Abstract
We describe here the genetic organization of the mec element downstream of the mecA gene in 34 different methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates carrying 13 of the most frequent polymorphisms of mecA and representing the major epidemic clones of MRSA. All polymorphisms carried three common genetic elements: the hypervariable region, a copy of IS431, and a unique 2-kb sequence (downstream constant segment, or dcs) for which no homologous sequences are found in data banks. Polymorphisms of the downstream region were shown to be caused by the presence of linearized plasmids flanked by insertion sequences (pUB110, pT181, and pI258) and the autonomous insertion sequence IS256.
The 2.1-kb mecA gene is part of a larger (40 to 60 kb) mec element of extraspecies origin which extends both upstream and downstream of the mecA determinant. mecA has a single ClaI digestion site, and therefore chromosomal ClaI digests of methicillin-resistant Staphylococcus aureus (MRSA) produce two mecA hybridizing fragments, one of which, including sequences downstream of mecA, exhibits considerable strain-to-strain variation in molecular size, ranging from 3.5 to 20 kb. The size of the second smaller mecA-hybridizing fragment, produced from the upstream side of mecA, is virtually invariant. Thus, most of the polymorphisms observed in the mecA region reflect variation in DNA downstream of mecA. Up to now, as many as 21 such mecA polymorphisms have been identified in various MRSA isolates (3, 12, 16, 18, 21, 26), and these polymorphisms have been used as an epidemiological marker and have also been the basis of speculation concerning the evolutionary origin of methicillin resistance in S. aureus (16).
The main purpose of the study described here was to determine the molecular basis of mecA polymorphisms by cloning and sequencing the downstream region of the mecA element in isolates representing the major current clonal types of MRSA.
MATERIALS AND METHODS
Bacterial strains, phage, and plasmids.
Escherichia coli host strains, vectors, and recombinant vectors are described in Table 1. MRSA clones and reference strains are listed in Table 2.
TABLE 1.
E. coli host strains, vectors, and recombinant vectors
| Host strain, vector, or recombinant vector | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli DH5α | Recipient strain for recombinant plasmids | Gibco BRL |
| E. coli XL1 Blue MRF′ | Recipient strain for recombinant lambda | Stratagene |
| pGEM-3Z | Ampr high-copy-number subcloning vector | Promega |
| pACYC177 | Ampr Kanr low-copy-number cloning vector | 23 |
| Lambda ZAP Express | Cloning vector | Stratagene |
| pDO-1 | pACYC177/6.2-kb ClaI-EcoRI fragment containing the 3′ end of mecA, the HVR, the IS431 left copy, and the aadD, ble, and rec genes of pUB110 | This study |
| λDO-2 | Lambda ZAP Express/6.3-kb BamHI-BamHI fragment containing the rep gene of pUB110 and the IS431 right copy | This study |
| pDO-3 | pGEM-3Z/3.5-kb PstI-BamHI fragment of pDO-1 insert | This study |
| pDO-4 | pACYC177/2.9-kb BamHI-BglII fragment of λDO-2 insert | This study |
| pDO-5 | pACYC177/3.4-kb BglII-BamHI fragment of λDO-2 insert | This study |
| pDO-6 | pACYC177/4.2-kb BamHI-ClaI fragment of the remaining sequence up to the first ClaI digestion site downstream of the mecA gene | This study |
TABLE 2.
