Abstract
The spurious acquisition and optimization of a furin cleavage site in the SARS-CoV-2 spike protein is associated with increased viral transmission and disease, and has generated intense interest in the development and application of therapeutic furin inhibitors to thwart the COVID-19 pandemic. This review summarizes the seminal studies that informed current efforts to inhibit furin. These include the convergent efforts of endocrinologists, virologists, and yeast geneticists that, together, culminated in the discovery of furin. We describe the pioneering biochemical studies which led to the first furin inhibitors that were able to block the disease pathways which are broadly critical for pathogen virulence, tumor invasiveness, and atherosclerosis. We then summarize how these studies subsequently informed current strategies leading to the development of small-molecule furin inhibitors as potential therapies to combat SARS-CoV-2 and other diseases that rely on furin for their pathogenicity and progression.
Keywords: furin, proprotein convertase, insulin, cancer, atherosclerosis, anthrax, HIV-1, SARS-CoV-2, protease inhibitor, α1-PDX
1. Introduction
When I penned the first comprehensive review on furin 20 years ago [1], the USA was reeling from the deliberate dissemination of Bacillus anthracis spores through the mail, and Asia was in turmoil from the emergence of the highly pathogenic H5N1 avian influenza virus which was capable of infecting humans. Caught ill-prepared, the USA resorted to dispatching swarms of biohazard containment teams to cordon off contaminated areas, whereas the Hong Kong government was forced to extinguish all the poultry in the region to prevent further zoonotic transfer. Perplexed by these calamitous events, I rhetorically asked “what next?” [1]. The answer: SARS-CoV-2. Exploiting a devious strategy long used by other pathogenic viruses, SARS-CoV-2 acquired a consensus furin site in its spike proprotein, greatly increasing viral transmission [2]. The recognition of furin as a virulence factor for disparate microbial pathogens ranging from Bacillus anthracis to SARS-CoV-2, and as a player in diseases ranging from atherosclerosis to cancer spurred the development of furin inhibitors as a novel broad-based therapeutic strategy. The goal of this short review is to summarize the key discoveries that led to the identification of furin as the prototypic member of the proprotein convertase (PC) family—a nearly 70-year endeavor—and, in turn, the generation of the first furin inhibitors capable of attenuating furin-dependent disease pathways. These pioneering studies laid the foundation for the generation of additional design strategies leading to the development of peptidomimetic and small-molecule furin inhibitors.
2. A Diabetic Dog, Sterile Yeast, and a Pox Virus Led to the Identification of Furin
As with many avenues in biomedical research, the path to experimental furin inhibitors began in 1921 with the Nobel-prize-winning work of Fred Banting, a medical instructor at the University of Western Ontario (now Western University), and Charles Best, an undergraduate assistant in John Macleod’s lab at the University of Toronto, who together conducted the seminal experiment that led to the discovery of insulin [3,4]. Working under John Macleod’s supervision, they perfected a two-step pancreatectomy method that allowed them to rescue glucose homeostasis by injecting a diabetic dog with an islet extract isolated from a healthy donor dog. Aided by the expertise of James Collip, a biochemist from the University of Alberta, the team soon purified insulin from islets and demonstrated its effectiveness in treating type I diabetics. The large-scale production of insulin isolated from feedlot animals by Ely Lily revolutionized the treatment of diabetes [5].
During the 1940s, Fred Sanger at Cambridge University perfected methods necessary to sequence proteins [6,7]. The ready supply of insulin produced by the pharmaceutical industry provided Sanger with the ideal protein to test this methodology. In a series of studies that led to Sanger’s first Nobel Prize, his team reported the primary structure of insulin and discovered that it is composed of two peptide chains, that is, a 30-amino acid B chain and a 21-amino acid A chain, covalently linked by a pair of interchain disulfide bridges [8,9]. The importance of these studies cannot be overstated, as Sanger not only reported the first protein sequence, but also suggested that each protein would have its own arrangement of amino acids, thus marking the birth of molecular biology [5].
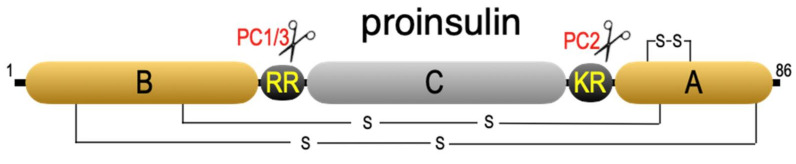
The question of how the two disulfide-linked insulin chains could assemble with such efficacy was puzzling. Attempts to join the two chains with the correct disulfide pairs failed miserably [10]. In 1967, Don Steiner at the University of Chicago solved this dilemma. Supplied with patient-derived β-cell tumors, Steiner conducted a pulse-chase study which demonstrated that insulin is synthesized as an approximately 10 kDa prohormone, proinsulin, which is subsequently converted to the disulfide-linked two-chain insulin hormone (Figure 1) [11]. Sequencing the proinsulin molecule revealed that the N-terminal B chain and C-terminal A chain sequences were joined by a connecting peptide (C-peptide) and flanked by doublets of basic amino acids (-ArgArg32- at the B/C junction and -LysArg65- at the C/A junction) [12,13].
Figure 1.
Proinsulin. Shown are the disulfide bonds and the sites cleaved by PC3 (PC1/3) and PC2.
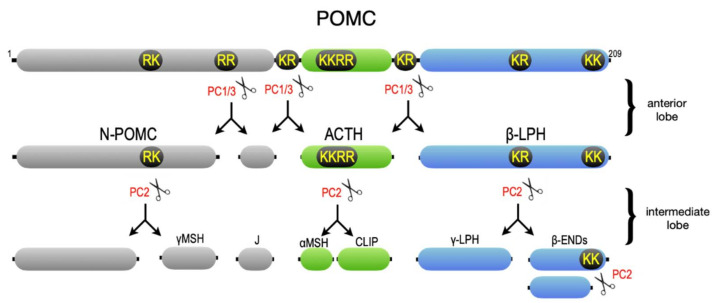
Intensive biochemical studies coupled with advances in cDNA cloning soon revealed that virtually all peptide hormones and neuropeptides are synthesized as prohormone molecules, many of which harbor sequences for multiple bioactive peptides, which are linked together by doublets or clusters of basic amino acids and are proteolytically excised in a tissue-specific manner [14,15]. One notable example is a study by Ed Herbert and his lab at the University of Oregon (and later at the Vollum Institute) that described the tissue-specific processing of POMC (proopiomelanocortin) in the pituitary gland (Figure 2) [16]. In the anterior lobe of the pituitary, POMC is processed at doublets of basic amino acids to generate ACTH (adrenocorticotropin), which stimulates cortisol release from the adrenal gland, and β-LPH (β-lipotropin), which was suggested to have lipolytic activity [17]. In the intermediate lobe, however, ACTH is cleaved at a cluster of basic amino acids to yield α-MSH (α-melanocyte-stimulating hormone), which modulates functions ranging from feeding to pigmentation. In addition, β-LPH is further cleaved at a doublet of basic amino acids to generate γ-LPH and β-endorphin, one of the principal opioid peptides. To ascertain the endoprotease “signatures” of these endocrine cells, Barbara Thorne in my lab analyzed a battery of POMC cleavage site mutants expressed in primary endocrine cells [18,19]. She suggested that just two endoproteases were broadly responsible for most prohormone processing steps in endocrine and neuroendocrine cells, including the activation of relatively simple prohormones such as insulin in the pancreas, as well as the tissue-specific processing of complex prohormones such as POMC in the pituitary.
