Summary
Liver resident macrophages Kupffer cells (KCs) and infiltrating Ly6Chi monocytes both contribute to liver tissue regeneration in various pathologies, but also to disease progression upon disruption of orderly consecutive regeneration cascades. Little is known about molecular pathways that regulate their differentiation, maintenance, or their inflammatory behavior during injury. Here we show that COMMD10-deficient KCs adopt liver-specific identity. Strikingly, COMMD10-deficiency in KCs and in other tissue resident macrophages impedes their homeostatic survival, leading to their continuous replacement by Ly6Chi monocytes. While COMMD10-deficiency in KCs mildly worsens acetaminophen-induced liver injury (AILI), its deficiency in Ly6Chi monocytes results in exacerbated and sustained hepatic damage. Monocytes display unleashed inflammasome activation and reduced type I interferon response and acquire ‘neutrophil-like’ and lipid associated macrophage differentiation fates. Collectively, COMMD10 appears indispensable for KC and other tissue resident macrophage survival and is an important regulator of Ly6Chi monocyte fate decisions and reparative behavior in the diseased liver.
Introduction
Most tissue-resident macrophages are established prenatally and self-maintain independently of bone marrow (BM) monocytes. Instructed by local cues, these cells adopt unique transcriptional modules that impart tissue-specific functional identity (Varol et al., 2015). In the liver, Kupffer cells (KCs) dominate the homeostatic macrophage pool. They reside in the sinusoidal vessels and in the space of Disse, interacting with hepatic stellate cells (HSC) and hepatocytes (Bonnardel et al., 2019), where they act as sentinels and perform specialized accessory functions involving iron and lipid metabolism (Scott and Guilliams, 2018). Upon depletion, resident KCs (ResKCs) are replaced by BM Ly6Chi monocyte-derived KCs (MoKCs) that adopt ResKCs functions during homeostasis and disease (Bonnardel et al., 2019; Remmerie et al., 2020; Sakai et al., 2019; Scott et al., 2016; Seidman et al., 2020; Tran et al., 2020).
Hepatic macrophages play a central role in the initiation, progression and resolution of various liver diseases (Krenkel and Tacke, 2017). KCs play a major role in initiating inflammation but have limited plasticity. In contrast, Ly6Chi monocytes and their-derived macrophage (MoMFs) descendants prevail in acute or chronic liver injury, display marked transcriptional variance and provide crucial functional plasticity (Krenkel et al., 2018; Ramachandran et al., 2019; Zigmond et al., 2014). A specific example of MoMFs is lipid-associated macrophages (LAMs) involved in progression of metabolic liver disease (Remmerie et al., 2020). Recent studies have uncovered binary developmental trajectories of monocytes in the BM. Neutrophil-like (NeuMo) and dendritic cell (DC)-like (DCMo) monocytes differentiate downstream of granulocyte-macrophage progenitors (GMP) and macrophage-dendritic cell progenitors (MDP), respectively (Weinreb et al., 2020; Yanez et al., 2017). While the impact of Ly6Chi monocyte bifurcation on liver diseases remains enigmatic, these findings challenge our current comprehension of macrophage heterogeneity in the diseased liver and stress the importance of identifying factors that govern macrophage and monocyte fate decisions.
The COMMD (copper metabolism MURR1 domain) family includes 10 evolutionarily conserved proteins. Functions of COMMD proteins are still being defined, but they seem to play non-redundant roles in regulating transcription and protein trafficking (Bartuzi et al., 2013; Burstein et al., 2005). COMMD1, the prototype of this family, negatively regulates NF-κB (Li et al., 2014; Maine et al., 2007) and hypoxia inducible factor-1 (HIF-1α) (Vonk et al., 2011) (van de Sluis et al., 2010). COMMD proteins are essential components of the COMMD/CCDC22/CCDC93 complex, which modulates endolysosome architecture and trafficking of transmembrane proteins (Bartuzi et al., 2016; Phillips-Krawczak et al., 2015). Embryonic lethality of COMMD10 deficiency has hampered the study of its function. Yet, utilizing conditional COMMD10 knockout mice we uncovered a role for COMMD10 in limiting inflammasome activation in Ly6Chi monocytes during experimental sepsis and colitis (Mouhadeb et al., 2018), and in supporting phagolysosomal biogenesis and maturation in KCs and BM-derived macrophages infected with Staphylococcus aureus (Ben Shlomo et al., 2019). These studies mark COMMD10 as a candidate mediator of monocyte and macrophage fate and immune responses.
Here, we uncover an important role for COMMD10 in homeostatic maintenance of KCs and other fetal tissue-resident macrophages. We also show that COMMD10 is involved in determining Ly6Chi monocyte differentiation fates and in restraining their inflammatory response in the injured liver.
Results COMMD10 is required for homeostatic maintenance of KCs
To target COMMD10 deficiency to KCs we crossed Clec4f+/cre mice (Scott et al., 2018) with Commd10fl/fl mice (Mouhadeb et al., 2018) (Clec4fΔCommd10). KCs were defined as CD45+CD11bloF4/80+Ly6G-Ly6C- cells, also expressing the KC markers TIM4, VSIG4, CLEC2 and CLEC4F (Figure S1A). Commd10 gene expression decreased by about 50% in Clec4fΔCommd10 KCs (Figure 1A). COMMD10-deficiency had no effect on KCs expression of CLEC4F (Figure 1B, S1B) or on their localization in the niche, where they intimately interacted with Desmin+ HSCs, hepatocytes and CD31+ LSECs (Figure 1C, S1C, S1D). Yet, KC counts were significantly lower in Clec4fΔCommd10 versus Commd10fl/fl livers (Figure 1D). Expression of TIM4, an apoptotic cell clearance receptor selectively expressed by ResKCs in the liver, was diminished in Clec4fΔCommd10 KCs at both protein (Figure 1E) and (Figure 1F) gene levels. Expression of CD163 and VSIG4, additional ResKC-associated cell surface receptors, was also altered in Clec4fΔCommd10 KCs with an increase in the fractions of CD163- and VSIG4- cells (Figure 1G, S1E) and VSIG4- counts, and reduction in CD163+ counts (Figure 1H). Similar results were obtained when Cx3cr1+/cre mice were used to delete COMMD10 in KCs (Cx3cr1ΔCommd10). Cx3cr1ΔCommd10 KCs expressed normal levels of CLEC4F (Figure S1F) but had diminished expression of TIM4 already at the perinatal stage (Figure S1G), later during maturation (Figure S1H) and intracellularly (Figure S1I). Cx3cr1ΔCommd10 KCs from older livers (24–30 weeks) displayed increasing numbers of TIM4+ cells, though the majority remained TIM4lo (Figure S1J). In contrast, TIM4+ KCs remained barely detectable even in older Clec4fΔCommd10 livers (24–30 weeks) (Figure 1I).
Figure 1. COMMD10-deficient ResKCs are replaced by MoKCs in the healthy liver.
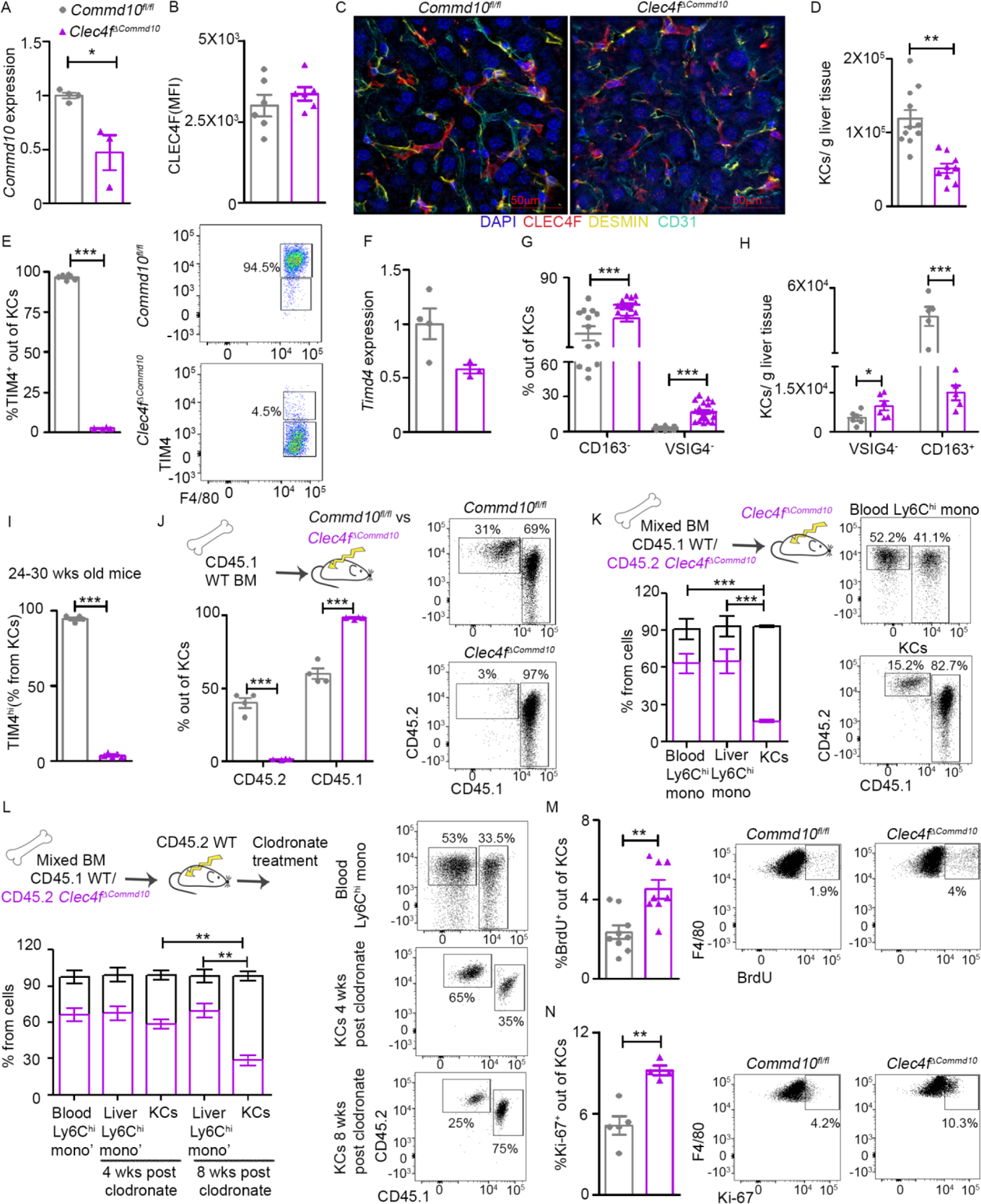
(A-J) KCs from Commd10fl/fl and Clec4fΔCommd10 healthy livers were compared. (A) RT-PCR gene expression of Commd10 (n>3). (B) Mean fluorescence intensity (MFI) of CLEC4F (n>6). (C) Confocal microscopy of livers showing expression of CLEC4F (red), Desmin (yellow), CD31 (cyan) and nuclei (blue), magnification: ×40. Bars=50 μm. (D) KC numbers normalized to liver mass (n>9). (E) Frequency of TIM4+ out of total KCs. (F) RT-PCR gene expression of Timd4 in KCs (n>3). (G) Frequency of CD163- and VSIG4- KCs (n>12). (H) Normalized cell counts of VSIG4- and CD163+ KCs (n>6). (I) Frequency of TIM4hi/total KCs in 24–30 wks old livers (n>4). (J-L) Quantification of % chimerism (J) in KCs at eight weeks post irradiation (n=4). (K) In blood and liver monocytes and KCs at eight weeks post irradiation (n=4). (L) In blood and liver monocytes and KCs as assessed 4/8 weeks post clodronate (n>4). (M) Quantification of BrdU+ cell frequency among KCs (n>7). (N) Quantification of Ki-67+ cell frequency among KCs (n>4). Data were analyzed by unpaired, two-tailed t-test and are presented as mean ± SEM with significance: *p < 0.05, **p < 0.01, ***p < 0.001. Experiments were repeated twice (A, B, H-J, M), once (C, E, F, K, L, N) or at least 3 times (D, G).
TIM4 is a discriminating marker between TIM4+ ResKCs and TIM4lo MoKCs (Remmerie et al., 2020; Tran et al., 2020). We next studied whether the persistently low expression of TIM4 by Clec4fΔCommd10 KCs is due to their replacement by MoKCs. Fetal-derived KCs are relatively radioresistant while MoKCs are radiosensitive (Soysa et al., 2019). Hence, if TIM4lo Clec4fΔCommd10 KCs are MoKCs, they would be expected to lose radioresistance. Clec4fΔCommd10 and Commd10fl/fl mice were lethally irradiated and reconstituted with congenic CD45.1+ WT BM graft. CD45.2+ radioresistant KCs constituted about 35% of KCs in reconstituted Commd10fl/fl livers but were barely detectable in Clec4fΔCommd10 livers (Figure 1J), confirming replacement of COMMD10-deficient KCs with radiosensitive MoKCs. Given the radiosensitivity of KCs in Clec4fΔCommd10 mice, we next subjected these mice to a competitive mixed BM chimerism experiment aiming to compare the fate of Clec4fΔCommd10 versus WT KCs in the same environment. Nine weeks following lethal irradiation and reconstitution with mixed CD45.1+ WT and CD45.2+ Clec4fΔCommd10 BM, circulating and liver Ly6Chi monocytes displayed mixed chimerism, with ~ 60% being of CD45.2+ Clec4fΔCommd10 origin. In contrast, only 20% of reconstituted KCs were of CD45.2+ Clec4fΔCommd10 origin (Figure 1K). This could be the result of COMMD10-deficiency hampering the ability of monocytes to reconstitute the KC niche and/or shortening MoKC survival. To examine this question, lethally irradiated CD45.2+ WT mice were reconstituted with mixed CD45.1+ WT and CD45.2+ Clec4fΔCommd10 BM, and eight weeks later clodronate liposomes were applied to effectively deplete the entire KC compartment (Figure S1K) (Soysa et al., 2019). Four weeks later, Ly6Chi monocytes and KCs displayed similar mixed chimerism, suggesting that COMMD10-deficiency had no effect on the ability of MoKCs to replenish the niche. However, eight weeks following KC depletion, although chimerism proportions of the Ly6Chi monocytes remained essentially the same, there was a significant reduction in the fraction of MoKCs of CD45.2+ Clec4fΔCommd10 origin, strongly indicating their shorter survival (Figure 1L). We next evaluated the proliferation rate of Clec4fΔCommd10 versus Commd10fl/fl KCs by measuring 5-bromo-2’-deoxyuridine (BrdU) incorporation. Clec4fΔCommd10 MoKCs showed a higher proliferation rate (Figure 1M). This was also evident by increased Ki-67 staining (Figure 1N). Altogether, these results show that absence of COMMD10 impedes KC survival.
