Abstract
Despite recent advancements in plant molecular biology and biotechnology, providing enough, and safe, food for an increasing world population remains a challenge. The research into plant development and environmental adaptability has attracted more and more attention from various countries. The transcription of some genes, regulated by transcript factors (TFs), and their response to biological and abiotic stresses, are activated or inhibited during plant development; examples include, rooting, flowering, fruit ripening, drought, flooding, high temperature, pathogen infection, etc. Therefore, the screening and characterization of transcription factors have increasingly become a hot topic in the field of plant research. BLH/BELL (BEL1-like homeodomain) transcription factors belong to a subfamily of the TALE (three-amino-acid-loop-extension) superfamily and its members are involved in the regulation of many vital biological processes, during plant development and environmental response. This review focuses on the advances in our understanding of the function of BLH/BELL TFs in different plants and their involvement in the development of meristems, flower, fruit, plant morphogenesis, plant cell wall structure, the response to the environment, including light and plant resistance to stress, biosynthesis and signaling of ABA (Abscisic acid), IAA (Indoleacetic acid), GA (Gibberellic Acid) and JA (Jasmonic Acid). We discuss the theoretical basis and potential regulatory models for BLH/BELL TFs’ action and provide a comprehensive view of their multiple roles in modulating different aspects of plant development and response to environmental stress and phytohormones. We also present the value of BLHs in the molecular breeding of improved crop varieties and the future research direction of the BLH gene family.
Keywords: BLH/BELL, transcription factors, plant development, environmental stress
1. Introduction
With the growth of the global population, humans are facing the challenges of increasing crop production and quality, while reducing land cultivation and responding to droughts, floods, pests and diseases due to global climate change [1]. Creating new plant varieties through biotechnology, to increase crop yields per unit area and improve their quality, is an effective way to solve the above challenges. It requires detailed knowledge of the underlying molecular mechanisms of plant development, senescence and stress responses and the identification of valuable genes and understanding of their actions, so they can be used to create new varieties through genetic engineering [2].
The specific expression of plant genes throughout the life cycle is transcriptionally regulated by transcription factors (TFs). There have been many reviews on the roles of important plant TF families, including NACs (NAM, ATAF and CUC) [3], MADS (MADS-box) [4], ERF (Ethylene Responsive Factor) [5] and TALE (Three-Amino-acid-Loop-Extension) [6]. The TALE TFs include two subfamilies, KNOX (Knotted-like Homeobox) and BLH (BEL1-like Homeobox), which play an important role in plant development and stress response [7,8,9,10]. With the rapid development of new generation sequencing technology, bioinformatics technology and molecular biology, BLH genes in different species have been cloned and verified. These play important roles in the regulation of plant development, hormone response and stress response [6,7,11,12,13]. Although several reviews of the TALE family have been published, they mainly focus on KNOX and there is a lack of review on the BLH subfamily [6]. A comprehensive review of BLH TFs in different species can contribute to a better understanding of the function of the BLH regulatory network and its role in plant development and stress response. Here, we review the current advances of the BLH family in different species and also explore their potential value in the molecular breeding of crops and their future research direction.
2. The BLH Family
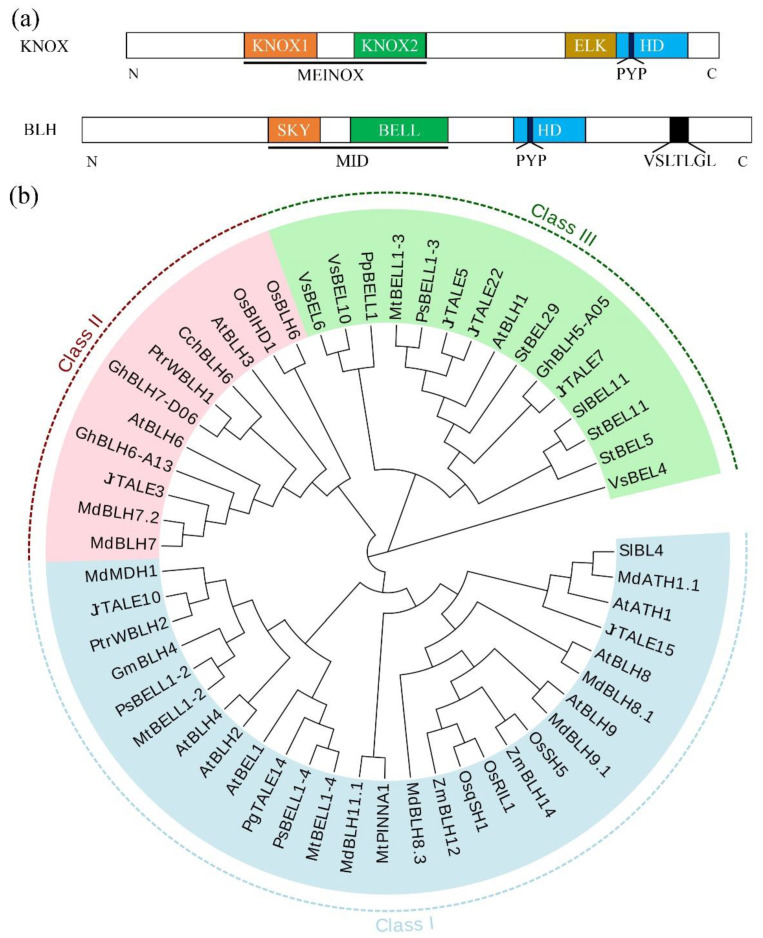
BLH and KNOX are characterized by three-amino-acid-loop-extensions and the conserved “PYP” (proline-tyrosine-proline) amino acid sequence in the homeodomain. However, they differ in their N-terminal and C-terminal regions (Figure 1a). KNOX proteins have a unique MEINOX domain, comprising KNOX 1 and 2 domains, and an ELK domain, while BLH proteins have an N-terminal conserved SKY domain and a BELL domain, upstream of the HD domain [6,14]. In addition, some BLH proteins contain a conserved C-terminal “VSLTLGL” sequence with unknown functions [15,16]. The SKY and BELL domains together make up the MID domain (also known as POX domain) (Figure 1a), which can interact with the MEINOX domain to form a homodimer or a heterodimer to regulate the growth and development of plants and response to stress [6,17,18]. HD domain is the DNA-binding domain [19]. In recent years, the BLH family has been identified in many species, and their functions have been deeply studied. The largest number of BLH family members is in Gossypium hirsutum, whereas only two members are found in Selaginella moellendorffii (Table 1). We also analyzed the kinship of BLH functional genes in different species. Phylogenetic tree analysis showed that these identified BLHs could be divided into three groups (class I, class II and class III) (Figure 1b). It can be found that BLHs involved in plant stress response are mainly concentrated in class I, while BLHs involved in plant development and hormone response are mainly concentrated in class II and class III (Table 1; Figure 1b).
Figure 1.
(a) Schematic diagram of the structure of KNOX and BLH proteins. KNOX proteins contain an ELK domain, an HD domain and an MEINOX domain which is composed of KNOX1 and KNOX2. BLH proteins contain an HD domain, a VSLTLGL domain and an MID domain which is composed of SKY and BELL. (b) Phylogenetic tree of 54 BLH homologous proteins that have been functionally identified. A total of 54 BLH protein sequences were aligned with ClustalX 2.1 program, and the phylogenetic tree was constructed by Neighbor-Joining method. All BLHs are divided into three class and each class is represented by different colors. Class I subfamily is represented by blue, class II subfamily is represented by pink and class III subfamily is represented by green. Gene prefixes represent different species. (Pp: Physcomitrium patens; Vs: Vandenboschia speciose; At: Arabidopsis thaliana; Sl: Solanum lycopersicum; St: Solanum tuberosum; Os: Oryza sativa L.; Zm: Zea mays L.; Gh: Gossypium hirsutum L.; Gm: Glycine max L.; Md: Malus domestica L.; Mt: Medicago truncatula; Ps: Pisum sativum L.; Cch: Camellia chekiangoleosa; Ptr: Populus trichocarpa; Jr: Juglans regia L.; Pg: Punica granatum L.).
Table 1.
