Abstract
Ninety-one ampicillin-resistant Shigella flexneri strains from Hong Kong and Shanghai were studied for production of β-lactamases. TEM-1-like and OXA-1-like enzymes were identified in 21 and 79% of the strains, respectively, by isoelectric focusing (IEF). No difference in the pattern of β-lactamase production was found between strains from Hong Kong and Shanghai. Four ribotypes were detected. Over 88% of OXA-producing strains had the same ribotype. All TEM-1-like strains harbored a plasmid which hybridized positively with the blaTEM probe. Total DNA from OXA-1-like strains failed to hybridize or only hybridized weakly with an OXA probe. The OXA resistance was not transferable. OXA-1-like enzymes exhibited substrate and inhibition profiles similar to that of OXA-1 and were shown to have a pI of 7.3 by further IEF using a narrow-range ampholine gel. The gene encoding the OXA-1-like enzyme from one isolate (CH-07) was cloned, sequenced, and found to differ from blaOXA-1 at codon 131 (AGA→GGA; Arg to Gly), resulting in the novel designation OXA-30. The predominance of OXA-type enzymes in ampicillin-resistant S. flexneri suggests host preference for specific β-lactamases.
Resistance to multiple antibiotics is common among clinical isolates of shigella. A previous study showed that 79% of the shigella isolates in our locality were resistant to ampicillin (26), among which 81% were also resistant to trimethoprim-sulfamethoxazole. Recently, nalidixic acid resistance was documented among 59.6% of Shigella flexneri isolates in Hong Kong (5). Although only one strain was reported to harbor ciprofloxacin resistance in that study, it is anticipated that resistance will develop quickly with widespread usage. Additionally, the use of quinolones in children, in whom bacillary dysentery most commonly occurs, should be avoided because of evidence of cartilage toxicity in the skeletally immature (9, 23). Alternative antimicrobial agents thus have to be sought.
Ampicillin-sulbactam has been suggested for the treatment of multidrug-resistant shigellosis because of its good in vitro activity. In a study of 129 shigella strains obtained between 1984 and 1987, only 2% were resistant to ampicillin-sulbactam (12). However, a recent study yielded contradictory results (5): most shigella isolates were resistant or only moderately susceptible to amoxicillin-clavulanate. Among members of the family Enterobacteriaceae, TEM-1 hyperproduction (25) and harboring of inhibitor-resistant TEM and OXA-1 β-lactamases have been reported to be mechanisms of resistance to β-lactamase–β-lactamase inhibitor combinations (14, 20). In order to elucidate the situation for shigella isolates in Hong Kong and Shanghai, β-lactamases from these isolates were characterized.
MATERIALS AND METHODS
Bacterial strains and screening for antimicrobial susceptibility.
S. flexneri isolates were obtained from inpatients with diarrheal disease in a teaching hospital in Hong Kong and from a public health laboratory in Shanghai. Screening for ampicillin resistance was performed by the disk diffusion method, and results were interpreted in accordance with the recommendations of the National Committee for Clinical Laboratory Standards (18). A total of 91 nonduplicate sporadic isolates of ampicillin-resistant S. flexneri (43 from Hong Kong and 48 from Shanghai) were collected from January to December 1994. Isolates were identified by standard microbiological techniques (9). Reference strains expressing the following plasmid-mediated enzymes were used: OXA-1, OXA-4, SHV-1, and TEM-1 (kindly provided by D. M. Livermore, Central Public Health Laboratory, London, United Kingdom, and G. A. Jacoby, Lahey Clinic, Burlington, Mass.).
Determination of MICs.
The MICs of the following agents were determined by the agar dilution method in accordance with National Committee for Clinical Laboratory Standards recommendations: ampicillin, cefuroxime, chloramphenicol, ceftazidime, cephalothin, cefotaxime, ceftriaxone, gentamicin, amikacin, tobramycin, tetracycline, nalidixic acid, ciprofloxacin, ofloxacin, sulfamethoxazole, streptomycin, spectinomycin, trimethoprim, mercury chloride, ampicillin-sulbactam, and amoxicillin-clavulanate (17). The combinations of ampicillin-sulbactam and amoxicillin-clavulanate were tested at a fixed ratio of 2:1. Antibiotic powders with known potency, including ampicillin-sulbactam (Pfizer Corporation, Hong Kong, China), amoxicillin-clavulanate (SmithKline Beecham, Hong Kong, China), ciprofloxacin (Bayer, Hong Kong, China), and ofloxacin (Hoechst AG, Hong Kong, China), were kindly provided by the manufacturers. All other antibiotics were purchased from Sigma (St. Louis, Mo.). Escherichia coli ATCC 25922 and ATCC 35218 were used as control strains for each test run.