COMMD10-deficient KCs acquire liver specific identity, but display increased stress and mitochondrial dysfunction
To examine the ability of COMMD10-deficient MoKCs to acquire KC-associated transcriptional programs we performed a MARS-seq transcriptome analysis of Clec4fΔCommd10 versus Commd10fl/fl KCs. There were 259 differentially expressed genes (DEGs) (≥1.5 fold, p.adj<0.05), among which 144 were upregulated and 115 were downregulated. Clec4fΔCommd10 KCs acquired a similar expression pattern of KC core genes and associated transcription factors (Figure 2A, B), with only 12% of ~70 KC-associated genes (Scott et al., 2018) being significantly downregulated (Table S1). Further supporting their recent ontogeny from Ly6Chi monocytes, they displayed higher expression of MoKC associated genes (Figure 2C). KCs specialize in lipid metabolism and iron recycling (Scott and Guilliams, 2018). With respect to lipid metabolism, Clec4fΔCommd10 KCs exhibited altered expression of several genes involved with lipid uptake (Cd36, Lpl), and cholesterol processing and export (Abca1, Apobec1, Apoc1) (Figure S2A). Yet, BODIPY staining for neutral lipids revealed no significant differences under homeostatic conditions (Figure S2B). Triglyceride levels were also identical in healthy Clec4fΔCommd10 versus Commd10fl/fl livers (Figure S2C). Some iron storage and transport genes were differentially expressed, with increased expression of Il6 and ferroportin 1 (Slc40a1), and reduced expression of ferritin light and heavy chains (Ftl1 and Fth1, respectively) and Marco (Figure S2D). Nevertheless, iron deposits were hardly detectable in healthy Clec4fΔCommd10 and Commd10fl/fl livers, suggesting normal physiological iron recycling (Figure S2E).
Figure 2: COMMD10-deficient KCs display an altered transcriptional profile involved with stress response.
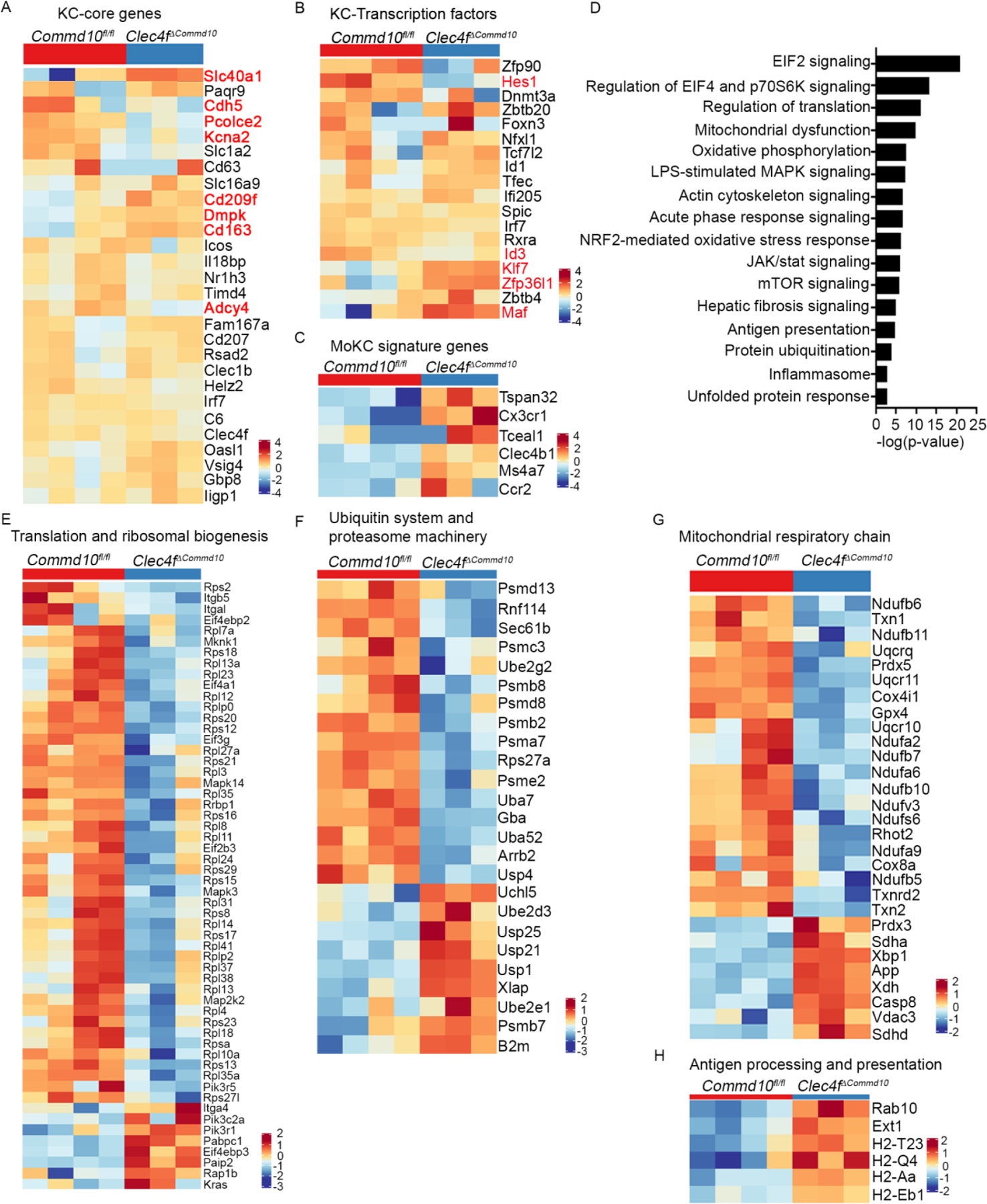
(A-C, E-H) MARS-RNAseq heat maps comparing between Commd10fl/fl and Clec4fΔCommd10 KCs and displaying expression of genes involved with (A) KC core signature and (B) KC transcription factors. Significantly varied genes are marked red. (C) MoKC signature, (E) Translation and ribosomal biogenesis, (F) Ubiquitin system and proteasome machinery, (G) Mitochondrial respiratory chain, (H) Antigen processing and presentation. (D) Top canonical pathways uncovered by ingenuity pathway analysis. Analysis was performed on differentially expressed genes (≥1.5-fold change, p < 0.05) (n>3).
In contrast, ingenuity pathway analysis (IPA) and MouseMine pathway and gene ontology analyses (p < 0.05, 1.5 FC) revealed among the top canonical pathways significant gene expression alterations involved with regulation of translation and EIF2/EIF4 signaling, as well as protein ubiquitination and degradation (Figure 2D). Accordingly, Clec4fΔCommd10 KCs displayed profound reduction in genes involved with translation initiation and ribosomal biogenesis (Figure 2E), and in genes belonging to the ubiquitin system and proteasome machinery (Figure 2F). Among the most significant pathways were also mitochondrial dysfunction and oxidative phosphorylation (Figure 2D), with reduction in many genes encoding for key enzymes and protein complexes mediating the mitochondrial respiratory chain and protecting cells from cytotoxicity elicited by oxidative stress (Figure 2G). Interestingly, Clec4fΔCommd10 KCs exhibited also increased expression of genes associated with interleukin-1 (IL-1) production and inflammasome (Figure S2F), and with antigen processing and presentation (Figure 2H), a feature that was also associated with MoKCs (Bonnardel et al., 2019; Seidman et al., 2020). Collectively, these results show that although COMMD10 is dispensable for the ability of MoKC to acquire KC identity, its deficiency is associated with transcriptional alterations that may facilitate their shortened survival.
COMMD10 is vital for the survival of diverse tissue resident macrophages
We next examined whether COMMD10 is important for the survival of other tissue resident macrophages. We utilized Lyz2+/cre mice to delete COMMD10 (LysMΔCommd10 mice) in large peritoneal macrophages (LPMs) and alveolar macrophages (AMs) (Abram et al., 2014). In the peritoneum, CD11b+MHCII−/+F4/80+ICAM2+ LPMs (Figure 3A) from LysMΔCommd10 and Commd10fl/fl mice were compared. Expression of Commd10 was diminished in LysMΔCommd10 LPMs, but expression of the LPM core transcription factor GATA-binding protein 6 (GATA6) was unaltered (Figure 3B), suggesting that COMMD10-deficiency does not alter their tissue-specific identity. There was reduced representation of LPMs in the LysMΔCommd10 peritoneum (Figure 3C, D). TIM4 expression in LPMs revealed three subsets: TIM4neg, TIM4low and TIM4hi. In LysMΔCommd10 peritoneum, TIM4hi were replaced with TIM4lo LPMs (Figure 3E). CD11c+SiglecF+F4/80+ AMs were isolated by bronchoalveolar lavage of Commd10fl/fl or LysMΔCommd10 mice. AM counts were similar, though LysMΔCommd10 AMs exhibited reduction in the expression of CD11c (Figure 3F, G). Revisiting the mixed BM chimerism approach, WT mice were lethally irradiated and reconstituted with mixed CD45.1+ WT and CD45.2+ LysMΔCommd10 BM. Nine weeks later, circulating Ly6Chi monocytes displayed mixed chimerism with near 60% being of CD45.2+ LysMΔCommd10 origin. In contrast, CD45.2+ LPMs and AMs dropped to 15% (Figure 3H). Similarly to COMMD10-deficient KCs, LysMΔCommd10 LPMs exhibited increased proliferation as depicted by BrdU incorporation (Figure 3I) and Ki67 staining (Figure 3J). Similar results were obtained using Cx3cr1ΔCommd10 mice, with reduction in LPM counts (Figure S3A), exchange of TIM4hi with TIM4lo LPMs in both young (8 weeks) and older (24–30 weeks) mice (Figure S3B), and a significant reduction in the fraction of CD45.2+ Cx3cr1ΔCommd10 versus CD45.1+ WT LPMs in a mixed BM chimerism experiment (Figure S3C).
Figure 3: Impeded survival of other COMMD10-deficient tissue-resident macrophages.
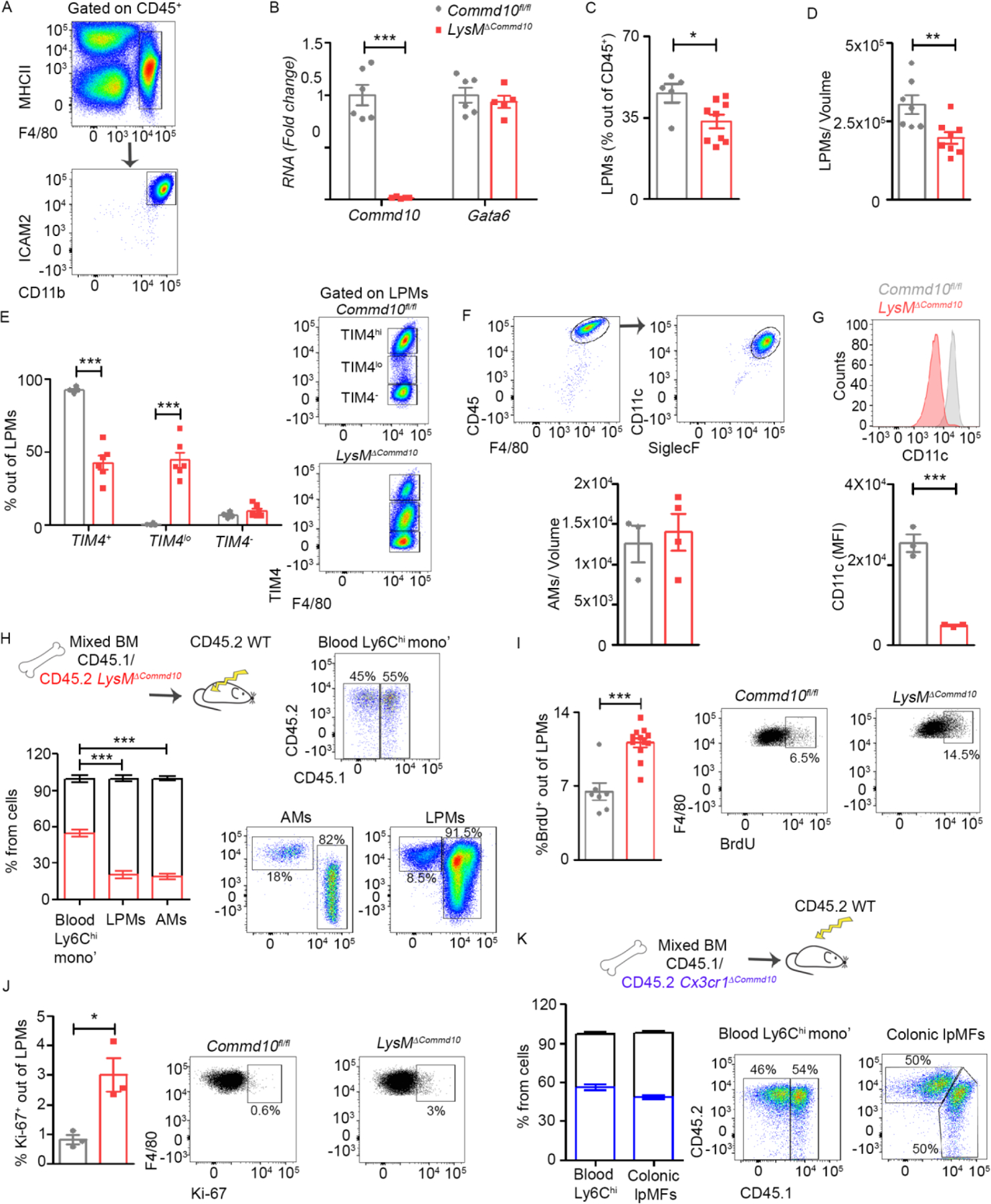
(A) Gating strategy of large peritoneal macrophages (LPMs) defined as CD45+F4/80+CD11b+ ICAM2+. (B-G) LPMs or alveolar macrophages (AMs) from Commd10fl/fl and LysMΔCommd10 mice were compared. (B) RT-PCR gene expression of Commd10 and Gata6 (n>5) (C) Fraction of LPMs among CD45+ cells. (D) LPM cell counts normalized to extracted volume (n>6). (E) Frequency of TIM4+, TIM4lo and TIM4- cells among LPMs (n>4). (F) Frequency of AMs (defined as CD45+F4/80+CD11C+SiglecF+) among CD45+ cells (n>3). (G) Mean fluorescence intensity (MFI) of CD11c expression by AMs (n=3). (H) Quantification of % chimerism in blood Ly6Chi monocytes, LPMs and AMs assessed in mixed CD45.1 WT/CD45.2 LysMΔCommd10 BM chimeras at eight weeks post irradiation (n>9). (I-J) Frequency of (I) BrdU+ cells (n>7), and (J) Ki-67+ cells (n>3), among total LPMs (n>3). (K) Quantification of % chimerism in blood Ly6Chi monocytes and lamina propria macrophages (lpMFs) as assessed in mixed CD45.1 WT/CD45.2 Cx3cr1ΔCommd10 BM chimeras at eight weeks post irradiation (n=7). Data were analyzed by unpaired, two-tailed t-test and are presented as mean ± SEM with significance: *p < 0.05, **p < 0.01, ***p < 0.001. Experiments were repeated twice (A, C, E, H, I, K) or once ( B, D, F, G, H, J).
