Abstract
To sense the transition from environment to host, bacteria use a range of environmental cues to control expression of virulence genes. Iron is tightly sequestered in host tissues and in the human pathogen enterohemorrhagic Escherichia coli (EHEC) iron-limitation induces transcription of the outer membrane haem transporter encoded by chuAS. ChuA expression is post-transcriptionally activated at 37°C by a FourU RNA thermometer ensuring that the haem receptor is only expressed under low iron, high temperature conditions that indicate the host. Here we demonstrate that expression of chuA is also independently regulated by the cAMP-responsive small RNA (sRNA) CyaR and transcriptional terminator Rho. These results indicate that chuAS expression is regulated at the transcription initiation, transcript elongation, and translational level. We speculate that additional sensing of the gluconeogenic environment allows further precision in determining when EHEC is at the gastrointestinal epithelium of the host. With previous studies, it appears that the chuAS transcript is controlled by eight regulatory inputs that control expression through six different transcriptional and post-transcriptional mechanisms. The results highlight the ability of regulatory sRNAs to integrate multiple environmental signals into a layered hierarchy of signal input.
Keywords: small RNA, Hfq, heme, iron, post-transcriptional, non-coding RNA, EHEC
Introduction
During infection, nutrients required by pathogenic bacteria are withheld by the host in a process known as nutritional immunity (Weinberg, 1975; Hood and Skaar, 2012). An example can be found in iron, a transition metal that is essential for pathogenic bacteria due to its role in essential physiological processes, such as respiration (Skaar, 2010; Evstatiev and Gasche, 2012). To deprive pathogens of this vital nutrient, the host sequesters iron in the high affinity iron binding molecules haem, ferritin and transferrin (Barber and Elde, 2014; Cornelissen, 2018). These ensure that the concentration of extracellular free iron in the host is too low to support bacterial growth and infection. To overcome this nutritional deficit, pathogens such as Haemophilus influenzae, Campylobacter jejuni, Pseudomonas aeruginosa, and uropathogenic Escherichia coli have evolved multiple systems that allow for the scavenging of iron, these include siderophores and haem uptake systems (Rossi et al., 2001; Torres et al., 2001; Ong et al., 2006; Ridley et al., 2006; Hagan and Mobley, 2009; Skaar, 2010; Fournier et al., 2011; Garcia et al., 2011; Morgenthau et al., 2013; Zambolin et al., 2016).
Enterohemorrhagic E. coli (EHEC) is an enteric pathogen that causes haemorrhagic colitis which can progress into potentially fatal hemolytic uremic syndrome. The primary reservoir of this pathogen are ruminants and transmission mainly occurs via the consumption of contaminated foodstuffs (Kaper et al., 2004; Gyles, 2007; Karpman and Ståhl, 2014). EHEC can express an outer membrane haem receptor from the iron regulated chu haem uptake operon (Torres and Payne, 1997). The chu locus is homologous to the shu locus found in Shigella, and contains genes that allow for haem uptake, transport, utilization and degradation. The locus consists of a bicistronic operon (chuAS) as well as two polycistronic operons (chuTWXY and chuUV). ChuA is a TonB-dependent outer membrane haem receptor that imports haem into the periplasm (Celia et al., 2016). The periplasmic binding protein ChuT binds to haem, and via the ABC transporter ChuUV, is shuttled into the cytoplasm. Haem is processed by the haem oxygenase ChuS that converts haem into biliverdin, carbon monoxide and free iron. This breakdown also prevents haem toxicity (Anzaldi and Skaar, 2010). Under anaerobic conditions, this role is taken up by the SAM-methyltransferase ChuW and the anaerobilin reductase ChuY (LaMattina et al., 2016, 2017).
The haem uptake operon is part of a larger regulatory network that allows EHEC to achieve iron homeostasis. Transcription of chuA is repressed by the iron-dependent transcriptional regulator Fur. In iron-rich conditions, Fur binds to iron which increases the affinity of Fur for its DNA-binding site ∼1000-fold (Bags and Neilands, 1987). Fur also regulates a Hfq-dependent small RNA (sRNA) RyhB that is central to maintaining iron homeostasis (Massé and Gottesman, 2002; Massé et al., 2007; Waters and Storz, 2009; Wagner and Romby, 2015). RyhB prevents the expression of non-essential proteins that require Fe as a cofactor such as the TCA cycle and respiration genes sdhC and fumAC, and iron storage genes such as bacterioferritin. Genes that contribute to iron uptake, such as the shikimate permease shiA and the colicin I receptor cirA, are positively regulated by this sRNA (Massé and Gottesman, 2002; Oglesby-Sherrouse and Murphy, 2013; Chareyre and Mandin, 2018).
Enterohemorrhagic Escherichia coli utilizes the Chu system when it is in an iron-poor, haem-rich host. For that reason, as well as the toxicity that accompanies haem and iron over-accumulation, it is necessary to precisely control expression of this operon. FourU RNA-thermometers are used in different bacterial pathogens, such as Shigella dysenteriae, Yersinia pseudotuberculosis, Vibrio cholerae, and Salmonella typhimurium, to regulate expression of their virulence factors in a temperature-dependent manner (Waldminghaus et al., 2007; Kouse et al., 2013; Weber et al., 2014; Righetti et al., 2016). A FourU RNA-thermometer was found in the chuA 5′UTR that blocks the ribosomal binding site (RBS) and is formed at temperatures below 37°C, indicating that the pathogen is outside of the host (Kouse et al., 2013). This limits chuA expression outside of the host where haem is unlikely to be available.
Identification of binding sites for the small RNA chaperone Hfq in EHEC identified the phage-encoded sRNA AsxR that positively regulates the haem oxygenase chuS by sponging interactions with the negatively regulating sRNA, FnrS (Tree et al., 2014). Further, analysis of the EHEC sRNA interactome revealed that the outer membrane haem receptor chuA was also negatively regulated by the EHEC-specific Hfq-binding sRNA Esr41 (Waters et al., 2017). Here, we show that translation of chuA is activated by the Crp-cAMP regulated sRNA CyaR in a temperature- and Rho-termination-independent manner. Our results suggest that EHEC employs an AND-OR logic gate that integrates information on iron availability, temperature and carbon availability to sense its location within the host for appropriate expression of chuA.
