Abstract
The prevalence and the genotypic and phenotypic characteristics of multinucleoside-resistant (MNR) human immunodeficiency virus type 1 (HIV-1) variants in Europe were investigated in a multicenter study that involved centers in nine European countries. Study samples (n = 363) collected between 1991 and 1997 from patients exposed to two or more nucleoside analogue reverse transcriptase inhibitors (NRTIs) and 274 control samples from patients exposed to no or one NRTI were screened for two marker mutations of multinucleoside resistance (the Q151M mutation and a mutation with a 2-amino-acid insertion at codon 69, T69S-XX). Q151M was identified in six of the study samples (1.6%), and T69S-XX was identified in two of the study samples (0.5%; both of them T69S-SS), but both patterns were absent among control samples. Non-NRTI (NNRTI)-related changes were observed in viral strains from two patients, which displayed the Q151M resistance pattern, although the patients were NNRTI naive. The patients whose isolates displayed multinucleoside resistance had received treatment with zidovudine and either didanosine, zalcitabine, or stavudine. Both resistance patterns conferred broad cross-resistance to NRTIs in vitro and a poor response to treatment in vivo. MNR HIV-1 is found only among multinucleoside-experienced patients. Its prevalence is low in Europe, but it should be closely monitored since it seriously limits treatment options.
Human immunodeficiency virus type 1 (HIV-1) has the ability to develop resistance to almost all clinically used antiretroviral drugs (11). Reduced sensitivity to nucleoside reverse transcriptase inhibitors (NRTIs), non-NRTIs (NNRTIs), and protease inhibitors (PIs) has been studied thoroughly and is linked to specific point mutations in the reverse transcriptase (RT) and protease (PR) genes, respectively (11, 16). Combinations of antiretroviral drugs have proved superior to monotherapy in delaying the emergence of resistant virus, but thus far no combination can guarantee the prevention of resistance development (17). A set of mutations (A62V, S68G, V75I, F77L, F116Y, and Q151M) that leads to high-level resistance to zidovudine, didanosine, zalcitabine, and stavudine was described previously (12–14). Since these mutations seem to accumulate in an ordered manner and the Q151M mutation is the first mutation to appear, Q151M can be considered a marker mutation (4, 13). The presence of Q151M by itself confers low-level resistance to these nucleoside analogues (4). A 6-bp insert between codons 68 and 70 of the RT gene, which is usually T69S-SS and which is often associated with zidovudine-related mutations, has been reported as a novel pattern of multinucleoside resistance (1, 7, 20). Despite the frequent use of NRTIs, the frequencies of both of these patterns appear to be relatively low. The prevalence of the Q151M resistance pattern has been reported to vary from 2.7 to 17% in patients receiving combination therapy with zidovudine and didanosine or zalcitabine, either sequentially or simultaneously (2, 5, 13), although no large international studies have ever been performed. In a preliminary report, the prevalence of the 6-bp insert at RT codon 69 in Spanish nucleoside-treated HIV-1 patients was estimated to be 0.8% (V. Soriano, M. Pérez-Olmeda, M. Gómez-Cano, C. Briones, and J. Gonzalez-Lahoz, Program abstr. Int. Conf. Discovery Clin. Dev. Antiretroviral Therapies, abstr. 49, p. 50, 1998).
A multicenter study was set up in an attempt to establish the prevalence in Europe of these two multinucleoside resistance patterns in the patients at highest risk, those with experience with multiple nucleoside analogues, compared to the prevalence of these patterns in single nucleoside-experienced or drug-naive patients. The variants that were identified were characterized genotypically and phenotypically, and the data were linked to the clinical outcome. Additionally, the frequency of the drug-specific NRTI-related mutations in HIV-1 RT was determined by a line probe assay (LiPA HIV-1 RT, Innogenetics, Ghent, Belgium).
MATERIALS AND METHODS
Patient population and study design.
In this multicenter study we had access to 755 samples, collected between 1991 and 1997, of HIV-1-infected patients from nine European countries. The participating centers were asked to select from their available samples on the basis of the following inclusion criteria: (i) the study samples had to be obtained from HIV-1-seropositive patients treated for at least 6 months with multiple NRTIs (irrespective of treatment with lamivudine) either sequentially or in combination and (ii) the control samples had to be obtained from HIV-1-seropositive patients who were either drug-naive or had been treated with only one NRTI. These inclusion criteria were based on the observation that previously these multinucleoside resistance patterns seemed to be linked to experience with at least two of the following nucleoside analogues: zidovudine, didanosine, zalcitabine, or stavudine (7, 13, 14, 21). On the basis of these criteria, 440 study samples (Denmark, n = 83; Germany, n = 10; Italy, n = 34; Luxembourg, n = 8; Spain, n = 67; Sweden, n = 45; Switzerland, n = 150; The Netherlands, n = 26; and Belgium, n = 15) were provided. For most countries, all readily available samples were collected, whereas for Switzerland a random selection of 150 samples was made, and for Sweden samples with a detectable viral load were selected. Plasma or serum samples, therapy history, and clinical data were provided by the participating centers. All amplified samples were screened for Q151M by a selective PCR (ARMS-151) or by a research version of the LiPA (LiPA-151). The results for the viral strains that scored a mixture of a wild-type and a mutant sequence by LiPA-151 were verified with an amplification-refractory mutation system (ARMS-151), and vice versa. In cases of ambiguous results, ARMS-151 was performed after cloning or a follow-up sample of the same patient was analyzed. To identify samples with an insert at position 69, LiPA HIV-1 RT was performed for all samples. Since the probes for positions 69 and 70 do not take into account a possible insertion at position 69 and since the strip is designed such that a template with a single mismatch fails to hybridize (15), the viral strains with an insertion at codon 69 are expected not to score at either the 70K wild-type or the 70R mutant lane. Thus, the RT genes of viral strains that scored negative for positions 69 and 70 were sequenced to verify the cause of hybridization failure and, eventually, to confirm the presence of an insert at this position.
RNA extraction and cDNA synthesis.
Viral RNA was extracted from plasma or serum with the QIAamp Viral RNA kit (QIAgen, Hilden, Germany), TRIzol (Life Technologies, Merelbeke, Belgium), or Nuclisens (Organon Teknika, Boxtel, The Netherlands), according to the manufacturer's instructions, followed by reverse transcription into cDNA with the Gene Amp RNA-PCR kit (Perkin-Elmer, Brussels, Belgium) with random hexamers.