ClaI::mecA vicinity organization features of MRSA epidemic clonesa
| Strain | Pattern | Organization featuresb | Observation/origin (reference) |
|---|---|---|---|
| HUC19 | I | ΔHVR + pUB110 + Ins117 | Iberian clone/Portugal (21) |
| HUC31 | I | ΔHVR + pUB110 + Ins117 | Iberian clone/Portugal (21) |
| BK1920 | I | ΔHVR + pUB110 + Ins117 | Iberian clone/USA (22) |
| BK59 | I | ΔHVR + pUB110 + Ins117 | mecA type I control strain (16) |
| BK2464 | I | pUB110 + Ins117 | New York clone/USA (17) |
| JP1 | I | pUB110 + Ins117 | Epidemic clone/Japanc |
| PL72 | I | pUB110 | Epidemic clone/Poland (18) |
| COL | II | Reference strain/RU collection (8) | |
| BB270 | II | (24) | |
| BK71 | II | mecA type II control strain (16) | |
| HUC191 | II | ΔHVR + Ins117 | Iberian clone related/Portugal (21) |
| DEN 2125 | II | Epidemic clone 1960s/Denmark (4) | |
| R35 | III | pT181 + pI258 | Reference strain/USA (13) |
| CPS22 | III | pT181 | Epidemic clone/Portugal (5) |
| CPS68 | III | pT181 | Epidemic clone/Portugal (5) |
| HU25 | III* | pI258 | Brazilian clone (29) |
| PER168 | IX | pT181 + pI258 | mecA type IX control strain (12) |
| PER222 | XI | pT181 + pI258 | mecA type XI control strain (12) |
| BK163 | III | pT181 + pI258 | mecA type III control strain (16) |
| HU106 | IX | pT181 + pI258 | Hungarian cloned (6) |
| HU41 | XI | pT181 + pI258 | Hungarian cloned (6) |
| HU101 | III | pT181 + pI258 | Hungarian cloned (6) |
| HU86 | III′ | pT181 + pI258 | Hungarian cloned (6) |
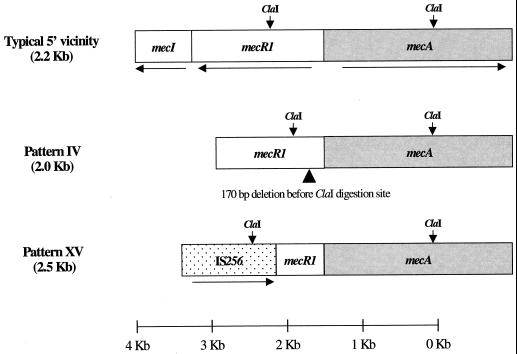
| BK2421 | IV | mecR1Δ170bp (5′)/pT181+pI258 (3′) | mecA type IV control strain (16) |
| LHH1 | IV | mecR1Δ170bp (5′)/pT181+pI258 (3′) | Epidemic clone/USA (7) |
| BK2529 | V | IS256 | mecA type V control strain (16) |
| BARGII17 | V | IS256 | Epidemic clone/USA (7) |
| HSA49 | V | IS256 | Sporadic clone/Portugal (10) |
| BK793 | VI | pI258 | mecA type VI control strain (16) |
| HU195 | VI | pI258 | Sporadic clone/Hungary (5) |
| HUC64 | VIII | ΔHVR + pUB110 + IS256 | Iberian clone related/Portugal (21) |
| IPO108 | XIII | ΔHVR + pUB110 + IS256 | Iberian clone related/Portugal (26) |
| VIC1 | XIV | ΔHVR + IS256 | Iberian clone related/Portugale |
| PL46 | XV | mecR1::IS256 (5′)/pT181 (3′) | Sporadic clone/Poland (18) |
The small differences among strains within pattern I or II (ΔHVR and Ins117) cannot be detected by the conventional gel electrophoresis settings used in ClaI::mecA pattern determination (5, 16). Polymorphisms IX, XI, III, and III′ differ primarily in the copy number of the short 40-bp sequence within the dru element: 11 copies in IX, 10 in XI, 9 in III, and 8 in III′ (see accession numbers in Materials and Methods).
Abbreviations: ΔHVR, truncated HVR, which has a deletion of 1,043 bp spanning all of the dru element; Ins117, 117-bp sequence flanked by two 15-bp direct repeats; mecR1Δ170bp, 170-bp deletion in the mecR1 coding region.
M. A. de Sousa et al., unpublished data.
D. Oliveira, P. Major, I. S. Sanches, A. Marton, and H. de Lencastre, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E170, 1998.
R. Sá-Leão, I. S. Sanches, F. Fonseca, and H. de Lencastre, Abstr. 1st Eur. Congr. Antimicrob. Agents Chemother., abstr. F164, 1996.
Media and growth conditions.
S. aureus strains were grown in tryptic soy broth or in tryptic soy agar (Difco Laboratories, Detroit, Mich.) with aeration at 37°C for 18 h. Luria-Bertani medium (Difco Laboratories) was used to propagate E. coli DH5α, and ampicillin (100 μg/ml) was added for selection and maintenance of plasmids. E. coli XL1-Blue MRF′ was the host strain for the Lambda ZAP Express vector. All manipulations were performed according to the manufacturer's recommendations (Stratagene Cloning Systems, La Jolla, Calif.).