Results
The Outer Membrane Haem Receptor chuA Is Regulated by CyaR and ChiX
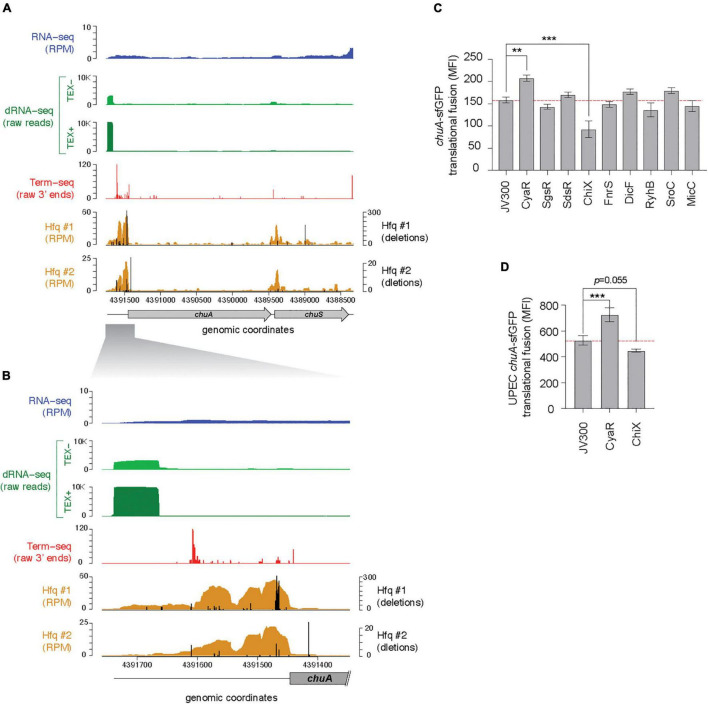
In previous work the binding sites of the small RNA chaperone Hfq were mapped using UV-crosslinking, denaturing purification and sequencing of Hfq-crosslinked RNA (termed CRAC) (Tree et al., 2014). Deletions introduced at the site of protein-RNA crosslinking during reverse transcription of UV-crosslinked RNAs can be used to identify the site of direct Hfq-RNA contact. Analysis of our previous Hfq-CRAC dataset reveals that Hfq binds strongly to the 5′UTR of the outer membrane haem receptor chuA (Figures 1A,B). Canonically, gene regulation through sRNAs occurs through direct base-pairing of the sRNA to the RBS within the 5′UTR of mRNA targets. Hfq-CRAC sequencing reads and contact-dependent deletions peaked at the RBS consistent with RyhB and Esr41 negative regulation at this site (Waters et al., 2017; Banerjee et al., 2020). A further Hfq-binding peak was located at position +160 to +200 suggesting that chuA may be subject to additional regulatory inputs within the 5′UTR (Figures 1A,B).
FIGURE 1.
The chuA 5′UTR is regulated by Hfq-dependent small RNAs. (A) RNA-sequencing data mapping to the chuAS operon. From top to bottom: total RNA-seq (dark blue; RPM), differential RNA-seq in samples untreated (light green) and treated (dark green) with TEX, Term-seq (red; raw 3′ end reads) (BioProject PRJNA601151), Hfq-binding sites from UV-crosslinking datasets (BioProject PRJNA197291). Sequencing deletions which indicate direct contact with Hfq are indicated in black. (B) Same as (A), but focusing on the 5′UTR of chuA. (C) Fluorescence measurements of the EHEC chuA-sfGFP fusion in the presence of predicted binding sRNAs. (D) Fluorescence measurements of the UPEC chuA-sfGFP fusion in the presence of CyaR or ChiX (***p < 0.001, **p < 0.01).
The chuA transcription start site is predicted to be ∼300 nt upstream of the chuA start codon (Nagy et al., 2001). To precisely map the transcription start site of chuA in EHEC str. Sakai, we analyzed our previously published differential RNA-seq (dRNA-seq) data, which enriches for primary transcripts using terminator exonuclease (TEX) (Sharma et al., 2010; Sy et al., 2020). This showed that the +1 site for chuA is at position 4,391,862, and that the 5′UTR is 291 nucleotides long, which concurs with previous predictions (Figure 1B; Nagy et al., 2001).
In previous analyses we found that sRNA-mRNA seed sites are often positioned within Hfq-bound reads recovered by Hfq-CRAC, and that the Hfq binding site can be used to restrict the sequence space for predicting sRNA–mRNA interactions (Tree et al., 2014; Wang et al., 2018). To identify sRNAs that may regulate chuA, IntaRNA (Busch et al., 2008; Mann et al., 2017) was used to predict interactions between the chuA 5′UTR and 44 known Hfq-dependent sRNAs (Li et al., 2013). Small RNAs that were predicted to interact within Hfq read peaks were retained. Through this analysis, we predicted that the sRNAs CyaR, SgrS, SdsR, ChiX, FnrS, RyhB, DicF, SroC, and MicC may have regulatory effects on chuA.
To confirm these predicted interactions the 5′UTR of chuA, starting from the +1 site up until the 15th codon of the coding sequence was fused to sfGFP by cloning into the plasmid pXG10SF (Corcoran et al., 2012). The fluorescence of this translational fusion was monitored in E. coli strain DH5α in the presence and absence of the candidate sRNAs expressed from constitutive expression plasmids (Figure 1C). This revealed that the Hfq-dependent sRNAs CyaR and ChiX have an activating and repressive effect on chuA, respectively.
CyaR Regulation of chuA Is Conserved in Uropathogenic Escherichia coli
Earlier work from the Murphy Lab indicated that chuA 5′UTR from EHEC and Shigella are more closely related than the 5′UTR from EHEC and uropathogenic E. coli (UPEC) (Kouse et al., 2013). We next examined whether CyaR and ChiX regulation are conserved in the divergent EHEC and UPEC 5′UTRs. In UPEC, haem acquisition using chuA is required for maximal colonization of the kidneys (Hagan and Mobley, 2009). The 5′UTRs of EHEC and UPEC (chuAEHEC and chuAUPEC) chuA share 83.4% identity (Nagy et al., 2001). The UPEC 5′UTR sequence has only 67.4% identity within the Hfq-binding site suggesting that chuAUPEC may not be regulated by Hfq-dependent sRNAs. To test whether chuAUPEC is post-transcriptionally regulated by CyaR and ChiX we constructed a translational fusion of chuAUPEC with sfGFP and monitored fluorescence in the presence or absence of CyaR and ChiX overexpression plasmids. The chuAUPEC fusion was activated by CyaR consistent with our results for chuAEHEC but was not significantly repressed by ChiX (Figure 1D). Notably, the chuAUPEC fusion was 55% more fluorescent than the chuAEHEC fusion. This suggests that while chuA in both pathotypes is subject to regulation by CyaR, the divergent 5′UTR sequence has de-repressed chuA expression in UPEC.
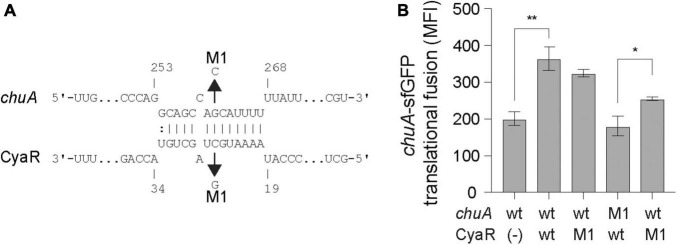
CyaR Activates chuA Translation Through Direct Base-Pairing
Both CyaR and ChiX are class II sRNAs that are known to modulate sRNA regulation by displacing sRNAs from Hfq when overexpressed (Moon and Gottesman, 2011; Santiago-Frangos et al., 2016). To determine whether CyaR and ChiX control chuA through direct RNA–RNA interactions with the chuA 5′UTR or indirectly through titration of Hfq, point mutations were made in both chuA and the sRNAs CyaR and ChiX to disrupt the predicted interaction sites. A predicted 3 nt compensatory mutation was made in the top scoring chuA–ChiX interaction as predicted by IntaRNA. The M1 mutation made in ChiX resulted in the de-repression of chuA-GFP expression. However, this effect was not observed when the compensatory chuA M1 mutation was introduced. This suggested that while the nucleotides mutated in ChiX contributed to chuA regulation, they did not interact with the predicted chuA interaction site. A second set of compensatory point mutants (M2) were made for the next highest scoring interaction that used the same ChiX seed region that was previously tested. Mutations in either chuA or ChiX alone resulted in the disruption of the chuA–ChiX interaction, but testing the mutants together did not restore the repression of chuA (Supplementary Figure 1). These results indicated that while ChiX affects translation of chuA, it is not due to a direct interaction with the chuA M1 or M2 site and may occur indirectly.
