Abstract
The fungicidal mechanism of a naturally occurring sesquiterpene dialdehyde, polygodial, was investigated in Saccharomyces cerevisiae. In an acidification assay, polygodial completely suppressed the glucose-induced decrease in external pH at 3.13 μg/ml, the same as the fungicidal concentration. Acidification occurs primarily through the proton-pumping action of the plasma membrane ATPase, Pma1p. Surprisingly, this ATPase was not directly inhibited by polygodial. In contrast, the two other membrane-bound ATPases in yeast were found to be susceptible to the compound. The mitochondrial ATPase was inhibited by polygodial in a dose-dependent manner at concentrations similar to the fungicidal concentration, whereas the vacuolar ATPase was only slightly inhibited. Cytoplasmic petite mutants, which lack mitochondrial DNA and are respiration deficient, were significantly less susceptible to polygodial than the wild type, as was shown in time-kill curves. A pet9 mutant which lacks a functional ADP-ATP translocator and is therefore respiration dependent was rapidly inhibited by polygodial. The results of these susceptibility assays link enzyme inhibition to physiological effect. Previous studies have reported that plasma membrane disruption is the mechanism of polygodial-induced cell death; however, these results support a more complex picture of its effect. A major target of polygodial in yeast is mitochondrial ATP synthase. Reduction of the ATP supply leads to a suppression of Pma1 ATPase activity and impairs adaptive responses to other facets of polygodial's cellular inhibition.
Systemic fungal infections are increasingly important causes of high morbidity and mortality (9, 18). Fungi are significantly affecting the growing population of patients with impaired immune systems due to AIDS, cancer chemotherapy, or immunosuppressive drugs. In addition to life-threatening systemic infection, superficial mycoses can clinically present as persistent infections that require continual treatment. Oropharyngeal and esophageal candidiasis occurs in over 70% of people with AIDS (46). With the increasing incidence of both systemic and superficial mycoses, it is critical that we develop new antifungal agents. Additionally, antifungal drug resistance has become an important problem and even further intensifies the need for new compounds (42, 54). Therefore, identifying promising cellular targets and understanding their physiological roles and basic biochemistry are critical for successful antifungal development (21, 36).
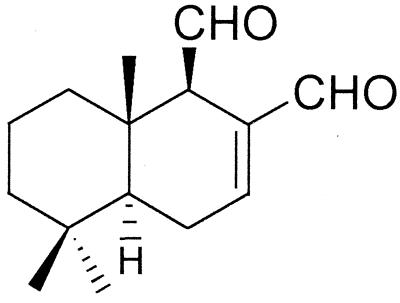
Polygodial (Fig. 1), a sesquiterpene dialdehyde, was originally isolated from the plant Polygonum hydropiper (3) and subsequently from Warburgia ugandensis and Warburgia stuhlmannii exhibiting insect antifeedant activity (28) and antimicrobial activity (48), as well as from Pseudowintera colorata exhibiting antimicrobial activity (34). Polygodial is a component of the “hot taste” in peppery spices of traditional Japanese cuisine (26). Unlike many other antifungal agents, polygodial has fungicidal activity against yeasts and filamentous fungi (29). The α,β-unsaturated aldehyde moiety in sesquiterpene dialdehydes is responsible for their antifungal activity (47).
FIG. 1.
Chemical structure of polygodial.
Polygodial is not mutagenic, as was determined by three variants of the Ames Salmonella test (2) and further confirmed by the mammal-based V79/HGPRT method (37). This is unique in that many other sesquiterpene dialdehydes possessing strong biological activity are mutagenic. In comparison with members of this group, polygodial exhibits the least cytotoxicity for compounds which have antifungal activity (2, 13).
Polygodial exhibits fungicidal activity against Saccharomyces cerevisiae, Candida albicans, and other fungal pathogens comparable to that of amphotericin B in standardized susceptibility tests (NCCLS) (31). Previous studies have reported that polygodial's fungicidal activity is the result of structural disruption of cell membranes. Radioactive monomer incorporation studies showed no selective inhibition of uptake in polymers of DNA, RNA, protein, or polysaccharide, as all uptake tapered off after 60 min (50). Electron microscopy (EM) showed generalized membrane disruption throughout the cell (29). Polygodial produced amounts of potassium leakage from yeast cells similar to those produced by amphotericin B and miconazole (56). This paper exposes another target of polygodial in yeast, specifically, mitochondrial ATP synthase.
MATERIALS AND METHODS
Chemicals.
Polygodial was isolated from the sprout of P. hydropiper (Polygonaceae) as described previously (28). Most chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.), including amphotericin B (concentrations of which were adjusted for the 80% purity of this compound), miconazole, sodium orthovanadate, and ammonium molybdate (USP quality). Sodium orthovanadate was prepared as described by Goodno (20), assuming a hydration number of 10, and then its actual concentration was verified by determining its absorbance at 265 nm from a sufficient dilution, through calculation with its extinction coefficient. Alamar Blue was purchased from Accumed International.
Microbial strains and media.
S. cerevisiae strains were purchased from the American Type Culture Collection (Manassas, Va.) or obtained as gifts. ATCC 7754 is wild-type Fleischmann bakers' yeast. ATCC 66089 is MATα ade1 his7 leu2-1 met8-1 pet9. W303-1a is MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100. Microbiological media were either 2.5% malt extract (ME) medium (BBL catalog no. 11885) or RPMI 1640 broth (Sigma catalog no. R-6504) buffered with 0.165 M MOPS (morpholinepropanesulfonic acid; Fisher Scientific, Santa Clara, Calif.) at pH 7. Adenine and uracil were added at 20 mg/liter to RPMI 1640 medium for W303-1a. Strains were subcultured on Sabouraud's dextrose agar medium (1% Bacto Peptone, 4% dextrose, 2% Bacto Agar) or maintained at −80°C in medium containing 20% glycerol. Cytoplasmic petite mutants of both strains 7754 and W303-1a were obtained by treatment with 25 μg of ethidium bromide per ml in 0.67% Difco yeast nitrogen base with 2% glucose, plus relevant nucleotides and amino acids for W303-1a. After the culture was grown to saturation, a small inoculum was transferred to ethidium bromide-free medium and the organisms were grown to saturation and streaked on YPD (1% yeast extract, 2% Bacto Peptone, 2% dextrose, 2% agar), which yielded petites entirely lacking mitochondrial DNA (mtDNA) ([rho0]) (15). Additionally, a spontaneously occurring cytoplasmic petite [rho−] was obtained from W303-1a. W303-1a exhibited a red phenotype on YPD plates, whereas colonies which are small and white are petite due to loss of mitochondrial function. On YPDG plates (1% yeast extract, 2% Bacto Peptone, 0.1% dextrose, 3% glycerol, 2% agar) wild-type colonies grew to normal size while petites gave rise to very small colonies, due to an inability to use glycerol as the primary carbon source, as is possible with respiratory-competent mitochondria. Petites were further confirmed as [rho−] by 4′,6-diamidino-2-phenylindole (DAPI) staining of DNA (22), and all petites displayed no visible mtDNA, unlike wild types. Petite strains were cultured in RPMI 1640 with MOPS (pH 7) plus 2% glucose, as were wild-type strains when they were directly compared.
Determination of MICs and MFCs.
Susceptibilities were determined with a slight modification of fungal microbroth dilution method M27-A of the NCCLS (39). A 2-fold dilution series of polygodial was prepared at 100-fold final strength in either dimethyl sulfoxide (DMSO; Sigma) or dimethylformamide (EM Science, Gibbstown, N.J.). The compound was diluted 25-fold in medium, and 50 μl was added to each well in a 96-well microtiter plate. Calcium was prepared at a fourfold concentration in medium, and 50 μl was added to the assay wells. Yeasts were subcultured by shaking at 30°C until early stationary phase, and 100 μl of an adjusted suspension at 2 × 103 CFU/ml was added to arrive at a final assay volume of 200 μl. The round-bottomed microplates were shaken gently on a vortexer with a microplate adapter and incubated at 30°C for 48 h. The MIC was the lowest concentration that demonstrated no visible growth. Minimum fungicidal concentrations (MFCs) were determined as follows. After the MIC was determined, a 5-μl aliquot was taken from each well and added to 200 μl of compound-free fresh medium. After 48 h of incubation, the MFC was determined as the lowest concentration of the test compound in which no recovery of microorganisms was observed. All assays were performed at least twice on separate occasions.
Time-kill studies.
