Summary
Post-translational modification of ribosomal proteins enables rapid and dynamic regulation of protein biogenesis. Site-specific ubiquitylation of 40S ribosomal proteins uS10 and eS10 plays a key role during ribosome-associated quality control. Distinct, and previously functionally ambiguous ubiquitylation events on the 40S proteins uS3 and uS5 are induced by diverse proteostasis stressors that impact translation activity. Here, we identify the ubiquitin ligase, RNF10, and the deubiquitylating enzyme, USP10, as the key enzymes that regulate uS3 and uS5 ubiquitylation. Prolonged uS3 and uS5 ubiquitylation results in 40S, but not 60S, ribosomal protein degradation in a manner independent of canonical autophagy. We show that blocking progression of either scanning or elongating ribosomes past the start codon triggers site-specific ribosome ubiquitylation events on uS5 and uS3. This study identifies and characterizes a distinct arm in the RQC pathway, initiation RQC (iRQC), that acts on 40S ribosomes during translation initiation to modulate translation activity and capacity.
Introduction
Translation is the critical process that decodes the genetic blueprint into functional proteins. While most translation events terminate in successful protein biogenesis, cis-acting features of the mRNA or nascent chain can result in abortive translation termination (Hinnebusch et al., 2016). Defects in either the emerging nascent polypeptide or translating mRNA can cause ribosomes to experience prolonged stalls during elongation which can subsequently result in 80S ribosome collisions and elicit a multifaceted ribosome-associated quality control (RQC) pathway (Inada, 2020; Joazeiro, 2019; Yip and Shao, 2021). Components of the RQC act to degrade the truncated nascent chain, destroy the associated mRNA, and recycle the ribosomal subunits (Joazeiro, 2019; Meydan and Guydosh, 2020). Current models suggest that ribosome collisions are the integral first signal necessary to recruit factors that facilitate downstream RQC activities (D’Orazio and Green, 2021; Ikeuchi et al., 2019; Juszkiewicz et al., 2018; Simms et al., 2017). Protein ubiquitylation plays two critical roles during mammalian RQC. The first involves conserved regulatory ribosomal ubiquitylation (RRub) of 40S proteins eS10 (RPS10) and uS10 (RPS20) mediated by the E3 ligase ZNF598 (Garzia et al., 2017; Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017). The second involves additional ligases, Listerin and the recently described CRL2/KLHDC10 and Pirh2 ligases, which are recruited to the 60S subunit, post-80S ribosome splitting, to catalyze nascent polypeptide chain ubiquitylation and subsequent degradation (Lyumkis et al., 2014; Shao and Hegde, 2014; Shao et al., 2013; Thrun et al., 2021). While these ubiquitylation events are well characterized, additional sites of ribosome ubiquitylation in mammals and other eukaryotes have been described that either play a direct role within the RQC (Ikeuchi et al., 2019; Saito et al., 2015) or operate outside of the RQC and have uncharacterized roles (Higgins et al., 2015; Montellese et al., 2020; Silva et al., 2015), suggesting ribosome ubiquitylation may regulate multiple steps during translation.
Dynamic feedback regulation between elongation and initiation meters ribosome traffic along mRNAs. Elevation in RQC activity due to increases in elongating ribosome collisions can indicate an overabundance of ribosome density on transcripts. Compensatory decreases in translation initiation rates can reduce ribosome collisions and RQC activity (Juszkiewicz et al., 2018). Further, recent studies have defined a collision-induced feedback loop that downregulates translation initiation. Following ribosome collisions, in a ZNF598-independent manner, the collision-sensor EDF1 recruits the translation repressors GIGYF2 and 4EHP to inhibit translation of stall-inducing transcripts (Juszkiewicz et al., 2020; Sinha et al., 2020). A separate study also demonstrated that the same translation repressors, GIGYF2 and 4EHP, when deleted, increased translation of a stall-inducing reporter (Hickey et al., 2020). These studies highlight the requirement for dynamic coordination between elongation and initiation rates to regulate elongation collision frequency. While elongating ribosome collisions and the corresponding RQC pathway have been well-established, a quality control pathway that acts on ribosomes during the initiation phase of translation has not been described.
Here we identify a surveillance pathway, iRQC, in which regulatory ribosomal ubiquitylation of distinct residues within the 40S proteins uS3 (RPS3) and uS5 (RPS2) promotes 40S subunit degradation. We identify and characterize the ubiquitin ligase RNF10, and the deubiquitylating enzyme, USP10 as the key ubiquitin pathway enzymes that regulate uS5 and uS3 ribosomal ubiquitylation. Loss of USP10 function or RNF10 overexpression resulted in enhanced uS5 and uS3 ubiquitylation in the absence of exogenous stressors. We show that prolonged uS3 and uS5 ubiquitylation induces selective degradation of 40S, but not 60S ribosomal proteins in a manner that is independent of the canonical autophagy pathway. Several pharmacological agents that act to repress translation initiation, including integrated stress response activators, also induce RNF10-dependent uS3 and uS5 ubiquitylation. These results indicate that stalled or otherwise defective scanning preinitiation complexes may be targeted by RNF10-dependent ubiquitylation to mediate 40S destruction. Our results establish parallel, but distinct, RQC pathways that act on ribosomes during the elongation (eRQC) or initiation (iRQC) phases of translation.
Results
RNF10 catalyzes uS5 and uS3 ubiquitylation
Previous studies have documented dynamic ubiquitylation of a variety of ribosomal proteins suggesting the ubiquitylation may be used to regulate ribosome function beyond the conical RQC pathway (Higgins et al., 2015; Montellese et al., 2020; Silva et al., 2015). We had previously demonstrated that either pharmacological translation inhibition or integrated stress response (ISR) activation results in robust uS3 (RPS3) and uS5 (RPS2) ubiquitylation (Higgins et al., 2015). Because these ubiquitylation events are not catalyzed by ZNF598 and do not function within the characterized RQC pathway (Garshott et al., 2020), how uS3 and uS5 ubiquitylation regulates ribosome function remained unknown.
In order to determine the molecular role(s) uS3 and uS5 ubiquitylation play during translation, we set out to identify the ubiquitin pathway enzymes that regulate uS3 and uS5 ubiquitylation. We utilized an siRNA-based loss-of-function screen targeting 18 known RNA-associated ubiquitin ligases and found that only depletion of RNF10 reproducibly prevented uS3 and uS5 ubiquitylation (Figures 1A and S1A–S1E). We then generated and verified RNF10 knockout cells using CRISPR-Cas9-based approaches and demonstrated that these cells completely lacked both ISR (DTT) or elongation inhibition (anisomycin, ANS)-induced uS3 and uS5 ubiquitylation (Figure 1B). To investigate the specificity of RNF10, we examined uS5, uS3, eS10, and uS10 ubiquitylation in 293 Flp-IN cells expressing inducible wild type RNF10. In the same manner that ZNF598 is specific in modifying eS10 and uS10 (Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017), RNF10 expression, in the absence of stress, resulted in enhanced uS3 and uS5 ubiquitylation but left eS10 and uS10 ubiquitylation largely unchanged (Figure 1C). Furthermore, in vitro ubiquitylation assays demonstrated that RNF10 maintains its ribosomal protein specificity when incubated with purified 40S subunits (Figures 1D and S1F). Collectively, these findings demonstrate that RNF10 is both necessary and sufficient to catalyze uS3 and uS5 ubiquitylation.
Figure 1. RNF10 catalyzes uS3 and uS5 ubiquitylation, See also Figure S1.
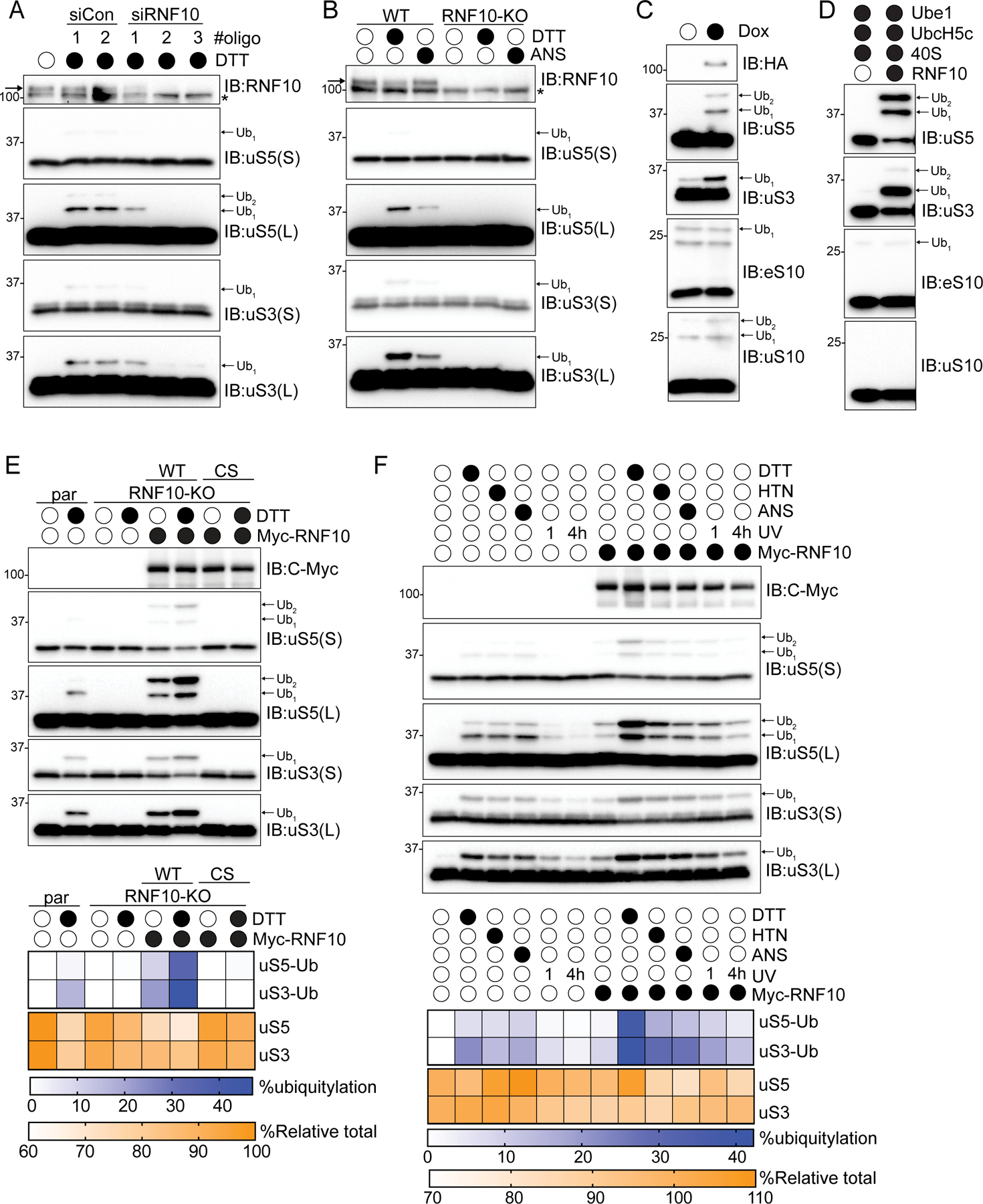
(A) Cell lysates from 293T cells transfected with either control siRNA oligos or three separate siRNA oligos targeting RNF10, followed by treatment with dithiothreitol (DTT) for 2 hours were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. * indicates non-specific background signal. Arrow indicates RNF10-specific immunoreactivity. For all blots the ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures, respectively.
(B) Cell extracts from parental 293T or RNF10 knockout (KO) cells were either untreated or treated with DTT or anisomycin (ANS) then analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
(C) HEK293-FlpIn cells expressing tet-inducible Flag-HA tagged RNF10 were treated with doxycycline (Dox) and cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
(D) In vitro ubiquitylation assay utilizing purified 40S ribosomal subunits and RNF10. Samples were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
(E) (top) RNF10 knockout (KO) cells were transfected with Myc-tagged wild type (WT) or inactive mutant (CS) RNF10 and parental 293T or RNF10-KO cells were either untreated or treated with DTT. Cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (bottom) Quantitative representation of percent ubiquitylated uS3 and uS5, and percent relative total abundance from immunoblots (bottom).
(F) (top) 293T cells with and without Myc-tagged wild type RNF10 expression were drug treated as indicated. UV indicates that cells were exposed to UV and were allowed to recover for 1 or 4 hours. Cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (bottom) Quantitative representation of percent ubiquitylated uS3 and uS5, and percent relative total abundance from immunoblots.
What is the fate of the ubiquitylated 40S? To begin to address this question, we utilized an RNF10 overexpression system where we transiently overexpressed either wild type RNF10, or a catalytically inactive mutant (C225S) in RNF10 knockout (KO) cells to enhance uS5 and uS3 ubiquitylation both at basal conditions and upon conditions that enhance uS3 and uS5 ubiquitylation. Expression of wild type, but not inactive RN10, rescued the ability to ubiquitylate uS3 and uS5 with or without DTT treatment (Figure 1E). Combining RNF10 overexpression with DTT treatment resulted in enhanced uS3 and uS5 ubiquitylation with more than 40% of total uS3 being ubiquitylated (Figures 1E,F). Notably, total uS3 and uS5 protein abundance was also reduced upon RNF10 overexpression with and without DTT treatment (Figures 1E,F). This result suggests the RNF10-catalyzed 40S ubiquitylation acts to reduce 40S protein levels. Utilizing a panel of ISR and elongation inhibitors, we observed that RNF10 overexpression further heightened uS3 and uS5 ubiquitylation while having no additional effect on eS10 or uS10 modification (Figures 1F and S1G). Because RNF10 overexpression, in the absence of stressors, induced uS5 and uS3 ubiquitylation, we hypothesized that the ribosomal species that is targeted by RNF10 is present under normal proliferative conditions and that the extent of ribosomal ubiquitylation may be limited by the cellular concentration of RNF10.
