Abstract
Background
The incidence of colorectal cancer (CRC) among individuals aged younger than 50 years has been increasing. As screening guidelines lower the recommended age of screening initiation, concerns including the burden on screening capacity and costs have been recognized, suggesting that an individualized approach may be warranted. We developed risk prediction models for early-onset CRC that incorporate an environmental risk score (ERS), including 16 lifestyle and environmental factors, and a polygenic risk score (PRS) of 141 variants.
Methods
Relying on risk score weights for ERS and PRS derived from studies of CRC at all ages, we evaluated risks for early-onset CRC in 3486 cases and 3890 controls aged younger than 50 years. Relative and absolute risks for early-onset CRC were assessed according to values of the ERS and PRS. The discriminatory performance of these scores was estimated using the covariate-adjusted area under the receiver operating characteristic curve.
Results
Increasing values of ERS and PRS were associated with increasing relative risks for early-onset CRC (odds ratio per SD of ERS = 1.14, 95% confidence interval [CI] = 1.08 to 1.20; odds ratio per SD of PRS = 1.59, 95% CI = 1.51 to 1.68), both contributing to case-control discrimination (area under the curve = 0.631, 95% CI = 0.615 to 0.647). Based on absolute risks, we can expect 26 excess cases per 10 000 men and 21 per 10 000 women among those scoring at the 90th percentile for both risk scores.
Conclusions
Personal risk scores have the potential to identify individuals at differential relative and absolute risk for early-onset CRC. Improved discrimination may aid in targeted CRC screening of younger, high-risk individuals, potentially improving outcomes.
The incidence of colorectal cancer (CRC) among individuals aged younger than 50 years (early-onset CRC) has been on the rise for the last several decades in the United States and several other countries (1-4). Early-onset CRC often presents at an advanced stage because of diagnostic delay and aggressive pathology (5), making earlier detection of susceptible individuals a high priority. In response to this increasing public health challenge, the American Cancer Society, the US Preventative Services Task Force, and the American College of Gastroenterology have recently made recommendations regarding lowering the screening age to younger than 50 years (6–8). However, other professional bodies still recommend a starting age for CRC screening at 50 years (9,10), whereas the US Multi-Society Task Force on Colorectal Cancer suggests a screening age of 45 years only for African Americans (11).
Although advocates for initiating screening at an earlier age propose that the benefits of life-years gained outweigh the concerns about unnecessary invasive procedures and associated costs, others suggest, given the extremely low absolute risk of cancer among persons younger than age 50 years, that more targeted approaches for individuals at higher risk are warranted, especially for the use of invasive methods such as colonoscopy (12,13). By using a combination of environmental and lifestyle risk factors and germline genetic variants, precision cancer screening may allow for improved risk discrimination and subsequent gains in the benefit-to-harm ratio compared with more traditional age-based screening regimens (14–18). To date, our risk prediction models for early-onset CRC have focused on genetic factors (16); thus, additional risk assessment incorporating environmental and lifestyle factors should be explored in conjunction with germline genetics.
In this study, we used data from 13 population-based studies, including 3486 cases and 3890 controls, to construct risk prediction models for early-onset CRC that incorporate a novel aggregate environmental risk score (ERS) and a recently expanded polygenic risk score (PRS) (15), now including 141 common genetic variants. We additionally evaluated the absolute risks of early-onset CRC across risk factor profiles of the ERS and PRS. The findings of this study may contribute towards identifying high-risk populations that may benefit from personalized preventive interventions for early-onset CRC.
Methods
Study Participants
Using data from 3 large consortia, the Colon Cancer Family Registry (CCFR), the Colorectal Transdisciplinary (CORECT) Study, and the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO), we included 13 cohort and case-control studies that both 1) evaluated genetic, lifestyle, and environmental factors known to be associated with CRC disease risk, and 2) included 20 or more early-onset CRC cases (<50 years of age at diagnosis of the first primary CRC) (Supplementary Table 1, available online) [see earlier publications for additional study information (16,19–21)]. The final study included 3486 early-onset cases, confirmed by medical record, pathology report, or death certificate. These were contrasted with 3890 controls aged younger than 50 years at recruitment who were ascertained using study-specific eligibility and matching criteria, if applicable, which predominantly involved age- and sex-matched participants. Study-specific participant recruitment occurred primarily between the 1990s and early 2010s, and participants were restricted to those of genetically defined European descent. Written informed consent was obtained from all participants, and the respective institutional review boards approved all research.
ERS Development
Lifestyle and environmental variables included self-reported anthropometric, dietary, lifestyle, and pharmacological risk factors. The data harmonization of these epidemiological variables used a multi-step data harmonization pipeline, reconciling each unique protocol and data-collection instrument (see the Supplementary Methods, available online, and previous publications) (19,20).
Missing data were addressed using sex- and study-specific mean imputation across the complete consortia dataset, as detailed in our previous publication (21). To develop the weighted sex-specific ERS for study participants, we applied sex-specific log-odds ratios from previously published multivariable logistic regression models developed for CRC, including 9748 CRC cases (>95% of which were late onset) and 10 590 age-matched controls ascertained using data from our consortium (19), with the referent level for each variable set at the category associated with the lowest risk for CRC. All variables were collected at the reference time of each respective study, defined as blood collection or participant recruitment for cohort studies, and approximately 1-2 years preceding participant recruitment for case-control studies. The models included the following independent variables: height, body mass index, educational attainment, history of type 2 diabetes, smoking status (ever vs never), alcohol consumption, aspirin use, nonsteroidal antiinflammatory drug use, use of menopausal hormones (women only), total energy consumption, sedentary lifestyle, and sex- and study-specific quartiles of smoking pack-years and dietary factors (intake of fiber, calcium, folate, processed meat, red meat, fruit, and vegetables). In addition, the models were adjusted for study, age, family history, and endoscopy history, defined as whether a participant underwent any sigmoidoscopy or colonoscopy screening before the study reference time (Supplementary Table 2, available online). We then multiplied the log-odds ratios by each participant’s value in our dataset for the corresponding risk factor, followed by summing across all risk factors to create a weighted risk score (19,20). The ERS was recoded as a percentile based on the distribution among control participants.
As a sensitivity analysis, we also produced an ERS with weights derived directly from the participants with early-onset CRC and their associated controls using ridge regression (22) to account for potential overfitting; 10-fold cross-validation (CV) was used for penalty parameter selection. Using this approach, we estimated log-odds ratios (ie, weights for the ERS) for all 16 lifestyle and environmental variables described above (Supplementary Table 3, available online). This model was adjusted for age, study, total energy consumption, and family history. Using these weights to construct an ERS, associations from multivariable logistic models with 10-fold CV between the ERS and early-onset CRC in this sensitivity analysis were comparable with those produced in the main analysis as indicated above, using the previously published log-odds ratios (Supplementary Table 4, available online). Furthermore, given that no participants from CCFR were used in the previously published study in which the external weights were derived (19), we carried out an additional sensitivity analysis restricting to the CCFR study after applying the externally derived weights for the ERS using the same methodology as above, which resulted in very comparable estimates compared with our main analysis (Supplementary Table 5, available online).
PRS Development
As previously described (16), we developed a PRS that included 141 single nucleotide polymorphisms (SNPs) that reached genome-wide statistical significance (P ≤ 5 × 10–8) in a previous large-scale CRC genome-wide association study (GWAS) as of January 2021 (15,23–43). The SNPs were imputed to the Haplotype Reference Consortium panel (44). Directly genotyped SNPs were coded as 0, 1, or 2 copies of the risk allele, whereas imputed SNPs were coded as imputed dosages representing the expected number of copies of the risk allele. To account for population substructure, all models including the PRS were adjusted for principal components of genetic ancestry. We developed the weighted PRS for 76 SNPs using previously published log-odds ratios from seminal GWAS publications among participants of European ancestry (15,23–43). For the 65 SNPs initially discovered in the GECCO and CORECT studies, using all available studies in our consortium (N = 118 673; approximately 10% aged <50 years), we estimated the log-odds ratios from a model fit with overall CRC (no age restrictions) as the outcome and the 141 SNPs as independent variables, adjusted for age, sex, principal components, and genotype platform; we then implemented a winner’s curse adjustment for these 65 SNPs (45). The weighted PRS was then estimated by multiplying the number of risk alleles for each SNP by their log-odds ratios (Supplementary Table 6, available online), followed by summing and recoding as a percentile based on cut points in the controls.