Time-kill curves were obtained by adding a 200-μl aliquot of an adjusted (107-CFU/ml) early-stationary-phase shaking subculture to 20 ml of fresh medium in a 250-ml flask, resulting in a starting culture of approximately 105 CFU/ml. The same medium conditions were used for subculture and assay cultures. After 30 min of shaking incubation in fresh medium, 200 μl of a 100-fold concentration of polygodial was added. For the calcium condition, calcium was added and 5 to 10 min of incubation transpired before polygodial was added. The flasks were incubated by shaking at 30°C for 48 h. Samples were taken at selected times, and serial dilutions were made in sterile saline before the samples were plated onto YPD plates. The plates were incubated for 2 days at 30°C before the number of CFU/ml was determined. Cells treated with polygodial grow on YPD plates even with a direct plating of 100 μl, so CFU per milliliter can be read down to 101. Results presented are the means of results of at least two independent assays.
Microscopy.
Nomarski differential interference contrast (DIC) microscopy was performed on a Nikon model TE300 inverted microscope equipped with a 100× Plan-Apo, 1.4 DIC objective and a charge-coupled device camera (Hamamatsu model C4742-95) controlled with Phase-3 (Philadelphia, Pa.) imaging system software. Fluorescence microscopy was performed with a 100× Plan-Fluor, 1.3 objective and DAPI filters.
Measurement of cellular leakage.
Cell leakage was assessed by measuring 260-nm-absorbing materials released to the medium, primarily representing nucleotides of which uracil-containing compounds exhibited the strongest absorbance. Cells cultured by shaking at 30°C to early stationary phase were washed two times and diluted to approximately 5 × 107 CFU/ml with cold MOPS buffer, pH 6.0. Cells were aliquoted to tubes, and compounds were added from 100-fold-concentration stocks. Cells were incubated stationary at 30°C, samples were taken at intervals and spun at 8,000 × g for 5 min in microcentrifuge tubes, and the supernatants were collected for analysis. Results presented are the means of values from at least two independent assays.
Measurement of medium acidification.
The glucose-induced medium acidification of S. cerevisiae was measured by a modified procedure based on the method of Haworth et al. (23). The strain was cultured with shaking in YPD broth overnight at 30°C and washed two times with cold distilled water. The cells were diluted to 107 CFU/ml with cold distilled water and kept on ice. The reaction mixture contained 2.7 ml of cells and 30 μl of compound dissolved in DMSO and was preincubated for 5 min at 30°C. A 20% glucose solution of 0.3 ml was added (final concentration, 2%) to induce medium acidification. After 10 min of incubation, the pH of the external medium was checked (Orion 8175 Ross semimicro electrode). This assay was performed in triplicate on separate occasions.
Isolation of membrane-bound ATPases.
S. cerevisiae was cultured to early stationary phase in a 50-liter fermentation batch, spun dry with a Sharples centrifuge, and stored at −80°C. Alternatively, cells were grown in 1-liter Fernbach flasks for collection during mid-log phase. A mixture of 100 ml of lysis buffer (50 mM Tris [pH 7.5], 1 mM EDTA, 2 mM Pefablock) plus 50 g of cells was homogenized with a nitrogen gas burst homogenizer (Avestin EmulsiFlex C5) by three or four passes at 25,000 to 30,000 lb/in2. The purification protocol is adapted from the method of Serrano (44). After pelleting of unbroken cells, the membrane-containing supernatant was centrifuged (25,000 × g for 20 min) to separate the membrane from the cytosol. The pellet was suspended in 50 mM Tris buffer (pH 7.5) containing 20% glycerol, 0.2 mM EDTA, and 2 mM Pefablock, mixed with a few strokes of a Dounce homogenizer (Weaton, loose pestle), and applied to a discontinuous sucrose gradient (2 parts 43% sucrose to 1 part 53% sucrose) at 55,000 × g for 3 h (Beckman SW25.1 swinging bucket rotor). Plasma membranes were recovered at the 43%-53% sucrose interface, and mitochondrial membranes were recovered at the glycerol–43% sucrose interface. A second sucrose gradient was performed on the glycerol–43% sucrose fraction. Sucrose fractions were stored at −80°C for long-term storage. For use, fractions were washed twice and resuspended with MTAE buffer (50 mM MES [morpholineethanesulfonic acid]-Tris, 1 mM ATP, 1 mM EDTA), which seems to stabilize the ATPases, permitting them to be stored at 4°C for several weeks without significant loss of activity.
Measurement of ATP hydrolysis.
The ATP hydrolysis assay was conducted in MTM buffer (100 mM MES-Tris buffer, 10 mM MgCl2) at optimal pHs for each of the three main membrane-bound ATPases of S. cerevisiae, namely, pH 5.7 for P-type ATPase, pH 7.0 for V-type ATPase, and pH 8.5 for F0F1-ATPase. MTM buffer was adjusted appropriately higher than its final pH to negate the pH drop caused by 10 mM ATP. A modified method of an ascorbate-ammonium molybdate assay was used to measure inorganic phosphate (35, 44). The linear range of phosphate detected was 1 to 40 nmol. Compounds were preincubated for 5 min, 10 mM ATP was added, and the reaction continued at 30°C for 15 min, at which point it was stopped with a solution containing sodium dodecyl sulfate (SDS), HCl, and ammonium molybdate (final concentrations, 0.5%, 0.2 M, and 0.5%, respectively). Next, 150 μl was transferred to a 96-well microplate, and then 50 μl of ascorbate was added (final concentration, 0.05%) with an eight-channel pipette to start the color development reaction simultaneously, which was monitored at 750 nm for 10 min until the absorbance reached its plateau. All assays were performed at least twice on each membrane preparation, and at least two independently isolated preparations were used. Protein concentrations were determined by the bicinchoninic acid protein assay (Pierce Co., Rockford, Ill.) with an SDS-based modification for more accurate assessment of membrane proteins (38), and bovine serum albumin was used as a standard.
RESULTS
Antifungal activity of polygodial.
Microbroth susceptibility assays were conducted with polygodial against S. cerevisiae, and the results are presented in Table 1. The MIC and MFC of polygodial were no more than twofold different, demonstrating its fungicidal activity. Calcium was previously reported to suppress membrane leakage induced by polygodial and presumably to protect against cell death (56). EDTA was reported to reverse the leakage protection. Therefore, both molecules were tested for their effects in the susceptibility assays. Unexpectedly, calcium did not protect the yeast from polygodial and actually enhanced the MIC but not the MFC in RPMI 1640 medium. The standard medium used in our lab is malt extract, but we also used RPMI 1640 buffered with MOPS at pH 7, which is the medium recommended for standardized susceptibility testing by the NCCLS (39). EDTA enhanced the MIC and MFC of polygodial in ME but killed the yeast by itself in RPMI 1640 at both concentrations tested. There was no observed difference between different types of calcium, as calcium chloride and calcium nitrate gave the same results (data not shown). The addition of calcium to RPMI 1640 medium resulted in high turbidity. In order to assess growth, we used the dye Alamar Blue, which permits clear distinction between viable and nonviable cells (16).
TABLE 1.
MICs and MFCs of polygodial with calcium and EDTA against S. cerevisiae
| Treatment | ME
|
RPMI 1640
|
||
|---|---|---|---|---|
| MICa | MFCb | MIC | MFC | |
| Polygodial | 1.56 | 3.13 | 3.13 | 6.25 |
| +1 mM EDTA | —c | 0.78 | — | — |
| +0.2 mM EDTA | 0.4 | 1.56 | — | — |
| +100 mM CaCl2 | 0.78 | 1.56 | 0.02 | 0.02 |
| +10 mM CaCl2 | 1.56 | 3.13 | 0.02 | 6.25 |
| +1 mM CaCl2 | 1.56 | 3.13 | 3.13 | 6.25 |
Micrograms per milliliter. Determined by the NCCLS microbroth dilution method M27-A.
Micrograms per milliliter. Determined by transferring 5 μl from the MIC wells to fresh media.
—, cells did not show growth in the absence of polygodial.
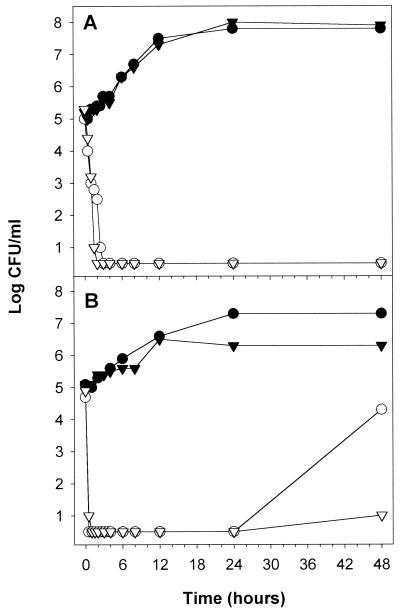
Time-kill curves were conducted with 6.25 μg of polygodial per ml (MFC) in the presence and absence of 10 mM calcium (Fig. 2). In both ME and RPMI 1640 media, lethality was rapid, with the number of CFU/ml dropping below 1 log unit within 0.5 to 3 h depending on the medium. Calcium exhibited no protective effect against the fungicidal activity of polygodial. In fact, calcium slowed the rebound of growth seen with RPMI 1640 at 48 h. Calcium was generally detrimental to growth, as seen in the MIC data also.
