Abstract
HuD, an RNA binding protein, plays a role in the regulation of gene expression in certain types of cells, including neuronal cells and pancreatic β-cells, via RNA metabolism. Its aberrant expression is associated with the pathogenesis of several human diseases. To explore HuD-mediated gene regulation, stable cells expressing short hairpin RNA against HuD were established using mouse neuroblastoma Neuro2a (N2a) cells, which displayed enhanced phenotypic characteristics of cellular senescence. Two approaches, RNA immunoprecipitation (RNA IP)-NanoString profiling and cytokine array, were used to subsequently identify a subset of putative HuD targets that act as senescence-associated secretory phenotype (SASP), including C-C motif ligand 2 (CCL2), CCL20, C-X-C motif chemokine ligand 2 (CXCL2), and interleukin-6 (IL-6). Here, we further demonstrated that HuD regulates the expression of CCL2, a SASP candidate upregulated in cells following HuD knockdown, by binding to the 3′-untranslated region (UTR) of Ccl2 mRNA. Downregulation of HuD increased the level of CCL2 in N2a cells and the brain tissues of HuD knockout (KO) mice. Exposure to γ-irradiation induced cellular senescence in N2a cells and HuD knockdown facilitated stress-induced cellular senescence. Our results reveal that HuD acts as a novel regulator of CCL2 expression, and its aberrant expression may contribute to cellular senescence by regulating SASP production.
Subject terms: Senescence, RNA quality control
Introduction
HuD (also known as ELAVL4) is an RNA binding protein belonging to human antigen Hu/ELAVL family. It regulates gene expression at the post-transcriptional level by affecting multiple aspects of RNA metabolism, including stability, translation, alternative splicing, and localization of target mRNAs [1–4]. HuD is predominantly expressed in the brain and plays an important role in brain function; however, it also acts as a pivotal regulator of gene expression in certain types of endocrine cells such as β cells in the islet of pancreas and small cells in the lung (reviewed in [5]). Aberrant expression of HuD is implicated in the pathogenesis of several diseases. For example, several mutations in HuD gene are found in patients with Parkinson’s disease [6, 7]. A differential expression of HuD has been reported in Alzheimer’s disease [8, 9], amyotrophic lateral sclerosis [10], schizophrenia [11], pancreatic neuroendocrine tumor [12], and type 2 diabetes [13]. Abnormal phenotypes associated with neural development and synaptic plasticity have also been reported in transgenic or knockout mice against HuD [14, 15]. Identification of target mRNAs is essential to understand HuD-mediated gene regulation. Several studies have shown that HuD interacts with various types of RNAs, including mRNA, long non-coding RNA (lncRNA), and circular RNA (circRNA), and regulates dynamic networks of gene expression [16–19]. However, additional efforts in target identification and functional studies may expand our knowledge of the role of HuD in health and disease.
Senescence is originally defined as a stable cell cycle arrest based on the limited capacity for proliferation [20, 21]. Currently, cellular senescence is considered as a response to a wide range of intrinsic and extrinsic stimuli, including genotoxic stress, oxidative stress, mitochondrial dysfunction, oncogene activation, exposure to irradiation, or various reagents [22, 23]. Irreversible cell cycle arrest is a common feature of senescent cells, and is coordinated by several cell cycle regulators; however, certain types of permanently differentiated cells such as neurons, hepatocytes, and adipocytes, also exhibit senescence-like phenotype, which indicates that senescence can occur independent of cell cycle arrest [22].
Senescent cells influence their surrounding environment by generating a senescence-associated secretory phenotype (SASP) [24]. The SASP consists of dynamic and heterogeneous components including cytokines, chemokines, extracellular matrix metalloproteases (MMPs), growth regulators, and angiogenic factors that initiate inflammation, wound healing, and growth responses in nearby cells [25–27]. Recent studies have extensively demonstrated the key role of SASP in various pathophysiological phenomena, including tumorigenesis, angiogenesis, and inflammation, thereby contributing to aging and diseases [23, 28]. Despite the biological significance of SASP, the intracellular signal networks regulating initiation and development of SASP, and the pathological relevance of its aberrant expression are not fully understood.
Herein, we proposed HuD as a novel factor regulating SASP expression in mouse neuroblastoma N2a cells. We found that HuD knockdown increased the levels of senescence-associated β-galactosidase, p16INK4a, and reactive oxygen species (ROS), and altered the profile of secreted proteins in the media of N2a cells. In this study, we sought to further identify novel targets of HuD and elucidate HuD-mediated regulatory mechanisms that potentially affect cellular senescence by modifying the microenvironment. Two approaches, RNA immunoprecipitation (RNA IP)-NanoString profiling and cytokine array, were used to subsequently identify a subset of putative HuD targets that function as SASP, including C-C motif ligand 2 (CCL2), CCL20, C-X-C motif chemokine ligand 2 (CXCL2), and interleukin-6 (IL-6). We further demonstrated that HuD regulates the expression of CCL2, a SASP candidate upregulated in HuD knockdown cells, by binding to the 3′-untranslated region (UTR) of Ccl2 mRNA. Downregulation of HuD not only increased the level of CCL2 in neuroblastoma cells and the brain of HuD knockout (KO) mice, but also increased the susceptibility of cells to stress-induced cellular senescence. Our data reveal that HuD acts as a novel regulator of CCL2 expression, and its aberrant expression may contribute to cellular senescence by regulating SASP production.