Statistical Analysis
Baseline participant characteristics between cases and controls were evaluated for comparability (Table 1). We used logistic regression to examine the association between the ERS and early-onset CRC, adjusting for reference age in years, sex, family history of CRC, total energy consumption, and study; models for PRS included additional adjustment for principal components, and genotype platform. ERS and PRS were modeled as continuous variables per 1 SD, transformed to the standard normal distribution (subsequently referred to as z-transformed), and as quartiles. Additionally, we evaluated for the presence of biological interaction between the 2 risk scores using the relative excess risk because of interaction, the proportion attributable to interaction, and the synergy index. Tenfold CV was used to evaluate model performance through the K-fold CV accuracy estimate because of the limited data sample. Relationships were explored by anatomic subsite (ie, proximal colon, distal colon, and rectum) using multinomial logistic regression and chi-squared tests for heterogeneity of associations across CRC subsites. We also used logistic regression to model combinations of ERS and PRS tertiles, adjusting for reference age in years, sex, family history of CRC, total energy consumption, principal components, study, and genotype platform.
Table 1.
Baseline participant characteristics of participants
| Characteristic | Cases | Controls |
|---|---|---|
| (n = 3486) | (n = 3890) | |
| Mean age (SD), y | 44.43 (7.39) | 44.52 (5.38) |
| Sex, No (%) | ||
| Female | 1818 (52.2) | 2043 (52.5) |
| Male | 1668 (47.8) | 1847 (47.5) |
| Disease site, No. (%) | ||
| Proximal colon | 891 (27.5) | — |
| Distal colon | 1056 (32.5) | — |
| Rectum | 1298 (40.0) | — |
| Family history, No. (%) | ||
| No | 2407 (76.5) | 2327 (87.3) |
| Yes | 741 (23.5) | 340 (12.7) |
| Combined risk scores | ||
| ERS | ||
| Quartile 1 | 828 (23.8) | 1019 (26.2) |
| Quartile 2 | 801 (23.0) | 1081 (27.8) |
| Quartile 3 | 915 (26.2) | 960 (24.7) |
| Quartile 4 | 942 (27.0) | 830 (21.3) |
| PRS | ||
| Quartile 1 | 640 (18.4) | 1209 (31.1) |
| Quartile 2 | 820 (23.5) | 1089 (28.0) |
| Quartile 3 | 920 (26.4) | 933 (24.0) |
| Quartile 4 | 1106 (31.7) | 659 (16.9) |
| Education, highest level completed, No. (%) | ||
| <High school graduate | 483 (13.9) | 622 (16.0) |
| High school graduate or completed GED | 762 (21.9) | 538 (13.8) |
| Some college or technical school | 1058 (30.3) | 1190 (30.6) |
| ≥College graduate | 1183 (33.9) | 1540 (39.6) |
| Mean height (SD), cm | 171.2 (9.8) | 170.8 (9.5) |
| Mean BMI (SD), kg/m2 | 27.2 (5.6) | 26.9 (5.2) |
| Red meat, No. (%), servings/d | ||
| Quartile 1a | 828 (24.7) | 1004 (26.4) |
| Quartile 2a | 843 (25.2) | 1234 (32.4) |
| Quartile 3a | 888 (26.5) | 998 (26.2) |
| Quartile 4a | 791 (23.6) | 573 (15.0) |
| Processed meat, No. (%), servings/d | ||
| Quartile 1a | 263 (13.7) | 385 (13.0) |
| Quartile 2a | 580 (30.1) | 1006 (34.0) |
| Quartile 3a | 698 (36.2) | 1296 (43.8) |
| Quartile 4a | 385 (20.0) | 272 (9.2) |
| Fruit, No. (%), servings/d | ||
| Quartile 1a | 1045 (31.3) | 1241 (33.0) |
| Quartile 2a | 1054 (31.5) | 1097 (29.1) |
| Quartile 3a | 711 (21.3) | 750 (19.9) |
| Quartile 4a | 531 (15.9) | 678 (18.0) |
| Vegetable, No. (%), servings/d | ||
| Quartile 1a | 801 (23.7) | 1173 (30.9) |
| Quartile 2a | 1271 (37.6) | 1101 (29.0) |
| Quartile 3a | 882 (26.1) | 878 (23.2) |
| Quartile 4a | 424 (12.6) | 639 (16.9) |
| Total fiber, No. (%), g/d | ||
| Quartile 1a | 354 (26.4) | 238 (27.1) |
| Quartile 2a | 331 (24.7) | 217 (24.7) |
| Quartile 3a | 309 (23.0) | 202 (23.0) |
| Quartile 4a | 348 (25.9) | 221 (25.2) |
| Total calcium intake, No. (%), mg/d | ||
| Quartile 1a | 298 (8.5) | 215 (5.5) |
| Quartile 2a | 1926 (55.2) | 2426 (62.4) |
| Quartile 3a | 1011 (29.0) | 1027 (26.4) |
| Quartile 4a | 251 (7.2) | 222 (5.7) |
| Total folate intake, No. (%), mcg/d | ||
| Quartile 1a | 787 (23.7) | 467 (12.4) |
| Quartile 2a | 1331 (40.1) | 2138 (56.7) |
| Quartile 3a | 646 (19.4) | 774 (20.5) |
| Quartile 4a | 559 (16.8) | 393 (10.4) |
| Sedentary lifestyle, No. (%) | ||
| No | 654 (78.9) | 1697 (82.2) |
| Yes | 175 (21.1) | 367 (17.8) |
| Pack-years of smoking, No. (%) | ||
| Never smoker | 1772 (55.9) | 2196 (63.2) |
| Quartile 1a | 395 (12.5) | 413 (11.9) |
| Quartile 2a | 401 (12.6) | 368 (10.6) |
| Quartile 3a | 376 (11.9) | 336 (9.7) |
| Quartile 4a | 226 (7.1) | 162 (4.7) |
| Alcohol use, No. (%), g/d | ||
| 0 | 1450 (43.1) | 1104 (28.7) |
| 1–28 | 1490 (44.3) | 2222 (57.9) |
| >28 | 424 (12.6) | 514 (13.4) |
| Aspirin use, No. (%) | ||
| No | 3090 (91.7) | 3520 (91.9) |
| Yes | 281 (8.3) | 312 (8.1) |
| NSAID use, No. (%) | ||
| No | 2967 (89.4) | 3115 (82.5) |
| Yes | 353 (10.6) | 661 (17.5) |
| Diabetes diagnosis, No. (%) | ||
| No | 3234 (95.5) | 3693 (97.4) |
| Yes | 154 (4.5) | 100 (2.6) |
Study and sex-specific quartiles. Note that the majority of lifestyle and environmental variables were modeled as ordinal sex- and study-specific quartiles throughout the analysis. BMI = body mass index; ERS = environmental risk score; GED = general educational development; NSAID = nonsteroidal antiinflammatory drug; PRS = polygenic risk score.
