandaei AER 14 is coordinated by a two-component regulon. J Bacteriol. 1997;179:2006–2013. doi: 10.1128/jb.179.6.2006-2013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Oshuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avidor B, Kletter Y, Abulafia S, Golan Y, Ephros M, Giladi M. Molecular diagnosis of cat scratch disease: a two-step approach. J Clin Microbiol. 1997;35:1924–1930. doi: 10.1128/jcm.35.8.1924-1930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellais S, Léotard S, Poirel L, Naas T, Nordmann P. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol Lett. 1999;171:127–132. doi: 10.1111/j.1574-6968.1999.tb13422.x. [DOI] [PubMed] [Google Scholar]
- 6.Bellais S, Poirel L, Naas T, Girlich D, Nordmann P. Genetic-biochemical analysis and distribution of the Ambler class A β-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob Agents Chemother. 2000;44:1–9. doi: 10.1128/aac.44.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloch K C, Nadarajah R, Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine (Baltimore) 1997;76:30–41. doi: 10.1097/00005792-199701000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Bush K. Metallo-beta-lactamases: a class apart. Clin Infect Dis. 1998;27(Suppl. 1):S48–S53. doi: 10.1086/514922. [DOI] [PubMed] [Google Scholar]
- 9.Bush K, Jacoby G A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carfi A, Pares S, Duee E, Galleni M, Duez C, Frère J-M, Dideberg O. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colding H, Bangsborg J, Fiehn N E, Bennekov T, Bruun B. Ribotyping for differentiating Flavobacterium meningosepticum isolates from clinical and environmental sources. J Clin Microbiol. 1994;32:501–505. doi: 10.1128/jcm.32.2.501-505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Concha N O, Rasmussen B A, Bush K, Herzberg O. Crystal structure of the wide-spectrum binuclear zinc metallo-β-lactamase from Bacteroides fragilis. Structure. 1996;4:823–836. doi: 10.1016/s0969-2126(96)00089-5. [DOI] [PubMed] [Google Scholar]
- 13.Cornish-Bowden A. Fundamentals of enzyme kinetics. Seattle, Wash: Portland Press, Inc.; 1995. Graphs of the Michaelis-Menten equation; pp. 30–37. [Google Scholar]
- 14.Da Silva G J, Leitão R. Emergence of carbapenem-hydrolyzing enzymes in Acinetobacter baumannii clinical isolates. J Clin Microbiol. 1999;37:2109–2110. doi: 10.1128/jcm.37.6.2109-2110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felici A, Amicosante G, Oratore A, Strom R, Ledent P, Joris B, Fanuel L, Frère J M. An overview of the kinetic parameters of class B β-lactamases. Biochem J. 1993;291:151–155. doi: 10.1042/bj2910151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser S L, Jorgensen J H. Reappraisal of the antimicrobial susceptibilities of Chryseobacterium and Flavobacterium species and methods for reliable susceptibility testing. Antimicrob Agents Chemother. 1997;41:2738–2741. doi: 10.1128/aac.41.12.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George R, Cochran C, Wheeler W. Epidemic meningitis of the newborn caused by Flavobacterium. Am J Dis Child. 1961;101:296–304. doi: 10.1001/archpedi.1961.04020040024005. [DOI] [PubMed] [Google Scholar]
- 18.Hansen J B, Olsen R H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978;135:227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewick R M, Hunkapiller M W, Le Hoo D, Dreyer W J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981;256:7990–7997. [PubMed] [Google Scholar]
- 20.Hussain M, Carlino A, Madonna M J, Lampen J O. Cloning and sequencing of the metallothioprotein β-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J Bacteriol. 1985;164:223–229. doi: 10.1128/jb.164.1.223-229.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito H, Arakawa Y, Ohsukan S, Wacharotayankun R, Kato N, Ohta M. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother. 1995;39:824–829. doi: 10.1128/aac.39.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyobe S, Yamada H, Minami S. Insertion of a carbapenemase gene cassette into an integron of a Pseudomonas aeruginosa plasmid. J Antimicrob Chemother. 1996;38:1114–1115. doi: 10.1093/jac/38.6.1114. [DOI] [PubMed] [Google Scholar]
- 23.Jones R N, Pfaller M A, Marshall S A, Hollis R J, Wilke W W. Antimicrobial activity of 12 broad-spectrum agents tested against 270 nosocomial blood stream infection isolates caused by non-enteric gram-negative bacilli: occurrence of resistance, molecular epidemiology, and screening for metallo-enzymes. Diagn Microbiol Infect Dis. 1997;29:187–192. doi: 10.1016/s0732-8893(97)81808-1. [DOI] [PubMed] [Google Scholar]
- 24.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 26.Laraki N, Galleni M, Thamm I, Riccio M L, Amicosante G, Frère J-M, Rossolini G M. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother. 1999;43:890–901. doi: 10.1128/aac.43.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laraki N, Franceschini N, Rossolini G M, Santucci P, Meunier C, de Pauw E, Amicosante G, Frère J-M, Galleni M. Biochemical characterization of the Pseudomonas aeruginosa 101/477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob Agents Chemother. 1999;43:902–906. doi: 10.1128/aac.43.4.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauretti L, Riccio M L, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini G M. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–1590. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim H M, Pene J J, Shaw R. Cloning, nucleotide sequence, and expression of the Bacillus cereus 5/B/6 β-lactamase II structural gene. J Bacteriol. 1988;170:2873–2878. doi: 10.1128/jb.170.6.2873-2878.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livermore D M. Acquired carbapenemases. J Antimicrob Chemother. 1997;39:673–676. doi: 10.1093/jac/39.6.673. [DOI] [PubMed] [Google Scholar]
