Abstract
Background
Cancer patients are at risk of secondary therapy–related myeloid neoplasms (t-MNs). Acquired blood-specific mutations in clonal hematopoiesis (CH)–associated genes are t-MN risk factors, and their occurrence associated with cancer therapy and age. Patients with ovarian cancer (OC) showed a particularly high prevalence of CH–associated gene mutations, which may additionally be explained by the high proportion of a hereditary disease cause in this cancer entity.
Methods
We performed a retrospective analysis of 448 OC patients enrolled in the AGO-TR1 study; 249 were enrolled at primary diagnosis and 199 at platinum-sensitive recurrence. Analyses included the most frequently altered CH–associated genes (ASXL1, DNMT3A, GNAS, JAK2, PPM1D, SF3B1, SH2B3, SRSF2, TET2, TP53). Results were analyzed according to the BRCA1/2 germline (gBRCA1/2) mutation status. All statistical tests were 2-sided.
Results
Advanced age at blood draw and a high number of prior platinum-based chemotherapy lines were risk factors to acquire CH–associated gene mutations, with gene-specific effects observed. Binomial logistic regression suggested increased probabilities for gBRCA1/2 mutation carriers to acquire CH-associated PPM1D and TP53 gene mutations (PPM1D: odds ratio = 4.30, 95% confidence interval = 1.48 to 12.46, P = .007; TP53: odds ratio = 6.20, 95% confidence interval = 0.98 to 53.9, P = .06). This observation was due to a statistically significantly increased number of platinum-based chemotherapy lines in gBRCA1/2 mutation carriers vs noncarriers (PPM1D: mean [SD] = 2.04 [1.27] vs 1.04 [0.99], P < .001; TP53: mean [SD] = 2.83 [1.33] vs 1.07 [1.01], P < .001). No interaction between platinum-based chemotherapy and gBRCA1/2 mutation status with the occurrence of CH–associated gene mutations was observed.
Conclusions
A positive gBRCA1/2 mutation status is not a risk factor to acquire CH–associated gene mutations. OC patients may benefit from monitoring CH–associated gene mutations, especially following carboplatin exposure. Future clinical studies are required to assess whether treatment regimen should be adapted according to individual t-MN risks.
Patients with cancer are at elevated risk of subsequent therapy-related myeloid neoplasms (t-MNs) such as acute myeloid leukemia and myelodysplastic syndrome. The recent MSK-IMPACT study analyzed blood-derived DNA from 24 146 patients with 56 different primary tumor types. In a subgroup of 10 138 patients with curated and detailed clinical data, older age at blood draw and cancer therapy before blood draw correlated with the presence of clonal hematopoiesis (CH), as shown by acquired mutations in CH–associated genes (1). In the MSK-IMPACT study, patients with ovarian cancer (OC) showed a particularly high prevalence of mutations in CH–associated genes, and it was hypothesized that cancer-specific differences may be due to interactions between mutations in specific genes and specific regimen of cancer therapy (1). Most OC patients received cytotoxic treatment regimen, which was also most common in the overall MSK-IMPACT study sample. A unique feature of OC is a high prevalence of pathogenic germline mutations in the BRCA1/2 (gBRCA1/2) cancer predisposition genes, which explain more than 10% of all OC cases irrespective of the patients’ cancer family history (2). BRCA1/2 mutation carriers with OC show a more favorable therapy response and survival than noncarriers with OC (3,4), which simultaneously may be associated with an increased risk to acquire CH–associated gene mutations. To assess whether the gBRCA1/2 mutation status modifies (either directly or indirectly) the association of drug treatment with the occurrence of CH–associated gene mutations, we performed a retrospective analysis of 448 OC patients enrolled in the observational AGO-TR1 study (NCT02222883). Our analyses included the most prevalently altered CH–associated genes ASXL1, DNMT3A, PPM1D, and TET2 along with GNAS, JAK2, SF3B1, SH2B3, SRSF2, and TP53 (5,6). The prevalence of CH-associated gene mutations was assessed according to age at blood draw, number of prior platinum-based chemotherapy lines, and gBRCA1/2 mutation status.
Methods
Study Sample
A total of 523 consecutive OC patients were enrolled in the AGO-TR1 study. The AGO-TR1 study protocol was approved by the Ethics Committee of the Landesaerztekammer Nordrhein (No. 2014340) and registered (NCT02222883, ClinicalTrials.gov). All patients were at least 18 years of age and gave their written informed consent before enrollment. Demographic data, disease characteristics, and family history of the overall study sample were previously described (7).