Results
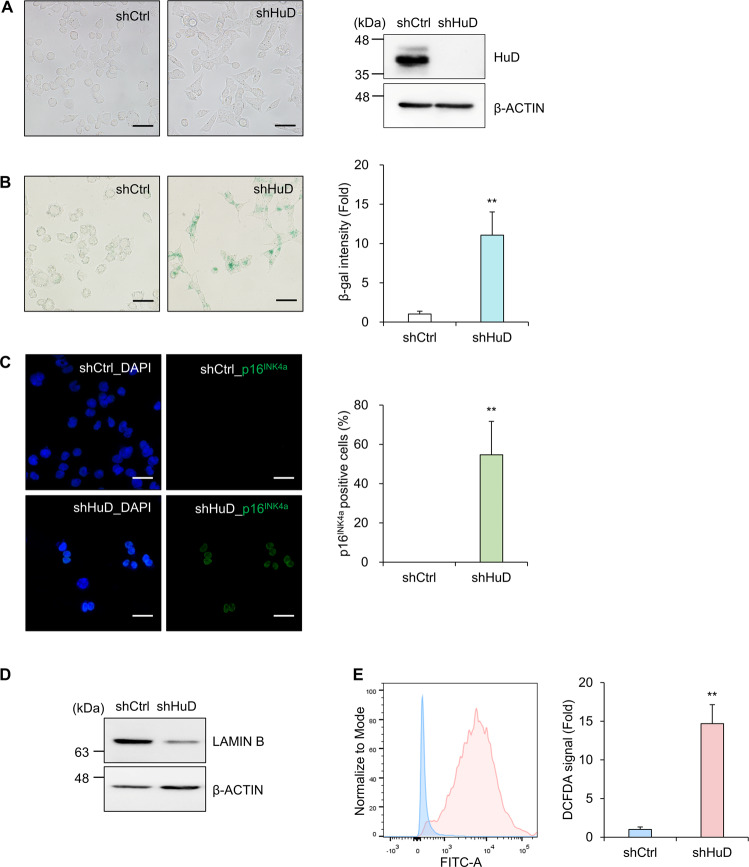
To identify the target mRNAs of HuD and explore HuD-mediated gene regulatory mechanisms, stable N2a cell lines expressing short hairpin RNA (shRNA) against HuD (shHuD) or control shRNA (shCtrl) were established via continuous selection using puromycin (Fig. 1A). Knockdown of HuD in these cell lines was confirmed by western blotting analysis as shown in Fig. 1A. N2a cells expressing shHuD plasmid (N2a_shHuD) were larger and flatter than control cells (N2a_shCtrl) and expressed higher levels of senescence-associated β-galactosidase (SA β-gal) (Fig. 1B). The number of p16INK4a-positive cells increased and the level of LAMIN B decreased in N2a_shHuD cells compared with N2a_shCtrl cells (Fig. 1C, D). In addition, N2a_shHuD cells carried higher levels of ROS than N2a_shCtrl cells (Fig. 1E). These results indicate that N2a_shHuD cells represent senescent phenotypes. Senescent cells are known to secrete various molecules, designated as SASP, and to affect gene expression through autocrine or paracrine pathways [24, 26]. Since HuD is one of pivotal factors in the post-transcriptional control of gene expression, we hypothesized that loss of HuD regulates the expression of SASP factors.
Fig. 1. Enhanced cellular senescence mediated via HuD downregulation.
A Stable cells expressing short hairpin RNA against HuD (shHuD) or control RNA (shCtrl) were established using mouse neuroblastoma Neuro2A (N2a) cells. Downregulation of HuD was assessed via western blotting analysis. β-ACTIN was used as a loading control. B Cells were stained with senescence-associated β-galactosidase (SA β-gal). The relative intensity of SA β-gal was assessed via densitometric analysis using Image J. C The level of p16INK4a was investigated via immunofluorescence microscopy and quantified by counting the number of p16INK4a-expressing cells. Green: p16INK4a, blue: nuclei. D The level of LAMIN B was assessed via western blotting analysis. β-ACTIN used as a loading control. E Cells were incubated with DCFDA, a fluorescent indicator of ROS and cellular ROS levels between N2a_shCtrl and N2a_shHuD cells were determined using flow cytometry. Images are representative, and data indicate the mean ± SEM from three independent analyses. Scale bar, 50 μm. The statistical significance of the data was analyzed via Student’s t-test; **p < 0.01.
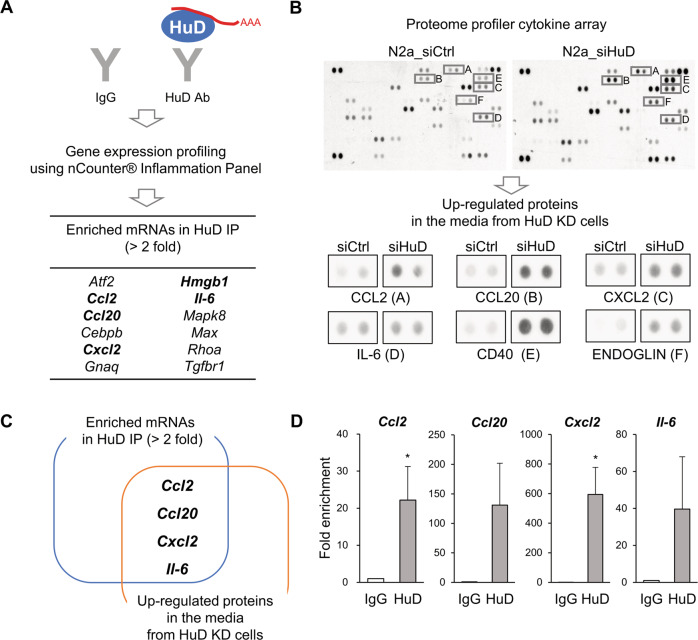
In this study, RNA IP-NanoString profiling and cytokine array were used to investigate the role of HuD in the expression of SASP in N2a cells. First, HuD-associating mRNAs were isolated from the RNP complexes containing HuD and analyzed via gene expression profiling using the NanoString nCounter® Inflammation Panel. Enriched mRNAs in HuD IPs were identified as putative target mRNAs of HuD and shown in Fig. 2A. In addition, the differential expression of secretory proteins affected by HuD was determined using the Proteome Profiler Mouse XL Cytokine Array (ARY028). Conditioned media were collected from N2a cells after transfection of either control siRNA or HuD siRNA and the secretory proteins were captured on nitrocellulose membrane and detected with biotinylated detection antibodies and visualized using chemiluminescence reagent according to the manufacturer’s instruction. Upregulated proteins in HuD knockdown cells are shown in Fig. 2B.
Fig. 2. Identification of molecular targets of HuD.
A The mRNAs that interact with HuD were isolated via RNA immunoprecipitation and analyzed via gene expression profiling using the NanoString nCounter® Inflammation Panel. Enriched mRNAs in HuD IP (>2-fold) compared to control IgG IP are listed in the table and senescence-associated secretory phenotype (SASP) mRNAs are shown in bold. B N2a cells were transiently transfected with control siRNA or HuD siRNA and the secretory proteins in the culture media were analyzed by western blotting using the Proteome Profiler Mouse XL cytokine array. The differential expression analysis of secretory proteins revealed six proteins, including CCL2, CCL20, CXCL2, IL-6, CD40, and ENDOGLIN, which were identified as upregulated ones in the media from HuD knockdown cells. C Four candidates that not only bind to HuD but also are upregulated by HuD knockdown were determined as putative targets of HuD; Ccl2, Ccl20, Cxcl2, and Il-6. D Interaction between HuD and its putative targets was validated by RNA IP followed by RT-qPCR using anti-HuD and control IgG antibodies. Gapdh mRNA was used for normalization. Data indicate the mean ± SEM from three independent analyses. The statistical significance of the data was analyzed via Student’s t-test; *p < 0.05.
These assays revealed four common putative candidates for HuD, including CCL2, CCL20, CXCL2, and IL-6, which are involved in chronic inflammatory response (Fig. 2C). The association between HuD and these target mRNAs was experimentally validated via RNA IP analysis followed by quantitative reverse transcription PCR (RT-qPCR) (Fig. 2D). Taken together, these results suggest the possibility that HuD may play a role in regulating the expression of these secretory proteins known as SASP factors. We further investigated whether HuD regulates the expression of CCL2, one of putative targets, in this study.
