Abstract
Despite the clear circumscription of tribe Sobralieae (Orchidaceae), its internal relationships are still dubious. The recently delimited genus Brasolia, based on previous Sobralia species, is now assumed to be paraphyletic, with a third genus, Elleanthus, nested in it. The morphology of these three genera is significantly different, indicating the necessity of new data for a better genera delimitation. Though morphology and molecular data are available, cytogenetics data for Sobralieae is restricted to two Sobralia and one Elleanthus species. Aiming to evaluate the potential of cytogenetic data for Brasolia-Elleanthus-Sobralia genera delimitation, we present chromosome number and genome size data for 21 and 20 species, respectively, and used such data to infer the pattern of karyotype evolution in these genera. The analysis allowed us to infer x = 24 as the base chromosome number and genome size of average 1C-value of 5.0 pg for the common ancestor of Brasolia-Elleanthus-Sobralia. The recurrent descending dysploidy in Sobralieae and the punctual genome upsize suggest a recent diversification in Sobralieae but did not allow differing between Brasolia and Sobralia. However, the basal position of tribe Sobralieae in the subfamily Epidendroideae makes this tribe of interest to further studies clarifying the internal delimitation and pattern of karyotype evolution.
Keywords: karyotype, phylogeny, nuclear DNA content, flow cytometry, dysploidy, phylogenomics, plant genetic diversity
1. Introduction
Since Ruiz and Pavón established the genus Sobralia (Orchidaceae) in 1794 [1], its species reached about 200 taxa distributed from southern Mexico to Bolivia and Brazil, with a remarkable species diversity in the Andes. The species are often terrestrial, rarely epiphytic, usually tall, or very tall (for example, stems of Sobralia luerorum Dodson can reach up to 3.5 m height), with slender, cane-like stems growing in clumps and strongly developed root systems. The inflorescence position and structure have served as a basis for the infrageneric classification of Sobralia [2], with five recognized sections [2,3].
The nominal section was characterized by lateral or rarely terminal inflorescences with branching, well-developed raceme and relatively small floral bracts compared to the size of the ovary [2]. The section Racemosae Brieger, despite terminal inflorescences, could differ from the former by its elongated and unbranched inflorescences with large floral bracts. Section Globosae Brieger is composed of small plants with narrow leaf blades, small flowers positioned in terminal and condensed inflorescences (shortened internodes hidden under the floral bracts) that successively produce a single flower at a time and elongate with successively produced floral bracts. Species from section Abbreviatae Brieger share terminal and condensed inflorescences with the previous section but, instead, present floral bracts forming a cone. The fifth section, Intermediae Brieger, was established for a single taxon—Sobralia fragrans Lindl.—to emphasize its elongated basal internode of the inflorescences. Dressler [3] enlarged this section, placing other species with small flowers and inflorescences (Figure 1).
Figure 1.
Diversity in Brasolia, Sobralia and Elleanthus flowers. (a) B. flava (Eric Hunt); (b) B. portillae (Lourens Grobler); (c) B. dichotoma (Marta Kolanowska); (d) S. fimbriata (Andreas Kay); (e) S. lancea (Andreas Kay); (f) S. bimaculata (Andreas Kay); (g) S. crocea (Andreas Kay); (h) S. decora (Przemyslaw Baranow); (i) S. rosea (K-Eric Hunt); (j) S. gloriosa (Andreas Kay); (k) E. brasiliensis (Ana Paula Moraes); (l) E. crinipes (Carlos Nunes). Scale bars = 10 mm.
Molecular phylogenetic studies [4,5] revealed that the nominal section of Sobralia is paraphyletic, indicating the necessity of taxonomic reassignment. Based on the morphological distinctness and the mentioned phylogenetic relations, the section Sobralia was elevated to the status of a separate genus named Brasolia (Rchb.f.) Baranow, Dudek and Szlach [4]. Dressler et al. [6] already had predicted the necessity of such reassignments, proposing to conserve the name Sobralia with Sobralia biflora Ruiz and Pav. as a nominal species to avoid transferring most of the genus representatives into Cyathoglottis Poepp. & Endl., the second-oldest generic name available for Sobralia. Thus, the former nominal section of the genus Sobralia now constitutes a separate genus renamed Brasolia, with the former section Abbreviatae being considered the Sobralia nominal section [4]. Nevertheless, the genus Elleanthus C. Presl was nested in the new Brasolia [4], an association previously suggested by Neubig et al. [5], suggesting further taxonomic realignments are necessary.
Despite the availability of molecular and morphological data, the cytogenetic information, which is known to significantly help to understand the relationship and delimitation among taxa [7], is limited in the tribe Sobralieae, with only three chromosome counts available until now: two Sobralia species, S. liliastrum Lindl. and S. sessilis Lindl., with 2n = 2x = 48, and one Elleanthus, E. brasiliensis (Lindl.) Rchb.f. with 2n = 2x = 50 [8]. Regarding the genome size, no data is available. The karyotype evolutionary history results from the successive accumulation of numerical and structural chromosome rearrangements (for a review, see [9,10]), making it mandatory to evaluate the cytogenetic data in a phylogenetic background. The family Orchidaceae Juss. is known for its large variation in both chromosome numbers, which varies more than 20-fold (from 2n = 10 to 2n = 240; [11,12]), and genome size, which varies more than 168-fold, the second-largest variability in Angiosperms [13]. It makes Orchidaceae an interesting clade for studies in karyotype evolution, and by inferring the historical karyotype and genome size modifications, it also gives clues about taxonomic delimitations [14,15]. However, the scare cytogenetic data in tribe Sobralieae prevents any further discussion on Brasolia and Sobralia delimitation, as well as the Sobralieae karyotype evolution.
Based on the statement that the more taxonomical data sources, the more realistic the classification becomes, we have chosen the karyotype evolution of Sobralieae as the next step to better understand the relationship between genera Brasolia, Elleanthus and Sobralia. Based on chromosome number and genome size for these three genera, we aim to answer the following questions: (1) Which are the main evolutionary mechanisms shaping karyotype and genome size in tribe Sobralieae? and (2) Based on the ancestral base chromosome number (x) and genome size (1C-value) for these three genera and tribe Sobralieae, is it possible to separate the genera? We gathered the somatic chromosome number for 21 species and the genome size for 20 species to answer these questions and interpreted all the data in a phylogenetic approach, based on Maximum Likelihood (ML) and Bayesian Inference (BI) analyses, both using nuclear ribosomal DNA (nrITS) and maturase K (matK) regions.
2. Results
2.1. Phylogenetic Analysis Suggested Brasolia as Paraphyletic
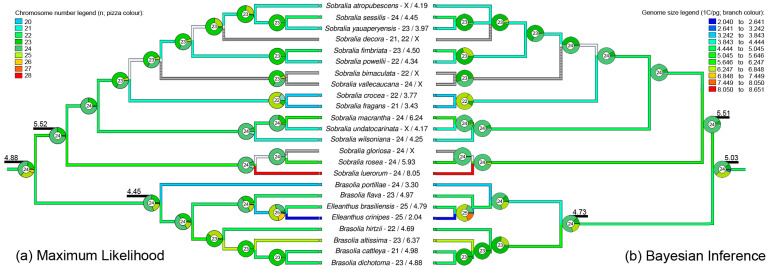
Both applied methodologies (ML and BI) revealed the same general results, with Sobralia as a monophyletic genus and Brasolia as paraphyletic with Elleanthus nested in it (Figure 2 and Figure S1). Despite the general similarity between both reconstruction methods, some differences could be detected, mainly inside Brasolia regarding the position of B. portillae and B. mandonii in relationship to the remainder Brasolia species (Figure S1). While this group was placed as a sister of remainder Brasolia and Elleanthus in ML, it was placed as a sister of B. flava and Elleanthus in BI. Considering the slight differences between ML and BI, we used both phylogenies to further steps. Both trees were pruned, removing species without chromosome number and genome size data, saving the tree topology for following analysis (Figure 2).
