Abstract
The ongoing global health crisis caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the virus which leads to Coronavirus Disease 2019 (COVID-19) has impacted not only the health of people everywhere, but the economy in nations across the world. While vaccine candidates and therapeutics are currently undergoing clinical trials, there is yet to be a proven effective treatment or cure for COVID-19. In this study, we have presented a synergistic computational platform, including molecular dynamics simulations and immunoinformatics techniques, to rationally design a multi-epitope vaccine candidate for COVID-19. This platform combines epitopes across Linear B Lymphocytes (LBL), Cytotoxic T Lymphocytes (CTL) and Helper T Lymphocytes (HTL) derived from both mutant and wild-type spike glycoproteins from SARS-CoV-2 with diverse protein conformations. In addition, this vaccine construct also takes the considerable glycan shield of the spike glycoprotein into account, which protects it from immune response. We have identified a vaccine candidate (a 35.9 kDa protein), named COVCCF, which is composed of 5 LBL, 6 HTL, and 6 CTL epitopes from the spike glycoprotein of SARS-CoV-2. Using multi-dose immune simulations, COVCCF induces elevated levels of immunoglobulin activity (IgM, IgG1, IgG2), and induces strong responses from B lymphocytes, CD4 T-helper lymphocytes, and CD8 T-cytotoxic lymphocytes. COVCCF induces cytokines important to innate immunity, including IFN-γ, IL4, and IL10. Additionally, COVCCF has ideal pharmacokinetic properties and low immune-related toxicities. In summary, this study provides a powerful, computational vaccine design platform for rapid development of vaccine candidates (including COVCCF) for effective prevention of COVID-19.
Keywords: Vaccine, SARS-CoV-2, Immunoinformatics, Molecular Dynamics
Introduction
The current Coronavirus Disease 2019 (COVID-19) pandemic has brought the world to a near standstill, with over 92 million cases worldwide and nearly 2 million deaths as of January 13, 2021. While some countries have been able to manage cases using a combination of stay-at-home orders, social distancing, and mask usage, the worldwide 7-day moving average for worldwide cases is currently over 730,000 per day (source: Worldometers.info), indicative of a need for effective prevention and/or treatment of COVID-19. As of January 13, 2021, the United States (U.S.) alone has more than 23 million confirmed cases, with a death toll approaching 400,000, accompanied by unprecedented social and economic consequences (coronavirus.jhu.edu). Recently, the FDA has approved two mRNA-based vaccines for emergency use(Baden et al. 2020; Polack et al. 2020); yet decreased stability of mRNA could lead to decreased potency due to immunostimulatory effects(Liu 2019). Peptide vaccines, on the other hand, have the advantage of enhanced stability, are readily synthesized, and lend well to repeated booster doses(Slingluff 2011).
Key to the interaction between Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the virus which causes COVID-19, and the human cell is the spike glycoprotein (S protein)(Tortorici and Veesler 2019), which in SARS-CoV-2 interacts with angiotensin-converting enzyme 2 (ACE2) via its receptor binding domain (RBD)(Hoffmann et al. 2020). The S protein is a 180 kDa homotrimer consisting of two subunits, S1 and S2, which mediate attachment to ACE2 and membrane fusion, respectively(Ou et al. 2020). The S1 subunit consists of an N-terminal domain (NTD) and the RBD, while the S2 subunit is composed of a fusion protein (FP), two heptad repeat domains (HR1 and HR2), a transmembrane domain (TM), and a cytoplasmic domain (CP)(Xia et al. 2020). In order to fuse its viral membrane with the host cell, the S protein must be activated at the S1/S2 boundary(Belouzard et al. 2012). This priming of the S protein is accomplished through the use of the cellular protein TMPRSS2(Hoffmann et al. 2020).
Because of this, the S protein has been a target for therapeutics(Martin and Cheng 2020; Xia et al. 2020), including vaccines(Amanat and Krammer 2020; Baden et al. 2020; Polack et al. 2020). However, key to the S protein’s ability to ward off an immune response is its considerable glycan shield(Watanabe et al. 2020a; Watanabe et al. 2020b). The glycosylation of the S glycoprotein creates somewhat of a barrier around the spike, preventing immune molecules from reaching the protein surface. Here, we have constructed a multi-epitope vaccine candidate using molecular dynamics simulations and immunoinformatics techniques while considering the impact of the glycan shield on the ability for a particular epitope to elicit an immune response (Figure 1). Selecting only epitopes which would be accessible in an immune response should improve the effectiveness of a vaccine. Our inclusion of multiple conformations for the spike glycoprotein, both in the wild-type as well as in 9 different mutated states, allowed for a broader reach with respect to B-cell epitope prediction; in fact, nearly 75% of the predicted epitopes were exclusive to the predictions from the mutated systems. Additionally, this method is superior to a sequence-based prediction; a preliminary test for linear B-lymphocyte epitopes over the 1,120 amino acid sequence available via crystal structure using BepiPred 2.0(Jespersen et al. 2017) yielded only 27 potential epitopes of length 6 or longer. This, along with careful consideration of the impact glycosylation will have on the ability for a particular epitope to elicit a useful immune response, allowed for the construction of our 331 amino acid vaccine construct, named COVCCF; the physiochemical properties and simulated immunogenicity indicate the potential for the induction of a strong immune response, which would also portend immunity toward the spike glycoprotein of SARS-CoV-2.
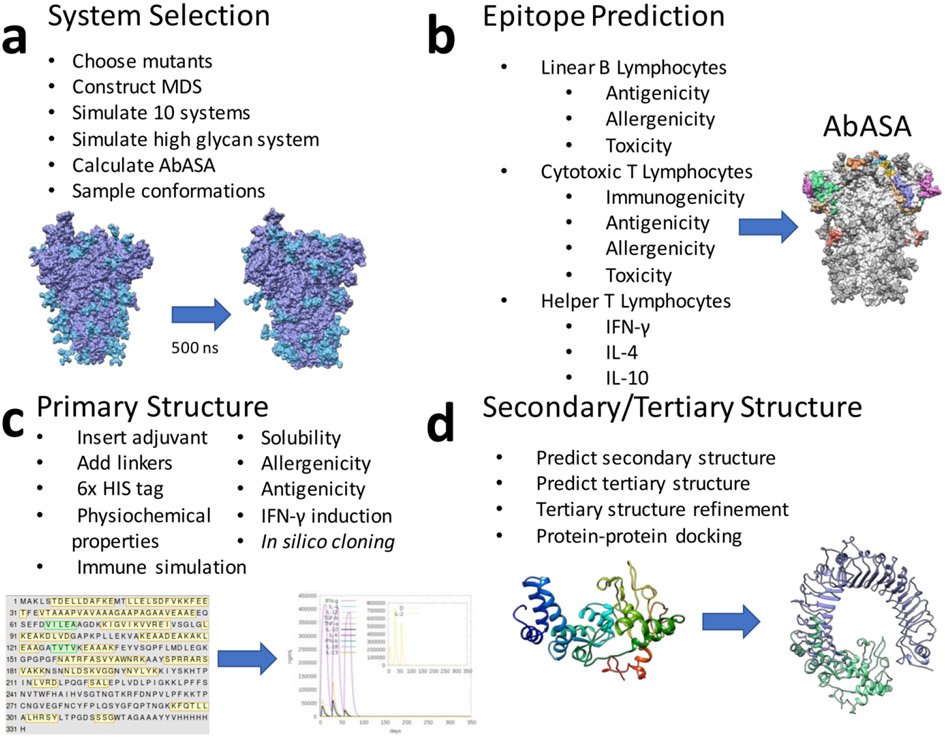
Figure 1: Overall workflow for project.
a) Selection of the systems used to generate conformations to be used in the linear B lymphocyte prediction. b) Epitope predictions, including linear B lymphocyte, cytotoxic T lymphocyte, and helper T lymphocytes. Predicted epitopes were assessed for multiple immune-relevant properties, as well as their ability to be accessed by a simulated antibody (antibody accessible surface area, AbASA). c) The final selected epitopes were linked together, along with an N-terminal adjuvant, using EAAAK, GPGPG, AAY, and KK linkers. This sequence was assessed for immune-relevant properties and simulated immune response. d) The secondary and tertiary structures were predicted and refined, and the final 3D structure was docked using protein-protein docking to toll-like receptor 2 and 4 (TLR2/4).
