Abstract
We aimed to investigate the link between serum metabolites, gut bacterial community composition, and clinical variables in Parkinson’s disease (PD) and healthy control subjects (HC). A total of 124 subjects were part of the study (63 PD patients and 61 HC subjects). 139 metabolite features were found to be predictive between the PD and Control groups. No associations were found between metabolite features and within-PD clinical variables. The results suggest alterations in serum metabolite profiles in PD, and the results of correlation analysis between metabolite features and microbiota suggest that several bacterial taxa are associated with altered lipid and energy metabolism in PD.
Subject terms: Parkinson's disease, Metabolomics, Next-generation sequencing
Introduction
Parkinson’ disease (PD) is the second most common neurodegenerative disorder and is associated with prominent gastrointestinal pathophysiological changes and symptoms1. In the gut of PD patients, there are signs of low-grade inflammation, increased permeability, and bacterial invasion, all of which may predispose to overexpression and accumulation of alpha-synuclein that subsequently may spread to the brain in a prion-like fashion. Indeed, recent research suggests that early gastrointestinal involvement may be a key determinant of PD subtypes and that in a significant group of patients the origin of PD may lie in the gut. Gut microbiota impact brain health through multiple pathways, including production of neuroactive metabolites and neurotransmitters, but also through interactions with the immune system and potentially by excreting aggregation-prone proteins. Compositional alterations of gut microbiota in PD have been robustly demonstrated across multiple cohorts2,3 and have been linked to motor- and non-motor symptoms as well as progression of the disease. However, the functional implications of these changes regarding microbiota-host interactions and PD pathology and progression are still poorly understood. Alterations of faecal and serum/plasma metabolites and inflammatory markers have been described in relation to gut microbiota4–6, but except for reproducible findings of reduced faecal short-chain fatty acid (SCFA) levels in PD7, shortlisting of relevant metabolites and pathways has been challenging and inconsistent. Multiomics analyses have linked faecal microbiota abundances to alterations of amino acid metabolites4, lipids, sulphur metabolism, bile acids8, and SCFAs9 in serum/plasma, and alterations of lipids, vitamins, amino acids, SCFAs, and other organic compounds in faeces of PD patients10–12. Thus, more research is needed to better understand how an altered microbial composition and metabolic activity may impact PD.
The Helsinki cohort has so far been analysed for microbiome correlations with clinical features2,13 and disease progression14. We have also studied the oral and nasal microbe communities15. Recently, the immune response and fecal SFCA levels were studied among the same individuals6. The current study was primarily designed to investigate, as broadly as possible, the existence of possible links between gut bacteria and metabolite features, using a data-driven, hypothesis-generating approach (as opposed to hypothesis-testing approach), using untargeted serum metabolomics, 16S rRNA bacterial marker gene data, and clinical symptoms in PD as compared to healthy control subjects (HC). A total of 124 subjects were part of the study, divided into 63 PD patients and 61 HC subjects14 (see supplementary file “Supplementary population data table” for details). The serum samples were collected as close to the collection timepoint of the stool samples as practically possible, with the following average difference between stool and serum collection (days mean ± SD): PD (1.27 ± 1.42) and HC (0.69 ± 2.20).
Results
Metabolome analysis
Data-driven metabolomics profiling of serum samples was undertaken, identifying a total of 7585 metabolite features. Support Vector Machines (SVM) using RBF kernel showed 81% classification accuracy using GC-MS metabolic profiles, and 77% and 72% classification accuracy was achieved using LC-MS positive and negative mode ionisation data, respectively (Table 1).
Table 1.
Confusion matrices.
| Predicted control | Predicted PD | ||
|---|---|---|---|
| GC-MS CCR = 81% | Actual control | 83% | 17% |
| Actual PD | 20% | 80% | |
| LC-MS Positive Mode CCR = 77% | Actual control | 75% | 25% |
| Actual PD | 21% | 79% | |
| LC-MS Negative Mode CCR = 72% | Actual control | 72% | 28% |
| Actual PD | 28% | 72% |
Confusion matrices for (a) LC-MS positive mode data, (b) LC-MS negative mode data, (c) GC-MS data. For each data, confusion matrix shows an average of 100 models tested by resampling. Each time 60% data were used as training set and 40% were used as test set. Average correct classification rate (CCR) is represented for each of the data. Upon permutation of class labels, LC-MS positive mode CCR dropped to 49%, LC-MS negative mode CCR dropped to 47% and GC-MS CCR dropped to 47%.
Selection of predictive metabolite features between Controls and PD subjects was carried out using SVM recursive feature elimination to select the top 10% of features from profiling experiments. The metabolite features that contributed the most to the classification of PD and control samples in the SVM model, selected via SVM-RFE, were called “key predictive” metabolites and the same terminology will be used throughout this paper. A total of 139 features (i.e. metabolites) were selected: 101 features from LC-MS data and 38 features from GC-MS data (Table 2). Pathway enrichment analysis using all the 7585 metabolomics features revealed significant changes in carnitine shuttle, vitamin E metabolism, glycerophospholipids, sphingolipids, fatty acids and aminoacyl-tRNA biosynthesis amongst 20 perturbed pathways (Table 3). These features were also putatively annotated based on accurate mass match at 5ppm using human metabolome database (HMDB v.4)16 and LipidMaps17 following Metabolite Standards Initiative (MSI)18 at Level 3. Age, gender, BMI, dietary components, and PD-related clinical variables like medications did not show any evidence of biologically meaningful effects on the selected 139 metabolite features, with the possible exception of a very minor effect from COMT-inhibitors (see “Supplementary Table 7 (MODELS).xlsx” and the ‘Methods’ section for details). Within the PD group, no association was established between clinical variables and all 7585 metabolite features after adjusting for time since motor symptom onset, age at sampling, and other known clinical covariates (see also “Supplementary Table 7 (MODELS).xlsx” and the ‘Methods’ section for details). All 7585 metabolite features were used for this within-PD analysis because irrespective of their lack of predictive potential for PD (unlike the selected 139), some could still have been associated with those clinical variables that are of interest only within the PD group.
Table 2.
Key predictive metabolite features.
| Peak number | Putative ID | Metabolite feature class | Avg P/C foldchange |
|---|---|---|---|
| 349 | GalCer(d18:1/23:0);GlcCer(d18:1/23:0) | Sphingolipid | 2.314 |
| 458 | 20:2-Glc-Campesterol | Sterol lipid | 2.035 |
| 1303 | MGDG(20:5(5Z,8Z,11Z,14Z,17Z)/18:3(9Z,12Z,15Z)) | Glycerolipid | 2.024 |
| 264 | 6-Keto-decanoylcarnitine | Fatty acyls | 1.979 |
| 199 | Palmitoleic acid | Fatty acyls | 1.958 |
| 793 | Tetrahydroaldosterone-3-glucuronide | Steroid and derivatives | 1.731 |
| 856 | FAHFA(18:1(9Z)/13-O-18:0) | Fatty acyls | 1.722 |
| 447 | Veranisatin C | Prenol lipids | 1.661 |
| 1160 | 5-Methyltetrahydropteroyltri-L-glutamate | Steroid and derivatives | 1.624 |
| 151 | PE(18:4(6Z,9Z,12Z,15Z)/18:1(9Z)) | Glycerophospholipid | 1.604 |
| 1005 | Unknown 2 | Unknown | 1.586 |
| 299 | Neoabietic acid | Isoprenoids | 1.567 |
| 429 | PC(16:0/18:1(6Z));PC(16:0/18:1(6E)) | Glycerophospholipid | 1.488 |
| 501 | Galbanic acid | Prenol lipids | 1.485 |
| 368 | Deca-4,6,8-triyne-1,1,2,3-tetraol | Artificial chemical | 1.403 |
| 280 | Citrulline | Carboxylic acid and derivatives | 1.377 |
| 191 | Sphinganine-phosphate | Sphingolipid | 1.356 |
| 398 | Unknown 1 | Unknown | 1.354 |
| 75 | N-stearoyl tyrosine | Carboxylic acid and derivatives | 1.346 |
| 32 | (-)-Jolkinol B | Chemical | 1.336 |
| 320 | 11-cis-Dehydroretinal;all-trans-Dehydroretinal | Prenol lipids | 1.319 |
| 144 | 1alpha,24,25,28-tetrahydroxyergocalciferol | Vitamin D2 derivative | 1.290 |
| 204 | Glycerol | Sugar alchohol | 1.289 |
| 362 | PC(22:4(7Z,10Z,13Z,16Z)/0:0) | Glycerophospholipid | 1.274 |
| 70 | (3S,5R,6S,7E,9x)-7-Megastigmene-3,6,9-triol 9-glucoside | Fatty acyl glycoside | 1.263 |
| 369 | PE-Cer(d15:2(4E,6E)/22:0(2OH)) | Glycerophospholipid | 1.255 |
| 415 | Estrone, 16alpha-hydroxy- | Steroid and derivatives | 1.253 |
| 1201 | PC(P-16:0/18:4(6Z,9Z,12Z,15Z)) | Glycerophospholipid | 1.248 |
| 1137 | PE(16:0/P-18:1(11Z)) | Glycerophospholipid | 1.247 |
| 461 | Sphingosine-1-phosphate;Sphingosine 1-phosphate | Phosphosphingolipids | 1.245 |
| 386 | Heptadecane, n- | Alkane | 1.242 |
| 1474 | PI-Cer(t20:0/22:0(2OH)) | Glycerophospholipid | 1.242 |
| 170 | PA(P-18:0/17:2(9Z,12Z)) | Glycerophospholipid | 1.230 |
| 36 | PG(16:1(9Z)/22:4(7Z,10Z,13Z,16Z)) | Glycerophospholipid | 1.229 |
| 792 | PS(19:0/0:0) | Glycerophospholipid | 1.219 |
| 439 | PI(16:0/20:1(11Z)) | Glycerophospholipid | 1.209 |
| 370 | 3-octadecylenic acid | Fatty acyls | 1.201 |
| 518 | PC(18:4(6Z,9Z,12Z,15Z)/18:1(11Z)) | Glycerophospholipid | 1.195 |
| 67 | 4-O-alpha-Cadinylangolensin | Flavonoids | 1.186 |
| 430 | PC(P-20:0/18:3(6Z,9Z,12Z)) | Glycerophospholipid | 1.185 |
| 406 | SM(d16:1/22:0) | Sphingolipid | 1.171 |
| 393 | Propionylcarnitine | Fatty acyls | 1.164 |
| 420 | PC(16:1(9Z)/0:0);PC(16:1(9E)/0:0) | Glycerophospholipid | 1.163 |
| 47 | PE(18:4(6Z,9Z,12Z,15Z)/15:1(9Z)) | Glycerophospholipid | 1.129 |
| 463 | PC(P-16:0/20:3(8Z,11Z,14Z)) | Glycerophospholipid | 1.100 |
| 243 | PS(20:3(8Z,11Z,14Z)/0:0) | Glycerophospholipid | 1.097 |
| 414 | PA(O-16:0/21:0) | Glycerophospholipid | 1.097 |
