Abstract
The androgen receptor (AR) is an important therapeutic target for treating prostate cancer (PCa). Moreover, there is an increasing need for understanding the AR-independent progression of tumor cells such as neuroendocrine prostate cancer (NEPC). Menin, which is encoded by multiple endocrine neoplasia type 1 (MEN1), serves as a direct link between AR and the mixed-lineage leukemia (MLL) complex in PCa development by activating AR target genes through histone H3 lysine 4 methylation. Although menin is a critical component of AR signaling, its tumorigenic role in AR-independent PCa cells remains unknown. Here, we compared the role of menin in AR-positive and AR-negative PCa cells via RNAi-mediated or pharmacological inhibition of menin. We demonstrated that menin was involved in tumor cell growth and metastasis in PCa cells with low or deficient levels of AR. The inhibition of menin significantly diminished the growth of PCa cells and induced apoptosis, regardless of the presence of AR. Additionally, transcriptome analysis showed that the expression of many metastasis-associated genes was perturbed by menin inhibition in AR-negative DU145 cells. Furthermore, wound-healing assay results showed that menin promoted cell migration in AR-independent cellular contexts. Overall, these findings suggest a critical function of menin in tumorigenesis and provide a rationale for drug development against menin toward targeting high-risk metastatic PCa, especially those independent of AR.
Keywords: androgen receptor, menin, menin inhibitor, metastasis, prostate cancer
INTRODUCTION
MEN1 is frequently mutated in multiple endocrine neoplasia type 1 and multiple types of tumors in different endocrine tissues, such as the parathyroid, pancreatic islets, and pituitary (Chandrasekharappa et al., 1997; Weber and Mulligan, 2017). MEN1 encodes menin, which is a protein of 610 amino acids, that serves as a scaffold to form complexes functioning in various pathways including transcription (Agarwal et al., 1999; Balogh et al., 2006; Kim et al., 2003; Matkar et al., 2013). By interacting with transcription factors and chromatin remodeling complexes, menin plays a critical role in regulating genes involved in development like HOX genes and cell cycle regulation, such as cyclin-dependent kinase inhibitors p18 and p27 (Milne et al., 2005; Yokoyama et al., 2005). Many mutations in MEN1 such as frameshift, nonsense, and missense mutations, result in premature termination and the loss of menin stability, leading to the development of tumors in an endocrine lineage (Dreijerink et al., 2017b; Feng et al., 2017a; Yang et al., 2013).
Alternatively, menin displays an oncogenic function in other cell types, such as blood cells. Mixed-lineage leukemia (MLL) is an extremely aggressive blood cancer characterized by MLL fusion oncogenes generated by the chromosomal translocation with genes involved in the transcription elongation (Slany, 2009). The most prominent binding partner of menin is MLL, which has a SET domain-derived methyltransferase activity toward histone H3 lysine 4 (H3K4). As an integral part of the MLL1/MLL2 complex, menin tethers MLL complexes to promote H3K4 trimethylation (H3K4me3) and gene activation (Hughes et al., 2004; Rao and Dou, 2015). In MLL-rearranged leukemia, menin recruits MLL fusion protein to target genes together with lens epithelium-derived growth factor. Menin-mediated chromatin remodeling via H3K4me3 and Dot1L-dependent H3K79me3 increases the transcription of HOXA/HOXB genes and the HOX cofactor MEIS1, blocking myeloid differentiation (Cierpicki and Grembecka, 2014; Liang et al., 2017; Thiel et al., 2010; Yokoyama et al., 2004). It is notable that the role of menin in tumor development may reside in its unique ability to interact with numerous transcription factors and chromatin remodelers in different cellular contexts. However, a better understanding of the pro-oncogenic functions of menin and the underlying molecular mechanisms within the protein-protein interaction network is further required.
In solid tumors, aberrantly expressed MEN1 is associated with hepatocellular carcinoma, at least in part, via the promotion of H3K4 methylation through MLL recruitment and the upregulation of many liver cancer genes including YAP1 (Kempinska et al., 2018; Xu et al., 2013). In breast cancer, menin interacts with estrogen receptor (ER) to activate ER target genes by recruitment of the MLL complex (Dreijerink et al., 2006; 2017a; Imachi et al., 2010). Aberrant H3K4 methylation is a crucial player of menin-mediated cell proliferation. Menin also binds to enhancers to activate genes regulating breast cancer proliferation.
Furthermore, menin and MLL participate in the androgen receptor (AR) pathway and play an essential role in the development of castration-resistance prostate cancer (CRPC) (Malik et al., 2015). CRPC develops by reactivated AR signaling via AR amplification or point mutations or the alteration of cofactors in response to androgen deprivation therapies. Menin directly interacts with the N-terminal domain (amino acids 469-559) of AR and recruits the MLL complex to AR target genes. Consequently, the menin-MLL complex increases H3K4me3 at target promoters, which is critical for AR signaling and AR-driven prostate cancer (PCa) development. Therefore, menin is a promising target for drug development in AR-dependent PCa (Grembecka et al., 2012; Huang et al., 2012; Xu et al., 2016).
However, the many incidences of AR-independent PCa growth indicate alternative AR-bypass mechanisms during disease progression. For example, neuroendocrine prostate cancer (NEPC) is highly aggressive and characterized by resistance to AR-targeted therapy due to its independence from AR signaling. Additionally, emerging lines of evidence suggest that menin plays diverse roles in tumorigenesis independently of MLL (Chou et al., 2020; Feng et al., 2017b; Gang et al., 2013). For instance, menin directly interacts with Myc in fibrosarcoma cells to upregulate Myc target genes in an MLL-independent manner (Wu et al., 2017).
In this study, we elucidated the role of menin in metastasis and proliferation in AR-independent tumorigenesis. The results of our study expand the potential therapeutic scope of existing limited therapeutic options, especially for AR-negative PCa.
MATERIALS AND METHODS
Cell culture
Human PCa DU145 cells from the American Type Culture Collection (ATCC, USA) were maintained in MEM media (Welgene, Korea) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Welgene) at 37°C with 5% CO2. LNCaP (ATCC) and 22RV1 (ATCC) cells were maintained in RPMI-1640 media (Welgene) supplemented with 10% FBS (Thermo Fisher Scientific), 100 U/ml penicillin, and 100 μg/ml streptomycin (Welgene) at 37°C with 5% CO2. PC3 (ATCC) cells were maintained in F12-K media (Welgene) supplemented with 10% FBS (Thermo Fisher Scientific), 100 U/ml penicillin, and 100 μg/ml streptomycin (Welgene) at 37°C with 5% CO2. LNCaP and 22RV1 cells are AR-positive, whereas DU145 and PC3 cells are AR-negative.
Trypan blue assay
A cell suspension aliquot was centrifuged at 800 rpm for 3 min, resuspended in 1 ml phosphate-buffered saline (PBS), and mixed with 0.4% trypan blue (Thermo Fisher Scientific) prior to loading a hemocytometer. The number of unstained (viable) and stained (nonviable) cells were counted using a binocular microscope. The percentage of unstained cells was calculated as follows: viable cells (%) = (number of viable cells per ml / number of cells per ml) × 100.
Cell proliferation assay
DU145 and LNCaP cells were seeded at 2,000 cells/well and 5,000 cells/well, respectively, in 96-well plates and treated with different concentrations of MI-503 for 96 h. Cell proliferation was determined using Cell Counting Kit-8 (CCK-8) from Dojindo Laboratories (USA) and CellTiter-Glo from Promega (USA). Twenty microliters of CCK-8 reagent were added to each well, and plates were incubated for 1-4 h at 37°C. Absorbance was measured at 490 nm using a multimode Synergy HTX reader (BioTek, USA). To detect viable cells, an ATP assay was performed with CellTiter-Glo. First, 100 μl of CellTiter-Glo reagent was added to each well, followed by incubation at room temperature for 10 min. The luminescence signal was recorded, and the half-maximal inhibitory concentration (IC50) was determined from the derived dose-response curve.
RNA interference
Cells were transfected with non-targeting control or specific siRNAs at concentrations ranging from 60 to 100 nM using Lipofectamine RNAiMAX (Thermo Fisher Scientific), according to the manufacturer’s instructions. After transfection, cells were incubated for 72 h. The efficiency of siRNA-mediated knockdown was determined by quantitative real-time polymerase chain reaction (RT-qPCR) and Western blotting. siRNA sequences are shown in Supplementary Table S1.
