Abstract
Inborn errors of metabolism (IEM) comprise a large class of recessive genetic diseases involving disorders of cellular metabolism that tend to be caused by missense mutations in which a single incorrect amino acid is substituted in the polypeptide chain. Cystathionine beta-synthase (CBS) deficiency is an example of an IEM that causes large elevations of blood total homocysteine levels resulting in phenotypes in several tissues. Current treatment strategies involve dietary restriction and vitamin therapy, but these are only partially effective and do not work in all patients. Over 85% of the described mutations in CBS deficient patients are missense mutations in which the mutant protein fails to fold into an active conformation. The ability of CBS to achieve an active conformation is affected by a variety of intracellular protein networks including the chaperone system and the ubiquitin/proteasome system, collectively referred to as the proteostasis network. Proteostasis modulators are drugs that perturb various aspects of these networks. In this article, we will review the evidence that modulation of the intracellular protein-folding environment can be used as a potential therapeutic strategy to treat CBS deficiency and discuss the pros and cons of such a strategy.
Keywords: homocystinuria, chaperone, protein folding, proteasome, sulfur metabolism, enzyme
Introduction: Missense Mutations and Inborn Errors of Metabolism
Inborn errors of metabolism comprise a large class of single gene genetic diseases involving disorders of cellular metabolism. The majority are caused by recessive loss-of-function mutations in genes that code for enzymes that catalyze biologically important reactions. Although each specific inborn error of metabolism is rare, taken together, they represent a significant source of birth defects. It is estimated that, on average, a genetically caused inborn error of metabolism occurs in 1/2500 births (Applegarth et al. 2000). Given the devastating nature of many of these diseases, novel treatment methods would be of great benefit both to patients and society as a whole.
The most common genetic alterations causing inborn errors are missense mutations that result in the production of proteins with single amino acid substitutions. Examination of the Human Gene Mutation Database indicates that approximately 2/3 of all mutant alleles found in inborn errors are of the missense variety. For example, missense mutations make up 62.4% of the alleles described in phenylketonuria patients, 65% of the alleles seen in ornithine transcarbamylase deficient patients, and 67% of the alleles seen in cystathionine beta-synthase (CBS) deficient patients (Stenson et al. 2003). Most disease-causing missense mutations do not target key catalytic residues in proteins but rather cause problems in protein stability and aggregation (DePristo et al. 2005). It is thought that missense mutations affect protein stability by preventing it from achieving its lowest-free energy native state. These trapped misfolded structural intermediates are either degraded or form large molecular weight aggregates (Bross et al. 1999). In theory, drugs that could reverse protein misfolding and promote proper folding of missense mutant proteins would be of great utility in the treatment of a wide variety of genetic diseases.
Cystathionine β-Synthase (CBS) Deficiency
A prototypical example of an inborn error of metabolism is CBS deficiency (Mudd et al. 2001). It is the most common inborn error of sulfur metabolism and is the cause of classical homocystinuria, a condition characterized by very high levels of plasma total homocysteine (tHcy). CBS deficiency is estimated to occur in about 1/100,000 births world-wide, although rates vary significantly in different countries (Weber Hoss et al. 2020). CBS catalyzes the condensation of homocysteine with serine to form cystathionine, which is the first step in the de novo production of cysteine. In healthy adults, tHcy concentration in plasma ranges from 5 to 15 μM, but untreated patients with CBS deficiency often have tHcy in excess of 200 μM. CBS-deficient patients suffer at an early age from various pathologies including thrombosis, osteoporosis, mental retardation, and dislocated lenses (Mudd et al. 1985). The major cause of mortality and morbidity in these patients is thrombosis. Current treatment strategies involve dietary restriction and vitamin therapy, but these are only partially effective and do not work in all patients (Morris et al. 2017; Morrison et al. 2021). There are several other treatment strategies under investigation including enzyme replacement (Majtan et al. 2018), gene therapy (Lee et al. 2019), and proteostasis modulation, which is the subject of this review.
