Abstract
The mechanisms responsible for macrolide resistance in Streptococcus pneumoniae mutants, selected from susceptible strains by serial passage in azithromycin, were investigated. These mutants were resistant to 14- and 15-membered macrolides, but resistance could not be explained by any clinically relevant resistance determinant [mef(A), erm(A), erm(B), erm(C), erm(TR), msr(A), mph(A), mph(B), mph(C), ere(A), ere(B)]. An investigation into the sequences of 23S rRNAs in the mutant and parental strains revealed individual changes of C2611A, C2611G, A2058G, and A2059G (Escherichia coli numbering) in four mutants. Mutations at these residues in domain V of 23S rRNA have been noted to confer erythromycin resistance in other species. Not all four 23S rRNA alleles have to contain the mutation to confer resistance. Some of the mutations also confer coresistance to streptogramin B (C2611A, C2611G, and A2058G), 16-membered macrolides (all changes), and clindamycin (A2058G and A2059G). Interestingly, none of these mutations confer high-level resistance to telithromycin (HMR-3647). Further, two of the mutants which had no changes in their 23S rRNA sequences had changes in a highly conserved stretch of amino acids (63KPWRQKGTGRAR74) in ribosomal protein L4. One mutant contained a single amino acid change (G69C), while the other mutant had a 6-base insert, resulting in two amino acids (S and Q) being inserted between amino acids Q67 and K68. To our knowledge, this is the first description of mutations in 23S rRNA genes or ribosomal proteins in macrolide-resistant S. pneumoniae strains.
The predominant forms of macrolide resistance in Streptococcus pneumoniae are mediated by mef(A), a gene encoding an efflux pump in the major facilitator superfamily, or by erm(B), an rRNA methylase. [Note that the mef(A) and mef(E) genes, originally named for the macrolide efflux determinants in Streptococcus pyogenes (6) and S. pneumoniae (44), respectively, have been classified into one group, mef(A) (32).] The prevalence of macrolide resistance varies geographically, being high in Japan (73%), Hong Kong (81.5%), France (47%), Italy (42%), and the United States (19 to 34%) (9; M. R. Jacobs, D. Felminghan, P. C. Appelbaum, and T. A. P. Group, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1044, 1999). In some countries, mef(A) predominates, while in others, erm(B) is the major resistance determinant (7, 19). Although the levels of prevalence of resistance to clarithromycin, erythromycin, and azithromycin are sometimes reported to be slightly different in surveillance studies, isolates containing either mef(A) or erm(B) should be regarded as coresistant to erythromycin, clarithromycin, and azithromycin regardless of the absolute MICs of these compounds (42; L. Brennan, J. Duignan, J. Petitpas, M. Anderson, W. Fu, J. Retsema, J. Rainville, D. Smyth, W. Su, and J. Sutcliffe, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-124, 1998). However, isolates containing mef(A) are susceptible to clindamycin and 16-membered macrolides while those containing erm(B) are resistant to these agents. Virtually all clinical isolates ofmacrolide-resistant S. pneumoniae that have been examined for macrolide resistance mechanisms have contained either mef(A) or erm(B), and occasional strains have contained both genes (7).
In a previous study we described the selection of azithromycin-resistant mutants from several macrolide-susceptible clinical strains of S. pneumoniae containing neither mef(A) nor erm(B) (29). Examination of these passage-derived mutants by PCR for mef(A) and erm(B) sequences showed that neither determinant accounted for the resistance. In this study we investigated the resistance mechanisms of these mutants by testing for the presence of other known macrolide resistance determinants, including three genes that encode macrolide phosphorylases (22, 26, 27; J. Cheng, T. Grebe, L. Wondrack, P. Courvalin, and J. Sutcliffe, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 837, 1999; K. O'Hara, T. Kawabe, K. Taniguchi, A. Nakamura, and T. Sawai, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-67, 1997), two genes that encode macrolide esterases, erm(TR) [now reclassified as erm(A) (32)], a newly described methylase found in Streptococcus pyogenes (38), and msr(A), which encodes an ABC-type transporter (36). Failing to find genes previously characterized to confer macrolide resistance in these mutants, we reasoned that mutations in the 23S rRNA alleles and/or in the ribosomal proteins L4 and L22 might account for resistance. A mutation in the ribosome seemed likely based on the precedents that rRNA mutations do exist in other species of bacteria (17, 18, 23, 24, 34, 35, 41, 45, 48, 49) and that mutations in L4 or L22 exist in erythromycin-resistant laboratory-derived mutants of Escherichia coli and Bacillus spp. (5, 30, 37, 39, 46, 50, 51).
MATERIALS AND METHODS
Bacterial strains.
The clinical isolates of S. pneumoniae used in the passage study and the mutants derived from them have been described previously (29). Briefly, the mutants were identified after they were subcultured 50 times in Mueller-Hinton broth (Difco) plus 5% lysed horse blood containing doubling dilutions of azithromycin. For each subsequent passage, an inoculum was taken from the tube with the MIC nearest in opacity to that of the antibiotic-free control. When the MIC increased fourfold, strains were subcultured in antibiotic-free medium for 10 serial passages. Strains for which MICs showed ≥4-fold increases were determined to be of the same clonal type as that of the parental strains by pulsed-field gel electrophoresis. S. pneumoniae R6 was used as a recipient in transformation experiments, and strain A9, a spontaneous streptomycin-resistant derivative of R6, served as a control (3).
MIC determinations.
MICs were determined with microtiter trays using Mueller-Hinton broth supplemented with 2.5% lysed horse blood according to the recommendations of the National Committee for Clinical Laboratory Standards (NCCLS) (25). All compounds were purchased from Sigma or made by published methods at Pfizer, Inc.
PCR primers and DNA sequencing.