| 423 | PC(P-20:0/18:2(9Z,12Z)) | Glycerophospholipid | 1.072 |
| 497 | SM(d17:1/24:1) | Sphingolipid | 1.056 |
| 1339 | PE(20:2(11Z,14Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | Glycerophospholipid | 1.056 |
| 509 | PI(O-16:0/13:0) | Glycerophospholipid | 1.049 |
| 511 | PC(18:3(9Z,12Z,15Z)/0:0) | Glycerophospholipid | 1.045 |
| 21 | 2-amino-2-deoxy-glucose | Glucose derivative | 1.043 |
| 500 | Butyrylcarnitine | Fatty acyls | 1.042 |
| 502 | PE(P-18:0/20:5(5Z,8Z,11Z,14Z,17Z)) | Glycerophospholipid | 1.038 |
| 160 | Methionine, N-formyl- | Amino acid derivative | 1.036 |
| 499 | 3-Deoxyvitamin D3 | Sterol lipid | 1.021 |
| 161 | Maltotriose | Oligosaccharides | 1.014 |
| 1338 | PG(P-20:0/20:1(11Z)) | Glycerophospholipid | 0.999 |
| 314 | Proline | Carboxylic acid and derivatives | 0.997 |
| 92 | Alanine, beta- | Carboxylic acid and derivatives | 0.996 |
| 459 | PC(P-18:0/20:5(5Z,8Z,11Z,14Z,17Z)) | Glycerophospholipid | 0.984 |
| 170 | Lactic acid, 3-imidazole- | Azoles | 0.975 |
| 467 | PE-Cer(d15:1(4E)/18:0) | Glycerophospholipid | 0.974 |
| 1434 | PI(15:0/22:0) | Glycerophospholipid | 0.959 |
| 348 | Cysteine, N-acetyl- | Drug | 0.955 |
| 68 | 3-Methyl-2-oxopentanoic-acid | Neurotoxin | 0.954 |
| 385 | Proline | Carboxylic acid and derivatives | 0.950 |
| 295 | Proline, 4-hydroxy-, trans- | Carboxylic acid and derivatives | 0.928 |
| 485 | SM(d18:1/21:0) | Sphingolipid | 0.928 |
| 1283 | PE(20:4(8Z,11Z,14Z,17Z)/20:4(8Z,11Z,14Z,17Z)) | Glycerophospholipid | 0.925 |
| 1391 | PS(19:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | Glycerophospholipid | 0.915 |
| 137 | Tridecane, n- | Alkane | 0.913 |
| 29 | Glucose, 2-amino-2-deoxy- | Glucose derivative | 0.912 |
| 498 | SM(d18:2/21:0) | Sphingolipid | 0.903 |
| 343 | Glycine, 2-phenyl- | Carboxylic acid and derivatives | 0.899 |
| 159 | Serine | Amino acid | 0.898 |
| 367 | PC(14:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | Glycerophospholipid | 0.896 |
| 65 | Dodecane | Alkane | 0.896 |
| 420 | Tartronic acid | Dicarboxylic acid | 0.893 |
| 11 | Dodecane | Alkane | 0.892 |
| 109 | Hydantoin, 5-methyl- | Allantoin metabolite | 0.892 |
| 47 | n-tricosane | Alkane | 0.888 |
| 845 | 2-methylbacteriohopane-32,33,34,35-tetrol | Prenol lipids | 0.886 |
| 247 | Proline, 4-hydroxy-, trans- | Carboxylic acid and derivatives | 0.885 |
| 474 | Acevaltrate | Carboxylic acid | 0.885 |
| 227 | Pentadecane, n- | Alkane | 0.883 |
| 355 | Glyceric acid | Sugar acids and derivatives | 0.876 |
| 126 | Methionine | Amino acid | 0.871 |
| 267 | Aniline, 3,4-dimethyl- | Xylidine isomer | 0.864 |
| 364 | Decane, n- | Alkane | 0.859 |
| 490 | PA(O-20:0/13:0) | Glycerophospholipid | 0.857 |
| 443 | GlcCer(d18:1(8Z)/21:0(2OH[R]));GlcCer(d18:1(8E)/21:0(2OH[R])) | Sphingolipid | 0.854 |
| 278 | 3-demethylubiquinone-9 | Prenol lipids | 0.852 |
| 323 | 3,3-Dibromo-2-n-hexylacrylic acid | Fatty acyls | 0.852 |
| 327 | Anandamide (20:5, n-3) | Fatty acid amide | 0.845 |
| 384 | Tetradecane, n- | Alkane | 0.845 |
| 134 | Unknown 4 | Unknown | 0.843 |
| 307 | Urea | Organic acids and derivatives | 0.842 |
| 387 | Benzaldehyde | Benzoids | 0.827 |
| 229 | 3,4-dimethyl-5-carboxyethyl-2-furanpentanoic acid | Furanoic fatty acids | 0.826 |
| 50 | Pyroglutamic acid | Carboxylic acid and derivatives | 0.816 |
| 433 | 25-hydroxy-1alpha-hydroxymethyl-23,23,24,24-tetradehydrocholecalciferol | Vitamin D metabolite | 0.815 |
| 379 | Galactose, 2-amino-2-deoxy-, D- | Glucose derivative | 0.800 |
| 1072 | OKHdiA-PS | Chemical | 0.797 |
| 360 | Heptadecane, n- | Alkane | 0.769 |
| 226 | Norvaline, DL- | Carboxylic acid and derivatives | 0.765 |
| 130 | 7,3’-Dihydroxy-4’-methoxy-8-methylflavan | Flavonoids | 0.752 |
| 188 | Unknown 3 | Unknown | 0.751 |
| 418 | Sorbitan stearate | Sorbitol derivative | 0.738 |
| 363 | 3-carboxy-4-methyl-5-pentyl-2-furanpropanoic acid | Furanoic fatty acids | 0.703 |
| 287 | Fuconic acid | Chemical | 0.690 |
| 361 | 3,4-dimethyl-5-carboxyethyl-2-furanhexanoic acid | Furanoic fatty acids | 0.690 |
| 399 | PE(O-20:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | Glycerophospholipid | 0.688 |
| 880 | DG(15:0/18:4(6Z,9Z,12Z,15Z)/0:0) | Fatty acyls | 0.684 |
| 390 | Fuconic acid | Chemical | 0.669 |
| 153 | Withaperuvin B | Steroid and derivatives | 0.655 |
| 1044 | OHOHA-PS | Chemical | 0.640 |
| 9 | OHOHA-PS | Chemical | 0.638 |
| 337 | C17 sphingosine-1-phosphocholine | Sphingolipid | 0.637 |
| 201 | Butenylcarnitine | Fatty acyls | 0.636 |
| 274 | (20S,24R)-20-fluoro-1alpha,24-dihydroxy-26,27-cyclovitamin D3 | Chemical | 0.634 |
| 311 | Sphingofungin A | Antifungal | 0.633 |
| 295 | Glycoursodeoxycholic acid | Steroid and derivatives | 0.610 |
| 177 | Glutarylcarnitine | Fatty acyls | 0.595 |
| 204 | 2,3-epoxyphylloquinone | Vitamin K derivative | 0.572 |
| 348 | Fuconic acid | Chemical | 0.560 |
| 276 | SM(d18:0/24:0) | Sphingolipid | 0.542 |
| 263 | Epigallocatechin 3-O-caffeate | Epigallocatechins | 0.536 |
| 115 | Hydroxybutyrylcarnitine | Fatty acyls | 0.522 |
| 5 | Palmitoleamide | Fatty amide | 0.519 |
| 365 | N-trans-Feruloyloctopamine | Cinnamic acids and derivatives | 0.515 |
| 313 | iodovulone I | Chemical | 0.485 |
| 321 | N2,N2-Dimethylguanosine | Purine nucleosides | 0.353 |
| 20 | Rubraflavone D | Flvonoids | 0.348 |
| 24 | Cycloheterophyllin | Pyranoflavonoids | 0.333 |
| 331 | cholesterol sulfate | Steroid and derivatives | 0.328 |
| 13 | Leukotriene D5 | Organooxygen compounds | 0.208 |
| 407 | PE(20:2(11Z,14Z)/0:0) | Glycerophospholipid | 0.105 |
Key predictive metabolite features between PD and Controls, organized in descending order of effect size. These top 10% metabolite features were selected after ranking them for their predictive power to distinguish between PD and HC. See ‘Methods’ section for details. The m/z features annotated as ‘Unknown’ had no accurate mass match or spectra match when compared to the library during database search. Mass spectra for these can be found in the Supplementary files as ”Supplementary Figure—Unknown X spectra”, with X corresponding to the respective unknown features 1, 2, 3, and 4 in the table.
Table 3.
Results from pathway analysis.
| Analytical Platform | Pathway name | Metabolite overlap | Pathway size | Adjusted p-value |
|---|---|---|---|---|
| LC-MS (pos mode) | Carnitine shuttle | 18 | 27 | 0.00934 |
| Vitamin E metabolism | 18 | 34 | 0.01417 | |
| Glycosphingolipid metabolism | 15 | 28 | 0.01585 | |
| N-Glycan Degradation | 5 | 6 | 0.01624 | |
| Porphyrin metabolism | 13 | 25 | 0.0209 | |
| Glycerophospholipid metabolism | 15 | 31 | 0.02649 | |
| Saturated fatty acids beta-oxidation | 8 | 15 | 0.03312 | |
| Linoleate metabolism | 9 | 18 | 0.03892 | |
| Squalene and cholesterol biosynthesis | 17 | 39 | 0.04848 | |
| LC-MS (neg mode) | De novo fatty acid biosynthesis | 13 | 18 | 0.00192 |
| Fatty acid activation | 12 | 17 | 0.00209 | |
| Hexose phosphorylation | 6 | 7 | 0.00292 | |
| Glycosphingolipid metabolism | 10 | 16 | 0.00384 | |
| Caffeine metabolism | 6 | 10 | 0.01308 | |
| Phosphatidylinositol phosphate metabolism | 4 | 6 | 0.02081 | |
| Fructose and mannose metabolism | 4 | 6 | 0.02081 | |
| Fatty Acid Metabolism | 4 | 6 | 0.02081 | |
| Starch and Sucrose Metabolism | 3 | 4 | 0.03001 | |
| Glycerophospholipid metabolism | 10 | 22 | 0.03784 | |
| GC-MS | Aminoacyl-tRNA biosynthesis | 6 | 48 | 0.00011601 |
| Pantothenate and CoA biosynthesis | 3 | 19 | 0.0037957 | |
| Valine, leucine and isoleucine biosynthesis | 2 | 8 | 0.007673 | |
| Phenylalanine metabolism | 2 | 10 | 0.012069 |
Results from pathway analysis for LC-MS and GC-MS data. Metabolite overlap shows the number of metabolites that overlap on the total metabolites on pathway indicated by pathway size. The p-values were adjusted for multiple comparisons as implemented within the Mummichog algorithm, as a penalisation process that takes into account the Cumulative Distribution Function (CDF) and the Expression Analysis Systematic Explorer (EASE).
Metabolome-microbiome correlation analysis
Correlation analysis between the selected 139 metabolite features and bacterial taxa at genus, family, and phylum levels was performed separately for the PD and Control groups to facilitate their contrast (Supplementary Tables 1–6). All results’ tables have been curated to obtain the best possible putative identification of the metabolite peak IDs at MSI level 3 identification level. Tables 2 and 5, as well as the two genus-level supplementary tables (Supplementary Tables 1 and 2) contain a metabolite “class” identification column (see ‘Methods’ section for details). All Supplementary Tables contain all identified taxa/metabolite correlation pairs (using the selected 139 metabolite features and all bacterial abundance data) that show posterior mean correlation values at or above 0.3 and at a 95% “confidence level” (see ‘Methods’ section for more details).
Table 5.
Mobile phases and gradient elution profile.
| Time (min) | Mobile Phase A (95:5 H2O:MeOH with 0.1% Formic acid) composition | Mobile Phase B (95:5 MeOH:H2O with 0.1% Formic acid) composition |
|---|---|---|
| 0 | 100 | 0 |
| 1 | 95 | 5 |
| 12 | 5 | 95 |
| 20 | 5 | 95 |
| 22 | 95 | 5 |
| 25 | 95 | 5 |
The within-PD analysis at genus level identified a total of 176 correlation pairs, while within-Controls analysis produced 202 pairs (Supplementary Tables 1 and 2, respectively). As can be seen, there is some overlap in the taxa and metabolites represented in the two groups, but overall there are substantial differences in the bacterial taxon-metabolite pair associations. To aid in the identification of possible links between metabolite classes and bacteria, we produced network figures for both within-PD and within-Controls results at genus level, using metabolite classes. (Figs. 1 and 2). At family level, the within-PD analysis identified a total of 67 correlation pairs, while the within-Controls analysis identified a total of 85 pairs (Supplementary Tables 3 and 4, respectively). Finally, the within-PD analysis at phylum level identified a total of 17 correlation pairs and the within-Controls analysis identified a total of 6 pairs (Supplementary Tables 5 and 6, respectively).