Targeted Next-Generation Sequencing (NGS)
Blood-derived DNA was available from all 523 patients enrolled in the AGO-TR1 study. Genomic DNA was isolated from venous blood samples collected between March and November 2015 by using a chemagic MSM instrument and the chemagic Prime DNA Blood 4k Kit H24 (PerkinElmer Chemagen Technology GmbH, Baesweiler, Germany). For DNA isolation from formalin-fixed and paraffin-embedded tumor samples, hematoxylin and eosin–stained 3-µm tissue sections were analyzed and tumor areas containing over 80% tumor nuclei were chosen for DNA isolation, which was performed as previously described (8). Tumor-derived DNA was available from 478 of the 523 patients enrolled in the AGO-TR1 study. A customized 48.48 amplicon-based gene panel (Access Array, Fluidigm, San Francisco, CA, USA) was used for target enrichment, which was suitable for the amplification of DNA isolated from both blood samples and formalin-fixed and paraffin-embedded tumor samples. Panel design was performed using the web-based D3 Assay Design tool (Fluidigm). The gene panel covered the entire coding regions and exon-flanking sequences (±2 nt) of 10 CH-associated genes, namely ASXL1 (MIM*612990, NM_015338), DNMT3A (MIM*602769, NM_175629), GNAS (MIM*139320, NM_000516.5), JAK2 (MIM*147796, NM_004972), PPM1D (MIM*605100, NM_003620), SF3B1 (MIM*605590, NM_012433), SH2B3 (MIM*605093, NM_005475), SRSF2 (MIM*600813, NM_003016), TET2 (MIM*612839, NM_001127208), and TP53 (MIM*191170, NM_000546). Overall, the gene panel covered 130 sequencing target regions (Supplementary Table 1, available online). NGS of the barcoded amplicons was performed by using a NextSeq 500 sequencing device and Mid-Output v2 kits (Illumina, San Diego, CA, USA). All DNA samples were centrally analyzed at the Center for Familial Breast and Ovarian Cancer, University Hospital Cologne, Germany. Raw BCL files were demultiplexed using bcl2fastq2 Conversion Software v2.19 (available at https://support.illumina.com). Sequence reads were mapped to the human reference genome assembly GRCh37, including decoy sequences (hs37d5), using BWA-MEM of Burrows-Wheeler Aligner v0.7.15 (9). Target-specific primer sequences were removed using BAMClipper v1.1 (10). Reads were filtered for reads mapped in proper pairs using samtools (11). Variant calling was performed using FreeBayes v1.0.0 (12) on a merged BAM file including RG-tagged reads from blood and tumor samples per sample ID. FreeBayes was run under specification of --min-mapping-quality 20, --min-base-quality 20, --min-coverage 1000, and --min-alternate-fraction 0.03, as well as use-duplicate-reads to account for the characteristics of amplicon sequencing.
Quality Control
All NGS analyses of blood- and tumor-derived DNA samples with an overall mean sequencing coverage greater than 1000× were included in this investigation; data of 448 patients met this quality criterion and were processed further. All sequencing targets with a mean sequencing coverage greater than 1000× in both blood- and tumor-derived DNA in the study sample of 448 patients were included; 11 sequencing targets were excluded. This allowed the comparative analysis of 119 sequencing targets in 448 patients (Supplementary Table 1, available online).
Variant Filtering
We excluded variants located within interspersed repeats and low complexity sequence regions as defined by RepeatMasker (13). Erroneous variant calls caused by technical artifacts were assumed to occur recurrently and to accumulate within the same sequencing run, with normally distributed variant fractions (VFs). Therefore, only variant calls with VFs reaching a modified Z-score greater than or equal to 3.5 (14) considering all VFs at the corresponding locus within the sequencing run were considered. All variants with a VF of at least 0.03 in the blood sample were analyzed further.
