Dr Barton Childs’ landmark 1999 text, Genetic Medicine – A Logic of Disease, led to the development of a new curriculum at Johns Hopkins School of Medicine entitled “Genes to Society.”1 A core part of contemporary medical school and pediatric residency training is individualized medicine. Trainees are now being equipped for an era of personalized, “precision” medicine. Childs said that, in the next century, medicine will be focused on the treatment of individuals rather than disease. This raises the question of how different are individuals at the genomic level? Early after the discovery of the genetic code, it was recognized that some 3 million single nucleotide polymorphisms could be used to distinguish individuals, based on their location within our unique DNA. Other sources of variation in the genome range from few-base-pair insertions and deletions and short tandem repeats to mega-base-pair variations owing to cytogenetic deletions, insertions, or aneuploidy. Once the human genome was sequenced, it became clear that more than 10% of the genome consists of copy number variations that also generate a unique signature for each individual.2 The fascinating observation that progression from simple organisms to higher-order organisms and ultimately humans was not associated with a significant increase in the number of genes, but rather, an increasing amount of DNA sequences unassociated with genes, so-called junk DNA. We now know that a substantial portion (≥30%) of this junk DNA is transcribed into noncoding RNA (ncRNA)—RNA that, instead of coding for proteins, serves a direct or indirect regulatory function for those genes that do code for proteins. To date, more than 18 000 distinct ncRNAs have been identified. In many cases, these ncRNAs serve as precursors to generate small inhibitory RNAs, which regulate target gene expression. Other ncRNAs bind proteins and serve as protein translocators and/or facilitate the formation of multiprotein complexes. Interestingly, the transcripts of 88% of single nucleotide polymorphisms that have been associated with different phenotypes are actually located within ncRNAs. These single nucleotide polymorphisms could result in different secondary structures, and are thereby likely responsible for differential functions of the ncRNA.
In 2012, the ENCODE project consortium revealed that as much as 80% of the human genome is actively transcribed.3 However, only about 2% of the genome is protein coding, suggesting the rest of the transcripts are ncRNA transcripts. This discovery led to great interest in unraveling the mechanism governing the biogenesis and function of ncRNAs. Although a vast majority of ncRNAs are transcribed at low levels, and may be transcriptional noise, without any functional role, recent advances of next-generation high-throughput sequencing coupled with functional analyses have yielded numerous discoveries of functional ncRNAs across species.4,5 The most abundant portion of ncRNAs are housekeeping ribosomal RNAs and transfer RNAs, which are well-known for their functional role in normal cellular processes for quite some time.4 Recent experiments have primarily focused on ncRNAs other than ribosomal RNAs and transfer RNAs. This minor fraction has been shown to play crucial roles in a myriad of physiologic processes, including genome integrity maintenance, innate immunity, neurodevelopment, and stem cell proliferation and differentiation, as well as diseases such as cancer. These new regulatory noncoding transcripts are traditionally divided into 2 major groups based on their size and an arbitrary cutoff: short noncoding RNAs (18–200 nucleotides) and long ncRNAs (lncRNAs) (>200 nucleotides). In addition, a novel class of ncRNAs was recently discovered, the circular RNAs (circRNAs), named because of the circular nature of the transcript generated from back-splicing of pre-mRNA and covalent linking of 3′ and 5′ ends.6–9 The size of circRNAs ranges from less than 200 to several thousand nucleotides (Figure 1).
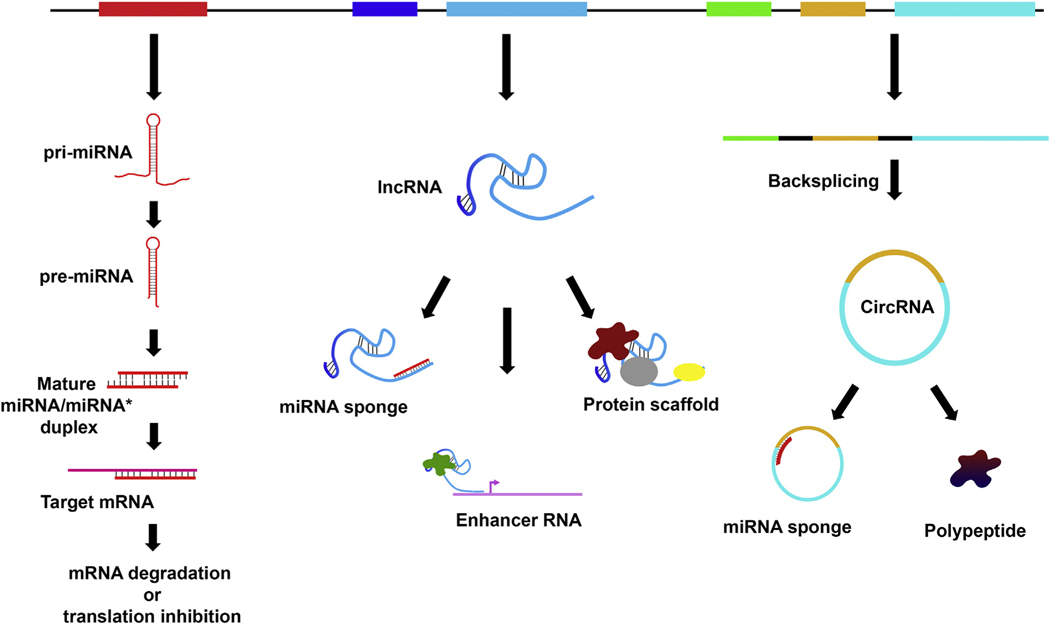
Figure 1.
A schematic illustrating various ncRNAs. The biogenesis mechanisms and functions of various ncRNAs are shown (miRNAs, left; lncRNAs, middle; circRNAs, right). pri-miRNA, primary microRNA.