Fig. 1. Network of within-PD correlations.
Network of within-PD correlations between bacterial genera and metabolite classes. Supplementary Tables 1 and 2 contain correlations with bins composed of more than one genus (or higher taxon), which were unclassified at genus level, but these bins were removed from Figs. 1 and 2 to aid in visualization and to focus on the identified genera. Edge thickness represents the strength of the correlation. Blue edges represent positive correlations, and Orange edges represent negative correlations. Green nodes represent metabolite classes and may contain more than one metabolite feature (hence why there may be multiple edges between two nodes), and Cyan nodes represent bacterial genera. Check Table 6 at the end of this article for the key to the abbreviations used in Figs. 1 and 2.
Fig. 2. Network of within-Controls’ correlations.
Network of within-Controls’ correlations between bacterial genera and metabolite classes. Supplementary Tables 1 and 2 contain correlations with bins composed of more than one genus (or higher taxon) which were unclassified at genus level, but these bins were removed from Figs. 1 and 2 to aid in visualization and to focus on the identified genera. Edge thickness represents the strength of the correlation. Blue edges represent positive correlations, and Orange edges represent negative correlations. Green nodes represent metabolite classes and may contain more than one metabolite feature (hence why there may be multiple edges between two nodes), and Cyan nodes represent bacterial genera. Check Table 6 at the end of this article for the key to the abbreviations used in Figs. 1 and 2.
To further focus the study, we trimmed the correlation pairs down to only those containing bacterial taxa that were (i) not unclassified at the target taxonomic level, (ii) differentially abundant between PD and Control groups at one or both of the two time points of sample collection in a previous study using the same subject data14, and (iii) taxa that were systematically reported previously in the PD microbiome literature as being differentially abundant between PD and Control groups3,14 (Table 4). The bacterial abundance data used in the present article corresponds to the second time point of sample collection in Aho et al.14. To aid in visualizing the relationships, a third within-PD network figure was produced, using genus-level data and metabolite classes as before, but limited to the trimmed correlation pairs (Fig. 3).
Table 4.
Selected results.
| Metabolite Peak ID | Metabolite MSI 3 ID | Class | p2.5 | post. mean corr. | p97.5 | Taxon Diff Abund Direction Consensus | |
|---|---|---|---|---|---|---|---|
| (Aho et al. 2019/Boertien et al. 2020) | |||||||
| Bacterial Genus: | |||||||
| Within-PD analysis: | |||||||
| Prevotella | X7253 | Proline | Carboxylic acid and derivatives | −0.5146 | −0.3307 | −0.1186 | Decreased in PD/same |
| Prevotella | X7287 | Tartronic acid | Dicarboxylic acid | −0.5025 | −0.3148 | −0.1066 | Decreased in PD/same |
| Prevotella | X7206 | N-formyl-methionine | Carboxylic acid and derivatives | −0.4957 | −0.3063 | −0.0929 | Decreased in PD/same |
| Roseburia | X499 | 3-Deoxyvitamin D3 | Sterol lipid | −0.5249 | −0.3348 | −0.1159 | Decreased in PD/same |
| Roseburia | X423 | PC(P-20:0/18:2(9Z,12Z)) | Glycerophospholipid | 0.1034 | 0.3070 | 0.4970 | Decreased in PD/same |
| Roseburia | X370 | 3-octadecylenic acid | Fatty acyls | 0.1231 | 0.3300 | 0.5208 | Decreased in PD/same |
| Lactobacillus | X6092 | 3,4-dimethyl-5-carboxyethyl-2-furanpentanoic acid | Furanoic fatty acids | −0.5270 | −0.3246 | −0.0991 | Increased in PD/mostly increased in PD |
| Lactobacillus | X7038 | PE(20:2(11Z,14Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | Glycerophospholipid | 0.0916 | 0.3260 | 0.5291 | Increased in PD/mostly increased in PD |
| Akkermansia | X7038 | PE(20:2(11Z,14Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | Glycerophospholipid | −0.5454 | −0.3572 | −0.1350 | NA/increased in PD |
| Akkermansia | X6564 | PS(19:0/0:0) | Glycerophospholipid | 0.1056 | 0.3305 | 0.5304 | NA/increased in PD |
| Within-Controls analysis: | |||||||
| Bifidobacterium | X7163 | 2-amino-2-deoxy-glucose | Hexoses | −0.5043 | −0.3109 | −0.0854 | Increased in PD/mostly increased in PD |
| Bacterial Family: | |||||||
| Within-PD analysis: | |||||||
| Bifidobacteriaceae | X7145 | PI-Cer(t20:0/22:0(2OH)) | Glycerophospholipid | 0.0948 | 0.3069 | 0.4977 | Increased in PD/mostly increased in PD |
| Pasteurellaceae | X6860 | PE(16:0/P-18:1(11Z)) | Glycerophospholipid | 0.1106 | 0.3521 | 0.5620 | Decreased in PD/NA |
| Lactobacillaceae | X32 | (−)-Jolkinol B | Chemical | −0.5073 | −0.3027 | −0.0664 | Increased in PD/mostly increased in PD |
| Lactobacillaceae | X7038 | PE(20:2(11Z,14Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | Glycerophospholipid | 0.0823 | 0.3074 | 0.5096 | Increased in PD/mostly increased in PD |
| Verrucomicrobiaceae | X7038 | PE(20:2(11Z,14Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | Glycerophospholipid | −0.5430 | −0.3579 | −0.1496 | NA/increased in PD |
| Verrucomicrobiaceae | X6564 | PS(19:0/0:0) | Glycerophospholipid | 0.1288 | 0.3413 | 0.5297 | NA/increased in PD |
| Erysipelotrichaceae | X7253 | Proline | Carboxylic acid or derivative | 0.1616 | 0.3715 | 0.5648 | NA/mostly increased in PD |
| Within-Controls analysis: | |||||||
| Bifidobacteriaceae | X7163 | 2-amino-2-deoxy-glucose | Hexoses | −0.5127 | −0.3171 | −0.1049 | Increased in PD/mostly increased in PD |
| Lactobacillaceae | X7183 | Beta-alanine | Carboxylic acid | −0.5079 | −0.3062 | −0.0822 | Increased in PD/mostly increased in PD |
| Lactobacillaceae | X7081 | PS(19:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | Glycerophospholipid | 0.1223 | 0.3725 | 0.5795 | Increased in PD/mostly increased in PD |
| Enterobacteriaceae | X485 | SM(d18:1/21:0) | Sphingolipid | 0.1140 | 0.3280 | 0.5281 | NA/mostly increased in PD |
| Enterobacteriaceae | X430 | PC(P-20:0/18:3(6Z,9Z,12Z)) | Glycerophospholipid | 0.1292 | 0.3299 | 0.5325 | NA/mostly increased in PD |
| Bacterial Phylum: | |||||||
| Within-PD analysis: | |||||||
| Verrucomicrobia | X7038 | PE(20:2(11Z,14Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | Glycerophospolipid | −0.5306 | −0.3414 | −0.1231 | NA/increased in PD |
| Verrucomicrobia | X6564 | PS(19:0/0:0) | Glycerophospolipid | 0.1064 | 0.3345 | 0.5302 | NA/increased in PD |
Selected results based on taxa previously reported in the literature at genus, family, and phylum levels. The last column presents a consensus on direction of effect based on previous reports: before the slash (/), we report the result obtained in Aho et al.14 using the same bacterial data as in the present study; after the slash, we report the consensus reported by Boertien et al.3 (see that study for details). NA means that no result for that taxon is available in that study14.
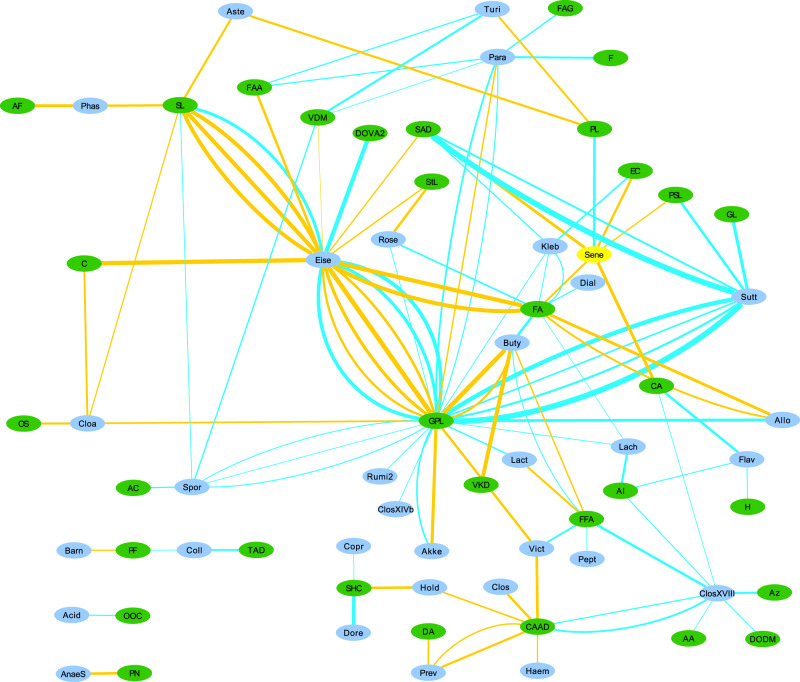
Fig. 3. Network of selected within-PD correlations.
Network of within-PD correlations between bacterial genera and metabolite classes using selected results (Table 4). Edge thickness represents the strength of the correlation. Blue edges represent positive correlations, and Orange edges represent negative correlations. Green nodes represent metabolite classes, and Cyan nodes represent bacterial taxa.
In the Helsinki cohort, six bacterial genera were previously reported as being differentially abundant (selected for the present article at an alpha threshold of statistical significance of 0.05 from the original 0.1), using one or more statistical methods14, between Control and PD groups at one or both time points, namely: Bifidobacterium, Roseburia, Prevotella, Blautia, Lactobacillus, and Clostridium XIVa. All these genera, except for Clostridium XIVa, have also been reported as being differentially abundant in previous publications contrasting a control group to PD patients14. Of these, Prevotella, Bifidobacterium, Roseburia, and Lactobacillus are also found to be correlated with one or more metabolite features in our genus-level analysis (Table 4).
Other differentially abundant bacterial genera reported previously in the PD microbiome literature3 besides those referred to in Aho et al.14 have also been found covarying with metabolite features in our dataset, and we also used that information for the purpose of focusing our study’s results. At genus level, Akkermansia, Bifidobacterium, Faecalibacterium, Prevotella, Lactobacillus, and Roseburia were reported multiple times in the literature, with only Akkermansia and Faecalibacterium not being reported in the Aho et al.14 study as being differentially abundant (Table 4).
In the Aho et al.14 study, seven bacterial families were reported as being differentially abundant between PD and Control groups at one or both time points, namely: Bifidobacteriaceae, Prevotellaceae, Rikenellaceae, Lachnospiraceae, Pasteurellaceae, Lactobacillaceae, and Puniceicoccaceae. All these families, except for Puniceicoccaceae, have also been reported as being differentially abundant in previous publications contrasting a control group to PD patients14. Three of them showed correlations with one or more metabolite features in our family-level analysis (Table 4). Boertien et al.3 also reported on the bacterial families most commonly found to be differentially abundant, namely Bifidobacteriaceae, Prevotellaceae, Lachnospiraceae, Lactobacillaceae, Verrucomicrobiaceae, Enterobacteriaceae, Erysipelotrichaceae, and Ruminococcaceae, with the last four families not being found to be differentially abundant at any time point in our cohort14 (Table 4).