Ribotyping.
Shigella chromosomal DNA was extracted for ribotyping (2). Approximately 5 μg of genomic DNA was digested overnight at 37°C with HincII (Amersham, Buckinghamshire, England) in accordance with the manufacturer's instructions. Digested total DNA and DIG (digoxigenin)-labeled marker VII consisting of EcoRI- and HindIII-digested λ DNA (Boehringer Mannheim, Mannheim, Germany) were separated in a 0.8% agarose gel by electrophoresis followed by transfer to nylon membrane (Amersham) by the Southern method. Probing was performed using a mixture of 16S and 23S rRNAs from E. coli (Roche, Mannheim, Germany) labeled by reverse transcription with DIG-labeled dUTP as described by the manufacturer. Ribotypes were designated with reference to the DIG-labeled molecular size marker VII.
IEF studies of β-lactamases.
Crude preparations of β-lactamases from isolates were obtained from sonic extracts prepared in 0.05 M phosphate buffer (pH 7.0). Primary isoelectric focusing (IEF) was performed using ampholine gel (Pharmacia, Hong Kong, China) from pI 3 to pI 10 (15). Further, confirmatory IEF was performed with a narrow-range ampholine gel from pI 5.5 to pI 8. The pI value of each enzyme was determined by spreading nitrocefin on the gel surface. The markers used included TEM-1, TEM-2, SHV-1, and OXA-1 for the primary IEF and OXA-1 and OXA-4 for the confirmatory IEF.
Plasmid and total DNA preparation for hybridization to β-lactamase gene probes.
Plasmids were isolated by a rapid method (10), and total DNA was prepared as described previously (2). Total DNA extracts were then digested with EcoRI. Isolated plasmids and enzyme-digested total DNA were subjected to agarose gel electrophoresis. The DNA was then transferred to a nylon membrane (Amersham) by the Southern method. Preparation of the blaTEM-1 and blaOXA-1 probes and DNA hybridization were performed as previously described (6, 8).
Transfer of ampicillin resistance by conjugation and transformation.
Conjugation was carried out by both simple broth mating and plate mating. A rifampin-resistant strain of E. coli (JP-995) was used as the recipient. Recipient and donor strains were separately inoculated into brain heart infusion broth (Oxoid, Hampshire, United Kingdom) and incubated at 37°C for 4 h. For broth mating, recipient and donor strains were mixed at a volume ratio of 1:1 for overnight incubation at 37°C. The next morning, 0.01 ml of the mixture was spread on a Mueller-Hinton (MH) agar plate containing rifampin (100 μg/ml) and ampicillin (100 μg/ml). As for plate mating, 0.2 ml of the donor and 1.8 ml of the recipient culture were mixed and passed through a filter with a diameter of 2.5 cm and a pore size of 0.45 μm. The filter was then placed on a prewarmed MH agar plate with the cells uppermost and incubated at 37°C for 4 h. The filter was then immersed in 2 ml of broth and the cells were resuspended. A 0.1-ml aliquot of the suspension was then plated on an ampicillin-rifampin selection plate. Transformation was conducted by a previously described method (22). Extracted plasmid DNA was mixed gently with competent cells, E. coli HB101 (ara-14, F− galK2 hsdS20 λ lacY1 leuB6 mtl-1 proA2 recA13 rpsL20 supE44 thi-1 xyl-5), and chilled on ice for 30 min. The cells were then heated to 42°C for 90 s and quickly placed in ice for 1 to 2 min. Brain heart infusion broth (400 μl) was added, and the cells were mixed gently. The mixture was incubated at 37°C for another 45 min, and 0.1 ml of the mixture was then plated on a selective MH plate (ampicillin at 50 μg/ml) to recover transformants.
β-Lactamase substrate and inhibition profiles.