Variants identified in blood-derived DNA were considered blood specific if 1) the variant position was covered at least 500× in the corresponding tumor-derived DNA; 2) the tumor VF did not exceed 0.10; and 3) the log2 ratio of the blood VF vs the tumor VF was greater than 1. Blood-specific variants were annotated with respect to the specified transcripts using SnpEff (15). Frameshift variants, nonsense variants, and variants located at the canonical splice sites ±2 bp were defined as protein-truncating variants (PTVs). Missense variants and in frame indels were defined as non-PTVs. Intronic variants outside the canonical splice sites and synonymous variants were excluded from this investigation. These analyses identified a total of 655 blood-specific variants in the overall study sample. All blood-specific variants were filtered for putative pathogenic effects in cancer development using OncoKB (16). Variants were annotated using the MafAnnotator utility of OncoKB Annotator v3.0.0 (https://github.com/oncokb/oncokb-annotator) without specification of a particular tumor type. Of the 655 blood-specific variants, 101 were classified as (likely) oncogenic according to oncoKB Annotator, subsequently referred to as CH-associated gene mutations.
Statistical Analysis
Analyses were conducted under R v3.6. Welch’s t test and Fisher’s exact test were used to assess the association between the patients’ age at blood draw, exposure to drug treatment, and gBRCA1/2 mutation status with the occurrence of CH-associated gene mutations. Benjamini-Hochberg adjustment was applied for multiple testing correction. All statistical tests were 2-sided, with P values less than or equal to .05 considered statistically significant. Association of counts of CH–associated gene mutations per individual with age and number of treatment lines was assessed via Spearman’s correlation using R’s cor.test() function. Binomial logistic regression was used to investigate the association of age at blood draw, exposure to drug treatment, and gBRCA1/2 mutation status with the occurrence of CH–associated gene mutations using R’s glm() utility. Models were fitted using iteratively reweighted least squares and served as input for age-dependent predictions of probabilities for the occurrence of CH–associated gene mutations in dependence to number of prior platinum-based chemotherapy lines and gBRCA1/2 mutation status using R’s predict() utility. P values were obtained applying 2-sided Wald tests. An interaction term between gBRCA1/2 mutation status and number of prior platinum-based chemotherapy lines was included in the binomial logistic regression model to assess whether gBRCA1/2 mutation status modifies the association of drug treatment with CH–associated gene mutations.
Results
CH–associated gene mutations were present in all 10 CH–related genes investigated and most prevalent in DNMT3A (n = 33) and PPM1D (n = 30), followed by TET2 (n = 12), ASXL1 (n = 8), TP53 (n = 7), JAK2 (n = 4), SRSF2 (n = 3), GNAS (n = 2), SF3B1 (n = 1), and SH2B3 (n = 1). All 101 CH–associated gene mutations are listed in Supplementary Table 2 (available online). VFs ranged from 0.03 (minimum cutoff) to 0.37, with a mean VF of 0.10 (Supplementary Table 2, available online). CH–associated mutations in the PPM1D gene, all PTVs, clustered in the terminal exon 6, in accordance with previous findings (17).
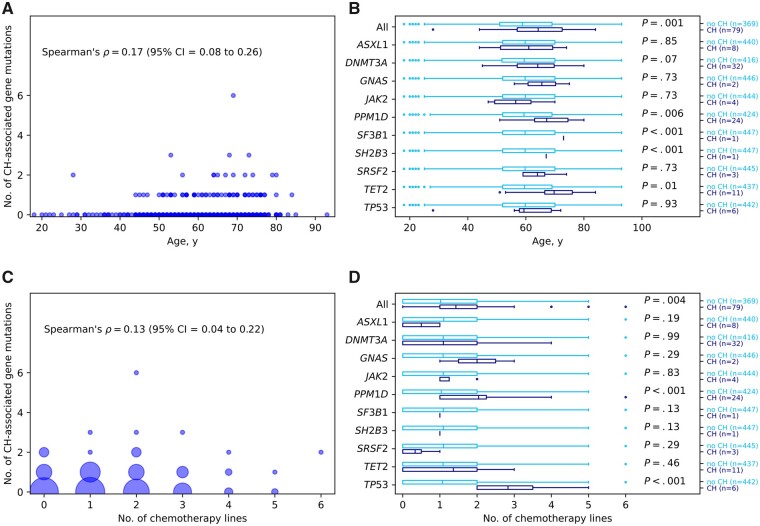
Demographic data of the 448 patients included in this investigation are presented in Supplementary Table 3 (available online). Of the 448 patients, 249 were enrolled in the AGO-TR1 study at primary OC diagnosis, and the remaining 199 patients at platinum-sensitive OC recurrence. On the patient level, 79 (17.6%) of the 448 OC patients carried at least 1 CH–associated gene mutation. Among these 79 patients, 64 patients carried 1, 11 patients carried 2, 3 patients carried 3, and 1 patient carried 6 CH–associated gene mutations (Figure 1, A). The occurrence of CH–associated gene mutations was statistically significantly associated with age at blood draw (Spearman’s rho = 0.17, P < .001; Figure 1, A). In the overall study sample of 448 patients, the mean age at blood draw was 59.8 years (range = 18-93 years, SD = 12.3 years). Patients with at least 1 CH–associated gene mutation in any of the investigated genes were statistically significantly older at the time of blood draw than patients without (79 vs 369 patients, mean age = 64.2 [SD = 10.4] vs 58.8 [SD = 12.5] years, Benjamini-Hochberg-adjusted Welch’s t test P = .001; Figure 1, B). Regarding the individual genes, the difference reached statistical significance for PPM1D (24 vs 424 patients, mean age = 67.2 [SD = 8.7] vs 59.3 [SD = 12.4] years, P = .006) as well as for TET2 (11 vs 437 patients, mean age = 69.7 [SD = 10.3] vs 59.5 [SD = 12.3] years, P = .01), SF3B1, and SH2B3. In the latter 2 genes, however, only a single CH-associated gene mutation was identified (Figure 1, B).
