Abstract
The improvement of cancer chemotherapy remains a major challenge, and thus new drugs are urgently required to develop new treatment regimes. Curcumin, a polyphenolic antioxidant derived from the rhizome of turmeric (Curcuma longa L.), has undergone extensive preclinical investigations and, thereby, displayed remarkable efficacy in vitro and in vivo against cancer and other disorders. However, pharmacological limitations of curcumin stimulated the synthesis of numerous novel curcumin analogs, which need to be evaluated for their therapeutic potential. In the present study, we calculated the binding affinities of 50 curcumin derivatives to known cancer-related target proteins of curcumin, i.e., epidermal growth factor receptor (EGFR) and nuclear factor κB (NF-κB) by using a molecular docking approach. The binding energies for EGFR were in a range of −12.12 (±0.21) to −7.34 (±0.07) kcal/mol and those for NF-κB ranged from −12.97 (±0.47) to −6.24 (±0.06) kcal/mol, indicating similar binding affinities of the curcumin compounds for both target proteins. The predicted receptor-ligand binding constants for EGFR and curcumin derivatives were in a range of 0.00013 (±0.00006) to 3.45 (±0.10) µM and for NF-κB in a range of 0.0004 (±0.0003) to 10.05 (±4.03) µM, indicating that the receptor-ligand binding was more stable for EGFR than for NF-κB. Twenty out of 50 curcumin compounds showed binding energies to NF-κB smaller than −10 kcal/mol, while curcumin as a lead compound revealed free binding energies of >−10 kcal/mol. Comparable data were obtained for EGFR: 15 out of 50 curcumin compounds were bound to EGFR with free binding energies of <−10 kcal/mol, while the binding affinity of curcumin itself was >−10 kcal/mol. This indicates that the derivatization of curcumin may indeed be a promising strategy to improve targe specificity and to obtain more effective anticancer drug candidates. The in silico results have been exemplarily validated using microscale thermophoresis. The bioactivity has been further investigated by using resazurin cell viability assay, lactate dehydrogenase assay, flow cytometric measurement of reactive oxygen species, and annexin V/propidium iodide assay. In conclusion, molecular docking represents a valuable approach to facilitate and speed up the identification of novel targeted curcumin-based drugs to treat cancer.
Keywords: bioinformatics, cancer, natural products, phytochemicals, synthetic derivatives, virtual drug screening
1. Introduction
Cancer is currently one of the leading causes of death worldwide. Incidence and death rates are increasing for several types of cancer [1,2]. More than 21 million cancer incidences have been predicted to occur in the year 2025 (https://www.statista.com/statistics/1031316/new-cancer-cases-forecast-worldwide/; accessed on 1 March 2022). Many advances have been made in the early diagnosis and treatment of cancer. However, lowering cancer mortality rates still remains a vital challenge [3]. Cancer chemotherapy involves using natural or synthetic chemicals to prevent or suppress cancer growth. As a matter of fact, more than half of all available anticancer drugs are derivatives of natural products or are compounds that mimic the modes of action of natural products [4]. The advantages of phytochemicals are their lower toxicity profiles and their ability to target multiple signaling pathways that prevent the rapid development of drug resistance [5,6,7]. A number of phytochemicals are known to address cancer-related target proteins, such as the epidermal growth factor receptor (EGFR) [8,9,10,11] or the nuclear factor-kappa B (NF-κB) [12,13,14] and others.
Curcumin is a naturally occurring compound derived from the rhizomes of Curcuma longa L. As a member of the ginger family, it has been commonly used as a spice for food preservation as well as in folk medicine. It possesses a wide array of functional characteristics, including antioxidant and anti-inflammatory [15] as well as antiviral, antibacterial, and antifungal properties [16]. Curcumin has been extensively investigated for its cellular and molecular modes of action against cancer [17,18], diabetes [19], neurological ailments [20], and osteoarthritis [21], and even entered several clinical trials [22,23]. Curcumin inhibits cancer cell proliferation [24,25], DNA repair along the p53-p21/GADD45A-cyclin/CDK-Rb/E2F-DNMT1 axis [26,27], metastasis by the NF-κB/c-JUN/MMP pathway [28], and the CXC-chemokine/NF-κB signaling pathway [29,30,31] as well as angiogenesis by the protein kinase C/NF-κB/AP-1 pathway [32,33]. EGFR is upregulated in several tumor types, including lung and colorectal tumors making EGFR an exquisite therapeutic target. Curcumin targets EGFR in lung cancer [34,35] and colorectal carcinoma [36,37] types leading to tumor cell killing. The transcription factor NF-κB is targeted by curcumin in a wide range of tumor types, including leukemia and lymphoma [38,39,40,41,42].
In contrast to conventional anticancer drugs that often exert severe side effects such as myelosuppression, mucositis, alopecia, nausea, vomiting, and others, curcumin displays only minimal toxicity [43,44,45]. However, the poor bioavailability of curcumin represents a major disadvantage for its clinical application [46]. Many efforts have been undertaken to improve its bioavailability using a variety of approaches, including innovative drug delivery systems (nanoparticles, liposomes, phospholipids, etc.) as well as the development of novel synthetic curcumin derivatives [47,48,49,50,51]. By synthesizing chemical libraries of curcumin derivatives and subjecting them to biological scrutiny, compounds with improved pharmacological features may be yielded.
Our study focuses on curcumin and a total of 50 curcumin compounds that were either reported by us [17] or mined in the PubChem database (https://pubchem.ncbi.nlm.nih.gov; accessed on 31 October 2021). We attempted to predict their activity using an in silico molecular docking approach. For this reason, we calculated the binding energies of these derivatives to two cancer-related proteins, i.e., EGFR and NF-κB. These proteins have been previously described as target proteins of curcumin [52].
The epidermal growth factor receptor (EGFR) is a tyrosine kinase in the cell membrane of many tumor types. This transmembrane receptor belongs to a gene family with three other members (HER2-4). The binding of extracellular ligands, i.e., epidermal growth factor (EGF) and transforming growth factor (TGFα), leads to dimerization, autophosphorylation, and downstream activation of signal transduction pathways. This ultimately leads to carcinogenesis, cell growth, metastasis, and inhibition of apoptosis [53], as well as the development of resistance to cytotoxic chemotherapy and radiotherapy [54]. Targeted therapies with small molecules (e.g., erlotinib, gefitinib, afatinib) or monoclonal antibodies (e.g., cetuximab, panitumumab) have significantly improved the treatment outcome of innumerable patients [55]. However, resistance to these targeted drugs has been emerging [56,57]. Thus, the search for new EGFR inhibitors has to continue [58]. In this context, phytochemicals gained interest as novel lead compounds [59].
The nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells (NF-κB) is a transcription factor that binds to specific DNA sequences (the 10 bp long κB motif) and thereby regulates the expression of downstream genes. This primarily involves genes that control the immune response, inflammation, cell proliferation, and apoptosis [60,61]. Since NF-κB activation is important for cancer development and progression [62], this transcription factor has become a target molecule for new drug development [63,64]. Phytochemicals represent an important reservoir for the identification of NF-κB inhibitors in cancer [65,66,67]. Interestingly, EGFR and NF-κB cooperate in tumor cells, amplifying their oncogenic signals [68]. Therefore, it is particularly useful to investigate inhibitors that inhibit both cancer-related proteins simultaneously to achieve a more effective antitumor effect.
The concept of the present investigation was to identify curcumin derivatives with better binding affinities to EGFR and NF-κB to improve tumor specificity and reduce side effects on normal organs. To validate the molecular docking data, we exemplarily tested the inhibitory effect of curcumin and two of the derivatives investigated by western blot experiments with these four proteins towards EGFR and NF-κB. Our results support the concept that novel synthetic curcumin derivatives with improved specificity to important cancer-related targets could be identified by combined in silico–in vitro drug screening approaches.
2. Results
2.1. Molecular Docking
We first performed molecular docking of 50 curcumin compounds (Figure S1) mined from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/; accessed on 31 October 2021) against EGFR and NF-κB. We intended to predict the potential activity of the synthetic derivatives by calculating their in silico binding activities to these proteins. It is noteworthy that the synthetic curcumin derivatives showed low binding energy values (i.e., higher affinities) to both target proteins.
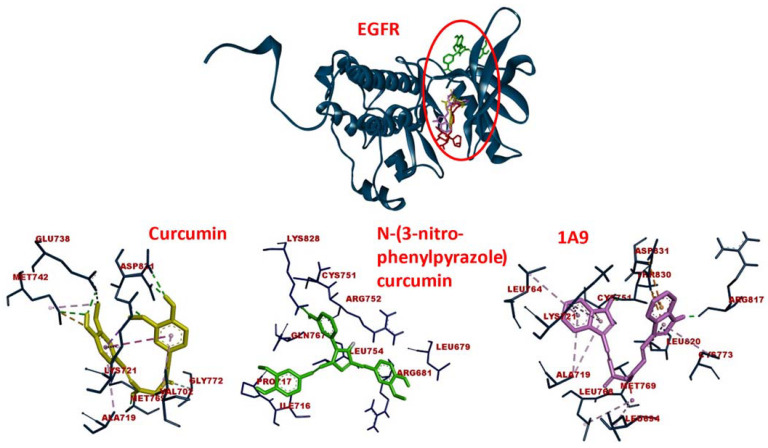
The binding energies for EGFR were in a range of −12.12 (±0.21) to −7.34 (±0.07) kcal/mol and those for NF-κB ranged from −12.97 (±0.47) to −6.24 (±0.06) kcal/mol, indicating similar binding affinities of the curcumin compounds for both target proteins (Table 1). Molecular dockings of curcumin and two selected curcumin derivatives to EGFR are shown in Figure 1. The three compounds were bound to the same domain but with different amino acids within this pharmacophore.
Table 1.