Figure 2.
Maximum likelihood (a) and Bayesian Inference (b) pruned phylogenies showing the ancestral reconstruction of base chromosome number on nodes and genome size (1C-value) on branches. The ancestral genome size is indicated on the node for Brasolia, Sobralia, and the ancestral of both genera. The colour legend for the base chromosome number is on the right, and for genome size, on the left.
2.2. Chromosome Number and Genome Size Varies in Sobralieae
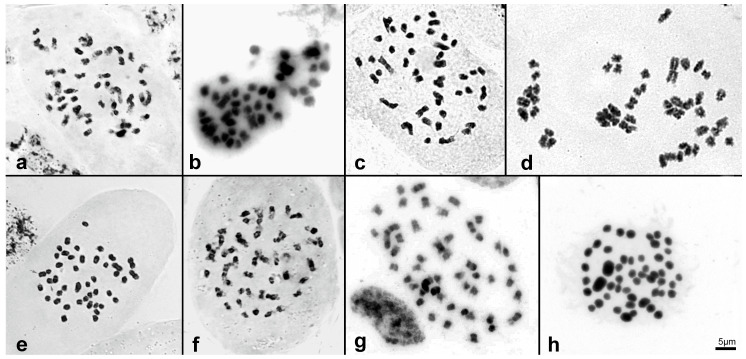
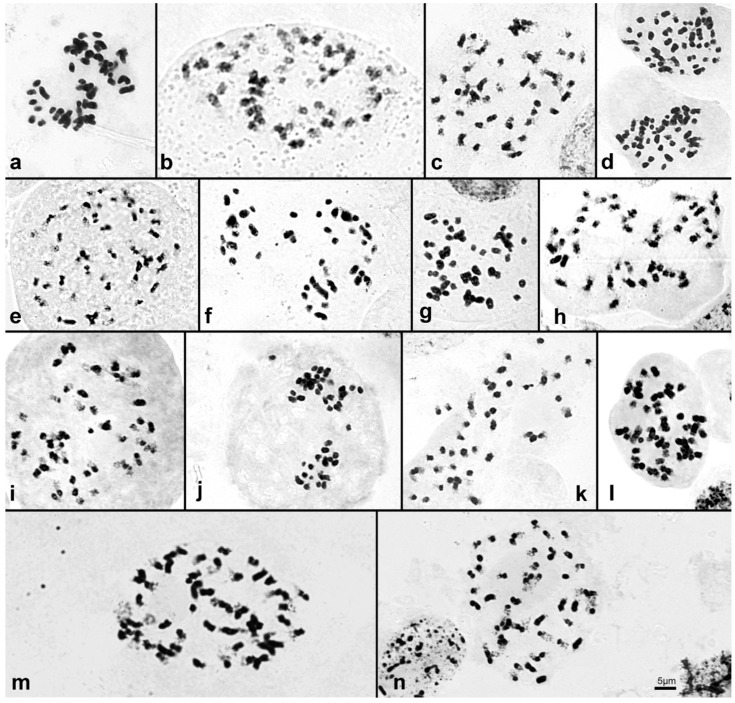
Chromosome numbers (2n) are provided for six Brasolia, two Elleanthus (Figure 3 and Figure S2), and 21 Sobralia species (Figure 4 and Figure S3), with two additional reports from the literature, totalizing chromosome numbers to 23 Sobralieae species (Table 1). Chromosome numbers varied from 2n = 40 to 2n = 48 in Brasolia and Sobralia, while both Elleanthus species presented 2n = 50 (Table 1).
Figure 3.
Chromosome complement of Brasolia and Elleanthus species. (a) B. altissima (2n = 46), (b) B. cattleya (2n = 42), (c) B. dichotoma (2n = 46), (d) B. flava (2n = 46), (e) B. hirtzii (2n = 44), (f) B. portillae (2n = 48), (g) E. brasiliensis (2n = 50), (h) E. crinipes (2n = 50). Bar in h correspond to 5 µm.
Figure 4.
Chromosome complement of Sobralia species. (a) S. bimaculata (2n = 44), (b) S. decora (2n = 42), (c) S. decora (2n = 44), (d) S. fimbriata (2n = 46), (e) S. macrantha (2n = 48), (f) S. powelli (2n = 44), (g) S. vallecaucana (2n = 48), (h) S. wilsoniana (2n = 48), (i) S. yauaperyensis (2n = 46), (j) S. crocea (2n = 44), (k) S. fragrans (2n = 42), (l) S. gloriosa (2n = 48), (m) S. luerorum (2n = 48), (n) S. rosea (2n = 48). Bar in h correspond to 5 µm.
Table 1.
Chromosome number and genome size in Sobralieae. The list of Brasolia, Elleanthus, and Sobralia species analyzed here and published on the literature is presented. For each species, the determined chromosome number is presented followed by its figure in text or reference on the literature. For each species analyzed here, the number of analyzed specimens in FCM is indicated in brackets. The genome size is presented in pg, followed by its standard deviation (SD), and Mega base-pairs (Mbp). The genome size (the 1C-value) is given in pg ± SD (Standard Deviation) and Mbp (Mega base-pairs). For each 1C-value, the sample Coefficient of Variation (CV) is presented and the genome size is classified according Soltis et al. [16].
| No. | Species [n. FCM Analyzed Specimens] | Chromosome Number | Figure/Reference | 1C DNA Content | CV Sample % |
Genome Size Category | |
|---|---|---|---|---|---|---|---|
| pg ± SD | Mbp | ||||||
| Brasolia | |||||||
| 01 | B. altissima [2] | 46 | Figure 3a | 6.37 ± 0.075 | 6157 | 3.65 | intermediate |
| 02 | B. cattleya [1] | 42 | Figure 3b | 4.98 ± 0.014 | 4876 | 3.95 | intermediate |
| 03 | B. dichotoma [1] | 46 | Figure 3c | 4.88 ± 0.005 | 4773 | 3.81 | intermediate |
| 04 | B. flava [2] | 46 | Figure 3d | 4.97 ± 0.028 | 4854 | 4.14 | intermediate |
| 05 | B. hirtzii [2] | 44 | Figure 3e | 4.69 ± 0.011 | 4582 | 3.76 | intermediate |
| 06 | B. portillae [3] | 48 | Figure 3f | 3.30 ± 0.016 | 3228 | 4.75 | small |
| Elleanthus | |||||||
| 07 | E. brasiliensis [3] | 50 | Figure 3g/FG10 [8] | 4.79 ± 0.075 | 4687 | 2.84 | intermediate |
| 08 | E. crinipes [3] | 50 | Figure 3h | 2.04 ± 0.078 | 1995 | 3.11 | small |
| Sobralia | |||||||
| Sect. Sobralia | |||||||
| 09 | S. atropubescens [1] | - | - | 4.19 ± 0.010 | 4098 | 5.08 | intermediate |
| 10 | S. bimaculata | 44 | Figure 3a | - | - | - | - |
| 11 | S. decora | 42, 44 | Figure 3b,c | - | - | - | - |
| 12 | S. fimbriata [1] | 46 | Figure 3d | 4.50 ± 0.019 | 4401 | 4.37 | intermediate |
| 13 | S. liliastrum | 48 | FG10 [8] | - | - | - | - |
| 14 | S. macrantha [2] | 48 | Figure 3e | 6.24 ± 0.121 | 6108 | 4.15 | intermediate |
| 15 | S. powelli [2] | 44 | Figure 3f | 4.34 ± 0.085 | 4248 | 4.45 | intermediate |
| 16 | S. sessilis [1] | 48 | FG10 [8] | 4.45 ± 0.095 | 4347 | 4.47 | intermediate |
| 17 | S. vallecaucana | 48 | Figure 3g | - | - | - | - |
| 18 | S. wilsoniana [2] | 48 | Figure 3h | 4.25 ± 0.025 | 4163 | 4.95 | intermediate |
| 19 | S. yauaperyensis [1] | 46 | Figure 4i | 3.97 ± 0.012 | 3878 | 4.52 | intermediate |
| Sect. Intermediae | |||||||
| 20 | S. crocea [2] | 44 | Figure 4j | 3.77 ± 0.005 | 3695 | 5.00 | intermediate |
| 21 | S. fragrans [1] | 42 | Figure 4k | 3.43 ± 0.025 | 3355 | 4.52 | small |
| Sect. Racemosae | |||||||
| 22 | S. gloriosa | 48 | Figure 4l | - | - | - | - |
| 23 | S. luerorum [2] | 48 | Figure 4m | 8.05 ± 0.103 | 7872 | 4.14 | intermediate |
| 24 | S. rosea [1] | 48 | Figure 4n | 5.93 ± 0.006 | 5800 | 3.48 | intermediate |
| Complex undatocarinata | |||||||
| 25 | S. undatocarinata [1] | - | - | 4.17 ± 0.013 | 4074 | 4.45 | intermediate |
The DNA content was estimated for 20 species (six Brasolia, two Elleanthus, and 12 Sobralia), ranging from 1C = 2.04 pg in E. crinipes to 1C = 8.05 pg in S. luerorum, reflecting a four-fold difference in genome size values (Table 1). According to the categorization proposed by Soltis et al. [16], three species can be considered as having small genomes (1C-value varying from 2.04 to 3.43 pg) and 17 as having intermediate genome sizes (1C-value varying from 3.77–8.05 pg; Table 1).
