Abstract
Microgliosis is a hallmark of many neurological diseases, including Alzheimer’s disease, stroke, seizure, traumatic brain and spinal cord injuries, and peripheral and optic nerve injuries. Recent studies have shown that the newly self-renewed microglia have specific neurological functions. However, the mechanism of adult microglia proliferation remains largely unclear. Here, with single-cell RNA sequencing, flow cytometry, and immunohistochemistry, we demonstrate that the sciatic nerve injury induced two distinct phases of microglia proliferation in mouse spinal cord, each with different gene expression profiles. We demonstrate that the transcription factor Myc was transiently upregulated in spinal cord microglia after nerve injury to mediate an early phase microglia proliferation. On the other hand, we reveal that the tumor-necrosis factor alpha-induced protein 3 (Tnfaip3) was downregulated to mediate the Myc-independent late-phase microglia proliferation. We show that cyclin dependent kinase 1, a kinase with important function in the M phase of the cell cycle, was involved only in the early phase. We reveal that although the early phase was neither necessary nor sufficient for the late phase proliferation, the late-phase suppressed the early phase microglia proliferation in the spinal cord. Finally, we demonstrate that the termination of spinal cord microglia proliferation required both Myc and Tnfaip3 to resume their baseline expression. Thus, we have delineated an interactive signaling network in the proliferation of differentiated microglia.
Subject terms: Mechanisms of disease, Transcriptomics
Introduction
Unlike other cells in central nervous-system (CNS) parenchyma, microglia originate from hematopoietic progenitors in yolk sac and migrate into CNS in early embryonic development1,2. Being the tissue macrophages in CNS, adult microglia are critical in keeping normal physiological function and in responding to pathological damages in CNS3. One character of adult microglia is that they can proliferate at homeostatic physiological condition and upon pathological stimulations4–9. Compared with embryonic microglia, which are highly proliferative1, adult microglia have a low rate of baseline proliferation to maintain their homeostatic population5–7.
In fact, microglia proliferation occurs in numerous neurological disorders, including Alzheimer’s disease5,10, Huntington’s disease11, amyotrophic lateral sclerosis (ALS)12, stroke13, traumatic brain and spinal cord injuries14,15, multiple sclerosis16,17, seizure18, Parkinson’s disease19, peripheral and optic nerve injuries7,9,20, neuropathic pain9,21, infection22,23, substance abuse24, irradiation25, and mental disorders26. In Alzheimer’s disease for example, increased microglia proliferation has been observed in both clinical and preclinical nervous system10; and the long-term in vivo single-cell imaging study suggests that proliferation of resident microglia is responsible for the increase of microglial numbers around amyloid deposits5. Similarly, the expansion of microglia population after peripheral or optic nerve injuries is not associated with infiltration of circulating monocytes20,27,28, indicating that microglia proliferation, rather than monocyte infiltration, is responsible for microglia population expansion after nerve injuries. Indeed, with a multicolor fluorescence fate-mapping system, microglia proliferation has been observed in facial nucleus after facial nerve injury7. In addition, several parabiotic studies suggest that there is little contribution of circulating monocytes to the expansion of microglia population in the mouse models of experimental autoimmune encephalitis (EAE)16, ALS28, and stroke13. These observations suggest that proliferation of adult microglia is a hallmark of many neurological diseases.
More importantly, adult microglia proliferation contributes to the development of neurological diseases. In a mouse neuropathic pain model, inhibition of microglia proliferation significantly reduces the development of neuropathic pain behavior21. Similarly, inhibition of microglia proliferation prevents the behavioral deficits and synaptic degeneration in a mouse model of Alzheimer’s disease10. Inhibition of microglia proliferation also prevents neuronal degeneration in prion disease23 and attenuates neuronal death in seizure18. On the other hand, increased microglia proliferation reduces lesion size and enhances functional recovery after spinal cord injury14. In fact, the newborn microglia generated from microglial proliferation have unique transcriptional profile, and they stimulate adult neurogenesis to facilitate the functional recovery after traumatic brain injury29. Thus, adult microglia proliferation not only is a hallmark but also has critical functions in the development of many neurological diseases.
Here we report that Myc and Tnfaip3 regulate the early phase and the late phase of microglia proliferation, respectively. We have delineated the interaction between these two pathways, and we have explored the mechanism of the termination of microglia proliferation.
Results
Microglia single-cell RNA sequencing (scRNA-Seq)
To investigate the mechanism of microglia proliferation, we applied a mouse sciatic nerve injury model, in which we and others have shown that the nerve injury stimulates microglia proliferation in the corresponding lumbar spinal cord9,21,27,30. As microglia are distinguished by Cx3cr1 expression1,31, with young adult Cx3cr1-creER-yfp/+ heterozygous mice32, we sorted YFP (+) cells by fluorescence-activated cell sorting (FACS) from the parenchyma of lumbar spinal cords at different timepoints after nerve injury for 10× Genomic nanoliter droplet-based scRNA-Seq33.
We analyzed our scRNA-Seq data with 10× Geneomic Loupe Cell Browser and found that virtually all FACS YFP (+) cells in the mouse lumbar spinal cord are microglia, because 99.97% of sequenced cells expressed Hexb (Supplementary Fig. S1), a gene specifically expressed in microglia34. Lumbar cord Cx3cr1-YFP (+) cells also expressed other microglia signature genes34 such as P2ry12 (99.7% of all cells), Tmem119 (99.7% of all cells), Csf1r (99.7% of all cells), and Fcrls (98.3% of all cells). Microglia are distinguished traditionally via flow cytometry as CD11bhighCD45intermediate markers; we analyzed the microglia from lumbar spinal cord of wild-type animals using flow cytometry (Supplementary Fig. S2) and found that 99% of cells labeled by CD11bhighCD45intermediate co-expressed CX3CR1, and 98.8% of CX3CR1 (+) cells were CD11bhighCD45intermediate (Supplementary Fig. S3).
As circulating monocytes also express Cx3cr1, along with monocyte-specific gene Ccr235, we analyzed our scRNA-Seq of Cx3cr1-YFP (+) cells and found minimal amount of Ccr2-expressing monocyte in lumbar cord in naive animals, or in animals 1 or 2 days after nerve injury (Supplementary Fig. S1), consistent with the previous report that there is minimal monocyte infiltration in the spinal cord after nerve injury27, and that proliferation of existing microglia is the source of microglia expansion after nerve injury9,21,27,30. We also found minimal Cd163 or Mrc1 expressing perivascular macrophages31 in mouse lumbar cord (Supplementary Fig. S1).
We then identified the cell-cycle state of each sequenced microglia cell by assessing its expression of specific cell-cycle marker genes36,37, followed by the visualization of these cells using linear-dimension reduction38. The scRNA-Seq principal component analysis (PCA) plots showed that most lumbar cord microglia in naive young adult Cx3cr1-creER-yfp/+ heterozygous mice were in gap-1 (G1) phase of the cell cycle, with a very small proportion of cells in synthesis (S) or gap-2 (G2)/mitosis (M) phases (Fig. 1a), consistent with previous reports of a low-baseline microglia proliferation5–7. One day after nerve injury, a substantial portion of microglia was in S phase (Fig. 1b), whereas a substantial G2/M-phase population was observed by 2 and 3 days after nerve injury (Fig. 1c, d). Thus, our scRNA-Seq results recapitulated the previously reported observations that peripheral nerve injury triggered microglia proliferation in the spinal cord9,21,27,30.
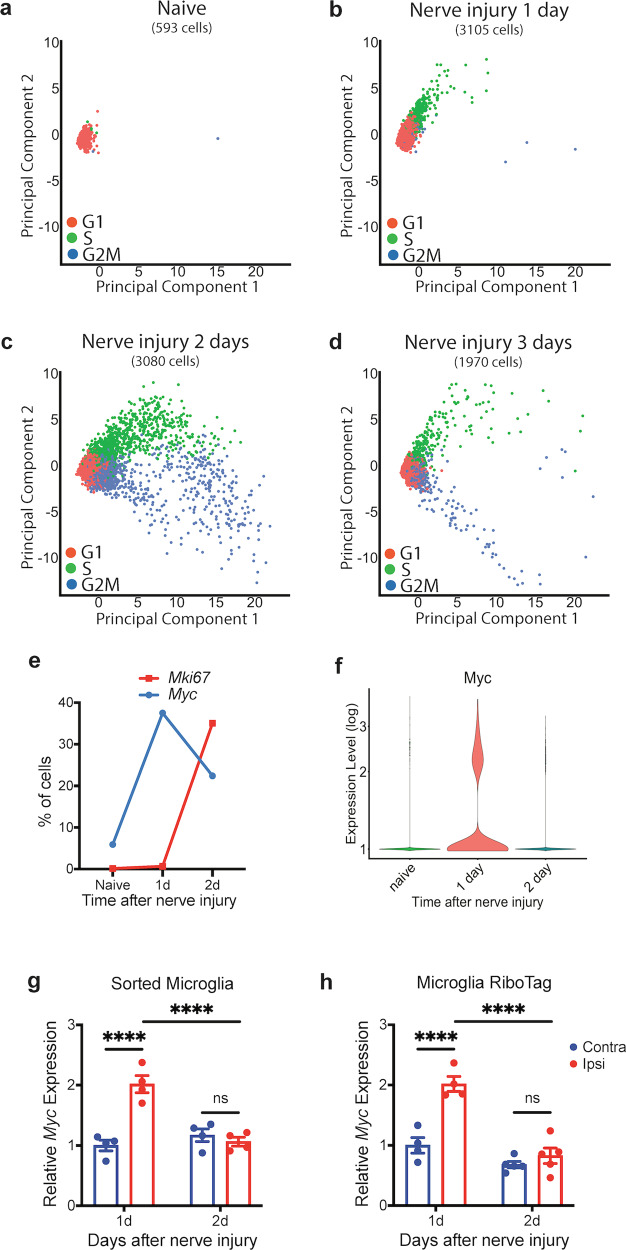
Fig. 1. scRNA-Seq of lumbar spinal cord microglia.