Mean binding energies and predicted binding constants were obtained by molecular docking for 50 curcumin compounds. Dockings were independently carried out three times with every 250,000 runs (mean values ± SD).
| Compounds | EGFR (Mean Binding Energy, kcal/mol) | EGFR (Mean pKi, µM) | NF-κB (Mean Binding Energy, kcal/mol) | NF-κB (Mean pKi, µM) | |
|---|---|---|---|---|---|
| 1 | Curcumin | −9.42 ± 0.09 | 0.13 ± 0.02 | −8.38 ± 0.16 | 0.73 ± 0.21 |
| 2 | Bisdemethoxycurcumin | −8.51 ± 0.02 | 0.58 ± 0.02 | −7.96 ± 0.05 | 1.46 ± 0.11 |
| 3 | Diacetylcurcumin | −10.72 ± 0.49 | 0.02 ± 0.02 | −9.67 ± 0.26 | 0.09 ± 0.04 |
| 4 | [18FP]-curcumin | −8.70 ± 0.21 | 0.43 ± 0.14 | −9.01 ± 0.07 | 0.25 ± 0.03 |
| 5 | Monodemethoxycurcumin | −9.04 ± 0.06 | 0.24 ± 0.02 | −8.07 ± 0.10 | 1.23 ± 0.20 |
| 6 | (E,E)-Bis(2-hydroxybenzylidene)acetone | −7.95 ± 0.01 | 1.49 ± 0.04 | −7.21 ± 0.06 | 5.19 ± 0.50 |
| 7 | Curcumin monoglucoside | −9.72 ± 0.76 | 0.11 ± 0.11 | −9.57 ± 0.34 | 0.11 ± 0.06 |
| 8 | Di-O-(2-thienoyl) curcumin | −11.71 ± 0.26 | 0.003 ± 0.002 | −10.71 ± 0.17 | 0.01 ± <0.01 |
| 9 | Cis-curcumin | −9.07 ± 0.30 | 0.24 ± 0.10 | −8.81 ± 0.33 | 0.39 ± 0.21 |
| 10 | Curcumin diglucoside | −9.10 ± 0.99 | 0.39 ± 0.35 | −10.35 ± 0.35 | 0.03 ± 0.02 |
| 11 | Tetrahydrocurcumin | −9.13 ± 0.17 | 0.21 ± 0.06 | −8.28 ± 0.21 | 0.90 ± 0.34 |
| 12 | Allyl curcumin | −10.07 ± 0.07 | 0.04 ± 0.01 | −9.15 ± 0.04 | 0.20 ± 0.01 |
| 13 | Monodemethylcurcumin | −9.96 ± 0.10 | 0.05 ± 0.01 | −8.16 ± 0.56 | 0.59 ± 0.12 |
| 14 | Didemethylcurcumin | −9.90 ± 0.07 | 0.06 ± 0.01 | −8.71 ± 0.25 | 0.49 ± 0.18 |
| 15 | Curcumin-4′-O-β-d-gentiotrioside | −7.85 ± 0,59 | 3.11 ± 0.23 | −7.51 ± 0.37 | 3.47 ± 2.05 |
| 16 | 4-Benzylidene curcumin | −10.62 ± 0.72 | 0.03 ± 0.03 | −9.10 ± 0.10 | 0.21 ± 0.04 |
| 17 | Monoglycinoyl curcumin | −12.12 ± 0.21 | 0.0013 ± 0.0006 | −10.07 ± 0.08 | 0.04 ± 0.01 |
| 18 | Ethyl curcumin | −10.09 ± 0.11 | 0.04 ± 0.01 | −9.23 ± 0.05 | 0.17 ± 0.01 |
| 19 | Curcumin dimer 1 | −11.64 ± 0.56 | 0.004 ± 0.003 | −11.11 ± 0.65 | 0.01 ± 0.01 |
| 20 | N-phenylpyrazole curcumin | −8.63 ± 0.19 | 0.49 ± 0.14 | −8.81 ± 0.01 | 0.35 ± 0.01 |
| 21 | Curcumin β-d-glucuronide | −10.36 ± 0.19 | 0.04 ± 0.02 | −10.07 ± 0.36 | 0.05 ± 0.03 |
| 22 | 3,4-Difluorobenzylidene curcumin | −10.08 ± 0.13 | 0.04 ± 0.01 | −9.30 ± 0.07 | 0.15 ± 0.02 |
| 23 | Di-O-(2-hydroxyethyl) curcumin | −9.60 ± 0.27 | 0.10 ± 0.05 | −8.97 ± 0.55 | 0.35 ± 0.27 |
| 24 | 4-(4-hydroxybenzylidene) curcumin | −10.55 ± 0.12 | 0.02 ± <0.01 | −8.98 ± 0.04 | 0.26 ± 0.02 |
| 25 | N-(3-nitrophenylpyrazole) curcumin | −10.50 ± 0.02 | 0.02 ± <0.01 | −9.57 ± 0.01 | 0.097 ± <0.01 |
| 26 | N-(4-flurophenylpyrazole) curcumin | −8.46 ± 0.19 | 0.65 ± 0.19 | −8.81 ± 0.09 | 0.35 ± 0.06 |
| 27 | N-(4-methoxyphenylpyrazole) curcumin | −8.49 ± 0.04 | 0.60 ± 0.03 | −8.95 ± 0.11 | 0.28 ± 0.05 |
| 28 | Di-O-chloropropionylethyl curcumin | −10.92 ± 0.25 | 0.01 ± <0.01 | −10.36 ± 0.59 | 0.03 ± 0.02 |
| 29 | Curcumin tri-adamantylaniniethylcarbonate | −10.75 ± 0.23 | 0.01 ± 0.01 | −12.97 ± 0.47 | 0.0004 ± 0.0003 |
| 30 | Curcumin dimer 2 | −11.08 ± 0.24 | 0.01 ± <0.01 | −12.05 ± 0.46 | 0.002 ± 0.001 |
| 31 | Curcumin tri-trithiadiazolaminoethylcarbonate | −10.35 ± 0.55 | 0.03 ± 0.02 | −10.97 ± 0.53 | 0.01 ± 0.01 |
| 32 | 4-(4-hydroxy-3-methoxybenzylidene) curcumin | −11.44 ± 0.16 | 0.004 ± 0.002 | −10.97 ± 0.53 | 0.01 ± 0.01 |
| 33 | Curcumin-β-d-glucuronide triacetate methyl ester | −10.06 ± 0.79 | 0.07 ± 0.06 | −10.35 ± 0.35 | 0.03 ± 0.02 |
| 34 | Curcumin 4′-O-β-d-gentiobiosyl 4″-O-β-d-glucoside | −9.54 ± 0.25 | 0.11 ± 0.05 | −10.23 ± 0.59 | 0.04 ± 0.04 |
| 35 | Tetrahydrocurcumin isoxazole | −9.43 ± 0.20 | 0.13 ± 0.04 | −8.60 ± 0.41 | 0.57 ± 0.32 |
| 36 | Hexahydrocurcumin | −9.25 ± 0.11 | 0.17 ± 0.03 | −8.41 ± 0.27 | 0.73 ± 0.35 |
| 37 | Bisdemethoxycurcumin isoxazole | −7.45 ± 0.01 | 3.45 ± 0.10 | −7.55 ± 0.10 | 2.96 ± 0.47 |
| 38 | Curcumin dimer 3 | −10.28 ± 0.22 | 0.03 ± 0.01 | −10.50 ± 0.77 | 0.01 ± 0.01 |
| 39 | Curcumin ED | −9.59 ± 0.08 | 0.10 ± 0.02 | −8.69 ± 0.14 | 0.43 ± 0.10 |
| 40 | Curcumin PE | −9.24 ± 0.12 | 0.17 ± 0.03 | −8.22 ± 0.22 | 0.99 ± 0.33 |
| 41 | Curcumin sulfate | −9.31 ± 0.16 | 0.15 ± 0.04 | −9.84 ± 0.08 | 0.06 ± 0.01 |
| 42 | Ferrocenyl curcumin | −8.34 ± 0.22 | 0.80 ± 0.29 | −7.10 ± 0.33 | 6.87 ± 3.92 |
| 43 | Di-O-deconyl curcumin | −10.57 ± 0.17 | 0.01 ± <0.01 | −11.58 ± 0.85 | 0.02 ± 0.01 |
| 44 | GNF-pf-2695 ((2E,5E)-2,5-bis[(3,4,5-trimethoxyphenyl) methylidene]cyclopentan-1-one) | −7.34 ± 0.07 | 4.22 ± 0.51 | −7.56 ± 0.15 | 2.93 ± 0.78 |
| 45 | Perfluoro curcumin | −7.55 ± 0.19 | 3.03 ± 0.86 | −6.86 ± 0.28 | 10.05 ± 4.03 |
| 46 | Keto-curcumin | −9.89 ± 0.41 | 0.07 ± 0.04 | −8.78 ± 0.15 | 0.37 ± 0.09 |
| 47 | Disalicyloyl curcumin | −7.67 ± 0.58 | 0.02 ± 0.02 | −6.24 ± 0.06 | 26.77 ± 3.36 |
| 48 | HO-3867 (3E,5E)-3,5-bis[(4-fluorophenyl)methylidene]-1-[(1-hydroxy-2,2,5,5-tetramethylpyrrol-3-yl)methyl]piperidin-4-one) | −9.73 ± 0.03 | 0.07 ± <0.01 | −7.79 ± 0.10 | 1.96 ± 0.33 |
| 49 | 1A6 ((1E,6E)-4-chloro-1,7-bis(3,4-dimethoxyphenyl)hepta-1,6-diene-3,5-dione) | −8.94 ± 0.06 | 0.28 ± 0.03 | −8.28 ± 0.21 | 0.90 ± 0.34 |
| 50 | 1A9 ((1E,6E)-4-chloro-1,7-di(1H-indol-3-yl)hepta-1,6-diene-3,5-dione) | −9.57 ± 0.02 | 0.10 ± 0.03 | −8.51 ± 0.11 | 0.58 ± <0.01 |
Figure 1.