USP10 antagonizes RNF10-dependent uS3 and uS5 ubiquitylation
Our demonstration that RNF10 overexpression can stimulate uS3 and uS5 ubiquitylation at steady state argued that robust deubiquitylating activity antagonizes RNF10 ribosomal ubiquitylation. We serendipitously identified USP10 as the deubiquitylating enzyme responsible for removing ubiquitin from uS3 and uS5 while identifying and characterizing deubiquitylating enzymes that antagonize ZNF598 (Figure S2A) (Garshott et al., 2020). A recent study also identified USP10 as a ribosomal deubiquitylating enzyme (Meyer et al., 2020). Consistent with this previous work, USP10 knockout (KO) cells display constitutively high levels of not only ubiquitylated eS10 and uS10, but also ubiquitylated uS3 and uS5 (Figure 2A) (Meyer et al., 2020). These modifications were not further induced upon translation elongation inhibition (harringtonine, HTN), suggesting that loss of USP10 results in maximal uS3 and uS5 ubiquitylation which cannot be further augmented by the stressors used here (Figure 2A). Similarly, treatment with ISR agonists or high dose elongation inhibitors did not further elevate uS3 and uS5 ubiquitylation in USP10-KO cells (Figure 2B). These observations suggest that excess levels of USP10 relative to RNF10 maintain low levels of ribosomal ubiquitylation at basal conditions. In agreement with this, exogenous USP10 overexpression resulted in a loss of observable ribosomal ubiquitylation that was largely dependent upon the deubiquitylating activity of USP10 (Figure 2A). Surprisingly, in USP10-KO cells, stress-induced uS3 and uS5 ubiquitylation decreased at later timepoints (Figure 2B). Because these cells lack the principle uS3 and uS5 deubiquitylating enzyme, the observed loss of ribosomal ubiquitylation was puzzling.
Figure 2. Persistent uS3 and uS5 ubiquitylation targets 40S ribosomal proteins for degradation, See also Figure S2.
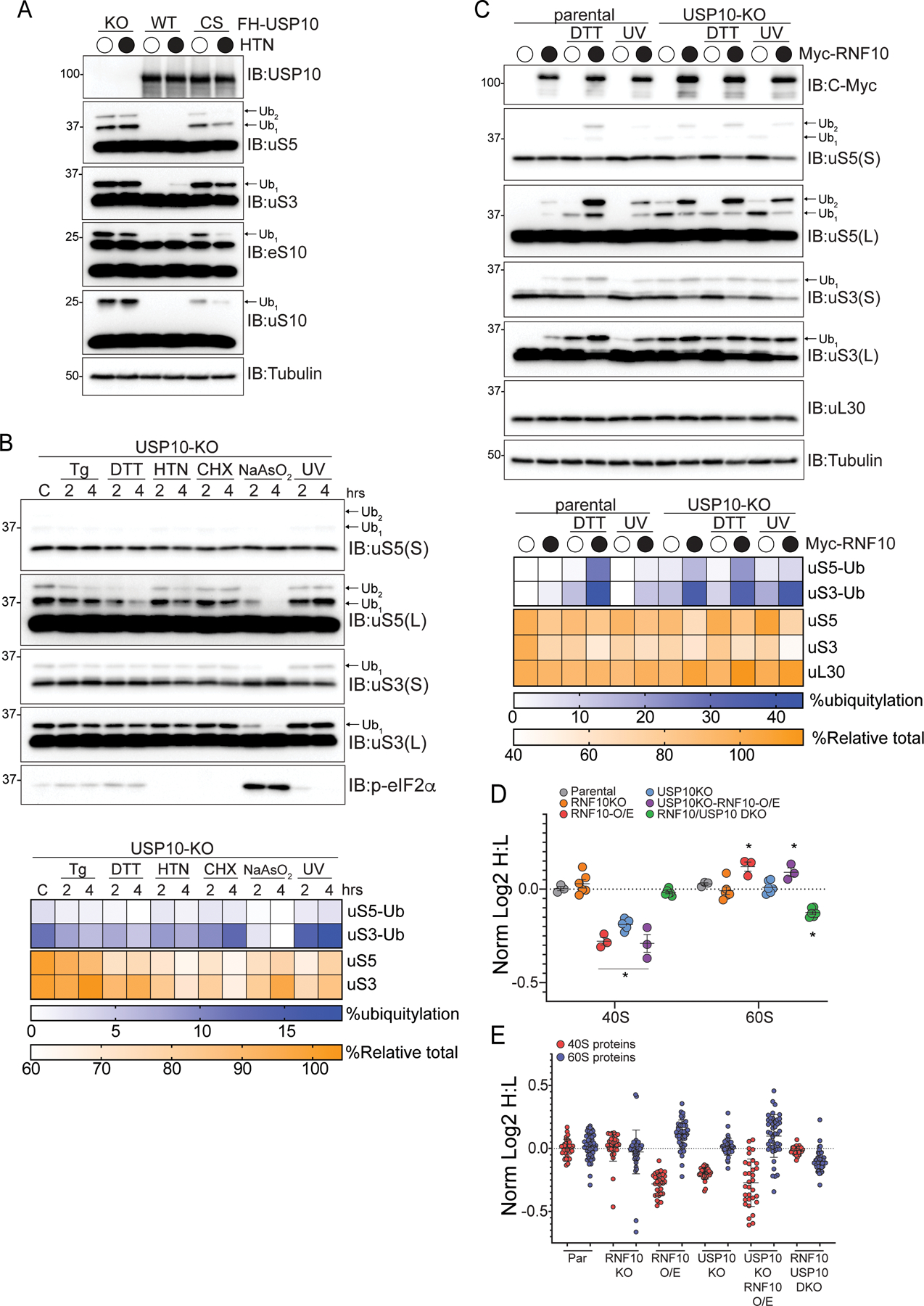
(A) 293T USP10-knockout (KO) cells constitutively expressing wild type (WT) or inactive mutant (CS) USP10 were treated with HTN and cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. For all blots the ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures, respectively.
(B) (top) USP10-KO cells were treated as indicated and cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (bottom) Percent ubiquitylated uS3 and uS5, and percent total relative abundance quantified from immunoblots
(C) (top) Parental 293T or USP10-KO cells expressing Myc-tagged wild type RNF10 were either untreated, treated as indicated and cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (bottom) Quantitative representation of uS3 and uS5 percent ubiquitylation, and percent relative total abundance for uS3, uS5 and uL30.
(D) The median normalized log2 SILAC ratio (H:L) for all quantified 40S and 60S ribosomal proteins comparing parental cells (light label) to cells of the indicated genotype (heavy label) with or without RNF10 overexpression (O/E). Each point represents a biological replicate, Bars denote mean value for replicate experiments with error bars displaying SEM. *=pvalue<0.05 by student’s t test compared to parental controls.
(E) The median normalized log2 SILAC ratio (H:L) for individual 40S and 60S ribosomal proteins comparing parental cells (light label) to cells of the indicated genotype with or without RNF10 overexpression (O/E). Bars denote mean and error bars denote SD.
RNF10-mediated ribosome ubiquitylation acts post-translationally to reduce 40S abundance
We surmised that the observed loss in uS5 and uS3 ubiquitylation upon protein synthesis inhibition was due to protein degradation in the absence of new protein production. To examine this possibility, we overexpressed RNF10 in the presence and absence of USP10, followed by ISR activation (Figure 2C). RNF10 overexpression in parental cells resulted in robust uS3 and uS5 ubiquitylation and an obvious reduction in overall uS3 and uS5 protein levels in DTT-treated cells overexpressing RNF10 (Figure 2C). This reduction of protein levels was evident when summing the intestines of both the ubiquitylated and unmodified forms of uS3 and uS5 (which is what is reported as total levels in all figures) indicating that the loss in total protein levels was not merely due to the increase in the amount of ubiquitylated uS3 and uS5. These observations were further enhanced when we overexpressed RNF10 in USP10 knockout cells, both at steady state and following stress induction (Figure 2C). Additionally, total protein levels for both eS10 and uS10 are also reduced, as observed via immunoblotting, upon RNF10 overexpression (Figures S2B,C). In contrast, levels of the 60S subunit protein uL30 (RPL7) were unchanged upon RNF10 overexpression in either parental or USP10-KO cells (Figures 2C and S2C). To examine if RNF10 expression suppressed ribosomal gene transcription, we measured uS3, eS6 (RSP6), or uL30 mRNA abundance in cells overexpressing RNF10. RNF10 overexpression did not decrease mRNA abundance, consistent with a post-transcriptional mechanism underlying the observed reduction in 40S protein levels (Figure S2D). Taken together, these results suggest that the abundance of 40S, but not 60S, ribosomal proteins is decreased upon conditions that result in constitutively high levels of uS3 and uS5 ubiquitylation.
To determine if the observed reduction in 40S protein levels occurred due to lower mRNA translation, we used a metabolic pulse labeling approach: heavy SILAC-labeled cells were switched to the light label and ribosomal protein synthesis was followed over time by quantitative proteomics. Global protein synthesis rates were unaltered in USP10 KO cells whereas a small but significant increase occurred in the rate of 40S, but not 60S, protein synthesis (Figure S2E). RNF10 overexpression suppressed global protein synthesis rates, consistent with observations that RNF10 overexpression reduces cellular proliferation rates. Despite this decrease in overall protein synthesis, 40S protein synthesis rates were increased in cells overexpressing RNF10 relative to 60S or total protein synthesis rates (Figure S2E). These results indicate that the observed selective reduction in 40S compared to 60S protein levels when uS3 and uS5 ubiquitylation is enhanced is not due to a decrease in 40S protein synthesis.
Constitutive uS5 and uS3 ubiquitylation results in 40S protein degradation
To examine if enhanced uS3 and uS5 ubiquitylation more broadly impacts overall 40S ribosomal protein abundance, we used SILAC-based quantitative proteomics to compare ribosome protein levels between parental cells and RNF10-KO, USP10-KO, USP10/RNF10 double knockout cells, or cells overexpressing RNF10. RNF10-KO cells had comparable 40S and 60S protein levels to parental cells, whereas RNF10 overexpression resulted in a ~17% reduction in 40S ribosomal protein abundance while modestly increasing 60S protein levels (Figure 2D; Table S1). Consistent with a previous report, cells lacking USP10 have reduced 40S protein levels (Meyer et al., 2020). Furthermore, 60S protein levels were unchanged relative to parental cells in USP10-KO cells. RNF10 overexpression in USP10-KO cells further reduced 40S protein levels while slightly increasing 60S protein abundance (Figure 2D). The observed decrease in 40S protein abundance in UPS10-KO cells was reversed in RNF10/USP10 double KO cells indicating that RNF10-dependent uS5 and uS3 ubiquitylation promotes 40S protein loss in USP10-KO cells. The abundance of the entire 40S subunit, rather than individual proteins, is reduced upon RNF10 overexpression or USP10 depletion suggesting that overall 40S protein stability is reduced by uS3 and uS5 ubiquitylation (Figure 2E; Table S1). These results are consistent with a model where ubiquitylated 40S ribosomal subunits that escape USP10-dependent ubiquitin removal can be targeted for degradation.
40S protein degradation is autophagy independent
Overall, our data indicate that RNF10 overexpression or loss of USP10 function results in 40S degradation. An autophagic mechanism seemed most plausible given that previous studies in S. cerevisiae have demonstrated that starvation conditions that inhibit mTOR signaling and stimulate autophagic flux result in enhanced ribosomal turnover by the autophagy pathway (Kraft et al., 2008). While mTOR-dependent degradation of ribosomes via autophagy does not appear to play a large role in regulating ribosomal abundance in mammalian cells (An and Harper, 2018; An et al., 2020), we directly evaluated if uS3 or uS5 ubiquitylation resulted in autophagy-dependent degradation of 40S ribosomal subunits. We first examined uS3 and uS5 protein levels upon RNF10 overexpression in parental or autophagy-deficient cells that are devoid of the critical ULK1 complex member, RB1CC1 (FIP200) (An et al., 2019). USP10 depletion or RNF10 overexpression alone or in combination resulted in the expected increase in uS3 and uS5 ubiquitylation and loss in 40S protein abundance in both parental and RB1CC1 knockout cells (Figure 3A). These results establish that cells deficient in canonical autophagy maintain the ability to degrade RNF10 targeted 40S proteins.
Figure 3. Enhanced ubiquitylation results in turnover of 40S ribosomal proteins in an autophagy-independent manner.
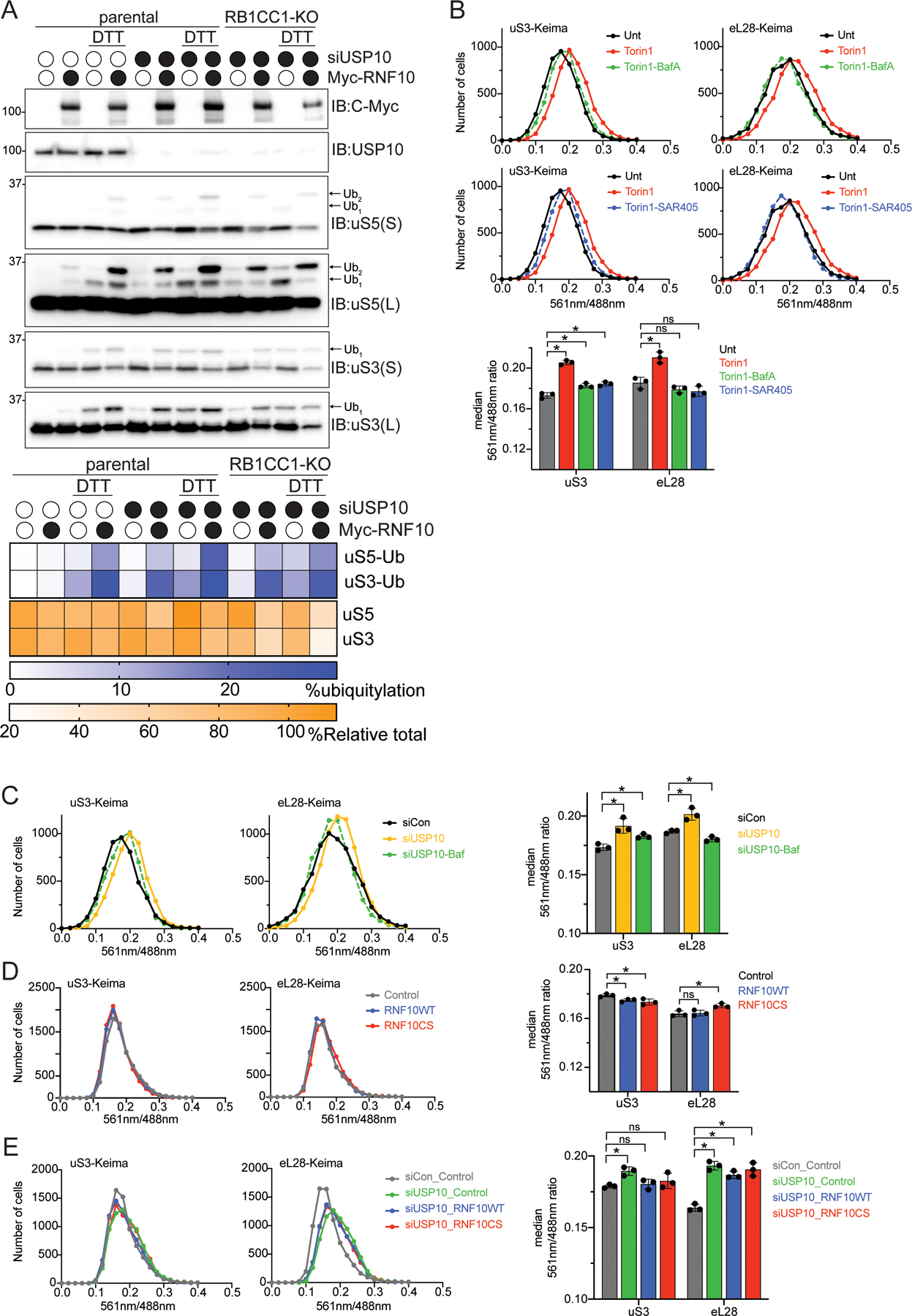
(A) (top) Cell extracts from parental 293T or RB1CC1-KO cells transfected with either a control siRNA oligo or siRNA oligo targeting USP10, followed by transfection with Myc-tagged wild type RNF10 treated as indicated were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (bottom) Quantitative representation of percent relative total abundance and uS3 and uS5 percent ubiquitylation.