TIM4 is also a marker of resident macrophages in the splenic marginal zone and red pulp (Fujiyama et al., 2019). Splenic macrophages were defined as CD11b+F4/80+CD64+ cells. Indeed, there was significant depletion of TIM4+ splenic macrophages in both young and older Cx3cr1ΔCommd10 mice (Figure S3D). The gut lpMF compartment also contains fetal TIM4+ macrophages that decline with age (Shaw et al., 2018). LpMFs, defined as CD45+CD11b+ CD64+MHCII+CX3CR1+ cells, exhibited a significant reduction in TIM4+ fetal lpMFs in the colons of Cx3cr1ΔCommd10 mice (Figure S3E). In contrast to LPMs, colonic lpMFs from the mixed BM chimera exhibited CD45.1+ WT and CD45.2+ Cx3cr1ΔCommd10 ratios similar to circulating Ly6Chi monocytes (Figure 3K), as expected given their constitutive renewal by these cells (Varol et al., 2009; Zigmond et al., 2012). Collectively, these results highlight COMMD10 as an essential factor in the maintenance of multiple tissue resident macrophages.
COMMD10-deficiency delays clearance of dying cells by KCs and LPMs
Being in direct contact with hepatocytes, KCs are the primary cells that sense and react to hepatic damage by initiating a restorative response (Krenkel and Tacke, 2017). Hence, we next sought to examine the effect of COMMD10 deficiency on KC survival and ability to recruit Ly6Chi monocytes and neutrophils to the injured liver, using AILI as a model. At AILI 24 h, plasma levels of the liver enzymes alanine and aspartate transaminases (ALT, AST) were similar in Clec4fΔCommd10 versus Commd10fl/fl mice (Figure 4A). In the Clec4fΔCommd10 livers histopathological score was mildly increased (Figure 4B), KC counts were still reduced (Figure 4C), and KCs were TIM4lo (Figure 4D). Ly6Chi monocytes and neutrophil cell numbers were lower (Figure 4E), yet expression of their recruitment chemokines as well as inflammatory cytokines was similar (Figure 4F). Despite the increased expression of genes involved with IL-1β production in Clec4fΔCommd10 KCs at steady state (Figure S2F), there was no evidence for increased inflammasome and caspase 1 activation in Clec4fΔCommd10 livers (Figure 4G). KCs are involved in clearing dying cells during AILI. In addition to there being fewer KCs, the reduction in TIM4 may compromise their ability to clear phosphatidylserine (PtdSer)-expressing apoptotic cells. To directly assess the effect of COMMD10 deficiency on the ability of KCs to clear dying cells, apoptotic CFSE-labeled thymocytes were injected into the portal vein. After 15 min, Commd10fl/fl KCs were already able to engulf apoptotic thymocytes, while these could not be detected in the Cx3cr1ΔCommd10 KCs (Figure 4H). TIM4 also mediates the recognition, engulfment, and clearance of PtdSer- expressing apoptotic cells by LPMs (Wong et al., 2010). Hence, the depletion of TIM4hi in COMMD10-deficient LPMs (Figure 3D, S3B) may compromise this function. Indeed, Cx3cr1ΔCommd10 LPMs displayed delayed uptake and clearance of engrafted with CFSE-labeled dying thymocytes (Figure 4I). This was further directly confirmed in ex vivo isolated Cx3cr1ΔCommd10 LPMs (Figure 4J). In agreement with the delayed clearance of dying cells by COMMD10-deficient KCs there was increased accumulation of TUNEL+ dying hepatocytes in Clec4fΔCommd10 livers (Figure 4K). Overall, COMMD10-deficiency in KCs interferes with their clearance of dying cells.
Figure 4: Clec4fΔCommd10 mice display mildly worse hepatic damage in response to acetaminophen-induced liver injury (AILI).
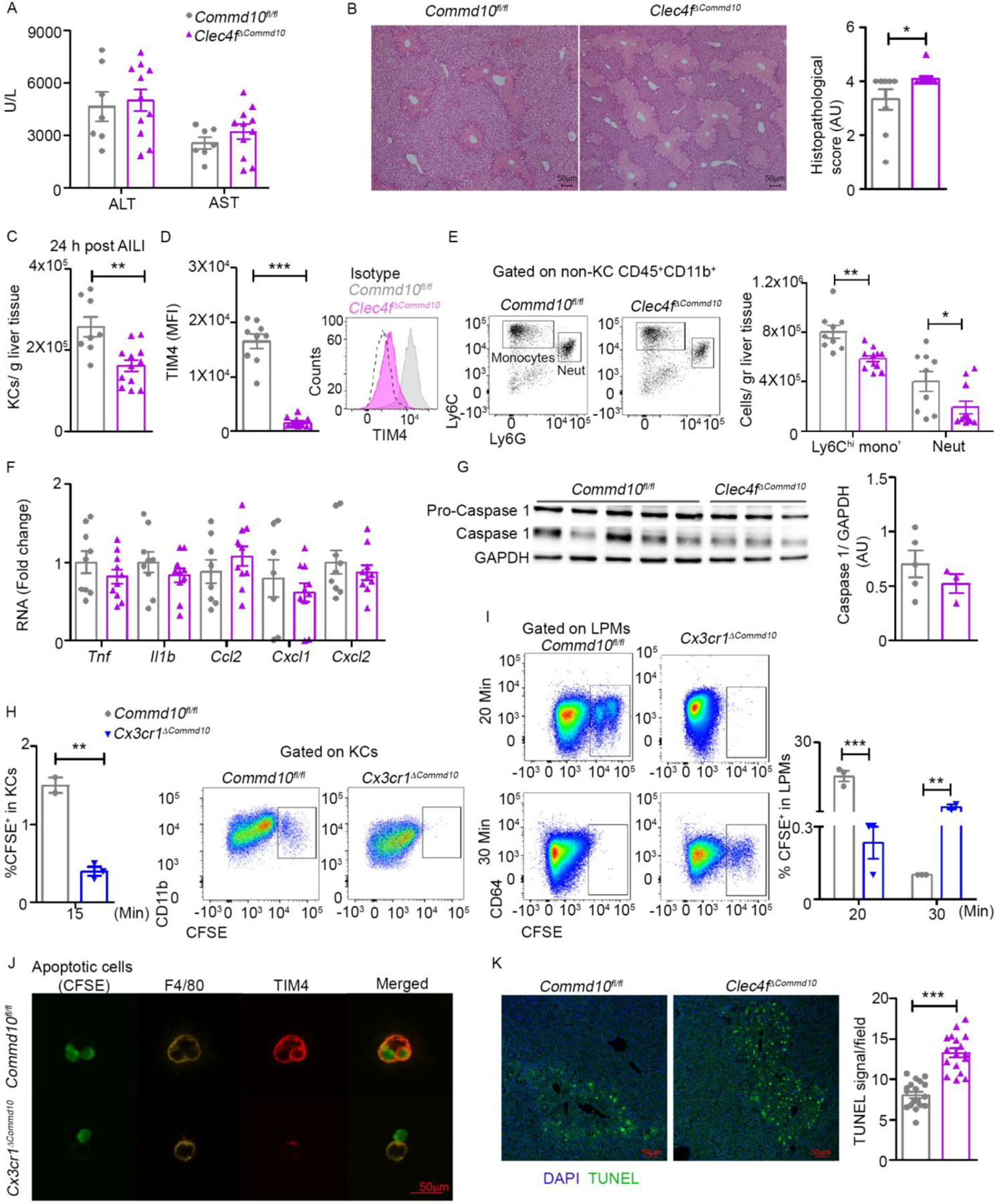
Commd10fl/fl and Clec4fΔCommd10 mice were injected with APAP (300 mg/kg) and sacrificed 24 h later. (A) ALT and AST serum levels (n>5). (B) Histopathological score and representative images of H&E stained liver sections. Magnification= x4. Bars=50µm (n>9). (C) Quantification of normalized KC counts (n>7). (D) Mean fluorescence intensity (MFI) of TIM4 (n>12) in KCs. (E) Quantification of normalized Ly6Chi monocyte and neutrophil counts (n>8). (F) Gene expression of inflammatory cytokines and chemokines quantified by qRT-PCR (n>8). (G) Immunoblot of total liver tissue protein showing caspase-1 activation. GAPDH was used as loading control (n>3). (H-I) Frequency of CFSE-labeled apoptotic thymocytes comparing Commd10fl/fl and Cx3cr1ΔCommd10 mice (H) In KCs 15 minutes post injection of 2×107 apoptotic cells into the portal vein (n>2). (I) In large peritoneal macrophages (LPMs) at 20 and 30 minutes post intraperitoneal injection of 2.5×106 apoptotic cells (n>3). (J) Confocal microscopy images showing expression of F4/80 and TIM4 in isolated LPMs 2 h following exposure to CFSE-labeled apoptotic cells at a ratio of 1:4. Magnification: ×40. Bars=50 μm. (K) Confocal images and quantification of average TUNEL signal per field in Commd10fl/fl and Clec4fΔCommd10 livers at AILI 24 h, magnification: ×10. Bars=50 μm (n=3, 20 images per group). Data were analyzed by unpaired, two-tailed t-test and are presented as mean ± SEM with significance: *p < 0.05, **p < 0.01, ***p < 0.001. Experiments repeated twice (A-F, J) or once (G- I).
Increased hepatic damage and inflammation is induced by COMMD10-deficiency in Ly6Chi monocytes
COMMD10-deficiency in Ly6Chi monocytes profoundly aggravates colonic damage and inflammation (Mouhadeb et al., 2018). Ly6Chi monocytes and their MoMF progenies play important roles in resolution of AILI (Graubardt et al., 2017; Zigmond et al., 2014), yet their inflammatory behavior must be strictly controlled to avoid collateral damage. Hence, we next endeavored to study the effect of COMMD10-deficiency in Ly6Chi monocytes on hepatic damage and inflammation during AILI. In LysMΔCommd10 mice, about 50% of liver-infiltrating Ly6Chi monocytes become COMMD10-deficient (Mouhadeb et al., 2018), and indeed there was a 50% reduction in Commd10 gene expression in liver infiltrating LysMΔCommd10 Ly6Chi monocytes sorted at AILI 24 h (Figure 5A). LysMΔCommd10 mice displayed profoundly increased hepatic damage as manifested by increased plasma levels of ALT/AST at AILI 24 h and 48 h (Figure 5B). Histopathological scoring revealed increased necrotic damage in LysMΔCommd10 livers with extended areas of panlobular and bridging necrosis (Figure 5C). There was profoundly higher accumulation of infiltrating Ly6Chi monocytes and neutrophils in LysMΔCommd10 livers (Figure 5D), and this was linked to elevated expression of various inflammatory genes, including TNFα, IL-1β and the Ly6Chi monocyte and neutrophil recruitment chemokines CCL2 and CXCL1/CXCL2, respectively (Figure 5E).
Figure 5: COMMD10-deficiency in Ly6Chi monocytes aggravates hepatic damage during AILI.
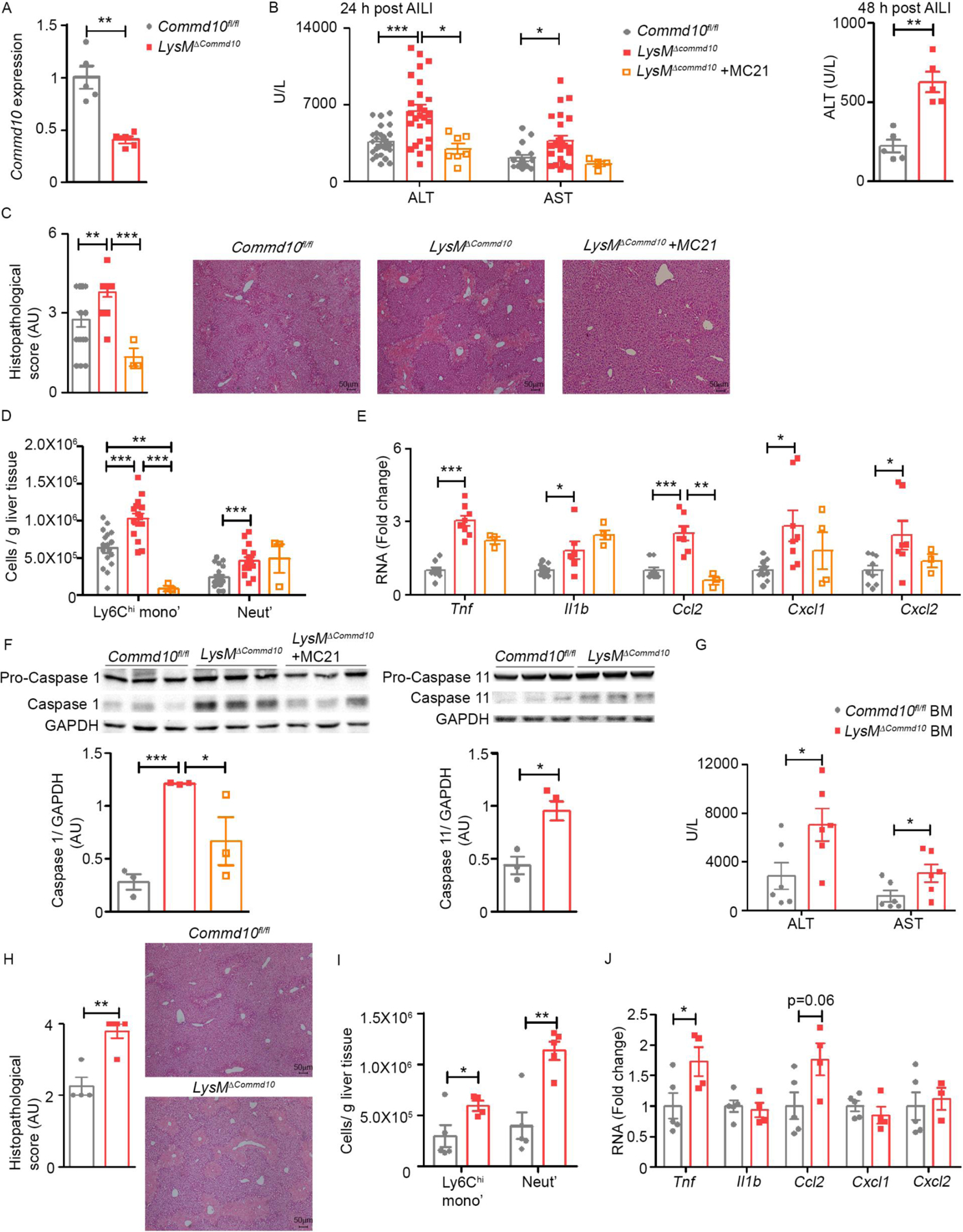
(A-F) Commd10fl/fl and LysMΔCommd10 were injected with APAP (300 mg/kg) and sacrificed 24 or 48 h later. Several LysMΔCommd10 mice were injected with MC-21 anti-mouse CCR2 antibody. (A) RT-PCR of Commd10 expression in liver Ly6Chi monocytes at 24 h of AILI (n=5). (B) ALT (24 and 48 h) and AST (24 h) serum levels (n>5). (C) Histopathological score of H&E stained liver sections at 24 h. Magnification= x4. Bars=50µm (n>14). (D) Normalized Ly6Chi monocyte and neutrophil counts at 24 h (n>17). (E) Gene expression of inflammatory cytokines and chemokines quantified by qRT-PCR (n>8). (F) Immunoblots of total liver tissue showing caspase-1 and caspase-11 activation at 24 h. GAPDH was used as loading control. (G-J) Ccr2−/− recipients were subjected to low dose irradiation (3 Gy) and transplanted with 5×106 BM cells from Commd10fl/fl or LysMΔCommd10 donors. Two weeks later they were subjected to AILI for 24 h. (G) ALT and AST serum levels (n>5). (H) Histopathological score of H&E stained liver sections. Magnification= x4. Bars=50µm (n>4). (I) Normalized counts of Ly6Chi monocyte and neutrophils. (J) Gene expression of inflammatory mediators quantified by qRT-PCR (n>4). Data were analyzed by unpaired, two-tailed t-test and are presented as mean ± SEM with significance: *p < 0.05, **p < 0.01, ***p < 0.001. Experiments repeated twice (A, F- J) or at least 3 times (B- E).