Molecular docking of curcumin-type compounds to EGFR. Top: The compounds were bound to the same domain of EGFR. Bottom: curcumin, N-(3-nitrophenylpyrazole) curcumin, and the derivative 1A9 were bound to different amino acids in this domain. The red circle indicates the binding site of the three compounds.
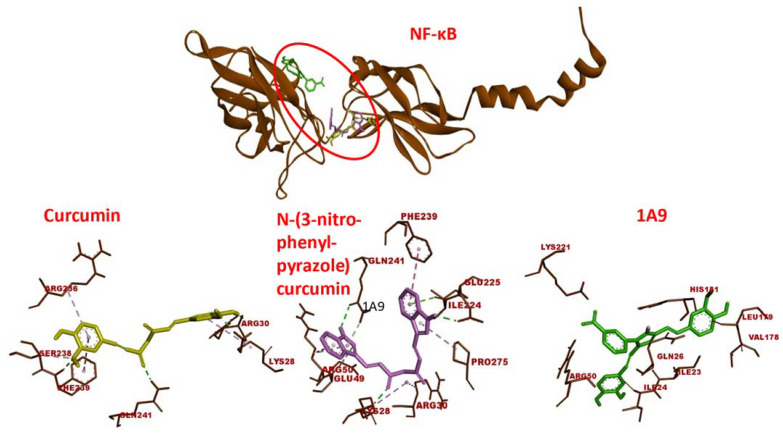
The predicted receptor-ligand binding constants for EGFR and curcumin derivatives were in a range of 0.00013 (±00.0006) to 3.45 (±0.10) µM and for NF-κB in a range of 0.0004 (±0.0003) to 10.05 (±4.03) µM, indicating that the receptor-ligand binding was more stable for EGFR than for NF-κB (Table 1). Molecular dockings of curcumin and two selected curcumin derivatives to NF-κB are shown in Figure 2. Like EGFR, the three compounds were bound to the same domain of NF-κB.
Figure 2.
Molecular docking of curcumin-type compounds to NF-κB. Top: The compounds were bound to the same domain of EGFR. Bottom: curcumin, N-(3-nitrophenylpyrazole) curcumin, and the derivative 1A9 were bound to different amino acids in this domain. The red circle indicates the binding site of the three compounds.
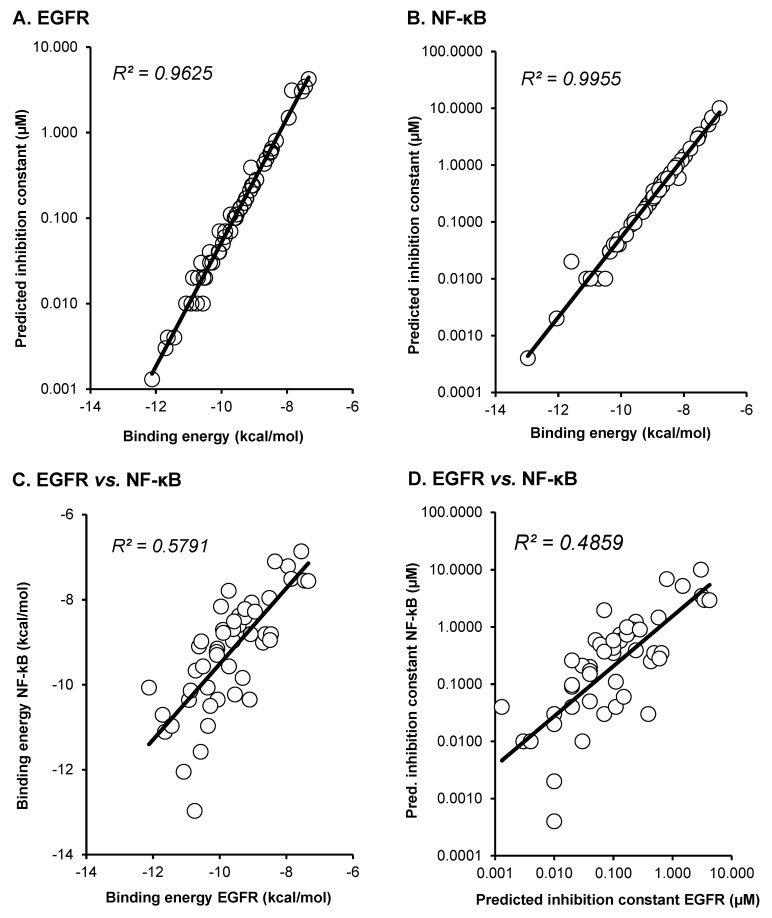
As a next step, we correlated the binding energies of the compounds for EGFR and NF-κB. By using Pearson correlation test, we found statistically significant relationships between binding energies and predicted inhibition constants of EGFR (p = 2.73 × 10−9; r = 0.715) and of (p = 4.39 × 10−7; r = 0.631) as well as of binding energies between EGFR and NF-κB (p = 4.32 × 10−5; r = 0.526) and predicted binding energies between both proteins (p = 2.53 × 10−8; r = 0.682). These results indicate that there may be a relationship between the “druggability” of the compounds being EGFR or NF-κB inhibitors. Compounds that were better bound to EGFR also showed a significant relationship to better bind to NF-κB and vice versa (Figure 3).
Figure 3.
Correlation of binding energies (kcal/mol) and predicted inhibition constants (pKi, µM) of 50 curcumin compounds were calculated using the Pearson correlation test. Correlation of binding energies and pKi values for (A) EGFR and (B) NF-κB. Correlation of (C) binding energies or (D) pKi values between EGFR and NF-κB.
2.2. Microscale Thermophoresis
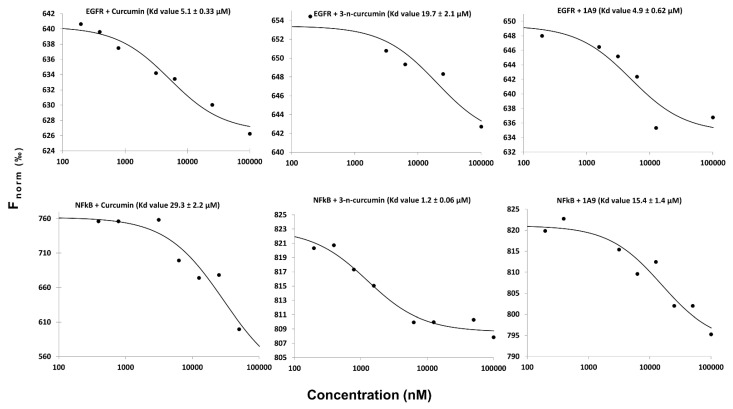
To verify the in silico predictions, we performed microscale thermophoresis. This is a biophysical assay to study the interactions between chemical ligands and their target proteins. For this reason, we used recombinant EGFR and NF-κB and assayed them with curcumin, N-(3-nitrophenylpyrazole) curcumin, and curcumin derivative 1A9. The equilibrium constants (KD) indicate that the three curcumin-type compounds were bound to EGFR and NF-κB (Figure 4).
Figure 4.
Analysis of the interaction between curcumin derivatives with recombinant EGFR and NF-κB by microscale thermophoresis (MST). The recombinant proteins were used at a concentration of 200 nM, while the concentration of curcumin, N-(3-nitrophenylpyrazole) curcumin, and the curcumin derivative 1A9 ranged from 100 to 100,000 nM. The migration of the fluorescent proteins was determined upon local heating using a Monolith NT.115Pico with 40% LED power and 80% MST power for EGFR and with 20% LED power and 20% MST power for NF-κB at room temperature.
2.3. Resazurin Assay
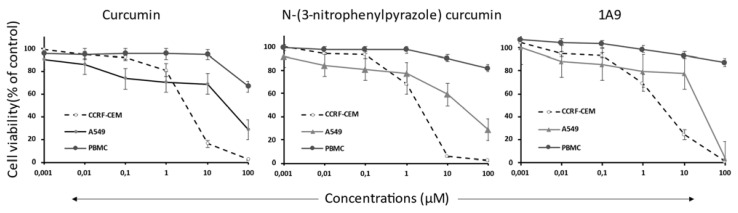
To study the effect of selected compounds on cell viability, we treated human CCRF-CEM leukemia and human A549 lung cancer cells with curcumin, N-(3-nitrophenylpyrazole) curcumin, and the curcumin derivative 1A9. CCRF-CEM and A549 cells were chosen as examples of hematopoietic and solid tumor cells. Peripheral blood mononuclear cells (PBMCs) were isolated from a healthy subject to compare the inhibitory effects of the curcumin compounds between tumor and normal cells. As shown in Figure 5, all three compounds inhibited the viability of CCRF-CEM and A549 cells in a dose-dependent manner, while normal PBMCs were not or only minimally inhibited. The dose-response curves were taken to calculate the 50% inhibition concentrations (IC50). CCRF-CEM leukemia cells were about one order of magnitude more sensitive to the compounds than A549 cells and N-(3-nitrophenylpyrazole) curcumin revealed a higher inhibitory activity than the other two compounds (Table 2).
Figure 5.
Cell viability dose-response curves of CCRF-CEM leukemia cells, A549 lung carcinoma cells, and healthy peripheral blood mononuclear cells treated with curcumin, 1A9, and N-(3-nitrophenylpyrazole) curcumin as determined by the resazurin assay. Cells were incubated with concentrations from 10−3 to 100 µM and incubated at 37 °C for 72 h. DMSO was used as vehicle control. Mean ± SD of three independent measurements.
Table 2.