DNA methods.
Standard DNA methodologies were used (2, 25). Restriction endonucleases (New England Biolabs, Beverely, Mass.), calf intestinal alkaline phosphatase, and T4 DNA ligase (Boehringer Mannheim, Indianapolis, Ind.) were used according to the manufacturer's instructions. Plasmid DNA was isolated using the Wizard Plus Minipreps or Wizard Plus Midipreps DNA purification systems (Promega, Madison, Wis.). Lambda DNA was isolated with the Qiagen Lambda Midi kit (Qiagen GmbH, Dusseldorf, Germany). Chromosomal DNAs for PCR were prepared as previously described (11). Southern hybridization, colony blotting, and plaque lifts were performed with the ECL random prime labeling and detection systems (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) according to the manufacturer's recommendations.
PCR.
PCR amplifications were performed in a DNA Thermal Cycler 480 (Perkin-Elmer Cetus [PEC], Branchburg, N.J.). The PCR mixture was prepared with a PCR reagent kit according to the manufacturer's recommendations (PEC). In each reaction, 40 pmol of each primer and 1 ng of plasmid or 5 ng of chromosomal template DNA were included. The amplifications were carried out at the following temperature profiles: 94°C for 4 min; 35 cycles of 94°C for 30 s, 47 to 53°C for 30 s, and 72°C for 1 to 4 min; and 72°C for 4 min and 4°C for the remainder of the reaction. For inverted PCR (IPCR), the chromosomal DNA was digested with ClaI; self-ligation and PCR amplification were performed essentially as previously described (9). Long-range PCR was performed by use of a GeneAmp XL kit (PEC) with a magnesium concentration of 1.1 mM, 40 pmol of each primer, and 5 ng of template DNA. The thermal profile was as follows: 94°C for 2 min, 37 cycles of 94°C for 30 s, 65°C for 7 min, 72°C for 10 min, and soaking at 4°C.
Cloning of the ClaI::mecA downstream vicinity of the Iberian MRSA clone (pattern I).
The recombinant plasmids and phage are listed in Table 1, and the digestion sites used are depicted in Fig. 1. The 6.0-kb ClaI-EcoRI fragment was cloned into the low-copy-number plasmid vector pACYC177 (recombinant plasmid pDO-1). The PstI-BamHI fragment of pDO-1 was then subcloned into pGEM-3Z (plasmid pDO-3). The 6.5-kb BamHI-BamHI fragment was cloned into bacteriophage λ to generate recombinant bacteriophage λDO-2. The DNA insert of λDO-2 was amplified by long-range PCR using the vector-based primers (T3-AAT TAA CCC TCA CTA AAG GG and T7-GTA ATA CGA CTC ACT ATA GGG C). The amplicon was digested with BamHI and BglII, and the two digests were ligated into pACYC177, creating the plasmids pDO-4 and pDO-5, respectively. The 3′ sequence of the AclI-BamHI fragment in the λDO-2 insert was used to design inverted primers (ABP3-AAT AAC TTG TGG ATA ACT GG and ABP4-TTG GTA ACT GTT TCT CAT GC), and IPCR was performed by use of AclI-digested chromosomal DNA as a template to obtain sequence data for the further downstream region. Finally, the BamHI-ClaI fragment was ligated with pACYC177 to produce pDO-6. The DNA inserts in plasmids pDO-1, pDO-3, pDO-4, pDO-5, and pDO-6 were sequenced by the primer walking strategy.
FIG. 1.
Molecular organization of the ClaI::mecA downstream polymorphisms. ClaI-mecA types are displayed on the left; between parentheses are the approximate sizes of the ClaI::mecA downstream vicinities in kilobases. ClaI digestion sites are indicated with vertical arrows. Coding sequences for each plasmid are indicated as described in the original references. ΔHVR, deleted HVR; dcs, downstream constant segment; Ins117, the 117-bp sequence flanked by two 15-bp direct repeats; orfX, insertion site of the mec element into the S. aureus chromosome as defined by Ito et al. (15). In pattern I organization the digestion sites used for cloning are indicated, as are the locations of primers used in the screening of the other vicinities (see Table 3 for primer sequences).