Methods
System Selection
Prior to epitope prediction, a total of 10 systems were simulated using the GROMACS 2020.1(Abraham et al. 2015) molecular dynamics package, including 9 mutated systems and wild type. The selected mutants (A852V, A930V, F797C, F970S, L752F, L861M, V615L, V860D, V860L) were selected(Koyama et al. 2020) based on conservation of the residue between SARS-CoV-2, SARS-CoV-1, and MERS-CoV. Each of the 9 mutations were added in silico using CHARMM-GUI(Jo et al. 2008; Lee et al. 2016) using PDB ID 6VSB(Wrapp et al. 2020) at the time of system creation. Following a processing step involving the addition of hydrogen atoms and completion of missing loops, a water box with edges at least 10 angstroms from any part of the protein was added. The systems were neutralized and brought to a total ionic strength of 0.15 M using sodium and chloride ions. Parameterization of the protein, ions, and TIP3P water molecules was accomplished using the CHARMM36m force field(Best et al. 2012). Each of these systems used the glycosylation state as in the crystal structure with no modifications. An additional wild type system with high mannose N-glycans was constructed in order to assess the change in the proteins immune accessibility due to the glycan shield.
Simulation Parameters
All systems were simulated using GROMACS 2020.1 on the AiMOS Supercomputer at the Rensselaer Polytechnic Institute Center for Computational Innovations in a three-step process. Initial minimization of the systems was run until changes in the potential energy of the system reached machine precision. Following minimization, an NVT equilibration step was completed with a 2 fs timestep for 500,000 steps using 400 kJ mol−1 nm−2 and 40 kJ mol−1 nm−2 positional restraints on the backbone and sidechains, respectively. A 500 ns production step was completed using the NPT ensemble with no position restraints and a 2 fs timestep.
Hydrogen atoms were constrained using the LINCS(Hess et al. 1997) algorithm. Temperature for the system was held at 300 K using a Nose-Hoover thermostat(Braga and Travis 2005) with a 1 ps coupling constant. For the production simulation, pressure was coupled isotropically using a Parrinello-Rahman barostat(Parrinello and Rahman 1981) with a 5.0 ps coupling constant and compressibility of 4.5e-05 bar−1 to maintain a pressure of 1 bar. The pair-list cutoff was constructed using the Verlet scheme(Verlet 1967) with a cutoff distance of 1.2 nm. Particle mesh Ewald electrostatics(Darden et al. 1993) were used to describe coulombic interactions with a 1.2 nm cutoff, while van der Waals forces were smoothly switched to using between 1.0 and 1.2 nm using a force-switch modifier to the cut-off scheme. Linear center of mass translation was removed every 100 steps for the entire system.
Linear B-cell Epitope Prediction
The above simulations were sampled at t = 0, 100, 200, 300, 400, and 500 nanoseconds for the prediction of linear B lymphocyte (LBL) epitopes, yielding a total of 60 structures. The use of these conformations allows for greater sampling of LBL epitopes when compared to the use of the protein sequence or a single PDB structure. These structure-based predictions were made using ElliPro(Ponomarenko et al. 2008) using the default minimum score of 0.5 and the default maximum distance of 6 angstroms. ElliPro implements three algorithms in its predictions: 1) an approximation of the shape of the protein as an ellipsoid; 2) calculation of the protrusion index for each residue, which is a quantification of the extent to which a residue protrudes from the surface of a protein based on the ellipsoid approximations; and 3) a clustering of neighboring residues based on protrusion index. While ElliPro is able to predict both linear and discontinuous epitopes, only linear epitopes are used in vaccine design(Nain et al. 2019). Since only structural epitopes were generated, only residues 27 through 1146 were included in any of the epitope predictions, as those are the only residues crystallized in the pdb used.
Cytotoxic T Lymphocytes (CTL) Epitope Prediction
Cytotoxic T lymphocyte (CTL) epitopes were predicted for each of the 10 sequences noted above using the NetCTL 1.2 server(Larsen et al. 2007). The default threshold for epitopes was retained at 0.75, and all 12 of the available MHC class I supertypes was used for the predictions. NetCTL uses artificial neural networks to predict major histocompatibility class (MHC) I binding and proteasomal cleavage, while TAP transport efficiency is predicted using a weight matrix. Additionally, the CTL epitopes were each evaluated for their immunogenicity using the MHC-1 immunogenicity tool on the IEDB server(Calis et al. 2013).
Helper T Lymphocytes (HTL) Epitope Prediction
Helper T cells help activate B cells to secrete antibodies and macrophages, and also help activate cytotoxic T cells, indicating their importance to adaptive immunity. Prediction of these HTL epitopes as peptides that bind MHC II molecules is therefore key to rational vaccine design(Nielsen et al. 2007). HTL epitopes of length 15 were predicted using the IEDB MHC-II binding predictions tool. The IEDB recommended prediction method was selected, which uses the consensus approach(Wang et al. 2010), combining NN-align(Nielsen and Lund 2009), SMM-align(Nielsen et al. 2007), CombLib(Sidney et al. 2008), and Sturniolo(Sturniolo et al. 1999) when possible, otherwise using NetMHCIIpan(Andreatta et al. 2015). The full HLA reference set was used for the prediction, and predictions with a percentile rank ≤ 2 were chosen; a lower percentile rank indicates a higher affinity.
Assessment of CTL/LBL Epitopes for Antigenicity, Allergenicity, and Toxicity
To ensure their ability to illicit an immune response, the antigenicity of each of the CTL and LBL epitopes was evaluated using the VaxiJen 2.0 server(Doytchinova and Flower 2007). VaxiJen uses an alignment-free approach based on auto cross covariance (ACC) transformation, a protein sequence mining method which has been applied to quantitative structure-activity relationships (QSAR) studies and protein classification(Wold et al. 1993). This application of ACC to the principal component analysis (PCA) of 29 properties of each of the 20 amino acids allows for the removal of irrelevant information, amplifying class-discriminating properties. Allergenicity of epitopes was determined using the AllerTOP 2.0 server(Dimitrov et al. 2014), which in addition to ACC uses a k-nearest neighbor algorithm based on a training set consisting of 2427 each of known allergens and non-allergens from different species. Toxicity of epitopes was predicted using the ToxinPred(Gupta et al. 2013) server, which uses the Support Vector Machine (SVM) algorithm, with a main dataset including 1805 sequences as positive training data and 3593 negative sequences from Swissprot(Luckheeram et al. 2012), and an independent dataset comprising of 303 positive and 300 negative sequences, also from Swissprot.
Identification of Cytokine-Inducing HTL epitopes
The ability of an HTL epitope to induce cytokines (specifically, interferon-gamma [IFN-γ], interleukin-4 [IL-4], and interleukin-10 [IL-10]) is key to a vaccine’s ability to illicit an effective immune response; the release of these cytokines helps in the activation of cytotoxic T-cells and other immune cells(Dhanda et al. 2013b). To determine the ability of our predicted HTL epitopes to induce these cytokines, we used the IFNepitope server(Dhanda et al. 2013b) (IFN-γ) using the motif and SVM hybrid approach with the IFN-gamma versus non IFN-gamma model; IL4pred server(Dhanda et al. 2013a) (IL-4) using the hybrid (SVM + motif) and the default SVM threshold of 0.2; and IL10pred server(Nagpal et al. 2017) (IL-10) using the SVM based method with the default SVM threshold of −0.3. These prediction servers, like ToxinPred above, use the SVM algorithm for their predictions.
Antibody-Accessible Surface Area Determination
To determine which predicted epitopes are most likely to be capable of eliciting a useful immune response, we determined the antibody-accessible surface area (AbASA) using a method similar to that outlined previously(Grant et al. 2020). Two calculations for AbASA were completed using the built in SASA tool in GROMACS 2020.1, selecting a probe size of 0.72 nanometers as opposed to the standard 0.14 nanometer probe used for a standard SASA calculation. The first calculation determined the AbASA for the bare protein, not accounting for glycosylation, while the second determined the AbASA for the protein while also taking the extensive glycosylation into account.
When selecting an epitope for COVCCF, a residue was deemed to be not antibody accessible if its AbASA was lower than 0.25 Å2 and therefore not included in the final vaccine candidate. As the spike glycoprotein is a homotrimer, an average of the AbASA across the three domains was used for this determination. Additionally, residues with a drop in AbASA when considering the glycan shield were inspected on a case-by-case basis with the knowledge that the 0.72 nm probe radius would only account for accessibility for an average loop in an antibody, and did not account for accessibility of an entire antibody. Regions which had a large change in AbASA were determined to be shielded, and predicted epitopes for these regions were not included in COVCCF.
Construction of the Multi-Epitope Vaccine Candidate Sequence
The CTL, HTL, and LBL epitopes which passed the above tests were used to generate the full vaccine sequence. In the event of overlap between epitopes, we did not duplicate the sequence. Epitopes were linked together using GPGPG, AAY, and KK linkers; GPGPG and AAY linkers were used to connect the HTL and CTL epitopes, while KK linkers were used for B-cell epitopes, allowing them to preserve their independent immunogenic properties(Gu et al. 2017). A 50 S ribosomal protein L7/L12 (locus RL7_MYCTU, UniProtKB ID: P9WHE3) was chosen as an adjuvant, and inserted on the N-terminus using an EAAAK linker.