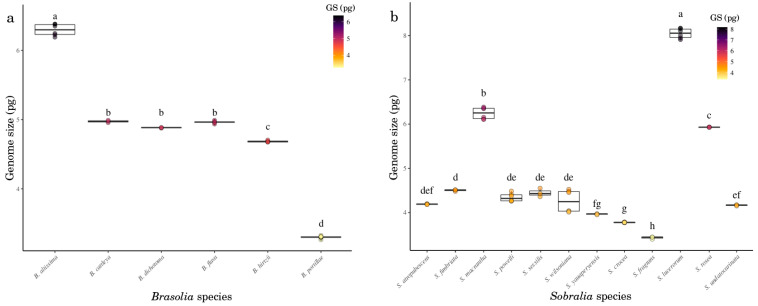
The genome size among Brasolia species ranged from 1C = 6.37 pg in B. altissima to 1C = 3.30 pg in B. portillae, representing a two-fold variation. According to 1C-value, Brasolia species could be organized into four groups (F = 3.719, p < 2 × 10−16; Figure 5a): the first represented by B. altissima with the largest genome size, the second group formed by B. cattleya-B. dichotoma-B. flava, the third composed by B. hirtzii, and the last group formed by B. portillae with the smallest genome size found in the genus (Figure 5a). The two Elleanthus species differ two-fold in their genome size, which enabled the identification of those two species based on the genome size (F = 5.622, p < 2 × 10−16; Table 1). Among the 12 Sobralia species, the estimated 1C-value presented a significant difference among species (F = 650, p < 2 × 10−16; Figure 5b), ranging from 1C = 3.43 pg in S. fragrans to 1C = 8.05 pg in S. luerorum (Table 1; Figure 5b). The three larger genome sizes were found in S. luerorum, S. macrantha, and S. rosea, while the two smaller ones were found in S. crocea and S. fragrans, while the remaining species formed a heterogeneous group without clear differentiation among species (Figure 5b).
Figure 5.
Brasolia (a) and Sobralia (b) genome sizes (1C-value). The 1C-value indicated by the same letter are not significantly different at p ≤ 0.05 according to the Duncan’s multiple range test.
2.3. Ancestral Reconstruction of Chromosome Number and Genome Size Indicate x = 25 and Small Genome Size for the Common Ancestor of Brasolia-Elleanthus-Sobralia
Regarding the ancestral reconstruction for chromosome number, for both trees, the ChromEvol suggested the same model as the best one: constant rate without duplication (Table 2). However, considering the ΔAIC < 2, no differences were found among constant rate, constant rate with demi-polyploidy and the best model for ML and BI phylogenies (Table 2). The Model Adequacy test confirmed the null model, constant rate without polyploidy, as the most adequate considering the variance and entropy tests (p = 0.13 and 0.21 for ML and p = 0.31 and 0.35 for IB, respectively) but not the parsimony (p = 0.00 for both ML and BI), suggesting that the chromosome number transitions are variable among clades. The best models highlight the importance of dysploidy variation as the main mechanism in chromosome evolution in Brasolia, Elleanthus and Sobralia.
Table 2.
ChromEvol model results to infer the ancestral base chromosome number and the chromosome number changes in the tribe Sobraliinae. The analysis was carried out for Maximum Likelihood and Bayesian Inference phylogenetic trees. For each tested ChromEvol model for each phylogenetic method, the Log-likelihood and AIC values are presented. The lowest AIC in each phylogenetic tree is indicated in bold and * indicates alternative model that does not differ from the lowest AIC.
| MODELS | Maximum Likelihood | Bayesian Inference | ||
|---|---|---|---|---|
| Log-Likelihood | AIC | Log-Likelihood | AIC | |
| Constante Rate | −51.76 | 109.5 * | −47.99 | 102 * |
| Constante Rate with demi-polyploid | −51.76 | 109.5 * | −47.99 | 102 * |
| Constante Rate with demi-polyploidy est | −51.76 | 111.5 | −47.99 | 104 |
| Constante Rate without polyploid (null model) | −51.76 | 107.5 | −47.99 | 99.99 |
| Linear rate | −50.92 | 111.8 | −47.71 | 105.4 |
| Linear rate with demi-polyploid | −50.92 | 111.8 | −47.71 | 105.4 |
| Linear rate with demi-polyploid est | −50.92 | 113.8 | −47.69 | 107.4 |
| Linear rate without polyploid (null model) | −50.92 | 109.8 | −47.71 | 103.4 |
The best model applied in both phylogenetic trees revealed similar results for most phylogenetic nodes. The base chromosome number x = 24 was suggested for the tribe Sobralieae, as well as for the genera Brasolia and Sobralia, while x = 25 was suggested for genus Elleanthus (Figure 2). The descending dysploidy was more frequently detected than ascending dysploidy, while no duplication was observed (Table S2). Successive descending dysploidies were observed in both phylogenetic trees reaching x = 23 in the large group comprehending S. atropubescens, S. sessilis, S. yauaperyensis, S. decora, S. fimbriata, S. powellii in IB phylogeny, but also grouping S. bimaculata, S. vallecaucana, S. crocea, and S. fragrans in ML phylogeny (Figure 2). Further descending dysploidies could be observed on S. crocea and S. fragrans, reaching x = 22, and B. cattleya and B. dichotoma, reaching x = 23 (ML and IB). Only one ascending dysploidy was detected in both trees, on the node of Elleanthus genus with x = 25 (Figure 2).
