Abstract
As a continuation of our earlier work against SARS-CoV-2, seven FDA-approved drugs were designated as the best SARS-CoV-2 nsp16-nsp10 2′-o-methyltransferase (2′OMTase) inhibitors through 3009 compounds. The in silico inhibitory potential of the examined compounds against SARS-CoV-2 nsp16-nsp10 2′-o-methyltransferase (PDB ID: (6W4H) was conducted through a multi-step screening approach. At the beginning, molecular fingerprints experiment with SAM (S-Adenosylmethionine), the co-crystallized ligand of the targeted enzyme, unveiled the resemblance of 147 drugs. Then, a structural similarity experiment recommended 26 compounds. Therefore, the 26 compounds were docked against 2′OMTase to reveal the potential inhibitory effect of seven promising compounds (Protirelin, (1187), Calcium folinate (1913), Raltegravir (1995), Regadenoson (2176), Ertapenem (2396), Methylergometrine (2532), and Thiamine pyrophosphate hydrochloride (2612)). Out of the docked ligands, Ertapenem (2396) showed an ideal binding mode like that of the co-crystallized ligand (SAM). It occupied all sub-pockets of the active site and bound the crucial amino acids. Accordingly, some MD simulation experiments (RMSD, RMSF, Rg, SASA, and H-bonding) have been conducted for the 2′OMTase—Ertapenem complex over 100 ns. The performed MD experiments verified the correct binding mode of Ertapenem against 2′OMTase exhibiting low energy and optimal dynamics. Finally, MM-PBSA studies indicated that Ertapenem bonded advantageously to the targeted protein with a free energy value of −43 KJ/mol. Furthermore, the binding free energy analysis revealed the essential amino acids of 2′OMTase that served positively to the binding. The achieved results bring hope to find a treatment for COVID-19 via in vitro and in vivo studies for the pointed compounds.
Keywords: SARS-CoV-2 nsp16-nsp10 2′-o-methyltransferase, FDA approved drugs, molecular fingerprints, structural similarity, molecular docking, MD simulations, MMPBSA
1. Introduction
The WHO, addressed on 16 February 2022, confirmed that the worldwide infections of COVID-19 were 414,525,183. Grievously, this number includes 5,832,333 deaths [1]. Although 10,227,670,521 vaccinations have been administered [1], the virus can still infect and spread widely [2]. Responding to these numbers, massive work is demanded from scientists all over the world to find a cure.
The regular process of new drug discovery is highly expensive and takes much time. The average required time for the complete development of a new drug is about 12 years, with a cost of 2.6 billion USD [3]. In contrast, drug repurposing or repositioning is a much faster technique in which the exploration of new pharmacological use for an old or existing drug occurs [4]. The strategy of drug repurposing was applied successfully in the discovery of anti-cancer [5], COVID-19 [6], anti-inflammatory [7], antibacterial [8], anti-parasitic [9], and anti-viral [10] drugs.
The tremendous applications of computational chemistry in drug discovery are due to different factors. First, the exploration of accurate 3D structures of different protein targets in the human body [11]. Second, the vast advancements in the fields of computer hardware and software [12]. Finally, the development of structure–activity relationship (SAR) principles [13]. Consequently, computational chemistry methods were applied to estimate various pharmacodynamic and pharmacokinetic parameters that relate the chemical structure of compounds to its activity and also to characterize the interaction of compounds with biological targets such as structure similarity [14], molecular fingerprints [15], QSAR [16], pharmacophores [17], homology models [18], molecular modeling [19], drug molecular design [20], rational drug design [21,22], molecular docking [23], MD simulations [24], absorption [25], distribution [26], metabolism [27], excretion [28], and toxicity properties [29], as well as physicochemical characterization [30] and DFT [31].
In this regard, our team employed the strategies of computer-based chemistry to discover the potential inhibitive effects of the secondary metabolites of Asteriscus hierochunticus [32], Monanchora sp. [33], Artemisia sublessingiana [34], and Artemisia sp. [35], as well as 69 isoflavonoids [36] against SARS-CoV-2. Additionally, we designed a multi-step in silico selection method to prime the most active inhibitor drugs against a SASRS-CoV-2 protein amongst a vast number of compounds. As an exemplification, amongst 310 natural metabolite and 69 semisynthetic compounds, the highest potential inhibitors against SARS-CoV-2 nsp10 [37] and the SARS-CoV-2 PLpro [38], respectively, were decided
In this research, a panel of 3009 FDA-approved compounds was retrieved from the internet [39] to be screened depending on various computational methods to distinguish the most potent SARS-CoV-2 nsp16-nsp10 2′-o-methyltransferase complex inhibitor.
The starting point in our study was (S-Adenosylmethionine, SAM), the co-crystallized ligand of the essential COVID-19 protein, 2′OMTase (PDB ID: (6W4H), that showed high binding affinity against it. Firstly, the selected compounds were subjected to two ligand-based computational techniques (molecular fingerprints and similarity) successively to select the most similar candidates to SAM. Then, several structure-based computational methods (molecular docking and MD simulations) were conducted to confirm the binding modes, energies, and dynamic behaviors of the singled-out candidates.
2. Results and Discussion
2.1. Filter Using Fingerprint
Molecular fingerprint is a ligand-based computational (in silico) computational technique. This approach can predict the biological activity of a molecule based on its chemical structure [40]. The scientific base of ligand-based calculations is influenced by the principles of target– structure–activity relationships (SAR). It can set a relationship between the measured bio response/s exerted by a molecule and its chemical structure. Accordingly, compounds with similar chemical structures are expected to exert similar activities [41].
A co-crystallized ligand is one that exerts an excellent binding affinity with the corresponding protein forming a crystallizable ligand–protein complex [42]. In accordance, the chemical structure of that ligand could be employed as a model to design and develop an inhibitor that can bind strongly to the target protein. The molecular fingerprints study was performed using Discovery Studio against SAM. The experiment examined the next variables: H-bond acceptor and donor [43], charge [44], hybridization [45], positive and negative ionizable [46], halogen, aromatic, or none of the above besides the ALogP of atoms and fragments.
In structural terms, the chemical structures of the examined molecules are encoded and transformed binary bit strings (sequences of 0′s and 1′s). Every bit corresponds to a “pre-defined/determined” structural descriptor or feature of substructure or fragment. If the examined molecule has that feature, the bit position that corresponds to this descriptor is set to 1 (ON). If it is absent, it is set to 0 (OFF) [47].
SA describes the number bits that were computed in the FDA-approved drugs and the SAM. SB identifies the number bits that were found in the FDA-approved drugs, but not SAM. SC refers to the number bits that were discovered in SAM, but not in the FDA-approved drugs.
The study (Table 1) favored 147 compounds. These compounds showed the highest fingerprint similarity with SAM.
Table 1.
Fingerprint similarity between the tested compounds and SAM.