Identification and characterization of BLH genes in different plant species.
| Species | Gene Number |
Characterized to Date |
Function | Reference |
|---|---|---|---|---|
| Physcomitrium patens | 4 | PpBELL1 | Sporophyte development | [20,21,22,23] |
| Selaginella moellendorffii | 2 | / | Unknown | [21] |
| Gnetum gnemon | 4 | / | Unknown | [24] |
| Vandenboschia speciosa | 11 | VsBELL4, VsBELL6, VsBELL10 | Gametophytic and the sporophytic developmental |
[25] |
| Arabidopsis thaliana | 13 |
BEL1, ATH1, BLH1, BLH2/SAW1, BLH3, BLH4/SAW2, BLH6, BLH8/PNF, BLH9/ PNY/BLR/RPL/VAN/LSN |
Meristems and inflorescence development; plant morphogenesis; cell wall; ovules; embryo; light signal; abiotic stress and hormone signaling | [6,7,11,13,26,27,28,29,30,31,32,33,34,35,36,37,38,39] |
| Solanum lycopersicum | 14 | SlBL4, SlBEL11 | Fruit chlorophyll; cell wall and hormone signaling |
[40,41,42] |
| Solanum tuberosum | 13 |
StBEL5, StBEL11, StBEL29 |
Phloem-mobile mRNA signals; yield; photoperiod and hormone signaling |
[15,16,43,44,45] |
| Daucus carota L. | 14 | / | Unknown | [20] |
| Oryza sativa L. | 14 | OsSH5/VBP1/RI, OsBIHD1, OsRIL1, qSH1, OsBLH6 | Seed shattering; regulation of inflorescence architecture and meristem maintenance; Secondary cell wall biosynthesis; morphological development; stress response |
[11,21,28,46,47,48,49,50] |
| Zea mays L. | 18 | ZmBEL12, ZmBEL14 | Meristem maintenance; leaf morphology; | [51] |
|
Gossypium hirsutum L. |
50 |
GhBLH5-A05, GhBLH6-A13 GhBLH7-D06 |
Secondary cell wall biosynthesis; morphological development; drought stress; virus response and hormone signaling | [52,53,54] |
| Glycine max L. | 34 | GmBLH4 | Morphological development; stress response; nodule development | [55,56] |
| Malus domestica L. | 19 | MdMDH1/MdBLH4.1, MdBLH7, MdBLH9.1, MdBLH8.1, MdBLH8.3, MdBLH11.1, MdATH1.1, MdBLH7.2 | plant morphogenesis; chlorophyll degradation; salt response; drought stress | [57,58,59] |
| Medicago truncatula | 14 | PINNA1, MtBELL1–2, MtBELL1–3, MtBELL1–4 | leaf morphology; nodulation | [60,61] |
| Pisum sativum L. | 14 | PsBELL1–2, PsBELL1–3, PsBELL1–4 | nodulation; hormone signaling | [60] |
| Brassica rapa L. | 14 | / | Unknown | [62] |
| Camellia chekiangoleosa | 12 | CchBLH6 | Fruit lignification | [63] |
| Populus trichocarpa | 20 | PtrWBLH1, PtrWBLH2 | Salt stress | [64] |
| Juglans regia L. | 17 | JrTALE3, JrTALE5, JrTALE7, JrTALE10, JrTALE15, JrTALE22 | Flower bud development | [65] |
| Punica granatum L. | 9 | PgTALE8, PgTALE14 | Ovule development; inflorescence development; | [66] |
| Cicer arietinum L. | 12 | / | Unknown | [55] |
| Lotus japonicus | 6 | / | Unknown | [55] |
3. The Function of BLHs in Regulating Plant Development
3.1. BLHs Regulate the Development of Plant Meristems
The apical meristem of higher plants produces different new tissues, such as roots, shoots, leaves, flowers and fruits [67]. The maintenance of meristem activity depends on the dynamic balance between cell division and cell differentiation, and BLH TFs play an important regulatory role in the development of meristems [68]. In Arabidopsis thaliana, ATH1 (ARABIDOPSIS THALIANA HOMEOBOX GENE1), BLH3 (BEL1-LIKE HOMEODOMAIN GENE3), PNF (POUND-FOOLISH) and PNY (PENNYWISE), as well as a series of PNY alleles VAN (VAAMANA), BLR (BELLRINGER), LSN (LARSON), BLH9 (BEL1-LIKE HOMEODOMAIN GENE9) and RPL (REPLUMLESS) encode the BLH proteins and were involved in the formation and maintenance of meristems [27,30,69,70,71]. ATH1 is expressed throughout the apical meristem and leaf primordia [72]. BLH3 is expressed in the central region of the apical meristem [13], and PNY is expressed in the central region of the apical meristem and inflorescence meristem [71]. BLR can inhibit the expression of PME5 (pectin methyl-esterase 5) in meristems to limit the demethylation of pectin in inflorescence meristems [73,74]. ATH1, PNY and PNF can interact with KNOX STM (SHOOT MERISTEMLESS) to form heterodimer and regulate the development of meristem [29,75,76].
VPB1 (verticillate primary branch 1) belonging to BLH family is expressed in the shoot tip meristem at the early stage of panicle development in rice. It not only maintains the activity of the inflorescence meristem, but also plays an important regulatory role in the development of primary branch meristem structures [28]. verticillate rachis (ri) and RI-LIKE1 (RIL1) are also rice BLH genes, which are homologous with PNY and PNF, respectively. They are expressed in restricted regions of the inflorescence meristem (IM) and primary branch meristem (PBM). The ri ril1 double mutant cannot normally establish and maintain the shoot apical meristem (SAM) during embryogenesis [11]. In maize, the double mutants of blh12 blh14 showed axillary meristem growth defects and could not produce normal tillers. On the other hand, Tsuda et al. (2017) found that BLH12/14 interacted with KNOTTED1 (KN1) and accumulated in the putative internode meristem of maize, while KN1 did not accumulate in the double mutant of blh12 blh14, suggesting that BLH transcription factors may be involved in the maintenance of meristems [51].
3.2. BLHs’ Regulation of Flower Development
The transition from vegetative growth to reproductive growth is a complex process, involving the regulation of many different genes and some TFs have been reported to regulate the expression of key flowering genes to ensure normal flowering at an appropriate time [77,78]. It was found that BLH TFs can regulate flowering-related genes expression in flowering plants [6].
The Arabidopsis, BLH family genes BLH3, BLH6, BLH8 (PNF), BLH9 (PNY/RPL/BLR) and ATH1 play an important role in flower development and are necessary for inflorescence formation [35,37,79,80,81]. Overexpression of BLH3 and BLH9 lead to early flowering, while overexpression of ATH1 and BLH6 lead to delayed flowering [29,32,33,34]. ATH1 is expressed in flower organs and its encoded protein inhibits flowering by regulating the expression of FLC (FLOWERING LOCUS C) [13,82]. ATH1, BLH3 and BLH6 can interact with OFP1 (OVATE FAMILY PROTEIN1) to regulate the inflorescence structure and flowering of plants [13,34,72]. The pny pnf double mutants can normally receive flowering induction signals, but show abnormal flowering phenotype [37,83]. AG (AGAMOUS) is the key regulator in the development of floral organs and meristems. BLR is necessary to prevent the ectopic expression of AG in floral meristem and can also directly inhibit the expression of AG. Bao et al. showed that BLR may recruit the corepressors of TFs into AG chromatin such as LEUNIG (LUG) and SEUSS (SEU), resulting in inhibiting the expression of AG and coordinating the development of floral organs [71]. In addition, PNY and PNF can interact with STM and participate in the recognition and formation of petals, stamens and pistil organs by regulating the expression of AP3 (APETALA3) and AG [39,71,84] (Figure 2). Previous studies has found that ATH1, PNY and PNF played both overlapping and opposing roles in inflorescence development and the ath1 pny pnf triple mutant showed normal flowering but abnormal flower organ development [29]. Recent studies have found that RPL, which is an allele of PNY, is very important in the normal development of the inflorescence in Brassicaceae. RPL can combine with specific TFs to jointly regulate the transcription of genes related to inflorescence development and promote the normal development of inflorescence structure [85]. The PgTALE8 from pomegranate is a homolog of ATH1, and recent studies have shown that it plays an important regulatory role in inflorescence development [66]. Guo et al. identified the walnut TALE family members and found that BLH genes (JrTALE3, JrTALE5, JrTALE7, JrTALE10, JrTALE15 and JrTALE22) were differentially expressed at different stages of flower bud development, which provided a basis for better understanding the transformation mechanism of the walnut flower bud [65]. TaqSH1 is a transcription factor of wheat BLH family, which is homologous with RPL in Arabidopsis. The overexpression of TaqSH1 in Arabidopsis affected the floral organ abscission in transgenic plants, and down-regulated the expression level of genes related to floral organ abscission [42].
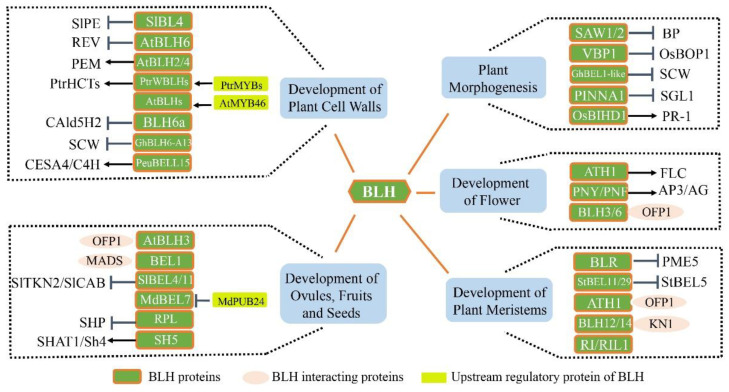
Figure 2.
Role of BLHs in plant development in different species. The composition of BLH in signaling pathways in plant development is divided into five aspects: development of plant meristems, development of flower, plant morphogenesis, development of plant cell walls and development of ovules, fruits and seeds. The main downstream genes are depicted in the diagram. Arrows indicate promotion and vertical lines indicate repression. Gene prefixes represent different species (At: Arabidopsis thaliana; Sl: Solanum lycopersicum; St: Solanum tuberosum; Gh: Gossypium hirsutum; Mt: Medicago truncatula; Ptr: Populus trichocarpa; Os: Oryza sativa; Gm: Glycine max L; Zm: Zea mays; Peu: Populus euphratica).