FIG. 2.
Time-kill curves of polygodial with (▾, ▿) and without (●, ○) 10 mM Ca2+ against S. cerevisiae. Results of assays with both malt extract (A) and RPMI 1640 plus MOPS (pH 7.0) (B) are presented. Results for controls are represented by filled symbols, and those for cells treated with polygodial (6.25 μg/ml) are represented by open symbols.
Cellular leakage effects.
Cell leakage can be assessed by measuring intracellular-component release to the medium from washed cells in buffer. MOPS buffer is synthetic and should not be metabolized by the cell and thus causes the cells to exist in stationary phase, so intracellular components are not effluxed. Cellular components which absorb at 260 nm represent one class of leakage components, primarily nucleotides of which uracil-containing compounds exhibit the strongest absorbance.
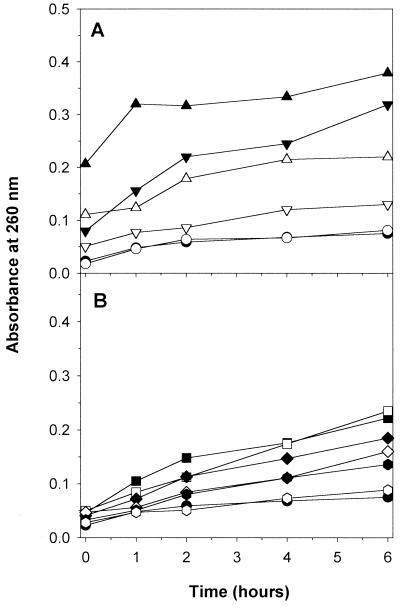
Figure 3 shows leakage from yeast cells over a 6-h period due to polygodial, amphotericin B, and miconazole. The concentrations used were at the MFC and at fourfold the MFC to account for a larger concentration of yeast cells. Polygodial exhibited a concentration-dependent effect on leakage, with 25 μg/ml resulting in larger amounts of leakage than 6.25 μg/ml, and both showed rapid effect. Calcium protected against leakage caused by polygodial and at both concentrations was suppressed approximately 50% at most time points. Miconazole caused much less leakage but was also suppressed by calcium. In comparison to polygodial, amphotericin B took 6 h to reach a similar amount of leakage, and calcium had only a small reduction effect. Therefore, levels of leakage of 260-nm-absorbing material from exposure to amphotericin B and polygodial appear different. By itself, polygodial absorbed at 260 nm at approximately the same amount as was seen with polygodial plus calcium at 0 h, for both concentrations. Twenty percent cell breakage resulted in values around 3 absorbance units, and we extrapolated the maximum potential absorbing material to be 15 absorbance units. Therefore, even after 6 h of polygodial treatment at high concentration, less than 3% of cellular material was lost.
FIG. 3.
Cellular leakage of 260-nm-absorbing materials. The following compounds were added to cells washed with MOPS buffer (50 mM, pH 6.0) and incubated for the specified times before the extracellular fractions were collected: DMSO (control) (●, ○), polygodial at 25 μg/ml (▴, ▵), and polygodial at 6.25 μg/ml (▾, ▿) (A), and amphotericin B at 6.25 μg/ml (■, □), amphotericin B at 1.56 μg/ml (⧫, ◊), and miconazole at 25 μg/ml ( , ) (B). Filled symbols represent the compound, and open symbols represent the compound plus 10 mM Ca2+.
Cell morphology.
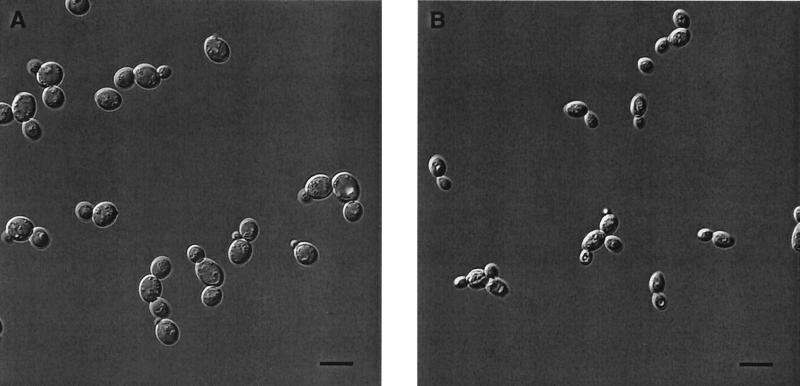
Nomarski DIC microscopy was performed to see what morphological changes occurred in live cells (Fig. 4). Cells were treated with 12.5 μg of polygodial per ml, the MFC for 106 CFU/ml in RPMI 1640 medium. After 3 h, the polygodial-treated cells exhibited significant shrinkage compared to controls, and the organization of subcellular organelles appears either disrupted or compacted. This disrupted organization apparently represents loss of ordered membranes of the secretory pathway as well as of the vacuole and mitochondria. Since various stages of budding were present, polygodial was not acting as a cell cycle inhibitor. In fact, these live cell images can be considered similar to electron micrographs taken previously (29), substantiating that the cell perturbations seen in the EM images are real and not artifacts of sample preparation. However, the prior assertion that breaks in the plasma membrane were visible in the EM micrographs is questionable, and the present images do not show major plasma membrane disruption or any evidence of cell lysis. Polygodial-treated cells did appear somewhat similar to cells with defects in the plasma membrane proton-pumping ATPase (51), which led us to investigate the possibility that polygodial inhibited this enzyme.
FIG. 4.
Morphological effect of polygodial on S. cerevisiae. DIC micrographs illustrate the difference between a control culture (A) grown in RPMI 1640 medium and cells treated with polygodial after 3 h at the MFC (B). The scale bar represents 10 μm.
Acidification and ATPase inhibition.
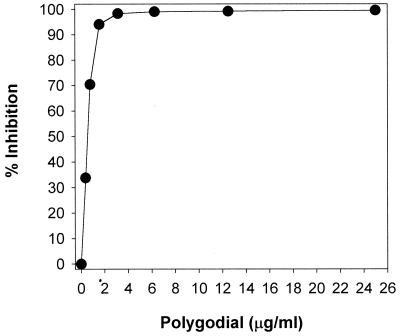
Acidification of the medium occurs primarily through the proton-pumping action of the plasma membrane ATPase, Pma1p, and was measured by comparing initial versus final pHs of washed yeast cells after 10 min (beginning with the addition of 2% glucose). pH was converted to proton concentration to calculate percent inhibition. Figure 5 shows that polygodial completely suppressed the glucose-induced decrease in external pH at 3.13 μg/ml, the same as the fungicidal concentration, which supports the idea of its mode of action being related to proton-pumping ability. Therefore, Pma1 ATPase was investigated as a likely target for polygodial. An alternative hypothesis, that the energy needed to drive Pma1 ATPase was being affected, was tested by investigating the mitochondrial ATPase.
FIG. 5.
The inhibitory effect of polygodial on medium acidification in S. cerevisiae. Acidification was induced by glucose (final concentration, 2%) and evaluated as the molar concentration of protons calculated from external pH. The inhibition ratio (percentage) was calculated as follows: (1 − [H+]inhibitor/[H+]inhibitor free) × 100.
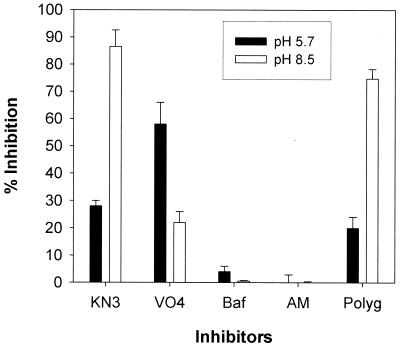
Partially purified membrane-bound ATPases were assayed for their ability to hydrolyze ATP. ATPase inhibitor profiles at pHs 5.7 and 8.5 were conducted to determine which ATPase activities were present (Fig. 6). In S. cerevisiae, the optimal pH for plasma membrane ATPase is 5.7 while that for mitochondrial ATPase is 8.5. Concentrations of selective inhibitors that maximally inhibit the ATPases in question were used (5, 19, 44). Potassium azide was used at 5 mM and inhibits the mitochondrial ATPase. Azide is commonly known as a respiration inhibitor affecting P450 enzymes but also directly inhibits both membrane-bound F0F1-ATPase and soluble F1-ATPase. Sodium orthovanadate was used at 100 μM and inhibits the plasma membrane ATPase, bafilomycin was used at 50 nM and inhibits vacuolar ATPase, ammonium molybdate was used at 0.2 mM and inhibits phosphatases, and polygodial was used at 50 μg/ml.