a–d scRNA-Seq PCA scatter plots of lumbar spinal cord YFP (+) cells of adult Cx3cr1-creER-yfp /+; +/+ heterozygous animals at different timepoints after sciatic nerve injury. The vast majority of microglia in naive animal were in G1 phase of the cell cycle (a), a substantial portion of microglia 1 day after nerve injury were in S phase of the cell cycle (b), and many microglia were in G2/M phase of the cell cycle 2 days (c) and 3 days (d) after nerve injury. Note that the S-phase cells appeared one day (b) before the appearance of G2/M-phase cells (c), and the distribution patterns of S phase cells were different between 1 day (b) and 2 days (c) after nerve injury. The distribution patterns of G2/M phase cells were also different between 2 days (c) and 3 days (d) after nerve injury. e Proportion of microglia cells expressing Myc or Mki67 from scRNA-Seq analysis. Myc was expressed in a higher proportion of microglia 1 day after nerve injury, and Mki67 was expressed in a higher proportion of microglia 2 days after nerve injury. f Violin plot of scRNA-Seq showing that a cluster of microglia transiently increased Myc expression on day 1 after nerve injury, and the expression returned to baseline by day 2. g, h qRT-PCR of sorted microglia from the lumbar spinal cord of adult Cx3cr1-creER-yfp/+; +/+ animals (g) and qRT-PCR of RiboTag enriched microglia mRNA from lumbar spinal cord (h). Myc was transiently upregulated in the ipsilateral lumbar cord microglia on day 1 after nerve injury, and the expression returned to baseline by day 2 after nerve injury. Analyses are two-way ANOVA with Sidak’s multiple-comparison test, n = 4–6, means ± SEM, ****P < 0.0001; ns, not significant.
Myc is transiently upregulated in spinal lumbar cord microglia after sciatic nerve injury
When we examined the single-cell expression of Mki67, which encodes proliferation marker Ki67, with 10× Genomic Loupe Cell Browser, we found that more microglia cells expressed Mki67 on day 2 after nerve injury (Fig. 1e), consistent with the previous reports that the proliferation of lumbar cord microglia begins 2 days after peripheral nerve injury9,39. We also found Myc transcripts, which encode the transcription factor MYC that is involved in regulating proliferation in many cell types40, in only a few microglia from naive animals, but in substantially more microglia 1 day after nerve injury (Fig. 1e), a time point preceding the induction of Mki67 expression. We found that Myc induction was transient, the highest at day 1, and resolved by day 2 after nerve injury (Fig. 1f).
To validate our scRNA-Seq finding of Myc upregulation in microglia shortly after nerve injury, we FACS-sorted YFP (+) microglia from the ipsilateral (injury) and contralateral (uninjury) sides of lumbar spinal cord of young adult Cx3cr1-creER-yfp/+ heterozygous mice 1 day and 2 days after nerve injury, and with quantitative RT-PCR (qRT-PCR) from these sorted cells, we confirmed that Myc was transiently upregulated in microglia of ipsilateral lumbar cord 1 day after nerve injury (Fig. 1g), when the expression of many microglia marker genes was unchanged (Supplementary Fig. S4). We also applied microglia RiboTag technique41 to investigate mRNA that specifically binds to microglia ribosomes, and we verified the transient upregulation of Myc in microglia of ipsilateral lumbar cord 1 day after nerve injury (Fig. 1h).
Two phases of microglia proliferation
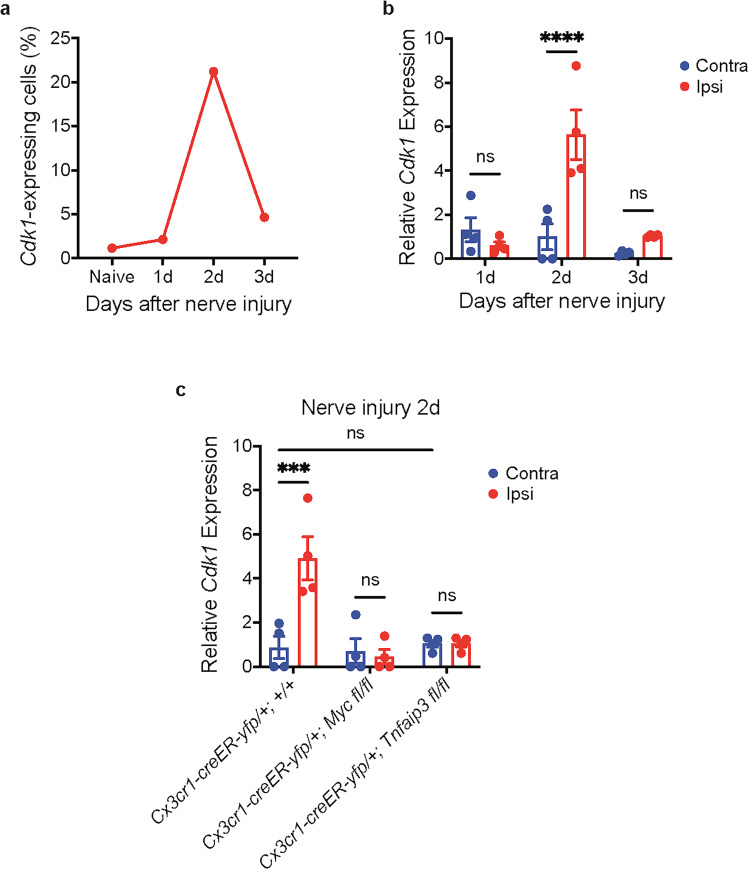
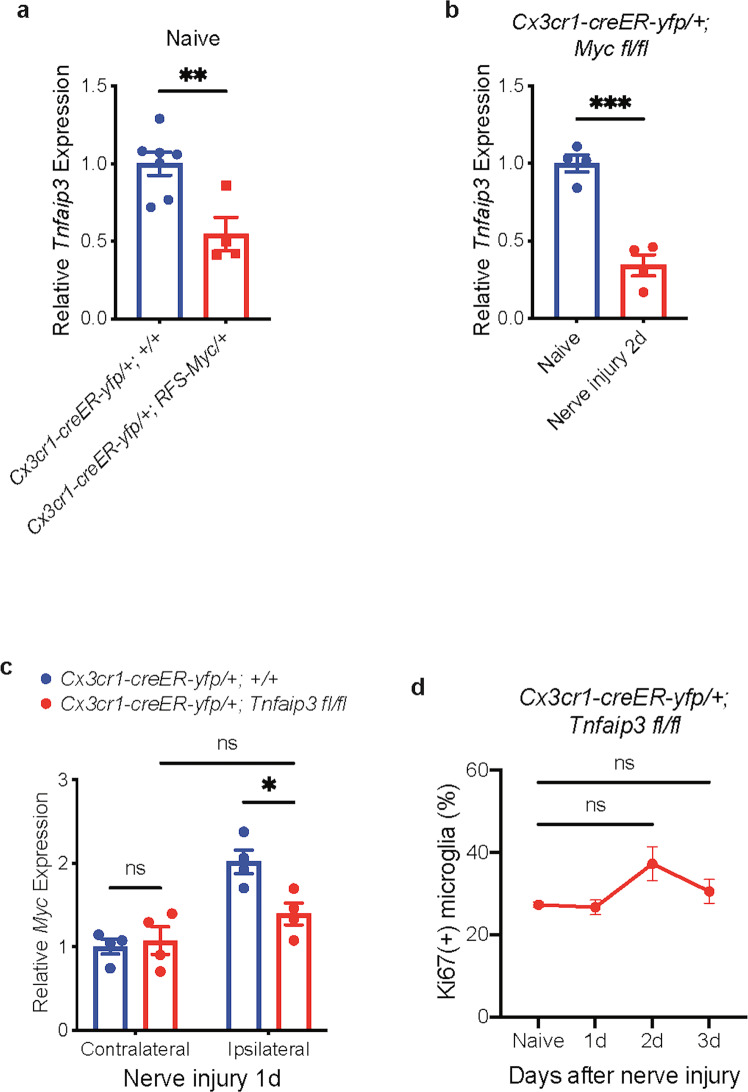
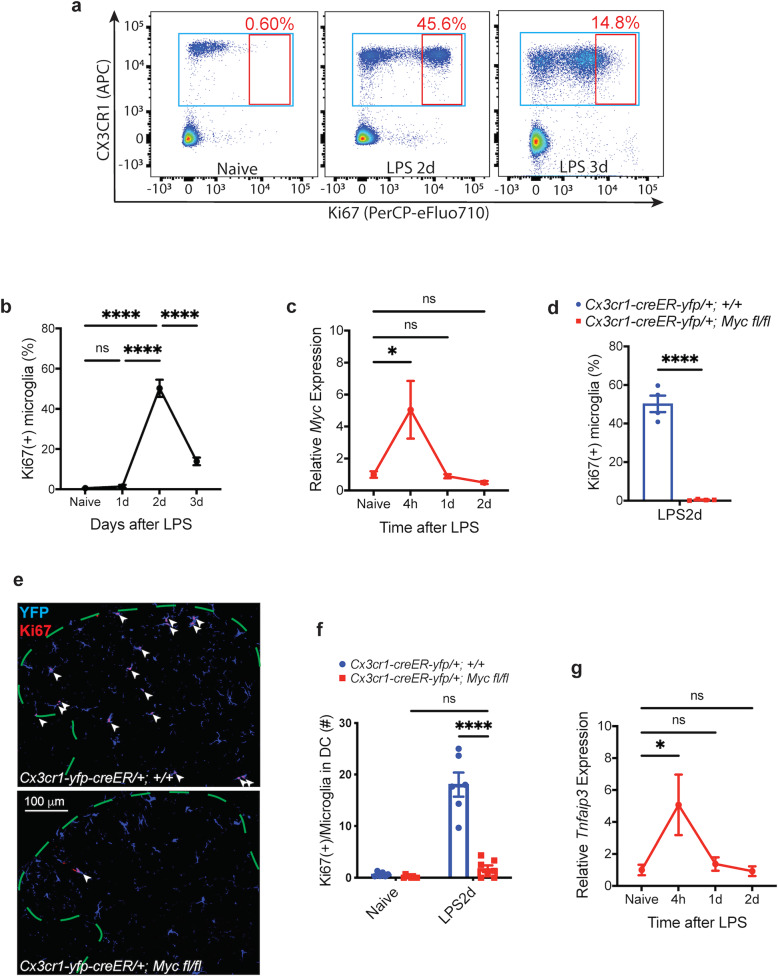