Prediction of Physiochemical Properties, Solubility, Allergenicity, Antigenicity, and IFN-γ induction
The full sequence for the multi-epitope vaccine construct COVCCF was tested for its allergenicity using the AllerTOP 2.0 server, and its antigenicity using the VaxiJen 2.0 server. The IFNepitope server was used to scan the full sequence for predicted IFN-γ inducing epitopes using the SVM based method for score prediction only. The ProtParam web server(Gasteiger et al.) allows for the prediction of various physiochemical properties, including amino acid composition, theoretical isoelectric point (pI), instability index, half-life (both in vivo and in vitro) aliphatic index, molecular weight, and grand average of hydropathicity (GRAVY). Solubility of the final protein sequence was predicted using the CamSol server(Sormanni et al. 2017; Sormanni et al. 2015), which allows for a pdb structure as input, taking into account the 3D conformation of the protein as opposed to only the sequence.
Prediction of Secondary Structure
The generated sequence for the full-length vaccine was submitted to the PSIPRED 4.0 server(Buchan and Jones 2019) to predict its secondary structure. PSIPRED uses a deep neural network architecture with two hidden layers and rectifier activations; the current version has a Q3 secondary structure prediction accuracy of 84.2%. The RaptorX Property server(Wang et al. 2016) was additionally employed as a second validation. RaptorX Property employs a new machine learning model, Deep Convolutional Neural Fields (DeepCNF), which implements both conditional neural fields (CNF) and deep convolutional neural networks (DCNN).
Tertiary Structure Prediction
Homology modeling of the final vaccine candidate was completed using the I-TASSER server(Yang et al. 2014). I-TASSER (Iterative Threading ASSEmbly Refinement) uses a three-step process to model the tertiary structure of a protein. First, the server tries to retrieve template proteins from the PDB library using LOMETS (Local Meta-Threading Server), which generates protein structure predictions by ranking and selecting models from multiple state of the art threading programs(Zheng et al. 2019). The second step involves assembling fragments excised from the PDB templates determined in step 1 using replica-exchange Monte Carlo simulations, with unaligned regions generated using ab initio modeling. The third step integrates spatial restraints to remove steric clashes, finally generating full atomic models. The secondary structure predictions generated by PSIPRED were submitted along with the primary structure of the multi-epitope vaccine candidate.
Tertiary Structure Refinement
The selected model generated by I-TASSER was further refined first using ModRefiner(Xu and Zhang 2011), followed by GalaxyRefine(Heo et al.). ModRefiner uses a two-step process, first a low-resolution step followed by a high-resolution step. The low-resolution step begins with a C-alpha trace of the initial structure, adding main chain atoms for an energy minimization. This structure is then passed to the high-resolution step, which adds side chain atoms and does a full atomic energy minimization, yielding a final model. GalaxyRefine begins by rebuilding all side chains, placing the highest probability rotamers starting from the core and extending to the surface, layer by layer. Upon reaching a steric clash, the next highest probability rotamer is selected. After being rebuilt, the model is refined using a two-step relaxation process, of which the lowest energy model is returned as model 1, and additional models are returned based on the closest clustered models.
Structural Validation
Multiple servers were implemented to validate the tertiary structure generated. ProSA-web(Wiederstein and Sippl 2007) gives an assessment of the overall model quality, displayed in the context of all X-ray and NMR structures, with a Z-score falling outside that of known structures generally implies errors in the structure. The ERRAT server(Colovos and Yeates 1993) was used to assess non-bonded interactions in comparison to high-resolution crystallographic structures. Finally, the MolProbity(Chen et al. 2010) server was used to generate a Ramachandran plot, which gives a visualization of energetically allowed and disallowed dihedral angles of amino acids, calculated based on the van der Waal radius of the side chain.
Prediction of LBL Epitopes in the Vaccine Protein
The presence of both linear and discontinuous B-cell epitopes was predicted using ElliPro as above. It has been estimated that greater than 90% of B-cell epitopes are discontinuous; that is, their segments are distant from each other in their primary structure, but are brought close to each other upon the folding of the protein(Barlow et al. 1986; Van Regenmortel 1996).
In Silico Cloning of Designed Vaccine Candidate
The vaccine protein sequence was submitted to the JAVA Codon Adaptation Tool (JCAT)(Grote et al.) to adapt the codon usage to E. coli K12. The options to avoid rho-independent transcription termination, prokaryote ribosome binding site, and restriction enzyme cleavage sites were selected. XhoI and XbaI restriction sites were added to the C and N termini of the sequence, respectively. The final nucleotide sequence was then cloned into the pET-30a (+) vector using the SnapGene 5.1.3 software.
Molecular Docking of the Vaccine Construct with TLR2/TLR4
The ability for COVCCF to generate an immune response depends on its ability to interact with immune receptors. Toll-like receptors 2 and 4 (TLR2, TLR4) are members of the TLR family, which play a role in pathogen recognition and activation of innate immunity. Therefore, the ability for COVCCF to interact with these receptors is key to the immune response. The adjuvant was selected as the region of interest, as it has been shown to be a TLR agonist(Khatoon et al. 2017). CPORT(de Vries and Bonvin 2011) was used to initially predict residues which could be involved in the protein-protein interaction. The results from this initial prediction were imported into the HADDOCK 2.4 server(Van Zundert et al. 2016) for data-driven protein-protein docking. HADDOCK (High Ambiguity Driven protein-protein DOCKing) uses a collection of python scripts which make use of crystal and NMR structures for structure calculations. The best structure from the best cluster was submitted to the PRODIGY (PROtein binDIng enerGY prediction) server(Xue et al. 2016) to predict the binding affinities of each of the protein-protein complexes. In addition to predicting a score, PRODIGY predicts a ΔG based on the following formula:
With the dissociation constant Kd calculated from ΔG using the standard formula.
Immune Simulation
The immunogenicity of COVCCF was further characterized using the C-ImmSim server(Rapin et al. 2010). C-ImmSim uses position-specific scoring matrix (PSSM) for immune epitope prediction and machine learning to predict immune interactions. It simulates hematopoietic stem cells in the bone marrow, T-cells in the thymus, and tertiary lymphatic organs, for their immune response. It has been determined computationally that an interval of several weeks between the prime (first) and boost (all subsequent) doses of a vaccine is required to obtain optimal antibody response(Castiglione et al. 2012). Therefore, the simulation was set to administer three injections at timesteps 1, 84, and 168, corresponding to time = 0, 4 weeks, and 8 weeks with a total of 1050 simulation steps. Each injection contained 1000 vaccine proteins, and all other parameters were set to their defaults. A further simulation with 12 injections setting 4 weeks apart was also carried out, which would simulate repeated exposure as typically seen in an endemic area, probing the clonal selection. The Simpson Index D, a measure of diversity, was interpreted from the plot.
Results
Generation of Conformations for Linear B Lymphocyte Prediction
Each of the simulated systems, including the 9 mutants, wild type, and the high mannose N-glycan substituted wild type system, were assessed for stability along the entire 500 nanosecond simulation using the RMSD of all backbone atoms after least squares fitting to the same using standard GROMACS(Abraham et al. 2015) tools (Supplementary Figure 1). A total of 5 μs of simulation time (10 systems at 500 nanoseconds each) was used for linear B lymphocyte prediction. No system was deemed to have any stability issues, so each system was sampled at its initial conformation (after equilibration but before production dynamics simulation) and every 100 nanoseconds of simulation, yielding a total of 6 conformations for each of the 9 mutant and 1 wild type system. The high mannose system was not sampled in this way and was processed separately.
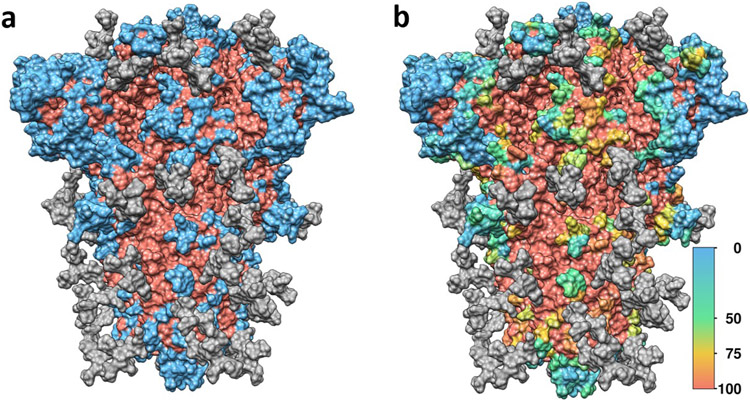
The high mannose system was further assessed for its antibody accessible surface area (AbASA). Using the built in GROMACS tool SASA, with a probe of size 0.72 nm, the surface area was determined for the protein alone (Figure 2a, Supplementary Figure 2) while ignoring the glycosylation, and again while taking the glycosylation into account (Supplementary Figure 3). The percent change in the AbASA was determined as the change in the AbASA when taking the glycosylation into account (Figure 2b). This change in accessibility to an immune response was used to dictate which of the predicted epitopes would be included in COVCCF; an epitope was rejected if there was a large change in the AbASA due to the glycosylation ( > 50% reduction), or if there were no residues with greater than 0.25 Å2 of accessible surface area, which would indicate a low likelihood of potential for immune interaction. Glycosylation of the spike glycoprotein creates a shield against immune response, reducing the AbASA for many surface residues by over 50%, protecting otherwise targetable epitopes.