LiPA protocols.
For the LiPA HIV-1 RT, amplification was performed as described by the manufacturer (15), and genotypic resistance at positions 41, 69, 70, 74, 184, and 215 in the RT gene was monitored. A research version of LiPA (LiPA-151) provided by Innogenetics (Ghent, Belgium), was used to score wild-type and mutant sequences at position 151 by using the same amplification product and protocol used for the LiPA HIV-1 RT which has a detection limit of 1,000 copies/ml (15). The performance of LiPA-151 was validated with the panel of plasmids as described by Stuyver et al. (15), as well as with several clinical samples with known DNA sequences, and the LiPA-151 showed 100% specificity for the wild-type and mutant templates.
Selective PCR for RT codon 151 (ARMS-151).
A selective PCR for detection of the Q151M mutation based on an ARMS was performed as described elsewhere (19). Starting with control HIV-1 RNA, the wild-type and mutant primers have a detection limit of 100 copies/ml with wild-type and mutant RNA templates, respectively. The discriminatory window (the difference in detection limit for the correct template and the detection limit caused by primer leakage) is at least 2 logs (19).
Sequencing of the RT and PR genes.
To amplify the PR gene or the RT gene, an outer PCR with primers AV150 and RT2 was followed by a nested PCR with primers RVP5 and RVP3 or M13USP-A35 and M13RSP-NE-(1)35, respectively (18). Sequencing was performed with primers USP, RSP, AV36, and AV44 for the RT gene and with primers RVP5 and RVP3 for the PR gene by use of dye terminator technology (ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit; Perkin-Elmer). The sequences were analyzed on an ABI Prism 310 sequencer (Perkin-Elmer) (19). The entire PR gene and the RT gene up to codon 255 were thus obtained. To screen for the presence of a 6-bp insert, 2 μl of the outer PCR product (obtained by the LiPA HIV-1 RT PCR or the ARMS-151 PCR) was used in a LiPA nested PCR. Primers RT-1 and RT-4 (15) were tailed with M13-USP and M13-RSP, respectively; the biotin group was omitted. Sequencing of the RT gene from amino acids 29 to 220 was performed as described above.
Cloning.
By using A35 and NE(35) (19) as the outer primers and the tailed RT-1 and RT-4 (15) as the inner primers (see the sequencing protocol described above), a 610-bp PCR fragment was inserted into a plasmid vector (TOPO TA cloning kit; Invitrogen, Groningen, The Netherlands) according to the instructions of the manufacturer. For each sample at least 10 colonies were screened by the ARMS-151 PCR with the inner primer pair.
Phenotypic drug resistance.
Phenotypic drug resistance was evaluated by the recombinant virus assay with the modifications described previously (18). A PCR with outer primers AV150 and RT2, which amplified the RT and PR genes, was followed by a nested PCR for amplification of the RT gene with primers IN5 and IN3 or the PR gene with primers RVP5 and RVP3 (18). The purified nested PCR products (Microcon-50; Amicon, Beverly, Mass.) were mixed with linearized plasmid pHIVΔRT-BstEII from which the RT gene was deleted (obtained from B. A. Larder, Glaxo Wellcome, Stevenage, United Kingdom) or plasmid pHIVΔPR-BstEII from which the PR gene was deleted (obtained from E. Blair, Glaxo Wellcome), and the plasmids were added to MT4 cells. After electroporation, the MT4 cells were maintained in supplemented RPMI 1640 medium until the virus-induced cytopathogenic effect (CPE) was microscopically detected. After titration for infectivity (10), a standardized virus input was used to infect MT4 cells in the presence of increasing concentrations of the drug to be tested. HIV-1 drug susceptibility was determined by an MT4–3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide-based CPE protection assay (9). The 50% effective concentration (EC50) was defined as the concentration of inhibitor required to inhibit the virus-induced CPE by 50%. Zidovudine was synthesized as described previously (3). Didanosine, zalcitabine, and stavudine were purchased from Sigma, Bornem, Belgium; lamivudine and abacavir were purchased from Glaxo Wellcome; adefovir and tenofovir were purchased from Gilead Sciences, Foster City, Calif.; nevirapine was purchased from Boehringer Ingelheim, Ridgefield, Conn.; delavirdine was purchased from Pharmacia & Upjohn, Kalamazoo, Mich.; efavirenz was purchased from Du Pont Pharmaceuticals, Wilmington, Del.; indinavir was purchased from Merck Research Laboratories, West Point, Pa.; saquinavir was purchased from Roche Products Limited, Welwyn Garden City, United Kingdom; ritonavir was purchased from Abbott Laboratories, Abbott Park, Ill.; and nelfinavir was purchased from Agouron Pharmaceuticals, La Jolla, Calif. EC50s were compared to the mean EC50s for a control wild-type strain (HIV-1 IIIB) and were expressed as fold resistance. The genotypes of the recombined virus stocks were verified by sequencing.
RESULTS
Prevalence of the Q151M mutation and the 6-bp insert at RT codon 69.
The RT genes in 82.5% (363 of 440) of the study samples and 87% (274 of 315) of the control samples could be amplified (Table 1). The detection limit of the PCRs used is about 1,000 RNA copies/ml. Viral load data could be collected for 42% of the study samples with PCR failure, and almost all of them had viral loads below 1,000 copies of RNA/ml, suggesting that low viral loads were the major cause of PCR failures. Six of 363 (1.6%) study samples were identified as containing viral strains that displayed the mutation Q151M (Table 1): in five samples as pure mutants and in one sample as a mixture with wild-type isolates (Table 2). No virus with the Q151M mutation was detected in the control samples (Table 1).
TABLE 1.
Treatments for patients from whom samples were obtained and prevalence of Q151M and T69S-SS mutations
| Group and treatmenta | No. of available samples | No. of samples from which virus was amplified | No. of samples containing virus with the following mutation:
|
|
|---|---|---|---|---|
| Q151M | T69S-SS | |||
| Study group | ||||
| ZDV-ddI | 81 | 57 | 0 | 0 |
| ZDV-ddC | 71 | 39 | 1 | 1 |
| ZDV-ddC-SQV | 11 | 11 | 0 | 0 |
| ZDV/ZDV-d4T | 11 | 11 | 1 | 0 |
| ZDV/ZDV-ddI | 36 | 33 | 2 | 0 |
| ZDV/ZDV-ddC | 25 | 20 | 0 | 0 |
| ZDV/ddI | 125 | 110 | 0 | 0 |
| ZDV/ddC | 6 | 4 | 0 | 0 |
| Other | 74 | 60 | 2 | 1 |
| Total study samples | 440 | 363 | 6 | 2 |
| Control group | ||||
| ZDV | 169 | 145 | 0 | 0 |
| ddI | 19 | 15 | 0 | 0 |
| Drug naive | 127 | 114 | 0 | 0 |
| Total control samples | 315 | 274 | 0 | 0 |
ZDV, zidovudine; ddI, didanosine; ddC, zalcitabine; SQV, saquinavir; d4T, stavudine. Slashes denote successive treatments.