CyaR is predicted to bind approximately 15 nt upstream of the chuA Shine–Dalgarno sequence. The interaction between CyaR and chuA results in a 1.3-fold activation of translation. Mutating CyaR at the M1 site reduced CyaR activation by a modest 8.1%, while the chuA point mutant completely abolished the interaction (Figure 2). Providing the CyaR-M1 mutant partially restore translational activation of chuA-M1, supporting a direct interaction between CyaR and the 5′UTR of chuA that activates translation.
FIGURE 2.
CyaR activates chuA translation through direct base-pairing. (A) IntaRNA prediction of the chuA–CyaR interaction. Compensatory point mutations predicted to disrupt the interaction are indicated by the arrows. (B) Fluorescence measurements of wild-type or mutant chuA-sfGFP translational fusions in the presence and absence of wild-type or mutant CyaR overexpression plasmid. Measurements are the mean median fluorescence intensity of three biological replicates (**p < 0.01, *p < 0.05).
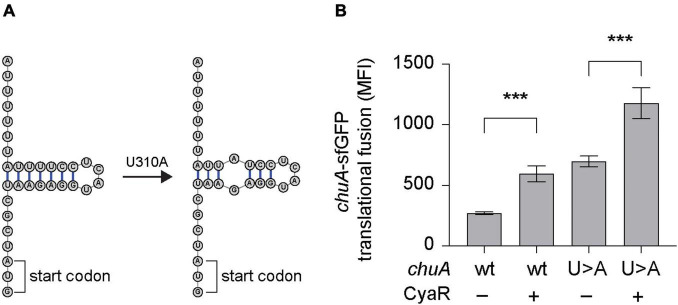
CyaR Activates ChuA Translation Independently of Temperature
Regulation of chuA occurs on both the transcriptional and translational level. Transcription of chuA is inhibited by Fur when cells are grown in iron-rich conditions (Torres and Payne, 1997), while the level of translation is dependent on the environmental temperature. At 25°C, translation is inhibited by the formation of a FourU RNA thermometer that occludes the RBS (Kouse et al., 2013). This inhibitory structure lies 15 nt downstream of the CyaR binding site. To determine whether CyaR-mediated activation of chuA translation occurs by inhibiting the formation of this RNA-thermometer, a U273A point mutation known to disrupt the formation of the FourU hairpin loop was made in the 5′UTR of the chuA-GFP translational fusion (Figure 3A; Kouse et al., 2013). Expression of wild-type and mutant chuA-sfGFP translational fusions were monitored in the presence and absence of CyaR overexpression plasmids at 25°C. Disruption of the RNA thermometer via the U273A point mutation resulted in a 2.6-fold increase in fluorescence compared to the wild-type, confirming the FourU temperature-dependent regulation of chuA translation. Disruption of this secondary structure however did not affect CyaR-mediated activation of chuA, as a 1.6-fold increase in fluorescence was observed when CyaR was provided with the U273A mutation in chuA (Figure 3B). This demonstrated that CyaR-mediated activation of chuA acts independently of post-transcriptional inhibition through the FourU RNA thermometer structure.
FIGURE 3.
CyaR activates chuA translation independently of a FourU RNA-thermometer. (A) Diagram indicating the point mutation made to disrupt the formation of the chuA FourU RNA thermometer. (B) Fluorescence measurements of wild-type and RNA-thermometer disrupted chuA-sfGFP translational fusions in the presence and absence of CyaR overexpression plasmid. Measurements are the mean median fluorescence intensity of three biological replicates (***p < 0.001).
The chuA 5′UTR Contains a Sequence That Inhibits Its Expression
While the activation of chuA translation by CyaR is not due to disruption of the RNA thermometer, in silico folding predictions on the 5′UTR of chuA indicated potential secondary structure throughout the 5′UTR, including some that occluded sequences downstream of the RBS (Supplementary Figure 2). To understand whether the activating chuA–CyaR interaction requires structured sequences within the chuA 5′UTR, we progressively removed sequence from the 5’ end in four truncations of the chuA-sfGFP translational fusion (Figure 4A). Predicted secondary structure and the presence of Hfq distal face binding motifs [(ARN)x] were used as guides in making truncates of the chuA 5′UTR (Soper and Woodson, 2008; Zhang et al., 2013; Schu et al., 2015). A search through the 5′UTR of chuA identified 10 (ARN)4 sites with one mismatch tolerated (ARN4m1). The first truncate (T1) begins at +57 nt and removes a section that forms two hairpins as well as 9 out of 10 ARN4m1 sites. The second (T2) and third (T3) begin at +116 and +177, respectively, and each one removes another hairpin from the overall predicted secondary structure while maintaining the RNA-thermometer. The second truncate T2 also removes the last ARN4m1 motif. The fourth truncate (T4) begins at +217 and leaves only the CyaR binding site, the RNA-thermometer, RBS and the start codon. In the absence of the CyaR overexpression plasmid, the chuA 5′UTR T1-T4 truncates caused 2.1-, 1.9-, 2.9-, and 9.2-fold increases in chuA translation, respectively. CyaR was able to activate truncates T1, T2, and T4 between 20 and 40% consistent with CyaR activation of the wild-type construct (Figure 4B). These results indicate that CyaR does not act through alleviation of an inhibitory secondary structure or a regulatory sequence in the upstream +1 to +217 nt region of the 5′UTR. Notably, removing the region between +177 and +217 nt (T3–T4) of the chuA 5′UTR resulted in a dramatic increase in chuA-GFP expression (9.2-fold) indicating that the T3–T4 region of the 5′UTR contains sequences or RNA structures that strongly repress expression.
FIGURE 4.
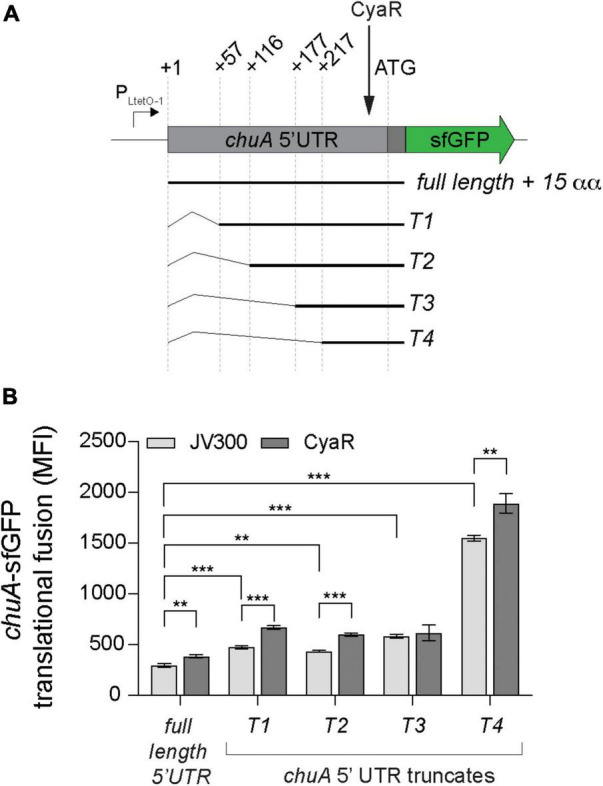
The chuA 5′UTR contains a sequence that disrupts its translation independently of CyaR. (A) Diagram indicating the positions of the different chuA-sfGFP truncates used. (B) Fluorescence measurements of full-length and truncated chuA-sfGFP constructs in the presence and absence of CyaR overexpression plasmid. The position of the CyaR binding site is indicated. Measurements are the mean median fluorescence intensity of three biological replicates (***p < 0.001, **p < 0.01).
