Abstract
Nuclear receptor coactivator 6 (NCOA6) is a transcriptional coactivator of nuclear receptors and other transcription factors. A general Ncoa6 knockout mouse was previously shown to be embryonic lethal, but we here generated liver-specific Ncoa6 knockout (Ncoa6 LKO) mice to investigate the metabolic function of NCOA6 in the liver. These Ncoa6 LKO mice exhibited similar blood glucose and insulin levels to wild type but showed improvements in glucose tolerance, insulin sensitivity, and pyruvate tolerance. The decrease in glucose production from pyruvate in these LKO mice was consistent with the abrogation of the fasting-stimulated induction of gluconeogenic genes, phosphoenolpyruvate carboxykinase 1 (Pck1) and glucose-6-phosphatase (G6pc). The forskolin-stimulated inductions of Pck1 and G6pc were also dramatically reduced in primary hepatocytes isolated from Ncoa6 LKO mice, whereas the expression levels of other gluconeogenic gene regulators, including cAMP response element binding protein (Creb), forkhead box protein O1 and peroxisome proliferator-activated receptor γ coactivator 1α, were unaltered in the LKO mouse livers. CREB phosphorylation via fasting or forskolin stimulation was normal in the livers and primary hepatocytes of the LKO mice. Notably, it was observed that CREB interacts with NCOA6. The transcriptional activity of CREB was found to be enhanced by NCOA6 in the context of Pck1 and G6pc promoters. NCOA6-dependent augmentation was abolished in cAMP response element (CRE) mutant promoters of the Pck1 and G6pc genes. Our present results suggest that NCOA6 regulates hepatic gluconeogenesis by modulating glucagon/cAMP-dependent gluconeogenic gene transcription through an interaction with CREB.
Keywords: cAMP response element-binding protein, gluconeogenesis, glucose-6-phosphatase, nuclear receptor coactivator 6, phosphoenolpyruvate carboxykinase
INTRODUCTION
Glucose is a major metabolic fuel for energy production in most organisms. Glucose homeostasis is therefore an important process and is maintained within a narrow range by various pathways of glucose metabolism, including glycogenesis, glycogenolysis, glycolysis and gluconeogenesis (Petersen et al., 2017). In the fed state, glycogen synthesis and glycolysis are dominant processes in the liver. In contrast, glycogen breakdown and gluconeogenesis mainly occur in a fasted state. Hepatic gluconeogenesis is the primary mechanism of endogenous glucose production during prolonged fasting or starvation because glycogen storage in the liver is rapidly depleted in the fasted state (Rui, 2014). Importantly in this regard, increased hepatic gluconeogenesis is considered to be a major contributor to the hyperglycemia observed in patients with type 2 diabetes, whereas glycogenolysis was found not to contribute (Cline et al., 1994; Magnusson et al., 1992). In addition, gluconeogenesis is a linking factor in the causal relationship between hepatic fat accumulation and hepatic insulin resistance (Samuel et al., 2004). Hence, a better understanding the regulation of gluconeogenesis is fundamentally critical to the development of new treatments for type 2 diabetes.
The main substrates for human gluconeogenesis are lactate, glycerol, and glucogenic amino acids (particularly alanine and glutamine). Pyruvate is first generated for gluconeogenesis from lactate or an α‐keto acid (e.g., α-ketoglutarate) derived from amino acid breakdown. Pyruvate is then transformed via carboxylation into oxaloacetate by pyruvate carboxylase (PC) in the mitochondria. After leaving the mitochondria via malate, oxaloacetate is converted to phosphoenolpyruvate by phosphoenolpyruvate carboxykinase 1 (PCK1). After five reverse steps of glycolysis, fructose-6‐phosphate is formed from fructose-1,6‐bisphosphate by fructose-1,6‐bisphosphatase 1 (FBP1). Fructose-6‐phosphate is then converted to glucose‐6‐phosphate by phosphoglucose isomerase. Glucose‐6‐phosphate is finally dephosphorylated by glucose‐6‐phosphatase (G6PC) to form free glucose. The reactions catalyzed by the PC, PCK1, FBP1, and G6PC enzymes are rate-limiting steps in gluconeogenesis. It is noteworthy that the gluconeogenic PC, PCK1 and G6PC genes are all directly activated by the cAMP response element binding protein (CREB) transcription factor (Thiel et al., 2005; Thonpho et al., 2010; Xing and Quinn, 1993). Latent nuclear CREB is phosphorylated by the cAMP/PKA pathway and then operates the gluconeogenic program through the transcriptional activation of the PC, PCK1 and G6PC genes (Benchoula et al., 2021).
Nuclear receptor coactivator 6 (NCOA6), also known as activating signal cointegrator-2 (ASC-2), nuclear receptor coregulator (NRC), peroxisome proliferator-activated receptor interacting protein (PRIP), nuclear receptor-activating protein 250 (RAP250), and thyroid hormone receptor-binding protein (TRBP), is a transcriptional coactivator of nuclear receptors and many other transcription factors (Caira et al., 2000; Ko et al., 2000; Lee et al., 1999; Mahajan and Samuels, 2000; Zhu et al., 2000). Ncoa6 null mouse embryos show growth retardation, hypoplastic heart development, defective placentation, and embryonic lethality (Antonson et al., 2003; Mahajan et al., 2004; Zhu et al., 2003). We previously reported using Ncoa6 heterozygous knockout mice that NCOA6 is involved in glucose homeostasis via the regulation of insulin secretion from pancreatic β-cells and hepatic insulin sensitivity (Kim et al., 2012; Yeom et al., 2006). In our present study, we generated liver-specific Ncoa6 knockout (Ncoa6 LKO) mice to enable us to study the metabolic function of hepatic NCOA6. Interestingly, we found from our analyses that pyruvate tolerance is improved in Ncoa6 LKO mice compared to wild type. This led us to investigate the molecular function of NCOA6 during the process of hepatic gluconeogenesis. Our results demonstrate that NCOA6 plays a crucial role as a gluconeogenic factor by stimulating the transcriptional activity of CREB towards the Pck1 and G6pc gene promoters.
MATERIALS AND METHODS
Mice
Ncoa6 LKO mice were generated by crossing homozygous floxed Ncoa6 (Ncoa6fl/fl) mice with hemizygous albumin-Cre mice (B6.Cg-Speer6-ps1Tg(Alb-cre)21Mgn/J, Stock No 003574; The Jackson Laboratory, USA). Ncoa6fl/+ mice were provided by Janardan K. Reddy from Northwestern University, Feinberg School of Medicine, Chicago, IL (Zhu et al., 2003). For genotyping, floxed Ncoa6 and Cre alleles were polymerase chain reaction (PCR) amplified from tail genomic DNA using the following primers: floxed Ncoa6 allele, 5′-GGC TCA TTT TCT AGC CCA TGA-3′ and 5′-AGG ACC AGC TCC TTG ACC ACC-3′; and Cre allele, 5′-CTG GTT ATG CGG CGG ATC CGA-3′ and 5′-GGC GCG AGT TGA TAG CTG GCT-3′. The genomic deletion of Ncoa6 exon 8 was confirmed by PCR amplification of liver genomic DNA using the floxed Ncoa6 allele primers. The mice were housed in a temperature-controlled facility with a 12-h light/12-h dark cycle and provided free access to water and regular rodent chow. All animal protocols were approved by the Institutional Animal Care and Use Committee of the Asan Institute for Life Sciences, Asan Medical Center (approval No. 2016-12-201).
Cell culture
Mouse primary hepatocytes were prepared from Ncoa6fl/fl and Ncoa6 LKO mice by collagenase digestion and Percoll density gradient centrifugation using a previously described procedure (Oh et al., 2015). After isolation, cells were incubated in M199 (Cat. No. 31100-035; Gibco, USA) supplemented with antibiotics (Cat. No. 15140-122; Gibco). HepG2 and 293T cells were maintained in DMEM (Cat. No. 12800-017; Gibco) supplemented with 10% fetal bovine serum (FBS) (Cat. No. 16000-044; Gibco) and antibiotics.