FIG. 6.
Effect of a panel of standard inhibitors and polygodial against membrane-bound ATPases in S. cerevisiae at pH 5.7 (filled bars) and pH 8.5 (open bars). Whole membranes were used, and the amount of enzyme activity ranged from 0.7 to 1.1 U (nanomoles of Pi per minute), with specific activity ranging from 50 to 90 U/mg. The inhibition ratio (percentage) was calculated as follows: {1 − [(I − C2)/(C1 − C2)] × 100}, where C1 is enzyme alone, I is enzyme plus inhibitor, and C2 is inhibitor without enzyme (10 mM ATP present in all). Inhibitors were used at concentrations known to give maximal inhibition of their selective enzyme. KN3, potassium azide at 5 mM; VO4, sodium orthovanadate at 100 μM; Baf, bafilomycin at 50 nM; AM, ammonium molybdate at 0.2 mM; Polyg, polygodial at 50 μg/ml. Values are means ± standard deviations.
At pH 5.7, vanadate inhibited around 60% while azide inhibited around 30%, showing significant mitochondrial ATPase activity even at pH 5.7. Polygodial showed less than 20% inhibition, which is less than that seen with azide. Standard assays for Pma1 ATPase inhibition include the use of azide to exclude mitochondrial ATPase contamination, so that only Pma1 ATPase activity remains. Viewing the results from this perspective, it is clear that there was no inhibition of Pma1 ATPase by polygodial. At pH 8.5 azide inhibition is near 85% while vanadate drops to near 20%. This residual inhibition by vanadate was not expected because Pma1 ATPase activity should not be present; however, this inhibition may be due to some phosphatase activity (B. Bowman, personal communication). Regardless, polygodial showed strong inhibition against mitochondrial ATPase (near 75%), at a level similar to that inhibited by azide. Bafilomycin and ammonium molybdate showed negligible inhibition at either pH, signifying that vacuolar ATPase and most phosphatases were not present.
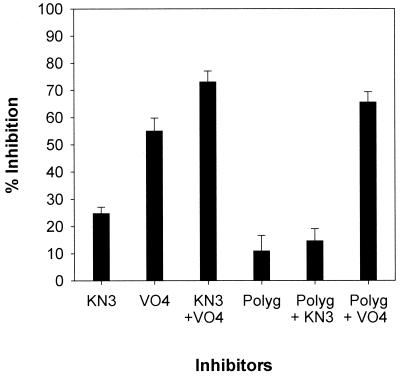
Combinations of inhibitors were assayed against plasma membrane ATPase at pH 5.7 (Fig. 7). Azide and vanadate inhibition were additive, indicating inhibition of different ATPases. The results with azide plus polygodial were not significantly different from results with polygodial or azide alone. Polygodial plus vanadate inhibited additively, similar to what occurred with azide plus vanadate. These results indicate that polygodial and azide inhibit the same ATPase.
FIG. 7.
Combinations of inhibitors against ATPases at pH 5.7. Whole membranes and inhibitors were used as described in the legend of Fig. 6. Values are means ± standard deviations.
Selective inhibition of isolated mitochondrial ATPase.
Further separation between membrane fractions of the plasma membrane and mitochondria was conducted by sucrose gradient centrifugation. SDS-polyacrylamide gels showed a large number of proteins still present in the 43-53 and G-43 fractions, many of them similar (data not shown). In general, pH had a greater effect in distinguishing ATPase activities than the sucrose gradient fraction, although specific activities were increased two- to fourfold and inhibition values had less error than whole membranes. Immunoblots with anti-F1β ATPase showed mitochondrial contamination of the 43-53 fraction, while those with anti-V-type ATPase (100 kDa; Molecular Probes) showed negligible amounts of vacuolar ATPases in the preparations.
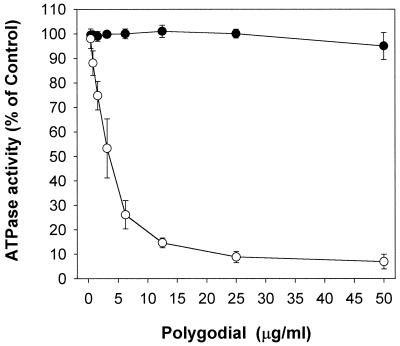
A dose-response curve is shown for polygodial against mitochondrial ATPase and plasma membrane ATPase in Fig. 8. The G-43 fraction plus vanadate was assayed at pH 8.5, and the 43-53 fraction plus azide was assayed at pH 5.7. Polygodial was tested at concentrations from 0.4 to 50 μg/ml and showed inhibition of the mitochondrial ATPase but no inhibition of the plasma membrane ATPase. There was no difference in mitochondrial ATPase inhibition between 5 and 30 min of preincubation. Polygodial's 50% inhibitory concentration (IC50) against mitochondrial ATPase was calculated by nonlinear regression to be 3.5 μg/ml (15 μM). These data clearly demonstrate that polygodial inhibits the mitochondrial ATPase.
FIG. 8.
Dose-response curves of polygodial comparing the plasma membrane ATPase (●) and mitochondrial ATPase (○) hydrolytic activities. Sucrose gradient membrane fractions were used, and the amount of enzyme activity ranged from 0.6 to 0.9 U (nanomoles of Pi per minute), with specific activity ranging from 100 to 200 U/mg of protein, depending on the fraction. ATP hydrolysis was calculated as follows: {[(I − C2)/(C1 − C2)] × 100}, where C1 is enzyme alone, I is enzyme plus inhibitor, and C2 is inhibitor without enzyme (10 mM ATP present in all). Values are means ± standard deviations.
Vacuolar ATPase was assayed at pH 7 when yeast cultures were harvested in mid-log phase. Immunoblots confirmed the presence of V-type ATPase. The G-43 fraction was tested, and polygodial at 50 μg/ml showed 15% more inhibition than azide alone, indicating inhibition of vacuolar ATPase as well. However, low inhibition at such a high concentration of polygodial is not likely relevant to polygodial's rapid fungicidal effect.
Polygodial's effect on respiration-deficient mutants.
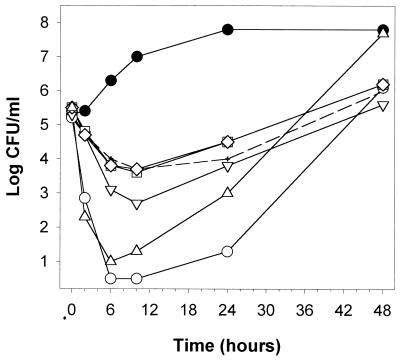
Polygodial-treated yeast displayed interesting morphology when they were observed on YPD plates, such as in the early time points of Fig. 2. There were dramatic differences in the sizes of colonies, ranging from the normal, large size to very small. Individual colonies were picked from these plates, subcultured twice in ME medium, and plated. Colonies which were originally large, medium, small, and very small all grew to roughly normal size on YPD plates. However, time-kill curves of these strains showed significant differences (Fig. 9). Strains which were originally medium and small were resistant to polygodial, compared to the large strain, which was slightly less resistant. The very small strain exhibited a curve more similar to that of the wild type. These strains were subsequently grown and then streaked on YPGD plates to test whether they might be respiration-deficient strains, commonly known as petite mutants. The large, medium, and small strains grew as petites, but the very small strain grew to the same large size as the wild type, thus correlating with the time-kill curves. They were further confirmed as cytoplasmic petites [rho−] by DAPI staining, which failed to show nonnuclear DNA present in these cells, whereas the wild-type and the very small strains showed mtDNA. Additionally, a cytoplasmic petite completely deficient in mtDNA ([rho0]) was generated by ethidium bromide treatment of wild-type yeast. The [rho0] time-kill curve was the same as those of the medium and small strains.
FIG. 9.
Time-kill curves of polygodial comparing wild-type S. cerevisiae ATCC 7754 to petite mutants, which are respiration deficient, in ME medium. Control cultures of each strain were not significantly different from the wild type (●). Polygodial at 3.13 μg/ml was exposed to the wild type (○) and to strains which were collected from another time-kill experiment as phenotypically large (▿), medium (□), small (◊), and very small (▵) strains. The [rho0] mutant is represented by dashed lines (+).
