Abstract
Mutant proteins with eight amino acid deletions in putative surface loops 2 and 3 of the imipenem-specific porin OprD of Pseudomonas aeruginosa failed to reconstitute imipenem susceptibility in an oprD-deficient background. The loop 3 deletion prevented the ability of imipenem to inhibit KCl conductance through the OprD channel, as previously shown for a loop 2 deletion. This suggests that both loops 2 and 3 have a role in imipenem binding to the OprD channel.
Porin OprD of Pseudomonas aeruginosa facilitates the uptake across the outer membrane of basic amino acids, small peptides that contain these amino acids, and their structural analogue imipenem. Indeed, prolonged imipenem treatment of patients with P. aeruginosa infections leads to imipenem-resistant mutants that either lack OprD due to an oprD gene mutation (9) or have strongly reduced OprD levels due to an nfxC-type mutation (mexT) which suppresses oprD expression at the same time as upregulation of the mexEF-oprN multidrug efflux operon (4, 8). Model membrane studies strongly indicate that OprD forms a channel that contains a binding site for imipenem and basic amino acids and peptides (6, 13); i.e., OprD is a so-called specific porin. Based on predictions of amphipathic β-strand regions and subsequent deletion mutagenesis, a model for the membrane topology of OprD was proposed. As for the crystallized nonspecific porins, e.g., OmpF of Escherichia coli (2, 3), it was proposed that OprD comprises a 16-strand transmembrane β-barrel structure, with the transmembrane β strands being interconnected by seven short turn sequences on the periplasmic side and by eight longer loop regions on the external surface. The general placement of six of the eight surface loops was confirmed by deletion mutagenesis, since deletions of four to eight amino acid residues were tolerated in loops 1, 2, 5, 6, 7, and 8 (5), a result consistent with studies for other porins which have demonstrated that deletions and insertions of amino acids are generally tolerated in the surface loops but prevent biogenesis of the porin protein when they occur in the transmembrane β strands (2, 12). However, the proposed loops 3 and 4 could not be confirmed since the deletion of amino acids 146 to 153 in proposed loop 3 was only poorly expressed in E. coli and was not expressed at all in P. aeruginosa, whereas the deletion of amino acids 196 to 199 in the originally proposed loop 4 was nonpermissive in either host. Study of the permissive deletion mutants showed that loop 2 was required for imipenem binding, while loops 5, 7, and 8 served to constrict the OprD channel entrance and prevent nonspecific passage of antibiotics (6). However, these results were somewhat contrary to investigations of other porins, since loop 3 generally inserts into the center of the porin channel, creating a constriction zone (2, 3, 11, 12) that determines ion selectivity for the nonspecific porins and, in part, the substrate binding site for the specific porins (11, 12). Therefore, we set out in the study described here to clarify the positioning and function of loops 2 and 3. With the recent release of the P. aeruginosa genome sequence (http://www.pseudomonas.com), we observed that OprD was the paradigm for a 19-member family of porins. The sequences of the 19 porins were multiply aligned and revealed a high level of similarity (ranging from 41 to 58% identity plus conservative changes compared to the sequence of OprD) but revealed seven regions in which large deletions or insertions had to be made to realign the proteins in the subsequent residues. It has been shown that related porin proteins from a given or different species tend to have large insertions or deletions that can be assigned to surface loops (7) and that for any given porin, including OprD, insertions or deletions of amino acids can be made exclusively at the position of the surface loops (2, 6, 12).
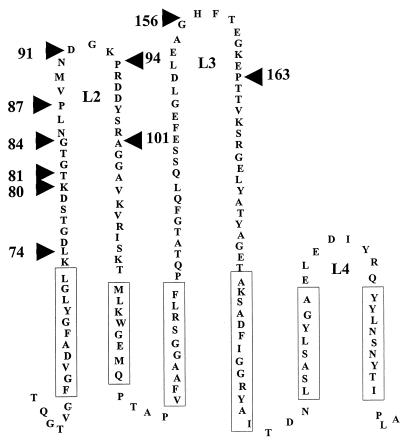
Consistent with these observations, the positions of OprD loops L1, L2, L5, L6, L7, and L8, as inferred by deletion mutagenesis (5), corresponded to six of the seven discordances in the sequence alignments. In contrast, loop L3 had been placed at the N-terminal side of the seventh misalignment and in fact largely corresponded to a region with quite a high degree of conservation. This, together with the nonpermissiveness of the previously made loop 3 deletion, led us to adjust the membrane topology model of OprD in this region (Fig. 1). We have shown in Fig. 1 only the topology model between loop 2 and loop 4 for clarity.
FIG. 1.