TABLE 2.
Characteristics and antiretroviral treatment for patients whose isolates displayed the Q151M or T69S-SS mutationa
| Mutation | Patient | Sex | Origin | Transmission route | Yr of first HIV detection | Date (mo/yr) of Q151M or T69S-SS detection | Viral load (log10 no. of RNA copies/ml | CD4 count (no. of cells/μl)b | Treatment duration (mo)c | Antiretroviral treatmentd |
|---|---|---|---|---|---|---|---|---|---|---|
| Q151M | IT2 | M | I | Homosexual | 1992 | 2/1993 | ND | 207 | 2 | ZDV-ddC |
| Q151M | IT1 | M | I | IVDU | 1988 | 10/1993 | 5.5 | 360 | 42 | ZDV/ZDV-ddI |
| Q151Q/M | SP1 | M | E | Homosexual | 1990 | 3/1996 | 5.3 | 50 | 8 | ZDV-ddC/ZDV-ddC-IDV |
| Q151M | SP2 | M | E | IVDU | 1986 | 2/1995 | 2.3 | 640 | 17 | ZDV/ZDV-d4T |
| Q151M | SW1 | M | S | Homosexual | 1983 | 3/1995 | 5 | 430 | 7 | ZDV/ZDV-ddI/ZDV-d4T-IDV/3TC-d4T-RTV-SQV |
| Q151M | SW2 | M | S | Homosexual | 1987 | 10/1995 | 4.2 | 6 | 34 | ZDV/ZDV-ddI/ddI/ZDV-ddI/ddI/ddI-3TC/ddI-3TC-IDV/ZDV-d4T-3TC-IDV/ZDV-d4T-3TC-SQV-RTV/d4T-3TC-SQV-RTV |
| T69S-SS | SW3 | M | S | Homosexual | 1989 | 8/1995 | 5.1 | 60 | 20 | ZDV/ZDV/ZDV-ddI/−/ZDV-ddI/ZDV-ddI-3TC/ZDV-ddI-3TC-IDV |
| T69S-SS | CHG | F | Z | Heterosexual | 1990 | 4/1995 | 3.8 | 293 | 51 | ZDV/ddI/−/ZDV-ddC/ZDV-3TC/d4T-3TC |
Abbreviations: M, male; F, female; I, Italy; E, Spain; S, Sweden; Z, Switzerland; IVDU, intravenous drug user; ND, not determined; ZDV, zidovudine; ddI, didanosine; ddC, zalcitabine; d4T, stavudine; 3TC, lamivudine; SQV, saquinavir; IDV, indinavir; RTV, ritonavir.
At the time that the Q151M or T69S-SS mutation was first identified; for viral load first available value is given.
Total duration of treatment with nucleoside analogues before Q151M or T69S-SS was identified retrospectively.
Slashes denote successive treatments, minus signs indicate no treatment, and hypens mark combination treatments. The presence of Q151M or T69S-SS is noted by underscores; the treatments that the patients were receiving at the time the original samples (that were included in the screening) were obtained are in boldface type.
All 637 samples from which RT genes were amplified were screened by the LiPA HIV-1 RT (363 study samples; 274 control samples). For 23 study samples and for 18 control samples, no score was obtained at positions 69 and 70. The RT genes in these 41 samples were sequenced. Two study samples with viral strains with a T69S-SS insert (Table 1) and one control sample with a viral strain with a T69S insert were identified. The viruses in the other 38 samples all had mismatches at positions 69 and 70 or at surrounding positions. One virus with the insert was obtained from a patient who was receiving zidovudine, didanosine, and lamivudine combination therapy; the other viral strain was from a patient who was receiving zidovudine and zalcitabine combination therapy. The control sample with the viral strain with the T69S mutation was from the latter patient but had been obtained in an earlier period, when the patient had been treated only with zidovudine.
Antiretroviral treatment and characteristics of patients with isolates with Q151M or T69S-SS mutations.
The treatment schedules for the six patients whose isolates had the Q151M mutation and the two patients whose isolates had the T69S-SS mutation are presented in Table 2. In all but one patient Q151M emerged when the patients were treated with zidovudine and a second NRTI. In one patient Q151M emerged during didanosine monotherapy following previous zidovudine exposure (Table 2). At the time that the Q151M mutation was first identified retrospectively, the average time of exposure to zidovudine was 15 months (range, 2 to 42 months), that to didanosine was 13.5 months (range, 6 to 24 months), that to zalcitabine was 5 months (range, 2 to 7 months), and that to stavudine was 12 months. When the Q151M mutation emerged, all except one of the patients had low CD4 counts and high viral loads.
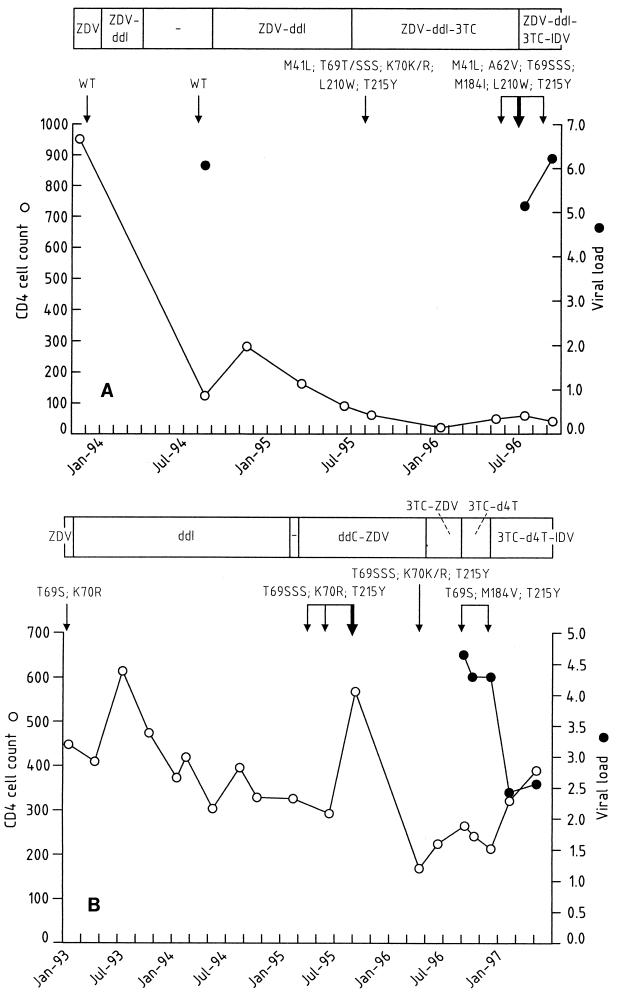
The results for the two patients whose isolates had the T69S-SS mutation are demonstrated in Fig. 1A and B. Isolates with the T69S-SS mutation were identified in these two patients 17 and 27 months after exposure to zidovudine, respectively. The corresponding exposure times to didanosine were 15 and 24 months.