IC50 values of CCRF-CEM leukemia and A549 lung carcinoma cells treated with curcumin, 1A9, and N-(3-nitrophenylpyrazole) curcumin as determined by the resazurin assay. The IC50 values (µM) were calculated from the dose-response curves shown in Figure 5. DMSO was used as vehicle control. Mean ± SD of three independent measurements.
| Cells | Compounds | ||
|---|---|---|---|
| Curcumin | N-(3-Nitrophenylpyrazole) Curcumin | 1A9 | |
| CCRF-CEM | 3.0 ± 0.4 | 1.9 ± 0.4 | 2.6 ± 0.3 |
| A549 | 29.4 ± 1.9 | 18.9 ± 1.4 | 23.3 ± 1.2 |
| PBMC | >100 | >100 | >100 |
2.4. LDH Assay
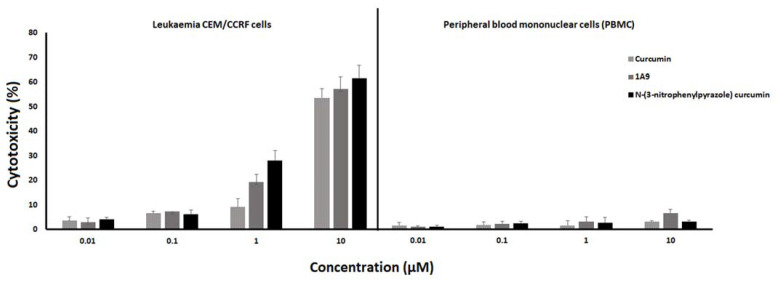
To assess the cytotoxicity, the LDH cell death assay was performed that measures the release of LDH from cells due to membrane damage. As shown in Figure 6, all three curcumin-type compounds led to a dose-dependent LDH release in a concentration range of 0.01 to 10 µM in CCRF-CEM leukemia cells. Only negligible LDH release was found in healthy peripheral blood mononuclear cells, indicating tumor-specific cytotoxic effects of the three curcumin compounds.
Figure 6.
Cytotoxicity in CCRF-CEM leukemia cells (left) and peripheral blood mononuclear cells (PBMCs) of a healthy donor (right) by curcumin, 1A9, and N-(3-nitrophenylpyrazole) curcumin as determined by the release of lactate dehydrogenase. Cells were incubated with concentrations from 0.01 to 10 µM and incubated at 37 °C for 48 h. DMSO was used as vehicle control. Mean ± SD of three independent measurements.
2.5. ROS Assay
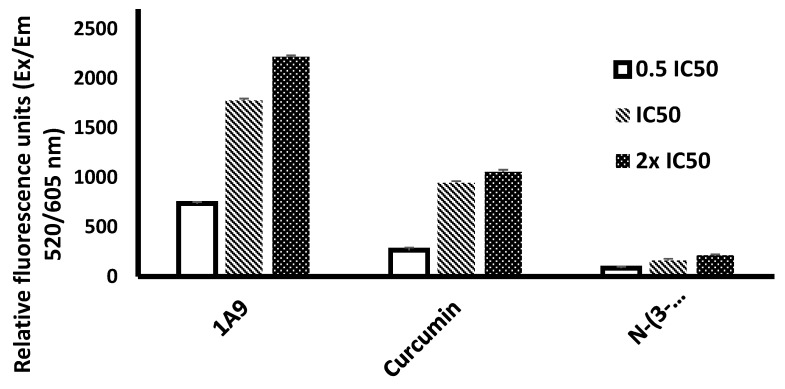
The generation of reactive oxygen species (ROS) upon exposure of CCRF-CEM cells with N-(3-nitrophenylpyrazole) curcumin or 1A9 was compared with the ROS generation by curcumin. As shown in Figure 7, dose-dependent effects were observed with 0.5×, 1×, and 2× IC50 concentrations of these three compounds. The strongest ROS generation was measured with 1A9, the lowest one with N-(3-nitrophenylpyrazole) curcumin. Curcumin produced intermediate ROS amounts.
Figure 7.
Generation of reactive oxygen species in CCRF-CEM cells by curcumin, N-(3-nitrophenylpyrazole) curcumin, and 1A9. Cells were incubated with 0.5×, 1×, 2×, or 4× IC50 and incubated at 37 °C for 24 h. DMSO was used as vehicle control. Mean ± SD of three independent measurements.
2.6. Annexin/PI Assay
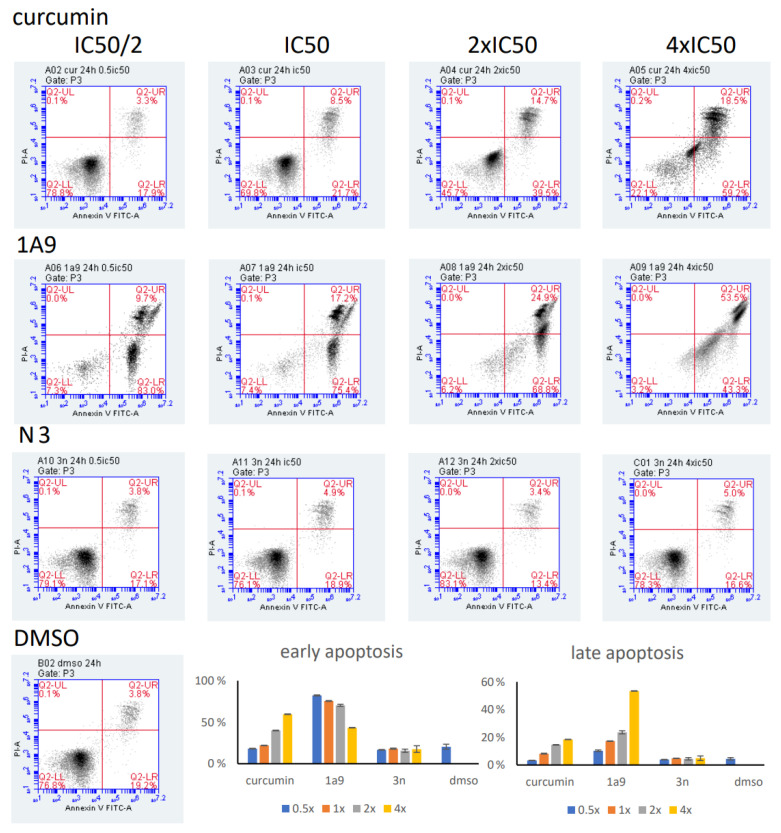
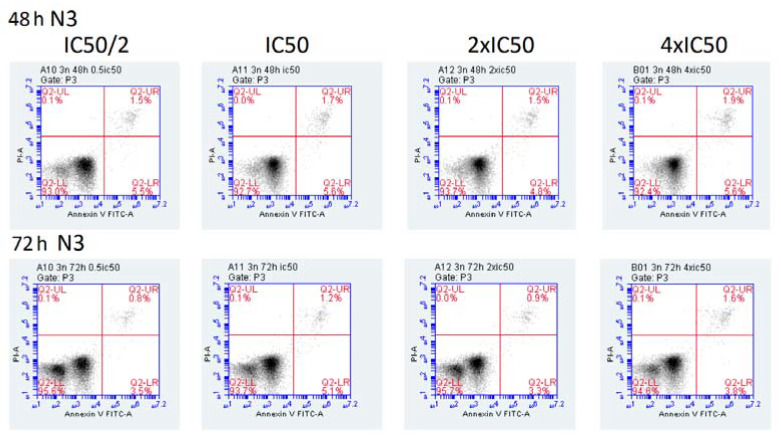
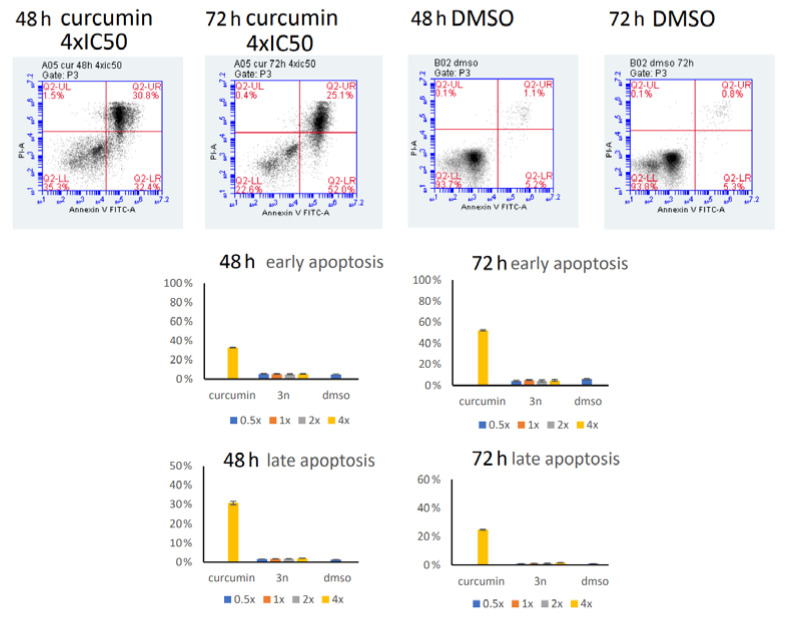
The induction of early and late apoptosis by the three curcumin compounds was determined by the annexin V/propidium iodide assay and flow cytometry. Incubation of CCRF-CEM cells at 37 °C for 24 h showed clear dose-dependent induction either of early or late apoptosis in a concentration range from 0.5× to 4× IC50 (Figure 8). The effects of 1A9 were even stronger: the highest fraction of apoptotic cells was observed already at the lowest concentration of 0.5× IC50. Higher concentrations led to a decrease in early apoptotic cells and an increase in late apoptotic cells, indicating a concentration-dependent switch from early to late apoptosis upon exposure with 1A9 at 37 °C for 24 h (Figure 8). Although N-(3-nitrophenylpyrazole) curcumin was cytotoxic in the LDH assay and inhibited viability in the resazurin assay, it did not induce early or late apoptosis after 24 h incubation (Figure 8). Even after prolonged incubation at 37 °C for 48 or 72 h, N-(3-nitrophenylpyrazole) curcumin did not induce apoptosis (Figure 9). The low percentages of apoptosis cells did not exceed the low rates of spontaneous apoptosis in DMSO-treated control cells.