With flow cytometry, we characterized the time course of spinal cord microglia proliferation after sciatic nerve injury by analyzing the percentage of Ki67 (+) microglia, and observed that microglia proliferation started at day 2, maintained through day 3, and then gradually reduced to the termination of proliferation by day 7 after nerve injury (Fig. 2a, b), consistent with previous reports from the immunohistochemistry (IHC) studies9,39. To study the function of Myc in microglia proliferation, we deleted Myc from adult microglia by crossing Mycflox mice42 with Cx3cr1-creER-yfp mice and then treated young adult Cx3cr1-creER-yfp/+; Myc fl/fl animals with tamoxifen. With flow cytometry, we found that Myc deletion from adult microglia completely prevented the early timepoint microglia proliferation 2 days after nerve injury (Fig. 2b; Supplementary Fig. S5), but microglia proliferation at later timepoints, as well as the termination of proliferation, remained (Fig. 2b). The same findings were echoed using IHC staining for Ki67 in the dorsal lumbar spinal cord: there was significantly less microglia proliferation in animals with Myc deletion specifically on day 2 after nerve injury, but the microglia proliferation 3 days after nerve injury was intact in Myc conditional-knockout (cKO) animals (Fig. 2c, d). Our IHC with EdU in lumbar dorsal cord further confirmed that Myc cKO mice had specific deficit in microglia proliferation 2 days, but not 3 days, after nerve injury (Supplementary Fig. S6).
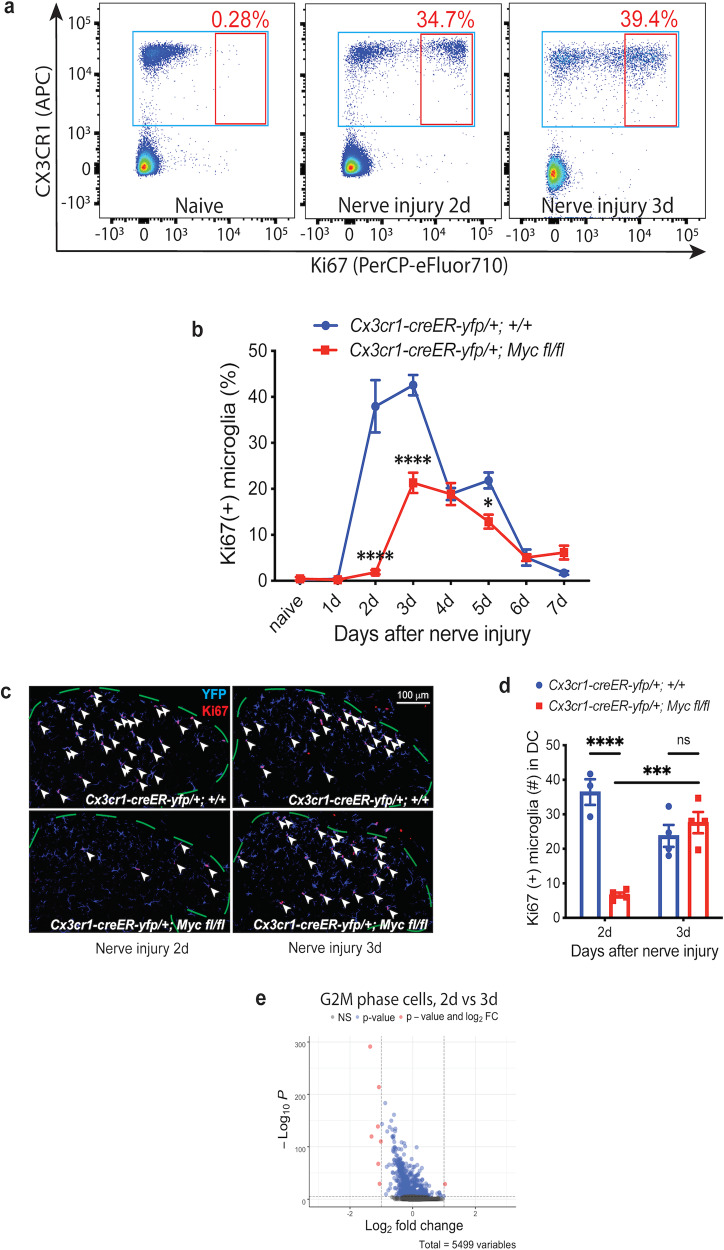
Fig. 2. Microglia proliferation after nerve injury consists of a Myc-dependent early phase and a Myc-independent late phase.
a Representative flow-cytometry scatter plots to assess microglia proliferation in lumbar spinal cord after sciatic nerve injury. The proportion of proliferating Ki67 (+) microglia in Cx3cr1-creER-yfp/+; +/+ animals increased 2 and 3 days after nerve injury. b Quantification of flow cytometry highlights the time course of nerve injury-induced microglia proliferation. In control Cx3cr1-creER-yfp/+; +/+ animals, microglia proliferation started at day 2, persisted through day 3, and then gradually reduced to termination by day 7 after nerve injury. In contrast, the early phase of microglia proliferation on day 2 after nerve injury was completely abolished in Cx3cr1-creER-yfp/+; Myc fl/fl cKO animals, but the late phase and the termination phase of microglia proliferation remained intact in cKO animals. c, d Representative Ki67 IHC staining (c) and the associated quantification (d) of lumbar dorsal cord microglia after nerve injury. Animals with Myc deletion from adult microglia had significantly less Ki67 (+) proliferating microglia in lumbar dorsal cord (DC) 2 days, but not 3 days, after nerve injury. The arrowheads point to the Ki67 (+) microglia, and the dashed lines outline the dorsal lumbar cord. Microglia are identified by YFP staining. e Differential expression volcano plot of pseudobulk scRNA-Seq analysis. The G2/M-phase microglia between day 2 and day 3 after nerve injury displayed distinct gene expression profiles. Analyses are two-way ANOVA with Sidak’s multiple-comparison test, n = 4–6, means ± SEM, *P < 0.05, ***P < 0.001, ****P < 0.0001, ns, not significant. The statistical analysis in b shows the difference between Myc cKO and control animals.
We also counted the total number of microglia in lumbar dorsal cord of naive animals and of animals at different timepoints after nerve injury and found that although Myc deletion in adult microglia did not change the total number of microglia in naive animals, it resulted in significantly less dorsal cord microgliosis after nerve injury (Supplementary Fig. S7). Consistent with our observation that the early timepoint microlgia proliferation 2 days after nerve injury was abolished in Myc cKO mice (Fig. 2b–d), the total number of dorsal cord microglia remained unchanged in Myc cKO mice 2 days after nerve injury, whereas it started to increase in control mice at that timepoint (Supplementary Fig. S7b).
The aforementioned results suggest that nerve injury-induced microglia proliferation has two phases, a Myc-dependent early phase on day 2 after nerve injury, and a Myc-independent late phase on day 3 after nerve injury (Fig. 2b). Indeed, the PCA plot distribution of the G2/M-phase microglia 2 days after nerve injury was different from that of the G2/M microglia 3 days after nerve injury (Fig. 1c, d). A further comparison of the G2/M microglia between 2 and 3 days after nerve injury using a pseudobulk differential expression analysis43 highlighted the differences in gene expression profiles between these two populations (Fig. 2e; Supplementary Table S1).
