Abstract
Thymic Epithelial Neoplasms (TENs) represent the most common tumors of the thymus gland. Epigenetic alterations are generally involved in initiation and progression of various cancer entities. However, little is known about the role of epigenetic modifications in TENs. In order to identify relevant studies, a literature review was conducted using the MEDLINE and LIVIVO databases. The search terms thymoma, thymic carcinoma, thymic epithelial neoplasm, epigenetics, DNA methylation, HDAC and miRNA were employed and we were able to identify forty studies focused on TENs and published between 1997 and 2021. Aberrant epigenetic alterations seem to be involved in the tumorigenesis of thymomas and thymic carcinomas, with numerous studies reporting on non-coding RNA clusters and altered gene methylation as possible biomarkers in different types of TENs. Interestingly, Histone Deacetylase Inhibitors have shown potent antitumor effects in clinical trials, thus possibly representing effective epigenetic therapeutic agents in TENs. Additional studies in larger patient cohorts are, nevertheless, needed to verify the clinical utility and safety of novel epigenetic agents in the treatment of patients with TENs.
Keywords: thymoma, thymic carcinoma, TEN, epigenetics, non-coding RNA, DNA methylation, HDACI
1. Introduction
The thymus is a secondary lymphoid gland located in the upper anterior mediastinum with its histologic architecture including the epithelium and the lymphoid component [1]. TENs represent a wide range of tumors deriving from the thymic epithelial cells that can be divided into thymomas and thymic carcinomas (TC). The World Health Organization (WHO) histopathological classification further subdivides thymomas into type A, type AB, type B1, type B2 and type B3 based on the morphology of epithelial tumor cells, the relative proportion of the lymphocytic component and the differentiation grade. TC are classified as type C TEN by the WHO [2,3,4].
Thymomas are considered as the most common neoplasms of the thymus gland, yet overall, thymomas are rare. The American Cancer Society estimates their incidence at 400 cases in the United States annually [5]. TCs account for 20% of all thymus cancers [5]. Based on patients diagnosed with thymus cancer between 2010 and 2016, the five-year relative survival rate is 71% for all surveillance, epidemiology, and end results (SEER) stages combined [6].
The majority of TEN are incidentally detected on chest X-ray or scan prior to symptom onset. Symptoms caused by thymus tumors either result from the compression of neighboring anatomical structures such as the airways and the superior vena cava, or are paraneoplastic, with myasthenia gravis (MG) representing the most common autoimmune disease associated with TEN [7].
Diagnostic evaluation of TEN includes, in addition to a physical examination, a computed tomography (CT) scan of the chest, usually combined with a positron emission tomography (PET) scan. magnetic resonance imaging (MRI) may be used to measure the tumor’s size. Definite diagnosis requires biopsy of the tumor mass [8]. Of note, the thymus gland physiologically disappears after puberty. As such, when children or infants present with thymic pathologies, the common diagnosis techniques like X-ray, CT or scintigraphy may not be always used, given their dangerous repercussions for future quality of life. Interestingly, when it comes to children, the roles of diet and especially obesity seems to play an important role in the altered morphology and function of TEN cells due to the infiltration and accumulation of adipocytes in tumor-bearing thymuses [9].
For patients with early-stage resectable TEN, thymectomy represents the first-line therapy. Patients with more advanced stage TEN may also be treated with radiation and/or chemotherapy after surgery, especially in cases of incomplete tumor resection [10].
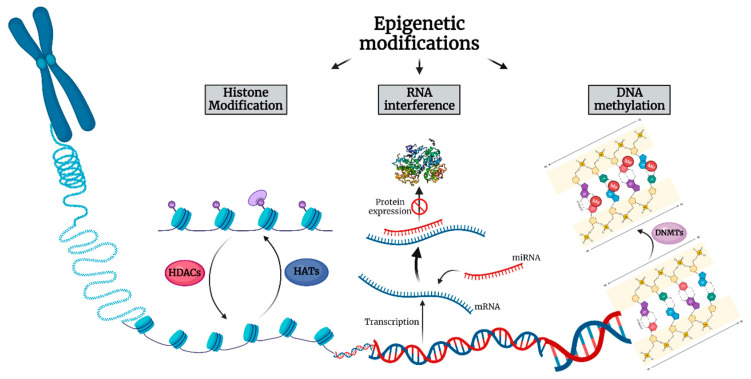
Cancer initiation and progression is now realized to involve genetic alterations along with epigenetic abnormalities. Epigenetics refers to the study of heritable changes in gene expression that enable cells to have distinct identities by regulating what genetic information can be accessed by cellular machinery without changing the primary DNA sequence [11]. Epigenetic modifications include DNA methylation, posttranslational modifications of histone proteins, nucleosome remodeling and non-coding RNAs, specifically microRNA (miRNA) expression (Figure 1) [12].
Figure 1.
The three main mechanisms of epigenetic alterations comprised of histone acetylation, protein expression obstruction through RNA interference and DNA methylation. HDAC: Histone deacetylase, HAT: Histone acetyltransferase, DNMT: DNA methyltransferase.
DNA methylation is regulated by a family of DNA methyltransferases (DNMTs) and involves the transfer of a methyl group onto the fifth carbon position of the cytosine to form 5-methylcytosine [13]. This heritable epigenetic mechanism regulates gene expression by recruiting proteins involved in gene repression or by inhibiting the binding of transcription factor(s) to DNA [14]. In cancer cells, global hypomethylation of the cancer genome, promoter hypermethylation of tumor suppressor genes and potentially direct mutagenesis of 5-methylcytosine-containing sequences through deamination of methylated cytosine can promote cancer initiation and evolution [15].
Post-translational modification of histone proteins by acetylation reduces the affinity of histones for the negatively charged DNA, thus enabling DNA strands to uncoil and transcription to occur [16]. The level of histone acetylation relies on the opposing actions of histone acetylases (HATs) and histone deacetylases (HDACs) [17]. HDACs catalyze the removal of acetyl groups on the NH2-terminal lysine residues of core nucleosomal histones, which generally results in transcriptional repression and the silencing of tumor-suppressor genes [18]. As a consequence, deregulation of histone acetylation can promote the development of human cancer, as shown by a great number of researchers who focused on revealing the link between histone acetylation/deacetylation and tumorigenesis [19,20,21].
The majority of the human genome is transcribed into non-coding RNAs [22]. miRNAs and long non-coding RNAs (lncRNAs) represent two major families of these non-protein-coding transcripts that can regulate fundamental cellular processes [23]. Dysregulation of miRNAs and lncRNAs is critical to cancer development and progression. Both miRNAs and lncRNAs have been reported to act as either oncogenes or tumor suppressors to regulate cancer biology via diverse molecular mechanisms, such as amplification, deletion, abnormal epigenetic or transcriptional regulation [24]. miRNAs are in particular associated with the hallmarks of neoplasia, ranging from angiogenesis induction to apoptosis resistance and metastasis activation [23].
