Abstract
A large proportion of the phenotypic variation in blood pressure (BP) appears to be inherited as a polygenic trait. This study examined the association between 12 single nucleotide polymorphisms (SNPs) in the guanine nucleotide binding protein beta polypeptide 3 (GNB3) and adducin 1 alpha (ADD1) genes and systolic (SBP), diastolic (DBP), and mean arterial (MAP) BP. A total of 3,142 individuals from 636 families were recruited from rural north China, and 2,746 met the eligibility criteria for analysis. BP measurements were obtained using a random-zero sphygmomanometer. Genetic variants were determined using SNPlex assays on an automated DNA Sequencer. A mixed linear model was used to estimate the association between each SNP and BP level. After Bonferroni correction, marker rs4963516 of the GNB3 gene remained significantly associated with DBP (corrected P values = 0.006, 0.007 and 0.002 for co-dominant, additive, and recessive models, respectively) and MAP (corrected P values = 0.02, 0.049, and 0.005, respectively). Compared to carriers of the major A allele, CC homozygotes had higher mean DBP (75.81 0.62 vs. 73.46 0.25 mmHg, P = 0.0002) and MAP (91.87 0.68 vs. 89.42 0.28 mmHg, P = 0.0004) after adjusting for covariates of age, gender, BMI, study site, and room temperature during BP measurement. In summary, these data support a role for the GNB3 gene in BP regulation in the Chinese population. Future studies aimed at replicating these novel findings are warranted.
Introduction
Blood pressure (BP) is a complex trait with the variation in BP levels being caused by multiple genetic and environmental factors. The genetic component is typically attributed to the additive effects of several genes, each having moderate effects on BP levels (Lifton and Jeunemaitre 1993). Approximately 30–60% of the BP variation in the population may be genetically determined (Levy et al. 2000; Melander 2001).
Many independent studies have confirmed an increased Na+/H+ exchanger activity in a subset of patients with hypertension (Siffert and Dusing 1995). Since both the guanine nucleotide binding protein (G-protein) and adducin aVect transport of Na+ ions (Bianchi 2005), associations of the G-protein β3 subunit (GNB3) and α-adducin (ADD1) genes and hypertension have been the focus of many studies (Cusi et al. 1997; Glorioso et al. 2002; Siffert et al. 1998). The effects of the GNB3 C825T and ADD1 Gly460Trp (rs4961) polymorphisms on BP variance or hypertension have been investigated extensively. The GNB3 825T allele has been associated with the expression of a shortened, functionally active splice variant of the G-protein β3 subunit and enhanced intracellular signal transduction. In a case–control study, the GNB3 825T allele was significantly associated with the risk of essential hypertension (Siffert et al. 1998). In 1997, an association of the ADD1 Gly460Trp variation with hypertension was first found (Cusi et al. 1997). Many subsequent studies have replicated the associations of the C825T and Gly460Trp polymorphisms with hypertension or BP-related phenotypes (Bianchi et al. 2005; Danoviz et al. 2006; Fava et al. 2007). However, inconsistencies among other association studies have also emerged (Ishikawa et al. 1998; Larson et al. 2000).
The current study was performed in a large, homogeneous sample of Han Chinese who took part in the Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study. The objective of the current investigation study was to examine the association between single nucleotide polymorphisms (SNPs) in the GNB3 and ADD1 genes and three quantitative BP traits: systolic (SBP), diastolic (DBP), and mean arterial (MAP) BP.