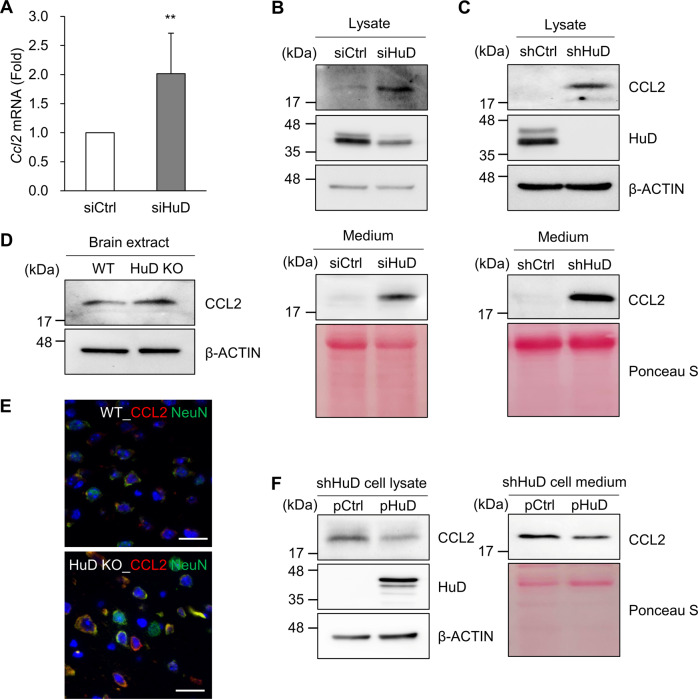
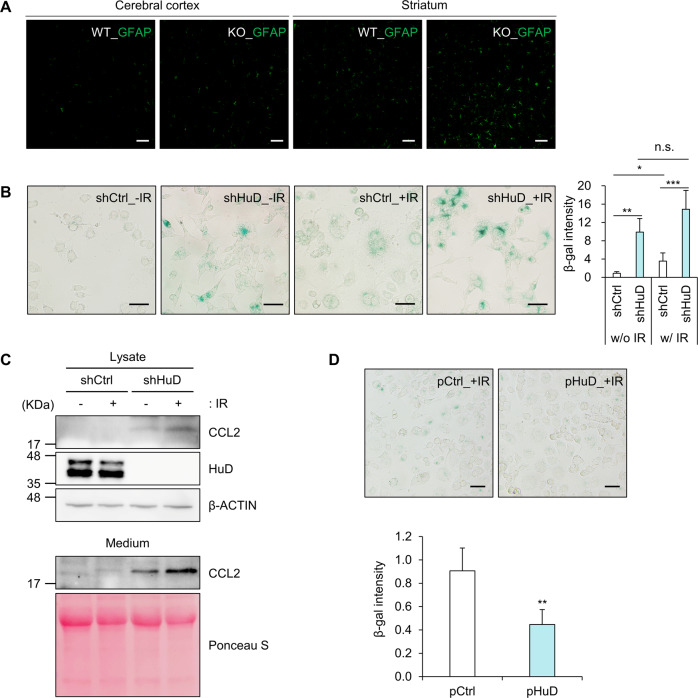
To determine whether HuD regulates CCL2 expression, the levels of Ccl2 mRNA and protein were assessed in N2a cells transfected with siRNAs. HuD knockdown moderately increased Ccl2 mRNA (Fig. 3A) and CCL2 protein in both lysates and media (Fig. 3B). The expression of CCL2 was also augmented in both lysates and media of stable N2a_shHuD cells (Fig. 3C), which indicates that HuD downregulation increases CCL2 expression in N2a cells. An increase of CCL2 by HuD knockdown was also observed in human neuroblastoma SH-SY5Y cells (Supplementary Fig. S1). In addition, the relative CCL2 level was investigated in the brain of HuD knockout (KO) mice or age-matched control brain via western blotting analysis and immunofluorescence microscopy. The results showed that CCL2 expression was increased in the brain of HuD KO mice compared with WT brain (Fig. 3D, E). Fluorescence for CCL2 was predominantly found in NeuN-positive (neuronal) cells and was also observed in some of GFAP-positive (astrocyte), OLIG2-positive (oligodendrocyte), or CD68-positive (macrophage) cells (Supplementary Fig. S2), which suggests that CCL2 increased in several types of cells in the brain of KO mice. To further determine whether HuD regulates CCL2 expression, the HuD level was restored in N2a_shHuD cells via ectopic expression of HuD and the relative expression of CCL2 was investigated. As shown in Fig. 3F, HuD overexpression downregulated CCL2 levels in both lysates and media of N2a_shHuD cells. Together, these results suggest that HuD plays a negative role in the regulation of CCL2 expression.
Fig. 3. Augmented expression of CCL2 by HuD knockdown.
A Following the transfection of N2a cells with either control siRNA or HuD siRNA, the expression of Ccl2 mRNA was determined via RT-qPCR. Gapdh mRNA was used for normalization. B, C The relative levels of CCL2 protein in both lysates (top) and culture media (bottom) were assessed via western blotting analysis in siRNA transfected cells (B) and stable cells (C). D, E Endogenous CCL2 levels in the brain of HuD KO mice and age-matched WT mice were assessed via western blotting analysis (D) and immunofluorescence microscopy (E). n = 3, Red: CCL2, green: NeuN, blue: nuclei. Scale bar, 20 μm. F N2a_shHuD cells were transiently transfected with plasmids (pHuD or control plasmid) and CCL2 levels in both lysates and media were determined by western blotting analysis. β-ACTIN blot and Ponceau S staining were used as loading controls. Images are representative, and data indicate the mean ± SEM from three independent experiments. The statistical significance of the data was analyzed via Student’s t-test; **p < 0.01.