RNA extraction and RT-qPCR
Total RNA was extracted using a NucleoSpin® RNA kit (Macherey-Nagel, USA), according to the manufacturer’s instructions. Approximately 2 μg of extracted RNA was used for cDNA synthesis with a cDNA synthesis kit (Thermo Fisher Scientific). RT-qPCR was performed using KAPA SYBR FAST Master Mix (KAPA Biosystem, USA) and a CFX96 Real-Time System (Bio-Rad Laboratories, USA) with specific primers (Supplementary Table S2). The relative quantification was calculated using the 2-ΔΔCT (Livak) method (Livak and Schmittgen, 2001) and the relative levels of mRNA were normalized to those of β-actin for each reaction.
Western blotting
Cells were harvested, washed twice with cold PBS, and lysed using a lysis buffer consisting of 25 mM Tris HCl (pH 8.0), 1 mM EDTA, 150 mM NaCl, and 0.5% NP40. After a 10-min incubation on ice, the cell lysate was centrifuged at 13,000 rpm at 4°C for 10 min. Equal amounts of protein from extracts were separated using SDS-PAGE and transferred to nitrocellulose membranes. The blots were blocked with 5% skim milk and incubated with primary antibodies, which were diluted in the range of 1:1,000 to 1:5,000. After washing with TBS-T (5 mM Tris HCl [pH 7.4], 150 mM NaCl, and 0.01% Tween 20), secondary antibodies were added at a dilution of 1:10,000 for a 1-h incubation at room temperature. Protein signals were detected using a western blot detection kit (AbFrontier, Korea). The following primary antibodies were used in this study: anti-β-actin (Merck Millipore, USA), anti-α-tubulin (AbFrontier), anti-menin (Bethyl Laboratories, USA), anti-cleaved-PARP (Cell Signaling Technology, USA), anti-AR (Santa Cruz Biotechnology, USA), and anti-Lamin B1 (Thermo Fisher Scientific).
Fluorescence-activated cell sorting (FACS) analysis
Cells grown in 6-well plates were treated with appropriate siRNAs or MI-503 for 48 h. Pelleted cells were washed thoroughly with PBS and resuspended in annexin V-FITC and propidium iodide (PI) at 50 μg/ml in binding buffer. After incubation at room temperature in the dark for 15 min, cells were detected using a Guava® easyCyte Flow Cytometer (Merck Millipore).
Immunofluorescence assay
Cells were fixed with 4% (w/v) paraformaldehyde, permeabilized with 0.5% Triton X-100 (Sigma, USA) in PBS, and blocked with 2% (w/v) bovine serum albumin in PBS. Cell nuclei were stained with DAPI (Thermo Fisher Scientific). Cells were examined under a LSM700 confocal microscope (Carl Zeiss, Germany), and images were processed using ZEISS ZEN Software.
mRNA sequencing
Total RNA was extracted using TRIzol® (Thermo Fisher Scientific) and checked for integrity using a RNA 6000 Nano ll kit and Bioanalyzer 2100 (Agilent, USA). A RNA library was prepared using a NEXTflex® Rapid Directional mRNA-Seq kit (Bio Scientific, USA). Briefly, mRNA was isolated from 400 ng of total RNA using RNA purification beads by polyA capture, followed by enzyme shearing. After cDNA synthesis, A-tailing and end repair were performed to ligate the cDNA to primers containing unique sequencing adaptors with an index for tracking Illumina reads. For each library, the total library size (~400 bp) was confirmed using a Bioanalyzer and High Sensitivity DNA Chips (Agilent) and quantified by RT-qPCR using a CFX96 Real-Time System (Bio-Rad Laboratories). After normalization, sequencing was performed using an Illumina NextSeq 500. Clusters of cDNA libraries were generated on a flow cell and sequenced generating 75-bp paired-end reads (2 × 75) using a NextSeq 500 and 150-Cycle Reagent Cartridge kit (Illumina, USA). Raw images were transformed by base-calling into sequence data and were stored in the FASTQ format.
Wound-healing assay
Cells were seeded in 6-well plates, incubated for 24 h, transfected with control or specific siRNAs using Lipofectamine 2000 (Thermo Fisher Scientific), and incubated for another 48 h until confluent. Cells were then scratched with a pin (SPL, Korea), and the detached cells were removed to create a cell-free zone as a thin wound. Cell migration was observed immediately and 24 h after wounding using a microscope.
Chromatin immunoprecipitation (ChIP) assay
DU145 cells were cross-linked with 1% formaldehyde at room temperature for 12.5 min, followed by quenching with 0.125 M glycine. Nuclei were prepared with Buffer A, containing 10 mM Tris HCl (pH 7.5), 10 mM KCl, 5 mM MgCl2, and 0.5% NP40, and resuspended in Buffer B, consisting of 50 mM Tris HCl (pH 7.9), 10 mM EDTA, and 0.5% SDS. The chromatin solution was prepared by sonication using a Bioruptor (Cosmo Bio, Japan). The supernatant of the sonicated nuclei was diluted tenfold in ChIP dilution buffer and used for immunoprecipitation with indicated antibodies. ChIP was performed by incubating the chromatin solution with 7.5 μg of anti-menin antibody (Bethyl Laboratories), 4 μg of H3K9 me3 antibody (Active Motif, USA), 2 μg of H3 antibody (Active Motif), or 2 μg of rabbit IgG (Thermo Fisher Scientific) as a control, followed by recovery using Protein A Sepharose beads and a 2-h incubation. Beads were washed sequentially with a low-salt wash buffer, high-salt wash buffer, LiCl wash buffer, and finally TE buffer (50 mM Tris HCl and 10 mM EDTA) twice. The precipitated DNA was resuspended in ChIP elution buffer. Cross-linking was reversed for 4 h at 65°C in the presence of RNase A. The precipitated DNA was then purified using a PCR purification kit (Qiagen, Germany) and measured using qPCR and specific ChIP-qPCR primers (Supplementary Table S3). The calculated input (%) was determined as follows: (immunoprecipitated DNA / input DNA) × 100, and represented the relative enrichment of immunoprecipitated DNA.
Analysis of The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets
The publicly available mRNA expression datasets from healthy controls and patients with corresponding cancers in GEO (https://www.ncbi.nlm.nih.gov/geo/) were analyzed. The gene expression levels of 497 patients with PCa from TCGA were log2 transformed and then analyzed. TCGA disease-free survival data and normalized mRNA expression levels (RSEM, batch normalized from Illumina HiSeq_RNASeqV2) were downloaded from the cBioPortal for Cancer Genomics (http://www.cbioportal.org). Kaplan–Meier analysis and log-rank tests were performed using the survival package in R.
Statistical analysis
Data were reported as mean from three biological replicates unless otherwise stated in figure legends. Error bars represent the SD. The Student’s t-test was used to compare two groups for statistical significance, and graphs were generated using GraphPad Prism v7.0 (GraphPad Software, USA). A P value < 0.05 was considered statistically significant.