Short ncRNAs are further categorized into several classes, including small nuclear RNAs, which are key components of the spliceosome and play an important role in pre-mRNA splicing, and small nucleolar RNAs, which regulate ribosomal RNA modification. Additional classes of short (21–30 nt) ncRNAs include small interfering RNAs (siRNAs), Piwi-interacting RNAs, and microRNAs (miRNAs or miRs), which are core components of RNA interference (RNAi), an evolutionarily conserved process that regulates gene expression in a sequence-specific manner. The siRNAs are derived from long double-stranded RNA precursors. They join and guide Argonaute protein-containing complexes to target mRNAs by complementary base pairing, leading to gene silencing via RNA degradation. Viral RNA replication intermediates (in the form of double-stranded RNAs) can be converted into siRNAs, which in turn can target viral RNA transcripts for degradation via RNAi.10–17 Thus, RNAi is a key constituent of antiviral defense mechanism in diverse host organisms. The miRNAs are transcribed as long stem-loop primary transcripts, which are processed into 22- to 24-nt segments.18–29 Similar to siRNAs, miRNAs are loaded into and guide Argonaute complexes to target mRNAs by imperfect base pairing between miRNAs and target mRNAs, and decrease protein output by mRNA destabilization and translation inhibition.30–32 Piwi-interacting RNAs are primarily produced in germ cells and control the expression of transposable elements, thereby contributing to the maintenance of germ-line genome integrity.33
The lncRNAs are characterized based on location and orientation of resulting transcript into long intergenic ncRNAs, natural antisense transcripts, enhancer RNAs, and bidirectional transcripts.5,34 LncRNAs are involved in functionally diverse mechanisms. They can serve as transcriptional repressors (eg, XIST), enhancers by promoting activating interactions between promoters and distal regulatory elements (eg, LUNAR1), miRNAs sponges (eg, TUG1), and hubs for protein-protein and protein-nucleic acid interactions (eg, SPRIGHTLY).5,34–39
The circRNAs are generated by back-splicing reactions during pre-mRNA processing. Emerging evidence has delineated diverse modes of action of circRNAs. For example, the mouse circRNA CDR1as/CiRS-7 shelters miR-7 and impacts brain development.9,40,41 In addition, the circRNA SRY plays a prominent role in male sex determination.9,40 Furthermore, select intron-containing circRNAs can interact with U1 small nuclear ribonucleoprotein particle and promote host gene transcription in the nucleus.42 Moreover, circRNAs can modulate gene expression by competing with linear splicing.43 Lastly, selective circRNAs can give rise to functional polypeptides, thereby expanding the complexity of the proteome.44,45
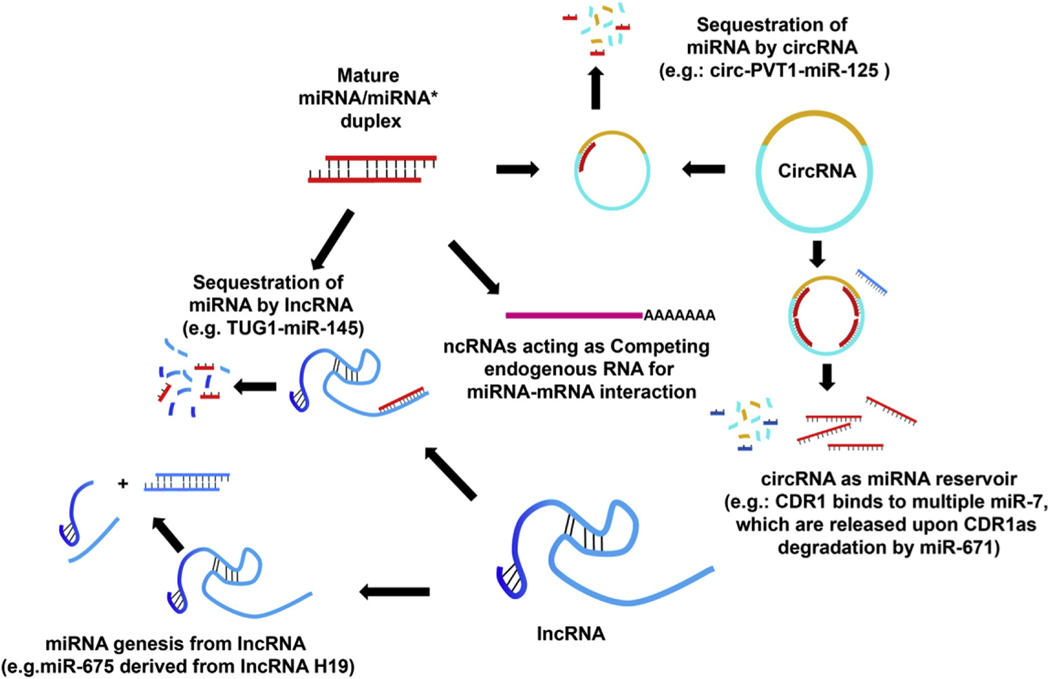
Cross-regulation among various classes of RNAs has been well-documented (Figure 2). For example, the lncRNA TUG1 can sequester and functionally inhibit the activity of a number of miRNAs.38,46–49 In addition, the lncRNA H19 serves as a source of precursors for miR-675 biogenesis.50–53 An elegant example of ncRNA cross-regulation is the miR-671-CDR1as-miR-7 axis: the circRNA CDR1as carries 1 near perfect binding site for miR-671 and dozens of imperfect binding sites for miR-7. Engagement of miR-671 with CDR1as results in CDR1as degradation, which leads to downregulation of miR-7 owing to loss of protection by CDR1as.9,40,41 Lastly, ncRNAs (lncRNAs and circRNAs) and protein-coding mRNAs can engage with and therefore compete for the same pool of miRNAs, resulting in cross-regulation. In fact, cross-regulation among various mRNAs and between mRNAs and lncRNAs via miRNA engagement provides support for this model.54
Figure 2.