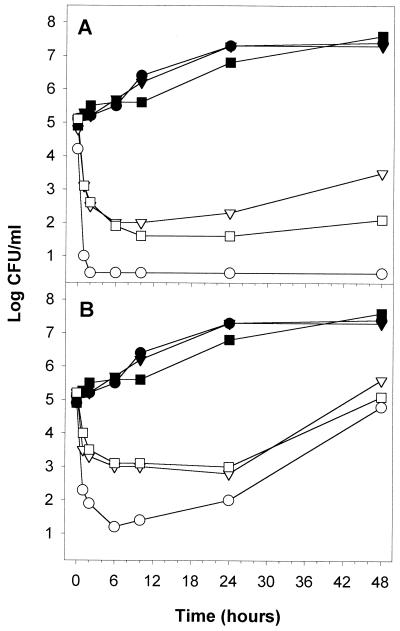
A common research strain, W303-1a, was also investigated to see if the polygodial-induced selection of [rho−] mutants is generally applicable. W303-1a is advantageous because it contains an adenine biosynthesis mutation, which causes colonies to appear red on YPD plates, versus petites, which appear small and white. A spontaneously occurring cytoplasmic petite of W303-1a was collected in this way; in addition, a [rho0] mutant was generated with ethidium bromide. Figure 10 shows the results of a time-kill curve of these strains with polygodial in RPMI 1640 plus MOPS (pH 7) supplemented with 2% glucose. At both 6.25 and 3.13 μg/ml, the cytoplasmic petites were more resistant than the W303-1a parent strain. At 6.25 μg/ml, the parent was rapidly killed to below 1 log CFU/ml within 1 h and remained nonviable at 48 h. In contrast, the petite mutants were reduced to 2 log CFU/ml after 6 h and then their numbers leveled off, the [rho−] mutant began to rebound, and the number of CFU per milliliter of the [rho0] mutant remained constant. With regard to the morphology of W303-1a parent colonies produced after 1 h of exposure, 95 to 100% of the population changed from large, red colonies to medium or small, white colonies, showing that the remaining viable population was petite. This percentage range remained constant throughout the 48-h assay.
FIG. 10.
Time-kill curves of polygodial comparing S. cerevisiae W303-1a to petite mutants, which are respiration deficient, in RPMI 1640 with MOPS (pH 7) plus 2% glucose. Results for controls are represented with filled symbols, and those for polygodial-treated cells are represented with open symbols as follows: ● and ○, W303-1a; ▾ and ▿, [rho−] strain; and ■ and □, [rho0] strain. Concentrations of polygodial were 6.25 μg/ml (A) and 3.13 μg/ml (B).
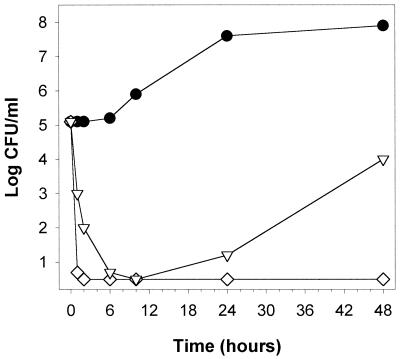
Polygodial's effect on a respiration-dependent mutant. One additional mutant was tested with polygodial. The pet9 strain is a nuclear petite strain caused by a point mutation in AAC2 (30) which eliminates functionality of the ADP-ATP carrier protein of the inner mitochondrial membrane (17). Therefore, ATP pools in the mitochondria and cytosol are separated and growth is possible only in the presence of fermentable sugars. ATP is also essential for various mitochondrial functions, and cells with a defective ADP-ATP translocator are known to be nonviable in the absence of oxygen (11), demonstrating their dependence on ATP produced via oxidative phosphorylation. Therefore, if polygodial targets ATP synthase in vivo, nonviable cells should result for the same reason. Figure 11 shows that polygodial rapidly inhibited the pet9 yeast. The pet9 strain's sensitivity to polygodial, in conjunction with [rho0] or [rho−] resistance, demonstrates causality between mitochondrial ATPase inhibition, reduction of cellular ATP supply, and fungicidal activity.
FIG. 11.
Time-kill curves of polygodial against the S. cerevisiae pet9 mutant, with a defective ADP-ATP translocator, in RPMI 1640 with MOPS (pH 7) plus 2% glucose. Shown are results with control cells (●) and cells treated with polygodial at 6.25 μg/ml (◊), and 3.13 μg/ml (▿).
DISCUSSION
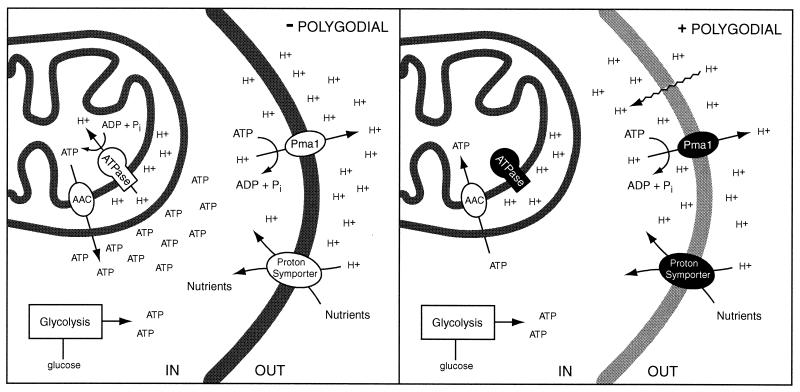
We propose the following model to explain the mechanism of fungicidal action by polygodial in S. cerevisiae (Fig. 12). Polygodial inhibits mitochondrial ATPase, eliminating a major source of ATP. It also causes plasma membrane disruption, which leads to an influx of protons. This influx results in the collapse of the proton motive force and inhibition of nutrient uptake coupled to this gradient. A decrease in internal pH which inhibits enzymes as they are shifted away from their pH optima also occurs. Cells can recover by pumping protons out of the cell via the plasma membrane ATPase, Pma1p, which consumes large amounts of ATP. The combination of ATP consumption stimulated by proton permeability and ATP synthase inhibition contributes to a decrease in ATP to levels below those necessary for normal metabolic functions, resulting in nonviable yeast.
FIG. 12.
Schematic representation of polygodial treatment on yeast. Polygodial inhibits mitochondrial ATPase, eliminating a major source of ATP (shading represents proteins with impaired functions). It also causes plasma membrane disruption, which leads to an influx of protons (wavy lines). This influx results in a collapse of the proton motive force and inhibition of nutrient uptake coupled to this gradient, followed by a decrease in internal pH, which inhibits enzymes as they are shifted off of their pH optima. Cells can recover by pumping protons out of the cell via the plasma membrane ATPase, Pma1p, which consumes large amounts of ATP. The combination of ATP consumption stimulated by proton permeability and ATP synthase inhibition contributes to a decrease in ATP to levels below those necessary for normal metabolic functions, resulting in nonviable yeast.
Either effect alone is not lethal to yeast. The existence of petite mutants shows that mitochondrial ATP production is not essential. The detrimental effects of proton permeability can be resisted with sufficient amounts of ATP, as evidenced with weak acid inhibitors, like sorbic acid, which require large concentrations to inhibit growth and are basically fungistatic (27). Weak acids penetrate the cell membrane in acidic media and then contribute their proton to the higher-pH intracellular environment. This process affects enzymes known to be sensitive to pH, such as phosphofructokinase (33), which can lead to reduced glycolytic ATP production. Additionally, the ATP pool can be consumed in a futile cycle of acid influx and ATPase-based export (7), thus inhibiting growth even though the cells remain metabolically active. Our previous report described polygodial's 64-fold potentiation of antifungal activity with sorbic acid, which presumably increased killing effectiveness due to inhibition of proton pumping by the plasma membrane ATPase, suppressing a key means of weak-acid resistance (27). In the context of the results presented here, this mechanism remains true, but with the cause of Pma1 ATPase inhibition being not due to direct enzyme inhibition but due to the lack of ATP as a substrate.
Amphotericin B is a well-known membrane-disruptive agent which has a direct membrane effect and is lethal to yeast. It forms stable pores in the membrane by complexing with ergosterol, which results in permeability to ions such as potassium and protons (53). Amphotericin B treatment causes greater membrane disruption than sorbates and results in lethality, but this requires sufficient time to occur. It took 10 h to decrease the count below 2 CFU/ml at the significant concentration of 4 μg/ml (32). At similar concentrations, polygodial completely killed the yeast within 2 h (Fig. 2), demonstrating an effect separate from, or in addition to, that of membrane disruption.
Previous reports supported that the plasma membrane was the target of polygodial (29, 50); therefore, further study was conducted. Surprisingly, polygodial-induced cell death was not suppressed by calcium addition. However, cellular leakage of 260-nm-absorbing materials was significantly suppressed by calcium, as was potassium leakage (56), making the correlation between leakage and cell death ambiguous. Additionally, only a small percentage of 260-nm-absorbing material was leaked, indicating that the cells were not lysed. This result is further supported by the fact that polygodial did not lyse erythrocytes at concentrations of up to 100 μg/ml, whereas amphotericin B lysed them at 6.25 μg/ml, which is similar to its MFC (31).