(B) HEK293 uS3 or eL28 (RPL28) Keima-tagged cells were treated as indicated and frequency distributions of the red (561nm) to green (488nm) ratio are plotted.
(C) HEK293 uS3 or eL28 Keima-tagged cells were transfected with either a control siRNA oligo (black line), siRNA targeting USP10 (yellow line) or in combination with Bafilomycin A (50nM, 1h) treatment (green line). Frequency distributions of the red (561nm) to green (488nm) ratio are plotted.
(D) HEK293 uS3 or eL28 Keima-tagged cells expressing either a control plasmid (grey line), RNF10 wild type (blue line) or the catalytic mutant (red line) 48 hours post transfection were collected and analyzed via FACS. Frequency distributions of the red (561nm) to green (488nm) ratio are plotted.
(E) HEK293 uS3 or eL28 Keima-tagged cells transfected with either a control siRNA oligo (grey line), or siRNA targeting USP10 and expressing either a control plasmid (green line), RNF10 wild type (blue line) or the catalytic mutant (red line) 48 hours post transfection were collected and analyzed via FACS (bottom). All bar graphs denote median red:green ratio from triplicate experiments. N=3, error bars denote SD of triplicate experiments. *=pvalue<0.05, ns = non-significant by unpaired student’s t test.
To confirm that RNF10-mediated ribosome ubiquitylation does not target 40S proteins for autophagy-dependent degradation, we utilized cell lines in which the genomic loci of uS3 or eL28 (RPL28) were tagged with the pH-sensitive fluorophore, Keima. Consistent with previous reports, inactivation of mTOR signaling enhanced both 40S and 60S flux to lysosomes as indicated by an increase in the red to green Keima fluorescence (Figure 3B) (An and Harper, 2018). This observed enhanced 40S and 60S flux through the autophagy pathway upon mTOR inhibition was inhibited by co-incubation of either SAR405 or Bafilomycin A, both of which inhibit autophagy by distinct mechanisms (Figure 3B). Similar to a previous report, transient knockdown of USP10 also resulted in enhanced autophagic flux of both Keima-tagged uS3 and eL28 which was reversed upon BafA treatment (Figure 3C) (Meyer et al., 2020). This result was inconsistent with our observation that 40S but not 60S protein levels were reduced in USP10 knockout cells. We note that the observed increase in 40S and 60S flux upon mTOR inhibition observed here, and previously, accounts for a 3% decrease in total ribosome abundance (An and Harper, 2020). The increase in 40S and 60S autophagic flux observed upon transient knockdown of USP10 was less than that observed upon mTOR inhibition suggesting that this level of enhanced flux would be insufficient to reduce ribosome abundance by the ~15% we measured using quantitative proteomics. It is possible that loss of USP10 activity results in a general increase in autophagy that is independent of the ubiquitin-mediated degradation of 40S proteins demonstrated here upon RNF10 overexpression or USP10 depletion. Consistent with this hypothesis, RNF10 overexpression alone, or combined with USP10 knockdown, did not result in an increase in either 40S or 60S ribosomal flux to the lysosome (Figures 3D,E), further suggesting that the canonical autophagy pathway is not responsible for the observed robust degradation of ubiquitylated 40S subunits.
RNF10 mediated uS5 ubiquitylation accelerates 40S protein turnover
In order to quantitatively examine 40S and 60S protein degradation, we utilized previously characterized cell lines in which the genomic uS3, uL24 (RPL26), or eL29 (RPL29) loci were tagged with Halo (hereafter called Ribo-Halo) (An et al., 2020). These Ribo-Halo cell lines enable evaluation of ribosomal protein degradation kinetics through fluorescent pulse-chase experiments using fluorescently labeled Halo ligands (Figure 4A). Ribo-Halo cells overexpressing a control protein (LRRC49), wild type RNF10, or inactive RNF10 were red-labeled with a tetramethylrhodamine (TMR) Halo ligand for 1 hour to mark the existing pool of uS3, uL24, or eL29. Following TMR labeling, excess label was washed out and the abundance of the TMR-labeled ribosomal pool was monitored over time by microscopy. Three days post-transfection, cells expressing wild type GFP-RNF10 but not inactive GFP-RNF10CS displayed a marked decrease in cellular uS3-Halo abundance while having no impact on uL24-Halo protein levels (Figure 4A). To directly evaluate uS3 protein turnover, we performed pulse-chase experiments upon RNF10 expression and quantified single-cell Ribo-Halo abundance by flow cytometry. Ribo-Halo abundance was initially measured 36 hours post transfection, and ribosome decay was observed following TMR washout for 24 hours. These experiments revealed an increased uS3 turnover rate in cells expressing wild type RNF10 that was not observed in cells expressing a control protein or inactive RNF10 (Figure 4B). Consistent with our proteomics results, RNF10 overexpression did not increase turnover of the 60S subunit protein eL29 (Figure 4B). Proteasome inhibition, but not autophagy inhibition, delayed the observed loss in uS3-Halo TMR signal 8 hours after TMR washout in cells with RNF10 overexpression (Figure 4C). These results indicate that constitutive uS3 and uS5 ubiquitylation enhances 40S, but not 60S, protein degradation.
Figure 4. RNF10-dependent uS5 ubiquitylation accelerates 40S protein turnover, See also Figure S3.
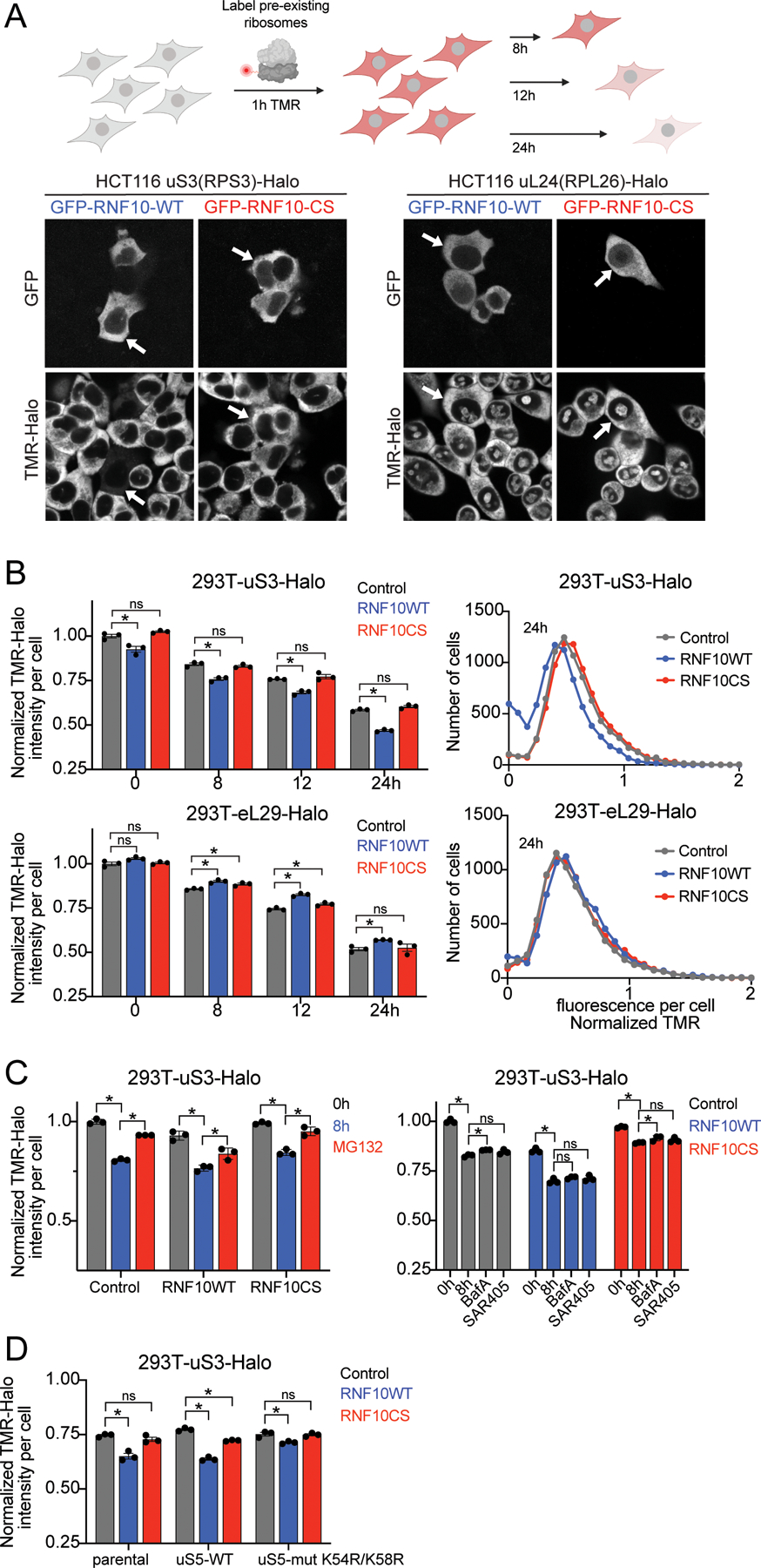
(A) Schematic of Ribo-Halo fluorescent pulse-chase assay (top). Microscopy images of HCT116 uS3 or uL24-Halo tagged cells expressing GFP-tagged wild type (WT) or inactive mutant (CS) RNF10. Ribosomes were labeled with TMR ligand for 2 hours prior to imaging. Arrows indicate the same cells across panels (bottom).
(B) The normalized (to control at 0h washout) TMR-fluorescence intensity for uS3 or eL29-Halo tagged cells expressing a control plasmid (grey bars), Myc-RNF10-WT (blue bars), or CS mutant (red bars) expression plasmid is depicted at the indicated time points post TMR washout (left). N=3, error bars denote SD of triplicate experiments. *=pvalue<0.05, ns = non-significant by multiple unpaired t tests compared to control protein. Frequency distribution of the normalized TMR signal at 24h is plotted (right).
(C) Normalized (to control at 0h washout) TMR-fluorescence intensities for uS3-Halo tagged cells expressing a control plasmid, Myc-RNF10-WT, or RNF10 mutant (CS) expression plasmid at time 0h (grey bars), 8h post TMR washout (blue bars) or with MG132 included during the 8h TMR washout (red bars) is depicted (left). TMR fluorescence intensities for cells expressing a control plasmid (grey bars), Myc-RNF10-WT (blue bars), or CS mutant (red bars) expression plasmid at 0 or 8h post TMR washout with or without BafA or SAR405 included in the TMR washout (right). N=3, error bars denote SD of triplicate experiments. *=pvalue<0.05 by unpaired student’s t test.
(D) TMR fluorescence intensities 12h post washout from parental 293T uS3-Halo tagged cells alone or with stable expression of wild type (WT) or K54R/K58R mutant (Mut) uS5 and transfected with a control plasmid (grey bars), GFP-RNF10-WT (blue bars), or GFP-RNF10-CS mutant (red bars) expression plasmids are depicted. The normalized (to control at 0h washout) TMR intensities are depicted. N=3, error bars denote SD of triplicate experiments. *=pvalue<0.05, ns = non-significant by unpaired student’s t tests compared to control protein.
Our previous studies delineated a hierarchical relationship among uS3 and uS5 ubiquitylation events such that eliminating uS3 ubiquitylation renders uS5 incompetent for ubiquitylation, whereas eliminating uS5 ubiquitylation did not prevent uS3 ubiquitylation (Garshott et al., 2020). Based on these results, we engineered uS3-Halo cell lines to express either wild type or a ubiquitylation deficient mutant version (K54R;K58R) of uS5. Consistent with previous results, stable expression of exogenous uS5 comprised 80% of total uS5 levels (Figure S3A). While uS5 ubiquitylation in cells expressing exogenous wild type uS5 remained intact following DTT treatment, uS5 ubiquitylation was absent upon expression of mutant uS5 (Figure S3A). We then performed Ribo-Halo pulse-chase experiments in the uS3-Halo cells containing wild type or ubiquitylation mutant uS5. Consistent with previous results, wild type RNF10 overexpression resulted in an 18% decrease in uS3-Halo levels 12 hours post TMR washout in cells expressing wild type uS5, compared to cells expressing a control protein, (Figure 4D). However, RNF10 overexpression failed to accelerate uS3 degradation in cells expressing the ubiquitylation deficient version of uS5 (Figure 4D). These experiments causally link RNF10-dependent enhanced 40S protein degradation to the observed increase in ribosome ubiquitylation and demonstrate that uS5 ubiquitylation is required for 40S turnover.
Translation initiation inhibition triggers 40S ribosomal ubiquitylation
Our previous observation that cells lacking uS5 or uS3 ubiquitylation sites retained RQC activity suggested that RNF10-mediated ubiquitylation targeted a distinct population of ribosomes than those targeted by ZNF598 during elongation collisions (Garshott et al., 2020). We had previously demonstrated that treating cells with a variety of translation elongation inhibitors effecting distinct steps during the elongation cycle induced uS3 and uS5 ubiquitylation (Higgins et al., 2015). We were particularly intrigued by our observation that Harringtonine, which blocks progression of 80S ribosomes at the start codon without impacting elongating or scanning ribosomes (Fresno et al., 1977), induces uS3 and uS5 ubiquitylation (Higgins et al., 2015). This result suggested that inhibition of either elongation immediately after start codon recognition, or a defect in the ability of scanning preinitiation complexes to transition to elongation competent 80S ribosomes triggered uS3 and uS5 ubiquitylation. Consistent with previous results, HTN treatment resulted in rapid and robust uS5 and uS3 ubiquitylation (Figures 5A) (Higgins et al., 2015). Treatment with either HTN or lactimidomycin (LTM), a functionally similar but mechanistically distinct compound (Lee et al., 2012; Schneider-Poetsch et al., 2010), induced uS5 and uS3 ubiquitylation that was detectable after 5 minutes and further increased over time (Figure 5B). We then utilized characterized inhibitors that impede the mRNA scanning step of translation initiation to examine if inhibiting progression of preinitiation complexes prior to start codon recognition induces ribosome ubiquitylation. Addition of rocaglates (RocA) (Iwasaki et al., 2016) or pateamine A (PatA) (Low et al., 2005), which inhibit the RNA helicase eIF4A and impair mRNA scanning, induced uS5 and uS3 ubiquitylation in a dose dependent manner (Figures 5C and S4A,B). Combined, these results suggest that impeding early events during the translation cycle generates a ribosomal subpopulation that is targeted by RNF10. One possibility is that terminally stalled preinitiation complexes that cannot transition into 80S elongation complexes are targeted for RNF10-dependend ribosome ubiquitylation. Alternatively, collisions between either multiple 43S preinitiation complexes scanning within the 5’UTR or between scanning preinitiation complexes and a stalled 80S trigger uS5 and uS3 ubiquitylation.