We estimated the discriminatory accuracy of the ERS and PRS by computing the covariate-adjusted area under the receiver operating characteristic curve (AUC), using the adjusted ROC function from the R Package ROCt. We computed the 95% confidence intervals (CIs) for the AUC estimates using 1000 bootstrap samples. Further, we evaluated the 5-year and 10-year absolute risks of developing early-onset CRC for selected risk profiles of the ERS and PRS, as previously detailed (14,19,20). Using age- and sex-specific population CRC incidence rates among non-Hispanic White individuals from the Surveillance, Epidemiology, and End Results (SEER) registry between 1992 and 2015 (Supplementary Table 7, available online) (46), we estimated the sex-specific baseline hazard function by multiplying the incidence rate with 1 minus the sex-specific population attributable risk, which was computed using the mean inverse exponential of risk scores among cases (47). In addition, we accounted for competing risks from death because of non-CRC causes in the absolute risk estimation using mortality rates from the National Center for Health Statistics (Supplementary Table 8, available online). The 95% confidence intervals for the absolute risks were obtained based on 1000 bootstrap replicates. All tests of statistical significance were 2-sided, and a P value less than .05 was considered statistically significant.
Results
ERS and PRS and Risk of Early-Onset CRC
A greater ERS value was linked to increased risk for early-onset CRC (odds ratio [OR] per SD = 1.14, 95% CI = 1.08 to 1.20) (Table 2); risks were 36% greater comparing the highest ERS quartile with the lowest (OR = 1.36, 95% CI = 1.16 to 1.58). A greater PRS value was also linked to increased risk for early-onset CRC (OR per SD = 1.59, 95% CI = 1.51 to 1.68); risks for early-onset CRC were 3.5-fold greater (OR = 3.50, 95% CI = 3.00 to 4.09) comparing the highest PRS quartile with the lowest. The 10-fold CV accuracy was greater than 0.70 across all models. ERS and PRS had independent predictive values; including both risk scores in a risk prediction model showed that effect estimates were largely unchanged compared with those from models including only one of the predictors. Furthermore, given that no participants from CCFR were included in the previously published study from which the external weights were derived (19), we carried out an additional sensitivity analysis restricting analysis to the CCFR study, using the same methodology as above. The results were strongly comparable for the CCFR (Supplementary Table 5, available online) and main analyses (Table 2).
Table 2.
Odds ratio of ERS and PRS associated with early-onset CRC risk using repeated 10-fold cross-validation
| Model | OR (95% CI) | P a | K-Fold cross-validation accuracy (SD) |
|---|---|---|---|
| Models with ERS as predictor | |||
| Model 1: ERS per 1 SDb | 1.14 (1.08 to 1.20) | <.001 | 0.721 (0.011) |
| Model 2: ERS by quartilec | 0.721 (0.013) | ||
| 1 | 1 (Referent) | — | — |
| 2 | 1.00 (0.86 to 1.16) | .97 | — |
| 3 | 1.22 (1.05 to 1.42) | .009 | — |
| 4 | 1.36 (1.16 to 1.58) | <.001 | — |
| Models with PRS as predictor | |||
| Model 3: PRS per 1 SDd | 1.59 (1.51 to 1.68) | <.001 | 0.720 (0.014) |
| Model 4: PRS by quartilee | 0.717 (0.016) | ||
| 1 | 1 (Referent) | — | — |
| 2 | 1.54 (1.32 to 1.80) | <.001 | — |
| 3 | 2.15 (1.84 to 2.51) | <.001 | — |
| 4 | 3.50 (3.00 to 4.09) | <.001 | — |
| Models with ERS and PRS as predictors | |||
| Model 5f: | 0.737 (0.014) | ||
| ERS per 1 SD | 1.12 (1.06 to 1.19) | <.001 | — |
| PRS per 1 SD | 1.59 (1.50 to 1.68) | <.001 | — |
| Model 6g: | 0.734 (0.011) | ||
| ERS by quartile | |||
| 1 | 1 (Referent) | — | — |
| 2 | 0.99 (0.85 to 1.16) | .91 | — |
| 3 | 1.24 (1.06 to 1.44) | .008 | — |
| 4 | 1.32 (1.12 to 1.54) | <.001 | — |
| PRS by quartile | |||
| 1 | 1 (Referent) | — | — |
| 2 | 1.50 (1.28 to 1.75) | <.001 | — |
| 3 | 2.06 (1.77 to 2.41) | <.001 | — |
| 4 | 3.52 (3.00 to 4.14) | <.001 | — |
2-sided P values per the Wald test. CI = confidence interval; CRC = colorectal cancer; ERS = environmental risk score; OR = odds ratio; PRS = polygenic risk score.
The model includes age, sex, total energy consumption, study, family history, and a continuous z-transformed ERS.
The model includes age, sex, total energy consumption, study, family history, and ERS in quartiles.
The model includes age, sex, genotype platform, family history, principal components, and a continuous z-transformed PRS.
The model includes age, sex, genotype platform, family history, principal components, and PRS in quartiles.
The model includes age, sex, total energy consumption, study, family history, principal components, genotype platform, and continuous z-transformed ERS and PRS.
The model includes age, sex, total energy consumption, study, family history, principal components, genotype platform, and ERS and PRS in quartiles.
When models were restricted by anatomic location, risks for early-onset disease according to the ERS were relatively consistent across sites, whereas the PRS showed greater risks for rectal (OR per SD = 1.67, 95% CI = 1.55 to 1.80) and distal colon cancer (OR per SD = 1.73, 95% CI = 1.60 to 1.87) compared with proximal colon cancer (OR per SD = 1.38, 95% CI = 1.27 to 1.50; P < .001, respectively) (Supplementary Table 9, available online).
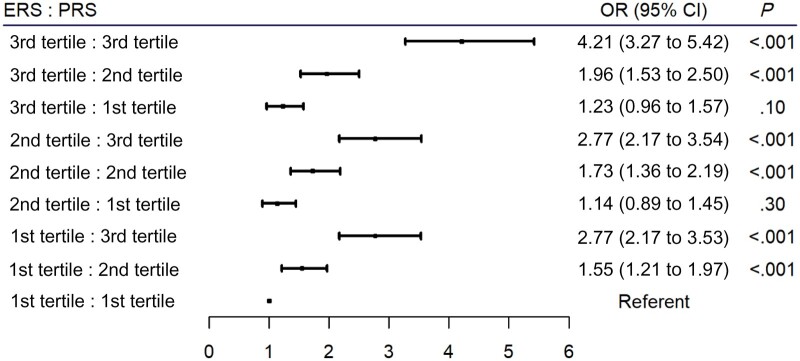
Evaluating the risks for early-onset CRC across varying risk profiles of the ERS and PRS demonstrated a clear trend in increasing risk for early-onset disease with increasing risk scores in both the ERS and PRS (Figure 1). Individuals with a risk profile characterized by the highest tertiles of both the ERS and PRS had a 4.2-fold greater risk (OR = 4.21, 95% CI = 3.27 to 5.42) for early-onset disease compared with those in the lowest tertiles for both measures. As indicated by the proportion attributable to interaction and the synergy index estimates, there is a possibility that modest positive interaction or more than additivity may be occurring between the ERS and PRS (Supplementary Table 10, available online).
Figure 1.
Odds ratio of different combinations of environmental risk score (ERS) and polygenic risk score (PRS) risk profiles across tertiles and associations with early-onset colorectal cancer (CRC) risk. Models were adjusted for age, sex, total energy consumption, study, family history, genotype platform, and principal components. The referent category is the first tertile for both the ERS and PRS. Two-sided P values per the Wald test. The error bars represent the 95% confidence intervals (CIs). OR = odds ratio.
Discriminatory Accuracy of the ERS and PRS
Covariate-adjusted AUC comparisons between risk prediction models for early-onset CRC showed greater risk discrimination with the PRS compared with the ERS (Table 3). The AUC estimate for the ERS was 0.536 (95% CI = 0.519 to 0.552), whereas the AUC for the PRS was 0.628 (95% CI = 0.613 to 0.644). When including both risk scores into a combined model, the AUC was 0.631 (95% CI = 0.615 to 0.647), suggesting limited additional contribution of the ERS, as currently constructed, to the overall AUC. Further, the combined model (PRS plus ERS) showed markedly improved discrimination for early-onset CRC compared with family history alone (AUC = 0.563, 95% CI = 0.555 to 0.571). Similar patterns were also observed when AUC estimates were stratified by sex.