Tnfaip3 is downregulated in spinal lumbar cord microglia after sciatic nerve injury
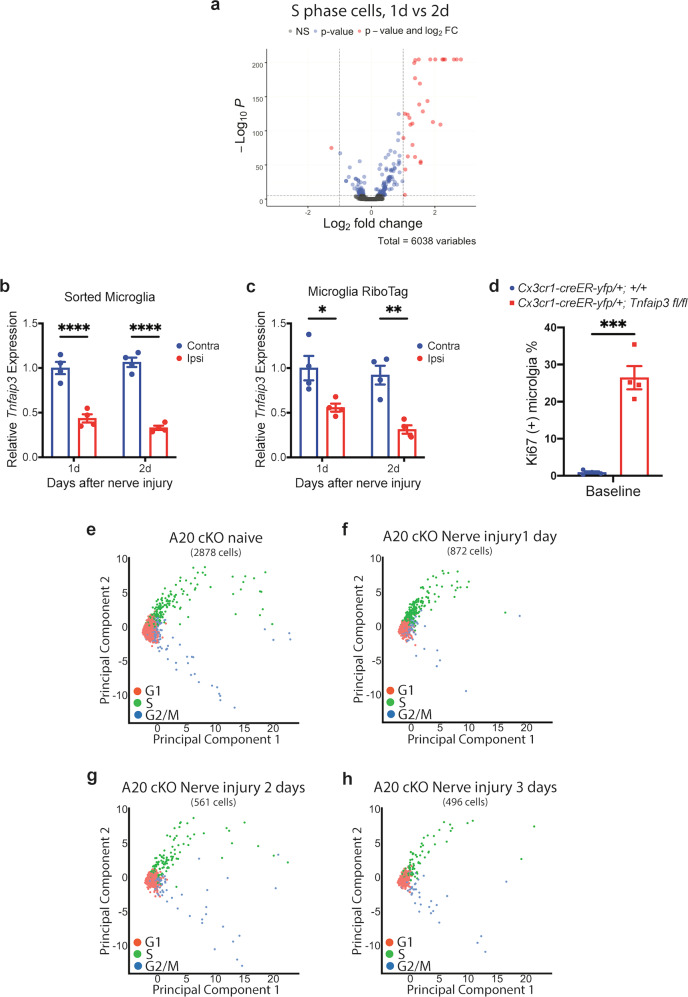
The PCA plot distribution of the S-phase microglia 1 day after nerve injury was also notably different from that of the S-phase microglia 2 days after nerve injury (Fig. 1b, c), which reflects the different gene expression profiles of the S-phase microglia between the two timepoints (Fig. 3a; Supplementary Table S2). To further investigate the mechanism behind such difference, we turned to tumor-necrosis factor alpha-induced protein 3 (Tnfaip3), commonly known as A20, as Tnfaip3 has been reported to inhibit microglia proliferation44. qRT-PCR of sorted microglia and microglia RiboTag revealed that Tnfaip3 was downregulated in microglia of ipsilateral lumbar spinal cord, both 1 and 2 days after nerve injury (Fig. 3b, c). Of note, on day 2 after nerve injury when Myc expression had returned to baseline (Fig. 1g, h), Tnfaip3 remained downregulated in microglia (Fig. 3b, c). With flow cytometry, we found that deleting Tnfaip3 from adult microglia, by crossing Tnfaip3flox mice45 with Cx3cr1-creER-yfp mice and then treating young adult Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl animals with tamoxifen, significantly enhanced baseline microglia proliferation without any nerve injury (Fig. 3d; Supplementary Fig. S8a). Our IHC studies also showed that A20 cKO mice have increased microglia proliferation (Supplementary Fig. S9) and increased microglia number (Supplementary Fig. S10) in the lumbar spinal dorsal cord without nerve injury. These results indicate that A20 inhibits microglia proliferation, and nerve injury induced A20 downregulation to promote microglia proliferation.
Fig. 3. Tnfaip3 is downregulated in microglia after nerve injury.
a Differential expression volcano plot of pseudobulk scRNA-Seq analysis. The S-phase microglia from day 1 after nerve injury displayed different gene expression profile from that of the S-phase microglia 2 days after nerve injury in Cx3cr1-creER-yfp/+; +/+ animals. b, c qRT-PCR of FACS-sorted microglia from lumbar spinal cord of Cx3cr1-creER-yfp/+; +/+ animals (b) and of RiboTag enriched microglia mRNA from lumbar spinal cord (c). Tnfaip3 was downregulated in the microglia of ipsilateral lumbar cord 1 and 2 days after nerve injury. d Quantification of proliferating Ki67 (+) microglia by flow cytometry. Naive uninjured Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl A20 cKO animals had significantly more baseline microglia proliferation than those of naive control Cx3cr1-creER-yfp/+; +/+ animals. e scRNA-Seq PCA scatter plot of naive Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl A20 cKO microglia. The distributions of G1, S, and G2/M phase cells were similar to that of Cx3cr1-creER-yfp/+; +/+ control animals 3 days after nerve injury as shown in Fig. 1d. f–h scRNA-Seq PCA scatter plot of Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl A20 cKO microglia 1 day (f), 2 days (g), and 3 days (h) after nerve injury. The distributions of G1, S, and G2/M-phase cells were similar to that of naive Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl A20 cKO animals (e). Analyses are two-way ANOVA with Sidak’s multiple comparison test (b, c) and unpaired two-tailed t-test (d), n = 4–6, means ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To further investigate the function of Tnfaip3 in microglia proliferation, we FACS-sorted YFP (+) cells from the parenchyma of lumbar spinal cords of young adult Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO mice at different timepoints after nerve injury for scRNA-Seq. We found that the microglia in naive Tnfaip3 cKO mice had a substantial proportion of S-phase and G2/M-phase cells (Fig. 3e), consistent with our flow cytometry and IHC results of enhanced microglia proliferation in naive Tnfaip3 cKO mice. Interestingly, the distribution of the S- and G2/M-phase cells in PCA plot of naive Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO mice without injury (Fig. 3e) was similar to that of Cx3cr1-creER-yfp/+; +/+ control mice 3 days after nerve injury (Fig. 1d), but was quite different from that of Cx3cr1-creER-yfp/+; +/+ animals 2 days after nerve injury (Fig. 1c).
Tnfaip3 mediates late phase of microglia proliferation
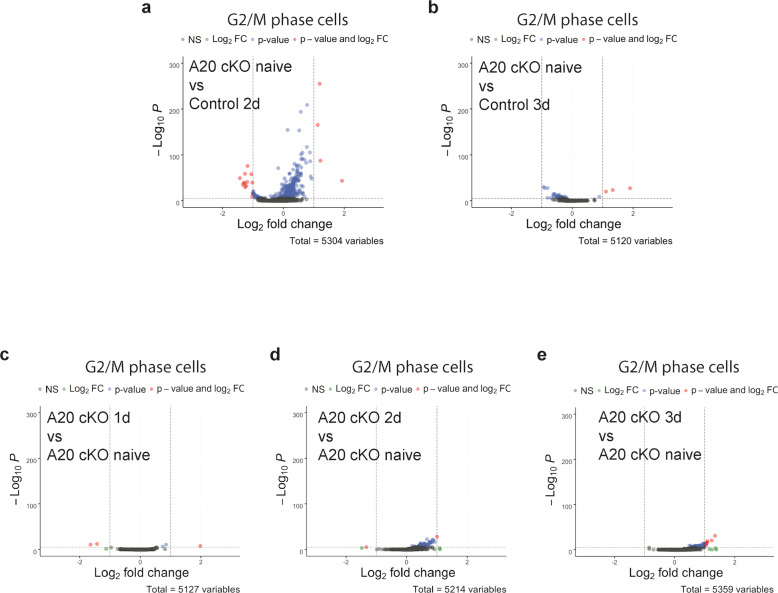
Because the G2/M microglia 2 days after nerve injury had different gene expression profile from the G2/M microglia 3 days after nerve injury (Fig. 2e), we compared the gene expression profile of the G2/M microglia from naive Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO mice without injury to the gene expression profiles of the G2/M microglia from control Cx3cr1-creER-yfp/+; +/+ mice 2 days and 3 days after nerve injury. We found that the G2/M microglia of naive Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO mice had distinct gene expression profile from the G2/M microglia of Cx3cr1-creER-yfp/+; +/+ mice 2 days after nerve injury (Fig. 4a; Supplementary Table S3). However, the naive Tnfaip3 cKO G2/M microglia shared a much similar gene expression profile with the G2/M microglia of Cx3cr1-creER-yfp/+; +/+ mice 3 days after nerve injury (Fig. 4b). These results suggested that the downregulation of Tnfaip3 in adult microglia results in increased microglia proliferation with a transcriptomic signature resembling that of the late-phase microglia proliferation 3 days after nerve injury.
Fig. 4. Differential expression volcano plots of pseudobulk scRNA-Seq showing that A20 naive cKO has similar G2/M microglia expression profile as control 3 days after nerve injury.
a The G2/M-phase microglia of naive uninjured Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl A20 cKO animals exhibited different gene expression profile from the G2/M phase microglia of control Cx3cr1-creER-yfp/+; +/+ animals 2 days after nerve injury. b The G2/M-phase microglia of naive uninjured Cx3cr1-creER-yfp/+; Tnfaip3 fl/flA20 cKO animals shared similar gene expression profile with the G2/M-phase microglia of control Cx3cr1-creER-yfp/+; +/+ animals 3 days after nerve injury. c, e The gene expression profile of the G2/M-phase microglia from Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl A20 cKO animals remained largely unchanged 1 day (c), 2 days (d), or 3 days (e) after nerve injury.
The early phase G2/M microglia do not appear in Tnfaip3 cKO mice after nerve injury
We found that the distributions of the S- and G2/M-phase cells in the PCA plot of Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO mice 1 day (Fig. 3f), 2 days (Fig. 3g), and 3 days (Fig. 3h) after nerve injury were all similar to that of naive Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO mice without injury (Fig. 3e) and that of Cx3cr1-creER-yfp/+; +/+ mice 3 days after nerve injury (Fig. 1d). Similarly, a comparison of the G2/M microglia transcriptome profiles between Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO mice 1 day (Fig. 4c), 2 days (Fig. 4d), and 3 days (Fig. 4e) after nerve injury and the naive Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO mice showed that the G2/M microglia transcriptome profiles in Tnfaip3 cKO remained relatively unchanged even after nerve injury. These results suggest that the early phase G2/M microglia 2 days after nerve injury in control animals did not appear in Tnfaip3 cKO mice after nerve injury.