Figure 5. Translational initiation inhibition induces ribosomal ubiquitylation, See also Figure S4.
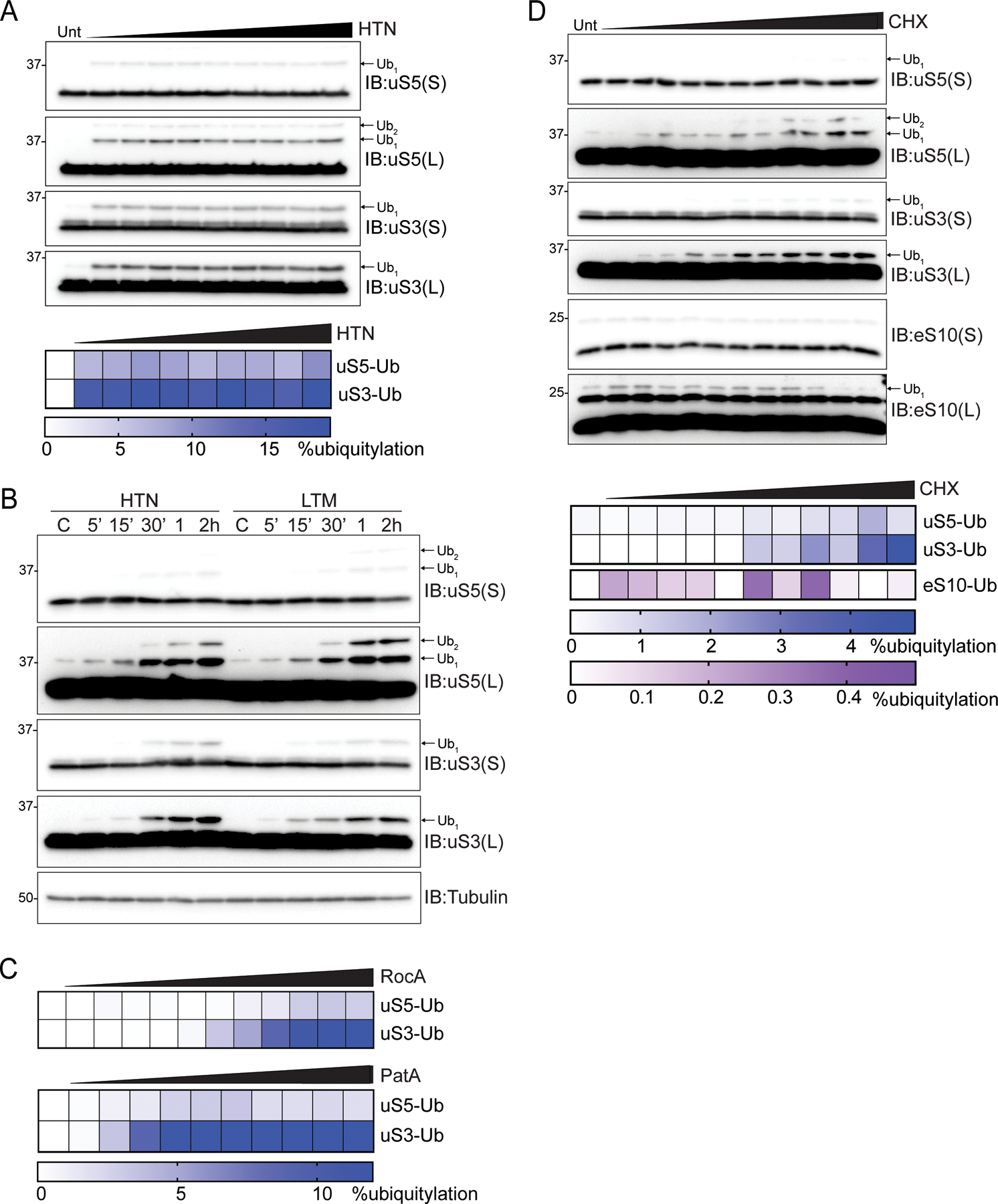
(A) (top) Cell extracts from 293T cells treated with increasing doses of HTN were analyzed by SDS-PAGE and immunoblotted (IB) with the indicated antibodies. For all blots, the ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures, respectively. (bottom) Percent ubiquitylated uS3 and uS5 quantified from immunoblots.
(B) Cell extracts from 293T cells treated with HTN or lactimidomycin (LTM) for the indicated times were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
(C) Quantification of uS3 or uS5 percent ubiquitylation from 293T cells treated with increasing doses of either rocaglates (RocA) or patamineA (PatA) from blots in S4A,B.
(D) (top) Cell extracts from 293T cells treated with increasing concentration of cycloheximide (CHX) were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (bottom) Quantitative representation of uS3, uS5, and eS10 percent ubiquitylation from immunoblots.
The demonstration that maximal elongation collisions and uS10 and eS10 ubiquitylation occur with low dose, rather than high dose treatment of elongation inhibitors was a critical result establishing that ribosome collisions are the key event leading to ribosomal ubiquitylation and RQC pathway activation (Juszkiewicz et al., 2018; Simms et al., 2017). To further examine potential differences between ZNF598-targeted elongation collisions and RNF10-targeted ribosome ubiquitylation events that occur upon translation initiation inhibition, we treated cells with increasing concentrations of translation elongation inhibitors. As expected, the ubiquitylation of eS10 ubiquitylation was induced at low CHX concentrations and diminished at high concentrations (Figure 5D). In contrast, uS3 and uS5 ubiquitylation increased with increasing CHX concentration and uS3 and uS5 ubiquitylation remained induced at the highest doses of CHX which failed to stimulate eS10 ubiquitylation (Figure 5D). This observation that uS3 and uS5 ubiquitylation is stimulated by high concentrations of elongation inhibitors suggests that inhibition of elongation shortly after start codon recognition, which would be more prevalent at higher inhibitor concentrations, elevates the abundance of the ribosomal population targeted by RNF10. Taken together, our results demonstrate that impeding progression of scanning or elongating ribosomes near start codons induces site-specific uS3 and uS5 ribosome ubiquitylation.
ISR activation similarly elicits uS5 and uS3 ubiquitylation in a ternary-complex concentration manner
We previously demonstrated that uS3 and uS5 ubiquitylation occurs upon activation of the integrated stress response (ISR) in an eIF2α phosphorylation-dependent manner (Higgins et al., 2015). These findings are distinct from those observed with HTN, as HTN treatment enhances uS3 and uS5 ubiquitylation in cells with compromised eIF2α phosphorylation (Higgins et al., 2015). ISR stimulated eIF2α phosphorylation inhibits translation initiation through depletion of the ternary complex (TC), which consists of methionyl-initiator tRNA (Met-tRNAi) and guanosine triphosphate (GTP)–bound eIF2 (Costa-Mattioli and Walter, 2020; Hinnebusch, 2014). It was initially puzzling why stressors that reduce translation initiation activity would result in uS3 and uS5 ubiquitylation if stalled preinitiation complexes or early elongating 80S ribosomes are needed to trigger ribosome ubiquitylation. One explanation would be that ISR activation also induces stalled preinitiation complexes or otherwise defective scanning ribosomes. We noticed that distinct ISR agonists increased uS3 and uS5 ubiquitylation to varying degrees, with those inducing low levels of eIF2α phosphorylation resulting in higher uS3 and uS5 ubiquitylation (Figures 6A and S3C). Notably, high concentration sodium arsenite (NaAsO2) treatment resulted in the greatest extent of eIF2α phosphorylation, but poorly stimulated uS3 or uS5 ubiquitylation (Figures 6A and S4C).
Figure 6. Moderate integrated stress response activation induces uS3 and uS5 ubiquitylation, See also Figure S4.
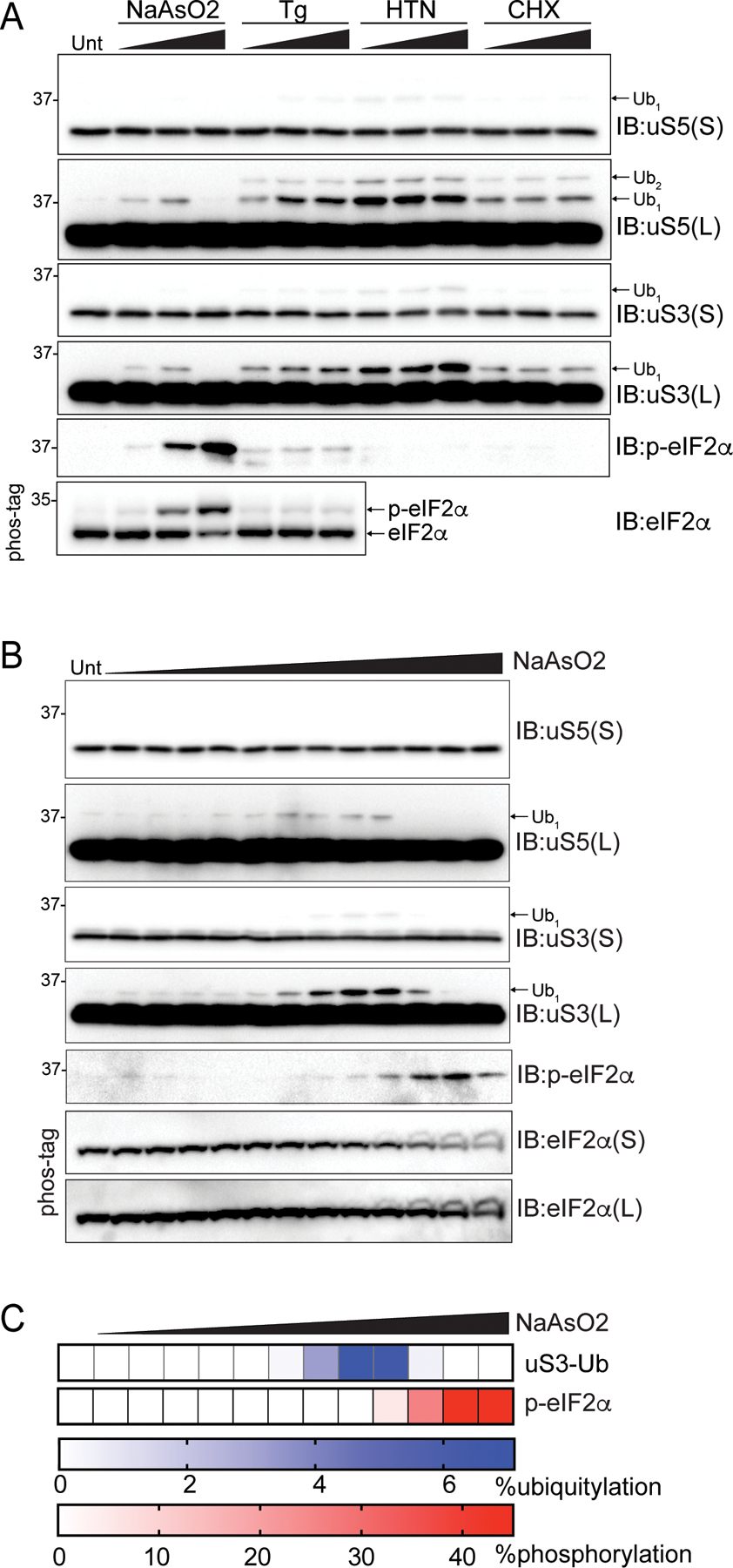
(A) Cell extracts from 293T cells treated as indicated were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. For all blots, the ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures, respectively.
(B) Cell extracts from 293T cells treated with increasing concentrations of sodium arsenite (NaAsO2) were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
(C) Quantification of uS3 percent ubiquitylation and eIF2α percent phosphorylation (from Phos-tag gels) following NaAsO2 treatment from B.
We reasoned that high stoichiometry eIF2α phosphorylation would reduce stalled preinitiation complexes by completely depleting GTP-bound ternary complexes and blocking translation initiation. In contrast, ISR agonists that induce low levels of eIF2α phosphorylation may allow for loading of scanning preinitiation complexes with GTP-bound ternary complexes that are unable to reacquire ternary complex due to reduced, but not ablated TC levels, upon encountering and translating upstream open reading frames (uORFs) (Hinnebusch et al., 2016). Thus, upon completion of uORF translation, scanning ribosomes that maintain engagement with mRNAs after uORF termination that cannot reacquire ternary complex may progress past downstream start codons and become terminally stalled. To test if low stoichiometry eIF2α phosphorylation induces a ribosomal population that is targeted for RNF10-depedent ubiquitylation, we treated cells with a range of sodium arsenite concentrations and quantified uS3 and uS5 ubiquitylation and eIF2α phosphorylation. Arsenite concentrations that induced less than 5% eIF2α phosphorylation resulted in maximal uS3 and uS5 ubiquitylation whereas conditions in which eIF2α was phosphorylated in excess of 40% did not induce ubiquitylation (Figures 6B,C). These results are consistent with our hypothesis that ISR activation resulting in low stoichiometry eIF2α phosphorylation results in elevated stalled preinitiation complexes that are targeted for ubiquitylation. Our results demonstrate that distinct, conserved ribosomal ubiquitylation events operate within separate RQC pathways which we classify as elongation RQC (eRQC) and initiation RQC (iRQC).
Sucrose gradient analysis of preinitiation collisions
We next sought to determine if uS3 and uS5 ubiquitylation is enriched within ribosomal populations that may contain elevated levels of stalled translation preinitiation complexes. We initially examined ribosome protein abundance across sucrose gradients from lysates treated with RNaseA. We compared untreated and HTN treated cells and observed that HTN treatment resulted in a noticeable broadening of the canonical 80S monosome peak, with a skew toward the lower density fractions (Figure 7A). Immunoblotting revealed that ribosomes with maximal HTN-induced uS3 and uS5 ubiquitylation migrated within fraction 5 which is at the front edge of the traditional monosome peak (Figure 7B). Abundant ubiquitylation within the 40S peak, which may also contain individual 43S preinitiation complexes, was also observed. Examination of endogenous RNF10 sedimentation within sucrose gradients revealed RNF10 to be present in fractions containing peak 40S ubiquitylation upon HTN treatment (Figure 7B). This result suggests that RNF10 associates with ribosomes in a manner that is stimulated upon conditions that enhance the abundance of stalled preinitiation complexes.