Despite statistics test could not differentiate Brasolia and Sobralia based on genome size (F = 0.488; p = 0.487), the ancestral reconstruction for genome size suggested a higher ancestral 1C-value for Sobralia (1C = 5.5 pg) compared to Brasolia (1C = 4.45–4.73 pg) (Figure 2). Some punctual amplifications could be observed inside each genus, even without any important chromosome number variation. For example, the species S. wilsoniana (1C = 4.25 pg), S. rosea (1C = 5.93 pg), S. macrantha (1C = 6.24 pg), and S. luerorum (1C = 8.05 pg) strongly vary among their 1C-values (Figure S1), although they present the same chromosome number 2n = 48 (n = 24; Figure 2). Inside Brasolia, genome size increase could be observed in B. altissima (1C = 6.37 pg, 2n = 46; Figure 2). A genome size reduction could be observed in S. crocea (1C = 3.77 pg, 2n = 44) and S. fragrans (1C = 3.43 pg, 2n = 42) and in B. portillae (1C = 3.3 pg, 2n = 48). An additional important genome size reduction was also observed in E. crinipes that presented half of the genome size (1C = 2.04 pg) compared with E. brasiliensis (1C = 4.79 pg), but with the same chromosome number (2n = 50 for both species; Figure 2).
Chromosome number presents no phylogenetic signal considering both phylogenetic trees, ML and BI, and tested indexes, Pagel’s lambda and Blomberg’s K suggesting that phylogenetic history has a minimal effect on both traits (Table 3). Otherwise, genome size seems to present phylogenetic signal considering both phylogenetic trees, ML and BI, and tested indexes, Pagel’s lambda (values very close to 1) and Blomberg’s K (values equal or superior to 1; see Table 3). Following the concept of phylogenetic signal, i.e., related species seems to present similar traits more than they resemble species drawn at random in this group, the phylogenetic signals for genome size suggested that this trait could reflect the evolutionary history of Brasolia-Elleanthus-Sobralia, being a potential informative trait to discuss the genera delimitation.
Table 3.
Pagel’s lambda and Blomberg’s K phylogenetic signals for chromosome number and genome size under Maximum Likelihood and Bayesian Inference.
| Index/Phylogenetic Reconstruction | Chromosome Number | Genome Size | ||
|---|---|---|---|---|
| Maximum Likelihood | Bayesian Inference | Maximum Likelihood | Bayesian Inference | |
| Pagel’s λ | 0.891 (p = 0.0003) | 0.885 (p = 0.0002) | 0.9875 (p = 4.83 × 10−7) | 0.9897 (p = 3.53 × 10−7) |
| Blomberg’s K | 0.276 (p = 0.003) | 0.523 (p = 0.001) | 1.00 (p = 0.001) | 2.20 (p = 0.001) |
3. Discussion
3.1. Phylogenetic Reconstruction
Both methodologies, ML and BI, presented the same general topology and agreed with previously published literature [4,5]. Baranow et al. [4] and Neubig et al. [5] noted that Brasolia species (along with Elleanthus) and Sobralia were placed on separate clades. It explicitly confirms the Dressler [6] attempt, followed by Baranow et al. [4], to place these two groups in two genera. The relationship between Brasolia and Elleanthus is hard to explain, especially considering the morphological distinctness between these two genera (compare Figure 1a–c with Figure 1d–j). The morphological differences between Brasolia and Elleanthus are much more visible than Sobralia and Brasolia morphological differences (compare Figure 1a–c with Figure 1d–j). Despite this, Sobralia and Brasolia formed two clades, both with high support. However, Elleanthus, even presenting apparent morphological differences from Brasolia, was placed nested on it in both phylogenies. Taking into account the present and previous phylogenetic studies, e.g., [4,5,6], one can suggest that Brasolia should be transferred to Elleanthus. However, from the practical point of view, it is not supported as the taxa are morphologically distinct, and the Elleanthus sampling is low. The Elleanthus taxonomic circumscription remains an open question for future studies in which a broader Sobralieae sampling should be mandatory.
3.2. Ancestral Base Chromosome Number Reconstruction Indicates Dysploidy as the Main Mechanism of Karyotype Evolution
There were just three available data regarding Sobralieae chromosome number [8] and no data about genome size. However, such data is now enlarged to 23 chromosome numbers and 20 genome sizes. The chromosome number reported to Sobralia [8], 2n = 48, was recurrent among species analyzed here, present in 38.1% of the counts. Furthermore, we confirm 2n = 50 to Elleanthus brasiliensis [8].
Our data showed a sizeable dysploid variation in both Brasolia and Sobralia, confirmed by the ancestral reconstruction analysis that suggested the descending dysploidy as the main mechanism of chromosome change in Sobralieae. Nevertheless, one should keep in mind that here dysploidy refers to chromosome number variation by one or few chromosomes and, in this sense, it includes both aneuploidy and dysploidy (for review see [9,17]). The importance of dysploidy in orchid chromosome evolution was already suggested in different orchid groups as subtribe Pleurothaliidinae (tribe Cymbidieae [18]) and the genera Christensonella Szlach., Mytnik, Górniak and Śmiszek [19], Heterotaxis Lindl. [16], Bifrenaria Lindl. and close genera [20], all from subtribe Maxillariinae, tribe Cymbidieae. In Christensonella and Heterotaxis, it was possible to map the point involved in the fusion-fission rearrangement causing the dysploid variation, confirming the occurrence of fission-fusion (i.e., dysploidy variation) associated with DAPI+ heterochromatic bands, reducing from 2n = 38 to 2n = 36 in the former genus [19] or with rDNA 45S, reducing from 2n = 42 to 2n = 40 in the last genus [15]. In tribe Sobralieae, we could confirm the importance of descending dysploidy with chromosome number reduction from 2n = 48 (n = 24) to 2n = 46, 44, 42 in Sobralia and Brasolia genera. However, based on the homogeneous chromosome morphology presented in the species analyzed here, it was impossible to infer the fission-fusion points or chromosome loss, causing the chromosome number reduction, making mandatory the use of in situ hybridization techniques.
The frequent occurrence of dysploidy in Sobralieae is in accordance with the diploidisation period following whole-genome duplication. Polyploidy is a central evolutionary mechanism in plants, but its occurrence is not random or constant throughout plant evolution [21,22,23]. Otherwise, it is suggested that polyploidy pulses occurred with high frequency at some geological periods as the Cretaceous-Paleogene transition (K-Pg; 66 Mya), Paleogene (Paleocene-Eocene transition; 56 Mya), and Neogene (at Late Miocene; 11 - 5 Mya), configuring periods of major stress (reviewed by [24]). Such concentration of polyploidy events in periods of geological history is confirmed by a high concentration of polyploid basal nodes in the Angiosperms phylogeny, representing some large extant plant families [23,24,25,26], which could be true for Orchidaceae with an estimated age of 90 million years ago (Mya; [27,28,29,30]). Furthermore, the suggested base chromosome number in Orchidaceae is x = 7 [8] or x = 10 [31], which suggests that most extant orchid species are polyploids. Such an assumption could be applied to the tribe Sobralieae (x = 24).
The tribe Sobralieae is placed on the base of the subfamily Epidendroideae (44.50–60.27 Mya; [30]), the largest Orchidaceae subfamily [32]. Sobralieae possibly diverged at 45 Mya on the Neotropics, with Brasolia, Elleanthus, and Sobralia diverging more recently, at ca. 7–10 Mya [27,28,30]. Based on the previously assumed base chromosome number of Orchidaceae, the tribe Sobralieae (x = 24) should be considered a hexaploid followed by descending dysploidy (x = 7 -> 14 -> 28 -> 26 -> 24) or tetraploid followed by ascending dysploidy (x = 10 -> 20 -> 22 -> 24). The genomic redundancy generated by polyploidy were “accommodated” in a diploid genome by the diploidisation period with descending dysploidy as one of the main chromosomal rearrangements observed during diploidisation [33]. The identification of descending dysploidy as the main mechanism of karyotype evolution in tribe Sobralieae is in accordance with the diploidisation period following polyploid pulses, possibly placed on the ancestral node of Orchidaceae. Since Brasolia and Sobralia are subject to the same karyotype evolution mechanism and because chromosome number has no phylogenetic signal, it is hard to use the karyotype to differentiate these groups. However, along the descending dysploidy, the elimination of repetitive sequences is another frequent event during the diploidisation period [34], affecting the genome size, which presents a phylogenetic signal.
