Graphical abstract
Keywords: Bacterial infection, Metallopeptidase, Protein inhibitor, Protein design, Therapeutic protein, Crystal structure
Abstract
Aureolysin, a secreted metallopeptidase (MP) from the thermolysin family, functions as a major virulence factor in Staphylococcus aureus. No specific aureolysin inhibitors have yet been described, making this an important target for the development of novel antimicrobial drugs in times of rampant antibiotic resistance. Although small-molecule inhibitors are currently more common in the clinic, therapeutic proteins and peptides (TPs) are favourable due to their high selectivity, which reduces off-target toxicity and allows dosage tuning. The greater wax moth Galleria mellonella produces a unique defensive protein known as the insect metallopeptidase inhibitor (IMPI), which selectively inhibits some thermolysins from pathogenic bacteria. We determined the ability of IMPI to inhibit aureolysin in vitro and used crystal structures to ascertain its mechanism of action. This revealed that IMPI uses the “standard mechanism”, which has been poorly characterised for MPs in general. Accordingly, we designed a cohort of 12 single and multiple point mutants, the best of which (I57F) inhibited aureolysin with an estimated inhibition constant (Ki) of 346 nM. Given that animals lack thermolysins, our strategy may facilitate the development of safe TPs against staphylococcal infections, including strains resistant to conventional antibiotics.
1. Introduction
Antibiotic resistance is a major global health burden, leading to hundreds of thousands of deaths every year and greatly increasing healthcare costs associated with the treatment of bacterial infections [1], [2], [3]. Resistance arises from selection pressure caused by the widespread abuse, overuse and misuse of antibiotics in humans, including premature treatment discontinuation [4], subtherapeutic dosing, and the distribution of counterfeit drugs [5]. Furthermore, ∼80% of all antimicrobials used in the USA are administered as prophylactics to farm animals to boost their health and productivity [6]. Once acquired, resistance is spread by horizontal gene transfer, often across species barriers, ultimately giving rise to multidrug-resistant strains [7]. The impact of antibiotic resistance is heightened by the lack of new drugs in the development pipeline, with only two new classes of antibiotics approved in the last 30 years: the oxazolidinones, which target protein synthesis, and the acidic lipopeptides, which target bacterial membranes [8], [9]. This lack of progress reflects decades of low returns compared with other drug classes, discouraging investment by the pharmaceutical industry [2], [7], [10] and thus posing a serious threat to public health [11]. There are few therapeutic options for the treatment of infections with “superbugs” such as Acinetobacter baumannii, Neisseria gonorrhoeae, Pseudomonas aeruginosa, Streptococcus pneumoniae and Staphylococcus aureus, which kill someone every 15 min in the USA [12], [13], [14]. Drug-resistant strains of S. aureus cause severe endocarditis, pneumonia, sepsis, and toxic shock syndrome [15]. Thus, there is an urgent need for the development of new classes of antibiotics to tackle such infections.
Microbial pathogenesis involves diverse pathways and mechanisms that lead to host colonisation and infection [16]. Virulence factors are secreted by the pathogen to facilitate this process, including peptidases that break down host defence proteins, regulate the availability of other secreted bacterial factors, and provide peptide nutrients for the pathogen. One example is the thermolysin family of bacterial metallopeptidases (MPs), also referred to as the M4 family according to the MEROPS database (www.ebi.ac.uk/merops) [17]. The archetype is Bacillus thermoproteolyticus thermolysin, which was the first endo-MP to be structurally resolved [18] and the founding member of the gluzincin clan of MPs [19], [20]. Related MPs produced by human pathogens include P. aeruginosa pseudolysin [21], vibriolysin from several Vibrio species [22], Burkholderia cenocepacia ZmpA/B [23], Enterococcus faecalis coccolysin [24], Legionella pneumophila Msp [25], Clostridium perfringens λ-toxin [26], and aureolysin from Staphylococcus epidermidis and S. aureus [16], [27], [28], [29].
Aureolysin was discovered in S. aureus strain V8 [30] and is the product of the aur gene, which is located on a monocistronic operon [31] and regulated by the alternative sigma factor σB and the staphylococcal accessory regulator SarA [31]. Aureolysin is prevalent in both pathogenic and commensal S. aureus strains [32], and peak abundance occurs during post-exponential growth and when the bacterial cells are phagocytosed by human neutrophils [33]. The enzyme accounts for ∼ 50% of the total peptidase activity in culture supernatants [28] and participates in the extracellular peptidase system of S. aureus by activating the V8-type serine peptidase SspA, which in turn activates the cysteine peptidase SspB [16]. Together with the cysteine peptidase ScpA, they constitute the four major extracellular peptidases of S. aureus [34] known as the “staphylococcal proteolytic cascade” [31]. Moreover, aureolysin recruits nutrients from host proteins [35] and contributes to staphylococcal infections by promoting hypervirulence and the transition from a sessile, biofilm-forming lifestyle to a mobile, invasive phenotype [36], [37]. It degrades the human antimicrobial peptide LL-37 [38] and complement protein C3, while releasing the chemoattractant C5a to prevent complement-mediated killing by neutrophils [39]. It also contributes to the intracellular survival of S. aureus in human macrophages [40]. Furthermore, aureolysin highjacks the blood coagulation and fibrinolytic systems by activating prothrombin [41] and inactivating the serpin-type serine peptidase inhibitors α1-proteinase inhibitor, α2-antiplasmin, and α1-antichymotrypsin by cleaving their “reactive-centre loops” (RCLs). This deregulates their targets such as neutrophil urokinase-type plasminogen activator, elastase, and plasmin [42], [43], [44], [45], [46]. Finally, aureolysin was shown to trigger osteoblast death and bone destruction in a murine model of osteomyelitis [29], which is a hallmark of S. aureus infection in humans [47]. Aureolysin is therefore a promising drug target given its role in the establishment and persistence of infection, which underpins its relevance for bacterial survival in vivo [48].
Aureolysin occurs as two isoforms (I and II) across distinct S. aureus strains that share 93% sequence identity [49]. It is exported as a 509-residue pre-pro-enzyme (UniProt access code [UP] P81177) comprising a 27-residue signal peptide for secretion, a 181-residue pro-domain (S28–E208; aureolysin residue numbering in subscript), and a 301-residue mature catalytic domain (CD, A209–E509; [27]) with 49% identity to thermolysin [28]. Once secreted, the zymogen is self-processed to yield the mature form [50], which (like other thermolysins [51]) prefers neutral pH and hydrophobic residues in the substrate P1′ position [28] (nomenclature of enzyme sub-site and substrate positions on the non-primed and primed sides of the active-site cleft according to [52], [53]). Typically for MPs, the enzyme is inhibited by the general metal chelators EDTA and o-phenanthroline, as well as the nonspecific pan-peptidase inhibitor α2-macroglobulin [28], but no specific small-molecule or protein inhibitors have yet been reported.
Small-molecule drugs are favoured in the clinic because they are often characterised by a long shelf life, oral bioavailability, efficient uptake by cells, and ease of manufacturing [54]. However, they generally have a small surface area for interaction with targets (usually large proteins), and this can limit their specificity and promote off-target effects. In contrast, therapeutic proteins (TPs) have larger surface areas, which result in higher selectivity, fewer toxic side effects, and tuneable dosage [54], often without harmful immune responses [55]. Although most TPs must be injected due to poor gastrointestinal absorption, various systems have been developed to overcome these limitations [56]. Recombinant TPs can also be redesigned to increase their specificity or efficacy. For example, defence proteins produced by one animal host against a class of bacterial virulence factors may be adapted to another host. Overall, this has increased the efficacy and potency of TPs, and they now account for ∼ 10% of the broader pharmaceutical market [54].
The MP inhibitor from Streptomyces nigrescens was the first M4 family inhibitor (MEROPS I36) shown to target thermolysin, pseudolysin and griselysin [57], [58], but its mechanism of action remains unknown. In contrast, the mature 68-residue inducible insect metallopeptidase inhibitor (IMPI) from the greater wax moth Galleria mellonella (MEROPS I8; UP P82176) is a potent inhibitor of thermolysin, pseudolysin, vibriolysin, bacillolysin, and Bacillus polymyxa peptidase, and, importantly, its mechanism is known [59], [60], [61], [62], [63]. Moreover, IMPI is currently under development for the therapy of ectopic infections caused by S. aureus to cure chronic wounds formulated in poloxamer hydrogels, which caused no side effects in the swine ear model [63], [64]. We therefore sought a protein inhibitor of aureolysin for further development as a TP by designing several IMPI mutants with the ability to block aureolysin, and determined their mechanisms of action by kinetic and structural analysis.
2. Materials and methods
Expression constructs — Plasmid pIMPI-WT contains the sequence of wild-type (wt) IMPI in its mature form (residues I20–S88, superscript numbering based on UP P82176) [62]. It is a modified pET–32a vector, with the IMPI sequence inserted at the BglII and XhoI restriction sites, preceded by an N-terminal His6–tagged thioredoxin fusion partner and a tobacco etch virus (TEV) peptidase recognition site, placing the peptide sequence G–M−S upstream of I20 in the final purified protein. We used pIMPI-WT to generate 13 mutants (T50N, T50Q, T50R, T50Y, I54M, I55R, I55W, I55Y, I57F, I57Y, R58E, T50Y + I55R and T50Y + I55R + I57F). T50N was used only as an intermediate to prepare T50Y and was not tested for activity. The mutants were generated by site-directed mutagenesis with overlapping primers (Table 1) using Phusion high fidelity DNA polymerase (Thermo Fisher Scientific) according to the manufacturer’s instructions. Template DNA was digested with DpnI (Thermo Fisher Scientific) and the product was used to transform competent Escherichia coli DH5α cells (Thermo Fisher Scientific). Plasmid DNA was purified using the E.Z.N.A. Plasmid DNA Mini Kit I (Omega Bio-Tek) and all constructs were verified by sequencing (Eurofins and Macrogen).
Table 1.