Figure 7. HTN induces 40S ubiquitylation in density gradient fractions with excess 40S relative to 60S ribosomal proteins, See also Figure S5.
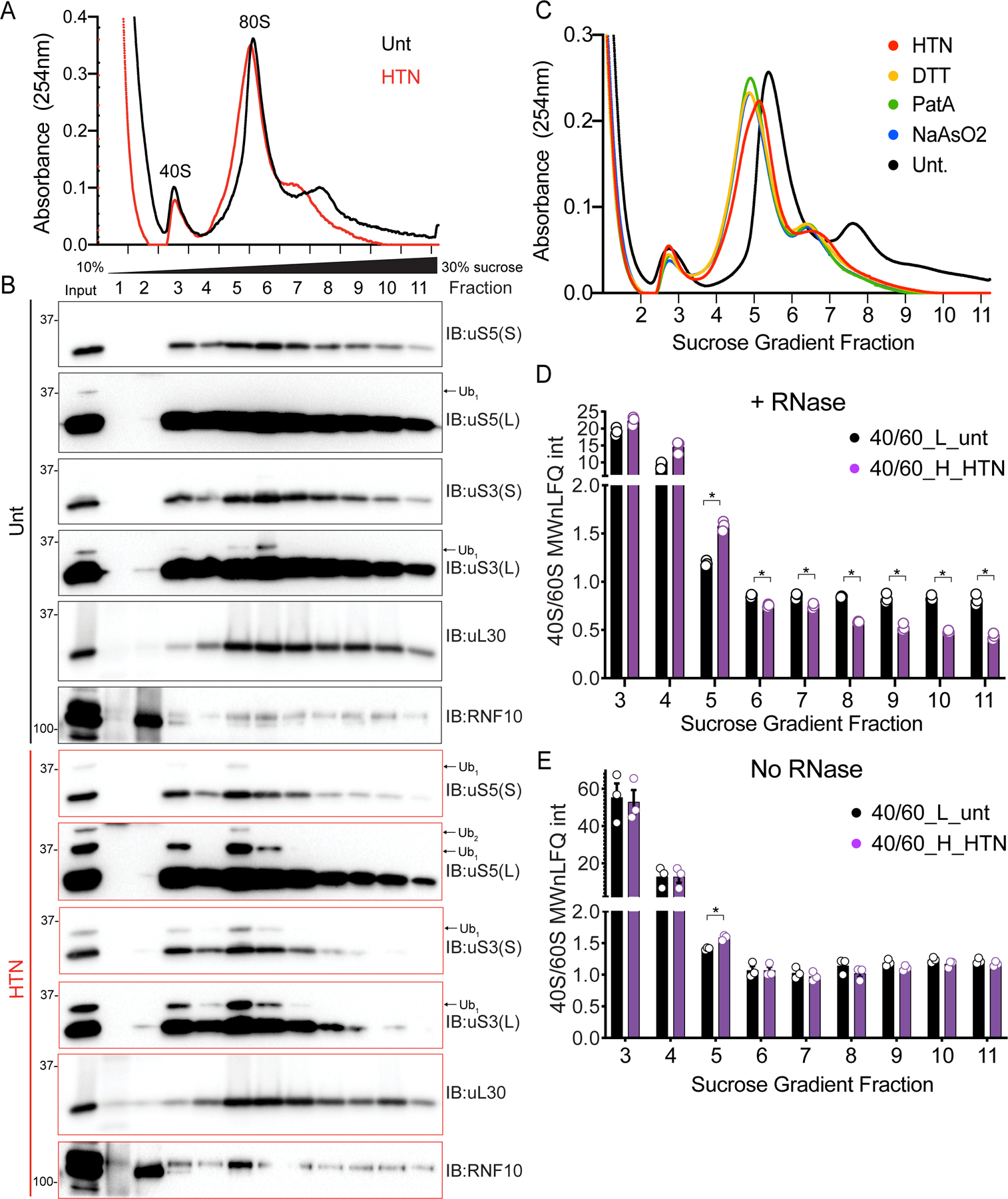
(A) RNaseA treated cell extracts from untreated (black line) or HTN treated (red line) 293T cells were fractionated on 10–30% sucrose gradients. The 254nm absorbance trace is depicted.
(B) Fractions (designated in A) were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures respectively.
(C) RNaseA treated cell lysates 293T cells treated as indicated were fractionated on 10–30% sucrose gradients. The relative 254nm absorbance trace is depicted.
(D,E) The ratio of the summed molecular weight (MW) normalized LFQ intensities 40S proteins:60S proteins from untreated, light labeled (black bars) or HTN treated, heavy labeled (purple bars) 293T cells. Cell extracts with either untreated (E) or treated (D) with RNaseA prior to density gradient centrifugation. Bars denote mean value for replicate experiments (n=3) with error bars displaying SEM. *=pvalue<0.05 by unpaired two-tailed Student’s t test.
In addition to HTN treatment, the widening and skewing of the monosome peak was observed in fractionated lysates from cells treated with DTT, PatA, or a moderate dose of NaAsO2, all of which induce uS5 and uS3 ubiquitylation (Figure 7C). While the monosome peak presumably contains predominately individual 80S complexes, this peak may also contain mRNAs with multiple loaded preinitiation complexes. According to this hypothesis, we should observe more 40S ribosome proteins relative to 60S proteins within the monosome peak under conditions that stimulate uS3 and uS5 ubiquitylation. We utilized SILAC-based quantitative proteomics to compare the abundance of 40S relative to 60S proteins across the sucrose gradient. Heavy-labeled HTN treated cells were mixed with untreated cells prior to lysis and density centrifugation. Directly comparing ribosome protein ratios revealed the expected increase in both 40S and 60S proteins in monosome-containing sucrose fractions in HTN-treated cells (Figures S5A–S5B; Table S3). We observed a significant increase in the 40S protein ratio compared to 60S in fraction 5, at the leading edge of the monosome peak, when comparing the summed and molecular weight normalized ion intensities from all 40S or 60S ribosomal proteins (Figure 7D; Table S3). This result is consistent with the hypothesis that HTN-induces stalled preinitiation complexes which migrate within the canonical monosome fraction in sucrose gradients. RNase treatment also resulted in noticeable deviation from the expected 40S:60S ratio in polysome-containing fractions (Figure 7D). Because RNaseA-mediated rRNA degradation may be impacting the integrity of 40S or 60S subunits, we repeated the sucrose gradient analysis without RNase treatment. We observed robust uS3 ubiquitylation throughout the broad HTN-induced monosome peak that was absent in untreated samples (Figures S5C–S5D). Further, we observed an increase in 40S protein abundance relative to 60S only in fraction 5 from HTN-treated cells (Figures 7E and S5E–S5F, Table S3). These results suggest that HTN increases the abundance of stalled preinitiation complexes, which stimulates iRQC pathway activation.
Discussion
We identify RNF10 and USP10 as the key ubiquitylation enzymes that regulate uS3 and uS5 ubiquitylation and demonstrate that persistent uS3 and uS5 ubiquitylation, surprisingly, triggers 40S but not 60S protein degradation. We further demonstrate that RNF10-dependent ubiquitylation is stimulated by a variety of distinct pharmacological agents that inhibit progression of ribosomes either during mRNA scanning, or shortly after the transition to elongating 80S ribosomes. One possible model is that terminally stalled preinitiation complexes, in isolation, are targeted by RNF10 to promote 40S degradation. However, and similar to early descriptions of how eRQC events are triggered, it is unclear how a terminally stalled preinitiation complex that requires iRQC activity can be differentiated from a slowly scanning or paused, but otherwise functional, preinitiation complex. This quandary was rectified by the demonstration that elongation collisions were the key trigger that stimulates eRQC pathway activation (Juszkiewicz et al., 2018; Simms et al., 2017). Intriguingly, the uS5 and uS3 ubiquitylation sites are positioned in the vicinity of the uS10 (RPS20) and eS10 (RPS10) ubiquitylation sites that are required for RQC events during elongation collisions (Figure S6A) (Ikeuchi et al., 2019; Juszkiewicz et al., 2018; Juszkiewicz and Hegde, 2017; Matsuo et al., 2017; Sundaramoorthy et al., 2017). As such, it is plausible that preinitiation complex collisions during the mRNA scanning phase of translation initiation trigger uS3 and uS5 ubiquitylation. However, despite similarities with elongation collisions, collisions between preinitiation complexes or between preinitiation complexes and stalled 80S ribosomes at the start codon would contain initiation factors which would likely constitute a unique collision interface.
Currently, our data cannot distinguish between whether isolated terminally stalled preinitiation complexes or preinitiation complex collisions trigger uS3 and uS5 ubiquitylation. Further biochemical evidence is needed to establish if preinitiation complex collisions occur within cells and if those collisions are targeted by RNF10. However, rapid rates of preinitiation complex loading and scanning relative to translation start would generate queues of potentially collided 43S preinitiation complexes within 5’UTRs. Evidence for such queueing has been demonstrated using in vitro translation systems and translation complex profile sequencing (TCP-seq) in yeast and human cells (Bohlen et al., 2020; Shirokikh et al., 2019; Sogorin et al., 2012; Wagner et al., 2020). Further, generating queues of preinitiation complexes using cycloheximide or insertion of an upstream open reading frame (uORF) resulted in alternative start codon utilization and translation recoding (Ivanov et al., 2018; Kearse et al., 2019). Combined, these studies suggest the possibility that preinitiation ribosome collisions occur.
Because RNF10-catalyzed 40S degradation appears to be autophagy-independent, 40S degradation, in a presumably proteasome-dependent manner, would require 40S disassembly prior to degradation. Thus, iRQC-dependent ribosomal degradation appears distinct from the proteasomal degradation of unassembled ribosomal proteins mediated by either Huwe1 or Ube2O (Nguyen et al., 2017; Sung et al., 2016; Yanagitani et al., 2017). We note that while proteasome inhibition does reduce 40S Ribo-Halo decay upon TMR washout (Figure 4C), it does so in cells expressing wild type and inactive RNF10 as well as those expressing a control protein. It is possible that the inhibition in cell cycle progression observed upon proteasome inhibition may result in slower cell growth and thus reduced dilution of the Ribo-Halo signal. As such, further experiments are needed to examine possible proteosome-mediated degradation of RNF10-catalyzed ubiquitylated 40S ribosome subunits.
Interestingly, in cells lacking USP10, uS5 and uS3 ubiquitylation reaches 20% of total uS5 and uS3 protein. These levels approach and surpass what has been observed for histone ubiquitylation, the most abundantly, and originally identified, ubiquitylated protein in the cell (Goldknopf and Busch, 1977). The large extent of ribosome ubiquitylation in USP10-KO cells also suggests that preinitiation complexes stall at a high frequency in unstressed, albeit rapidly dividing, cells and that USP10 rapidly reverses ubiquitylation of these stalled preinitiation complexes. The fact that uS3 and uS5 ubiquitylation is low in cells with USP10 argue that translation activity has evolved to allow for rapid translation initiation rates and the possible subsequent increase in stalled, and possibly collided preinitiation complexes by maintaining an excess of USP10 relative to RNF10 (Nusinow et al., 2020) (Figure S6B). Further, controlling the relative USP10:RNF10 ratio would set the threshold for the abundance of stalled scanning 40S ribosomes at steady state while enabling stress-sensitive stall responses. USP10 protein abundance is reported to be in 2-fold excess of RNF10 in HEK293 cells and is often in even greater excess, (e.g. 27-fold in HCT116 cells) in many cell lines and tissues with RNF10 abundance being below detection thresholds (Wang et al., 2015). This data suggests that, under normal growth conditions, deubiquitylation of stalled preinitiation complexes is favored over degradation in most cell types to avoid the energetically costly spurious degradation of 40S subunits. These observations also suggest that, with sufficient USP10 activity, stalled preinitiation complexes can eventually transition into elongating ribosomes (Figure S6B).
Similar to eRQC, the iRQC pathway appears to be conserved in single-celled eukaryotes as USP10 and RNF10 orthologs have been shown to regulate uS3 ubiquitylation in S. cerevisiae (Jung et al., 2017; Sugiyama et al., 2019). Not only are the enzymes conserved, but so too are the mechanistic requirements: yeast with inactivating mutations in the peptidyl transferase center of ribosomes that allow for scanning, but block elongation, trigger ribosomal RNA decay in a manner dependent upon uS3 ubiquitylation (Sugiyama et al., 2019). The previous study in S. cerevisiae concluded that damaged ribosomes were the target of ubiquitylation. It is possible that some of the pharmacological agents used within this study to stimulate uS5 and uS3 ubiquitylation result in damaged and nonfunctional ribosomes. However, given the breadth of stimuli used here, and the observation that loss of USP10 results in enhanced uS3 and uS5 ubiquitylation in the absence of stress, we propose that ribosomal ubiquitylation is triggered by preinitiation complex collisions or singular stalled preinitiation complexes. Interestingly, and completely opposite to what we observed during mammalian iRQC, the USP10 homolog in S. cerevisiae, Ubp3, has also been implicated in regulating 60S, but not 40S, ribosome degradation in an autophagy-dependent manner upon starvation (Kraft et al., 2008). Future studies are needed to disentangle starvation-induced ribophagy from iRQC-mediated 40S degradation as they appear to utilize overlapping components.
We propose that conserved ribosomal ubiquitylation acts as a cellular rheostat to dynamically regulate translation dynamics during conditions that enhance collision frequencies. Accumulating evidence suggests that elongation collisions not only trigger ribosomal subunit recycling, but also reduce translation initiation rates (Meydan and Guydosh, 2020; Vind et al., 2020). Our data suggest a possible model wherein stalled preinitiation complexes trigger ubiquitylation of specific 40S ribosomal proteins and that persistent uS5 and uS3 ubiquitylation results in 40S degradation. It is possible that iRQC could be utilized to globally reset translation initiation rates. As cellular proliferation rates change, for example during cellular differentiation, translation capacity and ribosome abundance may also be altered to match metabolic needs. Our demonstration that ISR-stimulating conditions also induce conditions that stimulate RNF10-dependent uS5 and uS3 ubiquitylation suggests that chronic stress signaling may also reset translation capacity. These findings describe a previously uncharacterized, and likely conserved, distinct ribosome-associated quality control pathway that can be utilized to regulate 40S ribosomal levels.
STAR METHODS
RESOURCE AVAILABILTY
Lead contact
Requests for resources or further information can be directed to the Lead Contact Eric J. Bennett (e1bennett@ucsd.edu).