In this review, we extensively investigated epigenetic alternations in TEN, incorporating research works that explored their role in carcinogenesis in both in vitro and in vivo studies. In particular, we searched for possible epigenetic biomarkers in TEN, as well as possible treatment options. The literature review was conducted using the MEDLINE and LIVIVO databases. The search terms thymoma, thymic carcinoma, thymic epithelial neoplasm, epigenetics, DNA methylation, HDAC and miRNA were employed and we were able to identify forty studies focused on TEN and published between 1997 and 2021.
2. Alterations of Non-Coding RNAs in Thymic Epithelial Neoplasms
Alterations of non-coding RNAs play an established role in TEN development and progression. A large number of studies have assessed the epigenetic abnormalities associated with alterations of non-coding RNAs in TEN patient tissue samples (Table 1). Ganci et al. performed miRNA expression profiling by microarray analysis of formalin-fixed paraffin embedded tissue in TENs versus normal thymic tissues and identified 87 miRNAs differently expressed between thymic tumor and normal samples, with selected miRNAs distinguishing the different TEN histotypes. miRNA-21-5p was found to be up- and miRNA-145-5p down-regulated in TEN, whereas miRNA-142-5p, miRNA-363-3p and miRNA-16-2-3p were shown to be predicted to target molecular pathways involved in thymic carcinogenesis, such as Baculoviral IAP Repeat Containing 3 (BIRC3), SCYA20 and MYC [25]. Wei et al., found miRNA-140, miRNA-450b, miRNA-542, miRNA-639, miRNA-3613 and miRNA-3913–1 to be positively associated and miRNA-1976 to be negatively associated with overall survival in thymomas. In terms of disease-free survival, let-7a-1, let-7a-2 and let-7a-3 miRNAs positively correlated with disease-free survival in thyomas, whereas miRNA-324 negatively correlated with it. Binary tree prediction discriminated TC from type A-B3 thymomas by identifying 91 miRNAs with differential expression between TC and non-type C thymomas [26]. Bellissimo et al. selected miRNA-21-5p, miRNA-148a-3p, miRNA-141-3p, miRNA-34b-5p, miRNA-34c-5p, and miRNA-455-5p to evaluate their expression in blood plasma and peripheral blood mononuclear cells and confirmed increased levels for miRNA-21-5p and miRNA-148a-3p. Plasma levels of miRNA-21-5p and miRNA-148a-3p were significantly reduced during follow-up after thymectomy [27]. In their study, Radovich et al. revealed 100% concordance between gene expression clusters and TEN histotype. A large miRNA cluster on chromosome 19q13.42 (C19MC) was shown to be significantly overexpressed in all A and AB TEN and virtually absent in the other thymomas and normal tissues, while overexpression of C19MC activated the oncogenic PI3K/AKT/mTOR pathway [28]. Enkner et al. suggested, on the other hand, that C19MC expression is silenced in TCs as a result of promoter methylation, and that the expression of miRNA cluster C14MC on chromosome 14q32 is decreased in TCs as compared to type A thymomas. Moreover, miRNA-21, miRNA-9-3 and miRNA-375 were up-, whereas miRNA-34b, miRNA-34c, miRNA-130a and miRNA-195 were down-regulated in TCs [29]. Ji et al. performed RNA sequence analysis and detected 65 differentially-expressed lncRNAs in thymomas, including AFAP1-AS1, LINC00324, ADAMTS9-AS1, VLDLR-AS1, LINC00968, and NEAT1 that have been validated with The Cancer Genome Atlas (TCGA) database. 1695 miRNAs were also reported to be overexpressed in thymomas. A network analysis of the lncRNA-mRNA-miRNA regulation axes identified a cluster of miRNAs upregulated in thymomas, triggering the expression of target protein-coding genes and disrupting various biological pathways, such as the PI3K/Akt/mTOR, FoxO, and Hypoxia-Inducible Factor (HIF)-1 signaling pathways [30]. Furthermore, the lncRNAs ADAMTS9-AS1, HSD52, LINC00968 and LINC01697 were described as potential markers in terms of accurate patient stratification in low versus high risk TEN, as well as in effective recurrence probability prediction [31]. Gene expression analysis by microarray in a cohort of fresh frozen TEN and normal tissues identified LINC00174 as up-regulated in TEN cases, with its expression positively correlating with a 5-genes signature. LINC00174 favored the expression of Syntabulin (SYBU), FEM1B, and Stearoyl-CoA Desaturase 5 (SCD5) genes by sponging miRNA-145-5p, thus affecting lipid metabolism and, consequently, TEN cell migration [32]. In their study, Wang et al., found LOXL1-AS1 and HSPA9 to be overexpressed in TEN and associated with poor prognosis. miR-525-5p expression was down-regulated, predicted patient prognosis and inhibited the expression of HSPA9 protein by targeting the 3′-untranslated region (UTR) of HSPA9 mRNA. LOXL1-AS1 promoted HSPA9 expression as a sponge targeting miR-525-5p. An in vivo tumor-burdened assay also showed that knockdown of miR-525-5p promoted tumorigenesis by stimulating the expression of HSPA9 [33]. In thymomatous MG, the aberrant decrease of miRNA-19b regulated thymic stromal lymphopoietin (TSLP) expression and contributed to T helper type 17 cells development [34]. Similarly, miRNA-20b was found to be down-regulated in patients with thymoma-associated MG and to target Nuclear Factor of Activated T-cells 5 (NFAT5) and Calmodulin Binding Transcription Activator 1 (CAMTA1), thus inhibiting T-cell proliferation and activation. Of note, the expression levels of miRNA-20b and NFAT5/CAMTA1 were inversely correlated in patients with thymomatous MG [35]. Yang et al., reported on the disparate role of the pseudogene RP11-424C20.2 and its parental gene UHRF1, and showed that high expression levels predicted better overall survival in TEN through the regulation of tumor-infiltrating immune cell levels, which is mediated at least partly through Interferon (IFN)-γ-mediated Clathrin Light Chain A (CLTA)-4 and Programmed Death-Ligand 1 (PD-L1) pathways. miRNA-378a-3p was down-, whereas miRNA-422a up-regulated in thymomas [36]. Analyses of 87 thymoma samples with complete MG information revealed distinct expression of immune-related genes and lncRNAs in thymoma with and without MG. The study found the methylation level of the immune-related lncRNAs AC004943.1, WT1-AS and FOXG1-AS1 to be significantly decreased in TEN and to correlate with their expression. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses revealed the functional pathways of the target genes of these immune-related lncRNAs with transcription coregulator activity and cell cycle representing the most enriched pathways for targets of lncRNA AC004943.1, and actin binding and axon guidance the most enriched pathways for targets of lncRNA WT1-AS, respectively [37]. Chen et al., described that the three lncRNAs: LINC00665, NR2F1-AS1 and RP11-285A1.1, as well as the four miRNAs hsa-miRNA-143, hsa-miRNA-141, hsa-miRNA-140 and hsa-miRNA-3199, were significantly associated with prognosis and overall survival of TEN [38].