COMMD10 negatively regulates both canonical and non-canonical inflammasome pathways in Ly6Chi monocytes, but not tissue resident macrophages, during endotoxic shock and experimental colitis (Mouhadeb et al., 2018). We therefore speculated that locally released death-associated molecular patterns (DAMPs) might incite hyper-inflammasome activation in liver infiltrating LysMΔCommd10 Ly6Chi monocytes. Indeed, there was increased caspase-1 and caspase-11 activation in the LysMΔCommd10 livers at AILI 24 h (Figure 5F).
To pinpoint the contribution of Ly6Chi monocytes (out of total myeloid cells) to the increased hepatotoxicity in LysMΔCommd10 livers, some of the LysMΔCommd10 mice were treated prior to AILI with the anti-CCR2 antibody MC-21 (Mack et al., 2001), which efficiently blocks the arrival of Ly6Chi monocytes to the injured liver (Graubardt et al., 2017; Zigmond et al., 2014) (Figure 5E). Their inducible ablation was sufficient to ameliorate hepatic damage and inflammation to that of Commd10fl/fl livers as depicted by normalization of plasma ALT/AST levels (Figure 5B), histopathological score (Figure 5C), expression of inflammatory genes such as Ccl2 (Figure 5E), and inflammasome activation (Figure 5F). In a complementary approach, we implemented BM chimerism in which Ccr2−/− recipients are subjected to low dose irradiation and transplanted with BM cells from CCR2+ donors. This mild irradiation spares KCs, and given the established reliance of Ly6Chi monocytes on CCR2, enforces liver engraftment by CCR2+ Ly6Chi monocytes of donor BM origin, while other resident and infiltrating immune cells remain largely of recipient origin (Tran et al., 2020). Indeed, two weeks after transplantation, Ly6Chi monocytes were 90% of donor origin, while only 15–20% of neutrophils and CD11b- lymphocytes and a minor fraction of KCs (<2%) were of donor origin in blood and liver at AILI 24 h (Figure S4A, B). Next, irradiated Ccr2−/−Cx3cr1gfp/+ mice were engrafted with BM from LysMΔCommd10 (BM LysMΔCommd10) or Commd10fl/fl (BM Commd10fl/fl) mice (Figure S4C), and two weeks later were subjected to AILI 24 h. About 90% of Ly6Chi monocytes in BM Commd10fl/fl and BM LysMΔCommd10 livers were of donor BM origin (Cx3cr1-gfp-) (Figure S4D). Importantly, BM LysMΔCommd10 mice had significantly increased plasma ALT/AST levels (Figure 5G), histopathological score (Figure 5H) and infiltration of Ly6Chi monocytes and neutrophils (Figure 5I). Moreover, expression of TNFα (Tnf) and CCl2 (Ccl2) increased in BM LysMΔCommd10 livers (Figure 5J). Collectively, these results illuminate an important role for COMMD10 in tuning the recruitment and inflammatory activity of Ly6Chi monocytes in AILI.
COMMD10-deficient Ly6Chi monocytes exhibit a reduction in type I interferon response genes
To gain molecular insight into the immunoregulatory role played by COMMD10, LysMΔCommd10 and Commd10fl/fl Ly6Chi monocytes were sorted from livers at AILI 24 h, and subjected to bulk-RNAseq analysis. Among 651 DEGs (≥1.5 fold, p.adj<0.05), 374 genes were significantly downregulated. The most pronounced GO functional enrichments (MouseMine) among downregulated genes were associated with type I interferon response and production (Figure 6A, B, Table S2). These included genes related to sensing (Tlr3, Tlr9, Ddx58), transcriptional activators of type I interferon response (Irf7, Stat1, Stat2), and many interferon stimulated genes. Many of these genes were significantly upregulated during the transition of Ly6Chi monocytes (AILI 24 h) into pro-restorative MoMFs at the resolution phase (AILI 72 h) (Figure 6C, Table S3). An additional profoundly reduced GO enrichment pathway was antigen processing and presentation of exogenous peptide antigen (Figure 6A). Indeed, LysMΔCommd10 Ly6Chi monocytes exhibited reduction in genes encoding for HLA class II alpha and beta chain paralogs, and molecules involved with transport of antigens (Figure 6D, Table S2). Functional enrichment analyses of 277 upregulated genes revealed pathways associated with abnormal response to injury, abnormal innate immunity, abnormal macrophage physiology, abnormal liver regeneration, extracellular matrix organization and glucose metabolic process (Figure 6A, Table S4). In this respect, LysMΔCommd10 Ly6Chi monocytes exhibited higher expression of inflammatory genes, including Ccl2 and Cxcl2, and genes encoding for extracellular matrix (ECM) core proteins and remodeling enzymes (Figure 6E).
Figure 6: COMMD10-deficient Ly6Chi monocytes exhibit altered transcriptional profile associated with neutrophil like monocytes (NeuMO).
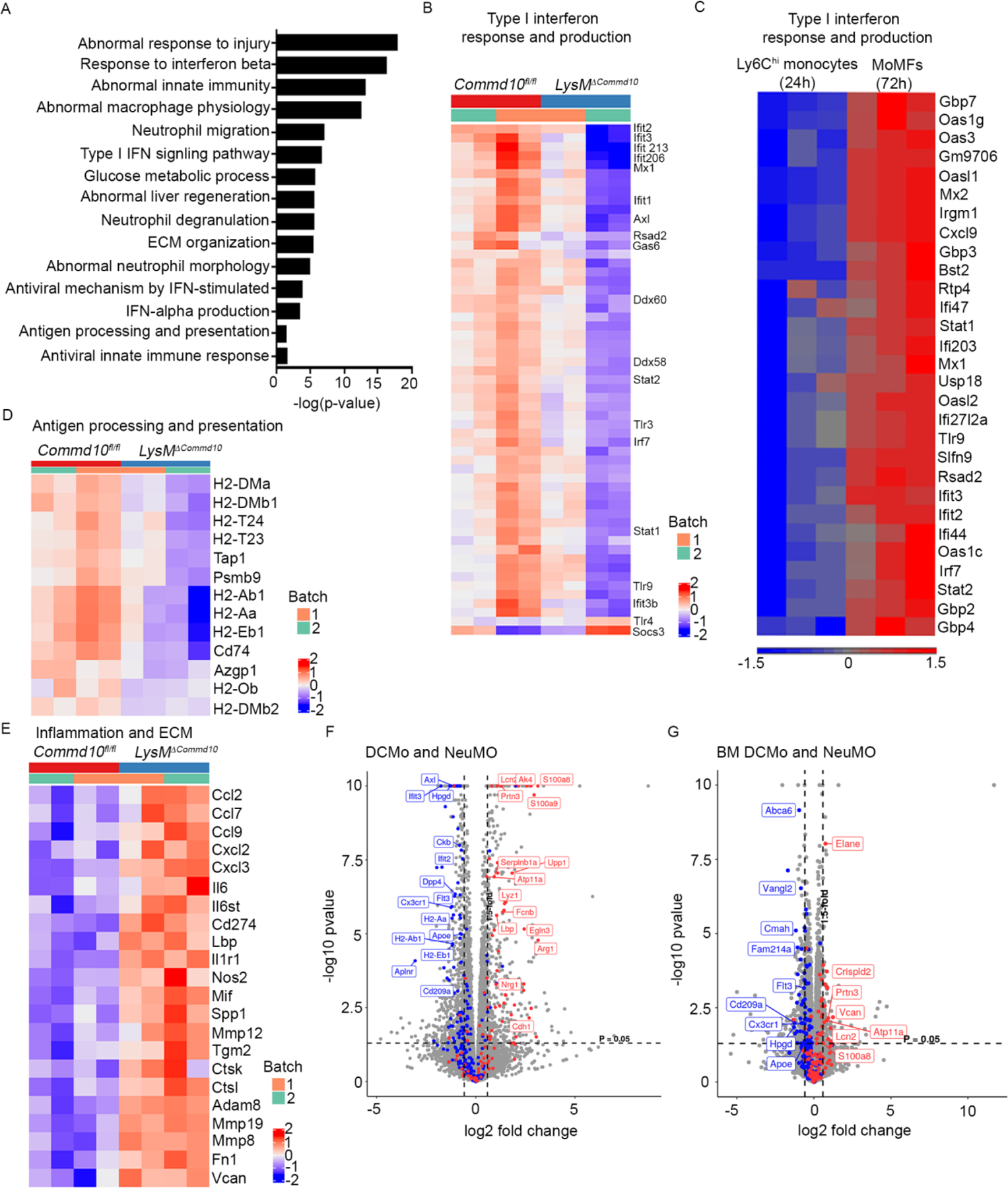
(A-B, D-F) Ly6Chi monocytes were sorted from Commd10fl/fl and LysMΔCommd10 livers at 24 h following AILI and subjected to RNAseq analysis. (A) Top pathways with significant gene expression alterations. (B, C) Heatmap analyses displaying variance of Type I interferon response genes in (B) Commd10fl/fl and LysMΔCommd10 Ly6Chi monocytes AILI 24 h, and (C) Sorted WT Ly6Chi monocytes (AILI 24 h) and descendent monocyte-derived macrophages (MoMFs) (AILI 72 h)(extracted from GSE55606). (D) Antigen processing and presentation genes (E) Inflammation and ECM remodeling genes. (F-G) Volcano plots comparing the expression pattern of NeuMO and DC like monocytes (DCMo) associated genes in Ly6Chi monocytes from (F) livers (AILI 24 h) and (G) from steady state bone marrow (BM) (G). Analysis was performed on differentially expressed genes (≥1.5-fold change, p< 0.05) (n>3).
COMMD10-deficiency promotes NeuMo fate of Ly6Chi monocytes
Among upregulated genes there was also enrichment for neutrophil associated pathways, such as neutrophil degranulation, abnormal neutrophil morphology and neutrophil migration (Figure 6A, Table S4). Given the recent discovery of neutrophil-like monocytes (NeuMo) and DC-like monocytes (DCMo) (Weinreb et al., 2020; Yanez et al., 2017), we speculated that COMMD10-deficieny in Ly6Chi monocytes might favor enrichment for NeuMO. Indeed, comparison with Yanez et al., NeuMo and DCMo gene signatures revealed in LysMΔCommd10 Ly6Chi monocytes a significant upregulation of about 40 NeuMo genes (e.g., S100a8, S100a9, Elane, Lcn2, and Prtn3), while about 80 DCMo genes were significantly reduced (Figure 6F, Table S5). Additional comparison with Weinreb et al. top 50 most significant NeuMo and DCMo signature genes revealed similar enrichment for NeuMo phenotype (Figure S5, Table S5). Further RNAseq analysis revealed that the differentiation bias of COMMD10-deficient Ly6Chi monocytes toward NeuMo fate initiates already in the steady state BM (Figure 6G). These results outline a role for COMMD10 in regulating Ly6Chi monocyte differentiation pathways.
Single cell RNAseq reveals distinct differentiation traits of COMMD10-deficient Ly6Chi monocytes
To further dissect the transcriptional changes evoked by COMMD10-deficiency we next performed single cell )SC) RNAseq analysis of purified Ly6Chi monocytes sorted from LysMΔCommd10 and Commd10fl/fl livers at AILI 24 h. Seurat (PC 1:18, resolution 0.5) was used to generate a uniform manifold approximation and projection (UMAP) revealing nine (0–8) significant (>100 cells) clusters (Figure 7A). Commd10fl/fl Ly6Chi monocytes mainly distributed in cluster 0, while LysMΔCommd10 Ly6Chi monocytes mainly distributed in cluster 1 (Figure 7B, C). Top marker genes of cluster 0 (Aif1, Itgb5, Cx3cr1, and Cd74) (Figure 7D, Figure S6A, Table S6) were reduced in LysMΔCommd10 Ly6Chi monocytes in the bulk RNAseq (Figure 7E), while top markers of cluster 1 (Arg1, Srgn, Adam8, Tgm2, F10, Ccl2, and Cxcl2) (Figure 7D, Figure S6B, Table S6) were significantly upregulated (Figure 7E). These results confirm a strong bias of COMMD10-deficient monocytes toward cluster 1 phenotype.
Figure 7: Higher representation of NeuMo and lipid associated macrophages (LAMs) among COMMD10-deficient Ly6Chi monocytes at AILI 24 h.
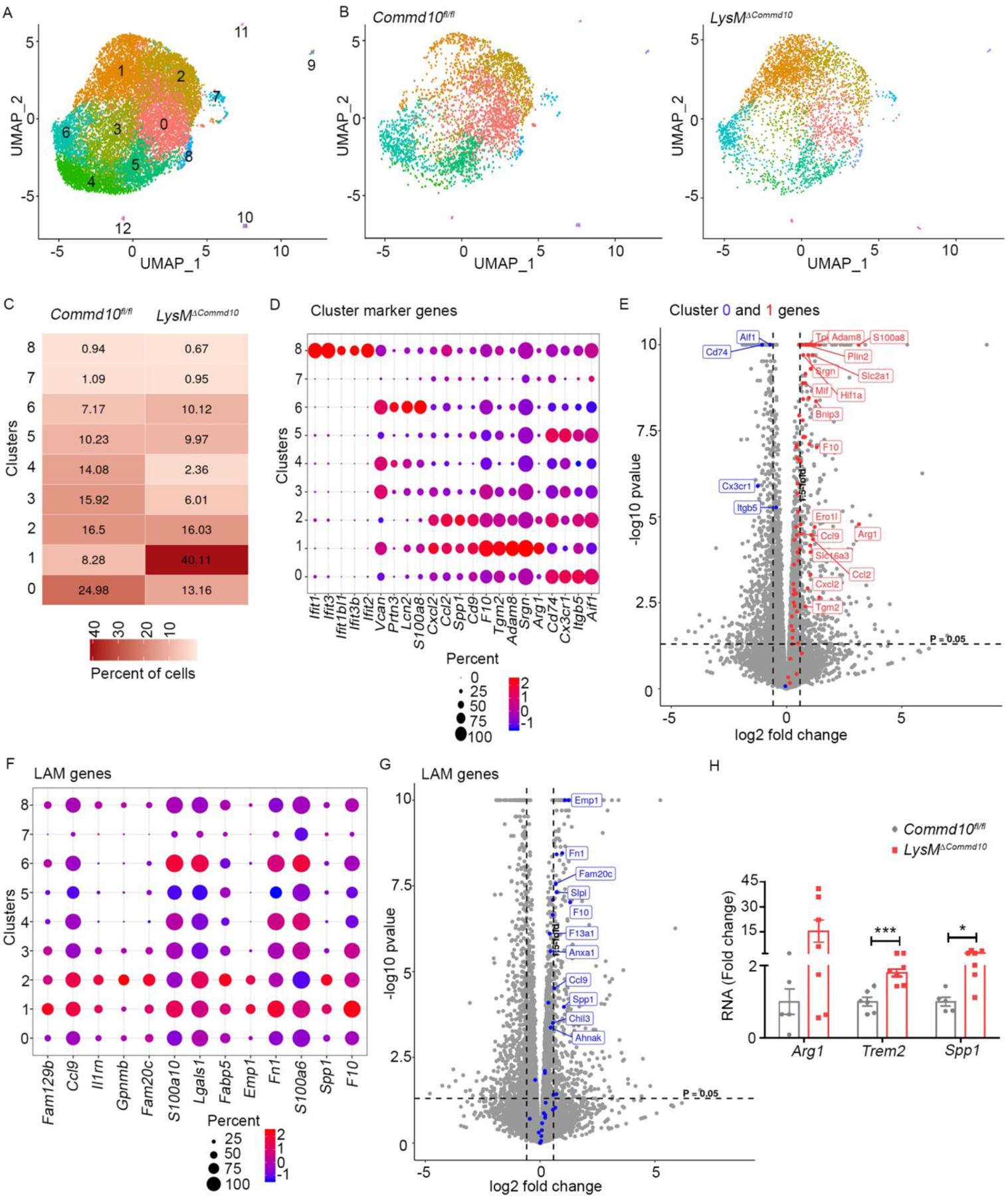
Ly6Chi monocytes sorted from LysMΔCommd10 and Commd10fl/fl livers (AILI 24 h) were compared by single cell RNAseq analysis. (A) Uniform manifold approximation and projection (UMAP) showing the distribution of Ly6Chi monocytes among 12 distinct clusters. (B) UMAPs displaying distinct patterns of cluster distribution. (C) Table displaying % of Ly6Chi monocytes in each of the main 0–8 clusters (> 100 cells). (D) Expression level and % of specific marker genes across the main clusters. (E) Volcano plot displaying the expression of cluster 0 (blue) and cluster 1 (red) top marker genes in LysMΔCommd10 versus Commd10fl/fl Ly6Chi monocytes (AILI 24 h), as depicted from bulk RNAseq analysis. (F) Expression levels and percentages of LAM genes across the main clusters. (G) Volcano plot displaying the expression of LAM genes among LysMΔCommd10 versus Commd10fl/fl Ly6Chi monocytes (AILI 24 h), as depicted from bulk RNAseq analysis. (H) RT-PCR gene expression analysis of LAM genes, comparing MoMFs sorted from Commd10fl/fl and LysMΔCommd10 livers (AILI 48 h) (n>6). Analysis was performed on differentially expressed genes (≥1.5-fold, p< 0.05) (n>3).