Cyclin-dependent kinase 1 (CDK1) in early phase microglia proliferation
Our results have demonstrated that the nerve injury-induced microglia proliferation had a Myc upregulation-mediated early phase and a Tnfaip3 downregulation-mediated late phase. To further explore the mechanism of microglia proliferation, we turned to CDK1, a catalytic subunit of the M phase-promoting factor that has crucial function in M phase of the cell cycle46. Our analysis of scRNA-Seq data of Cx3cr1-creER-yfp/+; +/+ mice with 10 × Genomic Loupe Cell Browser revealed that more microglia expressed Cdk1 on day 2 after nerve injury (Fig. 5a), a timepoint of early phase Myc-dependent microglia proliferation. However, the expression of Cdk1 reduced remarkably by 3 days after nerve injury (Fig. 5a), a timepoint of late-phase Myc-independent microglia proliferation (Fig. 2b).
Fig. 5. Signaling pathways in microglia proliferation.
a Proportion of microglia cells expressing Cdk1 from scRNA-Seq analysis. Cdk1 was expressed in a higher proportion of microglia 2 days after nerve injury. b qRT-PCR of sorted microglia from lumbar spinal cord of Cx3cr1-creER-yfp/+; +/+ animals. Cdk1 was upregulated in microglia from ipsilateral lumbar cord 2 days, but not 1 day or 3 days, after nerve injury. c qRT-PCR of sorted lumbar cord microglia. Cdk1 upregulation in microglia 2 days after nerve injury was prevented when Myc was deleted from adult microglia. Deleting Tnfaip3 from adult microglia did not induce Cdk1 upregulation. Deletion of Tnfaip3 also prevented Cdk1 upregulation 2 days after nerve injury. Analyses are two-way ANOVA with Sidak’s multiple-comparison test, n = 4–6, means ± SEM, ***P < 0.001, ****P < 0.0001; ns, not significant.
Our qRT-PCR of sorted microglia from Cx3cr1-creER-yfp/+; +/+ mice confirmed that Cdk1 was upregulated in the ipsilateral lumbar spinal cord 2 days after nerve injury, and its expression returned to baseline on day 3 after nerve injury (Fig. 5b). This result suggests that Cdk1 was induced only in the early phase microglia proliferation. In line with the previous report that Myc stimulates cell-cycle progression by activating CDK147, we found that deleting Myc from adult microglia (Cx3cr1-creER-yfp/+; Myc fl/fl) completely prevented the Cdk1 upregulation 2 days after nerve injury (Fig. 5c), indicating that upregulation of Cdk1 in microglia is downstream of Myc.
Along with our observation that Cdk1 was not upregulated in microglia 3 days after nerve injury (Fig. 5a, b), we found that deleting Tnfaip3 from adult microglia (Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl) did not cause Cdk1 upregulation (Fig. 5c). These results suggest that Cdk1 is specifically involved in the Myc-dependent early phase, but not in the Tnfaip3-mediated late phase, of microglia proliferation. We also found that the Cdk1 upregulation 2 days after nerve injury did not occur in Tnfaip3 cKO (Fig. 5c), consistent with our scRNA-seq analyses that the nerve injury-induced early phase microglia proliferation did not occur in Tnfaip3 cKO mice (Figs. 3e–h, 4).
Interactions between Myc and Tnfaip3 pathways
To study the potential interaction between Myc and Tnfaip3 pathways, we crossed Cx3cr1-creER-yfp mice with ROSA-Floxed-Stop (RFS)-Myc mice48, in which the treatment of young adult Cx3cr1-creER-yfp/+; RFS-Myc/+ mice with tamoxifen resulted in the upregulation of Myc in adult microglia (Supplementary Fig. S11). We found that Myc upregulation produced Tnfaip3 downregulation in microglia of naive animals without any injury (Fig. 6a), suggesting that Myc upregulation in microglia may contribute to the Tnfaip3 downregulation after nerve injury. However, microglia Tnfaip3 downregulation 2 days after nerve injury was intact in Cx3cr1-creER-yfp/+; Myc fl/fl cKO animals (Fig. 6b), indicating that Myc was not required for nerve injury-induced Tnfaip3 downregulation in microglia. On the other hand, although deleting Tnfaip3 from adult microglia in Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO did not change the baseline Myc expression in spinal cord microglia, it abolished the nerve injury-induced Myc upregulation in microglia of spinal cord one day after nerve injury (Fig. 6c), hinting that Tnfaip3 downregulation suppresses Myc upregulation in microglia after nerve injury. This result also explains our previous results that the Myc-dependent early phase microglia proliferation and Cdk1 upregulation 2 days after nerve injury did not occur in Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO mice (Figs. 3e–h, 4, 5c). Indeed, our flow cytometry (Fig. 6d; Supplementary Fig. S8b) study showed no changes in microglia proliferation beyond baseline levels in Tnfaip3 cKO mice after nerve injury. Our IHC studies confirmed that A20 cKO mice did not have increased microglia proliferation (Supplementary Fig. S12) or any increase in microglia number (Supplementary Fig. S13) in lumbar spinal dorsal cord after nerve injury.
Fig. 6. Interaction between Myc upregulation and Tnfaip3 downregulation in microglia.
a qRT-PCR of sorted lumbar cord microglia. Overexpression of Myc in microglia of adult naive Cx3cr1-creER-yfp/+; RFS-Myc/+ animals resulted in downregulation of Tnfaip3. b qRT-PCR of sorted lumbar cord microglia from Cx3cr1-creER-yfp/+; Myc fl/fl cKO animals. Deletion of Myc from adult microglia did not prevent Tnfaip3 downregulation 2 days after nerve injury. c qRT-PCR of sorted lumbar cord microglia. Deletion of Tnfaip3 from adult microglia prevented Myc upregulation 1 day after nerve injury. d Quantification of Ki67 flow cytometry. The proliferation of lumbar cord microglia in Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl cKO animals did not increase after nerve injury. Analyses are unpaired two-tailed t-test (a, b) and one-way ANOVA with Tukey’s multiple-comparison test (c, d), n = 4–6, means ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
Termination of induced microglia proliferation
Although it is important for microglia to expand their population by proliferation in response to various pathological stimulations, it is equally crucial for microglia to terminate their induced proliferation to prevent cancer-like uncontrollable cell growth. However, the mechanism for the termination of microglia proliferation is almost completely unknown. Even though the proliferative signal released by injured sensory neurons persists for at least several weeks9,49, the nerve injury-induced microglia proliferation only lasts for less than 7 days (Fig. 2b). When microglia proliferation has been terminated by day 7 after nerve injury, with qRT-PCR of sorted lumbar cord microglia, we found that the expression of Myc, Cdk1, and Tnfaip3 had returned to baseline (Fig. 7a). Moreover, our flow-cytometry study revealed that the persistent Myc upregulation (Cx3cr1-creER-yfp/+; RFS-Myc/+) or Tnfaip3 downregulation (Cx3cr1-creER-yfp/+; Tnfaip3 fl/fl) in adult microglia prevented the termination of microglia proliferation 7 days after nerve injury (Fig. 7b). Thus, the termination of nerve injury-induced microglia proliferation requires both Myc and Tnfaip3 to resume their baseline expression.
Fig. 7. The termination of microglia proliferation.