Plasmids and primers for overexpression.
| Plasmid | Forward primer | Reverse primer | Template |
|---|---|---|---|
| pIMPI-T50Na | CATATACAGAATAAAAATAACTGTCCC | GGGACAGTTATTTTTATTCTGTATATG | pIMPI-WT |
| pIMPI-T50Q | CATATACAGAATAAACAAAACTGTCCC | GGGACAGTTTTGTTTATTCTGTATATG | pIMPI-WT |
| pIMPI-T50R | CAGAATAAACGAAACTGTCCCATC | GATGGGACAGTTTCGTTTATTCTG | pIMPI-T50Q |
| pIMPI-T50Y | CATATACAGAATAAATATAACTGTCCC | GGGACAGTTATATTTATTCTGTATATG | pIMPI-T50N |
| pIMPI-I54M | CTGTCCCATGATTAATATAAGATGTAATGACAAGTGC | GCACTTGTCATTACATCTTATATTAATCATGGGACAG | pIMPI-WT |
| pIMPI-I55R | GTCCCATCCGTAATATAAGATGTAATG | CATTACATCTTATATTACGGATGGGAC | pIMPI-I55W |
| pIMPI-I55W | CAAACTGTCCCATCTGGAATATAAGATGTAATGAC | GTCATTACATCTTATATTCCAGATGGGACAGTTTG | pIMPI-WT |
| pIMPI-I55Y | CAAACTGTCCCATCTATAATATAAGATGTAATG | CATTACATCTTATATTATAGATGGGACAGTTTG | pIMPI-WT |
| pIMPI-I57F | CTGTCCCATCATTAATTTTAGATGTAATGACAAGTGC | GCACTTGTCATTACATCTAAAATTAATGATGGGACAG | pIMPI-WT |
| pIMPI-I57Y | CTGTCCCATCATTAATTATAGATGTAATGACAAGTGC | GCACTTGTCATTACATCTATAATTAATGATGGGACAG | pIMPI-WT |
| pIMPI-R58E | CATTAATATAGAATGTAATGACAAGTGC | GCACTTGTCATTACATTCTATATTAATG | pIMPI-WT |
| pIMPI-T50Y + I55R | CATATACAGAATAAATATAACTGTCCC | GGGACAGTTATATTTATTCTGTATATG | pIMPI-I55R |
| pIMPI-T50Y + I55R + I57F | CCCATCCGTAATTTTAGATGTAATGACAAGTGC | GCACTTGTCATTACATCTAAAATTACGGATGGG | pIMPI-T50Y + I55R |
This mutant was used as an intermediate to prepare T50Y and was not tested for activity. Only single nucleotides were exchanged in each reaction. For the double and triple mutants, a corresponding ancestral plasmid was used as the template.
Protein production and purification — The IMPI variants were expressed in E. coli BL21 (DE3) Origami2 cells (Novagen) transfected with the corresponding plasmid and grown on lysogeny broth (LB) agar supplemented with 100 µg/mL ampicillin. Single colonies were used to inoculate 25-mL LB starter cultures supplemented with 100 µg/mL ampicillin and 10 µg/mL tetracycline, and were incubated overnight at 37 °C under shaking. The starter cultures (1 mL) were used to inoculate 500 mL of the same medium, followed by cultivation under the same conditions until the OD550 reached 0.6. At this point, protein expression was induced with 0.2 mM isopropyl-β-D-1-thiogalactopyranoside (Thermo Fisher Scientific) and overnight incubation at 18 °C. Cells were harvested by centrifugation (3500 × g; 30 min; 4 °C), washed twice with cold buffer A (50 mM Tris·HCl, 250 mM sodium chloride, pH 8.0), and resuspended in the same buffer supplemented with 10 mM imidazole, the EDTA-free cOmplete protease inhibitor cocktail (Roche Life Sciences), and DNase I (Roche Life Sciences). Cells were lysed using a cell disrupter (Constant Systems) at a pressure of 135 MPa, and soluble protein was cleared by centrifugation (50,000 × g; 1 h; 4 °C) before passing the supernatant through a 0.22-µm filter (Merck Millipore). For immobilised-metal affinity chromatography (IMAC) [65], protein was captured on a nickel-Sepharose HisTrap HP column (Cytiva), previously washed and equilibrated with buffer A plus either 500 or 20 mM imidazole. Each IMPI construct was purified on a separate column to avoid cross-contamination. IMPI was washed and eluted using buffer A supplemented with either 20 or 300 mM imidazole. Protein-containing fractions were dialysed for 4 h at room temperature against a 50-fold excess volume of buffer B (50 mM Tris·HCl, 150 mM sodium chloride, 0.5 mM oxidised glutathione, 3 mM reduced glutathione, pH 8.0) and centrifuged (50,000 × g; 1 h; 4 °C) to remove precipitated protein. The inhibitors were dialysed overnight with His6-tagged TEV peptidase (produced in-house) at a peptidase:substrate ratio of 1:20 (w/w) in buffer A at room temperature to remove the fusion partner. After centrifugation (50,000 × g; 1 h; 4 °C) and 0.22-µm sterile filtration, the soluble fraction was loaded again onto the above HisTrap HP column for reverse IMAC. The flow-through fraction containing untagged inhibitor was collected, whereas TEV, thioredoxin and non-cleaved soluble IMPI aggregates bound to the column were eventually eluted using buffer A supplemented with 300 mM imidazole for column regeneration. The untagged IMPI was recovered after a second round of reverse IMAC, concentrated by exchange into buffer C (20 mM Tris·HCl, 150 mM sodium chloride, pH 8.0) using a HiPrep 26/10 desalting column (Cytiva), and polished by final size-exclusion chromatography (SEC) with buffer C in a Superdex 75 10/300 column (Cytiva) attached to an ÄKTA Purifier 10 apparatus (Cytiva).
Aureolysin isoform I was produced as previously described [66] with slight modifications. S. aureus V8-BC10 cells were streaked onto tryptic soy agar plates supplemented with 2.5 g/L glucose and 1% casein. A single colony, surrounded by a halo of digested casein, was then used to inoculate 20 mL of Bacto tryptic soy broth without dextrose (BD Biosciences) supplemented with 2.5 g/L glucose. This pre-inoculum was incubated for 8 h at 37 °C under shaking. The starter cultures were used to inoculate 4 L of the same medium, followed by overnight cultivation under the same conditions. The bacterial supernatant was cleared by centrifugation (7000 × g; 30 min; 4 °C) and passed through a 0.22-µm filter. Supernatant proteins were then precipitated in ammonium sulfate (80% saturation) with gentle stirring for 4 h at 4 °C, harvested by centrifugation (50,000 × g; 1 h; 4 °C), resuspended in buffer D (20 mM Tris⋅HCl, 10 mM calcium chloride, pH 7.8), and dialysed at 4 °C overnight against the same buffer. After centrifugation (50,000 × g; 1 h; 4 °C), the supernatant was loaded onto a 5-mL HiTrap Q FF anion exchange column (Cytiva) attached to an ÄKTA Pure 25 apparatus (Cytiva). The column was previously washed and equilibrated with buffer D, with or without 1 M sodium chloride. Protein bound to the column was washed extensively using buffer D supplemented with 50 mM sodium chloride, and eluted in a gradient of 50–750 mM sodium chloride in the same buffer. The purified aureolysin was polished by SEC in a Superdex 75 10/300 column with buffer E (20 mM Tris⋅HCl, 150 mM sodium chloride, 10 mM calcium chloride, 50 µM zinc chloride, pH 7.8).
Protein purity was assessed by SDS-PAGE on custom-made 14–20% glycine gels followed by staining with Coomassie Brilliant Blue (Sigma-Aldrich). Protein identities were confirmed by peptide mass fingerprinting (Suppl. Fig. 1) and N-terminal sequencing (Edman degradation) at the Protein Chemistry Service and the Proteomics Facility of the Centro de Investigaciones Biológicas (CIB-CSIC, Madrid, Spain). Ultrafiltration was carried out using Vivaspin 15 and Vivaspin 2 filter devices with Hydrosart membranes and a 2-kDa cut-off (Sartorius Stedim Biotech). Protein concentrations were determined using the BCA protein assay kit (Thermo Fisher Scientific) by comparison to a dilution series of bovine serum albumin.
Activity and inhibition assays — We tested the proteolytic and peptidolytic activity of aureolysin, thermolysin from B. thermoproteolyticus Rokko (Sigma-Aldrich), and ulilysin (produced according to [67], [68]) at 37 °C in 100-μL reactions containing buffer F (100 mM Tris·HCl, 150 mM sodium chloride, 10 mM calcium chloride, 50 µM zinc chloride, pH 7.5) in an Infinite M200 microplate fluorimeter (Tecan). As substrates, we used 10 μg/mL of the pig-skin gelatin fluorescein conjugate from the DQ Gelatin EnzCheck assay kit (λex = 485 nm, λem = 528 nm; Invitrogen, Thermo Fisher Scientific) or 20 μM FRET-4 (Abz-Y-G-K-R-V-F-K[dpn]–OH), an internally-quenched fluorogenic peptide (λex = 260 nm λem = 420 nm; GenScript).
Inhibition by wt-IMPI was measured using both substrates following the pre-incubation of the inhibitor (up to 200-fold molar excess) with 100 nM aureolysin, 10 nM thermolysin or 10 nM ulilysin for 1 h at room temperature. Inhibition by the IMPI mutants (T50Q, T50R, T50Y, I54M, I55R, I55W, I55Y, I57F, I57Y, R58E, T50Y + I55R, and T50Y + I55R + I57F) was measured using FRET-4 following the pre-incubation of each mutant (up to 100-fold molar excess) with 50 nM aureolysin for 1 h at room temperature. Reactions were carried out at 37 °C in buffer G (20 mM Tris·HCl, 150 mM sodium chloride, pH 7.5) in triplicate and the residual proteolytic activity was measured for 3 h. The activity of the inhibitors in the absence of peptidase was monitored for the same duration as a negative control. To determine the relative activity of the IMPI mutants compared to the wild type, initial cleavage velocities of the fluorogenic protein and peptide substrates, without (V0) and with (Vi) inhibitor, were determined from the slope of the linear range (R2 > 90%) of the fluorescence vs time curve, and (V0/Vi) was calculated using GRAPHPAD PRISM [69].