Primers were purchased from Sigma/Genosys Biotechnologies (The Woodlands, Tex.). Primers for erm(A), erm(B), erm(C), msr(A), mef(A), mph(A), mph(B), ere(A), and ere(B) have been described previously (43). Primers for erm(TR) have been designed for S. pyogenes and were used in this study (38). Primers for mph(C) are based on the sequence of a putative macrolide phosphorylase from Staphylococcus aureus clinical strains (22; Cheng et al., 39th ICAAC). The primers used to detect erm(TR) and mph(C) are listed in Table 1. Preliminary sequence data for 23S rRNA and for L4 and L22 ribosomal proteins in S. pneumoniae and E. coli were obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org). Primers were designed from these sequences and are listed in Table 1. All other L4 and L22 sequences were obtained courtesy of Incyte Pharmaceuticals (unpublished data, 1999). To locate mutations in 23S rRNAs, the genes were initially amplified from total genomic DNA as three overlapping contigs. Unique primers downstream of each 23S rRNA gene were designed from sequences beyond the 3′ end of 23S rRNA. The use of these primers enabled the peptidyl transferase region from each allele of 23S rRNA to be separately amplified and purified for DNA sequencing. For each primer pair, the concentration of MgCl2 was optimized and is listed in Table 1. All PCRs were subjected to 35 cycles of 94°C (1 min), either 54 or 44°C (1 min), 72°C (1 min), and then 72°C (10 min) for elongation. PCR products were purified using a QIAquick PCR purification kit (Qiagen Inc., Valencia, Calif.). Sequencing of purified DNA was performed on an ABI 373XL automated sequencing apparatus with stretch upgrade (PE Biosystems, Foster City, Calif.). Cycle sequencing using the BigDye Terminator TaqFS (BDT) chemistry (PE Biosystems) was performed according to the manufacturer's protocol with the following modifications: half-reaction BDT reaction mixtures (50% BDT, 50% ABI 5× buffer [PE Biosystems]) contained 5% dimethyl sulfoxide (Fisher Scientific, Fair Lawn, N.J.), and the cycle sequencing profile with a hot start was 95°C for 1 min (1 cycle); 98°C for 45 s, 50°C for 10 s, and 60°C for 4 min (1 cycle); and 98°C for 15 s, 50°C for 10 s, and 60°C for 4 min (29 cycles) on a PTC-225 DNA Engine Tetrad Thermal Cycler (MJ Research, Watertown, Mass.). Sequencing reaction mixtures were purified by following the manufacturer's protocol over 96-well gel filtration blocks (Edge BioSystems, Gaithersburg, Md.) and electrophoresed for 18 h at 2,500 V, 40 mA, on polyacrylamide gels made of 5.75% PAGE-Plus acrylamide (Amresco, Solon, Ohio)–6 M urea (Boehringer Mannheim, Indianapolis, Ind.) in 1× Tris-borate-EDTA (Roche Diagnostics Corp., Indianapolis, Ind.), with a 4% mobility file and ABI 100 base calling (PE Biosystems). Clones were assembled and edited using the ABI Factura and AutoAssembler programs (PE Biosystems). Sequence comparisons were carried out using the Lasergene sequence analysis software (DNASTAR, Inc., Madison, Wis.).
TABLE 1.
Primer descriptions and MgCl2 concentrations used in this study
| PCR primer pair | Nucleotide sequence
|
Product size (bp) | Optimum MgCl2 concn (mM) | Base start | Base end | |
|---|---|---|---|---|---|---|
| 5′ | 3′ | |||||
| 23S rRNA primers | ||||||
| 23S 5′ | 5′-GGTTAAGTTAATAAGGGCGC-3′ | 5′-TTTCGACTACGGATCTTAGC-3′ | 1,011 | 4 | 1 | 1011 |
| 23S center | 5′-CTGTTTGGGTGAGGGGTCC-3′ | 5′-ACCTGCATCTTCACAGGTAC-3′ | 1,060 | 1 | 900 | 2041 |
| 23S 3′ | 5′-CGGCGGCCGTAACTATAACG-3′ | 5′-TTGGATAAGTCCTCGAGCTATTAG-3′ | 1,003 | 1 | 1899 | 2902 |
| Unique 23S primers | ||||||
| DS_18 | 5′-CGGCGGCCGTAACTATAACG-3′ | 5′-GCCAGCTGAGCTACACCGCC-3′ | 1,900 | 2 | 1899 | +897a |
| DS_23 | 5′-CGGCGGCCGTAACTATAACG-3′ | 5′-TACACACTCACATATCTCTG-3′ | 1,200 | 2 | 1899 | +197a |
| DS_30 | 5′-CGGCGGCCGTAACTATAACG-3′ | 5′-TTTTACCACTAAACTACACC-3′ | 1,300 | 2 | 1899 | +297a |
| DS_91 | 5′-CGGCGGCCGTAACTATAACG-3′ | 5′-TACCAACTGAGCTATGGCGG-3′ | 1,900 | 3 | 1899 | +897a |
| Ribosomal protein primers | ||||||
| L4 | 5′-AAATCAGCAGTTAAAGCTGG-3′ | 5′-GAGCTTTCAGTGATGACAGG-3′ | 720 | 2 | −50b | 670b |
| L22 | 5′-GCAGACGACAAGAAAACACG-3′ | 5′-ATTGGATGTACTTTTTGACC-3′ | 420 | 2 | −41b | 380b |
| Erythromycin resistance primers | ||||||
| mph(C) | 5′-GGGAAATTGAACACAAACC-3′ | 5′-AATTCATCTGATACRCCATAAG-3′c | 500 | 3 | 1b | 520b |
| erm(TR) | 5′-ACAGAAAAACCCGAAAAATACG-3′ | 5′-TTGGATAATTTATCAAGATCAG-3′ | 679 | 1 | 5b | 731b |
Base relative to 3′ end of 23S rRNA.
Base relative to ATG.
R = A + G.
Southern hybridization.
S. pneumoniae genomic DNA was prepared as described previously (44). DNA was digested with restriction endonucleases from New England Biolabs (Beverly, Mass.) according to manufacturer's directions, and fragments were resolved by gel electrophoresis. DNA fragments were transferred to a Magnagraph nylon membrane (Micron Separations Inc., Westboro, Mass.). The 23S ribosomal DNA probe was a PCR product derived with the primer pair encompassing the 3′ end of the 23S rRNA gene (1,003 bp) (Table 1). The probe was purified, labeled, and detected by chemiluminescence with a digoxigenin DNA random prime labeling and detection kit (Boehringer Mannheim) according to manufacturer's directions. Hybridization was carried out at 60°C.
Transformation.
Synthetic competence-stimulating peptide I and the method of Havarstein et al. (15) were used to induce S. pneumoniae into a transformation-competent state. Briefly, cells were inoculated at a very low density (optical density at 550 nm [OD550], ∼0.005) and grown to early log phase (OD550, ∼0.02) in Todd-Hewitt medium plus 0.5% yeast extract. The cells were triggered to the competence state by the addition of NaOH (0.006 N final concentration), bovine serum albumin (0.2% final concentration), CaCl2 (1 mM final concentration), and synthetic competence-stimulating peptide I (0.2-μg/ml final concentration). After 15 min, DNA was added at a final concentration of 1 μg/ml to aliquots of cells and the mixture was incubated for an hour at 30°C. Cells with DNA were diluted fourfold and further incubated for 2 h at 37°C. They were then plated on Todd-Hewitt agar containing 5% sheep blood and appropriate antibiotics (0.3 to 0.5 μg of azithromycin per ml or 100 μg of streptomycin per ml) and incubated overnight at 37°C in 5% CO2. Cell suspensions were monitored for competence by determining the frequency of transformation of the streptomycin resistance gene from S. pneumoniae A9, a streptomycin-resistant mutant spontaneously derived from R6. Transformation frequencies for introduction of streptomycin resistance ranged from 0.1 to 0.5%.