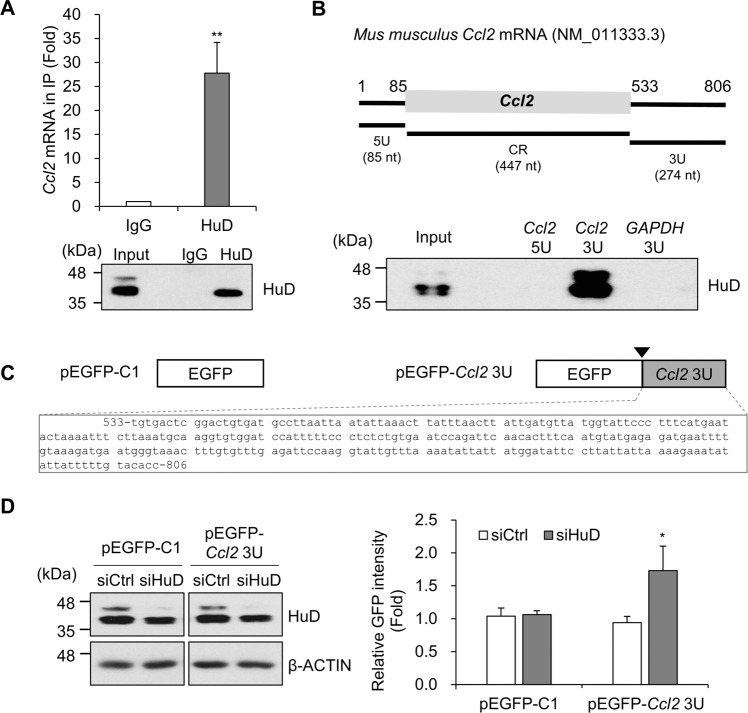
HuD is an RNA binding protein (RBP) known to interact with target mRNAs and affect their expressions [2, 5]. To understand HuD-mediated regulation of CCL2 expression, the association between HuD and Ccl2 mRNA was assessed by RNA IP analysis followed by RT-qPCR. Ccl2 mRNA was enriched in HuD IP (Fig. 4A), which indicates that the HuD-containing RNP complex binds to Ccl2 mRNA. Since RBPs usually affect the expression of target mRNAs via binding to their UTRs, the association between HuD and UTRs of Ccl2 mRNA was investigated by the pull-down assay using biotin-labeled probes and the results showed that HuD binds to 3′UTR of Ccl2 mRNA (Fig. 4B). To further confirm whether HuD regulates CCL2 expression by binding to its 3′UTR, the EGFP reporter plasmid containing the sequence of Ccl2 3′UTR (533–806 nt, pEGFP-Ccl2 3U) behind the coding region of EGFP was generated (Fig. 4C). The relative EGFP expression was analyzed after transfection of siHuD or control siRNA. As shown in Fig. 4D, HuD knockdown enhanced the fluorescence of EGFP-Ccl2 3U reporter plasmid, but not the level of EGFP control. These results suggest that HuD binds to 3′UTR of Ccl2 mRNA and downregulates its expression.
Fig. 4. Interaction between HuD and 3′UTR of Ccl2 mRNA.
A The interaction between HuD and Ccl2 mRNA was confirmed by RNA IP followed by RT-qPCR using anti-HuD and control IgG antibodies. Gapdh mRNA was used for normalization. B Top: A schematic of mouse Ccl2 mRNA (NM_011333.3). The UTRs of Ccl2 mRNA (5U and 3U) were transcribed in vitro using T7 RNA polymerase and biotin-labeled nucleotides. Bottom: The biotinylated transcripts (Ccl2 5U, Ccl2 3U, and GAPDH 3U) were incubated with the lysate from N2a cells. The proteins binding to the transcripts were pulled-down using streptavidin magnetic beads and analyzed via western blotting using HuD antibody. Biotinylated GAPDH 3U was used as a negative control. C Schematic diagram of EGFP reporters. The reporter plasmid (pEGFP-Ccl2 3U) was constructed by inserting the 3′UTR of Ccl2 mRNA (533–806 nt) into the pEGFP-C1. (▼: stop codon). D After sequential transfection with siRNAs and EGFP reporter plasmids, relative fluorescence of EGFP from each sample was measured by fluorescence microscopy and the levels of HuD and β-ACTIN were assessed by western blotting analysis. Images are representative, and data indicate the mean ± SEM from three independent experiments. The statistical significance of the data was analyzed via Student’s t-test; *p < 0.05; **p < 0.01.
CCL2 mediates astrocyte activation and promotes neuroinflammation during brain injury [29, 30]. Since augmented level of CCL2 was observed in the brain of HuD KO mice (Fig. 3D, E), astrocyte activation in the same brain tissues was assessed by immunofluorescence staining using an antibody to glial fibrillary acidic protein (GFAP), a marker for astrocytes. The intensity of GFAP-positive signal was enhanced in both regions of cerebral cortex and striatum in HuD KO mice (Fig. 5A), indicating enhanced astrocyte activation leading to altered brain microenvironment in HuD KO mice. This result suggests that HuD alters the brain microenvironment by regulating the expression of SASP molecules including CCL2 in HuD-expressing cells such as neurons.
Fig. 5. Sensitization of N2a cells via HuD knockdown in response to senescence inducer.
A Relative level of GFAP, an astrocyte marker, was analyzed in the cerebral cortex and striatum regions in the brain of HuD KO mice and their age-matched control mice by immunofluorescence microscopy. n = 3. B, C After exposure to γ-irradiation (5.5 Gy), the levels of SA β-gal (B) and CCL2 expression (C) were analyzed by β-gal staining and western blotting analysis. D N2a cells were transiently transfected with plasmids and exposed to γ-irradiation. The levels of SA β-gal were assessed by β-gal staining and densitometric analysis using Image J. Scale bar, 50 μm. Images are representative, and data indicate the mean ± SEM derived from three independent experiments. The statistical significance of the data was analyzed via Student’s t-test; *p < 0.05; **p < 0.01; ***p < 0.001; n.s., not significant.
The relative level of SA β-gal was elevated in N2a_shHuD cells compared to normal cells (Fig. 1B). This result led us to hypothesize that HuD downregulation promotes cellular senescence in response to extracellular stimuli. To determine whether N2a_shHuD cells are more sensitive to stress-induced cellular senescence, cells are exposed to γ-irradiation (IR), an inducer of senescence, and then relative levels of SA β-gal and CCL2 between N2a_shHuD cells and control cells were determined. In N2a_shHuD group, the intensity of SA β-gal as well as the expression of CCL2 were higher than in the control group after IR exposure (Fig. 5B, C), which suggests that N2a_shHuD cells were more vulnerable to IR. Transient knockdown of HuD also increased the expression of SA β-gal (Supplementary Fig. S3), while HuD overexpression decreased it (Fig. 5D) after exposure to IR. Taken together, these data suggest that HuD acts as a negative regulator of CCL2 expression and downregulation of HuD contributes to facilitating γ-irradiation-induced SA β-gal production.
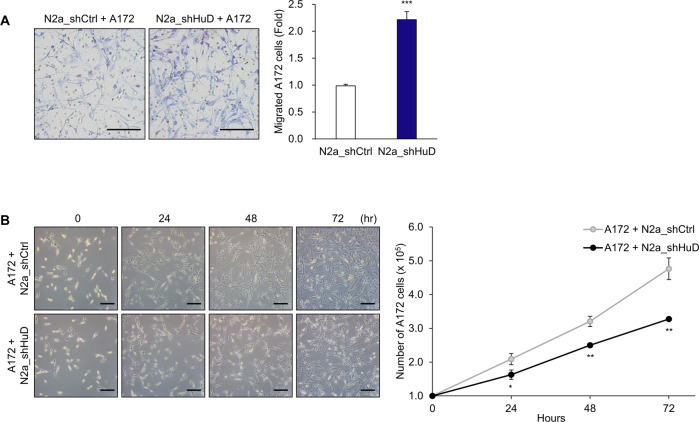
To further investigate whether HuD-mediated CCL2 regulation affects to neighbor cells via paracrine effects, A172 cells were co-cultured with N2a cells and the migration and growth of A172 cells were assessed by the transwell migration assay and cell counting. As shown in Fig. 6, co-culture with N2a_shHuD cells promoted migration of A172 cells (Fig. 6A) and decreased their growth (Fig. 6B) compared to co-culture with N2a_shCtrl cells. These data suggest that HuD has a potential to affect the migration and growth of nearby cells.