Complex formation and inhibitor cleavage detection — The complexes of aureolysin (at 100 μM) with wt-IMPI or the I57F-mutant were prepared by incubation in buffer H (50 mM Tris·HCl, 150 mM sodium chloride, pH 8.0) at a 1:2.5 M ratio for 30 min at room temperature. The complex was then disrupted by SEC in a Superdex 75 10/300 GL column (GE Healthcare) previously equilibrated in buffer H. The same amounts of aureolysin and inhibitor were processed separately as controls. IMPI cleavage was analysed by SDS-PAGE as above and mass spectrometry in a MALDI-TOF Autoflex III instrument (Bruker). Each sample was desalted using a C18 ZipTip (Millipore), mixed at a 1:1 ratio (v/v) with a matrix solution of 10 mg/mL sinapic acid in 50% acetonitrile, and spotted onto the plate using the dried-droplet method. Mass spectra were acquired in linear-mode geometry. Internal calibration was performed by correction of the average mass of the respective non-treated IMPI control sample (wt-IMPI: 7927.6 Da; I57F-IMPI: 7967.1 Da).
Crystallisation and diffraction data collection — Crystallisation conditions were screened at the joint IRB/IBMB Automated Crystallography Platform using the sitting-drop vapor diffusion method. A Freedom EVO robot (Tecan) prepared screening solutions and dispensed them into the reservoir wells of 96 × 2-well MRC crystallisation plates (Innovadyne Technologies). A Phoenix/RE robot (Art Robbins) pipetted crystallisation nanodrops containing 100 nL of each protein and reservoir solution into the shallow wells, and plates were incubated in steady-temperature crystal farms (Bruker) at 4 °C or 20 °C. Optimal aureolysin crystals complexed with either wt-IMPI or I57F-IMPI formed at 20 °C in solutions containing 5 mg/mL aureolysin and 2.9 mg/mL IMPI (peptidase:inhibitor molar ratio of 1:2.5) in 50 mM Tris·HCl pH 8.0, 150 mM sodium chloride, 1.6 mM calcium chloride, 8.3 μM zinc chloride, which was mixed with reservoir solution consisting of 0.1 M Bis-Tris pH 5.5, 25% (w/v) PEG 3350 or 0.1 M Bis-Tris pH 6.0, 31% (w/v) PEG 2000 MME. Crystals were cryoprotected with reservoir solution plus 10% ethylene glycol, harvested using round LithoLoops of 0.04–0.1 mm (Molecular Dimensions), and flash-vitrified in liquid nitrogen for data collection. X-ray diffraction data were recorded at 100 K on a Pilatus 6 M pixel detector (Dectris) at the XALOC beamline of the ALBA synchrotron (Cerdanyola, Catalonia, Spain) and on a Pilatus3 X 2 M detector (Dectris) at the ID23-2 beamline of the ESRF synchrotron (Grenoble, France). Diffraction data were processed with programs XDS [70] and XSCALE, and transformed with XDSCONV to MTZ-format for the PHENIX [71] and CCP4 [72] suites of programs. Statistics describing data collection and processing are provided in Table 2.
Table 2.
Crystallographic data.
| Dataset | Aureolysin/wt-IMPI | Aureolysin/I57F-IMPI (1) | Aureolysin/I57F-IMPI (2) |
|---|---|---|---|
| Beam line (synchrotron) | XALOC (ALBA) | ID23-2 (ESRF) | XALOC (ALBA) |
| Space group/complexes per a.u. a | P41/2 | P41212/1 | P41/2 |
| Twinning fraction α (−k,−h,−l) | 0.49 | 0.380 (estimated for P41) | 0.536 |
| Cell constants (a and c in Å) | 68.14, 166.18 | 68.80, 167.25 | 68.08, 166.69 |
| Wavelength (Å) | 0.97926 | 0.87313 | 0.97926 |
| Measurements/unique reflections | 874,126/64,323 | 326,552/26,086 | 398,888/99,152 |
| Resolution range (Å) (outermost shell) c | 52.7–1.85 (1.96–1.85) | 68.8–2.05 (2.17–2.05) | 68.1–1.60 (1.70–1.60) |
| Completeness (%)/Rmerged | 100.0 (99.8)/0.149 (2.772) | 100.0 (99.9)/0.193 (1.808) | 99.7 (99.4)/0.050 (1.069) |
| Rpime/CC(1/2) e | 0.042 (0.788)/0.999 (0.630) | 0.069 (0.712)/0.997 (0.641) | 0.029 (0.618)/0.999 (0.580) |
| Average intensity f | 14.7 (1.9) | 9.4 (1.6) | 14.2 (1.8) |
| B-Factor (Wilson) (Å2)/Aver. multiplicity | 42.2/13.6 (13.4) | 41.9/12.5 (13.4) | 34.4/4.0 (4.0) |
| Resolution range used for refinement (Å) | 52.7–1.85 | 68.1–1.60 | |
| Reflections used (test set) | 63,598 (724) | 98,470 (681) | |
| Crystallographic Rfactor (free Rfactor) d | 0.164 (0.219) | 0.158 (0.188) | |
| Non-H protein atoms/ionic ligands/ | 6467/6 Ca2+, 2 Zn2+ | 6322/6 Ca2+, 2 Zn2+ | |
| waters/non-ionic ligands per a.u. | 709/2 PEG, 3 EDO | 559/5 EDO | |
| Rmsd from target values | |||
| bonds (Å)/angles (°) | 0.008/1.64 | 0.008/1.76 | |
| Average B-factor (Å2) | 38.1 | 32.6 | |
| Protein contacts and geometry analysis b | |||
| Ramachandran favoured/outliers/all analysed | 686 (95.0%)/0/722 | 691 (95.5%)/1/723 | |
| Bond-length/bond-angle/chirality/planarity outliers | 0/3/0/2 | 0/2/0/3 | |
| Side-chain outliers | 22 (3.6%) | 15 (2.5%) | |
| All-atom clashes/clashscore b | 15/1.3 | 20/1.7 | |
| RSRZ outliers b/Fo:Fc correlation | 2 (0.3%)/0.97 (0.95) | 7 (1.0%)/0.98 (0.97) | |
| PDB access code | 7SKM | 7SKL |
Abbreviations: EDO, ethylene glycol; PEG, diethylene glycol; RSRZ, real-space R-value Z-score.
According to the wwPDB Validation Service (https://wwpdb-validation.wwpdb.org/validservice).
Values in parenthesis refer to the outermost resolution shell if not otherwise indicated.
Average intensity is 〈I/σ(I)〉 of unique reflections after merging according to XSCALE[70].
Structure solution and refinement — The structure of the complex of aureolysin and I57F-IMPI was solved by molecular replacement using PHASER [73] on a dataset initially processed as space group P41212 at 2.05 Å resolution (Table 2), with one complex per asymmetric unit (a.u.). The coordinates of the protein part of unbound aureolysin (Protein Data Bank [PDB] access code 1BQB [27]) and wt-IMPI in a complex with B. thermoproteolyticus thermolysin (PDB 3SSB [62]) were used as searching models. These calculations yielded unique solutions for the peptidase and inhibitor at Eulerian angles (in °) α = 13.7, β = 29.4, γ = 153.6 (fractional cell coordinates 0.019, 0.287, 0.972) and α = 166.5, β = 131.6, γ = 104.7 (fractional cell coordinates 0.751, 0.224, 0.191), respectively. The associated values for the translation functions after refinement were 15.6 and 34.0, and the final log-likelihood gain was 1316. The adequately rotated and translated molecules were refined using the REFINE protocol of PHENIX [74] and the BUSTER [75] program, including translation/libration/screw-motion (TLS) refinement. Unexpectedly, the free Rfactor stalled at ∼ 30% and the resulting Fourier maps were partially blurred, which together with the analysis of the intensity distribution with XTRIAGE [76] in PHENIX, and POINTLESS [77] in CCP4, indicated the presence of merohedral twinning following twin law (-k, -h, -l). At this point, a second dataset for the I57F-IMPI complex with a higher resolution (1.60 Å) became available, which was processed with the actual space group P41 (Table 2) and solved by Fourier synthesis after rigid-body refinement of the two copies of the partially refined complex structure in the a.u. The structure was manually rebuilt using COOT [78] and refined using REFMAC5 [79] considering twinning, as well as TLS and non-crystallographic symmetry (NCS) restraints. The final model included residues A209–E509, one zinc and three calcium ions of peptidase protomers A and C, as well as I20–I86 and I20–P84 of inhibitor moieties B and D, respectively, plus five ethylene glycol and 559 solvent molecules. Given that the structure of unbound aureolysin had originally been obtained before the gene sequence was available [27], [49], it contained five erroneous residues at positions 354, 361, 479, 492, and 493, which were corrected in the final model of the complex.
The structure of the wt-IMPI complex with aureolysin was solved at a resolution of 1.85 Å by Fourier synthesis after rigid-body refinement using the coordinates of the refined mutant complex structure. Model completion and refinement were carried out as described above. The final model comprised residues A209–E509 and A209–V508 of peptidase molecules A and C, plus one zinc and three calcium ions each, as well as I20–I86 and I20–K85 of inhibitor moieties B and D, respectively. Two diethylene glycol, three ethylene glycol, and 709 solvent molecules completed the model. Table 2 provides essential statistics on the final refined models, which were validated using the wwPDB validation service (https://validate-rcsb-1.wwpdb.org/validservice) and deposited at www.pdb.org (access codes 7SKL and 7SKM).
Miscellaneous — Structural superpositions were calculated with SSM [80] in COOT. Figures were prepared using CHIMERA [81]. Protein interfaces and intermolecular interactions were analysed using PDBEPISA [82] (www.ebi.ac.uk/pdbe/pisa) and verified by visual inspection. The interacting surface of a complex was taken as half the sum of the buried surface areas of either molecule.