Nucleotide sequencing.
DNA sequencing was performed at the Rockefeller University Protein/DNA Technology Center with an automated DNA sequencing system (Model 377; Perkin-Elmer Applied Biosystems, Foster City, Calif.). Sequence data were analyzed with the DNA Star software (Lasergene, Madison, Wis.). Homology searches were performed with the BLAST utility available through the National Center for Biotechnology Information website.
Nucleotide sequence accession number.
Nucleotide sequences were submitted to the DDBJ/EMBL/GenBank database under accession numbers AF142100 (mecR1 170-bp deletion; strain LHH1), AF142101 (hypervariable region [HVR]; strain HU106), AF142102 (HVR; strain HU41), AF142103 (HVR; strain HU101), and AF18195 (ClaI::mecA downstream vicinity; strain HUC19).
Previously published sequences.
Previously published sequences were obtained through the GenBank database website (http://www.ncbi.nlm.nih.gov/). The accession numbers are as follows: for pUB110, M19465; for pT181, J01764; for pI258, L29436; for IS431, X3818; and for IS256, M18086.
RESULTS AND DISCUSSION
Experimental strategy.
We first cloned and sequenced the 16-kb ClaI::mecA downstream region of the MRSA strain HUC19 (ClaI::mecA pattern I), a prototype of the widely spread Iberian clone (12, 21). Based on the sequence data obtained with HUC19, several primers were designed spanning the entire downstream region (Table 3; Fig. 1), and a representative collection of other MRSA clones was screened by PCR in order to detect regions of difference, revealed either by lack of amplification or by shifts in the amplified fragment sizes. When there was no amplification, long-range PCR was used to test for the presence of hypothetical insertion elements in the region. The flanking regions of such polymorphic fragments were then sequenced, and a homology search was performed.
TABLE 3.
Primers used for screening the mecA downstream vicinitya
| Primer | Sequence | Location |
|---|---|---|
| mecA P1 | ATC GAT GGT AAA GGT TGG C | 1 |
| HVR P1 | ATG TCC CAA GCT CCA TTT TG | 2213 |
| HVR P2 | TGG AGC TTG GGA CAT AAA TG | 2227 compl. |
| IS P1 | AAG GGA ATC TTC TGT ATG AAC | 2294 |
| IS P2 | TCA GTG TTC GCT TAA CTT GC | 2994 compl. |
| IS P3 | TTA CTT TAG CCA TTG CTA CC | 2744 compl. |
| IS P4 | CAG GTC TCT TCA GAT CTA CG | 2920 |
| MDV F1 | GCT TGG GTA ACT TAT CAT GG | 10146 |
| MDV F2 | CGC TGT TAA CAC TGT CTT ACC | 12127 |
| MDV F3 | CTT TAT CCG TAT CAC ATA GC | 14014 |
| MDV F4 | CTG CTT ATA TTC TTT CAT AGC | 9177 |
| MDV F5 | TAA CTT GTT CAC ACT GTT CC | 11124 |
| MDV R1 | AGA CAA CTT TAT GCA GGT CC | 10267 compl. |
| MDV R2 | AAT GGT GGA GGT ATT CGT GC | 12312 compl. |
| MDV R3 | ACG TGA TTA CAG GTA ACA CG | 14166 compl. |
| MDV R4 | TGT TAA TAA AGT CAA TCG C | 15890 compl. |
| MDV R5 | CAT GGC TAT GAT TTA GTA GC | 9326 compl. |
| MDV R6 | AGT TAT CCA CAA ATA CAC AGG | 11236 compl. |
Primer locations are based on the deduced sequence of the mecA downstream vicinity for strain HUC19 (accession number AF18195). Compl., complementary strand.
The MRSA collection (Table 2) tested included 34 strains with 13 different ClaI::mecA patterns (polymorphisms) covering most of the observed range of variability in the downstream vicinity. The most frequent polymorphisms (I, II, and III) were studied in several strains from different geographic origins. Less-frequent patterns were studied in two strains, the strain for which the pattern was originally described and a second strain from a different geographic origin. Strains sharing the same ClaI::mecA pattern were compared by sizes of PCR fragments and by nucleotide sequencing of the polymorphic regions.