Crude β-lactamase extracts were used for substrate and inhibition assays (4). The assays were performed spectrophotometrically by measuring the change in absorbance at the appropriate wavelength for each substrate. The wavelengths were 240 nm for benzylpenicillin, ampicillin, and carbenicillin; 260 nm for cephaloridine, cephalothin, and ceftazidime; 265 nm for oxacillin; and 500 nm for nitrocefin. β-Lactamase activity was determined in 1 ml of all of the substrates at 0.1 mM in 10 mM phosphate buffer, pH 7.0, except for ampicillin, of which a 1 mM solution was assayed. Substrate blanks were recorded for each reaction. One unit of activity was defined as the amount of enzyme that hydrolyzed 1 mol of benzylpenicillin per min per mg of total protein at room temperature. The rates of hydrolysis for each substrate were calculated relative to that of benzylpenicillin.
The IC50, the concentration of clavulanate required to inhibit enzymatic activity by 50%, was determined as follows. After 10 min of preincubation at room temperature of equal amounts of enzyme from each isolate with various amounts of clavulanate, enzyme hydrolytic activities were measured using nitrocefin as the substrate.
Cloning and sequencing.
Cloning was performed as previously described (22). The total DNA partially digested by EcoRI was ligated to the plasmid vector pMC1871 (Pharmacia), which was then transformed to CaCl2-treated E. coli HB101 using standard procedures (22). In the first cloning experiment, CaCl2-treated E. coli HB101 transformants were plated on MH agar supplemented with ampicillin (100 μg/ml) and tetracycline (30 μg/ml) to select for ampicillin-tetracycline-resistant clones harboring a recombinant plasmid containing the ampicillin resistance insert fragment. IEF of β-lactamases was performed for all transformants and an OXA-1 control strain. Enzymes with pIs similar to that of the parent strains and the OXA-1 strain (pI 7.3) were selected for further analysis. Plasmid DNA of the clones was extracted and analyzed by agarose gel electrophoresis. The plasmid with the lowest molecular weight was then selected for a subcloning experiment. Plasmid DNA from this low-molecular-weight clone was further digested completely by the BamHI enzyme, followed by self-ligation. The ligated products were again introduced into E. coli HB101 by transformation and selected by MH agar supplemented with ampicillin (100 μg/ml). The resulting clones were toothpicked and inoculated into two agar plates with either ampicillin (100 μg/ml) or ampicillin (100 μg/ml) with tetracycline (30 μg/ml). The clone with ampicillin resistance but tetracycline susceptibility was selected and subjected to IEF and sequencing as previously described (25). The initial sequencing primers were obtained from the vector (pMC1871) sequence. Further sequencing primers were designed after the preliminary sequencing data became available. Oligonucleotides used in sequencing experiments were synthesized by GIBCO BRL (New York, N.Y.). Both strands of the insert were sequenced.
PCR for specific detection of the OXA-1 gene family.
Total DNA extracts were used as templates in the PCR assay. Oligonucleotide primers used for the PCR assay were as follows: 5′-CCA AAG ACG TGG ATG (primer A) and 5′-GTT AAA TTC GAC CCC AAG TT (primer B) (GIBCO BRL). Primer A was known to be specific for blaOXA-1 (22), while a search of the GenBank database also confirmed the specificity of primer B for blaOXA-1. The predicted PCR product was a 540-bp intragenic fragment of blaOXA-1. Reactions were performed in a DNA thermal cycler (Bio-Rad, Hercules, Calif.) with 50-μl mixtures containing 2.5 U of Taq polymerase (Promega, Madison, Wis.); 1× buffer consisting of 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, and 0.01 μg of gelatin; 200 μM each deoxynucleoside triphosphate; and 2 μM each oligonucleotide primer. Thirty-five cycles were performed for each reaction, with the following temperature profile: 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min. All PCR products from the isolates were sequenced using the PCR primers described above.
Nucleotide sequence accession number.
The sequence of the gene blaOXA-30 has been given GenBank accession number AF255921.
RESULTS
Antibiotic susceptibility and IEF.