An increase in the cation bridging of membrane lipids is a reasonable supposition to account for leakage suppression, making it more difficult for the compound to enter the membrane. This suppression may not necessarily apply to penetration into the cell, however. Compounds which perturb lipid packing on a smaller scale would be suppressed by cations, as was seen with miconazole and polygodial. On the other hand, amphotericin B is more resistant to cation effect because it forms a stable transmembrane complex and causes leakage through a pore-like structure.
The strong synergistic effect of EDTA with polygodial is possibly due to abolishing most cation-lipid bridging, which would allow greater penetration of polygodial into the cell. EDTA may also disrupt ion flux, which is necessary for many membrane transport proteins as well as proper stress response (52), and holding captive a population of transportable ions would suppress these processes.
Since polygodial causes leakage of both 260-nm-absorbing compounds and potassium ions, it is reasonable to assume that protons have increased permeability also. In the presence of polygodial, protons transport down their concentration gradient into the cell. Changing the focus of thought from leakage, which is not relevant to cell death, to a consideration of polygodial-induced proton influx, which is physiologically relevant, permits a better understanding of polygodial's effect on the cell.
Microscopic observation provided a visual assessment of polygodial's effect on live, nonfixed yeast cells. There was significant change in morphology over time but nothing as dramatic as would be expected for a compound suspected of major membrane disruption. The cell wall can contribute to structural stability even if the cells are nonviable, which makes it difficult to distinguish if they would lyse without a cell wall, but the lack of erythrocyte lysis shows that they most likely would not. Therefore, we began to consider other possible modes of action. Similarity in morphology to plasma membrane ATPase mutants led us to investigate the possibility that polygodial inhibited this enzyme.
Yeast cells are observed to reduce their external pH upon addition of glucose, and this is known to be due to proton efflux of the plasma membrane proton-pumping ATPase Pma1 (12, 24, 45). The regulation of Pma1 ATPase by glucose at the molecular level is not yet fully understood, but the maintenance of proper internal pH is essential for enzyme activity and allows the cell to survive at a low environmental pH (33, 41). Acidification of the external medium is a complex process involving many cellular components. This phenomenon depends primarily on Pma1 ATPase, but the energy to run this pump requires large amounts of ATP, which comes from glycolysis coupled to oxidative phosphorylation, and requires sugar transporters as well. The cell actively modulates its proton pumping to maintain proper intracellular pH and to generate a proton motive force which drives most membrane transport via proton symport and antiport.
In an acidification assay, polygodial completely suppressed the glucose-induced decrease in external pH at 3.13 μg/ml, the same as the fungicidal concentration, which supports the idea of its mode of action being related to its proton-pumping ability. However, it is important to determine if this suppression of the decrease in external pH is a direct result of the inhibition of Pma1 ATPase or an indirect result through suppression of energy metabolism (44). There are examples of compounds, such as amiloride, that inhibit acidification but not Pma1 ATPase and that were found to affect glucose metabolism instead (23). Proton permeability may also contribute to some of the observed acidification inhibition.
Enzymatic characterization between the three major membrane-bound ATPases was conducted by using the optimal pH for each ATPase in conjunction with selective inhibitors. Surprisingly, Pma1 ATPase was not directly inhibited by polygodial. In contrast, the two other membrane-bound ATPases in yeast were found to be susceptible to the compound. Polygodial inhibited the mitochondrial ATPase strongly, whereas it inhibited the vacuolar ATPase only slightly. It is more challenging to separate vacuolar ATPase from the other two types, so detailed inhibition data are not available at this time. However, the slight inhibition seen in whole membranes at high polygodial concentrations is not likely of major significance to its fungicidal effect. Mitochondrial ATPase and vacuolar ATPase are evolutionarily related (4), so some degree of similar effects may be expected. Extending the preincubation time from 5 to 30 min did not affect polygodial's inhibition against any of the ATPases, indicating fast reactivity for those affected. Polygodial inhibition is most likely covalent in nature, as the dialdehyde moiety is reactive and will bind to primary amine and sulfhydryl groups.
Polygodial inhibited isolated F0F1-ATPase in a dose-dependent manner at concentrations similar to the MFC (IC50, 3.5 μg/ml), which supports the idea of mitochondrial ATPase being a physiologically relevant target of polygodial in S. cerevisiae. Pma1 ATPase is dependent on ATP levels such that a 20% drop inhibits proton pumping by 55% and a 50% drop inhibits proton pumping by 85% (43). Therefore, polygodial at the IC50 would lead to a decrease in ATP, which should inhibit Pma1 ATPase by about 90%, linking the ATPase inhibition to acidification inhibition.
Mitochondrial ATPase normally functions to produce ATP, but when free of the driving force of the electron transport chain, it can function in reverse and hydrolyze ATP. Measuring hydrolytic activity and distinguishing among the three major membrane-bound ATPases with known inhibitors is accepted procedure (5, 6, 19, 44). Other membrane-bound ATPases such as ABC transporters cannot function as ATP hydrolytic enzymes by themselves but require additional activating components to be functional.
Cytoplasmic petite mutants, which lack mtDNA and are respiration deficient, can occur spontaneously, and compromise up to a few percentages of most yeast cultures (15). The data above support the hypothesis that the rapid decrease in viability of wild-type cells represents a lethal effect on the majority population. Treatment of a commonly used research strain, W303-1a, with polygodial for only 1 h specifically selected for the survival of petites. It is reasonable to assume that polygodial asserts a selective pressure on the population, but it is possible that it favors the formation of petites also, as has been observed in strains with defective F1-ATPase, which were likely to lose their mtDNA (8). As the concentration of polygodial is increased, the [rho−] mutants are also increasingly killed, indicating increased membrane disruption or another effect of polygodial.
In S. cerevisiae, cytoplasmic petite mutants remain viable due to the presence of several nucleus-encoded proteins, including F1-ATPase and the ADP-ATP translocator (8). [rho0] or [rho−] mutants maintain their mitochondrial membrane potential by the electrogenic exchange of ADP for cytosolic ATP, hydrolysis via F1-ATPase, and the continuous cycling of these components. Inner membrane integrity also appears aided by a possible chaperone function of the F1 complex and the essential activities of Yme1p protease and the phosphatidylglycerolphosphate synthase Pgs1p (8). Proper membrane function in petite mutants is critical in maintaining the ability to process the hundreds of nucleus-encoded proteins transported into mitochondria via the TOM and TIM complexes and allows them to function properly (40). Since F1-ATPase is essential for the viability of [rho0] and [rho−] mutants, the partial lethality observed with polygodial may be due to suppression of the alternate F1-ATPase activities described above.
Mitochondria contain an ADP-ATP translocator which permits exchange of ATP and ADP across the inner mitochondrial membrane. Under normal conditions, mitochondrial ATP is produced and exported to the cytosol concomitant with ADP import to continue the cycle. Under anaerobic conditions, cytosolic ATP is produced and can be transported into the mitochondria by the ADP-ATP translocator. The pet9 strain is a nuclear petite strain caused by a point mutation in AAC2 (30) which basically eliminates the functionality of this carrier protein (17). Therefore, ATP pools in the mitochondria and cytosol are separated and growth is possible only in the presence of fermentable sugars. ATP is also essential for various mitochondrial functions, and cells with a defective ADP-ATP translocator are known to be nonviable in the absence of oxygen (11), demonstrating their dependence on ATP produced via oxidative phosphorylation.
In contrast to its effect in the cytoplasmic petite strains, polygodial showed rapid lethality in the pet9 mutant. Since this strain is solely dependent on respiration for its mitochondrial ATP supply, inhibition of ATP synthase will result in nonviable cells. This experiment most tightly links the enzyme inhibition of polygodial to its physiological effect. In conjunction with the other petite mutant time-kill curves, these data strongly support the theory that polygodial exerts a major portion of its fungicidal action through inhibition of ATP production.