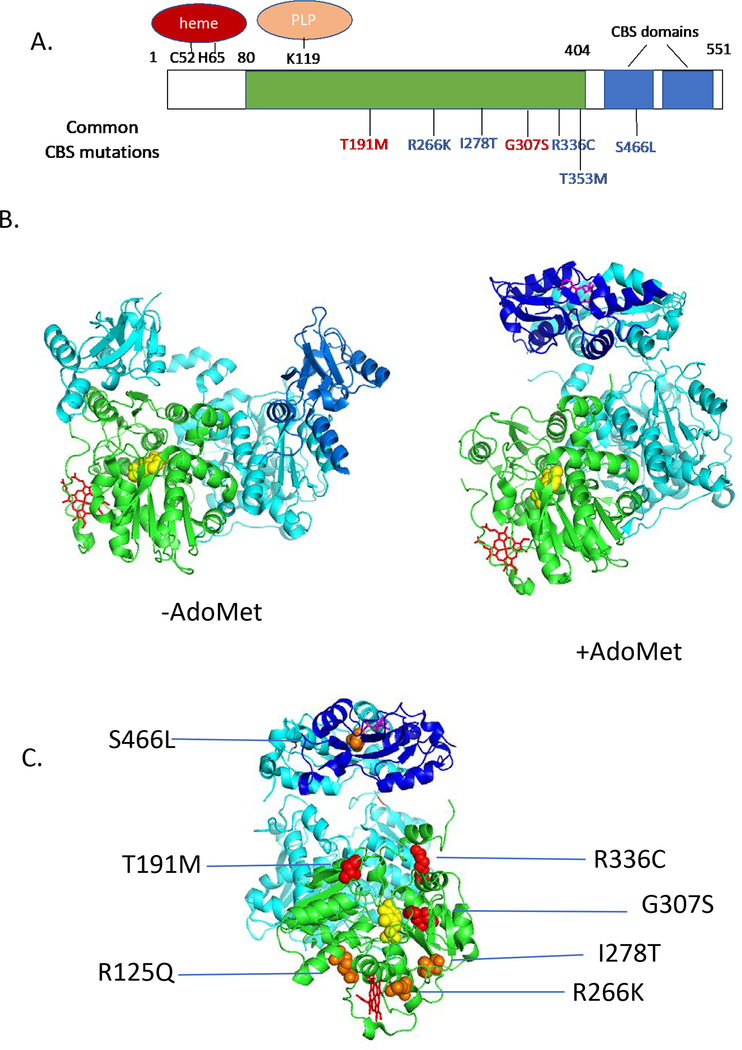
Human CBS (hCBS) encodes a 63 kDa protein that is 551 amino acids in length and is primarily found as a homotetramer (Kraus et al. 1978). The protein contains three functional domains: An N-terminal domain that binds heme (aa 1–80, a core domain (aa 80–404) that contains the catalytic site which binds pyridoxal phosphate (PLP) and shares similarity with other PLP-utilizing enzymes, and a C-terminal regulatory domain that is responsible for allosteric regulation by S-adenosylmethionine (aa 404–551) (Figure 1A). Heme does not appear to play a direct catalytic role in the hCBS reaction, as CBS proteins from lower eukaryotes lack this domain and function well. It has been proposed that heme may play a regulatory role in the enzyme, perhaps integrating hCBS activity with the cell intracellular redox state (Taoka et al. 2002; Taoka et al. 1998). However, there is some dispute as to whether this actually occurs in vivo, as the enzyme is still highly active when the heme is replaced by non-metal containing porphyrins, including biologically redox inert cobalt (Majtan et al. 2011; Smith et al. 2011). Rather, the data suggests that the heme moiety is more likely to play a role in protein stability, as others have proposed that the heme binding domains main role is to enhance folding and stability of the enzyme (Majtan et al. 2011; Majtan et al. 2008). The middle catalytic domain binds PLP at a specific lysine residue (K119). The PLP catalyzes the hCBS reaction by binding serine, forming an aminoacrylate intermediate, which is then attacked by homocysteine (Banerjee and Zou 2005). The C-terminal domain of the enzyme contains a repeated structural motif called the “CBS domain”, which is found in several classes of proteins including metabolic enzymes, calcium ion channels, and AMP-regulated protein kinases (Ignoul and Eggermont 2005). The CBS domain is always found in pairs in proteins and often forms structures that binds base-containing ligands, such as AMP, ATP, IMP, or in the case of CBS itself, S-adenosylmethionine.
Figure 1.
Human CBS structure and mutations. A. One-dimensional drawing showing homology domains of CBS. Green shows region similar to other PLP-containing enzymes, blue shows CBS domains. Above the rectangle shows location of key residues binding heme and PLP. Below show some patient derived mutations. Red mutations are “non-rescuable”, while blue mutations are rescuable. B. Structure of human CBS (Δ516–525) as determined in the presence and absence of AdoMet. One subunit of the dimer is shaded as in A., while the other is shaded in teal. PLP is shown in yellow, heme in red, and AdoMet in magenta. C. Patient derived mutants mapped onto AdoMet activated structure (rotated 90° on vertical axis compared to image in A.). Red mutations are non-responsive, while orange are pyridoxine-rescuable.
The three-dimensional structure of full length hCBS has not been determined, but the structure of a form missing a 9 amino acid loop region (Δ516–525) has been solved both in the presence and absence of AdoMet (Figure 2b) (Ereno-Orbea et al. 2013, 2014). The asymmetric unit of the crystal is a dimer and the relationship between the two molecules that make up the dimer changes dramatically when AdoMet is bound, allowing better access to the enzyme active-site by moving the regulatory domain out of the way.
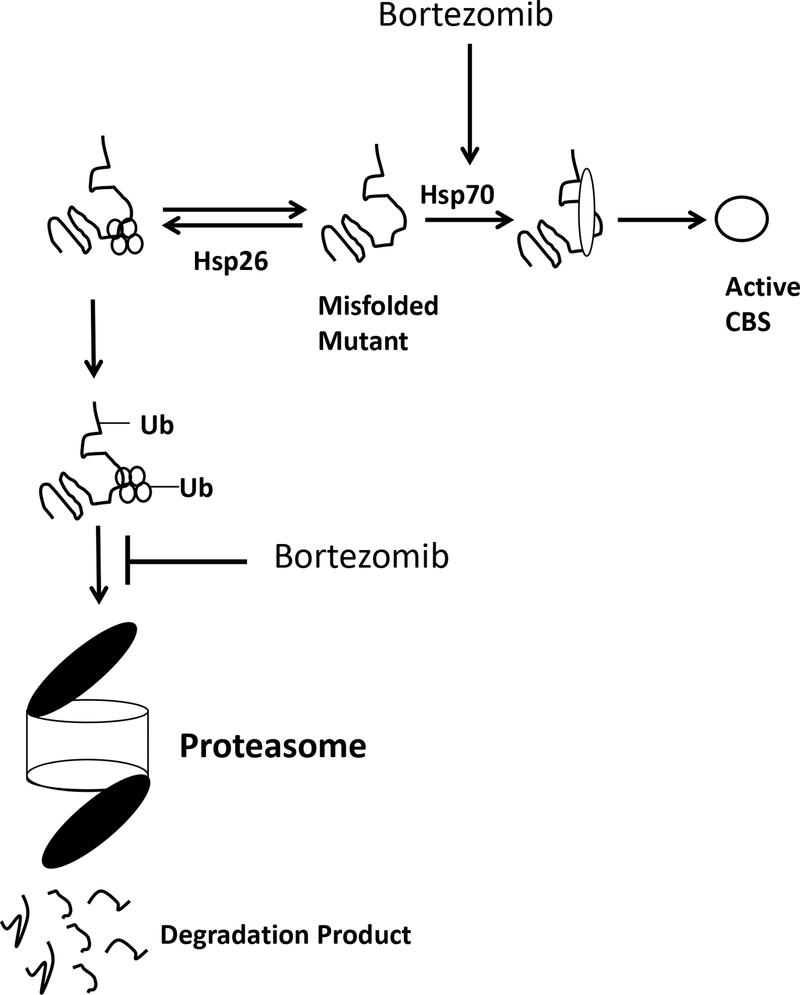
Figure 2.
Equilibrium model for hCBS folding in S. cerevisiae. Missense mutant proteins are acted on by two competing systems. If bound by small heat shock proteins, like Hsp26, missense mutant proteins are ubiquitinated and directed to the proteasome for degradation. If bound by Hsp70, mutant proteins are refolded into an active conformation. Proteasome inhibitors alter the equilibrium by both inhibiting the degradation arm, and stimulating the refolding arm by increasing Hsp70 levels.