Two types of β-lactamases were found in the shigella isolates with approximate pI values of 5.4 and 7.4, similar to TEM-1 and OXA-1 β-lactamases, respectively. The TEM-1-like β-lactamase was identified in 19 (21%) out of 91 isolates, and the remainder had an OXA-1-like enzyme. No isolate had both enzymes. All isolates were sensitive to cefuroxime, cefotaxime, ceftazidime, ceftriaxone, aztreonam, ciprofloxacin, ofloxacin, gentamicin, amikacin, and tobramycin but had variable susceptibility to trimethoprim, sulfamethoxazole, and nalidixic acid. All OXA-type isolates had a low level of resistance to ampicillin, whose MIC was 128 or 256 μg/ml, while TEM-type isolates were relatively highly resistant (MIC, 512 to ≥2,048 μg/ml). All OXA-type isolates were susceptible to cephalothin (MICs, 1 to 4 μg/ml) and mercury chloride (MICs, 4 to 8 μg/ml) but resistant to streptomycin, chloramphenicol, spectinomycin, and tetracycline. Variable susceptibility to chloramphenicol, mercury chloride, ampicillin-sulbactam, and amoxicillin-clavulanate was observed for TEM-type isolates. The MICs of ampicillin-sulbactam and amoxicillin-clavulanate for OXA-type isolates were in either the intermediate or the resistant range (Table 1). The results suggested that OXA-type isolates were not well inhibited by β-lactam–β-lactamase inhibitor combinations.
TABLE 1.
Comparison of MICs for OXA-type and TEM-type isolates of 91 S. flexneri isolates from Hong Kong and Shanghai
| Antibiotic(s) | OXA typea (n = 72)
|
TEM typeb (n = 19)
|
||||
|---|---|---|---|---|---|---|
| MIC range | MIC50 | MIC90 | MIC range | MIC50 | MIC90 | |
| Ampicillin | 128–256 | 128 | 256 | 512–≥2048 | 256 | ≥1,024 |
| Cephalothin | 1–4 | 4 | 4 | 2–64 | 4 | 32 |
| Cefuroxime | 1–4 | 2 | 4 | 1–2 | 1 | 2 |
| Cefotaxime | ≤0.015–0.06 | 0.03 | 0.06 | 0.03–0.06 | 0.03 | 0.06 |
| Ceftazidime | 0.06–0.25 | 0.06 | 0.12 | 0.06–0.25 | 0.06 | 0.12 |
| Ceftriaxone | ≤0.015–0.06 | 0.03 | 0.06 | ≤0.015–0.06 | 0.03 | 0.06 |
| Aztreonam | 0.03–0.06 | 0.03 | 0.06 | 0.03–0.25 | 0.03 | 0.25 |
| Chloramphenicol | 1–≥256 | 64 | 128 | 32–128 | 64 | 128 |
| Sulfamethoxazole | 8–≥4096 | 512 | ≥4,096 | 8–≥4,096 | 512 | ≥4,096 |
| Streptomycin | 32–≥256 | 128 | ≥256 | 64–≥256 | 128 | ≥256 |
| Spectinomycin | 32–≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 |
| Mercury chloride | 4–8 | 4 | 8 | 4–≥64 | 8 | ≥64 |
| Trimethoprim | 0.5–≥128 | ≥128 | ≥128 | 0.25–≥128 | ≥128 | ≥128 |
| Tetracycline | 64–≥512 | 128 | ≥512 | 64–≥512 | 128 | ≥512 |
| Nalidixic acid | 2–≥64 | ≥64 | ≥64 | 2–≥64 | 2 | ≥64 |
| Ciprofloxacin | 0.0075–0.25 | 0.0075 | 0.06 | 0.03–1 | 0.03 | 0.03 |
| Ofloxacin | 0.03–2 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 |
| Gentamicin | 1–2 | 1 | 2 | 1–2 | 2 | 2 |
| Amikacin | 2–8 | 4 | 8 | 2–8 | 4 | 8 |
| Tobramycin | 0.5–2 | 2 | 2 | 1–2 | 2 | 2 |
| Amoxicillin-clavulante | 16/8–32/16 | 16/8 | 32/16 | 8/4–32/16 | 8/4 | 32/16 |
| Ampicillin-sulbactam | 16/8–32/16 | 16/8 | 32/16 | 8/4–≥128/64 | 16/8 | 32/16 |
pI, approximately 7.4. MIC50 and MIC90, MICs for 50 and 90% of isolates, respectively. All values are in micrograms per milliliter.
pI, approximately 5.4.
Ribotyping.