3.3. BLHs’ Regulation of Plant Morphogenesis
In the stage of plant morphogenesis, new structures are formed from organ primordia, and the polar development pattern is gradually formed. In this process, the gene regulatory network is involved in the differentiation and expansion of cells, regulates the balance between morphogenesis and differentiation, and ultimately affects the size, shape and complexity of plant organs [86]. BLH TFs play an important role in the complex regulatory network of plant morphogenesis.
In Arabidopsis, the BLH family genes SAW1 (BLH2), SAW2 (BLH4), BLH6, PNY and VAN allele of PNY are involved in plant morphogenesis and play important regulatory roles [26,28,36]. The SAW1 and SAW2 genes are highly homologous, and overexpression of SAW1 results in smaller overall plant morphology and loss of leaf polarity. The saw1 saw2 double mutant results in sawtooth and revolute leaf margins, indicating these two genes act redundantly to affect leaf margin growth [36]. At the same time, both SAW1 and SAW2 can negatively regulate the expression of the KNOX family gene BP (BREVIPEDICELLUS) to regulate leaf edge morphogenesis [36,87]. The pny bp double mutant showed abnormal internode development and increased branching [28,88]. The van mutant showed a dwarf phenotype [27]. The blh6 mutant inhibited the effect of BP on the root phenotype. Genetic analysis of hybrid lines between BP overexpression and blh6 mutants proved that BLH6 was involved in the effect of BP on plant configuration [26]. Ectopic expression of apple BEL1-like gene MdMDH1 in Arabidopsis resulted in the dwarfing of transgenic plants and deformity of carpels and pods [58].
Rice BLH genes RI and RIL1 play an important role in the construction of the structure of rice inflorescence, where they regulate the structure of main inflorescence branches, the secondary branches and spikelets in rice. The inflorescence of ri ril1 double mutant showed an abnormal phenotype of increased branching [11]. VPB1 and OsBOP1 (BLADE-ON-PETIOLE1) are important regulators of inflorescence configuration [89]. VPB1 can negatively regulate the expression of OsBOP1 and affect the development of rice panicle configuration [28]. Overexpression of a rice BLH gene, OsBIHD1, in tobacco resulted in some abnormal morphological phenotypes in top buds and roots [46]. Overexpression of barley JuBEL2 in tobacco resulted in ectopic growth of transgenic plants. Some plants grow more branches and show abnormal bifurcation of leaf veins. When JuBEL2 was expressed in Arabidopsis, the transgenic plants formed more shoots compared with the wild type [90]. The blh12 blh14 double mutants of maize showed narrower leaves and failed to produce tillers or female inflorescences [51].
Overexpression of soybean GmBLH4 (BEL1-LIKE homeodomain 4) in Arabidopsis changed leaf phenotype and pod length [56]. The mutation of PINNA1 (PINNATE-LIKE PENTAFOLIATA1) of Medicago truncatula, a BLH family member, produced more compound leaves, which changed from a three leaf palm shape to a five leaf pinnate compound leaf shape [61].
3.4. BLHs’ Regulation of Plant Cell Wall Structure and Composition
Plant cell walls are mainly composed of cellulose, hemicellulose, pectin and lignin, and provide plants with important structural support and protection. The formation of the plant cell wall is a highly complex process in plant growth and development [91]. The synthesis of these components are regulated by a series of TFs in plants [92].
In Arabidopsis, BLH2, BLH4, BLH6 and BLR are involved in the formation and development of cell walls. Studies have proved that BLH2 and BLH4 are significantly expressed in seed mucus-secreting cells and affect the formation of the seed cell wall (mucus). In blh2 blh4 double mutants, the adhesion of mucus to seeds was significantly reduced [93]. In addition, BLH2 and BLH4 play a positive role in regulating the demethylation of HG (homogalacturonan) in seed mucus [31]. BLR participates in the formation of the cell wall by regulating the expression of PME5, a gene related to cell wall formation [94]. The homeodomain-leucine zipper (HD-ZIP) transcription factor REV (REVOLUTA) has been shown to be necessary for inter bundle fiber development [95]. BP and REV play a common role in the process of lignification, and the number of xylem cells in the secondary wall decreased in bp rev double mutant [94]. BLH6 negatively regulates the expression of secondary cell-wall-related genes by transcription inhibition. The secondary cell wall of the blh6 mutant was thicker than that of wild type. BLH6 also interacts with AtKNAT7 to participate in the formation of the secondary wall by inhibiting the expression of REV [32,96]. GhBLH6-A13 in cotton can also regulate the development of the secondary cell wall. Heterologous overexpression of GhBLH6-A13 in Arabidopsis significantly inhibited the synthesis of lignocellulose in the inter bundle fibers [52]. OsBLH6 is involved in the synthesis of the secondary cell wall and its overexpression enhanced the development of the stem secondary wall, while the lignin content was decreased in OsBLH6 knockout lines [49].
Lignin is one of the main components of the cell wall. Snapyl alcohol is an important precursor for lignin synthesis, and CAld5H2 (coniferaldehyde 5-hydroxylase) is the key enzyme catalyzing sinapyl alcohol biosynthesis [97]. BLH6a in poplar is a negative regulator of CAld5H2 and plays an important regulatory role in the synthesis of sinapyl alcohol [98]. PtrMYB021 and PtrMYB074 are transcriptional activators in the fiber of stem-differentiating xylem in Populus trichocarpa. PtrWBLH1 and PtrWBLH2 are the direct targets of PtrMYB021 and PtrMYB074, which can directly regulate 15 single molecule cellulose and cell wall cellulose biosynthesis genes. Similarly, AtMYB46 in Arabidopsis can directly regulate 17 transcription factors and 12 cell wall components, including AtBLH2, AtBLH3, AtBLH6 and AtBLH10 [64]. PeuBELL15 regulates the accumulation of glucan and lignin and promotes the expression of genes related to secondary vascular growth, cellulose synthase and lignin biosynthesis in Populus euphratica, such as CESA4 (Cellulose synthase 4), C4H (cinnamate-4-hydroxylase) and 4CL (4-coumarate: CoA ligase) [99]. CchBLH6 is a positive transcriptional regulator of lignin biosynthesis during the lignification of camellia fruit in Camellia chekiangoleosa [63] (Figure 2). In tomato, SlBL4 can directly repress the expression of SlPE (pectinesterase), resulting in reduced texture and cell wall thinning in tomato fruits [41].
3.5. BLHs’ Regulation of Fruit Development
There exist complex transcript regulatory networks during fruit development. BLH TFs are one of the important regulatory factors.
BEL1 is expressed and plays a regulatory role during ovules development of Arabidopsis. The bel1 mutant showed ovule with a defective embryo sac. On the other hand, BEL1 interact with MADS box protein complex to control the formation of the ovule integument [100,101,102,103]. The BLH gene PgTALE14 in pomegranate is also an important regulator of ovule development [66]. The RPL gene has been proved to be involved in the development of placental frame (replum), and RPL negatively regulates the expression of SHP to prevent the repeated use of the fate of valve marginal cells [30]. In rice, the BLH genes qSH1 and SH5 are involved in rice grain shedding by promoting the development of abscission zone and inhibiting lignin synthesis [47,50]. SHAT1 (shattering abortion 1) and SH4 (shattering 4) are the necessary regulatory factors for the formation of the abscission zone (AZ). Yoon et al. found that SH5 can induce the expression of SHAT1 and SH4 to promote the formation of abscission zone and seed dropping [50]. Overexpressing the barley JuBEL1 in tobacco resulted in the phenomenon of reduced male fertility [90]. Apple MdBEL7 was degraded by ubiquitination, with the U-box E3 ubiquitin ligase MdPUB24, resulting in the degradation of chlorophyll in apple during storage [57]. In tomato, SIBEL11 and SlBL4 have been shown to be involved in regulating the development of chloroplasts and chlorophyll synthesis in tomato fruit [40,41] (Figure 2). Plant developmental processes involving BLH proteins are summarized in Figure 2.
4. Functions of BLHs in the Response to the Environment
4.1. BLHs Involved in Light Responses
Light is an important regulatory signal during plant development. There are a large number of important regulatory factors involved in light signal transduction [104,105]. These regulatory factors cooperate with photoreceptor proteins, such as red and far-red sensing phytochromes, to participate in the growth and development of plants [106].
Studies have shown that ATH1 plays an important role in the regulation of light-induced gene expression and photomorphogenesis in Arabidopsis thaliana [107,108,109,110]. The expression of ATH1 is positively regulated by light. In the dark-grown cop1 det1 double mutants, the expression level of ATH1 increased, indicating that the expression of ATH1 is negatively regulated by COP1 (constitutive photomorphogenesis 1) and DET1 (de-etiolated 1). Genetic analysis shows that ATH1 may be an important downstream component of COP1 and DET1 signal transduction pathway [109] (Figure 3). Phytochrome A-mediated plant response has two stages, namely, the very low flux response (VLFR) and the high irradiation response (HIR). BLH1 can specifically regulate the HIR of phytochrome A. In addition, BLH5, a homolog of BLH1, showed the strongest similarity with BLH1 and blh5 mutation also reduces the HIR [111]. In potato, the movement of StBEL5 mRNA to stolons is induced by short-day photoperiods [43]. The promoter of StBEL5 contains many light response elements, which provides evidence for StBEL5 to play a role in photoperiod regulation. Studies have shown that the transcriptional activity of StBEL5 in leaves is not only induced by white light, but also induced by red and blue light, while the far-red light has no effect to StBEL5 expression [112]. The movement of StBEL11 and StBEL29 mRNAs was induced by short-day conditions [45]. The silencing of SlBEL11 or SlBL4 in tomato changes the expression level of many photosynthetic-related genes. This result shows that SlBEL11 and SlBL4 are also involved in the light signal response [40,41].