A schematic depicting cross-regulation among various classes of ncRNAs. On the one hand, miRNAs can bind and downregulate the expression of target lncRNAs; on the other hand, select lncRNAs can serve as miRNA precursors or sequestrate and functionally inhibit miRNAs. In addition, select circRNAs can inhibit or protect miRNAs by physically associating with miRNAs. Conversely, miRNAs that carry perfect complementarity to circRNAs can lead to target circRNA degradation. Last, both lncRNAs and circRNAs can serve as competing endogenous RNAs by sequestrating miRNAs, thereby modulating the availability of miRNAs to engage with and functionally inhibit target mRNAs.
Functionally, these ncRNAs species regulate cellular identity and function primarily via modulating transcriptional and post-transcriptional gene expression. Given the diverse physiological processes regulated by ncRNAs, it is not surprising that dysregulation of ncRNA expression can contribute to pathological conditions.55–57 There is now a growing body of evidence implicating ncRNAs in normal development, health, and dysfunctions. We summarize current knowledge on the role of ncRNAs in several diseases and the implications for therapy (Table). We note that extensive literature covering the topic of ncRNAs in pediatric diseases is still developing because the field of is at early stages. In particular, the pediatric translational research applications to date in the rapidly emerging basic science field of ncRNA remain fewer in number than among studies focused on diseases that primarily affect adults. The examples discussed herein are primarily based on studies involving cultured cells or animal models and focus heavily on basic science, and thus may not be specific to children. However, we believe that knowledge gained from these preclinical/early stage clinical studies discussed here will help us to better understand the molecular mechanisms underlying these diseases in the pediatric patient population and facilitate the development of novel diagnostic and therapeutic tools.
Table.
ncRNAs implicated in various diseases
| Diseases | ncRNA | Mechanism | References |
|---|---|---|---|
|
| |||
| β-thalassemia | miR-486-3p and miR-210 | Repressing BCL11A | 58 |
| miR-23a | Repressing KLF-2 | 59 | |
| miR-15a and miR-16-1 | Repressing MYB | 60 | |
| miR-27a | Repressing Sp1 | 59 | |
| HMI-LNCRNA | Repressing γ-globin | 61 | |
| DMD | miR-486 | Repressing PTEN and Foxo1a | 62 |
| miR-21 | Repressing PTEN and SPRY-1 | 63 | |
| miR-29a and miR-29c | Repressing COL3A1, FBN1 and YY1 | 63 | |
| linc-MD1 | Sequestering miR-133 and miR-135 | 64 | |
| circ-ZNF609 | Encoding a new protein | 44,45 | |
| Rett syndrome | miR-199a | Derepressing mTOR signaling | 65 |
| AK087060 | Dysregulation upon MeCP2 loss contributes to Rett syndrome | 66-68 | |
| AK081227 | Elevated expression correlates with downregulation of Gabrr2 | 68-70 | |
| Glioma | miR-9/9* | Repressing SOX2, PTCH1, FOXP1 and CAMTA1 | 71-73 |
| miR-17-92 cluster | repressing CFTG | 74,75 | |
| miR-17, miR-19a/b, miR-26a and miR-221/222 | Inhibiting PTEN signaling | 76-79 | |
| let-7a, miR-101, miR-124, miR-138, miR-214 and miR-708 | targeting EZH2-dependent epigenetic mechanisms | 80-85 | |
| miR-7 | Repressing EGFR | 86-88 | |
| miR-128 | Repressing EGFR, WEE1, MSI1, and RPS6KB1 | 89-91 | |
| miR-34a | Repressing CDK6 and CCND1 | 92 | |
| miRNA-100 | Repressing PLK1 | 93 | |
| H19 | Sequestering miR-140 and miR-29a | 94,95 | |
| HOTAIR | Sequestering miR-326 and interacting with EZH2 | 96,97 | |
| MEG3 | Activating p53 | 98 | |
| TUG1 | Interacting with EZH2, SUZ12 and YY1 | 99 | |
| GAS5 | Binding to miR-196a-5p and upregulating FOXO1 | 100 | |
| Medulloblastoma | miR-17-92 cluster | Positive effector of Shh-mediated proliferation | 101,102 |
| miR-10b | Expression positively correlated with BCL2 expression | 101,103 | |
| miR-21 | Repressing PDCD4 | 101,104 | |
| miR-124 | Repressing CDK6 and SCL16A1 | 105,106 | |
| miR-218 | Repressing NANOG RICTOR, CTSB and CDK6 | 107,108 | |
| miR-125b | Repressing SMO and LIFRα | 109,110 | |
| miR-326 | Repressing SMO | 109 | |
| MIR100HG | Sequestering miR-19a-3p, miR-19b-3p and miR-106a-5p and derepressing CDK6, MYCN, SNCAIP and KDM6A | 111 | |
| PVT1 | Stabilizing MYC | 112 | |
mTOR, mammalian target of rapamycin; PTEN, phosphatase and tensin homolog.