3.3. Genome Size Reconstruction: The Role of Genome Size in Species Evolution
The genera Brasolia and Sobralia did not differ in genome size, while Elleanthus differs from both genera, probably due to the reduced 1C-value of E. crinipes. The genome size seems not to have any association with chromosome number variation. For example, Sobralia species with 2n = 48 vary 2-fold in their genome size (1C-value from 4.34 pg to 8.08 pg), while Brasolia species with 2n = 46 vary from 4.97 to 6.37 pg, and between the two Elleanthus species, both with 2n = 50, the genome size varies more than 2-fold (from 2.04 to 4.79 pg). The difference observed in genome size is probably due to differences in repetitive sequences as transposable elements and satellite DNA families [34,35]. The homogenisation and elimination of such redundant sequences are characteristics of the diploidisation process [35] and probably shaped the genomes in alternative ways resulting in genome size differences, even with the same chromosome number [36].
The chromosome number and genome size are essential as reproductive barriers, ensuring the species’ integrity [37]. Here, the most significant difference in genome size, conserving the same chromosome number, was observed in Elleanthus. The two studied Elleanthus species, besides differing in genome size, also differ in their distribution. While E. brasiliensis ranges from French Guiana to southern Brazil and in the Brazilian Atlantic Rain Forest occurring from the sea level until 1000 m, E. crinipes is endemic to the highland forest areas in Brazilian Atlantic Rain Forest, occurring only above 800 m [38,39]. Despite the region of sympatry between the two species that present simultaneous blooming and partially share pollinators, no hybrids were described between these two species, with no fruit being formed after manual interspecific crossings [38]. Despite the two species presenting the same chromosome number with similar karyotype morphologies, the two-fold genome size difference should configure as a strong post-zygotic barrier preventing hybridisation, highlighting the importance of genome size as a reproductive barrier and its importance in speciation (reviewed [36]).
3.4. Genus Delimitation in Sobralieae: Future Perspectives
Despite the genera Brasolia and Sobralia being placed on two groups with high support in the molecular phylogeny [4,5], they present very similar morphology and could not be differentiated based on chromosome number and genome size, as suggested by the ancestral state reconstruction. However, Elleanthus, with a distinct morphology, was nested in Brasolia [4,5] and present study but also did not show a clear difference in chromosome number and genome size. Our limited sampling of Elleanthus prevents further discussion but highlights the importance of future studies in this genus.
The phylogenetic position of Sobralieae makes studies in this tribe of particular interest to better understand the evolutionary trends in the major subfamily of Orchidaceae. Furthermore, considering that Epidendroideae represents ca. 80% of orchids, better comprehending the diversification of this clade configures as one of the main objectives in orchid diversification and evolution studies [30,40] since such subfamily reflects the diversity of the whole family, and Sobralieae, placed at the base of subfamily Epidendroideae, is of particular interest.
4. Materials and Methods
4.1. Plant Material
Plant material for cytological and molecular studies was cultivated in two living orchids collections: of the University of Gdansk, Poland and Federal University of ABC (UFABC). All the experiments were performed in accordance with relevant guidelines and regulations.
4.2. Phylogenetic Analysis
For phylogenetic reconstruction we sampled 57 individuals from 53 species distributed in four genera: 11 Brasolia, four Elleanthus, 34 Sobralia, and four Neottia species (outgroup) (Table S1). We sampled species favoring those with available material for cytogenetic data (chromosome number and genome size), but that also represent all groups detected in previous studies [4,5].
Genomic DNA extraction was performed in two ways, both AX Sherlock kit (A&A Biotechnology, Gdansk, Poland) and CTAB 2× protocol [41] excluding β-mercaptoethanol were used. For both protocols, a fragment of plant tissue (~30 mg) and a ceramic sphere were placed in FastPrep tubes and the samples were homogenized. The final pellet was resuspended in 50 µL TE buffer. Two sequences were amplified: nrITS and matK. The nrITS sequences were amplified using two different pairs of primers: 101F and 102R [42] and 17SE and 26SE [43], while, for the matK gene, the primers 19F [44] and 1326R [45] were used for all samples. The PCRs, for both of the markers, were performed in a total volume of 25 µL, containing 1 µL temple DNA (~10–100 ng), 0.5 µL of 10 µM of each primer, 1.0 µL dNTP mix (0.2 mM of each dNTP), 2.5 µL 10× Hybrid buffer contains 15 mM MgCl2, 0.5 µL Hybrid DNA Polymerase (2U/µL) and water. The reaction parameters for the nrITS were: 94 °C, 4 min; 30× (94 °C, 45 s; 52 °C, 45 s; 72 °C, 1 min); 72 °C, 7 min. For the matK fragment, we set the following parameters: 95 °C, 3 min; 33× (94 °C, 45 s; 52 °C, 45 s; 72 °C, 2 min 30 s); 72 °C, 7 min. The products of PCR reaction were cleanup using Wizard SV Gel and a PCR Clean Up System (Promega, Madison, WI, USA), following the manufacturer’s protocol. Purified PCR products were sequenced by Macrogen (Seoul, South Korea), using the same primers mentioned above. DNA sequence chromatograms were examined/edited in FinchTV software (https://finchtv.software.informer.com/1.4/ accessed on 12 October 2021).
The phylogenetic reconstruction was based on the two obtained matrices, from nrITS and matK, aligned in MAFFT v. 7.453 software using progressive alignment and iterative refinement methods [46]. The aligned sequences were trimmed using BioEdit v. 7.2 [47] and concatenated using Mesquite v 3.61 [48]. The generated matrix was analyzed under Maximum Likelihood (ML) and Bayesian Inference (BI).
The ML analysis was performed using IQ-TREE2 [49], searching for the best-scoring ML tree under the following nucleotide substitution models: TNe+G4 to nrITS and K3Pu + F + G4 to matK, both chosen by ModelFinder applying the Bayesian Information Criterion (BIC) [50]. We used 100 bootstrapping to assess branch supports. The BI was performed using MrBayes v. 3.2.7a [51] with GTR+I+G as the selected evolutive model for both partitions. Four independent runs, one hot and three cold chains, were started from different random trees to ensure that individual runs had converged to the same result. We used 20 million generations per chain with a sampling frequency of 2000. Split frequencies below 0.01 were used to check for convergence and the effective sample size (ESS) for each run was checked in the Tracer v. 1.7.1 [52]. In this case, 25% of trees were excluded as burn-in. Saved trees were summarized in a majority rule consensus tree created with nodal confidence assessed by posterior probabilities (PP), which were strongly supported when equal to or higher than 0.95 [53,54]. The ML and BI trees were edited with FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/ accessed on 12 October 2021) and Adobe Photoshop CS5 (Adobe Systems, Inc., San Jose, CA, USA). Alignments and phylogenetic trees are available from TreeBase (http://treebase.org, submission number 28791, accessed on 12 October 2021).
4.3. Cytogenetics Analysis
For chromosome counting, between one to four specimens per species were analyzed. For each specimen, at least three slides were analyzed, with, at least, five metaphase plates each. Fresh root tips were pretreated with 0.002 M 8-hydroxyquinoline at 4 °C or 10 °C for 12 h. The material was then fixed in a mixture of 99.8% ethanol and glacial acetic acid (3:1, v:v) and stained according to standard methods with aceto-orcein [55], Feulgen’s stain [56] or DAPI (1 µg mL−1) for 30 min. Chromosomes in late prophase and/or metaphase were analyzed using a Nikon Eclipse E800 epifluorescence microscope equipped with differential interference contrast (DIC) optics and a Nikon DS-5Mc CCD camera (PRECOPTIC Co., Warszawa, Poland) or with a fully automated upright fluorescent microscope (Leica DM6000 B, KAWA.SKA Sp. z.o.o., Zalesie Górne, Poland). Photomicrographs were arranged and analyzed in Adobe Photoshop (Elements 11 and CS6; Adobe System Inc., San Jose, CA, USA).