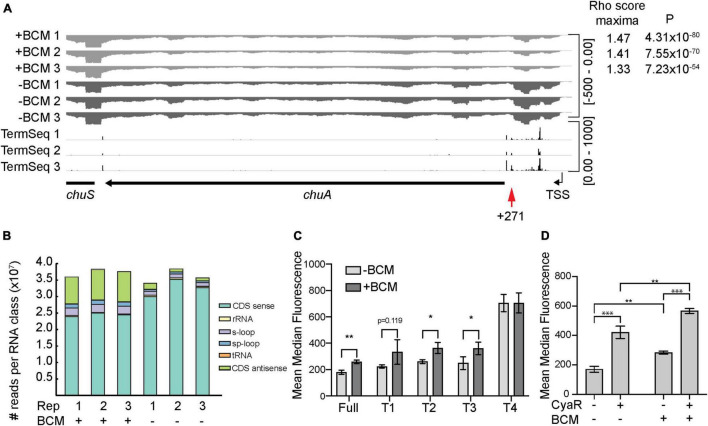
chuA Is Subject to Premature Transcription Termination by Rho
In E. coli, the average 5′UTR is approximately 25–35 nucleotides in length (Kim et al., 2012; Evfratov et al., 2017). The 5′UTR of chuA is 291 nucleotides, making it an unusually long UTR (Wyckoff et al., 1998; Kouse et al., 2013). In commensal E. coli, over half of all annotated genes with long 5′UTRs (defined as >80 nt) are prematurely terminated by the transcription termination factor Rho (Sedlyarova et al., 2016). Further, horizontally transferred genes have been shown to be more susceptible to regulation by Rho-dependent termination (Cardinale et al., 2008; Peters et al., 2012). By analyzing our previously published Term-seq data, we noticed that there were termination sites at positions +129, +271, and +297 within the chuA mRNA (Figure 5A). We did not identify Rho-independent (intrinsic) terminators within the chuA 5′UTR using ARNold (Figure 5A; Naville et al., 2011; Sy et al., 2020). Collectively, this suggested that transcription of the pathogenicity island encoded chuA may be prematurely terminated by Rho.
FIGURE 5.
The chuA 5′UTR is subject to termination by Rho. (A) RNA-seq coverage of chuA in EHEC O157:H7 strain Sakai treated and untreated with bicyclomycin. Rho scores for each replicate is shown. TSS is derived from differential RNA-seq data and is indicated by an arrow. Term-seq reads indicating chuA 3′ ends are also included. (B) Classification and number of reads per RNA class upon treatment of EHEC O157:H7 strain Sakai with bicyclomycin. (C) Fluorescence measurements of full-length and truncated chuA-sfGFP fusions in the presence and absence of induction with bicyclomycin. (D) Fluorescence measurements of chuA-sfGFP translational fusion in the presence and absence of CyaR overexpression plasmid and treatment with bicyclomycin. Measurements are the mean median fluorescence intensity of three biological replicates (***p < 0.001, **p < 0.01, *p < 0.05).
To identify Rho-termination sites in EHEC, we sequenced total RNA extracted from cells treated with the Rho-inhibiting antibiotic bicyclomycin (BCM) (Zwiefka et al., 1993). Rho-termination sites were identified by using the approach adopted by Adams et al. (2021) that previously identified termination sites in BCM-treated E. coli cells. Consistent with earlier studies we found that horizontally acquired regions and antisense RNAs are enriched among genes that are up-regulated by BCM treatment (Figure 5B; Cardinale et al., 2008; Peters et al., 2009). Read-through scores were calculated to assess Rho-dependent termination using previously published scripts (Adams et al., 2021). Read-through scores were maximal for all three replicates (±BCM) at positions +267–273 nt correlating well with the termination site at +271 nt identified by Term-seq. Read-through transcription at this site was up-regulated between 1.33- and 1.47-fold by BCM treatment indicating that Rho prematurely terminates transcription of chuA at position +271 (Figure 5A). To verify that chuA was subject to premature transcription termination by Rho, we measured the fluorescence of inducible full-length and truncated chuA-GFP translational fusions in the presence or absence of BCM (Figure 5A). Treatment with BCM resulted in a ∼1.4-fold increase in fluorescence for the full-length and T1–T3 chuA-GFP constructs. No significant change was observed for chuA truncate T4 (Figure 5C). Rho-dependent termination requires recruitment of the Rho at upstream Rho utilization sites (rut) and we speculate that the rut site for Rho-dependent termination at +271 is removed in the T4 truncate. Taken together, these results indicate that chuA is negatively regulated by Rho-dependent termination at position +271 and this repression requires the 30 nt between +177 and +217 nt of the chuA 5′UTR.
Small RNAs can regulate Rho-dependent termination by altering the accessibility of Rho utilization sites, either as a by-product of translation inhibition, or by directly binding to the rut site itself (Bossi et al., 2012; Sedlyarova et al., 2016; Chen et al., 2019; Silva et al., 2019). We hypothesized that CyaR may activate chuA translation by preventing premature Rho termination within the 5′UTR. To test this, we measured the fluorescence of our chuA-GFP translational fusion in the presence or absence of BCM and CyaR. CyaR was able to activate translation of chuA in the presence of BCM, indicating that CyaR activates chuA independently of Rho termination (Figure 5D). Collectively our data demonstrate that chuA expression is independently regulated by CyaR sRNA, Rho terminator, and the FourU RNA thermometer.
Expression of CyaR Is Correlated With chuA Expression in vivo
In order to obtain a more biologically relevant representation of the relationship between CyaR and chuA expression, we analyzed previously published RNA sequencing data of EHEC str. EDL933 obtained during colonization of a bovine rectum, intestine and rumen (Segura et al., 2018, 2021). Previously, these datasets have been used to show that genes relating to iron acquisition are differentially regulated in each of the gastrointestinal niches when compared to minimal M9 media. We compared the expression of both chuA and CyaR between each of the gastrointestinal niches and we find that chuA is most highly expressed in the rectum, followed by the rumen, then the intestine (Figure 6). These findings are consistent with induction of chuA by environmental signals at this site, including iron-limitation, temperature and a gluconeogenic environment (Snider et al., 2009; Bertin et al., 2013). We also observe this trend with CyaR expression, with the sRNA being maximally expressed in the rectum. This suggests that CyaR contributes to the maximal expression of chuA in the rectum, allowing for better colonization of this environmental niche.
FIGURE 6.

Expression of chuA and CyaR in bovine gastrointestinal niches. Log2FC expression of chuA (orange squares) and CyaR (blue circles) in rectum and rumen compared to intestine. Data from SRA accession SRP136076. Error values are standard errors calculated from DEseq2. Adjusted p-values are calculated from DEseq2.
Discussion
To successfully grow and colonize a host, pathogens utilize systems that allow them to retrieve trace minerals required for growth that are normally sequestered. Various pathotypes of E. coli, including EHEC and UPEC, utilize the ChuA outer membrane haem receptor to transport host derived haem. Transcription of chuA is repressed in the presence of iron by Fur (Torres and Payne, 1997), while translation is regulated by temperature through a FourU RNA-thermometer that occludes the RBS (Kouse et al., 2013). The combination of these two modes of regulation allows the pathogen to sense two signals associated with the host environment (low iron and high temperature) and activate expression of the haem receptor inside the host. Notably, each signal in isolation would generate many false positives when deciding on whether the cell had entered a mammalian host.