| Comp. | Similarity | SA | SB | SC | Comp. | Similarity | SA | SB | SC |
|---|---|---|---|---|---|---|---|---|---|
| SAM | 1 | 237 | 0 | 0 | 1670 | 0.57 | 257 | 214 | −20 |
| 4 | 0.497396 | 191 | 147 | 46 | 1694 | 0.5 | 191 | 145 | 46 |
| 42 | 0.597 | 138 | −6 | 99 | 1737 | 0.506944 | 146 | 51 | 91 |
| 50 | 0.651 | 157 | 4 | 80 | 1740 | 0.491582 | 146 | 60 | 91 |
| 51 | 0.581 | 137 | −1 | 100 | 1756 | 0.506912 | 220 | 197 | 17 |
| 56 | 0.665 | 171 | 20 | 66 | 1761 | 0.523404 | 246 | 233 | −9 |
| 58 | 0.491525 | 174 | 117 | 63 | 1766 | 0.54321 | 176 | 87 | 61 |
| 74 | 0.495652 | 171 | 108 | 66 | 1778 | 0.511299 | 181 | 117 | 56 |
| 91 | 0.496241 | 132 | 29 | 105 | 1792 | 0.50211 | 238 | 237 | −1 |
| 113 | 0.485714 | 170 | 113 | 67 | 1793 | 0.494792 | 285 | 339 | −48 |
| 130 | 0.490463 | 180 | 130 | 57 | 1802 | 0.56 | 237 | 186 | 0 |
| 152 | 0.624 | 143 | −8 | 94 | 1805 | 0.501433 | 175 | 112 | 62 |
| 158 | 0.5 | 189 | 141 | 48 | 1818 | 0.508475 | 210 | 176 | 27 |
| 186 | 0.644 | 150 | −4 | 87 | 1860 | 0.494024 | 124 | 14 | 113 |
| 189 | 0.5 | 122 | 7 | 115 | 1886 | 0.493478 | 227 | 223 | 10 |
| 190 | 0.492958 | 175 | 118 | 62 | 1911 | 0.490683 | 158 | 85 | 79 |
| 214 | 0.515723 | 164 | 81 | 73 | 1913 | 0.494033 | 207 | 182 | 30 |
| 241 | 0.717 | 160 | −14 | 77 | 1917 | 0.929 | 235 | 16 | 2 |
| 251 | 0.490956 | 190 | 150 | 47 | 1919 | 0.488701 | 173 | 117 | 64 |
| 272 | 0.508403 | 121 | 1 | 116 | 1927 | 0.489796 | 216 | 204 | 21 |
| 281 | 0.510806 | 260 | 272 | −23 | 1928 | 0.488636 | 215 | 203 | 22 |
| 304 | 0.488938 | 221 | 215 | 16 | 1932 | 0.50303 | 166 | 93 | 71 |
| 310 | 0.717 | 160 | −14 | 77 | 1949 | 0.505464 | 185 | 129 | 52 |
| 322 | 0.486154 | 158 | 88 | 79 | 1960 | 0.48995 | 195 | 161 | 42 |
| 380 | 0.514563 | 159 | 72 | 78 | 1993 | 0.522599 | 185 | 117 | 52 |
| 390 | 0.52862 | 157 | 60 | 80 | 1995 | 0.488998 | 200 | 172 | 37 |
| 404 | 0.535211 | 190 | 118 | 47 | 2002 | 0.49 | 147 | 63 | 90 |
| 428 | 0.498623 | 181 | 126 | 56 | 2009 | 0.511364 | 135 | 27 | 102 |
| 446 | 0.50641 | 158 | 75 | 79 | 2017 | 0.663 | 193 | 54 | 44 |
| 458 | 0.488136 | 144 | 58 | 93 | 2023 | 0.627 | 168 | 31 | 69 |
| 461 | 0.507837 | 162 | 82 | 75 | 2024 | 0.527378 | 183 | 110 | 54 |
| 470 | 0.491803 | 180 | 129 | 57 | 2031 | 0.57 | 147 | 21 | 90 |
| 515 | 0.501493 | 168 | 98 | 69 | 2036 | 0.487179 | 171 | 114 | 66 |
| 516 | 0.561 | 165 | 57 | 72 | 2042 | 0.664 | 172 | 22 | 65 |
| 539 | 0.519149 | 122 | −2 | 115 | 2174 | 0.488318 | 209 | 191 | 28 |
| 562 | 0.489496 | 233 | 239 | 4 | 2176 | 0.661 | 199 | 64 | 38 |
| 573 | 0.491049 | 192 | 154 | 45 | 2232 | 0.642 | 265 | 176 | −28 |
| 598 | 0.510504 | 243 | 239 | −6 | 2233 | 0.701 | 202 | 51 | 35 |
| 659 | 0.540816 | 159 | 57 | 78 | 2256 | 0.543662 | 193 | 118 | 44 |
| 663 | 0.492537 | 198 | 165 | 39 | 2268 | 0.538776 | 132 | 8 | 105 |
| 672 | 0.48913 | 135 | 39 | 102 | 2303 | 0.503597 | 210 | 180 | 27 |
| 679 | 0.501661 | 151 | 64 | 86 | 2306 | 0.494737 | 188 | 143 | 49 |
| 683 | 0.488798 | 240 | 254 | −3 | 2333 | 0.494595 | 183 | 133 | 54 |
| 711 | 0.566 | 137 | 5 | 100 | 2376 | 0.643 | 160 | 12 | 77 |
| 723 | 0.561 | 142 | 16 | 95 | 2396 | 0.491525 | 232 | 235 | 5 |
| 736 | 0.5 | 169 | 101 | 68 | 2410 | 0.513587 | 189 | 131 | 48 |
| 753 | 0.504425 | 228 | 215 | 9 | 2437 | 0.489189 | 181 | 133 | 56 |
| 771 | 0.486076 | 192 | 158 | 45 | 2467 | 0.503086 | 163 | 87 | 74 |
| 772 | 0.489703 | 214 | 200 | 23 | 2483 | 0.539185 | 172 | 82 | 65 |
| 781 | 0.487603 | 177 | 126 | 60 | 2488 | 0.542274 | 186 | 106 | 51 |
| 816 | 0.497297 | 184 | 133 | 53 | 2496 | 0.522099 | 189 | 125 | 48 |
| 821 | 0.493369 | 186 | 140 | 51 | 2501 | 0.496711 | 151 | 67 | 86 |
| 824 | 0.492958 | 175 | 118 | 62 | 2530 | 0.495468 | 164 | 94 | 73 |
| 874 | 0.553531 | 243 | 202 | −6 | 2532 | 0.491667 | 236 | 243 | 1 |
| 919 | 0.504032 | 125 | 11 | 112 | 2538 | 0.501887 | 133 | 28 | 104 |
| 1129 | 0.5 | 186 | 135 | 51 | 2581 | 0.486141 | 228 | 232 | 9 |
| 1179 | 0.488701 | 173 | 117 | 64 | 2585 | 0.524 | 131 | 13 | 106 |
| 1185 | 0.571 | 348 | 372 | −111 | 2612 | 0.504792 | 158 | 76 | 79 |
| 1187 | 0.510989 | 186 | 127 | 51 | 2618 | 0.489028 | 156 | 82 | 81 |
| 1249 | 0.497222 | 179 | 123 | 58 | 2717 | 0.555556 | 190 | 105 | 47 |
| 1274 | 0.502 | 251 | 263 | −14 | 2732 | 0.571 | 140 | 8 | 97 |
| 1315 | 0.514368 | 179 | 111 | 58 | 2751 | 0.490667 | 184 | 138 | 53 |
| 1391 | 0.494005 | 206 | 180 | 31 | 2786 | 0.562 | 140 | 12 | 97 |
| 1401 | 0.490446 | 154 | 77 | 83 | 2831 | 0.603 | 155 | 20 | 82 |
| 1411 | 0.491803 | 180 | 129 | 57 | 2853 | 0.52214 | 283 | 305 | −46 |
| 1444 | 0.495238 | 156 | 78 | 81 | 2861 | 0.522822 | 252 | 245 | −15 |
| 1458 | 0.5 | 166 | 95 | 71 | 2876 | 0.635 | 223 | 114 | 14 |
| 1478 | 0.558074 | 197 | 116 | 40 | 2877 | 0.519651 | 238 | 221 | −1 |
| 1587 | 0.485849 | 206 | 187 | 31 | 2879 | 0.7 | 168 | 3 | 69 |
| 1595 | 0.547414 | 127 | −5 | 110 | 2884 | 0.486425 | 215 | 205 | 22 |
| 1604 | 0.489189 | 181 | 133 | 56 | 2894 | 0.494279 | 216 | 200 | 21 |
| 1642 | 0.603 | 225 | 136 | 12 | 2907 | 0.488889 | 220 | 213 | 17 |
| 1651 | 0.586 | 309 | 290 | −72 | 2918 | 0.490028 | 172 | 114 | 65 |
| 1662 | 0.507576 | 134 | 27 | 103 | 2959 | 0.489362 | 230 | 233 | 7 |
SA: The number bits in both SAM and the test set. SB: The number bits in the test set, but not SAM. SC: The number bits in SAM but not the test set.
2.2. Molecular Similarity
The connection between chemical structures and biological activities of different compounds has always been an interesting area for research [48]. Consequently, the implementation of different molecular similarity strategies in drug design and development have been competently increased effectively [49]. Many descriptors have been considered in molecular similarity studies.
The examined descriptors are of a molecular type, such as molecular weight (M.W.) [50], hydrogen bond donors (HBA) [51], hydrogen bond acceptors (HBD) [52], partition coefficient (ALog p), which is the ratio of the concentration of a substance in the lipid phase to the concentration in the aqueous phase when the two concentrations are at equilibrium [53], number of rotatable bonds [54], number of rings, and aromatic rings [55], as well as the molecular fractional polar surface area (MFPSA) [56]. The examined compound is represented as a binary array (number of binary bits) to be computed.
The mentioned descriptors were calculated for the FDA-approved drugs then compared with the co-crystallized ligand of 2′OMTase (SAM) using Discovery studio software.
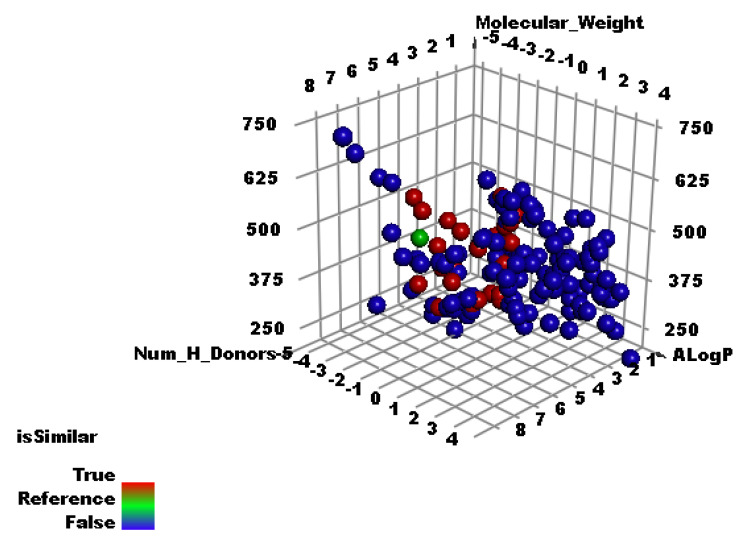
Figure 1 represented the co-crystalized ligand (SAM) (red ball), compounds with good similarities (green balls), and compounds with diminished similarities (blue balls). The degree of molecular likeness or similarity between two compounds depends on a similarity coefficient that is utilized to compute a quantitative score. That calculated score is equivalent to the degree of similarity and is based on the computed values of several structural descriptors. Similarity between two compounds is inversely proportional to the calculated distance between them in the descriptor space [57]. In this work, the distances between the several descriptors were computed to determine descriptor similarity among test compounds and SAM [58]. The computed distances describe the shortest distance between two points. Typed graph distances (Figure 1) show the overall similarity of behavior of the FDA-approved drugs compared to SAM. The study preferred 26 compounds among the most suitable 30 metabolites (Figure 1 and Figure 2, and Table 2).
Figure 1.
The molecular similarity of the examined compounds and SAM.
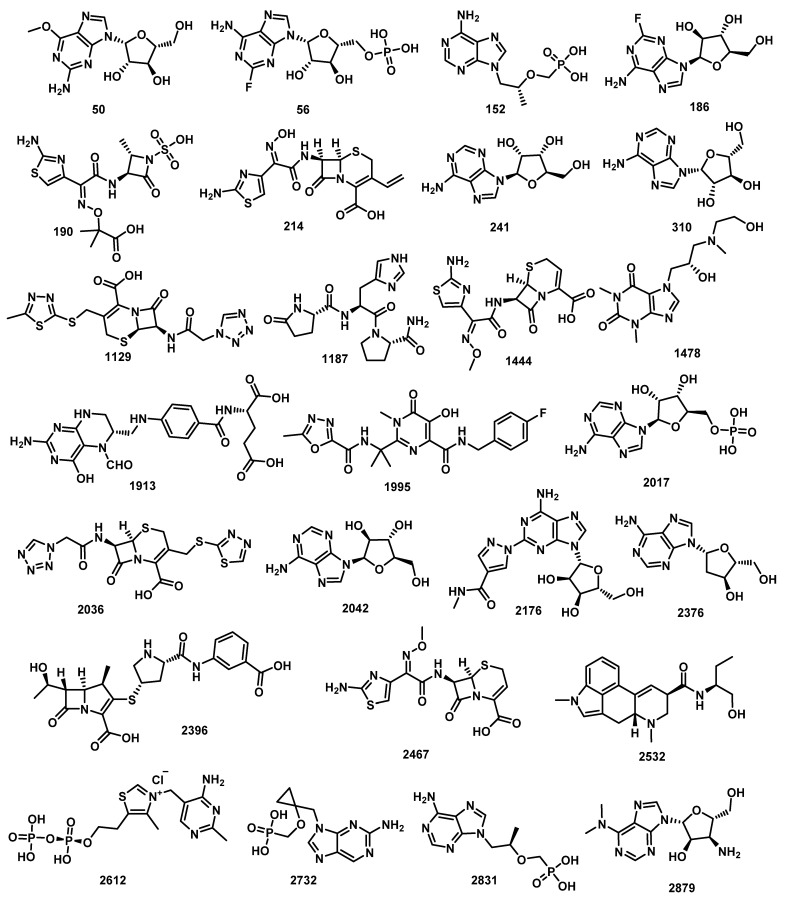
Figure 2.