There was also a higher fraction of COMMD10-deficient Ly6Chi monocytes in cluster 6 (Figure 7C), annotated by CellMarker as stage I neutrophils and characterized by their distinctive expression of NeuMo markers (S100a8, Lcn2, Prtn3, and Vcan) (Figure 7D, Figure S6C, Table S6). In fact, S100a8 and S100a9 were among the 10 most highly variable genes (Figure S6D). About 93 NeuMo signature genes were detected in this SC count, and hypergeometric testing revealed the most significant enrichment of these genes in clusters 6 (p=0.00038) and 1 (p=0.015), further highlighting the differentiation bias of LysMΔCommd10 monocytes toward NeuMo. Cluster 8 comprised a minor fraction of total monocytes, and was unique in its expression of type I interferon genes (Figure 7D, Table S6). In agreement with the reduced expression of type I interferon response genes in LysMΔCommd10 Ly6Chi monocytes (Figure 6B), there was a reduction in the fraction of cluster 8 among these cells (Figure 7C).
Osteopontin gene (Spp1) was among the 10 most highly variable genes (Figure S6D). Cluster 1 associated genes Spp1 and Cd9 were recently defined as markers of Ly6Chi monocyte-derived LAMs appearing in the metabolically diseased liver (Remmerie et al., 2020). Examination of top hepatic LAM markers (Remmerie et al., 2020) revealed that many of them appear in clusters 1, 2 and 6 (Figure 7F, Figure S6E), and were also significantly upregulated by LysMΔCommd10 Ly6Chi monocytes in the bulk RNAseq analysis (Figure 7G, Table S7). All 35 top LAM signature genes were present in this SC count, and hypergeometric testing revealed the most significant enrichment of these genes in clusters 1 (p= 2.9E-15), 2 (p= 5.7E-13) and 6 (p= 4.7E-13). Further qRT-PCR analysis confirmed the significant upregulation of Spp1, Trem2, and Arg1 in MoMFs sorted from LysMΔCommd10 versus Commd10fl/fl livers at AILI 48 h (Figure 7H). Altogether, these results outline an important role for COMMD10 in dictating Ly6Chi monocyte fate decisions in the injured liver.
Discussion
KC niche is composed of HSCs, hepatocytes, and liver sinusoidal endothelial cells. This ménage à trois conveys signals that imprint KC identity (Bonnardel et al., 2019; Sakai et al., 2019), yet the molecular pathways involved with homeostatic maintenance of KC survival remain largely unclear. Our findings highlight COMMD10 as a pivotal intracellular intersection molecule indispensable for KC survival. COMMD10-deficiency does not interfere with KC tissue specialization. Further, Clec4fΔCommd10 KCs normally express CLEC4F, and high-resolution imaging demonstrates their intact ability to closely interact with the aforementioned niche cells. KCs perform accessory lipid metabolic functions and iron recycling (Scott and Guilliams, 2018). Clec4fΔCommd10 mice exhibit normal KC lipid content, liver triglyceride levels and trace iron deposits. This suggests that KC-targeted COMMD10-deficiency has limited impact on homeostatic functions, despite altered expression of linked genes. However, COMMD10-deficiency leads to significant reduction in KC counts, and their entire replacement with TIM4- MoKCs. Clec4fΔCommd10 KCs remain TIM4- also in 6-month old livers. As reconstituting MoKCs renew TIM4 expression already 30 days following KC depletion (Scott et al., 2016), these results suggest that Clec4fΔCommd10 KCs do not survive long enough to upregulate TIM4 expression, obliging continuous renewal by Ly6Chi monocytes. Indeed, Clec4fΔCommd10 KCs express MoKC associated genes, and contain a larger fraction of VSIG4- and CD163- cells, probably preMoKCs at an earlier differentiation phase. Of note, MARS-RNAseq analysis did not reveal reduced expression of Timd4, Vsig4, and Cd163 genes in Clec4fΔCommd10 KCs. This may be due to the inferior accuracy of MARS-seq in comparison with RT-PCR and flow cytometry methods used to obtain the results above. BM chimerism experiments further reveal complete replacement of fetal radioresistant KCs with radiosensitive MoKCs in Clec4fΔCommd10 livers. Moreover, in mixed BM chimerism experiments, Clec4fΔCommd10 KCs evenly compete with their WT counterparts on the ability to replenish the KC niche following ablation, but later exhibit impaired survival. In agreement, Clec4fΔCommd10 KCs also exhibit higher homeostatic proliferation, originating from the constant need for replenishment. In agreement with these results, there is profound reduction in normalized counts of KCs in Clec4fΔCommd10 livers. This may be also a result of their increased fragility to the isolation protocol.
Clec4Fcre/+ mice used in our studies are a well-established tool to target KCs by virtue of their selective expression of the c-type lectin CLEC4F (Bonnardel et al., 2019; Scott et al., 2018). We show that Clec4FΔCommd10 KCs express the KC-associated CLEC4F protein similarly to control KCs, therefore allowing their efficient targeting in Clec4Fcre/+ mice. Cx3cr1+/cre mice are also used to target resident KCs. However, adult KCs are CX3CR1- and Cx3cr1cre/+ mice only target 25% of Ly6Chi monocytes (Yona et al., 2013). As it remains unknown to what extent MoKC express CX3CR1 during their differentiation pass, we speculated that the Cx3cr1cre/+ mice would be less efficient in targeting Commd10 gene deletion in MoKCs. Indeed, in contrast to the uniform loss of TIM4 expression in 6-months old Clec4FΔCommd10 KCs, there are about 30% of TIM4+ cells among Cx3cr1ΔCommd10 KCs. In addition, other liver resident macrophages such as CLEC4F-CX3CR1+ liver capsular macrophages (LCMs)(Sierro et al., 2017), will be targeted in Cx3cr1cre/+ mice, but not in Clec4Fcre/+ mice. Therefore, Clec4Fcre/+ mice were preferentially used in our studies to specifically target KCs.
Transcriptional analysis of Clec4fΔCommd10 KCs revealed a notable reduction in genes encoding for translation initiation- and ribosomal complex proteins, for ubiquitin and the proteasome protein complex, and for key proteins of the mitochondrial respiration chain. These transcriptional alterations allude to protein stress and mitochondrial dysfunction, and hint at the reason of Clec4fΔCommd10 KC premature death. Further defining the role of COMMD10 in macrophage homeostatic maintenance, we demonstrate that its deficiency impedes survival of other tissue resident macrophages. COMMD10-deficient LPMs display significantly reduced counts, replacement of TIM4hi with TIM4lo LPMs, and compensatory increased proliferation. Mixed BM chimerism further confirmed impaired survival of LysMΔCommd10 versus WT LPMs and AMs.
KCs are the main sentinel phagocytes located in juxtaposition to hepatocytes, and therefore initiate a reparative immune response while facing hepatocyte damage (Krenkel and Tacke, 2017). Despite a notable delay in clearance of dying cells, COMMD10-deficiency in KCs had moderate effects on hepatic damage. In contrast, COMMD10 deficiency in Ly6Chi monocytes profoundly aggravated hepatic damage and inflammation. In agreement with our previous studies during endotoxic shock and colitis (Mouhadeb et al., 2018), COMMD10-deficiency unleashes canonical and non-canonical inflammasome activation in liver infiltrating Ly6Chi monocytes in AILI. Moreover, LysMΔCommd10 Ly6Chi monocytes display upregulation of various inflammatory mediators and of ECM components and remodeling enzymes, which may contribute to the collateral damage. Specifically, their increased production of CCL2 may be linked to their increased accumulation in LysMΔCommd10 livers. In contrast, they dramatically reduce expression of genes associated with type I IFN production and response. During AILI, the release of DAMPs from damaged hepatocytes triggers activation of type I interferon pathways in liver immune cells (Araujo et al., 2018). Interestingly, similar type I IFN monocytes have been recently described in the metabolically diseased liver (Remmerie et al., 2020), during acute liver failure (Kolodziejczyk et al., 2020) and in injured peripheral nerves (Ydens et al., 2020), and hence may be functionally involved in regulating tissue inflammation and resolution.
Recent studies have indicated binary differentiation pathways of BM Ly6Chi monocytes yielding distinct NeuMo and DCMo-like monocyte progenies (Weinreb et al., 2020; Yanez et al., 2017). Based on the NeuMo and DCMo marker genes described in these two studies we show here that COMMD10-deficiency in Ly6Chi monocyte generates a clear bias towards NeuMo monocytes. This differentiation bias initiates already in the steady state BM. We also noticed a bias of LysMΔCommd10 Ly6Chi monocytes toward a cluster of cells characterized by the expression of markers associated with LAMs, a MoMF population accumulating in high damage areas in the metabolically diseased liver (Remmerie et al., 2020). We intend to pursue further the immunemetabolic functions of COMMD10 in these cells during chronic models of metabolic liver diseases.
Limitation of the study
Our findings highlight the impaired homeostatic survival of COMMD10-deficient KCs and suggest their continuous renewal by Ly6Chi monocytes. Yet, the BM chimerism approach used here does not directly examine the lineage of COMMD10-deficient KCs. Classical fate mapping studies, such as shielded liver irradiation, parabiosis, or transgenic approaches allowing discrimination between fetal or BM-derived macrophages, are required to directly prove the renewal of COMMD10-deficient KCs by Ly6Chi monocytes.