Polygodial's inhibition of mitochondrial ATPase explains a number of results from prior reports. In our previous paper (50), endogenous respiratory activity was assessed by measuring oxygen consumption in whole cells washed and resuspended in buffer, which therefore retained only small amounts of sugar. Respiration is strongly stimulated in such cells because the only method of producing sufficient ATP is oxidative phosphorylation. At MICs of polygodial, oxygen consumption initially increased more than 50%. However, polygodial neither stimulated nor inhibited major respiratory chain enzymes which were isolated from mitochondria (55). When these results are reviewed in light of mitochondrial ATPase inhibition, it is reasonable to assume that cellular regulatory mechanisms are required to detect decreasing ATP levels and then to stimulate electron transport to increase the proton gradient, and concomitantly ATP synthesis. The arrest of most biosynthetic processes as seen by inhibition of radioactive monomer incorporation after 60 min (50) is also explained by a rapid drop in cellular energy. Polygodial also lacks inhibition at concentrations up to 100 μg/ml against many gram-positive and gram-negative bacteria (29), which may be explained by the evolutionary divergence of ATP synthase between prokaryotes and eukaryotes. There was a previous report of cellular leakage of ATP from mammalian cells which showed 100% of ATP to be leaked from polygodial-treated ELD cells at 4.8 μg/ml (1). None of the other 14 sesquiterpene dialdehydes tested resulted in significant leakage without 1 or 2 orders of magnitude more compound. In light of our results, the leakage by polygodial was most likely an artifact of low intracellular ATP levels, and cells thus appeared to leak all of their ATP. Some degree of ATP leakage does occur as is indirectly corroborated by the results shown in Fig. 3, but probably at low levels more similar to those of the other sesquiterpene dialdehydes.
Our recent paper reported polygodial's antifungal activity against a variety of yeasts and filamentous fungi (31). There is substantial variation in MICs and MFCs. If mitochondrial ATPase is the primary target and is a strongly conserved protein, this variation is not likely due to differences in the target protein. The logP of polygodial, 3.02 (1), indicates that it can pass through membranes. However, it is at the high end of the range of logPs at which compounds are known to readily enter cells (10). Differences in the lipid compositions of various fungi may affect the ability of polygodial to reach its intracellular target, thus accounting for variable MIC data. Lipid differences may also influence the membrane disruptive effect of polygodial. Additionally, metabolizing polygodial may account for some variation. Muzigadial, a similar sesquiterpene dialdehyde, has been shown to be converted to a less potent hemiacetal by some fungal species (25), and there is some correlation to those species lacking antifungal activity in our assays (31). Finally, polygodial affecting another target may be the cause of some variation. In neuroblastoma cells, polygodial has been shown to inhibit muscarinic acetylcholine receptor signal transduction at concentrations similar to the MIC, but it was concluded that the inhibitory site is not the receptor itself but that it is perhaps at the G protein level or further down the signal transduction pathway (14). This possibility suggests that signaling mechanisms in yeast might be perturbed also.
The mitochondrial ATPase of S. cerevisiae is strongly supported as a major target of polygodial by the data presented above. It is a central protein in energy production, and inhibition would cause a significant decrease in ATP levels, which would indirectly inhibit the plasma membrane proton-pumping ATPase and account for inhibition of acidification. Low ATP levels cause significant perturbation of normal cellular functions and impair adaptive responses to other facets of polygodial's cellular inhibition.
Polygodial's inhibitory effect on ATP synthase will most likely preclude it from being used as a systemic antifungal agent. However, its apparent reduced ability to penetrate some fungal membranes indicates that it will not likely penetrate tough epithelial cells in human skin and might stay localized to exterior layers of cells such as in the mouth or vagina. It may make an effective topical antifungal agent, especially if used in combination, as the membrane disruptive effect in yeast cells will allow increased permeability of other agents (49). Further research in this direction may yield additional therapies to combat fungal infections.
ACKNOWLEDGMENTS
C.S.L. thanks S. H. Lee for his contributions to the early stages of this work and B. Glaeser for extensive discussion. C.S.L. also thanks J. Skoble for assistance with microscopy, P. Torrance for the W303-1a strain and helpful discussion, B. Lesch for fermentation runs, D. Baggott for the anti-F1β antibody, and J. Cope and J. Leveau for critical review of the manuscript.
REFERENCES
- 1.Andersson M, Bocchio F, Sterner O, Forsby A, Lewan L. Structure-activity relationships for unsaturated dialdehydes 7. The membrane toxicity of 15 sesquiterpenoids measured as the induction of ATP leakage in ELD cells. The correlation of the activity with structural descriptors by the multivariate PLS method. Toxicol In Vitro. 1993;7:1–6. doi: 10.1016/0887-2333(93)90106-f. [DOI] [PubMed] [Google Scholar]
- 2.Anke H, Sterner O. Comparison of the antimicrobial and cytotoxic activities of twenty unsaturated sesquiterpene dialdehydes from plants and mushrooms. Planta Med. 1991;57:344–346. doi: 10.1055/s-2006-960114. [DOI] [PubMed] [Google Scholar]
- 3.Barnes C, Loder J. The structure of polygodial, a new sesquiterpene dialdehyde from Polygonum hydropiperL. Aust J Chem. 1962;15:322–327. [Google Scholar]
- 4.Bowman B J, Dschida W J, Bowman E J. Vacuolar ATPase of Neurospora crassa: electron microscopy, gene characterization and gene inactivation/mutation. J Exp Biol. 1992;172:57–66. doi: 10.1242/jeb.172.1.57. [DOI] [PubMed] [Google Scholar]
- 5.Bowman B J, Mainzer S E, Allen K E, Slayman C W. Effects of inhibitors on the plasma membrane and mitochondrial adenosine triphosphatases of Neurospora crassa. Biochim Biophys Acta. 1978;512:13–28. doi: 10.1016/0005-2736(78)90214-6. [DOI] [PubMed] [Google Scholar]
- 6.Bowman E J, Bowman B J. Purification of vacuolar membranes, mitochondria, and plasma membranes from Neurospora crassaand modes of discriminating among the different H+-ATPases. Methods Enzymol. 1988;157:562–573. doi: 10.1016/0076-6879(88)57104-5. [DOI] [PubMed] [Google Scholar]
- 7.Brul S, Coote P. Preservative agents in foods: mode of action and microbial resistance mechanisms. Int J Food Microbiol. 1999;50:1–17. doi: 10.1016/s0168-1605(99)00072-0. [DOI] [PubMed] [Google Scholar]
- 8.Chen X J, Clark-Walker G D. Alpha and beta subunits of F1-ATPase are required for survival of petite mutants in Saccharomyces cerevisiae. Mol Gen Genet. 1999;262:898–908. doi: 10.1007/s004380051156. [DOI] [PubMed] [Google Scholar]
- 9.Dixon D M, McNeil M M, Cohen M L, Gellin B G, La Montagne J R. Fungal infections: a growing threat. Pub Health Rep. 1996;111:226–235. [PMC free article] [PubMed] [Google Scholar]
- 10.Domine D, Devillers J. A computer tool for simulating lipophilicity of organic molecules. Sci Comp Autom. 1998;1998(March):55–63. doi: 10.1021/js980101j. [DOI] [PubMed] [Google Scholar]
- 11.Drgon T, Sabová L, Nelson N, Kolarov J. ADP/ATP translocator is essential only for anaerobic growth of yeast Saccharomyces cerevisiae. FEBS Lett. 1991;289:159–162. doi: 10.1016/0014-5793(91)81059-h. [DOI] [PubMed] [Google Scholar]
- 12.Eraso P, Gancedo C. Activation of yeast plasma membrane ATPase by acid pH during growth. FEBS Lett. 1987;224:187–192. doi: 10.1016/0014-5793(87)80445-3. [DOI] [PubMed] [Google Scholar]
- 13.Forsby A, Andersson M, Lewan L, Sterner O. Structure-activity relationships for unsaturated dialdehydes, 4. The cytotoxicity of 22 sesquiterpenoid unsaturated dialdehydes, as determined by the neutral red absorption assay and by protein determination. Toxicol In Vitro. 1991;5:9–14. doi: 10.1016/0887-2333(91)90043-d. [DOI] [PubMed] [Google Scholar]