FIG. 1.
Evolution of CD4-cell count (number of cells per microliter), viral load (log10 number of copies of RNA per milliliter) and resistance-related genotype of the RT gene for patients SW3 (A) and CHG (B) who harbored viruses with the T69S-SS mutation. The times of retrieval of the original samples included in the screening, SW3-10125 and CHG-4815, respectively, are indicated by the bold arrow. Patient CHG had been treated with zidovudine (ZDV) since December 1990. ddI, didanosine; 3TC, lamivudine; d4T, stavudine; IDV, indinavir; WT, wild type.
Genotypic characterization of viruses carrying the Q151M or T69S-SS mutation.
For all patients whose viruses displayed the Q151M mutation, except PMAS, Q151M was associated with other mutation(s) characteristic of this resistance pattern, including A62V, S68G, V75I, F77L, and F116Y (Table 3). In the viruses of patients IT2 and SP1, no other resistance-related mutations in addition to the typical multinucleoside resistance mutations were identified. Viral strains from patients IT1 and SW2 showed several zidovudine-related mutations. For patient SW2, M184V (lamivudine-related) and Y115F (abacavir-related) mutations were additionally found in the absence of abacavir experience. Viruses from two patients displayed NNRTI-related changes, although the patients had not been treated with any NNRTI. Sequencing of PR (except for patient SP2) was performed using the same sample. Patient SW2 harbored virus with major PI-related mutations (V82A/V and L90L/M) (data not shown).
TABLE 3.
Resistance-related amino acid changes with respect to the HIV-1 HXB2 sequence detected by sequencing of the RT genes of strains harboring the Q151M or T69S-SS mutation
| Patient identification-sample codea | Amino acid change in the following codonb:
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M41 | A62 | D67 | S68 | T69 | K70 | V75 | F77 | A98 | K103 | Y115 | F116 | Q151 | V179 | Y181 | M184 | L210 | T215 | K219 | |
| IT1-31.8 | N | R | M | Q | |||||||||||||||
| IT1-31.8R | N | R | M | Q | |||||||||||||||
| IT2-31.15 | V/I | L | Y | M | |||||||||||||||
| IT2-31.15R | L | Y | M | ||||||||||||||||
| SP1-9612024 | I | L | Y | Q/Mc | |||||||||||||||
| SP1-9612024R | I | L | Y | Q/M | |||||||||||||||
| SP2-9502063 | G | Sd | M | C | |||||||||||||||
| SW1-10671 | V | G | I | L | Sd | Y | M | Id | |||||||||||
| SW1-10671R | V | G | I | L | Sd | Y | M | Id | |||||||||||
| SW2-11546 | V | N | G | Gd | I | L | F | M | V | Y/Fe | Q | ||||||||
| SW2-11546R | N | G | Gd | L | R | M | W | Y | Q | ||||||||||
| SW3-10125 | L | V | S-SS | I | W | Y | |||||||||||||
| SW3-10125R | L | V | S-SS | V/Ad | I | W | Y | ||||||||||||
| CHG-4815 | S-SS | R | Y | ||||||||||||||||
The strains with suffix R refer to the recombinant virus.
All amino acid changes related to resistance are given. Slashes denote mixtures. A, alanine; C, cysteine; D, aspartic acid; F, phenylalanine; G, glycine; I, isoleucine; K, lysine; L, leucine; M, methionine; N, asparagine; Q, glutamine; R, arginine; S, serine; T, threonine; V, valine; W, tryptophan; Y, tyrosine.
The Q/M mixture was detected by ARMS-151 and LiPA-151; by sequencing only Q151M was detected.
Polymorphisms other than the resistance-related changes at positions associated with resistance.
T215Y/F was detected by LiPA HIV-1 RT; T215F was scored by sequencing.
In the two patients whose viruses displayed the T69S-SS mutation, the RT genes of viral strains in preceding and follow-up samples were sequenced in order to follow the development of the 6-bp insert (Fig. 1). The viral strains from patient SW3 (Fig. 1A) carried no drug-related changes in any sample up to August 1994. At that time the patient had experience with zidovudine and didanosine. The T69S-SS insert and zidovudine-related changes were first identified when the treatment consisted of zidovudine, didanosine, and lamivudine. Later on during this regimen, the patient's virus also acquired the M184I mutation and the A62V mutation, a mutation usually seen in the context of the Q151M multinucleoside resistance pattern. The RT gene of the viral strain in the last sample analyzed (August 1996) showed the same genotype. Patient SW3 had CD4-cell counts that gradually decreased and high viral loads, clinical deterioration, and no response to a PI-containing therapy. The patient died in February 1997. After 2 years of zidovudine monotherapy (January 1993), the first sample from patient CHG analyzed (Fig. 1B) contained isolates with T69S and K70R mutations. The next isolates sequenced in samples obtained from April to September 1995 showed a T69S-SS insert together with the K70R and T215Y mutations. The patient was treated during this period with zidovudine and zalcitabine, after a period of didanosine monotherapy. In April 1996 the insert was still present. The therapy was later changed to zidovudine and lamivudine, with a reversion to T69S and K70, maintenance of the T215Y mutation, and acquisition of the M184V mutation. Subsequently, when the treatment consisted of lamivudine and stavudine, the RT genotype no longer changed and the insert remained absent. The patient remained asymptomatic, with high CD4 cell counts and a good response to lamivudine, stavudine, and indinavir therapy.