Figure 2: Antibody-accessible surface area (AbASA) of the spike glycoprotein.
a) Regions of the spike glycoprotein which have at least 0.25 Å2 of AbASA in blue, regions with less AbASA in red, and the glycosylation in gray. b) Percent change in AbASA due to glycosylation, with blue as no change, green as a 50% reduction in AbASA, and yellow a 75% reduction in AbASA. A value of 100% (red) was assigned for any residue with less than 0.25 Å2 of AbASA.
Identification of Antigenic Linear B-cell Epitopes
ElliPro(Ponomarenko et al. 2008) was used to predict the linear B-lymphocyte (LBL) epitopes for each of the 6 conformations for each of the nine mutant systems and the wild type. In total, this yielded 3,311 epitopes, of which 428 unique epitopes were found (Supplementary Data 1). Sequences were tested for allergenicity, antigenicity, and toxicity; the sequences which passed these tests were then aligned to the full-length sequence of the viral protein to determine which of the epitopes did not have significant impedance due to the glycosylation. The LBL epitopes included in the final construct are given in Table 1. A total of 5 LBL epitopes were chosen for COVCCF.
Table 1:
Predicted epitopes in the final vaccine construct.
| LBL Epitope | HTL Epitope | CTL Epitope |
|---|---|---|
| FEYVSQPFLMDLEGK (1) | ||
| FNATRFASVYAWNRK (2) | ||
| SPRRARSVA (3) | ||
| NSNNLDSKVGGNYNYLY (4) | SKVGGNYNY (4) | |
| IYSKHTPINLVRDLPQGFSALEPLVDLPIG (5) | ||
| LPFFSNVTWFHAIHVSGTNGTKRFDNPVLPF (6) | LPFFSNVTWFHAIHV (6) | HVSGTNGTK (6) |
| PFFSNVTWFHAIHVS (6) | VTWFHAIHV (6) | |
| YSKHTPINL (6) | ||
| TPCNGVEGFNCYFPLQSYGFQPTNG (7) | NGVEGFNCYFPLQSY (7) | |
| GVEGFNCYFPLQSYG (7) | ||
| FQTLLALHRSYLTPGDSSSGWTAGAAAYYV (8) | WTAGAAAYY (8) |
Note: Epitopes are numbered based on their order in the final vaccine construct. Epitopes with a shared sequence also share serial numbers; some LBL epitopes overlapped CTL/HTL epitopes, and some HTL epitopes overlapped CTL epitopes.
Identification of Cytotoxic T Lymphocyte Epitopes
Using all 10 sequences from the mutated and wild type proteins, a total of 3,844 non-unique epitopes were generated; 260 of these were unique (Supplementary Data 2). The epitopes which were predicted as immunogenic, antigenic, non-allergenic, and non-toxic were further assessed for their accessibility, yielding 6 total CTL epitopes (Table 1) included in the final construct. Epitopes which were either non-antigenic, allergenic, or toxic were not considered; accessibility was determined in the same fashion as for the LBL epitopes.
Identification of Helper T Lymphocyte Epitopes
As with the CTL prediction, all 10 sequences were submitted to the prediction server, with a total of 3,938 non-unique (and 272 unique) HTL epitopes (Supplementary Data 3). After predictions for their ability to induce cytokines, and assessment for antibody accessibility, 6 HTL epitopes were included in the final vaccine (Table 1). Epitopes which did not elicit a response from IFN-γ, IL-4, and IL-10 were not considered; accessibility was determined in the same fashion as for the LBL epitopes.
Construction of the Multi-Epitope Vaccine Candidate
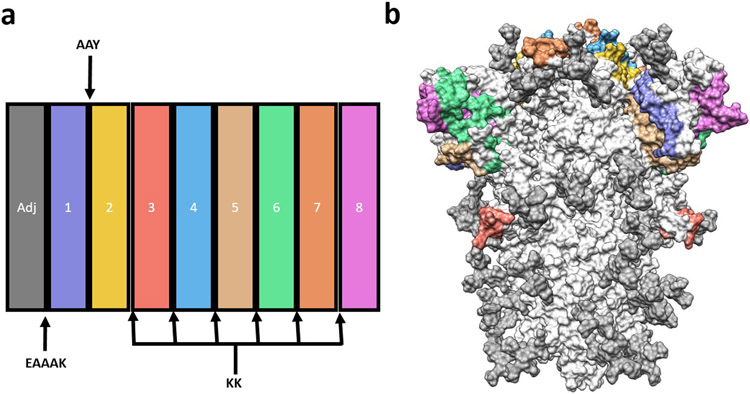
In total, 5 LBL, 6 HTL, and 6 CTL epitopes were selected for the final multi-epitope vaccine candidate COVCCF. These epitopes were determined to be antigenic, safe, and not impeded by the glycosylation of the spike glycoprotein. Epitopes for which there was overlap were not duplicated in the final construct and were instead merged. The 50S ribosomal L7/L12 (UniProt accession ID P9WHE3) was added as an adjuvant on the N-terminus of the construct and linked to the vaccine peptide using an EAAAK linker. This adjuvant was selected based on its strong activity as a TLR4 agonist(Khatoon et al. 2017). The GPGPG linker was chosen between the two HTL epitopes, an AAY linker between the HTL and CTL epitope, and KK linkers between the remaining epitopes as seen previously(Nain et al. 2019). A 6xHis tag was added to the C-terminus to aid in purification. The final vaccine candidate consists of 331 amino acids and 9 separate linked protein sequences (Figure 3a, b), the final order of which is outlined in Table 1.
Figure 3: Epitope selection for the multi-epitope vaccine construct.
a) Schematic representation of the final multi-epitope vaccine construct. Epitopes are labeled as in Table 1. b) Location of the selected epitopes on the spike glycoprotein. White residues indicate they are not in the final multi-epitope vaccine construct, with glycosylation in gray.
IFN-γ Inducing Epitope Prediction, Antigenicity and Allergenicity
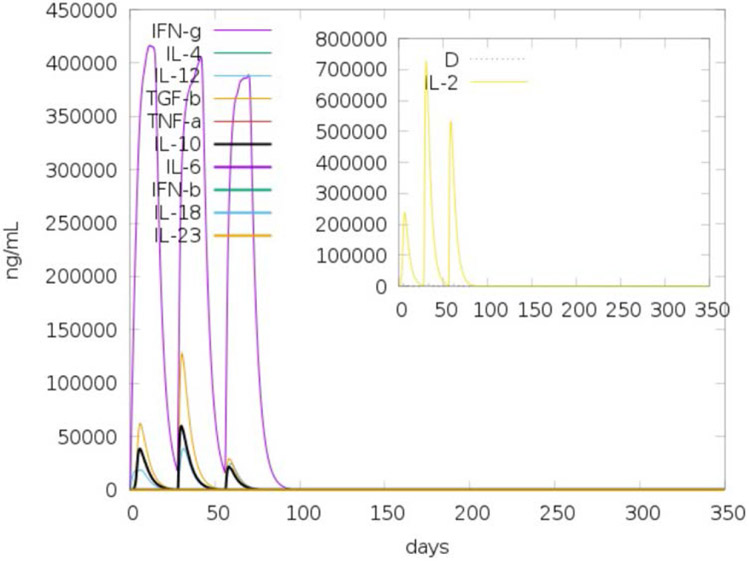
A total of 323 IFN-γ inducing epitopes were predicted using the scan function of the IFNepitope server(Dhanda et al. 2013b). Of these 323 predicted epitopes, 132 were predicted to have positive scores (Supplementary Data 4). These results are in line with the simulated levels predicted by C-ImmSim(Castiglione et al. 2012) (Figure 4). The prediction for antigenicity from VaxiJen 2.0(Doytchinova and Flower 2007) indicates it is antigenic under both a bacterial (0.5341) and viral model (0.4709) using the default threshold. COVCCF alone was also determined to be antigenic under both the bacterial (0.6180) and viral (0.5332) models using the default threshold of 0.4. Both the full construct and the vaccine peptide without adjuvant were predicted as non-allergenic using AllerTOP 2.0(Dimitrov et al. 2014). COVCCF is predicted to be non-allergenic, antigenic, and to elicit IFN-γ induction.
Figure 4: Cytokine levels induced by repeated injection of the vaccine construct.