In patients with CBS deficiency, at least 924 mutant alleles have been characterized with 85% of these being missense mutations (see http://cbs.lf1.cuni.cz/index.php). The most frequently observed missense mutations are: p.I278T (16.6%), p.T191M (16.1%), p.G307S (9.5%), p.R336C (9.0%), p.D444N (1.8%), p.R125Q (1.5%), p.E144K (1.4%), p.T353M (1.3%), and p.R266K (1%). An examination of the locations of patient derived missense mutations on the hCBS crystal structure shows that most of the mutations are located some distance from the PLP catalytic core of the enzyme (Figure 1C), suggesting that they do not directly interfere with the structure of the catalytic region.
In many cases, missense mutant hCBS enzymes have been functionally analyzed by either studying the protein in patient-derived fibroblast cell lines (Janosik et al. 2001b; Kluijtmans et al. 1999), or by expression of the mutant protein in heterologous expression systems such as E. coli, S. cerevisiae, or mammalian cell lines (Kozich and Kraus 1992; Kruger and Cox 1994; Maclean et al. 2002). The majority of missense mutant proteins that have been analyzed are either unstable, stable but form non-functional aggregates, or are simply catalytically impaired. However, it is important to point out that the behavior of the same mutant hCBS protein can vary significantly depending on the expression system. For example, p.S466L hCBS was first identified in a homocystinuric patient that presented later in life with unexplained thrombosis, implying that the mutation significantly reduces CBS activity (Gaustadnes et al. 2000). However, when expressed in either E. coli, S. cerevisiae, or Chinese hamster fibroblasts, the cell lysates exhibit near wild-type levels of enzyme activity even in the absence of AdoMet (Gupta et al. 2008; Maclean et al. 2002). Studies on the purified enzyme show that the S466L enzyme actually has enzyme activity as high as Adomet-stimulated purified wild-type CBS protein and, when AdoMet is added, it binds but does not raise enzyme activity (Janosik et al. 2001a). But, when human p.S466L is expressed in the liver of a transgenic mouse lacking endogenous mouse Cbs (Gupta et al. 2008), the enzyme is unstable and degraded resulting in the mice having less than 10% of the CBS activity of control transgenic mice expressing wild-type hCBS. This finding shows that the intracellular environment where a protein is produced can have a dramatic effect on the outcome of protein folding in certain circumstances.
Rescue with Pyridoxine
The first hint that certain hCBS mutant proteins might be “fixable” came from clinical studies that revealed that some CBS deficient patients responded to large doses (100–200 mg/day) of oral pyridoxine with a significant lowering of urinary homocysteine (Barber and Spaeth 1969). In families with multiple affected children, there was concordance of the pyridoxine response (Carson and Carre 1969), suggesting that it was determined genetically. Since pyridoxine is the precursor of PLP, the most obvious hypothesis was that certain mutations might cause the enzyme to bind PLP with reduced affinity, and that excess pyridoxine would alleviate this effect. However, despite many efforts, this has not been convincingly demonstrated. Addition of pyridoxine to tissue derived protein extracts from pyridoxine responsive patients results in either no or extremely small increases in hCBS activity (Fowler et al. 1978; Seashore et al. 1972). With the isolation of the hCBS gene, sequencing has identified several different mutations associated with pyridoxine-response in humans (Table 1), the most common being the p.I278T mutation (Hu et al. 1993; Kim et al. 1997; Sebastio 1995). However, expression of the p.I278T human CBS protein in either bacteria, yeast, human patient-derived fibroblasts, or mouse liver fail to exhibit any response to PLP addition (Chen et al. 2006; Kluijtmans et al. 1999; Kozich and Kraus 1992). Furthermore, titration experiments in which PLP is first stripped away from the purified enzyme, did not reveal any difference in the binding affinity between WT and p.I278T hCBS (Chen et al., 2006). When all of these studies are examined in total, the only strong conclusion that emerges is that pyridoxine-responsive alleles tend to have more residual enzyme activity than non-responsive ones. Thus, the exact mechanism by which pyridoxine lowers tHcy in humans is still unknown.
Table 1.
Pyridoxine-responsive CBS mutations*
| Mutation | Reference(s) | Notes |
|---|---|---|
| c.833T>C (p.I278T) | (Kluijtmans et al. 1999) (Gaustadnes et al. 2002) |
Most common allele. Vast majority of homozygous patients are reported as responsive. |
| c.797G>A (p.R266K) | (Kim et al. 1997) | Common in Norway. |
| c.1105C>T (p.R369C) | (Janoik et al. 2009) | Found as compound het in three patients. All were responsive. |
| c.456C>G (p.I152M) | (Kluijtmans et al. 1999) | Homozygous in Dutch patient. |
| c.1111G>A (p.V371M) | (Gaustadnes et al. 2002) | Found as compound het with E144K (non-responsive). |
| c.146C>T (p.49L) | (Poloni et al. 2018) | Found as compound het with two different non-responsive alleles. |
| c.676G>A (p.A226T) | (Kruger et al. 2003) | Found as compound het with T353M (non-responsive). |
| c.1616T>C (L539C) | (Aral et al. 1997) | Homozyous in a French patient. |
| c.1150G>A (K384E) | (Aral et al. 1997) | Homozygous in a French patient. |
| c.1330G>A (D444N) | (Kluijtmans et al. 1996) | Homozygous in a Dutch patient. |
| c.341C>T (A114V) | (de Franchis et al. 1999) | Compound het with three different alleles. |
| c.374G>A (R125Q) | (Sebastio 1995) | Partial response in homozygote. |
includes only alleles in which pyridoxine status has been determined for patients either homozygous for the mutation, or are a compound heterozygote with a known pyridoxine non-responsive allele.
