Abstract
Combinatorial peptide display on phage M13 protein pIII was used to discover peptide sequences that selectively bind to ErmC′ methyltransferase from Bacillus subtilis. One peptide, Ac-LSGVIAT-NH2, inhibited methylation in vitro with a 50% inhibitory concentration of 20 μM. Interestingly, the set of six peptides which inhibited ErmC′ stimulated ErmSF, a homologous methyltransferase from Streptomyces fradiae. Thus, Ac-LSGVIAT-NH2 may not act directly at the catalytic center of ErmC′, but may modulate its activity by binding at a structurally unrelated, but functionally linked, site.
The ErmC N-methyltransferase of Staphylococcus aureus and its close relative ErmC′ from Bacillus subtilis confer resistance to erythromycin and related macrolide antibiotics. These enzymes act by specifically methylating a single adenine residue in the peptidyl transferase center of bacterial 23S rRNA (for reviews, see references 7 and 8). The same methylation confers coresistance to structurally unrelated lincosamide and streptogramin type B antibiotics. The three groups are collectively known as the macrolide-lincosamide-streptogramin B (MLS) antibiotics, and the form of resistance based on the Erm group of methyltransferases is found in a wide range of pathogens. The three-dimensional structures of the ErmAM and ErmC′ methyltransferases have recently been solved by nuclear magnetic resonance (3, 9) and X-ray crystallography (1, 5), respectively. These developments bring us closer to rationally devising ligands that will selectively bind to Erm enzymes and possibly reduce the efficiency with which they confer MLS resistance.
Macrolide antibiotics have served as a mainstay of antimicrobial therapy for approximately the last half century, especially in instances where the recipient of the antibiotic was allergic to beta-lactam antibiotics. Attempts have been made to discover Erm methyltransferase inhibitors that maintain the effectiveness of macrolide antibiotics in the face of the increasing frequency of resistant isolates (2, 3). The latter work (3) was part of a series of major structural studies of Erm methyltransferases (1, 5, 9).
Nonpeptide ligands which displace an inhibitory peptide from its binding site on the enzyme might, themselves, have inhibitory activity. The use of inhibitory peptides might thus serve as a platform for the discovery of nonpeptide inhibitors of Erm enzymes. A proposed way to achieve this would be based on the discovery of test ligands to displace an inhibitory peptide from its association with its cognate Erm target. The displacement of an inhibitory ligand would also be easier to measure than methyltransferase activity.
Large-scale screening of inhibitory ligands by a direct assay of methyltransferase catalytic activity is cumbersome since it requires the separation of product (methyl-labeled 23S rRNA) from substrate (unreacted S-adenosyl-l-methionine). We report the use of combinatorial phage display to discover peptides that inhibit ErmC′ methylase activity and which might serve as displaceable ligands in a more efficient, broader assay for prospective methyltransferase inhibitors.
MATERIALS AND METHODS
Phage display.
Phage display was performed as described by Sparks et al. (6) by using a combinatorial peptide library specified by seven random codons, (NNK)7, where N denotes one of the four bases A, G, C, or T, and K denotes one of the two bases G or T (New England Biolabs). ErmC′ (1) was a gift of Abbott Pharmaceuticals, Inc. ErmC′ was originally derived from B. subtilis but was overexpressed in Escherichia coli from which it was purified. ErmSF was purified, and its activity was tested as described previously (4), along with that of ErmC′. The peptide specified by the combinatorial cassette was determined at the DNA level by the use of 5′ TAA GTG GAG CTT TCG TTC GAC 3′ as sequencing primer. This sequence is a part of the gene for M13 phage protein pIII on which the combinatorial peptide is displayed (6).
Effect of chemically synthesized peptides on Erm activity.
Peptides used for testing inhibitory activity were chemically synthesized (Research Genetics, Huntsville, Ala.) with N-acetyl- and C-amide modifications, respectively. The inhibitory (or stimulatory) activity of the chemically synthesized combinatorial peptides on the N-methyltransferase activity of ErmC′ and ErmSF was tested in triplicate at the stated concentrations. The complete system reaction mixture contained the following in a 50-μl total volume: 50 mM Tris HCl, pH 7.5; 4 mM MgCl2; 4 mM KCl; 40 mM dithiothreitol; 6 μM S-adenosyl-l-methionine, 75 Ci/mM; 10 U of RNasin; 207 ng of ErmC′ or ErmSF, as indicated; and 7.5 pmol of B. subtilis rRNA. Erythromycin-resistant (ermC) B. subtilis was used as the source of negative control rRNA. Incubation was at 37°C for 20 min. Reactions were phenol extracted, trichloroacetic acid-precipitated, filtered, and counted in a liquid scintillation spectrometer.
RESULTS
Peptides derived from phage display.