The genome size estimation was measured using fresh and young leaves of the selected Brasolia, Elleanthus, and Sobralia species. Depending on plant material availability, between one to three specimens per species were analyzed (Table 1), and each specimen was analyzed in triplicate (Table 1). Plant material was prepared as previously described [57], using 1 mL of Tris-MgCl2 nucleus isolation buffer [58] supplemented with 1% (w/v) polyvinylpyrrolidone (PVP-10) for Brasolia and Sobralia and using Ebihara buffer for Elleanthus [59]. The buffers were supplemented with ribonuclease A (50 mg mL−1) and propidium iodide (PI, 50 mg mL−1). For 25 accessions of Brasolia and Sobralia, Secale cereale cv. Dankowskie (2C = 16.19 pg; [60]) was used as an internal standard, whereas Pisum sativum cv. Set (2C = 9.11 pg; [61]) was used for S. luerorum accessions and Vicia faba cv. Inovec (2C = 26.90 pg; [60]) was used for Elleanthus species, avoiding sample and standard peaks overlapping. Genome size was estimated using a CyFlow SL Green (Partec GmbH, Münster, Germany) flow cytometer and a FACS Canto II cytometer (Becton Dickinson, San Jose, CA, USA), kindly made available by the Microbiology and Immunology Department of IBB-UNESP/Botucatu, Brazil, both equipped with a high-grade solid-state laser with green light emission at 532 nm. The 2C DNA content was measured in at least 5000 events using linear amplification. Histograms were evaluated using a FloMax program (Partec GmbH, Münster, Germany) and Flowing Software 2.5.1 (http://www.flowingsoftware.com/ accessed on 12 October 2021). The coefficient of variation (CV) of the G0/G1 peak of the studied species ranged between 2.84 and 5.00%. The nuclear DNA content of each species was calculated using the linear relationship between the ratio of the target species and the internal standard 2C peak positions on the histogram of fluorescence intensities. The 2C DNA contents (pg) were expressed in Mbp using the equation 1 pg = 978 Mbp [62]. The results of FCM estimation were analyzed using a one-way analysis of variance (ANOVA) following by Duncan’s test (p < 0.05) in R [63].
4.4. Chromosome Number and Genome Size Evolution
The phylogeny reconstructed here was pruned, removing species without information about chromosome number and genome size. The pruning used the function drop.tip from ‘ape’ package [64] in R [63]. The pruned phylogenetic tree was used to reconstruct the ancestral state for chromosome number, using ChromEvol v.2 [65,66] with posterior Adequacy test (Rice and Mayrose 2021) and genome size, using Trace Character History in Mesquite v 3.61 [60].
ChromEvol uses likelihood-based methods to evaluate the chromosome number changes along the phylogenetic branches, choosing the model that better performs among the eight available models. The inference considers four parameters: polyploidy (rate ρ), demi-duplication (i.e., a fusion of gametes of different ploidy, with rate μ), and dysploidy, which could be ascending (rate λ) or descending (rate δ). These parameters are combined and tested under two scenarios: constant and linear models. While the former estimates the rate of changes independently of the current chromosome number, the last considers the possibility of chromosome number change depending on the current chromosome number. Both model sets include a null model that assumes no polyploidization events. So, ultimately, the eight models (four constant models and four linear) were tested, allowing us to infer the main mechanism in chromosome number changes and the ancestral haploid number on the nodes of the phylogenetic tree. The best-fitted model was selected using the AIC and, considering the best-fitted model, we re-ran the analysis using 10,000 simulations. The difference among AIC values below two (ΔAIC < 2) were considered not distinguishable [67,68]. The model adequacy test [69] was performed online (http://chromevol.tau.ac.il/ accessed on 12 October 2021) to evaluate the selected model’s capability in generating the observed chromosome numbers.
Given the overall effect of phylogeny on a variation of chromosome number and genome size, we estimated the phylogenetic signal (i.e., the “tendency for related species to resemble each other more than they resemble species drawn at random from the tree”; [70,71] using Pagel’s lambda (λ; [72]). The index was estimated using phylosig from ‘phytools’ package [73] in R [63].
Acknowledgments
We want to express our gratitude to the curators of AMES, B, BM, COL, COAH, F, K, P, RPSC, U, US and W herbaria for the access to the collections they manage. We thank Carlos Nunes, John Romano and Bruce Rogers for the samples from their private collections used for the molecular study. We also want to thank the authors of the photos used to illustrate the studied species—Lourens Grobler, Eric Hunt, Marta Kolanowska, Andreas Kay and Carlos Nunes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23073948/s1.
Author Contributions
P.B., J.R. and A.P.M. designed the study; J.R., M.K. and J.B. performed chromosome analysis; M.D. provided molecular analysis; I.J. and M.R. conducted flow cytometric analysis; A.P.M. obtained chromosome data and genome size for Elleanthus, conducted phylogenetic analyses and ancestral reconstruction for chromosome number and genome size; J.R., P.B. and A.P.M. prepared the figures; D.S. provide conceptual advice; P.B., J.R. and A.P.M. wrote the manuscript; J.R. agrees to serve as the author responsible for contact and ensures communication. All authors have read and agreed to the published version of the manuscript.
Funding
The work was financially supported by University of Gdansk grants n. 531-D030-D243-20 and 531-D030-D847-21 to J.R., J.B. and M.K. and Brazilian National Council for Scientific and Technological Development (CNPq)—Research grant (proc. 312855/2021-4) to A.P.M.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Alignments and phylogenetic trees are available from TreeBase (http://treebase.org, submission number 28791, accessed on 12 October 2021).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ruiz H.J., Pavón J. In: Flora Peruvianae et Chilensis Prodromus. Ruiz H.J., Pavón J., editors. G. de Sancha; Madrid, Spain: 1794. [Google Scholar]
- 2.Brieger F.G. Subtribus Sobraliinae. In: Brieger F.G., Maatsch R., Senghas K., editors. Die Orchideen. Volume 13. Lieferung, VerlagPaul Parey; Berlin/Hamburg, Germany: 1983. pp. 780–800. [Google Scholar]
- 3.Dressler R.L. The major sections or groups within Sobralia, with four new species from Panama and Costa Rica, S. crispissima, S. gloriana, S. mariannae and S. nutans. Lankasteriana. 2002;5:9–15. doi: 10.15517/lank.v2i3.23088. [DOI] [Google Scholar]
- 4.Baranow P., Dudek M., Szlachetko D.L. Brasolia, a new genus highlighted from Sobralia (Orchidaceae) Plant Syst. Evol. 2017;303:853–871. doi: 10.1007/s00606-017-1413-z. [DOI] [Google Scholar]
- 5.Neubig K.M., Whitten W.M., Blanco M., Endara L., Williams N.H., Koehler S. Preliminary molecular phylogenetics of Sobralia and relatives (Orchidaceae: Sobralieae) Lankesteriana. 2011;11:307–317. doi: 10.15517/lank.v11i3.18286. [DOI] [Google Scholar]
- 6.Dressler R.L., Blanco M.A., Pupulin F., Neubig K.M. Proposal to conserve the name Sobralia (Orchidaceae) with a conserved type. Taxon. 2011;60:907–908. doi: 10.1002/tax.603030. [DOI] [Google Scholar]
- 7.de Resende K.F.M. Karyotype Evolution: Concepts and Applications. In: Bhat T., Wani A., editors. Chromosome Structure and Aberrations. Springer; New Delhi, India: 2017. pp. 181–200. [DOI] [Google Scholar]