Methods
Study population
The GenSalt study was conducted in a Han Chinese population at six field centers in Hebei, Henan, Shandong (two sites), Shaanxi, and Jiangsu provinces in rural north China. A community-based BP screening was carried out among residents aged 18–60 years in the study villages to identify potential probands. Their family members were invited to take part in this study. Those with a mean SBP between 130 and 160 mmHg and/or a DBP between 85 and 100 mmHg and no use of antihypertensive medications was identified as probands for this study. Detailed eligibility criteria for the study participants have been presented previously (GenSalt Collaborative Research Group. 2007). Both two-generation (probands, siblings, and their parents) and three-generation families (probands and their siblings, parents, spouses, and oVspring) were recruited for the study. Individuals who had secondary hypertension, clinical cardiovascular disease (CVD) or diabetes, current use of antihypertensive medications, pregnancy, or heavy alcohol consumption were excluded from the study. Of the 3,142 persons from 636 families recruited, 2,746 were included in the analysis. The 396 subjects who were excluded from analysis (in the parental generation) had experienced one or more of the following: on antihypertensive treatment, diagnosed with diabetes or elevated glucose levels, or had experienced heart attack, stroke, congestive heart failure or peripheral arterial disease or had kidney failure. Institutional Review Boards at all of the participating institutions approved the GenSalt study. Written informed consents were obtained from each participant.
Phenotype measurement
A standard questionnaire was administered by trained staV at the examination to collect information on family structure, demographic characteristics, personal and family medical history, and lifestyle risk factors. Three morning BP measurements were obtained according to a standard protocol on each of the 3 days of observation by trained and certified technicians using a random-zero sphygmomanometer (Perloff et al. 1993). BP was measured from the right arm of participants in the sitting position after 5 min of rest. In addition, participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 min prior to their BP measurements. Body weight and height were measured twice in light indoor clothing without shoes. Body mass index (BMI) was calculated as kilograms per square meter (kg/m2).
Candidate gene and SNP selection and genotyping
Two candidate genes from the intracellular messenger pathway, ADD1 and GNB3, were selected based on their potential biological effects on BP regulation. Tagger software was used to select tagSNPs from each gene. The r2 of 0.8 was used. Nine SNPs from the ADD1 gene and six SNPs from the GNB3 gene were chosen based on linkage disequilibrium (LD) structure in the Chinese population, as determined by the Chinese HapMap, or based on previously reported associations with BP. SNP genotyping was performed using SNPlex assays (Applied Biosystems) based on the oligonucleotide ligation assay (OLA) for capillary electrophoresis on an automated DNA Sequencer (ABI 3700 DNA Analyzer).
Of the 15 genotyped SNPs, 2 monomorphic SNPs and 1 with a low genotyping call rate (53.1%) were excluded from the analysis. Table 2 lists information for each SNP.
Table 2.
Characteristics of SNPs genotyped in ADD1 and GNB3 genes
| Candidate gene | SNP | Chromosome | Physical location | Alleles (major/minor) | Frequency (minor) | HW P value |
|---|---|---|---|---|---|---|
| ADDI | rs4690002 | 4 | 2878245 | C/T | 0.44 | 0.60 |
| rs12503220 | 4 | 2887147 | G/A | 0.14 | 0.70 | |
| rs1263359 | 4 | 2899031 | T/C | 0.43 | 0.93 | |
| rs17833172 | 4 | 2905519 | G/A | 0.04 | 0.21 | |
| rs3775067 | 4 | 2925627 | C/T | 0.34 | 0.77 | |
| rs4961 | 4 | 2943714 | T/G | 0.48 | 0.20 | |
| rs4963 | 4 | 2953769 | G/C | 0.48 | 0.54 | |
| GNB3 | rs4963516 | 12 | 6818288 | A/C | 0.32 | 0.12 |
| rs1129649 | 12 | 6818728 | T/C | 0.31 | 0.61 | |
| rs3213431 | 12 | 6818788 | T/C | 0.11 | 0.82 | |
| rs2301339 | 12 | 6824884 | G/A | 0.45 | 0.66 | |
| rs5446 | 12 | 6826722 | C/T | 0.20 | 0.68 |
Statistical analysis
ASPEX and GRR were used to check for potential misreported relationships in the GenSalt family pedigrees (Abecasis et al. 2001; Hinds and Risch 1999). The Mendelian consistency of the SNP genotype data was assessed by PLINK and PedCheck (O’Connell and Weeks 1998; Purcell et al. 2007). If Mendelian inconsistencies were found, genotypes for SNPs in those families were set to be missing. HWE for each SNP was examined using the PEDSTATS procedure, as implemented in Merlin (Abecasis et al. 2002). The genotyping call rate and MAF were also assessed for each SNP. Haploview software (version 4.0, http://www.broad.mit.edu/mpg/haploview) was used to estimate the extent of pairwise LD between SNPs.