Twenty-six compounds with good molecular similarity with the co-crystallized ligand (SAM) of 2′OMTase (PDB ID: (6W4H).
Table 2.
Molecular descriptors of the examined 26 compounds and SAM.
| Comp. | ALog p | MW | HBA | HBD | Rotatable Bonds | Rings | Aromatic Rings | MFPSA | Minimum Distance |
|---|---|---|---|---|---|---|---|---|---|
| SAM | −4.25 | 399.45 | 9 | 4 | 7 | 3 | 2 | 0.483 | 0 |
| 50 | −1.38 | 297.27 | 9 | 4 | 3 | 3 | 2 | 0.508 | 0.768 |
| 56 | −1.38 | 365.21 | 11 | 5 | 4 | 3 | 2 | 0.602 | 0.738 |
| 152 | −0.77 | 287.21 | 8 | 3 | 5 | 2 | 2 | 0.502 | 0.836 |
| 186 | −1.31 | 285.23 | 8 | 4 | 2 | 3 | 2 | 0.52 | 0.884 |
| 190 | −1.04 | 435.43 | 11 | 4 | 7 | 2 | 1 | 0.576 | 0.874 |
| 214 | −0.17 | 395.41 | 9 | 4 | 5 | 3 | 1 | 0.577 | 0.91 |
| 241 | −1.88 | 267.24 | 8 | 4 | 2 | 3 | 2 | 0.539 | 0.877 |
| 310 | −1.88 | 267.24 | 8 | 4 | 2 | 3 | 2 | 0.539 | 0.877 |
| 1129 | −2.81 | 476.49 | 11 | 1 | 7 | 4 | 2 | 0.624 | 0.896 |
| 1187 | −2.39 | 362.38 | 5 | 4 | 6 | 3 | 1 | 0.414 | 0.801 |
| 1444 | −0.64 | 383.4 | 9 | 3 | 5 | 3 | 1 | 0.56 | 0.874 |
| 1478 | −3.74 | 434.45 | 9 | 3 | 7 | 3 | 2 | 0.316 | 0.478 |
| 1913 | −3.05 | 511.5 | 12 | 5 | 9 | 3 | 2 | 0.545 | 0.67 |
| 1995 | −0.99 | 482.51 | 7 | 2 | 6 | 3 | 2 | 0.442 | 0.796 |
| 2017 | −2.16 | 365.24 | 12 | 6 | 4 | 3 | 2 | 0.655 | 0.856 |
| 2036 | −1.59 | 440.48 | 11 | 2 | 7 | 4 | 2 | 0.594 | 0.781 |
| 2042 | −2.09 | 285.26 | 9 | 5 | 2 | 3 | 2 | 0.589 | 0.874 |
| 2176 | −1.93 | 390.35 | 10 | 5 | 4 | 4 | 3 | 0.491 | 0.838 |
| 2376 | −1.32 | 269.26 | 8 | 4 | 2 | 3 | 2 | 0.54 | 0.917 |
| 2396 | −4.6 | 497.5 | 9 | 4 | 7 | 4 | 1 | 0.484 | 0.705 |
| 2467 | −2.12 | 405.39 | 9 | 2 | 5 | 3 | 1 | 0.628 | 0.87 |
| 2532 | −0.73 | 469.53 | 7 | 4 | 6 | 4 | 2 | 0.266 | 0.909 |
| 2612 | −1.98 | 460.77 | 10 | 4 | 8 | 2 | 2 | 0.572 | 0.594 |
| 2732 | −0.82 | 299.22 | 8 | 3 | 5 | 3 | 2 | 0.504 | 0.752 |
| 2831 | −0.98 | 305.23 | 9 | 4 | 5 | 2 | 2 | 0.55 | 0.76 |
| 2879 | −1.26 | 294.31 | 8 | 3 | 3 | 3 | 2 | 0.395 | 0.846 |
ALog p: lipid–water partition coefficient, MWt: molecular weight, HBA: hydrogen bond acceptor, HBD: hydrogen bond donor, Rotatable bonds: any single non-ring bond, attached to a non-terminal, non-hydrogen atom, Rings: non-aromatic rings, MFPSA: molecular fractional polar surface area, Minimum Distance: the shortest distance between a tested compound and the reference one.
2.3. Docking Studies
Docking studies of the tested compounds were conducted using the MOE (Molecular Operating Environment) software [58] to understand the proposed binding mode and the orientations of such compounds with the prospective target 2′OMTase (PDB ID: (6W4H)).
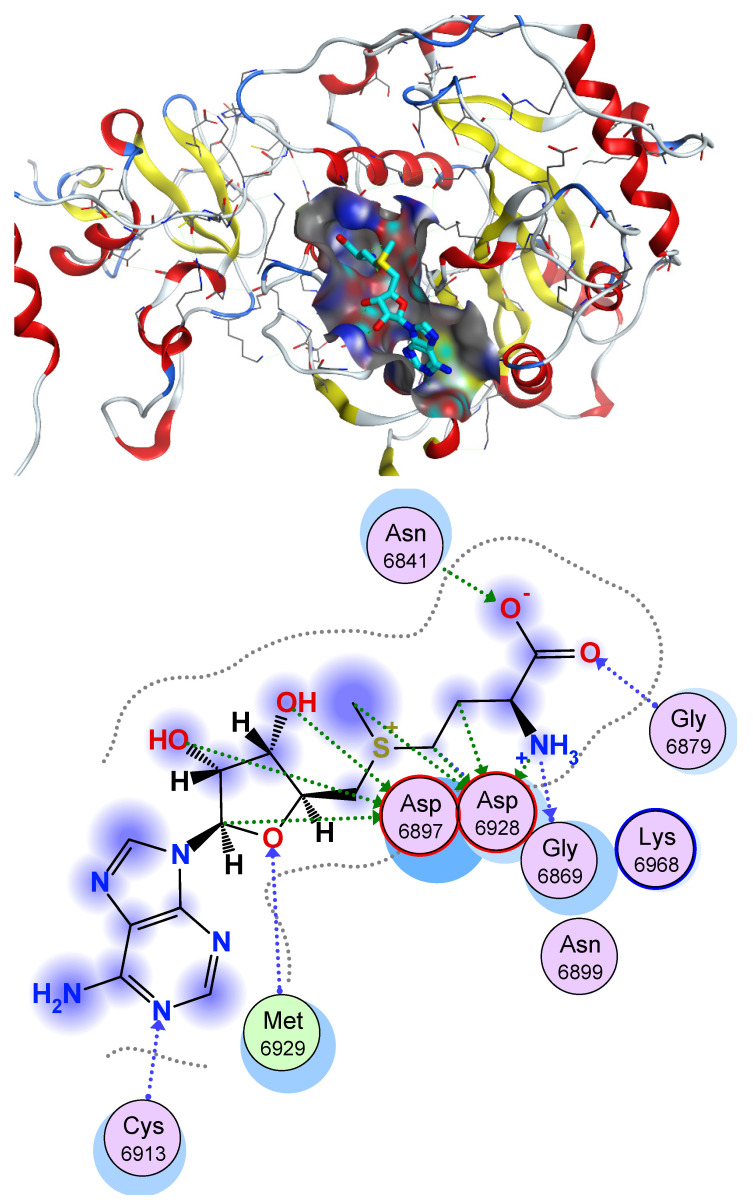
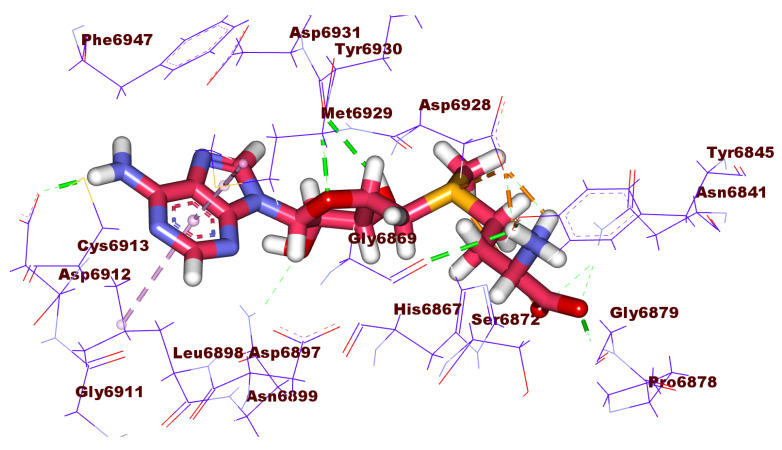
The active site of 2′OMTase consists of some crucial amino acids which can form hydrogen bonds with the active ligands. These amino acids include: Asn6841, Gly6879, Gly6869, Asp6928, Asp6897, Met6929, and Cys6913. In addition, there are some hydrophobic amino acids which can be incorporated in hydrophobic attractions with the active ligand and the hydrophobic amnio acids such as Leu6898 and Met6929 (Figure 3).
Figure 3.
Active site (3D and 2D) of 2′OMTase (PDB ID: (6W4H)).
The co-crystalized ligand s-adenosylmethionine (SAM) was used as a reference compound. First, the validation process was carried out to confirm the validity of the docking algorithm in obtaining accurate docking results. This was achieved by redocking the co-crystallized ligand (SAM) with 2′OMTase. The obtained low values of root mean square deviation (RMSD = 1.15 Å) between the native and redocked pose, in addition to the symmetrical superimposition in orientation between both the native (turquoise) and redocked (magenta) co-crystallized poses in Figure 4, guaranteed the valid performance of the docking protocol [36,38], in addition to the docking algorithm’s capability to obtain the reported binding mode of the co-crystalized ligand S-adenosylmethionine (SAM) [59].
Figure 4.
Alignment of the co-crystallized ligand (turquoise) and the docking pose (rose) of the same ligand (SAM) in the active site of 2′OMTase.
In comparing the tested compounds, the binding free energy (ΔG) between the docked molecules and the active site, and also the proper binding mode, were properly considered. The estimated (ΔG) (binding free energies) of the investigated drugs and the reference molecule (SAM) against the 2′OMTase are presented in Table 3.
Table 3.