Table 3.
Covariate-adjusted AUC comparisons between risk prediction models
| Model | All participants | Men | Women |
|---|---|---|---|
| AUC (95% CI) | AUC (95% CI) | AUC (95% CI) | |
| Model 1: Family historya | 0.563 (0.555 to 0,571) | 0.568 (0.558 to 0.580) | 0.558 (0.547 to 0.569) |
| Model 2: ERS per 1 SDb | 0.536 (0.519 to 0.552) | 0.546 (0.519 to 0.571) | 0.525 (0.494 to 0.543) |
| Model 3: PRS per 1 SDc | 0.628 (0.613 to 0.644) | 0.621 (0.592 to 0.651) | 0.633 (0.612 to 0.655) |
| Model 4: ERS and PRS per 1 SDd | 0.631 (0.615 to 0.647) | 0.629 (0.604 to 0.654) | 0.630 (0.607 to 0.652) |
The model includes family history as the predictor, adjusting for sex (for the model including all participants) and age. AUC = area under the receiver operating characteristic curve; CI = confidence interval; ERS = environmental risk score; PRS = polygenic risk score.
The model includes a z-transformed ERS as the predictor, adjusting for age, sex (for the model including all participants), total energy consumption, study, and family history.
The model includes a z-transformed PRS as the predictor, adjusting for age, sex (for the model including all participants), family history, genotype platform, and principal components.
The model includes z-transformed ERS and PRS as predictors, adjusting for age, sex (for the model including all participants), study, family history, total energy consumption, principal components, genotype platform, and a z-transformed ERS.
ERS and PRS and Absolute Risk of Early-Onset CRC
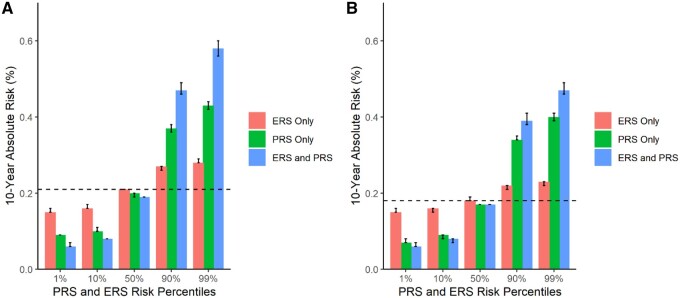
The absolute risk of early-onset CRC varied considerably given the ERS and PRS-dependent risk profile (Table 4; Figure 2). Also, absolute risks of early-onset CRC tended to be cumulative with respect to combined ERS and PRS scores. For example, the 10-year absolute risks of CRC for a 40-year-old at the 90th risk percentile of both the ERS and PRS were 0.47% (47 cases per 10 000) for men (Figure 2, A) and 0.39% (39 cases per 10 000) for women (Figure 2, B). In contrast, the 10-year absolute risks of CRC for a 40-year-old at the 10th risk percentile of both the ERS and PRS was 0.08% (8 cases per 10 000) for men and women. Compared with average 10-year absolute risks using data from SEER (21 cases per 10 000 men and 18 cases per 10 000 women), we can expect approximately 26 excess cases per 10 000 men and 21 excess cases per 10 000 women among 40-year-olds who score at the 90th percentile for both the ERS and PRS (estimated using data from Table 4). In addition, comparing average risks from SEER with those separately for the ERS and PRS at the 90th percentile, we can expect for men roughly 16 excess cases per 10 000 for the PRS and 6 excess cases for the ERS, whereas for females we can expect 16 excess cases per 10 000 for the PRS and 4 excess cases for the ERS (estimated using data from Table 4). Five-year risk differences comparing the 90th and 50th percentiles for both ERS and PRS for 40-year-olds resulted in 9 excess cases per 10 000 for men and 8 excess cases per 10 000 for women, whereas among 45-year-olds, excess cases in 5 years were 18 per 10 000 for men and 14 per 10 000 for women (estimated using data from Supplementary Table 11, available online).
Table 4.
Ten-year absolute risk estimates for early-onset CRC with variable risk factor profiles and starting ages
| ERS risk percentile | PRS risk percentile | Starting age of 30 y |
Starting Age of 40 years |
||
|---|---|---|---|---|---|
| Men | Women | Men | Women | ||
| %, (95% CI) | %, (95% CI) | %, (95% CI) | %, (95% CI) | ||
| Average riska | 0.06 (—) | 0.05 (—) | 0.21 (—) | 0.18 (—) | |
| ERS and PRS combinedb | |||||
| 1 | 1 | 0.02 (0.02 to 0.02) | 0.02 (0.02 to 0.02) | 0.06 (0.06 to 0.07) | 0.06 (0.06 to 0.07) |
| 10 | 10 | 0.02 (0.02 to 0.02) | 0.02 (0.02 to 0.02) | 0.08 (0.08 to 0.08) | 0.08 (0.07 to 0.08) |
| 50 | 50 | 0.05 (0.05 to 0.05) | 0.05 (0.05 to 0.05) | 0.19 (0.19 to 0.19) | 0.17 (0.17 to 0.17) |
| 90 | 90 | 0.13 (0.13 to 0.14) | 0.11 (0.11 to 0.12) | 0.47 (0.46 to 0.49) | 0.39 (0.38 to 0.41) |
| 99 | 99 | 0.16 (0.16 to 0.17) | 0.14 (0.13 to 0.14) | 0.58 (0.56 to 0.60) | 0.47 (0.46 to 0.49) |
| PRSc | |||||
| — | 1 | 0.03 (0.02 to 0.03) | 0.02 (0.02 to 0.02) | 0.09 (0.09 to 0.09) | 0.07 (0.07 to 0.08) |
| — | 10 | 0.03 (0.03 to 0.03) | 0.02 (0.02 to 0.03) | 0.10 (0.10 to 0.11) | 0.09 (0.08 to 0.09) |
| — | 50 | 0.05 (0.05 to 0.05) | 0.05 (0.05 to 0.05) | 0.20 (0.19 to 0.20) | 0.17 (0.17 to 0.17) |
| — | 90 | 0.10 (0.10 to 0.11) | 0.10 (0.10 to 0.10) | 0.37 (0.36 to 0.38) | 0.34 (0.34 to 0.35) |
| — | 99 | 0.12 (0.12 to 0.12) | 0.11 (0.11 to 0.12) | 0.43 (0.42 to 0.44) | 0.40 (0.39 to 0.41) |
| ERSd | |||||
| 1 | — | 0.04 (0.04 to 0.04) | 0.04 (0.04 to 0.04) | 0.15 (0.15 to 0.16) | 0.15 (0.15 to 0.16) |
| 10 | — | 0.05 (0.04 to 0.05) | 0.04 (0.04 to 0.05) | 0.16 (0.16 to 0.17) | 0.16 (0.15 to 0.16) |
| 50 | — | 0.06 (0.06 to 0.06) | 0.05 (0.05 to 0.05) | 0.21 (0.21 to 0.21) | 0.18 (0.18 to 0.19) |
| 90 | — | 0.07 (0.07 to 0.08) | 0.06 (0.06 to 0.06) | 0.27 (0.26 to 0.27) | 0.22 (0.21 to 0.22) |
| 99 | — | 0.08 (0.08 to 0.08) | 0.06 (0.06 to 0.07) | 0.28 (0.28 to 0.29) | 0.23 (0.22 to 0.23) |
Average risks in general population were calculated based on SEER incidence rates for men and women separately. CI = confidence interval; CRC = colorectal cancer; ERS = environmental risk score; PRS = polygenic risk score.
Adjusted for age, study, total energy consumption, family history, genotype platform, and principal components.
Adjusted for age, family history, genotype platform, and principal components.
Adjusted for age, study, total energy consumption, and family history.
Figure 2.