Phenotypic characterization of viruses carrying the Q151M or T69S-SS mutation.
The recombinant virus assay was successfully performed for six patients and failed for two patients. Genotypic and phenotypic data are summarized in Tables 3 and 4. Compared to the original patient virus, three recombinant viruses (indicated by R subscripts) showed drug-related alterations in their genotypes: IT2-31.15R, SP1-9612024R, and SW2-11546R (Table 3). The scoring of the polymorphic sites assured us that it was still the same patient viral strain and not a contaminant. All viruses were highly resistant to zidovudine, with extremely high levels of zidovudine resistance for SW1-10671R, which displayed the complete Q151M resistance pattern, and for SW2-11546R, which had the Q151M multinucleoside resistance pattern combined with zidovudine-related mutations, including T215Y. Both isolates were also resistant to lamivudine (greater than ninefold compared to a wild-type reference virus) in the absence of a lamivudine-related mutation at position 184. The Q151M resistance pattern is probably responsible for the lamivudine resistance. Compared to the phenotypic data for IT1-31.8R, with only Q151M as the characteristic amino acid change for this multinucleoside resistance pattern, isolates from all other patients had higher levels of resistance to zidovudine, didanosine, zalcitabine, and stavudine. Sample SW1-10671R also showed decreased sensitivity to abacavir (7-fold), tenofovir (6-fold), nevirapine (14-fold), and delavirdine (18-fold). Isolate SW2-11546R had decreased sensitivity to ritonavir (sixfold) (data not shown).
TABLE 4.
Susceptibility to RT inhibitors of strains with Q151M or T69S-SS
| Recombinant virus straina | Resistance pattern | 50% Inhibitory concn (μM)b
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZDV | ddI | ddC | d4T | 3TC | ABC | PMEA | PMPA | DLV | NVP | EFZ | ||
| IT1-31.8R | Q151M | 0.075 (19×) | 94 (5×) | 7.1 (2×) | 1.03 (5×) | 4.8 (1×) | 4.65 (1×) | 1.85 (0.3×) | 1.22 (0.3×) | 0.041 (4×) | 0.150 (3×) | 0.0022 (3×) |
| IT2-31.15R | Q151M | 0.150 (37×) | 121.6 (6×) | 29 (8×) | 3.44 (16×) | 11 (2×) | 20.2 (3×) | 3.1 (0.5×) | 2.3 (0.6×) | 0.041 4×) | 0.038 (1×) | 0.0010 (1×) |
| SP1-9612024R | Q151M | 0.187 (47×) | 116.9 (6×) | 21 (6×) | 2.46 (11×) | 18 (4×) | 20.9 (3×) | 4.37 (0.7×) | 3.693 (1×) | 0.021 (2×) | 0.075 (1×) | 0.0010 (1×) |
| SW1-10671R | Q151M | 4.494 (1124×) | >211.8 (>11×) | 37 (10×) | 7.14 (33×) | >44 (>9×) | 48.6 (7×) | 13.9 (2.2×) | 22.82 (6×) | 0.185 (18×) | 0.713 (14×) | 0.0029 (4×) |
| SW2-11546R | Q151M | 2.884 (721×) | 116.10 (6×) | 33 (9×) | 7.14 (33×) | >44 (>9×) | 29.7 (4×) | 11.19 (1.7×) | 14.98 (4×) | 0.041 (4×) | 0.014 (1×) | 0.0006 (1×) |
| SW3-10125R | T69S-SS | >7.491 (>1873×) | 22.46 (1×) | 31 (9×) | 3.48 (16×) | >66 (>14×) | 22.7 (3×) | 8.04 (1.2×) | 23.17 (6×) | 0.014 (1×) | 0.047 (1×) | 0.0006 (1×) |
| HIV-1 IIIbR | Wild type | 0.004 | 19.32 | 3.6 | 0.22 | 4.6 | 7.24 | 6.47 | 3.76 | 0.010 | 0.053 | 0.0008 |
The patient designation is followed by the sample code. The suffix R discriminates the original virus strain from the recombinant virus strain.
The results are the averages of two experiments performed in triplicate. The values for wild-type reference strain HIV-1 IIIbR are averages of 6 to 10 independent tests performed in triplicate. The fold resistance compared to the resistance of HIV-1 IIIbR is indicated in parentheses. Abbreviations: ZDV, zidovudine; ddI, didanosine; ddC, zalcitabine; d4T, stavudine; 3TC, lamivudine; ABC, abacavir; PMEA, adefovir; PMPA, tenofovir, DLV, delavirdine; NVP, nevirapine; EFZ, efavirenz.
Isolate SW3-10125R, which had the T69S-SS insert and several zidovudine- and lamivudine-related mutations (Table 3), showed high-level resistance to zidovudine, stavudine, and lamivudine (Table 4), with higher levels of resistance to zidovudine than those for the isolates that carried the Q151M mutation. Decreased sensitivities to zalcitabine and tenofovir were noted nine- and sixfold, respectively), while wild-type sensitivity to didanosine was seen, despite the didanosine experience. Genotyping of the recombinant virus showed no drug-related changes other than those present in the patient isolate (Table 3).
Other NRTI-related mutations in the study and control population.
For 456 of 637 amplifiable samples, LiPA HIV-1 RT gave interpretable results for all codons (71%). The detection failures for the different codons ranged from 6 to 10%: for codons 41, 69, 70, 74, 184, 214, and 215, 9, 7, 7, 6, 7, 10, and 9%, respectively.
Of the 274 control samples, 114 samples were obtained from antiretroviral drug-naive patients (from Denmark, Sweden, Spain, and Belgium) (Table 1). Viruses in 93 (82%) of the samples had wild-type genotypes at positions associated with resistance to nucleoside analogues. However, viruses in 21 (18%) of the samples displayed at least one resistance-related mutation associated with a loss of sensitivity to zidovudine, didanosine, zalcitabine, or lamivudine. Viral strains from 15 patients displayed the M41L, K70R, or T215Y/F mutation, or a combination of these mutations, which are related to zidovudine resistance. Viral strains from three patients displayed M184V, which is related to lamivudine, zalcitabine, and didanosine resistance. Three patients harbored virus with zidovudine mutations in combination with another mutation (T69K, L74V, or M184V).