Table 1.
Alterations of non-coding RNAs in TENs.
| Non-Coding RNA | Cell Lines/Tissue Samples | Methods | Main Results | References |
|---|---|---|---|---|
| 87 miRNAs | 54 thymic tumor samples, 12 normal counterparts |
Reverse transcription quantitative real-time Polymerase Chain Reaction (RT-qPCR), Immunohistochemistry |
|
[25] |
| 91 miRNAs | 17 type A, 38 type AB, 13 type B1, 31 type B2, 13 type B3, 11 type C thymomas |
KEGG pathway analysis of miRNA target genes |
|
[26] |
| miRNA-21-5p, miRNA-148a-3p, miRNA-141-3p, miRNA-34b-5p, miRNA-34c-5p, miRNA-455-5p | Peripheral blood samples from five patients with TEN |
MicroRNA expression analysis |
|
[27] |
| Large microRNA cluster on chr19q13.42 |
|
RNA-sequencing, qPCR, protein analyses, drug sensitivity experiments |
|
[28] |
| miRNA cluster C14MC on chromosome 14q32, miRNA-21, miRNA-9-3, miRNA-375, miRNA-34b, miRNA-34c, miRNA-130a, miRNA-195 |
18 type A, 19 type B3 thymomas, 35 TC |
Cancer gene panel sequencing, miRNA sequencing, fluorescence in situ hybridization (FISH), immunohistochemistry |
|
[29] |
| 65 differentially-expressed lncRNAs, 1695 miRNAs |
25 thymomas, 25 healthy thymus specimens |
lncRNA-miRNA-mRNA functional enrichment analyses |
|
[30] |
| lncRNAs ADAMTS9-AS1, HSD52, LINC00968, LINC01697 | TCGA | Statistics |
|
[31] |
| lncRNA LINC00174 |
|
Bioinformatics analysis of expression data from TCGA and Istituto Regina Elena thymoma cohorts, cDNA reverse transcriptase, RT-q PCR |
|
[32] |
| miR-525-5p |
|
RT-qPCR, dual luciferase reporter assay, cell counting kit 8 assay, flow cytometry, transwell assay, western blot analysis, tumor-burdened assay |
|
[33] |
| miRNA-19b | nine type A, 11 type B1, 20 type B2, 12 type B3 thymomas, 11 normal thymi | Luciferase reporter assay, qRT-PCR, western blot |
|
[34] |
| miRNA-20b |
|
qRT-PCR, MTT assay, cell cycle analysis, immunostaining, flow cytometric analysis, western blot |
|
[35] |
| RP11-424C20.2, miRNA-378a-3p, miRNA-422a |
DreamBase, TCGA | RP11-424C20.2 cellular localization prediction, GO and KEGG enrichment analysis, correlation of UHRF1 expression with immune infiltration analysis |
|
[36] |
| lncRNAs AC004943.1, WT1-AS, FOXG1-AS1 |
11 type A, 26 type AB, 10 type B1, 22 type B2, nine type B3 thymomas, 9 TC | Statistical analysis |
|
[37] |
|
16 samples of type A, 35 samples of type AB, 57 samples of type B thymomas, 11 samples of TC, two samples of normal tissue. |
GO and KEGG pathway annotation of DEmRNAs, overall survival analysis and establishment of the TEN-specific prognostic significance |
|
[38] |
3. Alterations of DNA Methylation in TENs
Numerous studies have investigated the effects of DNA methylation alterations in TEN (Table 2). Bioinformatic analysis of the TCGA dataset revealed 5155 and 6967 hyper- and hypo-methylated CpG sites in type A–B3 TEN and TC, respectively, of which 3600 were located within the gene promoter regions. One hundred and thirty-four genes were found to be silenced by promoter hypermethylation, while 174 mRNAs were up-regulated. Cox regression analysis showed significant association between the methylation levels of 187 sites and the overall survival in patients with TEN, with cg05784862 (KSR1), cg07154254 (ELF3), cg02543462 (ILRN), and cg06288355 (RAG1) representing independent prognostic factors [39]. Bi et al., attempted to examine the whole-genome DNA methylation status of TEN and identify differences in thymoma DNA methylation profiles. More than 10,000 CpGs were found to be differentially methylated between thymoma types A and B, with 36 genes having differentially methylated CpG sites in their promoter region. A Kyoto Encyclopedia of Genes and Genomes analysis identified focal adhesion and regulation of the actin cytoskeleton as the most enriched pathway of differentially methylated genes between tumors and healthy controls. Among the 29 genes that were hypomethylated with a high expression, zinc finger protein 396 and Fraser extracellular matrix complex subunit 1 had the largest area under the curve [40]. Another study, investigating whether a common polymorphism (×579G>T: rs1569686) in the promoter of the DNMT3B gene increases the risk to develop thymomatous MG, found a statistically significant association of the DNMT3B-579T allele and the TT homozygous genotype with the risk of TEN [41]. Coppedè et al. investigated MutL homolog 1 (MLH1), O(6)-methylguanine-DNA methyltransferase (MGMT), Cyclin Dependent Kinase Inhibitor (CDKN2A) and Ras association domain family 1 isoform A (RASSF1A) methylation levels in blood and tumor specimens from 69 patients with thymoma-associated MG and in the adjacent normal thymus available from 44 of them, and concluded that promoter methylation levels of these genes were neither increased in thymoma nor associated with the histopathological features of TEN [42]. On the contrary, Mokhtar et al. found MGMT methylation to be significantly more frequent in advanced TEN than in early thymomas [43]. Suzuki et al., also reported a significantly higher methylation index in TC than in thymomas, with the Secreted Protein Acidic and Rich in Cysteine (SPARC) genes being methylated in all examined TC samples [44]. The analysis of global methylation levels and of the