3. Results and discussion
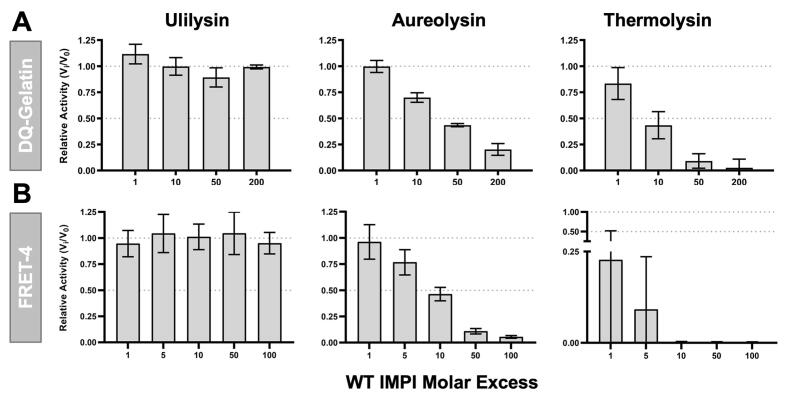
Assessment of wild-type IMPI as an aureolysin inhibitor and initial protein redesign — Wild-type IMPI was expressed in E. coli and recovered in a highly pure form (Fig. 1A-D; Suppl. Fig. 1). To assess its effect on aureolysin, which in turn was purified to homogeneity from cultures of S. aureus (Fig. 1E-H; Suppl. Fig. 1), the inhibitor was tested at molar ratios of 1:1 to 1:200 using a fluorogenic protein (Fig. 2A) and a fluorogenic peptide (Fig. 2B). We also tested thermolysin (the archetypal M4 family MP) and ulilysin, a metzincin MP from the pappalysin family (MEROPS M43B; [67], [68]) as controls. Thermolysin was efficiently inhibited as expected, whereas ulilysin was not inhibited at all, in agreement with IMPI being a specific inhibitor of M4 family MPs. Aureolysin was also inhibited in a dose-dependent manner, particularly when using the peptide substrate, although not to the same extent as thermolysin.
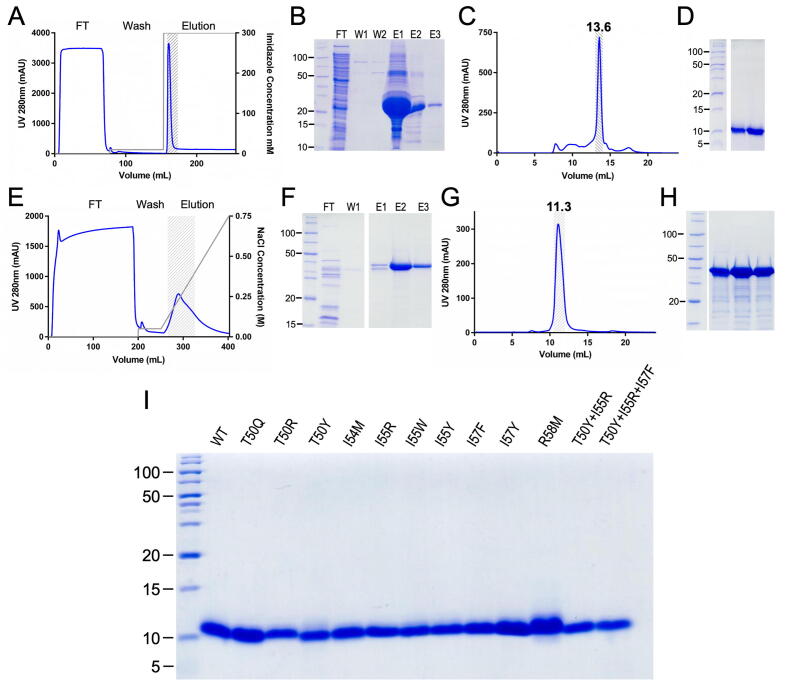
Fig. 1.
Protein production and purification. (A) Representative chromatogram and (B) SDS-PAGE analysis of the IMAC purification step of His6-thioredoxin-tagged wt-IMPI (expected molecular mass ∼ 25 kDa). FT, flow-through; W, wash step; E, elution step. (C) Chromatogram and (D) SDS-PAGE analysis of the SEC purification step of tag-depleted wt-IMPI (∼8 kDa), which migrated as a monomer (13.6 mL). (E) Representative chromatogram and (F) SDS-PAGE analysis of the anion-exchange chromatography purification step of aureolysin. (G) Chromatogram and (H) SDS-PAGE analysis of the SEC purification step of aureolysin. Despite the higher-than-expected molecular mass reported by SDS-PAGE (panels F,H), the protein is indeed mature aureolysin (expected mass ∼ 33 kDa), as confirmed by N-terminal sequencing, peptide-mass fingerprinting (Suppl. Fig. 1), and the retention volume in calibrated SEC (panel G; 11.3 mL) corresponding to ∼ 29 kDa. (I) 20% Glycine SDS-PAGE showing the purity of wt-IMPI and the 12 mutants (2–5 μg) analysed herein. All constructs behaved similarly to (A-D) during purification and yielded products of comparable purity and molecular mass.
Fig. 2.
Inhibitory activity of wild-type IMPI. (A) Residual fractional activity as Vi/V0 relative to the activity in the absence of inhibitor of (left) 10 nM ulilysin, (middle) 100 nM aureolysin, and (right) 10 nM thermolysin after incubation with wt-IMPI at several molar ratios using the DQ gelatin substrate. (B) As above, but using the internally quenched fluorescent FRET-4 peptide as the substrate.
We superposed the structure of unbound aureolysin [27] onto thermolysin in a complex with wt-IMPI [62] and hypothesised that replacing I57 (whose side chain interacts with the MP, see below) with a bulkier residue such as phenylalanine might achieve stronger inhibition. Accordingly, we produced the mutant I57F-IMPI as described above for wt-IMPI (Fig. 1I) and used it for further analysis.
Overall structure of the IMPI–aureolysin complex — We crystallised I57F-IMPI and wt-IMPI in complexes with aureolysin (Fig. 3A) and used molecular replacement to solve their tetragonal (P41) crystal structures, which contained two complexes per a.u. Structural solution and refinement (to 1.60 and 1.85 Å, respectively) was hindered by the presence of merohedral twinning in both crystals, with twinning fractions of 0.536 and 0.490, respectively (Table 2). Even so, the structures were refined to final free Rfactor values of 0.188 and 0.219, respectively, which are considered accurate. This was confirmed by the final Fourier maps (Fig. 3E). The two structures were practically indistinguishable upon superposition, so the following discussion focuses on the I57F-IMPI complex (protomers A and B) if not otherwise stated.
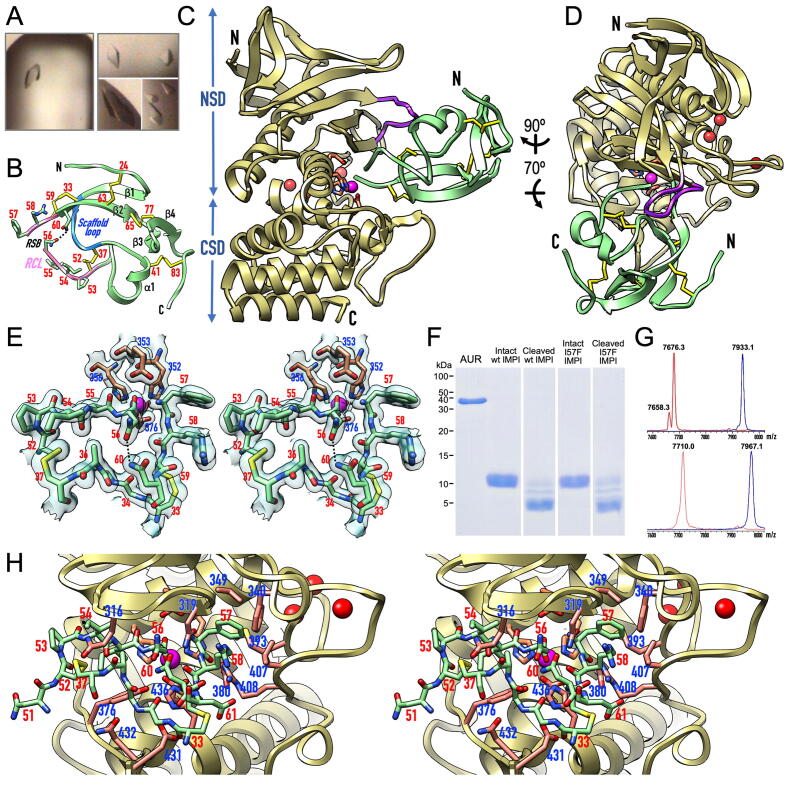
Fig. 3.