Glucose, glucagon, insulin, and pyruvate tolerance tests
For glucose or pyruvate tolerance testing, 10-week-old Ncoa6fl/fl and Ncoa6 LKO mice were intraperitoneally injected with 2 g/kg glucose or 2 g/kg pyruvate, respectively, after a 16 h overnight fast. For glucagon or insulin tolerance test, mice were intraperitoneally injected with 15 µg/kg glucagon (Cat. No. G2044; Sigma-Aldrich, USA) or 0.5 U/kg insulin (Cat. No. 12585-014; Thermo Fisher Scientific, USA), respectively, after a 6 h fast. Blood glucose concentrations were measured at 0, 30, 60, 90, and 120 min after injection using a One Touch Ultra Blood Glucose Monitoring System (LifeScan, USA).
Serum insulin and glucagon measurements
For insulin and glucagon measurements, serum was prepared by allowing whole blood to clot for 30 min followed by centrifugation at 1,000 × g, 4°C for 10 min. Serum insulin and glucagon levels were determined using an LBIS Mouse Insulin ELISA kit (Cat. No. 638-01489; Fujifilm Wako Shibayagi, Japan) and a Glucagon Enzyme Immunoassay kit (Cat. No. K4756; BioVision, USA), respectively, in accordance with the manufacturer’s instructions.
Glucose production assay
To assay glucose production, primary hepatocytes were plated into 6-well plates and incubated in M199 medium supplemented with antibiotics. The following day, cells were washed twice with 37°C pre-warmed phosphate-buffered saline (PBS) followed by the addition of 1 ml of glucose production buffer consisting of glucose-free DMEM (pH 7.4, without L-glutamine, phenol red, sodium pyruvate and sodium bicarbonate, Cat. No. D5030; Sigma-Aldrich) supplemented with 4 mM L-glutamine, 44 mM sodium bicarbonate, 20 mM sodium lactate, 2 mM sodium pyruvate, and 15 mM HEPES. The cells were then treated with 10 µM forskolin or DMSO and incubated at 37°C for 4 h. The glucose concentration in the glucose production buffer was measured using a Glucose (HK) Assay Kit (Cat. No. GAHK20-1KT; Sigma-Aldrich) in accordance with the manufacturer’s instructions.
RNA preparation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from mouse tissues or primary hepatocytes using TRIzol reagent (Cat. No. 15596018; Thermo Fisher Scientific) as per the manufacturer’s protocol. Purified total RNA (1 µg) was reverse-transcribed using M-MLV reverse transcriptase (M1705; Promega, USA). The transcript levels of each gene were analyzed by real-time qRT-PCR using the LightCycler 480 System (Roche, Switzerland) and SYBR Green PCR Master Mix (Cat. No. 04887352001; Roche). The 2-∆∆Ct method was used to calculate the relative transcript levels compared to internal control Rps29. The following primers were used for these qRT-PCR analyses: Rps29, 5′-CGC AAA TAC GGG CTG AAC A-3′ and 5′-GCC TAT GTC CTT CGC GTA CTG-3′; Ncoa6, 5′-GAA GAA ACC GCC TCG GAA GA-3′ and 5′-CCT CTA GAC CAG TTG GAC GAT TAT CT-3′; G6pc, 5′-CCA TGC AAA GGA CTA GGA ACA A-3′ and 5′-TAC CAG GGC CGA TGT CAA C-3′; Pck1, 5′-CCA CAG CTG CTG CAG AAC A-3′ and 5′-GAA GGG TCG CAT GGC AAA-3′; Pc, 5′-CCA CCT GGA TCC CCA ACT T-3′ and 5′-GCG TTC TCA TAG CCT ACC TGC TT-3′; Creb1, 5′-GGA GTG CCA AGG ATT GAA GA-3′ and 5′-CTG TCC ACT GCT AGT TTG GTA A-3′, Foxo1, 5′-CCA GCT CAA ATG CTA GTA CCA TCA-3′ and 5′- GTC CCC ATC TCC CAG GTC AT-3′; Pgc1a, 5′- AAG TGT GGA ACT CTC TGG AAC TG-3′ and 5′-GGG TTA TCT TGG TTG GCT TTA TG-3′.
Immunoblotting
For immunoblotting analysis, total cell lysates were first prepared from mouse livers and primary hepatocytes using lysis buffer containing 20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% NP-40, and protease inhibitor cocktail (Cat. No. 4693132001; Roche). Lysate proteins (50 µg) were separated by SDS-PAGE and transferred onto a PVDF membrane. After membrane blocking with 5% skim milk, the membrane was probed with primary antibodies against NCOA6 (Cat. No. NB200-335; Novus Biologicals, USA), PCK1 (Cat. No. sc-32879; Santa Cruz Biotechnology, USA), CREB (Cat. No. 9197; Cell Signaling Technology, USA), p-CREB (Cat. No. 9198S; Cell Signaling Technology), or α-tubulin (Cat. No. T9026; Sigma-Aldrich). The blots were then incubated with HRP-conjugated secondary antibodies and visualized using ECL substrate and the Chemi-Smart system (Vilber Lourmat, France). The intensities of the protein bands were determined using ImageJ software (NIH, USA).
Immunoprecipitation (IP)
293T cells were plated onto 100-mm dishes and transfected with 3xFLAG-hNCOA6 and HA-CREB Y134F (constitutively active CREB) using Lipofectamine 2000 (Cat. No. 11668019; Thermo Fisher Scientific), in accordance with the manufacturer’s instructions. After 24 h, the cells were washed with cold PBS and whole cell lysates were then prepared via the addition of lysis buffer. Aliquots of protein lysates (500 µg) were pre-cleared using protein G-agarose beads (Cat. No. sc-2002; Santa Cruz Biotechnology) and immunoprecipitated with anti-FLAG M2 affinity gel (Cat. No. A2220; Sigma-Aldrich) or anti-HA (Cat. No. MMS-101R; Covance, USA) antibody in conjunction with protein G-agarose at 4°C overnight. After IP, the beads were washed three times with IP buffer (20 mM Tris-Cl pH 7.5, 150 mM NaCl, and 0.1% NP-40) and then boiled for 5 min in SDS loading buffer to solubilize the proteins. The immunoprecipitated proteins were subsequently identified by immunoblotting analysis using anti-FLAG (Cat. No. F3165; Sigma-Aldrich) or anti-HA (Cat. No. MMS-101P; Covance) antibodies.
Construction of gluconeogenic gene promoter-luciferase reporters
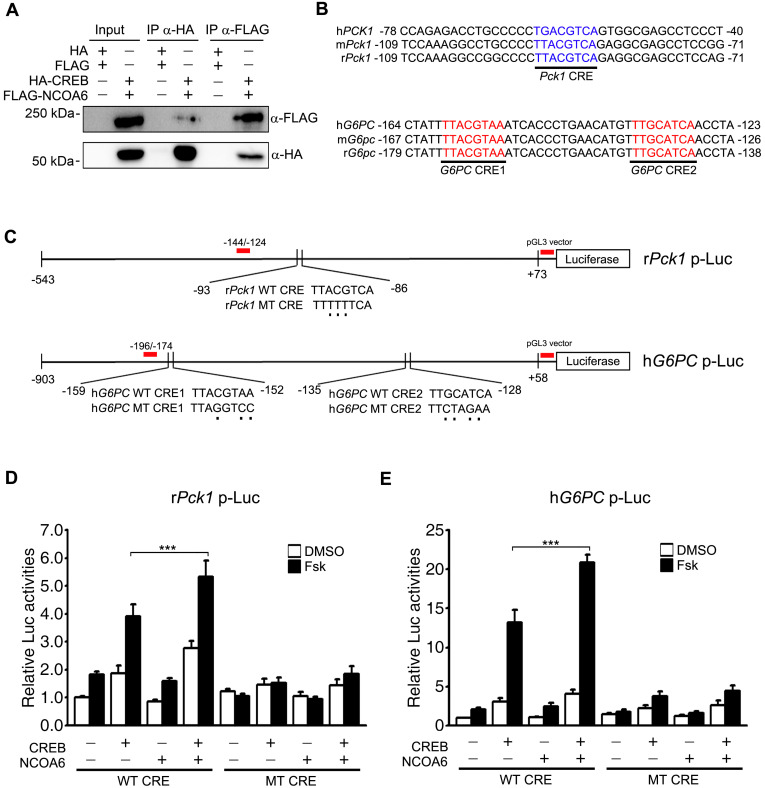
The human G6PC –903/+58 region was amplified from human genomic DNA by PCR using PrimeSTAR HS DNA polymerase with GC buffer (Cat. No. R044A; Takara, Japan) and the following primers: 5′-CGG GGT ACC TAA GAG ACA TGA GGC CAA-3′ and 5′-CCG CTC GAG GAA GAT GTC AGC AGA G-3′. The PCR product was digested with KpnI and XhoI and inserted upstream of the luciferase gene in the pGL3-Basic vector. A rat Pck1 –543/+73 Luc reporter was provided by JaeHun Cheong (Pusan National University, Korea). CRE mutant promoters of the G6PC and Pck1 genes were prepared via a PCR cloning strategy using the following primer pairs: G6PC CRE-1 mutant, 5′-TTC TAT TTT AGG TCC ATC ACC CTG AAC ATG-3′ and 5′-CTA GCT AGC ACT CTT CAT CTG AGG AGC-3′; G6PC CRE-2 mutant, 5′-ATG TTT CTA GAA ACC TAC TGG TGA TGC ACC-3′ and 5′-CCA GTA GGT TTC TAG AAA CAT GTT CAG GGT-3′; and Pck1 CRE mutant, 5′-CCC TTT TTT CAG AGG CGA GCC TCC-3′ and 5′- CTC TGA AAA AAG GGG CCG GCC TTT-3′.