promoter methylation status of the tumor suppressor genes (TSG) hMLH1, MGMT, p-16INK4a, RASSF1A, Fragile Histidine Triad Diadenosine Triphosphatase (FHIT), Anaphase-promoting complex subunit 1 (APC1A), Retinoic acid receptor β (RARB), Death-associated protein kinase (DAPK) and E-cadherin in 65 TEN samples revealed hypermethylation and decreased TSG expression in types B1 or higher thymomas. In comparison with early-stage TEN, global DNA methylation levels were found to be reduced, whereas DNMT1, DNMT3a, and DNMT3b expression was increased in advanced-stage thymomas [45]. The methylation status determination in a series of 13 resected thymoma samples by the sodium bisulfite-modification method followed by sequencing analysis revealed a significantly high frequency of methylation in the F-box and WD40 domain protein 7 (FBXW7) β-form promoter in types B1 or higher TEN [46]. Hirabayashi et al., examined 36 thymomas and four TC, and observed hypermethylation of the promoter region in the CDKN2 gene in four thymomas and one TC using a PCR-based assay, as well as genomic Southern hybridization [47]. Another relevant study examining aberrant DNA methylation of DAPK, p-16, MGMT and Hyperpigmentation, Progressive, 1 (HPP1) genes in 26 thymomas and 6 TC described the frequency of aberrant DNA methylation to correlate with the histopathologic TEN type and to be, consequently, higher in TC than thymoma [48]. Furthermore, Iaiza et al. found that the methyltransferase METTL3 is overexpressed in TEN and responsible for the induction of c-MYC expression through methylation and delocalization of the lncRNA MALAT1. Silencing of METTL3 combined with cisplatin or c-MYC inhibitor promotes apoptosis in TEN cells [49]. Bisulfite pyrosequencing in 46 TEN and 20 paired thymus tissues revealed higher promoter methylation of G protein subunit gamma 4 (GNG4), growth hormone secretagogue receptor (GHSR), homeobox D9 (HOXD9) and spalt like transcription factor 3 (SALL3) in TC than in thymoma. Higher promoter methylation of these four genes correlated with a significantly worse relapse-free survival in all TEN types [50]. In a subsequent study, the same study group speculated that DNA methylation of GHSR correlates with a shift from native to variant expression, thus inducing the tumorigenesis of thymoma, yet not TC [51]. Coppedè et al. investigated GHSR methylation levels in blood, tumor tissue, and adjacent healthy thymic tissue from 65 MG patients and observed hypermethylation in all WHO histological subtypes, but particularly at advanced disease stages [52]. The AutoImmune REgulator (AIRE) promoter was found to be hypomethylated in AIRE-negative TEN with variable individual patterns, thus indicating that the lack of expression does not result from the CpG-methylation mediated silencing of the AIRE gene promoter [53]. Ye et al., employed bisulphite sequencing to identify hypomethylation in the 5′ promoter region of the pro-opiomelanocortin (POMC) gene in five TC tumors resected from patients with ectopic adrenocorticotropic hormone (ACTH) syndrome and concluded that hypomethylation of this promoter correlates with POMC overexpression and the ectopic ACTH syndrome in TC [54]. The methylation-sensitive high-resolution melting technique for the analysis of DNA methylation levels of genes involved in one-carbon metabolism and DNA methylation in blood, tumor tissue, and healthy thymic epithelial cells from thymomatous MG patients showed significantly higher methylation of the methylenetetrahydrofolate reductase (MTHFR) promoter in thymomas, as well as some degrees of methylation of the DNMT3A gene in thymic tissue with respect to blood [55]. Masunaga et al., observed no methylation in the Phosphatase and Tensin homolog (PTEN) promoter region in TEN tumor cells [56]. Moreover, Deleted in liver cancer one (Dlc1) isoform 2 was described to undergo selective hypermethylation in oncogenic Kirsten rat sarcoma virus oncogene homolog (K-Ras) 2 induced TEN and significantly decrease the overall survival in Dlc1 gene trap mice [57]. Saito et al., showed that TC with Tet Methylcytosine Dioxygenase 2 (TET2) mutations had more hypermethylated genes and that hypermethylation correlated with down-regulation of gene expression [58]. Another study, examining DNA methylation by bisulfite pyrosequencing, revealed significantly higher methylation levels of glutamate decarboxylase 1 gene (GAD1) in TC tissues, with GAD1 methylation exhibiting high sensitivity and specificity for discriminating between TC and thymoma. More importantly, patients with TEN with high GAD1 DNA hypermethylation showed significantly shorter relapse-free survival rates [59]. Interestingly, the positive correlation between promoter DNA methylation and GAD1 expression in TEN was attributed to the inhibition of a Polycomb repressive complex 2 (PRC2) by the methylation of CCCTC-binding factor (CTCF) −3, with H3K27me3 levels markedly reduced at the GAD1 promoter. Through multiomic analysis of the TCGA, Yang et al. reported that KIT proto-oncogene ligand (KITLG) represents a new hallmark of type A and AB thymomas by inducing aberrant alterations of mRNA, miRNA and DNA methylation [60].
Table 2.
Alterations of DNA methylation in TENs.