To find peptides that bind to ErmC′ from B. subtilis, the random heptamer peptide library (NNK)7 displayed on phage M13 protein pIII was panned for the ability to bind to ErmC′-coated microtiter plate wells as described by Sparks et al. (6). Forty-eight phage clones were obtained which showed binding to the coated wells as determined by phage enzyme-linked immunosorbent assay. Of these, six that showed fivefold higher binding to ErmC′-coated wells, compared to bovine serum albumin-coated wells, were selected for further study. The respective active peptides were chemically synthesized (Research Genetics) by the solid-phase method and were tested for the ability to inhibit the catalytic activity of ErmC′.
Test for ability to inhibit ErmC′ methyltransferase catalytic activity.
All six peptides were tested in triplicate at 50 μg/ml (approximately 50 μM) for their ability to inhibit, in vitro, methylation of 23S rRNA by ErmC′. Results shown in Table 1 indicate that all six peptides were inhibitory to various degrees. As controls for the sequence specificity of the peptides, randomized peptides rnd 5 and rnd 6, based on the amino acid compositions of peptides 5 and 6, were synthesized and tested. The randomized peptides showed little if any inhibitory activity. Activity associated with some of the peptides seemed to show a high standard deviation. We believe that the phenol extraction step in the assay contributed a solid-phase component that traps methylated RNA along with denatured proteins to variable degrees in the different experimental test samples. Some samples showed a lower standard deviation, both in this set of experiments and in those described below.
TABLE 1.
Modulation of ErmC′ N-methyltransferase activity by peptides derived from phage display
| Peptide (50 μg/ml)a | cpm (avg) | Normalized % | SD (%) |
|---|---|---|---|
| Complete (with ErmC′) | 5,158 | 100 | |
| + peptide 1, TARVVLK | 2,577 | 61 | 39 |
| + peptide 2, YIGVETE | 1,548 | 33 | 11 |
| + peptide 3, SGFLGLQ | 2,078 | 48 | 28 |
| + peptide 4, LWVMKST | 2,309 | 52 | 25 |
| + peptide 5, YEVMMWG | 2,359 | 48 | 7 |
| + peptide 6, LSGVIAT | 1,488 | 35 | 24 |
| With resistant rRNA | 657 | 16 | 10 |
| No enzyme | 467 | 11 | 8 |
| Complete (with ErmC′) | 3,464 | 100 | |
| + rnd 5, WGEMMYV | 3,671 | 105 | 26 |
| + rnd 6, ATSVILG | 3,378 | 98 | 16 |
Peptides obtained by combinatorial phage display were chemically synthesized as the N-acetyl and C-amide, and their effect on methylation of 23S rRNA catalyzed by ErmC′ was tested in triplicate as described in Materials and Methods. Resistant rRNA was methylated in vivo, purified by phenol-extraction from ribosomes, and used as a negative control. The sequences of peptides rnd 5 and rnd 6 were based on a randomization of the sequences of peptides 5 and 6, respectively, and were used as negative controls. Two sets of experiments were performed. To facilitate comparison, the data were normalized, setting the unsupplemented complete reaction data to 100%.
Concentration dependence for inhibition of methylation.
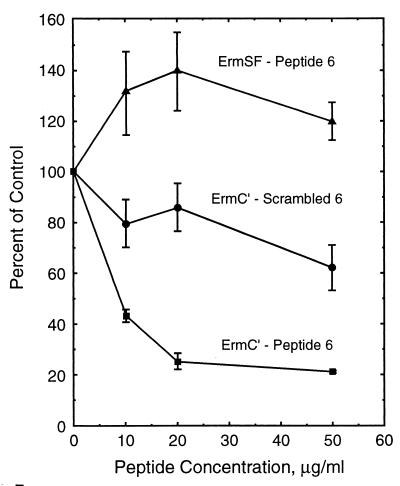
The concentration dependence of peptide 6 for inhibition of methylation by ErmC′ was tested along with ErmSF from Streptomyces fradiae, which was included for a comparison of enzyme specificity. The concentration dependence of rnd 6 acting on ErmC′ was also tested. The results shown in Fig. 1 indicate that peptide 6 inhibited methylation with a 50% inhibitory concentration between 3 and 10 μM, whereas rnd 6 had only a slight effect in this concentration range. Surprisingly, ErmSF activity was enhanced by peptide 6, whereas rnd had only a small effect on methylation catalyzed by ErmSF.
FIG. 1.
The results of a test of the action of peptide 6, Ac-LSGVIAT-NH2, and scrambled peptide 6 (referred to in Table 1 as rnd 6), Ac-ATSVILG-NH2, on methylation of 23S rRNA by ErmC′. The effect of peptide 6 on methylation of 23S rRNA by ErmSF was included for comparison. All values were determined in triplicate and were normalized to the unsupplemented control.
Test for ability to inhibit ErmSF methyltransferase catalytic activity.
We next asked whether other peptides would stimulate at 50 μg/ml (approximately 50 μM). The results shown in Table 2 indicate that individual peptides either stimulated ErmSF or had no effect. Also, rnd 5 and rnd 6 appeared to have no effect on ErmSF. Moreover, the ability of a peptide to inhibit ErmC′ did not appear to correlate with its ability to stimulate ErmSF.
TABLE 2.