Luciferase reporter assay
For the luciferase assays, HepG2 cells were seeded into 24-well plates in DMEM containing 10% FBS. A luciferase reporter and β-gal construct were cotransfected the next day with or without CREB and NCOA6 expression constructs using Lipofectamine 2000, in accordance with the manufacturer’s instructions. After a further 18 h, the cells were treated with 10 µM forskolin for 6 h and then lysed for the measurement of luciferase and β-gal activities. Luciferase activities were determined using a luminometer Centro LB 960 (Berthold Technologies, Germany) and normalized to β-galactosidase activities.
Chromatin immunoprecipitation (ChIP) of plasmid-bound proteins
ChIP assays were performed as described previously with minor modifications (Kim et al., 2015; Wolfe and Long, 2019). Briefly, 293T cells were cotransfected with 3xFLAG-hNCOA6, HA-CREB Y134F, and G6PC or Pck1 promoter constructs containing wild type or mutant CRE. The cells were incubated overnight and then fixed with 1% formaldehyde (Cat. No. F8775; Sigma-Aldrich) for 15 min at room temperature. The cells were next washed with cold PBS and lysed for 30 min at 4°C by resuspension in a buffer consisting of 50 mM Tris-Cl (pH 8.0), 1% SDS and 10 mM EDTA. Soluble chromatin was prepared by sonication (10 pulses of 10 s and 25% amplitude at 4°C using a VCX500 sonicator; Sonics & Materials, USA) and immunoprecipitated with antibodies against HA (Cat. No. MMS-101P; Covance), FLAG (Cat. No. A2220; Sigma-Aldrich) or IgG (Cat. No. ab46540; Abcam, UK). The final DNA extracts were analyzed by conventional PCR using the following primers: G6PC CRE forward (-196/-174), 5′- GCC GAT CAG GCT GTT TTT GTG TG-3′; Pck1 CRE forward (-144/-124), 5′-AGG TCA GTT CCA AAC CGT GCT-3′; and a commonly used reverse primer for the pGL3 vector, 5′-CGG TTC CAT CTT CCA GCG GAT A-3′.
Statistical analysis
Data are presented as the mean ± SE. Statistical analyses were performed using a two-tailed Student’s t-test or one-way ANOVA with SPSS software (ver. 19; IBM, USA). Differences with P values < 0.05 were considered statistically significant. Statistical results are indicated in the figures as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
RESULTS
Construction of liver-specific Ncoa6 knockout mice
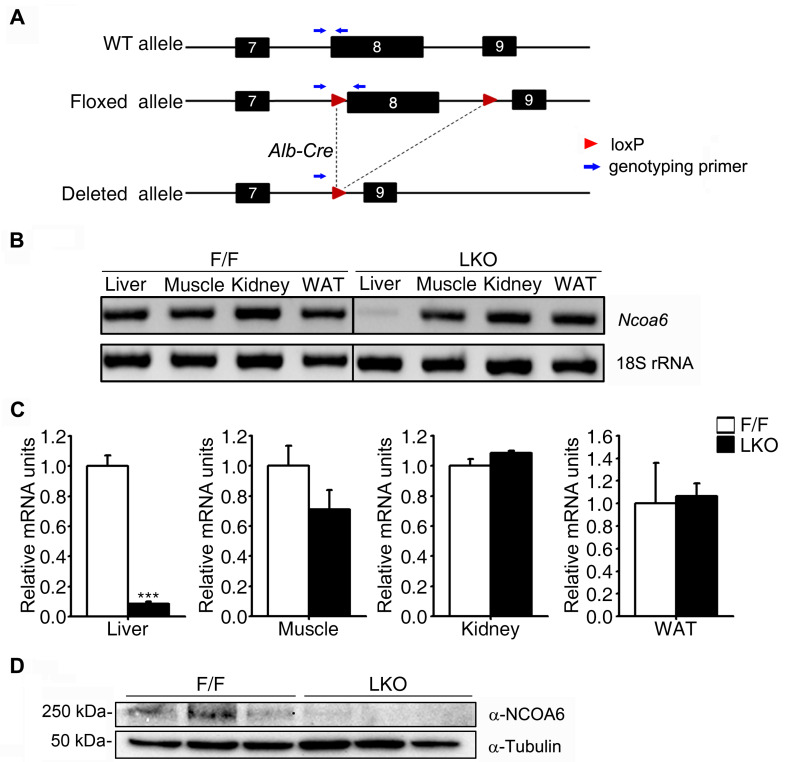
Since an Ncoa6 homozygous knockout in the mouse is embryonic lethal, we used heterozygous Ncoa6+/- and dominant-negative transgenic mice to study the physiological role of NCOA6 in the whole mouse body. Using these mouse models, we found that NCOA6 is an important transcriptional coactivator in many metabolic processes, including insulin secretion and insulin signaling (Kim et al., 2012; Yeom et al., 2006). In our present study, we generated liver-specific Ncoa6 knockout mice by crossing Ncoa6fl/fl and albumin-Cre mice (Fig. 1A). We then used these knockout animals to investigate the metabolic role of hepatic NCOA6 under more specific and clearly defined conditions. The liver-specific deletion of Ncoa6 exon 8 was validated by genomic conventional PCR (Fig. 1B). Consistently, the Ncoa6 transcript and NCOA6 protein levels were dramatically decreased in the liver tissue (Figs. 1C and 1D). The successful generation of an Ncoa6 LKO mouse was thus confirmed.
Fig. 1. Establishment of liver-specific Ncoa6 knockout mice.
(A) Schematic representation of the liver-specific knockout strategy used for the Ncoa6 gene, based on the Cre-LoxP system. Blue arrows denote the specific primer binding sites for genotyping. (B-D) The liver specific knockout of Ncoa6 was confirmed at the level of genomic DNA (gDNA), mRNA and protein. gDNA and mRNA were isolated from the liver, muscle, kidney and white adipose tissue (WAT) of Ncoa6fl/fl/Cre+/- (LKO) and Ncoa6fl/fl/Cre-/- (F/F) mice. The Ncoa6 gDNA levels were determined by conventional PCR (B). The Ncoa6 mRNA and NCOA6 protein levels were determined by qRT-PCR (C; n = 4) and western blotting (D; n = 3), respectively. mRNA data are presented as the mean ± SEM; ***P < 0.001 by t-test comparisons of the genotypes.