| DNA Methylation Sites | Cell Lines/Tissue Samples | Methods | Main Results | References |
|---|---|---|---|---|
| 5155 hyper-,6967 hypo-methylated CpG sites in type A–B3 TEN and TC | TCGA thymoma datasets with DNA methylation profiles of 124 tumor tissues and 2 matched adjacent normal tissues from 124 cases of thymoma, 54 thymomas, 46 TC |
Statistical analysis, pyrosequencing |
|
[39] |
| 10,000 CpG sites | 1 atypical type A, 1 type A, 1 type AB, 1 type B1, 1 type B2, 2 type B3 thymomas, 1 atypical TC | DNA isolation and bisulfite treatment, illumina 850K methylation microarray, statistical analysis |
|
[40] |
| DNMT3B-579T allele | Peripheral blood of 324 AChR+ MG patients, 735 healthy matched unrelated controls, 94 patients with thymoma | Genotyping analysis performed by means of PCR-RFLP techniques |
|
[41] |
| MLH1, MGMT, CDKN2A, RASSF1A | Blood samples and surgically resected thymomas from 69 patients with thymomatous MG and in the adjacent normal thymus available from 44 of them |
Methylation sensitive-high resolution melting |
|
[42] |
| MGMT | four type A, 12 type AB, 13 type B1, 7 type B2, eight type B3 thymomas, 23 TC | Nested methylation-specific PCR (MSP), immunohistochemical analysis |
|
[43] |
| SPARC | six thymoma, five TC, 22 non-small cell lung carcinoma samples | MSP, mutation assay |
|
[44] |
| hMLH1, MGMT, p-16INK4a, RASSF1A, FHIT, APC1A, RARB, DAPK, E-cadherin | 10 type A, 10 type AB, 12 type B1, nine type B2, seven type B3 thymomas, 17 TC | Real-time RT-PCR, nested MSP, immunohistologic analysis of global DNA methylation, measurement of global DNA methylation by an ELISA-like reaction |
|
[45] |
| FBXW7 β-form promoter | one type A, three type AB, five type B1, three type B2, one type B3 thymomas | Genomic sodium bisulfite sequencing analysis |
|
[46] |
| CDKN2 | 36 thymomas (non-invasive type, 16 cases; invasive type, 20 cases), three TC | Immunohistochemistry, PCR-SSCP, sequencing, PCR-based methylation assay, southern hybridization |
|
[47] |
| DAPK, p-16, MGMT, HPP1 | one type A, six type AB, 10 type B1, four type B2, five type B3 thymomas, six TC | Sodium bisulfite modification |
|
[48] |
| METTL3 |
|
Immunohistochemistry, clonogenic assays, real-time qRT-PCR analysis, lysis and immunoblotting analysis, polysome profiling and treatment with puromycin, m6A immunoprecipitation, FISH |
|
[49] |
| GNG4, GHSR, HOXD9, SALL3 | five type A, two type AB, four type B1, 10 type B2, and nine type B3 thymomas, 12 TC, four NECTT samples |
Bisulfite conversion of genomic DNA, bisulfite pyrosequencing |
|
[50] |
| GHSR | six type A, six type AB, eight type B1, 11 type B2, 10 type B3 thymomas, 13 TC, four NECTT samples |
RT-qPCR, immunohistochemistry |
|
[51] |
| GHSR | 12 type A, 12 type AB, five type B1, 22 type B2, eight type B3, five type B2-B3, one not specified thymoma samples |
Bisulfite modification, methylation- sensitive high-resolution melting analysis |
|
[52] |
| AIRE |
|
Reverse transcriptase quantitative RT-PCR, bisulfite genomic DNA sequencing, quick and quantitative chromatin immunoprecipitation |
|
[53] |
| POMC | three normal thymuses, one large cell lung cancer, five thymic carcinoid tumors | Immunohistochemistry, quantitation of POMC gene expression, bisulphite modification and sequencing |
|
[54] |
| MTHFR, DNMT3A | 13 type A, 13 type AB, five type B1, 23 type B2, eight type B3, five type B2/3, two non-specified thymoma samples | Bisulfite modification, methylation- sensitive–high-resolution melting analysis |
|
[55] |
| PTEN | three type A, eight type AB, 11 type B1, six type B2, five type B3 thymoma samples, four TC, two normal thymi | Immunohistochemistry, PCR direct sequencing, methylation-specific PCR, reverse transcription real-time PCR after target cell collection with laser microdissection |
|
[56] |
| Dlc1 |
|
Fluorescence microscopy, multiplex RT-PCR, western blotting, immunohistochemistry, microdissection, loss of heterozygosity analysis, DNA methylation study of Dlc1 promoter region, transendothelial migration and in vitro filipodia formation assay of the T lymphoma cells, treatment of cell culture with 5 azacytidine |
|
[57] |
| TET2 | nine squamous cell carcinomas, one undifferentiated carcinoma | Exome sequencing, analysis of mutational signature, genome copy number analysis and tumor content estimation, whole transcriptome sequencing, DNA methylation analysis, gene ontology analysis |
|
[58] |
| GAD1 | nine type A, 11 type AB, 19 type B1, 20 type B2, 14 type B3, 17 type C thymoma samples | Bisulfite conversion of genomic DNA, global methylation analysis, bisulfite pyrosequencing, RT-qPCR, immunohistochemistry |
|
[59] |
| KITLG |
|
RNA-seq data analysis, KITLG small-interfering RNA silencing, KITLG-overexpressing plasmid construction, real-time PCR, western blot analysis |
|
[60] |
4. Alterations of Histone Modifications in Thymic Epithelial Neoplasms
Several studies have examined the role of histone modification alterations in TEN by mainly focusing on modification of HDAC activity (Table 3). Arjonen et al., performed image-based high content drug screening to assess tumor cell specific responses and identified selective sensitivity of the TEN cells to HDAC Inhibitors (HDACI) Belinostat and Panobinostat [61]. A phase II study of Belinostat in 41 patients with recurrent or refractory advanced TEN demonstrated partial responses in two patients with thymoma, but no responses among patients with TC, with survival of patients with thymoma being significantly longer than that of patients with TC [62]. A phase I/II trial of Belinostat in combination with cisplatin, doxorubicin and cyclophosphamide in TEN, on the other hand, revealed objective response rates of 64% in thymomas and 21% in TC, respectively [63]. Bellissimo et al. found that treatment of TC1889 cells with the HDAC inhibitor Valproic Acid (VPA) led to miRNA-145-5p up-regulation and exhibited antitumor effects, such as reduction of cell viability, colony forming ability and migration capability. Interestingly, VPA also seemed to enhance the response of TEN cells to chemotherapy [64].
Table 3.
Alterations of histone modifications in TENs.