- 8.Felix L.P., Guerra M. Variation in chromosome number and the basic number of subfamily Epidendroideae (Orchidaceae) Bot. J. Linn. Soc. 2010;163:234–278. doi: 10.1111/j.1095-8339.2010.01059.x. [DOI] [Google Scholar]
- 9.Guerra M. Cytotaxonomy: The end of childhood. Plant Biosyst. 2012;146:703–710. doi: 10.1080/11263504.2012.717. [DOI] [Google Scholar]
- 10.Mayrose I., Lysak M.A. The Evolution of Chromosome Numbers: Mechanistic Models and Experimental Approaches. Genome Biol. Evol. 2021;13:evaa220. doi: 10.1093/gbe/evaa220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felix L.P., Guerra M. Cytogenetics and cytotaxonomy of some Brazilian species of Cymbidioid orchids. Genet. Mol. Biol. 2000;23:957–978. doi: 10.1590/S1415-47572000000400041. [DOI] [Google Scholar]
- 12.Yeh H.Y., Lin C.S., de Jong H., Chang S.B. Two reported cytotypes of the emergent orchid model species Erycina pusilla are two different species. Euphytica. 2017;213:957–978. doi: 10.1007/s10681-017-2026-x. [DOI] [Google Scholar]
- 13.Leitch I.J., Kahandawala I.M., Suda J., Hanson L., Ingrouille M.J., Chase M.W., Fay M.F. Genome size diversity in orchids: Consequences and evolution. Ann. Bot. 2009;104:469–481. doi: 10.1093/aob/mcp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moraes A.P., Souza-Chies T.T., Stiehl-Alves E.M., Burchardt P., Eggers L., Siljak-Yakovlev S., Brown S.C., Chauveau O., Nadot S., Bourge M., et al. Evolutionary trends in Iridaceae: New cytogenetic findings from the New World. Bot. J. Linn. Soc. 2015;177:27–49. doi: 10.1111/boj.12232. [DOI] [Google Scholar]
- 15.Moraes A.P., Simões A.O., Alayon D.I.O., De Barros F., Forni-Martins E.R. Detecting mechanisms of karyotype evolution in Heterotaxis (Orchidaceae) PLoS ONE. 2016;11:e0165960. doi: 10.1371/journal.pone.0165960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soltis D., Soltis P., Bennett M., Leitch I. Evolution of genome size in the angiosperms. Am. J. Bot. 2003;90:1596–1603. doi: 10.3732/ajb.90.11.1596. [DOI] [PubMed] [Google Scholar]
- 17.Guerra M. Chromosome numbers in plant cytotaxonomy: Concepts and implications. Cytogenet. Genome Res. 2008;120:339–350. doi: 10.1159/000121083. [DOI] [PubMed] [Google Scholar]
- 18.De Oliveira I.G., Moraes A.P., De Almeida E.M., de Assis F.N.M., Cabral J.S., De Barros F., Felix L.P. Chromosomal evolution in Pleurothallidinae (Orchidaceae: Epidendroideae) with an emphasis on the genus Acianthera: Chromosome numbers and heterochromatin. Bot. J. Linn. Soc. 2015;178:102–120. doi: 10.1111/boj.12273. [DOI] [Google Scholar]
- 19.Koehler S., Cabral J.S., Whitten W.M., Williams N.H., Singer R.B., Neubig K.M., Guerra M., Souza A.P., Amaral M.D.C.E. Molecular Phylogeny of the Neotropical Genus Christensonella (Orchidaceae, Maxillariinae): Species Delimitation and Insights into Chromosome Evolution. Ann. Bot. 2008;102:491–507. doi: 10.1093/aob/mcn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moraes A.P., Koehler S., Cabral J.S., Gomes S.S.L., Viccini L.F., Barros F., Felix L.P., Guerra M., Forni-Martins E.R. Karyotype diversity and genome size variation in Neotropical Maxillariinae orchids. Plant Biol. 2017;19:298–308. doi: 10.1111/plb.12527. [DOI] [PubMed] [Google Scholar]
- 21.Jiao Y. Double the Genome, Double the Fun: Genome Duplications in Angiosperms. Mol. Plant. 2018;11:357–358. doi: 10.1016/j.molp.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Magallón S., Sánchez-reyes L.L., Gómez-acevedo S.L. Thirty clues to the exceptional diversification of flowering plants. Ann. Bot. 2019;123:491–503. doi: 10.1093/aob/mcy182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin D.A. Did dysploid waves follow the pulses of whole genome duplications? Plant Syst. Evol. 2020;306:75. doi: 10.1007/s00606-020-01704-5. [DOI] [Google Scholar]
- 24.Van de Peer Y., Ashman T.L., Soltis P.S., Soltis D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell. 2021;33:11–26. doi: 10.1093/plcell/koaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanneste K., Baele G., Maere S., Van De Peer Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 2014;24:1334–1347. doi: 10.1101/gr.168997.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soltis P.S., Marchant D.B., Van de Peer Y., Soltis D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015;35:119–125. doi: 10.1016/j.gde.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Givnish T.J., Spalink D., Ames M., Lyon S., Hunter S.J., Zuluaga A., Iles W.J.D., Clements M.A., Arroyo M.T.K., Leebens-Mack J., et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B. 2015;282:20151553. doi: 10.1098/rspb.2015.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Givnish T.J., Spalink D., Ames M., Lyon S.P., Hunter S.J., Zuluaga A., Doucette A., Caro G.G., McDaniel J., Clements M.A., et al. Orchid historical biogeography, diversification, Antarctica and the paradox of orchid dispersal. J. Biogeogr. 2016;43:1905–1916. doi: 10.1111/jbi.12854. [DOI] [Google Scholar]
- 29.Li Y.-X., Li Z.-H., Schuiteman A., Chase M.W., Li J.-W., Huang W.-C., Hidayat A., Wu S.-S., Jin X.-H. Phylogenomics of Orchidaceae based on plastid and mitochondrial genomes. Mol. Phylogenet. Evol. 2019;139:106540. doi: 10.1016/j.ympev.2019.106540. [DOI] [PubMed] [Google Scholar]
- 30.Serna-Sánchez M.A., Pérez-Escobar O.A., Bogarín D., Torres-Jimenez M.F., Alvarez-Yela A.C., Arcila-Galvis J.E., Hall C.F., de Barros F., Pinheiro F., Dodsworth S., et al. Plastid phylogenomics resolves ambiguous relationships within the orchid family and provides a solid timeframe for biogeography and macroevolution. Sci. Rep. 2021;11:6858. doi: 10.1038/s41598-021-83664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carta A., Bedini G., Peruzzi L. A deep dive into the ancestral chromosome number and genome size of flowering plants. New Phytol. 2020;228:1097–1106. doi: 10.1111/nph.16668. [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Escobar O.A., Dodsworth S., Bogarín D., Bellot S., Balbuena J.A., Schley R.J., Kikuchi I.A., Morris S.K., Epitawalage N., Cowan R., et al. Hundreds of nuclear and plastid loci yield novel insights into orchid relationships. Am. J. Bot. 2021;108:1166–1180. doi: 10.1002/ajb2.1702. [DOI] [PubMed] [Google Scholar]
- 33.Baduel P., Bray S., Vallejo-Marin M., Kolář F., Yant L. The ‘Polyploid Hop’: Shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Front. Ecol. Evol. 2018;6:117. doi: 10.3389/fevo.2018.00117. [DOI] [Google Scholar]
- 34.Mandáková T., Lysak M.A. Post-polyploid diploidization and diversification through dysploid changes. Curr. Opin. Plant Biol. 2018;42:55–65. doi: 10.1016/j.pbi.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Garrido-Ramos M.A. Satellite DNA: An Evolving Topic. Genes. 2017;8:230. doi: 10.3390/genes8090230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellicer J., Hidalgo O., Dodsworth S., Leitch I.J. Genome size diversity and its impact on the evolution of land plants. Genes. 2018;9:88. doi: 10.3390/genes9020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rezende L., Suzigan J., Amorim F.W., Moraes A.P. Can plant hybridization and polyploidy lead to pollinator shift? Acta Bot. Bras. 2020;34:229–242. doi: 10.1590/0102-33062020abb0025. [DOI] [Google Scholar]
- 38.Nunes C.E., Amorim F.W., Mayer J.L., Sazima M. Pollination ecology of two species of Elleanthus (Orchidaceae): Novel mechanisms and underlying adaptations to hummingbird pollination. Plant Biol. 2016;18:15–25. doi: 10.1111/plb.12312. [DOI] [PubMed] [Google Scholar]
- 39.Udulutsch R.G. Elleanthus in Flora do Brasil. Jardim Botânico do Rio de Janeiro. [(accessed on 12 October 2021)];2020 Available online: http://floradobrasil.jbrj.gov.br/reflora/floradobrasil/FB86254.