Improved pyruvate tolerance in Ncoa6 LKO mice
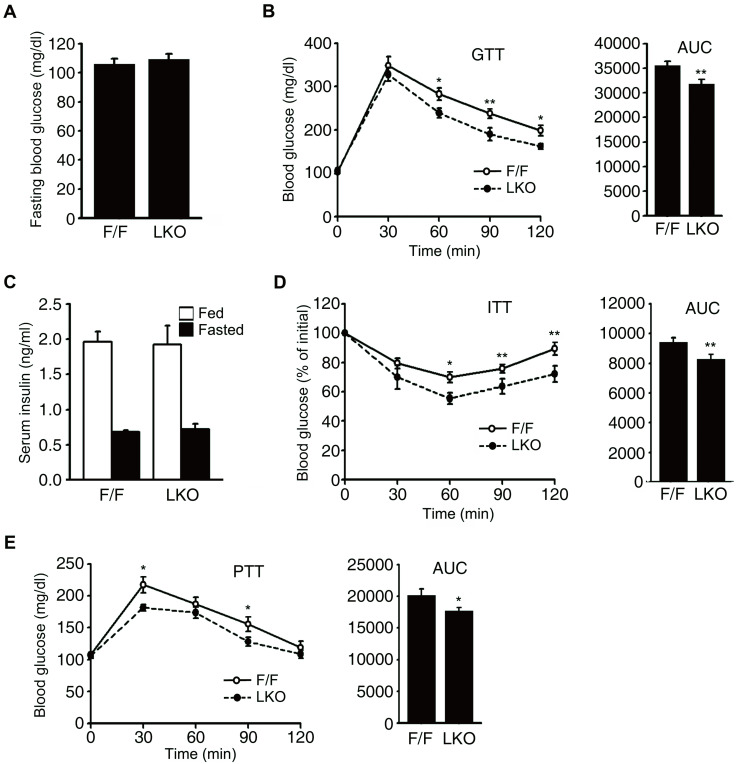
In a previous study of young (10 weeks old) Ncoa6+/- mice, the level of serum insulin was decreased and the insulin sensitivity was found to be increased compared to wild type mice, whereas the fasting blood glucose level and glucose tolerance were not significantly different (Kim et al., 2012). A normal glucose tolerance level in Ncoa6+/- mice was considered to be the result of a trade-off between a decrease in insulin secretion and an increase in insulin sensitivity. In our current analyses, we first analyzed the blood glucose and insulin levels and the glucose and insulin tolerance, of Ncoa6 LKO mice to compare their metabolic phenotypes with those of Ncoa6+/- mice. Ncoa6 LKO mice showed no difference in their fasting glucose level but displayed an improved glucose tolerance compared with Ncoa6fl/fl mice (Figs. 2A and 2B). The glucose area under the curve (AUC) is an index of whole glucose excursion after glucose loading. The AUC from the glucose tolerance test was found to be significantly decreased in the Ncoa6 LKO mice. As expected, the serum insulin levels of Ncoa6 LKO mice were not significantly different from Ncoa6fl/fl mice under either fed or fasting condition, because Ncoa6 LKO mice lack these NCOA6 expression only in liver tissue (Fig. 2C). Insulin sensitivity was found to be augmented in Ncoa6 LKO mice, consistent with the enhancement of hepatic insulin signaling in Ncoa6+/- mice (Fig. 2D).
Fig. 2. Glucose metabolism in Ncoa6 LKO mice.
(A) The blood glucose levels were measured in Ncoa6 LKO (n = 8) and WT (F/F, n = 9) mice after 16 h fasting. (B) Improved glucose tolerance in the Ncoa6 LKO mice. Intraperitoneal glucose tolerance tests were performed in 10-week-old mice (n = 9 for F/F, n = 8 for LKO) after a 16 h fast. (C) The serum insulin levels were measured in Ncoa6 LKO (n = 6 for fed, n = 7 for fasted) and WT mice (n = 7 for both fed and fasted) before and after a 16 h fast. (D) Enhanced insulin sensitivity in Ncoa6 LKO mice. Intraperitoneal insulin tolerance tests were performed in 10-week-old Ncoa6 LKO (n = 13) and WT mice (n = 10) after a 6 h fast. (E) Pyruvate tolerance was determined in 10-week-old Ncoa6 LKO (n = 11) and WT (n = 10) mice via an intraperitoneal injection with pyruvate (2 g/kg body weight) after overnight fasting for 16 h. AUC values for GTT, ITT, and PTT were calculated and are presented on the right of each graph. Data are presented as the mean ± SEM; *P < 0.05, **P < 0.01 by t-test comparisons of the genotypes. WT, wild type; GTT, glucose tolerance test; ITT, insulin tolerance test; PTT, pyruvate tolerance test.
We speculated that the improved glucose tolerance of Ncoa6 LKO mice was due to an increased hepatic insulin sensitivity. We next performed a pyruvate tolerance test to examine the gluconeogenic ability of Ncoa6 LKO mice. Gluconeogenesis occurs primarily in the liver, accounting for up to 80% of the total glucose production in healthy individuals during a prolonged fasting (Ekberg et al., 1999). To evaluate gluconeogenesis as the major hepatic glucose output, our mice were fasted for 16 h to deplete hepatic glycogen storage. Surprisingly, the pyruvate tolerance was significantly increased in Ncoa6 LKO mice (Fig. 2E), suggesting that glucose production from pyruvate is impaired in these animals.
Alteration of the gluconeogenic program in Ncoa6 LKO mice
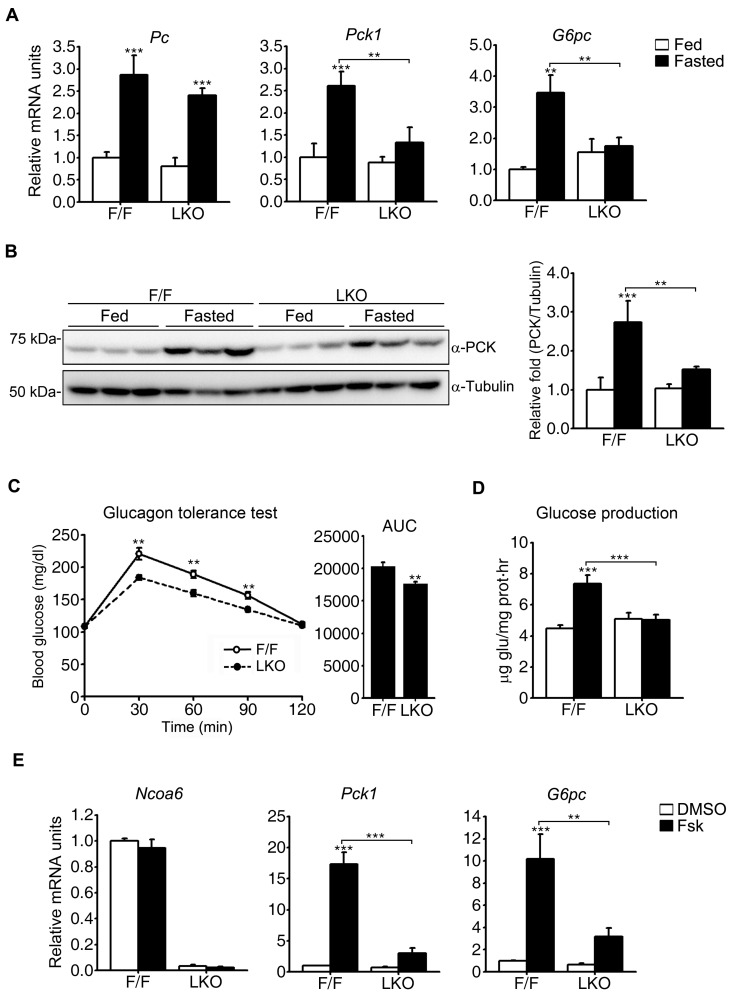
Gluconeogenesis is achieved via a series of enzymatic reactions to produce glucose from non-carbohydrate metabolites. The PC, PCK1, FBP1, and G6PC enzymes are unique to gluconeogenesis and considered to catalyze rate-limiting steps in this process. Among them, FBP1 is regulated allosterically by fructose 2,6-bisphosphate. In contrast, PC, PCK1, and G6PC are transcriptionally regulated by CREB. Since NCOA6 is known to be a transcriptional coactivator, we investigated the transcript levels of key gluconeogenic genes that are regulated by transcriptional mechanisms in our mouse model. The hepatic transcript levels of the Pc, Pck1, and G6pc genes were all increased by 24 h fasting in Ncoa6fl/fl mice, whereas the Pck1 and G6pc transcripts did not show a significant increase in fasted Ncoa6 LKO mice (Fig. 3A). The defective induction of the Pck1 gene was evident at the protein level following fasting in Ncoa6 LKO mice (Fig. 3B). It is well known that glucagon is markedly upregulated by fasting and is responsible for subsequent gluconeogenic gene induction. We thus performed glucagon tolerance tests to compare glucagon-stimulated glucose production between Ncoa6 LKO and Ncoa6fl/fl mice. As expected, the glucagon-stimulated increase in the blood glucose level was significantly diminished in Ncoa6 LKO mice (Fig. 3C).