| HDACI | Cell Lines/Tissue Samples/Patient Collective | Methods | Main Results | References |
|---|---|---|---|---|
| Belinostat, Panobinostat | Bronchial metastatic lesion of a 54-year old female patient with a metastatic thymoma with pleural invasion |
Mutation analysis, high content imaging drug screening |
|
[61] |
| Belinostat | 25 patients with thymoma, 16 patients with TC |
Pharmacodynamic analyses: Protein acetylation, peripheral blood mononuclear cell immune subsets, circulating angiogenic markers |
|
[62] |
| Belinostat | two patients with type B1, seven with type B2, three with type B3 thymoma, 14 patients with TC |
Monitoring for treatment-related adverse events, clinical laboratory tests, vital signs, physical examinations, 12-lead electrocardiograms, pharmacokinetic evaluations, multiparametric flow cytometry, CT scan |
|
[63] |
| VPA | 10 peritumoral thymi, 14 thymomas, 5 TC | Microarray hybridization, RT-qPCR, immunohistochemistry, transfection and treatment, lysate preparation and immunoblotting analysis, transwell migration assay, clonogenic assays, immunocytochemistry and morphological analysis, cell cycle analysis, ATPlite luminescence assay system, formaldehyde cross-linking and chromatin immunoprecipitation, plasmid construction and dual-luciferase reporter assay |
|
[64] |
5. Conclusions
The present review describes epigenetic alterations, searches for possible epigenetic biomarkers and explores possible novel treatment options in TEN. Alterations of non-coding RNAs seem to play an important role in TEN carcinogenesis and progression with numerous studies identifying up- or down-regulation of different miRNA/lncRNA clusters in thymomas and TC. Selected non-coding RNAs may distinguish different TEN histotypes, given their differential expression in normal thymi, thymomas and TC, target various molecular oncogenic pathways involved in TEN pathophysiology, such as the PI3K/AKT/mTOR, the FoxO, or the HIF-1 signaling pathway, as well as regulate immune cell proliferation and activation in thymomatous MG. Moreover, up- or down-regulation of non-coding RNAs either positively or negatively correlates with overall and disease specific survival in TEN, depending on each miRNA/lncRNA cluster. In terms of DNA methylation alterations, multiple CpG sites seem to be either hyper- or hypo-methylated in different histopathologic TEN types. Many of these CpG sites are located within the gene promoter regions, with DNA methylation alterations thus resulting in either the activation or silencing of important tumor suppressor and DNA repair genes associated with TEN tumorigenesis. Notably, MG-associated thymomas seem to represent a molecularly distinct subtype of TEN, given that hypermethylation of certain genes such as MLH1, MGMT, CDKN2A, and RASSF1A might be more frequent in TC than in thymomas [43,45,47,48], yet hypermethylation of these genes is neither frequent in thymomatous MG tissues nor does it correlate with the histopathologic features of the tumor [42]. Importantly, DNA methylation levels seem to significantly correlate with overall and relapse-free survival in patients with TEN, thus representing a significant prognostic factor. All in all, both non-coding RNAs and DNA methylation levels could be regarded as potential epigenetic biomarkers in terms of accurate patient stratification in low versus high risk TEN, effective recurrence probability prediction and, most importantly, overall patient survival, with a great number of studies incorporating hundreds of samples from datasets, such as the TCGA, and providing reliable and reproducible data that prove the role of epigenetic modifications in TEN pathogenesis and prognosis [31,36,39,60]. Interestingly, HDACIs seem to represent promising epigenetic therapeutic agents with potent antitumor effects in TEN. Belinostat has already been tested in a phase II clinical trial, showing promising results in a small patient collective with recurrent or refractory advanced thymomas [62], thus opening the way for further clinical studies to be conducted in larger patient collectives, with an aim to verify the clinical utility and safety of epigenetic modulators and develop novel treatments targeting different epigenetic alterations in TEN.
Author Contributions
Literature analysis and conceptualization, I.P., A.P. and S.T.; original draft preparation and writing, I.P.; art work, A.P.; review and supervision, K.V., S.T. and C.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Valavanis C., Stanc G.M., Baltayiannis N. Classification, histopathology and molecular pathology of thymic epithelial tumors: A review. J. BUON. 2021;26:1198–1207. [PubMed] [Google Scholar]
- 2.Marx A., Chan J.K.C., Chalabreysse L., Dacic S., Detterbeck F., French C.A., Hornick J.L., Inagaki H., Jain D., Lazar A.J., et al. The 2021 WHO Classification of Tumors of the Thymus and Mediastinum: What Is New in Thymic Epithelial, Germ Cell, and Mesenchymal Tumors? J. Thorac. Oncol. 2022;17:200–213. doi: 10.1016/j.jtho.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Marx A., Chan J.K., Coindre J.-M., Detterbeck F., Girard N., Harris N.L., Jaffe E.S., Kurrer M.O., Marom E.M., Moreira A.L., et al. The 2015 World Health Organization Classification of Tumors of the Thymus: Continuity and Changes. J. Thorac. Oncol. 2015;10:1383–1395. doi: 10.1097/JTO.0000000000000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marx A., Ströbel P., Badve S., Chalabreysse L., Chan J.K., Chen G., de Leval L., Detterbeck F., Girard N., Huang J., et al. ITMIG consensus statement on the use of the WHO histological classification of thymoma and thymic carcinoma: Refined definitions, histological criteria, and reporting. J. Thorac. Oncol. 2014;9:596–611. doi: 10.1097/JTO.0000000000000154. [DOI] [PubMed] [Google Scholar]
- 5.American Cancer Society . Key Statistics About Thymus Cancers. American Cancer Society; Atlanta, GA, USA: 2021. [Google Scholar]
- 6.American Cancer Society . Survival Rates for Thymus Cancer. American Cancer Society; Atlanta, GA, USA: 2021. [Google Scholar]
- 7.American Cancer Society . Signs and Symptoms of Thymus Cancers. American Cancer Society; Atlanta, GA, USA: 2021. [Google Scholar]
- 8.American Cancer Society . Tests for Thymus Cancer. American Cancer Society; Atlanta, GA, USA: 2021. [Google Scholar]
- 9.Lamas A., Lopez E., Carrio R., Lopez D.M. Adipocyte and leptin accumulation in tumor-induced thymic involution. Int. J. Mol. Med. 2016;37:133–138. doi: 10.3892/ijmm.2015.2392. [DOI] [PubMed] [Google Scholar]
- 10.American Cancer Society . Treatment of Thymus Cancers by Extent and Type of Tumor. American Cancer Society; Atlanta, GA, USA: 2021. [Google Scholar]