Figure 8.
Induction of apoptosis in CCRF-CEM cells upon exposure at 37 °C for 24 h with curcumin, N-(3-nitrophenylpyrazole) curcumin, and 1A9 as determined by the annexin V/propidium iodide assay and flow cytometry. Cells were incubated with 0.5×, 1×, 2×, or 4× IC50. DMSO was used as vehicle control. Mean ± SD of three independent measurements.
Figure 9.
Induction of apoptosis in CCRF-CEM cells upon exposure at 37 °C for 48 or 72 h with N-(3-nitrophenylpyrazole) curcumin as determined by the annexin V/propidium iodide assay and flow cytometry. Cells were incubated with 0.5×, 1×, 2×, or 4× IC50 N-(3-nitrophenylpyrazole) curcumin. Positive control: curcumin (4× IC50); negative control: DMSO. Mean ± SD of three independent measurements.
3. Discussion
In the present study, we investigated the in silico binding affinities of a total of 50 curcumin compounds to EGFR and NF-κB. The aim was to find novel derivatives with improved binding properties to these two cancer-related proteins as a starting point to develop curcumin-based drugs with improved pharmacological features.
EGFR plays a central role in the pathogenesis and progression of different carcinoma types. EGFR is overexpressed in many human carcinomas and is also involved in developing resistance to chemotherapy [54,56]. On average, 50–70% of lung, colon, and breast carcinoma express EGFR [69]. Inhibition of EGFR phosphorylation is caused by directly inhibitor effects of curcumin on the tyrosine kinase activity of EGFR as well as by curcumin-induced alterations of the physical plasma membrane properties influencing receptor dimerization [70]. Additionally, curcumin downregulated EGFR protein expression [36].
From previous investigations on the activity of curcumin derivatives towards EGFR [71], it is known that substitutions on the phenyl rings affected the extent of EGFR down-regulation. Moreover, methoxy or hydroxy substituents increased the compounds’ activity, while other alkyloxy groups did not. The activity was, in general, not influenced by methoxy, hydroxy, or halogen groups. Remarkably, N-(3-nitrophenylpyrazole) curcumin was bound better to EGFR than the halogenated curcumin derivatives investigated in this study, including difluorinated curcumin that is frequently described in the literature as a promising drug candidate for further drug development [72].
NF-κB regulates the expression of genes involved in many processes that play a key role in cancer biology, such as proliferation, migration, and apoptosis. NF-κB influences the expression of genes that are involved in a large number of physiological processes, including immune response, cell survival, differentiation, and proliferation. Curcumin has been described as a potent inhibitor of NF-κB activation [73]. Twenty out of 50 curcumin compounds showed binding energies to NF-κB smaller than −10 kcal/mol, while curcumin as a lead compound revealed free binding energies of >−10 kcal/mol. Comparable data were obtained for EGFR: 15 out of 50 curcumin compounds were bound to EGFR with free binding energies of <−10 kcal/mol, while the binding affinity of curcumin itself was >−10 kcal/mol. This indicates that the derivatization of curcumin may indeed be a promising strategy to improve targe specificity and to obtain more effective anticancer drug candidates.
Even though N-(3-nitrophenylpyrazole) curcumin was bound to EGFR and NF- κB, it inhibited cell viability (as shown by the resazurin assay) and was cytotoxic (as shown by the LDH assay) in a comparable manner as curcumin and 1A9, N-(3-nitrophenylpyrazole) curcumin did not induce ROS generation and did not induce apoptosis. This indicates that different curcumin derivatives may activate different downstream mechanisms despite identical upstream targets (EGFR, NF-κB). N-(3-nitrophenylpyrazole) curcumin apparently induced a ROS-independent non-apoptotic pathway of cell death. This is a novel finding that may have important therapeutic implications. It is well-known that tumor cells with defective apoptotic regulation exert resistance to the induction of cell death, resulting in tumor progression and resistance to anticancer drugs [74,75]. Having cytotoxic drugs at hand, such as N-(3-nitrophenylpyrazole) curcumin, may allow one to bypass apoptosis resistance and kill otherwise apoptosis-resistant tumors. It is now well-known that a plethora of different cell death mechanisms can be operative in tumor cells [76,77]. It is beyond the scope of the present investigation to clarify which alternative mechanisms of cell death are responsible for the cytotoxic activity of N-(3-nitrophenylpyrazole) curcumin. We will address this issue in the future.
4. Materials and Methods
4.1. Chemicals
Chemical structures for in silico analyses were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/; accessed on 31 October 2021). Curcumin and DMSO were of analytical grade and purchased from Sigma-Aldrich (Taufkirchen, Germany). The curcumin derivative 1A9 was synthesized as recently reported [17]. N-(3-nitrophenylpyrazole) curcumin was provided by Dr. Fadhil S. Kamounah (Department of Chemistry, University of Copenhagen, Denmark). Stock solutions (50 mM) were prepared in DMSO, stored at −20 °C, and diluted to the final concentration in fresh media before each experiment. The chemical structures of 50 curcumin compounds are shown in Supplementary Figure S1.
4.2. Docking
The X-ray crystallography-based three-dimensional protein structures of EGFR and NF-κB (PDB codes 1M17 and IVKX, respectively) were obtained from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do; accessed on 31 October 2021) and used as docking templates throughout the calculations. Two-dimensional structures of curcumin and its derivatives were energy-minimized and converted to 3D structures compatible for docking operation using the Chem 3D program. Molecular docking was then carried out with the AutoDock program 4.2.6 (The Scripps Research Institute, La Jolla, CA, USA) following a previously reported protocol [78,79]. Docking parameters were set to 250 runs and 250,000 energy evaluations for each cycle. Three independent cycles were performed, resulting in a total number of 750,000 calculations per compound and target protein. VMD (Visual Molecular Dynamics, Visual Molecular Dynamics, http://www.ks.uiuc.edu/Research/vmd/; accessed on 31 October 2021) was used as a visualization tool to illustrate further the binding modes obtained from docking.
4.3. Microscale Thermophoresis
In vitro protein binding assays were performed to validate the in silico interaction between EGFR and NF-κB). The recombinant proteins were obtained from commercial sources (EGFR cat. No. 10001-H08H, NF-κB cat. No. 12054-H09E, Sino Biological Europe GmbH, Eschborn, Germany). The proteins were labeled according to the Monolith™ NT.115 Protein Labeling Kit RED-NHS (NanoTemper Technologies GmbH, Munich, Germany). Varying concentrations of curcumin, N-(3-phenylpyrazole) curcumin, and the curcumin derivative 1A9 ranging from 100 to 100,000 nM were titrated with labeled EGFR or NF-κB. As previously described, the experiments were carried out using standard capillaries in the NanoTemper Monolith™ NT (NanoTemper Technologies GmbH, Munich, Germany) [80,81]. Microscale thermophoresis (MST) was performed with 40% LED power and 80% MST power for the labeled EGFR and with 20% LED power and 20% MST power for the labeled NF-κB.
4.4. Cell Culture
The A549 lung cancer cell line was obtained from the German Cancer Research Center (DKFZ, Heidelberg, Germany). The original source of the cell lines is the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultivated in a completed DMEM culture medium with GlutaMAX (Invitrogen, Germany) supplemented with 10% fetal bovine serum, l-glutamine (2 mM), and 1% of a 10,000 U/mL penicillin G and 10 mg/mL streptomycin at 37 °C in a humidified air incubator (95%) containing 5% CO2. All experiments were performed on cells in the logarithmic growth phase. CCRF-CEM cells were a gift from Dr. Axel Sauerbrey (University Hospital for Pediatrics, Univerity of Jena, Germany) and maintained in an RPMI medium supplemented with fetal bovine serum, glutamine, and penicillin as described above.
Peripheral blood from healthy donors was obtained from Transfusion Center (University Medical Center, Johannes Gutenberg University Mainz, Mainz, Germany). Human peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Histopaque-1077® (Sigma-Aldrich Co. LLC, Darmstadt, Germany), strictly following the manufacturer’s instructions. Isolated PBMCs were resuspended in warm Panserin 413 medium (PAN-Biotech, Aidenbach, Germany) supplemented with 2% phytohemagglutinin M (PHA-M, Life Technologies, Darmstadt, Germany) and then immediately used for experimentation.
4.5. Resazurin Cell Viability Assay
Viable cells reduce the non-fluorescent resazurin to the strongly-fluorescent resorufin. Suspension cells (1 × 104 cells/well) or adherent cells (5 × 103 cells/well, incubated overnight to allow attachment) were seeded in 96-well plates at a volume of 100 µL. Varying concentrations of curcumin, 1A9, or N-(3-nitrophenylpyrazole) curcumin were added to reach a total volume of 200 µL. After 72 h, 20 µL of 0.01% w/v resazurin (Sigma-Aldrich) was added to each well, and cells were incubated for another 4 h at 37 °C. The fluorescence of resorufin was measured at 544 nm (excitation) and 590 nm (emission) using an Infinite M2000 Pro™ plate reader (Tecan, Crailsheim, Germany). Each experiment was independently performed three times with six parallel measurements each. The effects on cell viability were assessed according to the percentage of untreated control and 50% inhibition concentrations (IC50) were calculated from dose-response curves using GraphPad Prism® v6.0 software (GraphPad Software Inc., San Diego, CA, USA).