Genetic rescue of mutant CBS enzymes
The first unambiguous evidence showing it was possible to restore significant enzymatic function to a patient-derived mutant hCBS protein came from studies using a yeast functional complementation assay (Kruger and Cox 1994). The basis of this assay is that wild-type hCBS can correct the cysteine auxotrophy in a yeast strain with a deletion of the endogenous yeast CBS gene (CYS4). This assay was effectively used to characterize the functional effects of over 20 unique patient-identified hCBS mutations (Kim et al. 1997; Kruger et al. 2003; Shan and Kruger 1998). The key discovery was the serendipitous finding that two different PCR-induced point mutation that prematurely truncated p.V168M hCBS at amino acid positions 409 or 411 could rescue the cysteine auxotrophy and restore substantial enzyme activity (Shan and Kruger 1998). The truncated enzyme was active, but no longer responded to stimulation of enzyme activity by addition of AdoMet. It was also shown that the truncating p.W409Stop allele could suppress at least eight other patient derived CBS alleles when combined in cis. In a follow-up study, specific missense mutations in the C-terminal domain were found to have similar effects (Shan et al. 2001). Taken together, these findings suggested that C-terminal domain of hCBS acts to inhibit enzyme function and that this inhibition is relieved by binding of AdoMet, consistent with the crystallographic data described above. In addition, these studies showed it was possible to “reactivate” a mutant hCBS protein by introducing a second mutation in the C-terminal region of the protein.
Chemical chaperone rescue
The ability to restore function of mutant hCBS by a cis-acting mutation, hinted that it also might be possible to restore function in trans via interaction with a small molecule (i.e. drug). In the course of screening for such molecules, the Kruger lab made a second serendipitous discovery, specifically that the solvent compound dimethylsulfoxide (DMSO) could partially rescue the function of yeast carrying the p.I278T mutation when added to the media at concentrations of 300 mM (Singh et al. 2007). DMSO is a member of a class of low molecular weight compounds called chemical chaperones, osmolytes that can affect the process of protein folding (Morello et al. 2000). The exact mechanism by which chemical chaperones affect protein-folding is unclear, although it is hypothesized that they help amino acid chains reach a stable conformation by stabilizing hydrophobic residues on the surface of proteins (Leandro and Gomes 2008). To test this idea further, Kruger and colleagues examined several other small osmolyte molecules including glycerol, trimethylamine-N-oxide, proline and sorbitol. All of the compounds could rescue at least one of the seven mutant hCBS proteins tested, although each mutant had their own unique “rescue” profile. The effects of chemical chaperone addition appeared to be direct, as the addition of osmolyte chaperones could increase specific activity of p.I278T CBS produced in an in vitro transcription/translation reaction (Singh et al. 2007).
Several other groups have now confirmed and extended these findings. The Kraus lab examined the effects of ethanol, TMAO, or DMSO on eight different patient-derived missense mutant CBS proteins expressed in E. coli (Majtan et al. 2010). They found that all eight mutations they tested could be rescued by addition of at least one of the three molecules tested. The mean amount of increased activity was 214%, ranging from a low of 35% to a high of 836%. Interestingly, while many of these mutant hCBS proteins had previously proved difficult to purify from E. coli, chaperone addition during growth reversed this effect. These chaperone-treated purified proteins also exhibited absorbance and thermostability profiles quite similar to wild-type hCBS. The Kozich lab examined 27 different human hCBS alleles expressed in E. coli for rescue with either the osmolytes betaine, taurine, and glycerol, or the heme precursor δ-aminolevulinic (ALA) acid (Kopecka et al. 2011). They found that fourteen of the mutant alleles responded by at least 30% increase in the amount of correctly assembled tetramers and enzymatic activity when either 0.5 mM ALA, 100 mM betaine, and/or 750 mM glycerol was added to the media. Examination of the characteristics of suppressible mutations showed that solvent exposed amino acids were more likely to be rescued than buried mutations. In a follow-up study, the Kozich lab showed that it was also possible to rescue some mutant human hCBS alleles expressed in mammalian cells (Melenovska et al. 2015). Specifically, they showed that addition of either 4-phenylbutyric acid (4-PBA), aminooxyacetic acid (AOAA), or heme arginate (HA) could increase specific activity of certain missense hCBS alleles. The most impressive increase was the combination of HA and the R125Q mutation, which resulted in a 900% increase in enzyme activity. The authors speculate that because R125Q is located near the heme binding pocket, addition of HA may help keep the pocket occupied, thus aiding protein folding. This idea is supported by studies indicating that heme is important for proper folding and stability of the hCBS molecule (Majtan et al. 2008; Oliveriusova et al. 2002).
Proteostasis Modulation and Proteasome inhibition
Large multi-domain proteins produced inside the cell often require help from other proteins in order to fold into the proper conformation. These helper proteins are known as molecular chaperones, key components of the cellular proteostasis network (Balch et al. 2008). Proteostasis is the concept that there are competing and integrated biological pathways within cells that control the biogenesis, folding, trafficking, and degradation of proteins (Powers et al. 2009). This network integrates both the chaperones that aid in stable protein folding, and the proteasome system of protein degradation for determining steady state protein levels (Balch et al. 2008). It has been proposed that modulation of the cellular proteostasis network may be useful in the correction of diseases associated with protein misfolding.
The first evidence that modulation of proteostasis might be useful in CBS deficiency came from studies in S. cerevisiae, where it was shown that yeast expressing p.I278T hCBS could be functionally rescued by addition of ethanol to the media (Singh and Kruger 2009). This rescue was associated with both an increase in enzyme activity and an increase in steady-state hCBS levels. Ethanol is a polar molecule and is normally not considered to be a chemical chaperone, but is known to induce the heat shock stress response in S. cerevisiae. The Kruger lab showed that addition of ethanol induced the Hsp70 and Hsp104 heat shock proteins and that deletion of the Hsp70 encoding gene SSA2, eliminated the ability of ethanol to rescue. They also showed that expression of p.I278T but not wild-type hCBS caused a dramatic decrease in steady-state Hsp26 levels and that this loss was eliminated when ethanol was added to the media. Additionally, deletion of HSP26 resulted in p.I278T rescue. A model to explain these observations is an equilibrium model for p.I278T hCBS in which there is a competition between the protein degradation pathway mediated by a Hsp26/ubiquitin/proteasome machine and Hsp70-mediated mediated protein folding pathway (Figure 2). A follow-up study showed that several other patient-derived hCBS missense mutation-containing proteins could be rescued by ethanol and/or deletion of the HSP26 gene (Singh et al. 2010).