Figure 1.
Associations between age at blood draw and number of prior platinum-based chemotherapy lines with the occurrence of clonal hematopoiesis (CH)–associated gene mutations. A) Number of CH–associated gene mutations per patient according to the age at blood draw. B) Boxplots for age at blood draw in years stratified by noncarriers (no CH) and carriers (CH) of CH–associated gene mutations and Benjamini-Hochberg-adjusted Welch’s t test P values. Mean lines in boxes correspond to mean values. C) Number of CH–associated gene mutations according to number of prior platinum-based chemotherapy lines. Marker sizes correspond to the number of observed samples. D) Boxplots for numbers of platinum-based chemotherapy lines received stratified by noncarriers (no CH) and carriers (CH) of CH–associated gene mutations and Benjamini-Hochberg-adjusted Welch’s t test P values. Mean lines in boxes correspond to mean values. CI = confidence interval.
Of the 448 patients, 303 patients received drug treatment at least 30 days before blood draw (mean age at blood draw = 60.3 years, range = 20-85 years, SD = 11.6 years); the remaining 145 patients did not receive drug treatment at least 30 days before blood draw (mean age at blood draw = 58.6 years, range = 18-93 years, SD = 13.7 years). All 303 patients received a carboplatin-based chemotherapy (Supplementary Table 3, available online); only 4 of the 303 patients received carboplatin and cisplatin. The standard dose in primary therapy was carboplatin AUC5 for 6 cycles, which was usually combined with taxane (Supplementary Table 3, available online). For the treatment of relapsed disease, multiple mostly platinum-based therapies are possible treatment options (Supplementary Table 3, available online).
The number of platinum-based chemotherapy lines at least 30 days before blood draw ranged from 1 to 6 (Figure 1, C; 169 patients received 1 platinum-based chemotherapy line, 97 received 2, 27 patients 3, and 10 patients received 4-6 platinum-based chemotherapy lines). Overall, the 79 patients carrying at least 1 CH-associated gene mutation in any of the investigated genes received statistically significantly more platinum-based chemotherapy lines starting at least 30 days before blood draw than the 369 patients without (mean = 1.43 [SD = 1.23] vs 1.02 [SD = 0.97], Benjamini-Hochberg-adjusted Welch’s t test P = .004, Figure 1, D). This difference was mainly driven by CH-associated gene mutations in the PPM1D gene (mean = 2.04 [SD = 1.27] vs 1.04 [SD = 0.99], P < .001, Figure 1, D) and the TP53 gene (mean = 2.83 [SD = 1.33] vs 1.07 [SD = 1.01], P < .001, Figure 1, D), which were exclusively identified in patients who received at least 1 platinum-based chemotherapy line starting at least 30 days before blood draw. In addition, CH–associated gene mutations in GNAS, JAK2, SF3B1, and SH2B3 were exclusively identified in patients who received prior platinum-based chemotherapy, though the differences in mean numbers of platinum-based chemotherapy lines received did not reach levels of statistical significance (Figure 1, D).