- 11.Centers for Disease Control and Prevention . What is Epigenetics? Centers for Disease Control and Prevention; Atlanta, GA, USA: 2020. [Google Scholar]
- 12.Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin B., Li Y., Robertson K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M., Costello J. DNA methylation: An epigenetic mark of cellular memory. Exp. Mol. Med. 2017;49:e322. doi: 10.1038/emm.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.McKenna N.J., O’Malley B.W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/S0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 18.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 19.Archer S.Y., Hodin R.A. Histone acetylation and cancer. Curr. Opin. Genet. Dev. 1999;9:171–174. doi: 10.1016/S0959-437X(99)80026-4. [DOI] [PubMed] [Google Scholar]
- 20.Ropero S., Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goutas D., Theocharis S., Tsourouflis G. Unraveling the Epigenetic Role and Clinical Impact of Histone Deacetylases in Neoplasia. Diagnostics. 2021;11:1346. doi: 10.3390/diagnostics11081346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palazzo A.F., Lee E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue M., Zhuo Y., Shan B. MicroRNAs, Long Noncoding RNAs, and Their Functions in Human Disease. Methods Mol. Biol. 2017;1617:1–25. doi: 10.1007/978-1-4939-7046-9_1. [DOI] [PubMed] [Google Scholar]
- 24.Grillone K., Riillo C., Scionti F., Rocca R., Tradigo G., Guzzi P.H., Alcaro S., Di Martino M.T., Tagliaferri P., Tassone P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020;39:117. doi: 10.1186/s13046-020-01622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganci F., Vico C., Korita E., Sacconi A., Gallo E., Mori F., Cambria A., Russo E., Anile M., Vitolo D., et al. MicroRNA expression profiling of thymic epithelial tumors. Lung Cancer. 2014;85:197–204. doi: 10.1016/j.lungcan.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Wei J., Liu Z., Wu K., Yang N., He Y., Chen G.G., Zhang J., Lin J. Identification of prognostic and subtype-specific potential miRNAs in thymoma. Epigenomics. 2017;9:647–657. doi: 10.2217/epi-2016-0161. [DOI] [PubMed] [Google Scholar]
- 27.Bellissimo T., Russo E., Ganci F., Vico C., Sacconi A., Longo F., Vitolo D., Anile M., Disio D., Marino M., et al. Circulating miR-21-5p and miR-148a-3p as emerging non-invasive biomarkers in thymic epithelial tumors. Cancer Biol. Ther. 2016;17:79–82. doi: 10.1080/15384047.2015.1108493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radovich M., Solzak J.P., Hancock B.A., Conces M.L., Atale R., Porter R.F., Zhu J., Glasscock J., Kesler K.A., Badve S.S., et al. A large microRNA cluster on chromosome 19 is a transcriptional hallmark of WHO type A and AB thymomas. Br. J. Cancer. 2016;114:477–484. doi: 10.1038/bjc.2015.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enkner F., Pichlhöfer B., Zaharie A.T., Krunic M., Holper T.M., Janik S., Moser B., Schlangen K., Neudert B., Walter K., et al. Molecular Profiling of Thymoma and Thymic Carcinoma: Genetic Differences and Potential Novel Therapeutic Targets. Pathol. Oncol. Res. 2017;23:551–564. doi: 10.1007/s12253-016-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji G., Ren R., Fang X. Identification and Characterization of Non-Coding RNAs in Thymoma. Med. Sci. Monit. 2021;27:e929727. doi: 10.12659/MSM.929727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su Y., Chen Y., Tian Z., Lu C., Chen L., Ma X. lncRNAs classifier to accurately predict the recurrence of thymic epithelial tumors. Thorac. Cancer. 2020;11:1773–1783. doi: 10.1111/1759-7714.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tito C., Ganci F., Sacconi A., Masciarelli S., Fontemaggi G., Pulito C., Gallo E., Laquintana V., Iaiza A., De Angelis L., et al. LINC00174 is a novel prognostic factor in thymic epithelial tumors involved in cell migration and lipid metabolism. Cell Death Dis. 2020;11:959. doi: 10.1038/s41419-020-03171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Huang H., Zhang X., Ma H. LOXL1AS1 promotes thymoma and thymic carcinoma progression by regulating miR5255pHSPA9. Oncol. Rep. 2021;45:117. doi: 10.3892/or.2021.8068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Wang Z., Chen Y., Xu S., Yang Y., Wei D., Wang W., Huang X. Aberrant decrease of microRNA19b regulates TSLP expression and contributes to Th17 cells development in myasthenia gravis related thymomas. J. Neuroimmunol. 2015;288:34–39. doi: 10.1016/j.jneuroim.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Xin Y., Cai H., Lu T., Zhang Y., Yang Y., Cui Y. miR-20b Inhibits T Cell Proliferation and Activation via NFAT Signaling Pathway in Thymoma-Associated Myasthenia Gravis. Biomed. Res. Int. 2016;2016:9595718. doi: 10.1155/2016/9595718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J., Zhang Y., Song H. A disparate role of RP11-424C20.2/UHRF1 axis through control of tumor immune escape in liver hepatocellular carcinoma and thymoma. Aging. 2019;11:6422–6439. doi: 10.18632/aging.102197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhuang J., Guan M., Liu M., Liu Y., Yang S., Hu Z., Lai F., He F. Immune-Related Molecular Profiling of Thymoma With Myasthenia Gravis. Front. Genet. 2021;12:756493. doi: 10.3389/fgene.2021.756493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen K., Bai L., Ji L., Wu L., Li G. Bioinformatics analysis of the key potential ceRNA biomarkers in human thymic epithelial tumors. Medicine. 2021;100:e26271. doi: 10.1097/MD.0000000000026271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S., Yuan Y., Xiao H., Dai J., Ye Y., Zhang Q., Zhang Z., Jiang Y., Luo J., Hu J., et al. Discovery and validation of DNA methylation markers for overall survival prognosis in patients with thymic epithelial tumors. Clin. Epigenet. 2019;11:38. doi: 10.1186/s13148-019-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi Y., Meng Y., Niu Y., Li S., Liu H., He J., Zhang Y., Liang N., Liu L., Mao X., et al. Genomewide DNA methylation profile of thymomas and potential epigenetic regulation of thymoma subtypes. Oncol. Rep. 2019;41:2762–2774. doi: 10.3892/or.2019.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coppedè F., Ricciardi R., Denaro M., De Rosa A., Provenzano C., Bartoccioni E., Baggiani A., Lucchi M., Mussi A., Migliore L. Association of the DNMT3B -579G>T polymorphism with risk of thymomas in patients with myasthenia gravis. PLoS ONE. 2013;8:e80846. doi: 10.1371/journal.pone.0080846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coppedè F., Ricciardi R., Lopomo A., Stoccoro A., De Rosa A., Guida M., Petrucci L., Maestri M., Lucchi M., Migliore L. Investigation of MLH1, MGMT, CDKN2A, and RASSF1A Gene Methylation in Thymomas From Patients With Myasthenia Gravis. Front. Mol. Neurosci. 2020;13:567676. doi: 10.3389/fnmol.2020.567676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mokhtar M., Kondo K., Namura T., Ali A.H., Fujita Y., Takai C., Takizawa H., Nakagawa Y., Toba H., Kajiura K., et al. Methylation and expression profiles of MGMT gene in thymic epithelial tumors. Lung Cancer. 2014;83:279–287. doi: 10.1016/j.lungcan.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki M., Chen H., Shigematsu H., Ando S., Iida T., Nakajima T., Fujisawa T., Kimura H. Aberrant methylation: Common in thymic carcinomas, rare in thymomas. Oncol. Rep. 2005;14:1621–1624. doi: 10.3892/or.14.6.1621. [DOI] [PubMed] [Google Scholar]