Proteasome inhibitors, such as bortezomib, are a class of drugs that are used clinically to treat humans with multiple myeloma and other cancers (Orlowski and Kuhn 2008). They work by inhibiting the proteasome which is the cellular machine that is responsible for the breakdown of misfolded proteins. Besides inhibiting the proteasome, they also induce Hsp70 in both yeast and mammalian cells (Kim and Li 1999; Lee and Goldberg 1998). Initially, the Kruger lab demonstrated that bortezomib could restore function to certain hCBS alleles in both S. cerevisiae and human fibroblasts (Singh et al. 2010). In the same study, they also demonstrated that a single i.v. injection of 750 μg/kg of bortezomib in a mouse expressing human p.I278T hCBS and deleted for endogenous mouse Cbs (Tg-I278T Cbs−/−) could increase steady state levels of p.I278T and increase liver hCBS activity 4.3-fold compared to untreated animals. In a later study, they showed that significantly more robust p.I278T hCBS rescue could be achieved by delivering bortezomib (490 μg/kg/day) subcutaneously using an osmotic pump (Gupta et al. 2013). In bortezomib-treated animals, median liver hCBS activity increased from 40.5 to 513 units, median kidney hCBS activity increased from 4.55 to 267 units, and median tHcy decreased from 329 to 75 μM. This was accompanied by a large increase in the steady-state level of p.I278T hCBS protein. In the same study, they also reported that a second proteasome inhibitor, ONX0914, given orally at 40 mg/kg/day had similar effects. One perplexing feature of these experiments, was that there was significant mouse to mouse variability with regards to the effectiveness of the treatment. Some of the treated mice had liver CBS activity levels similar to that observed in control mice expressing wild-type hCBS, while other mice showed no increase in activity. Microarray analysis showed that strong response was correlated with high levels of SerpinA6 mRNA in the liver, which encodes a corticosteroid binding protein that is secreted into the serum. How SerpinA6 levels and refolding of p.I278T hCBS are mechanistically related is still unknown.
Four additional missense mutant containing CBS proteins have been tested for rescue by various proteasome inhibitors in mice. Mice expressing p.S466L hCBS had their serum tHcy drop from a mean of 145 μM to 15 μM, and their liver CBS activity increase from 25 units to 1290 units, when treated with the oral proteasome inhibitor ONX0914 (Gupta et al. 2013). The p.R266K allele, which was originally found in several pyridoxine responsive patients from Norway (Kim et al. 1997), showed excellent response to bortezomib treatment, with serum tHcy dropping from 197 μM to 6.7 μM, their mean liver CBS activity increase from 41 to 1033 units. The p.R336C allele encodes a pyridoxine non-responsive allele which is the most common allele in Qatar, which has the highest rate of CBS deficiency in the world. The Tg-R336C Cbs−/− mouse model shows excellent rescue by bortezomib with mean liver CBS activity increasing from 134 to 1346 units (Gupta et al. 2019). The one allele that was not rescuable by proteasome inhibition in mice was p.G307S hCBS which is found primarily in non-pyridoxine responsive patients of Celtic ancestry (Gupta et al. 2018). Interestingly, this mutation, unlike the four suppressible mutations, is located near the entrance to the active-site of the enzyme (Figure 1c).
Translation to the clinic: Hurdles that must be overcome
Overall, the data described above indicates that it is possible to restore some level of CBS enzyme function to most mutant-containing hCBS proteins by manipulation of the intracellular protein-folding milieu. However, there are several obstacles that must be overcome to move these findings into the clinic. With regards to chemical chaperones, the biggest hurdle is simply achieving in vivo concentrations required to see an effect. All of the treatments that substantially increased CBS enzyme activity required chaperone concentrations in the range of 10–100 mM, which are probably well outside what is possible to achieve in human patients. To give some perspective, the most abundant amino acid in plasma is glutamine and this is found in the range of 400–600 μM. Proteasome inhibitors are potentially more promising in that they appear to function at concentrations that are typically used in the clinic. For example, a cancer patient undergoing bortezomib treatment typically will get an infusion of 1.3 mg/m2 twice a week, while the mice described above were receiving the equivalent of 1.47 mg/m2 per day for three or four days in a row. However, it must be stressed that bortezomib clearly has toxicity associated with it, which is why it is given in a specific dosing regimen involving two weeks on the drug followed by a week off the drug for four cycles, followed by five cycles in which the drug is only given once a week for two weeks followed by a 13-day rest period. Toxicities include fatigue, peripheral neuropathy, hypotension, cardiac toxicity, pulmonary toxicity, gastrointestinal toxicity, thrombocytopenia/neutropenia, and hepatoxicity. While these toxicities may be tolerable for cancer patients with a high risk of death, they are probably not going to be tolerable to patients with CBS deficiency who would have to be on treatment for a lifetime. One-way potential way to overcome this issue might be to use proteasome inhibitors at lower doses than are typically used in cancer patients. Whether a lower-dose window could be used to allow reactivation of mutant CBS protein, without serious toxicity is not currently known.
It should also be noted that proteasome inhibitors are not the only drugs that could be used to affect the liver proteostasis environment. There are a large number of different small molecules that have been shown to affect proteostasis, some of which exhibit minimal toxicity. These drugs include Ca2+ channel blockers, heat shock protein inducers, cyclooxygenase inhibitors, PPARγ agonists, and histone deacetylases (Kieran et al. 2004; Ong et al. 2010; Seemann et al. 2020; Yang et al. 2013). Future efforts should focus on screening these molecules using functional assays to determine if any of them might be effective in increasing residual CBS activity of mutant hCBS alleles.
Acknowledgments
Declarations
This work was funded by a grant from by grant DK101404 from the National Institutes of Health, and an appropriation from the Commonwealth of Pennsylvania. The author declares no conflict of interest.