For this study, we have also analysed our data at phylum level, unlike in Aho et al.14, and found correlations between various metabolites and the phyla Lentisphaerae, Verrucomicrobia, Synergistetes, and Tenericutes (Supplementary Tables 5 and 6). Some phyla recurrently found to be differentially abundant between PD and Control groups are Verrucomicrobia, Firmicutes, and Bacteroidetes3, although of those reported only Verrucomicrobia yielded correlations with metabolite features in our analysis (Table 4).
Discussion
Using mummichog approach, we have shown in this study that the identified serum metabolome differences in PD have functional significance on over 20 pathways, including carnitine shuttle, vitamin E metabolism, glycerophospholipids, sphingolipids, fatty acids, and aminoacyl-tRNA biosynthesis. In a separate study, we have demonstrated that carnitine shuttle, sphingolipids, and fatty acids pathways change in Parkinson’s sebum—these are within the 20 pathways enlisted above detected in serum in this study19.
The changes observed in pathways associated with sphingolipid metabolism may indicate a key shift in cell signalling and regulation. Dysregulation of sphingolipids is known to be associated with α-synucleinopathy20,21, changes in lysosomal metabolism, and in mitochondrial metabolism observed in PD22,23. Sphingolipids have shown strong associations with many neurodegenerative conditions as recently reviewed by Alessenko and Albi24. In addition to altered sphingolipid metabolism in plasma, metabolomics and lipidomics studies in PD have shown changes in ceramides, sphingosine and sphingosine-1-phosphate in particular25,26. It is not surprising, given that one of the top pathways where notable changes are seen in this study is linked to glycerophospholipid metabolism. Glycerophospholipids and sphingolipids are well-known as the ‘ying and yang’ of lipotoxicity in metabolic disease27. The dysregulation of these complementary and opposed forces in the metabolome leads to lipotoxicity seen in many metabolic diseases. It can thus be speculated that such lipotoxic insult may be one of the underlying pathophysiologies of PD, as measured in serum.
Decreased long-chain acylcarnitines due to insufficient β-oxidation has been shown to carry potential for early diagnosis of Parkinson’s28, especially 12–14 long chain acylcarnitines. In recent work studying the gut microbiome, Rosario et al.29 have shown the role of bacterial folate and homocysteine metabolism in PD. Higher numbers of bacterial mucin and host degradation enzymes were linked to the manifestation of PD. The contribution of bacterial folate metabolism to human metabolic regulation is not entirely clear. Folate is an essential vitamin B, that maintains methylation reactions. The liver, via many methylation reactions in post-translational modifications, regulates the synthesis of hormones, creatine, carnitine, and phosphatidylcholine30. If methylation capacity is compromised due to an alteration in folate metabolism, there may be impaired phosphatidylcholine synthesis along with shunted or disrupted carnitine shuttle observed in our results. Altered carnitine metabolism, fatty acids, and steroid metabolism were also observed in a metabolomics profiling study recently reported31. Several studies have reported decreased levels of carnitine and acylcarnitines in plasma from PD patients28,32,33; however, according to Jiménez-Jiménez et al.34 no changes were observed in acylcarnitine levels in plasma or cerebrospinal fluids of PD participants. Thus, there is no clear evidence of direction in which carnitines are expressed but there is much research evidence that suggests a link between perturbations in carnitine shuttle owing to protective mechanism of acylcarnitines leading to changes in other fatty acids and eventually the lipid make-up in PD. Researchers have shown molecules such as LDL and HDL to have direct association with sebum and related diseases such as acne35 and demonstrated that lipid metabolism is not just a diet processing effect, but a complex interaction that affects lipid uptake from the gut, biosynthesis in liver and sequestration in tissues including on skin36. These results in serum metabolome could be indicative of link between gut microbiome, serum metabolome and sebum metabolome.
Energy metabolism is highly regulated by facilitation of long chain fatty acid β-oxidation. Also, in serum from frail11 elderly participants without Parkinson’s, dysregulation of carnitine shuttle and vitamin E metabolism was observed when compared to similarly aged resilient individuals37. Thus, perturbation of carnitine shuttle and vitamin E, along with fatty acids in serum metabolome may indicate a significant change in energy metabolism during PD. Further, Vitamin E’s role as a protective factor against Parkinson’s has been extensively studied along with vitamin C for therapy of early onset PD38–41. Serum metabolome based on pathway analysis presented in this study indicates changes broadly in energy metabolism and lipid metabolism in PD. These disruptions have been recently reported in other biofluids such as sebum19 and CSF42 and the community is increasingly recognising the role of lipid dysregulation, well summarized in these reviews20,43,44. We used knowledge from our microbiome analysis to investigate if these metabolic shifts were entirely endogenous or were also partly contributed to by changing gut microbiome in Parkinson’s disease.
Regarding the correlations between bacterial taxa and metabolite features, and given that metabolite MSI 3 ID is a putative identification, we will mostly focus the following discussion at the level of metabolite classes. When mentioning specific bacterial taxa in terms of correlation results, we will report between brackets if the taxon is always (or usually) reported in the literature as being over- or underrepresented in PD (see the last column in the Table 4 for details), as well as the signal of the correlation detected in the present study.
At all bacterial taxonomic levels investigated in this study, the most commonly detected correlations were found with putative metabolites in the glycerophospholipid class and with lipids in general. This is the case both in our non-trimmed results (Supplementary Tables 1–6) and in the trimmed results focusing only on bacterial taxa found in the previous literature as being differentially abundant between Controls and PD cases (Table 4). Specifically, in the within-PD analysis and focusing the discussion on the trimmed results, we find correlations between various glycerophospholipids and Roseburia (decreased in PD; positive correlation), Lactobacillus (increased in PD; positive correlation), Akkermansia (increased in PD; one positive and one negative correlation), Bifidobacteriaceae (increased in PD; positive correlation), Pasteurellaceae (decreased in PD; positive correlation), Lactobacillaceae (increased in PD; positive correlation), Verrucomicrobiaceae (increased in PD; one positive and one negative correlation), and Verrucomicrobia (increased in PD; one positive and one negative correlation) (Table 4). Akkermansia, Verrucomicrobiaceae, and Verrucomicrobia share the same positive and negative correlations with two metabolite features. Lactobacillus and Lactobacillaceae correlate positively with the same metabolite feature.
With the exception of Roseburia and Pasteurellaceae, all these taxa are usually found to be overrepresented in PD, and are mostly positively correlated with various glycerophospholipids, which are also found in our analysis to be mostly overrepresented in PD (Table 2). This is not the case in the within-controls analysis, in which the only detected correlations with glycerophospholipids are with Lactobacillaceae (positive correlation with a different metabolite) and Enterobacteriaceae (also a positive correlation with a different metabolite) (Table 4). The genus-level network figures for PD and Controls (Figs. 1 and 2) also indicate the existence of possible alterations in bacterial metabolic dynamics in PD. Overall, these results suggest that these bacterial taxa, which have been found to be overabundant in PD in several studies, may be associated for the most part with an increase in glycerophospholipid abundance in PD.
All of these glycerophospholipids have an endogenous (human host) origin and are linked to cell signalling, lipid peroxidation, and lipid metabolism. These phospholipids are the main component of cell membranes in all known living systems, and play roles in various biological processes, including signal induction and acting as transporters. Interestingly, there are several genetic factors directly or indirectly related to glycerophospholipid metabolism, such as PLA2G620, that are associated with PD risk (PLA2G6 is the cause of early-onset PARK14-linked dystonia- parkinsonism45,46). Alpha-synuclein, characteristically found in aggregates within Lewy bodies in the brains of PD patients, directly binds to negatively charged phospholipids in the cells’ lipid membranes, and exhibits preferential binding to small lipid vesicles47. The binding of alpha-synuclein to lipid membranes can also lead to alterations in their bilayer structure that can induce the formation of those lipid vesicles48. Very importantly in the PD context, these interactions between lipid membranes and alpha-synuclein affect its rate of aggregation, and can lead to disruption of membrane integrity both in vitro and in vivo49. It has also been shown that the association of soluble alpha-synuclein with planar lipid bilayers results in the formation of aggregates and small fibrils50. Exposure to docosahexaenoic acid (DHA), which accounts for 60% of glycerophospholipid esterified fatty acids in the plasma membrane, gradually assembles alpha-synuclein into amyloid-like fibrils, with the notable feature that DHA itself becomes part of the aggregate51. Notably, alpha-synuclein gene expression is increased with elevated DHA intake, and the resulting oligomers are toxic to cells52,53. Alpha-synuclein also binds with specific phospholipids in mitochondrial membranes, modulating the efficiency of mitochondrial energy production54, with various mitochondrial phospholipids appearing to have an effect on alpha-synuclein toxicity20. Thus, the interaction between alpha-synuclein and various phospholipids and their metabolism may play an important role in PD pathogenesis, and gut microbiota may be implicated in these interactions.
We also detected correlations with other lipids in the within-PD analysis. Roseburia (decreased in PD) negatively correlates with a sterol lipid, probably of dietary origin. Roseburia is also positively correlated with a metabolite in the fatty acyl class, possibly also associated with diet. Lactobacillus (increased in PD) is negatively correlated with a furanoic fatty acid, which is associated with cell signalling, lipid peroxidation, lipid metabolism, and lipid transport metabolism, with dietary, human, and/or bacterial origin. Half of the metabolites from the fatty acyl class were found to be overrepresented in PD in the selected 139 metabolite features in our blood serum data (Table 2), but overall underrepresented in the PD sebum data from Sinclair et al.19.
In the within-controls analysis we detected a positive correlation between Enterobacteriaceae (increased in PD) and a sphingolipid, which is associated with membrane stabilization, lipid peroxidation, and lipid metabolism, of endogenous origin. In our study, this metabolite feature is slightly decreased in PD (Table 2) and no correlation is found between it and PD-linked bacterial taxa in the within-PD analysis.
Although the interpretation of these results regarding lipids in general is not suggestive of a particular pattern as in the case of glycerophospholipids, it is nevertheless interesting that virtually all correlations, positive or negative, with lipids are detected in the PD group, with most lipids detected in our study being overrepresented in PD relative to the control group (Table 2). Also interesting is that the majority of the identified correlations between metabolite features and the trimmed bacterial taxa list are with lipids, with relatively few other metabolite groups represented in the results (Table 4). The importance of this link between lipid metabolism and PD can’t be overstated: as mentioned earlier in the context of phospholipids, recent research shows that alpha-synuclein binds preferentially to specific lipid families and molecules, and that the latter promote alpha-synuclein interaction with synaptic membranes and affect alpha-synuclein oligomerization and aggregation. These same lipid-protein complexes also affect lipid metabolism by interfering with the catalytic activity of lipid enzymes in the cytoplasm and lipases in lysosomes. Lipid compositional alterations in PD have also been reported in brain and plasma, as well as linked to oxidative stress, inflammation, and progressive neurodegeneration through pro-inflammatory lipid mediators (see Alecu et al.20 for a full review on the role of lipids in PD). The link between lipids and bacteria in PD, if any, would probably consist of the bacterial modulation of lipid intake through diet and its differential effect on the bioavailability of those lipids in the host. The results of our study, by establishing associations between bacterial taxa found to be differentially abundant between Controls and the PD group and lipid metabolites present in serum that are themselves predictive of PD, suggests that such a scenario could have a role in PD pathology and development.