Genetic organization of the mecA downstream region in MRSA strain HUC19 carrying mecA polymorph I.
The ClaI::mecA downstream region of this strain was shown to be 15,897 bp, beginning at the ClaI digestion site internal to the mecA gene (Fig. 1). Directly downstream of mecA we found a copy of the HVR which, in relation to the HVR described by Ryffel et al. (24), had a deletion of 1,043 bp spanning the entire dru element. Adjoining the HVR downstream was a copy of the linearized plasmid pUB110 containing the kanamycin and neomycin resistance determinants flanked by two direct repeats of the insertion sequence IS431. The pUB110 linearization caused no interruption of any genes on the plasmid. The rep gene open reading frame contained a nonsense mutation and the rec gene showed a single amino acid substitution in comparison to the published pUB110 sequence (20), which may be a mechanism to stabilize the plasmid-borne antibiotic resistance markers in the MRSA chromosome. The inserted pUB110 was flanked by two external 16-bp inverted repeats, which belong to the ends of IS431, and by two 8-bp internal direct repeats (target sequence duplication). The remaining downstream sequence had some putative open reading frames. Within this region, a short 117-bp sequence flanked by two 15-bp direct repeats (Ins117) was identified.
Genetic organization of the downstream mecA region in MRSA carrying different mecA polymorphisms is shown in Fig. 1. Figure 2 shows two exceptional upstream patterns, one larger and the other smaller than the normal invariant upstream ClaI fragment.
FIG. 2.
Molecular organization of the ClaI::mecA upstream polymorphisms. ClaI-mecA types are displayed on the left; between parentheses are the approximate sizes of the ClaI::mecA upstream vicinities in kilobases. ClaI digestion sites are indicated with vertical arrows. Horizontal arrows represent coding sequences and their directions of transcription. The upstream ClaI::mecA fragments were studied by IPCR and sequencing of the amplified fragments.
dcs.
A unique 2-kb sequence, the downstream constant segment (dcs) (Fig. 1), was detected in each polymorph and was joined on the downstream side by a 6-kb sequence (Fig. 1, regions between orfX and ClaI), which showed high homology to a sequence in the methicillin-susceptible strain NCTC8325 (Advanced Center for Genome Technology, University of Oklahoma [http://www.genome.ou.edu]), while no sequence homologous to the 2-kb dcs was found in the NCTC8325 genome. Immediately next to the 3′ end of dcs, the same putative orfX—first identified in strain N315 (15)—was present in each strain, suggesting that this proposed integration site (15) is common in all MRSA strains.
Evolutionary relationships.
The variable length (up to 16 kb) of DNA extending from the 3′ end of mecA towards the S. aureus chromosomal sequence is composed of two types of genetic elements: conserved blocks (a copy of HVR either complete or carrying a 1043-bp deletion; an IS431 sequence; and dcs) and a variable number of different acquired genetic elements, the nature and arrangement of which can account for the genetic organization of the 3′ vicinity of mecA that is unique to the different polymorphisms of this region.
The simplest downstream organization (polymorphism II) may represent the structure of the earliest MRSA isolates, in which the conserved blocks of foreign DNA are contiguous and bridge the 3′ end of mecA with the beginning of the host chromosome. The downstream organization of other polymorphisms may have originated by the integration of plasmids through homologous recombination between the IS431 copy downstream to mecA and a copy of IS431 which has previously been transposed into the plasmid. Such a scenario has already been suggested in earlier studies (14, 19, 28) and agrees with our finding that all linearized plasmids were flanked by short 8-bp direct repeats, presumably formed when integration of the insertion sequence into the plasmids occurred. Moreover, this target sequence is the same for different strains with the same integrated plasmid, as is the insertion site, supporting the previously found site-specific insertion of IS431 into each plasmid (27, 28). The IS431 insertion sequence may have provided an efficient tool for the introduction of resistance determinants into the S. aureus chromosome.
Additional possible evolutionary relationships are also apparent. The genetic organization of polymorphism III* characteristic of the pandemic Brazilian clone of MRSA (strain HU25) may have originated from polymorphism III by deletion of pT181. The downstream regions of strains BK739 and HU195 (polymorphism VI) may have originated from the intramolecular recombination between the two IS431 copies flanking pT181 in a polymorphism III-like structure with pT181 and pI258 inserted in the mecA downstream vicinity. The potential role of IS431 in these types of genetic exchanges had already been proposed (14).