References
- Applegarth DA, Toone JR, Lowry RB (2000) Incidence of inborn errors of metabolism in British Columbia, 1969–1996. Pediatrics 105: e10. 10.1542/peds.105.1.e10. [DOI] [PubMed] [Google Scholar]
- Aral B, Coude M, London J, Aupetit J, Chasse JF, Zabot MT, Chadefaux-Vekemans B, Kamoun P (1997) Two novel mutations (K384E and L539S) in the C-terminal moiety of the cystathionine beta-synthase protein in two French pyridoxine-responsive homocystinuria patients. Human Mutation 9: 81–2. . [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319: 916–9. 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Banerjee R, Zou CG (2005) Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor protein. Archives of Biochemistry and Biophysics 433: 144–156. 10.1016/j.abb.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Barber GW, Spaeth GL (1969) The successful treatment of homocystinuria with pyridoxine. J Pediatr 75: 463–78. 10.1016/s0022-3476(69)80274-x. [DOI] [PubMed] [Google Scholar]
- Bross P, Corydon TJ, Andresen BS, Jorgensen MM, Bolund L, Gregersen N (1999) Protein misfolding and degradation in genetic diseases. Hum. Mutat. 14: 186–98. . [DOI] [PubMed] [Google Scholar]
- Carson NA, Carre IJ (1969) Treatment of homocystinuria with pyridoxine. A preliminary study. Arch Dis Child 44: 387–92. 10.1136/adc.44.235.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang L, Fazlieva R, Kruger WD (2006) Contrasting behaviors of mutant cystathionine beta-synthase enzymes associated with pyridoxine response. Hum Mutat 27: 474–82. 10.1002/humu.20320. [DOI] [PubMed] [Google Scholar]
- de Franchis R, Kraus E, Kozich V, Sebastio G, Kraus JP (1999) Four novel mutations in the cystathionine beta-synthase gene: effect of a second linked mutation on the severity of the homocystinuric phenotype. Hum Mutat 13: 453–7. . [DOI] [PubMed] [Google Scholar]
- DePristo MA, Weinreich DM, Hartl DL (2005) Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet 6: 678–87. 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- Ereno-Orbea J, Majtan T, Oyenarte I, Kraus JP, Martinez-Cruz LA (2013) Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration. Proc Natl Acad Sci U S A 110: E3790–9. 10.1073/pnas.1313683110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ereno-Orbea J, Majtan T, Oyenarte I, Kraus JP, Martinez-Cruz LA (2014) Structural insight into the molecular mechanism of allosteric activation of human cystathionine beta-synthase by S-adenosylmethionine. Proc Natl Acad Sci U S A 111: E3845–52. 10.1073/pnas.1414545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler B, Kraus J, Packman S, Rosenberg LE (1978) Homocystinuria. Evidence for three distinct classes of cystathionine beta-synthase mutants in cultured fibroblasts. Journal of Clinical Investigation 61: 645–53. 10.1172/JCI108976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaustadnes M, Rudiger N, Rasmussen K, Ingerslev J (2000) Intermediate and severe hyperhomocysteinemia with thrombosis: a study of genetic determinants. Thromb. Haemost. 83: 554–8. [PubMed] [Google Scholar]
- Gaustadnes M, Wilcken B, Oliveriusova J, McGill J, Fletcher J, Kraus JP, Wilcken DE (2002) The molecular basis of cystathionine beta-synthase deficiency in Australian patients: genotype-phenotype correlations and response to treatment. Hum Mutat 20: 117–26. 10.1002/humu.10104. [DOI] [PubMed] [Google Scholar]
- Gupta S, Gallego-Villar L, Wang L, Lee HO, Nasrallah G, Al-Dewik N, Haberle J, Thony B, Blom HJ, Ben-Omran T, Kruger WD (2019) Analysis of the Qatari R336C cystathionine beta-synthase protein in mice. J Inherit Metab Dis 42: 831–838. 10.1002/jimd.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kelow S, Wang L, Andrake M, Dunbrack RL Jr., Kruger WD (2018) Mouse modeling and structural analysis of the p.G307S mutation in human cystathionine beta-synthase (CBS) reveal effects on CBS activity but not stability. J Biol Chem 293: 13921–13931. 10.1074/jbc.RA118.002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Wang L, Anderl J, Slifker MJ, Kirk C, Kruger WD (2013) Correction of Cystathionine beta-Synthase Deficiency in Mice by Treatment with Proteasome Inhibitors. Hum Mutat 34: 1085–93. 10.1002/humu.22335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Wang L, Hua X, Krijt J, Kozich V, Kruger WD (2008) Cystathionine β-synthase p.S466L mutation causes hyperhomocysteinemia in mice. Hum Mutat 29: 1048–54. 10.1002/humu.20773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu FL, Gu Z, Kozich V, Kraus JP, Ramesh V, Shih VE (1993) Molecular basis of cystathionine beta-synthase deficiency in pyridoxine responsive and nonresponsive homocystinuria. Human Molecular Genetics 2: 1857–60. 10.1093/hmg/2.11.1857. [DOI] [PubMed] [Google Scholar]
- Ignoul S, Eggermont J (2005) CBS domains: structure, function, and pathology in human proteins. Am J Physiol Cell Physiol 289: C1369–78. 10.1152/ajpcell.00282.2005. [DOI] [PubMed] [Google Scholar]
- Janoik M, Sokolova J, Janosikova B, Krijt J, Klatovska V, Kozich V (2009) Birth Prevalence of Homocystinuria in Central Europe: Frequency and Pathogenicity of Mutation c.1105C > T (p.R369C) in the Cystathionine Beta-Synthase Gene. Journal of Pediatrics 154: 431–437. 10.1016/j.jpeds.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosik M, Kery V, Gaustadnes M, Maclean KN, Kraus JP (2001a) Regulation of human cystathionine beta-synthase by S-adenosyl-L-methionine: evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry 40: 10625–10633. 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- Janosik M, Oliveriusova J, Janosikova B, Sokolova J, Kraus E, Kraus JP, Kozich V (2001b) Impaired heme binding and aggregation of mutant cystathionine beta-synthase subunits in homocystinuria. Am J Hum Genet 68: 1506–13. 10.1086/320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieran D, Kalmar B, Dick JR, Riddoch-Contreras J, Burnstock G, Greensmith L (2004) Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nature Medicine 10: 402–5. 10.1038/nm1021. [DOI] [PubMed] [Google Scholar]