In total, 358 samples from the study arm and 145 samples from the control arm were from patients who were zidovudine experienced (Table 1), and the viruses in 77% of these samples had zidovudine-related mutations (M41L, K70R, and/or T215Y/F). Of the 267 samples from patients treated with didanosine (study and control samples together), 16% harbored viruses with the L74V mutation. Viral strains with the T69D mutation were found in 7.5% of the samples from patients with zalcitabine experience (n = 120), and isolates with the M184V mutation were found in 52% of the samples from patients with lamivudine experience (n = 21). Additionally, the M184V mutation occurred in 3.5 and 10% of the samples from patients treated with didanosine and zalcitabine, respectively, and caused decreased sensitivity to lamivudine.
DISCUSSION
This study was performed to monitor the prevalence of multinucleoside resistance in European HIV-1-infected patients, starting with 440 samples obtained from patients with multinucleoside experience (study samples) and 315 samples obtained from patients treated with no more than one NRTI (control samples). The samples were collected from patients in nine European countries between 1991 and 1997. We screened for the key mutations linked to the two currently known multinucleoside resistance patterns: (i) Q151M mutation linked to multinucleotide resistance pattern A62V, S68G, V75I, F77L, and F116Y (12, 14) by ARMS-151 or LiPA-151 and (ii) the 6-bp insert at position 69 in the RT gene which, together with zidovudine-related mutations, confers multinucleoside resistance (1, 7, 20), as detected by LiPA HIV-1 RT and by sequencing of samples with negative score for codons 69 and 70. Additionally, we were able to measure the prevalence of nucleoside analogue resistance in our sample groups as scored by LiPA HIV-1 RT.
Among the study samples (n = 363), the Q151M mutation was identified in six samples (1.6%) and the 6-bp insert at position 69 (in all isolates a T69S-SS mutation) was identified in two samples (0.5%). No multinucleotide resistance of either pattern was observed for isolates from the control samples. These are the first values of the overall prevalence of multinucleoside resistance reported for such a large set of European patients, making it the most accurate European estimate available. Although no restriction on viral load level was made as an inclusion criterion, the low rates of PCR failures for the viruses in both the study (17.5%) and control (13%) samples suggest that most of the samples included in this study have viral loads above the detection limit of the PCRs used (about 1,000 RNA copies/ml). All samples were collected retrospectively and dated from the period from 1991 to 1997. The suboptimal regimens that these patients received, which consisted of sequential monotherapies or bitherapies with nucleoside analogues, were generally not able to inhibit completely viral replication, creating an ideal situation for the emergence of drug-resistant virus variants. Previously, the frequency of the Q151M mutation in two studies of small populations (116 and 150 patients, respectively) of multinucleoside-experienced patients (with at least 6 months of experience) from a limited number of European countries was reported to be in the range of 2 to 3.5%, while no Q151M mutation was observed in patients with no NRTI experience or experience with a single NRTI (2, 13). The acquisition of Q151M requires 2 base changes (Q151L and Q151K), with a poor replicative capacity of the possible intermediate variants, and this can contribute to the low prevalence of Q151M (6). Kavlick et al. (5), however, described a 17% prevalence of the Q151M mutation in U.S. patients with long-term experience (more than 36 months) with zidovudine and didanosine. The possible linkage between the longer exposure to NRTIs (36 months in the study by Kavlick et al. [5]) and the high prevalence of the Q151M mutation needs further investigation, since in the future the duration of combination treatments that include NRTIs will undoubtedly increase. On the other hand, the introduction of HAART (highly active antiretroviral therapy), which results in sustained low viral loads in a large proportion of patients, may be responsible for a decrease in the prevalence of multinucleoside resistance. We could not draw conclusions on the evolution of the prevalence of multinucleoside resistance over time since there was not an equal distribution of the samples over the different years of collection (1991 through 1997). Since the strategies for the treatment of HIV infection in Europe have changed fundamentally, it is difficult to extrapolate the prevalence measured in this study to the current prevalence of multinucleoside resistance and that in the near future. Few data are available about the prevalence of the 6-bp insert at position 69. A preliminary report (Soriano et al., Program abstr. Int. Conf. Discovery Clin. Dev. Antiretroviral Therapies, 1998) of a study from Spain in which the same strategy of sequencing of samples with a negative score for codons 69 and 70 by LiPA was used mentions a prevalence of 0.8% in patients who failed didanosine- or stavudine-containing therapy. This is similar to what we report here for the overall prevalence in Europe. An underestimation of the prevalence by this strategy is possible, since isolates that contain inserts in a mixture with the wild type will be missed.
All six patients whose isolates were found to have the Q151M mutation had been treated with zidovudine and either didanosine, zalcitabine, or stavudine when the Q151M mutation was first observed. This indicates that combinations with zidovudine and stavudine can also select for the Q151M mutation-induced resistance pattern, an observation that was not reported before. The isolate from one patient showed the Q151M mutation 2 months after the patient started antiretroviral treatment (zidovudine and zalcitabine) (Table 2). Since no sample from an earlier date was available from this patient, it cannot be excluded that the Q151M mutation was already present in the period when the patient was antiretroviral drug naive, possibly as a result of the transmission of a virus that carried the Q151M mutation. The presence of the Q151M mutation in a recently infected naive patient was reported in a French study that screened for baseline resistance (D. Descamps, D. Costagliola, G. Glaude, C. Buffet-Janvresse, V. Calvez, G. Collin, J. Cottalorda, F. Ferchal, M. Harzic, J. Izopet, B. Masquelier, A. Ruffault, A. Schmuck, C. Tamalet, A. Yvon, F. Brun-Vézinet, and the French ANRS Antiretroviral Resistance Study Group, Int. Workshop Drug Resistance Treatment Strategies, Antivir. Ther. 4[Suppl. 1]:abstr. 123, 1999). Previous treatment with zidovudine and either didanosine, zalcitabine, or stavudine is, however, not a prerequisite for the development of the Q151M mutation. In a preliminary report Race et al. (E. Race, F. Ferchal, E. Dam, V. Picard, A. Maillard, V. Obry, M. M. Sombardier, G. Chêne, J. M. Molina, and F. Clavel, Int. Workshop Drug Resistance Treatment Strategies, Antivir. Ther. 4[Suppl. 1], abstr. 118, 1999) identified the Q151M mutation in isolates from patients who showed virologic escape during treatment with the combination of didanosine and stavudine without other antiretroviral exposure. In vitro, selection of the Q151M mutation seems to be possible with a single NRTI, either zidovudine, didanosine, zalcitabine, or stavudine (E. Fontaine, J. M. Plesseria, C. Lambert, T. Staub, V. Arendt, R. Hemmer, F. Schneider, A.-M. Vandamme, and J. C. Schmit, Int. Workshop Drug Resistance Treatment Strategies, Antivir. Ther. 4[Suppl. 1], abstr. 37, 1999). Similar to the Q151M resistance pathway, the 6-bp insert in our patients appeared during treatment that contained zidovudine and didanosine or zalcitabine. Since in clinical practice the majority of HIV-1-infected patients are zidovudine experienced, the prerequisite for zidovudine experience in the development of multinucleoside resistance is difficult to study.