In the overall study sample of 448 OC patients, 92 patients (20.7%) carried pathogenic gBRCA1/2 mutations (Supplementary Table 3, available online). The occurrence of at least 1 CH–associated gene mutation was not statistically significantly associated with a positive gBRCA1/2 mutation status, neither overall (19 occurrences in 92 gBRCA1/2-positive vs 60 in 356 gBRCA1/2-negative patients, Fisher’s exact test P = .44) nor in the subgroup of 303 patients who received platinum-based chemotherapy at least 30 days before blood draw (17 in 70 vs 45 in 233, P = .40).
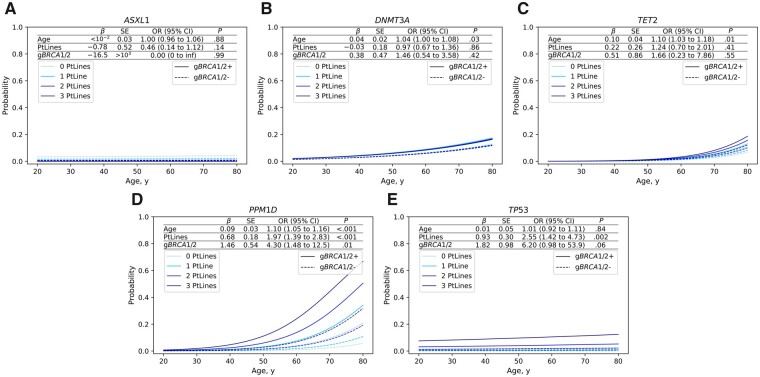
To visualize potential gene-specific effects of the age at blood draw, the number of prior platinum-based chemotherapy lines, and the gBRCA1/2 mutation status on the age-dependent occurrence of CH–associated gene mutations, we used a binomial logistic regression model for the most commonly affected genes: ASXL1, DNMT3A, PPM1D, TET2, and TP53 (Figure 2, A–E). CH–associated gene mutations in the ASXL1 gene did not associate with any of the potential risk factors investigated here (Figure 2, A). CH–associated gene mutations in the ASXL1 gene have recently been shown to be enriched in current or former smokers, which may explain the lack of a statistically significant association in our study that did not stratify for smoking behavior (1). CH–associated gene mutations in the DNMT3A and TET2 genes were associated with age at blood draw, but not with the number of prior platinum-based chemotherapy lines or gBRCA1/2 mutation status (Figure 2, B and C). CH–associated gene mutations in the PPM1D gene were associated with all 3 factors: age at blood draw, number of prior platinum-based chemotherapy lines, and BRCA1/2 mutations status (age: odds ratio [OR] =1.10, 95% confidence interval [CI] = 1.05 to 1.16, P < .001; chemotherapy lines: OR = 1.97, 95% CI = 1.39 to 2.83, P < .001; gBRCA1/2: OR = 4.30, 95% CI = 1.48 to 12.46, P = .007; Figure 2, D). CH–associated gene mutations in the TP53 gene were associated with the number of prior platinum-based chemotherapy lines, but not statistically significantly with BRCA1/2 mutation status and age at blood draw (age: OR = 1.01, 95% CI = 0.92 to 1.11, P = .84; chemotherapy lines: OR = 2.55, 95% CI = 1.42 to 4.73, P = .002; gBRCA1/2: OR = 6.20, 95% CI = 0.98 to 53.9, P = .06; Figure 2, E).
Figure 2.
Risk prediction for clonal hematopoiesis (CH)–associated gene mutations based on binomial logistic regression dependent on age at blood draw (age), number of prior platinum-based chemotherapy lines (PtLines), and BRCA1/2 germline mutation status (gBRCA1/2). Coefficients (β), standard errors (SE), odds ratios (OR), 95% confidence intervals (CI), and P values (P, 2-sided Wald test) as obtained from fitting a binomial logistic regression model for the observation of CH–associated gene mutations in ASXL1 (A), DNMT3A (B), TET2 (C), PPM1D (D), and TP53 (E).
Testing for interaction between the number of platinum-based chemotherapy lines and BRCA1/2 mutation status with the occurrence of CH–associated gene mutations as an outcome revealed no statistically significant association for either PPM1D (interaction OR = 1.02, 95% CI = 0.49 to 2.20, P = .95) or TP53 (interaction OR = 1.69, 95% CI = 0.52 to 7.19, P = .42) (Supplementary Table 4, available online). However, a positive gBRCA1/2 mutation status was statistically significantly associated with younger age at onset (mean age at diagnosis = 52.5 [SD = 8.9] vs 59.8 [SD = 12.8] years, Welch’s t test P < .001; Supplementary Table 3, available online) along with a higher number of platinum-based chemotherapy lines at least 30 days before blood draw (mean = 1.39 [SD = 1.15] vs 1.01 [SD = 0.98], Welch’s t test P = .004), which may explain the increased probability to acquire CH–associated gene mutations at younger ages (Figure 2).