Structure of the IMPI–aureolysin complex. (A) Tetragonal protein crystals of the aureolysin–wt-IMPI (left) and aureolysin–I57F-IMPI complexes (right). (B) Ribbon-type plot of I57F-IMPI depicting the four β-strands (β1–β4) and the single helix (α1) of the structure, as well as the five disulfide bridges (with numbered cysteine residues). The scaffold loop is shown in blue, and the reactive-centre loop (RCL) is shown in pink with numbered residues (sticks). The cleaved reactive-site bond (RSB), N-terminus, and C-terminus are labelled. Hydrogen bond N56Oδ1–N60Nδ2 is needed to maintain the position of the P1 residue in place. (C) Ribbon-type plot of the complex between I57F-IMPI (green ribbon, disulfide bonds as yellow sticks) and aureolysin (pale gold ribbon, catalytic zinc and structural calcium cations shown as magenta and red spheres, respectively) viewed along the active-site cleft (vertically rotated 90° counterclockwise away from the traditional “standard orientation” of MPs [53]). The side chains of the zinc-binding MP residues and the general/base acid glutamate are further shown as sticks for reference (carbons in salmon). The N-termini and C-termini are labelled, the characteristic flap is in purple, and the NSD and CSD of the peptidase are indicated. (D) Rotated view of (C). (E) Close-up in cross-eye stereo showing the RCL and scaffold loop of I57F-IMPI (green carbons) and the zinc site of aureolysin (carbons in salmon) superposed with the final 1.60-Å (2mFobs-DFcalc)-type Fourier map as a semi-transparent surface contoured at 1 σ in a similar view to (D). The RSB is cleaved, selective inhibitor and MP residues are numbered in red and blue, respectively. Hydrogen bond N56Oδ1–N60Nδ2 is shown as a dashed line. (F) In vitro proof that binding and inhibition of aureolysin by wt- and I57F-IMPI involves the cleavage of the inhibitor at the RSB (N56–I57) within the RCL as shown by SDS-PAGE analysis of the respective SEC fractions. (G) Mass spectra showing analysis of the cleavage of (top) intact wt-IMPI (blue spectrum; 7933.1 Da) giving rise to the cleaved inhibitor (red spectrum; 7676.3 Da) and (bottom) intact I57F-IMPI (blue spectrum; 7967.1 Da) to yield the cleaved inhibitor (red spectrum; 7710.0 Da). Incubation of both intact species with aureolysin leads to the removal of the N-terminal tag-segment G-M−S (–275 Da) and the addition of a water molecule (+18 Da) due to RSB cleavage. For wt-IMPI, a small fraction of tag-depleted noncleaved inhibitor was detected (7658.3 Da). (H) Close-up in stereo of (D), further rotated 25° downwards and 25° leftwards, giving insight into the interactions between I57F-IMPI (sticks with green carbons, residue numbers in red) and aureolysin (sticks with carbons in salmon, residue numbers in blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The structure of wt-IMPI has been reported in a complex with thermolysin [62]. Briefly, it has a spearhead shape (Fig. 3B), whose tip contains a “reactive-site bond” (RBS; N56–I/F57) within a RCL (C52–C59). The latter is anchored to a subjacent “scaffold loop” (C33–C37) via two disulfide bonds, which are part of a set of five that confer structural rigidity. The regular secondary structures of IMPI comprise four β-strands (β1–β4) and one α-helix (α1).
The structure of the aureolysin CD is known for its unbound form [27]. It conforms to that of the thermolysin family and consists of an N-terminal subdomain (NSD; A209–A363, Fig. 3C) featuring an N-terminal β-barrel grafted into a frontal five-stranded mixed β-sheet whose lowermost strand forms the “upper-rim” of the active-site cleft (N322–I326; Fig. 3C,D). This element binds substrates in an extended conformation as an antiparallel β-ribbon. The NSD also contains a “backing helix” and an “active-site helix”, encompassing the characteristic motif of the zincin MPs, H352-E-X-X-H356 [83], [84]. The two histidine residues are ligands of the catalytic zinc, and the glutamate is the general base/acid of the cleavage reaction [85] (Fig. 3E). The main distinctive structural element of the aureolysin NSD compared to other thermolysins is a “flap” (N312–N321) that precedes the upper-rim strand and protrudes from the surface above the cleft (Fig. 3C,D).
The C-terminal subdomain (CSD; N364–E509; Fig. 3C) starts with the characteristic “glutamate helix” of gluzincins [19], [20], which contains the third zinc-binding protein ligand (E376; Fig. 3E). It is followed by a long “irregular segment” (D388–G434) that shapes the bottom of the active-site cleft on its primed side, including the hydrophobic S1′ pocket. This pocket confers substrate specificity upon aureolysin and other M4 family MPs, as well as most other MP families [53]. Moreover, the irregular segment embraces three calcium-binding sites, which stabilise the structure [28]. The removal of these ions using chelators therefore causes irreversible inactivation [28], [86]. The CSD also contains a C-terminal four-helix bundle arranged as a Greek-key motif. Remarkably, the aureolysin CSD lacks the conspicuous β-ribbon that protrudes from the last turn of the first of these α-helices in thermolysin.
In the complex, I57F-IMPI inserts like a wedge into the active-site cleft of the peptidase (Fig. 3C,D) and interacts via interfaces of 865 and 849 Å2 (ΔiG = –5.2 and –4.5 kcal/mol [82]) in complexes A/B and C/D, respectively. This involves 24 hydrogen bonds and salt bridges, plus two metallorganic bonds, as well as hydrophobic interactions between five inhibitor and 10 peptidase residues (Table 3). The main participating elements are the RCL and scaffold loop of the inhibitor, as well as the flap, upper-rim strand, S1′-pocket shaping residues, and the initial and final stretches of the irregular segment. Diverging from the thermolysin complex, superposition of the aureolysin complexes with wt-IMPI and I57F-IMPI revealed a much smaller spread in the relative orientation between inhibitor and peptidase. The maximum deviation at the cleft-distal site of the inhibitor was ∼ 4°/1.8 Å across the four complexes of the two structures, compared to ∼ 10°/4.8 Å for the two thermolysin complexes in the a.u. [62].
Table 3.
Interactions at the I57F-IMPI–aureolysin interface.
| Hydrogen bonds/salt bridges (<3.7 Å) | Hydrophobic interactions (<4 Å) |
|---|---|
| Y31O – K430Nζ 3.10/2.79 Å | |
| E32Oε2 – D431N 2.75/2.82 Å | |
| A36O – Q317Nε2 –/2.72 Å | E32 – K430 |
| D38N – Q317Oε1 3.11/– Å | I54 – I326 |
| D38N – Q317Nε2 –/3.48 Å | I55 – H356 |
| Q47Nε2 - Q317Nε2 3.48/– | I55 – Y367 |
| I55N – W325O 2.92/2.86 Å | F57 – F340 |
| I55O – W325N 2.87/2.92 Å | F57 – L343 |
| N56O – E353Oε1 2.67/2.83 Å | F57 – V349 |
| N56O – E353Oε2 3.23/3.10 Å | F57 – H352 |
| N56OT – H436Nε2 3.15/3.08 Å | F57 – M396 |
| N56OT – Y367Oη 3.69/3.45 Å | F57 – L407 |
| N56Nδ2 – A323O 2.82/3.00 Å | R58 – F340 |
| F57N – N322Oδ1 3.09/3.25 Å | R58 – L407 |
| F57N – A323O 3.27/3.14 Å | |
| F57N – E353Oε2 2.90/3.01 Å | |
| F57O – R408Nη1 2.84/2.75 Å | |
| F57O – R408Nη2 2.83/2.76 Å | |
| R58N – N322Oδ1 3.51/3.45 Å | |
| R58Nε – N321O 2.85/2.47 Å | |
| R58Nη1 – N321O 2.74/– Å | |
| R58O – N322Nδ2 2.88/2.83 Å | |
| N60Oδ1 – N322Nδ2 3.11/2.80 Å | |
| K62Nζ – Q317Oε1 –/2.66 Å | |
| Ionic interactions | |
| N56OT – Zn999 2.11/2.08 Å | |
| N56O – Zn999 2.61/2.38 Å |
The first residue/atom belongs to IMPI, the second to aureolysin. The two values for each bond correspond to complexes between protomers A/B and C/D, respectively.
Finally, superposition of IMPI-bound aureolysin with the unbound structure [27] revealed negligible differences between the NTS and CTS. This contrasts with thermolysin, where a 5° relative rotation of the two subdomains distinguishes between the unbound and bound forms [87]. Similar relative motion was proposed for P. aeruginosa elastase and Bacillus cereus neutral proteinase [27]. Aureolysin therefore does not appear to undergo the closing hinge motion when binding ligands or substrates, in contrast to other M4 family MPs.
IMPI inhibits aureolysin via the standard mechanism — The IMPI RCL runs across the peptidase cleft in the direction of the substrate, blocking S4-S1′ with residues P53–I/F57 (Fig. 3H). Remarkably, the RSB was cleaved in the crystals (Fig. 3E), which was verified in vitro by incubating both wt-IMPI and I57F-IMPI with aureolysin. Indeed, both forms were cleaved at N56–I/F57 (Fig. 3F,G). This feature causes the terminal carboxylate oxygen of the P1 residue, N56OT, to bind the catalytic zinc and contribute to a distorted tetrahedral coordination sphere together with protein ligands H352Nε2, H356Nε2, and E376Oε2 (all 2.02–2.11 Å apart in the various structures). N56OT replaces the two solvent molecules found in the unbound structure [27] and further contacts H436Nε2 (3.08–3.15 Å), which is equivalent to H231 of thermolysin (thermolysin residues are shown in italics with subscript numbers for clarity). Together with Y157, equivalent to Y376 in aureolysin, this residue helps to stabilise the tetrahedral reaction intermediate [85]. Moreover, the other carboxylate oxygen of N56 is very close to the general base/acid glutamate (N56O–E353Oε1; 2.60–2.67 Å), indicating that one of them must be protonated. On the primed side of the cleft, P1′ residue I/F57 is bound via its α-amino group to E353Oε2 (2.90–3.01 Å) and the upper-rim main-chain carbonyl of A323 (3.14–3.27 Å) as well as the side-chain carboxamide of N322 (3.09–3.25 Å; Fig. 3E,H).
The inhibition mode described above agrees with the “standard mechanism” or “canonical mechanism” of peptidase inhibition [88], [89]. Remarkably, in standard-mechanism inhibitors (which mostly target serine endopeptidases), the RSB is cleaved very slowly because the cleavage reaction is kinetically unfavourable, so the intact complexes have half-lives of several years [90]. This has been verified by many crystal structures with intact RSBs [91]. In contrast, IMPI represents a unique case of a standard-mechanism MP inhibitor occurring as a cleaved inhibitor, first in its thermolysin complex [62] and now here with aureolysin, whose 69-residue structure is kept rigid through five disulfide bonds that are evenly distributed across the structure.