Levels were modeled in C-ImmSim based on three injections given 4 weeks apart. D in the inset plot is the danger signal (dotted line).
Physiochemical and Solubility Properties
The physiochemical properties of COVCCF are outlined in Table 2. COVCCF is predicted to have a molecular weight of 35.9 kDa, with a theoretical isoelectric point of 8.75, indicating a slightly basic protein. The half-life is predicted to be 30 hours in mammalian reticulocytes, > 20 hours in yeast, and > 10 hours in E. coli. The predicted instability index of 27.57 indicates a stable protein (> 40 indicates instability), while the aliphatic index of 79.09 indicates thermostability; a larger aliphatic index indicates higher stability. The predicted grand average of hydropathicity is −0.237, which indicates the protein is hydrophilic; this value is calculated as an average over the entire protein of the hydropathicity of each amino acid, where hydrophilic amino acids have a negative value and hydrophobic amino acids have a positive value. The solubility score as determined by CamSol(Sormanni et al. 2015) is 0.788 based on the sequence, with a corrected score of 1.994. Altogether, COVCCF has ideal solubility and physiochemical properties.
Table 2:
Predicted physiochemical properties of COVCCF.
| Property | Result |
|---|---|
| Amino acid count | 331 |
| Molecular weight | 35906.93 Da |
| Chemical formula | C1633H2526N432O471S5 |
| Predicted pI | 8.75 |
| Estimated half-life: | |
| Mammalian reticulocytes | 30 hours |
| Yeast Cells | > 20 hours |
| E. coli | > 10 hours |
| Instability Index | 27.57 |
| Aliphatic Index | 79.89 |
| Grand average of hydropathicity (GRAVY) | −0.237 |
| Solubility | 1.994 |
Secondary Structure Prediction
The final vaccine sequence was predicted to be 42.6% alpha helix, 9.4% beta sheet, and 48.0% coil by PSIPRED 4.0(Buchan and Jones 2019) (Supplementary Figure 4), while RaptorX property(Wang et al. 2016) predicted 37%, 6%, and 56%, respectively. 57% of residues were predicted to be solvent exposed, 24% medium exposed, and 18% buried; a total of 58 residues (17%) were predicted as disordered by RaptorX (Supplementary Figure 5). The secondary structure predictions were exported from PSIPRED for use in the tertiary structure modeling.
Tertiary Structure Prediction, Refinement, and Validation
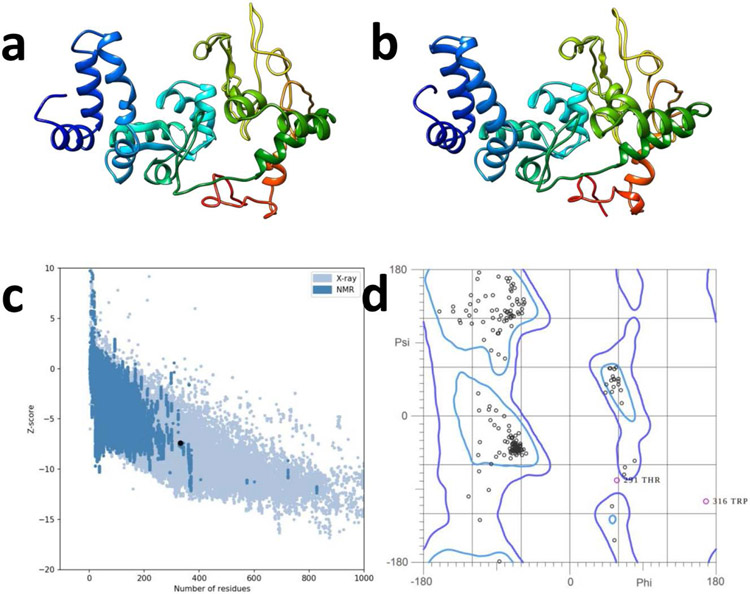
Five models were predicted using the I-TASSER webserver(Yang et al. 2014) based on alignments predicted by various threading programs. Z-scores for template alignments ranged from 0.84 to 5.61, with 1rquA, 3qtdA, 1dd3A, 1dd4A, and 2ftc as the top 5 ranking templates. Model 1 (Figure 5a) was selected for further refinement. A local installation of ModRefiner(Xu and Zhang 2011) was used for the initial refinement of the model in the two-step process outlined in the methods. The model refined using ModRefiner was then submitted to a local installation of GalaxyRefine(Heo et al.), where 10 models were generated for further assessment. The ERRAT server was used to assess the generated model, with model 1 (Figure 5b) having the highest quality factor of 81.013. Furthermore, ProSA-web(Wiederstein and Sippl 2007) was additionally used for validation, indicating a Z-score of −7.41, well within the range of native proteins of comparable size (Figure 5c). The Ramachandran plot indicated 92.7% of residues were in favored regions, 99.4% were in favored or allowed regions, and only two residues were in outlier regions (Figure 5d). These results indicate our predicted structure is likely to be close to the actual 3D structure.
Figure 5: Construction and refinement of the multi-epitope vaccine construct.
a) Final 3D model of COVCCF after modeling with I-TASSER. b) Refined model after refinement with ModRefiner and GalaxyRefine. c) Validation of structure with ProSA-web, indicating the structural properties are in line with other proteins of similar size (Z-score −7.41). d) Ramachandran plot indicating 92.7% of residues are in favored regions, and 2 residues are in outlier regions.
Prediction of LBL Epitopes in Final Vaccine Construct
A total of 8 discontinuous and 13 linear epitopes were found in COVCCF. The discontinuous epitopes ranged in length from 10 to 46 amino acids, encompassing a total of 213 of the 331 residues in the protein construct. The 13 linear epitopes were non-overlapping and encompassed 186 residues. Scores ranged from 0.508 to 0.832 for the linear epitopes, and 0.558 to 0.809 for the discontinuous epitopes. This result indicated COVCCF has the ability to induce an immune response not only from the selected epitopes, but from conformational discontinuous epitopes based on its 3D structure.
Protein-Protein Docking to TLR2 and TLR4
Protein-protein docking was performed using the HADDOCK 2.4 webserver(Van Zundert et al. 2016) with a data-driven approach. First, CPORT(de Vries and Bonvin 2011) was implemented to determine the predicted residues in a protein-protein interaction. Residues from the adjuvant were selected as part of the interaction with both toll-like receptors, since it has been shown as able to induce an immune response(Khatoon et al. 2017). In the vaccine, residues F32, V34, T35, A36, A38, P39, V42, A43, A45, G46, A48, P49, and A50 were selected to drive the docking, while in the adjuvant alone residues T35, A36, A38, P39, A41, V42, A43, A45, G46, A47, and P49 were chosen. In TLR4, residues I48, D50, N51, L52, P53, F54, S55, D60, P65, H68, G70, S71, Y72, S73, F75, S76, P78, D84, S86, D95, Q99, and S100 were chosen, and residues R63, T65, S85, G87, Y109, Y111 were chosen for TLR2. CPORT predicted many more residues as active, but they were not chosen in an effort to narrow the results (Supplementary Data 5). Surrounding residues were not entered as input in HADDOCK; instead, the default selection for passive residue selection was used, defining a 6.5 angstrom radius around active residues as the passive region.
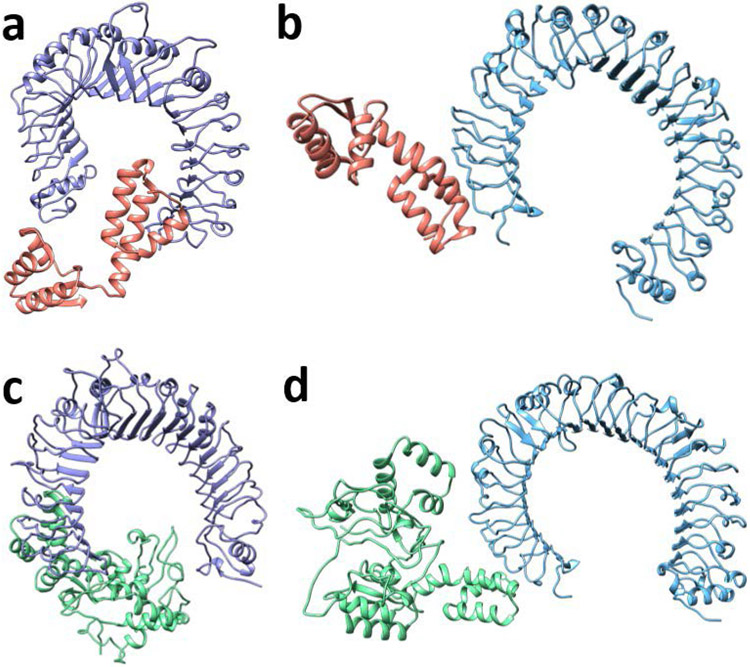
The predicted scores for each of the resulting structures (Figure 6a-d) are outlined in Table 3. The best predicted binding poses for both the TLR2 and TLR4 constructs indicated the vaccine construct induces a conformational change in the adjuvant region which is beneficial to the interaction; the Kd for the vaccine-TLR complexes is comparatively lower in both cases. This is likely due to a predicted increase in the number of interfacial contacts between the complexes. For example, in the TLR4 complexes, four of the six interfacial contact types increased (charged-charged from 3 to 4, charged-polar from 5 to 10, charged-apolar from 5 to 10, and apolar-apolar from 23 to 27), one remained the same (polar-polar at 2) and one decreased (polar-apolar from 13 to 12). Altogether, the docking results indicate that COVCCF will be able to bind TLR2/TLR4 and induce an immune response.