Figure 2.
POMC. Shown are the processing steps that occur in the pituitary anterior lobe and neurointermediate lobe. The sites cut by proprotein convertases PC1/3 and PC2 are shown. For a recent review, see [20].
Understanding how diverse proproteins are activated required the identification of the proteases that catalyze these reactions. Capitalizing on methodologies developed by Sanger, Don Steiner found that exposure of proinsulin to mild trypsin digestion excised the mature insulin hormone. This led to the suggestion that proinsulin is processed in the mildly acidic late secretory pathway compartments of the cell by the sequential activities of a trypsin-like endoprotease, frequently followed by trimming of the residual basic amino acids by a carboxypeptidase B-like activity to generate the mature peptide [21]. For the next quarter-century, numerous researchers sought to isolate the relevant endoproteases by conventional biochemical methods. A visit to the local abattoir and days in the cold room fractionating tissues yielded an overwhelming number of candidate activities, each capable of cutting protein or peptide substrates at basic amino acids under mildly acidic conditions. Unfortunately, these studies were lacking in a genetic approach to conclusively identify the bona fide converting endoproteases. As John Hutton (then at Cambridge University) lamented, “many candidates have been put forward—and eventually shot down” [22].
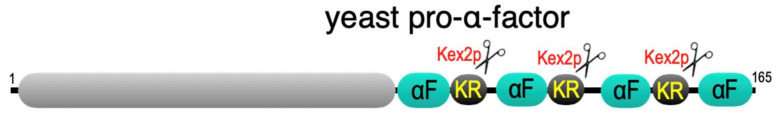
The first breakthrough using a genetic approach was made by Ira Herskowitz’s lab, then at the University of Oregon, who found that, similar to many mammalian neuropeptide precursors, the yeast mating pheromone, α-factor, is encoded by a tandem of four copies of the tridecapeptide encased in a larger prohormone, pro-α-mating factor [23]. Each α-factor sequence is flanked on one side by a linker segment containing a -LysArg- doublet of basic amino acids, suggesting that yeast may express a prohormone processing endoprotease (Figure 3). Jeremy Thorner’s lab at UC Berkeley capitalized on these findings and demonstrated that yeast Kex2 encodes a protease required for cleavage of the -LysArg- sites in pro-α-mating factor and pro-killer toxin [24]. They also raised the possibility that the Kex2 protein (Kex2p) may be the prototype for the mammalian prohormone convertases.
Figure 3.
Yeast pro-α-mating factor. Shown are the four cryptic α-factor (αF) peptides and the flanking -LysArg- cleavage sites, which are processed by Kexp2.
Leveraging advances in viral vector technology, we showed that, when expressed in mammalian cells, yeast Kex2p can correctly process POMC to sets of peptides generated in the pituitary gland (including γ-LPH and β-endorphin), demonstrating that Kex2p is functionally similar, and thus likely structurally similar, to the long-sought-after mammalian proprotein convertases [25,26]. A database search conducted by Robert Fuller, now at the University of Michigan, identified the first such human Kex2p homologue, an obscure open reading frame coined “furin” [27]. The Van de Ven team had serendipitously discovered the furin locus in 1986, designated fur (fes/fps upstream region), which they predicted would encode a membrane receptor [28]. In a series of papers beginning in 1990, Sean Molloy and Pat Bresnahan in my lab reported that furin is a trans-Golgi network (TGN) membrane-localized, calcium-dependent serine endoprotease that cuts the neurotrophin precursor pro-β-NGF at the multibasic site -Arg-X-Lys/Arg-Arg↓, which is a decisive step modulating neuron survival [29,30,31,32]. The number of proproteins identified as being cleaved by furin now exceeds 150 [33,34]. These range from TGF-β family members essential for embryogenesis to receptors, cell adhesion proteins, serum proteins, and other proteases involved in disease pathways ranging from cancer to cardiovascular disease, as well as a number of microbial proproteins critical for pathogen virulence [1,33,35,36,37,38].
The identification of furin as the first human proprotein convertase guided PCR strategies to identify additional members of the PC family [39]. As predicted by our earlier work [18,19], Laurel Thomas and Barbara Thorne, in collaboration with Don Steiner’s lab, demonstrated that PC2 and PC3 (also called PC1) are a common core of neuroendocrine proprotein convertases that cleave POMC to the complex sets of peptides found in the pituitary as well as proinsulin to insulin [40,41] (for a detailed description of the proprotein convertase family see [1,42]).
3. Furin Essentials
Furin’s broad role in proprotein activation is attributed to changes in its gene expression, as well as its highly regulated intracellular trafficking itinerary. For example, elevated furin expression negatively correlates with disease outcome in several cancers, increasing metastasis while reducing immune-cell infiltration [1,37,43,44]. Bioinformatic analyses identified FURIN polymorphisms as risk factors for diabetes, cardiovascular disease, obesity, and all-cause mortality [45,46,47]. Importantly, these furin-associated risk factors, together with furin’s key role in SARS-CoV-2 pathogenesis (as described below), may underpin the increased vulnerability of susceptible patients (including obese and diabetic patients) to negative outcomes from COVID-19 [48,49].
In cells, furin localizes to the TGN and traffics between this processing compartment and two other processing compartments: the cell surface and early endosomes [1]. In the TGN/biosynthetic pathway, furin activates many substrates, including TGF-βs, receptors, and viral envelope glycoproteins. Cell-surface furin activates cellular proteins involved in cell migration and tumor metastasis as well as several pathogen proteins, including anthrax protective antigen (PA), proaerolysin, and Clostridium septicum α-toxin [1,50,51]. The mildly acidic pH of early endosomal compartments enables profurin to complete its own autoproteolytic activation steps in a compartment-specific manner and is exploited by pathogens to activate A/B-type bacterial toxins, including Pseudomonas exotoxin A and Shiga toxin [1,52,53]. The trafficking of furin between these various processing compartments is mediated by sequences in its cytoplasmic domain, which bind several clathrin adaptors (i.e., AP-1, AP-2, and AP-4) and contain a pair of serine residues that are reversibly phosphorylated by the actions of protein kinase CK2 and specific isoforms of protein phosphatase 2A (PP2A) [1,54,55]. Phosphorylated furin binds the sorting protein PACS-1, which localizes furin to either the TGN or to a peripheral cycling loop between early endosomes and the cell surface [56,57]. Movement from early endosomes to the TGN requires the PP2A-dependent dephosphorylation of furin [55]. Furin molecules localized to the cell surface are tethered by the actin-binding protein filamin-A (ABP-280) [58]. Interestingly, in cancer cells hypoxia induces both furin expression and its redistribution to the plasma membrane, where it tethers to filamin-A and activates MT1-MMP/MM2P-dependent cell invasion [50,51].