Our findings support the thesis that the incorporation of the mec element into the S. aureus species was a rare event (1, 16). If the opposite were true, one would expect more extensive and diverse structural variation in the mecA downstream vicinity, other than the presence or absence of mobile elements.
ACKNOWLEDGMENTS
We thank Ana Madalena Ludovice for guidance and advice given during the initial phase of the study and Alexander Tomasz for stimulating discussions and critical revision of the manuscript.
Partial support for this study was provided by projects PRAXIS XXI/2/2.1/BIO/1154/95 and PRAXIS XXI/2/2.2/SAU/1295/95 (Portugal) awarded to H. de Lencastre and by a grant from the Irene Diamond Foundation to A. Tomasz, head of the Laboratory of Microbiology, The Rockefeller University, New York, N.Y. D.C. Oliveira was supported by grant BD/4162/96 from Fundação para a Ciência e Tecnologia, Lisbon, Portugal.
REFERENCES
- 1.Archer G L, Niemeyer D M. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 1994;2:343–347. doi: 10.1016/0966-842x(94)90608-4. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: Green Publishing Associates and John Wiley & Sons; 1992. [Google Scholar]
- 3.Corso A, Santos Sanches I, Aires de Sousa M, Rossi A, de Lencastre H. Spread of a methicillin-resistant and multiresistant epidemic clone of Staphylococcus aureus in Argentina. Microb Drug Resist. 1998;4:277–288. doi: 10.1089/mdr.1998.4.277. [DOI] [PubMed] [Google Scholar]
- 4.de Lencastre H, Chung M, Westh H. Archaic clones of methicillin-resistant Staphylococcus aureus (MRSA): molecular and microbiological properties of isolates from the 1960s in Denmark. Microb Drug Resist. 2000;6:1–10. doi: 10.1089/mdr.2000.6.1. [DOI] [PubMed] [Google Scholar]
- 5.de Lencastre H, Couto I, Santos I, Melo-Cristino J, Torres-Pereira A, Tomasz A. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur J Clin Microbiol Infect Dis. 1994;13:64–73. doi: 10.1007/BF02026129. [DOI] [PubMed] [Google Scholar]
- 6.de Lencastre H, Severina E P, Milch H, Thege M K, Tomasz A. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin Microbiol Infect. 1997;3:289–296. doi: 10.1111/j.1469-0691.1997.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 7.de Lencastre H, Severina E P, Roberts R B, Kreiswirth B N, Tomasz A. Testing the efficacy of a molecular surveillance network: methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VREF) genotypes in six hospitals in the metropolitan New York City area. The BARG Initiative Pilot Study Group. Bacterial Antibiotic Resistance Group. Microb Drug Resist. 1996;2:343–351. doi: 10.1089/mdr.1996.2.343. [DOI] [PubMed] [Google Scholar]
- 8.de Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lencastre H, Wu S W, Pinho M G, Ludovice A M, Filipe S, Gardete S, Sobral R, Gill S, Chung M, Tomasz A. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb Drug Resist. 1999;5:163–175. doi: 10.1089/mdr.1999.5.163. [DOI] [PubMed] [Google Scholar]
- 10.de Sousa M A, Sanches I S, Ferro M L, Vaz M J, Saraiva Z, Tendeiro T, Serra J, de Lencastre H. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J Clin Microbiol. 1998;36:2590–2596. doi: 10.1128/jcm.36.9.2590-2596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Sousa M A, Sanches I S, van Belkum A, van Leeuwen W, Verbrugh H, de Lencastre H. Characterization of methicillin-resistant Staphylococcus aureus isolates from Portuguese hospitals by multiple genotyping methods. Microb Drug Resist. 1996;2:331–341. doi: 10.1089/mdr.1996.2.331. [DOI] [PubMed] [Google Scholar]