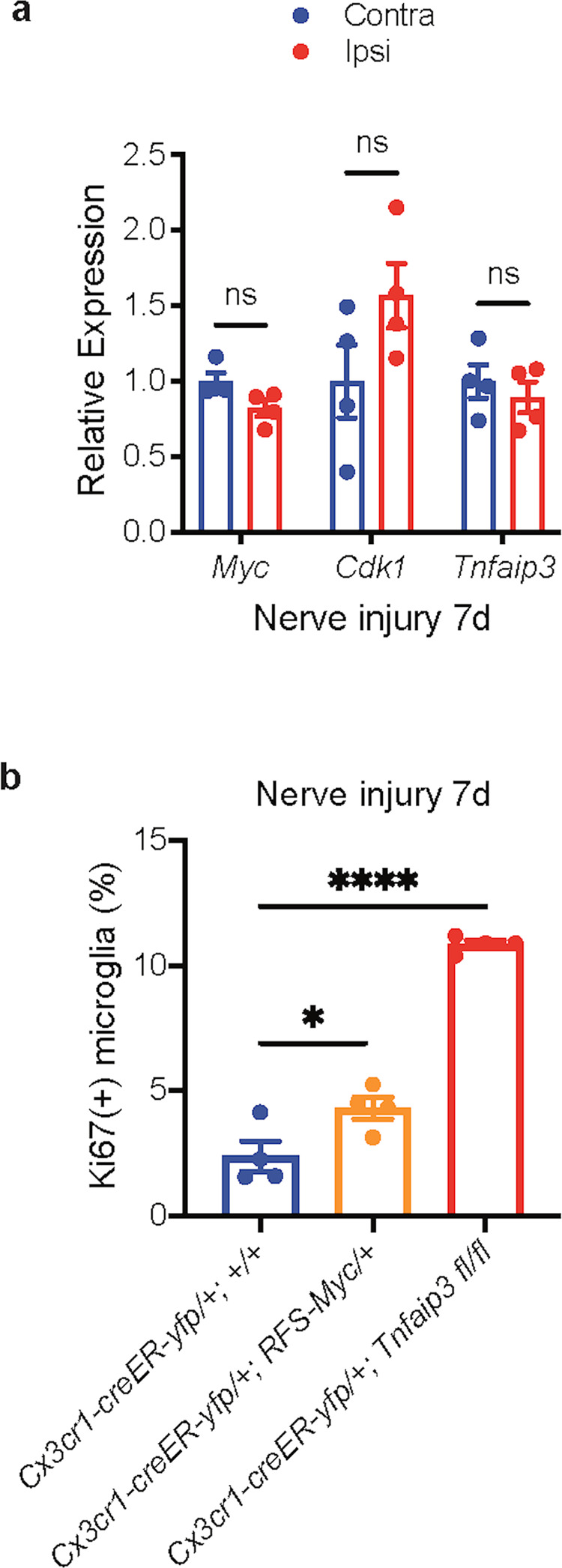
a qRT-PCR of sorted lumbar cord microglia from Cx3cr1-creER-yfp/+; +/+ animals. The expression levels of Myc, Cdk1, and Tnfaip3 of lumbar cord microglia returned to control contralateral levels 7 days after nerve injury. b Quantification of Ki67 flow cytometry. Overexpression of Myc in adult microglia or deletion of Tnfaip3 from adult microglia prevented the termination of lumbar cord microglia proliferation 7 days after nerve injury. Analyses are one-way ANOVA with Tukey’s multiple-comparison test (b) or two-way ANOVA with Sidak’s multiple-comparison test (a), n = 4–6, means ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
The Myc upregulation-mediated early phase is not sufficient to induce Tnfaip3 downregulation-mediated late-phase proliferation
To further investigate the mechanism of microglia proliferation, we turned to lipopolysaccharide (LPS), an endotoxin from Gram-negative bacteria that can produce microglia proliferation in mouse brain by a single peritoneal injection of 0.5–2.5 mg/kg22. With flow cytometry, we found that a single peritoneal injection of 2.5 mg/kg LPS induced microglia proliferation in mouse spinal cord (Fig. 8a, b) and multiple brain regions (Supplementary Fig. S14a) 2 days after the treatment. With CCR2-RFP mice, in which all the peripheral monocytes are labeled with RFP35, we found no RFP+ monocyte infiltration into the spinal cord after LPS injection (data not shown), suggesting that the expansion of microglia population after LPS treatment was the result of microglia proliferation. In agreement with the nerve-injury model, our qRT-PCR of sorted microglia showed that Myc was also transiently upregulated in the microglia of spinal cord (Fig. 8c) and various brain regions (Supplementary Fig. S14b) after LPS treatment. Our flow cytometry (Fig. 8d) and IHC studies (Fig. 8e, f; Supplementary Fig. S15) on animals with Myc deletion from adult microglia (Cx3cr1-creER-yfp/+; Myc fl/fl) revealed that Myc is necessary for LPS-induced microglia proliferation in spinal cord, as well as in many brain regions (Supplementary Fig. S14c).
Fig. 8. Myc-dependent early phase microglia proliferation is not sufficient to induce late-phase microglia proliferation.
a, b Ki67 flow-cytometry scatter plots (a) and the associated quantification (b). Ki67 (+) proliferating microglia increased transiently in the spinal cord of Cx3cr1-creER-yfp/+; +/+ animals 2 days after LPS treatment. c qRT-PCR of sorted lumbar cord microglia from Cx3cr1-creER-yfp/+; +/+ animals. Myc was transiently upregulated in microglia 4 h after LPS treatment, and the expression returned to baseline level by 1 day. d Quantification of Ki67 flow cytometry. Myc deletion in adult microglia abolished the LPS-induced lumbar cord microglia proliferation. e, f Representative Ki67 IHC images (e) and the associated quantification (f). Animals with Myc deletion from adult microglia had significantly less Ki67 (+) proliferating microglia in lumbar dorsal cord (DC) 2 days after LPS. The arrowheads point to the Ki67 (+) microglia, and the dashed lines outline the dorsal lumbar cord. Microglia are identified by YFP staining. g qRT-PCR of sorted lumbar cord microglia from Cx3cr1-creER-yfp/+; +/+ animals. Tnfaip3 was transiently upregulated in lumbar cord microglia 4 h after LPS treatment, and the expression returned to baseline level by 1 day, without any Tnfaip3 downregulation. Analyses are one-way ANOVA with Tukey’s multiple-comparison test (b, c, g) or two-way ANOVA with Sidak’s multiple-comparison test (f), and unpaired two-tailed t-test (d), n = 4–6, means ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
However, the LPS-induced microglia proliferation was short-lived and lasted for only one day (Fig. 8a, b), unlike the prolonged microglia proliferation seen after nerve injury (Fig. 2b). In addition, contrary to the nerve-injury model, we found that LPS treatment did not induce any Tnfaip3 downregulation (Fig. 8g), which we showed to mediate the late-phase of proliferation. Thus, the transient Myc-dependent microglia proliferation induced by LPS was not followed by Tnfaip3-mediated late-phase microglia proliferation. In other words, the Myc upregulation-mediated early phase of microglia proliferation by itself is not sufficient to induce the late phase proliferation. Given that the Myc upregulation-mediated early phase of proliferation is not required for the late phase proliferation either (Fig. 2b), our results demonstrate that the early phase of microglia proliferation is neither necessary nor sufficient for the late phases of microglia proliferation.
Discussion
Microglia proliferation has an early phase, a late phase, and a termination phase
In a nerve-injury-induced microglia proliferation model, we confirmed the previous reports9,39 that microglia proliferation starts on day 2, maintains on day 3, and gradually reduces to termination by day 7 after nerve injury (Fig. 2b). We also found that the early phase of microglia proliferation 2 days after nerve injury did not occur when Myc was deleted from adult microglia, with relatively intact late phase of microglia proliferation 3 days after nerve injury and the termination phase of microglia proliferation 6–7 days after nerve injury (Fig. 2b). Thus, the nerve-injury-induced microglia proliferation has a Myc-dependent early phase and a Myc-independent late phase.
The proliferating microglia at different timepoints after nerve injury have different gene expression profiles
Our observation that microglia proliferation has distinct early and late phases is further supported by our scRNA-seq study. By analyzing the expression of cell-cycle marker genes36,37, we assigned each sequenced microglia cell into specific cell cycles. We found that the G2M microglia cells in the early phase (2 days after nerve injury) had different gene expression profile from the G2M cells in the late phase (3 days after nerve injury) (Fig. 2e). Similarly, we found that the S-microglia cells 1 day after nerve injury were also different from the S cells 2 days after nerve injury (Fig. 3a).
Myc–CDK1 pathway mediates the early phase of microglia proliferation
CDK1 is a serine/threonine kinase that plays a crucial role in driving the M phase in mitosis, and its activity is increased only in the M phase of the cell cycle46. Both of our scRNA-Seq and qRT-PCR results showed that Cdk1 was upregulated specifically in microglia of Myc-dependent early phase (day 2 after nerve injury), but not in microglia of Myc-independent late phase (day 2 after nerve injury) (Fig. 5a, b), further supporting our observations that the early phase and the late phase of microglia proliferation are distinct with different mechanisms. Moreover, we found that the Cdk1 upregulation itself was Myc-dependent in microglia (Fig. 5c). Thus, our results demonstrate that the Myc–CDK1 pathway mediates the early phase of microglia proliferation. Further studies are needed to investigate how Myc regulates Cdk1 expression in microglia and how CDK1 regulates the early phase of microglia proliferation.
A20 mediates the late phase of microglia proliferation
A20, encoded by Tnfaip3 gene, is a key player in ubiquitin-dependent signals, and it participates in regulating diverse signaling pathways50. Our results that deletion of Tnfaip3 from adult microglia resulted in microglia proliferation in naive animals (Fig. 3d) and that Tnfaip3 was downregulated in microglia after nerve injury (Fig. 3b, c) suggested that A20 inhibits microglia proliferation, and that nerve injury suppressed the inhibitory effect of A20 to promote microglia proliferation. Importantly, our scRNA-seq analysis showed that the proliferating G2/M microglia in naive Tnfaip3 cKO without injury was similar to the G2/M microglia of the late-phase proliferation 3 days after nerve injury, but different from the G2/M microglia of the early phase 2 days after nerve injury (Fig. 4), suggesting that suppression of A20 mediates the late-phase microglia proliferation. A20 has been implicated in both promoting and inhibiting proliferation in a cell-type-specific context51,52, but how A20 regulates the late phase of microglia proliferation remains to be further studied.
The early phase is neither necessary nor sufficient for the late phases of microglia proliferation
Although the early phase precedes the late-phase microglia proliferation, our results demonstrated that the late-phase proliferation occurred independently from the early phase, further supporting that the mechanisms for the early phase and the late phase of microglia proliferation are different. In Myc cKO animals, Tnfaip3 was downregulated in microglia to the level similar as in control animals after nerve injury (Figs. 6b, 3b), and the late-phase microglia proliferation occurred without the precedent early phase proliferation (Fig. 2b), suggesting that the early phase was not required for the development of the late-phase microglia-proliferation. On the other hand, in LPS-induced microglia proliferation model, the Myc-dependent transient microglia proliferation 2 days after LPS treatment was not followed by Tnfaip3-downregulation-mediated late-phase proliferation (Fig. 8b), suggesting that the early phase was not sufficient for the development of the late-phase microglia proliferation either.