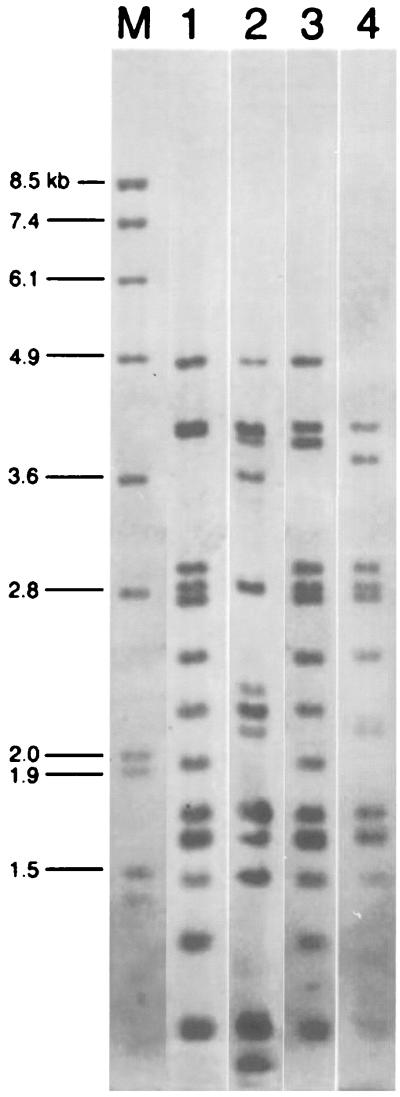
Four different ribotypes were obtained after HincII digestion of total DNA (Fig. 1). Ribotype 1 comprised 69 strains (76%) from both Shanghai and Hong Kong, including both TEM and OXA isolates. Ribotype 3 was found in only one TEM-type isolate from Hong Kong. Ribotypes 2 (6 of 91, 6.6%) and 4 (15 of 91, 16.5%) were found in OXA isolates from both Shanghai and Hong Kong.
FIG. 1.
Ribotyping of 91 strains of S. flexneri. Ribotype patterns 1 to 4 are shown with the marker in lane M (DIG-labeled, EcoRI- and HindIII-digested λ DNA).
Transferability of β-lactam resistance.
The β-lactam resistance of isolates harboring the TEM-1-like β-lactamase was found to be conjugatively transferable. Only one plasmid was found in each recipient. The plasmids from the transconjugants have molecular sizes ranging from approximately 90 to 120 kb. On the contrary, the β-lactam resistance of isolates with the OXA-1-like β-lactamase was not transferable by mating nor by transformation.
Southern hybridization of β-lactamase genes and affirmative IEF.
All TEM-type β-lactamases comigrated with the TEM-1 control, and plasmids from strains harboring these enzymes hybridized positively with the blaTEM probe. Plasmid DNA from strains with the OXA-type β-lactamase did not hybridize with the OXA probe. Further hybridization using HincII-digested total DNA as the template revealed a very weak band in some strains. Other strains yielded negative hybridization results. Narrow-range ampholine gel electrophoresis revealed pI values of 7.3 for the OXA-type enzymes and 7.4 for the reference enzymes (Fig. 2).
FIG. 2.
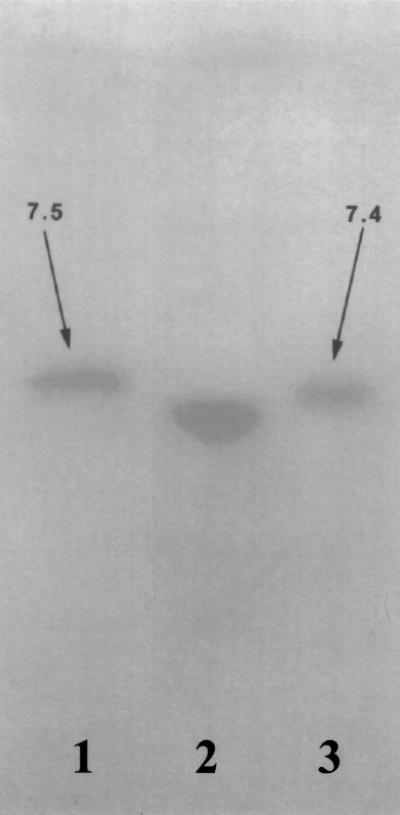
IEF showing the pIs of three β-lactamases. Lanes 1, OXA-4 (pI 7.5); 2, an OXA-1-like enzyme (OXA-30) from a strain of S. flexneri (pI 7.3); 3, OXA-1 (pI 7.4).
Substrate and inhibition profiles of the pI 7.3 β-lactamase.