Discussion
In this clinical cohort of 448 OC patients, we demonstrated a high prevalence of CH–associated gene mutations, affecting approximately 1 in 6 patients. Of note, integration of OncoKB annotation for CH classification may represent a conservative CH calling approach, which may lead to missing some less common CH–associated gene mutations.
In patients with t-MN, CH–associated gene mutations were most prevalent in the ASXL1, DNMT3A, PPM1D, TET2, and TP53 genes, whereas CH–associated gene mutations in the PPM1D and TP53 genes were statistically significantly enriched in patients with t-MN compared with de novo MN (18). The MSK-IMPACT study demonstrated that cancer therapy with radiation, platinum (especially carboplatin), and topoisomerase II inhibitors preferentially select for CH–related mutations in DNA damage response genes, including TP53 and PPM1D (1). Concordant with these findings, we identified CH–associated gene mutations in the TP53 and PPM1D genes exclusively in patients who received at least 1 line of platinum-based chemotherapy before blood draw. CH–associated gene mutations in the PPM1D and the TP53 genes were identified in 28 of the 303 patients (9.2%) who received carboplatin-based regimen, of which 2 patients carried CH–associated gene mutations in both genes. These 28 patients received first chemotherapy line 92 days to 11.5 years before blood draw (mean = 3.4 years, SD = 2.8 years; data not shown), with a mean number of 2.14 prior chemotherapy lines (range = 1-6, SD = 1.30). For the remaining 275 of the 303 patients who received the carboplatin-based regimen before blood draw but did not show CH-associated gene mutations in the TP53 and PPM1D genes, the time between first chemotherapy and blood draw ranged from 31 days to 16.1 years (mean = 2.5 years, SD = 2.5 years), with a mean number of 1.56 chemotherapy lines (range = 1-5, SD = 0.77).
Our results point towards a rather indirect association between gBRCA1/2 mutations and a higher probability to accumulate CH–associated gene mutations in the PPM1D, TP53, and probably other genes that accumulate CH–associated gene mutations in response to chemotherapy, which may have implications for the clinical management of patients with gBRCA1/2-associated hereditary cancers such as high-grade serous OC and triple-negative breast cancer (7,19). In the multivariable analysis, we did not observe a statistically significant interaction between gBRCA1/2 mutation status and the number of platinum-based chemotherapy lines with the occurrence of CH–associated gene mutations in PPM1D and TP53 genes, respectively, though minor effects cannot be excluded. In our study sample, gBRCA1/2 mutation carriers received a statistically significantly higher number of platinum-based chemotherapy lines before blood draw than noncarriers, which is most likely due to a more favorable therapy response and survival benefit observed for gBRCA1/2 mutation carriers (3,4).
There had been a concern that gBRCA1/2 mutation carriers may be more prone to t-NM after administration of PARP inhibitors such as niraparib or olaparib: the ENGOT -OV16/NOVA and SOLO2/ENGOT-Ov21 trials revealed increased t-MN rates in the niraparib and the olaparib arms, respectively, vs the placebo arms (20,21). A follow-up investigation of the SOLO2/ENGOT-Ov21 trial, however, revealed that PARP inhibition did not increase the t-MN risk vs placebo (22). Rather, a trend was seen for a higher t-MN incidence with an increasing number of prior platinum-based chemotherapy lines. These results support our findings of an indirect association with gBRCA1/2 gene mutations due to higher numbers of prior platinum-based chemotherapy lines.
Monitoring of patients after chemotherapy exposure with blood draws at defined time intervals and subsequent analysis for CH–associated gene mutations may allow optimized clinical management that considers the patients’ individual t-MN risk. Future clinical studies are required to assess the potential necessity to adapt the choice of treatment regimen according to the individual t-MN risk. For example, the MSK-IMPACT study suggested a lower CH risk following cisplatin or oxaliplatin than following carboplatin.