- 14.Forsby A, Nathanson M, Hasanvan H, Walum E. Polygodial inhibits carbachol-induced muscarinic acetylcholine receptor signal transduction. Toxicol In Vitro. 1998;12:585–585. doi: 10.1016/s0887-2333(98)00041-1. [DOI] [PubMed] [Google Scholar]
- 15.Fox T D, Folley L S, Mulero J J, McMullin T W, Thorsness P E, Hedin L O, Costanzo M C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 16.Franzblau S G, Witzig R S, McLaughlin J C, Torres P, Madico G, Hernandez A, Degnan M T, Cook M B, Quenzer V K, Ferguson R M, Gilman R H. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosisisolates by using the microplate Alamar Blue assay. J Clin Microbiol. 1998;36:362–366. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gawaz M, Douglas M G, Klingenberg M. Structure-function studies of adenine nucleotide transport in mitochondria. II. Biochemical analysis of distinct AAC1 and AAC2proteins in yeast. J Biol Chem. 1990;265:14202–14208. [PubMed] [Google Scholar]
- 18.Georgopapadakou N H, Walsh T J. Human mycoses: drugs and targets for emerging pathogens. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 19.Goffeau A, Dufour J P. Plasma membrane ATPase from the yeast Saccharomyces cerevisiae. Methods Enzymol. 1988;157:528–533. doi: 10.1016/0076-6879(88)57101-x. [DOI] [PubMed] [Google Scholar]
- 20.Goodno C C. Myosin active-site trapping with vanadate ion. Methods Enzymol. 1982;85:116–123. doi: 10.1016/0076-6879(82)85014-3. [DOI] [PubMed] [Google Scholar]
- 21.Groll A H, De Lucca A J, Walsh T J. Emerging targets for the development of novel antifungal therapeutics. Trends Microbiol. 1998;6:117–124. doi: 10.1016/s0966-842x(97)01206-7. [DOI] [PubMed] [Google Scholar]
- 22.Hasek J, Streiblová E. Fluorescence microscopy methods. Methods Mol Biol. 1996;53:391–405. doi: 10.1385/0-89603-319-8:391. [DOI] [PubMed] [Google Scholar]
- 23.Haworth R S, Cragoe E J, Jr, Fliegel L. Amiloride and 5-(N-ethyl-N-isopropyl) amiloride inhibit medium acidification and glucose metabolism by the fission yeast Schizosaccharomyces pombe. Biochim Biophys Acta. 1993;1145:266–272. doi: 10.1016/0005-2736(93)90298-e. [DOI] [PubMed] [Google Scholar]
- 24.Haworth R S, Lemire B D, Crandall D, Cragoe E J, Jr, Fliegel L. Characterisation of proton fluxes across the cytoplasmic membrane of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1991;1098:79–89. [PubMed] [Google Scholar]
- 25.Jurgens T M, Hufford C D, Clark A M. The metabolism of muzigadial by microorganisms. Xenobiotica. 1992;22:569–577. doi: 10.3109/00498259209053120. [DOI] [PubMed] [Google Scholar]
- 26.Kubo I, Ganjian I. Insect antifeedant terpenes, hot-tasting to humans. Experientia. 1981;37:1063–1064. doi: 10.1007/BF02085009. [DOI] [PubMed] [Google Scholar]
- 27.Kubo I, Lee S H. Potentiation of antifungal activity of sorbic acid. J Agric Food Chem. 1998;46:4052–4055. [Google Scholar]
- 28.Kubo I, Lee Y W, Pettei M, Pilkiewicz F, Nakanishi K. Potent army worm antifeedants from the East African Warburgiaplants. Chem Comm. 1976;24:1013–1014. [Google Scholar]
- 29.Kubo I, Taniguchi M. Polygodial, an antifungal potentiator. J Nat Prod. 1988;51:22–29. doi: 10.1021/np50055a002. [DOI] [PubMed] [Google Scholar]
- 30.Lawson J E, Gawaz M, Klingenberg M, Douglas M G. Structure-function studies of adenine nucleotide transport in mitochondria. I. Construction and genetic analysis of yeast mutants encoding the ADP/ATP carrier protein of mitochondria. J Biol Chem. 1990;265:14195–14201. [PubMed] [Google Scholar]
- 31.Lee S H, Lee J R, Lunde C S, Kubo I. In vitro antifungal susceptibilities of Candida albicansand other fungal pathogens to polygodial, a sesquiterpene dialdehyde. Planta Med. 1999;65:204–208. doi: 10.1055/s-1999-13981. [DOI] [PubMed] [Google Scholar]
- 32.Liao R S, Rennie R P, Talbot J A. Assessment of the effect of amphotericin B on the vitality of Candida albicans. Antimicrob Agents Chemother. 1999;43:1034–1041. doi: 10.1128/aac.43.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madshus I H. Regulation of intracellular pH in eukaryotic cells. Biochem J. 1988;250:1–8. doi: 10.1042/bj2500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCallion R F, Cole A L, Walker J R L, Blunt J W, Munro M H G. Antibiotic compounds from New Zealand plants, II: polygodial, an anti-Candida agent from Pseudowintera colorata. Planta Med. 1982;44:134–138. doi: 10.1055/s-2007-971422. [DOI] [PubMed] [Google Scholar]
- 35.Monk B C, Kurtz M B, Marrinan J A, Perlin D S. Cloning and characterization of the plasma membrane H+-ATPase from Candida albicans. J Bacteriol. 1991;173:6826–6836. doi: 10.1128/jb.173.21.6826-6836.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monk B C, Perlin D S. Fungal plasma membrane proton pumps as promising new antifungal targets. Crit Rev Microbiol. 1994;20:209–223. doi: 10.3109/10408419409114555. [DOI] [PubMed] [Google Scholar]
- 37.Morales P, Andersson M, Lewan L, Sterner O. Structure-activity relationships for unsaturated dialdehydes 6. The mutagenic activity of 11 compounds in the V79/HGPRT assay. Mut Res. 1992;268:315–321. doi: 10.1016/0027-5107(92)90237-v. [DOI] [PubMed] [Google Scholar]
- 38.Morton R E, Evans T A. Modification of the bicinchoninic acid protein assay to eliminate lipid interference in determining lipoprotein protein content. Anal Biochem. 1992;204:332–334. doi: 10.1016/0003-2697(92)90248-6. [DOI] [PubMed] [Google Scholar]
- 39.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 40.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 41.Ramos S, Balbin M, Raposo M, Valle E, Pardo L A. The mechanism of intracellular acidification induced by glucose in Saccharomyces cerevisiae. J Gen Microbiol. 1989;135:2413–2422. doi: 10.1099/00221287-135-9-2413. [DOI] [PubMed] [Google Scholar]
- 42.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candidaspecies to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serrano R. Effect of ATPase inhibitors on the proton pump of respiratory-deficient yeast. Eur J Biochem. 1980;105:419–424. doi: 10.1111/j.1432-1033.1980.tb04516.x. [DOI] [PubMed] [Google Scholar]
- 44.Serrano R. H+-ATPase from plasma membranes of Saccharomyces cerevisiae and Avena sativaroots: purification and reconstitution. Methods Enzymol. 1988;157:533–544. doi: 10.1016/0076-6879(88)57102-1. [DOI] [PubMed] [Google Scholar]
- 45.Serrano R, Kielland-Brandt M C, Fink G R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature. 1986;319:689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- 46.Sternberg S. The emerging fungal threat. Science. 1994;266:1632–1634. doi: 10.1126/science.7702654. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi M, Adachi T, Oi S, Kimura A, Katsumura S, Isoe S, Kubo I. Structure-activity relationship of the Warburgiasesquiterpene dialdehydes. Agric Biol Chem. 1984;48:73–78. [Google Scholar]
- 48.Taniguchi M, Chapya A, Kubo I, Nakanishi K. Screening of East African plants for antimicrobial activity I. Chem Pharm Bull. 1978;26:2910–2913. [Google Scholar]
- 49.Taniguchi M, Yano Y, Motoba K, Tanaka T, Oi S, Haraguchi H, Hashimoto K, Kubo I. Polygodial-induced sensitivity to rifampicin and actinomycin D of Saccharomyces cerevisiae. Agric Biol Chem. 1988;52:1881–1884. [Google Scholar]
- 50.Taniguchi M, Yano Y, Tada E, Ikenishi K, Oi S, Haraguchi H, Hashimoto K, Kubo I. Mode of action of polygodial, an antifungal sesquiterpene dialdehyde. Agric Biol Chem. 1988;52:1409–1414. [Google Scholar]
- 51.Vallejo C G, Serrano R. Physiology of mutants with reduced expression of plasma membrane H+-ATPase. Yeast. 1989;5:307–319. doi: 10.1002/yea.320050411. [DOI] [PubMed] [Google Scholar]
- 52.van der Rest M E, Kamminga A H, Nakano A, Anraku Y, Poolman B, Konings W N. The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol Rev. 1995;59:304–322. doi: 10.1128/mr.59.2.304-322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warnock D W. Amphotericin B: an introduction. J Antimicrob Chemother. 1991;28(Suppl. B):27–38. doi: 10.1093/jac/28.suppl_b.27. [DOI] [PubMed] [Google Scholar]
- 54.White T C, Marr K A, Bowden R A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yano Y, Taniguchi M, Tada E, Tanaka T, Oi S, Haraguchi H, Hashimoto K, Kubo I. Potentiation by polygodial of the respiration inhibiting activity of maesanin in Candida utilis. Agric Biol Chem. 1989;53:1525–1530. [Google Scholar]
- 56.Yano Y, Taniguchi M, Tanaka T, Oi S, Kubo I. Protective effects of Ca2+ on cell membrane damage by polygodial in Saccharomyces cerevisiae. Agric Biol Chem. 1991;55:603–604. [Google Scholar]