RESULTS
Inhibition of menin represses the growth of both AR-positive and AR-negative PCa cells
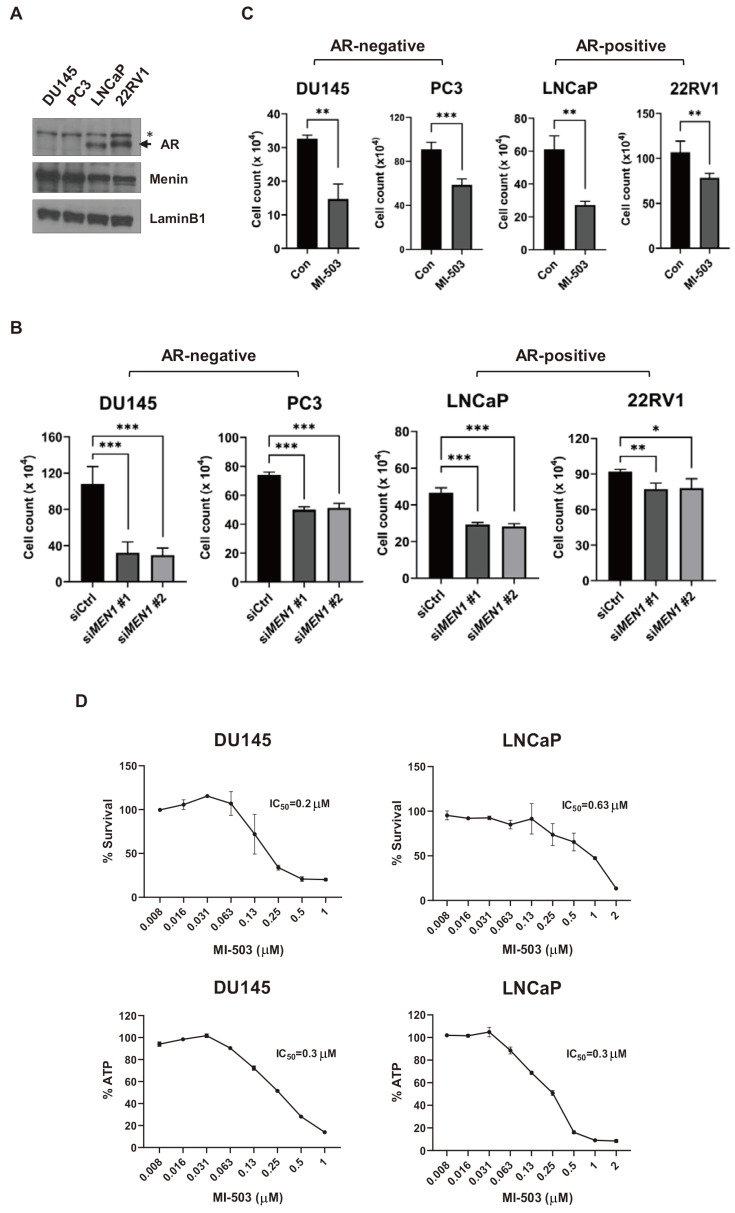
Menin is a critical component of AR signaling, but its tumorigenic role in AR-negative PCa cells remains unknown. To elucidate the activity of menin in the AR-independent pathway, we first investigated whether the inhibition of menin affects the growth of AR-negative and AR-positive PCa cells. In this regard, the DU145, PC3, LNCaP, and 22RV1 cell lines were used as a preclinical model of PCa for comparison. DU145 and PC3 are AR-negative and androgen unresponsive PCa cells, whereas LNCaP and 22RV1 are AR-positive and androgen responsive PCa cells. These cell lines showed comparable levels of menin expression (Fig. 1A). Cells were treated with control or two independent MEN1 siRNAs. The level of MEN1 mRNA was reduced upon MEN1 knockdown, and the depletion of menin by specific siRNAs was confirmed by Western blotting (Supplementary Fig. S1). Although the knockdown efficiency was relatively low in PC3 and 22RV1 cells for unknown reasons, the two MEN1 siRNAs significantly suppressed the cell growth of both cell types (Fig. 1B). These results suggest a potential role of menin in the tumorigenic growth of PCa cells independent of AR. In addition, by treating cells with 1 μM of MI-503, which is a potent and selective inhibitor of the menin-MLL interaction, we found that MI-503 significantly suppressed the growth of PCa cells, regardless of the presence or absence of AR (Fig. 1C). Moreover, we determined the IC50 of MI-503 with DU145 and LNCaP cells using two colorimetric methods to quantify the number of metabolically active cells. As expected, cell viability was dose-dependently reduced by MI-503 with a similar IC50 in the two cell types (Fig. 1D). Our results indicate that menin plays a pro-tumorigenic role in both AR-positive and AR-negative PCa cells. Given the reproducibility and efficiency of RNAi-mediated knockdown of MEN1, we selected the DU145 and LNCaP cell lines as representatives of AR-negative and AR-positive PCa for further studies.
Fig. 1. Inhibition of menin represses the growth of both AR-positive and AR-negative PCa cells.
(A) Western blot assay of AR and menin in PCa cells. Lamin B1 (70 kDa) was used as a loading control. The AR protein band is indicated by an arrow. An asterisk indicates nonspecific bands. (B) The proliferation of DU145, PC3, LNCaP, and 22RV1 cells upon MEN1 knockdown for 48 h as measured by Trypan blue assay. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Cell proliferation analysis of DU145, PC3, LNCaP, and 22RV1 cells treated with 1 μM of MI-503 for 48 h. **P < 0.01; ***P < 0.001. Con, control. (D) Proliferation of DU145 and LNCaP cells treated with different concentrations of MI-503 for 96 h. The IC50 value was measured using CCK-8 (upper) and CellTiter-Glo (below) methods.
Inhibition of menin induces apoptosis in PCa cells
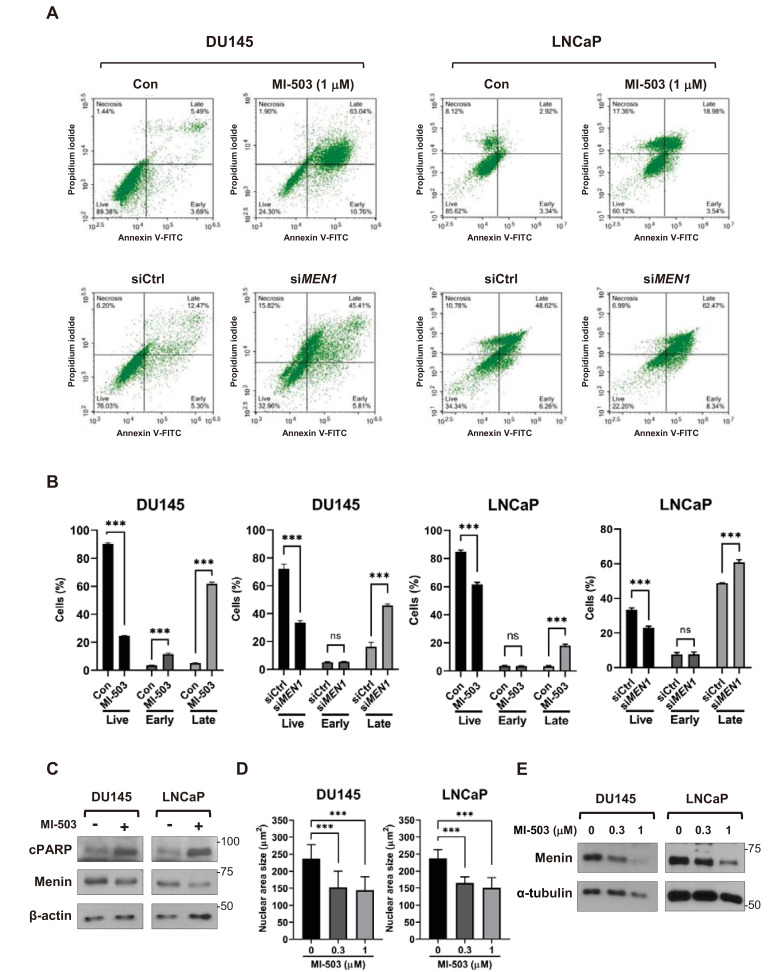
The inhibition of menin increases apoptosis in Rb-deficient thyroid carcinoma and MLL-rearranged leukemia cells (Grembecka et al., 2012; Matoso et al., 2008; Shin et al., 2016). Considering the inhibitory effect of menin depletion on cell growth, we also compared the effect of menin inhibition on apoptotic cell death. FACS analysis with annexin V-FITC/PI double staining showed a significant increase in apoptotic signals, indicating that cell apoptosis was induced by the transfection of MEN1 siRNA or the treatment of MI-503 in DU145 and LNCaP cells (Figs. 2A and 2B). Western blotting of the cleaved fragment of poly-ADP ribose polymerase (cPARP), a marker of apoptosis, also indicated that apoptosis was significantly increased in MI-503-treated PCa cells (Fig. 2C). In addition, the size of cell nuclei, another indicator of apoptosis, was shown to shrink upon MI-503 treatment, according to confocal microscopy (Fig. 2D, Supplementary Fig. S2A). MI-503 reduces menin stability, by promoting its degradation through the ubiquitin/proteasome pathway (Wu et al., 2019). Here, we also observed a substantial decrease in the level of menin protein after treatment with MI-503 without attenuation of MEN1 mRNA (Fig. 2E, Supplementary Fig. S2B). These data indicate that the depletion of menin is the primary cause of apoptosis. Therefore, menin likely contributes to the tumorigenicity of AR-positive and AR-negative PCa cells by mediating antiapoptotic and pro-survival effects, where the necessity of MLL remains unclear.
Fig. 2. Inhibition of menin induces apoptosis in PCa cells.
(A and B) FACS analysis of DU145 and LNCaP cells treated with 1 μM of MI-503 (48 h) or siRNAs (48 h). Cells were stained with Annexin V and PI and analyzed by FACS. n = 3, ***P < 0.001; ns, not significant. Con, control. (C) Western blot analysis of DU145 and LNCaP cells treated with MI-503 (1 μM) using specific antibodies as indicated. β-actin was used as a loading control. (D) DAPI staining of MI-503-treated PCa cells. The nuclei of DU145 (n = 50) and LNCaP (n = 61) cells after treatment with MI-503 were DAPI-stained to measure the size of the nuclei. ***P < 0.001. (E) Western blot assay of menin in DU145 and LNCaP cells treated with 0.3 μM or 1 μM of MI-503, respectively. α-tubulin was used as a loading control.