In the AGO-TR1 study sample, pathogenic germline mutations in non-BRCA1/2 OC predisposition genes were observed (7). Their prevalence, however, was too low to perform meaningful calculations. A further limitation of our study is the focus on 10 CH–associated genes only, that is, additional genes that accumulate CH–associated gene mutations following chemotherapy such as CHEK2 were not considered. A stringent minimum VF of 0.03 was chosen due to the NGS target enrichment used, which is more error-prone than hybrid capture techniques. The AGO-TR1 trial did not assess t-MN as an endpoint.
Funding
This work was supported by the Federal Institute for Drugs and Medical Devices (grant/award number: V-16698/68502/2016–2020).
Notes
Role of the funder: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements: We thank all patients for participating in this study. Computations were partially run on the German Research Foundation (DFG)–funded high-performance computing cluster CHEOPS, which is provided and maintained by the computing center of the University of Cologne (RRZK).
Disclosures: F.H.: Travel grants: AstraZeneca, Tesaro, Roche; Honoraria: Roche, AstraZeneca; Clovis, PharmaMar; Advisory: Roche; P.H.: Consulting or Advisory Role: AstraZeneca, Roche/Genentech, Tesaro, Clovis, PharmaMar Lilly, Sotio; Research Funding: AstraZeneca (Inst); Travel, Accommodations, Expenses: Medac; R.K.S.: Honoraria: AstraZeneca; Consulting or Advisory Role: AstraZeneca; Research Funding: AstraZeneca (Inst); E.H.: Honoraria: AstraZeneca, Consulting or Advisory Role: AstraZeneca, Research Funding: AstraZeneca (Inst).
Author contributions: Conceptualization, K.W.L., C.E., R.K.S., P.H., E.H.; Methodology, K.W.L., C.E., J.H., N.W.L., E.P.R., E.H., R.K.S.; Software, C.E.; Formal Analysis, K.W.L., C.E.; Investigation, K.W.L., C.E.; Resources, A.R., K.B., C.J., T.W.P.S., L.H., K.P., S.K., F.M., F.H., P.H.; Data Curation, K.W.L., C.E., A.R., E.H.; Writing—Original Draft, K.W.L., C.E., R.K.S., E.H.; Writing—review & editing, K.W.L., C.E., A.R., K.M., K.B., C.J., J.H., D.D., J.B., T.W.P.S., L.H., K.P., S.M., N.W.L., E.P.R., S.K., F.M., F.H., J.C.S., R.K.S., P.H., E.H.; Visualization, C.E.; Supervision, R.K.S., E.H.; Project Administration, R.K.S., E.H.; Funding Acquisition, R.K.S.
Supplementary Material
Data Availability
All relevant data are shown in the main manuscript and the Supplementary Materials.
Contributor Information
Konstantin Weber-Lassalle, Center for Familial Breast and Ovarian Cancer, Center for Integrated Oncology (CIO), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, Germany.
Corinna Ernst, Center for Familial Breast and Ovarian Cancer, Center for Integrated Oncology (CIO), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, Germany.
Alexander Reuss, Coordinating Center for Clinical Trials, Philipps-University Marburg, Marburg, Germany.
Kathrin Möllenhoff, Institute of Medical Statistics and Computational Biology, Faculty of Medicine, University of Cologne, Cologne, Germany.
Klaus Baumann, Department of Gynecology, Medical Center Ludwigshafen, Ludwigshafen, Germany.
Christian Jackisch, Department of Gynecology and Obstetrics, Sana Klinikum Offenbach, Offenbach, Germany.
Jan Hauke, Center for Familial Breast and Ovarian Cancer, Center for Integrated Oncology (CIO), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, Germany.
Dimo Dietrich, Department of Otorhinolaryngology, Head and Neck Surgery, University Medical Center Bonn (UKB), Bonn, Germany.
Julika Borde, Center for Familial Breast and Ovarian Cancer, Center for Integrated Oncology (CIO), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, Germany.
Tjoung-Won Park-Simon, Department of Gynecology & Gynecologic Oncology, Medizinische Hochschule Hannover, Hannover, Germany.
Lars Hanker, Department of Gynecology and Obstetrics, University of Schleswig-Holstein, Lübeck, Germany.
Katharina Prieske, Department of Gynecology and Gynecologic Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Sandra Schmidt, Center for Familial Breast and Ovarian Cancer, Center for Integrated Oncology (CIO), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, Germany.
Nana Weber-Lassalle, Center for Familial Breast and Ovarian Cancer, Center for Integrated Oncology (CIO), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, Germany.