Systematic analyses of the furin-dependent cleavage of two bona fide furin substrates (i.e., anthrax PA and virulent H5N8 influenza virus HA proteins), each containing defined amino acid changes surrounding their cleavage sites, identified -Arg-X-X-Arg-↓ as the minimal site required for efficient processing by furin [30,59,60,61]. Subsequent crystallography and enzymology studies reinforced these findings, identifying the residues in the catalytic domain that bind the critical Arg residues and demonstrating that favorable amino acids at the P2 and P6 subsites can compensate for less-favorable ones at P4 [62,63].
4. Pathogenic Viruses from Bird Flu to SARS-CoV-2 Acquire Furin Cleavage Sites to Increase Tropism
Pioneering studies in the 1980s by Robert Webster at St. Jude Research and Hans-Dieter Klenk at the University of Marburg converged on furin as a key factor regulating viral tropism [64]. They determined that the pathogenicity of avian influenza viruses correlated with the cleavability of its fusion protein precursor HA0 to generate the fusion-competent HA1–HA2 complex. Avirulent avian influenza viruses, which lack a consensus furin site in HA0, cause a localized infection in the intestinal tract. Mutation of the HA0 cleavage site to a consensus furin site enables the virus to be activated by ubiquitously expressed furin, allowing the virus to spread systemically throughout the bird [65]. Analysis of the H5N1 influenza virus, which caused the deadly flu outbreak in Hong Kong in 1997, revealed that just two mutations were required to generate the lethal virus, including the generation of a tandem furin site in the cleavage junction between HA1 and HA2 (-ArgGluArgArgArgLysLysArg-↓), which increases cleavability [66]. Increases in viral tropism resulting from the acquisition of a furin site have been reported for other pathogenic viruses [1,36].
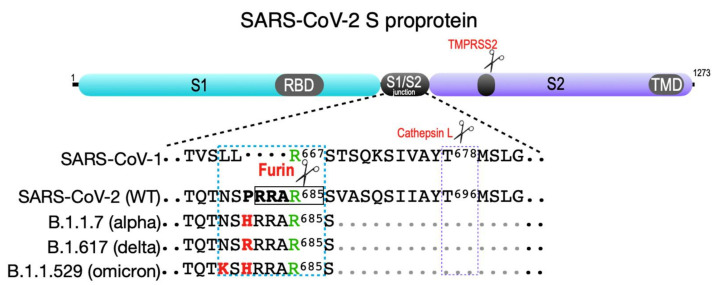
SARS-CoV-2 illustrates in real-time how acquiring and then optimizing the furin cleavage site can have devastating consequences on morbidity and mortality. Both SARS-CoV and SARS-CoV-2 rely on the cleavage of their spike (S) proproteins by the cell surface protease TMPRSS2 to expose the receptor-binding domain (RBD) that contacts the primary virus receptor, ACE2 (Figure 4). However, TMPRSS2 cleavage does not fully expose the SARS-CoV-2 RBD [67]. To overcome this block, SARS-CoV-2 acquired a suboptimal furin site (-TNSP681RRAR↓S686-(furin cleavage site is underlined)) at the SARS-CoV-2 S1/S2 junction, which directs furin-dependent cleavage of the S proprotein to fully unmask the RBD, allowing it to efficiently bind ACE2 [67]. In addition, the exposed basic amino acids at the cleaved S1 C-terminus are not removed by a carboxypeptidase but instead bind the co-receptor neuropilin-1, further augmenting viral transmission [68,69]. The B.1.1.7 (alpha) variant contains a P681 → H change (-TNRH681RRAR↓S686-), which increases cleavability by furin [70,71]. This strategically placed His residue likely becomes protonated following the transit of the S protein to late secretory pathway compartments, providing a positive charge that triggers cleavage by furin in endosomal compartments [53]. The highly transmissible B.1.617.2 (delta) variant contains a P681 → R change (-TNSR681RRAR↓S686-), creating a permanent positive charge at residue 681 and further increasing furin-dependent infectivity, possibly in multiple cellular compartments [70,71]. Intriguingly, the recently described B.1.1.529 (omicron) variant appears to have “doubled-down” on the endosomal processing by furin through the addition of both the P681 → H and N679 → K changes (-TK679RH681RRAR↓S686-). This double change reinstates reliance on the pH-sensitive His681 as initially observed with B.1.1.7, but likely further increases cleavage efficiency in endosomes through the addition of the positive charge at Lys679—a nefarious mimicry of the efficacious compartment-specific autoactivation pathway employed by furin [53]. These possibilities await experimental testing, and may illuminate the endomembrane itinerary used by SARS-CoV-2 to optimize or limit virus assembly and transmissibility. Intuitively, one would predict that an Ala684 → Arg change (-TNSPRRR684R↓S686-) would maximize transmission efficiency. While this change increases syncytia formation in vitro, it nonetheless impedes virus entry [72]. Similar findings were reported for HIV-1 gp160, which also maintains an unfavorable acidic amino acid at the P3 site (-ArgGluLysArg-↓) [73]. Thus, maintenance of a suboptimal furin site may best support subsequent conformational changes in an envelope-protein-specific manner for optimal infectivity.
Figure 4.
SARS-CoV-2 spike protein. Shown are the S1 and S2 segments in the SARS family S proprotein that flank the S1/S2 cleavage site junction as well as the ACE2 receptor-binding domain (RBD) in S1 and the transmembrane domain (TMD) in S2. The S2′ TMPRSS2 cleavage site is common to all SARS coronaviruses (violet box). The SARS-CoV-1 S1/S2 junction is cut by cathepsin L at Thr678 (see [74]). SARS-CoV-2 contains a four-amino-acid insertion (PRAA684), which converts the trypsin-sensitive Arg685 residue (green) to the P1 site cut by furin (RRAR685, boxed). The cyan box also shows the B.1.1.7 (alpha variant) furin site containing the P681 → H change; the more transmissible B.1.617.2 (delta variant) furin site, which contains the P681 → R change; and the recently reported B.1.1.529 (omicron variant), which contains both P681 → H and N679 → K changes.