Ten-year absolute risk estimates for early-onset colorectal cancer (CRC) with varying risk factor profiles for a 40-year-old individual. Dashed lines indicate the average 10-year absolute risks of early-onset CRC estimated using Surveillance, Epidemiology, and End Results for a 40-year-old person: A) 0.21% in men and B) 0.18% in women. The environmental risk score (ERS) and polygenic risk score (PRS) combined model adjusted for age, study, total energy consumption, family history, genotype platform, and principal components. The ERS-only model adjusted for age, study, total energy consumption, and family history. The PRS-only model adjusted for age, family history, genotype platform, and principal components.
Discussion
In this study, we demonstrated that greater values of the ERS and PRS were linked to greater risk for early-onset CRC. The discriminatory capacity of the scores, as measured by the covariate-adjusted AUC, was greatest for the PRS, with limited improvement after additional incorporation of the ERS. Similarly, analysis of 5-year and 10-year absolute risks showed that the excess of expected cases varied considerably, with greatest risk stratification stemming from the combined risk scores, although only moderately greater than when considering the PRS alone. However, the absolute number of cases expected was relatively modest even in high-risk categories, largely driven by the overall low rates of CRC at ages younger than 50 years. With screening recommendations increasingly beginning to consider including younger age groups (6–11), concerns need to be recognized regarding societal costs, including increased burden on screening capacity by diverting resources away from higher-risk, older populations to younger, low-risk groups, and furthering disparities in CRC (13,48). Therefore, it is important to evaluate more targeted screening approaches compared with traditional age-based models.
This study is the first to our knowledge to implement a risk score integrating lifestyle, environmental, and genetic factors in early-onset CRC, which complements similar efforts for cohorts consisting predominantly of late-onset disease. Some of these late-onset studies relied either on lifestyle and environmental factors (20,49,50) or on genetics only (51). Previous research in our consortia, using 19 lifestyle and environmental factors and 63 common genetic variants, found similar increases in risk of predominantly late-onset CRC per equivalent increase in the ERS or PRS, with improved case-control discrimination for the combined measures compared with using family history alone (AUC = 0.63 vs 0.53) (19). However, we show here that the PRS contributes most importantly to case-control discrimination for early-onset CRC (AUC: family history alone = 0.563; plus ERS = 0.536; plus PRS = 0.628; plus both risk scores = 0.631). The weaker performance of the ERS in early-onset disease may be due to the lesser importance of certain lifestyle and environmental CRC risk factors that have been generally identified in older people and, most provocatively, indicates the need for further research specifically in the early-onset setting to identify novel lifestyle risk factors for CRC and potentially other cancers in this age group (52). Furthermore, as prediction models move to implementation, it will be important to track changes in exposure prevalence and time-dependent risks.
Additional insight into developing risk prediction models for early-onset CRC can be gleaned from models developed for advanced colorectal neoplasia (adenoma and cancer) in individuals aged younger than 50 years, as recently reported from Korea (53,54), with analysis of established CRC risk factors (53) and clinical factors including H. pylori (54), the latter of which was previously linked to CRC in adults younger than 55 years of age (55). Further opportunities for refinement of risk prediction in early-onset CRC include incorporating information on childhood radiation exposures, antibiotic use, and the microbiome (56,57). Simulation studies suggest that risk-stratified CRC screening may be cost-effective compared with age-based uniform screening if AUC estimates for the PRS are approximately 0.65 or greater (58), pointing to the potential for targeted CRC prevention with improved understanding of the causes of CRC in those younger than 50 years of age.
Our study has the unique strengths of a large sample of cases and controls aged younger than 50 years, in which we leveraged 13 cohort and case-control studies with participants stemming from heterogeneous populations that underwent rigorous harmonization of risk factors (19,20). The study also used data from individuals of European ancestry, thus limiting generalizability to other racially and ethnically diverse populations. The risk factors in the ERS could be strengthened in future studies. The environmental risk factors in our study were self-reported, which could lead to misclassification, although research suggests that self-reported lifestyle and dietary factors are fairly reliable (59,60). In addition, because risk factors were evaluated after cancer diagnosis in case-control studies, data may have been vulnerable to recall bias and may not entirely reflect the most relevant period of exposure for CRC carcinogenesis, particularly for early-life exposures, which were not systematically captured in these studies. Further, imputation to account for missing data can lead to biased estimates, although our prior work with these data showed robustness of estimates to missingness (21). Another limitation related to our study is that we were unable to account for genetic mutations related to hereditary cancer syndromes (61–65) or variants specifically linked to early-onset CRC, given the absence of GWAS specific for early-onset CRC (16).
In conclusion, we showed that an ERS developed from lifestyle and environmental risk factors and a PRS developed with 141 genetic variants provide risk stratification for early-onset CRC. Absolute risks for developing early-onset CRC varied substantially across the various risk profiles of both the ERS and PRS, although the excess number of cases in higher risk strata remained modest, largely due to the relatively low incidence of CRC in young age groups. Additionally, moderate improvement of the predictive performance for the combined risk scores vs the PRS alone indicated that risk stratification of young individuals may be more easily achieved using the PRS alone, although future improvement of the ERS may argue for its eventual utility as well. These risk scores provide an important step toward developing personalized screening regimens targeting individuals younger than 50 years of age who are at increased risk of early-onset CRC (17,18).
Funding
This work was funded by the National Cancer Institute under R03CA21577502, awarded to RBH, and through the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) funded by the National Cancer Institute, National Institutes of Health, US Department of Health and Human Services (U01CA164930, R01CA201407), awarded to UP. This research was funded in part through the NIH/NCI Cancer Center Support Grants, P30CA016087, P30CA015704, and P20CA252728, and training grant T32HS026120 from the Agency for Healthcare Research and Quality. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality.
The Colon Cancer Family Registry (CCFR, www.coloncfr.org) is supported in part by funding from the National Cancer Institute (NCI), National Institutes of Health (NIH) (award U01 CA167551). Support for case ascertainment was provided in part from the Surveillance, Epidemiology, and End Results (SEER) Program and the following US state cancer registries: AZ, CO, MN, NC, NH; and by the Victoria Cancer Registry (Australia) and Ontario Cancer Registry (Canada). The CCFR Set-1 (Illumina 1M/1M-Duo) and Set-2 (Illumina Omni1-Quad) scans were supported by NIH awards U01 CA122839 and R01 CA143247 (to GC). The CCFR Set-3 (Affymetrix Axiom CORECT Set array) was supported by NIH award U19 CA148107 and R01 CA81488 (to SBG). The CCFR Set-4 (Illumina OncoArray 600K SNP array) was supported by NIH award U19 CA148107 (to SBG) and by the Center for Inherited Disease Research (CIDR), which is funded by the NIH to the Johns Hopkins University, contract number HHSN268201200008I. Additional funding for the OFCCR/ARCTIC was through award GL201-043 from the Ontario Research Fund (to BWZ), award 112746 from the Canadian Institutes of Health Research (to TJH), through a Cancer Risk Evaluation (CaRE) Program grant from the Canadian Cancer Society (to SG), and through generous support from the Ontario Ministry of Research and Innovation. The SFCCR Illumina HumanCytoSNP array was supported in part through NCI/NIH awards U01 CA074794 (to JDP) and U24 CA074794 and R01 CA076366 (to PAN). The content of this manuscript does not necessarily reflect the views or policies of the NCI, NIH or any of the collaborating centers in the Colon Cancer Family Registry (CCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government, any cancer registry, or the CCFR.
CRCGEN: Colorectal Cancer Genetics & Genomics, Spanish study was supported by Instituto de Salud Carlos III, co-funded by FEDER funds –a way to build Europe– (grants PI14-613 and PI09-1286), Agency for Management of University and Research Grants (AGAUR) of the Catalan Government (grant 2017SGR723), and Junta de Castilla y León (grant LE22A10-2). Sample collection of this work was supported by the Xarxa de Bancs de Tumors de Catalunya sponsored by Pla Director d’Oncología de Catalunya (XBTC), Plataforma Biobancos PT13/0010/0013 and ICOBIOBANC, sponsored by the Catalan Institute of Oncology.