Modulation of ErmSF N-methyltransferase activity by peptides derived from phage display
| Peptide (50 μg/ml)a | cpm (avg) | Normalized % | SD (%) |
|---|---|---|---|
| Complete (with ErmSF) | 2,798 | 100 | |
| + peptide 1 | 5,330 | 229 | 125 |
| + peptide 2 | 4,243 | 191 | 107 |
| + peptide 3 | 6,183 | 271 | 121 |
| + peptide 4 | 3,365 | 139 | 106 |
| + peptide 5 | 4,959 | 213 | 108 |
| + peptide 6 | 5,059 | 190 | 54 |
| With resistant rRNA | 584 | 22 | 5 |
| no enzyme | 553 | 20 | 9 |
| Complete (with ErmSF) | 7,224 | 100 | |
| + rnd 5 | 6,808 | 97 | 20 |
| + rnd 6 | 6,910 | 97 | 16 |
Peptides obtained by combinatorial phage display were chemically synthesized as the N-acetyl and C-amide, and their effect on methylation of 23S rRNA catalyzed by ErmSF was tested in triplicate as in Table 1. Two sets of experiments were performed. To facilitate comparison, the data were normalized, setting the unsupplemented complete reaction data to 100%.
DISCUSSION
Drug discovery programs generally seek inhibitors, rather than potentiators, of enzyme function, and published studies appear to favor the search for inhibitors of the catalytic center of the target protein. S-adenosyl homocysteine inhibits the catalytic activity of methyltransferases by binding to the active center. S-adenosyl homocysteine was unable, at concentrations up to 1 mM, to compete with phage carrying peptide 6 for binding to ErmC′ (data not shown). These observations suggest that peptide 6 does not act at the active site of ErmC′ methyltransferase. In the present set of experiments, we have shown that all of the combinatorial peptides selected for binding to ErmC′ inhibited ErmC′ methyltransferase activity, albeit to different degrees, but also stimulated ErmSF activity. These findings suggest that one might be able to screen for agents that potentiate the activity of a given enzyme by initially screening for inhibitors of a homolog.
In summary, peptides have been obtained with the help of phage-displayed peptides which inhibit the activity of ErmC′ but stimulate the activity of a homolog, ErmSF. The results of these studies may serve as a model for the discovery of enzyme potentiators as well as an efficient assay for inhibitors of the enzymatic activity of Erm methyltransferases.
ACKNOWLEDGMENTS
We thank Brian K. Kay for introducing us to phage display and Abbott Pharmaceuticals, Inc. for a gift of purified ErmC′.
REFERENCES
- 1.Bussiere D E, Muchmore S W, Dealwis C G, Schluckebier G, Nienaber V L, Edalji R P, Walter K A, Ladror U S, Holzman T F, Abad-Zapatero C. Crystal structure of ErmC′, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry. 1998;37:7103–7112. doi: 10.1021/bi973113c. [DOI] [PubMed] [Google Scholar]
- 2.Clancy J, Schmieder B J, Petitpas J W, Manousos M, Williams J A, Faiella J A, Girard A E, McGuirk P R. Assays to detect and characterize synthetic agents that inhibit the ErmC methyltransferase. J Antibiot. 1995;48:1273–1279. doi: 10.7164/antibiotics.48.1273. [DOI] [PubMed] [Google Scholar]
- 3.Hajduk P J, Dinges J, Schkeryantz J M, Janowick D, Kaminski M, Tufano M, Augeri D J, Petros A, Nienaber V, Zhong P, Hammond R, Coen M, Beutel B, Katz L, Fesik S W. Novel inhibitors of Erm methyltransferases from NMR and parallel synthesis. J Med Chem. 1999;42:3852–3859. doi: 10.1021/jm990293a. [DOI] [PubMed] [Google Scholar]
- 4.Kovalic D, Giannattasio R B, Jin H J, Weisblum B. 23S rRNA domain V, a fragment that can be specifically methylated in vitro by the ErmSF (TlrA) methyltransferase. J Bacteriol. 1994;176:6992–6998. doi: 10.1128/jb.176.22.6992-6998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schluckebier G, Zhong P, Stewart K D, Kavanaugh T J, Abad-Zapatero C. The 2.2 Å structure of the rRNA methyltransferase ErmC′ and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J Mol Biol. 1999;289:277–291. doi: 10.1006/jmbi.1999.2788. [DOI] [PubMed] [Google Scholar]
- 6.Sparks A, Adey N, Cwirla S, Kay B K. Chapter 13. In: Kay B K, Winter J, McCafferty J, editors. Phage display of peptides and proteins: a laboratory manual. San Diego, Calif: Academic Press; 1996. pp. 227–253. [Google Scholar]
- 7.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weisblum B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother. 1995;39:797–805. doi: 10.1128/aac.39.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu L, Petros A M, Schnuchel A, Zhong P, Severin J M, Walter K, Holzman T F, Fesik S W. Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide-streptogramin antibiotic resistance. Nat Struct Biol. 1997;4:483–489. doi: 10.1038/nsb0697-483. [DOI] [PubMed] [Google Scholar]