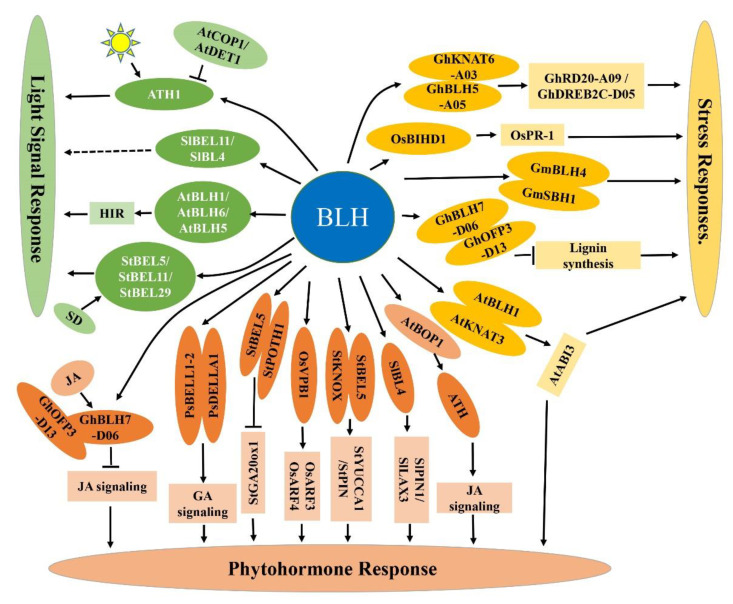
Figure 3.
Role of BLHs in light, stress and hormone signaling pathways. The composition of BLH signal pathway is divided into three parts, which is represented by three different colors. Light signals are described in green, stress signals in yellow and hormone signals in orange. The main downstream genes and downstream signaling pathways are described in rectangular boxes. BLH protein and its interacting proteins were described by oval box. Arrows indicate promotion and vertical lines indicate repression. Gene prefixes represent different species (At: Arabidopsis thaliana; Sl: Solanum lycopersicum; St: Solanum tuberosum; Gh: Gossypium hirsutum; Ps: Medicago truncatula; Os: Oryza sativa; Gm: Glycine max L.).
4.2. BLHs Involved in Plant Resistance to Stress
Plant adaptation to environmental stress mainly depends on the complex molecular regulatory network, including stress signal perception and transmission, stress-response-related gene expression and metabolite synthesis [106]. Studying the molecular mechanism of plant signal transmission under stress is of great significance for breeding stress-resistant crops [113]. BLH genes plays an important regulatory role in the plant abiotic stress response. In Arabidopsis, BLH1 is involved in salt stress response. During seed germination and early seedling development, the sensitivity of the blh1 mutant to salt decreased, while the sensitivity of a BLH1 overexpressing line to salt increased [114]. Study has shown that BLH8 is involved in the abiotic stress response. The blh8 mutant specifically showed a leaf chlorosis phenotype under ion stress (especially Na+ and K+), but the plant roots are unaffected [115]. Overexpression of rice OsBIHD1 in tobacco significantly increased the sensitivity of transgenic plants to salt and oxidative stress, showing the cell membrane damage phenotype under oxidative stress [46]. Apple MdBLH4.1, MdBLH9.1, MdBLH8.1, MdBLH8.3 and MdBLH11.1 and poplar PtTALE5 (homologous gene of Arabidopsis BLH7) showed significant changes in gene expression under different concentrations of NaCl and MdATH1.1, and MdBLH7.2 showed a significant decrease in expression under mannitol treatment [59,116]. Cotton GhBLH5-A05 is a positive regulator of drought stress. Overexpressing GhBLH5-A05 in Arabidopsis and cotton resulted in stronger drought tolerance. Study has also showed that GhBLH5-A05 could interact with GhKNAT6-A03 to promote the expression of drought stress response genes GhRD20-A09 and GhDREB2C-D05, so as to enhance the tolerance of cotton to drought stress [54] (Figure 3). When soybean GmBLH4 was overexpressed in Arabidopsis under HTH (high temperature and high humidity), the stress, seed vigor and germination rate of transgenic lines were significantly higher than those of wild type. The KNOX protein GmSBH1 plays an important regulatory role in soybean response to HTH stress environments. The study also shows that GmBLH4 interacts with GmSBH1, resulting in the formation of a complex to jointly regulate the response of soybean to HTH stress [56].
BLH genes are also involved in the response of plants to biological stress. A large number of studies have shown that the accumulation of lignin and plant hormone signals are directly related to the resistance of plants to Verticillium [117]. The cotton GhBLH7-D06 negatively regulates the resistance of cotton to Verticillium. Silencing GhBLH7-D06 by VIGS (virus-induced gene silencing) induced the expression of lignin synthesis and hormone-signal-related genes, which enhanced the resistance of cotton to Verticillium [53]. The promoter of the StBEL5 gene contains trauma response elements, such as the W, WUN and G-boxes, indicating that the expression of StBEL5 may be induced by trauma. Chatterjee et al. analyzed the GUS activity of StBEL5 transgenic plants and found that the transcription of StBEL5 was activated by trauma and pests [112]. Rice OsBIHD1 plays an important role in the resistance response of Magnaporthe grisea, and the expression of OsBIHD1 in rice seedlings increased significantly after inoculation with Magnaporthe grisea [48]. Overexpression of OsBIHD1 in tobacco resulted in activation of the expression of defense-related genes PR-1, which enhanced the disease resistance of rice to viruses and the oomycete pathogen [46] (Figure 3).
5. BLHs Involved in Phytohormones Biosynthesis and Signaling
Plant hormones are an important part of the complex molecular network regulating plant growth and development, and transcriptional regulation plays an important role in plant hormone-mediated signal transduction pathways [118]. BLH transcription factors have been shown to participate in plant hormone-mediated growth and development, especially abscisic acid (ABA), indole-3-acetic acid (IAA), gibberellin acid (GA) and jasmonic acid (JA).
5.1. BLHs Involved in ABA Response
In Arabidopsis, BLH1 is involved in ABA-mediated seed germination and seedling development. Overexpression of BLH1 promotes the expression of ABA response genes ABI3 (abscisic acid insensitive 3) and ABI5 in transgenic plants. The complex formed by the interaction between BLH1 and KNAT3 can activate the expression of ABI3, to enhance the ABA response in plants and jointly regulate seed germination and seedling growth [114,119].
5.2. BLHs’ Regulation of IAA Response
BP is a potential interaction partner of BLH6, and their expression regions overlap partially in roots. The overexpression of BP affects the redistribution of auxin in plants, resulting in the abnormal growth of plant lateral roots. These results show that BLH6 and BP are involved in the plant hormone-dependent root regulation network [26]. The BEL1 transcription factor plays an important role in the ovule hormone network, especially the auxin and cytokinin signaling pathways. In addition, PIN1 is an auxin efflux-promoting factor [120], and the ectopic expression of PIN1 caused the abnormal ovule development in the bel1 mutant [121]. The auxin response factor ETT (ETTIN/ARF3) is important for the establishment of pistil apical polarity, which ensures the normal synthesis and distribution of auxin in plant organs [122,123]. SPT (SPATULA) and IND (INDEHISCENT) are important bHLH family regulators of pistil development [124,125,126]. Recent research results show that ETT, IND, BP, RPL and SEU jointly regulate the distribution of auxin in different organs [85]. In rice, VPB1 regulates the arrangement of panicles and branches, and the distribution or content of auxin in the vpb1 mutant changed. This resulted in decreased activity of the inflorescence meristem, and finally led to the disorder of initiation and arrangement of branch meristems. Transcriptome analysis showed that VPB1 affected the expression of hormone signaling pathway genes, such as ARF3, ARF4 and ARF12. This result showed that VPB1 plays a role in plants, mainly by regulating meristem development-related genes, to affect the activity of inflorescence primordium and subsequent differentiation [28].
The phloem transport of StBEL5 mRNA is related to auxin synthesis and signal transduction. StBEL5 cooperates with KNOX to induce the expression of auxin biosynthetic gene YUCCA1 and transport gene PIN [43] (Figure 3). Silencing tomato SlBL4 enhanced the sensitivity of plants to IAA, and exogenous IAA hormone treatment inhibited the early abscission phenotype of flower stems of SlBL4 RNAi plants. Transcriptome analysis shows that SlBL4 regulates the expression of many genes related to auxin signal transduction, and experimental analysis shows that SlBL4 can activate the transcription of auxin transport-related genes PIN1 and LAX3 [42] (Figure 3).
5.3. BLHs’ Regulation of GA Response
DELLA protein plays an important role in the gibberellin regulatory network. Research has found that PsBELL1–2 interacts with PsDELLA1 to regulate the nodulation process in Pisum sativum. The study also showed that PsBELL1–2 could interact with PsKNOX9, and both may be regulated by NIN, which is one of the most important regulatory factors of nodule organogenesis and infection [60]. In potato, StBEL5 interacts with POTH1 (potato homeobox 1) to regulate potato development by regulating GA and cytokinin levels. Ga20ox1 encodes a key enzyme in the gibberellin biosynthesis pathway. Overexpression of StBEL5 in potato reduces the GA20ox1 mRNA level at the stolon tip and increases the cytokinin level in the shoot [15]. The StBEL5-POTH1 protein complex can bind to the promoter of GA20ox1 and negatively regulate its expression [124] (Figure 3). In cotton, GhBEL1, GhBLH1 and GhBLH6 also participate in the gibberellin signal regulation network, but the specific mechanism is not clear [52].