Binding free energies (calculated ΔG in kcal/mol) of the examined compounds and ligand SAM against 2′OMTase.
| Comp. | Name | ΔG [kcal/mol] |
|---|---|---|
| SAM | S-Adenosylmethionine | −21.52 |
| 50 | Arranon (Nelarabine) | −13.84 |
| 56 | Fludara (Fludarabine) | −15.53 |
| 152 | Tenofovir (PMPA) | −13.58 |
| 186 | Fludarabine | −14.19 |
| 190 | Azactam (aztreonam) | −14.88 |
| 214 | Cefdinir (cefdinir) | −15.41 |
| 241 | Adenosine | −14.09 |
| 310 | VIRA-A (vidarabine) | −14.10 |
| 1129 | Cefazolin | −16.66 |
| 1187 | Protirelin | −18.68 |
| 1444 | Ceftizoxime | −10.99 |
| 1478 | Xanthinol Nicotinate | −16.19 |
| 1913 | Calcium folinate | −19.09 |
| 1995 | Raltegravir | −21.07 |
| 2017 | Adenosine 5′-monophosphate | −15.34 |
| 2036 | Ceftezole | −15.21 |
| 2042 | Vidarabine | −13.41 |
| 2176 | Regadenoson | −18.54 |
| 2376 | 2′-Deoxyadenosine | −13.16 |
| 2396 | Ertapenem | −20.73 |
| 2467 | Ceftizoxime | −13.63 |
| 2532 | Methylergometrine | −20.46 |
| 2612 | Thiamine pyrophosphate hydrochloride | −18.03 |
| 2732 | Besifovir | −13.47 |
| 2831 | Tenofovir | −14.36 |
| 2879 | Puromycin aminonucleoside | −15.33 |
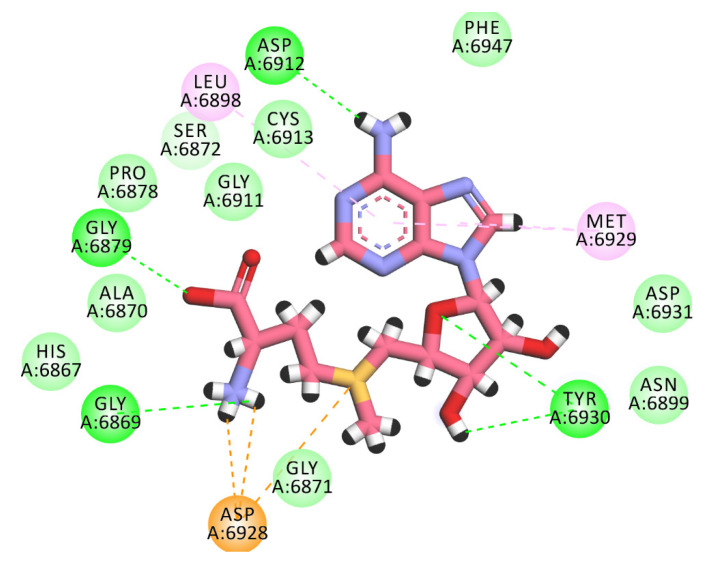
The predicted binding mode of the redocked ligand (SAM) yielded an affinity value of −21.52 kcal/mol. It interacted with its 6-amino-purin moiety and formed one hydrogen bond with Asp6912, in addition to hydrophobic interactions with Leu6898 and Met6929. Moreover, the di hydroxy tetrahydrofuran moiety formed two hydrogen bonds with Tyr6930, and the sulfur atom was involved in electrostatic interaction with Asp6928. Additionally, the terminal NH2 group was found to form one hydrogen bond with Gly6869, and two electrostatic interactions with Asp6928. Finally, the terminal carboxylic group formed one hydrogen bond with Gly6879 (Figure 5).
Figure 5.
3D and 2D binding mode of the redocked ligand (SAM) in the active site of the target protein.
From the tested compounds, seven members showed good binding mode with high binding energy. These compounds are 1187 (Protirelin), 1913 (Calcium folinate), 1995 (Raltegravir), 2176 (Regadenoson), 2396 (Ertapenem), 2532 (Methylergometrine), and 2612 (Methylergometrine).
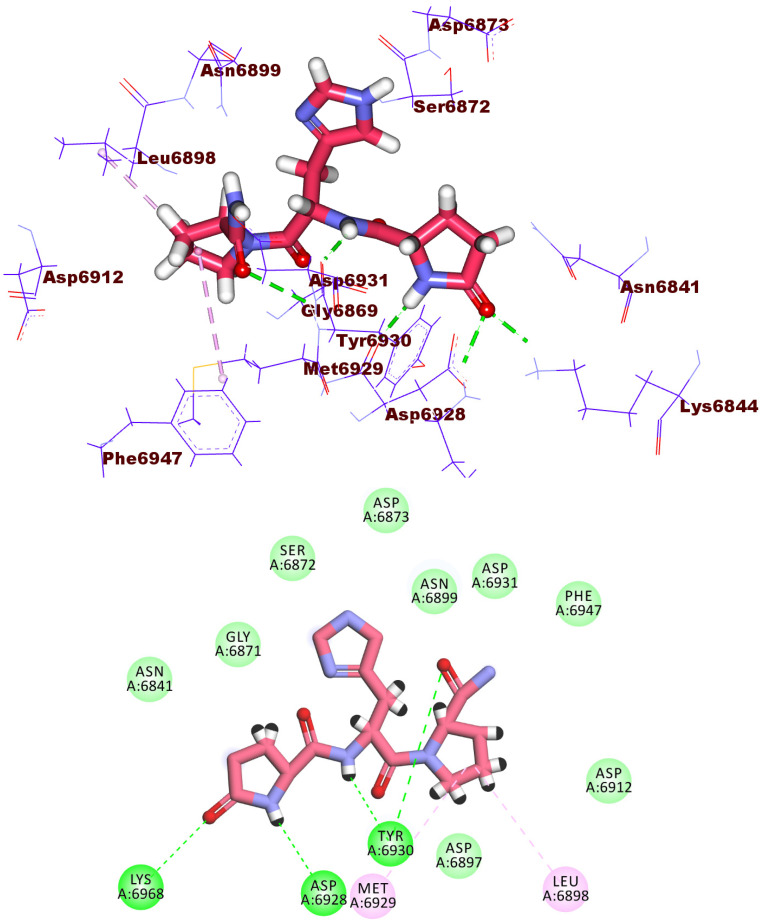
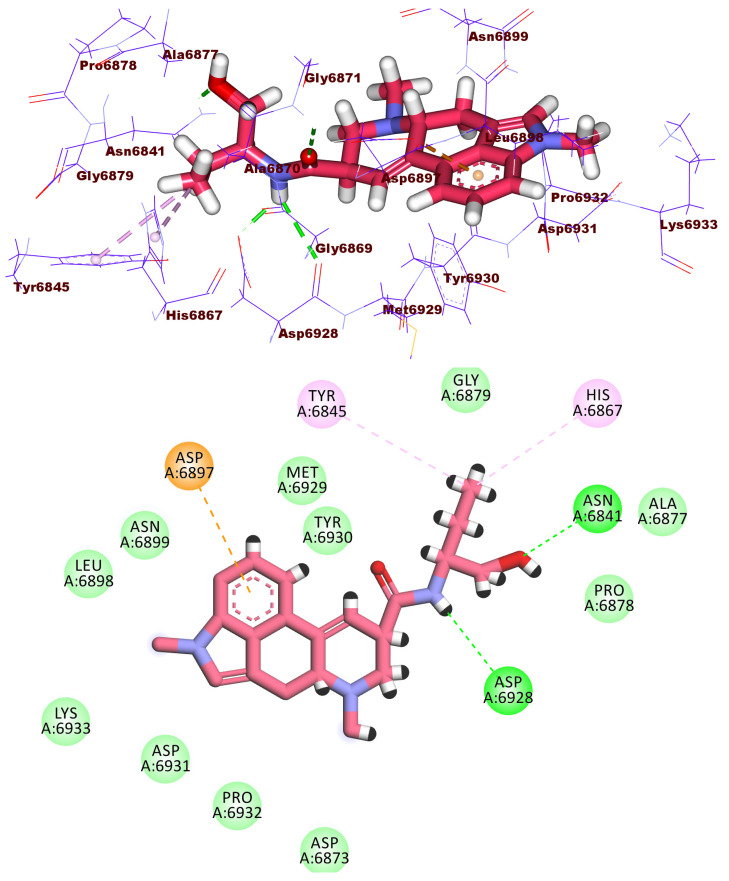
Compound 1187 has a docking score of −18.68 kcal/mol and formed four hydrogen bonds with the crucial amino acids in the active site of the 2′OMTase enzyme. The pyrrolidin-2-one moiety formed two hydrogen bonds with Asp6928 and Lys6968 via its NH and C=O groups, respectively. Furthermore, the NH group of the central amide moiety formed one hydrogen bond with Tyr6930. Moreover, the (S)-pyrrolidine-2-carboxamide moiety formed one hydrogen with Tyr6930 and two hydrophobic bonds with Met6929 and Leu6898 (Figure 6).
Figure 6.
3D and 2D binding mode of compound 1187 in the active site of the target protein.
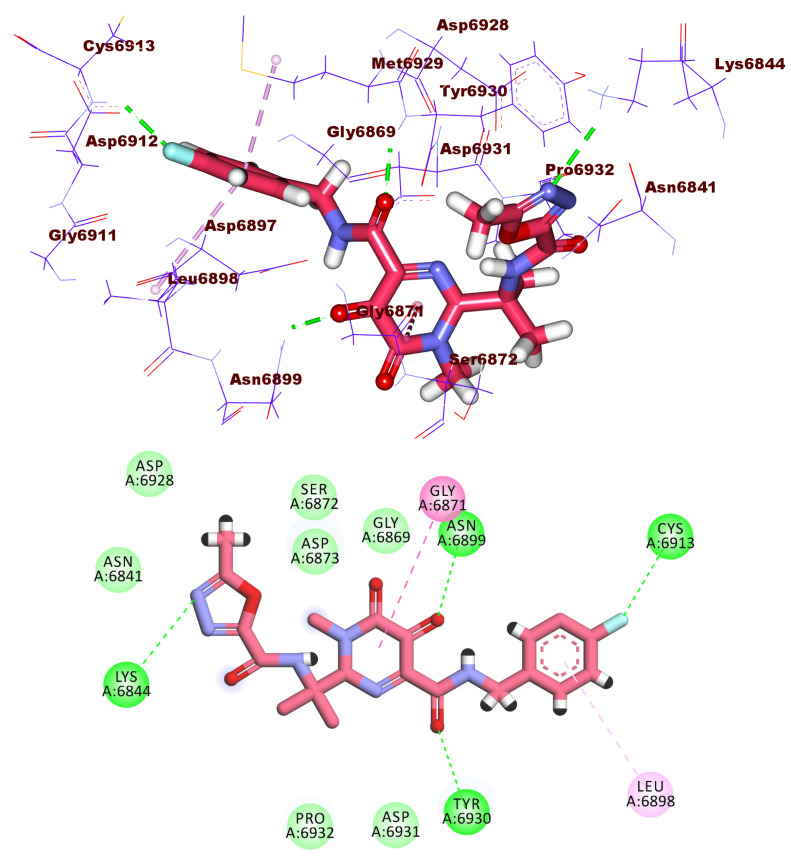
Compound 1913 has a docking score of −19.09 kcal/mol, forming six hydrogen bonds within the active site. The 2-amino-4-hydroxy-7,8-dihydropteridine-5(6H)-carbaldehyde moiety formed three hydrogen bonds with Cys6913, Gly6911, and Asp6912 via its NH2, OH groups, and the hetero nitrogen atom at 3-position. In addition, the glutamic acid moiety formed three hydrogen bonds with Asn6841, Gly6879, and Gly6871. Moreover, a carboxylate group of glutamic acid moiety formed one electrostatic interaction with Asp6873 (Figure 7).