Finally, in the aureolysin complexes, the cleaved RSB is poised for rejoining, which is another functional requisite of the standard mechanism [91]. This is indicated by the proximity and orientation of the α-amino group of I/F57 relative to the carboxylate carbon of N56, which are ideally situated for a nucleophilic attack. Indeed, the angle I/F57N–N56C–N56OT, where N56OT is the oxygen that is not bound to the general base/acid glutamate, is ∼ 110° on average over all four I57F-IMPI and wt-IMPI complexes, thus in good agreement with the value postulated for a productive Bürgi-Dunitz geometrical reaction coordinate (105 ± 5° [92]). This is supported by the ability of cleaved wt-IMPI to rejoin in vitro following the addition of catalytic amounts of thermolysin [62].
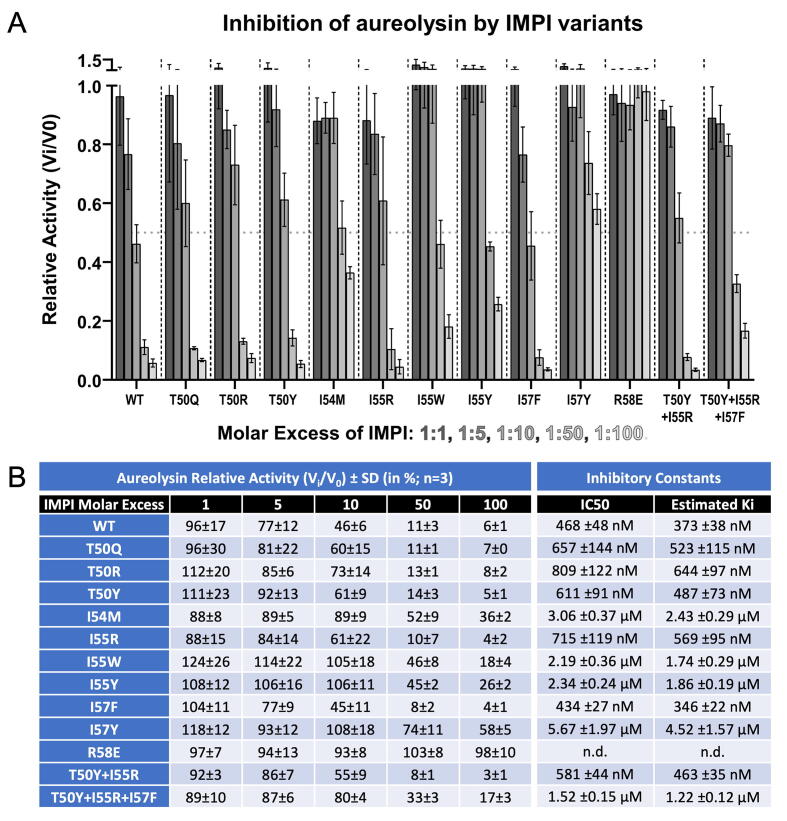
Redesign of IMPI — Based on the IMPI–aureolysin crystal structures described above, we identified positions 50, 54, 55, 57 and 58 of the RCL as ideal for mutagenesis and constructed 11 single, double and triple point mutants in addition to the wt-IMPI and I57F-IMPI variants (T50Q, T50R, T50Y, I54M, I55R, I55W, I55Y, I57Y, R58E, T50Y + I55R, and T50Y + I55R + I57F). All variants were produced and purified as efficiently as described above for wt-IMPI (Fig. 1I), and were compared to wt-IMPI for their ability to inhibit aureolysin at molar ratios of 1:1 to 1:100 using the fluorogenic peptide FRET-4 as the substrate (Fig. 4A,B). R58E did not affect peptidase activity. We tested the mutant with thermolysin, which revealed ∼ 200-fold weaker inhibition than the wild type (Suppl. Fig. 2). We thus conclude that the mutant was properly folded, as suggested by its behaviour during purification, but functionally impaired and thus unable to block thermolysins. The rest of the cohort of mutants achieved the concentration-dependent inhibition of aureolysin. They could be assigned to two groups, one similar to the wild type, with residual activities of 3–8% at the highest molar ratio (Fig. 4B), whereas and the others showing weaker inhibition, with residual activities of 17–58% (Fig. 4B). The derived IC50 values enabled us to estimate Ki values of 346–644 nM for the first group and 1220–4520 nM for the second group (Fig. 4B). Notably, mutant I57F (from the initial stage of the project, see above) achieved the highest inhibition among all variants tested (Ki = 346 nM) and would thus provide a suitable lead for further development.
Fig. 4.
Inhibitory activity of the IMPI mutants. (A) Relative fractional activity as Vi/V0 of 50 nM aureolysin after incubation with IMPI mutants, relative to the wild type at molar ratios 1:1, 1:5, 1:10, 1:50 and 1:100 with FRET-4 (at 20 μM) as the substrate. Experiments were performed at least in triplicate, and error bars show standard deviations. (B) Tabular representation of the relative aureolysin activity data (in %) shown in (A). Average half-maximal inhibitory concentrations (IC50) were determined using a four-parameter sigmoidal fit in GRAPHPAD (see Suppl. Fig. 2), and the inhibitor constant Ki was estimated usinf the equation Ki = IC50/([S]/KM + 1) [93].
Corollary — Aureolysin plays multiple roles during S. aureus infections and is a promising target for the development of novel antimicrobials. We tested the M4-specific inhibitor IMPI, and found that it inhibited the peptidase using the standard mechanism, best described for serine endopeptidases, based on the analysis of crystal structures. We therefore designed a cohort of point mutants, with I57F emerging as the strongest inhibitor. This is, to our knowledge, the first report of a TP candidate that can inhibit one of the major proteolytic virulence factors of S. aureus. The only other protein-based inhibitor with this ability is the general pan-peptidase inhibitor α2-macroglobulin, which has a molecular mass of ∼720 kDa and a broad spectrum of targets, making it unsuitable for therapeutic applications. Cell-based and disease challenge studies are now required to confirm the potential of I57F-IMPI as a TP for the treatment of S. aureus infections.
Author contributions
F.X.G.R. and A.V. conceived, supervised, and funded the project; S.R.M. produced and purified all proteins, prepared the mutants, performed in vitro studies with U.E. and P.C., analysed kinetics and mass spectrometry data with U.E., and crystallised proteins with assistance from U.E. and T.G.; S.R.M., U.E. and A.R.-B. collected diffraction data, and U.E. performed initial data analysis; E.M. performed biocomputational calculations; F.X.G.R. solved and refined crystal structures; and F.X.G.R. and A.V. wrote the manuscript with contributions from all authors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to Laura Company, Xandra Kreplin and Joan Pous from the joint IBMB/IRB Automated Crystallography Platform and the Protein Purification Service at IBMB for assistance during purification and crystallisation experiments, and Carme Quero from the Institut de Química Avançada de Catalunya (IQAC-CSIC) is thanked for assistance with mass spectrometry. We also acknowledge the kind gift of S. aureus strain V8-BC10 (for aureolysin production) from Jan Potempa, Jagiellonian University of Kraków, Poland. The authors would also like to thank the ESRF and ALBA synchrotrons for beamtime allocation and the beamline staff for assistance during diffraction data collection. This study was supported in part by Spanish and Catalan public and private bodies that provided funding to the Proteolysis Lab (grants PID2019-107725RG-I00 from MCIN/AEI/10.13039/501100011033, 2017SGR3 from the National Government of Catalonia, and 201815 from Fundació “La Marató de TV3”). S.M.E. acknowledges grant BES2016-076877 from the Spanish State Agency for Research (MCIN/AEI/10.13039/501100011033) and the European Social Fund “ESF invests in your future”. U.E. acknowledges a “Beatriu-de-Pinós” COFUND fellowship from the National Government of Catalonia (2018BP00163). A.V. and P.C. acknowledge funding from the German Federal Ministry for Education and Research (BMBF) through project “4-In” (Inhalable Virulence-Inhibitors from Insects for the Therapy of lung infections, ref. 16GW0137K). The authors thank Richard M. Twyman for editing the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.01.001.
Contributor Information
Ulrich Eckhard, Email: ueccri@ibmb.csic.es.
Andreas Vilcinskas, Email: andreas.vilcinskas@agrar.uni-giessen.de.
F. Xavier Gomis-Rüth, Email: xgrcri@ibmb.csic.es.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.WHO (2014) Antimicrobial resistance: global report on surveillance 2014. . Geneva: WHO. p. 257.