5. The First-Generation Furin Inhibitors
The identification of furin as the principal cellular endoprotease that cleaves viral envelope glycoproteins prompted the development of furin inhibitors able to block viral pathogenicity. These initial studies relied on two markedly different strategies [75,76]. Elliot Shaw at the Friedrich Miescher Institute, together with Hans-Dieter Klenk, developed an irreversible peptidyl chloromethyl ketone (CMK) active site furin inhibitor. Their strategy was based on Shaw’s work from the 1960s which demonstrated that attaching the substrate P1 amino acid to the CMK warhead produced inhibitors selective for different classes of serine proteases by reacting with the active site histidine: Tos-Phe-CH2Cl (TPCK) inhibits chymotrypsin and Tos-Lys-CH2Cl (TLCK) inhibits trypsin [77,78,79]. Incorporating furin’s consensus cleavage sequence, Shaw generated furin-directed CMKs (e.g., dec-Arg-Glu-Lys-Arg-CH2Cl) [76]. A decanoyl group was added to increase membrane permeability. Treatment of cells with dec- Arg-Glu-Lys-Arg -CMK blocked the processing of HIV-1 gp160 and the production of infectious HIV-1 as well as several other pathogenic viruses, including paramyxovirus, pathogenic avian influenza viruses, and SARS-CoV-2 [60,76,80,81]. Unfortunately, the furin-directed CMKs lack specificity and are low nanomolar inhibitors of all proprotein convertases, reducing their utility [82].
In an alternative strategy, my lab engineered a variant of the serpin α1-antitrypsin that is highly selective for furin called α1-antitrypsin Portland (α1-PDX) [75]. Circulating α1-antitrypsin (α1-AT) is the primary inhibitor of neutrophil elastase, protecting the lungs during the acute-phase response to tissue injury. Mechanistically, α1-AT functions as a suicide substrate [83]. This globular serpin contains a metastable 19-amino acid reactive center loop (RCL) that corresponds to the P15-P4′ residues spanning the target enzyme cleavage site. The formation of the protease—α1-AT acyl intermediate invokes a massive conformational rearrangement of the RCL, which traps the bound protease in a stable complex. The RCL can be engineered to have high target specificity, as demonstrated by a unique case in the 1970s, where a boy from Pittsburgh was diagnosed with a fatal posttraumatic bleeding disorder caused by a Met358 → Arg mutation at the P1 site in the α1-AT RCL (Ala-Ile-Pro-Met358 → Ala-Ile-Pro-Arg358) [84,85]. This single amino acid change in this α1-AT mutant, called α1-antitrypsin Pittsburgh (α1-PIT), had two profound effects. It switched the serpin from an elastase inhibitor to a potent thrombin inhibitor, explaining the fatal acute-phase bleeding disorder and highlighting the target specificity that can be accommodated by the RCL. In addition, α1-PIT was associated with circulating proalbumin, which requires cleavage at an -ArgArg- site, leading the authors to speculate that α1-PIT encounters and inhibits the proalbumin convertase during transport through the hepatocyte secretory pathway [86].
Eric Anderson and Francois Jean, now at the University of British Columbia, determined that α1-PIT was a weak inhibitor of furin [75,82]. However, changing the RCL to the minimal Arg-X-X-Arg consensus furin site (Ala-Ile-Pro-Arg358 → Arg-Ile-Pro-Arg358) generated an engineered serpin, α1-PDX, which is a highly selective subnanomolar inhibitor of furin, which no longer recognizes elastase or thrombin. A1-PDX can be expressed from the nucleus, or it can be generated in bacteria and either applied to cells in vitro or systemically delivered to mice. Using these delivery approaches, α1-PDX blocks the furin-dependent activation of several pathogenic viruses, including HIV-1, measles virus, and HCMV, as well as the furin-dependent activation of Pseudomonas exotoxin A [82,87,88]. α1-PDX is effective in cancer models, inhibiting tumor cell invasiveness in vitro and tumor metastasis in vivo [43,89,90,91]. Finally, systemic administration of the recombinant α1-PDX reduced atherosclerotic progression in vivo, in part by inhibiting the furin-dependent activation of MT1-MMP/MMP2 [92]. The primary obstacle to the development of α1-PDX, or any form of α1-AT, as a potential therapeutic has been the lack of an expression system that can generate the physiologically stable α1-AT protein. However, new advances in formulating α1-AT and its variants have overcome this challenge [93]. Together with recent work suggesting that donor-purified α1-AT inhibits the TMPRSS2-dependent cell entry of SARS-CoV-2 [94], these advancements raise the exciting possibility that combinations of recombinant α1-AT and α1-PDX may be a potential treatment for SARS-CoV-2 [93,95].
6. The Promise of Small-Molecule Furin Inhibitors as Broad-Based Therapeutics
The successes achieved by using peptidyl CMKs or α1-PDX to inhibit furin have guided additional approaches toward the development of therapeutic furin inhibitors [96]. Leveraging Elliott Shaw’s success with peptidyl CMKs and the furin crystal structure, several cell-penetrant peptide inhibitors, including substrate-based inhibitors containing the H5N1 cleavage site and poly-D-arginine-based peptides, have been developed and shown to prevent the processing of anthrax PA in cellulo and to protect mice from anthrax toxemia in vivo [97,98,99,100,101]. In addition, structure–activity relationship (SAR) analyses have been performed to improve molecular stability and cell penetrance, yielding various peptidomimetic inhibitors containing a decarboxylated P1 Arg, a replacement of the P2 and P4 Arg by canavanine, an addition of P5 Arg mimetics, or the addition of azaβ3 moieties to the N- and C-termini, and leading to nanomolar furin inhibitors that prevent pathogen activation [102,103,104,105,106,107]. Importantly, the discovery of small-molecule furin inhibitors, notably a series of 2,5-dideoxystreptamine derivatives with nanomolar potency that protect cells from anthrax PA toxicity, represents a critical next step in the realization of therapeutic furin inhibitors [108,109,110]. A recent study suggesting the bioavailable small-molecule furin inhibitor BOS-981 blocks cleavage of the SARS-CoV-2 S1/S2 site and reduces viral titer further supports this approach [111]. The next few years should reveal which of these approaches may lead us closer to the realization of a broad-based therapeutic that selectively targets furin to combat multiple diseases.
Acknowledgments
We would like to thank T. Brosenitsch, S. Villar-Pazos, and Y. Yang for critically reading the manuscript.