Materials availability
All reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
RESOURCE TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Rabbit monoclonal anti-RPS2 (uS5) | Bethyl Laboratories | Cat# A303-794A; RRID:AB_11218192 |
| Rabbit monoclonal anti-RPS3 (uS3) | Bethyl Laboratories | Cat# A303-840A; RRID:AB_2615588 |
| Rabbit polyclonal anti-RPS10 (eS10) | ABclonal | Cat# A6056; RRID:AB_2766730 |
| Rabbit monoclonal anti-RPS20 (uS10_ | Abcam | Cat# ab133776; RRID:AB_2714148 |
| Rabbit monoclonal anti-p-eIF2α | Cell Signaling Technology | Cat# 3398S; RRID:AB_2096481 |
| Rabbit polyclonal anti-RPL7 (uL30) | Bethyl Laboratories | Cat# A300-741A; RRID:AB_2301241 |
| Rabbit polyclonal anti-USP10 | Abcam | Cat# ab72486; RRID:AB_1271412 |
| Mouse monoclonal α-Tubulin | Cell Signaling Technology | Cat# 3873; RRID:AB_1904178 |
| Mouse monoclonal c-Myc | Santa Cruz Biotechnology | Cat# sc-40 AC; RRID:AB_2857941 |
| Mouse monoclonal anti-tubulin | Cell Signaling Technology | Cat# 3873S; RRID:AB_1904178 |
| Anti-Rabbit IgG (H+L), HRP Conjugate antibody | Promega | Cat# W4011; RRID:AB_430833 |
| Anti-Mouse IgG (H+L), HRP Conjugate antibody | Promega | Cat# W4021; RRID:AB_430834 |
| Lipofectamine RNAiMax | Thermo-Fisher | Cat# 13778030 |
| Lipofectamine 2000 | THermo-Fisher | Cat# 11668019 |
| Mirus TransIT 293 | Mirus Bio llc | Cat# MIR 2700 |
| Protease inhibitor cocktail tablet | Roche | Cat# 11836170001 |
| Immun-Blot® PVDF Membrane | BioRad | Cat# 1620177 |
| Albumin Standard | Thermo Scientific | Cat# 23209 |
| Clarity™ Western ECL Substrate | BioRad | Cat# 170-5061 |
|
| ||
| Chemicals | ||
|
| ||
| DL-Dithiothreitol | ACROS organics | Cat# 165680050 |
| Anisomycin | Fisher Scientific | Cat# 50995788 |
| Harringtonine | LKT labs | Cat# H0169 |
| Thapsigargin | Tocris Biosciences | Cat# 1138 |
| Cycloheximide | MP Biomedicals | Cat# 100183 |
| Sodium arsenite solution | Fluka Analytical | Cat# 35000-1L-R |
| Torin1 | Tocris Biosciences | Cat# 4247 |
| PatamineA | Gift from Jeremy Pelletier’s lab | |
| MG-132 | Enzo Life Sciences | Cat# BML-PI102 |
| Rocaglamide | MedChemExpress | Cat# HY19356 |
| Lactimidomycin | Millipore Sigma | Cat# 5062910001 |
| Bafilomycin A | Sigma | Cat# B1793 |
| SAR405 | APExBio | Cat# A8883 |
| Doxycycline hydrochloride | Fisher Scientific | Cat# BP2653-5 |
| Ammonium persulfate (≥ 98%) | Sigma | Cat# A3678 |
| Fetal Bovine Serum | VWR | Cat# 97068-085 |
| Trypsin | Sigma-Aldrich | Cat# T1426 |
| N-Ethylmaleimide | Sigma-Aldrich | Cat# E3876 |
| Puromycin | Mediatech (Corning) | Cat# 61-385-RA |
| RNaseA | Thermo Scientific | Cat# EN0531 |
| SUPERase-In™ | Invitrogen | Cat# AM2696 |
| Turbo™ DNase | Invitrogen | Cat# AM2238 |
| Trizol | Life Technologies | Cat# 15596026 |
| Dimethyl sulfoxide | Corning | Cat# 25-950-CQC |
| β-mercaptoethanol | J.T. Baker | Cat# 4049-00 |
| Triton-X 100 | Sigma | Cat# T8787 |
| EDTA | Amersco | Cat# E177 |
| Trichloroacetic acid solution | Sigma | Cat# T0699 |
| Thermostable Inorganic | New England | Cat# M0296S |
| Pyrophosphatase | Biolabs | |
| ATP solution | AMSBIO | Cat# 2121-100 |
| Creatine kinase | Biovision | Cat# 76289-160 |
| Creatine phosphate | Sigma Aldrich | Cat# 10621714001 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| BCA Protein Assay | Thermo Scientific (Pierce) | Cat# 23225 |
| Bradford Assay; protein assay dye reagent concentrate | BioRad | Cat# 500-0006 |
| HaloTag®TMR ligand | Promega | Cat# G8251 |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| 293T | ATCC | Cat# CRL-3216 |
| HCT116 | ATCC | Cat# CCL-247 |
| HEK293 | ATCC | Cat# CRL-1573 |
| 293Flp-In T-REx | Thermo Fisher | Cat# R78007 |
| 293T-USP10KO | This paper | N/A |
| 293T-USP21KO | Garshott et al. 2020 | N/A |
| 293T-OTUD3KO | Garshott et al. 2020 | N/A |
| HCT116-uS3-Halo | An et al. 2020 | N/A |
| HCT116-uL26-Halo | This paper | N/A |
| 293T-uS3-Halo | An et al. 2020 | N/A |
| 293T-eL29-Halo | An et al. 2020 | N/A |
| HEK293-uS3-Keima | An et al. 2018 | N/A |
| HEK293-eL28-Keima | An et al. 2018 | N/A |
| HEK293T-RB1CC1KO | An et al. 2020 | N/A |
| 293FlpIn-FRT-Flag-HA-RNF10wt | This paper | N/A |
| 293FlpIn-RNF10KO | This paper | N/A |
| 293FlpIn-RNF10/USP10-DKO | This paper | N/A |
| 293T-Flag-HA-USP10wt | This paper | N/A |
| 293T-Flag-HA-USP10CS | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCMV-N-Myc-RNF10wt | This paper | N/A |
| pCMV-N-Myc-RNF10CS | This paper | N/A |
| pCMV-N-Myc-LRRC49 | This paper | N/A |
| pCMV-N-GFP-RNF10wt | This paper | N/A |
| pCMV-N-GFP-RNF10CS | This paper | N/A |
| pCMV-N-Myc-GFP | This paper | N/A |
|
| ||
| Sequence-Based Reagents | ||
|
| ||
| See Supplemental Experimental Procedures for siRNA reagents | ||
|
| ||
| Software and Algorithms | ||
|
| ||
| FlowJo (v10.6.2) | BD biosciences | |
| Prism 9.0 | GraphPad software | |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All HEK293, HEK293T, HCT116 and 293Flp-In cells were grown in DMEM (high glucose, pyruvate and L-Glutamine) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and maintained in a 5% CO2 humidified incubator.
METHOD DETAILS
Plasmids
Using Gateway cloning (Invitrogen) all protein coding regions were cloned into Myc- of GFP-tagged CMV expression vectors. Mutations were introduced using QuickChange site-directed mutagenesis utilizing PCR-based approaches (primers 5’ to 3’: RNF10-C225S, CATGAAGTGCCATCTTCCCCAATATGCCTCTATC). Template DNA was digested by Dpn1 followed by transformation of the mutated plasmids into TOP10 E. coli cells. Plasmids were confirmed by sequencing and screened for expression by immunoblotting.
Treatments, transfections and siRNA
Prior to harvesting cells were treated with either 1uM Tg, 5mM DTT, 2ug/ml HTN, 100ug/ml CHX, 500uM NaAsO2, 150nM Torin1, 50–100nM Bafilomycin A, 1uM SAR405, 10uM MG132 or were exposed to 0.02J/cm2 UV radiation using a SpectorlinkerTM XL-1000 (Spectronics).
Lentiviral transduction was used to generate stable cells lines expressing Flag-HA tagged USP10. Using Mirus TransIT 293 transfection reagent cells were transfected with five helper plasmids pHAGE-GAG-POL; pHAGE-VSVG; pHAGE-tat1b; pHAGE-rev and pHAGE-Flag-HA-USP10 (wild type or catalytic mutant), followed by the addition of fresh media after 24 hours. The supernatant was filtered using a 0.45 mm sterile syringe filter and mixed with 2ul of 6mg/ml polybrene. The viral mixture was then added to cells seeded at 50% confluency and infected for 24hours. Stable expression clones were selected with 1ug/ml Puromycin.
The Flp-In™ system (Thermo Fisher) through single locus integration and hygromycin selection was used to generate stable doxycycline inducible cell lines expressing Flag-HA-tagged proteins. Flp-In 293 cells were transfected with Flp-In expression vectors for RNF10 using TransIT 293 transfection reagent (Mirus) according to manufacturer guidelines. Cells were seeded at 60% confluency, transfected for 24 hours followed by selection of stable expression clones with 100ug/mL Hygromycin. Treatment with 2ug/mL doxycycline for 16 hours prior to harvesting was used to induce protein expression.
All transient transfections were carried out using Lipofectamine 2000 (Thermo Fisher) and all siRNA knockdown transfections were performed using Lipofectamine RNAiMAX (Thermo Fisher) according to manufacturer instructions. A list of all RNAi oligonucleotides used in this study can be found in table below.
Immunoblotting
For all immunoblot analysis, cell pellets were resuspended in urea denaturing lysis buffer (8M urea, 50mM Tris-Cl, pH 8.0, 75mM NaCl, 1mM NaV, 1mM NaF, 1mM β-glycerophosphate, 40mM NEM in the presence of EDTA-free protease inhibitor cocktail) and kept on ice during preparation. Cell lysates were sonicated for 10 s (output of 3W on a membrane dismembrator model 100 (Fisher Scientific) with a microtip probe then centrifuged for 10 min at 15,000rpm at 4°C. Lysate protein concentrations were measured by BCA Protein Assay (23225, Thermo Scientific Pierce). Laemmli sample buffer with β-mercaptoethanol was then added to cell lysates and heated at 95°C for 10 min. Samples were then cooled to room temperature and centrifuged briefly. Lysates were resolved on 12% Tris-glycine SDS -PAGE gels, followed by transfer to PVDF membranes (1620177, BioRad) using Bjerrum semi-dry transfer buffer (48mMTris Base, 39mM Glycine-free acid, 0.0375% SDS, 20% MeOH, pH 9.2) and a semi-dry transfer apparatus (Bio-Rad Turbo Transfer) for 30 min at 25V. Immunoblots were blocked with 5% blotting grade nonfat dry milk (APEX Bioresearch) in TBST for 1 hour. Primary antibodies were diluted in 5% BSA and rocked overnight. Immunoblots were developed using Clarity Western ECL Substrate (1705061, BioRad) and imaged on a Bio-Rad Chemi-Doc XRS+ system. All blots were processed using Imagelab (BioRad) software, with final images prepared in Adobe Illustrator. All plots were prepared using GraphPad Prism 9.0.
Phos-Tag SDS-PAGE
For Phos-tag analysis, cell pellets were resuspended in 500ul of lysis buffer (8M urea, 50mM Tris-Cl, pH 8.0, 75mM NaCl, 1mM NaV, 1mM NaF, 1mM β-glycerophosphate in the presence of EDTA-free protease inhibitor cocktail). Lysates were sonicated for 10s (as described above) followed by centrifugation for 10 min at 15,000rpm at 4°C. 125ul of TCA was added to each sample, then incubated on ice for 2h at 4°C. Protein was collected by spinning tube in microcentrifuge at 15,000 rpm for 30min at 4°C. The TCA protein pellet was washed with 200ul cold acetone, followed by centrifugation at 15,000 rpm for 10min at 4°C. The acetone wash step was repeated two more times. Pellets were left to dry for 30min at room temperature to evaporate any remaining acetone, then resuspended in 50ul 8M urea/20mM DTT. Protein concentrations were measured by Bradford Assay (protein assay dye reagent concentrate, 500–0006, BioRad). Laemmli sample buffer with β-mercaptoethanol was then added to protein samples and heated at 95°C for 10 min. Samples were resolved on 12.5% SuperSepTM Phos-tagTM gels (198–17981, Fujifilm), followed by Zn2+ ion elimination. Gel was soaked in 1X transfer buffer (25mM Tris, 192mM Glycine, 10% v/v methanol) with 10mM EDTA for 20min with gentle agitation. This step was repeated three times with buffer exchanges, followed by 10min without EDTA. Wet-tank transfer to PVDF membranes using Towbin transfer buffer (25mM Tris, 192mM Glycine, 20% v/v methanol) was done overnight (16h) at 30V. Immunoblots were blocked, developed, and imaged as described above.
Sucrose density gradient fractionation
Cell pellets were lysed in 500 ul of lysis buffer (20mM Tris-Cl, pH 8.0, 150mM NaCl, 15mM MgCl2, 1% Triton-X 100, 40U Turbo DNase I, 40mM NEM, 1mM DTT, EDTA-free protease inhibitor cocktail in DEPC treated water) followed by vigorous pipetting and incubated on ice for 15min. The cell lysates were centrifuged at 15,000 rpm for 10min at 4°C and the supernatant was transferred to a new microcentrifuge tube. Total RNA concentration of each lysate was determined using a nanodrop (Thermo Scientific). 500ug of total RNA was digested with 3.5ug/ml of RNaseA for 15min at 25°C on a thermomixer (Eppendorf) at 500rpm. The digestion was stopped with 166.5U of SUPERaseIn RNase Inhibitor. Samples were fractionated over a 10–30% sucrose gradient containing 150ug/ml cycloheximide (prepared on Gradient Master 108 (Biocomp): 1min 54s, 81.5 degrees, 16rpm). Samples were centrifuged at 41,000rpm for 2 hr at 4°C in an SW41i rotor. 1ml fractions were collected using a PGFip piston gradient fractionator (Biocomp). Protein fractions were precipitated overnight with 10% TCA at 4°C, followed by three ice-cold acetone washes. Pellets were dried in Vacufuge plus (Eppendorf) at room temperature for 5 min. Pellets were then resuspended in Laemmli sample buffer with β-mercaptoethanol, heated at 95°C for 10 min.