Further correlations with putative metabolites in other classes have also been detected in our study, in particular in the hexoses class and carboxylic acid or derivatives class. In the hexoses case, two negative correlations for the same metabolite feature were found for Bifidobacterium and Bifidobacteriaceae (both taxon levels increased in PD; Table 4). These two correlations are only found in the within-controls analysis. The metabolite is probably endogenous in origin and is involved in sugar metabolism shunts, diverting a proportion of glucose from the main glycolytic path and returning metabolites at the level of triose phosphate and fructose 6-phosphate.
Finally, a positive correlation in the within-PD analysis is found between a metabolite feature in the carboxylic acid or derivatives class and the bacterial family Erysipelotrichaceae, which is mostly found in previous studies to be overrepresented in PD. This metabolite feature is tentatively identified as proline and may have a microbiome or endogenous source. Four different metabolite features found to be predictive of PD in our data are tentatively identified as proline (Table 2), and in all cases show a slight decrease in mean abundance in PD. Interestingly, L-proline can act as a weak agonist for glycine and glutamate receptors55, like NMDA, AMPA, and kainite. Both glutamate and glycine are neurotransmitters. Although that is not the case in our data, proline has been reported previously as being overrepresented in PD31 and is known to be linked to protein metabolism and structure, cell differentiation, conceptus growth and development56, and gut microbiota community re-equilibration in cases of dysbiosis, with L-proline dietary supplementation being known to affect gut microbial composition and gut concentrations of several bacterial metabolites57. One of the detected correlations with Prevotella also involves (putative) proline. Prevotella and Prevotellaceae, when detected in PD studies, are usually found decreased in PD. In this study it correlates negatively with proline, which is found to be slightly underrepresented in the PD group.
In conclusion, we designed our study to investigate, as broadly as possible, the existence of associations between bacteria and metabolite abundances in PD. To this end, we used a data-driven, hypothesis-generating approach based on untargeted metabolomics data. It is our hope that the results can a posteriori be used to build more focused studies of either an observational or experimental nature to explore our putative findings. Circa 7500 serum metabolite features were detected by gas chromatography and liquid chromatography using untargeted metabolomics. Of these, 139 were deemed to be particularly predictive for PD status. For the most part they are related to lipid metabolism, and are also mostly overrepresented in PD. The evidence for metabolic differences in PD is related to carnitine shuttle, vitamin E metabolism, glycerophospholipids, sphingolipids, and fatty acids, suggesting alterations in PD related to energy and lipid metabolism. Our results indicate that the abundance of several gut bacterial genera (e.g. Prevotella, Bifidobacteria, Akkermansia, Lactobacillus, Roseburia) correlates with the abundance of several of these metabolite features, and thus may be implicated in these metabolic alterations in PD.
One limitation of this study, like in many other metabolomics clinical studies, could be the variation introduced by drug usage. We have analysed possible associations between the metabolite features against several medications (e.g. LEDD score) and found no clear indication of drug effects. However, without a targeted metabolomics study, it is hard to rule out such effects. We may have these drugs or their breakdown products in our data, but we cannot target them specifically due to the unknown masses of all possibly relevant drug metabolites. We hope that untargeted metabolomics studies like the present one could, with their broad hypothesis-generation approach, serve as a basis for future targeted metabolomics studies that could better deal with potential sources of variation such as drugs or their metabolic products.
Methods
Study subjects, clinical data, and sampling
This study was conducted in accordance with the Declaration of Helsinki and was approved by the ethics committee of the Hospital District of Helsinki and Uusimaa. All participants provided written informed consent.
The present study uses bacterial abundance data that was used in a previously published study by Aho et al.14. The study subjects and associated clinical data in the present study is similar to the data referred to previously in that study as “follow-up” timepoint, with minor changes specific to the present study: of the original 128 subjects, 61 control subjects and 63 PD patients were used in the present study, i.e. 3 control subjects and 1 PD subject less than in the original cohort (C75, C82, C123, and P119). This difference in sample numbers was due to insufficient metabolite data available to perform the study.
For DNA sequencing, the stool samples were collected at home by the study subjects using tubes containing DNA stabilizer (PSP Spin Stool DNA Plus Kit, STRATEC Molecular), which were stored for a maximum of three days in a freezer until transport. At the clinic, the received samples were stored at −80 °C and later transferred to the sequencing centre, where they were also stored at −80 °C until further processing14.
For serum samples, blood was drawn at the study visit and, after processing, immediately transferred to −20 °C and subsequently to −80 °C. Samples were shipped overnight on dry ice from Helsinki to Manchester for analysis.
Sample preparation and metabolomics methods
Metabolomics sample preparation
Untargeted metabolite profiling was performed on serum samples that were collected from participants and stored at −80 °C prior to analysis. Complementary coverage of metabolites was obtained using ultra-high performance liquid chromatography mass spectrometry (UHPLC-MS) and gas chromatography mass spectrometry (GC-MS). The procedures were adapted from the Dunn58 and Begley59 protocols as summarized here:
Metabolites were extracted from the serum samples by individually adding 400 µL of cold methanol to 200 µL of serum. This was followed by vortexing and centrifugation (17,500 × g) to yield a metabolite rich supernatant that was split into two aliquots and lyophilised for 12 h. Resultant metabolite pellet was stored at −80 °C until analysis. A pooled QC standard was also generated by combining 20 µL aliquots of each sample into a pooled vial with subsequent 200 µL aliquots from the pool, being extracted identical to each sample.
LC-MS method parameters
Processed metabolite pellets were defrosted at 4 °C and subsequently reconstituted in 100 µL of 95:5 H2O:MeOH (v/v). UHPLC-MS analysis was performed using an Accela UHPLC with cooled auto sampler system coupled to an electrospray LTQ-Orbitrap XL hybrid mass spectrometer (ThermoFisher, Bremen, Germany). Analysis was carried out in positive and negative ESI modes while samples were completely randomised to negate for any bias. The mobile phases and gradient elution profile were as tabulated in Table 5. From each sample vial, 10 µL of the extract was injected onto a Hypersil GOLD UHPLC C18 column (length 100 mm, diameter 2.1 mm, particle size 1.9 µm, Thermo-Fisher Ltd. Hemel Hempsted, UK) held at a constant temperature of 50 °C with a solvent flow rate of 400 µL min−1.
Prior to analysis, LTQ-Orbitrap XL was calibrated according to manufacturer’s instructions using caffeine (20 µg mL−1), the tetrapeptide MRFA (1 µg ml−1) and Ultramark 1621 (0.001%) in an aqueous solution of acetonitrile (50%), methanol (25%) and acetic acid (1%). The data acquisition was performed in centroid mode with 30 K mass resolution and scan rate of 400 ms per scan. The masses were measured between 100 and 1200 m/z range with source gases set at sheath gas = 40 arb units, aux gas = 0 arb units, sweep gas = 5 arb units. The ESI source voltage was set to 3.5 V, and capillary ion transfer tube temperature set at 275 °C.
LC-MS data processing
Xcaliber software (v.3.0; Thermo-Fisher Ltd. Hemel Hempsted, UK) was used as the operating software for the Thermo LTQ-Orbitrap XL mass spectrometer. Data processing was initiated by the conversion of the standard UHPLC.raw files into the.mzML format using Proteowizard60. Subsequently, peak picking was carried out in RStudio61 using the XCMS62 package for data deconvolution (http://masspec.scripps.edu/xcms/xcms.php). The output data was a matrix of mass spectral features with accurate m/z and retention time pairs. Any missing values after deconvolution were replaced using k-nearest neighbours algorithm. Peaks with relative standard deviation of more than 20% within pooled QCs were removed. The remaining data was normalised with total ion count to account for injection to injection signal variations, log10 transformed, and pareto scaled prior to statistical analysis.
GC-MS method parameters
Analysis of serum samples was also carried out on a Agilent 7250 GC-Time-of-Flight mass spectrometer coupled to a Gerstel-MPS autosampler. Two step derivatization of metabolite pellets thawed at 4 °C was carried out as described in the Begley protocol59. The source temperature was set to 230 °C and quad temperature was at 150 °C. The total run time was 25 min for 10 µL sample injected each time. The sample was injected in split mode with 20:1 split ratio and split flow of 20 mL per minute. Agilent CP8944 VF-5 ms column was used for separation (30 m × 250 µm × 0.25 µm). With a 5 min solvent delay at the start of run, gradient elution method was used to elute and separate analytes from serum. The oven temperature was ramped from 70 °C to 300 °C with an increase of 14 °C per minute. At 300 °C the temperature was held for 4 min before dropping back to starting conditions.
GC-MS data processing
The raw data files were in Agilent.D format that were converted to.mzML format using Proteowizard60. Peak picking was carried out in RStudio61 using an in-house script for the eRah63 package for GC-MS peak picking and deconvolution. The peaks were annotated using eRah’s MassBank library. Any missing values after deconvolution were replaced using k-nearest neighbours algorithm. Peaks with relative standard deviation of more than 20% within pooled QCs were removed. The remaining data was normalised with total ion count to account for injection to injection signal variations, log10 transformed, and pareto scaled prior to statistical analysis.
All metabolites successfully annotated within both the LC-MS and GC-MS analysis were assessed and scored at MSI level 3 putative identification according to rules set out by the Chemical Analysis Working Group of the Metabolite Standards Initiative18.
Sample preparation and DNA sequencing
DNA extraction was performed according to the manufacturer’s instructions. We then amplified the V3-V4 regions of the bacterial 16 S rRNA gene, using two technical replicates (25 μL reactions) per biological sample, and a mixture of the universal bacterial primers 341F1–4 (5′ CCTACGGGNGGCWGCAG 3′) and 785R1–4 (5′ GACTACHVGGGTATCTAATCC 3′) with partial Illumina TruSeq adapter sequences added to the 5′ ends (F1; ATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT, F2; ATCTACACTCTTTCCCTACACGACGC TCTTCCGATCTgt, F3; ATCTACACTCTTTCCCTACACGACGCTCTTCCGA TCTagag, F4; ATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTtagtgt and R1; GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT, R2; GTGACT GGAGTTCAGACGTGTGCTCTTCCGATCTa, R3; GTGACTGGAGTTCAGACGT GTGCTCTTCCGATCTtct, R4; GTGACTGGAGTTCAGACGTGTGCTCTTCCGA TCTctgagtg)14. The small letters in the above sequences are additional nucleotides introduced for purposes of mixing in the sequencing process. We performed two-step PCRs followed by quantification, pooling, and purification14. The resulting PCR products were then sequenced with Illumina MiSeq (v3 600 cycle kit), with 325 bases and 285 bases for the forward and reverse reads, respectively.
Bioinformatics and statistical data analysis
Statistical analysis of metabolomics data
In this untargeted profiling study, we detected a total of 7585 features combined between LC-MS positive ionisation mode, LC-MS negative ionisation mode and GC-MS data. After processing the raw data and applying QC based relative standard deviation filtering, in LC-MS positive ionisation mode 5897 features remained, and in LC-MS negative ionisation mode data 1260 features remained. In GC-MS data 428 features were retained for statistical analysis. As described earlier in this article, these features were scaled and transformed prior to any statistical analysis.
(i) Classification by disease status
Individual dataset from LC-MS positive mode, LC-MS negative mode, and GC-MS were first analysed separately and then standardised combined data was used to generate a model based on the whole metabolome measured by 7585 features in total. Individual datasets from each technique were analysed using partial least squares discriminant analysis (PLS-DA) to classify between PD and control using the Metaboanalyst package in R. The models were validated by 60:40 split of data with 250 bootstraps and also by comparing correct classification rates with permuted models (Supplementary Table 7 (MODELS)).