Fig. 3. Alterations in the gluconeogenic enzyme expression levels in Ncoa6 LKO mice.
(A) Decreased induction of gluconeogenic gene transcripts in the Ncoa6 LKO liver following fasting. Liver RNAs were isolated from Ncoa6 LKO and WT mice after a 24 h fast (n = 6 for each group). The mRNA levels of Pc, Pck1, and G6pc were determined by real-time qRT-PCR. (B) Attenuated induction of PCK protein in the Ncoa6 LKO liver after fasting. Protein expression was analyzed in the liver of ad libitum fed mice and 16 h fasted mice (n = 7 for each group) by western blot analysis. Intensities of the protein bands were measured using the ImageJ program. (C) Glucagon tolerance of Ncoa6 LKO mice (n = 10 for F/F, n = 14 for LKO). Ten-week-old mice were injected intraperitoneally with glucagon (15 µg/kg) after 6 h of fasting. Blood samples were prepared at 0, 30, 60, 90, and 120 min after the glucagon injection. AUC values are shown on the right. (D) Effects of NCOA6 on the glucose production level in primary hepatocytes. Glucose production was compared between Ncoa6 LKO and WT mouse primary hepatocytes after a 4 h forskolin treatment using glucose free media supplemented with lactate and sodium pyruvate (n = 4 independent experiments). (E) Alteration of Pck1 and G6pc transcript induction in Ncoa6 LKO primary hepatocytes following a 4 h treatment of these cells with forskolin (n = 5 independent experiments). Transcript levels were analyzed by real-time qRT-PCR. Data are presented as the mean ± SEM; **P < 0.01, ***P < 0.001, determined using one-way ANOVA. Fsk, forskolin.
We next examined the effects of NCOA6 on the autonomous glucose production of hepatocytes induced by forskolin, a potent activator of adenylate cyclase. Glucagon-receptor complex activates adenylate cyclase to generate second messenger cAMP, which in turn stimulates the PKA-CREB pathway. Thus, forskolin treatments mimic fasting signals to induce gluconeogenesis. In contrast to the normal glucose output of Ncoa6fl/fl hepatocytes, forskolin-stimulated glucose production was abolished in Ncoa6 LKO hepatocytes (Fig. 3D). In addition, and consistent with the observations in the whole mouse liver, the forskolin-stimulated induction of Pck1 and G6pc mRNA was dramatically decreased in primary hepatocytes isolated from Ncoa6 LKO mice (Fig. 3E). Taken together, these results suggest that hepatic NCOA6 is required for the transcriptional induction of gluconeogenic enzyme genes, including Pck1 and G6pc, by fasting signals in a cell-autonomous manner.
Gluconeogenesis regulators and CREB phosphorylation in Ncoa6 LKO mice
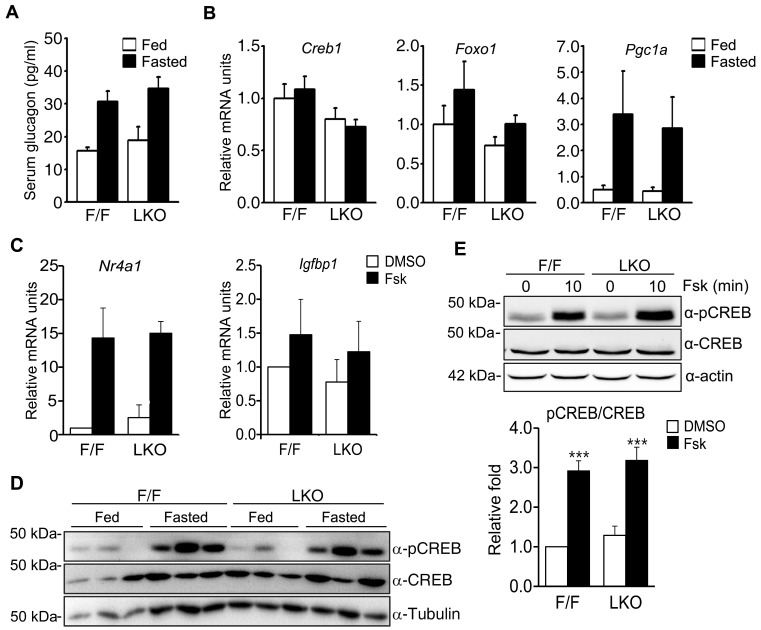
In addition to the enzymes that catalyze each step of the gluconeogenesis pathway from pyruvate to glucose, other gluconeogenic regulatory factors were examined for quantitative changes caused by fasting in Ncoa6 LKO mice in comparison with Ncoa6fl/fl mice. These analyses aimed to identify factors responsible for the gluconeogenic defects of Ncoa6 LKO mice. We first examined glucagon, which is a peptide hormone produced by pancreatic α-cells and stimulates gluconeogenesis largely through transcriptional regulation. The serum glucagon levels were increased by fasting in both Ncoa6fl/fl and Ncoa6 LKO mice at a comparable level (Fig. 4A). We next determined the mRNA levels of transcriptional regulators involved in glucose production, which included two transcription factors, CREB and forkhead box protein O1 (FOXO1), and a transcriptional coactivator, peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α). Notably however, Ncoa6 LKO mice did not show significant differences from Ncoa6fl/fl mice in the hepatic transcript levels of Creb1, Foxo1, or Pgc1a under either fed or fasted conditions (Fig. 4B). Additionally we compared the effects of an NCOA6 deficiency on the hepatic transcript levels of CREB- and FOXO1-specific target genes because the Pck1 and G6pc genes are targeted by both of these transcription factors. By contrast, Nr4a1 (nuclear receptor subfamily 4 group A member 1) and Igfbp1 (insulin-like growth factor binding protein 1) are specific target genes of CREB and FOXO1, respectively. However, Ncoa6 LKO hepatocytes did not exhibit any significant alteration in the transcript levels either Nr4a1 or Igfbp1 irrespective of forskolin treatment (Fig. 4C).
Fig. 4. Gluconeogenic factors and CREB phosphorylation in the Ncoa6 LKO mouse.
(A) Serum glucagon levels in the Ncoa6 LKO (n = 6 for fed, n = 7 for fasted) and WT mice (n = 7 for both fed and fasted) before and after 16 h of fasting. (B) mRNA levels of gluconeogenic transcriptional factors in the liver of Ncoa6 LKO and WT mice in a fed or 24 h fasted state (n = 6 for each group). Hepatic mRNAs were analyzed by real-time qRT-PCR. (C) Transcript levels of the CREB target gene Nr4a1 and FOXO1 target gene Igfbp1 in the primary hepatocytes of Ncoa6 LKO (n = 4) and WT mice (n = 3). (D) Induced phosphorylation of CREB by 16 h of fasting in the livers of Ncoa6 LKO and WT mice (n = 3 for each group). (E) Forskolin-induced phosphorylation of CREB in primary hepatocytes of Ncoa6 LKO and WT mice. Protein levels were determined by western blot analyses using anti-CREB, anti-pCREB (Ser133) or anti-tubulin antibodies (n = 5 independent experiments). The intensities of the protein bands were measured using the ImageJ program. Data are presented as the mean ± SEM; ***P < 0.001, determined using one-way ANOVA. Fsk, forskolin.