Partial membrane topology model of P. aeruginosa outer membrane protein OprD showing the portion of the protein from β strands 3 to 8 and the surface loops 2, 3, and 4. The boundaries of the insertions made in this study are shown by arrows. Only that portion of the model that was predicted to be altered compared to the sequence of the previously published model (5) is presented. Numbering of amino acids refers to the mature OprD protein.
To test this revised model in part, we constructed an extensive series of plasmid-encoded OprD derivatives with deletions in proposed loop 2 and one deletion in loop 3, using the same methods described previously (5). Plasmids were constructed as follows. Deletions were introduced into the oprD wild-type gene by PCR-mediated mutagenesis as described previously (5). Detailed information about the oligonucleotide primers and PCR protocols used is available from the authors on request. Plasmid pXH3 was constructed by excision of the oprD gene from pXH2 (5) with BamHI and SstI and cloning of the gene into the corresponding sites of pUCP18. Plasmids pHP2 (5) and pHP21-23 (this report) are derivatives of pXH2, while pXH12 is a derivative of pXH3 (this report). All OprD deletion mutants were transformed into P. aeruginosa mutant H729 oprD::Kmr (5) and/or P. aeruginosa H846 oprD::Gmr-xylE (10). All mutant proteins were expressed at the same level as the wild-type OprD, as demonstrated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (data not shown) and Western immunoblotting with a polyclonal antibody (5) specific for OprD (Fig. 2). All OprD variants remained normally (5, 6) heat modifiable except for the loop 2 mutant OprDΔ94-101, which was not apparently heat modifiable.
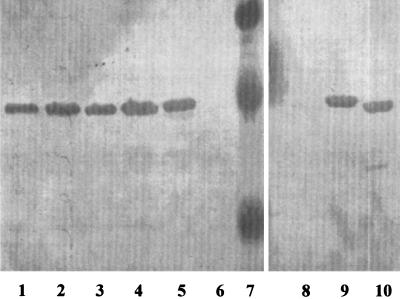
FIG. 2.
Western immunoblot probed with antiserum specific for OprD (5) of P. aeruginosa strain H729 oprD::Kmr containing the following plasmids or standards: pHP23 (lane 1), pHP22 (lane 2), pHP21 (lane 3), pHP2 (lane 4), pXH2 (lane 5), vector control pUCP18 (lanes 6 and 8), chromogenic molecular weight standards (lane 7), pXH3 (lane 9), and pXH12 (lane 10).
Strains carrying the wild-type oprD gene expressed from either plasmid pXH2 (oprD cloned in the same orientation as the lac promoter) or plasmid pXH3 (opposite orientation relative to that of the lac promoter) reversed the imipenem resistance due to the loss of OprD in strain H729 oprD::Kmr, becoming 4- to 16-fold more susceptible to imipenem relative to the susceptibilities of the vector controls. In contrast, all three new deletions in the proposed loop 2, covering residues 80 to 101, had only a twofold increase in imipenem susceptibility (i.e., MICs eightfold higher than that observed for the wild-type gene), exactly as observed for the OprDΔ84-91 mutant previously constructed in this laboratory (6) (Table 1). In contrast, deletions in loops L1, L5, L6, L7, and L8 were previously shown to demonstrate no significant changes in ability to reconstitute imipenem susceptibility. Expression of the deletion in putative loop 3, OprDΔ156-163, completely failed to reconstitute imipenem susceptibility (Table 1). As controls we could demonstrate that there were no changes in the MIC of ciprofloxacin (Table 1), ceftazidime, gentamicin, polymyxin B, tetracycline, or chloramphenicol caused by these mutations.
TABLE 1.
Influence of loop deletions on the ability of OprD variants to reconstitute imipenem susceptibility to a P. aeruginosa oprD::Kmr mutant
| Plasmid | Cloned gene | MIC (μg/ml)a
|
|
|---|---|---|---|
| Imipenem | Ciprofloxacin | ||
| pUCP19 | None | 8 | 0.03 |
| pXH2 | OprD | 0.5 | 0.03 |
| pHP21 | OprDΔ74-81b | 4 | 0.03 |
| pHP22 | OprDΔ80-87b | 4 | 0.03 |
| pHP2 | OprDΔ84-9b | 4 | 0.03 |
| pHP23 | OprDΔ94-101b | 4 | 0.03 |
| pUCP18 | None | 8 | 0.06 |
| pXH3 | OprD | 2 | 0.06 |
| pXH12 | OprDΔ156-163c | 8 | 0.06 |
No changes in the MIC of ceftazidime, gentamicin, polymyxin B, tetracycline, or chloramphenicol were observed.
Putative loop 2 deletion.
Putative loop 3 deletion.