Figure 7.
3D and 2D binding mode of compound 1913 in the active site of the target protein.
With a docking score of −21.07 kcal/mol, compound 1995 fit well into the active site of the 2′OMTase enzyme and formed four hydrogen bonds. The fluorobenzene formed one hydrogen bond with Cys6913 and one hydrophobic interaction with Leu6898. The carbonyl group of the amide moiety formed one hydrogen bond with Tyr6930. The carbonyl group of 5-hydroxy-3-methylpyrimidin-4(3H)-one moiety formed one hydrogen bond with Asn6899. In addition, the 5-hydroxy-3-methylpyrimidin-4(3H)-one moiety formed hydrophobic bond with Gly6871. The NH group of 2-methyl-1,3,4-oxadiazole moiety formed one hydrogen bond with Lys6844 (Figure 8).
Figure 8.
3D and 2D binding mode of compound 1995 in the active site of the target protein.
Compound 2176 showed a binding energy of −18.54 kcal/mol. This compound formed four hydrogen bonds in the active site of the target protein. The ribose sugar moiety formed three hydrogen bonds with Gly6879, Ala6870, and Gly6871. Furthermore, the NH group of the 9H-purin-6-amine moiety formed a hydrogen bond with Ty6930. Moreover, the N-methyl-1H-pyrazole-4-carboxamide moiety was incorporated in hydrophobic interaction with Met6929 and one electrostatic interaction with Asp6897 (Figure 9).
Figure 9.
3D and 2D binding mode of compound 2176 in the active site of the target protein.
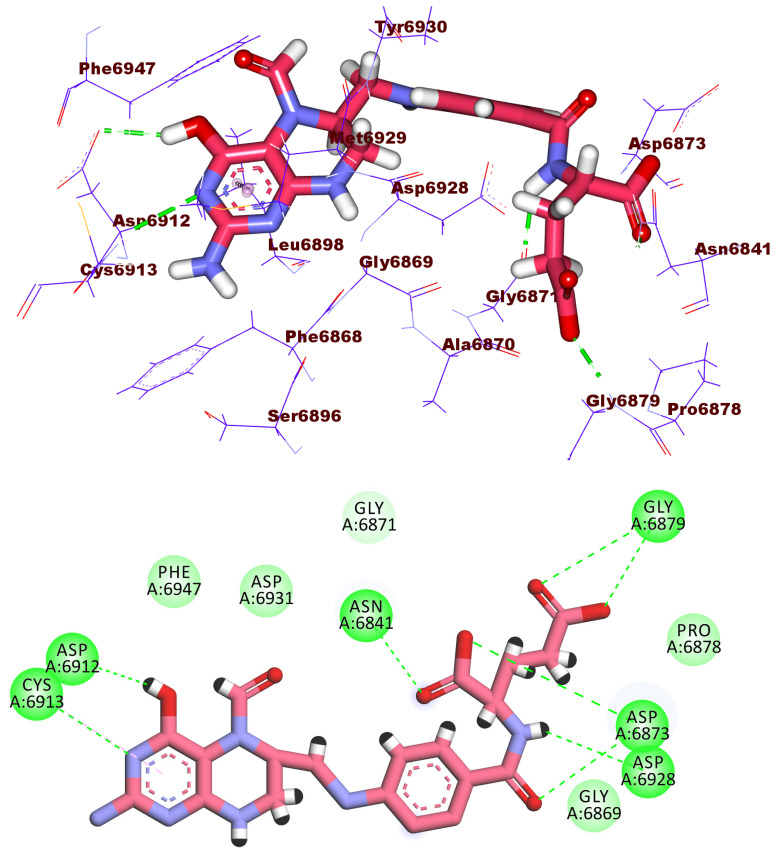
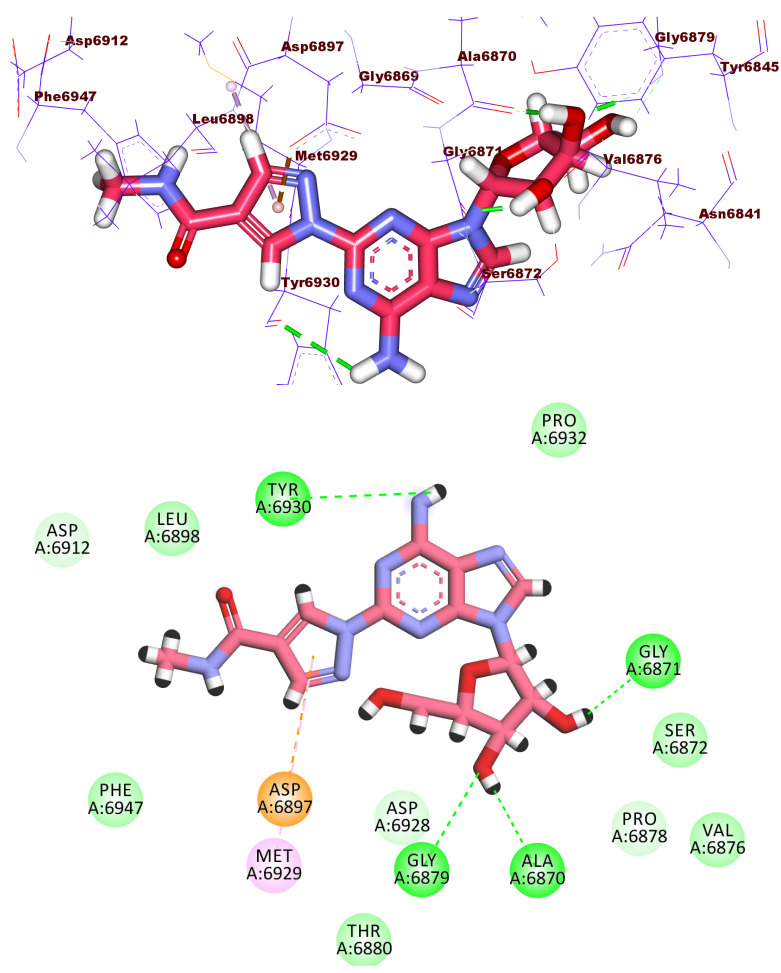
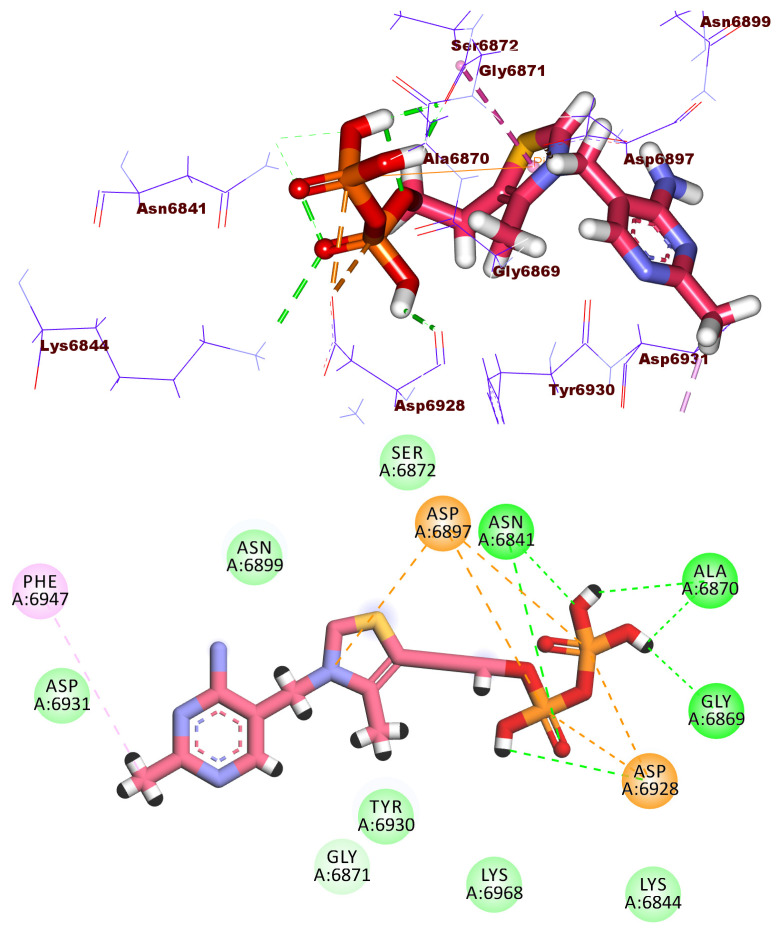
Compound 2396 (Ertapenem) has a docking score of −20.73 kcal/mol and created five hydrogen bonds with the crucial amino acids in the active site of the 2′OMTase enzyme. The benzoic acid moiety formed one hydrogen bond with Cys6913 via its carboxylic group, and two hydrophobic interactions with Met6929 and Leu6898. Furthermore, the NH group formed another hydrogen bond with Asp6897. Moreover, the carboxylate group at 2-position of 1-azabicyclo[3.2.0]hept-2-ene moiety formed one hydrogen and one electrostatic bond with Asp6873. The terminal hydroxyl group formed two hydrogen bonds with Asp6928 and Gly6869 (Figure 10). Although Ertapenem showed a binding energy less than Raltegravir, it showed an ideal binding mode like that of the co-crystallized ligand (SAM). It occupied all sub pockets of the active site and bound the crucial amino acids. Accordingly, it was used for further in silico testing via MD simulations.
Figure 10.
3D and 2D binding mode of compound 2396 in the active site of the target protein.