By using crude β-lactamase extracts from 10 of the OXA-type isolates and two OXA-1 reference strains, it was shown that the relative hydrolysis rate of oxacillin was approximately 150% for the reference enzyme (hydrolysis of penicillin G was taken as 100%), while that from the study strains ranged from 98 to 189%. With the same extracts, the IC50s of clavulanate were 2.5 and 2.8 mM for the two reference OXA-1 β-lactamases while the study enzyme yielded values ranging from 2 to 3.2 mM (Table 2).
TABLE 2.
Comparison of the enzyme kinetic parameters of a novel, OXA-type β-lactamase (OXA-30) from S. flexneri with those of OXA-1 and SHV-1
| Straina | β-Lactamase produced | Rate of relative hydrolysis (Vmax)b of:
|
IC50 of CLAc | |||||
|---|---|---|---|---|---|---|---|---|
| PEN | AMP | CAR | LOR | CEF | OXA | |||
| DMO-1 | OXA-1 | 100 | 542 | 72 | 17 | 5.2 | 147 | 2.5 |
| pMON301 | OXA-1 | 100 | 370 | 65 | 15 | 5.2 | 147 | 2.8 |
| pMON38 | SHV-1 | 100 | 161 | 5 | 38 | 7.2 | <1 | <0.1 |
| CH-03 | OXA-30 | 100 | 407 | 40 | 35 | 1.3 | 137 | 2.5 |
| CH-40 | OXA-30 | 100 | 565 | 46 | 18 | 1 | 189 | 3.0 |
| SF-12 | OXA-30 | 100 | 475 | ND | 14 | 5.2 | 110 | 2.0 |
| SF-13 | OXA-30 | 100 | 646 | 72 | 20 | 5.2 | 98 | 2.2 |
Control strains were from D. M. Livermore (DMO-1) and G. A. Jacoby (pMON301 and pMON38). Clinical strains of S. flexneri were from Shanghai (CH-03 and CH-40) and Hong Kong (SF-12 and SF-13).
Abbreviations: PEN, benzylpenicillin; AMP, ampicillin; CAR, carbenicillin; LOR, cephaloridine; CEF, cephalothin; OXA, oxacillin. ND, not detected.
CLA, clavulanate.
Sequence homology of insertion gene to Tn2603.
A 2.5-kb DNA fragment from S. flexneri CH-07 was found to confer resistance to ampicillin and induce production of the pI 7.3 β-lactamase when cloned into E. coli HB101. Sequencing of this fragment revealed a portion of tnpA (transponase), tnpR (resolvase or recombinase), res (resolution site), and blaOXA. The result was compared with Tn2603 (the transposon encoding the blaOXA-1 gene), and over 99% nucleotide sequence homology was found. The blaOXA gene differed from blaOXA-1 by one base at codon 131, AGA→GGA (Arg to Gly). In addition to the structural mutation, one cytosine base insertion within the partial sequence of tnpA was identified (data not shown).
PCR for specific detection of the OXA-1-like gene and hybridization of PCR products to the blaOXA-1 probe.
PCR amplified products were 540 bp in length, which was consistent with the predicted size. All isolates harboring the OXA-1-like β-lactamase showed positive results in the PCR assay. Except for the strain with the OXA-4 enzyme, which has 99% DNA homology with OXA-1, there was no nonspecific amplification among reference strains with TEM-1, SHV-1, OXA-2, OXA-3, OXA-5, OXA-6, OXA-7, OXA-10, OXA-11, PSE-1, PSE-3, and PSE-4 β-lactamases. Upon hybridization with the blaOXA-1 fragment probe, all amplicons showed positive results, confirming the specificity of the PCR assay. Sequencing of all PCR products revealed the same point mutation as described in the previous paragraph.
DISCUSSION
This study revealed that resistance to ampicillin in most S. flexneri strains is associated with an OXA-type β-lactamase (OXA-30), instead of TEM-1, which is ubiquitous in the closely related bacterium E. coli (16). Since our isolates were obtained from sporadic cases and four different ribotypes were obtained among these isolates, clonal spread of specific β-lactamases is an unlikely explanation for the predominance of OXA types among the isolates. Moreover, the predominance of OXA-type β-lactamase in S. flexneri has also been reported from other geographical areas, including Denmark (24) and Tanzania (19). In the former study, 17 of 18 isolates that were obtained in 1989 primarily from patients with a history of travel to the Middle East, Asia, and Africa were reported to produce a β-lactamase with properties similar to those of OXA-1. In the latter, ampicillin resistance in 75% of the S. flexneri strains collected in 1997 was associated with the presence of an OXA-1-type β-lactamase. Taken together, these findings suggest the probable occurrence of host preference for OXA-type β-lactamase in S. flexneri.