5.4. BLHs’ Regulation of JA Response
The disease resistance response of plants involves a complex signal transduction network, regulated by a series of signal molecules. Cotton GhBLH7-D06 negatively regulates cotton resistance to Verticillium and jasmonic acid (JA) and can induce the expression of GhBLH7-D06. Silencing GhBLH7-D06 can significantly increase the expression level of genes related to JA biosynthesis and signal transduction genes, such as GhLOX1-A08, GhLOX2-A05 and GhLOX3-A09, and enhance the resistance of cotton to Verticillium [53]. BLADE-ON-PETIOLE1 (BOP1) is a lateral organ boundary protein and it can directly activate ATH1 under the action of cofactors, to increase the content of JA in plants by promoting the expression of JA biosynthetic genes [69] (Figure 3).
The essential role played by BLH proteins in many aspects of plant growth and development, environmental and plant hormone signaling, is summarized in Figure 3. At present, research on BLHs and their interacting networks is incomplete and needs further exploration.
6. Conclusions and Prospects
6.1. BLHs Are Important Candidate Genes for Molecular Breeding
The BLH gene family has a variety of functions in plant development and stress response. Many BLHs can be used as high-quality candidate genes for molecular breeding. For example, ZmBLH12 and ZmBLH14 play an important regulatory role in maintaining the development of axillary meristem in maize. Proper expression regulation of ZmBLH12 and ZmBLH14 may be helpful to cultivate maize of ideal plant type [51]. In potatoes, StBEL5 is a candidate gene for cultivating potato varieties with a short growth cycle, which is of great significance to alleviate the world’s food problems [15]. In addition, overexpression of SlBL4 improves the firmness of tomatoes, which helps to reduce the loss caused by tomato transportation [41]. Populus pilosa BLH family genes PtrWBLH1/2 and Populus euphratica PeuBELL15 are important regulators in the expression of plant primary and secondary cell wall-related genes [64,99]. Through the regulation and expression of these genes, we can improve and select improved wood varieties, with rapid growth and high wood biomass, so as to promote the high-quality and efficient utilization of wood. In Medicago truncatula, gene editing of PINNA1 may be helpful for cultivating forage varieties with higher leaf yield, which has important economic value [61]. In addition to the regulation of plant growth and development, BLHs can also be used as candidate genes in the cultivation of plant varieties resistant to biotic and abiotic stress. Screening varieties with high expression of GhBLH5-A05 and low expression of GhBLH7-D06 and GhBLH6 in cotton may hasten the production of varieties with drought resistance and Verticillium resistance [53]. In addition, due to the specificity of promoter expression, some BLHs are expressed in specific locations, such as ATH1, PNY and OsBLH6 [13,49,71]. These promoters can be utilized to direct transgenes expression, in the right place, at the right time, to precisely regulate the relevant phenotype, maximize the positive influence and minimize any adverse effects of the gene.
6.2. The Post-Translational Modification of BLHs Remains to Be Further Studied
In addition to transcriptional regulation, post-translational modifications play an important role in the regulatory network of transcription factors. Recent studies have shown that an E3 ubiquitin ligase (MdPUB24) in apple promotes the degradation of MdBEL7 and regulates the greening process of the apple fruit [57]. However, there are few studies on the post-translational modification of BLHs. How a large number of BLHs with different functions in plants are modified has not been reported. Whether BLHs are regulated by E3 ubiquitin ligases other than PUB needs to be further explored. Whether there are other protein post-translational modifications in BLHs also needs further research. With the continuous advancement of scientific research technology, combining different developmental stages or stress conditions of plants through proteomics methods will help us to identify potential modified proteins of BLHs more quickly. Understanding the post-translational modification process of BLHs will help us better study the functions of the BLH gene family and lay the foundation for the construction of the complex regulatory networks of BLHs.
6.3. The BLHs Regulatory Network Requires Further Study
BLHs are an important class of TFs. To date, only a few target genes regulated by BLHs have been analyzed. For example, BLH1 in Arabidopsis directly targets the AtABI3 promoter and increases its expression to regulate seed germination [114]. SlBEL4 and SlBEL11 directly inhibit the expression of genes related to chloroplast development and negatively regulate chloroplast development in tomato fruits [40,41]. OsVPB1 in rice regulates the inflorescence structure of rice by directly inhibiting the expression of OsBOP1 [28]. However, there are still many downstream genes directly regulated by BLHs that have not been identified or reported. These target genes of BLHs need to be studied in much greater detail. In addition, the regulatory principle and mechanism of BLHs in plant tissues require further clarification. Our understanding of the regulatory pathways is not deep enough, and the regulation at different levels and interactions between proteins are unclear. The identification of BLHs’ interacting proteins is essential to explain how these genes affect the development of different organs. In the future, it will be necessary to combine genomics, transcriptomics and molecular genetics to study the function of BLHs in crop growth and development. Bioinformatics and system analysis in different species are of great significance for the discovery of new BLHs and function prediction. With the advancement of high-throughput sequencing technology and genomics research, a large number of target genes of BLHs are expected to be identified and the regulatory network of the BLHs are also expected to be clarified in detail. This will be of great significance for future crop genetics and breeding.
Acknowledgments
The authors would like to thank Donald Grierson (The University of Nottingham) for critical reading and editing of this manuscript.
Author Contributions
X.N. and D.F. designed the review and wrote the whole manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32072277 and 31571898) to D.F.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sonko E., Florkowski W.J., Agodzo S., Antwi-Agyei P. Subsistence farmer knowledge of strategies alleviating food insecurity in the context of climate change in the lower river region of the Gambia. Food Secur. 2020;12:607–624. doi: 10.1007/s12571-020-01024-z. [DOI] [Google Scholar]
- 2.Scheben A., Wolter F., Batley J., Puchta H., Edwards D. Towards CRISPR/Cas crops—Bringing together genomics and genome editing. New Phytol. 2017;216:682–698. doi: 10.1111/nph.14702. [DOI] [PubMed] [Google Scholar]
- 3.Bian Z., Gao H., Wang C. Nac Transcription Factors as Positive or Negative Regulators during Ongoing Battle between Pathogens and Our Food Crops. Int. J. Mol. Sci. 2021;22:81. doi: 10.3390/ijms22010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreni L., Kater K. Mads Reloaded: Evolution of the Agamous Subfamily Genes. New Phytol. 2014;201:717–732. doi: 10.1111/nph.12555. [DOI] [PubMed] [Google Scholar]
- 5.Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. Ap2/Erf Family Transcription Factors in Plant Abiotic Stress Responses. Biochim. Biophys. Acta—Gene Regul. Mech. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Hamant O., Pautot V. Plant development: A TALE story. Comptes Rendus Biol. 2010;333:371–381. doi: 10.1016/j.crvi.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Bellaoui M., Pidkowich M.S., Samach A., Kushalappa K., Kohalmi S.E., Modrusan Z., Haughn C. The Arabidopsis BELL1 and KNOX TALE Homeodomain Proteins Interact through a Domain Conserved between Plants and Animals. Plant Cell. 2001;13:2455–2470. doi: 10.1105/tpc.010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bürglin T.R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hay A., Tsiantis M. KNOX genes: Versatile regulators of plant development and diversity. Development. 2010;137:3153–3165. doi: 10.1242/dev.030049. [DOI] [PubMed] [Google Scholar]
- 10.Modrusan Z., Reiser L., Feldmann K.A., Fischer R.L., Haughn G.W. Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell. 1994;6:333–349. doi: 10.2307/3869754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda T., Tanaka W., Toriba T., Suzuki C., Maeno A., Tsuda K., Shiroishi T., Kurata T., Sakamoto T., Murai M. BELL1-like homeobox genes regulate inflorescence architecture and meristem maintenance in rice. Plant J. 2019;98:465–478. doi: 10.1111/tpj.14230. [DOI] [PubMed] [Google Scholar]
- 12.Kanrar S., Onguka O., Smith H.M. Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta. 2006;224:1163–1173. doi: 10.1007/s00425-006-0298-9. [DOI] [PubMed] [Google Scholar]