Esther Pohl-Rescigno, Center for Familial Breast and Ovarian Cancer, Center for Integrated Oncology (CIO), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, Germany.
Stefan Kommoss, Department Gynecology & Gynecologic Oncology, University of Tübingen, Tübingen, Germany.
Frederik Marmé, Center for Tumor Disease, Department of Gynecology, University of Heidelberg, Heidelberg, Germany.
Florian Heitz, Department of Gynecology & Gynecologic Oncology, Kliniken Essen-Mitte (KEM) Evang, Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany; Department for Gynecology, Charité – Universitätsmedizin Berlin, Freie Universität Berlin, Humboldt-Universität zu Berlin, Berlin, Germany; Berlin Institute of Health, Berlin, Germany.
Julia C Stingl, Institute of Clinical Pharmacology, University Hospital of Rheinisch-Westfälische Technische Hochschule Aachen, Aachen, Germany.
Rita K Schmutzler, Center for Familial Breast and Ovarian Cancer, Center for Integrated Oncology (CIO), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, Germany.
Philipp Harter, Department of Gynecology & Gynecologic Oncology, Kliniken Essen-Mitte (KEM) Evang, Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany.
Eric Hahnen, Center for Familial Breast and Ovarian Cancer, Center for Integrated Oncology (CIO), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, Germany.
References
- 1. Bolton KL, Ptashkin RN, Gao T, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet. 2020;52(11):1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arts-de Jong M, de Bock GH, van Asperen CJ, et al. Germline BRCA1/2 mutation testing is indicated in every patient with epithelial ovarian cancer: a systematic review. Eur J Cancer. 2016;61:137–145. [DOI] [PubMed] [Google Scholar]
- 3. Bolton KL, Chenevix-Trench G, Goh C, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307(4):382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20(3):764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harter P, Hauke J, Heitz F, et al. Prevalence of deleterious germline variants in risk genes including BRCA1/2 in consecutive ovarian cancer patients (AGO-TR-1). PLoS One. 2017;12(10):e0186043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hauke J, Hahnen E, Schneider S, et al. Deleterious somatic variants in 473 consecutive individuals with ovarian cancer: results of the observational AGO-TR1 study (NCT02222883). J Med Genet. 2019;56(9):574–580. [DOI] [PubMed] [Google Scholar]
- 9. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Au CH, Ho DN, Kwong A, et al. BAMClipper: removing primers from alignments to minimize false-negative mutations in amplicon next-generation sequencing. Sci Rep. 2017;7(1):1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012;1207.3907. [q-bio.GN].
- 13. Smit A, Hubley R, Green P. RepeatMasker Open-3.0. http://www.repeatmasker.org. Accessed May 20, 2021.
- 14. Iglewicz B, Hoaglin D. How to detect and handle outliers. Milwaukee: ASQC Quality Press; 1993. [Google Scholar]
- 15. Cingolani P, Platts A, Wang Le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. J Clin Oncol Precis Oncol. 2017;2017;(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kahn JD, Miller PG, Silver AJ, et al. PPM1D truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood. 2018;132(11):1095–1105. doi: 10.1182/blood-2018-05-850339:blood-2018-05-850339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsu JI, Dayaram T, Tovy A, et al. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell. 2018;23(5):700–713.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hahnen E, Hauke J, Engel C, Neidhardt G, Rhiem K, Schmutzler RK. Germline mutations in triple-negative breast cancer. Breast Care (Basel). 2017;12(1):15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mirza MR, Benigno B, Dørum A, et al. Long-term safety in patients with recurrent ovarian cancer treated with niraparib versus placebo: results from the phase III ENGOT-OV16/NOVA trial. Gynecol Oncol. 2020;159(2):442–448. [DOI] [PubMed] [Google Scholar]
- 21. Poveda A, Floquet A, Ledermann JA, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a final analysis of a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2021;22(5):620–631. [DOI] [PubMed] [Google Scholar]
- 22. Korach J, Turner S, Milenkova T, et al. Incidence of myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) in patients (pts) with a germline (g) BRCA mutation (m) and platinum-sensitive relapsed ovarian cancer (PSR OC) receiving maintenance olaparib in SOLO2: Impact of prior lines of platinum therapy. J Clin Oncol. 2018;36(15_suppl):5548–5548. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are shown in the main manuscript and the Supplementary Materials.