- 12.Dominguez M A, de Lencastre H, Linares J, Tomasz A. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J Clin Microbiol. 1994;32:2081–2087. doi: 10.1128/jcm.32.9.2081-2087.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubin D T, Matthews P R, Chikramane S G, Stewart P R. Physical mapping of the mec region of an American methicillin-resistant Staphylococcus aureus strain. Antimicrob Agents Chemother. 1991;35:1661–1665. doi: 10.1128/aac.35.8.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inglis B, Matthews P R, Stewart P R. Induced deletions within a cluster of resistance genes in the mec region of the chromosome of Staphylococcus aureus. J Gen Microbiol. 1990;136:2231–2239. doi: 10.1099/00221287-136-11-2231. [DOI] [PubMed] [Google Scholar]
- 15.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 17.Kreiswirth B N, Lutwick S M, Chapnick E K, Gradon J D, Lutwick L I, Sepkowitz D V, Eisner W, Levi M H. Tracing the spread of methicillin-resistant Staphylococcus aureus by Southern blot hybridization using gene-specific probes of mec and Tn554. Microb Drug Resist. 1995;1:307–313. doi: 10.1089/mdr.1995.1.307. [DOI] [PubMed] [Google Scholar]
- 18.Leski T, Oliveira D, Trzcinski K, Sanches I S, de Sousa M A, Hryniewicz W, de Lencastre H. Clonal distribution of methicillin-resistant Staphylococcus aureus in Poland. J Clin Microbiol. 1998;36:3532–3539. doi: 10.1128/jcm.36.12.3532-3539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews P R, Inglis B, Stewart P R. Clustering of resistance genes in the mec region of the chromosome of Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers, Inc.; 1990. pp. 69–83. [Google Scholar]
- 20.McKenzie T, Hoshino T, Tanaka T, Sueoka N. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986;15:93–103. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira D, Sanches I S, Tamayo M, Ribeiro G, Mato R, Costa D, de Lencastre H. Virtually all MRSA infections in the largest Portuguese hospital are caused by two internationally spread multiresistant strains: the “Iberian” and the “Brazilian” clones of MRSA. Clin Microbiol Infect. 1998;4:373–384. doi: 10.1111/j.1469-0691.1998.tb00081.x. [DOI] [PubMed] [Google Scholar]
- 22.Roberts R B, Tennenberg A M, Eisner W, Hargrave J, Drusin L M, Yurt R, Kreiswirth B N. Outbreak in a New York City teaching hospital burn center caused by the Iberian epidemic clone of MRSA. Microb Drug Resist. 1998;4:175–183. doi: 10.1089/mdr.1998.4.175. [DOI] [PubMed] [Google Scholar]
- 23.Rose R E. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988;16:356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryffel C, Bucher R, Kayser F H, Berger-Bachi B. The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to the Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J Bacteriol. 1991;173:7416–7422. doi: 10.1128/jb.173.23.7416-7422.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Sanches I S, Saraiva Z C, Tendeiro T C, Serra J M, Dias D C, de Lencastre H. Extensive intra-hospital spread of a methicillin-resistant staphylococcal clone. Int J Infect Dis. 1998;3:26–31. doi: 10.1016/s1201-9712(98)90091-1. [DOI] [PubMed] [Google Scholar]
- 27.Stewart P R, Dubin D T, Chikramane S G, Inglis B, Matthews P R, Poston S M. IS257 and small plasmid insertions in the mec region of the chromosome of Staphylococcus aureus. Plasmid. 1994;31:12–20. doi: 10.1006/plas.1994.1002. [DOI] [PubMed] [Google Scholar]
- 28.Sugiyama M, Yuasa K, Bhuiyan M Z, Iwai Y, Masumi N, Ueda K. IS431mec-mediated integration of a bleomycin-resistance gene into the chromosome of a methicillin-resistant Staphylococcus aureus strain isolated in Japan. Appl Microbiol Biotechnol. 1996;46:61–66. doi: 10.1007/s002530050783. [DOI] [PubMed] [Google Scholar]
- 29.Teixeira L A, Lourenco M C, Figueiredo A M. Emergence of a methicillin-resistant Staphylococcus aureus clone related to the Brazilian epidemic clone III:B:A causing invasive disease among AIDS patients in a Brazilian hospital. Microb Drug Resist. 1996;2:393–399. doi: 10.1089/mdr.1996.2.393. [DOI] [PubMed] [Google Scholar]