RESULTS
Passaged mutants do not contain known erythromycin resistance determinants.
Genomic DNAs from all of the resistant isolates and parental strains were isolated and subjected to PCR analysis using primers specific for macrolide esterases [ere(A) and ere(B)], phosphotransferases [mph(A), mph(B), and mph(C)], an ABC-binding transporter [msr(A)], and rRNA methylases [erm(TR), erm(A), and erm(C)] (38, 43; Cheng et al., 39th ICAAC). None of the isolates had a PCR product specific to any of the known resistance determinants described for enterics, staphylococci, or streptococci (data not shown). These results, in conjunction with our previous data [the absence of mef(A) and erm(B)], suggested that these isolates contained a novel mechanism(s) of resistance.
Copy number determination of 23S rRNA.
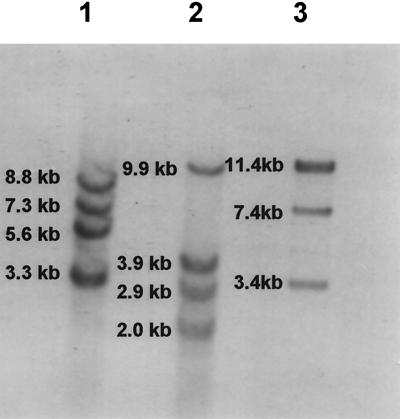
Since the resistant isolates were negative for all known macrolide resistance determinants, we reasoned that the isolates could have mutations at the level of the ribosome. Alteration in 23S rRNA seemed a possibility, and we initially sought to determine the number of 23S rRNA alleles present in S. pneumoniae. The copy number of 23S rRNA was uncertain, as there had been reports that pneumococcal strains contained either six (10; P. Matsushima and R. H. Baltz, Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996, abstr. H-63, 1996) or four (4) copies of 23S rRNA. Search of the TIGR database for S. pneumoniae with the published 23S rRNA sequence from staphylococci yielded six contigs: a single contig (spn_23) with sequence beyond the expected 5′ and 3′ ends of a canonical rRNA operon (16S-23S-5S rRNA), three copies with homology to a portion of the 23S rRNA sequence but each possessing unique 3′ ends, and two contigs with sequence homology to internal regions of 23S rRNA (http://www.tigr.org). The last two, being internal, were not considered to be additional individual copies. To determine the number of alleles of 23S rRNA, genomic DNA was restricted with 18 enzymes; digestion with ApaI or ClaI revealed four bands, while the majority of enzymes gave less than four bands following digestion and Southern blot analysis with a probe specific for 23S rRNA (data not shown). Since some of the fragment sizes were large, we employed a double-restriction enzyme strategy to help distinguish if multiple alleles were present on one fragment. I-CeuI from New England Biolabs is an intron-encoded protein from the chloroplast large rRNA gene of Chlamydomonas eugametos (20). This enzyme has a 26-bp recognition sequence that is unique and conserved only within 23S ribosomal genes. Restriction with I-CeuI was coupled with EcoRI, which cuts outside of 23S rRNA. After enzyme digestion with I-CeuI and EcoRI, Southern blot analysis with a probe specific for 23S rRNA revealed four bands in each of three different isolates of S. pneumoniae (Fig. 1). Four 23S-rRNA-specific bands were observed at 8.8, 7.3, 5.6, and 3.3 kb in isolate 6T0, and bands of 9.9, 3.9, 2.9, and 2.0 kb were observed in isolate 8T0; a doublet band at 11.4 kb, with singlet fragments of 7.4 and 3.4 kb, were seen in R6. These results are consistent with the presence of four copies of 23S rRNA genes in S. pneumoniae.
FIG. 1.
Determination of the number of 23S rRNA genes in pneumococci. Genomic DNAs from three strains of pneumococci were digested with I-CeuI and EcoRI. Lane 1, strain 6T0; lane 2, strain 8T0; lane 3, strain R6.
Analysis of 23S rRNA and L4 and L22 sequences in passaged mutants.
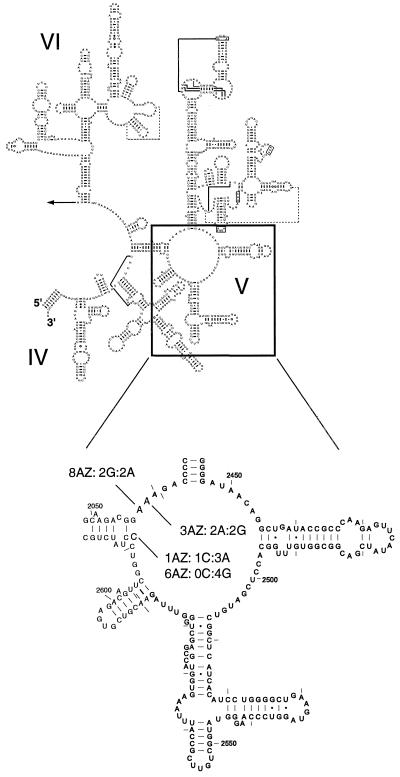
DNAs from parental strains and macrolide-resistant passaged isolates were amplified using three pairs of primers specific to overlapping regions of 23S rRNA in pneumococci (Table 1). Although there was some strain-to-strain variation in 23S rRNA sequences among susceptible isolates, the only change between susceptible and resistant pairs was seen in the peptidyl transferase region within domain V (E. coli nucleotides 2042 to 2628) of the 23S rRNA. Four of the six mutants showed changes in this region (Fig. 2). Two resistant strains, 3Az and 8Az, showed changes in A2059G and A2058G, respectively (Table 2). Resistance to 14-membered (erythromycin, clarithromycin), 15-membered (azithromycin), and 16-membered (spiramycin) macrolides was noted for both of these mutants, and resistance to lincosamides (lincomycin and clindamycin) was also noted for strain 8Az, according to NCCLS guidelines (where available). For strain 3Az, the MIC of streptogramin B was not increased and, although there was decreased susceptibility to the ketolide telithromycin, the MIC of the ketolide remained low. Conversion of A2058 to guanine in 8Az provided a significant increase in the MICs of 14- and 15-membered macrolides and more modest increases in the MICs of spiramycin, lincosamides, streptogramin B, and telithromycin. C2611 in two other strains, 1Az and 6Az, was changed to adenine and guanine, respectively. Changes to adenine in 1Az conferred low-level resistance to the 14-, 15-, and 16-membered macrolides, with slight and large increases in the MICs of telithromycin and streptogramin B, respectively. The C2611G change in 6Az resulted in notable resistance to 14- and 15-membered macrolides and streptogramin B but less resistance to lincosamides and spiramycin. Further, there was a 130-fold increase in the MIC of telithromycin.