The late phase suppresses the early phases of microglia proliferation
In contrast, our results showed that the Tnfaip3-downregulation-mediated late-phase proliferation inhibited the Myc-upregulation-mediated early phase proliferation. We found that genetic deletion of Tnfaip3 from adult microglia completely blocked the nerve-injury-induced upregulation of Myc (Fig. 6c) or Cdk1 (Fig. 5c) in microglia. Moreover, our scRNA-seq results revealed that the early phase-specific G2/M microglia cells never appeared in Tnfaip3 cKO after nerve injury (Fig. 4). Thus, the late-phase microglia proliferation not only occurred independently from the early phase, but also suppressed the early phase proliferation.
Termination of microglia proliferation
We and others have previously demonstrated that peripheral nerve injury induces the injured dorsal root ganglion (DRG) sensory neurons to express colony-stimulating factor 1 (CSF1)9,49, a cytokine critical for microglia development1, and CSF1 is subsequently transported from the injured DRG neurons to the spinal cord to stimulate microglia proliferation9. Interestingly, although the nerve-injury-induced CSF1 expression in the injured DRG neurons lasts for at least several weeks9,49, the nerve-injury-induced spinal cord microglia proliferation only lasts for less than one week39 (Fig. 2b). We now show that when microglia proliferation was terminated at 7 days after nerve injury, the expression of both Myc and Tnfaip3 in microglia had returned to baseline (Fig. 7a), and this resumption of baseline expression of Myc and Tnfaip3 was necessary for the termination of nerve-injury-induced microglia proliferation (Fig. 7b). How Myc and Tnfaip3 resume their baseline expression needs to be further studied.
Mechanisms of microglia proliferation
In summary, our results suggest that mature differentiated microglia proliferate with two distinct phases mediated by two different mechanisms (Supplementary Fig. S16). Nerve injury leads to transient Myc upregulation in microglia, which in turn induces Cdk1 upregulation to mediate an early phase of microglia proliferation. In parallel, nerve injury also induces sustained Tnfaip3 downregulation in microglia, which mediates the late-phase microglia proliferation. Although the early phase proliferation is neither necessary nor sufficient for the late-phase proliferation, Myc upregulation enhances Tnfaip3 downregulation, whereas Tnfaip3 downregulation suppresses Myc upregulation and the subsequent Cdk1 upregulation. However, because the Myc upregulation after nerve injury is transient, such transient Myc upregulation is unlikely to produce sustained Tnfaip3 downregulation for late-phase microglia proliferation. The termination of microglia proliferation requires both Myc and Tnfaip3 to resume their baseline expression levels. Thus, we have delineated the signaling network in the distinct phases of microglia proliferation. To our knowledge, this is the first report on the separate phases of proliferation in fully differentiated cells.
Materials and methods
Animal lines
Animal experiments were approved by UCSF Institutional Animal Care and Use Committee and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory animals. C57BL/6 wild-type, Cx3cr1-creER-yfp32 and RiboTag53 mice were purchased from Jackson Laboratory. Mycflox, Tnfaip3flox, and ROSA-Floxed-Stop (RFS)-Myc mice were described previously42,45,48,54. The experiments were conducted with roughly equal number of male and female young/adult animals.
Surgeries and treatments
Sciatic-nerve transection and ligation was performed as previously described9. LPS (E. coli O127:B8, Sigma) was administered as a single 2.5 mg/kg intraperitoneal dose. Tamoxifen (Sigma) was injected as 100 mg/kg per animal for five consecutive days for recombination. All the experiments were performed 7 days after the first dose of tamoxifen injection.
Microglia isolation
Mice were anesthetized with Avertin (Sigma) 25 mg per animal by intraperitoneal injection. After perfusion of 1× HBSS (Hank’s Balanced Salt Solution, Gibco), brain or spinal cord were collected freshly, with meningeal membrane removed. All the fresh samples were homogenized with 1× HBSS in Dounce homogenizer (DWK Life Science). The cell suspensions were then filtered through 70 mm cell strainer and washed with 1× HBSS, followed by mixture with Percoll (Sigma) gradient in HBSS with the final Percoll concentration of 33%. Samples were then centrifuged at 800 relative centrifugal force (RCF) for 20 min with no brake. Myelin and supernatant were removed by vacuum aspiration. Pellets, which contain isolated microglia, were collected and resuspended in 1 × HBSS.
Fluorescence-activated cell sorting (FACS)
Freshly isolated microglia were sorted by FACS machine (BD FACSAria2) using 85um nozzle. After size selection and doublet exclusion, YFP (+) cells were sorted (1) directly into lysis buffer (QIAGEN), and RNA was isolated by RNeasy Plus Micro Kit (QIAGEN) following the manufacturer’s protocol for RNA collection or (2) into optimized serum-free media for single-cell RNA sequencing.
Flow cytometry
Isolated microglia samples were first treated with Fc blocker (BD Pharmingen), then stained with surface CX3CR1-APC (1:500, Biolegend) and LIVE/DEAD Fixable Near-IR Dead Cell Staining Kit (1:2000, Invitrogen). The cells were then washed, fixed, and permeabilized with FoxP3/Transcription factor staining set (ThermoFisher), and then stained with Ki67–PerCP–eFluor710 (1:2000, Invitrogen). Flow cytometry was performed with a BD FACS Canto II and the results were analyzed by Flowjo software v10.6.1. Our gating strategy is similar as previously reported55 and presented in Supplementary Fig. S3.
Immunohistochemistry
Immunohistochemistry was performed as previously described9, with the following antibodies: GFP (Abcam), Ki67 (Abcam), and CD11b (Abcam). Confocal images were collected with a Carl Zeiss LSM 700 microscope and were processed with Fiji/ImageJ (NIH).
RiboTag immunoprecipitation
RiboTag immunoprecipitation was performed as previously described41. In short, the tissue of interest was dissected into homogenization buffer with 3% weight per volume ratio. Homogenized samples were centrifuged at 10,000 RCF for 10 min, and the supernatant was carefully separated for immunoprecipitation. The sample was incubated with anti-HA antibody (1:500, Biolegend) for 4 h. Protein A/G magnetic beads (Thermofisher Scientific) were prewashed with homogenization buffer, then added to the sample, and incubated overnight. The beads were collected by magnetic separation and then washed in high-salt buffer before being resuspended in lysis buffer for RNA isolation using RNeasy Plus Micro Kit (QIAGEN).
Single-cell sequencing
Microglia sample preparation was performed as in our FACS experiments. Lumbar spinal cords of three animals were pooled per group, and the cells were harvested from naive animals and from animals 24 h, 48 h, or 72 h after nerve injury. Cell pellets were resuspended in optimized serum-free media previously described for improved microglia viability56: human recombinant TGF-β2 (2 ng/mL, Peprotech), murine IL34 (100 ng/mL, R&D Systems), and cholesterol (1.5 µg/mL, Avanti Polar Lipids). Single-cell libraries were generated using Chromium Single Cell 3′ Reagent Kit v3 (10× Genomics) by the UCSF Genomics Core57. The libraries were sequenced using Illumina NovaSeq 6000 according to the following protocol: read 1 (26 cycles, 16-bp cell barcode, 10-bp UMI), i7 (8 cycles, Illumina i7 sample index), and read 2 (91 cycles, transcript insert). The raw data have been uploaded to GEO (GSE 163948). The secure token for the reviewers is “irodugcozfudrmx”.
scRNA-Seq data analysis
scRNA-Seq analysis was performed using RStudio (v1.3)58 with the Seurat package (V3.2.1)38,59. In brief, cells were filtered based on genes per cell, UMIs per cell, and percentage of mitochondrial gene reads per cell. The data were log-normalized and scaled. The scRNA-Seq from each condition was integrated to remove batch effects. Each cell was assigned a score, based on its expression of G2/M- and S-phase markers, linear-dimension reduction was performed subsequently via principal component analysis on cell-cycle genes to visualize the scRNA-Seq data36,37. To perform the pseudobulk differential expression comparison analysis43, two clusters of cells of interest were first selected and differentially expressed genes were compared based on the negative binomial distribution model and visualized as volcano plots. The scRNA-Seq data were also analyzed by 10× Genomic Loupe Cell Browser.
Quantitative reverse-transcription polymerase-chain reaction
Total RNA was extracted with RNeasy Plus Micro Kit (QIAGEN) and cDNA was synthesized with SuperScript kit (Invitrogen). qRT-PCR was performed as previously described9 with Power iTag SYBR Green SuperMix (Bio-Rad) and specific primers in CFX384 Real-Time System (Bio-Rad). All primers were designed from the NCBI Primer-BLAST program. β-actin was used as the internal control for all the samples. The primer pairs used are
M b-actin F: CCACACCCGCCACCAGTTCG
M b-actin R: TACAGCCCGGGGAGCATCGT
cMyc 3′-F: AAAACAACGAAAAGGCCCCC
cMyc 3′-R: TTCAGAGGTGAGCTTGTGCT
qRT-PCR was also performed using the following TaqMan Gene Expression Assay probes with the TaqMan Universal PCR Master Mix and performed in CFX384 Real-Time System (Bio-Rad) according to the manufacturer’s suggested protocol (Thermo Fisher):
Actin: Mm02619580_g1
Myc: Mm00487804_m1
Tnfaip3: Mm00437121_m1
Cdk1: Mm00772472_m1
Hexb: Mm01282432_m1
Cx3cr1: Mm02620111_s1
Csf1r: Mm01266652_m1
P2ry12: Mm01950543_s1
Statistical analysis
The measurements were taken from distinct samples. Two-tailed Student’s t-test was used to compare means of two groups, and one-way or two-way ANOVA tests were used for multiple comparisons with GraphPad Prism 9.0.1 (GraphPad Software, San Diego). Data are presented as means ± SEM. The data distribution was assumed to be normal and variances were assumed to be equal across the groups, but this was not formally tested.