Gene ontology (GO) analysis of differentially expressed genes (DEGs) in menin-depleted PCa cells
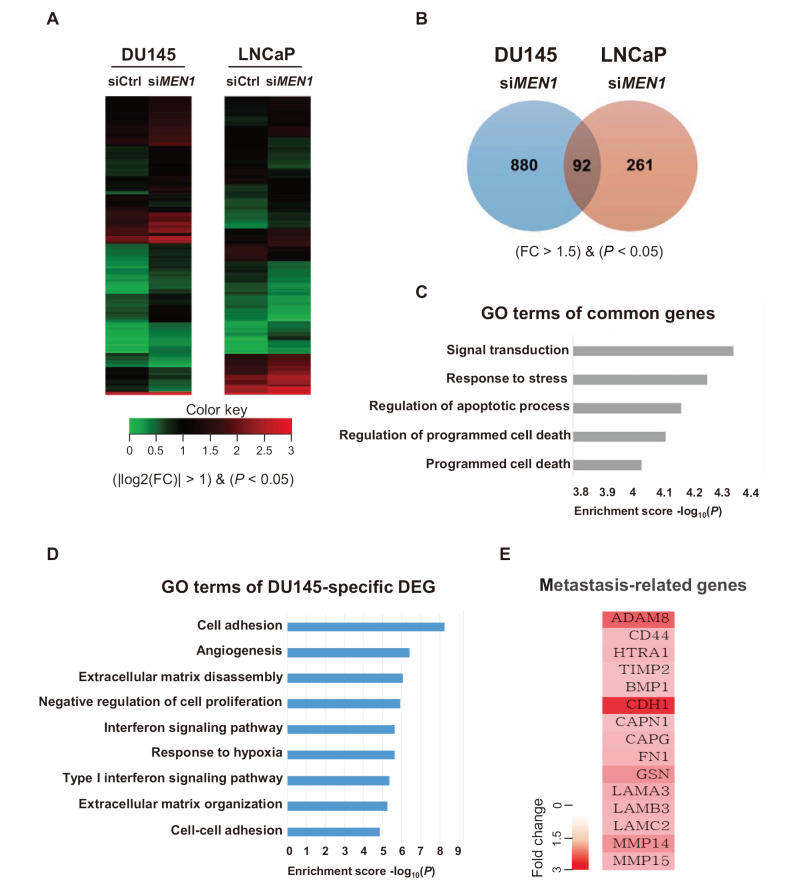
To assess the effect of menin depletion on gene expression, we performed gene expression analysis with DU145 and LNCaP cells transfected with MEN1 siRNAs. RNA purified from menin-depleted cells was subjected to mRNA-sequencing (RNA-seq), followed by genome-wide transcriptome analysis (Fig. 3A). A total of 19,869 genes were analyzed in DU145 and LNCaP cells, and genes with a fold change (FC) of more than 1.5 were further considered (P < 0.05). A total of 972 genes were significantly up or downregulated in menin-depleted DU145 cells, whereas the expression of 353 genes was significantly changed in LNCaP cells (Fig. 3B). Among these genes, 92 were commonly affected in both DU145 and LNCaP cells. Further GO analysis of these common genes using DAVID Bioinformatics resources 6.8 revealed that many of them were significantly associated with several anti-growth pathways, including signal transduction, stress response, and programmed cell death (Fig. 3C). These data suggest that the common downstream targets of menin are linked to the apoptotic pathway in AR-dependent and AR-independent PCa cells.
Fig. 3. GO analysis of DEGs with menin-depleted PCa cells.
(A) Heatmaps of DEGs from siMEN1-treated DU145 and LNCaP cells. (B) The expression of 972 and 353 genes was altered in MEN1 knockdown DU145 and LNCaP cells, respectively. The expression profiles of 92 genes were common in DU145 and LNCaP cells. Genes whose expression was changed by MEN1 knockdown (>1.5-fold) are shown. (C) A diagram showing the GO of 92 genes ordered by P value. The biological functions of genes with the five lowest P values are shown. (D) Biological process analysis was performed using DAVID to select upregulated genes (P < 0.05, FC > 2). (E) Representative genes affected by menin and associated with the metastatic pathway.
We further dissected the role of menin in AR-negative cells by analyzing the 880 DU145-specific DEGs. Surprisingly, our GO analysis showed that the genes perturbed in DU145 cells, are significantly associated with biological functions indicative of metastasis, such as cell adhesion, angiogenesis, negative regulation of cell proliferation, and extracellular matrix disassembly/organization (Fig. 3D). Representative genes upregulated by menin depletion are listed in Fig. 3E. Together, these data demonstrate that menin plays an unexpected role in enhancing the metastatic potential in low or null AR PCa cells.
Menin is highly represented in metastatic cancer and linked to cell migration
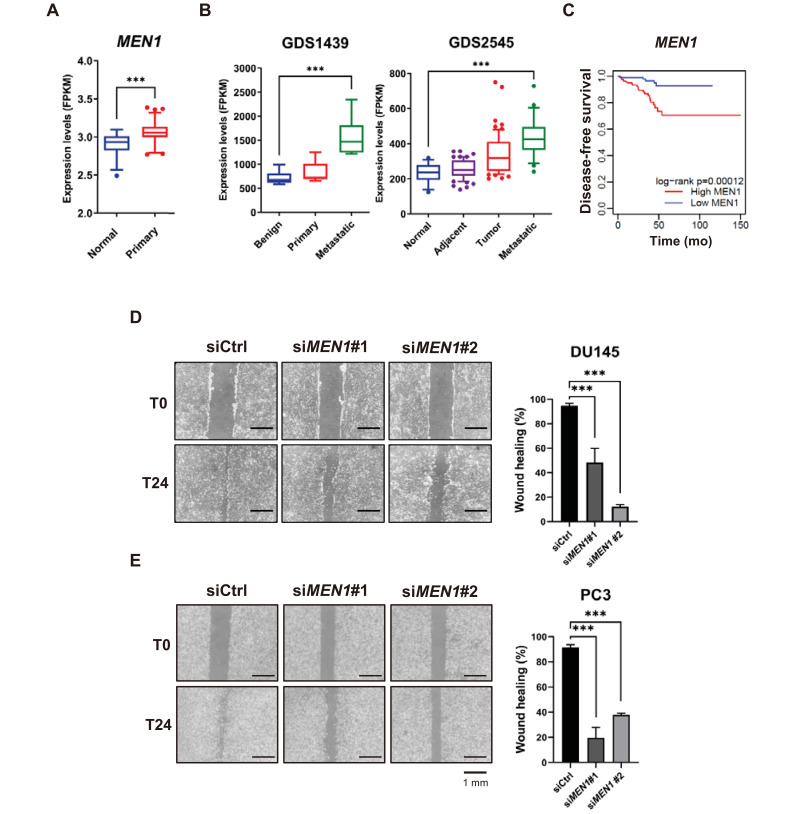
To further explore the potential role of menin in metastatic PCa, we evaluated the expression profiles of MEN1 from TCGA and two available GEO datasets of patients with PCa (GDS1439 and GDS2545). We determined that menin expression is significantly higher in prostate tumors than in normal tissues (Fig. 4A). Moreover, the MEN1 mRNA profile is further elevated in metastatic and hormone refractory tumors compared with normal tissues or clinically localized primary tumors (Fig. 4B). These results are consistent with recent findings (Malik et al., 2015). In addition, our Kaplan–Meier survival analysis of TCGA datasets showed that menin is related to poor prognosis of patients with PCa (log-rank = 0.00012) (Fig. 4C).
Fig. 4. Menin is highly represented in metastatic cancer and linked to cell migration.
(A) The relative expression of MEN1 in the TCGA cohort (normal = 52, primary prostate tumor = 497). ***P < 0.001. (B) MEN1 expression profiles from the GEO datasets. Raw values were obtained from GDS1439 (benign n = 6, primary n = 7, and metastatic n = 6) and GDS2545 (normal n = 18, adjacent n = 63, tumor n = 65, and metastatic n = 25). ***P < 0.001. (C) Kaplan–Meier disease-free survival analysis of patients with prostate adenocarcinoma and high (n = 156, red line) or low (n = 176, blue line) MEN1 expression. P values were determined using the log-rank test. (D and E) DU145 (D) and PC3 (E) cells were scratched 48 h after siMEN1 transfection and the migration efficiency was analyzed 24 h after scratching. The percentage of wound healing was defined as the extent of relative wound closure (n = 3). Scale bars = 1 mm. ***P < 0.001.