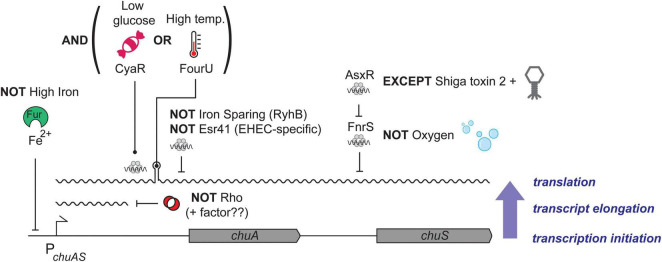
The natural order imposed by transcription and post-transcriptional regulation creates an AND-logic gate, where low iron levels AND high temperature are required for expression of chuA in the host. Here we have shown that chuA is controlled by two additional post-transcriptional signals: repression through Rho termination and activation through CyaR. CyaR acts independently of both Rho and the FourU thermometer potentially creating a post-transcriptional OR-logic gate. The effect of the FourU thermometer and CyaR are additive, and either regulator can activate independently of the other. In Boolean terms it appears that chuA uses an AND-OR logic gate where expression requires low iron AND (high temperature OR CyaR) (Figure 7).
FIGURE 7.
Model of chuAS regulation. Regulation of chuAS expression occurs at the transcriptional, transcript elongation, and translational levels (indicated blue, right). The dependence of each layer on the previous creates regulatory AND gates. (Bottom layer) Repression of chuAS transcription by Fe-loaded Fur is relieved under iron-limited conditions allowing transcription initiation. (Middle layer) Elongating RNAP is prematurely terminated by Rho within the 5′UTR of chuA. An additional factor (potentially another sRNA) is likely to modulate Rho termination and allow read-through transcription. (Top layer) Translation of the chuAS transcript is activated by the cAMP-responsive sRNA CyaR or temperatures ł37°C that melt the FourU thermometer. Melting of the chuAS mRNA FourU element can be overridden under iron-sparing conditions by the sRNA RyhB or the EHEC-specific sRNA Esr41 that repress translation from the exposed RBS. The downstream haem oxygenase encoded by chuS is repressed under aerobic conditions by the sRNA FnrS, except in EHEC where repression is relieved by the sRNA sponge AsxR carried by the Shiga toxin 2-encoding bacteriophage.
Transcription of CyaR is activated by the global regulator Crp when cyclic-AMP levels are high, such as in the absence of glucose (De Lay and Gottesman, 2009). EHEC colonizes the colon where the primary source of carbon is mucins. Bacteroides thetaiotaomicron (Bt) can cleave mucins to release sugars that are utilized by EHEC (Xu et al., 2003; Fischbach and Sonnenburg, 2011). During colonization of the gastrointestinal mucus layer and epithelium, EHEC is likely to encounter varying niches that include both glycolytic and gluconeogenic environments (Miranda et al., 2004). Sensing variation in the availability of sugars and oxygen availability have been suggested to be key regulatory signals that allow EHEC to determine location in the gastrointestinal tract and proximity to the epithelium (Carlson-Banning and Sperandio, 2016). Close to the intestinal epithelium, the relative absence of microorganisms that metabolize and release sugars from mucins creates at gluconeogenic environment (Conway and Cohen, 2015). Lower levels of fucose, and higher levels of succinate reduces the level of the fucose-sensing two-component system FusKR that induces expression of Cra. In gluconeogenic environments, Cra can enhance binding of the cAMP receptor protein Crp to its targets (Ryu et al., 1995). Activation of Cra can induce expression of genes required for T3SS and adhesion to the epithelium (Njoroge et al., 2012; Curtis et al., 2014; Carlson-Banning and Sperandio, 2016). Expression of Crp can also be activated in the presence of Cra (Zhang et al., 2014). We propose that the gluconeogenic environment encountered at the gastrointestinal tract provides an activating signal for CyaR that has been incorporated into chuA regulation as an additional signal that indicates host colonization and haem availability. Consistent with this hypothesis, we find that both CyaR and chuA transcription is activated in bovine rectal samples (Figure 6).
The mechanism of CyaR activation of chuA expression remains unclear although we have ruled out three probable mechanisms: CyaR acts through direct base-pairing with the chuA 5′UTR rather than general titration of Hfq, CyaR does not disrupt the FourU RNA thermometer to override temperature-dependence, and CyaR does not inhibit premature Rho termination of chuA. While these results have uncovered two additional post-transcriptional regulators of chuA (CyaR and Rho) they also suggest that there exists an additional repressive element in the chuA 5′UTR that is overcome by CyaR binding. We note that our chuA-GFP translational fusion reports on multiple post-transcriptional processes including translation efficiency and mRNA stability, and that CyaR may be modulating either of these to affect chuA expression.
Our Term-seq data indicates that the chuA 5′UTR is terminated at positions +129, +271, and +297 with position +271 nt maximally sensitive to treatment with BCM. Using truncates of the chuA 5′UTR we have also found that the sequence between +177 and +217 nt (T3 and T4) represses chuA expression almost 10-fold. We suggest that Rho associates with the nascent chuA transcript between +177 and +217 nt to terminate transcription at +271 nt. Small RNAs can promote or occlude rut sites to control Rho termination in response to regulatory signals (Sedlyarova et al., 2016; Silva et al., 2019) and it remains plausible that Rho termination is modulated by yet another regulatory signal.
The chuA encoded haem receptor is transcribed as a bicistronic transcript with the haem oxygenase chuS. In previous work we showed that chuS expression is repressed by the sRNA FnrS that is induced under anaerobic conditions, consistent with ChuS requiring oxygen for activity. FnrS repression is over-ridden by the sRNA sponge AsxR transcribed from the Shiga toxin 2 encoding bacteriophage Sp5 (Tree et al., 2014). Collectively, expression of chuAS is subject to an impressive level of post-transcriptional regulation that provides complex integration of environmental signals beyond low iron (transcriptional regulation by Fur). These post-transcriptional signals appear to provide a much more accurate interpretation of the environment to indicate whether haem transport is required (i.e., whether EHEC is at the gastrointestinal epithelium). The logic of the regulatory circuit that controls chuAS expression is outlined in Figure 7.
Post-transcriptional regulation appears ideally suited to this layering of logic gates because the natural order (dependence) imposed by transcriptional and post-transcriptional signals creates AND gates. There are at least three ordered layers of AND gates in the chuA transcript that need to be satisfied before the next layer of signals are incorporated. Fur-dependent transcription, Rho-dependent termination, and post-transcriptional control through sRNAs or the FourU thermometer. Together these form an elegant set of AND, OR, and NOT gates that interpret the environment and genetic background of the host (the later through the EHEC-specific sRNAs AsxR and Esr41) (Figure 7). The extensive post-transcriptional logic of the chuAS operon suggests that layered post-transcriptional regulation can create complex regulatory logic gates to integrate and interpret environmental signals.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains, oligonucleotides and plasmids used for this study are listed in Supplementary Table 1. E. coli was routinely grown at 37°C in liquid Luria-Bertani (LB) broth or on solid LB-agar plates. Bacterial media was supplemented with ampicillin (100 μg/mL) or chloramphenicol (34 μg/mL) where appropriate. For inhibition of Rho termination, bicyclomycin (BioAustralis, 50 μg/mL) was added to exponential phase (OD600 = 0.6) cultures for 30 min before being assayed.
In silico Prediction of Interacting sRNAs
A list of published sRNAs present in enterohemorrhagic E. coli was taken from the Bacterial Small Regulatory RNA Database (Li et al., 2013). sRNAs listed as not being Hfq-binding were filtered out, and the sequences of the remaining sRNAs were input into IntaRNA (Busch et al., 2008; Wright et al., 2014; Mann et al., 2017) to search for interactions with the 5′UTR of chuA. sRNAs that were predicted to bind to regions of the chuA 5′UTR that were not Hfq-binding were removed from consideration.