Role of ncRNAs in Pediatric Diseases
β-Thalassemia is a recessive inherited disease affecting hundreds of thousands of individuals worldwide, with symptomatic onset in childhood. Before birth, the predominant hemoglobin is α2γ2. In the last trimester, fetal γ globin synthesis decreases and adult β globin synthesis increase. A hallmark of β-thalassemia is a decrease in or absence of β globin synthesis, necessary for the predominant adult hemoglobin (α2β2), resulting in anemia and ineffective erythropoiesis. Current treatments include chelation therapy, blood transfusion, and bone marrow transplantation. Recently, a novel strategy has been explored, which involves reactivation of gene encoding fetal γ globin, to compensate for the shortage of β globin. A suite of miRNAs that functionally inhibit the γ globin gene transcriptional repressors have been identified and validated: miR-486–3p and miR-210 (which target BCL11A mRNA), miR-23a (against KLF-2), miR-15a and miR-16–1 (against MYB), and miR-27a (against Sp1).58–60 Experimental approaches that enhance the activities of these miRNAs are expected to induce the γ globin gene, thereby harboring potential as a new approach to treating β-thalassemia. The lncRNAs have also been implicated in β-thalassemia.61,113,114 For example, the nuclear lncRNA HMI-LNCRNA generated from the HBS1L-MYB enhancer region displays significantly higher levels of expression in erythroblasts derived from cultured adult peripheral blood cells, which express more β globin, compared with erythroblasts from cultured cord blood cells, which express more γ globin. Notably, downregulation of HMI-LNCRNA in HUDEP-2 cells, which express mostly β globin, significantly reactivates γ globin expression and promotes erythroid maturation. Thus, HMI-LNCRNA might be a potential therapeutic target for γ globin induction treatment in β-thalassemia.
Duchenne muscular dystrophy (DMD) is a lethal neuromuscular disease and is the most common muscular dystrophy affecting children. DMD is characterized by a rapid progression of muscle degeneration caused by mutations in the dystrophin gene. Several miRNAs have been implicated in DMD. For example, the muscle-enriched miRNA miR-486 is markedly decreased in the muscles of dystrophin-deficient mice and in DMD patient samples. Mechanistically, miR-486 suppresses the expression of phosphatase and tensin homolog (PTEN) and Foxo1a, negative regulators of phosphoinositide-3-kinase (PI3K)/Akt signaling, which regulates muscle hypertrophy and growth, as well as dedicator of cytokinesis 3 (DOCK3), thereby playing a key role in myotube survival.62,115 In addition, miR-21 expression is significantly increased in DMD samples, and correlates with a significant reduction in the expression of miR-21 target transcripts including PTEN and SPRY-1 (Sprouty homolog 1), whereas miR-29a and miR-29c are significantly decreased in Duchenne muscle and myoblasts, accompanied by a concordant increase in miR-29 target transcripts, including COL3A1, FBN1, and YY1.63 Several additional muscle-enriched miRNAs, such as miR-1, miR-133, and miR-206 display an increase in the serum of DMD patients and/or in muscle tissues of mouse DMD models, suggesting that they may serve as biomarkers for DMD. When it comes to lncRNAs, it has been shown that the lncRNA linc-MD1 can operate as a miRNA sponge by sequestering miR-133 and miR-135, thereby modulating the expression of Maml1, Mef2c, Myog, and Mhc, which regulate muscle-specific gene expression. In the muscle of DMD patients the level of linc-MD1 is greatly reduced, whereas linc-MD1 overexpression can rescue the defective myogenic differentiation and restore the normal expression of the aforementioned linc-MD1-regulated genes.64 Last, a recent study provided evidence that the circRNA circ-ZNF609 can be translated into a functional protein that modulates myogenesis, thereby adding circRNAs to the list of regulatory RNAs in muscle development.44,45
Rett syndrome is a neurodevelopmental disorder associated with mutations in the MeCP2 gene encoding methyl-CpG binding protein 2. It has been reported that the MeCP2 protein associates with the miRNA biogenesis machinery and is required for appropriate post-translational processing of a suite of miRNAs.65 Among these MeCP2-regulated miRNAs is miR-199a, which suppresses the expression of inhibitory factors of mammalian target of rapamycin (mTOR) signaling that has been implicated in Rett syndrome. Besides miRNA regulation, MeCP2 can also modulate lncRNA expression. MeCP2 loss in the mouse brain is associated with upregulation of 2 lncRNAs, AK081227 and AK087060.116 In particular, elevated expression of AK087060 in MeCP2 knockout mouse brain correlates with an increase in the expression of its host gene Arhgef26 encoding a Rho guanine nucleotide exchange factor that contributes to axon patterning.66,67 Thus, it is possible that dysregulation of AK087060 and Arhgef26 upon MeCP2 loss in mouse brain contributes to Rett syndrome phenotypes. In contrast, elevated expression of AK081227 is associated with downregulation of gamma-aminobutyric acid (GABA) receptor subunit rho 2 (Gabrr2). Because dysfunction in GABAergic inhibitory neurotransmission is associated with many neurodevelopmental disorders, including Rett syndrome, and that the expression of another GABA receptor subunit member (GABRB3) is reduced in Rett syndrome, it is likely that AK081227 and Gabrr2 are candidates to be altered in Rett syndrome.69,70,117,118 Lastly, the observation that most circRNAs are enriched in the brain suggests a functional relevance in neurodevelopment. In fact, the circRNA CDR1as/CiRS-7 plays a key role in brain development, at least in part, by associating with and stabilizing miR-7.9,40,41 We envision that advances in circRNA study will continue to provide insights regarding the role of circRNAs in normal neurodevelopment and neuro-logic diseases, such as Rett syndrome.
Glioma is a cancer originating in glial cells that are primarily involved in nourishment and upkeep of neighboring neurons.119 Gliomas are the most common brain tumor in children. The most frequent form, low-grade gliomas, are generally not associated with poor prognosis whereas high-grade glioma are often fatal.120 Various differentially regulated miRNAs and lncRNAs have been identified in gliomas.