In order to investigate the classification of samples into PD and control, support vector machines (SVM) were used on 7585 metabolite features. Using Python via Orange user interface, SVM models were generated for this analysis. The data were pre-treated as described earlier in this article. The data were then split into train (60% data) and test (40% data), and resampling was repeated 100 times. The SVM model was generated with RBF-kernel, cost (C) was set to 1.5, and regression loss epsilon was set at 0.10. We accounted for potential effects of hypercholesterolaemia, gender, age at sampling, BMI, defecations per week, weekly stool characteristics average, clinical scores (GDS15, MMSE, NMSQ, NMSS, RBDSQ, Rome-III constipation score, SDQ, Wexner total, Progression score, and Rome-III IBS criteria) as well as 68 dietary and lifestyle related variables as described in Supplementary table 7 (MODELS). The top 10% variables were selected after ranking them by ReliefF algorithm, and models were regenerated to rule out the possibility of an effect due to a large number of metabolite features.
(ii) Key predictive metabolite feature selection
To select metabolite features (i.e., features predictive of PD status) contributing to the SVM models, the mSVM-RFE algorithm was used64. The iterative algorithm worked backwards from an initial set of features consisting of all the metabolite features in the dataset. At each iterative round, firstly a simple linear SVM was fitted, then features were ranked based on their weights in the SVM solution and lastly, the algorithm eliminated the feature with the lowest weight. In order to stabilize these feature rankings, at each iteration cross validation resampling was used. By using k-fold cross validation (k = 10) multiple SVM-RFE iterations were carried out. From the resultant ranked feature list, the top 10% of the features were selected (the 139 key predictive metabolite features) for further interpretation as they contributed the most towards SVM models.
(iii) Effects of PD medications, UPDRS-III score, and time since onset of motor symptoms on key metabolite features selected
During the selection of the key predictive metabolite features, potential effects from clinical variables like PD medication could not be taken directly into account in those models, since they are restricted to the PD group. However, such an approach would leave open the question regarding possible effects of those variables on the metabolite predictors. Thus, to investigate the potential effects of PD medications, UPDRS-III score, and time since motor symptoms on PD vs HC classification (i.e. the 139 metabolite features), and to investigate associations of drug dosages to the key metabolite features, partial least squares and logistic regression were carried out within-PD for continuous and categorical responses, respectively. The variables that had continuous scaled values were subjected to partial least squares regression with 20 PCs, for 1000 iterations. The variables that had categorical values were subjected to logistic regression with ridge penalization with cost value of 1. All partial least squares and logistic regression models were validated by leave-one-out cross-validation (LOOCV). Partial least squares R2 and root mean squared error (RMSE) and logistic regression classification accuracy were used for model evaluation (Supplementary Table 7 (MODELS)). The top 10% of variables were selected after ranking them by ReliefF algorithm, and models were regenerated to rule out the possibility of an effect due to a large number of metabolite features. Our results indicate that the choice of 139 metabolite features was not affected by PD medication use or dosage. One possible exception could be COMT inhibitors, which may have a weak effect and may be associated with the PD metabolome in serum: using the 139 key metabolites, we were able to classify between a PD subject with and without COMT inhibitor treatment with an accuracy of 63%, which is low. Thus, these results suggest by extension that drug use did not affect the choice of the 139 key predictive metabolite features in any substantial way during the HC vs PD predictive feature analysis. Supplementary Table 7 (MODELS) includes information about variables and adjusted variables, for each model.
(iv) Effects of clinical variables on metabolome within PD
We investigated potential effects and association of clinical variables within the PD cohort. All metabolite features (not just the key predictive metabolite features) were regressed against clinical features of Parkinson’s viz. GDS-1565 (depression), MMSE66 (cognition), NMSS67 (non-motor symptoms), RBDSQ68 (REM-sleep behaviour disorder), Rome-III constipation score69, Rome-III IBS status, Wexner score70 (constipation), SCS-PD71 (drooling), SDQ72 (dysphagia), UPDRS II-III, Hoeh & Yahr scale, and progression category from Aho et al.14, while adjusting for model covariates (see Supplementary Table 7 (MODELS)). All RBDSQ and Progression categories were adjusted for age at sampling and time since motor onset. SCS-PD, SDQ, UPDRS-II, UPDRS-III and Hoehn & Yahr were also adjusted for LEDD. UPDRS-III was additionally adjusted for beta blockers. Wexner score and Rome-III IBS status and Rome-III constipation scores were also adjusted for anticholinergic medication, constipation medication, opioids, and tricyclic medications, as well as dietary fibre intake. SCS-PD was additionally adjusted for anticholinergic and tricyclics. GDS-15 was additionally adjusted for SSRI medications. MMSE was additionally adjusted for anticholinergic and tricyclic medications.
Partial least squares and logistic regression was carried out for continuous and categorical responses, respectively. The features that had continuous scaled values were subjected to partial least squares regression with 20 PCs, for 1000 iterations. The features that had categorical values were subjected to logistic regression with ridge penalization with cost value of 1. All partial least squares and logistic regression models were validated by splitting data into 60:40 training: test sets and resampling for 100 times. Partial least square R2 and root mean squared error (RMSE) and logistic regression classification accuracy were used for model evaluation (Supplementary Table 7 (MODELS)). Top 10% of variables (759 variables) were selected after ranking them by ReliefF algorithm, and models were regenerated to rule out the possibility of an effect due to a large number of metabolite features.
(v) Pathway analysis
In this data driven approach, we have interrogated data generated from an untargeted profiling study. It is often impractical to identify each peak in a metabolomics profiling study as it could contain upwards to 5000 features in a single sample. To identify them accurately, the only option is to purchase commercial standards and perform MS/MS analysis in both samples and standards and then match fragmentation spectra. This could be relevant when performing a targeted analysis with a defined set of metabolites. Computationally predicted m/z based identification alone is not adequate for pathway analysis due to multiple metabolite matches to single m/z. Thus, we have employed mummichog analysis that does not depend on identification of metabolites and then mapping on pathways. Instead, mummichog leverages the collective power in the organisation of metabolic networks. If a list of m/z values truly reflects a biological activity, the true metabolites that are represented by these m/z values should show enrichment on a local structure of a metabolic network. If the measured m/z matches to a falsely represented metabolite, the distribution will be observed randomly. The overall significance of mapping and pathway enrichment is estimated by ranking the p-values from the real data among the p-values from permutation data to adjust for type I error, along with penalisation. Thus, a robust functional metabolic network gives us insight into our data more than identifying a handful of features. Mummichog73 (v.1.0.9) pathway analysis was used to predict network activity from pre-processed UHPLC-MS metabolomics data. The full metabolite data set consisting of 5897 and 1260 features from LC-MS positive and negative mode, respectively, was used as an input. Pathway enrichment analysis was performed on annotated 428 GC-MS features using MetaboAnalystR74.
Data pre-processing for 16S rRNA gene sequence data
The raw sequence data amounted to a total of 34,701,899 reads. In brief, primers were removed from the reads using cutadapt before further processing14. We then used mothur to pair, quality trim, taxonomically classify, and finally cluster the reads into OTUs, following mothur’s Standard Operating Procedure (SOP) for MiSeq. The following customizations were made to the SOP parameters: insert = 40 and deltaq = 10 in make.contigs; maxlength = 450 in the first screen.seqs step; start = 6428 and end = 23,440 in the second screen.seqs step; diffs = 4 in pre.cluster. Singleton sequences were also removed with split.abunds (cutoff = 1) before running classify.seqs. The reference databases used were the full-length SILVA alignment release 128 for align.seqs and the RDP 16S rRNA reference (PDS) version 16 for classify.seqs. The final, processed data set (without sequencing blanks) consisted of 18,867 278 reads14.
Metabolome-microbiome correlation analysis
For correlation analysis between metabolites and bacterial taxa at genus, family, and phylum levels, we used the fido75 package (v.0.1.13; the package was formerly known as stray) for the R Statistical Programming Software76 (v.3.6.0). fido provides a framework for inferring multinomial logistic-normal models which can account for zeros and compositional constraints, as well as sampling and technical variation present in sequence count data75. For the present study we used the function orthus from fido which enables joint modelling of multivariate count data (e.g. 16S rRNA gene amplicon sequence data) and multivariate Gaussian data (e.g. metabolomics data on the log-scale).
For samples j ∈ {1, …, N} we denote by Yj the observed D1 -dimensional vector of sequence counts, Zj the standardized (i.e. Z-transformed) and log10 -transformed P -dimensional vector of observed metabolite abundances, and Xj a Q -dimensional vector of covariates. Using this notation, the orthus likelihood model is given by
| 1 |
with priors and Inverse Wishart and with denoting the inverse additive log-ratio (ALR) transform with respect to the D -th taxa77. Of note, the ultimate inference is invariant to the chosen ALR transform. This represents a joint linear model over the latent relative abundances of microbial taxa and metabolite abundances. For computational scalability this model was inferred using the multinomial-Dirichlet Bootstrap approximation to the marginal posterior density p (π | Y) that is available in fido. The multinomial-Dirichlet bootstrap approximates the true marginal posterior density using the posterior of a Bayesian multinomial-Dirichlet model centred at the maximum a posteriori (map) estimate of π. In brief, this is accomplished as follows: for each sample j, the marginal posterior distribution p (πj | Y) is approximated as the posterior of a Bayesian multinomial Dirichlet model where . Here, the Dirichlet parameters αj were all taken to be 0.5; this can be thought of as a probabilistic equivalent to using a pseudo-count of 0.5 yet also producing quantified uncertainty due to multivariate counting. The prior parameters were chosen as
| 2 |
and v = D + P + 9. Finally, we set the prior
| 3 |
where G is the (D − 1) × D ALR contrast matrix given by . This choice of Θ, Γ, , and v represents the weak prior belief that the correlation between the absolute abundance of taxa is, on average, small. This prior is closely related to the sparse penalization used by SparCC78. Using this model, priors, and inference, we sampled 2000 independent samples from the posterior distribution p (Λ, Σ | Y, Z, X).
Variable selection for these metabolite feature/microbiota correlation models was performed as follows: we selected the relevant variables based on multivariate analyses of the communities’ Bray-Curtis dissimilarity measure using PERMANOVA, a semi-parametric approach that does not assume multivariate normality. These analyses were performed separately for the within-Controls and within-PD groups. First, all clinical and technical variables of interest were analysed on their own in a univariable model (i.e. a model with a single explanatory variable). Given that the amount of variance explained in microbiome models is always very low, the choice of variables at this step was based solely on achieving statistical significance equal to or lower than 0.05. The variables that passed this alpha threshold of significance were then combined into a multivariable model (i.e. a model with more than one explanatory variable) using marginal testing. Those that retained significance at 0.05 or less in this full model were considered for the Bayesian covariance models. Of the latter, only those variables in common between the metabolite-based variable selection (see section iii above) and the microbiota-based variable selection were used for the Bayesian covariance models. Thus, for the three within-Parkinson’s covariance models (i.e. using only PD subjects at three taxonomic levels), the models were adjusted for COMT inhibitor medication use. For the three within-Controls covariance models we used intercept-only models.
The matrix Σ represents a ([D − 1] + Q) × ([D − 1] + Q) covariance matrix encoding all possible covariances between ALR coordinates and metabolites. For model interpretation and inference, each posterior sample of the upper (D − 1) × Q submatrix was transformed to a D × Q matrix representing the covariance between microbial composition (now represented with respect to centred log-ratio coordinates, or CLR) and metabolite abundances. Covariances were transformed to correlations using the function cov2cor in the R programming language. For the purposes of this study, we considered only those correlations that had a posterior mean equal to or larger than 0.3 and that had a 95% credible region not including zero. Conditioned on our chosen priors, this decision boundary can be thought of as limiting our results to correlations which we believe (with at least 95% certainty) are non-zero.