- 13.Cole M., Nolte C., Werr W. Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 2006;34:1281–1292. doi: 10.1093/nar/gkl016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar R. Ph.D. Thesis. University of British Columbia; Vancouver, BC, Canada: Dec, 2006. The Roles of BEL1-Like Proteins in Organ Morphogenesis in Arabidopsis thaliana. [Google Scholar]
- 15.Chen H., Rosin F.M., Prat S., Hannapel D.J. Interacting Transcription Factors from the Three-Amino Acid Loop Extension Superclass Regulate Tuber Formation. Plant Physiol. 2003;132:1391–1404. doi: 10.1104/pp.103.022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma P., Lin T., Grandellis C., Yu M., Hannapel D.J. The BEL1-like family of transcription factors in potato. J. Exp. Bot. 2014;65:709–723. doi: 10.1093/jxb/ert432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J.H., Lin H., Joo S., Goodenough U. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell. 2008;133:829–840. doi: 10.1016/j.cell.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 18.Magnani E., Hake S. KNOX lost the OX: The Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell. 2008;20:875–887. doi: 10.1105/tpc.108.058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao C., Banerjee A.K., Hannapel D.J. The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 2004;38:276–284. doi: 10.1111/j.1365-313X.2004.02048.x. [DOI] [PubMed] [Google Scholar]
- 20.Feng Q., Wang G.L., Tong L., Wang Y.H., Xu Z.S., Xiong A.S. Genome-wide identification, expansion, and evolution analysis of homeobox genes and their expression profiles during root development in carrot. Funct. Integr. Genom. 2018;18:685–700. doi: 10.1007/s10142-018-0624-x. [DOI] [PubMed] [Google Scholar]
- 21.Mukherjee K., Brocchieri L., Bürglin T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009;26:2775–2794. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison J. Development and Genetics in the Evolution of Land Plant Body Plans. Philos. Trans. R. Soc. B Biol. Sci. 2017;372:1713. doi: 10.1098/rstb.2015.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horst N., Katz A., Pereman I., Decker E.L., Ohad N., Reski R. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat. Plants. 2016;2:15209. doi: 10.1038/nplants.2015.209. [DOI] [PubMed] [Google Scholar]
- 24.Becker A., Bey M., Bürglin T., Saedler H., Theissen G. Ancestry and diversity of BEL1-like homeobox genes revealed by gymnosperm (Gnetum gnemon) homologs. Dev. Genes Evol. 2002;212:452–457. doi: 10.1007/s00427-002-0259-7. [DOI] [PubMed] [Google Scholar]
- 25.Mercedes R.E., Mohammed B., Rubén M., Manuel G.R. Identification and Characterization of TALE Homeobox Genes in the Endangered Fern Vandenboschia speciosa. Genes. 2017;8:275. doi: 10.3390/genes8100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Souček P., Hanáček P., Mazura P., Reinöhl V. Interaction among BREVIPEDICELLUS, BLH6 and auxin in roots of Arabidopsis thaliana. Russ. J. Plant Physiol. 2017;64:386–397. doi: 10.1134/S1021443717030189. [DOI] [Google Scholar]
- 27.Bhatt A.M., Etchells J.P., Canales C., Lagodienko A., Dickinson H. VAAMANA—A BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene. 2004;328:103–111. doi: 10.1016/j.gene.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 28.Li M., Fu D., Xu T., Wu C. VPB1 Encoding BELL-like Homeodomain Protein Is Involved in Rice Panicle Architecture. Int. J. Mol. Sci. 2021;22:7909. doi: 10.3390/ijms22157909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutjens B., Bao D., Van Eck-Stouten E., Brand M., Smeekens S., Proveniers M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 2009;58:641–654. doi: 10.1111/j.1365-313X.2009.03809.x. [DOI] [PubMed] [Google Scholar]
- 30.Roeder A., Ferrándiz C., Yanofsky M.F. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 2003;13:1630–1635. doi: 10.1016/j.cub.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y., Wang Y., Wang X., Pei S., Kong Y., Hu R., Zhou G. Transcription factors BLH2 and BLH4 regulate demethylesterification of homogalacturonan in seed mucilage. Plant Physiol. 2020;183:96–111. doi: 10.1104/pp.20.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., You S., Taylor-Teeples M., Li W.L., Schuetz M., Brady S.M., Douglas C.J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 Interact and Regulate Secondary Cell Wall Formation via Repression of REVOLUTA. Plant Cell. 2014;26:4843. doi: 10.1105/tpc.114.128322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith H.M., Hake S. The Interaction of Two Homeobox Genes, BREVIPEDICELLUS and PENNYWISE, Regulates Internode Patterning in the Arabidopsis Inflorescence. Plant Cell. 2003;15:1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L., Zhang X., Ju H., Chen J., Ying C. OVATE FAMILY PROTEIN1 interaction with BLH3 regulates transition timing from vegetative to reproductive phase in Arabidopsis. Biochem. Biophys. Res. Commun. 2016;470:492–497. doi: 10.1016/j.bbrc.2016.01.135. [DOI] [PubMed] [Google Scholar]
- 35.Kanrar S., Bhattacharya M., Arthur B., Courtier J., Smith H.M. Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J. 2008;54:924–937. doi: 10.1111/j.1365-313X.2008.03458.x. [DOI] [PubMed] [Google Scholar]
- 36.Kumar R., Kushalappa K., Godt D., Pidkowich M.S., Pastorelli S., Hepworth S.R., Haughn G.W. The Arabidopsis BEL1-LIKE HOMEODOMAIN Proteins SAW1 and SAW2 Act Redundantly to Regulate KNOX Expression Spatially in Leaf Margins. Plant Cell. 2007;19:2719–2735. doi: 10.1105/tpc.106.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith H., Campbell B.C., Hake S. Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 2004;14:812–817. doi: 10.1016/j.cub.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 38.Pagnussat G.C., Yu H.J., Sundaresan V. Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell. 2007;19:3578–3592. doi: 10.1105/tpc.107.054890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu L., Patibanda V., Smith H.M. A novel role of BELL1-like homeobox genes, PENNYWISE and POUND-FOOLISH, in floral patterning. Planta. 2009;229:693–707. doi: 10.1007/s00425-008-0867-1. [DOI] [PubMed] [Google Scholar]
- 40.Meng L., Fan Z., Zhang Q., Wang C., Gao Y., Deng Y., Zhu B., Zhu H., Jianye C., Shan W. BEL1-LIKE HOMEODOMAIN 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. Plant J. 2018;94:1126–1140. doi: 10.1111/tpj.13924. [DOI] [PubMed] [Google Scholar]
- 41.Yan F., Gao Y., Pang X., Xu X., Zhu N., Chan H., Hu G., Wu M., Yuan Y., Li H. BEL1-LIKE HOMEODOMAIN4 regulates chlorophyll accumulation, chloroplast development, and cell wall metabolism in tomato fruit. J. Exp. Bot. 2020;71:5549–5561. doi: 10.1093/jxb/eraa272. [DOI] [PubMed] [Google Scholar]
- 42.Yan F., Gong Z., Hu G., Ma X., Bai R., Yu R., Zhang Q., Deng W., Li Z., Wuriyanghan H. Tomato SlBL4 plays an important role in fruit pedicel organogenesis and abscission. Hortic. Res. 2021;8:1–15. doi: 10.1038/s41438-021-00515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hannapel D.J., Sharma P., Lin T. Phloem-mobile messenger RNAs and root development. Front. Plant Sci. 2013;4:257. doi: 10.3389/fpls.2013.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banerjee A.K., Lin T., Hannapel D.J. Untranslated Regions of a Mobile Transcript Mediate RNA Metabolism. Plant Physiol. 2009;151:1831–1843. doi: 10.1104/pp.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghate T.H., Sharma P., Kondhare K.R., Hannapel D.J., Banerjee A.K. The mobile RNAs, StBEL11 and StBEL29, suppress growth of tubers in potato. Plant Mol. Biol. 2017;93:563–578. doi: 10.1007/s11103-016-0582-4. [DOI] [PubMed] [Google Scholar]
- 46.Luo H., Song F., Zheng Z. Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. J. Exp. Bot. 2005;56:2673–2682. doi: 10.1093/jxb/eri260. [DOI] [PubMed] [Google Scholar]
- 47.Konishi S., Izawa T., Lin S., Ebana K., Fukuta Y., Sasaki T., Yano M. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 48.Luo H., Song F., Goodman R.M., Zheng Z. Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biol. 2005;7:459–468. doi: 10.1055/s-2005-865851. [DOI] [PubMed] [Google Scholar]
- 49.Hirano K., Kondo M., Aya K., Miyao A., Sato Y., Antonio B.A., Namiki N., Nagamura Y., Matsuoka M. Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol. 2013;54:1791–1802. doi: 10.1093/pcp/pct122. [DOI] [PubMed] [Google Scholar]
- 50.Yoon J., Cho L.H., Kim S.L., Choi H., Koh H.J., An G. The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 2014;79:717–728. doi: 10.1111/tpj.12581. [DOI] [PubMed] [Google Scholar]
- 51.Tsuda K., Abraham-Juarez M.J., Maeno A., Dong Z., Aromdee D., Meeley R., Shiroishi T., Nonomura K., Hake S. KNOTTED1 Cofactors, BLH12 and BLH14, Regulate Internode Patterning and Vein Anastomosis in Maize. Plant Cell. 2017;29:1105–1118. doi: 10.1105/tpc.16.00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma Q., Wang N., Hao P., Sun H., Yu S. Genome-wide identification and characterization of TALE superfamily genes in cotton reveals their functions in regulating secondary cell wall biosynthesis. BMC Plant Biol. 2019;19:432. doi: 10.1186/s12870-019-2026-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Ma Q., Wang N., Ma L., Lu J., Wei H. The Cotton BEL1-Like Transcription Factor GhBLH7-D06 Negatively Regulates the Defense Response against Verticillium dahliae. Int. J. Mol. Sci. 2020;21:7126. doi: 10.3390/ijms21197126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J. Ph.D. Thesis. Central China Normal University; Wuhan, China: 2021. Study on the Roles of Drought Stress-Responsive Genes GhBLH5-A05 and GhHDT4D in Cotton Response to Drought Stress. [Google Scholar]