FIG. 2.
Secondary structures of domains IV to VI of E. coli 23S rRNA and mutations in azithromycin-passaged mutants. The locations and heterozygous nature of the mutations in peptidyl transferase region of 23S rRNA (domain V) are shown for the four mutants. The two-dimensional sequence is courtesy of Gutell et al. (R. R. Gutell, S. Subashchandran, M. Schnare, Y. Du, N. Lin, L. Madabusi, K. Muller, N. Pande, N. Yu, Z. Shang, S. Date, D. Konings, V. Schweiker, B. Weiser, and J. J. Cannone, http://www.rna.icmb.utexas.edu).
TABLE 2.
Phenotypic analysis of passage-derived strains with mutations in 23S rRNA
| Straina | MIC (μg/ml) ofb:
|
Mutation | Heterozygosity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AZI | ERY | CLAR | SPIR | LINC | CLIN | STREP B | TELI | PEN | |||
| Parent 1T0 | 0.20 | 0.05 | 0.02 | 0.39 | 0.20 | 0.05 | 1.56 | 0.006 | 0.01 | None | 4C:0A |
| Mutant 1Az | 3.12 | 0.78 | 0.78 | 1.56 | 0.39 | 0.20 | 50 | 0.01 | 0.02 | C2611A | 1C:3A |
| Parent 6T0 | 0.10 | 0.05 | 0.02 | 0.10 | 1.56 | 0.05 | 0.78 | 0.003 | 0.01 | None | 4C:0G |
| Mutant 6Az | 12.5 | >100 | 100 | 0.78 | 6.25 | 0.20 | >100 | 0.39 | 0.01 | C2611G | 0C:4G |
| Parent 3T0 | 0.02 | 0.02 | 0.01 | 0.10 | 0.20 | 0.02 | 1.56 | 0.003 | 0.02 | None | 4A:0G |
| Mutant 3Az | >200 | 12.5 | 3.12 | >100 | 1.56 | 0.20 | 1.56 | 0.01 | 0.02 | A2059G | 2A:2G |
| Parent 8T0 | 0.10 | 0.05 | 0.05 | 0.39 | 0.20 | 0.10 | 3.12 | 0.01 | 0.05 | None | 4A:0G |
| Mutant 8Az | >200 | >100 | 100 | 6.25 | 25 | 3.12 | 12.5 | 0.10 | 0.05 | A2058G | 2A:2G |
| ATCC 49619 | 0.12 | 0.030 | 0.03 | 0.20 | 0.39 | ≤0.20 | 3.12 | 0.01 | 0.39 | NDc | ND |
Strains designated nT0 are the parental, macrolide-susceptible strains; strains designated as nAz are the mutants that were characterized following passage in azithromycin (29). ATCC 49619 is the control strain suggested by the NCCLS (25).
AZI, azithromycin; ERY, erythromycin; CLAR, clarithromycin; SPIR, spiramycin; LINC, lincomycin; CLIN, clindamycin; STREP B, streptogramin B; TELI, telithromycin (HMR-3647); PEN, penicillin G. The 14-membered macrolides used are erythromycin and clarithromycin, the 15-membered macrolide is azithromycin, the 16-membered macrolide is spiramycin, the lincosamides are lincomycin and clindamycin, and the ketolide is telithromycin.
ND, not done.
Examination of the sequencing traces covering domain V showed that some of the mutants (1Az, 3Az, and 8Az) had a mixture of bases at the altered residue. Using primers designed to amplify each of the four 23S rRNA genes (Table 1), we confirmed that the nature of the mutations was heterozygous (Fig. 2 and Table 2). 3Az and 8Az contained two alleles of A2059G and A2058G, respectively. 1Az had three alleles of C2611A. 6Az was confirmed to be homozygous, with all four 23S rRNA alleles containing C2611G. We also examined the mutants for any changes in the ribosomal proteins L4 and L22 based on the observation that laboratory strains of E. coli or Bacillus selected for resistance to erythromycin had changes in the sequence of either L4 or L22. In all four mutants, both the L4 and L22 sequences were wild type, having the same sequence that resides in the TIGR database (http://www.tigr.org) for S. pneumoniae type 4.
Two of the mutants, 4Az and 5Az, had no changes in their 23S rRNA sequences. However, when these two strains were examined for changes in protein L4 or L22, both mutants were discovered to contain mutations in L4. These changes were in a highly conserved region of the protein (32 amino acids), where there are zero to three amino acid changes among gram-positive pathogens and five to seven amino acid differences from those of gram-negative pathogens (Table 3). 4Az differed from its sensitive parent by 2 bases (G211T and A213C, S. pneumoniae numbering), leading to a single amino acid change from glycine to cysteine at position 69. 5Az had a 6-bp in-frame insertion, leading to the addition of 2 amino acids, serine and glutamine, between amino acids glutamine67 and lysine68 (Table 4). In general, the MICs of 14-, 15-, and 16-membered macrolides and streptogramin B were increased ≥4-fold, but the MICs of the ketolide telithromycin and the lincosamides were increased not more than 2-fold (Table 4). In some cases, the increase in resistance to 14- and 15-membered macrolides would not be sufficient to reveal these isolates as macrolide resistant, according to NCCLS guidelines (25).
TABLE 3.
Conservation of L4 sequences among gram-positive and gram-negative bacteriaa
| Organism | Position of beginning residue | Amino acid sequence | Position of ending residue |
|---|---|---|---|
| Streptococcus pneumoniae | 59 | G G G R K P W R Q K G T G R A R Q G S I R S P Q W R G G G V V F | 90 |
| Enterococcus faecalis | 59 | G G G R K P W R Q K G T G R A R Q G S I R S P Q W R G G G V V F | 90 |
| Streptococcus pyogenes | 59 | G G G R K P W R Q K G T G R A R Q G S I R S P Q W R G G G V V F | 90 |
| Staphylococcus aureus | 59 | G G G R K P W K Q K G T G R A R Q G T I R A P Q W R G G G I V F | 90 |
| Staphylococcus epidermidis | 50 | G G G R K P W R Q K G T G R A R Q G T I R A P Q W R G G G V V F | 81 |
| Streptococcus haemolyticus | 50 | G G G R K P W R Q K G T G R A R Q G T I R A P Q W R G G G I V F | 81 |
| Escherichia coli | 54 | G S G K K P W R Q K G T G R A R S G S I K S P I W R S G G V T F | 85 |
| Enterobacter cloacae | 54 | G S G K K P W R Q K G T G R A R S G S I K S P I W R S G G V T F | 85 |
| Klebsiella pneumoniae | 54 | G S G K K P W R Q K G T G R A R S G S I K S P I W R S G G V T F | 85 |
| Pseudomonas aeruginosa | 54 | G G G K K P W R Q K G T G R A R A G T I R S P I W R S G G T T F | 85 |
| Morganella morganii | 54 | G S G K K P W R Q K G T G R A R S G S I K S P I W R S G G I T F | 85 |
| Proteus mirabilis | 54 | G S G K K P W R Q K G T G R A R S G S I K S P I W R S G G G T F | 85 |
| Yersinia pestis | 54 | G S G K K P W R Q K G T G R A R A G S V K S P I W R S G G V T F | 85 |
Numbers at either end of the sequences refer to the amino acid positions of L4 in the individual species. The source of the S. pneumoniae and E. coli sequences were the TIGR website; the remainder of the sequences were courtesy of Incyte Pharmaceuticals. Boldface letters indicate residues that differ from the S. pneumoniae L4 sequence.