Descriptive statistics were calculated for each of the analysis variables and covariates for the combined analysis sample. Associations between SNP genotypes and BP traits were examined under codominant, additive, dominant and recessive genetic models using a mixed linear model. A sandwich estimator was used to account for the non-independence of family members. This method assumes the same degree of dependency among family members. The raw BP variables were analyzed with multiple predictors that included a given SNP plus age, gender, BMI, room temperature of BP measurement, and study site. The P values were adjusted for the total number of analyses (N = 12 corresponding to the 12 SNPs) using the Bonferroni correction. Additionally, we estimated the mean effect size and 95% confidence interval (CI) of each genotype using a mixed linear model. These analyses were conducted using SAS statistical software (version 9.1; SAS Institute Inc).
Results
On average, participants were 47.4 years of age, had a BMI of 23.0 kg/m2 and mean SBP, DBP, and MAP of 121.9, 73.7, and 89.8 mmHg, respectively. There were 1,427 (52.0%) male subjects in the present study. Table 1 presents these descriptive statistics for all participants.
Table 1.
Characteristics of 2,746 study participants
| V ariable | Mean ± SD or percentage | Range |
|---|---|---|
| Age (years) | 47.4 ± 16.1 | (15.0, 93.0) |
| Male (%) | 52.0 | - |
| BMI (kg/m2) | 23.0 ± 3.2 | (14.3, 37.8) |
| BP (mmHg) | ||
| Systolic | 121.9 ± 19.0 | (80.9, 222) |
| Diastolic | 73.7 ± 10.5 | (38.9, 115.1) |
| Mean arterial pressure | 89.8 ± 12.1 | (58.7, 146.5) |
BMI body mass index, BP blood pressure
MAF and HWE were examined for each SNP (Table 2). Table 3 shows the raw P values for the association between each SNP and SBP, DBP and MAP adjusted for age, gender, BMI, study site and room temperature of BP measurement. For additional details of the mean effect size and 95% CI, see Supplementary Table 1.
Table 3.
The association between SNPs and blood pressure level
| Gene | SNP | Codominant P value | Additive P value | Dominant P value | Recessive P value |
|---|---|---|---|---|---|
| Systolic blood pressure | |||||
| ADDI | rs4690002 | 0.96 | 0.77 | 0.77 | 0.87 |
| rs12503220 | 0.59 | 0.32 | 0.30 | 0.74 | |
| rs1263359 | 0.26 | 0.27 | 0.82 | 0.10 | |
| rs17833172 | 0.81 | 0.61 | 0.69 | 0.62 | |
| rs3775067 | 0.19 | 0.07 | 0.14 | 0.15 | |
| rs4961 | 0.54 | 0.38 | 0.77 | 0.27 | |
| rs4963 | 0.36 | 0.22 | 0.58 | 0.15 | |