- 31.Massidda O, Rossolini G M, Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among metallo-β-lactamases. J Bacteriol. 1991;173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumura N, Minami S, Watanabe Y, Iyobe S, Mitsuhashi S. Role of permeability in the activities of β-lactams against gram-negative bacteria which produce a group 3 β-lactamase. Antimicrob Agents Chemother. 1999;43:2084–2086. doi: 10.1128/aac.43.8.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medeiros A A. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl. 1):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard. NCCLS document M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 35.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum beta-lactamase from Pseudomonas aeruginosa and comparison with class A beta-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38:71–78. doi: 10.1128/aac.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palissa H, von Dohren H, Kleinkauf H, Ting H H, Baldwin J E. Beta-lactam biosynthesis in a gram-negative eubacterium: purification and characterization of isopenicillin N synthase from Flavobacterium sp. strain SC 12.154. J Bacteriol. 1989;171:5720–5728. doi: 10.1128/jb.171.10.5720-5728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payne D J, Cramp R, Bateson J H, Neale J, Knowles D. Rapid identification of metallo- and serine beta-lactamases. Antimicrob Agents Chemother. 1994;38:991–996. doi: 10.1128/aac.38.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pickett M J. Methods for identification of flavobacteria. J Clin Microbiol. 1989;27:2309–2315. doi: 10.1128/jcm.27.10.2309-2315.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poirel L, Guibert M, Girlich D, Naas T, Nordmann P. Cloning, sequence analyses, expression and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob Agents Chemother. 1999;43:769–776. doi: 10.1128/aac.43.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poirel L, Naas T, Guibert M, Chaibi E B, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo J-D, Nordmann P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–897. doi: 10.1128/aac.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasmussen B A, Bush K. Carbapenem-hydrolyzing β-lactamases. Antimicrob Agents Chemother. 1997;41:223–232. doi: 10.1128/aac.41.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen B A, Gluzman Y, Tally F P. Cloning and sequencing of the class B β-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1990;34:1590–1592. doi: 10.1128/aac.34.8.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossolini G M, Franceschini N, Lauretti L, Caravelli B, Riccio M L, Galleni M, Frère J-M, Amicosante G. Cloning of Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaACME) encoding an extended-spectrum class A β-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER β-lactamases. Antimicrob Agents Chemother. 1999;43:2193–2199. doi: 10.1128/aac.43.9.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossolini G M, Franceschini N, Riccio M L, Mercuri P S, Perilli M, Galleni M, Frère J-M, Amicosante G. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: a new molecular class B β-lactamase showing a broad substrate profile. Biochem J. 1998;332:145–152. doi: 10.1042/bj3320145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saino Y, Kobayashi F, Inoue M, Mitsuhashi S. Purification and properties of inducible penicillin β-lactamase isolated from Pseudomonas maltophilia. Antimicrob Agents Chemother. 1982;22:564–570. doi: 10.1128/aac.22.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanschagrin F, Dufresne J, Levesque R C. Molecular heterogeneity of the L-1 metallo-β-lactamase family from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1998;42:1245–1248. doi: 10.1128/aac.42.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segatore B, Massidda O, Satta G, Setacci D, Amicosante G. High specificity of cphA-encoded metallo-beta-lactamase from Aeromonas hydrophila AE036 for carbapenems and its contribution to beta-lactam resistance. Antimicrob Agents Chemother. 1993;37:1324–1328. doi: 10.1128/aac.37.6.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J Clin Microbiol. 1996;34:2909–2913. doi: 10.1128/jcm.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, Kato N, Ohta M. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob Agents Chemother. 1996;40:349–353. doi: 10.1128/aac.40.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegman-Igra Y, Schwartz D, Soferman G, Konforti N. Flavobacterium group IIb bacteremia: report of a case and review of Flavobacterium infections. Med Microbiol Immunol. 1987;176:103–111. doi: 10.1007/BF00200682. [DOI] [PubMed] [Google Scholar]
- 53.Swofford D L. PAUP (version 3.0): phylogenetic analysis using parsimony. Champaign, Ill: Illinois Natural History Survey; 1989. [Google Scholar]
- 54.Tarentino A L, Quinones G, Schrader W P, Changchien L M, Plummer T H., Jr Multiple endoglycosidase (Endo) F activities expressed by Flavobacterium meningosepticum. Endo F1: molecular cloning, primary sequence, and structural relationship to Endo H. J Biol Chem. 1992;267:3868–3872. [PubMed] [Google Scholar]
- 55.Ullah J H, Walsh T R, Taylor I A, Emery D C, Verma C S, Gamblin S J, Spencer J. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 Å resolution. J Mol Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 56.Vandamme P, Bernardet J F, Segers P, Kersters K, Holmes B. New perspectives in the classification of the flavobacteria: description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int J Syst Bacteriol. 1994;44:827–831. [Google Scholar]
- 57.Walsh T R, Hall L, Assinder S J, Nichols W W, Cartwright S J, MacGowan A P, Bennett P M. Sequence analysis of the L-1 metallo β-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta. 1994;1218:199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 58.Wang Z, Fast W, Benkovic S J. On the mechanism of the metallo-β-lactamase from Bacteroides fragilis. Biochemistry. 1999;38:10013–10023. doi: 10.1021/bi990356r. [DOI] [PubMed] [Google Scholar]