Construction of GFP-Translational Fusions and sRNA Expression Vectors for Testing sRNA–mRNA Interactions
Plasmids pXG10SF containing the full-length and truncated chuA-GFP translational fusion and pZE12 carrying candidate sRNAs were cloned according to method described in Urban and Vogel (2007). Briefly, the 5′UTR of chuA and candidate sRNAs were amplified from genomic DNA using primers identified in Supplementary Table 1. The chuA-GFP translational fusion was inserted using NheI-NsiI restriction cloning. Candidate sRNAs were inserted by first linearizing pZE12-luc using primers pLlacO-B and pLlacO-D. The linearized amplicon, as well as the candidate sRNA amplicon were digested using XbaI and ligated together using T4 DNA ligase.
Mutations were made in the chuA or sRNA sequences by using the Quikchange XL mutagenesis kit (Agilent) according to the manufacturer’s instructions. Primers for mutagenesis were designed using the Quikchange primer design program.1
Confirmation of sRNA–chuA Interactions Using the sfGFP 2-Plasmid System
The expression of sfGFP was monitored and quantified with and without candidate sRNAs using a BD FACSCanto II or a BD LSRFortessa™ Special Order Research Product cell analyzer. Fluorescence was measured using a 530/30 nm bandpass filter. Forward scatter (FSC) and side scatter (SSC) were also measured to gate the bacterial population. For each sample, at least 100,000 events were recorded. Data was analyzed using FlowJo software (BD) and statistics were calculated using Prism 8 (GraphPad) to obtain each sample’s mean median fluorescence intensity (mean MFI). p-Values were calculated using a standard two-tailed Student’s t-test.
Plasmids expressing the wild-type or mutant chuA-GFP translational fusion and those expressing candidate sRNAs were co-transformed into E. coli DH5α or TOP10F′. Three colonies from each transformation were purified and grown overnight in LB broth. Overnight cultures are subcultured 1/100 in 0.22 μm filtered LB broth and grown to exponential phase (OD600 = 0.6). Expression of the chuA-GFP translational fusions were induced in TOP10F′ using 200 nM of anhydrotetracycline. These are diluted fivefold in 0.22 μm-filtered PBS, then read on the flow cytometer as described above. Plasmid pJV300 expressing a scrambled sRNA, and pXG1 and pXG0 plasmids expressing GFP and Lux, respectively, were used as controls.
RNA Secondary Structure Prediction
The secondary structure for the 5′UTR of chuA was predicted using the mfold2 and RNAfold webservers (Zuker, 2003; Lorenz et al., 2011). Figures were drawn on RNAStructure version 6.0.1 (Reuter and Mathews, 2010). ARN motifs were detected using custom scripts previously used in Tree et al. (2014).
RNA Sequencing of Bicyclomycin-Treated Cells
Three single colonies of E. coli O157:H7 str. Sakai stx- were grown for 16 h in LB broth. The following day, these subcultured 1/100 in MEM-HEPES supplemented with 0.1% glucose and 250 nM Fe(NO3)3. At an OD600 of 0.75, cultures were split into two (treated and untreated), and 50 μg/mL of bicyclomycin was added to the treated sample. Cultures were incubated for a further 15 min, followed by addition of RNA stop solution (5% water-saturated phenol in ethanol). RNA was extracted using guanidinium thiocyanate-phenol as previously described in Tollervey and Mattaj (1987). Genomic DNA was digested using RQ1 DNase (Promega) and RNA was cleaned using another phenol-chloroform extraction. Total RNA was ribodepleted using the Zymo-Seq RiboFree Total RNA library kit and libraries were prepared using the Illumina NextSeq 500/550 Mid-Output Kit. Samples were 2 × 75 bp paired-end sequenced on an Illumina NextSeq 500 platform. Library preparation and sequencing were performed by the Ramaciotti Centre for Genomics in the University of New South Wales, Sydney, NSW, Australia.
RNA Sequencing Analysis and Identification of Rho-Dependent Transcripts
RNA sequencing data for in vivo analysis was taken from SRA accession number SRP136076. Alignment and DEseq analysis of RNA sequencing output was done using the “align,” “coverage,” “gene-quanti,” and “deseq” modules of READemption v1.0 (Förstner et al., 2014) using default settings.
To identify Rho-terminated transcripts, fastq files were re-aligned to the E. coli O157:H7 str. Sakai genome (accession number: NC_002695.2) using BWA-MEM (v0.7.17) (Li and Durbin, 2010). BAM files were generated using samtools (v.1.10) (Li et al., 2009) and 3′ end read counts were calculated using the genomecov tool from bedtools (v2.27.1) (Quinlan and Hall, 2010). Rho readthrough scores were calculate using the program RhoTerm-Peaks using a window size of 250 nt (Adams et al., 2021).
Data Availability Statement
RNA sequencing datasets are deposited at the NCBI Gene Expression Omnibus (GEO) under accession numbers GSE143631, GSE46118, and GSE197379.
Author Contributions
BS and JT conceived and designed the experimental work, analyzed the data, and wrote the manuscript. BS performed the experimental work. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
BS was supported by a Research Training Program Scholarship from the Australian Government. JT and BS were supported by funding from the National Health and Medical Research Council (GNT1161161). Flow cytometry in this work was performed at the Mark Wainwright Analytical Centre of the University of New South Wales (UNSW) Sydney, which is part funded by the Research Infrastructure Programme of UNSW Sydney.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.821196/full#supplementary-material
ChiX indirectly represses chuA translation. (A,B) (Left) IntaRNA prediction of the chuA–ChiX interaction. Compensatory point mutations predicted to disrupt the interaction are indicated by the arrows. (Right) Fluorescence measurements of wild-type or mutant chuA-sfGFP translational fusions in the presence and absence of wild-type or mutant ChiX overexpression plasmid. Measurements are the mean median fluorescence intensity of three biological replicates.
Secondary structure of the chuA 5′UTR as predicted by RNAfold. The +1 site, CyaR binding site, FourU RNA thermometer and sites where truncations were made are indicated.
Primers, bacterial strains, and plasmids used in this work.