Oncogenic miRNAs that promote glioma pathogenesis include miR-9/9* that regulates SOX2, PTCH1, FOXP1, and CAMTA1, the miR-17–92 cluster targeting CFTG, and miR-17, miR-19a/b, miR-26a, and miR-221/222 that inhibit tumor suppressive PTEN signaling.71–79 Tumor suppressor miRNAs, such as let-7a, miR-101, miR-124, miR-138, miR-214, and miR-708 are involved in inhibiting glioma/glioblastoma growth, particularly by targeting EZH2-dependent epigenetic mechanisms.80–85 Other tumor suppressor miRNAs in high-grade glioma include miR-7 (targeting EGFR), miR-128 (targeting EGFR, WEE1, MSI1, and RPS6KB1), miR-34a (targeting CDK6 and CCND1), and miRNA-100 (targeting PLK1).86–93
LncRNAs are also found to be associated with IDH1/2 mutation status and glioma grade, thereby highlighting their potential diagnostic and prognostic significance.121 Importantly, select lncRNAs have been analyzed for potential functional role in gliomas. For example, the lncRNA H19 is upregulated in gliomas where it acts as oncogene by sequestering miR-140 and miR-29a, thereby relieving their inhibitory effect on key oncogenes such as CDK6, CASH2, MDR, and so on.94,95 Another lncRNA, HOTAIR, can activate the fibroblast growth factor (FGF), PI3K/AKT, and MEK pathways and promote glioma proliferation and metastasis by serving as a miR-326 sponge, and regulate cell cycle progression in glioma via interaction with EZH2.96,97 Of note, HOTAIR has also been shown to be activated by epigenetic regulator BRD4 in glioma genesis, a well-known oncogene across cancer types.122 In addition, the lncRNA MEG3 inhibits cell proliferation via p53 activation.98 Furthermore, the lncRNA TUG1 maintains glioma stem cells through interactions with PRC2 components (EZH2 and SUZ12) and transcription factor YY1, thereby epigenetically suppressing multiple neuronal differentiation-associated genes.99 Moreover, the lncRNA GAS5 suppresses glioma stem cell proliferation, migration, and invasion by binding to the oncogenic miR-196a-5p and upregulating the downstream FOXO1.100 Lastly, XIST is another oncogenic lncRNA that modulates epigenetic pathways and promotes cell proliferation and migration in gliomas.123
Medulloblastoma represents the most common malignant pediatric brain tumor, localized in cerebellum. Recent genetic and epigenetic studies have characterized medulloblastoma into four clinical and molecular subgroups, namely, wingless (WNT), sonic hedgehog (SHH), group 3, and group 4 (reviewed in124). As of now, miRNAs and lncRNAs are the predominant ncRNA species that have been investigated in medulloblastoma pathogenesis (reviewed in101). Both miRNAs and lncRNAs show subgroup specific expression pattern, thereby highlighting subgroup-specific molecular mechanism regulated by these ncRNA species.
Several miRNA such as miR-17–92 cluster, miR-10b and miR-21, have been shown to promote medulloblastoma proliferation and/or metastasis in vitro and in vivo.101–104 Conversely, miR-124, miR-218, miR-125b, and miR-326 are examples of tumor suppressor miRNAs found downregulated in medulloblastomas.105–110,125,126 Some of these candidates also represent potential therapeutic targets. For example, the miR-17–92 cluster, which was found to be associated with SHH medulloblastoma, promotes tumor development in vivo.127 Consequently, complete knockout or locked nucleic acid (LNA) based inhibition of the miR-17–92 cluster reduced tumor growth and improved survival in SHH medulloblastoma mice.
The lncRNAs in medulloblastoma have received comparatively little attention, as of yet. A recent genome-wide lncRNA analysis highlighted subgroup-specific lncRNA expression in medulloblastoma patients and proposed a diagnostic and prognostic model based on lncRNAs.128 Several other in vitro studies also highlight functional role of lncRNAs in medulloblastoma. The lncRNA MIR100HG was found to act as oncogene in group 4 tumors where it sponged miR-19a-3p, miR-19b-3p, and miR-106a-5p, thereby derepressing their targets, including CDK6, MYCN, SNCAIP, and KDM6A, and promoting proliferation.111 However, surprisingly, overexpression of MIR100HG in the group 3 cell line downregulated their proliferation. PVT1 is another oncogenic lncRNA in medulloblastoma, particularly group 3 patients, where it is frequently found fused to oncogene MYC.129 One consequence of the resulting fusion transcript is stabilization of MYC mRNA.112
CircRNAs are also gaining interest in medulloblastoma research. It has been shown that various cancer types display distinct circRNA signatures and that circRNAs may serve as tumor biomarkers.130,131 In addition, Lv et al recently identified and validated 33 circRNAs that are dysregulated in medulloblastoma.132 Interestingly, 2 circRNAs (circ-SKA3 and circ-DTL) seem to modulate expression of corresponding host genes and impact the proliferation, migration, and invasion of tumor cells. Considering that circRNA research is still at infancy, we expect that rapid progress in this field will solidify the notion that circRNAs can modulate and perhaps drive the development and progression of various cancer types, such as medulloblastoma.
Conclusions and Future Directions
We have summarized several physiologic processes regulated by ncRNAs, and provide examples of a wide variety of diseases resulting from ncRNA dysregulation. Although the examples discussed herein may not be strictly specific to children, similar, if not identical, underlying molecular mechanisms operate in both adults and children. We envision that the exciting field of transcriptomics and ncRNA research, which encompasses both basic science and translational studies, will continue to benefit from rapid advances in the development of next generation sequencing technology and bioinformatics tools. This will facilitate the elucidation of the molecular mechanism underlying the function and regulation of ncRNAs in physiological and pathological settings, and provide insights into the development of ncRNA-based diagnostic and therapeutic strategies against pediatric diseases.
Acknowledgments
Publication of this supplement was supported by the Johns Hopkins All Children’s Foundation.