| GNB3 | rs4963516 | 0.05 | 0.12 | 0.60 | 0.02 |
| rs1129649 | 0.29 | 0.13 | 0.12 | 0.49 | |
| rs3213431 | 0.30 | 0.16 | 0.23 | 0.24 | |
| rs2301339 | 0.86 | 0.93 | 0.80 | 0.69 | |
| rs5446 | 0.27 | 0.47 | 0.90 | 0.11 | |
| Diastolic blood pressure | |||||
| ADDI | rs4690002 | 0.78 | 0.56 | 0.78 | 0.49 |
| rs12503220 | 0.88 | 0.80 | 0.72 | 0.78 | |
| rs1263359 | 0.96 | 0.99 | 0.88 | 0.86 | |
| rs17833172 | 0.50 | 0.86 | 0.73 | 0.28 | |
| rs3775067 | 0.32 | 0.33 | 0.16 | 0.82 | |
| rs4961 | 0.66 | 0.68 | 0.42 | 0.83 | |
| rs4963 | 0.56 | 0.46 | 0.29 | 0.94 | |
| GNB3 | rs4963516 | 0.0005 † | 0.0006 † | 0.02 | 0.0002 † |
| rs1129649 | 0.13 | 0.046 | 0.10 | 0.12 | |
| rs3213431 | 0.33 | 0.52 | 0.74 | 0.14 | |
| rs2301339 | 0.57 | 0.53 | 0.94 | 0.30 | |
| rs5446 | 0.79 | 0.67 | 0.56 | 0.83 | |
| Mean arterial pressure | |||||
| ADDI | rs4690002 | 0.86 | 0.60 | 0.74 | 0.60 |
| rs12503220 | 0.77 | 0.52 | 0.47 | 0.99 | |
| rs1263359 | 0.77 | 0.60 | 0.87 | 0.47 | |
| rs17833172 | 0.89 | 0.72 | 0.68 | 0.80 | |
| rs3775067 | 0.29 | 0.14 | 0.12 | 0.54 | |
| rs4961 | 0.79 | 0.51 | 0.53 | 0.68 | |
| rs4963 | 0.57 | 0.29 | 0.36 | 0.45 | |
| GNB3 | rs4963516 | 0.002 † | 0.004 † | 0.10 | 0.0004 † |
| rs1129649 | 0.15 | 0.05 | 0.08 | 0.19 | |
| rs3213431 | 0.26 | 0.29 | 0.45 | 0.12 | |
| rs2301339 | 0.70 | 0.69 | 0.93 | 0.43 | |
| rs5446 | 0.73 | 0.51 | 0.65 | 0.46 | |
Original P <0.05 is indicated in bold italic
After Bonferroni correction, P value < 0.05
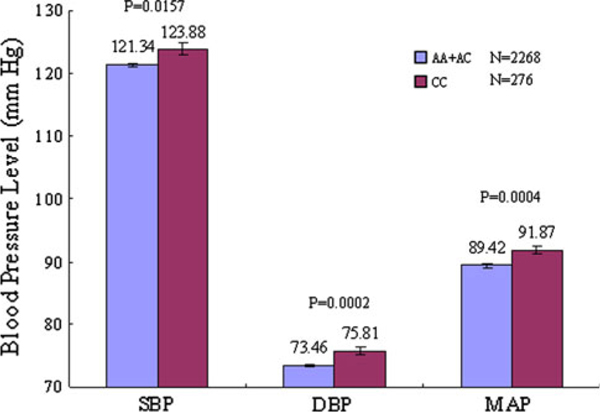
Marker rs4963516 of the GNB3 gene was significantly associated with SBP, DBP or MAP in at least one of the genetic models tested. After Bonferroni correction, marker rs4963516 remained significantly associated with DBP (corrected P values = 0.006, 0.007, and 0.002 for the codominant, additive, and recessive genetic models, respectively) and MAP (corrected P values = 0.02, 0.049, and 0.005 for the codominant, additive, and recessive genetic models, respectively). Although associations of this variant were significant under all three genetic models, the recessive model provided the best fit based on assessment of the P values and mean BP levels. Compared to the individuals carrying the major A allele, participants with rs4963516 CC genotype had higher levels of DBP (75.81 ± 0.62 vs. 73.46 ± 0.25 mmHg, P value = 0.0002) and MAP (91.87 ± 0.68 vs. 89.42 ± 0.28 mmHg, P value = 0.0004). Although not statistically significant, a similar trend was observed for mean SBP level for this variant (Fig. 1).
Fig. 1.