- 45.Chen C., Yin B., Wei Q., Li D., Hu J., Yu F., Lu Q. Aberrant DNA methylation in thymic epithelial tumors. Cancer Investig. 2009;27:582–591. doi: 10.1080/07357900802620869. [DOI] [PubMed] [Google Scholar]
- 46.Gu Z., Mitsui H., Inomata K., Honda M., Endo C., Sakurada A., Sato M., Okada Y., Kondo T., Horii A. The methylation status of FBXW7 beta-form correlates with histological subtype in human thymoma. Biochem. Biophys. Res. Commun. 2008;377:685–688. doi: 10.1016/j.bbrc.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 47.Hirabayashi H., Fujii Y., Sakaguchi M., Tanaka H., Yoon H.E., Komoto Y., Inoue M., Miyoshi S., Matsuda H. p16INK4, pRB, p53 and cyclin D1 expression and hypermethylation of CDKN2 gene in thymoma and thymic carcinoma. Int. J. Cancer. 1997;73:639–644. doi: 10.1002/(SICI)1097-0215(19971127)73:5<639::AID-IJC5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 48.Hirose Y., Kondo K., Takizawa H., Nagao T., Nakagawa Y., Fujino H., Toba H., Kenzaki K., Sakiyama S., Tangoku A. Aberrant methylation of tumour-related genes in thymic epithelial tumours. Lung Cancer. 2009;64:155–159. doi: 10.1016/j.lungcan.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 49.Iaiza A., Tito C., Ianniello Z., Ganci F., Laquintana V., Gallo E., Sacconi A., Masciarelli S., De Angelis L., Aversa S., et al. METTL3-dependent MALAT1 delocalization drives c-Myc induction in thymic epithelial tumors. Clin. Epigenet. 2021;13:173. doi: 10.1186/s13148-021-01159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kishibuchi R., Kondo K., Soejima S., Tsuboi M., Kajiura K., Kawakami Y., Kawakita N., Sawada T., Toba H., Yoshida M., et al. DNA methylation of GHSR, GNG4, HOXD9 and SALL3 is a common epigenetic alteration in thymic carcinoma. Int. J. Oncol. 2020;56:315–326. doi: 10.3892/ijo.2019.4915. [DOI] [PubMed] [Google Scholar]
- 51.Tegshee B., Kondo K., Soejima S., Muguruma K., Tsuboi M., Kajiura K., Kawakami Y., Kawakita N., Toba H., Yoshida M., et al. GHSR methylation-dependent expression of a variant ligand and receptor of the ghrelin system induces thymoma tumorigenesis. Oncol. Lett. 2021;22:793. doi: 10.3892/ol.2021.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coppedè F., Stoccoro A., Nicolì V., Gallo R., De Rosa A., Guida M., Maestri M., Lucchi M., Ricciardi R., Migliore L. Investigation of GHSR methylation levels in thymomas from patients with Myasthenia Gravis. Gene. 2020;752:144774. doi: 10.1016/j.gene.2020.144774. [DOI] [PubMed] [Google Scholar]
- 53.Kont V., Murumägi A., Tykocinski L.-O., Kinkel S.A., Webster K.E., Kisand K., Tserel L., Pihlap M., Ströbel P., Scott H., et al. DNA methylation signatures of the AIRE promoter in thymic epithelial cells, thymomas and normal tissues. Mol. Immunol. 2011;49:518–526. doi: 10.1016/j.molimm.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 54.Ye L., Li X., Kong X., Wang W., Bi Y., Hu L., Cui B., Li X., Ning G. Hypomethylation in the promoter region of POMC gene correlates with ectopic overexpression in thymic carcinoids. J. Endocrinol. 2005;185:337–343. doi: 10.1677/joe.1.05963. [DOI] [PubMed] [Google Scholar]
- 55.Lopomo A., Ricciardi R., Maestri M., De Rosa A., Melfi F., Lucchi M., Mussi A., Coppedè F., Migliore L. Gene-Specific Methylation Analysis in Thymomas of Patients with Myasthenia Gravis. Int. J. Mol. Sci. 2016;17:2121. doi: 10.3390/ijms17122121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masunaga A., Omatsu M., Kunimura T., Uematsu S., Kamio Y., Kitami A., Miyagi Y., Hiroshima K., Suzuki T. Expression of PTEN and its pseudogene PTENP1, and promoter methylation of PTEN in non-tumourous thymus and thymic tumours. J. Clin. Pathol. 2017;70:690–696. doi: 10.1136/jclinpath-2016-204220. [DOI] [PubMed] [Google Scholar]
- 57.Sabbir M.G., Prieditis H., Ravinsky E., Mowat M.R. The role of Dlc1 isoform 2 in K-Ras2(G12D) induced thymic cancer. PLoS ONE. 2012;7:e40302. doi: 10.1371/journal.pone.0040302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saito M., Fujiwara Y., Asao T., Honda T., Shimada Y., Kanai Y., Tsuta K., Kono K., Watanabe S., Ohe Y., et al. The genomic and epigenomic landscape in thymic carcinoma. Carcinogenesis. 2017;38:1084–1091. doi: 10.1093/carcin/bgx094. [DOI] [PubMed] [Google Scholar]
- 59.Soejima S., Kondo K., Tsuboi M., Muguruma K., Tegshee B., Kawakami Y., Kajiura K., Kawakita N., Toba H., Yoshida M., et al. GAD1 expression and its methylation as indicators of malignant behavior in thymic epithelial tumors. Oncol. Lett. 2021;21:483. doi: 10.3892/ol.2021.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Z., Liu S., Wang Y., Chen Y., Zhang P., Liu Y., Zhang H., Tao Z., Xiong K. High expression of KITLG is a new hallmark activating the MAPK pathway in type A and AB thymoma. Thorac. Cancer. 2020;11:1944–1954. doi: 10.1111/1759-7714.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arjonen A., Mäkelä R., Härmä V., Rintanen N., Kuopio T., Kononen J., Rantala J.K. Image-based ex vivo drug screen to assess targeted therapies in recurrent thymoma. Lung Cancer. 2020;145:27–32. doi: 10.1016/j.lungcan.2020.04.036. [DOI] [PubMed] [Google Scholar]
- 62.Giaccone G., Rajan A., Berman A., Kelly R.J., Szabo E., Lopez-Chavez A., Trepel J., Lee M.-J., Cao L., Espinoza-Delgado I., et al. Phase II study of belinostat in patients with recurrent or refractory advanced thymic epithelial tumors. J. Clin. Oncol. 2011;29:2052–2059. doi: 10.1200/JCO.2010.32.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas A., Rajan A., Szabo E., Tomita Y., Carter C.A., Scepura B., Lopez-Chavez A., Lee M.-J., Redon C.E., Frosch A., et al. A phase I/II trial of belinostat in combination with cisplatin, doxorubicin, and cyclophosphamide in thymic epithelial tumors: A clinical and translational study. Clin. Cancer Res. 2014;20:5392–5402. doi: 10.1158/1078-0432.CCR-14-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bellissimo T., Ganci F., Gallo E., Sacconi A., Tito C., De Angelis L., Pulito C., Masciarelli S., Diso D., Anile M., et al. Thymic Epithelial Tumors phenotype relies on miR-145-5p epigenetic regulation. Mol. Cancer. 2017;16:88. doi: 10.1186/s12943-017-0655-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.