Author Disclosures
Supported by the National Institutes of Health (1R21AI131099, 1R03AI131068, and 1R01AI140049 [to R.Z.]; 1R21CA202197, 1R03CA172847, and 1R03CA165184 [to R.J.P.]), Florida Department of Health (Bankhead-Coley Cancer Research Program 5BC08 [to R.J.P.]), and Johns Hopkins University (to R.Z. and R.J.P.).
Glossary
- circRNA
Circular RNA
- DMD
Duchenne muscular dystrophy
- GABA
Gamma-aminobutyric acid
- lncRNA
Long ncRNA
- miRNA
MicroRNAs
- ncRNA
Noncoding RNA
- SHH
Sonic hedgehog
- siRNA
Small interfering RNA
References
- 1.Childs B. Genetic medicine: a logic of disease. Baltimore: Johns Hopkins University Press; 1999. [Google Scholar]
- 2.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature 2006;444:444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hombach S, Kretz M. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol 2016;937:3–17. [DOI] [PubMed] [Google Scholar]
- 5.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009;23:1494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–8. [DOI] [PubMed] [Google Scholar]
- 10.Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science 2006;312:452–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 2006;20:2985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol 2006;7:590–7. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science 2013;342: 231–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, et al. Antiviral RNA interference in mammalian cells. Science 2013;342:235–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, et al. An endogenous small interfering RNA pathway in Drosophila. Nature 2008;453:798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, et al. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell 2009;138:340–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong XP, Kurthkoti K, Chang KY, Lichinchi G, De N, Schneemann A, et al. Core small nuclear ribonucleoprotein particle splicing factor SmD1 modulates RNA interference in Drosophila. Proc Natl Acad Sci U S A 2013;110:16520–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901–6. [DOI] [PubMed] [Google Scholar]
- 19.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–54. [DOI] [PubMed] [Google Scholar]
- 20.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004;432:235–40. [DOI] [PubMed] [Google Scholar]
- 21.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003;425:415–9. [DOI] [PubMed] [Google Scholar]
- 22.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature 2004;432: 231–5. [DOI] [PubMed] [Google Scholar]
- 23.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol 2005;3:e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001;293: 834–8. [DOI] [PubMed] [Google Scholar]
- 25.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 2005;3:e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, et al. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 2009;15:1886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 2005;19: 1674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong XP, Vogler G, Kurthkoti K, Samsonova A, Zhou R. SmD1 Modulates the miRNA pathway independently of its pre-mRNA splicing function. PLoS Genet 2015;11:e1005475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell 2008;32:592–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther 2016;1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010;466: 835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 2005;309:1573–6. [DOI] [PubMed] [Google Scholar]
- 33.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 2008;322:1387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM, Li CW, et al. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res 2012;4:127–50. [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 2016;73: 2491–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008;322: 750–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 2014;158:593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu C, Li L, Xie F, Guo S, Liu F, Dong N, et al. LncRNA TUG1 sponges miR-204–5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc Res 2018;114:168–79. [DOI] [PubMed] [Google Scholar]
- 39.Lee B, Sahoo A, Marchica J, Holzhauser E, Chen X, Li JL, et al. The long noncoding RNA SPRIGHTLY acts as an intranuclear organizing hub for pre-mRNA molecules. Sci Adv 2017;3:e1602505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient micro-RNA sponges. Nature 2013;495:384–8. [DOI] [PubMed] [Google Scholar]
- 41.Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017;357:eaam8526. [DOI] [PubMed] [Google Scholar]
- 42.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256–64. [DOI] [PubMed] [Google Scholar]
- 43.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55–66. [DOI] [PubMed] [Google Scholar]
- 44.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 2017;66:22–37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell 2017;66:9–21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi L, Tian C, Sun L, Cao F, Meng Z. The lncRNA TUG1/miR-145–5p/FGF10 regulates proliferation and migration in VSMCs of hypertension. Biochem Biophys Res Commun 2018;501:688–95. [DOI] [PubMed] [Google Scholar]
- 47.Zeng B, Ye H, Chen J, Cheng D, Cai C, Chen G, et al. LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget 2017;8: 113650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Liao S, Li H, Chen Y, Yu J. Long Non-coding RNA TUG1 sponges Mir-145a-5p to regulate microglial polarization after oxygen-glucose deprivation. Front Mol Neurosci 2019;12:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou H, Sun L, Wan F. Molecular mechanisms of TUG1 in the proliferation, apoptosis, migration and invasion of cancer cells. Oncol Lett 2019;18:4393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012;14:659–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu G, Xiang T, Wu QF, Wang WX. Long Noncoding RNA H19-Derived miR-675 enhances proliferation and invasion via RUNX1 in gastric cancer cells. Oncol Res 2016;23:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675–3p and miR-675–5p to promote skeletal muscle differentiation and regeneration. Genes Dev 2014;28:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo H, Wang J, Liu D, Zang S, Ma N, Zhao L, et al. The lncRNA H19/miR-675 axis regulates myocardial ischemic and reperfusion injury by targeting PPARalpha. Mol Immunol 2019;105:46–54. [DOI] [PubMed] [Google Scholar]
- 54.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med 2018;24:257–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861–74. [DOI] [PubMed] [Google Scholar]
- 57.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome 2008;19:454–92. [DOI] [PubMed] [Google Scholar]