TABLE 4.
Phenotypic analysis of passage-derived strains with mutations in ribosomal protein L4
| Straina | MIC (μg/ml) of b:
|
Mutation | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ERY | CLAR | AZI | JOSA | CLIN | STREP B | TELI | PEN | ||
| 4T0 | 0.02 | 0.01 | 0.05 | 0.20 | 0.10 | 0.78 | 0.003 | 0.01 | 63KPWRQKGTGRAR74 |
| 4Az | 0.05 | 0.05 | 0.39 | 1.56 | 0.20 | 3.12 | 0.006 | 0.02 | 63KPWRQKCTGRAR74 |
| 5T0 | 0.05 | 0.02 | 0.10 | 0.20 | 0.05 | 0.78 | 0.006 | 0.02 | 63KPWRQKGTGRAR74 |
| 5Az | 0.20 | 0.20 | 1.56 | 0.78 | 0.05 | 3.12 | 0.01 | 0.02 | 63KPWRQSQKGTGRAR74 |
Strains designated nT0 are the parental, macrolide-susceptible strains; strains designated nAz are the mutants that were characterized following passage in azithromycin (29).
AZI, azithromycin; ERY, erythromycin; CLAR, clarithromycin; JOSA, josamycin; CLIN, clindamycin; STREP B, streptogramin B; TELI, telithromycin (HMR-3647); PEN, penicillin G. The 14-membered macrolides used were erythromycin and clarithromycin, the 15-membered macrolide was azithromycin, the 16-membered macrolide was josamycin, the lincosamide was clindamycin, and the ketolide was telithromycin.
Effects of mutations on growth physiology.
Mutations in ribosomal proteins and alterations to 23S rRNA have been shown to alter growth rate and to confer temperature sensitivity to growth (5, 30, 39, 46, 50). To determine if any of the mutants were temperature sensitive, each mutant was evaluated for the ability to form colonies on four media (brain heart infusion, Todd-Hewitt, Trypticase soy, and Mueller-Hinton) supplemented with 5% sheep red blood cells at three different temperatures (25, 35, and 42°C) (data not shown). The mutants grew well on each medium at all temperatures with the exception of mutant 5Az. This mutant did not form colonies on any of the media at 25°C. Further, neither 5Az nor its parent, 5T0, were able to sustain growth at 42°C on any medium. As a confirmation of the phenotype, 5Az and 5T0 were grown in Todd-Hewitt medium containing 0.5% yeast extract at 35°C for ∼1.5 generations and then shifted to either 25 or 42°C. Growth was monitored every hour by measuring the number of CFU on Todd-Hewitt broth–blood agar plates at 35°C. Both 5T0 and 5Az stopped increasing in cell number upon the temperature shift to 25°C, and the cell number remained constant for 8 h. However, parent 5T0 was capable of forming colonies on plates incubated at 25°C whereas 5Az was not. Upon being shifted to 42°C, both strains lost viability. 5Az had a longer doubling time (110 min versus 60 min) at 35°C than that of its isogenic parent. Thus, it appears that the insertion mutation (SQ) in 5Az resulted in a protein that impacted the growth rate at 35°C and viability at 25°C.
Transformation of L4 mutations into R6.
To determine if the mutations in L4 were sufficient to confer a macrolide resistance phenotype, the sequences for L4 from strains 4Az and 5Az were amplified. Purified PCR products were introduced into R6 by transformation, and azithromycin-resistant transformants were selected on Todd-Hewitt broth–blood agar containing 0.5 μg of azithromycin per ml. Transformants were selected, streaked onto selective medium, and phenotyped. The L4 sequences from MS transformants were amplified, and the PCR products were purified and sequenced. All transformants had the same MS phenotypes and mutations in L4 as their parental version, 4Az or 5Az (Table 4).
DISCUSSION
The S. pneumoniae parental strains and the mutants obtained by exposure to azithromycin did not contain any known mechanism of resistance that has been described for clinical strains, namely, rRNA methylases, efflux genes, esterases, or phosphorylases. Since these resistance determinants are generally acquired, this finding is not totally unexpected. Mutations in 23S rRNA conferring erythromycin resistance have been described for clinical strains of Mycobacterium avium (24), Mycobacterium intracellulare (23), Mycobacterium chelonae (49), Mycobacterium abscessus (49), Brachyspira (Serpulina) hyodysenteriae (18), Helicobacter pylori (17, 41, 45, 48), and Proprionibacterium spp. (34, 35). These mutations have been mapped to the peptidyl transferase region, specifically, G2057, A2058, or A2059 in the E. coli numbering scheme. Changes of A2058G have also been seen in chloroplasts from either Chlamydomonas reinhardtii or Nicotiana plumbaginafolia, mitochondria from Saccharomyces cerevisiae, and laboratory mutants of E. coli (50) and Mycoplasma pneumoniae (21). In addition, the A2058G mutation was selected in Streptomyces ambofaciens, a spiramycin producer, by another 16-membered macrolide, chalcomycin (31). Methylation at A2058 by one of many erythromycin methylases is a major mechanism of resistance that is widespread in many bacterial species, including macrolide producer strains (32, 50). The A2059G transition in N. plumbaginafolia chloroplast rRNA has been described previously (50). Four of the six passage-derived strains in our study contain mutations in A2058, A2059, and C2611, a residue that pairs with G2507 in the secondary structure of 23S rRNA (13). The mutations C2611U and -G have been described for laboratory-derived mutants of E. coli, as chloroplast mutations in Chlamydomonas moewusei or C. reinhardtii, and as mitochondrial mutations in S. cerevisiae (50). The conversion of C2611A is described for the first time for the S. pneumoniae mutant 1Az.