Supplementary information
Acknowledgements
The authors thank Drs. Allan Basbaum and Lewis Lanier for reading the previous version of the paper. Z.G. is supported by NINDS R01NS121144.
Author contributions
W.T. supervised, performed experiments, and analyzed data, and P.P.S. performed single-cell sequencing, most studies on Tnfaip3, and edited the paper. J.L. performed IHC, X.G. performed most qRT-PCR, J.C. performed part of the flow cytometry, and G.K. participated in some of the flow cytometry. A.G. initiated the whole project. A.M. contributed to A20 study, and Z.G. designed the experiments, analyzed the data, wrote the paper, and supervised all aspects of the study.
Data availability
Microglia single-cell sequencing data are available at GEO (GSE 163948), with reviewers’ token of “irodugcozfudrmx”.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Wulin Tan, Po-Yi Paul Su.
Supplementary information
The online version contains supplementary material available at 10.1038/s41421-022-00377-3.
References
- 1.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 3.Salter MW, Beggs S. Sublime microglia: expanding roles for the guardians of the CNS. Cell. 2014;158:15–24. doi: 10.1016/j.cell.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Rossi F, Lewis C. Microglia’s heretical self-renewal. Nat. Neurosci. 2018;21:455–456. doi: 10.1038/s41593-018-0123-3. [DOI] [PubMed] [Google Scholar]
- 5.Fuger P, et al. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat. Neurosci. 2017;20:1371–1376. doi: 10.1038/nn.4631. [DOI] [PubMed] [Google Scholar]
- 6.Reu P, et al. The lifespan and turnover of microglia in the human brain. Cell Rep. 2017;20:779–784. doi: 10.1016/j.celrep.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tay TL, et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 2017;20:793–803. doi: 10.1038/nn.4547. [DOI] [PubMed] [Google Scholar]
- 8.Askew K, et al. Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep. 2017;18:391–405. doi: 10.1016/j.celrep.2016.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan Z, et al. Injured sensory neuron-derived CSF1 induces microglial proliferation and DAP12-dependent pain. Nat. Neurosci. 2016;19:94–101. doi: 10.1038/nn.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olmos-Alonso A, et al. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer’s-like pathology. Brain. 2016;139:891–907. doi: 10.1093/brain/awv379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraft AD, Kaltenbach LS, Lo DC, Harry GJ. Activated microglia proliferate at neurites of mutant huntingtin-expressing neurons. Neurobiol. Aging. 2012;33:e17–e33. doi: 10.1016/j.neurobiolaging.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spiller KJ, et al. Microglia-mediated recovery from ALS-relevant motor neuron degeneration in a mouse model of TDP-43 proteinopathy. Nat. Neurosci. 2018;21:329–340. doi: 10.1038/s41593-018-0083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, et al. Proliferation of parenchymal microglia is the main source of microgliosis after ischaemic stroke. Brain. 2013;136:3578–3588. doi: 10.1093/brain/awt287. [DOI] [PubMed] [Google Scholar]
- 14.Bellver-Landete V, et al. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun. 2019;10:518. doi: 10.1038/s41467-019-08446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritzel RM, et al. Old age increases microglial senescence, exacerbates secondary neuroinflammation, and worsens neurological outcomes after acute traumatic brain injury in mice. Neurobiol. Aging. 2019;77:194–206. doi: 10.1016/j.neurobiolaging.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FM. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat. Neurosci. 2011;14:1142–1149. doi: 10.1038/nn.2887. [DOI] [PubMed] [Google Scholar]
- 17.Jordao, M. J. C. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science, 363, (6425):eaat7554. 10.1126/science.aat7554 (2019). [DOI] [PubMed]
- 18.Feng L, et al. Microglial proliferation and monocyte infiltration contribute to microgliosis following status epilepticus. Glia. 2019;67:1434–1448. doi: 10.1002/glia.23616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu X, et al. Macrophage antigen complex-1 mediates reactive microgliosis and progressive dopaminergic neurodegeneration in the MPTP model of Parkinson’s disease. J. Immunol. 2008;181:7194–7204. doi: 10.4049/jimmunol.181.10.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan TF, Liang YX, Peng B, Lin B, So KF. Local proliferation is the main source of rod microglia after optic nerve transection. Sci. Rep. 2015;5:10788. doi: 10.1038/srep10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu N, et al. Spinal microgliosis due to resident microglial proliferation is required for pain hypersensitivity after peripheral nerve injury. Cell Rep. 2016;16:605–614. doi: 10.1016/j.celrep.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shankaran M, et al. Measurement of brain microglial proliferation rates in vivo in response to neuroinflammatory stimuli: application to drug discovery. J. Neurosci. Res. 2007;85:2374–2384. doi: 10.1002/jnr.21389. [DOI] [PubMed] [Google Scholar]
- 23.Gomez-Nicola D, Fransen NL, Suzzi S, Perry VH. Regulation of microglial proliferation during chronic neurodegeneration. J. Neurosci. 2013;33:2481–2493. doi: 10.1523/JNEUROSCI.4440-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nixon K, Kim DH, Potts EN, He J, Crews FT. Distinct cell proliferation events during abstinence after alcohol dependence: microglia proliferation precedes neurogenesis. Neurobiol. Dis. 2008;31:218–229. doi: 10.1016/j.nbd.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han W, et al. Cranial irradiation induces transient microglia accumulation, followed by long-lasting inflammation and loss of microglia. Oncotarget. 2016;7:82305–82323. doi: 10.18632/oncotarget.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehmann ML, Cooper HA, Maric D, Herkenham M. Social defeat induces depressive-like states and microglial activation without involvement of peripheral macrophages. J. Neuroinflammation. 2016;13:224. doi: 10.1186/s12974-016-0672-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tashima R, et al. Bone marrow-derived cells in the population of spinal microglia after peripheral nerve injury. Sci. Rep. 2016;6:23701. doi: 10.1038/srep23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 29.Willis EF, et al. Repopulating microglia promote brain repair in an IL-6-dependent manner. Cell. 2020;180:833–846. doi: 10.1016/j.cell.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Guimaraes RM, et al. Frontline science: blood-circulating leukocytes fail to infiltrate the spinal cord parenchyma after spared nerve injury. J. Leukoc. Biol. 2019;106:541–551. doi: 10.1002/JLB.HI1118-458R. [DOI] [PubMed] [Google Scholar]
- 31.Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 32.Parkhurst CN, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda T, et al. Novel Hexb-based tools for studying microglia in the CNS. Nat. Immunol. 2020;21:802–815. doi: 10.1038/s41590-020-0707-4. [DOI] [PubMed] [Google Scholar]
- 35.Saederup N, et al. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS ONE. 2010;5:e13693. doi: 10.1371/journal.pone.0013693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scialdone A, et al. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 2016;535:289–293. doi: 10.1038/nature18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohno K, et al. Temporal kinetics of microgliosis in the spinal dorsal horn after peripheral nerve injury in rodents. Biol. Pharm. Bull. 2018;41:1096–1102. doi: 10.1248/bpb.b18-00278. [DOI] [PubMed] [Google Scholar]
- 40.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haimon Z, et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 2018;19:636–644. doi: 10.1038/s41590-018-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/S1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voet S, et al. A20 critically controls microglia activation and inhibits inflammasome-dependent neuroinflammation. Nat. Commun. 2018;9:2036. doi: 10.1038/s41467-018-04376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tavares RM, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalous, J., Jansova, D. & Susor, A. Role of cyclin-dependent kinase 1 in translational regulation in the M-phase. Cells, 9, 1568. 10.3390/cells9071568 (2020). [DOI] [PMC free article] [PubMed]
- 47.Garcia-Gutierrez L, et al. Myc stimulates cell cycle progression through the activation of Cdk1 and phosphorylation of p27. Sci. Rep. 2019;9:18693. doi: 10.1038/s41598-019-54917-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 2011;71:925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okubo M, et al. Macrophage-colony stimulating factor derived from injured primary afferent induces proliferation of spinal microglia and neuropathic pain in rats. PLoS ONE. 2016;11:e0153375. doi: 10.1371/journal.pone.0153375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malynn BA, Ma A. A20: A multifunctional tool for regulating immunity and preventing disease. Cell Immunol. 2019;340:103914. doi: 10.1016/j.cellimm.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das T, Chen Z, Hendriks RW, Kool M. A20/Tumor necrosis factor alpha-induced protein 3 in immune cells controls development of autoinflammation and autoimmunity: lessons from mouse models. Front. Immunol. 2018;9:104. doi: 10.3389/fimmu.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S, et al. Up-regulated A20 promotes proliferation, regulates cell cycle progression and induces chemotherapy resistance of acute lymphoblastic leukemia cells. Leuk. Res. 2015;39:976–983. doi: 10.1016/j.leukres.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 53.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl Acad. Sci. USA. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greten FR, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 55.Martin, E., El-Behi, M., Fontaine, B. & Delarasse, C. Analysis of microglia and monocyte-derived macrophages from the central nervous system by flow cytometry. J. Vis. Exp. 22, 55781.10.3791/55781 (2017). [DOI] [PMC free article] [PubMed]
- 56.Bohlen CJ, et al. Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron. 2017;94:759–773. doi: 10.1016/j.neuron.2017.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.RStudio. R Studio: Integrated Development for R. RStudio, http://www.rstudio.com (2020).
- 59.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microglia single-cell sequencing data are available at GEO (GSE 163948), with reviewers’ token of “irodugcozfudrmx”.