Identification of a tnpR (resolvase) sequence upstream of blaOXA-30 suggests that the genes are encoded in a transposon. The gene blaOXA-30 is probably encoded not on the transposon encoding OXA-1 (Tn2603) but on that encoding OXA-4 (Tn1409). This is inferred from the known resistance patterns of the transposons. While both transposons confer resistance to sulfamethoxazole, spectinomycin, streptomycin, chloramphenicol, and trimethoprim, only Tn2603 yields resistance to mercury (19). In previous studies, ampicillin resistance determinants in shigella were also reported to be within mobile genetic elements such as plasmids and integrons (13, 19). Yet, it is intriguing that ampicillin resistance mediated by OXA-30 was not transferable by conjugation and transformation. Within the tnpA region from S. flexneri strain CH-07, one cytosine base insertion was found. This might have caused a frameshift mutation in the genes required for recombination, leading to loss of transferability. Further studies to validate this hypothesis are warranted. Hybridization results of this study suggest that the transposon encoding blaOXA-30 is probably located in the chromosome. As previously reported for chromosomally encoded SHV-1 in Klebsiella pneumoniae (1, 11), the hybridization signal for blaOXA-30 was weak in most of the S. flexneri isolates in the current study. In the study by Burman et al., OXA-type β-lactamase-producing E. coli also showed negative results upon hybridization with the blaOXA-1 probe (3).
The gene blaOXA-30 differs from blaOXA-1 by having one mutation at codon 131, AGA→GGA (arginine to glycine). The functional significance of this amino acid substitution appears to be minimal, as the Vmax and IC50 of OXA-30 are similar to those of OXA-1. According to the general consensus, this enzyme was named distinctly from OXA-1 (K. Bush, personal communication, 1996). Due to the instability of OXA-type β-lactamases, reporting of the relative Vmax is more appropriate than that of Km values (H. Chardon, S. Farzaneh, R. Labia, V. Jarlier, M. H. Nicolas, G. Paul, C. Poyart, D. Sirot, and J. Sirot, Letter, J. Antimicrob. Chemother. 36:267–269, 1995. IEF is highly reproducible. It will retain its usefulness as a screening test because it allows a large number of samples to be tested concurrently. However, this test alone may not yield an exact identification due to the number of β-lactamases with similar pIs. The definitive pI may even be missed, as was the case with OXA-30 initially, if a broad-range ampholine gel is used. Ultimately, a molecular approach such as PCR and sequencing may prove to be the most convenient method for further study of the molecular epidemiology of blaOXA-30.
In summary, a novel OXA-30 β-lactamase from S. flexneri was reported. The OXA-type β-lactamase is more prevalent than TEM-1 in ampicillin-resistant S. flexneri. This finding, together with similar data from other countries, suggests a host specificity for blaOXA-1-like-genes in S. flexneri. Further studies on the prevalence of OXA-1-like β-lactamases in S. flexneri and other bacteria in different geographical localities should be pursued.
ACKNOWLEDGMENTS
This study was supported by a research grant from the University Research Committee, The University of Hong Kong. The work described here was performed at the Department of Microbiology, The University of Hong Kong.