Future Directions
COMMD proteins, and specifically COMMD10, are poorly defined mechanistically. To define the mechanistic link between COMMD10-deficiency and altered resident macrophage cell survival, it would be important to develop tools that allow investigation of the COMMD10 interactome. Our comprehension of Ly6Chi monocyte heterogeneity, of how COMMD10 mediates Ly6Chi monocyte differentiation, and of whether the biased differentiation fates of COMMD10-deficient monocytes are responsible for the aggravated necro-inflammatory response during liver injury, remains limited. Establishing methods that enable phenotypic and functional discrimination between distinct Ly6Chi monocyte-derived effector cells will help clarify these questions.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chen Varol (chenv@tlvmc.gov.il).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All Single-cell RNA-seq, Bulk RNA-seq and MARS-RNA-seq data have been deposited at GEO and are publicly available. Accession numbers are listed in the key resources table. Original Western blot images were deposited in Mendeley, and associated link appears in the key resources table. Original microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC/Cy7 anti mouse CD45 (30-F11) | BioLegend | Cat #103116; RRID: AB_312981 |
| PE/Cy7 anti mouse CD11b (M1/70) | BioLegend | Cat #101216; RRID: AB_312799 |
| PerCP/Cy5.5 anti mouse Ly6C (HK1.4) | BioLegend | Cat#128012; RRID: AB_1659241 |
| PB anti mouse Ly6G (1A8) | BioLegend | Cat#127612; RRID: AB_2251161 |
| PE anti mouse CD115 (AF598) | BioLegend | Cat#135505; RRID: AB_1937254 |
| APC anti mouse TIM4 (RMT4–54) | BioLegend | Cat#130008; RRID: AB_2271648 |
| APC anti mouse CD163 (TNKUPJ) | ThermoFisher Scientific | Cat#12–1631-80; RRID: AB_2716923 |
| PE anti mouse F4/80 (REA126) | Miltenyi | Cat#130–116-499; RRID: AB_2727574 |
| APC anti mouse VSIG4 (NLA14) | ThermoFisher Scientific | Cat#25–5752-82; RRID: AB_2637431 |
| Unconjugated anti mouse CLEC4F | R&D Systems | Cat#AF2784; RRID: AB_2081339 |
| PE/Cy7 anti mouse CD64 (X54–5/7.1) | BioLegend | Cat#139314; RRID: AB_2563904 |
| PE anti mouse CLEC2 (17D9) | BioLegend | Cat#146104; RRID: AB_2562383 |
| APC anti mouse CD45.1 (A20) | BioLegend | Cat#110713; RRID: AB_313502 |
| CFL 647 donkey anti goat IgG | Santa Cruz Biotechnology | sc-362285 |
| Alexa flour 488 anti mouse CX3CR1 (SA011F11) | BioLegend | Cat#149022; RRID: AB_2565705 |
| Alexa flour 647 anti mouse SiglecF (E50–2440 (RUO)) | BD | BD562680; RRID: AB_2687570 |
| PE anti mouse F4/80 (BM8) | BioLegend | Cat#123110; RRID: AB_893486 |
| PE/Cy7 anti mouse TIM4 (RMT4–54) | BioLegend | Cat#130009; RRID: AB_2565718 |
| Alexa flour 647 anti mouse CD102 (3C4 (MIC2/4)) | BioLegend | Cat#105611; RRID: AB_2122183 |
| Alexa flour 488 donkey anti rat IgG (H+L) | ThermoFisher Scientific | A21208; RRID: AB_2535794 |
| Ki-67 eFluor 660 (SolA15) | Invitrogen | Cat#50–5698-82; RRID: AB_2574235 |
| Anti-Desmin | Abcam | ab15200; RRID: AB_301744 |
| Alex flour 555 goat anti rabbit IgG (H+L) | Abcam | ab150078; RRID: AB_2722519 |
| Anti CD31 (MEC 13.3) | BD | BD550274; RRID: AB_393571 |
| PB anti mouse CD11c (N418) | BioLegend | Cat#117321; RRID: AB_755987 |
| MC-21 anti mouse CCR2 | Jung Lab (Weizmann Institute of Science) (Mack et al., 2001) | Clone: MC-21 |
| Chemicals, Peptides and Recombinant Proteins | ||
| APC BRDU FLIW KIT | BD | BD552598 |
| APAP | Merck | A7085 |
| BODIPY 493/503 | ThermoFisher Scientific | D3922 |
| CFSE | Merck | Cat#65–0850 |
| Collagenase VIII from Clostridium histolyticum | Merck | C5138–500MG |
| Dexamethasone | Merck | D4902–25MG |
| Dnase I (Deoxyribonuclease I from bovine pancreas) | Merck | 10104159001 DN25–100MG |
| DPBS | Biological Industries | 02–023-1A |
| DTT (Dithiothreitol) | Formidum | 3483–12-3 |
| EDTA (Ethylenediaminetetraacetic acid disodium salt dihydrate) | Merck | E5134 |
| Fetal Bovine Serum | Biological industries | 04–001-1A |
| Fixation/Permeablization Kit | BD | BD554714; RRID: AB_2869008 |
| HBSS −/− | gibco | 2183113 |
| Clodronate | Liposoma | C-005 |
| Paraformaldehyde 32% | Electron microscopy sciences | 15714 |
| Protease inhibitor | Merck | P8340 |
| RIPA buffer | Cell Signaling | C-9806S |
| Tissue-Tek O.C.T | Scigen | 23–730-625 |
| TRIzol® reagent | Merck | T9424–100ML |
| Critical Commercial Assays | ||
| cDNA reverse transcription kit | Applied Biosystems | 4368814 |
| miRNeasy Mini Kit | QIAGEN | 217084 |
| SYBR Green PCR Master Mix kit | Applied Biosystems | 4309155 |
| TUNEL staining | Promega | G3250 |
| Deposited Data | ||
| KCs steady state: Bulk MARS-Seq | This paper | GEO: GSE183493 |
| Ly6Chi monocytes AILI 24 h: Bulk RNAseq | This paper | GEO: GSE183494 |
| BM Ly6Chi monocytes: Bulk RNAseq | This paper | GEO: GSE183495 |
| Ly6Chi monocytes AILI 24 h: Single cell RNAseq | This paper | GEO: GSE183367 |
| RNAseq SuperSeries bundle of all four datasets | This paper | GEO: GSE183756 |
| RAW Western blot images (refer to Figures 4G, 5F) | This paper | Mendeley, DOI: 10.17632/kfst2hy224.1 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Commd10fl/fl | Generated by our lab, (Mouhadeb et al., 2018) | N/A |
| Mouse: Cx3cr1cre/+ (Cx3cr1tm1.1(cre)Jung) | Jung Lab (Weizmann Institute of Science) (Yona et al., 2013) | Stock No: 025524 |
| Mouse: Lyz2cre/+ (B6.129P2-Lyz2tm1(cre)Ifo) | The Jackson Laboratories | Stock No: 004781 |
| Mouse: Clec4fcre/+ | Guilliams Lab (Ghent University) (Scott et al., 2018) | N/A |
| Mouse: CD45.1 (B6.SJL-Ptprca Pepcb/Boy) | The Jackson Laboratories | Stock No: 002014 |
| Mouse: CD45.2 C57BL/6JOlaHsd male mice | Envigo (Israel) | Stock No: 000664 |
| Mouse: Ccr2−/−Cx3cr1gfp/+ | Jung Lab (Weizmann Institute of Science) (Zigmond et al., 2012) | N/A |
| Oligonucleotides | ||
| Commd10 FWD: GAGAGCCCCAGCATGAAGAA | Merck | N/A |
| Commd10 REV: AATCCGGCTGAGAAGTCGTG | Merck | N/A |
| Rplp0 FWD: TCCAGCAGGTGTTTGACAAC | Merck | N/A |
| Rplp0 REV: CCATCTGCAGACACACACT | Merck | N/A |
| Arg1 FWD:CAGAAGAATGGAAGAGTCAG | Merck | N/A |
| Arg1 REV:CAGATATGCAGGGAGTCACC | Merck | N/A |
| Trem2 FWD: GGCCCATGCCAGCGTGTGGT | Merck | N/A |
| Trem2 REV: CCAGAGATCTCCAGCATC | Merck | N/A |
| Spp1 FWD: AGCAAGAAACTCTTCCAAGCAA | Merck | N/A |
| Spp1 REV: TGAGATTCGTCAGATTCATCCG | Merck | N/A |
| More Oligonucleotides appear in Figure S7 | ||
| Sofwere and Algorithms | ||
| FlowJo 10.7.1 | Flow jo | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism 5 | GraphPad Software | https://www.graphpad.com/ |
| ImageJ v1.51j | NIH | https://imagej.nih.gov/ij/ |
| StepOne v2.3 | StepOne software | https://www.thermofisher.com/il/en/home/technicalresources/software-downloads/StepOne-and-StepOnePlus-Real-Time-PCR-System.html |
| ZEN 2011 | Zeiss | https://www.zeiss.com/microscopy/int/products/microscopesoftware/zen-lite.html |
| Partek Genomics Suite | Partek | https://www.partek.com/partek-genomics-suite/ |
| Cutadapt | DOI:10.14806/ej.17.1.200 | https://cutadapt.readthedocs.io/en/stable/ |
| Ensembl | DOI:10.1093/nar/gkaa942 | https://www.ensembl.org/index.html |
| DESeq2 | Bioconductor, (Love et al., 2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| snakemake | (Koster and Rahmann, 2012) | https://snakemake.readthedocs.io/en/stable/ |
| STAR | (Dobin et al., 2013) | http://code.google.com/p/rna-star/. |
| MouseMine | (Motenko et al., 2015) | http://www.mousemine.org/mousemine/begin.do |
| CellMarker | (Zhang et al., 2019) | http://biocc.hrbmu.edu.cn/CellMarker/ |
| Ingenuity Pathway Analysis | QIAGEN Bioinformatics | https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice strain
Animal experiments were performed with male adult mice (C57BL/6 background). Animals were maintained in specific pathogen-free animal facility, and experiments were performed according to protocols approved by the Animal Care Use Committee of the Sourasky Medical Center. Mice were housed with 12-h light cycles and a constant temperature of 22°C. LysMΔCommd10, Cx3cr1ΔCommd10, and Clec4fΔCommd10 mice, and their individual littermate Commd10fl/fl controls, were generated by crossing Lyz2cre/+ (B6.129P2-Lyz2tm1(cre)Ifo, Stock No: 004781, The Jackson Laboratory), Cx3cr1cre/+ (Yona et al., 2013), and Clec4fcre/+ (Scott et al., 2018) mice with Commd10fl/fl mice (Mouhadeb et al., 2018), respectively. Ccr2−/−Cx3cr1gfp/+ (Zigmond et al., 2012) were provided by the Jung laboratory. For chimera mice experiments, six-week-old CD45.1 (B6.SJL-Ptprca Pepcb/Boy, Stock No: 002014, The Jackson Laboratory) and CD45.2 C57BL/6JOlaHsd male mice (Envigo, Israel) were used.
METHOD DETAILS
Acetaminophen-induced liver injury (AILI)
Mice were fasted overnight for 12 h prior to intraperitoneal (i.p.) administration of 300 mg/kg acetaminophen (APAP). Water was returned concomitantly with APAP administration and chow 2 h later.
Quantification of hepatic damage
Liver samples were obtained at 24/48 h after AILI, fixed (4% paraformaldehyde), paraffinembedded, sectioned, and stained with H&E of liver sections. Blinded pathologic evaluation was performed by a liver pathologist. Necrosis was scored as 0 (no necrosis), 1 (spotty necrosis), 2 (confluent, zone 3 necrosis), 3 (confluent, zone 2 plus 3 necrosis), or 4 (panlobular necrosis). Bridging necrosis was scored as 0 (absent) or 1 (present). Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were measured using a ADVIA Chemistry XPT analyzer.
MC-21-mediated ablation of Ly6Chi monocytes
When monocyte ablation was required, mice received an i.p. injection of 400 μL conditioned media of anti-mouse CCR2 mAb (clone MC-21)(Mack et al., 2001)(about 29 μg Ab/mL). Injections were performed at 12 h prior to APAP challenge.
Isolation of hepatic non-parenchymal cells
Isolation of hepatic non-parenchymal cells was performed as previously described (Ben Shlomo et al., 2019; Graubardt et al., 2017; Zigmond et al., 2014). In brief, mice were anesthetized and perfused livers were collected, cut into small fragments, and incubated with 5 mL digestion buffer composed of 5% fetal bovine serum, 0.5 mg/mL collagenase VIII from Clostridium histolyticum, 0.1 mg/mL Deoxyribonuclease I from bovine pancreas in Dulbecco’s phosphate-buffered saline with calcium and magnesium (PBS+/+), in a shaker-incubator at 250 rpm, 37°C for 45 min. Samples were then subjected to three cycles of washing with Dulbecco’s phosphate-buffered saline without calcium and magnesium (PBS−/−) at 400 rpm, 4°C for 5 min from which the supernatant was kept, omitting the parenchymal cell pellet. Subsequently, the supernatant was centrifuged at 1,400 rpm, 4°C for 5 min and the cell pellet was lysed for erythrocytes by 2 min incubation with Ammonium-Chloride-Potassium (ACK) buffer composed of 0.15 M NH4Cl and 0.01 M KHCO3 and washed with PBS−/−.
Isolation of peritoneal macrophages
Mice were anesthetized and resident peritoneal macrophages were obtained by peritoneal lavage with ice-cold PBS from Commd10fl/fl, Cx3cr1ΔCommd10 or LysMΔCommd10 mice. Subsequently, the supernatant was centrifuged at 1,400 rpm, 4°C for 5 min and the pellet was stained.
Isolation of colonic lamina propria macrophages
Colonic lamina propria phagocytes were isolated as previously described (Zigmond et al., 2012). In brief, colons were extracted, extra-intestinal fat tissue was carefully removed and colons were then flushed of their luminal content with cold PBS−/−, opened longitudinally, and cut into 3–5 mm pieces. Epithelial cells and mucus were removed by 40 min incubation with 5 ml HBSS−/− (without Ca2+ and Mg2+) containing 5% FBS, 2 mM EDTA, and 1 mM DTT at 37°C shaking at 250 rpm. Colon pieces were then incubated with shaking at 37°C, 250 rpm for 45 min with 5 ml digestion buffer [5% FCS, 1 mg/ml collagenase VIII in PBS+/+]. The cell suspension was then filtered through 200 μM wire mesh and washed with PBS−/−.
Isolation of alveolar macrophages
Mice were anesthetized and the trachea on the ventral side of the neck was exposed. A 22 G needle attached to a 1 ml syringe containing PBS−/− was inserted through the tracheal wall into the lumen just below the larynx. The PBS−/− was inserted into the lung and withdrew into the syringe. The supernatant was centrifuged at 1,400 rpm, 4°C for 5 min.
Isolation of spleen macrophages
The spleen was perfused with 1ml of digestion buffer composed of 5% fetal bovine serum, 0.5 mg/ml, collagenase VIII from Clostridium histolyticum, 0.1 mg/mL Deoxyribonuclease I from bovine pancreas in PBS+/+ and incubated in 37°C for 40 min. The supernatant was centrifuged at 1,400 rpm, 4°C for 5 min and the cell pellet was lysed for erythrocytes by 2 min incubation with (ACK) and washed with PBS−/−.
Isolation of blood and BM Ly6Chi monocytes
Blood peripheral blood mononuclear and polymorphonuclear cells were isolated using the BD FACS Lysing Solution, according to the manufacturer’s instructions. For isolation of BM cells, femurs and tibia were carefully dissected and flashed with cold PBS-/−.
Protein immunoblotting
Total protein from liver was extracted in ice cold RIPA buffer containing protease inhibitors. Proteins were detected by immunoblotting using standard techniques. Antibodies used: caspase-11 (sc-374615), GAPDH (sc-47724), from Santa Cruz; caspase-1(AG-20B-0042) from Adipogen. Blots were incubated with HRP-conjugated secondary antibodies and subjected to chemiluminescent detection using the MicroChemi imaging system (DNR Bio-Imaging Systems, Israel).
Liver triglyceride measuring
Liver tissues were harvest and homogenized in 5% NP-40 using homogenizer. The lysates were heated to 100°C for 5 minutes and then cooled down to room temperature twice. The lysates were measured using a ADVIA Chemistry XPT analyzer.
Staining for iron deposits in the liver
Liver samples were obtained at steady state, fixed (4% paraformaldehyde), paraffin-embedded, sectioned, and stained with Perls’s stain for Iron of liver sections. Blinded pathologic evaluation was performed by a liver pathologist.
Quantitative Real-Time PCR (RT-PCR)
Total RNA was extracted from tissues with TRIzol® reagent, and from sorted Ly6Chi monocytes or Kupffer cells using the miRNeasy Mini Kit. RNA was reverse transcribed with a high-capacity cDNA reverse transcription kit. All PCR reactions were performed with SYBR Green PCR Master Mix kit and Applied Biosystems 7300 Real-Time PCR machine. Quantification of PCR signals of each sample was performed by the ΔCt method normalized to Rplp0 housekeeping gene. Oligonucleotide sequences appear in the Key Resources Table and Figure S7.
Flow cytometry analysis
Cells (0.5–5 X 106) isolated from liver, peritoneum, alveolar space, spleen, colon, BM, or peripheral blood were stained with appropriate antibodies (listed in the key resource table) at 40C in the dark for 20 min and were analyzed with BD FACSCanto™ II (BD Bioscience). Flow cytometry analysis was performed using FlowJo™ software (Ashland, OR, Becton, Dickinson & Company USA). Cell sorting was performed using the BD FACSAria™ FUSION cell sorter (BD Bioscience).
Confocal microscopy
Perfused livers were fixed using 4% PFA for 24 h, dehydrated in 30% sucrose, and subsequently embedded in OCT freezing media. About 13 µm sections were sliced with cryostat (Thermo fisher) and blocked with a blocking buffer containing 2% BSA for 1h. Sections were stained with directly conjugated antibodies or appropriate primary and secondary antibodies 1h or overnight, respectively. For detection of in situ cell death (TUNEL staining), staining was performed using the supplier’s protocol (Promega). Sections were mounted with fluorescence mounting medium containing DAPI. Images were taken with ZEISS Confocal Microscope LSM700 (MicroImaging GmbH, ZEISS, Germany). Image processing was performed with ZEN 2011 SP7 software. TUNEL quantification was calculated by subtraction of the background from each slide and an average was performed.