- 58.Gasparello J, Fabbri E, Bianchi N, Breveglieri G, Zuccato C, Borgatti M, et al. BCL11A mRNA targeting by miR-210: a possible network regulating gamma-globin gene expression. Int J Mol Sci 2017;18:2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma Y, Wang B, Jiang F, Wang D, Liu H, Yan Y, et al. A feedback loop consisting of microRNA 23a/27a and the beta-like globin suppressors KLF3 and SP1 regulates globin gene expression. Mol Cell Biol 2013;33:3994–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sankaran VG, Menne TF, Scepanovic D, Vergilio JA, Ji P, Kim J, et al. MicroRNA-15a and −16–1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc Natl Acad Sci U S A 2011;108: 1519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma J, Liu F, Du X, Ma D, Xiong L. Changes in lncRNAs and related genes in beta-thalassemia minor and beta-thalassemia major. Front Med 2017;11:74–86. [DOI] [PubMed] [Google Scholar]
- 62.Small EM, O’Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci U S A 2010;107:4218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zanotti S, Gibertini S, Curcio M, Savadori P, Pasanisi B, Morandi L, et al. Opposing roles of miR-21 and miR-29 in the progression of fibrosis in Duchenne muscular dystrophy. Biochim Biophys Acta 2015;1852:1451–64. [DOI] [PubMed] [Google Scholar]
- 64.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147: 358–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsujimura K, Irie K, Nakashima H, Egashira Y, Fukao Y, Fujiwara M, et al. miR-199a Links MeCP2 with mTOR signaling and its dysregulation leads to Rett syndrome phenotypes. Cell Rep 2015;12:1887–901. [DOI] [PubMed] [Google Scholar]
- 66.Kolpak AL, Jiang J, Guo D, Standley C, Bellve K, Fogarty K, et al. Negative guidance factor-induced macropinocytosis in the growth cone plays a critical role in repulsive axon turning. J Neurosci 2009;29: 10488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo D, Standley C, Bellve K, Fogarty K, Bao ZZ. Protein kinase Calpha and integrin-linked kinase mediate the negative axon guidance effects of Sonic hedgehog. Mol Cell Neurosci 2012;50:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petazzi P, Sandoval J, Szczesna K, Jorge OC, Roa L, Sayols S, et al. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol 2013;10:1197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Medrihan L, Tantalaki E, Aramuni G, Sargsyan V, Dudanova I, Missler M, et al. Early defects of GABAergic synapses in the brain stem of a MeCP2 mouse model of Rett syndrome. J Neurophysiol 2008;99:112–21. [DOI] [PubMed] [Google Scholar]
- 70.Coghlan S, Horder J, Inkster B, Mendez MA, Murphy DG, Nutt DJ. GABA system dysfunction in autism and related disorders: from synapse to symptoms. Neurosci Biobehav Rev 2012;36:2044–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ben-Hamo R, Zilberberg A, Cohen H, Efroni S. hsa-miR-9 controls the mobility behavior of glioblastoma cells via regulation of MAPK14 signaling elements. Oncotarget 2016;7:23170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen X, Yang F, Zhang T, Wang W, Xi W, Li Y, et al. MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J Exp Clin Cancer Res 2019;38:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, et al. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9-mediated suppression of SOX2. Cancer Res 2011;71:3410–21. [DOI] [PubMed] [Google Scholar]
- 74.Lu S, Wang S, Geng S, Ma S, Liang Z, Jiao B. Increased expression of microRNA-17 predicts poor prognosis in human glioma. J Biomed Biotechnol 2012;2012:970761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ernst A, Campos B, Meier J, Devens F, Liesenberg F, Wolter M, et al. De-repression of CTGF via the miR-17–92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 2010;29:3411–22. [DOI] [PubMed] [Google Scholar]
- 76.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev 2009;23:1327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jia Z, Wang K, Zhang A, Wang G, Kang C, Han L, et al. miR-19a and miR-19b overexpression in gliomas. Pathol Oncol Res 2013;19:847–53. [DOI] [PubMed] [Google Scholar]
- 78.Tokudome T, Sasaki A, Tsuji M, Udaka Y, Oyamada H, Tsuchiya H, et al. Reduced PTEN expression and overexpression of miR-17–5p, −19a-3p, −19b-3p, −21–5p, −130b-3p, −221–3p and −222–3p by glioblastoma stem-like cells following irradiation. Oncol Lett 2015;10:2269–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quintavalle C, Garofalo M, Zanca C, Romano G, Iaboni M, del Basso De Caro M, et al. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu. Oncogene 2012;31:858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang XR, Luo H, Li HL, Cao L, Wang XF, Yan W, et al. Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro-oncology 2013;15:1491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo P, Lan J, Ge J, Nie Q, Mao Q, Qiu Y. miR-708 acts as a tumor suppressor in human glioblastoma cells. Oncol Rep 2013;30:870–6. [DOI] [PubMed] [Google Scholar]
- 82.Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget 2010;1:710–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 2012;61:278–89. [DOI] [PubMed] [Google Scholar]
- 84.Qiu S, Huang D, Yin D, Li F, Li X, Kung HF, et al. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochim Biophys Acta 2013;1832:1697–707. [DOI] [PubMed] [Google Scholar]
- 85.Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell 2009;36:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 2008;68:3566–72. [DOI] [PubMed] [Google Scholar]
- 87.Liu Z, Liu Y, Li L, Xu Z, Bi B, Wang Y, et al. MiR-7–5p is frequently downregulated in glioblastoma microvasculature and inhibits vascular endothelial cell proliferation by targeting RAF1. Tumour Biol 2014;35: 10177–84. [DOI] [PubMed] [Google Scholar]
- 88.Katakowski M, Zheng X, Jiang F, Rogers T, Szalad A, Chopp M. MiR-146b-5p suppresses EGFR expression and reduces in vitro migration and invasion of glioma. Cancer Invest 2010;28:1024–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, Dugas JC, Gill RM, Huillard E, et al. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene 2012;31:1884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wuchty S, Arjona D, Li A, Kotliarov Y, Walling J, Ahn S, et al. Prediction of associations between microRNAs and gene expression in glioma biology. PLoS One 2011;6:e14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shi ZM, Wang J, Yan Z, You YP, Li CY, Qian X, et al. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS One 2012;7:e32709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 2008;582:1564–8. [DOI] [PubMed] [Google Scholar]