- Kim CE, Gallagher PM, Guttormsen AB, Refsum H, Ueland PM, Ose L, Folling I, Whitehead AS, Tsai MY, Kruger WD (1997) Functional modeling of vitamin responsiveness in yeast: a common pyridoxine-responsive cystathionine beta-synthase mutation in homocystinuria. Hum Mol Genet 6: 2213–21. 10.1093/hmg/6.13.2213. [DOI] [PubMed] [Google Scholar]
- Kim D, Li GC (1999) Proteasome inhibitors lactacystin and MG132 inhibit the dephosphorylation of HSF1 after heat shock and suppress thermal induction of heat shock gene expression. Biochem Biophys Res Commun 264: 352–8. 10.1006/bbrc.1999.1371. [DOI] [PubMed] [Google Scholar]
- Kluijtmans LA, Boers GH, Kraus JP, van den Heuvel LP, Cruysberg JR, Trijbels FJ, Blom HJ (1999) The molecular basis of cystathionine beta-synthase deficiency in Dutch patients with homocystinuria: effect of CBS genotype on biochemical and clinical phenotype and on response to treatment. Am. J. Hum. Genet. 65: 59–67. 10.1086/302439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluijtmans LA, Boers GH, Stevens EM, Renier WO, Kraus JP, Trijbels FJ, van den Heuvel LP, Blom HJ (1996) Defective cystathionine beta-synthase regulation by S-adenosylmethionine in a partially pyridoxine responsive homocystinuria patient. Journal of Clinical Investigation 98: 285–9. 10.1172/JCI118791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecka J, Krijt J, Rakova K, Kozich V (2011) Restoring assembly and activity of cystathionine beta-synthase mutants by ligands and chemical chaperones. Journal of Inherited Metabolic Disease 34: 39–48. 10.1007/s10545-010-9087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich V, Kraus JP (1992) Screening for mutations by expressing patient cDNA segments in E. coli: homocystinuria due to cystathionine beta-synthase deficiency. Human Mutation 1: 113–23. 10.1002/humu.1380010206. [DOI] [PubMed] [Google Scholar]
- Kraus J, Packman S, Fowler B, Rosenberg LE (1978) Purification and properties of cystathionine beta-synthase from human liver. Evidence for identical subunits. J. Biol. Chem. 253: 6523–8. [PubMed] [Google Scholar]
- Kruger WD, Cox DR (1994) A yeast system for expression of human cystathionine beta-synthase: structual and functional conservation of the human and yeast genes. Proc. Natl. Acad. Sci. USA 91: 6614–6618. 10.1073/pnas.91.14.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger WD, Wang L, Jhee KH, Singh RH, Elsas LJ 2nd (2003) Cystathionine beta-synthase deficiency in Georgia (USA): correlation of clinical and biochemical phenotype with genotype. Hum. Mutat. 22: 434–41. 10.1002/humu.10290. [DOI] [PubMed] [Google Scholar]
- Leandro P, Gomes CM (2008) Protein misfolding in conformational disorders: rescue of folding defects and chemical chaperoning. Mini Rev Med Chem 8: 901–11. 10.2174/138955708785132783. [DOI] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL (1998) Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol Cell Biol 18: 30–8. 10.1128/MCB.18.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Gallego-Villar L, Grisch-Chan HM, Haberle J, Thony B, Kruger WD (2019) Treatment of Cystathionine beta-Synthase Deficiency in Mice Using a Minicircle-Based Naked DNA Vector. Hum Gene Ther 30: 1093–1100. 10.1089/hum.2019.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean KN, Gaustadnes M, Oliveriusova J, Janosik M, Kraus E, Kozich V, Kery V, Skovby F, Rudiger N, Ingerslev J, Stabler SP, Allen RH, Kraus JP (2002) High homocysteine and thrombosis without connective tissue disorders are associated with a novel class of cystathionine beta-synthase (CBS) mutations. Hum. Mutat. 19: 641–655. 10.1002/humu.10089. [DOI] [PubMed] [Google Scholar]
- Majtan T, Freeman KM, Smith AT, Burstyn JN, Kraus JP (2011) Purification and characterization of cystathionine beta-synthase bearing a cobalt protoporphyrin. Arch Biochem Biophys 508: 25–30. 10.1016/j.abb.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majtan T, Jones W Jr., Krijt J, Park I, Kruger WD, Kozich V, Bassnett S, Bublil EM, Kraus JP (2018) Enzyme Replacement Therapy Ameliorates Multiple Symptoms of Murine Homocystinuria. Mol Ther 26: 834–844. 10.1016/j.ymthe.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majtan T, Liu L, Carpenter JF, Kraus JP (2010) Rescue of cystathionine beta-synthase (CBS) mutants with chemical chaperones: purification and characterization of eight CBS mutant enzymes. Journal of Biological Chemistry 285: 15866–73. 10.1074/jbc.M110.107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majtan T, Singh LR, Wang L, Kruger WD, Kraus JP (2008) Active cystathionine beta-synthase can be expressed in heme-free systems in the presence of metal-substituted porphyrins or a chemical chaperone. J Biol Chem 283: 34588–95. 10.1074/jbc.M805928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melenovska P, Kopecka J, Krijt J, Hnizda A, Rakova K, Janosik M, Wilcken B, Kozich V (2015) Chaperone therapy for homocystinuria: the rescue of CBS mutations by heme arginate. J Inherit Metab Dis 38: 287–94. 10.1007/s10545-014-9781-9. [DOI] [PubMed] [Google Scholar]
- Morello JP, Petaja-Repo UE, Bichet DG, Bouvier M (2000) Pharmacological chaperones: a new twist on receptor folding. Trends Pharmacol. Sci. 21: 466–9. 10.1016/s0165-6147(00)01575-3. [DOI] [PubMed] [Google Scholar]