Figure 6: Protein-protein docking results of adjuvant or vaccine construct with TLR2 or TLR4.
Results from HADDOCK for the a) adjuvant and TLR2, b) adjuvant and TLR4, c) vaccine and TLR2, and d) vaccine and TLR4.
Table 3:
Results from protein-protein docking in HADDOCK and binding affinity predictions in PRODIGY.
| Cluster Score (±) | Z-Score | Best Structure Score |
PRODIGY Kd Prediction |
|
|---|---|---|---|---|
| Adjuvant - TLR2 | −76.7 (5.0) | −1.4 | −83.422 | 1.3E-07 |
| Vaccine - TLR2 | −84.6 (1.4) | −1.7 | −86.138 | 3.3E-08 |
| Adjuvant - TLR4 | −81.7 (2.9) | −1.4 | −85.447 | 2.6E-07 |
| Vaccine - TLR4 | −84.3 (2.3) | −1.9 | −87.720 | 2.2E-07 |
Note: The cluster score and Z-score are the aggregate scores for all proteins within the best cluster, while the best structure score is for the structure with the lowest HADDOCK score. The PRODIGY prediction is for the predicted best structure by HADDOCK score. TLR: toll-like receptor.
In Silico Codon Optimization
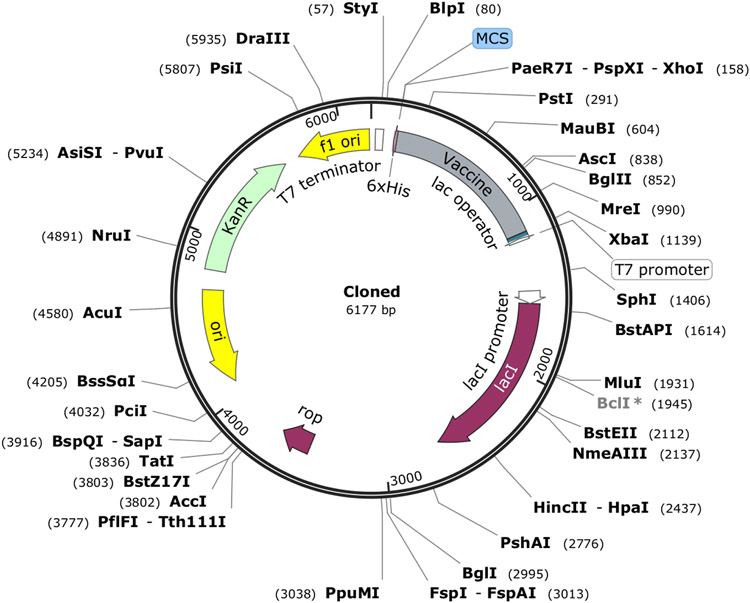
The Java Codon Adaptation Tool(Grote et al.) was used to optimize codon usage of the vaccine construct, to be expressed in E. coli (K12). This optimization would allow for maximal protein expression. A 993 base pair sequence was generated with a Codon Adaptation Index (CAI) value of 0.916, and a GC content of 50.25%, which compares favorably with the 50.73% GC content in the chosen E. coli strain. The sequence of the recombinant plasmid was then inserted in a pET30a (+) vector using SnapGene software (www.snapgene.com, Figure 7). These results indicate our protein would likely be easily cloneable in a common vector, the K12 strain of E. coli.
Figure 7:
In silico cloning of the vaccine construct using the pET30a (+) expression vector. The vaccine insertion is denoted with a gray bar. The His-tag is located at the C-terminal end. MCS: multiple cloning site; KanR: kanamycin resistance protein.
Immune Simulation Indicates Strong Secondary and Tertiary Response
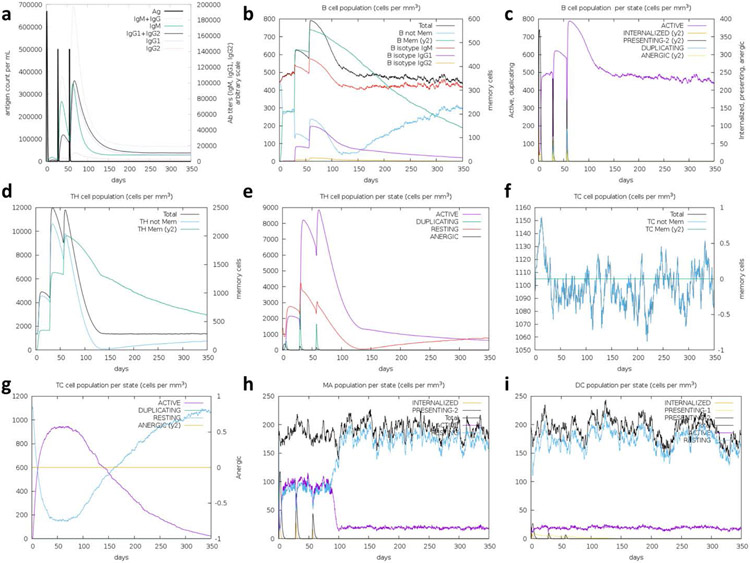
The immune simulations carried out on the C-ImmSim(Castiglione et al. 2012) server gave results consistent with an actual immune response, highlighted by the increased secondary response when compared to the primary response. High levels of immunoglobulin activity (IgM, IgG1, IgG2) in the secondary and tertiary response were matched with a corresponding decrease in antigen concentration (Ag, Figure 8a). B-cell population also increased with each injection (Figure 8b, c), while a corresponding T-helper and cytotoxic T-cell response was evident as well (Figure 8d-g). Additionally, macrophage activity was increased, while dendritic cell activity remained consistent during exposure (Figure 8h, i). Taken together, these results support the immunogenicity of COVCCF.
Figure 8: Immune simulation results from C-ImmSim.
Injections were simulated to occur at T = 0, 4, and 8 weeks. a) Antigen and immunoglobulin levels, subdivided per isotype; B lymphocytes by b) total count and c) population per entity state (active, presenting, internalized, duplicating, or anergic); CD4 T-helper lymphocytes by d) count and e) population per entity state; CD8 T-cytotoxic lymphocytes by f) count and g) population per entity state; total count of h) macrophages and i) dendritic cells. Supplementary Figures 6-14 include high-resolution copies of these figures.
Discussion
While there are currently two mRNA vaccines approved for emergency use by the FDA, it is clear more research into possible candidates must be continued. Additionally, other vaccine candidates are approaching endpoints in their clinical trials. However, as of January 13th, the 7-day moving average for cases is pushing toward 750,000 per day; it is clear that further work toward the discovery of more vaccine candidates must be done. Multiple vaccine candidates could allow for faster distribution and implementation. Additionally, novel candidates may come with fewer or reduced side effects, which could also help with implementation in those who are worried about the severity of the side effects in the mRNA vaccines(Wadman 2020). Here, we have used computational techniques, including molecular dynamics simulations and immunoinformatics techniques, to design a multi-epitope vaccine candidate which appears capable of eliciting an immune response. The full multi-epitope vaccine sequence was predicted to contain 132 IFN-γ positive epitopes; this line up well with the predicted induction of IFN-γ from C-ImmSim, which predicted levels over 400,000 ng/mL for both the primary and secondary doses (Figure 3). The immune simulation indicated results consistent with an expected immune response to a vaccine, based on the general increase in the immune response upon secondary and tertiary doses of vaccine. Protein-protein docking comparisons between the full vaccine construct with the adjuvant alone indicated a stronger interaction with both TLR2 and TLR4 in the vaccine than the adjuvant alone, indicating a potential shift in conformation which allows for better binding. This stronger interaction could indicate a quicker immune response to the vaccine candidate.