The level of systolic (SBP), diastolic (DBP), and mean arterial pressure (MAP) according to SNP rs4963516 genotypes of the GNB3 gene adjusted by age, gender, BMI, study site and BP measurement room temperature. Values are means with standard error bars
Discussion
In the present study, we identified a novel genetic variant in the GNB3 gene that may be an important predictor of resting BP. Marker rs4963516 in the GNB3 gene was significantly associated with DBP and MAP. Participants homozygous for the minor C allele had higher DBP and MAP compared to those who were heterozygous or homozygous for the major A allele.
The widespread constitutive cellular abnormalities in sodium handling observed in rats and hypertensive patients, both characterized by a common defect of renal tubular function, were suggestive of an adducin protein alteration in the actin cytoskeleton (Bianchi et al. 2005). ADD1 has been a target for candidate gene studies of BP-related phenotypes because of its role in modulating the re-absorption of sodium in renal tubular cells (Manunta et al. 2007). Many studies have investigated the effect of the ADD1 Gly460Trp polymorphism on BP traits or essential hypertension, and found that the Trp substitution was associated with the increased BP level (Fava et al. 2007; Shioji et al. 2004). While several studies have reported higher BP levels among participants with the 460Trp variant, the current study did not find such an association. The discrepant findings reported here may reflect population heterogeneity, as the LD structure of the Han Chinese population examined here may be diVerent to that of the Caucasian populations examined in the majority of previous studies (Cusi et al. 1997; Grant et al. 2002).
Previous studies have shown that the Na+/H+ exchanger isoform-1 activity was enhanced in hypertensive patients, suggesting the involvement of G-protein. Hence, alterations that aVect G-protein function or expression may modulate BP homeostasis (Farfel et al. 1999). In this study, participants with the CC genotype of the GNB3 marker rs4963516 had significantly higher DBP and MAP compared to those who were homozygous or heterozygous for the A allele. This novel finding has not been reported previously.
Numerous studies have examined the association between the C825T variant of the GNB3 gene and BP-related phenotypes and identified a significantly increased risk for hypertension or higher BP levels associated with the variant T allele (Benjafield et al. 1998; Schunkert et al. 1998). However, our previous study, and studies in Chinese and other populations such with Finnish, Japanese and African ancestry did not report this association (Huang et al. 2003; Larson et al. 2000; Li et al. 2005; Shioji et al. 2004; Snapir et al. 2001). Although the current study did not examine C825T specifically, the measured rs2301339 variant of GNB3 is in perfect LD with the C825T polymorphism (D’ and r2 = 1.0) in the HapMap Chinese (CHB) and Caucasian (CEU) populations. However, this SNP was not associated with SBP, DBP or MAP in this study. In contrast, the maker rs4963516 in the GNB3 gene that was significant in the current study was in weak LD with rs2301339 (r2 = 0.247) and thus is likely to represent a new signal.
For the pair-wise LD analysis using the latest Chinese HapMap data, there is only one maker, rs5440, in moderate LD with rs4963516 (D’ = 0.83 and r2 = 0.60). Marker rs4963516 lies in the 5’-flanking region of the GNB3 gene. Additionally, it has been reported as a variation of intron 13 in the leprecan-like 2 (LEPREL2) gene. It should be noted that the GNB3 and LEPREL2 genes overlap on chromosome 12. The SNP rs5440 is located at position −476 bp upstream from the transcription-initiation site of GNB3 gene. We explored the possible functions of rs4963516 and rs5440 in post hoc analysis using TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess), a web tool for predicting transcription factor binding sites in DNA sequences. TESS analysis showed that rs4963516 lies in a recognition sequence of the transcription element c-Jun. The variant rs5440 also lies in a recognition sequence of the transcription element Elk-1 (ETS like gene 1). This placement suggests that rs4963516 may act as a transcriptional regulatory element contributing to the development and occurrence of hypertension. Certainly, functional studies will be important to determine whether this SNP regulates the transcription of the GNB3 or LEPREL2 gene, and whether these changes may be associated with BP-related traits. Whether the SNP rs5440 in moderate LD with rs4963516 is a causative polymorphism deserves further functional study.