Author Contributions
Conceptualization, G.T.; writing—original draft preparation, G.T.; writing—review and editing, G.T., F.C. and A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by NIH grants DK114855 and NS123649 (to G.T.) and FRQNT grant 291453 (to F.C.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thomas G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell. Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson B.A., Xie X., Bailey A.L., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best C.H. The internal secretion of the pancreas. J. Am. Med. Assoc. 1935;105:270–274. doi: 10.1001/jama.1935.92760300002008. [DOI] [Google Scholar]
- 4.Rosenfeld L. Insulin: Discovery and controversy. Clin. Chem. 2002;48:2270–2288. doi: 10.1093/clinchem/48.12.2270. [DOI] [PubMed] [Google Scholar]
- 5.Ward C.W., Lawrence M.C. Landmarks in insulin research. Front. Endocrinol. 2011;2:76. doi: 10.3389/fendo.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanger F. The free amino groups of insulin. Biochem. J. 1945;39:507–515. doi: 10.1042/bj0390507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanger F. Sequences, sequences, and sequences. Annu. Rev. Biochem. 1988;57:1–28. doi: 10.1146/annurev.bi.57.070188.000245. [DOI] [PubMed] [Google Scholar]
- 8.Ryle A.P., Sanger F., Smith L.F., Kitai R. The disulphide bonds of insulin. Biochem. J. 1955;60:541–556. doi: 10.1042/bj0600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanger F. Chemistry of insulin; determination of the structure of insulin opens the way to greater understanding of life processes. Science. 1959;129:1340–1344. doi: 10.1126/science.129.3359.1340. [DOI] [PubMed] [Google Scholar]
- 10.Cahill G.F., Jr. Insulin and proinsulin. N. Engl. J. Med. 1970;283:762. doi: 10.1056/NEJM197010012831412. [DOI] [PubMed] [Google Scholar]
- 11.Steiner D.F., Cunningham D., Spigelman L., Aten B. Insulin biosynthesis: Evidence for a precursor. Science. 1967;157:697–700. doi: 10.1126/science.157.3789.697. [DOI] [PubMed] [Google Scholar]
- 12.Clark J.L., Steiner D.F. Insulin biosynthesis in the rat: Demonstration of two proinsulins. Proc. Natl. Acad. Sci. USA. 1969;62:278–285. doi: 10.1073/pnas.62.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chance R.E., Ellis R.M., Bromer W.W. Porcine proinsulin: Characterization and amino acid sequence. Science. 1968;161:165–167. doi: 10.1126/science.161.3837.165. [DOI] [PubMed] [Google Scholar]
- 14.Douglass J., Civelli O., Herbert E. Polyprotein gene expression: Generation of diversity of neuroendocrine peptides. Annu. Rev. Biochem. 1984;53:665–715. doi: 10.1146/annurev.bi.53.070184.003313. [DOI] [PubMed] [Google Scholar]
- 15.Bicknell A.B. The tissue-specific processing of pro-opiomelanocortin. J. Neuroendocrinol. 2008;20:692–699. doi: 10.1111/j.1365-2826.2008.01709.x. [DOI] [PubMed] [Google Scholar]
- 16.Herbert E., Uhler M. Biosynthesis of polyprotein precursors to regulatory peptides. Cell. 1982;30:1–2. doi: 10.1016/0092-8674(82)90002-2. [DOI] [PubMed] [Google Scholar]
- 17.Richter W.O., Schwandt P. Lipolytic potency of proopiomelanocorticotropin peptides in vitro. Neuropeptides. 1987;9:59–74. doi: 10.1016/0143-4179(87)90033-3. [DOI] [PubMed] [Google Scholar]
- 18.Thorne B.A., Thomas G. An in vivo characterization of the cleavage site specificity of the insulin cell prohormone processing enzymes. J. Biol. Chem. 1990;265:8436–8443. doi: 10.1016/S0021-9258(19)38907-0. [DOI] [PubMed] [Google Scholar]
- 19.Thorne B.A., Viveros O.H., Thomas G. Expression and processing of mouse proopiomelanocortin in bovine adrenal chromaffin cells. A model system to study tissue-specific prohormone processing. J. Biol. Chem. 1991;266:13607–13615. doi: 10.1016/S0021-9258(18)92743-2. [DOI] [PubMed] [Google Scholar]
- 20.Harno E., Gali Ramamoorthy T., Coll A.P., White A. POMC: The Physiological Power of Hormone Processing. Physiol. Rev. 2018;98:2381–2430. doi: 10.1152/physrev.00024.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubenstein A.H., Steiner D.F. Proinsulin. Annu. Rev. Med. 1971;22:1–18. doi: 10.1146/annurev.me.22.020171.000245. [DOI] [PubMed] [Google Scholar]
- 22.Marx J. How peptide hormones get ready for work. Science. 1991;252:779–780. doi: 10.1126/science.1674172. [DOI] [PubMed] [Google Scholar]
- 23.Kurjan J., Herskowitz I. Structure of a yeast pheromone gene (MF alpha): A putative alpha-factor precursor contains four tandem copies of mature alpha-factor. Cell. 1982;30:933–943. doi: 10.1016/0092-8674(82)90298-7. [DOI] [PubMed] [Google Scholar]
- 24.Julius D., Brake A., Blair L., Kunisawa R., Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- 25.Thomas G., Herbert E., Hruby D.E. Expression and cell type—Specific processing of human preproenkephalin with a vaccinia recombinant. Science. 1986;232:1641–1643. doi: 10.1126/science.3754979. [DOI] [PubMed] [Google Scholar]
- 26.Thomas G., Thorne B.A., Thomas L., Allen R.G., Hruby D.E., Fuller R., Thorner J. Yeast KEX2 endopeptidase correctly cleaves a neuroendocrine prohormone in mammalian cells. Science. 1988;241:226–230. doi: 10.1126/science.3291117. [DOI] [PubMed] [Google Scholar]
- 27.Fuller R.S., Brake A.J., Thorner J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science. 1989;246:482–486. doi: 10.1126/science.2683070. [DOI] [PubMed] [Google Scholar]
- 28.Roebroek A.J., Schalken J.A., Bussemakers M.J., van Heerikhuizen H., Onnekink C., Debruyne F.M., Bloemers H.P., Van de Ven W.J. Characterization of human c-fes/fps reveals a new transcription unit (fur) in the immediately upstream region of the proto-oncogene. Mol. Biol. Rep. 1986;11:117–125. doi: 10.1007/BF00364823. [DOI] [PubMed] [Google Scholar]
- 29.Bresnahan P.A., Leduc R., Thomas L., Thorner J., Gibson H.L., Brake A.J., Barr P.J., Thomas G. Human fur gene encodes a yeast KEX2-like endoprotease that cleaves pro-beta-NGF in vivo. J. Cell Biol. 1990;111:2851–2859. doi: 10.1083/jcb.111.6.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molloy S.S., Bresnahan P.A., Leppla S.H., Klimpel K.R., Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 1992;267:16396–16402. doi: 10.1016/S0021-9258(18)42016-9. [DOI] [PubMed] [Google Scholar]
- 31.Molloy S.S., Thomas L., VanSlyke J.K., Stenberg P.E., Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: Localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee R., Kermani P., Teng K.K., Hempstead B.L. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- 33.Osman E.E.A., Rehemtulla A., Neamati N. Why All the Fury over Furin? J. Med. Chem. 2021;65:2747–2784. doi: 10.1021/acs.jmedchem.1c00518. [DOI] [PubMed] [Google Scholar]