Fig. 6. Alterations in migration and growth of A172 cells co-cultured with N2a_shHuD cells.
A A172 cells (upper chamber) and N2a cells (lower chambers) were co-cultured for 48 h and migration of A172 cells were determined by staining of migrated cells on the transwell membrane. B A172 cells (lower chamber) and N2a cells (upper chambers) were co-cultured and the number of A172 cells was analyzed by cell counting at each time point. Images are representative, and data indicate the mean ± SEM derived from three independent experiments. Scale bar, 200 μm. The statistical significance of the data was analyzed via Student’s t-test; *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
HuD is an RBP belonging to Hu antigen family and acts as a pivotal regulator of gene expression at the post-transcriptional level [2, 5]. Differential expression of HuD protein or mutations involving HuD gene is implicated in the pathogenesis of several diseases such as Alzheimer’s disease, Parkinson’s disease, cancer, and diabetes [6, 8, 9, 12, 31]. Although HuD has been recognized as an essential factor regulating RNA metabolism, the detailed mechanisms of its expression, molecular targets, and disease relevance remain to be further elucidated. In this study, we established stable N2a cells expressing shRNA against HuD to explore HuD-mediated gene regulation and observed enhanced levels of SA β-gal and ROS in HuD knockdown cells. We identified an SASP molecule, CCL2, as a novel target of HuD and demonstrated that HuD negatively regulated CCL2 expression by binding to its 3′UTR. HuD knockdown increased CCL2 expression and the level of SA β-gal in response to irradiation. In addition, we showed HuD-mediated CCL2 regulation affects the migration and growth of A172 cells via paracrine effects. These results suggest that HuD regulates the expression of SASP molecules, including CCL2, and downregulation of HuD sensitizes the cells to extracellular stimuli, thereby leading to cellular senescence.
Several phenotypic characteristics of senescence were observed, including accumulation of SA β-gal and p16INK4a, increased production of ROS, and downregulation of LAMIN B, in N2a cells expressing shRNA against HuD (Fig. 1). In addition, we showed that HuD altered the expression of several secreted proteins such as CCL2, CCL20, IL-6, and CXCL2, suggesting a novel role of HuD in regulating the SASP expression. These results suggest the possibility that HuD not only regulates intracellular gene expression but also mediates cell-to-cell communication. Although downregulation of HuD has been shown in senescent astrocytes [32] using the SeneQuest (http://Senequest.net) provided by the International Cell Senescence Association (ICSA) [23], HuD expression during cellular senescence remains to be elucidated. We observed no significant changes in HuD level after short-term exposure to irradiation (data not shown). However, a follow-up study investigating the changes in HuD expression in response to senescence inducers may provide an important clue to understand the HuD-mediated cell-to-cell crosstalk during cellular senescence.
We previously reported that HuD knockdown induces mitochondrial dysfunction (enhanced mitochondrial fission, reductions in ATP synthesis, mitochondrial membrane potential, and oxygen consumption rate) by regulating Mfn2 mRNA translation [31]. In addition, several recent articles demonstrate that mitochondrial dysfunction increases SASP production and induces chronic inflammation and/or cellular senescence [33–36]. These suggest that mitochondrial dysfunction is one of important factors facilitating cellular senescence. However, activation of DNA damage response, increased intracellular ROS level, and enhanced oxidative stress response observed in N2a_shHuD cells (data not shown) may act as the positive feedback mechanism in accelerating cellular senescence. Since it is not easy to completely define the role of HuD in the causal relationship between cellular senescence and SASP generation, in this study, we tried to focus on the HuD-mediated SASP regulation. Further studies may be enabled to elucidate a fine mechanism of HuD in the regulation of cellular senescence and age-related diseases.
CCL2 is a proinflammatory chemokine that facilitates the accumulation of immune cells at inflammatory sites [37]. The level of CCL2 is upregulated in several tumors and correlates with poor prognosis of patients with cancers of breast, stomach, and liver [38–40]. CCL2-overexpressing mice exhibit metabolic dysregulation and premature death with accelerating aging [41, 42]. In addition, CCL2 promotes the activation of astrocytes and microglia in the brain [29, 30]. Therefore, understanding of the detailed mechanism of CCL2 expression is important for the development of therapeutic strategies targeting inflammatory response. CCL2 expression is largely controlled via the transcription factor nuclear factor κ B (NFκB) signaling; however, other factors, such as p53 and forkhead box K1 (FOXK1), also regulate the transcription of Ccl2 [43–45]. Several microRNAs, including miR-33, miR-124, miR-206, and miR-374, negatively regulate CCL2 expression [46–49], while HuR positively regulates it in response to tumor necrosis factor α (TNFα) [50]. Here, we demonstrated that HuD knockdown upregulated CCL2 expression in both mouse neuroblastoma N2a cells and human neuroblastoma SH-SY5Y cells, suggesting a potential role of HuD as a negative regulator of CCL2 expression (Figs. 3 and S1). Although we observed a moderate, but significant, increase in Ccl2 mRNA level via HuD knockdown (Fig. 3A), the detailed mechanism underlying the decreased expression of CCL2 mediated via HuD needs to be further elucidated.
Several RBPs, including HuD, AU-rich element protein 1 (AUF1), tristetraprolin (TTP), T cell intracellular antigen 1 (TIA1), WIG1, and CUG-binding protein 1 (CUGBP1), are implicated in the regulation of cellular senescence (reviewed in [51]). Loss of HuR has been reported to promote cellular senescence in several cell types, suggesting that HuR plays a suppressive role in senescence [52–56]. Cheng et al. [57] recently reported the roles of HuB and HuD in the regulation of telomerase activity. Interestingly, downregulation of HuB and HuD together increased cell growth and delayed cellular senescence of human neuroblastoma SH-SY5Y cells. Here, we demonstrated that knockdown of HuD promoted cellular senescence and SASP expression in mouse neuroblastoma N2a cells. We did not investigate whether telomerase activity was altered by HuD downregulation or whether HuD regulates SASP expression cooperatively or competitively with other Hu family proteins in our system. Therefore, it is difficult to determine the cause of this difference in senescent phenotypes after HuD regulation. Additional experiments are required to fully understand the regulatory mechanisms of gene expression by Hu family of proteins among different cell types or species.
In conclusion, we propose a novel function of HuD in the regulation of SASP expression. We demonstrated that downregulation of HuD increased the level of SASPs, such as CCL2, CCL20, CXCL2, and IL-6, and sensitized the cells to senescence inducers, thereby promoting cellular senescence. Our results suggest that HuD acts as a novel regulator of CCL2 expression, and its aberrant expression induces cellular senescence by altering SASP production. Further studies investigating the relative level of HuD in response to senescence inducers altered cell-to-cell communication mediated by SASPs in HuD-deficient models, and their underlying mechanisms may provide additional insight into HuD-mediated gene regulation during cellular senescence or neuroinflammation.