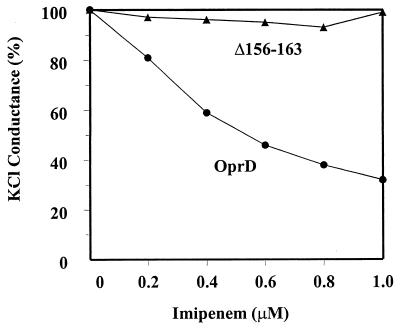
These data suggested a possible role for loop 3 in imipenem uptake, in agreement with the finding of a role for loop 3 in maltodextrin uptake in LamB (12) and phosphate uptake in OprP (12). To confirm this, we purified OprD and OprDΔ156-163 as described previously (6) and studied the function of both proteins in the model membrane planar bilayer system. Consistent with previous results, we found that OprD had a single-channel conductance in 1 M KCl of 22.1 ± 0.6 pS and observed the blockage of K+Cl− conductance through OprD by progressive addition of imipenem (Fig. 3) (6). From the latter data, a Kd for imipenem binding of 0.35 μM could be extrapolated, similar to the value described previously (6). In contrast, the mutant OprDΔ156-163 porin demonstrated a slightly lower single-channel conductance of 12.6 ± 0.4 pS, and K+Cl− conductance was not substantially inhibited by the addition of imipenem. These data were consistent with loop 3 having a role in imipenem passage through OprD, but given that we observed similar results for the loop 2 deletion, OprDΔ84-91, we could not state that loop 3 is involved in direct imipenem binding or whether it stabilizes the binding site. In an attempt to address this issue, using site-specific mutagenesis, we made the loop 2 mutations D74N, D78N, D90N, D95N, and D96N and the loop 3 mutations E146Q, E148Q, E151N, E153Q, H156L, E159Q, K161M, and E162Q, either in pairs or in combinations of up to six mutations. None of these had any impact on the ability of OprD to reconstitute full imipenem susceptibility.
FIG. 3.
Inhibition by imipenem of macroscopic KCl conductance through native OprD and the loop 3 deletion mutant Δ156-163 expressed from pXH12. Macroscopic conductance inhibition experiments were performed exactly as described previously (6). Each point represents the average of four to seven measurements.
These studies have indicated a role for loop 3 in imipenem passage through OprD and have helped to adjust the membrane topology model. In addition, they confirmed and extended our previous, somewhat surprising result in demonstrating a role for loop 2 in imipenem binding (6). It must be pointed out, however, that these deletion mutants would be expected to destabilize or alter the positioning of the entire loop, and thus provide only general information about the involvement of loops 2 and 3 in imipenem passage through the OprD channel. To provide a more detailed picture, we are currently engaged in trying to crystallize and solve the structure of OprD.
Acknowledgments
This research was supported by the Medical Research Council of Canada (MRC). R.E.W.H. was the recipient of an MRC Distinguished Scientist Award. M.M.O. received fellowships from the Canadian Cystic Fibrosis Foundation and the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in laboratory medicine. Baltimore, Md: The Williams & Wilkins Co.; 1991. pp. 72–78. [Google Scholar]
- 2.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 3.Cowan S W, Garavito R M, Jansonius J N, Jenkins J A, Karlsson R, König N, Pai E F, Pauptit R A, Rizkallah P J, Rosenbusch J P. The structure of OmpF porin in a tetragonal crystal form. Structure. 1995;3:1041–1050. doi: 10.1016/s0969-2126(01)00240-4. [DOI] [PubMed] [Google Scholar]
- 4.Fukuda H, Hosaka M, Iyobe S, Gotoh N, Nishino T, Hirai K. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:790–792. doi: 10.1128/AAC.39.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H, Jeanteur D, Pattus F, Hancock R E W. Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosa porin OprD. Mol Microbiol. 1995;16:931–941. doi: 10.1111/j.1365-2958.1995.tb02319.x. [DOI] [PubMed] [Google Scholar]
- 6.Huang H, Hancock R E W. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J Bacteriol. 1996;178:3085–3090. doi: 10.1128/jb.178.11.3085-3090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeanteur D, Lakey J H, Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 8.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 9.Lynch M J, Drusano G L, Mobley H L T. Emergence of resistance to imipenem in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1987;31:1892–1896. doi: 10.1128/aac.31.12.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ochs M M, McCusker M P, Bains M, Hancock R E W. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob Agents Chemother. 1999;43:1085–1090. doi: 10.1128/aac.43.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schirmer T, Keller T A, Wang Y F, Rosenbusch J P. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 12.Sukhan A, Hancock R E W. Insertion mutagenesis of the Pseudomonas aeruginosa phosphate-specific porin OprP. J Bacteriol. 1995;177:4914–4920. doi: 10.1128/jb.177.17.4914-4920.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trias J, Nikaido H. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem. 1990;265:15680–15684. [PubMed] [Google Scholar]