- 93.Li C, Gao Y, Zhang K, Chen J, Han S, Feng B, et al. Multiple roles of microRNA-100 in human cancer and its therapeutic potential. Cell Physiol Biochem 2015;37:2143–59. [DOI] [PubMed] [Google Scholar]
- 94.Zhao H, Peng R, Liu Q, Liu D, Du P, Yuan J, et al. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch Biochem Biophys 2016;610:1–7. [DOI] [PubMed] [Google Scholar]
- 95.Jia P, Cai H, Liu X, Chen J, Ma J, Wang P, et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett 2016;381:359–69. [DOI] [PubMed] [Google Scholar]
- 96.Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget 2015;6: 21934–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015;6:537–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem 2012;113:1868–74. [DOI] [PubMed] [Google Scholar]
- 99.Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun 2016;7:13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao X, Liu Y, Zheng J, Liu X, Chen J, Liu L, et al. GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochim Biophys Acta Mol Cell Res 2017;1864:1605–17. [DOI] [PubMed] [Google Scholar]
- 101.Joshi P, Katsushima K, Zhou R, Meoded A, Stapleton S, Jallo G, et al. The therapeutic and diagnostic potential of regulatory non-coding RNAs in medulloblastoma. Neuro-Oncology Adv 2019;1:vdz023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res 2009;69: 3249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pal R, Greene S. microRNA-10b is overexpressed and critical for cell survival and proliferation in medulloblastoma. PLoS One 2015;10: e0137845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grunder E, D’Ambrosio R, Fiaschetti G, Abela L, Arcaro A, Zuzak T, et al. MicroRNA-21 suppression impedes medulloblastoma cell migration. Eur J Cancer 2011;47:2479–90. [DOI] [PubMed] [Google Scholar]
- 105.Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol 2008;90:1–7. [DOI] [PubMed] [Google Scholar]
- 106.Li KK, Pang JC, Ching AK, Wong CK, Kong X, Wang Y, et al. miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum Pathol 2009;40:1234–43. [DOI] [PubMed] [Google Scholar]
- 107.Thiebes KP, Nam H, Cambronne XA, Shen R, Glasgow SM, Cho HH, et al. miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1-Lhx3. Nat Commun 2015;6:7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Venkataraman S, Birks DK, Balakrishnan I, Alimova I, Harris PS, Patel PR, et al. MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J Biol Chem 2013;288:1918–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J 2008;27:2616–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Salm F, Dimitrova V, von Bueren AO, Cwiek P, Rehrauer H, Djonov V, et al. The phosphoinositide 3-kinase p110alpha isoform regulates leukemia inhibitory factor receptor expression via c-Myc and miR-125b to promote cell proliferation in medulloblastoma. PLoS One 2015;10: e0123958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Laneve P, Po A, Favia A, Legnini I, Alfano V, Rea J, et al. The long noncoding RNA linc-NeD125 controls the expression of medulloblastoma driver genes by microRNA sponge activity. Oncotarget 2017;8: 31003–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014;512:82–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morrison TA, Wilcox I, Luo HY, Farrell JJ, Kurita R, Nakamura Y, et al. A long noncoding RNA from the HBS1L-MYB intergenic region on chr6q23 regulates human fetal hemoglobin expression. Blood Cells Mol Dis 2018;69:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lai K, Jia S, Yu S, Luo J, He Y. Genome-wide analysis of aberrantly expressed lncRNAs and miRNAs with associated co-expression and ceRNA networks in beta-thalassemia and hereditary persistence of fetal hemoglobin. Oncotarget 2017;8:49931–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alexander MS, Casar JC, Motohashi N, Vieira NM, Eisenberg I, Marshall JL, et al. MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J Clin Invest 2014;124:2651–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Petazzi P, Sandoval J, Szczesna K, Jorge OC, Roa L, Sayols S, et al. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol 2013;10:1197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM. 15q11–13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum Mol Genet 2007;16:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet 2005;14:483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fangusaro J. Pediatric high grade glioma: a review and update on tumor clinical characteristics and biology. Front Oncol 2012;2:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Blionas A, Giakoumettis D, Klonou A, Neromyliotis E, Karydakis P, Themistocleous MS. Paediatric gliomas: diagnosis, molecular biology and management. Ann Transl Med 2018;6:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reon BJ, Anaya J, Zhang Y, Mandell J, Purow B, Abounader R, et al. Expression of lncRNAs in low-grade gliomas and glioblastoma multiforme: an in silico analysis. PLoS Med 2016;13:e1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN, et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A 2015;112:8326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J, et al. Long Non-coding RNA XIST Promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. J Cancer 2017;8: 4106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 2012;123:465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Miele E, Po A, Begalli F, Antonucci L, Mastronuzzi A, Marras CE, et al. beta-arrestin1-mediated acetylation of Gli1 regulates Hedgehog/Gli signaling and modulates self-renewal of SHH medulloblastoma cancer stem cells. BMC Cancer 2017;17:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS, et al. MicroRNA profiling in human medulloblastoma. Int J Cancer 2009;124:568–77. [DOI] [PubMed] [Google Scholar]
- 127.Murphy BL, Obad S, Bihannic L, Ayrault O, Zindy F, Kauppinen S, et al. Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer Res 2013;73:7068–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Joshi P, Perera RJ. In silico analysis of long non-coding RNAs in medulloblastoma and its subgroups. bioRxiv 2019783092. [DOI] [PubMed] [Google Scholar]
- 129.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012;488:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA. Cancer Cell 2019;176:869–81.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018;37:555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lv T, Miao YF, Jin K, Han S, Xu TQ, Qiu ZL, et al. Dysregulated circular RNAs in medulloblastoma regulate proliferation and growth of tumor cells via host genes. Cancer Med 2018;7:6147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]