- 2.Sabtu N., Enoch D.A., Brown N.M. Antibiotic resistance: what, why, where, when and how? Br Med Bull. 2015;116:105–113. doi: 10.1093/bmb/ldv041. [DOI] [PubMed] [Google Scholar]
- 3.Bengtsson-Palme J., Kristiansson E., Larsson D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev. 2018;42(fux053) doi: 10.1093/femsre/fux053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odenholt I., Gustafsson I., Löwdin E., Cars O. Suboptimal antibiotic dosage as a risk factor for selection of penicillin-resistant Streptococcus pneumoniae: in vitro kinetic model. Antimicrob Agents Chemother. 2003;47(2):518–523. doi: 10.1128/AAC.47.2.518-523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delepierre A., Gayot A., Carpentier A. Update on counterfeit antibiotics worldwide; public health risks. Med Mal Infect. 2012;42(6):247–255. doi: 10.1016/j.medmal.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Van Boeckel T.P., Brower C., Gilbert M., Grenfell B.T., Levin S.A., Robinson T.P., et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA. 2015;112(18):5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livermore D. Can better prescribing turn the tide of resistance? Nat Rev Microbiol. 2004;2(1):73–78. doi: 10.1038/nrmicro798. [DOI] [PubMed] [Google Scholar]
- 8.Diekema D.J., Jones R.N. Oxazolidinone antibiotics. Lancet. 2001;358(9297):1975–1982. doi: 10.1016/S0140-6736(01)06964-1. [DOI] [PubMed] [Google Scholar]
- 9.Strieker M., Marahiel M.A. The structural diversity of acidic lipopeptide antibiotics. Chembiochem : Eur J Chem Biol. 2009;10(4):607–616. doi: 10.1002/cbic.200800546. [DOI] [PubMed] [Google Scholar]
- 10.Projan S.J. Why is big Pharma getting out of antibacterial drug discovery? Curr Opin Microbiol. 2003;6(5):427–430. doi: 10.1016/j.mib.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Norrby SR, Nord CE, Finch R, ESCMID f (2005) Lack of development of new antimicrobial drugs: a potential serious threat to public health. Lancet Infect Dis 5:115-119. [DOI] [PubMed]
- 12.Livermore D.M. The need for new antibiotics. Clin Microbiol Infect. 2004;10(Suppl 4):1–9. doi: 10.1111/j.1465-0691.2004.1004.x. [DOI] [PubMed] [Google Scholar]
- 13.Hawkey P.M., Warren R.E., Livermore D.M., McNulty C.A.M., Enoch D.A., Otter J.A., et al. Treatment of infections caused by multidrug-resistant Gram-negative bacteria: report of the British Society for Antimicrobial Chemotherapy/Healthcare Infection Society/British Infection Association Joint Working Party. J Antimicrob Chemother. 2018;73(suppl_3):iii2–iii78. doi: 10.1093/jac/dky027. [DOI] [PubMed] [Google Scholar]
- 14.Nelson R.E., Hatfield K.M., Wolford H., Samore M.H., Scott R.D., Reddy S.C., et al. National estimates of healthcare costs associated with multidrug-resistant bacterial infections among hospitalized patients in the United States. Clin Infect Dis. 2021;72(Supplement_1):S17–S26. doi: 10.1093/cid/ciaa1581. [DOI] [PubMed] [Google Scholar]
- 15.David M.Z., Daum R.S. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-García S., Rodríguez-Martínez S., Cancino-Diaz M.E., Cancino-Diaz J.C. Extracellular proteases of Staphylococcus epidermidis: roles as virulence factors and their participation in biofilm. APMIS. 2018;126(3):177–185. doi: 10.1111/apm.12805. [DOI] [PubMed] [Google Scholar]
- 17.Rawlings N.D., Bateman A. How to use the MEROPS database and website to help understand peptidase specificity. Protein Sci. 2021;30:83–92. doi: 10.1002/pro.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews B.W., Jansonius J.N., Colman P.M., Schoenborn B.P., Dupourque D. Three-dimensional structure of thermolysin. Nature. 1972;238:37–41. doi: 10.1038/newbio238037a0. [DOI] [PubMed] [Google Scholar]
- 19.Hooper N.M. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 20.Cerdà-Costa N., Xavier Gomis-Rüth F. Architecture and function of metallopeptidase catalytic domains. Prot Sci. 2014;23(2):123–144. doi: 10.1002/pro.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galdino A.C.M., de Oliveira M.P., Ramalho T.C., de Castro A.A., Branquinha M.H., Santos A.L.S. Anti-virulence strategy against the multidrug-resistant bacterial pathogen Pseudomonas aeruginosa: pseudolysin (elastase B) as a potential druggable target. Curr Protein Pept Sci. 2019;20(5):471–487. doi: 10.2174/1389203720666190207100415. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi S.-I. Extracellular proteolytic enzymes produced by human pathogenic vibrio species. Front Microbiol. 2013;4:339. doi: 10.3389/fmicb.2013.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kooi C., Subsin B., Chen R., Pohorelic B., Sokol P.A. Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infect Immun. 2006;74(7):4083–4093. doi: 10.1128/IAI.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makinen P.L., Makinen K.K. The Enterococcus faecalis extracellular metalloendopeptidase (EC 3.4.24.30; coccolysin) inactivates human endothelin at bonds involving hydrophobic amino acid residues. Biochem Biophys Res Commun. 1994;200(2):981–985. doi: 10.1006/bbrc.1994.1546. [DOI] [PubMed] [Google Scholar]
- 25.Sahney N.N., Summersgill J.T., Ramírez J.A., Miller R.D. Inhibition of oxidative burst and chemotaxis in human phagocytes by Legionella pneumophila zinc metalloprotease. J Med Microbiol. 2001;50:517–525. doi: 10.1099/0022-1317-50-6-517. [DOI] [PubMed] [Google Scholar]
- 26.Okabe A., Matsushita O. In: Handbook of Proteolytic Enzymes. Rawlings N.D., Salvesen G.S., editors. Academic Press; Oxford: 2013. Chapter 113 - Lambda toxin (Clostridium perfringens) pp. 561–563. [Google Scholar]
- 27.Banbula A., Potempa J., Travis J., Fernandez-Catalén C., Mann K., Huber R., et al. Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72Å. resolution. Structure. 1998;6(9):1185–1193. doi: 10.1016/s0969-2126(98)00118-x. [DOI] [PubMed] [Google Scholar]
- 28.Potempa J., Shaw L.N. In: Handbook of Proteolytic Enzymes. Rawlings N.D., Salvesen G.S., editors. Academic Press; Oxford: 2013. Chapter 114 - Aureolysin; pp. 563–569. [Google Scholar]
- 29.Cassat J., Hammer N., Campbell J.P., Benson M., Perrien D., Mrak L., et al. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe. 2013;13(6):759–772. doi: 10.1016/j.chom.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arvidson S., Holme T., Lindholm B. The formation of extracellular proteolytic enzymes by Staphylococcus aureus. Acta Pathol Microbiol Scand B Microbiol Immunol. 1972;80:835–844. doi: 10.1111/j.0365-5563.1973.tb00009.x. [DOI] [PubMed] [Google Scholar]
- 31.Shaw L., Golonka E., Potempa J., Foster S.J. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology. 2004;150:217–228. doi: 10.1099/mic.0.26634-0. [DOI] [PubMed] [Google Scholar]
- 32.Dubin G. Extracellular proteases of Staphylococcus spp. Biol Chem. 2002;383:1075–1086. doi: 10.1515/BC.2002.116. [DOI] [PubMed] [Google Scholar]
- 33.Burlak C., Hammer C.H., Robinson M.-A., Whitney A.R., McGavin M.J., Kreiswirth B.N., et al. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol. 2007;9(5):1172–1190. doi: 10.1111/j.1462-5822.2006.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elmwall J., Kwiecinski J., Na M., Ali A.A., Osla V., Shaw L.N., et al. Galectin-3 Is a target for proteases involved in the virulence of Staphylococcus aureus. Infect Immun. 2017;85(7) doi: 10.1128/IAI.00177-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehman M.K., Nuxoll A.S., Yamada K.J., Kielian T., Carson S.D., Fey P.D., et al. Protease-Mediated Growth of Staphylococcus aureus on Host Proteins Is opp3 Dependent. mBio. 2019;10(2) doi: 10.1128/mBio.02553-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martí M., Trotonda M.P., Tormo-Más M.Á., Vergara-Irigaray M., Cheung A.L., Lasa I., et al. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 2010;12(1):55–64. doi: 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Gimza B.D., Jackson J.K., Frey A.M., Budny B.G., Chaput D., Rizzo D.N., et al. Unraveling the Impact of Secreted Proteases on Hypervirulence in Staphylococcus aureus. mBio. 2021;12(1) doi: 10.1128/mBio.03288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sieprawska-Lupa M., Mydel P., Krawczyk K., Wójcik K., Puklo M., Lupa B., et al. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob Agents Chemother. 2004;48(12):4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laarman A.J., Ruyken M., Malone C.L., van Strijp J.A.G., Horswill A.R., Rooijakkers S.H.M. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol. 2011;186(11):6445–6453. doi: 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 40.Kubica M, Guzik K, Koziel J, Zarebski M, Richter W et al. (2008) A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS one 3:e1409. [DOI] [PMC free article] [PubMed]
- 41.Pietrocola G., Nobile G., Rindi S., Speziale P. Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Front Cell Infect Microbiol. 2017;7:166. doi: 10.3389/fcimb.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potempa J., Watorek W., Travis J. The inactivation of human plasma α1-proteinase inhibitor by proteinases from Staphylococcus aureus. J Biol Chem. 1986;261:14330–14334. [PubMed] [Google Scholar]
- 43.Potempa J., Dubin A., Watorek W., Travis J. An elastase inhibitor from equine leukocyte cytosol belongs to the serpin superfamily. Further characterization and amino acid sequence of the reactive center. J Biol Chem. 1988;263(15):7364–7369. [PubMed] [Google Scholar]
- 44.Potempa J., Fedak D., Dubin A., Mast A., Travis J. Proteolytic inactivation of α1-anti-chymotrypsin. Sites of cleavage and generation of chemotactic activity. J Biol Chem. 1991;266:21482–21487. [PubMed] [Google Scholar]
- 45.Potempa J., Wunderlich J.K., Travis J. Comparative properties of three functionally different but structurally related serpin variants from horse plasma. Biochem J. 1991;274(Pt 2):465–471. doi: 10.1042/bj2740465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beaufort N., Wojciechowski P., Sommerhoff C., Szmyd G., Dubin G., Eick S., et al. The human fibrinolytic system is a target for the staphylococcal metalloprotease aureolysin. Biochem J. 2008;410(1):157–165. doi: 10.1042/BJ20070650. [DOI] [PubMed] [Google Scholar]