CREB phosphorylation is important for the recruitment of coactivators to promoter regions and for the subsequent activation of gluconeogenic gene transcription. We therefore compared the levels of CREB phosphorylation induced by fasting in the Ncoa6fl/fl and Ncoa6 LKO mice. Ncoa6 LKO mice exhibited fasting-induced phosphorylation of CREB to a similar level as Ncoa6fl/fl mice (Fig. 4D). We then examined forskolin-stimulated CREB phosphorylation in primary hepatocytes and found that CREB was phosphorylated by forskolin treatment to a similar degree in both Ncoa6fl/fl and Ncoa6 LKO hepatocytes (Fig. 4E). These results indicated that the alterations of gluconeogenic activity by an NCOA6 deficiency are not due to changes in the levels of serum glucagon or of intracellular gluconeogenic transcriptional factors such as CREB, FOXO1, and PGC-1α.
NCOA6 as a transcriptional coactivator of CREB
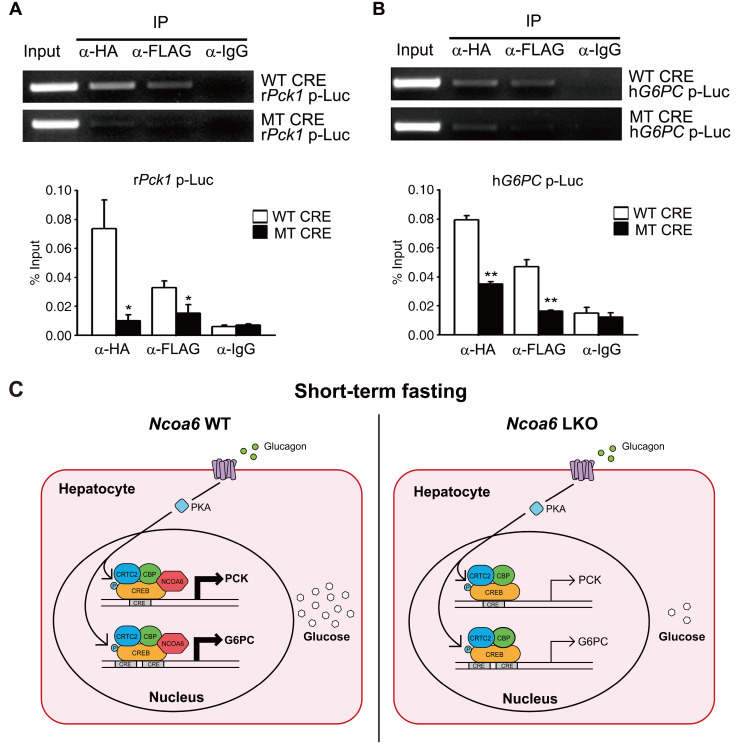
Our current results had demonstrated that the transcriptional activation of Pck1 and G6pc by gluconeogenic signaling is impaired in Ncoa6 LKO mice and in the primary hepatocytes derived from these animals (Fig. 3). Both the Pck1 and G6pc genes are activated by the binding of CREB to CRE in response to glucagon or forskolin. However, there has been no evidence of any interaction between CREB and NCOA6 to date. We here examined the interaction between HA-CREB and FLAG-NCOA6 following the cotransfection of these constructs into 293T cells. CREB and NCOA6 were successfully co-immunoprecipitated in both directions, i.e., using either anti-HA or anti-FLAG antibodies (Fig. 5A). NCOA6 is a bona fide coactivator of many transcription factors, in addition to nuclear hormone receptors. We thus investigated the coactivator function of NCOA6 in the context of CRE-containing promoters of gluconeogenic biosynthetic genes (Figs. 5B and 5C). The transcriptional activities of CREB were further enhanced at the promoters of both Pck1 and G6pc by NCOA6 (Figs. 5D and 5E). Notably, these additional activation events were abolished by using CRE mutant promoters of Pck1 and G6pc genes (Figs. 5D and 5E). The CRE-dependent transactivation of NCOA6 was also confirmed in the context of a synthetic CRE-Luc reporter (Supplementary Fig. S1). NCOA6 alone had no effect on reporter activity, and a further activation only occurred when it was cotransfected with CREB. We next determined the extent of the recruitment of CREB and NCOA6 to the CRE regions of Pck1 and G6pc genes using a modified ChIP assay and promoter-Luc reporters. Both CREB and NCOA6 were found to be recruited to the CREs of the Pck1 and G6pc gene promoters (Figs. 6A and 6B). Moreover, these recruitments were markedly reduced in the CRE mutants of Pck1 and G6pc promoter-Luc reporters (Figs. 6A and 6B). Taken together, these results suggest that NCOA6 regulates gluconeogenesis by enhancing the transcriptional activity of CREB at the Pck1 and G6pc genes (Fig. 6C).
Fig. 5. Transcriptional activation of the Pck1 and G6PC genes by NCOA6.
(A) Interactions between CREB and NCOA6. 293T cells were transfected with HA-CREB and FLAG-NCOA6 and lysates were immunoprecipitated from these cells using anti-HA or anti-FLAG antibody. The coimmunoprecipitates were then analyzed by western blotting with anti-HA or anti-FLAG antibody. (B) Sequence alignment of the CRE-containing promoter regions of the Pck1 or G6pc gene among human, mouse, and rat. (C) Schematic diagram of the rPck1 or hG6PC promoter-Luc reporter containing WT or MT CRE. The red bars indicate the loci of the ChIP primers used in Figs. 6A and 6B. (D and E) Effects of NCOA6 on the CREB-mediated transcriptional activation of the rPck1 (D) and hG6PC (E) promoter (n = 5 independent experiments). WT or MT CRE-containing Pck1 or G6PC promoter-Luc reporters were cotransfected into HepG2 cells with or without HA-CREB, FLAG-NCOA6 and actin-β-galactosidase. Luciferase activities were then measured with a luminometer and normalized using β-galactosidase activities. Data are presented as the mean ± SEM; ***P < 0.001, determined using one-way ANOVA. IP, immunoprecipitation; WT, wild type; MT, mutant; Fsk, forskolin.
Fig. 6. Recruitment of CREB and NCOA6 to the CRE regions of the Pck1 and G6PC promoter.
(A and B) 293T cells were cotransfected with a Pck1 (A) or G6PC (B) promoter-Luc reporter, HA-CREB and FLAG-NCOA6. Modified ChIP assays were then performed using anti-HA, anti-FLAG or IgG antibodies, followed by analysis using conventional PCR (n = 4 independent experiments). The intensities of the PCR bands were measured using the ImageJ program. Data are presented as the mean ± SEM; *P < 0.05, **P < 0.01, determined by t-test. WT, wild type; MT, mutant; IP, immunoprecipitation. (C) Hypothetical model for the gluconeogenic transcription of the Pck1 and G6pc genes mediated via NCOA6.
DISCUSSION
The blood glucose levels are maintained within a very narrow range by a complex network of metabolic organs. In simple terms however, this control represents a balance between glucose uptake by peripheral tissues and glucose release by the liver. Hence, hepatic gluconeogenesis is a very important process for regulating glucose homeostasis in vivo, especially during prolonged fasting. In addition, gluconeogenesis is aberrantly stimulated in type 2 diabetes and its elevation is associated with high fasting glucose and excessive postprandial hyperglycemia in this disease (Basu et al., 2005; Chevalier et al., 2006; Wajngot et al., 2001). Further elucidating the regulatory mechanisms of gluconeogenesis will therefore be critical not only for a fuller understanding of physiology of glucose homeostasis, but also for developing novel therapeutic agents for type 2 diabetes. In our present study, we demonstrate that the coactivator NCOA6 affects glucose production in the mouse by regulating the transcription of the Pck1 and G6pc genes, both of which express rate-limiting enzymes in the gluconeogenic process. However, as shown in Fig. 2A, the 16 h fasting glucose level in Ncoa6 LKO mice did not differ from that in their wild type counterparts. The hepatic fasting response maintains glucose homeostasis through a feedforward mechanism that operates in two stages, depending on the duration of fasting (Altarejos and Montminy, 2011; Zhang et al., 2014). During short-term fasting, CREB acts as a major transcription factor for the expression of gluconeogenic enzymes and transcription factors, and then under conditions of prolonged fasting, PGC-1α, NR4A1, and FOXO1 participate in the maintenance of glucose homeostasis. Further to this, our present findings indicated that NCOA6 does not affect the transcription of the gluconeogenic FOXO1 target gene Igfbp1, but selectively contributes to the transcription of gluconeogenic CREB target enzyme genes including Pck1 and G6pc. Hence, in a long-term fasting state, it would be expected that the relative contribution of NCOA6 to the entire gluconeogenesis process decreases, and that the differences caused by an NCOA6 deficiency during short-term fasting would be offset by the homeostasis mechanism.