The Q151M multinucleoside resistance pattern and the classical zidovudine resistance pathway with T215Y/F as a marker mutation were previously described as two distinct mutational pathways, were seldom identified in viruses from the same patient, and did not occur in the same HIV-1 strain (5, 13). We observed one patient (patient SW2) who harbored virus with both the Q151M and the T215Y/F mutations as the complete mutant population, indicating that both mutational pathways are compatible (data not shown). It is possible, however, that the presence of both mutations is the result of recombination events after the separate development of both patterns. In this patient, the accumulation of the multinucleoside resistance pattern and of NRTI- and PI-related mutations still results in a fully competent virus capable of replicating to high viral loads.
The viruses from two patients with the Q151M mutation displayed NNRTI-related changes in the absence of NNRTI experience (Tables 2 and 3). In addition, the recombinant virus isolate from patient SW2 had acquired NNRTI-related mutation K103R, which was absent in the original patient virus. Cross-talk between the Q151M-related mutations which cluster together next to the active site opposite the NNRTI binding pocket and the NNRTI-related mutations has been reported before (A.-M. Vandamme et al., Sixth Eur. Conf. Clin. Aspects Treat. HIV Infect., Hamburg, Germany, 1997), with the difference being that the NNRTI-related mutations had been acquired under NNRTI drug pressure. As we observed in patient SW1 (Table 4), NNRTI-related changes and decreased sensitivity to NNRTIs in patients with the Q151M pattern and without any NNRTI experience have been mentioned in a preliminary report (Race et al., Int. Workshop on Drug Resistance Treatment Strategies, Antivir. Ther. 4[Suppl. 1], abstr. 118, 1999). This phenomenon could have detrimental implications by further reducing the therapy options in the presence of the multinucleoside resistance pattern.
According to a previous report (13), phenotyping of viruses with several mutations of the Q151M mutation pathway revealed important cross-resistance to zidovudine, didanosine, zalcitabine, and stavudine and partial cross-resistance to lamivudine. We confirmed this cross-resistance and found that these viruses maintain normal or only slightly reduced susceptibility to abacavir, tenofovir, and adefovir; one isolate, however, was resistant to abacavir and tenofovir (Table 4). Phenotyping of one of the isolates with the T69S-SS mutation revealed high-level resistance to zidovudine, stavudine, and lamivudine and decreased sensitivity to zalcitabine and tenofovir. Winters et al. (20) also found that viruses with variable 6-bp inserts in combination with the T215Y mutation and without adefovir-associated mutations had reduced susceptibility to adefovir. The implications for abacavir, adefovir, and tenofovir therapy cannot yet be drawn from the few available data.
Except for the one patient with stable low viral loads and high CD4 counts during zidovudine and stavudine combination therapy reported above, the patients whose viruses had the Q151M pattern had poor responses to NRTI-containing regimens. Indinavir was included as part of triple-drug therapy with NRTIs (patients SW1, SP1, and SW2), resulting in a sustained drop in viral load only for patient SP1. We can only speculate about a reversion of Q151Q/M toward the wild type since samples obtained later were not analyzed. The change to dual PI therapy (saquinavir and ritonavir) had only a transient effect on the viral load for patient SW1; for patient SW2 the viral load remained above 4.5 logs. Genotyping was performed during indinavir therapy for the viral strain from patient SW2 and revealed major PI mutations. Thus, use of the combination of NRTIs with a PI(s) to treat patients with the Q151M multinucleoside resistance pattern does not guarantee suppression of viral replication, as it can result in the accumulation of PI-related mutations.
For the viruses from two patients with the T69S-SS insert in combination with zidovudine-related mutations, we observed two different evolutions (Fig. 1) both in the stability of the genetic rearrangement and in the clinical evolution of the patients. Patient SW3 was already in a later stage of the disease at the time that the virus acquired the insert, and the maintenance of the T69S-SS insert was associated with a progressive deterioration and no therapy response even after the inclusion of indinavir in the therapeutic regimen. The virus from patient CHG lost the T69S-SS insert, after it was present for at least 12 months, with a reversion to the baseline genotype (T69S). This patient remained asymptomatic and had a good response to therapy (lamivudine, stavudine, and indinavir) after the insert was lost. Even though we identified only two patients whose viral strains had the T69S-SS insert, we observe that this multinucleoside resistance pattern does not always predict the failure of therapy and a bad clinical outcome.
For viruses from samples from antiretroviral drug-naive patients (n = 114) the prevalence of baseline resistance mutations to NRTIs was 18%. The viruses mostly displayed resistance to zidovudine, followed by resistance to lamivudine. If K70R is considered a naturally occurring polymorphism rather than a resistance-related mutation (8), the prevalence of baseline resistance drops to 8.5%. Mutations in viruses from antiretroviral drug-naive patients were mostly present as a mixture with the wild-type population. In viruses from NRTI-experienced patients we observed a high prevalence of zidovudine- and lamivudine-related mutations and a low prevalence of zalcitabine- and didanosine-related mutations.
In conclusion, both multinucleotide resistance patterns, the Q151M resistance pattern and the 6-bp insert at codon 69, display very similar characteristics. They both appear at low prevalences in Europe (<2% each) and in patients with long-term zidovudine experience and either didanosine, zalcitabine, or stavudine experience (on average, 11 months for patients whose isolates carried the Q151M mutation and 21 months for patients whose isolates had the T69S-SS insert). These mutations have not been observed in patients with no NRTI experience or experience with a single NRTI. Both resistance patterns are associated with phenotypic cross-resistance to zidovudine, stavudine, lamivudine, and zalcitabine and a poor response to NRTI therapy. Genotypically, viruses with the Q151M or T69S-SS resistance pattern can display other mutations related to NRTI, NNRTI, or PI resistance, including the classical zidovudine resistance mutations such as T215Y. The reasons why one of both multinucleoside resistance patterns occur and why most patients with multinucleoside experience do not develop either of these resistance patterns are issues that remain to be clarified.