- 55.Bhattacharjee A., Ghangal R., Garg R., Jain M. Genome-Wide Analysis of Homeobox Gene Family in Legumes: Identification, Gene Duplication and Expression Profiling. PLoS ONE. 2015;10:e0119198. doi: 10.1371/journal.pone.0119198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tao Y., Chen M., Shu Y.J., Zhu Y.J., Wang S. Identification and functional characterization of a novel BEL1-LIKE homeobox transcription factor GmBLH4 in soybean. Plant Cell. 2018;134:331–344. doi: 10.1007/s11240-018-1419-4. [DOI] [Google Scholar]
- 57.Wei Y., Jin J., Xu Y., Liu W., Wang A. Ethylene ctivated MdPUB24 mediates ubiquitination of MdBEL7 to promote chlorophyll degradation in apple fruit. Plant J. 2021;108:169–182. doi: 10.1111/tpj.15432. [DOI] [PubMed] [Google Scholar]
- 58.Dong Y.H., Yao J.L., Atkinson R.G., Putterill J.J., Morris B.A., Gardner R.C. MDH1: An apple homeobox gene belonging to the BEL1 family. Plant Mol. Biol. 2000;42:623–633. doi: 10.1023/A:1006301224125. [DOI] [PubMed] [Google Scholar]
- 59.Li H., Zhao Q., Wang H., Dong Q., Xu Y. Genome-Wide Identification, Expression Profiling and Protein-Protein Interaction Properties of the BEL-Like Homeodomain Gene Family in Apple. Phyton-Int. J. Exp. Bot. 2022;91:315–331. doi: 10.32604/phyton.2022.016951. [DOI] [Google Scholar]
- 60.Dolgikh A.V., Rudaya E.S., Dolgikh E.A. Identification of BELL Transcription Factors Involved in Nodule Initiation and Development in the Legumes Pisum sativum and Medicago truncatula. Plants. 2020;9:1808. doi: 10.3390/plants9121808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He L., Liu Y., He H., Liu Y., Chen J. A molecular framework underlying the compound leaf pattern of Medicago truncatula. Nat. Plants. 2020;6:511–521. doi: 10.1038/s41477-020-0642-2. [DOI] [PubMed] [Google Scholar]
- 62.Khan N., Hu C.M., Khan W.A., Wang W., Ke H., Huijie D., Zhishuo Z., Hou X. Genome-wide Identification, Classification, and Expression Pattern of Homeobox Gene Family in Brassica rapa under Various Stresses. Sci. Rep. 2018;8:16265. doi: 10.1038/s41598-018-34448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan C., Hu Z., Nie Z., Li J., Yin H. CcBLH6, a bell-like homeodomain-containing transcription factor, regulates the fruit lignification pattern. Planta. 2021;253:90. doi: 10.1007/s00425-021-03610-7. [DOI] [PubMed] [Google Scholar]
- 64.Chen H., Wang J.P., Liu H., Li H., Lin Y.C.J., Shi R., Yang C., Gao J., Zhou C., Li Q. Hierarchical Transcription Factor and Chromatin Binding Network for Wood Formation in Populus trichocarpa. Plant Cell. 2019;31:602–626. doi: 10.1105/tpc.18.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo C., Niu J., Quan S., Zhang Z., Kang C., Liu J. Genome-Wide Identification, Characterization and Expression Profile of Tale Gene Family in Juglans Regia L. Sci. Hortic. 2020;297:110945. doi: 10.1016/j.scienta.2022.110945. [DOI] [Google Scholar]
- 66.Wang Y., Zhao Y., Yan M., Zhao H., Yuan Z. Genome-Wide Identification and Expression Analysis of TALE Gene Family in Pomegranate (Punica granatum L.) Agronomy. 2020;10:829. doi: 10.3390/agronomy10060829. [DOI] [Google Scholar]
- 67.Aida M., Masao T. Genetic Control of Shoot Organ Boundaries. Curr. Opin. Plant Biol. 2006;9:72–77. doi: 10.1016/j.pbi.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 68.Grandi V., Veronica G., Martin M.K. Uncovering Genetic and Molecular Interactions among Floral Meristem Identity Genes in Arabidopsis thaliana. Plant J. 2012;69:881–893. doi: 10.1111/j.1365-313X.2011.04840.x. [DOI] [PubMed] [Google Scholar]
- 69.Khan M., Ragni L., Tabb P., Salasini B.C., Chatfield S., Datla R., Lock J., Kuai X., Despres C., Proveniers M. Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for meristem maintenance and flowering in Arabidopsis thaliana1. Plant Physiol. 2015;169:2166–2186. doi: 10.1104/pp.15.00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ung N., Lal S., Smith H. The Role of PENNYWISE and POUND-FOOLISH in the Maintenance of the Shoot Apical Meristem in Arabidopsis. Plant Physiol. 2011;156:605–614. doi: 10.1104/pp.110.171462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bao X., Franks R.G., Levin J.Z., Liu Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell. 2004;16:1478–1489. doi: 10.1105/tpc.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L., Sun L., Zhang X., Zhang S., Xie D., Liang C., Huang W., Fan L., Fang Y., Chang Y. OFP1 interaction with ATH1 regulates stem growth, flowering time and flower basal boundary formation in Arabidopsis. Genes. 2018;9:399. doi: 10.3390/genes9080399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Byrne M.E., Groover A.T., Fontana J.R., Martienssen R.A. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development. 2003;130:3941–3950. doi: 10.1242/dev.00620. [DOI] [PubMed] [Google Scholar]
- 74.Peaucelle A., Louvet R., Johansen J.N., Salsac F., Morin H., Fournet F., Belcram K., Gillet F., Hofte H., Laufs P. The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development. 2011;138:4733–4741. doi: 10.1242/dev.072496. [DOI] [PubMed] [Google Scholar]
- 75.Byrne M.E., Simorowski J., Martienssen R.A. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development. 2002;129:1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- 76.Belles-Boix E., Hamant O., Witiak S.M., Morin H., Traas J., Pautot V. KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell. 2006;18:1900–1907. doi: 10.1105/tpc.106.041988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spanudakis E., Stephen J. The Role of Micrornas in the Control of Flowering Time. J. Exp. Bot. 2014;2:365–380. doi: 10.1093/jxb/ert453. [DOI] [PubMed] [Google Scholar]
- 78.Sagheer A., Lu C.Q., Wu J.Q., Wei Y.L., Gao J., Jin J.P., Zheng C.Y., Zhu G.F., Yang F.X. Transcriptional Cascade in the Regulation of Flowering in the Bamboo Orchid Arundina graminifolia. Biomolecules. 2021;6:771. doi: 10.3390/biom11060771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arnaud N., Pautot V. Ring the BELL and tie the KNOX: Roles for TALEs in gynoecium development. Front. Plant Sci. 2014;5:93. doi: 10.3389/fpls.2014.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khan M., Tabb P., Hepworth S.R. BLADE-ON-PETIOLE1 and 2 regulate Arabidopsis inflorescence architecture in conjunction with homeobox genes KNAT6 and ATH1. Plant Signal. Behav. 2012;7:788–792. doi: 10.4161/psb.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang L.M. ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture. J. Exp. Bot. 2012;63:1423–1433. doi: 10.1093/jxb/err376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Proveniers M., Rutjens B., Brand M., Smeekens S. The Arabidopsis TALE homeobox gene ATH1 controls floral competency through positive regulation of FLC. Plant J. 2007;52:899–913. doi: 10.1111/j.1365-313X.2007.03285.x. [DOI] [PubMed] [Google Scholar]
- 83.Andrés F., Romera-Branchat M., Martínez-Gallegos R., Patel V., Schneeberger K., Jang S., Altmüller J., Nürnberg P., Coupland G. Floral Induction in Arabidopsis by FLOWERING LOCUS T Requires Direct Repression of BLADE-ON-PETIOLE Genes by the Homeodomain Protein PENNYWISE. Plant Physiol. 2015;169:2187–2199. doi: 10.1104/pp.15.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ragni L., Belles-Boix E., Gunl M., Pautot V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis Inflorescences. Plant Cell. 2008;20:888–900. doi: 10.1105/tpc.108.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simonini S., Stephenson P., Østergaard L. A molecular framework controlling style morphology in Brassicaceae. Development. 2018;145:158105. doi: 10.1242/dev.158105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poethig R.S., Sussex I.M. The Cellular Parameters of Leaf Development in Tobacco: A Clonal Analysis. Planta. 1985;2:170–184. doi: 10.1007/BF00395039. [DOI] [PubMed] [Google Scholar]
- 87.Blein T., Hasson A., Laufs P. Leaf development: What it needs to be complex. Curr. Opin. Plant Biol. 2010;13:75–82. doi: 10.1016/j.pbi.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 88.Zhao M., Yang S., Chen C.Y., Li C., Shan W., Lu W., Cui Y., Liu X., Wu K. Arabidopsis BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate KNAT2 and KNAT6 Expression in Control of Inflorescence Architecture. PLoS Genet. 2015;11:e1005125. doi: 10.1371/journal.pgen.1005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Madiha K., Xu H.S., Hepworth S.R. Blade-on-Petiole Genes: Setting Boundaries in Development and Defense. Plant Sci. 2014;215:157–171. doi: 10.1016/j.plantsci.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 90.Müller J., Wang Y., Franzen R., Santi L., Salamini F., Rohde W. In vitro interactions between barley TALE homeodomain proteins suggest a role for protein–protein associations in the regulation of Knox gene function. Plant J. 2001;27:13–23. doi: 10.1046/j.1365-313x.2001.01064.x. [DOI] [PubMed] [Google Scholar]