SILAC LC-MS-MS analysis
Cells were grown in a media containing dialyzed FBS (FB03, Omega Scientific) and either light (K0) lysine and arginine (R0) or 13C615N2-labeled (K8) lysine and (R10) arginine (Cambridge Isotopes). Cells were harvested and mixed 1:1 by cell count and were processed for mass spectrometry as described previously (Markmiller et al., 2019). Briefly, cells were lysed using 8M urea lysis buffer with 40mM fresh NEM and lysates were quantified for protein content using the BCA assay. 20μg of total cell extract was diluted to a final urea concentration of 1M and then digested overnight with trypsin (V5111, Promega) at a 1:100 (enzyme:protein) ratio. The digests were reduced with 1mM DTT for 30 min and then alkylated with 10mM NEM in a dark for 30min. The digests were desalted using Stage-Tip method and analyzed by LC-MS/MS as described below. Mixed SILAC lysates were fractionated over sucrose gradients as described. Fractions were TCA precipitated, followed by resuspension in 50mM ammonium bicarbonate and digested overnight with 500ng/ul of trypsin (V5111, Promega) at 37°C. Digests were reduced, alkylated and desalted as described above.
All the samples (1ug digested peptides) were analyzed in triplicate by LC-MS/MS using a Q-Exactive mass spectrometer (Thermo Scientific, San Jose, CA) with the following conditions. A fused silica microcapillary column (100 mmID, 20 cm) packed with C18 reverse-phase resin (XSELECT CSH 130 C18 2.5 mm, Waters Co., Wilford, MA) using an in-line nano-flow EASY-nLC 1000 UHPLC (Thermo Scientific) was used to resolve the peptides. Peptides were eluted over a 45 min 2%–30% ACN gradient, a 5 min 30%–60% ACN gradient, a 2 min 60%–95% gradient, with a final 8 min isocratic step at 0% ACN for a total run time of 60 min at a flow rate of 250 nl/min. All gradient mobile phases contained 0.1% formic acid. MS/MS data were collected in a data dependent fashion using a top 10 method with a full MS mass range from 300–1750 m/z, 70,000 resolution, and an AGC target of 3e6. MS2 scans were triggered when an ion intensity threshold of 1e5 was reached with a maximum injection time of 60 ms. Peptides were fragmented using a normalized collision energy setting of 25. A dynamic exclusion time of 20 s was used, and the peptide match setting was disabled. Singly charged ions, charge states above 8 and unassigned charge states were excluded.
The resultant RAW files were analyzed using Andromeda/MaxQuant (version 1.6.12.0) using the combined UniProt reviewed only database for Homo sapiens (Dec 2020). The default parameters were used and ‘match between the runs’ and ‘requantify’ options were enabled in the MaxQuant settings. The proteingroups output table was imported into Microsoft Excel for subsequent data analysis. Normalized SILAC ratios and LFQ intensities were used for data analysis.
Purification of RNF10
Cells were seeded at 50% confluency in ten 10cm plates one day prior to transfection of a N-Flag-TEV-RNF10 expression plasmid using the calcium phosphate method. 20ug of total DNA was mixed with 2M CaCl2 in distilled water. The mixture was added in a dropwise manner to equal volumes 2XHBS (280mM NaCl, 10mM KCl, 1.5mM Na2HPO4, 12mM glucose and 50mM HEPES pH 7.05) solution at room temperature with continuous mixing, followed by incubation at room temperature for 30 minutes. Transfection mixture was added to each plate and incubated overnight at 37°C. 48 hours post transfection cells were collected by scrapping into cold 1X PBS and pelleted at 1,000 rpm for 5min at 4°C. Cells were lysed in 2mL of lysis buffer (50mM HEPES, pH 7.4, 100mM KAc, 5mM MgAc2, 0.5% NP40, 1 mM DTT (made fresh) and 1X EDTA-free Complete protease inhibitor cocktail) and incubated on ice for 20min. Lysates were clarified by centrifugation at 15,000 rpm for 10min at 4°C. 200ul of clarified lysate was added to a 1:1 slurry of pre-equilibrated (in lysis buffer with 0.1% NP40) anti-Flag M2 resin (A2220, Sigma) and incubated with rotation for 2 hours at 4°C. Resin was collected by centrifugation at 3,000 rpm for 1min at 4°C, while flow through was saved in a new tube. Resin was washed three times in 1ml of IP buffer (50mM HEPES, pH 7.4, 100mM KAc, 5mM MgAc2, 0.1% NP40, 1mM DTT (made fresh) and 1X EDTA-free Complete protease inhibitor cocktail) for 2min with rotation, followed by centrifugation. Resin was then washed three times with 1ml of high salt buffer (50mM HEPES, pH 7.4, 400mM KAc, 5mM MgAc2, 0.1% NP40, 1mM DTT), followed by three washes with 1ml of elution buffer (50mM HEPES, pH 7.4, 100mM KAc, 5mM MgAc2, 1mM DTT). Following elution, 100U of His-TEV protease (Z03030–1K, GenScript) was added to the 1:1 slurry of resin in elution buffer and incubated at room temperature for 30min. Resin was washed with an additional 100ul of elution buffer and then pooled with the first elution. 50ul of pre-equilibrated NiNTA agarose resin (30210, Qiagen) was added to the pooled elution fractions and incubated with rotation for 1h at 4°C. Cleared elution was collected by centrifugation, followed by silver stain and immunoblotting for confirmation of protein purification.
In vitro ubiquitylation assay
All in vitro ubiquitylation reactions were carried out for 60min at 37°C. Single reactions consisted of 400nM recombinant human His6-Ubiquitin E1 enzyme Ube1 (E-304, BostonBiochem), 2uM recombinant human UbcH5c/UBE2D3 protein (E2–627, BostonBiochem), 200uM recombinant human ubiquitin no K (UM-NOK, BostonBiochem), 125nM 40S ribosomes (Purified from Hap1 cells, gift from Jody Puglisi and Alex Johnson, Stanford University), 50mM Tris-Cl pH 7.5, 50mM MgCl2, 20mM ATP, 6U/ml pyrophosphatase, 35U/ml creatine kinase and 100mM creatine phosphate, and 8uM RNF10. Reactions were inactivated with Laemmli buffer, then incubated for 10min at 95°C. Proteins were resolved by 12% SDS-PAGE and visualized by immunoblotting.
Generation of knockout and knockin cell lines
Using CRISPR/Cas9 genome engineering USP10 and RNF10 knockout was done in 293Flp-In and 293T cells. Three individual guide RNAs were designed for each gene using CHOPCHOP website (https://chopchop.cbu.uib.no). RNF10: 5’-GCCGGCGAGTCTAAACCCAA-3’, 5’- GCCACGTTAGACTCGGGAAG-3’, 5’- CCGTTGATGCCGCTGAGCTC-3’, USP10: 5’- GACTCCTCGATCTTCAGTTG-3’, 5’- CTTACCTCAACTGAAGATCG-3’ and 5’- GCCTGGGTACTGGCAGTCGA-3. Cells were transfected with the pSpCas9(BB)-2a-GFP plasmid containing individual guide RNAs using lipofectamine 2000. 48 hours post transfection, GFP positive cells were either single cell sorted on a BD FACSAria Fusion (BD BioSciences) cell sorter, or pooled cell sorts were clonally isolated by limiting dilution method. Cells were validated for loss of USP10 and RNF10 by immunoblotting and sequencing. For HaloTag7 knock-in, guide RNA (gRNA) targeting the C-terminal region of human RPL26 gene was designed using the CHOPCHOP website (http://chopchop.cbu.uib.no/). The guide sequence for RPL26 gene (5′- GAAACCATTGAGAAGATGC-3′) was assembled into a pX459 plasmid. Donor vector was constructed by assembling a HaloTag7 transgene with upstream and downstream homology arms (650 nucleotide each) into a digested pSMART plasmid by Gibson assembly. Wild type HCT116 cells were transfected with donor and gRNA vectors (1 to 1 ratio) by Lipofectamine 3000 (Invitrogen). Five days after the transfection, the pool of transfected cells was treated with 100 nM Halo-TMR ligand for 1h, followed by washing three times. Fluorescence-positive cells were sorted into 96-well plates by flow cytometry (MoFlo Astrios EQ, Beckman Coulter). Three weeks later, the expanded single-cell colonies were screened for the integration of the HaloTag7 transgene by immunoblotting with α-RPL26.
Ribo-Halo microscopy
HCT116 Ribo-HaloTag7 cells were transfected with either GFP-RNF10 WT or CS expression plasmid (2 ug/dish) using lipofectamine 3000 (Invitrogen). 24 hours post transfection, the cells were plated onto 35 mm-glass bottom dishes (No. 1.5, 14 mm glass diameter, MatTek) pre-treated with poly-L-lysine. 48 hours later, Halo-TMR containing medium (50 nM) was added to the cells and incubated for 1 hour. The medium was removed, and the cells were washed with warm DMEM for two times. DMEM was replaced by FluoroBrite™DMEM (Thermo Fisher) before the live cell imaging. The cells were imaged using a Yokogawa CSU-X1 spinning disk confocal with Spectral Applied Research Aurora Borealis modification on a Nikon Ti motorized microscope equipped with a Nikon Plan Apo 60×/1.40 N.A objective lens. Pairs of images for TMR and GFP fluorescence were collected sequentially using 100 mW 488 nm and 100 mW 561 solid state lasers attenuated and controlled with an AOTF (Spectral Applied Research LMM-5), and emission collected with a 525/50 nm or 620/60 nm filter (Chroma Technologies), respectively. Confocal images were acquired with the Hamamatsu ORCA-ER cooled CCD camera and MetaMorph software. The images were analyzed using FiJi software.
Flowcytometry analysis for Ribo-Halo labeling
Ribo-Halo cells were seeded at 40% confluency in 12-well plates one day prior to transient transfections. 36 hours post transfection cells were treated with 100nM TMR-ligand (G8251, Promega) for 1–2 hours. After TMR-labeling, cells were washed with fresh warmed DMEM without the Halo-ligand three times with 10min incubations in between washes. Fresh warm DMEM was added to cells and cells were collected at various time points post washout. Cells were trypsinized then collected in fresh media. Following a short 3min centrifugation at 3,500rpm, cell pellets were resuspended in 800ul of FACS buffer (2% FBS in 1× DPBS) and passed through a nitex nylon mesh (Genesee Scientific). Samples were analyzed by flow-cytometry on a BD LSRFortessaTM X-20 cell analyzer (BD Biosciences). FACS data was analyzed using FlowJo (v10.6.2).
qPCR analysis
For qPCR analysis, cells were plated at 50–60% confluency prior to lipofectamine based transfection, as described previously. 48 hours post transfection cells were collected in TRIzol and RNA was isolated using Direct-zol RNA miniprep kit (11–331, Zymo Research). Using 2ug RNA template, cDNA was synthesized is SuperScript III First Strand Synthesis system (18080–051, Invitrogen). Five standards were prepared by making four-fold dilutions of a sample pool. cDNA samples were each diluted 1:5 in water prior to plating. 8ul of each standard or sample was plated into a 96-well thermocycler plate, followed by 12ul of master mix containing SYBR green super mix (1725121, BioRad) and primers for gene of interest. The following primers were used in this study: RPS3: 5’-CAGAACAGAAGGGTGGGAAG-3’, 5’- GCAACATCCAGACTCCAGAATA-3’, RPS6: 5’-GAGCGTTCTCAACTTGGTTATTG-3’, 5’-GCGGATTCTGCTAGCTCTTT-3’, RPL7: 5’-GGCGAGGATGGCAAGAAA-3’, 5’-CTTTGGGCTCACTCCATTGATA-3’, GAPDH: 5’-AACGGGAAGCTTGTCATCAATGGAAA-3’, 5’-GCAGGAGGCAGCTGATGATCTT-3’. The following PCR conditions were run on a C1000 Thermo Cycler (BioRad): 50°C for 10min, 95°C for 15min, 95°C for 10s, 60°C for 30s (repeat for 40 cycles). All relative quantifications were calculated using the delta delta Ct method.
QUANTIFICATION AND STATISTICAL ANALYSIS
All FACS-based assays were performed in triplicate (n = 3) as biologically distinct samples. The median 561nm/488nm ratio and SD were calculated. Transient overexpression experiments were compared to a transfection control. Immunoblot quantification of the relative % ubiquitylation and % phosphorylation was calculated by normalization of the individual intensities for each concentration to that of the no treatment control. Significance (p value) was calculated using an unpaired two-tailed Student’s t test using GraphPad Prism 9.0.
Supplementary Material
Table S1. Proteomic analysis of ribosome abundance in cells with altered iRQC activity, Related to Figure 2.
Log2 SILAC ratios (H-indicated genotype:L-parental) for all measured ribosomal proteins from parental, RNF10-KO, parental with RNF10 overexpression (OE), USP10-KO, USP10-KO with RNF10 OE, or USP10/RNF10 double KO (DKO) cells across replicate experiments. Data represented in Figures 2D and 2E.
Table S2. Proteomic analysis of sucrose gradient fractions with and without HTN, Related to Figures 7 and S5.
Log2 SILAC ratios (H-HTN:L-unt) or label free quantification (LFQ) ion intensities for all measured ribosomal proteins within individual sucrose gradient fractions from untreated or RNase treated cell lysates across three replicate experiments. Data represented in Figures 7D and 7E and S5A–S5F.
Acknowledgments
We thank Jody Puglisi and Alex Johnson (Stanford University) for technical advice and for providing purified ribosomal submits. We also acknowledge Julie Monda, Xuezhen Ge, and Steven Wasserman (UCSD) for editing assistance during manuscript preparation. The graphical abstract and Figure 4A were created with BioRender.com. This research was supported by funding from the National Institutes for Health (DP2GM119132 (E.J.B.), R01GM136994 (E.J.B.), R01AG011085 (J.W.H.), R01NS083524 (J.W.H.), T32GM007240 (D.M.G.), and the National Science Foundation (DGE-1650112 (D.M.G.)).
Footnotes
Declaration of interests
E.J.B. serves on the scientific advisory board of Plexium Inc.. J.W.H. is a consultant and founder of Caraway Therapeutics and is a founding board member of Interline Therapeutics.