To determine if one or more alleles encoding 23S rRNA contained a mutation conferring macrolide resistance, we first had to ascertain how many 23S rRNA operons S. pneumoniae contained. The literature was confusing, with one reference reporting results consistent with four copies of 23S rRNA (4) but with others claiming six alleles (10; Matsushima and Baltz, Abstr. 96th Gen. Meet. Am. Soc. Microbiol., 1996). We reexamined the number of 23S rRNA alleles and found in three different isolates that four copies of 23S rRNA were present. Other clinical species with 23S rRNA mutations have either one, two, or three copies of the 23S rRNA operon. For those species containing either one copy (Mycobacterium, Brachyspira) or two copies (Helicobacter, Mycoplasma pneumoniae), a change in one of the copies has been shown to be sufficient to confer erythromycin resistance. Although Proprionibacterium avidum contains one copy and Proprionibacterium granulosum contains two copies, Proprionibacterium acnes contains three alleles of 23S rRNA (34). However, there was no evidence of heterozygosity in Proprionibacterium granulosum or Proprionibacterium acnes isolates that were macrolide resistant. A mutation in one of the seven alleles of rRNA in E. coli was recessive, with erythromycin resistance being conferred only when point mutations were provided in trans on a multicopy plasmid (40). It was Streptomyces ambofaciens, with one of four rRNA alleles carrying A2058G, that provided the precedent that changes in one of the four alleles could be sufficient to confer macrolide resistance (31).
Using a strategy that allowed us to amplify each allele independently, we found that all but one of the 23S rRNA mutants were heterozygous. Two mutants, 3Az and 8Az, had changes of guanine for A2059 and A2058, respectively, in two of their four alleles. The resistance phenotype was notably different for these two mutants; change at position 2058 resulted in a stronger phenotype to macrolide, lincosamide, and streptogramin B antibiotics, while the change at position 2059 did not confer resistance to streptogramin B. Interestingly, erythromycin-resistant Proprionibacterium acnes mutants with A2059G were homozygous at all three loci and highly resistant to both 14- and 16-membered macrolides (34) whereas the pneumococcal isolate carrying half of its alleles as 2059G was not as highly resistant to 14-membered macrolides as to the 15-membered macrolide azithromycin and the 16-membered macrolide spiramycin. Clarithromycin-resistant H. pylori clinical isolates with the mutations A2058G, A2058C, and A2059G have been described previously (17, 41, 45, 48). Heterozygosity was evident in some isolates, but the majority contained both alleles with their respective mutations. The MICs of clarithromycin were generally higher for isolates with A2058 mutations than for those with A2059 mutations. Cross-resistance to other 14-membered macrolides was not seen in every case. In B. hyodysenteriae, resistance to tylosin, erythromycin, and clindamycin was associated with an A-to-T transversion mutation in nucleotide position 2058 (18). Susceptible strains subcultured on agar containing tylosin yielded resistant mutants with an A-to-G transition at 2058.
Two passage-derived mutants had changes to either adenine or guanine at C2611, a residue important in maintaining the stem preceding the single-stranded portion of the peptidyl transferase region containing positions A2058 and A2059 (Fig. 2) (13). The C2611A and C2611G mutants contained either three alleles of adenine or four alleles of guanine. Strain 6Az, containing the homozygous transversion, appeared to be more resistant to 14- and 15-membered macrolides, telithromycin, and streptogramin B than strain 1Az (ratio of A to C at 2611, 3:1). However, whether the higher level of resistance was due to homozygosity of a purine or the presence of guanine rather than adenine at this residue is not known.
The 50S subunit of bacterial ribosomes is composed of 23S and 5S rRNAs and more than 30 proteins (11). An affinity-labeling study with two photoreactive erythromycin analogs identified a strong interaction of both compounds with the proteins L22 and L15, while only one of two derivatives labeled proteins L2 and L4 (2). L4 has been reported to interact largely with the 5′ end (domains I and II) of 23S rRNA (28, 33). Erythromycin-resistant mutants of E. coli or Bacillus stearothermophilus have been described to have mutations in L4 (5, 30, 37, 50, 51) or, with E. coli or Bacillus subtilis, mutations in protein L22 (5, 39, 46, 50). The presence of erythromycin resistance mutations in either the L4 or L22 ribosomal protein is consistent with the interpretation that these proteins are also in contact with or near the peptidyl transferase region in domain V since erythromycin footprints at A2058 and A2059. The only mutations conferring macrolide resistance that have been identified in ribosomal DNA sequences are those in E. coli; the L4 mutation was a point mutation that changed Lys63 to Glu in L4, while the L22 mutation was a deletion of 3 amino acids, Met82, Lys83, and Arg84 (5). Both mutations appear to be in regions predicted to interact with RNA, suggesting that the mutations in the ribosomal proteins may act indirectly to alter 23S rRNA conformation (47). Interestingly, when the E. coli L4 and L22 proteins carrying the mutations were evaluated in ribosome footprinting experiments, they were found to profoundly alter the conformation of 23S rRNA as assessed by chemical modifications in domains II, III, and V (12), not previously described. Notably, there were no detectable effects at or near A2058 in domain V.
It is interesting that a number of passages (31 to 45 passages for 1Az, 3Az, 4Az, and 5Az; 13 and 17 passages for 8Az and 6Az, respectively) appeared to be necessary before stable mutations in either 23S rRNA or L4 occurred. However, since the 23S rRNA genes in S. pneumoniae are identical in sequence, it is likely that a mutation had to occur in only one copy and that, by homologous recombination, other alleles were converted. We have not yet isolated a laboratory mutant or seen in a clinical strain a mutational change in one 23S rRNA allele conferring macrolide resistance. However, only passaged isolates for which MICs of azithromycin exhibited a ≥4-fold change were examined in the previous study (29). It is possible that one of four changes confers a weaker phenotype and was not examined. As for L4 gene copy number, there appears to be only one L4 sequence per species (Incyte Pharmaceuticals, unpublished data, 1999; TIGR website). The mutations that we detected in L4 were either a change at amino acid 69 from glycine (GGA) to cysteine (TGC) in 4Az or a 6-nucleotide insert (AGTCAA) in 5Az. It is not clear how the 2-nucleotide change occurred in 4Az, but TGA would encode a stop triplet and GGC would be silent, as it still encodes glycine. The 6-nucleotide insert may result from slippage, as the preceding 6 nucleotides (CGTCAA) are nearly identical to those in the insert (TIGR website).
Like 5Az, the E. coli strain with a mutation in L4 did not form colonies at low temperatures (5). The inability of the E. coli L4 mutant to form colonies at 20 to 25°C was associated with a 50S subunit assembly defect at the reduced temperature (5, 14). In addition, the E. coli ribosomes containing the altered L4 bound erythromycin poorly and purified ribosomes from the mutant had reduced peptidyltransferase activities (5, 51). No mutations were found in L22 in this study, but two previously unidentified mutations in L4 were defined in two independently passage-derived isolates. Interestingly, neither L4 mutation conferred cross-resistance to telithromycin or the lincosamides, clindamycin, or lincomycin by NCCLS guidelines (where available). We have not examined subunit assembly in the S. pneumoniae mutants or the ability of altered L4 proteins to bind erythromycin.