Our study has several important strengths. First, to our knowledge this is the most comprehensive study of BP variation in relation to tag-SNPs and previously reported SNPs in the ADD1 and GNB3 genes. Second, the study participants are relatively homogeneous with regard to genetic background (all Han Chinese) and environmental risk factors (i.e. common diets and physical activity levels). Thus, confounding effects due to these exposures should be minimal. Finally, rigorous quality control procedures were employed during measurement of BP, other study covariates, and genotypes, again reducing sources of error in the data analysis.
In conclusion, our results support a role for the GNB3 gene in BP regulation. In particular, we identified a significant association between variant rs4963516 of the GNB3 gene and BP level. Since this finding may have important clinical implications, future studies aimed at replicating these novel results will be of interest.
Supplementary Material
Acknowledgments
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by a cooperative agreement project Grant (U01HL072507) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD.
Appendix: GenSalt Collaborative Research Group
Tulane University Health Sciences Center, New Orleans, USA: Jiang He (PI), Lydia A. Bazzano, Chung-Shiuan Chen, Jing Chen, Tanika N. Kelly, L. Lee Hamm, Paul Muntner, Kristi Reynolds, Paul K. Whelton, Wenjie Yang, and Qi Zhao.
Washington University School of Medicine, St. Louis, USA: DC Rao (PI), Matthew Brown, Charles Gu, Treva Rice, Karen Schwander, and Shiping Wang.
Chinese Academy of Medical Sciences, Beijing, China: Dongfeng Gu (PI), Jie Cao, Jichun Chen, Xiufang Duan, Jianfeng Huang, Jinghan Huang, Jianxin Li, De-Pei Liu, Donghua Liu, Enchun Pan, Wei Yang, and Xigui Wu. Shandong Academy of Medical Sciences, Shandong, China: Fanghong Lu (PI), Shikuan Jin, Qingjie Meng, Fan Wu, and Yingxin Zhao; Shandong Center for Diseases Control and Prevention, Shandong, China: Jixiang Ma (PI), Weika Li, and Jiyu Zhang; Zhengzhou University: Dongsheng Hu (PI), Yaxin Ding, Hongwei Wen, Meixi Zhang, and Weidong Zhang; Xinle Traditional Chinese Medicine Hospital, Hebei, China: Xu Ji (PI), Rongyan Li, Haijun Zu; Nanjing University of Medical Sciences, Jiangsu, China: Cailiang Yao (PI), Yongchao Li, Chong Shen, and Jiayi Zhou; Xi’an Jiaotong University, Shanxi, China: Jianjun Mu (PI), Enrang Chen, Qinzhou Huang, and Man Wang.
Chinese National Human Genome Center at Beijing: Zhi-Jian Yao (PI), Shufeng Chen, Dongfeng Gu, Hongfan Li, Laiyuan Wang, Penghua Zhang, Qi Zhao.
University of Texas Health Sciences Center at Houston: James E. Hixson (PI) and Lawrence C. Shimmin.
National Heart, Lung, and Blood Institute: Cashell E. Jaquish.
Footnotes
For the GenSalt Collaborative Research Group.
Electronic supplementary material The online version of this article (doi:10.1007/s00439-010-0834-3) contains supplementary material, which is available to authorized users.
Contributor Information
Shufeng Chen, Division of Population Genetics, Department of Evidence Based Medicine, Cardiovascular Institute, Fu Wai Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, 167 Beilishi Road, Beijing 100037, China.
Hongwei Wang, Division of Population Genetics, Department of Evidence Based Medicine, Cardiovascular Institute, Fu Wai Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, 167 Beilishi Road, Beijing 100037, China.
Xiangfeng Lu, Division of Population Genetics, Department of Evidence Based Medicine, Cardiovascular Institute, Fu Wai Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, 167 Beilishi Road, Beijing 100037, China.