- 34.Tian S., Huang Q., Fang Y., Wu J. FurinDB: A database of 20-residue furin cleavage site motifs, substrates and their associated drugs. Int. J. Mol. Sci. 2011;12:1060–1065. doi: 10.3390/ijms12021060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren K., Jiang T., Zheng X.L., Zhao G.J. Proprotein convertase furin/PCSK3 and atherosclerosis: New insights and potential therapeutic targets. Atherosclerosis. 2017;262:163–170. doi: 10.1016/j.atherosclerosis.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Braun E., Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019;8:e1073. doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaaks P., Bernasconi M. The proprotein convertase furin in tumour progression. Int. J. Cancer. 2017;141:654–663. doi: 10.1002/ijc.30714. [DOI] [PubMed] [Google Scholar]
- 38.Roebroek A.J., Umans L., Pauli I.G., Robertson E.J., van Leuven F., Van de Ven W.J., Constam D. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 1998;125:4863–4876. doi: 10.1242/dev.125.24.4863. [DOI] [PubMed] [Google Scholar]
- 39.Steiner D.F., Smeekens S.P., Ohagi S., Chan S.J. The new enzymology of precursor processing endoproteases. J. Biol. Chem. 1992;267:23435–23438. doi: 10.1016/S0021-9258(18)35852-6. [DOI] [PubMed] [Google Scholar]
- 40.Smeekens S.P., Montag A.G., Thomas G., Albiges-Rizo C., Carroll R., Benig M., Phillips L.A., Martin S., Ohagi S., Gardner P., et al. Proinsulin processing by the subtilisin-related proprotein convertases furin, PC2, and PC3. Proc. Natl. Acad. Sci. USA. 1992;89:8822–8826. doi: 10.1073/pnas.89.18.8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas L., Leduc R., Thorne B.A., Smeekens S.P., Steiner D.F., Thomas G. Kex2-like endoproteases PC2 and PC3 accurately cleave a model prohormone in mammalian cells: Evidence for a common core of neuroendocrine processing enzymes. Proc. Natl. Acad. Sci. USA. 1991;88:5297–5301. doi: 10.1073/pnas.88.12.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seidah N.G., Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 2012;11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 43.Bassi D.E., Lopez De Cicco R., Mahloogi H., Zucker S., Thomas G., Klein-Szanto A.J. Furin inhibition results in absent or decreased invasiveness and tumorigenicity of human cancer cells. Proc. Natl. Acad. Sci. USA. 2001;98:10326–10331. doi: 10.1073/pnas.191199198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He Z., Khatib A.M., Creemers J.W.M. Loss of the proprotein convertase Furin in T cells represses mammary tumorigenesis in oncogene-driven triple negative breast cancer. Cancer Lett. 2020;484:40–49. doi: 10.1016/j.canlet.2020.05.001. [DOI] [PubMed] [Google Scholar]
- 45.Consortium C.A.D., Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T.L., Thompson J.R., Ingelsson E., Saleheen D., Erdmann J., et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fitzgerald K. Furin Protease: From SARS CoV-2 to Anthrax, Diabetes, and Hypertension. Perm. J. 2020;24:20.187. doi: 10.7812/TPP/20.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh S., Vivar J., Nelson C.P., Willenborg C., Segre A.V., Makinen V.P., Nikpay M., Erdmann J., Blankenberg S., O’Donnell C., et al. Systems Genetics Analysis of Genome-Wide Association Study Reveals Novel Associations Between Key Biological Processes and Coronary Artery Disease. Arterioscler. Thromb. Vasc. Biol. 2015;35:1712–1722. doi: 10.1161/ATVBAHA.115.305513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drucker D.J. Diabetes, obesity, metabolism, and SARS-CoV-2 infection: The end of the beginning. Cell Metab. 2021;33:479–498. doi: 10.1016/j.cmet.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su Y., Yuan D., Chen D.G., Ng R.H., Wang K., Choi J., Li S., Hong S., Zhang R., Xie J., et al. Multiple Early Factors Anticipate Post-Acute COVID-19 Sequelae. Cell. 2022;185:881–895.e20. doi: 10.1016/j.cell.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arsenault D., Lucien F., Dubois C.M. Hypoxia enhances cancer cell invasion through relocalization of the proprotein convertase furin from the trans-Golgi network to the cell surface. J. Cell Physiol. 2012;227:789–800. doi: 10.1002/jcp.22792. [DOI] [PubMed] [Google Scholar]
- 51.McMahon S., Grondin F., McDonald P.P., Richard D.E., Dubois C.M. Hypoxia-enhanced expression of the proprotein convertase furin is mediated by hypoxia-inducible factor-1: Impact on the bioactivation of proproteins. J. Biol. Chem. 2005;280:6561–6569. doi: 10.1074/jbc.M413248200. [DOI] [PubMed] [Google Scholar]
- 52.Anderson E.D., VanSlyke J.K., Thulin C.D., Jean F., Thomas G. Activation of the furin endoprotease is a multiple-step process: Requirements for acidification and internal propeptide cleavage. EMBO J. 1997;16:1508–1518. doi: 10.1093/emboj/16.7.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feliciangeli S.F., Thomas L., Scott G.K., Subbian E., Hung C.H., Molloy S.S., Jean F., Shinde U., Thomas G. Identification of a pH sensor in the furin propeptide that regulates enzyme activation. J. Biol. Chem. 2006;281:16108–16116. doi: 10.1074/jbc.M600760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones B.G., Thomas L., Molloy S.S., Thulin C.D., Fry M.D., Walsh K.A., Thomas G. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 1995;14:5869–5883. doi: 10.1002/j.1460-2075.1995.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molloy S.S., Thomas L., Kamibayashi C., Mumby M.C., Thomas G. Regulation of endosome sorting by a specific PP2A isoform. J. Cell Biol. 1998;142:1399–1411. doi: 10.1083/jcb.142.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molloy S.S., Anderson E.D., Jean F., Thomas G. Bi-cycling the furin pathway: From TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 1999;9:28–35. doi: 10.1016/S0962-8924(98)01382-8. [DOI] [PubMed] [Google Scholar]