This study characterized mutations in azithromycin-derived isolates; however, similar results can be obtained with mutants derived from passage with erythromycin or clarithromycin (8; W. Fu, M. Anderson, S. Williams, A. Tait-Kamradt, J. Sutcliffe, and J. Retsema, Prog. Abstr. 5th Int. Conf. Macrolides Azalides Streptogramins Ketolides Oxazolidinones, abstr. 07–10, 2000). Although some may question the utility of laboratory-derived mutants to predict the nature and frequency of resistance in clinical strains, laboratory experiments with nonfermentation-derived antibiotics like quinolones have identified mutations in gyrase, topoisomerase, and efflux genes that have been subsequently observed in clinical strains (16). It seems likely that, when there are resistance determinants that have mobility via plasmids or transposons, experiments in the laboratory might not predict the frequency of resistance or even the nature of resistance in the clinic. However, laboratory-derived mutants can be predictive for mutations observed in clinical strains, as has been noted for isolates that are linezolid resistant (E. faecium) (S. M. Swaney, D. L. Shinabarger, R. D. Schaadt, J. H. Bock, J. L. Slightom, and G. E. Zurenko, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-104, 1998; G. E. Zurenko, W. M. Todd, B. Hafkin, B. Meyers, C. Kauffman, J. Bock, J. Slightom, and D. Shinabarger, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 0848, 1999) or everninomicin resistant (S. pneumoniae) (1; P. Adrian, C. Mendrick, D. Loebenberg, K. J. Shaw, K. P. Klugman, and R. S. Hare, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 0845, 1999; P. M. McNicholas, P. A. Mann, D. J. Najarian, L. Miesel, T. A. Black, R. S. Hare, and K. J. Shaw, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 0846, 1999). Perhaps not surprisingly, we have identified some clinical strains of pneumococci that have either L4 or 23S rRNA mutations (A. Tait-Kamradt, T. Davies, L. Brennan, F. Depardieu, P. Courvalin, J. Duignan, J. Petitpas, L. Wondrack, M. Jacobs, P. Appelbaum, and J. Sutcliffe, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. LB-8, 1999). Although the frequency of these mutations is unknown, the presence of clinical strains with similar mutations does serve to verify that what we have seen in the laboratory is of some predictive value in the clinic.
In summary, we established that mutations in either 23S rRNA alleles or ribosomal protein L4 were responsible for the macrolide resistance in S. pneumoniae isolates passaged with azithromycin. Furthermore, we determined that there are four copies of 23S rRNA in S. pneumoniae and that macrolide resistance is conferred when at least two or more alleles carry a mutation in A2058, A2059, or C2611, important residues for binding of macrolides and maintaining the conformation of the peptidyl transferase region within 23S rRNA. The application of these findings in characterizing macrolide resistance in clinical isolates will enhance our understanding of the nature and transmission of clones of macrolide-resistant pneumococci.
ACKNOWLEDGMENTS
Sequencing of the S. pneumoniae genome was accomplished with support from TIGR, The National Institute of Allergy and Infectious Diseases, and the Merck Genome Research Institute.
We are also indebted to Incyte Pharmaceuticals for allowing the publication of portions of the L4 sequences from many bacterial species. We thank Robert Kessler for reviewing the manuscript and Paul Miller for enthusiastic support of the study. We also thank all members of the DNA sequencing lab for their consummate patience and careful work.
REFERENCES
- 1.Adrian P V, Zhao W, Black T A, Shaw K J, Hare R S, Klugman K P. Mutations in ribosomal protein L16 conferring reduced susceptibility to evernimicin ( SCH27899): implications for mechanism of action. Antimicrob Agents Chemother. 2000;44:732–738. doi: 10.1128/aac.44.3.732-738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arevalo M A, Tejedor F, Polo F, Ballesta J P. Protein components of the erythromycin binding site in bacterial ribosomes. J Biol Chem. 1988;263:58–63. [PubMed] [Google Scholar]
- 3.Avery O T, MacLeod C M, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a deoxyribonucleic acid fraction isolated from Pneumococcus type III. 1944. Mol Med. 1995;1:344–365. [PMC free article] [PubMed] [Google Scholar]
- 4.Bacot C M, Reeves R H. Novel tRNA gene organization in the 16S-23S intergenic spacer of the Streptococcus pneumoniae rRNA gene cluster. J Bacteriol. 1991;173:4234–4236. doi: 10.1128/jb.173.13.4234-4236.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chittum H S, Champney W S. Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr Microbiol. 1995;30:273–279. doi: 10.1007/BF00295501. [DOI] [PubMed] [Google Scholar]
- 6.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 7.Corso A, Severina E P, Petruk V F, Mauriz Y R, Tomasz A. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb Drug Resist. 1998;4:325–327. doi: 10.1089/mdr.1998.4.325. [DOI] [PubMed] [Google Scholar]
- 8.Davies T A, Dewasse B E, Jacobs M R, Appelbaum P C. In vitro development of resistance to telithromycin (HMR 3647), four macrolides, clindamycin, and pristinamycin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2000;44:414–417. doi: 10.1128/aac.44.2.414-417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doern G V, Brueggemann A B, Huynh H, Wingert E. Antimicrobial resistance with Streptococcus pneumoniae in the United States, 1997–98. Emerg Infect Dis. 1999;5:757–765. doi: 10.3201/eid0506.990603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasc A M, Kauc L, Barraille P, Sicard M, Goodgal S. Gene localization, size, and physical map of the chromosome of Streptococcus pneumoniae. J Bacteriol. 1991;173:7361–7367. doi: 10.1128/jb.173.22.7361-7367.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green R, Noller H F. Ribosomes and translation. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 12.Gregory S T, Dahlberg A E. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23S ribosomal RNA. J Mol Biol. 1999;289:827–834. doi: 10.1006/jmbi.1999.2839. [DOI] [PubMed] [Google Scholar]
- 13.Gutell R R, Gray M W, Schnare M N. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993;21:3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guthrie C, Nashimoto H, Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc Natl Acad Sci USA. 1969;63:384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper D C. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin Infect Dis. 1998;27(Suppl. 1):S54–S63. doi: 10.1086/514923. [DOI] [PubMed] [Google Scholar]