ACKNOWLEDGMENTS
We thank Ria Swinnen and Inge Aerts (Rega Institute, Leuven, Belgium) for fine editorial assistance and Kristien Erven and Cindy Heens (Rega Institute, Leuven, Belgium) and Nadia Van der Cruyssen (Innogenetics, Ghent, Belgium) for excellent technical assistance.
This work was partially supported by the Biomedical Research Programme of the European commission (EC BIOMED2 grant BMH4-CT-95-1634), the Flemish Fonds voor Wetenschappelijk Onderzoek (FWO grant G.0104.98), and the Belgian Geconcerteerde Onderzoeksacties (GOA 95/5). All the participating centers are part of the European Network for the Virologic Evaluation of the International Trials for New Anti-HIV Therapies, sponsored by the European Commission (EC BIOMED2 grant BMH4-CT-96-0409).
REFERENCES
- 1.de Jong J J, Goudsmit J, Lukashov V V, Hillebrand M E, Baan E, Huismans R, Danner S A, ten Veen J H, de Wolf F, Jurriaans S. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS. 1999;13:75–80. doi: 10.1097/00002030-199901140-00010. [DOI] [PubMed] [Google Scholar]
- 2.Gómez-Cano M, Rubio A, Puig T, Perez-Olmeda M, Ruiz L, Soriano V, Pineda J A, Zamora L, Xaus N, Clotet B, Leal M. Prevalence of genotypic resistance to nucleoside analogues in antiretroviral-naive and antiretroviral-experienced HIV-infected patients in Spain. AIDS. 1998;12:1015–1020. [PubMed] [Google Scholar]
- 3.Horwitz J P, Chua J, Noel M. Nucleosides. V. The monomesylates of 1-(2′-deoxy-β-d-lyxofuranoxyl) thymine. J Org Chem. 1964;29:2076–2078. [Google Scholar]
- 4.Iversen A K N, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kavlick M F, Wyvill K, Yarchoan R, Mitsuya H. Emergence of multidideoxynucleoside-resistant human immunodeficiency virus type 1 variants, viral sequence variation, and disease progression in patients receiving antiretroviral chemotherapy. J Infect Dis. 1998;98:1506–1513. doi: 10.1086/515324. [DOI] [PubMed] [Google Scholar]
- 6.Kosalaraksa P, Kavlick M F, Maroun V, Le R, Mitsuya H. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an in vitro competitive HIV-1 replication assay. J Virol. 1999;73:5356–5363. doi: 10.1128/jvi.73.7.5356-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larder B, Bloor S, Kemp D, Hertogs K, Desmet R L, Miller V, Sturmer M, Staszweski S, Ren J, Stammers D K, Stuart D I, Pauwels R. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Najera I, Richman D D, Olivares I, Rojas J M, Peinado M A, Perucho M, Najera R, Lopez-Galindez C. Natural occurrence of drug resistance mutations in the reverse transcriptase of human immunodeficiency virus type 1 isolates. AIDS Res Hum Retrovir. 1994;10:1479–1488. doi: 10.1089/aid.1994.10.1479. [DOI] [PubMed] [Google Scholar]
- 9.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Herdewijn P, Desmyter J, De Clercq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 10.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 11.Schinazi R F, Larder B, Mellors J W. Mutations in retroviral genes associated with drug resistance: 1999–2000 update. Int Antivir News. 1999;7:46–69. [Google Scholar]
- 12.Schmit J-C, Cogniaux J, Hermans P, Van Vaeck C, Sprecher S, Van Remoortel B, Witvrouw M, Balzarini J, Desmyter J, De Clercq E, Vandamme A-M. Multiple drug resistance to nucleoside analogues and nonnucleoside reverse transcriptase inhibitors in an efficiently replicating human immunodeficiency virus type 1 patient strain. J Infect Dis. 1996;174:962–968. doi: 10.1093/infdis/174.5.962. [DOI] [PubMed] [Google Scholar]
- 13.Schmit J-C, Van Laethem K, Ruiz L, Hermans P, Sprecher S, Sonnerborg A, Leal M, Harrer T, Clotet B, Arendt V, Lissen E, Witvrouw M, Desmyter J, De Clercq E, Vandamme A-M. Multiple dideoxynucleoside analogue-resistant (MddNR) HIV-1 strains isolated from patients from different European countries. AIDS. 1998;12:2007–2015. doi: 10.1097/00002030-199815000-00012. [DOI] [PubMed] [Google Scholar]
- 14.Shirasaka T, Kavlick M F, Ueno T, Gao W Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R F, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandamme A-M, Van Laethem K, De Clercq E. Managing resistance to anti-HIV drugs. An important consideration for effective disease management. Drugs. 1999;57:337–361. doi: 10.2165/00003495-199957030-00006. [DOI] [PubMed] [Google Scholar]
- 17.Vandamme A-M, Van Vaerenbergh K, De Clercq E. Anti-human immunodeficiency virus drug combination strategies. Antivir Chem Chemother. 1998;9:187–203. doi: 10.1177/095632029800900301. [DOI] [PubMed] [Google Scholar]
- 18.Vandamme A-M, Witvrouw M, Pannecouque C, Balzarini J, Van Laethem K, Schmit J-C, Desmyter J, De Clercq E. Evaluating clinical isolates for their phenotypic and genotypic resistance against anti-HIV drugs. In: Kinchington D, Schinazi R F, editors. Methods in cellular and molecular medicine: antiviral chemotherapy. Totowa, N.J: The Humana Press Inc.; 1999. [DOI] [PubMed] [Google Scholar]
- 19.Van Laethem K, Van Vaerenbergh K, Schmit J-C, Sprecher S, Hermans P, De Vroey V, Schuurman R, Harrer T, Witvrouw M, Van Wijngaerden E, Stuyver L, Van Ranst M, Desmyter J, De Clercq E, Vandamme A-M. Phenotypic assays and sequencing are less sensitive than point mutation assays for the detection of resistance in mixed HIV-1 genotypic populations. J Acquir Immune Defic Syndr. 1999;22:107–118. doi: 10.1097/00126334-199910010-00001. [DOI] [PubMed] [Google Scholar]
- 20.Winters M A, Coolley K L, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]