De-Pei Liu, National Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, China.
Jing Chen, Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA, USA; Department of Medicine, Tulane University School of Medicine, New Orleans, LA, USA.
Cashell E. Jaquish, National Heart, Lung, and Blood Institute, National Institute of Health, Bethesda, MD, USA
Dabeeru C. Rao, Washington University in St. Louis School of Medicine, St. Louis, MO, USA
James E. Hixson, University of Texas School of Public Health, Houston, TX, USA
Tanika N. Kelly, Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA, USA
Liping Hou, Division of Population Genetics, Department of Evidence Based Medicine, Cardiovascular Institute, Fu Wai Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, 167 Beilishi Road, Beijing 100037, China.
Laiyuan Wang, Division of Population Genetics, Department of Evidence Based Medicine, Cardiovascular Institute, Fu Wai Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, 167 Beilishi Road, Beijing 100037, China.
Jianfeng Huang, Division of Population Genetics, Department of Evidence Based Medicine, Cardiovascular Institute, Fu Wai Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, 167 Beilishi Road, Beijing 100037, China.
Chung-Shiuan Chen, Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA, USA.
Treva K. Rice, Washington University in St. Louis School of Medicine, St. Louis, MO, USA
Paul K. Whelton, Loyola University Medical Center, Maywood, IL, USA
Jiang He, Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA, USA; Department of Medicine, Tulane University School of Medicine, New Orleans, LA, USA.
Dongfeng Gu, Division of Population Genetics, Department of Evidence Based Medicine, Cardiovascular Institute, Fu Wai Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, 167 Beilishi Road, Beijing 100037, China; Chinese National Human Genome Center, Beijing, China.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlinrapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Benjafield AV, Jeyasingam CL, Nyholt DR, Griffiths LR, Morris BJ (1998) G-protein beta3 subunit gene (GNB3) variant in causation of essential hypertension. Hypertension 32:1094–1097 [DOI] [PubMed] [Google Scholar]
- Bianchi G (2005) Genetic variations of tubular sodium reabsorption leading to “primary” hypertension: from gene polymorphism to clinical symptoms. Am J Physiol Regul Integr Comp Physiol 289:R1536–R1549 [DOI] [PubMed] [Google Scholar]
- Bianchi G, Ferrari P, Staessen JA (2005) Adducin polymorphism: detection and impact on hypertension and related disorders. Hypertension 45:331–340 [DOI] [PubMed] [Google Scholar]
- Cusi D, Barlassina C, Azzani T, Casari G, Citterio L, Devoto M, Glorioso N, Lanzani C, Manunta P, Righetti M, Rivera R, Stella P, TroVa C, Zagato L, Bianchi G (1997) Polymorphisms of alpha-adducin and salt sensitivity in patients with essential hypertension. Lancet 349:1353–1357 [DOI] [PubMed] [Google Scholar]
- Danoviz ME, Pereira AC, Mill JG, Krieger JE (2006) Hypertension, obesity and GNB 3 gene variants. Clin Exp Pharmacol Physiol 33:248–252 [DOI] [PubMed] [Google Scholar]
- Farfel Z, Bourne HR, Iiri T (1999) The expanding spectrum of G protein diseases. N Engl J Med 340:1012–1020 [DOI] [PubMed] [Google Scholar]
- Fava C, Montagnana M, Almgren P, Rosberg L, Guidi GC, Berglund G, Melander O (2007) Association between adducin-1 G460W variant and blood pressure in Swedes is dependent on interaction with body mass index and gender. Am J Hypertens 20:981–989 [DOI] [PubMed] [Google Scholar]