- 17.Hulten K, Gibreel A, Skold O, Engstrand L. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob Agents Chemother. 1997;41:2550–2553. doi: 10.1128/aac.41.11.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson M, Fellstrom C, Heldtander M U, Johansson K E, Franklin A. Genetic basis of macrolide and lincosamide resistance in Brachyspira (Serpulina) hyodysenteriae. FEMS Microbiol Lett. 1999;172:255–260. doi: 10.1111/j.1574-6968.1999.tb13476.x. [DOI] [PubMed] [Google Scholar]
- 19.Latini L, Ronchetti M P, Merolla R, Guglielmi F, Bajaksouzian S, Villa M P, Jacobs M R, Ronchetti R. Prevalence of mefE, erm and tet(M) genes in Streptococcus pneumoniae strains from central Italy. Int J Antimicrob Agents. 1999;13:29–33. doi: 10.1016/s0924-8579(99)00097-7. [DOI] [PubMed] [Google Scholar]
- 20.Liu S L, Hessel A, Sanderson K E. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci USA. 1993;90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucier T S, Heitzman K, Liu S K, Hu P C. Transition mutations in the 23S rRNA of erythromycin-resistant isolates of Mycoplasma pneumoniae. Antimicrob Agents Chemother. 1995;39:2770–2773. doi: 10.1128/aac.39.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuoka M, Endou K, Kobayashi H, Inoue M, Nakajima Y. A plasmid that encodes three genes for resistance to macrolide antibiotics in Staphylococcus aureus. FEMS Microbiol Lett. 1998;167:221–227. doi: 10.1111/j.1574-6968.1998.tb13232.x. [DOI] [PubMed] [Google Scholar]
- 23.Meier A, Kirschner P, Springer B, Steingrube V A, Brown B A, Wallace R J, Jr, Bottger E C. Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob Agents Chemother. 1994;38:381–384. doi: 10.1128/aac.38.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nash K A, Inderlied C B. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob Agents Chemother. 1995;39:2625–2630. doi: 10.1128/aac.39.12.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 26.Noguchi N, Emura A, Matsuyama H, O'Hara K, Sasatsu M, Kono M. Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2′-phosphotransferase I in Escherichia coli. Antimicrob Agents Chemother. 1995;39:2359–2363. doi: 10.1128/aac.39.10.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noguchi N, Katayama J, O'Hara K. Cloning and nucleotide sequence of the mphB gene for macrolide 2′-phosphotransferase II in Escherichia coli. FEMS Microbiol Lett. 1996;144:197–202. doi: 10.1111/j.1574-6968.1996.tb08530.x. [DOI] [PubMed] [Google Scholar]
- 28.Ostergaard P, Phan H, Johansen L B, Egebjerg J, Ostergaard L, Porse B T, Garrett R A. Assembly of proteins and 5 S rRNA to transcripts of the major structural domains of 23 S rRNA. J Mol Biol. 1998;284:227–240. doi: 10.1006/jmbi.1998.2185. [DOI] [PubMed] [Google Scholar]
- 29.Pankuch G A, Jueneman S A, Davies T A, Jacobs M R, Appelbaum P C. In vitro selection of resistance to four beta-lactams and azithromycin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2914–2918. doi: 10.1128/aac.42.11.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pardo D, Rosset R. Genetic studies of erythromycin resistant mutants of Escherichia coli. Mol Gen Genet. 1974;135:257–268. doi: 10.1007/BF00268620. [DOI] [PubMed] [Google Scholar]
- 31.Pernodet J L, Boccard F, Alegre M T, Blondelet-Rouault M H, Guerineau M. Resistance to macrolides, lincosamides and streptogramin type B antibiotics due to a mutation in an rRNA operon of Streptomyces ambofaciens. EMBO J. 1988;7:277–282. doi: 10.1002/j.1460-2075.1988.tb02810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts M C, Sutcliffe J, Courvalin P, Jensen L B, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823–2830. doi: 10.1128/aac.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohl R, Nierhaus K H. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci USA. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross J I, Eady E A, Cove J H, Jones C E, Ratyal A H, Miller Y W, Vyakrnam S, Cunliffe W J. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob Agents Chemother. 1997;41:1162–1165. doi: 10.1128/aac.41.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross J I, Eady E A, Cove J H, Ratyal A H, Cunliffe W J. Resistance to erythromycin and clindamycin in cutaneous propionibacteria is associated with mutations in 23S rRNA. Dermatology. 1998;196:69–70. doi: 10.1159/000017871. [DOI] [PubMed] [Google Scholar]
- 36.Ross J I, Farrell A M, Eady E A, Cove J H, Cunliffe W J. Characterisation and molecular cloning of the novel macrolide-streptogramin B resistance determinant from Staphylococcus epidermidis. J Antimicrob Chemother. 1989;24:851–862. doi: 10.1093/jac/24.6.851. [DOI] [PubMed] [Google Scholar]
- 37.Schnier J, Gewitz H S, Behrens S E, Lee A, Ginther C, Leighton T. Isolation and characterization of Bacillus stearothermophilus 30S and 50S ribosomal protein mutations. J Bacteriol. 1990;172:7306–7309. doi: 10.1128/jb.172.12.7306-7309.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seppala H, Skurnik M, Soini H, Roberts M C, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–262. doi: 10.1128/aac.42.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharrock R A, Leighton T, Wittmann H G. Macrolide and aminoglycoside antibiotic resistance mutations in the Bacillus subtilis ribosome resulting in temperature-sensitive sporulation. Mol Gen Genet. 1981;183:538–543. doi: 10.1007/BF00268778. [DOI] [PubMed] [Google Scholar]
- 40.Sigmund C D, Ettayebi M, Borden A, Morgan E A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- 41.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamm R K, Graham D Y, Ghoneim A T, Tanaka S K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutcliffe J. Resistance to macrolides mediated by efflux mechanisms. Curr Opin Investig Drugs. 1999;1:403–412. [Google Scholar]
- 43.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tipper D J, Johnson C W, Ginther C L, Leighton T, Wittmann H G. Erythromycin resistant mutations in Bacillus subtilis cause temperature sensitive sporulation. Mol Gen Genet. 1977;150:147–159. doi: 10.1007/BF00695395. [DOI] [PubMed] [Google Scholar]
- 47.Urlaub H, Kruft V, Bischof O, Muller E C, Wittmann-Liebold B. Protein-rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J. 1995;14:4578–4588. doi: 10.1002/j.1460-2075.1995.tb00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace R J, Jr, Meier A, Brown B A, Zhang Y, Sander P, Onyi G O, Bottger E C. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother. 1996;40:1676–1681. doi: 10.1128/aac.40.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wittmann H G, Stoffler G, Apirion D, Rosen L, Tanaka K, Tamaki M, Takata R, Dekio S, Otaka E. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol Gen Genet. 1973;127:175–189. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]