Clearance of apoptotic cells, in vivo assay
To generate apoptotic cells, thymocytes isolated from 6- to 8-weeks-old C57BL/6 mice were labeled with CFSE (Merck), and then incubated for 16 hr in 1 mM dexamethasone (Merck), washed several times with PBS, spun through a FCS cushion to eliminate the dexamethasone, and resuspended in 1% FCS in RPMI or PBS. Peritoneal macrophages were isolated from Commd10fl/fl and Cx3cr1ΔCommd10 mice, 20 or 30 minutes after intraperitoneal injection of 2 X 107 apoptotic thymocytes. Resident peritoneal macrophages were obtained by peritoneal lavage with ice-cold PBS. KCs were isolated from the livers of Commd10fl/fl or Cx3cr1ΔCommd10 mice, 15 minutes after injection to the portal vein of 2 X 107 CFSE-labeled apoptotic thymocytes. KCs were isolated as described above.
Clearance of apoptotic cells, in vitro assay
Resident macrophages were isolated from the peritoneum of Commd10fl/fl and Cx3cr1ΔCommd10 mice by peritoneal lavage with ice-cold 4% FCS in PBS−/− and plated on coverslips in 24-well plates for immunofluorescent confocal microscopy. Macrophages were allowed to rest for 2 hours at 37 0C at 5% CO2 before starting experiments. Macrophages were incubated with apoptotic cells in 1% FCS in RPMI for 2 hours at a ratio of 1:5 at 37 0C. Afterwards, unbound cells were gently washed away with ice-cold PBS. For immunofluorescence microscopy, macrophages on coverslips were stained as indicated and mounted on glass slides. Images were taken with ZEISS Confocal Microscope (MicroImaging GmbH, ZEISS, Germany). Processing was performed with ZEN 2011 software.
Total body irradiation and BM transplantation
Ccr2−/− mice (C57BL/6 background) were exposed to 3Gy total body irradiation. BM cells were harvested from donor mice the day after by gently flushing their femurs, and 5 × 106 cells were intravenously injected into each of recipient mice. A two-week recovery period was sufficient to ensure donor BM engraftment and blood monocyte reconstitution.
WT C57BL/6, Commd10fl/fl or Clec4fΔCommd10 mice were exposed to 9 Gy total body irradiation. Mice were reconstituted the day after by intravenous administration of 5 × 106 BM cells from congenic CD45.1 WT BM, or of 5 × 106 mixed 1:1 CD45.1 WT/CD45.2 Clec4fΔCommd10, CD45.1 WT / CD45.2 LysMΔCommd10 or CD45.1 WT / CD45.2 Cx3cr1ΔCommd10 BM cells. An eight to nine weeks recovery period was sufficient to ensure donor BM engraftment and reconstitution of macrophage compartments in the liver, peritoneum, alveolar space or colonic lamina propria.
Clodronate liposome mediated depletion of KCs
To deplete KCs, mice were injected intravenous (10ul/gr body weight) of the liposome suspension (Liposoma, the Netherlands) to the lateral tail vain. The mice were sacrificed 4/8 weeks post injection.
Extraction of microarray data
Date was extracted from our previously established Affymetrix GeneChip database deposited at the National Center for Biotechnology Information Gene Expression Omnibus public database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE55606 (Zigmond et al., 2014). Ly6Chi monocytes were sorted at the inflammatory peak of the necrotic phase, 24 h post-AILI (n=3). Their Ly6Clo MoMF descendants were sorted at the recovery phase, 72 h post-AILI (n=3). Heat maps were performed using Partek Genomics Suite software.
Bulk RNAseq
RNAseq and bioinformatics were performed as a service at The Crown Genomics and The Mantoux Bioinformatics institutes of the Nancy and Stephen Grand Israel National Center for Personalized Medicine (GINCPM), Weizmann Institute of Science. All analyses were performed by Dr. Dayana Yahalomi. Sequencing: Libraries were prepared using the GINCPM-mRNA-seq protocol (Weizmann Institute of Science). PolyA fraction (mRNA) was purified from 500 ng of total RNA following by fragmentation and the generation of double-stranded cDNA. Then, end repair, A base addition, adapter ligation and PCR amplification steps were performed. Libraries were evaluated by Qubit (Thermo fisher scientific) and TapeStation (Agilent). Sequencing libraries were constructed with barcodes to allow multiplexing of samples. Around 20 million single-end 60-bp reads were sequenced per sample on Illumina SR 60 v4, High Output, one lane.
Bioinformatics: Poly-A/T stretches and Illumina adapters were trimmed from the reads using Cutadapt; and resulting reads shorter than 30bp were discarded. Reads were mapped to the M. musculus reference genome GRCm38, supplied with gene annotations downloaded from Ensembl (Version 92) (and with End-To-End option and outFilterMismatchNoverLmax was set to 0.04). Expression levels for each gene were quantified using htseq-count (Anders et al., 2015), using the gtf above. Differentially expressed genes were identified using DESeq2 (Love et al., 2014), with the betaPrior, cooks Cutoff and independent Filtering parameters set to False. Raw P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg. Pipeline was run using snakemake (Koster and Rahmann, 2012).
Bulk MARS-Seq
A bulk adaptation of the MARS-Seq protocol (Jaitin et al., 2014; Keren-Shaul et al., 2019) was used to generate RNA-Seq libraries for expression profiling. Briefly, mRNA was purified from ~10,000 cells of each sample using Dynabeads mRNA Direct purification kit (ThermoFisher), barcoded during reverse transcription and pooled. Following Agencourct Ampure XP beads cleanup (Beckman Coulter), the pooled samples underwent second strand synthesis and were linearly amplified by T7 in vitro transcription. The resulting RNA was fragmented and converted into a sequencing-ready library by tagging the samples with Illumina sequences during ligation, RT, and PCR. Libraries were quantified by Qubit and TapeStation as well as by qPCR for mouse ActB gene. Sequencing was done on a Nextseq 75 cycles high output kit (Illumina).
Bioinformatics:
Poly-A/T stretches and Illumina adapters were trimmed from the reads using Cutadapt; resulting reads shorter than 30bp were discarded. Remaining reads were mapped onto 3’ UTR regions (1000 bases) of M. musculus, mm10 genome according to Refseq annotations, using STAR (Dobin et al., 2013) with End-To-End option and outFilterMismatchNoverLmax was set to 0.05). Deduplication was carried out by flagging all reads that were mapped to the same gene and had the same UMI. Counts for each gene were quantified using htseq-count (Anders et al., 2015), using the gtf above and corrected for UMI saturation. Differentially expressed genes (DEGs) were identified using DESeq2 (Love et al., 2014), with the betaPrior, cooksCutoff and independentFiltering parameters set to False. Raw P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg. Pipeline was run using snakemake (Koster and Rahmann, 2012). MouseMine bioinformatics tool (Motenko et al., 2015) and Ingenuity pathway analysis (IPA) (QIAGEN) was used to determine pathway and gene ontology (GO) enrichments among the DEGs.
Single cell RNAseq
Cells were counted and diluted to a final concentration in PBS supplemented with 0.04% BSA. Cellular suspension was loaded onto Next GEM Chip G and then ran on a Chromium Controller instrument to generate GEM emulsion (10x Genomics). Single-cell 3’ RNA-seq libraries were generated according to the manufacturer’s protocol (10x Genomics Chromium Single Cell 3’ Reagent Kit User Guide v3/v3.1 Chemistry). Final libraries were quantified using NEBNext Library Quant Kit for Illumina (NEB) and high sensitivity D1000 TapeStation (Agilent). Libraries were pooled according to targeted cell number, aiming for ~50,000 reads per cell. Pooled libraries were sequenced on a NovaSeq 6000 instrument using an SP 100 cycles reagent kit (Illumina).
Bioinformatics:
Cell Ranger v3 with default parameters was used for alignment, filtering, barcode counting, and UMI counting. The Seurat package in R was used for downstream analysis and visualization. Log normalization was used to normalized the reads. Dimension reduction was done using PCA. Clustering was done using KNN graph and visualization and non-linear reduction was done using UMAP. Marker genes were found by preforming differential expression based on the non-parametric Wilcoxon rank sum test (Seurat default). CellMarker (http://biocc.hrbmu.edu.cn/CellMarker/)(Zhang et al., 2019) was used to annotate each cluster according to the top 10 marker genes. Cell type enrichment was carried out using hypergeometric test. Each cluster annotation was determined according to cell type with the best p value.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical differences between two groups were determined according to unpaired two-tailed student’s t-test. Statistical differences between three groups and above were determined using one way ANOVA with Tukey post-tests. All experiments were analyzed using Prism 7 (GraphPad Software). Graphical data was shown as mean values with error bars indicating SEM. Significance was defined if p-value was less than 0.05 as following: * p< 0.05; ** p< 0.01; *** p< 0.001. Statistical details of experiments can be found in figure legends.
Supplementary Material
Table S1: Expression of KC associated core genes in Commd10fl/fl vs. Clec4fΔCommd10 KCs (Related to Figure 2).
Significantly downregulated genes are marked in red.
Table S2: Functional enrichment pathways extracted from downregulated genes between Commd10fl/fl vs. LysMΔCommd10 Ly6Chi monocytes at 24 h post-APAP (Related to Figure 6A, B, D, E).
Table S3: Upregulation of type I interferon response genes upon differentiation of Ly6Chi monocytes (AILI 24 h) into pro-restorative MoMFs (AILI 72 h) (Related to Figure 6C).
Table S4: Functional enrichment pathways extracted from upregulated genes between Commd10fl/fl vs. LysMΔCommd10 Ly6Chi monocytes at 24 h post-AILI (Related to Figure 6A).
Table S5: Variable expression of NeuMo and DCMo signature genes in LysMΔCommd10 versus Commd10fl/fl Ly6Chi monocytes (Related to Figure 6F, G, S5).
NeuMo and DCMo gene signatures were extracted from (Weinreb et al., 2020; Yanez et al., 2017)
Table S6: Single cell RNAseq analysis of sorted Commd10fl/fl versus LysMΔCommd10 Ly6Chi monocytes at AILI 24 h (Related to Figure 7D, E, G, S6A-C).
Table S7: Biased expression of top hepatic lipid associated macrophages (LAM) markers by LysMΔCommd10 Ly6Chi monocytes (Related to Figure 7G).
Top score (above 2) LAM signature genes were extracted from (Remmerie et al., 2020)
Acknowledgements
The authors acknowledge support from the Israel Science Foundation (grants # 1745/19 to CV and # 331/18 to NG), the Azrieli Foundation Canada-Israel (to CV), and the NIH (NIAID) R01 AI134987 (to HSG).
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Abram CL, Roberge GL, Hu Y, and Lowell CA (2014). Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J Immunol Methods 408, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo AM, Antunes MM, Mattos MS, Diniz AB, Alvarenga DM, Nakagaki BN, Carvalho E, Lacerda VAS, Carvalho-Gontijo R, Goulart J, et al. (2018). Liver Immune Cells Release Type 1 Interferon Due to DNA Sensing and Amplify Liver Injury from Acetaminophen Overdose. Cells 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuzi P, Billadeau DD, Favier R, Rong S, Dekker D, Fedoseienko A, Fieten H, Wijers M, Levels JH, Huijkman N, et al. (2016). CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat Commun 7, 10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuzi P, Hofker MH, and van de Sluis B (2013). Tuning NF-kappaB activity: a touch of COMMD proteins. Biochim Biophys Acta 1832, 2315–2321. [DOI] [PubMed] [Google Scholar]
- Ben Shlomo S, Mouhadeb O, Cohen K, Varol C, and Gluck N (2019). COMMD10-Guided Phagolysosomal Maturation Promotes Clearance of Staphylococcus aureus in Macrophages. iScience 14, 147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnardel J, T’Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, Vanneste B, De Prijck S, Nedospasov SA, Kremer A, et al. (2019). Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 51, 638–654 e639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, Maine GN, Wilkinson JC, Mayo MW, and Duckett CS (2005). COMMD proteins, a novel family of structural and functional homologs of MURR1. The Journal of biological chemistry 280, 22222–22232. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama S, Nakahashi-Oda C, Abe F, Wang Y, Sato K, and Shibuya A (2019). Identification and isolation of splenic tissue-resident macrophage sub-populations by flow cytometry. Int Immunol 31, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubardt N, Vugman M, Mouhadeb O, Caliari G, Pasmanik-Chor M, Reuveni D, Zigmond E, Brazowski E, David E, Chappell-Maor L, et al. (2017). Ly6C(hi) Monocytes and Their Macrophage Descendants Regulate Neutrophil Function and Clearance in Acetaminophen-Induced Liver Injury. Front Immunol 8, 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, and Amit I (2014). Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, Kenigsberg E, Jaitin DA, David E, Paul F, Tanay A, and Amit I (2019). MARS-seq2.0: an experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat Protoc 14, 1841–1862. [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Federici S, Zmora N, Mohapatra G, Dori-Bachash M, Hornstein S, Leshem A, Reuveni D, Zigmond E, Tobar A, et al. (2020). Acute liver failure is regulated by MYC- and microbiome-dependent programs. Nat Med [DOI] [PubMed] [Google Scholar]
- Koster J, and Rahmann S (2012). Snakemake--a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Krenkel O, Puengel T, Govaere O, Abdallah AT, Mossanen JC, Kohlhepp M, Liepelt A, Lefebvre E, Luedde T, Hellerbrand C, et al. (2018). Therapeutic inhibition of inflammatory monocyte recruitment reduces steatohepatitis and liver fibrosis. Hepatology 67, 1270–1283. [DOI] [PubMed] [Google Scholar]
- Krenkel O, and Tacke F (2017). Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 17, 306–321. [DOI] [PubMed] [Google Scholar]
- Li H, Chan L, Bartuzi P, Melton SD, Weber A, Ben-Shlomo S, Varol C, Raetz M, Mao X, Starokadomskyy P, et al. (2014). Copper metabolism domain-containing 1 represses genes that promote inflammation and protects mice from colitis and colitis-associated cancer. Gastroenterology 147, 184–195 e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack M, Cihak J, Simonis C, Luckow B, Proudfoot AE, Plachy J, Bruhl H, Frink M, Anders HJ, Vielhauer V, et al. (2001). Expression and characterization of the chemokine receptors CCR2 and CCR5 in mice. J Immunol 166, 4697–4704. [DOI] [PubMed] [Google Scholar]