DACHS: This work was supported by the German Research Council (BR 1704/6–1, BR 1704/6–3, BR 1704/6–4, CH 117/1–1, HO 5117/2–1, HE 5998/2–1, KL 2354/3–1, RO 2270/8–1 and BR 1704/17–1), the Interdisciplinary Research Program of the National Center for Tumor Diseases (NCT), Germany, and the German Federal Ministry of Education and Research (01KH0404, 01ER0814, 01ER0815, 01ER1505A and 01ER1505B).
DALS: National Institutes of Health (R01 CA48998 to M. L. Slattery).
EPIC: The coordination of EPIC is financially supported by the European Commission (DGSANCO) and the International Agency for Research on Cancer. The national cohorts are supported by Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF), Deutsche Krebshilfe, Deutsches Krebsforschungszentrum and Federal Ministry of Education and Research (Germany); the Hellenic Health Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRCItaly and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); ERC-2009-AdG 232997 and Nordforsk, Nordic Centre of Excellence programme on Food, Nutrition and Health (Norway); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPICOxford) (United Kingdom).
Kentucky: This work was supported by the following grant support: Clinical Investigator Award from Damon Runyon Cancer Research Foundation (CI-8); NCI R01CA136726.
LCCS: The Leeds Colorectal Cancer Study was funded by the Food Standards Agency and Cancer Research UK Programme Award (C588/A19167).
MECC: This work was supported by the National Institutes of Health, US Department of Health and Human Services (R01 CA81488 to SBG and GR).
NCCCS I & II: We acknowledge funding support for this project from the National Institutes of Health, R01 CA66635 and P30 DK034987.
NFCCR: This work was supported by an Interdisciplinary Health Research Team award from the Canadian Institutes of Health Research (CRT 43821); the National Institutes of Health, US Department of Health and Human Services (U01 CA74783); and National Cancer Institute of Canada grants (18223 and 18226). The authors wish to acknowledge the contribution of Alexandre Belisle and the genotyping team of the McGill University and Génome Québec Innovation Centre, Montréal, Canada, for genotyping the Sequenom panel in the NFCCR samples. Funding was provided to Michael O. Woods by the Canadian Cancer Society Research Institute.
Harvard cohort (NHS): NHS is supported by the National Institutes of Health (R01 CA137178, P01 CA087969, UM1 CA186107, R01 CA151993, R35 CA197735, K07CA190673, and P50 CA127003).
OFCCR: The Ontario Familial Colorectal Cancer Registry was supported in part by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under award U01 CA167551 and award U01/U24 CA074783 (to SG). Additional funding for the OFCCR and ARCTIC testing and genetic analysis was through and a Canadian Cancer Society CaRE (Cancer Risk Evaluation) program grant and Ontario Research Fund award GL201-043 (to BWZ), through the Canadian Institutes of Health Research award 112746 (to TJH), and through generous support from the Ontario Ministry of Research and Innovation.
UK Biobank: This research has been conducted using the UK Biobank Resource under Application Number 8614.
Notes
Role of the funder: The funders had no role in the design of the study, the writing of the manuscript, the decision to submit the manuscript for publication, and the collection, analysis, and interpretation of the data.
Disclosures: The authors have no conflicts of interest to report and assume full responsibility for all aspects of this study.
Author contributions: Conceptualization: ANA, RBH, UP, LH; Formal analysis: ANA, JJ, YL; Investigation: ANA, JJ; Methodology: ANA, RBH, UP, LH, JJ, DAC, PSL, MD; Supervision: RBH, UP, LH, DAC; Writing—original draft: ANA, RBH, UP, LH, DAC; Writing—review & editing: ANA, JJ, YL, MT, TAH, DTB, HB, GC, ATC, JC-C, JCF, SG, SBG, MJG, FG, MH, MAJ, TOK, LLM, LL, VM, PAN, RP, PSP, GR, LCS, JKL, MLS, MS, AKW, MOW, NM, PTC, Y-RS, IL-V, EFPP, YC, AZ-J, PSL, MD, DAC, LH, UP, RBH; Funding acquisition: ANA, RBH, UP.
Acknowledgements: Participating studies would like to acknowledge the respective contributors. The Colon CFR graciously thanks the generous contributions of their study participants, dedication of study staff, and the financial support from the US National Cancer Institute, without which this important registry would not exist. The authors would like to thank the study participants and staff of the Seattle Colon Cancer Family Registry and the Hormones and Colon Cancer study (CORE Studies). The Darmkrebs: Chancen der Verhütung durch Screening study would like to thank all participants and cooperating clinicians, and Ute Handte-Daub and Utz Benscheid for excellent technical assistance. For EPIC, where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization. The Kentucky study would like to acknowledge and thank the staff at the Kentucky Cancer Registry. The Leeds Colorectal Cancer Study would like to acknowledge the contributions of Jennifer Barrett, Robin Waxman, Gillian Smith and Emma Northwood in conducting this study. North Carolina Colon Cancer Studies I & II would like to thank the study participants, and the NC Colorectal Cancer Study staff. For the Harvard cohort (Nurses' Health Study), the study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We would like to thank the participants and staff of the NHS for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. Lastly, the authors would like to thank the study participants and staff of the Hormones and Colon Cancer and Seattle Cancer Family Registry studies (CORE Studies).
Supplementary Material
Data Availability
The data underlying this article were accessed from the Fred Hutchinson Cancer Center (https://research.fredhutch.org/peters/en/genetics-and-epidemiology-of-colorectal-cancer-consortium.html). The derived data generated in this research will be shared on reasonable request to the corresponding author with permission of the Fred Hutchinson Cancer Center.
Contributor Information
Alexi N Archambault, Division of Epidemiology, Department of Population Health, New York University School of Medicine, New York, NY, USA.
Jihyoun Jeon, Department of Epidemiology, University of Michigan, Ann Arbor, MI, USA.
Yi Lin, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Minta Thomas, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Tabitha A Harrison, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
D Timothy Bishop, Leeds Institute of Medical Research, St. James’s University of Leeds, Leeds, UK.
Hermann Brenner, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany; Division of Preventive Oncology, German Cancer Research Center (DKFZ) and National Center for Tumor Diseases (NCT), Heidelberg, Germany; German Cancer Consortium (DKTK), German Cancer Research Center (DKFZ), Heidelberg, Germany.
Graham Casey, Center for Public Health Genomics, University of Virginia, Charlottesville, VA, USA.
Andrew T Chan, Division of Gastroenterology, Massachusetts General Hospital, Boston, MA, USA; Channing Division of Network Medicine, Brigham and Women’s Hospital, Boston, MA, USA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital, Boston, MA, USA; Broad Institute of Harvard and MIT, Cambridge, MA, USA; Department of Epidemiology, Harvard T.H. Chan School of Public Health, Harvard University, Boston, MA, USA; Department of Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health, Harvard University, Boston, MA, USA.
Jenny Chang-Claude, Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany; University Medical Centre Hamburg-Eppendorf, University Cancer Centre Hamburg (UCCH), Hamburg, Germany.
Jane C Figueiredo, Department of Medicine, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Steven Gallinger, Lunenfeld Tanenbaum Research Institute, Mount Sinai Hospital, University of Toronto, Toronto, ON, Canada.
Stephen B Gruber, Center for Precision Medicine, City of Hope National Medical Center, Duarte, CA, USA.
Marc J Gunter, Nutrition and Metabolism Section, International Agency for Research on Cancer, World Health Organization, Lyon, France.
Feng Guo, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Michael Hoffmeister, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Mark A Jenkins, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Melbourne, Victoria, Australia.
Temitope O Keku, Center for Gastrointestinal Biology and Disease, University of North Carolina, Chapel Hill, NC, USA.
Loïc Le Marchand, Epidemiology Program, University of Hawaii Cancer Center, Honolulu, HI, USA.