Materials and methods
Cell culture, transfection of plasmids and small interfering RNAs
Mouse neuroblastoma Neuro2a (N2a) and human glioblastoma A172 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) and RPMI 1640 (Capricorn Scientific, Ebsdorfergrund, Germany) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics and incubated at 37 °C in the presence of 5% CO2. Stable cells expressing shRNAs were established by transfection of shHuD plasmid (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or control plasmid under puromycin (Invitrogen™, Waltham, MA, USA) selection. Enhanced green fluorescent protein (EGFP) reporter was prepared by cloning the 3′UTR sequence of Ccl2 mRNA (533–806, 274 nt) into the pEGFP-C1 (BD Bioscience, Franklin Lakes, NJ, USA) vector. HuD overexpression plasmids (pHuD) were received as a gift from Prof. Alessandro Quattrone [18]. Transfection of small interfering RNAs (HuD siRNA (siHuD) and control siRNA (siCtrl)) (Genolution Pharmaceuticals, Inc., Seoul, South Korea) or plasmids were achieved using Lipofectamine™ 2000 (Invitrogen™) according to the manufacturer’s instructions.
RNA analysis
Total RNA was isolated from whole cells using RNAiso Plus (Takara Bio, Inc., Shiga, Japan) and cDNA was synthesized by reverse transcription using ReverTra® Ace qPCR RT kit (Toyobo Co., Ltd, Osaka, Japan). The level of transcripts was determined via quantitative PCR (qPCR) using the SensiFAST™ SYBR Hi-ROX kit (Meridian Bioscience, Inc., Cincinnati, OH, USA), gene-specific primers (Supplementary Table S1), and StepOnePlus™ Real-Time PCR System (Applied Biosystems™, Waltham, MA, USA). Data were processed using the ΔΔCT method for comparison between control and experimental groups. Gapdh mRNA was used for normalization.
For RNA immunoprecipitation, ribonucleoprotein (RNP) complexes were immunoprecipitated from the cell lysates using Protein A bead (Invitrogen™) incubated with anti-HuD or control IgG antibody (Santa Cruz Biotechnology, Inc.) [58]. The immunoprecipitated RNP complexes were sequentially incubated with DNase I and proteinase K. The isolated RNAs from the complexes were used for further analysis including RT-qPCR and gene profiling using the nCounter® Analysis Systems (NanoString Technologies, Inc., Seattle, WA, USA). Gene expression profiling in the complexes was accomplished by using the nCounter® Inflammation Panel. Data were analyzed using nSolver software according to the instruction.
Biotin pull down assay
To synthesize biotinylated transcripts, DNA fragments corresponding to the 5′UTR and 3′UTR of Ccl2 mRNA (NM_011333.3) were generated using forward primers containing T7 RNA polymerase binding sequence (5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′). After purification of the PCR products, biotinylated transcripts were synthesized using the MaxiScript T7 kit (Invitrogen™) and biotin-CTP (Enzo Life Sciences, Inc., Farmingdale, NY, USA). Cell lysates were incubated with the purified biotinylated transcripts for 30 min at room temperature. The RNA–protein complexes were isolated using streptavidin-coupled Dynabeads (Invitrogen™). Proteins were isolated from the complex and subjected to western blotting analysis using HuD antibody [58]. 3′UTR of Gapdh mRNA was used as a negative control for the binding assay.
Western blotting analysis
Whole-cell lysates were prepared using RIPA buffer (Biosesang, Inc., Seongnam, South Korea) containing protease inhibitor cocktail (Roche, Basel, Switzerland). The samples were separated by SDS polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Burlington, MA, USA), incubated with primary antibodies including CCL2 (Abcam Plc., Cambridge, UK), HuD, LAMIN B, GFP (Santa Cruz Biotechnology, Inc.), and β-ACTIN (Genetex, Inc., Irvine, CA, USA) at 4 °C overnight, and further incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Sigma-Aldrich, Burlington, MA, USA). Chemiluminescence was detected with the Clarity Western ECL Substrate (Bio-Rad, Inc., Hercules, CA, USA) using the ChemiDoc Imaging Systems (Bio-Rad, Inc.).
Cytokine array
Conditioned media collected from the cell culture were centrifuged to remove cell debris and used for the analysis of secretory proteins. Cytokine array was performed using the Proteome Profiler Mouse XL Cytokine Array Kit (Cat. No. ARY028, R&D Systems, Inc., Minneapolis, MN, USA) according to the manufacturer’s instructions. In brief, after transfection of N2a cells with siCtrl or siHuD, the conditioned media from each cell were collected to 50 mL tube and concentrated by centrifugation using Amicon® Ultracentrifugal filter (Millipore). The concentrated media were incubated the nitrocellulose membrane containing 111 different anti-mouse cytokine antibodies from the Mouse XL Cytokine Array kit. Captured proteins on the membranes were further incubated with detection antibodies and visualized using chemiluminescent detection reagents.
Measurement of reactive oxygen species
The intracellular ROS level was determined using a general oxidative stress indicator, CM-H2DCFDA (DCFDA) (Invitrogen™). Cells were incubated with DCFDA reagent at 37 °C for 30 min and washed with Hank’s balanced salt solution (HBSS) (Gibco™, Waltham, MA, USA). The fluorescence signal was analyzed by detecting the DCF intensity using the FACSCanto™ II Flow Cytometry System (BD Bioscience).
Cell staining and fluorescence microscopy
Cells were fixed with 4% FA solution for immunofluorescence microscopy and brain tissues embedded with paraffin (50 weeks, female mice) were deparaffinized for fluorescence immunohistochemistry. After permeabilization with Triton X-100, cells were sequentially incubated with blocking solution and primary antibodies raised against p16INK4a (Santa Cruz Biotechnology, Inc.), CCL2 (Abcam Plc.), NeuN (Abcam Plc.) and GFAP (Sigma-Aldrich) at 4 °C overnight, and further incubated with secondary antibodies conjugated with Alexa Flour® 488 or Alexa Flour® 555 (Abcam Plc.). DAPI (4′,6-diamidino-2-phenylindole) solution (Invitrogen™) was used to stain the nuclei. Fluorescence signals were observed and imaged using the ZEISS Axio Imager M1 microscope (Carl Zeiss, Oberkochen, Germany).
For detection of senescence-associated β-galactosidase (SA β-gal), cells were fixed with 4% FA and incubated with the SA β-gal solution containing 40 mM citric acid/sodium phosphate (pH 6.0), 150 mM NaCl, 2 mM MgCl2, 5 mM potassium ferricyanide and potassium ferrocyanide, and 1 mg/mL X-gal (BEAMS Biotechnology, Seongnam, South Korea) at 37 °C for 16 h in the dark. After washing with PBS, stained cells were observed under the IX70w microscope (Olympus Corp., Tokyo, Japan). The intensity of β-gal was derived by dividing the value of β-gal expression quantified using ImageJ software [59], by the total number of cells.