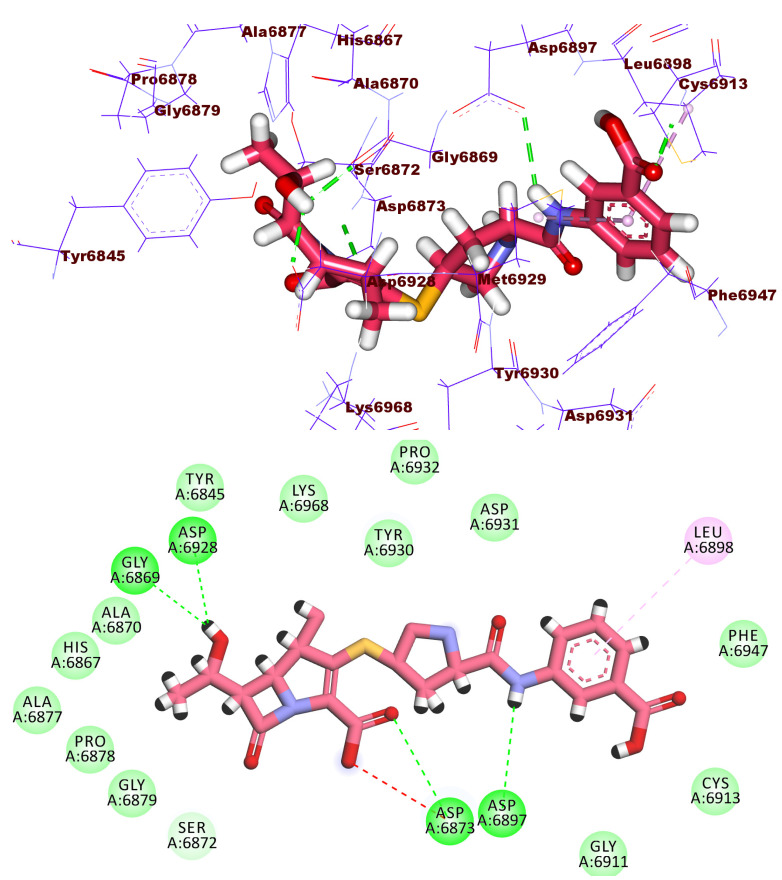
The binding mode of compound 2532 (affinity value of −20.46 kcal/mol), which is extremely close to ligand SAM, revealed that the amide group formed two hydrogen bonds with fundamental amino acids Asp6928 and Gly6871. In addition, the OH group formed another hydrogen bond with the amino acid Asn6841. Furthermore, the terminal ethyl moiety was incorporated in hydrophobic interaction with His6867 and Tyr6845. The phenyl ring formed an electrostatic attraction with Asp6897 (Figure 11).
Figure 11.
3D and 2D binding mode of compound 2532 in the active site of the target protein.
As demonstrated in Figure 12, compound 2612 had a high potential binding affinity (ΔG = −18.03 kcal/mol) with 2′OMTase enzyme active site. The strong binding affinity is assumed to be due to the formation of four hydrogen bonds in addition to many hydrophobic and electrostatic attractions. The terminal diphosphate moiety formed four hydrogen bonds with Gly6871, and Asn6841. It also formed three electrostatic attractions with Asp6928. In addition, the 4-methylthiazol-3-ium moiety formed a hydrophobic interaction with Gly6871 and an electrostatic attraction with Asp6897. Furthermore, the 2-methylpyrimidin-4-amine moiety was incorporated in a hydrophobic attraction with Phe6947.
Figure 12.
3D and 2D binding mode of compound 2612 in the active site of the target protein.
2.4. Molecular Dynamic Simulation
Molecular dynamics (MD) simulations studies can be applied to examine almost every kind of biomacromolecule (protein, nucleic acid, or carbohydrate) of biological significance [60]. The MD experiments can afford abundant information regarding the dynamic structural of the studied system [61]. Additionally, it contributes large amounts of energetic data. Such data are essential to understand the structure–function relationship of the examined ligand, its target protein, as well as the protein–ligand interactions. Correspondingly, MD studies could be a vital guide in the drug design and discovery processes [62].
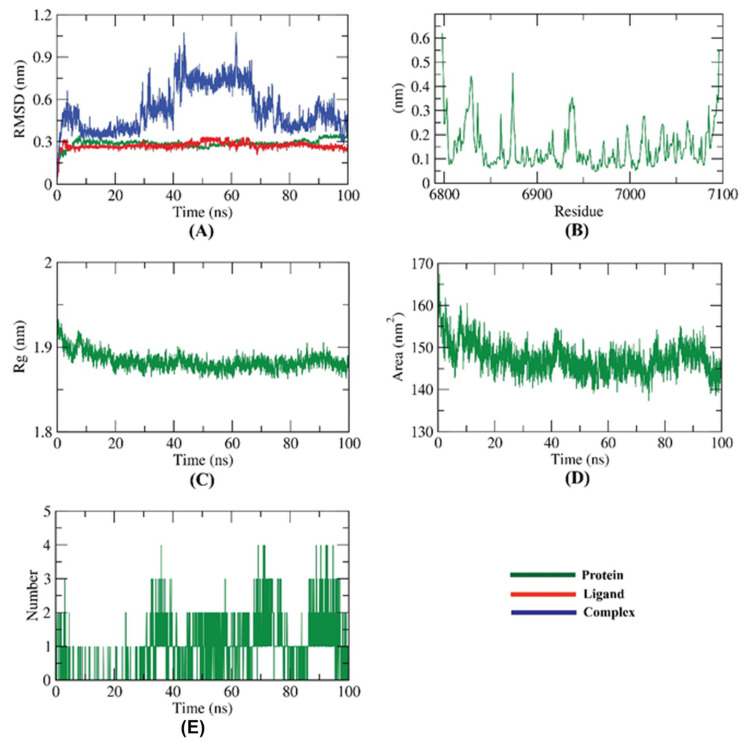
The dynamic, as well as conformational shifts of backbone atoms of 2′OMTase, Ertapenem in addition to 2′OMTase—Ertapenem complex, were estimated through the calculation of the root mean square deviation (RMSD) to distinguish the stability of the examined molecules before and after binding. RMSD investigation demonstrates both of conformational and dynamics changes [63] that occur after binding. Excitingly, the 2′OMTase—Ertapenem complex demonstrated low RMSD values with slight fluctuations from 40–70 ns~ and was stabilized later until the end of the study (Figure 13A) Fortunately, this slight fluctuation did not affect the integrity of the 2′OMTase—Ertapenem complex as the next experiments (Rg and SASA) did not record major changes in this time. However, the H-binding showed that the number of H-bonds decreased in this period (40–70 ns), from 3–4 bonds to 2 bonds. The study demonstrated that the number of H-bonds became 3–4 again after 70 ns. The flexibility of the evaluated complex was measured in the terms of RMSF to identify the fluctuated region of 2′OMTase during the 100 ns of the simulation. Favorably, the binding of Ertapenem does not make 2′OMTase very flexible (Figure 13B). The compactness of the 2′OMTase—Ertapenem complex was investigated by the computation of radius of gyration (Rg) of the evaluated enzyme. Complementarily, the Rg exhibited was noticed to be of lower value during the 100 ns of the experiment compared to the starting time (Figure 13C). In a similar manner, SASA (solvent accessible surface area) denotes the interaction between 2′OMTase—Ertapenem complex, and the surrounding solvents was measured. SASA value is an excellent indicator to the conformational changes that occurred during the simulation experiment because of binding interactions. Of note, the surface area of 2′OMTase (Figure 13D) displayed a considerable reduction in SASA values through the simulation time compared to the starting point. Finally, hydrogen bonding, as an essential factor in the binding of 2′OMTase—Ertapenem complex, was estimated. The greatest number of H-bonds that formed between 2′OMTase—Ertapenem complex was up to three H-bonds (Figure 13E).
Figure 13.
Results of M D simulations of 2′OMTase—Ertapenem complex; (A) RMSD, (B) RMSF, (C) Rg, (D) SASA, and (E) H-bonding.
2.5. Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) Studies
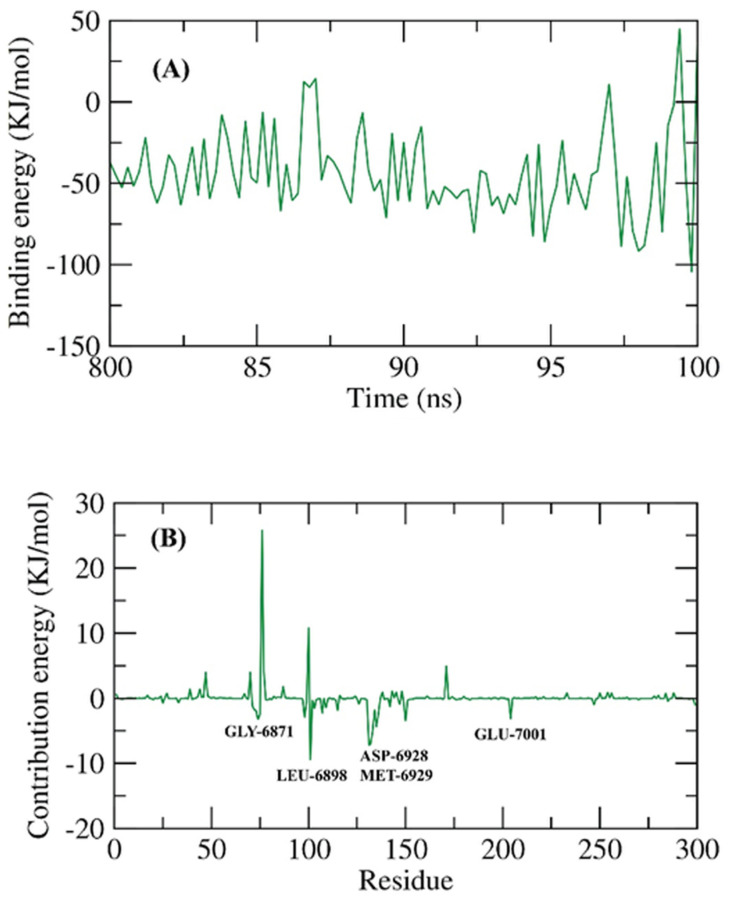
The binding free energy of 2′OMTase—Ertapenem complex was investigated in the last 20 ns of the MD run with an interval of 100 ps from the produced MD trajectories. The MM/PBSA method was utilized with the MmPbSaStat.py script to compute the average free binding energy as well as its standard deviation/error. Interestingly, as shown in Figure 14A, Ertapenem demonstrated a low binding free energy with a value of −43 KJ/mol (equivalent to −10.28 kcal/mol) with 2′OMTase. The share of the different amino acid residues of 2′OMTase in respect of the binding energy compared to the binding with Ertapenem. Total binding free energy decomposing of the 2′OMTase—Ertapenem complex into per residue share energy was achieved. The following amino acid residues of 2′OMTase, GLY-6871, LEU-6898, ASP-6928, MET-6829, and GLU-7001, contributed the binding energy with values that are more than −3 KJ/mol (Figure 14B).
Figure 14.
MM-PBSA study of 2′OMTase—Ertapenem; (A): total binding free energy, (B): analyzed binding free energy per amino acid residue.