4.6. Lactate Dehydrogenase (LDH) Assay
The LDH release from the cells was determined using an LDH-cytotoxicity assay kit (catalog no. ab65393, Abcam, Berlin, Germany). CEM/CCRF leukemia cells and peripheral blood mononuclear cells (PBMCs) were seeded in 96-well plates at a density of 4 × 104 cells/well. Varying concentrations ranging from 0.01 to 10 µM of curcumin, N-(3-nitrophenylpyrazole) curcumin, or 1A9 were added to the cells and incubated for 48 h. At the end of the incubation period, the plates were gently shaken to ensure LDH was evenly distributed in the culture medium. Afterward, cells were precipitated at 600× g for 10 min and 10 μL/well of the clear supernatant medium were transferred to optically clear 96-well plates. The LDH reaction mix (100 μL/well) was added and incubated for 30 min at room temperature. The absorbance of LDH was measured with an Infinite M2000 Pro™ plate reader (Tecan, Crailsheim, Germany) using a 450 (440–490) nm filter.
4.7. Reactive Oxygen Species (ROS) Detection Assay
Intracellular reactive oxygen species (ROS) were determined using the cellular reactive oxygen species detection assay kit (catalog No. ab186027, Abcam, Berlin, Germany). Briefly, 4 × 104 CEM/CCRF leukemia cells/well were seeded in 96-well plates and incubated with 100 µL/well of ROS red working solution for 1 h in an incubator at 37 °C in a 5% CO2 atmosphere. Afterward, cells were treated with curcumin, N-(3-nitrophenylpyrazole) curcumin, or 1A9. For each compound, varying concentrations (0.5× IC50, IC50, 2× IC50) were used. After incubation for 24 h, the ROS induction was monitored at Ex/Em = 520/605 nm using an Infinite M2000 Pro™ plate reader (Tecan, Crailsheim, Germany).
4.8. Annexin V/Propidium Iodide (PI) Assay
CCRF-CEM cells (1 × 106 cells/mL) were seeded in 6-well plates and different concentrations of curcumin, 1A9, or N-(3-nitrophenylpyrazole) curcumin (0.5×, 1×, 2×, 4× IC50) were applied at 37 °C for 24 h. The cells were washed and re-suspended in 1 mL cold PBS and 500 μL 1× binding buffer, respectively, followed by incubation with 5 µL annexin V/PI and 10 µL of PE (50 mg/mL) (Thermo Fisher Scientific, Dreieich, Germany) in the dark for 15 min. The histograms were measured using an Accuri C6 flow cytometer (Becton Dickinson, Heidelberg, Germany). Four different cell populations were determined: viable cells, annexin V positive/PI negative cells (cells that are in early apoptosis), annexin V negative/PI-positive cells (cells that are necrotic), and annexin V positive/PI-positive cells (cells that are in late apoptosis or already dead).
5. Conclusions
In conclusion, we speculated that synthetic derivatives bear potential for cancer treatment, especially as EGFR inhibitors. Some synthetic derivatives are bound to and inhibit their target proteins in a comparable or even better manner than their parent lead compound, curcumin. The chemistry of curcumins is a fascinating area of research. It should be kept in mind that curcumin is not only active against cancer but also against many diseases such as metabolic syndrome, ulcerative colitis and inflammatory diseases in general, neurodegenerative diseases, infectious diseases, etc. [82,83,84,85,86]. It is well in the scope of expectations that some of the curcumin derivatives described in the present study may also be valuable drug candidates for other diseases than cancer. We believe they may be valuable candidates in the further drug discovery process.
Acknowledgments
M.-E.F.H. gratefully acknowledges the financial support from the Alexander von Humboldt Foundation “Georg Foster Research Fellowship for Experienced Researchers”. R.Y. is funded by the Theophrastus Foundation, Germany. M.E.M.S., O.K. and E.-J.S. are funded by intramural funds at the Johannes Gutenberg University Mainz, Germany. We are grateful for a Ph.D. stipend from the German Academic Exchange Service (DAAD) to E.O.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/article/10.3390/ijms23073966/s1.
Author Contributions
T.E. and B.B. designed the study. R.Y., A.D. and M.D. performed molecular docking experiments and calculations. M.E.M.S., E.O., R.Y., E.-J.S., A.D., M.D. and O.K. carried out the experiments. S.J.T. and F.S.K. synthesized the compounds. M.-E.F.H. optimized the compounds and revised the structures for molecular docking. T.E. supervised the work and provided the facilities for the study. M.E.M.S. and T.E. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
M.-E.F.H. gratefully acknowledges the financial support from the Alexander von Humboldt Foundation “Georg Foster Research Fellowship for Experienced Researchers”. M.E.M.S., O.K. and E.J.S. are funded by intramural funds at the Johannes Gutenberg University Mainz, Germany. We are grateful for a Ph.D. stipend from the German Academic Exchange Service (DAAD) to E.O.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available on request.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Toyoda Y., Tabuchi T., Nakayama T., Hojo S., Yoshioka S., Maeura Y. Past trends and future estimation of annual breast cancer incidence in Osaka, Japan. Asian Pac. J. Cancer Prev. 2016;17:52. [PubMed] [Google Scholar]
- 3.Mattiuzzi C., Lippi G. Current cancer epidemiology. J. Epidemiol. Glob. Health. 2019;9:217–222. doi: 10.2991/jegh.k.191008.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 5.Kuete V., Efferth T. African flora has the potential to fight multidrug resistance of cancer. BioMed Res. Int. 2015;2015:914813. doi: 10.1155/2015/914813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Efferth T., Saeed M.E.M., Kadioglu O., Seo E.J., Shirooie S., Mbaveng A.T., Nabavi S.M., Kuete V. Collateral sensitivity of natural products in drug-resistant cancer cells. Biotechnol. Adv. 2020;38:107342. doi: 10.1016/j.biotechadv.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Efferth T., Kadioglu O., Saeed M.E.M., Seo E.-J., Mbaveng A.T., Kuete V. Medicinal plants and phytochemicals against multidrug-resistant tumor cells expressing ABCB1, ABCG2, or ABCB5: A synopsis of 2 decades. Phytochem. Rev. 2021;20:7–53. doi: 10.1007/s11101-020-09703-7. [DOI] [Google Scholar]
- 8.Sertel S., Plinkert P.K., Efferth T. Natural products derived from traditional Chinese medicine as novel inhibitors of the epidermal growth factor receptor. Comb. Chem. High Throughput Screen. 2010;13:849–854. doi: 10.2174/138620710793360266. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Q., Kretschmer N., Bauer R., Efferth T. Shikonin and its derivatives inhibit the epidermal growth factor receptor signaling and synergistically kill glioblastoma cells in combination with erlotinib. Int. J. Cancer. 2015;137:1446–1456. doi: 10.1002/ijc.29483. [DOI] [PubMed] [Google Scholar]
- 10.Saeed M.E.M., Rahama M., Kuete V., Dawood M., Elbadawi M., Sugimoto Y., Efferth T. Collateral sensitivity of drug-resistant ABCB5- and mutation-activated EGFR overexpressing cells towards resveratrol due to modulation of SIRT1 expression. Phytomedicine. 2019;59:152890. doi: 10.1016/j.phymed.2019.152890. [DOI] [PubMed] [Google Scholar]
- 11.Lee H.Y.J., Meng M., Liu Y., Su T., Kwan H.Y. Medicinal herbs and bioactive compounds overcome the drug resistance to epidermal growth factor receptor inhibitors in non-small cell lung cancer. Oncol. Lett. 2021;22:646. doi: 10.3892/ol.2021.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamdoun S., Efferth T. Ginkgolic acids inhibit migration in breast cancer cells by inhibition of NEMO sumoylation and NF-κB activity. Oncotarget. 2017;8:35103–35115. doi: 10.18632/oncotarget.16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Liu R.H. Potential mechanisms of action of dietary phytochemicals for cancer prevention by targeting cellular signaling transduction pathways. J. Agric. Food. Chem. 2018;66:3260–3276. doi: 10.1021/acs.jafc.7b04975. [DOI] [PubMed] [Google Scholar]
- 14.Dawood M., Ooko E., Efferth T. Collateral sensitivity of parthenolide via NF-κB and HIF-α inhibition and epigenetic changes in drug-resistant cancer cell lines. Front. Pharmacol. 2019;10:542. doi: 10.3389/fphar.2019.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menon V.P., Sudheer A.R. Antioxidant and anti-inflammatory properties of curcumin. Adv. Exp. Med. Biol. 2007;595:105–125. doi: 10.1007/978-0-387-46401-5_3. [DOI] [PubMed] [Google Scholar]
- 16.Moghadamtousi S.Z., Kadir H.A., Hassandarvish P., Tajik H., Abubakar S., Zandi K. A review on antibacterial, antiviral, and antifungal activity of curcumin. BioMed Res. Int. 2014;2014:186864. doi: 10.1155/2014/186864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ooko E., Alsalim T., Saeed B., Saeed M.E., Kadioglu O., Abbo H.S., Titinchi S.J., Efferth T. Modulation of P-glycoprotein activity by novel synthetic curcumin derivatives in sensitive and multidrug-resistant T-cell acute lymphoblastic leukemia cell lines. Tox. Appl. Pharmacol. 2016;305:216–233. doi: 10.1016/j.taap.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Liu L., Fu Y., Zheng Y., Ma M., Wang C. Curcumin inhibits proteasome activity in triple-negative breast cancer cells through regulating p300/miR-142-3p/PSMB5 axis. Phytomedicine. 2020;78:153312. doi: 10.1016/j.phymed.2020.153312. [DOI] [PubMed] [Google Scholar]