Transwell migration assay and cell counting
Transwell migration assay was performed using a Falcon® Permeable Support for 24-well Plate with 8.0 μm Transparent Membrane (Corning Inc., NY, USA). N2a shCtrl and shHuD cells were cultured in a lower chamber, while A172 cells were added in an upper chamber. After incubating A172 cells with RPMI media containing 1% FBS for 48 h, the membrane was stained with a Diff Quit kit (Sysmex Asia Pacific Pte Ltd, Singapore) and observed under a ZEISS Axio Imager A1 microscope (Carl Zeiss). For cell counting assay, A172 cells were seeded in the lower chamber of a Falcon® 6-well TC-treated Polystyrene Permeable Support Companion Plate and N2a cells were added in upper chambers. The number of A172 cells were determined by cell counting with a hemocytometer under the Leica DM IL LED microscope (Leica Microsystems Ltd, Wetzlar, Germany).
Statistical analysis
Data were expressed as mean ± SEM of three independent experiments. The statistical significance of the data was analyzed via Student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgments
The authors appreciate SY Yang for her technical assistance and Professor JH Lee for providing A172 cells.
Author contributions
SR and EKL conceived and designed the experiments. SR, MJ, CK, HK, SH, and SC performed the experiments and analyzed the data. SMJ and EKL discussed the experiments and provided advice. SR and EKL wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the Basic Science Research Programs through the National Research Foundation of Korea (NRF) funded by the Minister of Education and the Minister of Science and ICT (2020R1F1A1048425 and 2021R1A2C1004128).
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data may be available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by Professor Massimiliano Agostini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-022-04792-y.
References
- 1.Bronicki LM, Jasmin BJ. Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. RNA. 2013;19:1019–37. doi: 10.1261/rna.039164.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perrone-Bizzozero N, Bird CW. Role of HuD in nervous system function and pathology. Front Biosci (Sch Ed) 2013;5:554–63. doi: 10.2741/S389. [DOI] [PubMed] [Google Scholar]
- 3.Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, et al. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–33. doi: 10.1016/0092-8674(91)90184-Z. [DOI] [PubMed] [Google Scholar]
- 4.Simone LE, Keene JD. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr Opin Genet Dev. 2013;23:35–43. doi: 10.1016/j.gde.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung M, Lee EK. RNA-binding protein hud as a versatile factor in neuronal and non-neuronal systems. Biology (Basel) 2021;10:361. doi: 10.3390/biology10050361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noureddine MA, Qin XJ, Oliveira SA, Skelly TJ, van der Walt J, Hauser MA, et al. Association between the neuron-specific RNA-binding protein ELAVL4 and Parkinson disease. Hum Genet. 2005;117:27–33. doi: 10.1007/s00439-005-1259-2. [DOI] [PubMed] [Google Scholar]
- 7.DeStefano AL, Latourelle J, Lew MF, Suchowersky O, Klein C, Golbe LI, et al. Replication of association between ELAVL4 and Parkinson disease: the GenePD study. Hum Genet. 2008;124:95–99. doi: 10.1007/s00439-008-0526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subhadra B, Schaller K, Seeds NW. Neuroserpin up-regulation in the Alzheimer’s disease brain is associated with elevated thyroid hormone receptor-beta1 and HuD expression. Neurochem Int. 2013;63:476–81. doi: 10.1016/j.neuint.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang MJ, Abdelmohsen K, Hutchison ER, Mitchell SJ, Grammatikakis I, Guo R, et al. HuD regulates coding and noncoding RNA to induce APP->Abeta processing. Cell Rep. 2014;7:1401–9. doi: 10.1016/j.celrep.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dell’Orco M, Sardone V, Gardiner AS, Pansarasa O, Bordoni M, Perrone-Bizzozero NI, et al. HuD regulates SOD1 expression during oxidative stress in differentiated neuroblastoma cells and sporadic ALS motor cortex. Neurobiol Dis. 2021;148:105211. doi: 10.1016/j.nbd.2020.105211. [DOI] [PubMed] [Google Scholar]
- 11.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–51. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim C, Jeong DE, Heo S, Ji E, Rho JG, Jung M, et al. Reduced expression of the RNA-binding protein HuD in pancreatic neuroendocrine tumors correlates with low p27(Kip1) levels and poor prognosis. J Pathol. 2018;246:231–43. doi: 10.1002/path.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C, Lee H, Kang H, Shin JJ, Tak H, Kim W, et al. RNA-binding protein HuD reduces triglyceride production in pancreatic beta cells by enhancing the expression of insulin-induced gene 1. Biochim Biophys Acta. 2016;1859:675–85. doi: 10.1016/j.bbagrm.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci USA. 2005;102:4625–30. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolognani F, Tanner DC, Merhege M, Deschenes-Furry J, Jasmin B, Perrone-Bizzozero NI. In vivo post-transcriptional regulation of GAP-43 mRNA by overexpression of the RNA-binding protein HuD. J Neurochem. 2006;96:790–801. doi: 10.1111/j.1471-4159.2005.03607.x. [DOI] [PubMed] [Google Scholar]
- 16.Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010;38:117–30. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheckel C, Drapeau E, Frias MA, Park CY, Fak J, Zucker-Scharff I, et al. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. Elife. 2016;5:e10421. doi: 10.7554/eLife.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tebaldi T, Zuccotti P, Peroni D, Kohn M, Gasperini L, Potrich V, et al. HuD is a neural translation enhancer acting on mtorc1-responsive genes and counteracted by the Y3 small non-coding RNA. Mol Cell. 2018;71:256–70e210. doi: 10.1016/j.molcel.2018.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dell’Orco M, Oliver RJ, Perrone-Bizzozero N. HuD binds to and regulates circular RNAs derived from neuronal development- and synaptic plasticity-associated genes. Front Genet. 2020;11:790. doi: 10.3389/fgene.2020.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 21.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–36. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 22.Herranz N, Gil J. Mechanisms and functions of cellular senescence. J Clin Invest. 2018;128:1238–46. doi: 10.1172/JCI95148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, et al. Cellular senescence: defining a path forward. Cell. 2019;179:813–27. doi: 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21:1424–35. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borodkina AV, Deryabin PI, Giukova AA, Nikolsky NN. “Social life” of senescent cells: what is SASP and why study it? Acta Nat. 2018;10:4–14. doi: 10.32607/20758251-2018-10-1-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basisty N, Kale A, Jeon OH, Kuehnemann C, Payne T, Rao C, et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020;18:e3000599. doi: 10.1371/journal.pbio.3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birch J, Gil J. Senescence and the SASP: many therapeutic avenues. Genes Dev. 2020;34:1565–76. doi: 10.1101/gad.343129.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joly-Amado A, Hunter J, Quadri Z, Zamudio F, Rocha-Rangel PV, Chan D, et al. CCL2 overexpression in the brain promotes glial activation and accelerates Tau pathology in a mouse model of tauopathy. Front Immunol. 2020;11:997. doi: 10.3389/fimmu.2020.00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, De, Koninck Y. Spatial and temporal relationship between monocyte chemoattractant protein-1 expression and spinal glial activation following peripheral nerve injury. J Neurochem. 2006;97:772–83. doi: 10.1111/j.1471-4159.2006.03746.x. [DOI] [PubMed] [Google Scholar]