3. Method
3.1. Molecular Similarity Detection
Discovery studio 4.0 software was used (see method part in Supplementary Materials).
3.2. Fingerprint Studies
Discovery studio 4.0 software was used (see method part in Supplementary Materials).
3.3. Docking Studies
Docking studies were performed against target enzymes using Discovery studio software [64] (see method part in Supplementary Materials).
3.4. Molecular Dynamics Simulation
The system was prepared using the web-based CHARMM-GUI [65,66,67] interface utilizing CHARMM36 force field [68] and NAMD 2.13 [69] package. The TIP3P explicit solvation model was used (see Supplementary Materials).
3.5. MM-PBSA Studies
The g_mmpbsa package of GROMACS was utilized to calculate the MM/PBSA (See Supplementary Materials).
4. Conclusions
Seven FDA-approved compounds (Protirelin, (1187), Calcium folinate (1913), Raltegravir (1995), Regadenoson (2176), Ertapenem (2396), Methylergometrine (2532), and Thiamine pyrophosphate hydrochloride (2612), out of 3009 were elected as the strongest 2′OMTase inhibitors. The selection of compounds was based on a multistep in silico study. The utilized studies included molecular fingerprints and structure similarity studies against SAM, the co-crystallized ligand of the targeted enzyme in addition to molecular docking studies. Ertapenem (2396) was subjected to MD simulation studies (RMSD, RMSF, Rg, SASA, and H-bonding) for 100 ns, confirming the excellent binding. These encouraging results could be a step to discover an effective cure against COVID-19 through further in vitro and in vivo studies for the pointed candidates.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27072287/s1, The method of Fingerprints; Molecular Similarity; Docking; Molecular dynamics; and MMPBSA studies.
Author Contributions
Conceptualization, I.H.E. and A.M.M.; Funding acquisition, B.A.A.; Project administration, I.H.E. and A.M.M.; Software, I.H.E., M.S.A., A.M.S. and E.B.E.; Writing—review and editing, E.B.E., B.A.A., A.-A.M.M.E.-A. and A.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Princess Nourah Bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R142), Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data is contained in the published article.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compoundsre not available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO WHO Coronavirus (COVID-19) Dashboard. [(accessed on 19 January 2022)]. Available online: https://covid19.who.int/
- 2.Wang Q., Yang L., Jin H., Lin L. Vaccination against COVID-19: A systematic review and meta-analysis of acceptability and its predictors. Prev. Med. 2021;150:106694. doi: 10.1016/j.ypmed.2021.106694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan H.S., Shan H., Dahoun T., Vogel H., Yuan S. Advancing drug discovery via artificial intelligence. Trends Pharmacol. Sci. 2019;40:592–604. doi: 10.1016/j.tips.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 5.Sleire L., Førde H.E., Netland I.A., Leiss L., Skeie B.S., Enger P.Ø. Drug repurposing in cancer. Pharmacol. Res. 2017;124:74–91. doi: 10.1016/j.phrs.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Singh T.U., Parida S., Lingaraju M.C., Kesavan M., Kumar D., Singh R.K. Drug repurposing approach to fight COVID-19. Pharmacol. Rep. 2020;72:1479–1508. doi: 10.1007/s43440-020-00155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong J., Bang M. Anti-inflammatory strategies for schizophrenia: A review of evidence for therapeutic applications and drug repurposing. Clin. Psychopharmacol. Neurosci. 2020;18:10. doi: 10.9758/cpn.2020.18.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konreddy A.K., Rani G.U., Lee K., Choi Y. Recent drug-repurposing-driven advances in the discovery of novel antibiotics. Curr. Med. Chem. 2019;26:5363–5388. doi: 10.2174/0929867325666180706101404. [DOI] [PubMed] [Google Scholar]
- 9.Shirley D.-A., Sharma I., Warren C.A., Moonah S. Drug repurposing of the alcohol abuse medication disulfiram as an anti-parasitic agent. Front. Cell. Infect. Microbiol. 2021;11:165. doi: 10.3389/fcimb.2021.633194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trivedi J., Mohan M., Byrareddy S.N. Drug repurposing approaches to combating viral infections. J. Clin. Med. 2020;9:3777. doi: 10.3390/jcm9113777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westbrook J.D., Burley S.K. How structural biologists and the Protein Data Bank contributed to recent FDA new drug approvals. Structure. 2019;27:211–217. doi: 10.1016/j.str.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimme S., Schreiner P.R. Computational chemistry: The fate of current methods and future challenges. Angew. Chem. Int. Ed. 2018;57:4170–4176. doi: 10.1002/anie.201709943. [DOI] [PubMed] [Google Scholar]
- 13.Amin S., Banerjee S., Singh S., Qureshi I.A., Gayen S., Jha T. First structure–activity relationship analysis of SARS-CoV-2 virus main protease (Mpro) inhibitors: An endeavor on COVID-19 drug discovery. Mol. Divers. 2021;25:1827–1838. doi: 10.1007/s11030-020-10166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ranjan S., Devarapalli R., Kundu S., Saha S., Deolka S., Vangala V.R., Reddy C.M. Isomorphism: Molecular similarity to crystal structure similarity’in multicomponent forms of analgesic drugs tolfenamic and mefenamic acid. IUCrJ. 2020;7:173–183. doi: 10.1107/S205225251901604X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baidya A.T., Ghosh K., Amin S.A., Adhikari N., Nirmal J., Jha T., Gayen S. In silico modelling, identification of crucial molecular fingerprints, and prediction of new possible substrates of human organic cationic transporters 1 and 2. New J. Chem. 2020;44:4129–4143. doi: 10.1039/C9NJ05825G. [DOI] [Google Scholar]
- 16.Shi Y. Support vector regression-based QSAR models for prediction of antioxidant activity of phenolic compounds. Sci. Rep. 2021;11:8806. doi: 10.1038/s41598-021-88341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Idris M.O., Yekeen A.A., Alakanse O.S., Durojaye O.A. Computer-aided screening for potential TMPRSS2 inhibitors: A combination of pharmacophore modeling, molecular docking and molecular dynamics simulation approaches. J. Biomol. Struct. Dyn. 2021;39:5638–5656. doi: 10.1080/07391102.2020.1792346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y., Li M. A new computer model for evaluating the selective binding affinity of phenylalkylamines to T-Type Ca2+ channels. Pharmaceuticals. 2021;14:141. doi: 10.3390/ph14020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eissa I.H., Ibrahim M.K., Metwaly A.M., Belal A., Mehany A.B., Abdelhady A.A., Elhendawy M.A., Radwan M.M., ElSohly M.A., Mahdy H.A. Design, molecular docking, in vitro, and in vivo studies of new quinazolin-4 (3H)-ones as VEGFR-2 inhibitors with potential activity against hepatocellular carcinoma. Bioorgan. Chem. 2021;107:104532. doi: 10.1016/j.bioorg.2020.104532. [DOI] [PubMed] [Google Scholar]
- 20.Zhanzhaxina A., Suleimen Y., Metwaly A.M., Eissa I.H., Elkaeed E.B., Suleimen R., Ishmuratova M., Akatan K., Luyten W. In vitro and in silico cytotoxic and antibacterial activities of a diterpene from cousinia alata schrenk. J. Chem. 2021;2021:5542455. doi: 10.1155/2021/5542455. [DOI] [Google Scholar]
- 21.Jalmakhanbetova R., Elkaeed E.B., Eissa I.H., Metwaly A.M., Suleimen Y.M. Synthesis and molecular docking of some grossgemin amino derivatives as tubulin inhibitors targeting colchicine binding site. J. Chem. 2021;2021:5586515. doi: 10.1155/2021/5586515. [DOI] [Google Scholar]
- 22.El-Adl K., El-Helby A.-G.A., Ayyad R.R., Mahdy H.A., Khalifa M.M., Elnagar H.A., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M. Design, synthesis, and anti-proliferative evaluation of new quinazolin-4 (3H)-ones as potential VEGFR-2 inhibitors. Biorgan. Med. Chem. 2021;29:115872. doi: 10.1016/j.bmc.2020.115872. [DOI] [PubMed] [Google Scholar]
- 23.Imieje V.O., Zaki A.A., Metwaly A.M., Eissa I.H., Elkaeed E.B., Ali Z., Khan I.A., Falodun A. Antileishmanial derivatives of humulene from Asteriscus hierochunticus with in silico tubulin inhibition potential. Rec. Nat. Prod. 2021;16:150–171. [Google Scholar]
- 24.Rafi M.O., Al-Khafaji K., Tok T.T., Rahman M.S. Computer-based identification of potential compounds from Salviae miltiorrhizae against Neirisaral adhesion A regulatory protein. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1856189. [DOI] [PubMed] [Google Scholar]
- 25.Parmar D.R., Soni J.Y., Guduru R., Rayani R.H., Kusurkar R.V., Vala A.G., Talukdar S.N., Eissa I.H., Metwaly A.M., Khalil A. Discovery of new anticancer thiourea-azetidine hybrids: Design, synthesis, in vitro antiproliferative, SAR, in silico molecular docking against VEGFR-2, ADMET, toxicity, and DFT studies. Biorgan. Chem. 2021;115:105206. doi: 10.1016/j.bioorg.2021.105206. [DOI] [PubMed] [Google Scholar]
- 26.El-Adl K., Sakr H.M., Yousef R.G., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M., ElSohly M.A., Abulkhair H.S., Eissa I.H. Discovery of new quinoxaline-2 (1H)-one-based anticancer agents targeting VEGFR-2 as inhibitors: Design, synthesis, and anti-proliferative evaluation. Biorgan. Chem. 2021;114:105105. doi: 10.1016/j.bioorg.2021.105105. [DOI] [PubMed] [Google Scholar]
- 27.Suleimen Y.M., Metwaly A.M., Mostafa A.E., Elkaeed E.B., Liu H.-W., Basnet B.B., Suleimen R.N., Ishmuratova M.Y., Turdybekov K.M., Van Hecke K. Isolation, Crystal Structure, and In Silico Aromatase Inhibition Activity of Ergosta-5, 22-dien-3β-ol from the Fungus Gyromitra esculenta. J. Chem. 2021;2021:5529786. doi: 10.1155/2021/5529786. [DOI] [Google Scholar]
- 28.Yousef R.G., Sakr H.M., Eissa I.H., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M., ElSohly M.A., Abulkhair H.S., El-Adl K. New quinoxaline-2 (1 H)-ones as potential VEGFR-2 inhibitors: Design, synthesis, molecular docking, ADMET profile and anti-proliferative evaluations. New J. Chem. 2021;45:16949–16964. doi: 10.1039/D1NJ02509K. [DOI] [PubMed] [Google Scholar]