To test a potential link between menin and metastasis, we next investigated whether menin affects tumor cell motility in AR-negative cells. We performed a scratch wound-healing assay to evaluate the migration capability of DU145 and PC3, following transfection with control or MEN1 siRNAs. As shown in the images of the scratched areas, we found that MEN1 knockdown reduced migration at the edge of the exposed scratch regions compared with control siRNA in both DU145 and PC3, indicating that cell migration of AR-negative PCa cells was markedly reduced by menin inhibition (Figs. 4D and 4E). To quantify the effect of MEN1 knockdown, the percentage of wound closure (% wound healing) after 24 h was evaluated and clearly showed that menin depletion caused a significant inhibition of cell migration. In contrast, LNCaP and 22RV1 showed a much slower migration rate and longer incubation over the next 24 to 48 h led to cell death in MEN1 KD conditions prior to wound closure (Supplementary Fig. S3). Collectively, both clinical correlation data and in vitro migration assay strongly support the involvement of menin in enhanced metastasis of PCa cells in an AR-independent manner.
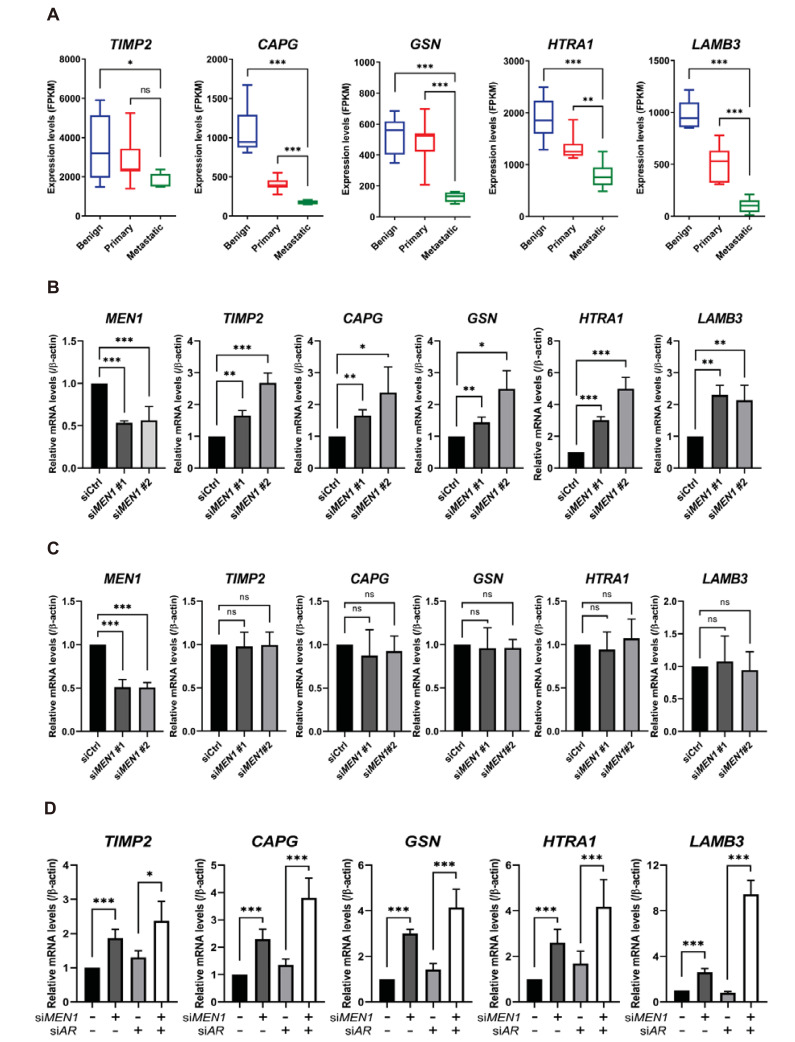
Menin regulates metastasis-related genes in AR-negative DU145 cells
Cell migration is an essential process involved in cancer metastasis and is driven by a complex transcriptional program. To explore the AR-independent function of menin in metastasis, we further focused on DU145-specific DEGs that showed connectivity to metastasis. Some metastasis-related genes, including TIMP2, CAPG, GSN, HTRA1, and LAMB3, were elevated by MEN1 knockdown (Fig. 3E). These genes are particularly intriguing given that they are involved in the regulation or process of metastasis (Fujita et al., 2001; Huang et al., 2018; Mullany et al., 2011; Reis et al., 2012; Ross et al., 2003; Zhang et al., 2019). We first examined their expression profiles using the same GEO data set (GDS1439). Interestingly, the expression of TIMP2, CAPG, GSN, HTRA1, and LAMB3 was substantially reduced in the metastatic cancer tissue (Fig. 5A), demonstrating a negative correlation with menin. To validate this observation and our transcriptome data, we performed RT-qPCR to detect their mRNA levels upon siRNA treatment. As shown in Fig. 5B, we found that menin was a critical player in the suppression of TIMP2, CAPG, GSN, HTRA1, and LAMB3 in DU145 cells. In contrast, their mRNA levels were barely affected by MEN1 knockdown in LNCaP cells (Fig. 5C). These data demonstrate that menin is involved in the tight regulation of metastasis genes in DU145 cells. Although the DU145 cell line is considered AR-negative, several studies have reported that it expresses a detectable level of AR mRNA (Alimirah et al., 2006). To further clarify our observation, we compared the mRNA levels of AR in DU145 and LNCaP cells by RT-qPCR. As a result, we noticed that DU145 cells indeed retained a residual amount of AR mRNA, but the level was extremely low compared with LNCaP cells (Supplementary Fig. S4A). Next, to rule out any possibility of AR-dependency, we depleted residual AR mRNA using an AR-specific siRNA and compared the extent of suppression by menin. As shown in Fig. 5D, both MEN1 siRNAs alone or cotreatment with MEN1 and AR siRNAs efficiently derepressed metastasis genes. In addition, given the pro-metastatic contribution of menin in PC3 cells (Fig. 4E), we analyzed the effect of menin depletion on the same set of genes in PC3 cells and found that their mRNA levels were largely unaltered or marginally downregulated upon MEN1 knockdown (Supplementary Fig. S4B). These data indicate that distinct gene expression programs are operating in two different AR-negative cell lines. Taken together, these results show a specific role for menin in the metastasis of AR-negative cells, at least in part, by regulating a group of genes linked to metastatic pathways.
Fig. 5. Menin downregulates metastasis-associated genes in AR-negative DU145 cells.
(A) The expression of metastasis-associated genes in the GEO dataset (GDS1439). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. (B) Validation of the expression of metastasis-related genes identified from transcriptome data using RT-qPCR. DU145 cells were treated with a control or two independent MEN1-specific siRNAs. *P < 0.05; **P < 0.01; ***P < 0.001. (C) LNCaP cells were treated with a control or MEN1-specific siRNAs. ***P < 0.001; ns, not significant. (D) DU145 cells were treated with siRNAs targeting MEN1 or AR, as indicated. Bars show the mean mRNA level and SD. *P < 0.05; ***P < 0.001.
Menin downregulates TIMP2 and plays a unique role in PCa cell metastasis
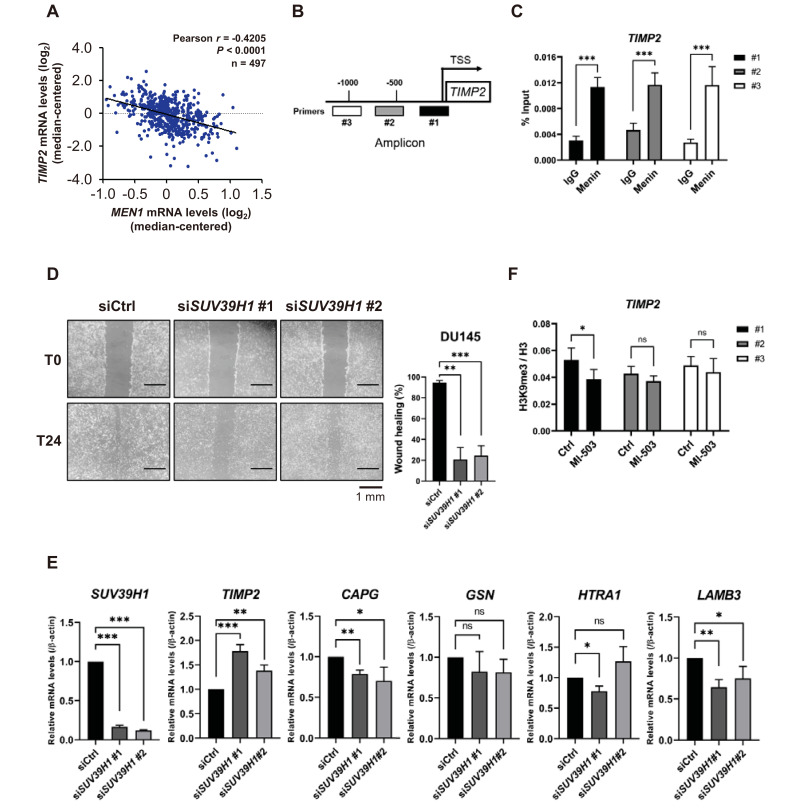
To gain further insights into the function of menin in metastatic pathways, we focused on TIMP2 regulation. TIMP2 is a known natural inhibitor of matrix metalloproteinases (MMPs) and is critical for suppressing tumor cell migration and extracellular matrix (ECM) remodeling through tight regulation of the proteolytic activity of MMPs (Reis et al., 2012; Ross et al., 2003). In agreement with our RT-qPCR data, a Pearson’s statistical analysis of 497 samples of patients with prostate tumors from the TCGA dataset showed a significant negative correlation between MEN1 and TIMP2 (r = -0.4205, P < 0.0001) (Fig. 6A). These data support the idea that menin enhances the metastatic potential of tumor cells by downregulating TIMP2. We further examined TIMP2 regulation by investigating the occupancy of menin at the TIMP2 promoter using the ChIP assay. The location of three amplicons is indicated within the TIPM2 promoter (Shin and Kim, 2012; Yi et al., 2017) (Fig. 6B). Our ChIP-qPCR analysis showed that menin was highly enriched at these three TIMP2 promoter loci (Fig. 6C), suggesting its direct involvement in the transcriptional regulation of TIMP2.