- GenSalt Collaborative Research Group (2007) GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens 21:639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorioso N, Filigheddu F, Cusi D, TroVa C, Conti M, Natalizio M, Argiolas G, Barlassina C, Bianchi G (2002) alpha-Adducin 460Trp allele is associated with erythrocyte Na transport rate in North Sardinian primary hypertensives. Hypertension 39:357–362 [DOI] [PubMed] [Google Scholar]
- Grant FD, Romero JR, Jeunemaitre X, Hunt SC, Hopkins PN, Hollenberg NH, Williams GH (2002) Low-renin hypertension, altered sodium homeostasis, and an alpha-adducin polymorphism. Hypertension 39:191–196 [DOI] [PubMed] [Google Scholar]
- Hinds D, Risch M (1999) The ASPEX package: aVected sib-pair exclusion mapping. 88: [cited 2008 May 20]. ftp://lahmed.stan-ford.edu/pub/aspex/doc/usage.html [Google Scholar]
- Huang X, Ju Z, Song Y, Zhang H, Sun K, Lu H, Yang Z, Jose PA, Zhou G, Wang M, Wang W, Feng S, Hui R (2003) Lack of association between the G protein beta3 subunit gene and essential hypertension in Chinese: a case-control and a family-based study. J Mol Med 81:729–735 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Katsuya T, Sato N, Nakata Y, Takami S, Takiuchi S, Fu Y, Higaki J, Ogihara T (1998) No association between alpha-adducin 460 polymorphism and essential hypertension in a Japanese population. Am J Hypertens 11:502–506 [DOI] [PubMed] [Google Scholar]
- Larson N, Hutchinson R, Boerwinkle E (2000) Lack of association of 3 functional gene variants with hypertension in African Americans. Hypertension 35:1297–1300 [DOI] [PubMed] [Google Scholar]
- Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH (2000) Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the Framingham heart study. Hypertension 36:477–483 [DOI] [PubMed] [Google Scholar]
- Li B, Ge D, Wang Y, Zhao W, Zhou X, Gu D, Chen R (2005) G protein beta 3 subunit gene variants and essential hypertension in the northern Chinese Han population. Ann Hum Genet 69:468–473 [DOI] [PubMed] [Google Scholar]
- Lifton RP, Jeunemaitre X (1993) Finding genes that cause human hypertension. J Hypertens 11:231–236 [DOI] [PubMed] [Google Scholar]
- Manunta P, Citterio L, Lanzani C, Ferrandi M (2007) Adducin polymorphisms and the treatment of hypertension. Pharmacogenomics 8:465–472 [DOI] [PubMed] [Google Scholar]
- Melander O (2001) Genetic factors in hypertension–what is known and what does it mean? Blood Press 10:254–270 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PerloV D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ (1993) Human blood pressure determination by sphygmomanometry. Circulation 88:2460–2470 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H, Hense HW, Doring A, Riegger GA, Siffert W (1998) Association between a polymorphism in the G protein beta3 subunit gene and lower renin and elevated diastolic blood pressure levels. Hypertension 32:510–513 [DOI] [PubMed] [Google Scholar]
- Shioji K, Kokubo Y, Mannami T, Inamoto N, Morisaki H, Mino Y, Tagoi N, Yasui N, Iwaii N (2004) Association between hypertension and the alpha-adducin, beta1-adrenoreceptor, and G-protein beta3 subunit genes in the Japanese population; the Suita study. Hypertens Res 27:31–37 [DOI] [PubMed] [Google Scholar]
- Siffert W, Dusing R (1995) Sodium-proton exchange and primary hypertension. An update. Hypertension 26:649–655 [DOI] [PubMed] [Google Scholar]
- Siffert W, Rosskopf D, Siffert G, Busch S, Moritz A, Erbel R, Sharma AM, Ritz E, Wichmann HE, Jakobs KH, Horsthemke B (1998) Association of a human G-protein beta3 subunit variant with hypertension. Nat Genet 18:45–48 [DOI] [PubMed] [Google Scholar]
- Snapir A, Heinonen P, Tuomainen TP, Lakka TA, Kauhanen J, Salonen JT, Scheinin M (2001) G-protein beta3 subunit C825T polymorphism: no association with risk for hypertension and obesity. J Hypertens 19:2149–2155 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.