Glucose homeostasis is essentially controlled by two opposing hormones, insulin and glucagon. In our previous study, we demonstrated that NCOA6 inhibits hepatic insulin signaling through the induction of the insulin signaling inhibitors SOCS1 and SOCS3, and hence that hepatic insulin sensitivity is enhanced by a decreased expression of NCOA6 (Kim et al., 2012). Here, we have demonstrated that NCOA6 is required to deliver glucagon signals to induce the transcription of gluconeogenic genes by stimulating CREB activity, and therefore that gluconeogenesis is impaired by an Ncoa6 knockout in fasting conditions. We speculate from this that the modulation of hepatic NCOA6 activity could potentially repress the increase in the blood glucose level after feeding and block the hyperglycemia that arises due to increased gluconeogenesis under diabetic conditions. NCOA6 could therefore be further investigated as a promising therapeutic target for the treatment of type 2 diabetes.
NCOA6 is known to function as a coactivator not only for nuclear hormone receptors, but also for other transcription factors such as AP-1, NFκB, and SRF. In our present analyses, CREB was found for the first time to be a transcription factor that interacts and cooperates with NCOA6 to control gluconeogenesis. As the expression of both the G6pc and Pck1 genes is regulated by many transcription factors including FOXO1, HNF4α, and glucocorticoid receptor, in addition to CREB, it will be worth investigating the possible association of these other transcription factors with NCOA6 in terms of gluconeogenesis in a future study. FOXO1, HNF4α, and glucocorticoid receptor are all involved in the activation of gluconeogenesis during fasting. Therefore, the activity of other gluconeogenic transcription factors may be impaired in the Ncoa6 LKO mouse. In this regard, it is noteworthy that the transcriptional activity of HNF4α was found previously to be enhanced by NCOA6 cotransfection in a synthetic promoter reporter assay using HeLa cells (Yeom et al., 2006).
CREB is well known to be modified posttranslationally in the course of the gluconeogenic program. First, CREB is phosphorylated at Ser133 through the cAMP-PKA-CREB pathway by a fasting signal. CREB binding protein (CBP), one of the known coactivators of CREB, is then recruited to the phosphorylated CREB and acetylates Lys91, Lys94, and Lys136 within the CREB activation domain. The resulting doubly modified phospho (Ser133)-acetyl (Lys136) CREB protein then further potentiates CBP recruitment to it (Paz et al., 2014). In this context, it will be very interesting in a future study to determine the point at which NCOA6 participates in the CREB activation process and how these known modifications (phosphorylation and acetylation) of CREB affect NCOA6 recruitment to it. It would then be possible to obtain information about the crosstalk among CREB, CBP, and NCOA6 by investigating the binding of specific domains and particular modification sites of CREB with NCOA6. NCOA6 recruitment to Pck1 and G6pc promoters would be enhanced under the CREB modification conditions in which its binding to NCOA6 is stronger. Conversely, CREB-NCOA6 binding may affect the recruitment of other coactivators including CBP.
Interestingly, the Pc gene, which encodes the enzyme that catalyzes the first committed step of the gluconeogenesis pathway, showed fasting-induced increases in its mRNA level in the Ncoa6 LKO liver, in contrast to the Pck1 and G6pc genes. PC is a mitochondrial enzyme that performs anaplerotic carboxylation from pyruvate to oxaloacetate. On the other hand, both PCK1 and G6PC are cytosolic enzymes that determine the overall rate of the gluconeogenic process. There are two isoforms of the PCK enzyme, cytosolic PCK1 (PEPCK-C) and mitochondrial PCK2 (PEPCK-M), which are encoded by two different nuclear genes (Nordlie and Lardy, 1963). While PCK1 expression can be strongly stimulated by glucagon and inhibited by insulin, PCK2 is constitutively expressed without hormonal control (Hanson and Patel, 1994; Modaressi et al., 1998). Thus, PCK2 expression would not be altered during the transient increase of glucose production under fasting conditions. In conclusion therefore, NCOA6 appears to take part selectively in the transcription of cytosolic gluconeogenic genes, but not in the expression of mitochondrial enzymes under fasting conditions. Further studies are required to elucidate the differential regulation of these cytosolic and mitochondrial enzymes, even though they are all involved in the gluconeogenesis pathway.
The regulatory regions of the Pc, Pck1, and G6pc genes all have CREs. Jitrapakdee et al. (1997; 2001) demonstrated that the rat Pc gene has two promoter regions, i.e., a proximal and distal promoter. The proximal promoter (P1) of rat Pc is active in the liver and adipose tissue, while the distal promoter (P2) is active in the pancreatic islets. The CRE sequence is present in the proximal promoter region of the rat Pc gene. In addition, CREB reportedly binds to the -1639/-1631 CRE of the mouse Pc gene and transactivates the CRE-containing 1.95 kb 5' flanking sequence of mouse Pc (Thonpho et al., 2010). As mentioned above, the Pc, Pck1, and G6pc genes are all activated by CREB. However, NCOA6 is involved in the fasting-induced transcription of Pck1 and G6pc selectively, but not in the transcription of the Pc gene, even though NCOA6 and CREB were shown in our present study to interact with each other using IP experiments with transfected 293T cells. This suggests that the interaction between CREB and NCOA6 is not the only condition necessary for the expression of the Pck1 and G6pc genes by NCOA6. The characteristics of the CRE-adjacent sequences of the Pc gene could affect the recruitment of relevant factors to the promoter region. The interactions between CREB and other transcription factors may also alter the binding and action of NCOA6. Therefore, it is reasonable to assume that NCOA6 does not always act as a major coactivator for CREB, that is, NCOA6 is not always involved in the transcription of all CREB target genes. Nevertheless, our present findings have revealed the significance of the interaction between NCOA6 and CREB in gluconeogenesis. To further our understanding of the transcriptional effects of NCOA6 on overall gluconeogenic regulation, it would be helpful in the future to analyze the gene profile when NCOA6 is recruited after a fasting signal, and also to analyze changes in the hepatic transcript levels under conditions of an Ncoa6 KO or overexpression.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
We thank the GEAR Core Lab core facility at the ConveRgence mEDIcine research cenTer (CREDIT), Asan Medical Center for the use of their shared equipment, services and expertise. This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (MSIT) (NRF-2018R1A2B6007013, NRF-2021R1H1A2095350); and by a grant (2021IL0038) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea.