- 57.Wan L., Molloy S.S., Thomas L., Liu G., Xiang Y., Rybak S.L., Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/S0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 58.Liu G., Thomas L., Warren R.A., Enns C.A., Cunningham C.C., Hartwig J.H., Thomas G. Cytoskeletal protein ABP-280 directs the intracellular trafficking of furin and modulates proprotein processing in the endocytic pathway. J. Cell Biol. 1997;139:1719–1733. doi: 10.1083/jcb.139.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klimpel K.R., Molloy S.S., Thomas G., Leppla S.H. Anthrax toxin protective antigen is activated by a cell surface protease with the sequence specificity and catalytic properties of furin. Proc. Natl. Acad. Sci. USA. 1992;89:10277–10281. doi: 10.1073/pnas.89.21.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stieneke-Grober A., Vey M., Angliker H., Shaw E., Thomas G., Roberts C., Klenk H.D., Garten W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker J.A., Molloy S.S., Thomas G., Sakaguchi T., Yoshida T., Chambers T.M., Kawaoka Y. Sequence specificity of furin, a proprotein-processing endoprotease, for the hemagglutinin of a virulent avian influenza virus. J. Virol. 1994;68:1213–1218. doi: 10.1128/jvi.68.2.1213-1218.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henrich S., Cameron A., Bourenkov G.P., Kiefersauer R., Huber R., Lindberg I., Bode W., Than M.E. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat. Struct. Biol. 2003;10:520–526. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- 63.Krysan D.J., Rockwell N.C., Fuller R.S. Quantitative characterization of furin specificity. Energetics of substrate discrimination using an internally consistent set of hexapeptidyl methylcoumarinamides. J. Biol. Chem. 1999;274:23229–23234. doi: 10.1074/jbc.274.33.23229. [DOI] [PubMed] [Google Scholar]
- 64.Webster R.G., Rott R. Influenza virus A pathogenicity: The pivotal role of hemagglutinin. Cell. 1987;50:665–666. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- 65.Zambon M.C. The pathogenesis of influenza in humans. Rev. Med. Virol. 2001;11:227–241. doi: 10.1002/rmv.319. [DOI] [PubMed] [Google Scholar]
- 66.Hatta M., Gao P., Halfmann P., Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 67.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Anton-Plagaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lubinski B., Frazier L.E., Phan M.V.T., Bugembe D.L., Tang T., Daniel S., Cotton E., Jaimes J.A., Whittaker G.R. Spike protein cleavage-activation mediated by the SARS-CoV-2 P681R mutation: A case-study from its first appearance in variant of interest (VOI) A.23.1 identified in Uganda. bioRxiv. 2021 doi: 10.2139/ssrn.3966642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peacock T.P., Sheppard C.M., Brown J.C., Goonawardane N., Zhou J., Whiteley M., Consortium P.V., de Silva T.I., Barclay W.S. The SARS-CoV-2 variants associated with infections in India, B.1.617, show enhanced spike cleavage by furin. bioRxiv. 2021 doi: 10.1101/2021.05.28.446163. [DOI] [Google Scholar]
- 72.Hoffmann M., Kleine-Weber H., Pohlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Binley J.M., Sanders R.W., Master A., Cayanan C.S., Wiley C.L., Schiffner L., Travis B., Kuhmann S., Burton D.R., Hu S., et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 2002;76:2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bosch B.J., Bartelink W., Rottier P.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson E.D., Thomas L., Hayflick J.S., Thomas G. Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed alpha 1-antitrypsin variant. J. Biol. Chem. 1993;268:24887–24891. doi: 10.1016/S0021-9258(19)74548-7. [DOI] [PubMed] [Google Scholar]
- 76.Hallenberger S., Bosch V., Angliker H., Shaw E., Klenk H.D., Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 77.Powers J.C., Asgian J.L., Ekici O.D., James K.E. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 78.Schoellmann G., Shaw E. Direct evidence for the presence of histidine in the active center of chymotrypsin. Biochemistry. 1963;2:252–255. doi: 10.1021/bi00902a008. [DOI] [PubMed] [Google Scholar]
- 79.Shaw E. [58] Site-specific reagents for chymotrypsin, trypsin, and other serine proteases. Methods Enzymol. 1972;25:655–660. doi: 10.1016/S0076-6879(72)25062-5. [DOI] [PubMed] [Google Scholar]
- 80.Garten W., Hallenberger S., Ortmann D., Schafer W., Vey M., Angliker H., Shaw E., Klenk H.D. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 1994;76:217–225. doi: 10.1016/0300-9084(94)90149-X. [DOI] [PubMed] [Google Scholar]
- 81.Tang T., Jaimes J.A., Bidon M.K., Straus M.R., Daniel S., Whittaker G.R. Proteolytic Activation of SARS-CoV-2 Spike at the S1/S2 Boundary: Potential Role of Proteases beyond Furin. ACS Infect. Dis. 2021;7:264–272. doi: 10.1021/acsinfecdis.0c00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jean F., Stella K., Thomas L., Liu G., Xiang Y., Reason A.J., Thomas G. alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: Application as an antipathogenic agent. Proc. Natl. Acad. Sci. USA. 1998;95:7293–7298. doi: 10.1073/pnas.95.13.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huntington J.A. Shape-shifting serpins—Advantages of a mobile mechanism. Trends Biochem. Sci. 2006;31:427–435. doi: 10.1016/j.tibs.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 84.Lewis J.H., Iammarino R.M., Spero J.A., Hasiba U. Antithrombin Pittsburgh: An alpha1-antitrypsin variant causing hemorrhagic disease. Blood. 1978;51:129–137. doi: 10.1182/blood.V51.1.129.129. [DOI] [PubMed] [Google Scholar]
- 85.Owen M.C., Brennan S.O., Lewis J.H., Carrell R.W. Mutation of antitrypsin to antithrombin. alpha 1-antitrypsin Pittsburgh (358 Met leads to Arg), a fatal bleeding disorder. N. Engl. J. Med. 1983;309:694–698. doi: 10.1056/NEJM198309223091203. [DOI] [PubMed] [Google Scholar]
- 86.Brennan S.O., Owen M.C., Boswell D.R., Lewis J.H., Carrell R.W. Circulating proalbumin associated with a variant proteinase inhibitor. Biochim. Biophys. Acta. 1984;802:24–28. doi: 10.1016/0304-4165(84)90029-1. [DOI] [PubMed] [Google Scholar]
- 87.Jean F., Thomas L., Molloy S.S., Liu G., Jarvis M.A., Nelson J.A., Thomas G. A protein-based therapeutic for human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA. 2000;97:2864–2869. doi: 10.1073/pnas.050504297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Watanabe M., Hirano A., Stenglein S., Nelson J., Thomas G., Wong T.C. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J. Virol. 1995;69:3206–3210. doi: 10.1128/jvi.69.5.3206-3210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mercapide J., Lopez De Cicco R., Bassi D.E., Castresana J.S., Thomas G., Klein-Szanto A.J.P. Inhibition of furin-mediated processing results in suppression of astrocytoma cell growth and invasiveness. Clin. Cancer Res. 2002;8:1740–1746. doi: 10.1158/1078-0432.CCR-19-0489. [DOI] [PubMed] [Google Scholar]