- 91.Burton R.A., Michael J.G., Geoffrey B.F. Heterogeneity in the Chemistry, Structure and Function of Plant Cell Walls. Nat. Chem. Biol. 2010;10:724–732. doi: 10.1038/nchembio.439. [DOI] [PubMed] [Google Scholar]
- 92.Wang H.Z., Utku A., Jin N., Michael G.H., Fang C., Richard A.D. Mutation of WRKY Transcription Factors Initiates Pith Secondary Wall Formation and Increases Stem Biomass in Dicotyledonous Plants. Proc. Natl. Acad. Sci. USA. 2010;51:22338–22343. doi: 10.1073/pnas.1016436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Francoz E., Ranocha P., Burlat V., Dunand C. Arabidopsis seed mucilage secretory cells: Regulation and dynamics. Trends Plant Sci. 2015;20:515–524. doi: 10.1016/j.tplants.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 94.Etchells J.P., Moore L., Wen Z.J., Prescott H., Dickinson H.G. A role for BELLRINGER in cell wall development is supported by loss-of-function phenotypes. BMC Plant Biol. 2012;12:212. doi: 10.1186/1471-2229-12-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. Radial Patterning of Arabidopsis Shoots by Class Iii Hd-Zip and Kanadi Genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 96.Liu Y., Douglas C.J. A role for OVATE FAMILY PROTEIN1 (OFP1) and OFP4 in a BLH6-KNAT7 multi-protein complex regulating secondary cell wall formation in Arabidopsis thaliana. Plant Signal. Behav. 2015;10:e1033126. doi: 10.1080/15592324.2015.1033126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang P.Y. Ph.D. Thesis. North Carolina State University; Raleigh, NC, USA: 2012. Towards a Systems Approach for Lignin Biosynthesis in Populus Trichocarpa: Functional Redundancy of the Two 5-Hydroxylases and Kinetic Modeling of the Monolignol Biosynthetic Pathway. [Google Scholar]
- 98.Wang Q., Dai X., Pang H., Cheng Y., Li Q. BEL1-like Homeodomain Protein BLH6a Is a Negative Regulator of CAld5H2 in Sinapyl Alcohol Monolignol Biosynthesis in Poplar. Front. Plant Sci. 2021;12:761291. doi: 10.3389/fpls.2021.761291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han X., An Y., Zhou Y., Liu C., Xia X. Comparative transcriptome analyses define genes and gene modules differing between two Populus genotypes with contrasting stem growth rates. Biotechnol. Biofuels. 2020;13:139. doi: 10.1186/s13068-020-01758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brambilla V., Battaglia R., Colombo M., Masiero S., Bencivenga S., Kater M.M., Colombo L. Genetic and Molecular Interactions between BELL1 and MADS Box Factors Support Ovule Development in Arabidopsis. Plant Cell. 2007;19:2544–2556. doi: 10.1105/tpc.107.051797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reiser L., Modrusan Z., Margossian L., Samach A., Ohad N., Haughn G.W., Fischer R.L. The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell. 1995;83:735–742. doi: 10.1016/0092-8674(95)90186-8. [DOI] [PubMed] [Google Scholar]
- 102.Robinson-Beers K., Gasser P. Ovule Development in Wild-Type Arabidopsis and Two Female-Sterile Mutants. Plant Cell. 1992;4:1237–1249. doi: 10.2307/3869410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ray A., Robinson-Beers K., Ray S., Baker S.C., Lang J.D., Preuss D., Gasser M. Arabidopsis floral homeotic gene BELL (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG) Proc. Natl. Acad. Sci. USA. 1994;91:5761–5765. doi: 10.1073/pnas.91.13.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen M., Joanne C., Christian F. Light Signal Transduction in Higher Plants. Annu. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 105.Ma L. Light Control of Arabidopsis Development Entails Coordinated Regulation of Genome Expression and Cellular Pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang G.T., Ma S.L., Bai L.P., Zhang L., Ma H., Jia P., Liu J., Zhong M., Guo Z.F. Signal Transduction During Cold, Salt, and Drought Stresses in Plants. Mol. Biol. Rep. 2012;39:969–987. doi: 10.1007/s11033-011-0823-1. [DOI] [PubMed] [Google Scholar]
- 107.Ejaz M., Bencivenga S., Tavares R., Bush M., Sablowski R. ARABIDOPSIS THALIANA HOMEOBOX GENE 1 controls plant architecture by locally restricting environmental responses. Proc. Natl. Acad. Sci. USA. 2021;118:e2018615118. doi: 10.1073/pnas.2018615118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gomez-Mena C., Sablowski R. ARABIDOPSIS THALIANA HOMEOBOX GENE1 Establishes the Basal Boundaries of Shoot Organs and Controls Stem Growth. Plant Cell. 2008;20:2059–2072. doi: 10.1105/tpc.108.059188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Quaedvlieg N., Dockx J., Rook F., Weisbeek P., Smeekens S. The Homeobox Gene ATH1 of Arabidopsis Is Derepressed in the Photomorphogenic Mutants cop1 and det1. Plant Cell. 1995;7:117–129. doi: 10.1105/tpc.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu L.I., Yang Y.J., Yan L.I., Wang J., Xiao X.J., Guo X.H., Tang D.Y., Liu X.M. Protein identification and mRNA analysis of phytochrome-regulated genes in Arabidopsis under red light. Sci. China Ser. C Life Sci. 2009;52:371–380. doi: 10.1007/s11427-009-0045-0. [DOI] [PubMed] [Google Scholar]
- 111.Staneloni R.J., Rodriguez-Batiller M.J., Legisa D., Scarpin M.R., Agalou A., Cerdan P.D., Meijer A.H., Ouwerkerk P.B.F., Casal J.J. Bell-like homeodomain selectively regulates the high-irradiance response of phytochrome A. Proc. Natl. Acad. Sci. USA. 2009;106:13624–13629. doi: 10.1073/pnas.0906598106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chatterjee M., Banerjee A.K., Hannapel D.J. A BELL1-like gene of potato is light activated and wound inducible. Plant Physiol. 2007;145:1435–1443. doi: 10.1104/pp.107.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu J.-K. Salt and Drought Stress Signal Transduction in Plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim D., Cho Y.H., Ryu H., Kim Y., Kim T.H., Hwang I. BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 2013;75:755–766. doi: 10.1111/tpj.12236. [DOI] [PubMed] [Google Scholar]
- 115.Park H.C., Park J.Y., Baek D.W., Yun D.J. Functional characterization of Arabidopsis thaliana BLH 8, BEL1-Like Homeodomain 8 involved in environmental stresses. J. Plant Biotechnol. 2011;38:162–168. doi: 10.5010/JPB.2011.38.2.162. [DOI] [Google Scholar]
- 116.Zhao K., Zhang X., Cheng Z., Yao W., Li R., Jiang T., Zhou B. Comprehensive analysis of the three-amino-acid-loop-extension gene family and its tissue-differential expression in response to salt stress in poplar. Plant Physiol. Bioch. 2019;136:1–12. doi: 10.1016/j.plaphy.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 117.Reusche M., Thole K., Janz D., Truskina J., Rindfleisch S., Drubert C., Polle A., Lipka V., Teichmann T. Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. Plant Cell. 2012;24:3823–3837. doi: 10.1105/tpc.112.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nemhauser J.L., Hong F.X., Chory J. Different Plant Hormones Regulate Similar Processes through Largely Nonoverlapping Transcriptional Responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 119.Hoth S., Morgante M., Sanchez J.P., Hanafey M.K., Tingey S.V., Chua N.H. Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 2002;115:4891–4900. doi: 10.1242/jcs.00175. [DOI] [PubMed] [Google Scholar]
- 120.Keek P., Skpa P., Libus J., Naramoto S., Tejos R., Friml J., Zaímalová E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009;10:1–11. doi: 10.1186/gb-2009-10-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bencivenga S., Simonini S., Benkova E., Colombo L. The Transcription Factors BEL1 and SPL Are Required for Cytokinin and Auxin Signaling During Ovule Development in Arabidopsis. Plant Cell. 2012;24:2886–2897. doi: 10.1105/tpc.112.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sessions A.R., Nemhauser J.L., Mccoll A., Roe J.L., Zambryski P.C. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- 123.Simonini S., Bencivenga S., Trick M., Ostergaard L. Auxin-Induced Modulation of ETTIN Activity Orchestrates Gene Expression in Arabidopsis. Plant Cell. 2017;29:1864–1882. doi: 10.1105/tpc.17.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Girin T., Paicu T., Stephenson P., Fuentes S., Stergaard L. INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell. 2011;23:3641–3653. doi: 10.1105/tpc.111.090944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Heisler M., Atkinson A., Bylstra Y.H., Walsh R., Smyth D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development. 2001;128:1089–1098. doi: 10.1242/dev.128.7.1089. [DOI] [PubMed] [Google Scholar]
- 126.Liljegren S.J., Roeder A., Kempin S.A., Gremski K., Østergaard L., Guimil S., Reyes D.K., Yanofsky M.F. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;116:843–853. doi: 10.1016/S0092-8674(04)00217-X. [DOI] [PubMed] [Google Scholar]