Fig. 6. Menin downregulates TIMP2 and plays a unique role in PCa cell metastasis.
(A) Pearson’s correlation analysis between MEN1 and TIMP2 mRNA levels in prostate primary tumor tissues (n = 497). (B) A diagram of the TIMP2 promoter, showing the locations of ChIP primers. TSS, transcription start site. (C) ChIP was performed using a chromatin solution prepared from DU145 cells treated with either rabbit IgG or anti-menin antibody and analyzed by RT-qPCR with the indicated ChIP primers. Results are shown as the percentage (%) input. ***P < 0.001. (D) DU145 cells were scratched 48 h after siSUV39H1 transfection and the migration efficiency was analyzed 24 h after scratching. The percentage of wound healing was defined as the extent of relative wound closure (n = 3). Scale bars = 1 mm. **P < 0.01; ***P < 0.001. (E) The mRNA level of metastasis-related genes upon SUV39H1 knockdown in DU145 cells. Bars show the mean mRNA level and SD. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. (F) DU145 cells were treated with 1 μM of MI-503 for 48 h. ChIP was performed using rabbit IgG, anti-H3, or anti-H3K9 me3 antibody. H3K9 me3 enrichment was normalized by H3 enrichment. *P < 0.05; ns, not significant.
To explore the molecular mechanisms of menin-dependent TIMP2 regulation, we examined SUV39H1, which is a H3K9 methyltransferase responsible for heterochromatin formation and transcriptional suppression (Kudithipudi et al., 2017; Lachner et al., 2001). Previously, we reported that menin mediates the recruitment of SUV39H1 to repress target genes through H3K9me3 (Yang et al., 2013). Furthermore, Feng et al. (2017b) recently showed that menin and DAXX facilitate the SUV39H1-mediated suppression of membrane metallo-endopeptidase in murine β cells. Considering the negative relationship between menin and selected metastasis genes, we tested the potential engagement of SUV39H1 in their regulation, by examining the mRNA profiles of SUV39H1 in TCGA and GEO. Although TCGA data showed a marginal difference between normal and primary prostate tumor tissues, an additional evaluation of GDS1439 indicated that SUV39H1 was substantially higher in metastatic PCa tissues (Supplementary Figs. S5A and S5B). Furthermore, Kaplan–Meier survival analysis revealed that patients with higher expression of SUV39H1 have shorter overall survival (Supplementary Fig. S5C). We then investigated the effect of SUV39H1 on cell migration by performing a wound-healing assay using two independent SUV39H1 siRNAs. SUV39H1 knockdown diminished the migration potential of DU145 cells, indicating that the elevated expression of SUV39H1 is associated with prostate tumor metastasis (Fig. 6D).
Next, we explored any potential link between menin and SUV39H1 in the regulation of metastasis-related genes. We determined whether the expression of TIMP2, CAPG, GSN, HTRA1, and LAMB3 was also affected by SUV39H1. Interestingly, our RT-qPCR data showed that CAPG, GSN, HTRA1, and LAMB3 were either minimally affected or downregulated by SUV39H1 knockdown. In contrast, the expression of TIMP2 was significantly increased by SUV39H1 knockdown (Fig. 6E). Moreover, treatment of the menin inhibitor MI-503 resulted in a decreased level of H3K9me3 at the transcription start site (TSS) of the TIMP2 promoter (Fig. 6F, amplicon #1), implying a potential link between menin and SUV39H1 in the coregulation of TIMP2 expression. In summary, our data showed that menin plays a distinct role in PCa metastasis by targeting metastasis-related genes in an AR-independent manner.
DISCUSSION
NEPC is a subtype of PCa with poor prognosis. NEPC can arise from resistance to AR-targeted hormonal therapy or de novo development of small-cell carcinoma of the prostate (SCCP) (Beltran et al., 2014; Epstein et al., 2014; Feldman and Feldman, 2001). Our current understanding of the AR-independent development of NEPC or SCCP is insufficient and limits available clinical strategies, which typically rely on platinum-based chemotherapy (Mohler et al., 2019).
Menin is strongly associated with the tumorigenesis of various cancers, functioning as a direct coactivator of many prognostic transcriptional factors through the tethering of MLL complexes to chromatin. Thus, the menin-MLL interface is a valuable target for anticancer therapy (Grembecka et al., 2012; Huang et al., 2012; Xu et al., 2016). Menin and MLL are also involved in AR signaling as coactivators to promote transcriptional activation of AR target genes in prostate tumors. In this study, using TCGA and GEO analysis, we showed that the level of menin was significantly elevated in PCa and even higher in metastatic cancers. These findings suggest that a high level of menin is related to metastasis and the negative prognosis of patients with PCa. Furthermore, we observed that treating AR-negative cells with MEN1-specific siRNAs impaired cell growth as effectively as in AR-positive cells. The quantitative determination of IC50 of MI-503 also confirmed that the inhibition and depletion of menin have a similar detrimental effect on both cell types. These results show that menin has additional pro-oncogenic and metastasis-promoting functions, independent of the AR signaling axis.
In particular, the analysis of DU145-specific DEGs revealed the downstream target genes of menin as associated with metastasis. MMPs and tissue inhibitors of metalloproteinases (TIMPs) are well-known players of tissue homeostasis and controllers of tumor behavior in various cancer types (Reis et al., 2012; Ross et al., 2003). MMPs are responsible for the degradation of the ECM to facilitate malignant tumor invasion and metastasis. For example, MMP2 cleaves the basal components of ECM, such as type IV collagen and gelatin. MMPs are mainly regulated at the posttranslational level by TIMPs, which prevent the substrate interaction of MMPs by binding proximally to their catalytic domain. Therefore, the decreased expression of TIMPs may enhance MMP activity, thereby promoting tumor growth and invasiveness.
Importantly, we showed that menin is directly associated with the TIMP2 promoter and negatively regulated its transcription, in part, via SUV39H1. H3K9me3 at the TSS of TIMP2 was reduced upon MI-503 treatment. However, as noticeable changes were not observed on the other tested loci of the TIMP2 promoter in our data, we assume that there are additional epigenetic factors involved in menin-dependent TIMP2 regulation. Indeed, TIMP2 is under epigenetic regulation of the enhancer of zeste homolog 2 (EZH2) in ovarian and PCa (Shin and Kim, 2012; Yi et al., 2017). In addition, menin independently cooperates with EZH2 to silence many genes via an indirect mechanism (Gherardi et al., 2017; Mishra et al., 2018; Thiel et al., 2013). In this view, it is likely that menin contributes to epigenetic silencing of TIMP2 and other metastasis genes via SUV39H1 and/or EZH2, although further analysis is required.
The AR signaling axis is a critical part of tumor growth and metastatic development of PCa (Ko et al., 2015; Lin et al., 2018). However, unexpectedly, our migration assay showed that menin depletion affected the migratory potential of AR-negative DU145 and PC3 cells. At the molecular level, menin was linked to the regulation of metastasis-associated genes in DU145 cells, indicating that menin promotes metastatic behavior of tumor cells through the regulation of metastasis genes in an AR-independent manner. However, the underlying mechanisms and affected genes are expected to be distinct for each cell line. For example, genes like CDH11, FYN, and SPARCL1 are not on the list of the 972 DEGs identified in DU145 cells, although they are involved in the metastatic behavior of PC3 cells (Chu et al., 2008; Gururajan et al., 2015; Xiang et al., 2013). These differences may be attributed to biological distinctions between tumor cells.