- 19.Heshmati J., Moini A., Sepidarkish M., Morvaridzadeh M., Salehi M., Palmowski A., Mojtahedi M.F., Shidfar F. Effects of curcumin supplementation on blood glucose, insulin resistance and androgens in patients with polycystic ovary syndrome: A randomized double-blind placebo-controlled clinical trial. Phytomedicine. 2021;80:153395. doi: 10.1016/j.phymed.2020.153395. [DOI] [PubMed] [Google Scholar]
- 20.Seo E.J., Efferth T., Panossian A. Curcumin downregulates expression of opioid-related nociceptin receptor gene (OPRL1) in isolated neuroglia cells. Phytomedicine. 2018;50:285–299. doi: 10.1016/j.phymed.2018.09.202. [DOI] [PubMed] [Google Scholar]
- 21.Hsiao A.F., Lien Y.C., Tzeng I.S., Liu C.T., Chou S.H., Horng Y.S. The efficacy of high- and low-dose curcumin in knee osteoarthritis: A systematic review and meta-analysis. Complement. Ther. Med. 2021;63:102775. doi: 10.1016/j.ctim.2021.102775. [DOI] [PubMed] [Google Scholar]
- 22.Saghatelyan T., Tananyan A., Janoyan N., Tadevosyan A., Petrosyan H., Hovhannisyan A., Hayrapetyan L., Arustamyan M., Arnhold J., Rotmann A.R., et al. Efficacy and safety of curcumin in combination with paclitaxel in patients with advanced, metastatic breast cancer: A comparative, randomized, double-blind, placebo-controlled clinical trial. Phytomedicine. 2020;70:153218. doi: 10.1016/j.phymed.2020.153218. [DOI] [PubMed] [Google Scholar]
- 23.Zhang T., He Q., Liu Y., Chen Z., Hu H. Efficacy and safety of curcumin supplement on improvement of insulin resistance in people with type 2 diabetes mellitus: A systematic review and meta-analysis of randomized controlled trials. Evid.-Based Complement. Altern. Med. 2021;2021:4471944. doi: 10.1155/2021/4471944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sertel S., Eichhorn T., Bauer J., Hock K., Plinkert P.K., Efferth T. Pharmacogenomic determination of genes associated with sensitivity or resistance of tumor cells to curcumin and curcumin derivatives. J. Nutr. Biochem. 2012;23:875–884. doi: 10.1016/j.jnutbio.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Ye M.X., Li Y., Yin H., Zhang J. Curcumin: Updated molecular mechanisms and intervention targets in human lung cancer. Int. J. Mol. Sci. 2012;13:3959–3978. doi: 10.3390/ijms13033959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong Y., Ma W., Liu X., Zu Y., Fu Y., Wu N., Liang L., Yao L., Efferth T. Cytotoxic activity of curcumin towards CCRF-CEM leukemia cells and its effect on DNA damage. Molecules. 2009;14:5328–5338. doi: 10.3390/molecules14125328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong R., Wu X., Liu Y., Liu Y., Zhou J., Jiang X., Zhang L., He X., Ma L. Curcumin-induced DNA demethylation in human gastric cancer cells is mediated by the DNA-damage response pathway. Oxid. Med. Cell. Longev. 2020;2020:2543504. doi: 10.1155/2020/2543504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachmeier B., Nerlich A.G., Iancu C.M., Cilli M., Schleicher E., Vené R., Dell’Eva R., Jochum M., Albini A., Pfeffer U. The chemopreventive polyphenol Curcumin prevents hematogenous breast cancer metastases in immunodeficient mice. Cell Physiol. Biochem. 2007;19:137–152. doi: 10.1159/000099202. [DOI] [PubMed] [Google Scholar]
- 29.Killian P.H., Kronski E., Michalik K.M., Barbieri O., Astigiano S., Sommerhoff C.P., Pfeffer U., Nerlich A.G., Bachmeier B.E. Curcumin inhibits prostate cancer metastasis in vivo by targeting the inflammatory cytokines CXCL1 and -2. Carcinogenesis. 2012;33:2507–2519. doi: 10.1093/carcin/bgs312. [DOI] [PubMed] [Google Scholar]
- 30.Kronski E., Fiori M.E., Barbieri O., Astigiano S., Mirisola V., Killian P.H., Bruno A., Pagani A., Rovera F., Pfeffer U., et al. miR181b is induced by the chemopreventive polyphenol curcumin and inhibits breast cancer metastasis via down-regulation of the inflammatory cytokines CXCL-1 and -2. Mol. Oncol. 2014;8:581–595. doi: 10.1016/j.molonc.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz de Porras V., Bystrup S., Martínez-Cardús A., Pluvinet R., Sumoy L., Howells L., James M.I., Iwuji C., Manzano J.L., Layos L., et al. Curcumin mediates oxaliplatin-acquired resistance reversion in colorectal cancer cell lines through modulation of CXC-Chemokine/NF-κB signalling pathway. Sci. Rep. 2016;6:24675. doi: 10.1038/srep24675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norooznezhad F., Rodriguez-Merchan E.C., Asadi S., Norooznezhad A.H. Curcumin: Hopeful treatment of hemophilic arthropathy via inhibition of inflammation and angiogenesis. Expert Rev. Hematol. 2020;13:5–11. doi: 10.1080/17474086.2020.1685867. [DOI] [PubMed] [Google Scholar]
- 33.Bhandarkar S.S., Arbiser J.L. Curcumin as an inhibitor of angiogenesis. Adv. Exp. Med. Biol. 2007;595:185–195. doi: 10.1007/978-0-387-46401-5_7. [DOI] [PubMed] [Google Scholar]
- 34.Shafiee M., Mohamadzade E., Shahid Sales S., Khakpouri S., Maftouh M., Parizadeh S.A., Hasanian S.M., Avan A. Current status and perspectives regarding the therapeutic potential of targeting EGFR pathway by curcumin in lung cancer. Curr. Pharm. Des. 2017;23:2002–2008. doi: 10.2174/1381612823666170123143648. [DOI] [PubMed] [Google Scholar]
- 35.Wan Mohd Tajuddin W.N.B., Lajis N.H., Abas F., Othman I., Naidu R. Mechanistic understanding of curcumin’s therapeutic effects in lung cancer. Nutrients. 2019;11:2989. doi: 10.3390/nu11122989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pabla B., Bissonnette M., Konda V.J. Colon cancer and the epidermal growth factor receptor: Current treatment paradigms, the importance of diet, and the role of chemoprevention. World J. Clin. Oncol. 2015;6:133–141. doi: 10.5306/wjco.v6.i5.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz de Porras V., Layos L., Martínez-Balibrea E. Curcumin: A therapeutic strategy for colorectal cancer? Semin. Cancer Biol. 2021;73:321–330. doi: 10.1016/j.semcancer.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Thomas R.K., Sos M.L., Zander T., Mani O., Popov A., Berenbrinker D., Smola-Hess S., Schultze J.L., Wolf J. Inhibition of nuclear translocation of nuclear factor-kappaB despite lack of functional IkappaBalpha protein overcomes multiple defects in apoptosis signaling in human B-cell malignancies. Clin. Cancer Res. 2005;11:8186–8194. doi: 10.1158/1078-0432.CCR-05-0224. [DOI] [PubMed] [Google Scholar]
- 39.Zambre A.P., Kulkarni V.M., Padhye S., Sandur S.K., Aggarwal B.B. Novel curcumin analogs targeting TNF-induced NF-kappaB activation and proliferation in human leukemic KBM-5 cells. Bioorg. Med. Chem. 2006;14:7196–7204. doi: 10.1016/j.bmc.2006.06.056. [DOI] [PubMed] [Google Scholar]
- 40.Mackenzie G.G., Queisser N., Wolfson M.L., Fraga C.G., Adamo A.M., Oteiza P.I. Curcumin induces cell-arrest and apoptosis in association with the inhibition of constitutively active NF-kappaB and STAT3 pathways in Hodgkin’s lymphoma cells. Int. J. Cancer. 2008;123:56–65. doi: 10.1002/ijc.23477. [DOI] [PubMed] [Google Scholar]
- 41.Shehzad A., Lee J., Lee Y.S. Curcumin in various cancers. Biofactors. 2013;39:56–68. doi: 10.1002/biof.1068. [DOI] [PubMed] [Google Scholar]
- 42.Zhu G., Shen Q., Jiang H., Ji O., Zhu L., Zhang L. Curcumin inhibited the growth and invasion of human monocytic leukaemia SHI-1 cells in vivo by altering MAPK and MMP signalling. Pharm. Biol. 2020;58:25–34. doi: 10.1080/13880209.2019.1701042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song X., Zhang M., Dai E., Luo Y. Molecular targets of curcumin in breast cancer (Review) Mol. Med. Rep. 2019;19:23–29. doi: 10.3892/mmr.2018.9665. [DOI] [PubMed] [Google Scholar]
- 44.Akbari S., Kariznavi E., Jannati M., Elyasi S., Tayarani-Najaran Z. Curcumin as a preventive or therapeutic measure for chemotherapy and radiotherapy induced adverse reaction: A comprehensive review. Food Chem. Toxicol. 2020;145:111699. doi: 10.1016/j.fct.2020.111699. [DOI] [PubMed] [Google Scholar]
- 45.Ashrafizadeh M., Zarrabi A., Hashemi F., Moghadam E.R., Hashemi F., Entezari M., Hushmandi K., Mohammadinejad R., Najafi M. Curcumin in cancer therapy: A novel adjunct for combination chemotherapy with paclitaxel and alleviation of its adverse effects. Life Sci. 2020;256:117984. doi: 10.1016/j.lfs.2020.117984. [DOI] [PubMed] [Google Scholar]
- 46.Liu W., Zhai Y., Heng X., Che F.Y., Chen W., Sun D., Zhai G. Oral bioavailability of curcumin: Problems and advancements. J. Drug Target. 2016;24:694–702. doi: 10.3109/1061186X.2016.1157883. [DOI] [PubMed] [Google Scholar]