References
- Adams P. P., Baniulyte G., Esnault C., Chegireddy K., Singh N., Monge M., et al. (2021). Regulatory roles of Escherichia coli 5’ UTR and ORF-internal RNAs detected by 3’ end mapping. eLife 10:e62438. 10.7554/eLife.62438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzaldi L. L., Skaar E. P. (2010). Overcoming the heme paradox: Heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78 4977–4989. 10.1128/IAI.00613-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bags A., Neilands J. B. (1987). Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26 5471–5477. 10.1021/bi00391a039 [DOI] [PubMed] [Google Scholar]
- Banerjee R., Weisenhorn E., Schwartz K. J., Myers K. S., Glasner J. D., Perna N. T., et al. (2020). Tailoring a global iron regulon to a uropathogen. mBio 11:e00351-20. 10.1128/mBio.00351-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber M. F., Elde N. C. (2014). Escape from bacterial iron piracy through rapid evolution of transferrin. Science 346 1362–1366. 10.1126/science.1259329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin Y., Chaucheyras-Durand F., Robbe-Masselot C., Durand A., de la Foye A., Harel J., et al. (2013). Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157: H7 in bovine intestinal content. Environ. Microbiol. 15 610–622. 10.1111/1462-2920.12019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Schwartz A., Guillemardet B., Boudvillain M., Figueroa-Bossi N. (2012). A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev. 26 1864–1873. 10.1101/gad.195412.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Richter A. S., Backofen R. (2008). IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24 2849–2856. 10.1093/bioinformatics/btn544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale C. J., Washburn R. S., Tadigotla V. R., Brown L. M., Gottesman M. E., Nudler E. (2008). Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320 935–938. 10.1126/science.1152763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson-Banning K. M., Sperandio V. (2016). Catabolite and oxygen regulation of enterohemorrhagic Escherichia coli virulence. mBio 7:e01852-16. 10.1128/mBio.01852-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celia H., Noinaj N., Zakharov S. D., Bordignon E., Botos I., Santamaria M., et al. (2016). Structural insight into the role of the Ton complex in energy transduction. Nature 538 60–65. 10.1038/nature19757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chareyre S., Mandin P. (2018). Bacterial iron homeostasis regulation by sRNAs. Microbiol. Spectr. 6 1–15. 10.1128/microbiolspec.RWR-0010-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Morita T., Gottesman S. (2019). Regulation of transcription termination of small RNAs and by small RNAs: molecular mechanisms and biological functions. Front. Cell. Infect. Microbiol. 9:201. 10.3389/fcimb.2019.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Cohen P. S. (2015). Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol Spectr. 3:10.1128/microbiolspec.MBP-0006-2014. 10.1128/9781555818883.ch16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran C. P., Podkaminski D., Papenfort K., Urban J. H., Hinton J. C. D., Vogel J. (2012). Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 84 428–445. 10.1111/j.1365-2958.2012.08031.x [DOI] [PubMed] [Google Scholar]
- Cornelissen C. N. (2018). Subversion of nutritional immunity by the pathogenic Neisseriae. Pathog. Dis. 76:ftx112. 10.1093/femspd/ftx112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M. M., Hu Z., Klimko C., Narayanan S., Deberardinis R., Sperandio V. (2014). The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16 759–769. 10.1016/j.chom.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N., Gottesman S. (2009). The crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 191 461–476. 10.1128/JB.01157-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evfratov S. A., Osterman I. A., Komarova E. S., Pogorelskaya A. M., Rubtsova M. P., Zatsepin T. S., et al. (2017). Application of sorting and next generation sequencing to study 5’-UTR influence on translation efficiency in Escherichia coli. Nucleic Acids Res. 45 3487–3502. 10.1093/nar/gkw1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evstatiev R., Gasche C. (2012). Iron sensing and signalling. Gut 61 933–952. 10.1136/gut.2010.214312 [DOI] [PubMed] [Google Scholar]
- Fischbach M. A., Sonnenburg J. L. (2011). Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10 336–347. 10.1016/j.chom.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstner K. U., Vogel J., Sharma C. M. (2014). READemption-a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics 30 3421–3423. 10.1093/bioinformatics/btu533 [DOI] [PubMed] [Google Scholar]
- Fournier C., Smith A., Delepelaire P. (2011). Haem release from haemopexin by HxuA allows Haemophilus influenzae to escape host nutritional immunity. Mol. Microbiol. 80 133–148. 10.1111/j.1365-2958.2011.07562.x [DOI] [PubMed] [Google Scholar]
- Garcia E. C., Brumbaugh A. R., Mobley H. L. T. (2011). Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect. Immun. 79 1225–1235. 10.1128/iai.01222-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyles C. L. (2007). Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85 E45–E62. 10.2527/jas.2006-508 [DOI] [PubMed] [Google Scholar]
- Hagan E. C., Mobley H. L. T. (2009). Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 71 79–91. 10.1111/j.1365-2958.2008.06509.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. I., Skaar E. P. (2012). Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10 525–537. 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J. B., Nataro J. P., Mobley H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2 123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- Karpman D., Ståhl A.-L. (2014). Enterohemorrhagic Escherichia coli pathogenesis and the host response. Microbiol. Spectr. 2 1–15. 10.1128/microbiolspec.EHEC-0009-2013 [DOI] [PubMed] [Google Scholar]
- Kim D., Hong J. S. J., Qiu Y., Nagarajan H., Seo J. H., Cho B. K., et al. (2012). Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet. 8:e1002867. 10.1371/journal.pgen.1002867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouse A. B., Righetti F., Kortmann J., Narberhaus F., Murphy E. R. (2013). RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS One 8:e63781. 10.1371/journal.pone.0063781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMattina J. W., Delrossi M., Uy K. G., Keul N. D., Nix D. B., Neelam A. R., et al. (2017). Anaerobic heme degradation: ChuY is an anaerobilin reductase that exhibits kinetic cooperativity. Biochemistry 56 845–855. 10.1021/acs.biochem.6b01099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMattina J. W., Nix D. B., Lanzilotta W. N. (2016). Radical new paradigm for heme degradation in Escherichia coli O157:H7. Proc. Natl. Acad. Sci. U.S.A. 113 12138–12143. 10.1073/pnas.1603209113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Huang D., Cheung M. K., Nong W., Huang Q., Kwan H. S. (2013). BSRD: a repository for bacterial small regulatory RNA. Nucleic Acids Res. 41 233–238. 10.1093/nar/gks1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R., Bernhart S. H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P. F., et al. (2011). ViennaRNA Package 2.0. Algorithms Mol. Biol. 6 122–128. 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M., Wright P. R., Backofen R. (2017). IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 45 W435–W439. 10.1093/nar/gkx279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E., Gottesman S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99 4620–4625. 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E., Salvail H., Desnoyers G., Arguin M. (2007). Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10 140–145. 10.1016/j.mib.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Miranda R. L., Conway T., Leatham M. P., Chang D. E., Norris W. E., Allen J. H., et al. (2004). Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect. Immun. 72 1666–1676. 10.1128/IAI.72.3.1666-1676.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K., Gottesman S. (2011). Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 82 1545–1562. 10.1111/j.1365-2958.2011.07907.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenthau A., Pogoutse A., Adamiak P., Moraes T. F., Schryvers A. B. (2013). Bacterial receptors for host transferrin and lactoferrin: molecular mechanisms and role in host-microbe interactions. Future Microbiol. 8 1575–1585. 10.2217/fmb.13.125 [DOI] [PubMed] [Google Scholar]
- Nagy G., Dobrindt U., Kupfer M., Emody L., Karch H., Hacker J. (2001). Expression of hemin receptor molecule chuA is influenced by rfaH in uropathogenic E. coli strain 356. Infect. Immun. 69 1924–1928. 10.1128/IAI.69.3.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naville M., Ghuillot-Gaudeffroy A., Marchais A., Gautheret D. (2011). ARNold: a web tool for the prediction of rho-independent transcription terminators. RNA Biol. 8 10–13. 10.4161/rna.8.1.13346 [DOI] [PubMed] [Google Scholar]