Li Li, Department of Family Medicine, University of Virginia, Charlottesville, VA, USA.
Victor Moreno, Oncology Data Analytics Program, Catalan Institute of Oncology-IDIBELL, L'Hospitalet de Llobregat, Barcelona, Spain; CIBER Epidemiología y Salud Pública (CIBERESP), Madrid, Spain; Department of Clinical Sciences, Faculty of Medicine, University of Barcelona, Barcelona, Spain; ONCOBEL Program, Bellvitge Biomedical Research Institute (IDIBELL), L'Hospitalet de Llobregat, Barcelona, Spain.
Polly A Newcomb, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; School of Public Health, University of Washington, Seattle, WA, USA.
Rish Pai, Department of Laboratory Medicine and Pathology, Mayo Clinic Arizona, Scottsdale, AZ, USA.
Patrick S Parfrey, Faculty of Medicine, Memorial University, St John’s, NL, Canada.
Gad Rennert, Department of Community Medicine and Epidemiology, Lady Davis Carmel Medical Center, Haifa, Israel; Ruth and Bruce Rappaport Faculty of Medicine, Technion-Israel Institute of Technology, Haifa, Israel; Clalit National Cancer Control Center, Haifa, Israel.
Lori C Sakoda, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Division of Research, Kaiser Permanente Northern California, Oakland, CA, USA.
Jeffrey K Lee, Division of Research, Kaiser Permanente Northern California, Oakland, CA, USA.
Martha L Slattery, Department of Internal Medicine, University of Utah, Salt Lake City, UT, USA.
Mingyang Song, Division of Gastroenterology, Massachusetts General Hospital, Boston, MA, USA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital, Boston, MA, USA; Department of Nutrition, Harvard T.H. Chan School of Public Health, Harvard University, Boston, MA, USA.
Aung Ko Win, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Melbourne, Victoria, Australia.
Michael O Woods, Discipline of Genetics, Memorial University of Newfoundland, St John’s, NL, Canada.
Neil Murphy, Section of Nutrition and Metabolism, International Agency for Research on Cancer, Lyon, France.
Peter T Campbell, Department of Population Science, American Cancer Society, Atlanta, GA, USA.
Yu-Ru Su, Biostatistics Unit, Kaiser Permanente Washington Health Research Institute, Seattle, WA, USA.
Iris Lansdorp-Vogelaar, Department of Public Health, Erasmus University Medical Center, Rotterdam, the Netherlands.
Elisabeth F P Peterse, Department of Public Health, Erasmus University Medical Center, Rotterdam, the Netherlands.
Yin Cao, Division of Public Health Sciences, Department of Surgery, Washington University School of Medicine, St Louis, MO, USA; Washington University School of Medicine, Alvin J. Siteman Cancer Center, St Louis, MO, USA; Division of Gastroenterology, Department of Medicine, Washington University School of Medicine, St Louis, MO, USA.
Anne Zeleniuch-Jacquotte, Division of Epidemiology, Department of Population Health, New York University School of Medicine, New York, NY, USA.
Peter S Liang, Department of Medicine, New York University School of Medicine, New York, NY, USA.
Mengmeng Du, Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Douglas A Corley, Division of Research, Kaiser Permanente Northern California, Oakland, CA, USA.
Li Hsu, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Department of Biostatistics, University of Washington, Seattle, WA, USA.
Ulrike Peters, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Department of Epidemiology, University of Washington School of Public Health, Seattle, WA, USA.
Richard B Hayes, Division of Epidemiology, Department of Population Health, New York University School of Medicine, New York, NY, USA.
References
- 1. Siegel RL, Torre LA, Soerjomataram I, et al. Global patterns and trends in colorectal cancer incidence in young adults. Gut. 2019;68(12):2179–2185. doi: 10.1136/gutjnl-2019-319511:gutjnl-2019-319511. [DOI] [PubMed] [Google Scholar]
- 2. Feletto E, Yu XQ, Lew J-B, et al. Trends in colon and rectal cancer incidence in Australia from 1982 to 2014: analysis of data on over 375,000 cases. Cancer Epidemiol Biomarkers Prev. 2019;28(1):83–90. [DOI] [PubMed] [Google Scholar]
- 3. Brenner DR, Ruan Y, Shaw E, et al. Increasing colorectal cancer incidence trends among younger adults in Canada. Prev Med. 2017;105:345–349. [DOI] [PubMed] [Google Scholar]
- 4. Siegel RL, Jemal A, Ward EM. Increase in incidence of colorectal cancer among young men and women in the United States. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1695–1698. [DOI] [PubMed] [Google Scholar]
- 5. Yeo H, Betel D, Abelson JS, et al. Early-onset colorectal cancer is distinct from traditional colorectal cancer. Clin Colorectal Cancer. 2017;16(4):293–299.e6. [DOI] [PubMed] [Google Scholar]
- 6.The Lancet Gastroenterology Hepatology. USPSTF recommends expansion of colorectal cancer screening. Lancet Gastroenterol Hepatol. 2021;6(1):1. [DOI] [PubMed] [Google Scholar]
- 7. Wolf AMD, Fontham ETH, Church TR, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin. 2018;68(4):250–281. [DOI] [PubMed] [Google Scholar]
- 8. Shaukat A, Kahi CJ, Burke CA, et al. ACG clinical guidelines: colorectal cancer screening 2021. Am J Gastroenterol. 2021;116(3):458–479. [DOI] [PubMed] [Google Scholar]
- 9. Qaseem A, Crandall CJ, Mustafa RA, et al. ; Clinical Guidelines Committee of the American College of Physicians. Screening for colorectal cancer in asymptomatic average-risk adults: a guidance statement from the American College of Physicians. Ann Intern Med. 2019;171(9):643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Provenzale D, Ness RM, Llor X, et al. NCCN guidelines insights: colorectal cancer screening, version 2.2020. J Natl Compr Canc Netw. 2020;18(10):1312–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rex DK, Boland CR, Dominitz JA, et al. Colorectal cancer screening: recommendations for physicians and patients from the U.S. Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2017;153(1):307–323. [DOI] [PubMed] [Google Scholar]
- 12. Corley DA, Peek RM Jr. When should guidelines change? A clarion call for evidence regarding the benefits and risks of screening for colorectal cancer at earlier ages. Gastroenterology. 2018;155(4):947–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang PS, Allison J, Ladabaum U, et al. Potential intended and unintended consequences of recommending initiation of colorectal cancer screening at age 45 years. Gastroenterology. 2018;155(4):950–954. [DOI] [PubMed] [Google Scholar]
- 14. Hsu L, Jeon J, Brenner H, et al. Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO). A model to determine colorectal cancer risk using common genetic susceptibility loci. Gastroenterology. 2015;148(7):1330–1339.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huyghe JR, Bien SA, Harrison TA, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51(1):76–87. doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Archambault AN, Su YR, Jeon J, et al. Cumulative burden of colorectal cancer-associated genetic variants is more strongly associated with early-onset vs late-onset cancer. Gastroenterology. 2020;158(5):1274–1286.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hull MA, Rees CJ, Sharp L, et al. A risk-stratified approach to colorectal cancer prevention and diagnosis. Nat Rev Gastroenterol Hepatol. 2020;17(12):773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robertson DJ, Ladabaum U. Opportunities and challenges in moving from current guidelines to personalized colorectal cancer screening. Gastroenterology. 2019;156(4):904–917. [DOI] [PubMed] [Google Scholar]
- 19. Jeon J, Du M, Schoen RE, et al. ; Colorectal Transdisciplinary Study and Genetics and Epidemiology of Colorectal Cancer Consortium. Determining risk of colorectal cancer and starting age of screening based on lifestyle, environmental, and genetic factors. Gastroenterology. 2018;154(8):2152–2164.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, O'Connell K, Jeon J, et al. Combined effect of modifiable and non-modifiable risk factors for colorectal cancer risk in a pooled analysis of 11 population-based studies. BMJ Open Gastroenterol. 2019;6(1):e000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Archambault AN, Lin Y, Jeon J, et al. Nongenetic determinants of risk for early-onset colorectal cancer. JNCI Cancer Spectr. 2021;5(3):pkab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 23. Al-Tassan NA, Whiffin N, Hosking FJ, et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci Rep. 2015;5:10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tomlinson IP, Webb E, Carvajal-Carmona L, et al. ; The CORGI Consortium. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40(5):623–630. [DOI] [PubMed] [Google Scholar]