- 47.Anand P., Kunnumakkara A.B., Newman R.A., Aggarwal B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 48.Vyas A., Dandawate P., Padhye S., Ahmad A., Sarkar F. Perspectives on new synthetic curcumin analogs and their potential anticancer properties. Curr. Pharm. Des. 2013;19:2047–2069. [PMC free article] [PubMed] [Google Scholar]
- 49.Ooko E., Kadioglu O., Greten H.J., Efferth T. Pharmacogenomic characterization and isobologram analysis of the combination of ascorbic acid and curcumin-two main metabolites of Curcuma longa-in cancer cells. Front. Pharmacol. 2017;8:38. doi: 10.3389/fphar.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karimpour M., Feizi M.A.H., Mahdavi M., Krammer B., Verwanger T., Najafi F., Babaei E. Development of curcumin-loaded gemini surfactant nanoparticles: Synthesis, characterization and evaluation of anticancer activity against human breast cancer cell lines. Phytomedicine. 2019;57:183–190. doi: 10.1016/j.phymed.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 51.Moballegh Nasery M., Abadi B., Poormoghadam D., Zarrabi A., Keyhanvar P., Khanbabaei H., Ashrafizadeh M., Mohammadinejad R., Tavakol S., Sethi G. Curcumin delivery mediated by bio-based nanoparticles: A review. Molecules. 2020;25:689. doi: 10.3390/molecules25030689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin J.K. Molecular targets of curcumin. Adv. Exp. Med. Biol. 2007;595:227–243. doi: 10.1007/978-0-387-46401-5_10. [DOI] [PubMed] [Google Scholar]
- 53.Sigismund S., Avanzato D., Lanzetti L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018;12:3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan G.E., Efferth T. Broad-spectrum cross-resistance to anticancer drugs mediated by epidermal growth factor receptor. Anticancer Res. 2019;39:3585–3593. doi: 10.21873/anticanres.13505. [DOI] [PubMed] [Google Scholar]
- 55.Singh D., Attri B.K., Gill R.K., Bariwal J. Review on EGFR inhibitors: Critical updates. Mini Rev. Med. Chem. 2016;16:1134–1166. doi: 10.2174/1389557516666160321114917. [DOI] [PubMed] [Google Scholar]
- 56.Yan G., Saeed M.E.M., Foersch S., Schneider J., Roth W., Efferth T. Relationship between EGFR expression and subcellular localization with cancer development and clinical outcome. Oncotarget. 2019;10:1918–1931. doi: 10.18632/oncotarget.26727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kadioglu O., Saeed M.E.M., Mahmoud N., Azawi S., Mrasek K., Liehr T., Efferth T. Identification of novel drug resistance mechanisms by genomic and transcriptomic profiling of glioblastoma cells with mutation-activated EGFR. Life Sci. 2021;284:119601. doi: 10.1016/j.lfs.2021.119601. [DOI] [PubMed] [Google Scholar]
- 58.Efferth T. Signal transduction pathways of the epidermal growth factor receptor in colorectal cancer and their inhibition by small molecules. Curr. Med. Chem. 2012;19:5735–5744. doi: 10.2174/092986712803988884. [DOI] [PubMed] [Google Scholar]
- 59.Kadioglu O., Cao J., Saeed M.E., Greten H.J., Efferth T. Targeting epidermal growth factor receptors and downstream signaling pathways in cancer by phytochemicals. Target. Oncol. 2015;10:337–353. doi: 10.1007/s11523-014-0339-4. [DOI] [PubMed] [Google Scholar]
- 60.Taniguchi K., Karin M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 61.Hoesel B., Schmid J.A. The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 63.Gilmore T.D., Herscovitch M. Inhibitors of NF-kappaB signaling: 785 and counting. Oncogene. 2006;25:6887–6899. doi: 10.1038/sj.onc.1209982. [DOI] [PubMed] [Google Scholar]
- 64.Gaptulbarova K.A., Tsyganov M.M., Pevzner A.M., Ibragimova M.K., Litviakov N.V. NF-kB as a potential prognostic marker and a candidate for targeted therapy of cancer. Exp. Oncol. 2020;42:263–269. doi: 10.32471/exp-oncology.2312-8852.vol-42-no-4.15414. [DOI] [PubMed] [Google Scholar]
- 65.Ríos J.L., Recio M.C., Escandell J.M., Andújar I. Inhibition of transcription factors by plant-derived compounds and their implications in inflammation and cancer. Curr. Pharm. Des. 2009;15:1212–1237. doi: 10.2174/138161209787846874. [DOI] [PubMed] [Google Scholar]
- 66.Luqman S., Pezzuto J.M. NFkappaB: A promising target for natural products in cancer chemoprevention. Phytother. Res. 2010;24:949–963. doi: 10.1002/ptr.3171. [DOI] [PubMed] [Google Scholar]
- 67.Soukhtanloo M., Mohtashami E., Maghrouni A., Mollazadeh H., Mousavi S.H., Roshan M.K., Tabatabaeizadeh S.A., Hosseini A., Vahedi M.M., Jalili-Nik M., et al. Natural products as promising targets in glioblastoma multiforme: A focus on NF-κB signaling pathway. Pharmacol. Rep. 2020;72:285–295. doi: 10.1007/s43440-020-00081-7. [DOI] [PubMed] [Google Scholar]
- 68.Shostak K., Chariot A. EGFR and NF-κB: Partners in cancer. Trends Mol. Med. 2015;21:385–393. doi: 10.1016/j.molmed.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 69.Normanno N., De Luca A., Bianco C., Strizzi L., Mancino M., Maiello M.R., Carotenuto A., De Feo G., Caponigro F., Salomon D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 70.Starok M., Preira P., Vayssade M., Haupt K., Salome L., Rossi C. EGFR Inhibition by curcumin in cancer cells: A dual mode of action. Biomacromolecules. 2015;16:1634–1642. doi: 10.1021/acs.biomac.5b00229. [DOI] [PubMed] [Google Scholar]
- 71.Wada K., Lee J.Y., Hung H.Y., Shi Q., Lin L., Zhao Y., Goto M., Yang P.C., Kuo S.C., Chen H.W., et al. Novel curcumin analogs to overcome EGFR-TKI lung adenocarcinoma drug resistance and reduce EGFR-TKI-induced GI adverse effects. Bioorg. Med. Chem. 2015;23:1507–1514. doi: 10.1016/j.bmc.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Momtazi A.A., Sahebkar A. Difluorinated curcumin: A promising curcumin analogue with improved anti-tumor activity and pharmacokinetic profile. Curr. Pharm. Des. 2016;22:4386–4397. doi: 10.2174/1381612822666160527113501. [DOI] [PubMed] [Google Scholar]
- 73.Hackler L., Jr., Ozsvari B., Gyuris M., Sipos P., Fabian G., Molnar E., Marton A., Farago N., Mihaly J., Nagy L.I., et al. The curcumin analog C-150, influencing NF-kappaB, UPR and Akt/Notch pathways has potent anticancer activity in vitro and in vivo. PLoS ONE. 2016;11:e0149832. doi: 10.1371/journal.pone.0149832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carneiro B.A., El-Deiry W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong R.S. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reed J.C. Bcl-2: Prevention of apoptosis as a mechanism of drug resistance. Hematol. Oncol. Clin. N. Am. 1995;9:451–473. doi: 10.1016/S0889-8588(18)30104-7. [DOI] [PubMed] [Google Scholar]
- 77.Gottesman M.M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 78.Kadioglu O., Saeed M.E., Valoti M., Frosini M., Sgaragli G., Efferth T. Interactions of human P-glycoprotein transport substrates and inhibitors at the drug binding domain: Functional and molecular docking analyses. Biochem. Pharmacol. 2016;104:42–51. doi: 10.1016/j.bcp.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 79.Abdelfatah S., Böckers M., Asensio M., Kadioglu O., Klinger A., Fleischer E., Efferth T. Isopetasin and S-isopetasin as novel P-glycoprotein inhibitors against multidrug-resistant cancer cells. Phytomedicine. 2021;86:153196. doi: 10.1016/j.phymed.2020.153196. [DOI] [PubMed] [Google Scholar]
- 80.Lu X., Yan G., Dawood M., Klauck S.M., Sugimoto Y., Klinger A., Fleischer E., Shan L., Efferth T. A novel moniliformin derivative as pan-inhibitor of histone deacetylases triggering apoptosis of leukemia cells. Biochem. Pharmacol. 2021;194:114677. doi: 10.1016/j.bcp.2021.114677. [DOI] [PubMed] [Google Scholar]
- 81.Naß J., Abdelfatah S., Efferth T. The triterpenoid ursolic acid ameliorates stress in Caenorhabditis elegans by affecting the depression-associated genes skn-1 and prdx2. Phytomedicine. 2021;88:153598. doi: 10.1016/j.phymed.2021.153598. [DOI] [PubMed] [Google Scholar]
- 82.Kumar S., Ahuja V., Sankar M.J., Kumar A., Moss A.C. Curcumin for maintenance of remission in ulcerative colitis. Cochrane Database Syst. Rev. 2012;10:CD008424. doi: 10.1002/14651858.CD008424.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bukhari S.N., Franzblau S.G., Jantan I., Jasamai M. Current prospects of synthetic curcumin analogs and chalcone derivatives against Mycobacterium tuberculosis. Med. Chem. 2013;9:897–903. doi: 10.2174/1573406411309070002. [DOI] [PubMed] [Google Scholar]
- 84.Monroy A., Lithgow G.J., Alavez S. Curcumin and neurodegenerative diseases. BioFactors. 2013;39:122–132. doi: 10.1002/biof.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cicero A.F., Colletti A. Role of phytochemicals in the management of metabolic syndrome. Phytomedicine. 2016;23:1134–1144. doi: 10.1016/j.phymed.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 86.Hernandez M., Wicz S., Corral R.S. Cardioprotective actions of curcumin on the pathogenic NFAT/COX-2/prostaglandin E2 pathway induced during Trypanosoma cruzi infection. Phytomedicine. 2016;23:1392–1400. doi: 10.1016/j.phymed.2016.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on request.