- 47.Lew D.P., Waldvogel F.A. Osteomyelitis. Osteomyelitis. Lancet. 2004;364(9431):369–379. doi: 10.1016/S0140-6736(04)16727-5. [DOI] [PubMed] [Google Scholar]
- 48.Jusko M., Potempa J., Kantyka T., Bielecka E., Miller H.K., Kalinska M., et al. Staphylococcal proteases aid in evasion of the human complement system. J Innate Immun. 2014;6(1):31–46. doi: 10.1159/000351458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuomanen E.I., Sabat A., Kosowska K., Poulsen K., Kasprowicz A., Sekowska A., et al. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect Immun. 2000;68(2):973–976. doi: 10.1128/iai.68.2.973-976.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nickerson N.N., Joag V., McGavin M.J. Rapid autocatalytic activation of the M4 metalloprotease aureolysin is controlled by a conserved N-terminal fungalysin-thermolysin-propeptide domain. Mol Microbiol. 2008;69(6):1530–1543. doi: 10.1111/j.1365-2958.2008.06384.x. [DOI] [PubMed] [Google Scholar]
- 51.Adekoya O.A., Sylte I. The thermolysin family (M4) of enzymes: therapeutic and biotechnological potential. Chem Biol Drug Des. 2009;73(1):7–16. doi: 10.1111/j.1747-0285.2008.00757.x. [DOI] [PubMed] [Google Scholar]
- 52.Schechter I., Berger A. On the size of active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 53.Gomis-Rüth F.X., Botelho T.O., Bode W. A standard orientation for metallopeptidases. Biochim Biophys Acta. 2012;1824(1):157–163. doi: 10.1016/j.bbapap.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 54.Craik D.J., Fairlie D.P., Liras S., Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81(1):136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 55.Dingermann T. Recombinant therapeutic proteins: production platforms and challenges. Biotechnol J. 2008;3(1):90–97. doi: 10.1002/biot.200700214. [DOI] [PubMed] [Google Scholar]
- 56.Bruno B.J., Miller G.D., Lim C.S. Basics and recent advances in peptide and protein drug delivery. Ther Deliv. 2013;4(11):1443–1467. doi: 10.4155/tde.13.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oda K., Koyama T., Murao S. Purification and properties of a proteinaceous metallo-proteinase inhibitor from Streptomyces nigrescens TK-23. Biochim Biophys Acta. 1979;571(1):147–156. doi: 10.1016/0005-2744(79)90235-3. [DOI] [PubMed] [Google Scholar]
- 58.Seeram S.S., Hiraga K., Oda K. Resynthesis of reactive site peptide bond and temporary inhibition of Streptomyces metalloproteinase inhibitor. J Biochem. 1997;122(4):788–794. doi: 10.1093/oxfordjournals.jbchem.a021824. [DOI] [PubMed] [Google Scholar]
- 59.Wedde M., Weise C., Kopacek P., Franke P., Vilcinskas A. Purification and characterization of an inducible metalloprotease inhibitor from the hemolymph of greater wax moth larvae, Galleria mellonella. Eur J Biochem. 1998;255(3):535–543. doi: 10.1046/j.1432-1327.1998.2550535.x. [DOI] [PubMed] [Google Scholar]
- 60.Clermont A., Wedde M., Seitz V., Podsiadlowski L., Lenze D., Hummel M., et al. Cloning and expression of an inhibitor of microbial metalloproteinases from insects contributing to innate immunity. Biochem J. 2004;382(1):315–322. doi: 10.1042/BJ20031923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wedde M., Weise C., Nuck R., Altincicek B., Vilcinskas A. The insect metalloproteinase inhibitor gene of the lepidopteran Galleria mellonella encodes two distinct inhibitors. Biol Chem. 2007;388:119–127. doi: 10.1515/BC.2007.013. [DOI] [PubMed] [Google Scholar]
- 62.Arolas J.L., Botelho T.O., Vilcinskas A., Gomis-Rüth F.X. Structural evidence for standard-mechanism inhibition in metallopeptidases from a complex poised to resynthesize a peptide bond. Angew Chem Int Ed Engl. 2011;50(44):10357–10360. doi: 10.1002/anie.201103262. [DOI] [PubMed] [Google Scholar]
- 63.Eisenhardt M., Schlupp P., Höfer F., Schmidts T., Hoffmann D., Czermak P., et al. The therapeutic potential of the insect metalloproteinase inhibitor against infections caused by Pseudomonas aeruginosa. J Pharm Pharmacol. 2019;71(3):316–328. doi: 10.1111/jphp.13034. [DOI] [PubMed] [Google Scholar]
- 64.Eisenhardt M., Dobler D., Schlupp P., Schmidts T., Salzig M., Vilcinskas A., et al. Development of an insect metalloproteinase inhibitor drug carrier system for application in chronic wound infections. J Pharm Pharmacol. 2015;67(11):1481–1491. doi: 10.1111/jphp.12452. [DOI] [PubMed] [Google Scholar]
- 65.Block H., Maertens B., Spriestersbach A., Brinker N., Kubicek J., et al. Immobilized-metal affinity chromatography (IMAC): a review. Methods Enzymol. 2009;463:439–473. doi: 10.1016/S0076-6879(09)63027-5. [DOI] [PubMed] [Google Scholar]
- 66.Sabat A.J., Wladyka B., Kosowska-Shick K., Grundmann H., van Dijl J.M., Kowal J., et al. Polymorphism, genetic exchange and intragenic recombination of the aureolysin gene among Staphylococcus aureus strains. BMC Microbiol. 2008;8(1) doi: 10.1186/1471-2180-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tallant C., García-Castellanos R., Seco J., Baumann U., Gomis-Rüth F.X. Molecular analysis of ulilysin, the structural prototype of a new family of metzincin metalloproteases. J Biol Chem. 2006;281(26):17920–17928. doi: 10.1074/jbc.M600907200. [DOI] [PubMed] [Google Scholar]
- 68.Huesgen P.F., Lange P.F., Rogers L.D., Solis N., Eckhard U., Kleifeld O., et al. LysargiNase mirrors trypsin for protein C-terminal and methylation-site identification. Nat Methods. 2015;12(1):55–58. doi: 10.1038/nmeth.3177. [DOI] [PubMed] [Google Scholar]
- 69.Motulsky H., Christopoulos A. Oxford University Press; New York: 2004. Fitting models to biological data using linear and nonlinear regression. A practical guide to curve fitting; p. 352. [Google Scholar]
- 70.Kabsch W. XDS. Acta Crystallogr sect D. 2010;66(2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr sect D. 2010;66(2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr sect D. 2011;67(4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J Appl Crystallogr. 2007;40(4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr sect D. 2019;75(10):861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smart O.S., Womack T.O., Flensburg C., Keller P., Paciorek W., Sharff A., et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr sect D. 2012;68(4):368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zwart P.H., Grosse-Kunstleve R.W., Adams P.D. In: CCP4 Newsletter on Protein Crystallography. Remacle F., editor. Daresbury Laboratory; Daresburt, Warrington (UK): 2005. Xtriage and Fest: automatic assessment of X-ray data and substructure structure factor estimation; pp. 27–35. [Google Scholar]
- 77.Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr sect D. 2011;67(4):282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Casañal A., Lohkamp B., Emsley P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 2020;29(4):1055–1064. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kovalevskiy O., Nicholls R.A., Long F., Carlon A., Murshudov G.N. Overview of refinement procedures within REFMAC5: utilizing data from different sources. Acta Crystallogr sect D. 2018;74(3):215–227. doi: 10.1107/S2059798318000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr sect D. 2004;60(12):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 81.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27(1):14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 83.McKerrow J.H. Human fibroblast collagenase contains an amino acid sequence homologous to the zinc-binding site of Serratia protease. J Biol Chem. 1987;262(13):5943. doi: 10.1016/S0021-9258(18)45517-2. [DOI] [PubMed] [Google Scholar]
- 84.Bode W., Gomis-Rüth F.X., Stöcker W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ´metzincinś. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- 85.Matthews B.W. Structural basis of the action of thermolysin and related zinc peptidases. Acc Chem Res. 1988;21(9):333–340. [Google Scholar]
- 86.Wasylewski Z., Stryjewski W., Waśniowska A., Potempa J., Baran K. Effect of calcium binding on conformational changes of staphylococcal metalloproteinase measured by means of intrinsic protein fluorescence. Biochim Biophys Acta. 1986;871(2):177–181. doi: 10.1016/0167-4838(86)90171-8. [DOI] [PubMed] [Google Scholar]
- 87.Hausrath A.C., Matthews B.W. Thermolysin in the absence of substrate has an open conformation. Acta Crystallogr sect D. 2002;58(6):1002–1007. doi: 10.1107/s090744490200584x. [DOI] [PubMed] [Google Scholar]
- 88.Laskowski M., Kato I. Protein inhibitors of proteinases. Annu Rev Biochem. 1980;49(1):593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 89.Bode W., Huber R. Natural protein proteinase inhibitors and their interaction with proteinases. Eur J Biochem. 1992;204(2):433–451. doi: 10.1111/j.1432-1033.1992.tb16654.x. [DOI] [PubMed] [Google Scholar]
- 90.Ascenzi P., Bocedi A., Bolognesi M., Spallarossa A., Coletta M., et al. The bovine basic pancreatic trypsin inhibitor (Kunitz inhibitor): a milestone protein. Curr Protein Pept Sci. 2003;4:231–251. doi: 10.2174/1389203033487180. [DOI] [PubMed] [Google Scholar]
- 91.Laskowski M., Qasim M.A. What can the structures of enzyme-inhibitor complexes tell us about the structures of enzyme substrate complexes? Biochim Biophys Acta. 2000;1477(1-2):324–337. doi: 10.1016/s0167-4838(99)00284-8. [DOI] [PubMed] [Google Scholar]
- 92.Bürgi H.B., Dunitz J.D., Shefter E. Geometrical reaction coordinate. II. Nucleophilic addition to a carbonyl group. J Am Chem Soc. 1973;95:5065–5067. [Google Scholar]
- 93.Cer R.Z., Mudunuri U., Stephens R., Lebeda F.J. IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009;37(Web Server):W441–W445. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.García-Castellanos R., Marrero A., Mallorquí-Fernández G., Potempa J., Coll M., Gomis-Rüth F.X. Three-dimensional structure of MecI : Molecular basis for transcriptional regulation of staphylococcal methicillin resistance. J Biol Chem. 2003;278(41):39897–39905. doi: 10.1074/jbc.M307199200. [DOI] [PubMed] [Google Scholar]
- 95.Weiss M.S. Global indicators of X-ray quality. J Appl Cryst. 2001;34:130–135. [Google Scholar]
- 96.Karplus P.A., Diederichs K. Linking crystallographic model and data quality. Science. 2012;336(6084):1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.