Key to this work was the implementation of the glycan shield for the selection of epitopes to be included in the final vaccine construct COVCCF. We believe it to be important to design a vaccine which would be capable of creating an immune response which would be effective against the SARS-CoV-2 spike glycoprotein in both its unglycosylated and fully glycosylated states; this unfortunately means not including epitopes which, though they may elicit a strong immune response, would generate antibodies which would not be capable of reaching their intended target. An example of this is a predicted LBL epitope from A701 through I720, predicted in all nine mutant systems and the wild type. However, while there are residues in this region which have some antibody-accessible surface area in a non-glycosylated protein, glycosylation of residues N709 and N1074 almost completely abolish this accessibility. As an example, S704 has 11.4 Å2 of AbASA when glycosylation is not accounted for (using a probe size of 0.72 nm), but is reduced to 1.04 Å2 of AbASA when glycosylation is accounted for. While there is a limitation to blindly following the probe size of 0.72 nm as the only criteria, the knowledge that this probe size only accounts for an averaged loop radius, and not the size of the loop-containing antibody, was taken into account when selecting epitopes where some interference from glycosylation was evident.
In addition, as opposed to taking a sequence-based approach to the prediction of LBL epitopes, we chose to include conformational changes which could uncover more epitopes, which may not be found using only the amino acid sequence. Additionally, we included multiple mutated systems, with the hopes of improving our chances of discovering epitopes which may not be discovered when using only the wild-type structure. To do this, we used 500 nanosecond molecular dynamics simulations of 10 different systems, which included 9 mutated systems and the wild-type system. This expanded our predictions in more than one way; for example, the initial equilibrated system which was to be submitted to unrestrained molecular dynamics simulation generated 51 LBL epitopes, while a combination of this conformation with conformations at 100, 200, 300, 400, and 500 nanoseconds of simulation yielded 120 unique LBL epitopes. Further adding to our predictions, the 9 mutated systems added another 309 unique LBL epitopes not predicted in the 6 conformations used for the wild-type system. In fact, only one out of the five LBL epitopes included in the final vaccine construct was predicted in any of the conformations of the wild-type system. It should be noted that although each of the 10 systems was used for predictions of CTL and HTL epitopes as well, since only the primary structure of the protein is used for these predictions, it was not expected that there would be a significant increase in the number of epitopes due to the inclusion of the mutants.
The final multi-epitope vaccine construct (COVCCF) consisted of antigenic, non-toxic, non-allergenic, and antibody accessible B-cell and T-cell epitopes; in addition, multiple helper T-cell epitopes, all of which were determined to induce cytokines important to innate immunity, such as IFN-γ, IL4, and IL10, were included. Our 35.9 kDa protein was predicted to be soluble upon overexpression in an E. coli host, with a theoretical pI of 8.75, implying its best stability would be in a slightly basic environment. The instability index indicates a protein that is likely to be stable in a test tube; a protein with an instability index (II) greater than 40 is not predicted to be stable, whereas the II of COVCCF is 27.57. Additionally, the aliphatic index is a positive factor for the increase of thermostability, for which our vaccine construct was scored at 79.09. Finally, the negative value for the grand average of hydropathy (GRAVY), −0.237, indicates a hydrophilic protein, allowing it to properly interact with water molecules. The in vivo half-life was predicted using the “N-end rule”; the “N-end rule” relates the half-life of a protein to the identity of the N-terminal residue, which for this protein is a methionine. Outside of an N-terminal valine, this yields the highest predicted half-life for the vaccine construct, which is a measure of how long it would take for half of the amount of protein in the cell to disappear, based on host.
Further functional validation of the final multi-epitope vaccine candidate would be required prior to implementation, the first step of which is screening for immunoreactivity using serological analysis(Gori et al. 2013). Expression of the protein in a suitable host would be required for this, with E. coli the preferred choice for recombinant protein expression. Both the value for the codon adaptation index (0.916) and GC content (50.25%) were favorable, indicating the probability of high expression in the selected E. coli (strain K12) vector. Validation of effectiveness in an animal model is also a possibility, with studies in ferrets indicating rapid transmission and indicate efficacy in identification of therapeutic applications(Kim et al. 2020; Park et al. 2020).
A key limitation to this study is the lack of inclusion of mutants in the SARS-CoV-2 spike glycoprotein which have proven to have much higher prevalence in the general population. Specifically, the D614G mutation, which in certain locations has become the predominant species(Daniloski et al. 2020), was not included. Selection of mutants for simulation studies was completed before the prevalence of this mutation had been demonstrated. It is not known if this would impact the selection of LBL epitopes for the final candidate, though CTL and HTL epitope selection would largely be unaffected since both are sequence based as opposed to conformation based in this study.
It is becoming clear that control of SARS-CoV-2, the virus which causes COVID-19, will require multiple vaccines in order to generate enough herd immunity to prevent overwhelming the capabilities of hospitals around the world. Our integration of molecular dynamics simulations and immunoinformatics techniques has allowed us to generate a potential multi-epitope vaccine, coding for multiple LBL, CTL, and HTL epitopes. The inclusion of 9 mutated spike glycoproteins, along with long timescale molecular dynamics simulations, allowed for greater sampling of conformations for the spike glycoprotein; this sampling improved our ability to select LBL epitopes which are immunogenic, antigenic, and are not shrouded by the glycan shield. The final 331 amino acid peptide vaccine, COVCCF, with its ideal physiochemical properties and ability to initiate an immune response, could be a first step in preventing the further spread of COVID-19. If broadly applied, the synergistic computational approaches presented here can be utilized to design vaccines for other emerging infectious diseases as well.
Supplementary Material
Acknowledgements
We acknowledge support from the Oak Ridge Leadership Computing Facility, the Ohio Super Computer Center, and the IBM ResearchAI Hardware Center, and the Center for Computational Innovation at Rensselaer Polytechnic Institute for computational resources for computing resources.
Funding:
This work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) under Award Number R00 HL138272 and the National Institute of Aging under Award Number R01AG066707 and 3R01AG066707-01S1. This work was supported, in part, by the VeloSano Pilot Program (Cleveland Clinic Taussig Cancer Institute).
Footnotes
Code availability. All codes written for and used in this study are available from the corresponding author upon reasonable request.
Supplementary information is available in the online version of the paper.
Competing interests. The authors declare that they have no conflict of interest. The content of this publication does not necessarily reflect the views of the Cleveland Clinic.
Data availability.
All predicted epitopes are available from Supplemental Data S1-S5. All other data are available from the corresponding author upon reasonable request.
References
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. 2015. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1-2:19–25. [Google Scholar]
- Amanat F, Krammer F. 2020. Sars-cov-2 vaccines: Status report. Cell Press. p. 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta M, Karosiene E, Rasmussen M, Stryhn A, Buus S, Nielsen M. 2015. Accurate pan-specific prediction of peptide-mhc class ii binding affinity with improved binding core identification. Immunogenetics. 67(11-12):641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB et al. 2020. Efficacy and safety of the mrna-1273 sars-cov-2 vaccine. New England Journal of Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DJ, Edwards MS, Thornton JM. 1986. Continuous and discontinuous protein antigenic determinants. Nature. 322(6081):747–748. [DOI] [PubMed] [Google Scholar]
- Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 4(6):1011–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD. 2012. Optimization of the additive charmm all-atom protein force field targeting improved sampling of the backbone ï†, ïˆ and side-chain ï‡(1) and ï‡(2) dihedral angles. Journal of chemical theory and computation. 8(9):3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga C, Travis KP. 2005. A configurational temperature nosé-hoover thermostat. The Journal of Chemical Physics. 123(13):134101–134101. [DOI] [PubMed] [Google Scholar]
- Buchan DWA, Jones DT. 2019. The psipred protein analysis workbench: 20 years on. Web Server issue Published online. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calis JJA, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, Keşmir C, Peters B. 2013. Properties of mhc class i presented peptides that enhance immunogenicity. PLoS Computational Biology. 9(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglione F, Mantile F, De Berardinis P, Prisco A. 2012. How the interval between prime and boost injection affects the immune response in a computational model of the immune system. Computational and mathematical methods in medicine. 2012:842329–842329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. Molprobity: All-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography. 66(1):12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C, Yeates TO. 1993. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Science. 2(9):1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z, Guo X, Sanjana NE. 2020. The d614g mutation in sars-cov-2 spike increases transduction of multiple human cell types. bioRxiv.2020.2006.2014.151357-152020.151306.151314.151357. [Google Scholar]
- Darden T, York D, Pedersen L. 1993. Particle mesh ewald: An n ·log( n ) method for ewald sums in large systems. The Journal of Chemical Physics. 98(12):10089–10092. [Google Scholar]