Footnotes
AUTHOR CONTRIBUTIONS
S.W.K. conceived the experiments, wrote the manuscript, and secured funding. G.S.O. and S.R.K. wrote the initial draft of the manuscript and analyzed the data. G.S.O., S.R.K., E.S.L., J.Y., M.K.S., H.K.R., and D.S.K. performed the experiments.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Altarejos J.Y., Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonson P., Schuster G.U., Wang L., Rozell B., Holter E., Flodby P., Treuter E., Holmgren L., Gustafsson J.Å. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol. Cell. Biol. 2003;23:1260–1268. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R., Chandramouli V., Dicke B., Landau B., Rizza R. Obesity and type 2 diabetes impair insulin-induced suppression of glycogenolysis as well as gluconeogenesis. Diabetes. 2005;54:1942–1948. doi: 10.2337/diabetes.54.7.1942. [DOI] [PubMed] [Google Scholar]
- Benchoula K., Parhar I.S., Madhavan P., Hwa W.E. CREB nuclear transcription activity as a targeting factor in the treatment of diabetes and diabetes complications. Biochem. Pharmacol. 2021;188:114531. doi: 10.1016/j.bcp.2021.114531. [DOI] [PubMed] [Google Scholar]
- Caira F., Antonson P., Pelto-Huikko M., Treuter E., Gustafsson J.Å. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J. Biol. Chem. 2000;275:5308–5317. doi: 10.1074/jbc.275.8.5308. [DOI] [PubMed] [Google Scholar]
- Chevalier S., Burgess S.C., Malloy C.R., Gougeon R., Marliss E.B., Morais J.A. The greater contribution of gluconeogenesis to glucose production in obesity is related to increased whole-body protein catabolism. Diabetes. 2006;55:675–681. doi: 10.2337/diabetes.55.03.06.db05-1117. [DOI] [PubMed] [Google Scholar]
- Cline G.W., Rothman D.L., Magnusson I., Katz L.D., Shulman G.I. 13C-nuclear magnetic resonance spectroscopy studies of hepatic glucose metabolism in normal subjects and subjects with insulin-dependent diabetes mellitus. J. Clin. Invest. 1994;94:2369–2376. doi: 10.1172/JCI117602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekberg K., Landau B.R., Wajngot A., Chandramouli V., Efendic S., Brunengraber H., Wahren J. Contributions by kidney and liver to glucose production in the postabsorptive state and after 60 h of fasting. Diabetes. 1999;48:292–298. doi: 10.2337/diabetes.48.2.292. [DOI] [PubMed] [Google Scholar]
- Hanson R.W., Patel Y.M. Phosphoenolpyruvate carboxykinase (GTP): the gene and the enzyme. Adv. Enzymol. Relat. Areas Mol. Biol. 1994;69:203–281. doi: 10.1002/9780470123157.ch6. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S., Booker G.W., Cassady A.I., Wallace J.C. The rat pyruvate carboxylase gene structure. Alternate promoters generate multiple transcripts with the 5'-end heterogeneity. J. Biol. Chem. 1997;272:20522–20530. doi: 10.1074/jbc.272.33.20522. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S., Petchamphai N., Sunyakumthorn P., Wallace J.C., Boonsaeng V. Structural and promoter regions of the murine pyruvate carboxylase gene. Biochem. Biophys. Res. Commun. 2001;287:411–417. doi: 10.1006/bbrc.2001.5599. [DOI] [PubMed] [Google Scholar]
- Kim G.H., Lee K.J., Oh G.S., Yoon J., Kim H.W., Kim S.W. Regulation of hepatic insulin sensitivity by activating signal co-integrator-2. Biochem. J. 2012;447:437–447. doi: 10.1042/BJ20120861. [DOI] [PubMed] [Google Scholar]
- Kim G.H., Oh G.S., Yoon J., Lee G.G., Lee K.U., Kim S.W. Hepatic TRAP80 selectively regulates lipogenic activity of liver X receptor. J. Clin. Invest. 2015;125:183–193. doi: 10.1172/JCI73615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko L., Cardona G.R., Chin W.W. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6212–6217. doi: 10.1073/pnas.97.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Anzick S.L., Choi J.E., Bubendorf L., Guan X.Y., Jung Y.K., Kallioniemi O.P., Kononen J., Trent J.M., Azorsa D., et al. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 1999;274:34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- Magnusson I., Rothman D.L., Katz L.D., Shulman R.G., Shulman G.I. Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J. Clin. Invest. 1992;90:1323–1327. doi: 10.1172/JCI115997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M.A., Das S., Zhu H., Tomic-Canic M., Samuels H.H. The nuclear hormone receptor coactivator NRC is a pleiotropic modulator affecting growth, development, apoptosis, reproduction, and wound repair. Mol. Cell. Biol. 2004;24:4994–5004. doi: 10.1128/MCB.24.11.4994-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M.A., Samuels H.H. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol. Cell. Biol. 2000;20:5048–5063. doi: 10.1128/MCB.20.14.5048-5063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modaressi S., Brechtel K., Christ B., Jungermann K. Human mitochondrial phosphoenolpyruvate carboxykinase 2 gene. Structure, chromosomal localization and tissue-specific expression. Biochem. J. 1998;333(Pt 2) doi: 10.1042/bj3330359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlie R.C., Lardy H.A. Mammalian liver phosphoneolpyruvate carboxykinase activities. J. Biol. Chem. 1963;238:2259–2263. doi: 10.1016/S0021-9258(19)67962-7. [DOI] [PubMed] [Google Scholar]
- Oh G.S., Lee G.G., Yoon J., Oh W.K., Kim S.W. Selective inhibition of liver X receptor α-mediated lipogenesis in primary hepatocytes by licochalcone A. Chin. Med. 2015;10:8. doi: 10.1186/s13020-015-0037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz J.C., Park S., Phillips N., Matsumura S., Tsai W.W., Kasper L., Brindle P.K., Zhang G., Zhou M.M., Wright P.E., et al. Combinatorial regulation of a signal-dependent activator by phosphorylation and acetylation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17116–17121. doi: 10.1073/pnas.1420389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M.C., Vatner D.F., Shulman G.I. Regulation of hepatic glucose metabolism in health and disease. Nat. Rev. Endocrinol. 2017;13:572–587. doi: 10.1038/nrendo.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L. Energy metabolism in the liver. Compr. Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel V.T., Liu Z.X., Qu X., Elder B.D., Bilz S., Befroy D., Romanelli A.J., Shulman G.I. Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J. Biol. Chem. 2004;279:32345–32353. doi: 10.1074/jbc.M313478200. [DOI] [PubMed] [Google Scholar]
- Thiel G., Al Sarraj J., Stefano L. cAMP response element binding protein (CREB) activates transcription via two distinct genetic elements of the human glucose-6-phosphatase gene. BMC Mol. Biol. 2005;6:2. doi: 10.1186/1471-2199-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thonpho A., Sereeruk C., Rojvirat P., Jitrapakdee S. Identification of the cyclic AMP responsive element (CRE) that mediates transcriptional regulation of the pyruvate carboxylase gene in HepG2 cells. Biochem. Biophys. Res. Commun. 2010;393:714–719. doi: 10.1016/j.bbrc.2010.02.067. [DOI] [PubMed] [Google Scholar]
- Wajngot A., Chandramouli V., Schumann W.C., Ekberg K., Jones P.K., Efendic S., Landau B.R. Quantitative contributions of gluconeogenesis to glucose production during fasting in type 2 diabetes mellitus. Metabolism. 2001;50:47–52. doi: 10.1053/meta.2001.19422. [DOI] [PubMed] [Google Scholar]
- Wolfe K.B., Long D.T. Chromatin immunoprecipitation (ChIP) of plasmid-bound proteins in Xenopus egg extracts. Methods Mol. Biol. 2019;1999:173–184. doi: 10.1007/978-1-4939-9500-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Quinn P.G. Involvement of 3',5'-cyclic adenosine monophosphate regulatory element binding protein (CREB) in both basal and hormone-mediated expression of the phosphoenolpyruvate carboxykinase (PEPCK) gene. Mol. Endocrinol. 1993;7:1484–1494. doi: 10.1210/me.7.11.1484. [DOI] [PubMed] [Google Scholar]
- Yeom S.Y., Kim G.H., Kim C.H., Jung H.D., Kim S.Y., Park J.Y., Pak Y.K., Rhee D.K., Kuang S.Q., Xu J., et al. Regulation of insulin secretion and β-cell mass by activating signal cointegrator 2. Mol. Cell. Biol. 2006;26:4553–4563. doi: 10.1128/MCB.01412-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Yin R., Yang X. O-GlcNAc: a bittersweet switch in liver. Front. Endocrinol. 2014;5:221. doi: 10.3389/fendo.2014.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Kan L., Qi C., Kanwar Y.S., Yeldandi A.V., Rao M.S., Reddy J.K. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J. Biol. Chem. 2000;275:13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- Zhu Y.J., Crawford S.E., Stellmach V., Dwivedi R.S., Rao M.S., Gonzalez F.J., Qi C., Reddy J.K. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J. Biol. Chem. 2003;278:1986–1990. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.