- Njoroge J. W., Nguyen Y., Curtis M. M., Moreira C. G., Sperandio V. (2012). Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. mBio 3:e00280-12. 10.1128/mBio.00280-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby-Sherrouse A. G., Murphy E. R. (2013). Iron-responsive bacterial small RNAs: variations on a theme. Metallomics 5 276–286. 10.1039/c3mt20224k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. T., Shan Ho J. Z., Ho B., Ding J. L. (2006). Iron-withholding strategy in innate immunity. Immunobiology 211 295–314. 10.1016/j.imbio.2006.02.004 [DOI] [PubMed] [Google Scholar]
- Peters J. M., Mooney R. A., Grass J. A., Jessen, Tran F., Landick R. (2012). Rho and NusG suppress pervasive antisense transcription in E. coli. Genes Dev. 26 2621–2633. 10.1101/gad.196741.112.The [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M., Mooney R. A., Kuan P. F., Rowland J. L., Keles S., Landick R. (2009). Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 106 15406–15411. 10.1073/pnas.0903846106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter J. S., Mathews D. H. (2010). RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11:129. 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley K. A., Rock J. D., Li Y., Ketley J. M. (2006). Heme utilization in Campylobacter jejuni. J. Bacteriol. 188 7862–7875. 10.1128/JB.00994-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righetti F., Nuss A. M., Twittenhoff C., Beele S., Urban K., Will S., et al. (2016). Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U.S.A. 113 7237–7242. 10.1073/pnas.1523004113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M. S., Fetherston J. D., Létoffé S., Carniel E., Perry R. D., Ghigo J. M. (2001). Identification and characterization of the hemophore-dependent heme acquisition system of Yersinia pestis. Infect. Immun. 69 6707–6717. 10.1128/IAI.69.11.6707-6717.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S., Ramseier T. M., Michotey V., Saier M. H., Garges S. (1995). Effect of the FruR regulator on transcription of the pts operon in Escherichia coli. J. Biol. Chem. 270 2489–2496. 10.1074/jbc.270.6.2489 [DOI] [PubMed] [Google Scholar]
- Santiago-Frangos A., Kavita K., Schu D. J., Gottesman S., Woodson S. A. (2016). C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. U.S.A. 113 E6089–E6096. 10.1073/pnas.1613053113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu D. J., Zhang A., Gottesman S., Storz G. (2015). Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 34 2557–2573. 10.15252/embj.201591569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlyarova N., Shamovsky I., Bharati B. K., Epshtein V., Chen J., Gottesman S., et al. (2016). sRNA-mediated control of transcription termination in E. coli. Cell 167 111–121.e13. 10.1016/j.cell.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura A., Bertin Y., Durand A., Benbakkar M., Forano E. (2021). Transcriptional analysis reveals specific niche factors and response to environmental stresses of enterohemorrhagic Escherichia coli O157:H7 in bovine digestive contents. BMC Microbiol. 21:284. 10.1186/s12866-021-02343-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura A., Bertoni M., Auffret P., Klopp C., Bouchez O., Genthon C., et al. (2018). Transcriptomic analysis reveals specific metabolic pathways of enterohemorrhagic Escherichia coli O157:H7 in bovine digestive contents. BMC Genomics 19:766. 10.1186/s12864-018-5167-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C. M., Hoffmann S., Darfeuille F., Reignier J., Findeiß S., Sittka A., et al. (2010). The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464 250–255. 10.1038/nature08756 [DOI] [PubMed] [Google Scholar]
- Silva I. J., Barahona S., Eyraud A., Lalaouna D., Figueroa-Bossi N., Massé E., et al. (2019). SraL sRNA interaction regulates the terminator by preventing premature transcription termination of rho mRNA. Proc. Natl. Acad. Sci. U.S.A. 116 3042–3051. 10.1073/pnas.1811589116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6:e1000949. 10.1371/journal.ppat.1000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider T. A., Fabich A. J., Conway T., Clinkenbeard K. D. (2009). E. coli O157:H7 catabolism of intestinal mucin-derived carbohydrates and colonization. Vet. Microbiol. 136 150–154. 10.1016/j.vetmic.2008.10.033 [DOI] [PubMed] [Google Scholar]
- Soper T. J., Woodson S. (2008). The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14 1907–1917. 10.1261/rna.1110608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy B. M., Lan R., Tree J. J. (2020). Early termination of the Shiga toxin transcript generates a regulatory small RNA. Proc. Natl. Acad. Sci. U.S.A. 117 25055–25065. 10.1073/pnas.2006730117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Mattaj I. W. (1987). Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 6 469–476. 10.1002/j.1460-2075.1987.tb04777.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A. G., Payne S. M. (1997). Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23 825–833. 10.1046/j.1365-2958.1997.2641628.x [DOI] [PubMed] [Google Scholar]
- Torres A. G., Redford P., Welch R. A., Payne S. M. (2001). TonB-dependent systems of uropathogenic Escherichia coli: Aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69 6179–6185. 10.1128/IAI.69.10.6179-6185.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree J. J., Granneman S., McAteer S. P., Tollervey D., Gally D. L. (2014). Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol. Cell 55 199–213. 10.1016/j.molcel.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J. H., Vogel J. (2007). Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35 1018–1037. 10.1093/nar/gkl1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. G. H., Romby P. (2015). “Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it,” in Advances in Genetics, eds Friedmann T., Dunlap J. C., Goodwin S. F. (Waltham, MA: Academic Press; ). 10.1016/bs.adgen.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Waldminghaus T., Heidrich N., Brantl S., Narberhaus F. (2007). FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 65 413–424. 10.1111/j.1365-2958.2007.05794.x [DOI] [PubMed] [Google Scholar]
- Wang D., McAteer S. P., Wawszczyk A. B., Russell C. D., Tahoun A., Elmi A., et al. (2018). An RNA-dependent mechanism for transient expression of bacterial translocation filaments. Nucleic Acids Res. 46 3366–3381. 10.1093/nar/gky096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. S., Storz G. (2009). Regulatory RNAs in bacteria. Cell 136 615–628. 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters S. A., McAteer S. P., Kudla G., Pang I., Deshpande N. P., Amos T. G., et al. (2017). Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E. EMBO J. 36 374–387. 10.15252/embj.201694639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G. G., Kortmann J., Narberhaus F., Klose K. E. (2014). RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 111 14241–14246. 10.1073/pnas.1411570111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg E. D. (1975). Nutritional Immunity: host’s attempt to withhold iron from microbial invaders. J. Am. Med. Assoc. 231 39–41. 10.1093/ajcn/30.9.1485 [DOI] [PubMed] [Google Scholar]
- Wright P. R., Georg J., Mann M., Sorescu D. A., Richter A. S., Lott S., et al. (2014). CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 42 119–123. 10.1093/nar/gku359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff E. E., Duncan D., Torres A. G., Mills M., Maase K., Payne S. M. (1998). Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28 1139–1152. 10.1046/j.1365-2958.1998.00873.x [DOI] [PubMed] [Google Scholar]
- Xu J., Bjursell M. K., Himron J., Deng S., Carmichael L. K., Chaing H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299 2074–2076. 10.1126/science.1080029 [DOI] [PubMed] [Google Scholar]
- Zambolin S., Clantin B., Chami M., Hoos S., Haouz A., Villeret V., et al. (2016). Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Nat. Commun. 7:11590. 10.1038/ncomms11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Schu D. J., Tjaden B. C., Storz G., Gottesman S. (2013). Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J. Mol. Biol. 425 3678–3697. 10.1016/j.jmb.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Aboulwafa M., Saier M. H. (2014). Regulation of crp gene expression by the catabolite repressor/activator, Cra, in Escherichia coli. J. Mol. Microbiol. Biotechnol. 24 135–141. 10.1159/000362722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiefka A., Kohn H., Widger W. R. (1993). Transcription termination factor rho: the site of bicyclomycin inhibition in Escherichia coli. Biochemistry 32 3564–3570. 10.1021/bi00065a007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ChiX indirectly represses chuA translation. (A,B) (Left) IntaRNA prediction of the chuA–ChiX interaction. Compensatory point mutations predicted to disrupt the interaction are indicated by the arrows. (Right) Fluorescence measurements of wild-type or mutant chuA-sfGFP translational fusions in the presence and absence of wild-type or mutant ChiX overexpression plasmid. Measurements are the mean median fluorescence intensity of three biological replicates.
Secondary structure of the chuA 5′UTR as predicted by RNAfold. The +1 site, CyaR binding site, FourU RNA thermometer and sites where truncations were made are indicated.
Primers, bacterial strains, and plasmids used in this work.
Data Availability Statement
RNA sequencing datasets are deposited at the NCBI Gene Expression Omnibus (GEO) under accession numbers GSE143631, GSE46118, and GSE197379.