REFERENCES
- 1.Arlet G, Philippon A. Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB) FEMS Microbiol Lett. 1991;66:19–25. doi: 10.1016/0378-1097(91)90414-6. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Morre D D, Seidman J G, Smith J, editors. Current protocols in molecular biology. Boston, Mass: John Wiley & Sons; 1995. [Google Scholar]
- 3.Burman L G, Haeggman S, Kuistila M, Tullus K, Huovinen P. Epidemiology of plasmid-mediated β-lactamases in enterobacteria in Swedish neonatal wards and relation to antimicrobial therapy. Antimicrob Agents Chemother. 1992;36:989–992. doi: 10.1128/aac.36.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K, Sykes R B. Methodology for the study of β-lactamases. Antimicrob Agents Chemother. 1986;30:6–10. doi: 10.1128/aac.30.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu Y W, Houang E T, Lyon D J, Ling J M, Ng T K, Cheng A F. Antimicrobial resistance in Shigella flexneri and Shigella sonnei in Hong Kong, 1986 to 1995. Antimicrob Agents Chemother. 1998;42:440–443. doi: 10.1128/aac.42.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooksey R C, Clark N C, Thornsberry C. A gene probe for TEM type β-lactamases. Antimicrob Agents Chemother. 1985;28:154–156. doi: 10.1128/aac.28.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray L D. Escherichia, Salmonella, Shigella, and Yersinia. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C.: American Society for Microbiology; 1995. pp. 450–456. [Google Scholar]
- 8.Huovinen S, Huovinen P, Jacoby G A. Detection of plasmid-mediated β-lactamases with DNA probes. Antimicrob Agents Chemother. 1988;32:175–179. doi: 10.1128/aac.32.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingham B, Brentnall E, Dale E A, McFadzean J A. Arthopathy induced by antibacterial fused N-alkyl-4-pyridone-3-carboxylic acids. Toxicol Lett. 1977;1:21–26. [Google Scholar]
- 10.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung M, Shannon K, French G. Rarity of transferable β-lactamase production by Klebsiella species. J Antimicrob Chemother. 1997;39:737–745. doi: 10.1093/jac/39.6.737. [DOI] [PubMed] [Google Scholar]
- 12.Ling J, Kam K M, Lam A W, French G L. Susceptibilities of Hong Kong isolates of multiply resistant Shigella spp. to 25 antimicrobial agents, including ampicillin plus sulbactam and new 4-quinolones. Antimicrob Agents Chemother. 1988;32:20–23. doi: 10.1128/aac.32.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling J M, Shaw P C, Kam K M, Cheng A F, French G L. Molecular studies of plasmids of multiply-resistant Shigella spp. in Hong Kong. Epidemiol Infect. 1993;110:437–446. doi: 10.1017/s095026880005086x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthew M, Harris A M. Identification of β-lactamases by analytical isoelectric focusing: correlation with bacterial taxonomy. J Gen Microbiol. 1976;94:55–67. doi: 10.1099/00221287-94-1-55. [DOI] [PubMed] [Google Scholar]
- 16.Medeiros A A. Plasmid-determined β-lactamases. In: Bryan L E, editor. Handbook of experimental pharmacology. Berlin, Germany: Springer-Verlag; 1989. pp. 101–128. [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 3rd ed. 1997. Approved standard M7-A3. National Committee for Clinical Laboratory Standards, Villanova, Pa. [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. 5th ed. 1997. Approved standard M2-A5. National Committee for Clinical Laboratory Standards, Villanova, Pa. [Google Scholar]
- 19.Navia M M, Capitano L, Ruiz J, Vargas M, Urassa H, Schellemberg D, Gascon J, Vila J. Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J Clin Microbiol. 1999;37:3113–3117. doi: 10.1128/jcm.37.10.3113-3117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicolas-Chanoine M H. Inhibitor-resistant β-lactamases. J Antimicrob Chemother. 1997;40:1–3. doi: 10.1093/jac/40.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Ouellette M, Paul G C, Philippon A M, Roy P H. Oligonucleotide probes (TEM-1, OXA-1) versus isoelectric focusing in β-lactamase characterization of 114 resistant strains. Antimicrob Agents Chemother. 1988;32:397–399. doi: 10.1128/aac.32.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritish E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Schaad U B, Wedgwood K J. Nalidixic acid in children: retrospective matched controlled study for cartilage toxicity. Infection. 1987;15:165–168. doi: 10.1007/BF01646040. [DOI] [PubMed] [Google Scholar]
- 24.Schumacher H, Nir M, Mansa B, Grassy A. β-Lactamases in Shigella. APMIS. 1992;100:954–956. [PubMed] [Google Scholar]
- 25.Siu L K, Ho P L, Yuen K Y, Wong S S T, Chau P Y. Transferable hyperproduction of TEM-1 β-lactamase in Shigella flexneri due to a point mutation in the Pribnow box. Antimicrob Agents Chemother. 1997;41:468–470. doi: 10.1128/aac.41.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsang R S W, Yung R W H. Relative frequency and antimicrobial susceptibility of gastrointestinal bacterial pathogens. Lab Med. 1991;22:793–797. [Google Scholar]