- 90.Scamuffa N., Siegfried G., Bontemps Y., Ma L., Basak A., Cherel G., Calvo F., Seidah N.G., Khatib A.M. Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J. Clin. Investig. 2008;118:352–363. doi: 10.1172/JCI32040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma Y.C., Fan W.J., Rao S.M., Gao L., Bei Z.Y., Xu S.T. Effect of Furin inhibitor on lung adenocarcinoma cell growth and metastasis. Cancer Cell Int. 2014;14:43. doi: 10.1186/1475-2867-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yakala G.K., Cabrera-Fuentes H.A., Crespo-Avilan G.E., Rattanasopa C., Burlacu A., George B.L., Anand K., Mayan D.C., Corliano M., Hernandez-Resendiz S., et al. FURIN Inhibition Reduces Vascular Remodeling and Atherosclerotic Lesion Progression in Mice. Arterioscler. Thromb. Vasc. Biol. 2019;39:387–401. doi: 10.1161/ATVBAHA.118.311903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McNulty M.J., Silberstein D.Z., Kuhn B.T., Padgett H.S., Nandi S., McDonald K.A., Cross C.E. Alpha-1 antitrypsin deficiency and recombinant protein sources with focus on plant sources: Updates, challenges and perspectives. Free Radic. Biol. Med. 2021;163:10–30. doi: 10.1016/j.freeradbiomed.2020.11.030. [DOI] [PubMed] [Google Scholar]
- 94.Wettstein L., Weil T., Conzelmann C., Muller J.A., Gross R., Hirschenberger M., Seidel A., Klute S., Zech F., Prelli Bozzo C., et al. Alpha-1 antitrypsin inhibits TMPRSS2 protease activity and SARS-CoV-2 infection. Nat. Commun. 2021;12:1726. doi: 10.1038/s41467-021-21972-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takeda K., Kim S.H., Joetham A., Petrache I., Gelfand E.W. Therapeutic benefits of recombinant alpha1-antitrypsin IgG1 Fc-fusion protein in experimental emphysema. Respir. Res. 2021;22:207. doi: 10.1186/s12931-021-01784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Couture F., Kwiatkowska A., Dory Y.L., Day R. Therapeutic uses of furin and its inhibitors: A patent review. Expert Opin. Ther. Pat. 2015;25:379–396. doi: 10.1517/13543776.2014.1000303. [DOI] [PubMed] [Google Scholar]
- 97.Cameron A., Appel J., Houghten R.A., Lindberg I. Polyarginines are potent furin inhibitors. J. Biol. Chem. 2000;275:36741–36749. doi: 10.1074/jbc.M003848200. [DOI] [PubMed] [Google Scholar]
- 98.Kacprzak M.M., Peinado J.R., Than M.E., Appel J., Henrich S., Lipkind G., Houghten R.A., Bode W., Lindberg I. Inhibition of furin by polyarginine-containing peptides: Nanomolar inhibition by nona-D-arginine. J. Biol. Chem. 2004;279:36788–36794. doi: 10.1074/jbc.M400484200. [DOI] [PubMed] [Google Scholar]
- 99.Lewandowska-Goch M.A., Kwiatkowska A., Lepek T., Ly K., Navals P., Gagnon H., Dory Y.L., Prahl A., Day R. Design and Structure-Activity Relationship of a Potent Furin Inhibitor Derived from Influenza Hemagglutinin. ACS Med. Chem. Lett. 2021;12:365–372. doi: 10.1021/acsmedchemlett.0c00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Remacle A.G., Gawlik K., Golubkov V.S., Cadwell G.W., Liddington R.C., Cieplak P., Millis S.Z., Dejardins R., Routhier S., Yuan X.W., et al. Selective and potent furin inhibitors protect cells from anthrax without significant toxicity. Int. J. Biochem. Cell Biol. 2010;42:987–995. doi: 10.1016/j.biocel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sarac M.S., Peinado J.R., Leppla S.H., Lindberg I. Protection against anthrax toxemia by hexa-D-arginine in vitro and in vivo. Infect. Immun. 2004;72:602–605. doi: 10.1128/IAI.72.1.602-605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Becker G.L., Lu Y., Hardes K., Strehlow B., Levesque C., Lindberg I., Sandvig K., Bakowsky U., Day R., Garten W., et al. Highly potent inhibitors of proprotein convertase furin as potential drugs for treatment of infectious diseases. J. Biol. Chem. 2012;287:21992–2003. doi: 10.1074/jbc.M111.332643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Becker G.L., Sielaff F., Than M.E., Lindberg I., Routhier S., Day R., Lu Y., Garten W., Steinmetzer W. Potent inhibitors of furin and furin-like proprotein convertases containing decarboxylated P1 arginine mimetics. J. Med. Chem. 2010;53:1067–1075. doi: 10.1021/jm9012455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3:e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gagnon H., Beauchemin S., Kwiatkowska A., Couture F., D’Anjou F., Levesque C., Dufour F., Desbiens A.R., Vaillancourt R., Bernard S., et al. Optimization of furin inhibitors to protect against the activation of influenza hemagglutinin H5 and Shiga toxin. J. Med. Chem. 2014;57:29–41. doi: 10.1021/jm400633d. [DOI] [PubMed] [Google Scholar]
- 106.Hardes K., Ivanova T., Thaa B., McInerney G.M., Klokk T.I., Sandvig K., Kunzel S., Lindberg I., Steinmetzer W. Elongated and Shortened Peptidomimetic Inhibitors of the Proprotein Convertase Furin. ChemMedChem. 2017;12:613–620. doi: 10.1002/cmdc.201700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Van Lam van T., Heindl M.R., Schlutt C., Bottcher-Friebertshauser E., Bartenschlager R., Klebe G., Brandstetter H., Dahms S.O., Steinmetzer W. The Basicity Makes the Difference: Improved Canavanine-Derived Inhibitors of the Proprotein Convertase Furin. ACS Med. Chem. Lett. 2021;12:426–432. doi: 10.1021/acsmedchemlett.0c00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiao G.S., Cregar L., Wang J., Millis S.Z., Tang C., O’Malley S., Johnson A.T., Sareth S., Larson J., Thomas G. Synthetic small molecule furin inhibitors derived from 2,5-dideoxystreptamine. Proc. Natl. Acad. Sci. USA. 2006;103:19707–197012. doi: 10.1073/pnas.0606555104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Komiyama T., Coppola J.M., Larsen M.J., van Dort M.E., Ross B.D., Day R., Rehemtulla A., Fuller R.S. Inhibition of furin/proprotein convertase-catalyzed surface and intracellular processing by small molecules. J. Biol. Chem. 2009;284:15729–15738. doi: 10.1074/jbc.M901540200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramos-Molina B., Lick A.N., Blanco E.H., Posada-Salgado J.A., Martinez-Mayorga K., Johnson A.T., Jiao G.S., Lindberg I. Identification of potent and compartment-selective small molecule furin inhibitors using cell-based assays. Biochem. Pharmacol. 2015;96:107–118. doi: 10.1016/j.bcp.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Essalmani R., Jain J., Susan-Resiga D., Andréo U., Evagelidis A., Derbali R.M., Huynh D.N., Dallaire F., Laporte M., Delpal A., et al. Furin cleaves SARS-CoV-2 spike-glycoprotein at S1/S2 and S2′ for viral fusion/entry: Indirect role of TMPRSS2. bioRxiv. 2020 doi: 10.1101/2020.12.18.423106. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.