In summary, our study shows an unexpected role of menin in the metastasis of PCa that does not depend on AR signaling. In light of our findings, menin may be a future target for small molecule inhibitors and act as a prognostic biomarker for PCa metastasis.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (NRF-2019R1A5A2027340, 2021R1F1A1049941, and 2019R1A2C2084716 to E.J.C. and 2012R1A3A2048767 to H.D.Y.).
Footnotes
AUTHOR CONTRIBUTIONS
T.K., K.J., E.K., K.Y., J.H.P., and J.C. performed the experiments, analyzed the data, wrote the manuscript. J.C. and J.H.K. performed bioinformatics. H.D.Y. and H.S.K. provided reagents. E.J.C. conceived the study design and wrote the manuscript. T.K. and K.J. contributed equally to this work.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Agarwal S.K., Guru S.C., Heppner C., Erdos M.R., Collins R.M., Park S.Y., Saggar S., Chandrasekharappa S.C., Collins F.S., Spiegel A.M., et al. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell. 1999;96:143–152. doi: 10.1016/S0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- Alimirah F., Chen J.M., Basrawala Z., Xin H., Choubey D. DU-145 and PC-3 human prostate cancer cell lines express androgen receptor: implications for the androgen receptor functions and regulation. FEBS Lett. 2006;580:2294–2300. doi: 10.1016/j.febslet.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Balogh K., Racz K., Patocs A., Hunyady L. Menin and its interacting proteins: elucidation of menin function. Trends Endocrinol. Metab. 2006;17:357–364. doi: 10.1016/j.tem.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Beltran H., Tomlins S., Aparicio A., Arora V., Rickman D., Ayala G., Huang J.T., True L., Gleave M.E., Soule H., et al. Aggressive variants of castration-resistant prostate cancer. Clin. Cancer Res. 2014;20:2846–2850. doi: 10.1158/1078-0432.CCR-13-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharappa S.C., Guru S.C., Manickam P., Olufemi S.E., Collins F.S., EmmertBuck M.R., Debelenko L.V., Zhuang Z.P., Lubensky I.A., Liotta L.A., et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- Chou C.W., Tan X., Hung C.N., Lieberman B., Chen M.Z., Kusi M., Mitsuya K., Lin C.L., Morita M., Liu Z.J., et al. Menin and menin-associated proteins coregulate cancer energy metabolism. Cancers (Basel) 2020;12:2715. doi: 10.3390/cancers12092715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu K., Cheng C.J., Ye X.C., Lee Y.C., Zurita A.J., Chen D.T., Yu-Lee L.Y., Zhang S., Yeh E.T., Hu M.C.T., et al. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol. Cancer Res. 2008;6:1259–1267. doi: 10.1158/1541-7786.MCR-08-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cierpicki T., Grembecka J. Challenges and opportunities in targeting the menin- MLL interaction. Future Med. Chem. 2014;6:447–462. doi: 10.4155/fmc.13.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreijerink K.M.A., Groner A.C., Vos E.S.M., Font-Tello A., Gu L., Chi D., Reyes J., Cook J., Lim E., Lin C.Y., et al. Enhancer-mediated oncogenic function of the Menin tumor suppressor in breast cancer. Cell Rep. 2017a;18:2359–2372. doi: 10.1016/j.celrep.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreijerink K.M.A., Mulder K.W., Winkler G.S., Hoppener J.W.M., Lips C.J.M., Timmers H.T.M. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- Dreijerink K.M.A., Timmers H.T.M., Brown M. Twenty years of menin: emerging opportunities for restoration of transcriptional regulation in MEN1. Endocr. Relat. Cancer. 2017b;24:T135–T145. doi: 10.1530/ERC-17-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J.I., Amin M.B., Beltran H., Lotan T.L., Mosquera J.M., Reuter V.E., Robinson B.D., Troncoso P., Rubin M.A. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am. J. Surg. Pathol. 2014;38:756–767. doi: 10.1097/PAS.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman B.J., Feldman D. The development of androgen-independent prostate cancer. Nat. Rev. Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- Feng Z.J., Ma J., Hua X.X. Epigenetic regulation by the menin pathway. Endocr. Relat. Cancer. 2017a;24:T147–T159. doi: 10.1530/ERC-17-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z.J., Wang L., Sun Y.M., Jiang Z.Z., Domsic J., An C.Y., Xing B.W., Tian J.J., Liu X.H., Metz D.C., et al. Menin and Daxx interact to suppress neuroendocrine tumors through epigenetic control of the membrane metallo-endopeptidase. Cancer Res. 2017b;77:401–411. doi: 10.1158/0008-5472.CAN-16-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H., Okada F., Hamada J., Hosokawa M., Moriuchi T., Koya R.C., Kuzumaki N. Gelsolin functions as a metasatsis suppressor in B16-BL6 mouse melanoma cells and requirement of the carboxyl-terminus for its effect. Int. J. Cancer. 2001;93:773–780. doi: 10.1002/ijc.1413. [DOI] [PubMed] [Google Scholar]
- Gang D., Hongwei H., Hedai L., Ming Z., Qian H., Zhijun L. The tumor suppressor protein menin inhibits NF-kappa B-mediated transactivation through recruitment of Sirt1 in hepatocellular carcinoma. Mol. Biol. Rep. 2013;40:2461–2466. doi: 10.1007/s11033-012-2326-0. [DOI] [PubMed] [Google Scholar]
- Gherardi S., Ripoche D., Mikaelian I., Chanal M., Teinturier R., Goehrig D., Cordier-Bussat M., Zhang C.X., Hennino A., Bertolino P. Menin regulates Inhbb expression through an Akt/Ezh2-mediated H3K27 histone modification. Biochim. Biophys. Acta Gene Regul. Mech. 2017;1860:427–437. doi: 10.1016/j.bbagrm.2017.02.003. [DOI] [PubMed] [Google Scholar]
- Grembecka J., He S.H., Shi A.B., Purohit T., Muntean A.G., Sorenson R.J., Showalter H.D., Murai M.J., Belcher A.M., Hartley T., et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururajan M., Cavassani K.A., Sievert M., Duan P., Lichterman J., Huang J.M., Smith B., You S.Y., Nandana S., Chu G.C.Y., et al. SRC family kinase FYN promotes the neuroendocrine phenotype and visceral metastasis in advanced prostate cancer. Oncotarget. 2015;6:44072–44083. doi: 10.18632/oncotarget.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Gurung B., Wan B.B., Matkar S., Veniaminova N.A., Wan K., Merchant J.L., Hua X.X., Lei M. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482:542–546. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Chi Y.Y., Qin Y., Wang Z.L., Xiu B.Q., Su Y.H., Guo R., Guo L., Sun H.F., Zeng C.J., et al. CAPG enhances breast cancer metastasis by competing with PRMT5 to modulate STC-1 transcription. Theranostics. 2018;8:2549–2564. doi: 10.7150/thno.22523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C.M., Rozenblatt-Rosen O., Milne T.A., Copeland T.D., Levine S.S., Lee J.C., Hayes D.N., Shanmugam K.S., Bhattacharjee A., Biondi C.A., et al. Menin associates with a trithorax family histone methyltransferase complex and with the Hoxc8 locus. Mol. Cell. 2004;13:587–597. doi: 10.1016/S1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Imachi H., Murao K., Dobashi H., Bhuyan M.M., Cao X.Y., Kontani K., Niki S., Murazawa C., Nakajima H., Kohno N., et al. Menin, a product of the MENI gene, binds to estrogen receptor to enhance its activity in breast cancer cells: possibility of a novel predictive factor for tamoxifen resistance. Breast Cancer Res. Treat. 2010;122:395–407. doi: 10.1007/s10549-009-0581-0. [DOI] [PubMed] [Google Scholar]
- Kempinska K., Malik B., Borkin D., Klossowski S., Shukla S., Miao H.Z., Wang J.Y., Cierpicki T., Grembecka J. Pharmacologic inhibition of the menin-MLL interaction leads to transcriptional repression of PEG10 and blocks hepatocellular carcinoma. Mol. Cancer Ther. 2018;17:26–38. doi: 10.1158/1535-7163.MCT-17-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee J.E., Cho E.J., Liu J.O., Youn H.D. Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res. 2003;63:6135–6139. [PubMed] [Google Scholar]