- de Vries SJ, Bonvin AMJJ. 2011. Cport: A consensus interface predictor and its performance in prediction-driven docking with haddock. PLoS ONE. 6(3):e17695–e17695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Gupta S, Vir P, Raghava GPS. 2013a. Prediction of il4 inducing peptides. Clinical and Developmental Immunology. 2013:263952–263952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Vir P, Raghava GPS. 2013b. Designing of interferon-gamma inducing mhc class-ii binders. Biology Direct. 8(1):30–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I, Bangov I, Flower DR, Doytchinova I. 2014. Allertop v.2 - a server for in silico prediction of allergens. Journal of Molecular Modeling. 20(6). [DOI] [PubMed] [Google Scholar]
- Doytchinova IA, Flower DR. 2007. Vaxijen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 8(1):4–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the expasy server. [DOI] [PubMed] [Google Scholar]
- Gori A, Longhi R, Peri C, Colombo G. 2013. Peptides for immunological purposes: Design, strategies and applications. Amino Acids. p. 257–268. [DOI] [PubMed] [Google Scholar]
- Grant OC, Montgomery D, Ito K, Woods RJ. 2020. Analysis of the sars-cov-2 spike protein glycan shield: Implications for immune recognition. bioRxiv.2020.2004.2007.030445-032020.030404.030407.030445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote A, Hiller K, Scheer M, Mü Nch R, Nö Rtemann B, Hempel DC, Jahn D. Jcat: A novel tool to adapt codon usage of a target gene to its potential expression host. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Sun X, Li B, Huang J, Zhan B, Zhu X. 2017. Vaccination with a paramyosin-based multi-epitope vaccine elicits significant protective immunity against trichinella spiralis infection in mice. Frontiers in Microbiology. 8(AUG). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GPS. 2013. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE. 8(9):e73957–e73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo L, Park H, Seok C. Galaxyrefine: Protein structure refinement driven by side-chain repacking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. 1997. Lincs: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 18(12):1463–1472. [Google Scholar]
- Hoffmann M, Kleine-Weber H, Krueger N, Mueller MA, Drosten C, Poehlmann S. 2020. The novel coronavirus 2019 (2019-ncov) uses the sars-coronavirus receptor ace2 and the cellular protease tmprss2 for entry into target cells. Cold Spring Harbor Laboratory. p. 2020.2001.2031.929042-922020.929001.929031.929042. [Google Scholar]
- Jespersen MC, Peters B, Nielsen M, Marcatili P. 2017. Bepipred-2.0: Improving sequence-based b-cell epitope prediction using conformational epitopes. Nucleic Acids Research. 45(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W. 2008. Charmm-gui: A web-based graphical user interface for charmm. Journal of Computational Chemistry. 29(11):1859–1865. [DOI] [PubMed] [Google Scholar]
- Khatoon N, Pandey RK, Prajapati VK. 2017. Exploring leishmania secretory proteins to design b and t cell multi-epitope subunit vaccine using immunoinformatics approach. Scientific Reports. 7(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang JH, Kim EJ, Lee S, Casel MAB et al. 2020. Infection and rapid transmission of sars-cov-2 in ferrets. Cell Host and Microbe. 27(5):704–709.e702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Weeraratne D, Snowdon JL, Parida L. 2020. Emergence of drift variants that may affect covid-19 vaccine development and antibody treatment. Pathogens. 9(5):324–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. 2007. Large-scale validation of methods for cytotoxic t-lymphocyte epitope prediction. BMC Bioinformatics. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y et al. 2016. Charmm-gui input generator for namd, gromacs, amber, openmm, and charmm/openmm simulations using the charmm36 additive force field. Journal of Chemical Theory and Computation. 12(1):405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MA. 2019. A comparison of plasmid dna and mrna as vaccine technologies. Vaccines. 7(2):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckheeram RV, Zhou R, Verma AD, Xia B. 2012. Cd4 + t cells: Differentiation and functions. Clinical and Developmental Immunology. 2012:12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WR, Cheng F. 2020. Repurposing of fda-approved toremifene to treat covid-19 by blocking the spike glycoprotein and nsp14 of sars-cov-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal G, Usmani SS, Dhanda SK, Kaur H, Singh S, Sharma M, Raghava GPS. 2017. Computer-aided designing of immunosuppressive peptides based on il-10 inducing potential. Scientific Reports. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nain Z, Abdullah F, Rahman MM, Karim MM, Khan MSA, Sayed SB, Mahmud S, Rahman SMR, Sheam MM, Haque Z et al. 2019. Proteome-wide screening for designing a multi-epitope vaccine against emerging pathogen elizabethkingia anophelis using immunoinformatic approaches. Journal of Biomolecular Structure and Dynamics. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Lund O. 2009. Nn-align. An artificial neural network-based alignment algorithm for mhc class ii peptide binding prediction. BMC Bioinformatics. 10:296–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Lund O. 2007. Prediction of mhc class ii binding affinity using smm-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 8:238–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J et al. 2020. Characterization of spike glycoprotein of sars-cov-2 on virus entry and its immune cross-reactivity with sars-cov. Nature Communications. 11(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Yu KM, Kim YI, Kim SM, Kim EH, Kim SG, Kim EJ, Casel MAB, Rollon R, Jang SG et al. 2020. Antiviral efficacies of fda-approved drugs against sars-cov-2 infection in ferrets. mBio. 11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M, Rahman A. 1981. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics. 52(12):7182–7190. [Google Scholar]
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C et al. 2020. Safety and efficacy of the bnt162b2 mrna covid-19 vaccine. New England Journal of Medicine. 383(27):2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, Peters B. 2008. Ellipro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 9:514–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapin N, Lund O, Bernaschi M, Castiglione F. 2010. Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 5(4):e9862–e9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, Peters B. 2008. Quantitative peptide binding motifs for 19 human and mouse mhc class i molecules derived using positional scanning combinatorial peptide libraries. Immunome Research. 4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingluff CL Jr. 2011. The present and future of peptide vaccines for cancer: Single or multiple, long or short, alone or in combination? Cancer journal (Sudbury, Mass). 17(5):343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormanni P, Amery L, Ekizoglou S, Vendruscolo M, Popovic B. 2017. Rapid and accurate in silico solubility screening of a monoclonal antibody library. Scientific Reports. 7(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormanni P, Aprile FA, Vendruscolo M. 2015. The camsol method of rational design of protein mutants with enhanced solubility. Journal of Molecular Biology. 427(2):478–490. [DOI] [PubMed] [Google Scholar]
- Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP, Sinigaglia F et al. 1999. Generation of tissue-specific and promiscuous hla ligand databases using dna microarrays and virtual hla class ii matrices. Nature Biotechnology. 17(6):555–561. [DOI] [PubMed] [Google Scholar]
- Tortorici MA, Veesler D. 2019. Structural insights into coronavirus entry. Academic Press Inc. p. 93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Regenmortel MHV. 1996. Mapping epitope structure and activity: From one-dimensional prediction to four-dimensional description of antigenic specificity. Methods: A Companion to Methods in Enzymology. 9(3):465–472. [DOI] [PubMed] [Google Scholar]
- Van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, Van Dijk M, De Vries SJ, Bonvin AMJJ. 2016. The haddock2.2 web server: User-friendly integrative modeling of biomolecular complexes. Journal of Molecular Biology. 428(4):720–725. [DOI] [PubMed] [Google Scholar]
- Verlet L 1967. Computer "Experiments" On classical fluids. I. Thermodynamical properties of lennard-jones molecules. Physical Review. 159(1):98–103. [Google Scholar]
- Wadman M 2020. Public needs to prep for vaccine side effects. Science. 370(6520):1022. [DOI] [PubMed] [Google Scholar]
- Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. 2010. Peptide binding predictions for hla dr, dp and dq molecules. BMC Bioinformatics. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li W, Liu S, Xu J. 2016. Raptorx-property: A web server for protein structure property prediction. Nucleic Acids Research. 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. 2020a. Site-specific glycan analysis of the sars-cov-2 spike. Science.eabb9983–eabb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Berndsen ZT, Raghwani J, Seabright GE, Allen JD, Pybus OG, McLellan JS, Wilson IA, Bowden TA, Ward AB et al. 2020b. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nature Communications. 11(1):2688–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ. 2007. Prosa-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research. 35:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Jonsson J, Sjörström M, Sandberg M, Rännar S. 1993. Dna and peptide sequences and chemical processes multivariately modelled by principal component analysis and partial least-squares projections to latent structures. Analytica Chimica Acta. 277(2):239–253. [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. 2020. Cryo-em structure of the 2019-ncov spike in the prefusion conformation. Science (New York, NY). 367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, Du L, Liu S et al. 2020. Inhibition of sars-cov-2 (previously 2019-ncov) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Research. 30(4):343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhang Y. 2011. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophysical Journal. 101(10):2525–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue LC, Rodrigues JP, Kastritis PL, Bonvin AM, Vangone A. 2016. Prodigy: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics (Oxford, England). 32(23):3676–3678. [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2014. The i-tasser suite: Protein structure and function prediction. Nature Publishing Group. p. 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhang C, Wuyun Q, Pearce R, Li Y, Zhang Y. 2019. Lomets2: Improved meta-threading server for fold-recognition and structure-based function annotation for distant-homology proteins. Nucleic Acids Research. 47:429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All predicted epitopes are available from Supplemental Data S1-S5. All other data are available from the corresponding author upon reasonable request.