- 31.Hong Y, Tak H, Kim C, Kang H, Ji E, Ahn S, et al. RNA binding protein HuD contributes to beta-cell dysfunction by impairing mitochondria dynamics. Cell Death Differ. 2020;27:1633–43. doi: 10.1038/s41418-019-0447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limbad C, Oron TR, Alimirah F, Davalos AR, Tracy TE, Gan L, et al. Astrocyte senescence promotes glutamate toxicity in cortical neurons. PLoS ONE. 2020;15:e0227887. doi: 10.1371/journal.pone.0227887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiley CD, Velarde MC, Lecot P, Liu S, Sarnoski EA, Freund A, et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 2016;23:303–14. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vizioli MG, Liu T, Miller KN, Robertson NA, Gilroy K, Lagnado AB, et al. Mitochondria-to-nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev. 2020;34:428–45. doi: 10.1101/gad.331272.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riley JS, Tait SW, Mitochondrial DNA. in inflammation and immunity. EMBO Rep. 2020;21:e49799. doi: 10.15252/embr.201949799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zlotorynski E. Defective mitochondria ignite the SASP. Nat Rev Mol Cell Biol. 2020;21:179. doi: 10.1038/s41580-020-0228-x. [DOI] [PubMed] [Google Scholar]
- 37.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–26. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueno T, Toi M, Saji H, Muta M, Bando H, Kuroi K, et al. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin Cancer Res. 2000;6:3282–9. [PubMed] [Google Scholar]
- 39.Tao LL, Shi SJ, Chen LB, Huang GC. Expression of monocyte chemotactic protein-1/CCL2 in gastric cancer and its relationship with tumor hypoxia. World J Gastroenterol. 2014;20:4421–7. doi: 10.3748/wjg.v20.i15.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Yao W, Yuan Y, Chen P, Li B, Li J, et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–67. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 41.Luciano-Mateo F, Cabre N, Baiges-Gaya G, Fernandez-Arroyo S, Hernandez-Aguilera A, Elisabet Rodriguez-Tomas E, et al. Systemic overexpression of C-C motif chemokine ligand 2 promotes metabolic dysregulation and premature death in mice with accelerated aging. Aging. 2020;12:20001–23. doi: 10.18632/aging.104154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez-Gallego E, Riera-Borrull M, Hernandez-Aguilera A, Marine-Casado R, Rull A, Beltran-Debon R, et al. Ubiquitous transgenic overexpression of C-C chemokine ligand 2: a model to assess the combined effect of high energy intake and continuous low-grade inflammation. Mediators Inflamm. 2013;2013:953841. doi: 10.1155/2013/953841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng X, Xu M, Yuan C, Yin L, Chen X, Zhou X, et al. Transcriptional regulation of increased CCL2 expression in pulmonary fibrosis involves nuclear factor-kappaB and activator protein-1. Int J Biochem Cell Biol. 2013;45:1366–76. doi: 10.1016/j.biocel.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Hacke K, Rincon-Orozco B, Buchwalter G, Siehler SY, Wasylyk B, Wiesmuller L, et al. Regulation of MCP-1 chemokine transcription by p53. Mol Cancer. 2010;9:82. doi: 10.1186/1476-4598-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakatsumi H, Matsumoto M, Nakayama KI. Noncanonical pathway for regulation of CCL2 expression by an mTORC1-FOXK1 axis promotes recruitment of tumor-associated macrophages. Cell Rep. 2017;21:2471–86. doi: 10.1016/j.celrep.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Wei M, Xie Q, Zhu J, Wang T, Zhang F, Cheng Y, et al. MicroRNA-33 suppresses CCL2 expression in chondrocytes. Biosci Rep. 2016;36:e00332. doi: 10.1042/BSR20160068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60:1294–304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- 48.Zhang G, Wang J, Yao G, Shi B. Downregulation of CCL2 induced by the upregulation of microRNA-206 is associated with the severity of HEV71 encephalitis. Mol Med Rep. 2017;16:4620–6. doi: 10.3892/mmr.2017.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z, Guo Z, Dong J, Sheng S, Wang Y, Yu L, et al. miR-374a regulates inflammatory response in diabetic nephropathy by targeting MCP-1 expression. Front Pharmacol. 2018;9:900. doi: 10.3389/fphar.2018.00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan J, Ishmael FT, Fang X, Myers A, Cheadle C, Huang SK, et al. Chemokine transcripts as targets of the RNA-binding protein HuR in human airway epithelium. J Immunol. 2011;186:2482–94. doi: 10.4049/jimmunol.0903634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim C, Kang D, Lee EK, Lee JS. Long noncoding RNAs and RNA-binding proteins in oxidative stress, cellular senescence, and age-related diseases. Oxid Med Cell Longev. 2017;2017:2062384. doi: 10.1155/2017/2062384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M. Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol. 2001;21:5889–98. doi: 10.1128/MCB.21.17.5889-5898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hashimoto M, Tsugawa T, Kawagishi H, Asai A, Sugimoto M. Loss of HuR leads to senescence-like cytokine induction in rodent fibroblasts by activating NF-kappaB. Biochim Biophys Acta. 2014;1840:3079–87. doi: 10.1016/j.bbagen.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 54.Lee JH, Jung M, Hong J, Kim MK, Chung IK. Loss of RNA-binding protein HuR facilitates cellular senescence through posttranscriptional regulation of TIN2 mRNA. Nucleic Acids Res. 2018;46:4271–85. doi: 10.1093/nar/gky223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hassibi S, Baker J, Donnelly L, Barnes P. The RNA binding protein HuR regulates the senescence-associated secretory phenotype under conditions of oxidative stress. Eur Respir J. 2019;54:PA2374. [Google Scholar]
- 56.Shao ZX, Ni LB, Hu SL, Xu TZ, Meftah Z, Yu ZP, et al. RNA-binding protein HuR suppresses senescence through Atg7 mediated autophagy activation in diabetic intervertebral disc degeneration. Cell Prolif. 2021;54:e12975. doi: 10.1111/cpr.12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng XL, Gu XP, Xia TJ, Ma ZL, Yang ZZ, Feng HL, et al. HuB and HuD repress telomerase activity by dissociating HuR from TERC. Nucleic Acids Res. 2021;49:2848–58. doi: 10.1093/nar/gkab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim C, Kim W, Lee H, Ji E, Choe YJ, Martindale JL, et al. The RNA-binding protein HuD regulates autophagosome formation in pancreatic beta cells by promoting autophagy-related gene 5 expression. J Biol Chem. 2014;289:112–21. doi: 10.1074/jbc.M113.474700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data may be available from the corresponding author on reasonable request.