- Ko C.J., Huang C.C., Lin H.Y., Juan C.P., Lan S.W., Shyu H.Y., Wu S.R., Hsiao P.W., Huang H.P., Shun C.T., et al. Androgen-induced TMPRSS2 activates matriptase and promotes extracellular matrix degradation, prostate cancer cell invasion, tumor growth, and metastasis. Cancer Res. 2015;75:2949–2960. doi: 10.1158/0008-5472.CAN-14-3297. [DOI] [PubMed] [Google Scholar]
- Kudithipudi S., Schuhmacher M.K., Kebede A.F., Jeltsch A. The SUV39H1 protein lysine methyltransferase methylates chromatin proteins involved in heterochromatin formation and VDJ recombination. ACS Chem. Biol. 2017;12:958–968. doi: 10.1021/acschembio.6b01076. [DOI] [PubMed] [Google Scholar]
- Lachner M., O'Carroll N., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Liang K.W., Volk A.G., Haug J.S., Marshall S.A., Woodfin A.R., Bartom E.T., Gilmore J.M., Florens L., Washburn M.P., Sullivan K.D., et al. Therapeutic targeting of MLL degradation pathways in MLL-rearranged leukemia. Cell. 2017;168:59–72.e13. doi: 10.1016/j.cell.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.Y., Jan Y.J., Kuo L.K., Wang B.J., Huo C., Jiang S.S., Chen S.C., Kuo Y.Y., Chang C.R., Chuu C.P. Elevation of androgen receptor promotes prostate cancer metastasis by induction of epithelial-mesenchymal transition and reduction of KAT5. Cancer Sci. 2018;109:3564–3574. doi: 10.1111/cas.13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Malik R., Khan A.P., Asangani I.A., Cieslik M., Prensner J.R., Wang X.J., Iyer M.K., Jiang X., Borkin D., Escara-Wilke J., et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat. Med. 2015;21:344–352. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkar S., Thiel A., Hua X.X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem. Sci. 2013;38:394–402. doi: 10.1016/j.tibs.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoso A., Zhou Z.X., Hayama R., Flesken-Nikitin A., Nikitin A.Y. Cell lineage-specific interactions between Men1 and Rb in neuroendocrine neoplasia. Carcinogenesis. 2008;29:620–628. doi: 10.1093/carcin/bgm207. [DOI] [PubMed] [Google Scholar]
- Milne T.A., Hughes C.M., Lloyd R., Yang Z.H., Rozenblatt-Rosen O., Dou Y.L., Schnepp R.W., Krankel C., LiVolsi V.A., Gibbs D., et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Ayasolla K., Kumar V., Lan X.Q., Vashistha H., Aslam R., Hussain A., Chowdhary S., Shoshtari S.M., Paliwal N., et al. Modulation of apolipoprotein L1-microRNA-193a axis prevents podocyte dedifferentiation in high-glucose milieu. Am. J. Physiol. Renal Physiol. 2018;314:F832–F843. doi: 10.1152/ajprenal.00541.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler J.L., Antonarakis E.S., Armstrong A.J., D'Amico A.V., Davis B.J., Dorff T., Eastham J.A., Enke C.A., Farrington T.A., Higano C.S., et al. Prostate cancer, version 2.2019, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 2019;17:479–505. doi: 10.6004/jnccn.2019.0023. [DOI] [PubMed] [Google Scholar]
- Mullany S.A., Moslemi-Kebria M., Rattan R., Khurana A., Clayton A., Ota T., Mariani A., Podratz K.C., Chien J., Shridhar V. Expression and functional significance of HtrA1 loss in endometrial cancer. Clin. Cancer Res. 2011;17:427–436. doi: 10.1158/1078-0432.CCR-09-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R.C., Dou Y.L. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer. 2015;15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis S.T., Antunes A.A., Pontes J., de Sousa-Canavez J.M., Dall'Oglio M.F., Piantino C.B., da Cruz J.A.S., Morais D.R., Srougi M., Leite K.R.M. Underexpression of MMP-2 and its regulators, TIMP2, MT1-MMP and IL-8, is associated with prostate cancer. Int. Braz. J. Urol. 2012;38:167–174. doi: 10.1590/S1677-55382012000200004. [DOI] [PubMed] [Google Scholar]
- Ross J.S., Kaur P., Sheehan C.E., Fisher H.A.G., Kaufman R.A., Kallakury B.V.S. Prognostic significance of matrix metalloproteinase 2 and tissue inhibitor of metalloproteinase 2 expression in prostate cancer. Mod. Pathol. 2003;16:198–205. doi: 10.1097/01.MP.0000056984.62360.6C. [DOI] [PubMed] [Google Scholar]
- Shin M.H., He Y.L., Marrogi E., Piperdi S., Ren L., Khanna C., Gorlick R., Liu C.Y., Huang J. A RUNX2-mediated epigenetic regulation of the survival of p53 defective cancer cells. PLoS Genet. 2016;12:e1005884. doi: 10.1371/journal.pgen.1005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y.J., Kim J.H. The role of EZH2 in the regulation of the activity of matrix metalloproteinases in prostate cancer cells. PLoS One. 2012;7:e30393. doi: 10.1371/journal.pone.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slany R.K. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel A.T., Blessington P., Zou T., Feather D., Wu X.J., Yan J.Z., Zhang H., Liu Z.G., Ernst P., Koretzky G.A., et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel A.T., Feng Z.J., Pant D.K., Chodosh L.A., Hua X.X. The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBP alpha and differentiation in MLL-AF9 leukemia. Haematologica. 2013;98:918–927. doi: 10.3324/haematol.2012.074195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Mulligan L.M. Happy 20th anniversary MEN1: from positional cloning to gene function restoration. Endocr. Relat. Cancer. 2017;24:E7–E11. doi: 10.1530/ERC-17-0346. [DOI] [PubMed] [Google Scholar]
- Wu G.W., Yuan M.Q., Shen S.Q., Ma X.Y., Fang J.W., Zhu L.B., Sun L.C., Liu Z.J., He X.P., Huang D., et al. Menin enhances c-Myc-mediated transcription to promote cancer progression. Nat. Commun. 2017;8:15278. doi: 10.1038/ncomms15278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Doepner M., Hojnacki T., Feng Z.J., Katona B.W., He X., Ma J., Cao Y., Busino L., Zhou F.X., et al. Disruption of the menin-MLL interaction triggers menin protein degradation via ubiquitin-proteasome pathway. Am. J. Cancer Res. 2019;9:1682–1694. [PMC free article] [PubMed] [Google Scholar]
- Xiang Y.Z., Qiu Q.C., Jiang M., Jin R.J., Lehmann B.D., Strand D.W., Jovanovic B., DeGraff D.J., Zheng Y., Yousif D.A., et al. SPARCL1 suppresses metastasis in prostate cancer. Mol. Oncol. 2013;7:1019–1030. doi: 10.1016/j.molonc.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Li S.H., Zheng R., Gao S.B., Ding L.H., Yin Z.Y., Lin X., Feng Z.J., Zhang S., Wang X.M., et al. Menin promotes hepatocellular carcinogenesis and epigenetically up-regulates Yap1 transcription. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17480–17485. doi: 10.1073/pnas.1312022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Yue L.Y., Wang Y.L., Xing J., Chen Z.F., Shi Z., Liu R.F., Liu Y.C., Luo X.M., Jiang H.L., et al. Discovery of novel inhibitors targeting the menin-mixed lineage leukemia interface using pharmacophore- and docking-based virtual screening. J. Chem. Inf. Model. 2016;56:1847–1855. doi: 10.1021/acs.jcim.6b00185. [DOI] [PubMed] [Google Scholar]
- Yang Y.J., Song T.Y., Park J., Lee J., Lim J., Jang H., Kim Y.N., Yang J.H., Song Y., Choi A., et al. Menin mediates epigenetic regulation via histone H3 lysine 9 methylation. Cell Death Dis. 2013;4:e583. doi: 10.1038/cddis.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X.Q., Guo J.F., Guo J., Sun S., Yang P., Wang J.J., Li Y., Xie L.S., Cai J., Wang Z.H. EZH2-mediated epigenetic silencing of TIMP2 promotes ovarian cancer migration and invasion. Sci. Rep. 2017;7:3568. doi: 10.1038/s41598-017-03362-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A., Somervaille T.C.P., Smith K.S., Rozenblatt-Rosen O., Meyerson M., Cleary M.L. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Yokoyama A., Wang Z., Wysocka J., Sanyal M., Aufiero D.J., Kitabayashi I., Herr W., Cleary M.L. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Pan Y.Z., Cheung M., Cao M., Yu C., Chen L., Zhan L., He Z.W., Sun C.Y. LAMB3 mediates apoptotic, proliferative, invasive, and metastatic behaviors in pancreatic cancer by regulating the PI3K/Akt signaling pathway. Cell Death Dis. 2019;10:230. doi: 10.1038/s41419-019-1320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.