Covariance modelling was performed for bacterial genus, family, and phylum levels, using only metabolite abundance data at “Peak ID” level. This means that, although several of their corresponding MSI level 3 putative identifications were nominally the same, these were not merged before analysis, because they have different retention times and there is non-negligible uncertainty in their identification. After the correlations were calculated, we broadly assigned metabolite class information to these metabolites for Table 4 and Table 2 to aid in interpretation. These class assignments were then used to produce the Cytoscape79 network visualisations (v.3.8.0), both because classes simplify the networks and because they are more plausible than the putative MSI level 3 identifications. These classes were assigned by searching each putative identification of a metabolite feature against the Human Metabolome Database16 (HMDB) and the Kyoto Encyclopedia of Genes and Genomes80 (KEGG) entry (Table 6).
Table 6.
Key to Figures 1 and 2.
| Metabolite class key | Metabolite feature class | Metabolite class key | Metabolite feature class |
|---|---|---|---|
| AA | Amino acid | GL | Glycerolipid |
| AC | Acylcarnitines | GuD | Glucose derivative |
| AF | Antifungal | GPL | Glycerophospholipid |
| Al | Alkane | H | Hexoses |
| Ar | Arylamine | NT | Neurotoxin |
| ArC | Artificial chemical | OAAD | Organic acids and derivatives |
| Az | Azoles | OOC | Organooxygen compounds |
| C | Chemical | OS | Oligosaccharides |
| CA | Carboxylic acid | PF | Pyranoflavonoids |
| CAAD | Carboxylic acid and derivatives | PL | Prenol lipids |
| DA | Dicarboxylic acid | PN | Purine nucleosides |
| DODM | Drug or drug metabolite | PSL | Phosphosphingolipids |
| DOVA2 | Derivative of Vitamin A2 | SAD | Steroid and derivatives |
| EC | Epigallocatechins | SD | Sorbitol derivative |
| F | Flavonoids | SHC | Saturated hydrocarbon |
| FA | Fatty acyls | SL | Sphingolipid |
| FAA | Fatty acid amide | StL | Sterol lipid |
| FAG | Fatty acyl glycoside | TAD | Tyrosine and derivatives |
| FFA | Furanoic fatty acids | VDM | Vitamin D metabolite |
| GD | Galactose derivative | VKD | Vitamin K derivative |
| Genus key | Genus | Genus key | Genus |
|---|---|---|---|
| Acid | Acidaminococcus | Hold | Holdemanella |
| Akke | Akkermansia | Howa | Howardella |
| Allo | Alloprevotella | Inte | Intestinimonas |
| AnaeS | Anaerostipes | Kleb | Klebsiella |
| AnaeT | Anaerotruncus | Lach | Lachnospira |
| Asac | Asaccharobacter | Lact | Lactobacillus |
| Aste | Asteroleplasma | Mega | Megasphaera |
| Barn | Barnesiella | Odor | Odoribacter |
| Bifi | Bifidobacterium | Para | Parasutterella |
| Buty | Butyrivibrio | ParaP | Paraprevotella |
| Cate | Catenibacterium | Pept | Peptococcus |
| Cloa | Cloacibacillus | Phas | Phascolarctobacterium |
| Clos | Clostridium_sensu_stricto | Prev | Prevotella |
| ClosXIVb | Clostridium_XlVb | Porp | Porphyromonas |
| ClosXVIII | Clostridium_XVIII | Pseu | Pseudoflavonifractor |
| Coll | Collinsella | Romb | Romboutsia |
| Copr | Coprobacter | Rose | Roseburia |
| Desu | Desulfovibrio | Rumi | Ruminococcus |
| Dial | Dialister | Rumi2 | Ruminococcus2 |
| Dore | Dorea | Sene | Senegalimassilia |
| E/S | Escherichia/Shigella | Stre | Streptococcus |
| Eise | Eisenbergiella | Spor | Sporobacter |
| Euba | Eubacterium | Succ | Succiniclasticum |
| Faec | Faecalicoccus | Sutt | Sutterella |
| Flav | Flavonifractor | Turi | Turicibacter |
| Gemm | Gemmiger | Veil | Veillonella |
| Haem | Haemophilus | Vict | Victivallis |
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This study was funded by the Michael J. Fox Foundation for Parkinson’s Research, the Academy of Finland (295724, 310835), the Finnish Medical Foundation, and the Hospital District of Helsinki and Uusimaa (UAK1014004, UAK1014005, TYH2018224, TYH2020335). D.K.T. thanks Michael J Fox Foundation and Parkinson’s UK for supporting research leading to contribution to this work, and Prof. Perdita Barran for her invaluable input throughout this project. We thank Dr. Velma Aho for supporting this project by curating metadata from the Helsinki cohort, and for the R script for collapsing taxonomic levels.
Author contributions
F.S. conceived the study. P.A.B.P., D.K.T., I.C.D., L.P., P.A., and F.S. developed and organized the study, and collected samples, data, and measurements. P.A.B.P. and D.K.T. analysed the data and are “co-first authors”, having contributed equally to this manuscript. J.S. provided statistical analysis support. L.P. was responsible for the design and execution of the amplicon sequencing. P.A.B.P., D.K.T., J.S., and F.S. drafted the manuscript. All authors read, reviewed, edited, and approved the final manuscript. All authors have contributed substantially for this study and meet all four authorship criteria required by the present journal.
Data availability
The 16S rRNA gene sequence raw data is available at the European Nucleotide Archive, with the accession number PRJEB27564. The metabolomics data is available at MetaboLights, with the accession number MTBLS4332. The clinical data is also available upon direct request from the corresponding authors of the present article. This is due to European patient confidentiality laws, and may require signing a Data Usage Agreement, depending on the specifics of the request.
Code availability
Code for the bacterial taxa-metabolite correlation analyses is available as R scripts’ files (Supplementary files “Metabolomics.SERVER.SCRIPT.PD_ONLY.Selected.CORR.final” and “Metabolomics.SERVER.SCRIPT.CONTROLS_ONLY.Selected.CORR.final”). For metabolomics-only data analysis, deconvolution R scripts using XCMS and eRah, Matlab code for Partial Least Squares Discriminant Analysis (PLS-DA), R code for metabolite correlations, and Python code for Support Vector Machine (SVM) are available in the GitHub repository at github.com/drupadt/.
Competing interests
P.A.B.P., L.P., P.A. and F.S. have patents issued (FI127671B & US10139408B2) and pending (US16/186,663 & EP3149205) that are assigned to NeuroBiome Ltd. F.S. is founder and CEO of NeuroInnovation Oy and NeuroBiome Ltd., is a member of the scientific advisory board and has received consulting fees and stock options from Axial Biotherapeutics. Authors J.S., I.C.D. and J.S. declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pedro A. B. Pereira, Drupad K. Trivedi.
Contributor Information
Pedro A. B. Pereira, Email: pedro.pereira@helsinki.fi
Filip Scheperjans, Email: filip.scheperjans@hus.fi.
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-022-00300-3.
References
- 1.Skjærbæk C, Knudsen K, Horsager J, Borghammer P. Gastrointestinal dysfunction in Parkinson’s disease. J. Clin. Med. 2021;10:3 493. doi: 10.3390/jcm10030493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheperjans, et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015;30:350–8. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 3.Boertien JM, Pereira PAB, Aho VTE, Scheperjans F. Increasing comparability and utility of gut microbiome studies in Parkinson’s disease: a systematic review. J. Parkinsons Dis. 2019;9:S297–S312. doi: 10.3233/JPD-191711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirstea, et al. Microbiota composition and metabolism are associated with gut function in Parkinson’s disease. Mov. Disord. 2020;35:1208–1217. doi: 10.1002/mds.28052. [DOI] [PubMed] [Google Scholar]
- 5.Tan, et al. Gut microbial ecosystem in parkinson disease: new clinicobiological insights from multi-omics. Ann. Neurol. 2021;89:546–559. doi: 10.1002/ana.25982. [DOI] [PubMed] [Google Scholar]
- 6.Aho, et al. Relationships of gut microbiota, short-chain fatty acids, inflammation, and the gut barrier in Parkinson’s disease. Mol. Neurodegener. 2021;16:6. doi: 10.1186/s13024-021-00427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unger, et al. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat. Disord. 2016;32:66–72. doi: 10.1016/j.parkreldis.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Hertel, et al. Integrated analyses of microbiome and longitudinal metabolome data reveal microbial-host interactions on sulfur metabolism in Parkinson’s disease. Cell Rep. 2019;29:1767–1777.e8. doi: 10.1016/j.celrep.2019.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin C, Lim Y, Lim H, Ahn TB. Plasma short-chain fatty acids in patients with Parkinson’s disease. Mov. Disord. 2020;35:1021–1027. doi: 10.1002/mds.28016. [DOI] [PubMed] [Google Scholar]
- 10.Vascellari, et al. Gut microbiota and metabolome alterations associated with Parkinson’s disease. mSystems. 2020;5:5 e00561–20. doi: 10.1128/mSystems.00561-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan, et al. Gut microbial ecosystem in Parkinson disease: new clinicobiological insights from multi-omics. Ann. Neurol. 2021;89:546–559. doi: 10.1002/ana.25982. [DOI] [PubMed] [Google Scholar]
- 12.Yan, et al. Alterations of gut microbiota and metabolome with Parkinson’s disease. Micro. Pathog. 2021;160:105187. doi: 10.1016/j.micpath.2021.105187. [DOI] [PubMed] [Google Scholar]
- 13.Mertsalmi, et al. More than constipation—bowel symptoms in Parkinson’s disease and their connection to gut microbiota. Eur. J. Neurol. 2017;24:1375–1383. doi: 10.1111/ene.13398. [DOI] [PubMed] [Google Scholar]
- 14.Aho, et al. Gut microbiota in Parkinson’s disease: temporal stability and relations to disease progression. EBioMedicine. 2019;44:691–707. doi: 10.1016/j.ebiom.2019.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira, et al. Oral and nasal microbiota in Parkinson’s disease. Parkinsonism Relat. Disord. 2017;38:61–67. doi: 10.1016/j.parkreldis.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 16.Wishart, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sud, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35(Database issue):D527–32. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumner, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinclair, et al. Metabolomics of sebum reveals lipid dysregulation in Parkinson’s disease. Nat. Commun. 2021;12:1592. doi: 10.1038/s41467-021-21669-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alecu I, Bennett SAL. Dysregulated lipid metabolism and its role in α-synucleinopathy in Parkinson’s disease. Front Neurosci. 2019;13:328. doi: 10.3389/fnins.2019.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin G, Wang L, Marcogliese PC, Bellen HJ. Sphingolipids in the pathogenesis of Parkinson’s disease and Parkinsonism. Trends Endocrinol. Metab. 2019;30:106–117. doi: 10.1016/j.tem.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Hallett PJ, Engelender S, Isacson O. Lipid and immune abnormalities causing age-dependent neurodegeneration and Parkinson’s disease. J. Neuroinflammation. 2019;16:153. doi: 10.1186/s12974-019-1532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xicoy H, Wieringa B, Martens GJM. The role of lipids in Parkinson’s disease. Cells. 2019;8:27. doi: 10.3390/cells8010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alessenko AV, Albi E. Exploring sphingolipid implications in neurodegeneration. Front Neurol. 2020;11:437. doi: 10.3389/fneur.2020.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, et al. Integrated metabolomics and proteomics analysis reveals plasma lipid metabolic disturbances in patients with Parkinson’s disease. Front Mol. Neurosci. 2020;13:80. doi: 10.3389/fnmol.2020.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Kruining, et al. Sphingolipids as prognostic biomarkers of neurodegeneration, neuroinflammation, and psychiatric diseases and their emerging role in lipidomic investigation methods. Adv. Drug Deliv. Rev. 2020;159:232–244. doi: 10.1016/j.addr.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Cuenca S, Pellegrinelli V, Campbell M, Oresic M, Vidal-Puig A. Sphingolipids and glycerophospholipids—The “ying and yang” of lipotoxicity in metabolic diseases. Prog. Lipid Res. 2017;66:14–29. doi: 10.1016/j.plipres.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Saiki, et al. Decreased long-chain acylcarnitines from insufficient β-oxidation as potential early diagnostic markers for Parkinson’s disease. Sci. Rep. 2017;7:7328. doi: 10.1038/s41598-017-06767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosario, et al. Systematic analysis of gut microbiome reveals the role of bacterial folate and homocysteine metabolism in Parkinson’s disease. Cell Rep. 2021;34:108807. doi: 10.1016/j.celrep.2021.108807. [DOI] [PubMed] [Google Scholar]