- 40.Freudenstein J.V., Chase M.W. Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: Progressive specialization and diversification. Ann. Bot. 2015;115:665–681. doi: 10.1093/aob/mcu253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 42.Douzery E.J.P., Pridgeon A.M., Kores P., Linder H.P., Kurzweil H., Chase M.W. Molecular Phylogenetics of Disae (Orchidaceae): A contribution from nuclear ribosomal ITS sequences. Am. J. Bot. 1999;86:887–899. doi: 10.2307/2656709. [DOI] [PubMed] [Google Scholar]
- 43.Sun Y., Skinner D.Z., Liang G.H., Hulbert S.H. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoret. Appl. Genet. 1994;89:26–32. doi: 10.1007/BF00226978. [DOI] [PubMed] [Google Scholar]
- 44.Molvary M., Kores P.J., Chase M.W. Polyphyly of mycoheterotrophic orchids and functional influences on floral and molecular characters. In: Morrison D.A., Wilson K.L., editors. Monocots: Systematic and Evolution. CSIRO Publishing; Collingwood, Australia: 2000. pp. 441–448. [Google Scholar]
- 45.Cuénoud P., Savolainen V., Chatrou L.W., Powell M., Grayer R.J., Chase M.W. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB and matK DNA sequences. Am. J. Bot. 2002;89:132–144. doi: 10.3732/ajb.89.1.132. [DOI] [PubMed] [Google Scholar]
- 46.Katoh K., Standley D.M. MAFFT Multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 48.Maddison W.P., Maddison D.R. Mesquite: A Modular System for Evolutionary Analysis. 2019. [(accessed on 12 October 2021)]. Version 3.61. Available online: http://www.mesquiteproject.org.
- 49.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: New models and efficient methodes for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalyaanamoorthy S., Quang M.B., Wong T., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronquist F., Teslenko M., van der Mark P., Huelsenbeck J. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambaut A., Drummond A., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian Phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cummings M.P., Handley S.A., Myers D.S., Reed D.L., Rokas A., Winka K. Comparing Bootstrap and Posterior Probability Values in the Four-Taxon Case. Syst. Biol. 2003;52:477–487. doi: 10.1080/10635150390218213. [DOI] [PubMed] [Google Scholar]
- 54.Simmons M.P., Pickett K.M., Miya M. How Meaningful Are Bayesian Support Values? Mol. Biol. Evol. 2004;21:188–199. doi: 10.1093/molbev/msh014. [DOI] [PubMed] [Google Scholar]
- 55.Jensen W.A. In: Botanical Histochemistry: Principles and Practice. Jensen W.A., editor. W.H. Freeman & Co.; San Francisco, CA, USA: 1962. pp. 1–408. [Google Scholar]
- 56.Fukui K., Nakayama S. In: Plant Chromosomes: Laboratory Methods. Fukui K., Nakayama S., editors. CRC Press; Boca Raton, FL, USA: 1996. pp. 1–288. [Google Scholar]
- 57.Jedrzejczyk I., Sliwinska E. Leaves and seeds as materials for flow cytometric estimation of the genome size of 11 Rosaceae woody species containing DNA-staining inhibitors. J. Bot. 2010;2010:930895. doi: 10.1155/2010/930895. [DOI] [Google Scholar]
- 58.Rewicz A., Rewers M., Jędrzejczyk I., Rewicz T., Kołodziejek J., Jakubska-Busse A. Morphology and genome size of Epipactis helleborine (L.) Crantz (Orchidaceae) growing in anthropogenic and natural habitats. PeerJ. 2018;6:e5992. doi: 10.7717/peerj.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ebihara A., Ishikawa H., Matsumoto S., Lin S.J., Iwatsuki K., Takamiya M., Watano Y., Ito M. Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. Am. J. Bot. 2005;92:1535–1547. doi: 10.3732/ajb.92.9.1535. [DOI] [PubMed] [Google Scholar]
- 60.Doležel J., Greilhuber J., Lucretti S., Meister A., Lysák M.A., Nardi L., Obermayer R. Plant genome size estimation by flow cytometry: Inter-laboratory comparison. Ann. Bot. 1998;82:17–26. doi: 10.1093/oxfordjournals.aob.a010312. [DOI] [Google Scholar]
- 61.Doležel J., Bartos J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pellicer J., Powell R.F., Leitch I.J. The application of flow cytometry for estimating genome size, ploidy level, endopolyploidy, and reproductive modes in plants. In: Besse P., editor. Molecular Plant Taxonomy. Springer; New York, NY, USA: 2021. [DOI] [PubMed] [Google Scholar]
- 63.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2021. [(accessed on 12 October 2021)]. Available online: https://www.R-project.org/ [Google Scholar]
- 64.Paradis E., Schliep K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 65.Mayrose I., Barker M.S., Otto S. Probabilistic models of chromosome number evolution and the inference of polyploidy. Syst. Biol. 2010;59:132–144. doi: 10.1093/sysbio/syp083. [DOI] [PubMed] [Google Scholar]
- 66.Glick L., Mayrose I. ChromEvol: Assessing the pattern of chromosome number evolution and the inference of polyploidy along a phylogeny. Mol. Biol. Evol. 2014;31:1914–1922. doi: 10.1093/molbev/msu122. [DOI] [PubMed] [Google Scholar]
- 67.Burnham K.P., Anderson D.R., editors. Model Selection and Multimodel Inference. Springer; New York, NY, USA: 2002. pp. 49–97. [DOI] [Google Scholar]
- 68.Burnham K.P., Anderson D.R. Multimodel Inference: Understanding AIC and BIC in Model Selection. Sociol. Methods Res. 2004;33:261–304. doi: 10.1177/0049124104268644. [DOI] [Google Scholar]
- 69.Rice A., Mayrose I. Model adequacy tests for probabilistic models of chromosome number evolution. New Phytol. 2021;229:3602–3613. doi: 10.1111/nph.17106. [DOI] [PubMed] [Google Scholar]
- 70.Blomberg S.P., Garland T. Tempo and mode in evolution: Phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 2002;15:899–910. doi: 10.1046/j.1420-9101.2002.00472.x. [DOI] [Google Scholar]
- 71.Münkemüller T., Lavergne S., Bzeznik B., Dray S., Jombart T., Schiffers K., Thuiller W.W. How to measure and test phylogenetic signal. Methods Ecol. Evol. 2012;3:743–756. doi: 10.1111/j.2041-210X.2012.00196.x. [DOI] [Google Scholar]
- 72.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 73.Revell L.J., Mahler D.L., Peres-Neto P.R., Redelings B.D. A new phylogenetic method for identifying exceptional phenotypic diversification. Evolution. 2012;66:135–146. doi: 10.1111/j.1558-5646.2011.01435.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Alignments and phylogenetic trees are available from TreeBase (http://treebase.org, submission number 28791, accessed on 12 October 2021).