- 29.Amer H.H., Alotaibi S.H., Trawneh A.H., Metwaly A.M., Eissa I.H. Anticancer activity, spectroscopic and molecular docking of some new synthesized sugar hydrazones, Arylidene and α-Aminophosphonate derivatives. Arab. J. Chem. 2021;14:103348. doi: 10.1016/j.arabjc.2021.103348. [DOI] [Google Scholar]
- 30.Husain A., Farooqui A., Khanam A., Sharma S., Mahfooz S., Shamim A., Akhter F., Alatar A.A., Faisal M., Ahmad S. Physicochemical characterization of C-phycocyanin from Plectonema sp. and elucidation of its bioactive potential through in silico approach. Cell. Mol. Biol. 2021;67:68–82. doi: 10.14715/cmb/2021.67.4.8. [DOI] [PubMed] [Google Scholar]
- 31.Mohammed S.O., El Ashry E.S.H., Khalid A., Amer M.R., Metwaly A.M., Eissa I.H., Elkaeed E.B., Elshobaky A., Hafez E.E. Expression, Purification, and Comparative Inhibition of Helicobacter pylori Urease by Regio-Selectively Alkylated Benzimidazole 2-Thione Derivatives. Molecules. 2022;27:865. doi: 10.3390/molecules27030865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imieje V.O., Zaki A.A., Metwaly A.M., Mostafa A.E., Elkaeed E.B., Falodun A. Comprehensive In Silico Screening of the Antiviral Potentialities of a New Humulene Glucoside from Asteriscus hierochunticus against SARS-CoV-2. J. Chem. 2021;2021:5541876. doi: 10.1155/2021/5541876. [DOI] [Google Scholar]
- 33.El-Demerdash A., Metwaly A.M., Hassan A., El-Aziz A., Mohamed T., Elkaeed E.B., Eissa I.H., Arafa R.K., Stockand J.D. Comprehensive virtual screening of the antiviral potentialities of marine polycyclic guanidine alkaloids against SARS-CoV-2 (COVID-19) Biomolecules. 2021;11:460. doi: 10.3390/biom11030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jalmakhanbetova R.I., Suleimen Y.M., Oyama M., Elkaeed E.B., Eissa I., Suleimen R.N., Metwaly A.M., Ishmuratova M.Y. Isolation and In Silico Anti-COVID-19 Main Protease (Mpro) Activities of Flavonoids and a Sesquiterpene Lactone from Artemisia sublessingiana. J. Chem. 2021;2021:5547013. doi: 10.1155/2021/5547013. [DOI] [Google Scholar]
- 35.Suleimen Y.M., Jose R.A., Suleimen R.N., Arenz C., Ishmuratova M., Toppet S., Dehaen W., Alsfouk A.A., Elkaeed E.B., Eissa I.H., et al. Isolation and In Silico Anti-SARS-CoV-2 Papain-Like Protease Potentialities of Two Rare 2-Phenoxychromone Derivatives from Artemisia spp. Molecules. 2022;27:1216. doi: 10.3390/molecules27041216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alesawy M.S., Abdallah A.E., Taghour M.S., Elkaeed E.B., H Eissa I., Metwaly A.M. In Silico Studies of Some Isoflavonoids as Potential Candidates against COVID-19 Targeting Human ACE2 (hACE2) and Viral Main Protease (Mpro) Molecules. 2021;26:2806. doi: 10.3390/molecules26092806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eissa I.H., Khalifa M.M., Elkaeed E.B., Hafez E.E., Alsfouk A.A., Metwaly A.M. In Silico Exploration of Potential Natural Inhibitors against SARS-Cov-2 nsp10. Molecules. 2021;26:6151. doi: 10.3390/molecules26206151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alesawy M.S., Elkaeed E.B., Alsfouk A.A., Metwaly A.M., Eissa I. In Silico Screening of Semi-Synthesized Compounds as Potential Inhibitors for SARS-CoV-2 Papain-like Protease: Pharmacophoric Features, Molecular Docking, ADMET, Toxicity and DFT Studies. Molecules. 2021;26:6593. doi: 10.3390/molecules26216593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.FDA-Approved Drug Library. [(accessed on 19 November 2021)]. Available online: https://www.selleckchem.com/screening/fda-approved-drug-library.html.
- 40.Briem H., Kuntz I.D. Molecular similarity based on DOCK-generated fingerprints. J. Med. Chem. 1996;39:3401–3408. doi: 10.1021/jm950800y. [DOI] [PubMed] [Google Scholar]
- 41.Vidal D., Garcia-Serna R., Mestres J. Chemoinformatics and Computational Chemical Biology. Springer; Berlin/Heidelberg, Germany: 2011. Ligand-based approaches to in silico pharmacology; pp. 489–502. [DOI] [PubMed] [Google Scholar]
- 42.Hassell A.M., An G., Bledsoe R.K., Bynum J.M., Carter H.L., Deng S.-J., Gampe R.T., Grisard T.E., Madauss K.P., Nolte R.T. Crystallization of protein–ligand complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007;63:72–79. doi: 10.1107/S0907444906047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu H., He Q.-X., Wang J., Hu Y., Wang Y.-Q., Lin Z.-H. In silico design of novel benzohydroxamate-based compounds as inhibitors of histone deacetylase 6 based on 3D-QSAR, molecular docking, and molecular dynamics simulations. New J. Chem. 2020;44:21201–21210. doi: 10.1039/D0NJ04704J. [DOI] [Google Scholar]
- 44.Ieritano C., Campbell J.L., Hopkins W.S. Predicting differential ion mobility behaviour in silico using machine learning. Analyst. 2021;146:4737–4743. doi: 10.1039/D1AN00557J. [DOI] [PubMed] [Google Scholar]
- 45.Taha M., Ismail N.H., Ali M., Rashid U., Imran S., Uddin N., Khan K.M. Molecular hybridization conceded exceptionally potent quinolinyl-oxadiazole hybrids through phenyl linked thiosemicarbazide antileishmanial scaffolds: In silico validation and SAR studies. Biorgan. Chem. 2017;71:192–200. doi: 10.1016/j.bioorg.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Opo F.A.D.M., Rahman M.M., Ahammad F., Ahmed I., Bhuiyan M.A., Asiri A.M. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci. Rep. 2021;11:4049. doi: 10.1038/s41598-021-83626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durant J.L., Leland B.A., Henry D.R., Nourse J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002;42:1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 48.Maggiora G., Vogt M., Stumpfe D., Bajorath J. Molecular similarity in medicinal chemistry: Miniperspective. J. Med. Chem. 2014;57:3186–3204. doi: 10.1021/jm401411z. [DOI] [PubMed] [Google Scholar]
- 49.Bender A., Glen R.C. Molecular similarity: A key technique in molecular informatics. Org. Biomol. Chem. 2004;2:3204–3218. doi: 10.1039/b409813g. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan K.M., Enoch S.J., Ezendam J., Sewald K., Roggen E.L., Cochrane S. An adverse outcome pathway for sensitization of the respiratory tract by low-molecular-weight chemicals: Building evidence to support the utility of in vitro and in silico methods in a regulatory context. Appl. Vitr. Toxicol. 2017;3:213–226. doi: 10.1089/aivt.2017.0010. [DOI] [Google Scholar]
- 51.Altamash T., Amhamed A., Aparicio S., Atilhan M. Effect of hydrogen bond donors and acceptors on CO2 absorption by deep eutectic solvents. Processes. 2020;8:1533. doi: 10.3390/pr8121533. [DOI] [Google Scholar]
- 52.Wan Y., Tian Y., Wang W., Gu S., Ju X., Liu G. In silico studies of diarylpyridine derivatives as novel HIV-1 NNRTIs using docking-based 3D-QSAR, molecular dynamics, and pharmacophore modeling approaches. RSC Adv. 2018;8:40529–40543. doi: 10.1039/C8RA06475J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turchi M., Cai Q., Lian G. An evaluation of in-silico methods for predicting solute partition in multiphase complex fluids—A case study of octanol/water partition coefficient. Chem. Eng. Sci. 2019;197:150–158. doi: 10.1016/j.ces.2018.12.003. [DOI] [Google Scholar]
- 54.Escamilla-Gutiérrez A., Ribas-Aparicio R.M., Córdova-Espinoza M.G., Castelán-Vega J.A. In silico strategies for modeling RNA aptamers and predicting binding sites of their molecular targets. Nucleosides Nucleotides Nucleic Acids. 2021;40:798–807. doi: 10.1080/15257770.2021.1951754. [DOI] [PubMed] [Google Scholar]
- 55.Kaushik A.C., Kumar A., Bharadwaj S., Chaudhary R., Sahi S. Bioinformatics Techniques for Drug Discovery. Springer; Berlin/Heidelberg, Germany: 2018. Ligand-Based Approach for In-silico Drug Designing; pp. 11–19. [Google Scholar]
- 56.Zhang H., Ren J.-X., Ma J.-X., Ding L. Development of an in silico prediction model for chemical-induced urinary tract toxicity by using naïve Bayes classifier. Mol. Divers. 2019;23:381–392. doi: 10.1007/s11030-018-9882-8. [DOI] [PubMed] [Google Scholar]
- 57.Sheridan R.P., Kearsley S.K. Why do we need so many chemical similarity search methods? Drug Discov. Today. 2002;7:903–911. doi: 10.1016/S1359-6446(02)02411-X. [DOI] [PubMed] [Google Scholar]
- 58.MOE MOE User Guide. [(accessed on 1 October 2021)]. Available online: https://www.easa.europa.eu/sites/default/files/dfu/B01.UG_.CAO_.00024-008%20User%20Guide%20for%20Maintenance%20Organisation%20Exposition.PDF.
- 59.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Brunzelle J., Satchell K.J. High-resolution structures of the SARS-CoV-2 2′-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 2020;13:eabe1202. doi: 10.1126/scisignal.abe1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sousa S.F., Fernandes P.A., Ramos M.J. Protein–ligand docking: Current status and future challenges. Proteins Struct. Funct. Bioinform. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 61.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X., Shi D., Zhou S., Liu H., Liu H., Yao X. Molecular dynamics simulations and novel drug discovery. Expert Opin. Drug Discov. 2018;13:23–37. doi: 10.1080/17460441.2018.1403419. [DOI] [PubMed] [Google Scholar]
- 63.Kuzmanic A., Zagrovic B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys. J. 2010;98:861–871. doi: 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Protein Data Bank. 2021. [(accessed on 5 January 2022)]. Available online: https://www.rcsb.org/structure/4OW0.
- 65.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 66.Brooks B.R., Brooks C.L., III., Mackerell A.D., Jr., Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Best R.B., Zhu X., Shim J., Lopes P.E., Mittal J., Feig M., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is contained in the published article.