- Morris AA, Kozich V, Santra S, Andria G, Ben-Omran TI, Chakrapani AB, Crushell E, Henderson MJ, Hochuli M, Huemer M, Janssen MC, Maillot F, Mayne PD, McNulty J, Morrison TM, Ogier H, O’Sullivan S, Pavlikova M, de Almeida IT, Terry A, Yap S, Blom HJ, Chapman KA (2017) Guidelines for the diagnosis and management of cystathionine beta-synthase deficiency. J Inherit Metab Dis 40: 49–74. 10.1007/s10545-016-9979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison T, Bosch F, Landolt MA, Kozich V, Huemer M, Morris AAM (2021) Homocystinuria patient and caregiver survey: experiences of diagnosis and patient satisfaction. Orphanet J Rare Dis 16: 124. 10.1186/s13023-021-01764-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd SH, Levy HL, Kraus JP (2001) Disorders in transsulfuration. In: Scriver CR, Beaudet A, Sly W, Valle D (eds) The Metabolic Basis of Inherited Disease. McGraw-Hill, New York, pp 2007–2056. [Google Scholar]
- Mudd SH, Skovby F, Levy HL, Pettigrew KD, Wilcken B, Pyeritz RE, Andria G, Boers GH, Bromberg IL, Cerone R, et al. (1985) The natural history of homocystinuria due to cystathionine beta-synthase deficiency. Am. J. Hum. Genet. 37: 1–31. [PMC free article] [PubMed] [Google Scholar]
- Oliveriusova J, Kery V, Maclean KN, Kraus JP (2002) Deletion mutagenesis of human cystathionine beta-synthase. Impact on activity, oligomeric status, and S-adenosylmethionine regulation. J Biol Chem 277: 48386–94. 10.1074/jbc.M207087200. [DOI] [PubMed] [Google Scholar]
- Ong DS, Mu TW, Palmer AE, Kelly JW (2010) Endoplasmic reticulum Ca2+ increases enhance mutant glucocerebrosidase proteostasis. Nat Chem Biol 6: 424–32. 10.1038/nchembio.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski RZ, Kuhn DJ (2008) Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res 14: 1649–57. 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- Poloni S, Sperb-Ludwig F, Borsatto T, Weber Hoss G, Doriqui MJR, Embirucu EK, Boa-Sorte N, Marques C, Kim CA, Fischinger Moura de Souza C, Rocha H, Ribeiro M, Steiner CE, Moreno CA, Bernardi P, Valadares E, Artigalas O, Carvalho G, Wanderley HYC, Kugele J, Walter M, Gallego-Villar L, Blom HJ, Schwartz IVD (2018) CBS mutations are good predictors for B6-responsiveness: A study based on the analysis of 35 Brazilian Classical Homocystinuria patients. Mol Genet Genomic Med 6: 160–170. 10.1002/mgg3.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE (2009) Biological and chemical approaches to diseases of proteostasis deficiency. Annual Review of Biochemistry 78: 959–91. 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Seashore MR, Durant JL, Rosenberg LE (1972) Studies of the mechanism of pyridoxine-responsive homocystinuria. Pediatr Res 6: 187–96. 10.1203/00006450-197203000-00007. [DOI] [PubMed] [Google Scholar]
- Sebastio G, Sperandeo MP, Panico M, de Franchis R, Kraus JP, Andria G (1995) The molecular basis of homocystinuria due to cystathionine beta synthase deficiency in italian families, and report of four novel mutations. Am. J. Hum. Genet 56: 1324–1333. [PMC free article] [PubMed] [Google Scholar]
- Seemann S, Ernst M, Cimmaruta C, Struckmann S, Cozma C, Koczan D, Knospe AM, Haake LR, Citro V, Brauer AU, Andreotti G, Cubellis MV, Fuellen G, Hermann A, Giese AK, Rolfs A, Lukas J (2020) Proteostasis regulators modulate proteasomal activity and gene expression to attenuate multiple phenotypes in Fabry disease. Biochem J 477: 359–380. 10.1042/BCJ20190513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Dunbrack RL Jr., Christopher SA, Kruger WD (2001) Mutations in the regulatory domain of cystathionine beta synthase can functionally suppress patient-derived mutations in cis. Hum. Mol. Genet. 10: 635–43. 10.1093/hmg/10.6.635. [DOI] [PubMed] [Google Scholar]
- Shan X, Kruger WD (1998) Correction of disease-causing CBS mutations in yeast. Nat Genet 19: 91–3. 10.1038/ng0598-91. [DOI] [PubMed] [Google Scholar]
- Singh LR, Chen X, Kozich V, Kruger WD (2007) Chemical chaperone rescue of mutant human cystathionine β-synthase. Mol Genet Metab 91: 335–42. 10.1016/j.ymgme.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LR, Gupta S, Honig NH, Kraus JP, Kruger WD (2010) Activation of mutant enzyme function in vivo by proteasome inhibitors and treatments that induce Hsp70. PLoS Genet 6: e1000807. 10.1371/journal.pgen.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LR, Kruger WD (2009) Functional rescue of mutant human cystathionine β-synthase by manipulation of Hsp26 and Hsp70 levels in Saccharomyces cerevisiae. J. Biol. Chem. 284: 4238–45. 10.1074/jbc.M806387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AT, Majtan T, Freeman KM, Su Y, Kraus JP, Burstyn JN (2011) Cobalt cystathionine beta-synthase: a cobalt-substituted heme protein with a unique thiolate ligation motif. Inorg Chem 50: 4417–27. 10.1021/ic102586b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN (2003) Human Gene Mutation Database (HGMD): 2003 updat e. Hum. Mutat 21: 577–81. 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Taoka S, Lepore BW, Kabil O, Ojha S, Ringe D, Banerjee R (2002) Human cystathionine beta-synthase is a heme sensor protein. Evidence that the redox sensor is heme and not the vicinal cysteines in the CXXC motif seen in the crystal structure of the truncated enzyme. Biochemistry 41: 10454–61. 10.1021/bi026052d. [DOI] [PubMed] [Google Scholar]
- Taoka S, Ohja S, Shan X, Kruger WD, Banerjee R (1998) Evidence for heme-mediated redox regulation of human cystathionine beta-synthase activity. Journal of Biological Chemistry 273: 25179–84. 10.1074/jbc.273.39.25179. [DOI] [PubMed] [Google Scholar]
- Weber Hoss GR, Sperb-Ludwig F, Schwartz IVD, Blom HJ (2020) Classical homocystinuria: A common inborn error of metabolism? An epidemiological study based on genetic databases. Mol Genet Genomic Med 8: e1214. 10.1002/mgg3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Huntoon K, Ksendzovsky A, Zhuang Z, Lonser RR (2013) Proteostasis modulators prolong missense VHL protein activity and halt tumor progression. Cell Rep 3: 52–9. 10.1016/j.celrep.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]