- 25. Tomlinson IP, Carvajal-Carmona LG, Dobbins SE, et al. ; EPICOLON Consortium. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7(6):e1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Broderick P, Carvajal-Carmona L, Pittman AM, et al. ; CORGI Consortium. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39(11):1315–1317. [DOI] [PubMed] [Google Scholar]
- 27. Dunlop MG, Dobbins SE, Farrington SM, et al. ; COIN Collaborative Group. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44(7):770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zeng C, Matsuda K, Jia WH, et al. Identification of susceptibility loci and genes for colorectal cancer risk. Gastroenterology. 2016;150(7):1633–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tomlinson I, Webb E, Carvajal-Carmona L, et al. ; CORGI Consortium. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39(8):984–988. [DOI] [PubMed] [Google Scholar]
- 30. Zhang B, Jia WH, Matsuda K, et al. ; Colon Cancer Family Registry (CCFR). Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet. 2014;46(6):533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang M, Gu D, Du M, et al. Common genetic variation in ETV6 is associated with colorectal cancer susceptibility. Nat Commun. 2016;7:11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Houlston RS, Webb E, Broderick P, et al. ; CoRGI Consortium. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40(12):1426–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Orlando G, Law PJ, Palin K, et al. Variation at 2q35 (PNKD and TMBIM1) influences colorectal cancer risk and identifies a pleiotropic effect with inflammatory bowel disease. Hum Mol Genet. 2016;25(11):2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang H, Burnett T, Kono S, et al. ; GECCO consortium members. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat Commun. 2014;5(1):4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu Y, Kweon SS, Tanikawa C, et al. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology. 2019;156(5):1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Whiffin N, Hosking FJ, Farrington SM, et al. ; Swedish Low-Risk Colorectal Cancer Study Group. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum Mol Genet. 2014;23(17):4729–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Houlston RS, Cheadle J, Dobbins SE, et al. ; COINB Collaborative Group. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42(11):973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Law PJ, Timofeeva M, Fernandez-Rozadilla C, et al. ; PRACTICAL consortium. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat Commun. 2019;10(1):2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmit SL, Edlund CK, Schumacher FR, et al. Novel common genetic susceptibility loci for colorectal cancer. J Natl Cancer Inst. 2019;111(2):146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jia WH, Zhang B, Matsuo K, et al. ; Colon Cancer Family Registry (CCFR). Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat Genet. 2013;45(2):191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schumacher FR, Schmit SL, Jiao S, et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat Commun. 2015;6(1):7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peters U, Jiao S, Schumacher FR, et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology. 2013;144(4):799–807.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmit SL, Edlund CK, Schumacher FR, et al. Novel common genetic susceptibility loci for colorectal cancer. J Natl Cancer Inst. 2019;11(2):146–157. doi: 10.1093/jnci/djy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCarthy S, Das S, Kretzschmar W, et al. ; Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhong H, Prentice RL. Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics. 2008;9(4):621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surveillance Explorer, and End Results (SEER) Program (www.seer.cancer.gov) SEERStat Database: Incidence - SEER Research Data, 13 Registries, Nov 2019 Sub (1992-2017) - Linked to County Attributes - Time Dependent (1990-2017) Income/Rurality, 1969-2018 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2020, based on the November 2019 submission.
- 47. Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for White females who are being examined annually. J Natl Cancer Inst. 1989;81(24):1879–1886. [DOI] [PubMed] [Google Scholar]
- 48. Bretthauer M, Kalager M, Weinberg DS. From colorectal cancer screening guidelines to headlines: beware! Ann Intern Med. 2019;170(10):734. [DOI] [PubMed] [Google Scholar]
- 49. Erben V, Carr PR, Holleczek B, et al. Strong associations of a healthy lifestyle with all stages of colorectal carcinogenesis: results from a large cohort of participants of screening colonoscopy. Int J Cancer. 2019;144(9):2135–2143. [DOI] [PubMed] [Google Scholar]
- 50. Carr PR, Weigl K, Jansen L, et al. Healthy lifestyle factors associated with lower risk of colorectal cancer irrespective of genetic risk. Gastroenterology. 2018;155(6):1805–1815.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jenkins MA, Win AK, Dowty JG, et al. Ability of known susceptibility SNPs to predict colorectal cancer risk for persons with and without a family history. Fam Cancer. 2019;18(4):389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scannell Bryan M, Argos M, Andrulis IL, et al. Germline variation and breast cancer incidence: a gene-based association study and whole-genome prediction of early-onset breast cancer. Cancer Epidemiol Biomarkers Prev. 2018;27(9):1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jung YS, Park CH, Kim NH, et al. Impact of age on the risk of advanced colorectal neoplasia in a young population: an analysis using the predicted probability model. Dig Dis Sci. 2017;62(9):2518–2525. [DOI] [PubMed] [Google Scholar]
- 54. Park YM, Kim HS, Park JJ, et al. A simple scoring model for advanced colorectal neoplasm in asymptomatic subjects aged 40-49 years. BMC Gastroenterol. 2017;17(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Epplein M, Pawlita M, Michel A, et al. Helicobacter pylori protein-specific antibodies and risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2013;22(11):1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Akimoto N, Ugai T, Zhong R, et al. Rising incidence of early-onset colorectal cancer - a call to action. Nat Rev Clin Oncol. 2021;18(4):230–243. doi: 10.1038/s41571-020-00445-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hofseth LJ, Hebert JR, Chanda A, et al. Early-onset colorectal cancer: initial clues and current views. Nat Rev Gastroenterol Hepatol. 2020;17(6):352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Naber SK, Kundu S, Kuntz KM, et al. Cost-effectiveness of risk-stratified colorectal cancer screening based on polygenic risk: current status and future potential. JNCI Cancer Spectr. 2020;4(1):pkz086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hu FB, Satija A, Rimm EB, et al. Diet assessment methods in the Nurses’ Health Studies and contribution to evidence-based nutritional policies and guidelines. Am J Public Health. 2016;106(9):1567–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wolf AM, Hunter DJ, Colditz GA, et al. Reproducibility and validity of a self-administered physical activity questionnaire. Int J Epidemiol. 1994;23(5):991–999. [DOI] [PubMed] [Google Scholar]
- 61. Jasperson KW, Tuohy TM, Neklason DW, et al. Hereditary and familial colon cancer. Gastroenterology. 2010;138(6):2044–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Giráldez MD, López-Dóriga A, Bujanda L, et al. ; Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Susceptibility genetic variants associated with early-onset colorectal cancer. Carcinogenesis. 2012;33(3):613–619. [DOI] [PubMed] [Google Scholar]
- 63. Pinto C, Veiga I, Pinheiro M, et al. MSH6 germline mutations in early-onset colorectal cancer patients without family history of the disease. Br J Cancer. 2006;95(6):752–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. de Voer RM, Hahn MM, Mensenkamp AR, et al. Deleterious germline BLM mutations and the risk for early-onset colorectal cancer. Sci Rep. 2015;5:14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pearlman R, Frankel WL, Swanson B, et al. ; Ohio Colorectal Cancer Prevention Initiative Study Group. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2017;3(4):464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article were accessed from the Fred Hutchinson Cancer Center (https://research.fredhutch.org/peters/en/genetics-and-epidemiology-of-colorectal-cancer-consortium.html). The derived data generated in this research will be shared on reasonable request to the corresponding author with permission of the Fred Hutchinson Cancer Center.