- Maine GN, Mao X, Komarck CM, and Burstein E (2007). COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. The EMBO journal 26, 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motenko H, Neuhauser SB, O’Keefe M, and Richardson JE (2015). MouseMine: a new data warehouse for MGI. Mamm Genome 26, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhadeb O, Ben Shlomo S, Cohen K, Farkash I, Gruber S, Maharshak N, Halpern Z, Burstein E, Gluck N, and Varol C (2018). Impaired COMMD10-Mediated Regulation of Ly6C(hi) Monocyte-Driven Inflammation Disrupts Gut Barrier Function. Front Immunol 9, 2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Krawczak CA, Singla A, Starokadomskyy P, Deng Z, Osborne DG, Li H, Dick CJ, Gomez TS, Koenecke M, Zhang JS, et al. (2015). COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Molecular biology of the cell 26, 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, Portman JR, Matchett KP, Brice M, Marwick JA, et al. (2019). Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmerie A, Martens L, Thone T, Castoldi A, Seurinck R, Pavie B, Roels J, Vanneste B, De Prijck S, Vanhockerhout M, et al. (2020). Osteopontin Expression Identifies a Subset of Recruited Macrophages Distinct from Kupffer Cells in the Fatty Liver. Immunity 53, 641–657 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai M, Troutman TD, Seidman JS, Ouyang Z, Spann NJ, Abe Y, Ego KM, Bruni CM, Deng Z, Schlachetzki JCM, et al. (2019). Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity 51, 655–670 e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, and Guilliams M (2018). The role of Kupffer cells in hepatic iron and lipid metabolism. J Hepatol 69, 1197–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, T’Jonck W, Martens L, Todorov H, Sichien D, Soen B, Bonnardel J, De Prijck S, Vandamme N, Cannoodt R, et al. (2018). The Transcription Factor ZEB2 Is Required to Maintain the Tissue-Specific Identities of Macrophages. Immunity 49, 312–325 e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, Lippens S, Abels C, Schoonooghe S, Raes G, et al. (2016). Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun 7, 10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman JS, Troutman TD, Sakai M, Gola A, Spann NJ, Bennett H, Bruni CM, Ouyang Z, Li RZ, Sun X, et al. (2020). Niche-Specific Reprogramming of Epigenetic Landscapes Drives Myeloid Cell Diversity in Nonalcoholic Steatohepatitis. Immunity [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw TN, Houston SA, Wemyss K, Bridgeman HM, Barbera TA, Zangerle-Murray T, Strangward P, Ridley AJL, Wang P, Tamoutounour S, et al. (2018). Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J Exp Med 215, 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro F, Evrard M, Rizzetto S, Melino M, Mitchell AJ, Florido M, Beattie L, Walters SB, Tay SS, Lu B, et al. (2017). A Liver Capsular Network of Monocyte-Derived Macrophages Restricts Hepatic Dissemination of Intraperitoneal Bacteria by Neutrophil Recruitment. Immunity 47, 374–388 e376. [DOI] [PubMed] [Google Scholar]
- Soysa R, Lampert S, Yuen S, Douglass AN, Li W, Pfeffer K, and Crispe IN (2019). Fetal origin confers radioresistance on liver macrophages via p21(cip1/WAF1). J Hepatol 71, 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran S, Baba I, Poupel L, Dussaud S, Moreau M, Gelineau A, Marcelin G, Magreau-Davy E, Ouhachi M, Lesnik P, et al. (2020). Impaired Kupffer Cell Self-Renewal Alters the Liver Response to Lipid Overload during Non-alcoholic Steatohepatitis. Immunity [DOI] [PubMed] [Google Scholar]
- van de Sluis B, Mao X, Zhai Y, Groot AJ, Vermeulen JF, van der Wall E, van Diest PJ, Hofker MH, Wijmenga C, Klomp LW, et al. (2010). COMMD1 disrupts HIF-1alpha/beta dimerization and inhibits human tumor cell invasion. The Journal of clinical investigation 120, 2119–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C, Mildner A, and Jung S (2015). Macrophages: development and tissue specialization. Annual review of immunology 33, 643–675. [DOI] [PubMed] [Google Scholar]
- Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, Luche H, Fehling HJ, Hardt WD, Shakhar G, and Jung S (2009). Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity 31, 502–512. [DOI] [PubMed] [Google Scholar]
- Vonk WI, Bartuzi P, de Bie P, Kloosterhuis N, Wichers CG, Berger R, Haywood S, Klomp LW, Wijmenga C, and van de Sluis B (2011). Liver-specific Commd1 knockout mice are susceptible to hepatic copper accumulation. PloS one 6, e29183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C, Rodriguez-Fraticelli A, Camargo FD, and Klein AM (2020). Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Valdez PA, Tan C, Yeh S, Hongo JA, and Ouyang W (2010). Phosphatidylserine receptor Tim-4 is essential for the maintenance of the homeostatic state of resident peritoneal macrophages. Proc Natl Acad Sci U S A 107, 8712–8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez A, Coetzee SG, Olsson A, Muench DE, Berman BP, Hazelett DJ, Salomonis N, Grimes HL, and Goodridge HS (2017). Granulocyte-Monocyte Progenitors and Monocyte-Dendritic Cell Progenitors Independently Produce Functionally Distinct Monocytes. Immunity 47, 890–902 e894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ydens E, Amann L, Asselbergh B, Scott CL, Martens L, Sichien D, Mossad O, Blank T, De Prijck S, Low D, et al. (2020). Profiling peripheral nerve macrophages reveals two macrophage subsets with distinct localization, transcriptome and response to injury. Nat Neurosci 23, 676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng C, Luo T, Xu L, Liao G, Yan M, et al. (2019). CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res 47, D721–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E, Samia-Grinberg S, Pasmanik-Chor M, Brazowski E, Shibolet O, Halpern Z, and Varol C (2014). Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. J Immunol 193, 344–353. [DOI] [PubMed] [Google Scholar]
- Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, et al. (2012). Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37, 1076–1090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Expression of KC associated core genes in Commd10fl/fl vs. Clec4fΔCommd10 KCs (Related to Figure 2).
Significantly downregulated genes are marked in red.
Table S2: Functional enrichment pathways extracted from downregulated genes between Commd10fl/fl vs. LysMΔCommd10 Ly6Chi monocytes at 24 h post-APAP (Related to Figure 6A, B, D, E).
Table S3: Upregulation of type I interferon response genes upon differentiation of Ly6Chi monocytes (AILI 24 h) into pro-restorative MoMFs (AILI 72 h) (Related to Figure 6C).
Table S4: Functional enrichment pathways extracted from upregulated genes between Commd10fl/fl vs. LysMΔCommd10 Ly6Chi monocytes at 24 h post-AILI (Related to Figure 6A).
Table S5: Variable expression of NeuMo and DCMo signature genes in LysMΔCommd10 versus Commd10fl/fl Ly6Chi monocytes (Related to Figure 6F, G, S5).
NeuMo and DCMo gene signatures were extracted from (Weinreb et al., 2020; Yanez et al., 2017)
Table S6: Single cell RNAseq analysis of sorted Commd10fl/fl versus LysMΔCommd10 Ly6Chi monocytes at AILI 24 h (Related to Figure 7D, E, G, S6A-C).
Table S7: Biased expression of top hepatic lipid associated macrophages (LAM) markers by LysMΔCommd10 Ly6Chi monocytes (Related to Figure 7G).
Top score (above 2) LAM signature genes were extracted from (Remmerie et al., 2020)
Data Availability Statement
All Single-cell RNA-seq, Bulk RNA-seq and MARS-RNA-seq data have been deposited at GEO and are publicly available. Accession numbers are listed in the key resources table. Original Western blot images were deposited in Mendeley, and associated link appears in the key resources table. Original microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC/Cy7 anti mouse CD45 (30-F11) | BioLegend | Cat #103116; RRID: AB_312981 |
| PE/Cy7 anti mouse CD11b (M1/70) | BioLegend | Cat #101216; RRID: AB_312799 |
| PerCP/Cy5.5 anti mouse Ly6C (HK1.4) | BioLegend | Cat#128012; RRID: AB_1659241 |
| PB anti mouse Ly6G (1A8) | BioLegend | Cat#127612; RRID: AB_2251161 |
| PE anti mouse CD115 (AF598) | BioLegend | Cat#135505; RRID: AB_1937254 |
| APC anti mouse TIM4 (RMT4–54) | BioLegend | Cat#130008; RRID: AB_2271648 |
| APC anti mouse CD163 (TNKUPJ) | ThermoFisher Scientific | Cat#12–1631-80; RRID: AB_2716923 |
| PE anti mouse F4/80 (REA126) | Miltenyi | Cat#130–116-499; RRID: AB_2727574 |
| APC anti mouse VSIG4 (NLA14) | ThermoFisher Scientific | Cat#25–5752-82; RRID: AB_2637431 |
| Unconjugated anti mouse CLEC4F | R&D Systems | Cat#AF2784; RRID: AB_2081339 |
| PE/Cy7 anti mouse CD64 (X54–5/7.1) | BioLegend | Cat#139314; RRID: AB_2563904 |
| PE anti mouse CLEC2 (17D9) | BioLegend | Cat#146104; RRID: AB_2562383 |
| APC anti mouse CD45.1 (A20) | BioLegend | Cat#110713; RRID: AB_313502 |
| CFL 647 donkey anti goat IgG | Santa Cruz Biotechnology | sc-362285 |
| Alexa flour 488 anti mouse CX3CR1 (SA011F11) | BioLegend | Cat#149022; RRID: AB_2565705 |
| Alexa flour 647 anti mouse SiglecF (E50–2440 (RUO)) | BD | BD562680; RRID: AB_2687570 |
| PE anti mouse F4/80 (BM8) | BioLegend | Cat#123110; RRID: AB_893486 |
| PE/Cy7 anti mouse TIM4 (RMT4–54) | BioLegend | Cat#130009; RRID: AB_2565718 |
| Alexa flour 647 anti mouse CD102 (3C4 (MIC2/4)) | BioLegend | Cat#105611; RRID: AB_2122183 |
| Alexa flour 488 donkey anti rat IgG (H+L) | ThermoFisher Scientific | A21208; RRID: AB_2535794 |
| Ki-67 eFluor 660 (SolA15) | Invitrogen | Cat#50–5698-82; RRID: AB_2574235 |
| Anti-Desmin | Abcam | ab15200; RRID: AB_301744 |
| Alex flour 555 goat anti rabbit IgG (H+L) | Abcam | ab150078; RRID: AB_2722519 |
| Anti CD31 (MEC 13.3) | BD | BD550274; RRID: AB_393571 |
| PB anti mouse CD11c (N418) | BioLegend | Cat#117321; RRID: AB_755987 |
| MC-21 anti mouse CCR2 | Jung Lab (Weizmann Institute of Science) (Mack et al., 2001) | Clone: MC-21 |
| Chemicals, Peptides and Recombinant Proteins | ||
| APC BRDU FLIW KIT | BD | BD552598 |
| APAP | Merck | A7085 |
| BODIPY 493/503 | ThermoFisher Scientific | D3922 |
| CFSE | Merck | Cat#65–0850 |
| Collagenase VIII from Clostridium histolyticum | Merck | C5138–500MG |
| Dexamethasone | Merck | D4902–25MG |
| Dnase I (Deoxyribonuclease I from bovine pancreas) | Merck | 10104159001 DN25–100MG |
| DPBS | Biological Industries | 02–023-1A |
| DTT (Dithiothreitol) | Formidum | 3483–12-3 |
| EDTA (Ethylenediaminetetraacetic acid disodium salt dihydrate) | Merck | E5134 |
| Fetal Bovine Serum | Biological industries | 04–001-1A |
| Fixation/Permeablization Kit | BD | BD554714; RRID: AB_2869008 |
| HBSS −/− | gibco | 2183113 |
| Clodronate | Liposoma | C-005 |
| Paraformaldehyde 32% | Electron microscopy sciences | 15714 |
| Protease inhibitor | Merck | P8340 |
| RIPA buffer | Cell Signaling | C-9806S |
| Tissue-Tek O.C.T | Scigen | 23–730-625 |
| TRIzol® reagent | Merck | T9424–100ML |
| Critical Commercial Assays | ||
| cDNA reverse transcription kit | Applied Biosystems | 4368814 |
| miRNeasy Mini Kit | QIAGEN | 217084 |
| SYBR Green PCR Master Mix kit | Applied Biosystems | 4309155 |
| TUNEL staining | Promega | G3250 |
| Deposited Data | ||
| KCs steady state: Bulk MARS-Seq | This paper | GEO: GSE183493 |
| Ly6Chi monocytes AILI 24 h: Bulk RNAseq | This paper | GEO: GSE183494 |
| BM Ly6Chi monocytes: Bulk RNAseq | This paper | GEO: GSE183495 |
| Ly6Chi monocytes AILI 24 h: Single cell RNAseq | This paper | GEO: GSE183367 |
| RNAseq SuperSeries bundle of all four datasets | This paper | GEO: GSE183756 |
| RAW Western blot images (refer to Figures 4G, 5F) | This paper | Mendeley, DOI: 10.17632/kfst2hy224.1 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Commd10fl/fl | Generated by our lab, (Mouhadeb et al., 2018) | N/A |
| Mouse: Cx3cr1cre/+ (Cx3cr1tm1.1(cre)Jung) | Jung Lab (Weizmann Institute of Science) (Yona et al., 2013) | Stock No: 025524 |
| Mouse: Lyz2cre/+ (B6.129P2-Lyz2tm1(cre)Ifo) | The Jackson Laboratories | Stock No: 004781 |
| Mouse: Clec4fcre/+ | Guilliams Lab (Ghent University) (Scott et al., 2018) | N/A |
| Mouse: CD45.1 (B6.SJL-Ptprca Pepcb/Boy) | The Jackson Laboratories | Stock No: 002014 |
| Mouse: CD45.2 C57BL/6JOlaHsd male mice | Envigo (Israel) | Stock No: 000664 |
| Mouse: Ccr2−/−Cx3cr1gfp/+ | Jung Lab (Weizmann Institute of Science) (Zigmond et al., 2012) | N/A |
| Oligonucleotides | ||
| Commd10 FWD: GAGAGCCCCAGCATGAAGAA | Merck | N/A |
| Commd10 REV: AATCCGGCTGAGAAGTCGTG | Merck | N/A |
| Rplp0 FWD: TCCAGCAGGTGTTTGACAAC | Merck | N/A |
| Rplp0 REV: CCATCTGCAGACACACACT | Merck | N/A |
| Arg1 FWD:CAGAAGAATGGAAGAGTCAG | Merck | N/A |
| Arg1 REV:CAGATATGCAGGGAGTCACC | Merck | N/A |
| Trem2 FWD: GGCCCATGCCAGCGTGTGGT | Merck | N/A |
| Trem2 REV: CCAGAGATCTCCAGCATC | Merck | N/A |
| Spp1 FWD: AGCAAGAAACTCTTCCAAGCAA | Merck | N/A |
| Spp1 REV: TGAGATTCGTCAGATTCATCCG | Merck | N/A |
| More Oligonucleotides appear in Figure S7 | ||
| Sofwere and Algorithms | ||
| FlowJo 10.7.1 | Flow jo | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism 5 | GraphPad Software | https://www.graphpad.com/ |
| ImageJ v1.51j | NIH | https://imagej.nih.gov/ij/ |
| StepOne v2.3 | StepOne software | https://www.thermofisher.com/il/en/home/technicalresources/software-downloads/StepOne-and-StepOnePlus-Real-Time-PCR-System.html |
| ZEN 2011 | Zeiss | https://www.zeiss.com/microscopy/int/products/microscopesoftware/zen-lite.html |
| Partek Genomics Suite | Partek | https://www.partek.com/partek-genomics-suite/ |
| Cutadapt | DOI:10.14806/ej.17.1.200 | https://cutadapt.readthedocs.io/en/stable/ |
| Ensembl | DOI:10.1093/nar/gkaa942 | https://www.ensembl.org/index.html |
| DESeq2 | Bioconductor, (Love et al., 2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| snakemake | (Koster and Rahmann, 2012) | https://snakemake.readthedocs.io/en/stable/ |
| STAR | (Dobin et al., 2013) | http://code.google.com/p/rna-star/. |
| MouseMine | (Motenko et al., 2015) | http://www.mousemine.org/mousemine/begin.do |
| CellMarker | (Zhang et al., 2019) | http://biocc.hrbmu.edu.cn/CellMarker/ |
| Ingenuity Pathway Analysis | QIAGEN Bioinformatics | https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis/ |