- 30.da Silva RP, Kelly KB, Al Rajabi A, Jacobs RL. Novel insights on interactions between folate and lipid metabolism. Biofactors. 2014;40:277–83. doi: 10.1002/biof.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao, et al. Comprehensive metabolic profiling of Parkinson’s disease by liquid chromatography-mass spectrometry. Mol. Neurodegener. 2021;16:4. doi: 10.1186/s13024-021-00425-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao, et al. Potential biomarkers of Parkinson’s disease revealed by plasma metabolic profiling. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018;1081-1082:101–108. doi: 10.1016/j.jchromb.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 33.Crooks, et al. Carnitine levels and mutations in the SLC22A5 gene in Faroes patients with Parkinson’s disease. Neurosci. Lett. 2018;675:116–119. doi: 10.1016/j.neulet.2018.03.064. [DOI] [PubMed] [Google Scholar]
- 34.Jiménez-Jiménez, et al. Cerebrospinal fluid carnitine levels in patients with Parkinson’s disease. J. Neurol. Sci. 1997;145:183–5. doi: 10.1016/s0022-510x(96)00259-6. [DOI] [PubMed] [Google Scholar]
- 35.Utami OC, Kurniawati Y, Diba S, Saleh MI. Correlation between serum lipid profile and acne vulgaris severity. J. Phys.: Conf. Ser. 2019;1246:012066. [Google Scholar]
- 36.Camera E, Picardo M. Lipids in serum and sebum, Pathogenesis and Treatment of Acne and Rosacea (Springer, 2013)
- 37.Rattray, et al. Metabolic dysregulation in vitamin E and carnitine shuttle energy mechanisms associate with human frailty. Nat. Commun. 2019;10:5027. doi: 10.1038/s41467-019-12716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fariss MW, Zhang JG. Vitamin E therapy in Parkinson’s disease. Toxicology. 2003;189:129–46. doi: 10.1016/s0300-483x(03)00158-6. [DOI] [PubMed] [Google Scholar]
- 39.Etminan M, Gill SS, Samii A. Intake of vitamin E, vitamin C, and carotenoids and the risk of Parkinson’s disease: a meta-analysis. Lancet Neurol. 2005;4:362–5. doi: 10.1016/S1474-4422(05)70097-1. [DOI] [PubMed] [Google Scholar]
- 40.Schirinzi, et al. Dietary Vitamin E as a protective factor for Parkinson’s Disease: clinical and experimental evidence. Front Neurol. 2019;10:148. doi: 10.3389/fneur.2019.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikam S, Nikam P, Ahaley SK, Sontakke AV. Oxidative stress in Parkinson’s disease. Indian J. Clin. Biochem. 2009;24:98–101. doi: 10.1007/s12291-009-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernández-Irigoyen J, Cartas-Cejudo P, Iruarrizaga-Lejarreta M, Santamaría E. Alteration in the cerebrospinal fluid lipidome in Parkinson’s disease: a post-mortem pilot study. Biomedicines. 2021;9:491. doi: 10.3390/biomedicines9050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fanning S, Selkoe D, Dettmer U. Parkinson’s disease: proteinopathy or lipidopathy? NPJ Parkinsons Dis. 2020;6:3. doi: 10.1038/s41531-019-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erskine, et al. Lipids, lysosomes and mitochondria: insights into Lewy body formation from rare monogenic disorders. Acta Neuropathol. 2021;141:511–526. doi: 10.1007/s00401-021-02266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, et al. Phospholipase PLA2G6, a Parkinsonism-associated gene, affects Vps26 and Vps35, retromer function, and ceramide levels, similar to α-synuclein gain. Cell Metab. 2018;28:605–618.e6. doi: 10.1016/j.cmet.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 46.Belarbi, et al. Glycosphingolipids and neuroinflammation in Parkinson’s disease. Mol. Neurodegener. 2020;15:59. doi: 10.1186/s13024-020-00408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu M, Li J, Fink AL. The association of alpha-synuclein with membranes affects bilayer structure, stability, and fibril formation. J. Biol. Chem. 2003;278:40186–97. doi: 10.1074/jbc.M305326200. [DOI] [PubMed] [Google Scholar]
- 48.Madine J, Doig AJ, Middleton DA. A study of the regional effects of alpha-synuclein on the organization and stability of phospholipid bilayers. Biochemistry. 2006;45:5783–92. doi: 10.1021/bi052151q. [DOI] [PubMed] [Google Scholar]
- 49.Rawat A, Langen R, Varkey J. Membranes as modulators of amyloid protein misfolding and target of toxicity. Biochim Biophys. Acta Biomembr. 2018;1860:1863–1875. doi: 10.1016/j.bbamem.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jo E, McLaurin J, Yip CM, St George-Hyslop P, Fraser PE. alpha-Synuclein membrane interactions and lipid specificity. J. Biol. Chem. 2000;275:34328–34. doi: 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- 51.Broersen K, van den Brink D, Fraser G, Goedert M, Davletov B. Alpha-synuclein adopts an alpha-helical conformation in the presence of polyunsaturated fatty acids to hinder micelle formation. Biochemistry. 2006;45:15610–6. doi: 10.1021/bi061743l. [DOI] [PubMed] [Google Scholar]
- 52.De Franceschi, et al. Molecular insights into the interaction between alpha-synuclein and docosahexaenoic acid. J. Mol. Biol. 2009;394:94–107. doi: 10.1016/j.jmb.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 53.De Franceschi, et al. Structural and morphological characterization of aggregated species of α-synuclein induced by docosahexaenoic acid. J. Biol. Chem. 2011;286:22262–74. doi: 10.1074/jbc.M110.202937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ludtmann, et al. Monomeric alpha-synuclein exerts a physiological role on brain ATP synthase. J. Neurosci. 2016;36:10510–10521. doi: 10.1523/JNEUROSCI.1659-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henzi V, Reichling DB, Helm SW, MacDermott AB. L-proline activates glutamate and glycine receptors in cultured rat dorsal horn neurons. Mol. Pharm. 1992;41:793–801. [PubMed] [Google Scholar]
- 56.Wu, et al. Proline metabolism in the conceptus: implications for fetal growth and development. Amino Acids. 2008;35:691–702. doi: 10.1007/s00726-008-0052-7. [DOI] [PubMed] [Google Scholar]
- 57.Ji Y, Guo Q, Yin Y, Blachier F, Kong X. Dietary proline supplementation alters colonic luminal microbiota and bacterial metabolite composition between days 45 and 70 of pregnancy in Huanjiang mini-pigs. J. Anim. Sci. Biotechnol. 2018;9:18. doi: 10.1186/s40104-018-0233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dunn, et al. Human serum metabolome (HUSERMET) consortium. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011;6:1060–83. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 59.Begley, et al. HUSERMET Consortium, Goodacre R, Kell DB. Development and performance of a gas chromatography-time-of-flight mass spectrometry analysis for large-scale nontargeted metabolomic studies of human serum. Anal. Chem. 2009;81:7038–46. doi: 10.1021/ac9011599. [DOI] [PubMed] [Google Scholar]
- 60.Chambers, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–20. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.RStudio Team. RStudio: Integrated Development Environment for R. Boston, MA. Available from: http://www.rstudio.com/ (2015).
- 62.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–87. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 63.Domingo-Almenara, et al. eRah: a computational tool integrating spectral deconvolution and alignment with quantification and identification of metabolites in GC/MS-based metabolomics. Anal. Chem. 2016;88:9821–9829. doi: 10.1021/acs.analchem.6b02927. [DOI] [PubMed] [Google Scholar]
- 64.Duan KB, Rajapakse JC, Wang H, Azuaje F. Multiple SVM-RFE for gene selection in cancer classification with expression data. IEEE Trans. Nanobioscience. 2005;4:228–34. doi: 10.1109/tnb.2005.853657. [DOI] [PubMed] [Google Scholar]
- 65.Kurlowicz L, Greenberg SA. The geriatric depression scale (GDS) Am. J. Nurs. 2007;107:67–68. [Google Scholar]
- 66.Ombaugh TN, McDowell I, Kristjansson B, Hubley AM. Mini-mental state examination (MMSE) and the modified MMSE (3MS): a psychometric comparison and normative data. Psychological Assess. 1996;8:48–59. [Google Scholar]
- 67.Chaudhuri KR, Martinez-Martin P. Quantitation of non-motor symptoms in Parkinson’s disease. Eur. J. Neurol. 2008;15(Suppl 2):2–7. doi: 10.1111/j.1468-1331.2008.02212.x. [DOI] [PubMed] [Google Scholar]
- 68.Stiasny-Kolster, et al. The REM sleep behavior disorder screening questionnaire-a new diagnostic instrument. Mov. Disord. 2007;22:2386–93. doi: 10.1002/mds.21740. [DOI] [PubMed] [Google Scholar]
- 69.Rome Foundation. Guidelines-Rome III diagnostic criteria for functional gastrointestinal disorders. J. Gastrointestin Liver Dis. 2006;15:307–12. [PubMed] [Google Scholar]
- 70.Agachan F, Chen T, Pfeifer J, Reissman P, Wexner SD. A constipation scoring system to simplify evaluation and management of constipated patients. Dis. Colon Rectum. 1996;39:681–5. doi: 10.1007/BF02056950. [DOI] [PubMed] [Google Scholar]
- 71.Lloret, et al. Validation of a new scale for the evaluation of sialorrhea in patients with Parkinson’s disease. Mov. Disord. 2007;22:107–11. doi: 10.1002/mds.21152. [DOI] [PubMed] [Google Scholar]
- 72.Lam, et al. Simple clinical tests may predict severe oropharyngeal dysphagia in Parkinson’s disease. Mov. Disord. 2007;22:640–4. doi: 10.1002/mds.21362. [DOI] [PubMed] [Google Scholar]
- 73.Li, et al. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chong J, Xia J. MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics. 2018;34:4313–4314. doi: 10.1093/bioinformatics/bty528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Silverman JD, Roche K, Holmes ZC, David LA, Mukherjee S. Bayesian Multinomial Logistic Normal Models through Marginally Latent Matrix-T Processes. J. Mach. Learn. Res. 2022;23:1–42. [Google Scholar]
- 76.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (2020).
- 77.Aitchison J. The Statistical Analysis of Compositional Data. London, New York: Monographs on statistics and applied probability, Chapman and Hall; 1986. [Google Scholar]
- 78.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shannon, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequence raw data is available at the European Nucleotide Archive, with the accession number PRJEB27564. The metabolomics data is available at MetaboLights, with the accession number MTBLS4332. The clinical data is also available upon direct request from the corresponding authors of the present article. This is due to European patient confidentiality laws, and may require signing a Data Usage Agreement, depending on the specifics of the request.
Code for the bacterial taxa-metabolite correlation analyses is available as R scripts’ files (Supplementary files “Metabolomics.SERVER.SCRIPT.PD_ONLY.Selected.CORR.final” and “Metabolomics.SERVER.SCRIPT.CONTROLS_ONLY.Selected.CORR.final”). For metabolomics-only data analysis, deconvolution R scripts using XCMS and eRah, Matlab code for Partial Least Squares Discriminant Analysis (PLS-DA), R code for metabolite correlations, and Python code for Support Vector Machine (SVM) are available in the GitHub repository at github.com/drupadt/.