References
- An H, and Harper JW (2018). Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol 20, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, and Harper JW (2020). Ribosome Abundance Control Via the Ubiquitin-Proteasome System and Autophagy. J Mol Biol 432, 170–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Ordureau A, Korner M, Paulo JA, and Harper JW (2020). Systematic quantitative analysis of ribosome inventory during nutrient stress. Nature 583, 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Ordureau A, Paulo JA, Shoemaker CJ, Denic V, and Harper JW (2019). TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol Cell 74, 891–908 e810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen J, Fenzl K, Kramer G, Bukau B, and Teleman AA (2020). Selective 40S Footprinting Reveals Cap-Tethered Ribosome Scanning in Human Cells. Mol Cell 79, 561–574 e565. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, and Walter P (2020). The integrated stress response: From mechanism to disease. Science 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio KN, and Green R (2021). Ribosome states signal RNA quality control. Mol Cell 81, 1372–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno M, Jimenez A, and Vazquez D (1977). Inhibition of translation in eukaryotic systems by harringtonine. European journal of biochemistry 72, 323–330. [DOI] [PubMed] [Google Scholar]
- Garshott DM, Sundaramoorthy E, Leonard M, and Bennett EJ (2020). Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T, et al. (2017). The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun 8, 16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf IL, and Busch H (1977). Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proceedings of the National Academy of Sciences of the United States of America 74, 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey KL, Dickson K, Cogan JZ, Replogle JM, Schoof M, D’Orazio KN, Sinha NK, Hussmann JA, Jost M, Frost A, et al. (2020). GIGYF2 and 4EHP Inhibit Translation Initiation of Defective Messenger RNAs to Assist Ribosome-Associated Quality Control. Mol Cell 79, 950–962 e956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R, Gendron JM, Rising L, Mak R, Webb K, Kaiser SE, Zuzow N, Riviere P, Yang B, Fenech E, et al. (2015). The Unfolded Protein Response Triggers Site-Specific Regulatory Ubiquitylation of 40S Ribosomal Proteins. Mol Cell 59, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2014). The scanning mechanism of eukaryotic translation initiation. Annual review of biochemistry 83, 779–812. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, and Sonenberg N (2016). Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R, and Inada T (2019). Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T (2020). Quality controls induced by aberrant translation. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Shin BS, Loughran G, Tzani I, Young-Baird SK, Cao C, Atkins JF, and Dever TE (2018). Polyamine Control of Translation Elongation Regulates Start Site Selection on Antizyme Inhibitor mRNA via Ribosome Queuing. Mol Cell 70, 254–264 e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, and Ingolia NT (2016). Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534, 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CAP (2019). Mechanisms and functions of ribosome-associated protein quality control. Nature reviews. Molecular cell biology 20, 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Kim HD, Yang HW, Kim HJ, Jang CY, and Kim J (2017). Modulating cellular balance of Rps3 mono-ubiquitination by both Hel2 E3 ligase and Ubp3 deubiquitinase regulates protein quality control. Exp Mol Med 49, e390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, and Hegde RS (2018). ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol Cell 72, 469–481 e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, and Hegde RS (2017). Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination. Mol Cell 65, 743–750 e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Slodkowicz G, Lin Z, Freire-Pritchett P, Peak-Chew SY, and Hegde RS (2020). Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Goldman DH, Choi J, Nwaezeapu C, Liang D, Green KM, Goldstrohm AC, Todd PK, Green R, and Wilusz JE (2019). Ribosome queuing enables non-AUG translation to be resistant to multiple protein synthesis inhibitors. Genes Dev 33, 871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, and Peter M (2008). Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10, 602–610. [DOI] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, and Qian SB (2012). Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proceedings of the National Academy of Sciences of the United States of America 109, E2424–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, and Liu JO (2005). Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20, 709–722. [DOI] [PubMed] [Google Scholar]
- Lyumkis D, Oliveira dos Passos D, Tahara EB, Webb K, Bennett EJ, Vinterbo S, Potter CS, Carragher B, and Joazeiro CA (2014). Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proceedings of the National Academy of Sciences of the United States of America 111, 15981–15986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Fulzele A, Higgins R, Leonard M, Yeo GW, and Bennett EJ (2019). Active Protein Neddylation or Ubiquitylation Is Dispensable for Stress Granule Dynamics. Cell Rep 27, 1356–1363 e1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, et al. (2017). Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun 8, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meydan S, and Guydosh NR (2020). A cellular handbook for collided ribosomes: surveillance pathways and collision types. Current genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Garzia A, Morozov P, Molina H, and Tuschl T (2020). The G3BP1-Family-USP10 Deubiquitinase Complex Rescues Ubiquitinated 40S Subunits of Ribosomes Stalled in Translation from Lysosomal Degradation. Mol Cell 77, 1193–1205 e1195. [DOI] [PubMed] [Google Scholar]
- Montellese C, van den Heuvel J, Ashiono C, Dorner K, Melnik A, Jonas S, Zemp I, Picotti P, Gillet LC, and Kutay U (2020). USP16 counteracts mono-ubiquitination of RPS27a and promotes maturation of the 40S ribosomal subunit. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Prado MA, Schmidt PJ, Sendamarai AK, Wilson-Grady JT, Min M, Campagna DR, Tian G, Shi Y, Dederer V, et al. (2017). UBE2O remodels the proteome during terminal erythroid differentiation. Science 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER 3rd, Kalocsay M, Jane-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, et al. (2020). Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 180, 387–402 e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Horikawa W, and Ito K (2015). Inhibiting K63 polyubiquitination abolishes no-go type stalled translation surveillance in Saccharomyces cerevisiae. PLoS Genet 11, e1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, and Liu JO (2010). Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nature chemical biology 6, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, and Hegde RS (2014). Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol Cell 55, 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, von der Malsburg K, and Hegde RS (2013). Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol Cell 50, 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokikh NE, Dutikova YS, Staroverova MA, Hannan RD, and Preiss T (2019). Migration of Small Ribosomal Subunits on the 5’ Untranslated Regions of Capped Messenger RNA. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GM, Finley D, and Vogel C (2015). K63 polyubiquitination is a new modulator of the oxidative stress response. Nat Struct Mol Biol 22, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, and Zaher HS (2017). Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol Cell 68, 361–373 e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NK, Ordureau A, Best K, Saba JA, Zinshteyn B, Sundaramoorthy E, Fulzele A, Garshott DM, Denk T, Thoms M, et al. (2020). EDF1 coordinates cellular responses to ribosome collisions. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogorin EA, Shirokikh NE, Ibragimova AM, Vasiliev VD, Agalarov S, and Spirin AS (2012). Leader sequences of eukaryotic mRNA can be simultaneously bound to initiating 80S ribosome and 40S ribosomal subunit. Biochemistry (Mosc) 77, 342–345. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Li S, Kato M, Ikeuchi K, Ichimura A, Matsuo Y, and Inada T (2019). Sequential Ubiquitination of Ribosomal Protein uS3 Triggers the Degradation of Non-functional 18S rRNA. Cell Rep 26, 3400–3415 e3407. [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, and Bennett EJ (2017). ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol Cell 65, 751–760 e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MK, Porras-Yakushi TR, Reitsma JM, Huber FM, Sweredoski MJ, Hoelz A, Hess S, and Deshaies RJ (2016). A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrun A, Garzia A, Kigoshi-Tansho Y, Patil PR, Umbaugh CS, Dallinger T, Liu J, Kreger S, Patrizi A, Cox GA, et al. (2021). Convergence of mammalian RQC and C-end rule proteolytic pathways via alanine tailing. Mol Cell 81, 2112–2122 e2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vind AC, Genzor AV, and Bekker-Jensen S (2020). Ribosomal stress-surveillance: three pathways is a magic number. Nucleic Acids Res 48, 10648–10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Herrmannova A, Hronova V, Gunisova S, Sen ND, Hannan RD, Hinnebusch AG, Shirokikh NE, Preiss T, and Valasek LS (2020). Selective Translation Complex Profiling Reveals Staged Initiation and Co-translational Assembly of Initiation Factor Complexes. Mol Cell 79, 546–560 e547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, and von Mering C (2015). Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15, 3163–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Juszkiewicz S, and Hegde RS (2017). UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357, 472–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip MCJ, and Shao S (2021). Detecting and Rescuing Stalled Ribosomes. Trends in biochemical sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Proteomic analysis of ribosome abundance in cells with altered iRQC activity, Related to Figure 2.
Log2 SILAC ratios (H-indicated genotype:L-parental) for all measured ribosomal proteins from parental, RNF10-KO, parental with RNF10 overexpression (OE), USP10-KO, USP10-KO with RNF10 OE, or USP10/RNF10 double KO (DKO) cells across replicate experiments. Data represented in Figures 2D and 2E.
Table S2. Proteomic analysis of sucrose gradient fractions with and without HTN, Related to Figures 7 and S5.
Log2 SILAC ratios (H-HTN:L-unt) or label free quantification (LFQ) ion intensities for all measured ribosomal proteins within individual sucrose gradient fractions from untreated or RNase treated cell lysates across three replicate experiments. Data represented in Figures 7D and 7E and S5A–S5F.
Data Availability Statement
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
RESOURCE TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Rabbit monoclonal anti-RPS2 (uS5) | Bethyl Laboratories | Cat# A303-794A; RRID:AB_11218192 |
| Rabbit monoclonal anti-RPS3 (uS3) | Bethyl Laboratories | Cat# A303-840A; RRID:AB_2615588 |
| Rabbit polyclonal anti-RPS10 (eS10) | ABclonal | Cat# A6056; RRID:AB_2766730 |
| Rabbit monoclonal anti-RPS20 (uS10_ | Abcam | Cat# ab133776; RRID:AB_2714148 |
| Rabbit monoclonal anti-p-eIF2α | Cell Signaling Technology | Cat# 3398S; RRID:AB_2096481 |
| Rabbit polyclonal anti-RPL7 (uL30) | Bethyl Laboratories | Cat# A300-741A; RRID:AB_2301241 |
| Rabbit polyclonal anti-USP10 | Abcam | Cat# ab72486; RRID:AB_1271412 |
| Mouse monoclonal α-Tubulin | Cell Signaling Technology | Cat# 3873; RRID:AB_1904178 |
| Mouse monoclonal c-Myc | Santa Cruz Biotechnology | Cat# sc-40 AC; RRID:AB_2857941 |
| Mouse monoclonal anti-tubulin | Cell Signaling Technology | Cat# 3873S; RRID:AB_1904178 |
| Anti-Rabbit IgG (H+L), HRP Conjugate antibody | Promega | Cat# W4011; RRID:AB_430833 |
| Anti-Mouse IgG (H+L), HRP Conjugate antibody | Promega | Cat# W4021; RRID:AB_430834 |
| Lipofectamine RNAiMax | Thermo-Fisher | Cat# 13778030 |
| Lipofectamine 2000 | THermo-Fisher | Cat# 11668019 |
| Mirus TransIT 293 | Mirus Bio llc | Cat# MIR 2700 |
| Protease inhibitor cocktail tablet | Roche | Cat# 11836170001 |
| Immun-Blot® PVDF Membrane | BioRad | Cat# 1620177 |
| Albumin Standard | Thermo Scientific | Cat# 23209 |
| Clarity™ Western ECL Substrate | BioRad | Cat# 170-5061 |
|
| ||
| Chemicals | ||
|
| ||
| DL-Dithiothreitol | ACROS organics | Cat# 165680050 |
| Anisomycin | Fisher Scientific | Cat# 50995788 |
| Harringtonine | LKT labs | Cat# H0169 |
| Thapsigargin | Tocris Biosciences | Cat# 1138 |
| Cycloheximide | MP Biomedicals | Cat# 100183 |
| Sodium arsenite solution | Fluka Analytical | Cat# 35000-1L-R |
| Torin1 | Tocris Biosciences | Cat# 4247 |
| PatamineA | Gift from Jeremy Pelletier’s lab | |
| MG-132 | Enzo Life Sciences | Cat# BML-PI102 |
| Rocaglamide | MedChemExpress | Cat# HY19356 |
| Lactimidomycin | Millipore Sigma | Cat# 5062910001 |
| Bafilomycin A | Sigma | Cat# B1793 |
| SAR405 | APExBio | Cat# A8883 |
| Doxycycline hydrochloride | Fisher Scientific | Cat# BP2653-5 |
| Ammonium persulfate (≥ 98%) | Sigma | Cat# A3678 |
| Fetal Bovine Serum | VWR | Cat# 97068-085 |
| Trypsin | Sigma-Aldrich | Cat# T1426 |
| N-Ethylmaleimide | Sigma-Aldrich | Cat# E3876 |
| Puromycin | Mediatech (Corning) | Cat# 61-385-RA |
| RNaseA | Thermo Scientific | Cat# EN0531 |
| SUPERase-In™ | Invitrogen | Cat# AM2696 |
| Turbo™ DNase | Invitrogen | Cat# AM2238 |
| Trizol | Life Technologies | Cat# 15596026 |
| Dimethyl sulfoxide | Corning | Cat# 25-950-CQC |
| β-mercaptoethanol | J.T. Baker | Cat# 4049-00 |
| Triton-X 100 | Sigma | Cat# T8787 |
| EDTA | Amersco | Cat# E177 |
| Trichloroacetic acid solution | Sigma | Cat# T0699 |
| Thermostable Inorganic | New England | Cat# M0296S |
| Pyrophosphatase | Biolabs | |
| ATP solution | AMSBIO | Cat# 2121-100 |
| Creatine kinase | Biovision | Cat# 76289-160 |
| Creatine phosphate | Sigma Aldrich | Cat# 10621714001 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| BCA Protein Assay | Thermo Scientific (Pierce) | Cat# 23225 |
| Bradford Assay; protein assay dye reagent concentrate | BioRad | Cat# 500-0006 |
| HaloTag®TMR ligand | Promega | Cat# G8251 |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| 293T | ATCC | Cat# CRL-3216 |
| HCT116 | ATCC | Cat# CCL-247 |
| HEK293 | ATCC | Cat# CRL-1573 |
| 293Flp-In T-REx | Thermo Fisher | Cat# R78007 |
| 293T-USP10KO | This paper | N/A |
| 293T-USP21KO | Garshott et al. 2020 | N/A |
| 293T-OTUD3KO | Garshott et al. 2020 | N/A |
| HCT116-uS3-Halo | An et al. 2020 | N/A |
| HCT116-uL26-Halo | This paper | N/A |
| 293T-uS3-Halo | An et al. 2020 | N/A |
| 293T-eL29-Halo | An et al. 2020 | N/A |
| HEK293-uS3-Keima | An et al. 2018 | N/A |
| HEK293-eL28-Keima | An et al. 2018 | N/A |
| HEK293T-RB1CC1KO | An et al. 2020 | N/A |
| 293FlpIn-FRT-Flag-HA-RNF10wt | This paper | N/A |
| 293FlpIn-RNF10KO | This paper | N/A |
| 293FlpIn-RNF10/USP10-DKO | This paper | N/A |
| 293T-Flag-HA-USP10wt | This paper | N/A |
| 293T-Flag-HA-USP10CS | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCMV-N-Myc-RNF10wt | This paper | N/A |
| pCMV-N-Myc-RNF10CS | This paper | N/A |
| pCMV-N-Myc-LRRC49 | This paper | N/A |
| pCMV-N-GFP-RNF10wt | This paper | N/A |
| pCMV-N-GFP-RNF10CS | This paper | N/A |
| pCMV-N-Myc-GFP | This paper | N/A |
|
| ||
| Sequence-Based Reagents | ||
|
| ||
| See Supplemental Experimental Procedures for siRNA reagents | ||
|
| ||
| Software and Algorithms | ||
|
| ||
| FlowJo (v10.6.2) | BD biosciences | |
| Prism 9.0 | GraphPad software | |


